- 1Department of ophthalmology, JiuJiang City Key Laboratory of Cell Therapy, Jiujiang No. 1 People’s Hospital, JiuJiang, Jiangxi, China
- 2Department of Otolaryngology, The Seventh Affiliated Hospital, Sun Yat-sen University, ShenZhen, Guangdong, China
Purpose: This study aimed to develop a hybrid deep learning model for classifying multiple fundus diseases using ultra-widefield (UWF) images, thereby improving diagnostic efficiency and accuracy while providing an auxiliary tool for clinical decision-making.
Methods: In this retrospective study, 10,612 UWF fundus images were collected from the JiuJiang No. 1 People’s Hospital and the Seventh Affiliated Hospital, Sun Yat-sen University between 2020 and 2025, covering 16 fundus diseases, including normal fundus, nine common eye diseases, and six rare retinal conditions. The model employed DenseNet121 as a feature extractor combined with an XGBoost classifier. Gradient-weighted Class Activation Mapping (Grad-CAM) was used to visualize the model’s decision-making process. Performance was evaluated on validation and external test sets using accuracy, recall, precision, F1 score, and AUC-ROC. The model’s diagnostic accuracy was also compared with that of junior and intermediate ophthalmologists.
Results: The model demonstrated exceptional diagnostic performance. For common diseases such as retinal vein occlusion, age-related macular degeneration, and diabetic retinopathy, AUC values exceeded 0.975, with accuracy rates above 0.980. For rare diseases, AUC values were above 0.970, and accuracy rates surpassed 0.998. Grad-CAM visualizations confirmed that the model’s focus areas aligned with clinical pathological features. Compared to ophthalmologists, the model achieved significantly higher accuracy across all diagnostic tasks.
Conclusion: The proposed deep learning model can automatically identify and classify multiple ophthalmic diseases using UWF images. It holds promise for enhancing clinical diagnostic efficiency, assisting ophthalmologists in optimizing workflows, and improving patient care quality.
Introduction
With the rapid growth of the global elderly population, many age-related health issues have become increasingly prominent, among which the incidence of retinal diseases has significantly increased (Wong et al., 2014; Flaxman et al., 2017). As a vital element in vision formation, the retina directly impacts visual acuity. Therefore, the prompt, efficient, and precise detection of retinal lesions is of utmost importance. Traditional fundus photography is confined to the central 30–60 degrees of the retina, allowing identification of only the posterior pole (Shoughy et al., 2015). In contrast, Ultra-Wide-Field (UWF) fundus imaging offers a comprehensive view of nearly the entire retina with a 200° field of view, covering both the posterior pole and peripheral areas (Nagiel et al., 2016; Silva et al., 2012). This extensive coverage provides richer clinical information, aiding in the early diagnosis and timely intervention of conditions such as pathologic myopia, retinal vein occlusion, retinal detachment, retinal holes, diabetic retinopathy, and age-related macular degeneration (Tan et al., 2019; Ohsugi et al., 2017; Masumoto et al., 2019; Zhang et al., 2021; Horie and Ohno-Matsui, 2022). However, despite the fact that UWF technology can provide a broader coverage of the retina, doctors still face many challenges in identifying lesion features in these images. For instance, in the diagnosis and management of diabetic retinopathy, UWF fundus imaging can detect peripheral lesions that are difficult to find with traditional methods, which helps to more accurately assess the severity of the disease and formulate treatment strategies (Ashrafkhorasani et al., 2024; Silva et al., 2013). However, due to the complexity and large amount of information in UWF fundus images, doctors need to possess higher professional skills and experience when interpreting these images (Ghasemi Falavarjani et al., 2017).
In recent years, with the rapid development of artificial intelligence (AI) technology, its application in ophthalmic imaging examinations has become increasingly widespread, especially achieving remarkable progress in fields such as optical coherence tomography (OCT) (Gan et al., 2023a; Wei et al., 2023), fundus photography (Li et al., 2024; Rajalakshmi et al., 2018), and fluorescein fundus angiography (FFA) (Long et al., 2025; Wei et al., 2025). It is capable of rapidly and precisely identifying and analyzing the characteristics of fundus lesions, thereby enabling early screening, diagnosis stratification, and disease monitoring for various fundus diseases, which substantially enhances diagnostic efficiency and accuracy (Grassmann et al., 2018; Li et al., 2023). In previous studies, Nguyen et al. (2024) proposed a deep learning-based approach for retinal disease diagnosis using ultra-wide-field fundus images, achieving high accuracy with models like ResNet152. Despite their promising results, their work was limited to binary classification (normal vs. abnormal). In contrast, DenseNet121, with its unique dense connection mechanism and feature reuse capability, demonstrates significant advantages in handling complex medical image tasks. Its dense connection structure not only enhances feature propagation and reduces the problem of gradient vanishing, but also significantly improves parameter utilization efficiency and reduces model complexity through feature reuse, while enhancing the model’s generalization ability (Zhang et al., 2025). XGBoost, with its powerful classification performance and nonlinear combination ability, can further optimize classification results (Ye et al., 2022; Dong et al., 2018).
Therefore, this study proposes a hybrid deep learning model based on DenseNet121 and XGBoost for the intelligent diagnosis of 16 retinal conditions (including normal fundus, nine common eye diseases, and six rare diseases) in ultra-wide-field fundus images. The model innovatively combines the multi-scale feature extraction advantages of DenseNet121s dense connection architecture with XGBoost’s ensemble learning capabilities for handling imbalanced data (Gan et al., 2023b). By leveraging the complementary strengths of deep convolutional features and gradient boosting decision trees, it effectively addresses addressed the performance bottleneck of traditional end-to-end CNN models in classifying rare diseases.
Methods
Data collection
This retrospective study collected ultra-widefield (UWF) fundus photographs from patients who visited the JiuJiang No. 1 People’s Hospital between January 2020 and January 2025. The dataset was randomly partitioned into training (80%) and validation (20%) sets. An additional external test set was compiled from UWF images obtained from the Seventh Affiliated Hospital, Sun Yat-sen University during the same period. The study protocol received approval from the institutional review boards of both participating hospitals and complied with the Declaration of Helsinki. As all patient data were anonymized and contained no personally identifiable information, the ethics committees waived the requirement for individual informed consent. The study design is illustrated in Figure 1.
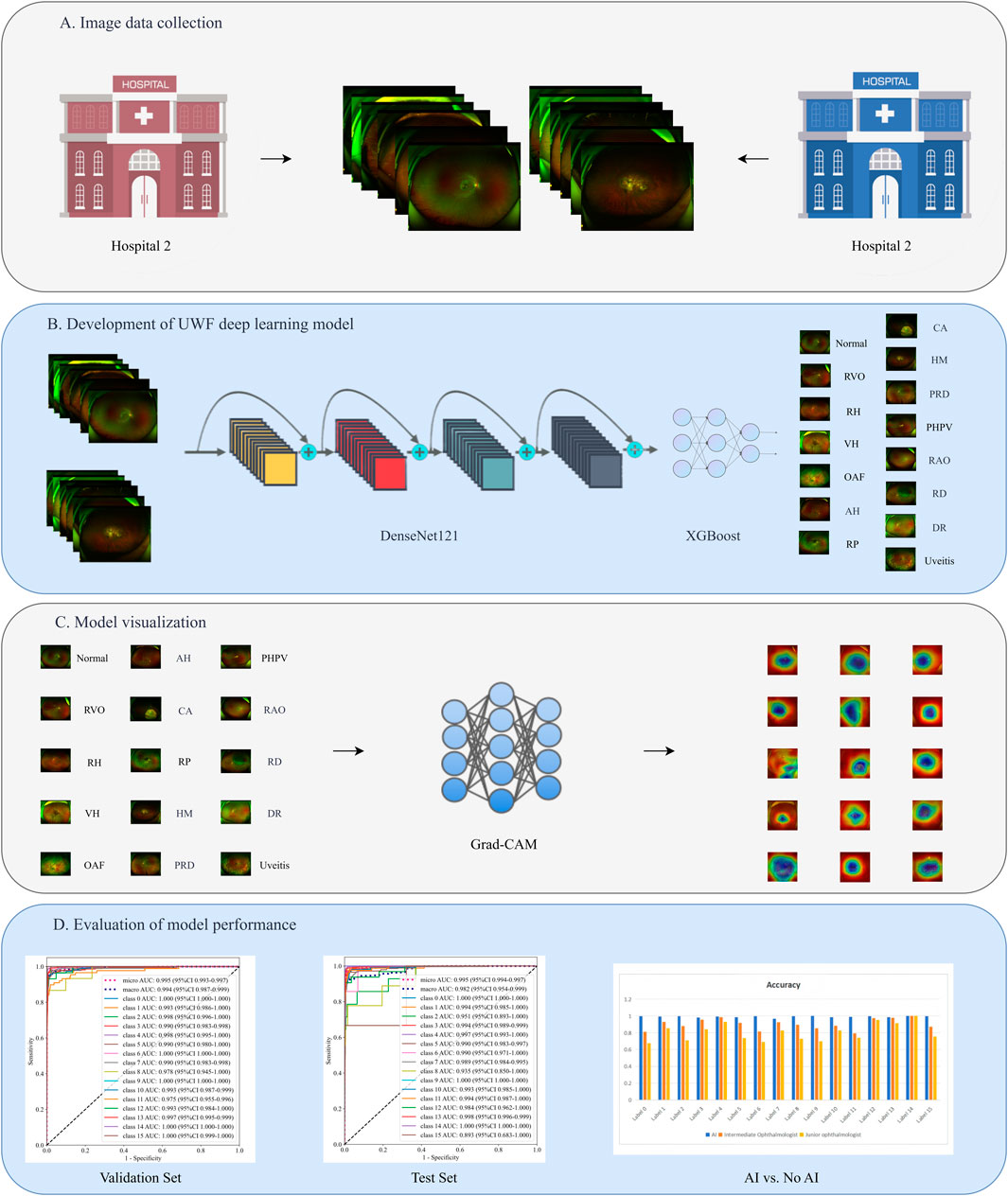
Figure 1. The study design flowchart. (A) Image data collection. (B) Development of UWF deep learning model. (C) Model visualization. (D) Evaluation of model performance.
Image preprocessing
Initial quality control excluded images with poor resolution, excessive blurring, or duplicate content. Diagnostic labeling followed a rigorous two-stage process: 1) preliminary classification by an independent ophthalmologist using comprehensive clinical data from electronic medical records, and 2) validation by a senior retinal specialist who reviewed all ambiguous cases to ensure diagnostic consistency. Based on clinical diagnoses, images were categorized into 16 classes: Retinal Artery Occlusion (RAO, 0), Retinal Vein Occlusion (RVO, 1), Age-related Macular Degeneration (AMD, 2), Diabetic Retinopathy (DR, 3), Retinal Detachment (RD, 4), High Myopia (HM, 5), Uveitis (6), Vitreous Hemorrhage (VH, 7), Asteroid Hyalosis (AH, 8), Choroidal Atrophy (CA, 9), Peripheral Retinal Degeneration (PRD, 10), Retinal Hole (RH, 11), Retinitis Pigmentosa (RP, 12), Normal fundus (13), Ocular Albinism Fundus (OAF, 14), and Persistent Hyperplastic Primary Vitreous (PHPV, 15).
Development of deep learning model
We employed DenseNet121, pretrained on ImageNet, for feature extraction. The model’s dense connection architecture facilitates feature propagation and reuse, enhancing its ability to capture both local and global image features. All images were resized to 224 × 224 pixels to meet network input requirements. To improve model generalization and robustness, we implemented data augmentation techniques including random flipping, rotation (±30°), and contrast-limited adaptive histogram equalization (CLAHE) (Wu et al., 2022).
To address class imbalance, we applied stratified sampling to maintain consistent class distributions across training, validation, and test sets. Rare disease samples underwent 20-fold augmentation (rotation, horizontal flipping, CLAHE), yielding 300 effective training samples per rare disease category. The model incorporated Focal Loss in DenseNet121 to reduce majority class weighting, while the XGBoost classifier used scale_pos_weight (reciprocal of class frequency) to further mitigate imbalance effects.
Computational implementation
Model training and evaluation were conducted on a workstation equipped with an NVIDIA RTX 4090 GPU (24 GB VRAM), Intel Core i9-13900K CPU, and 64 GB DDR5 RAM, achieving an average inference speed of 0.07 ± 0.01 s per image. Training parameters included:
• Batch size: 32
• Initial learning rate: 0.001 (reduced by factor of 0.1 every 50 epochs)
• Optimizer: Adam (β1 = 0.9, β2 = 0.999)
• Training epochs: 150 (with early stopping if validation loss failed to improve for 15 consecutive epochs)
High-level features were extracted from the “features.norm5” layer prior to global average pooling. These features underwent dimensionality reduction via principal component analysis (PCA) before being classified by a tuned XGBoost model (Gan et al., 2023a).
Model visualization
After the completion of model training, we employ the Gradient-weighted Class Activation Mapping (Grad-CAM) (Selvaraju et al., 2024) technique to visualize the decision-making basis of the model. Specifically, through backpropagation, we compute the gradient of the target category with respect to the feature map of the last convolutional layer in DenseNet121. These gradients are then multiplied element-wise by the corresponding feature maps and aggregated via weighted summation to generate a normalized activation map. Subsequently, this activation map is transformed into a heatmap and overlaid onto the original UWF fundus image, thereby visually highlighting the regions of interest that the model focuses on. Then, a specialist in retinal diseases from a tertiary hospital evaluated the consistency between the heat map-marked regions and the corresponding clinical pathological features. This process provides valuable insights into the model’s decision-making process, offering interpretable visualization support.
Comparison of diagnostic performance between ophthalmologists and AI models
Diagnostic performance was compared between the AI model and:
1. Junior ophthalmologists (<5 years’ experience, completed residency training)
2. Intermediate ophthalmologists (5–10 years’ experience, attending physicians)
Evaluations used a standardized electronic platform allowing image parameter adjustment but no clinical context. A single-blind design ensured objective assessment, with clinicians unaware of ground truth labels.
Statistical analysis
All analyses were performed using Python 3.9.7. Model performance was assessed using:
1. Accuracy: Overall classification correctness
2. Recall: True positive detection rate
3. Precision: Positive predictive value
4. F1 score: Harmonic mean of recall and precision
5. AUC-ROC: Classification discrimination across thresholds
ROC curves visualized true-positive vs. false-positive tradeoffs, while confusion matrices detailed per-class prediction errors. Comparative analyses with clinician performance provided practical insights into clinical applicability.
Results
Dataset characteristics
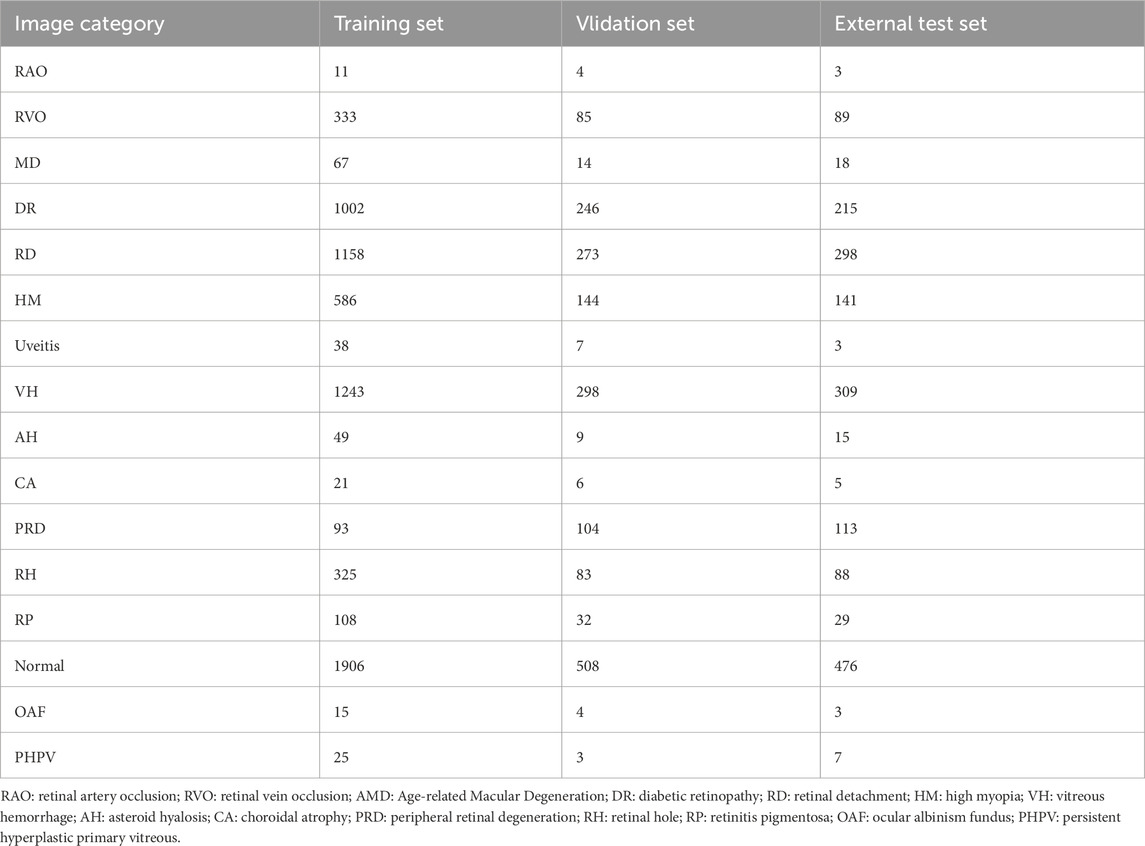
The dataset of this study contains a total of 10,612 ultra-wide-angle fundus images, covering 16 different types of retinal images, which were divided into a training set (6,980 images), a validation set (1,820 images), and an external test set (1,812 images). The dataset includes normal retinal images as well as images of 9 common eye diseases (RVO, AMD, DR, RD, HM, Uveitis, VH, PRD, RH) and 6 rare retinal diseases (RAO, AH, CA, RP, OAF, PHPV). The distribution of images of each category in the training set, validation set, and external test set are detailed in Table 1.
Evaluation of model diagnostic performance
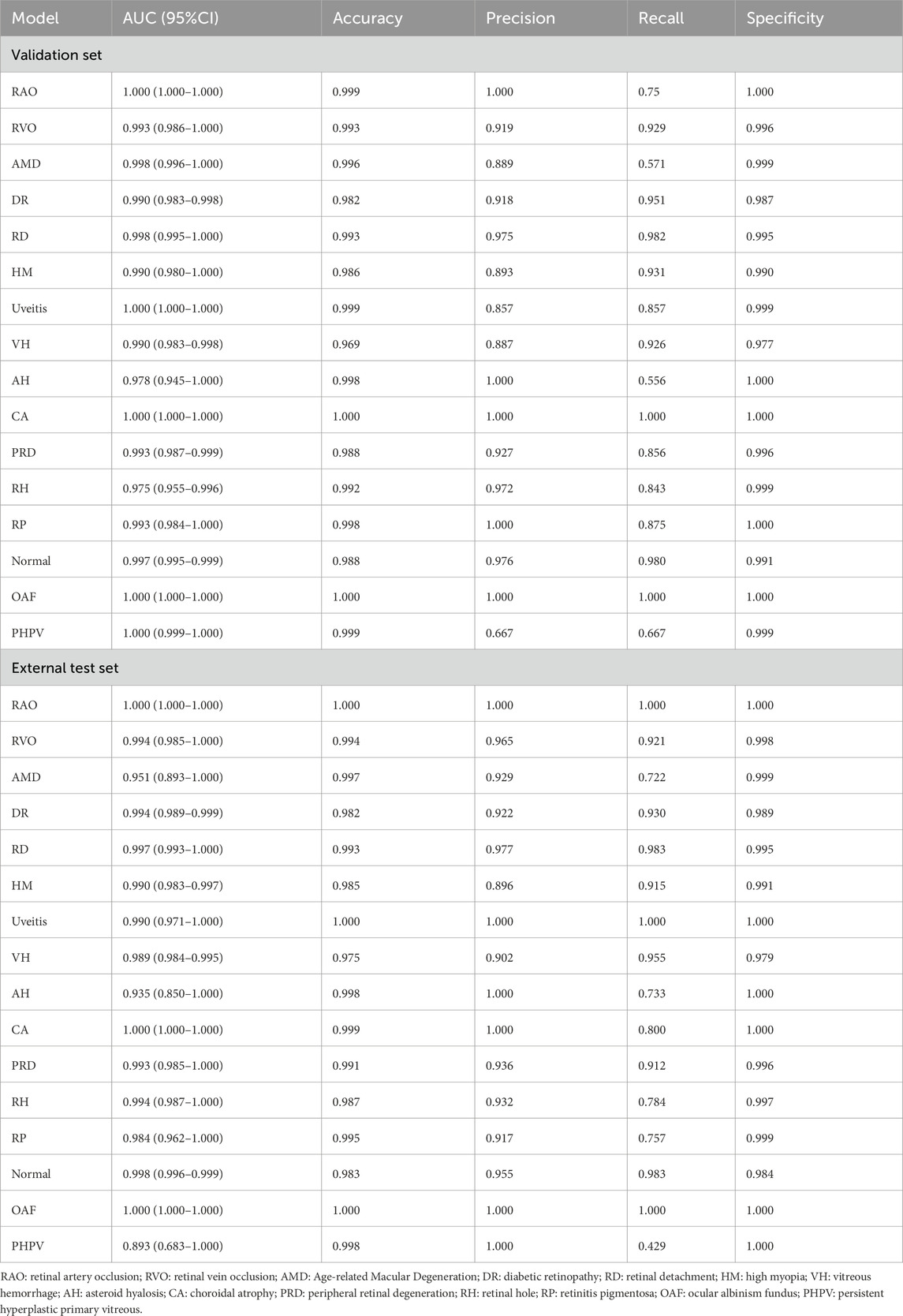
Our deep learning model has exhibited exceptional performance in diagnosing nine common eye diseases (RVO, AMD, DR, RD, HM, Uveitis, VH, PRD, and RH). On the validation set, the AUC values for all diseases exceeded 0.975, with accuracy rates consistently above 0.980. Notably, the AUC values for RVO, AMD, DR, RD, HM, Uveitis, PRD, and RH reached or exceeded 0.990, while the accuracy rates for AMD, RD, Uveitis, and RH surpassed 0.990. These results underscore the model’s high precision and robust discriminative ability in identifying these prevalent ocular conditions. On the external test set, the model maintained strong performance, achieving AUC values above 0.950 and accuracy rates exceeding 0.970. This further validates its reliability and stability in real-world applications.
For six rare retinal diseases, the model also demonstrated superior diagnostic capabilities. On the validation set, the AUC values for these diseases were all above 0.970, with accuracy rates reaching as high as 0.998. On the external test set, the AUC values remained above 0.893, and the accuracy rates exceeded 0.991. Despite the relatively small sample size of rare diseases, which may pose challenges to the model’s generalization, its performance remained consistently high. This highlights the model’s significant potential and reliability in diagnosing rare retinal diseases. The detailed diagnostic performance indicators of the model are presented in Table 2, and the ROC curves and confusion matrices for different types of retinal diseases are shown in Figure 2.
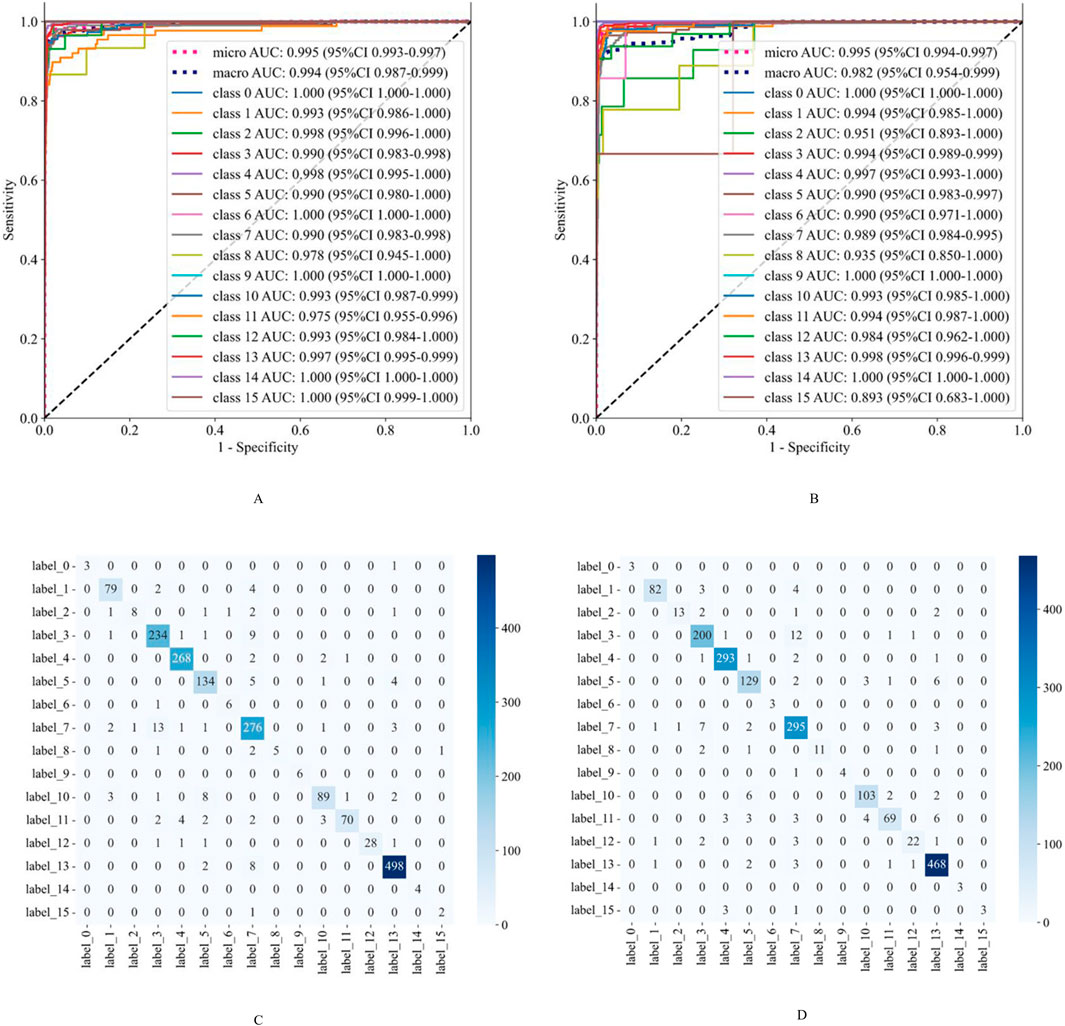
Figure 2. The classification performance evaluation results of the model on the validation set and the external test set. (A,B) are the ROC curves of the model on the validation set and the external test set, respectively. (C,D) are the confusion matrices of the model on the validation set and the external test set, respectively. The rows of the confusion matrix represent the actual categories, and the columns represent the predicted categories. Class 0: Retinal Artery Occlusion (RAO); Class 1: Retinal Vein Occlusion (RVO); Class 2: Age-related Macular Degeneration (AMD); Class 3: Diabetic Retinopathy (DR); Class 4: Retinal Detachment (RD); Class 5: High Myopia (HM); Class 6: Uveitis; Class 7: Vitreous Hemorrhage (VH); Class 8: Asteroid Hyalosis (AH); Class 9: Choroidal Atrophy (CA); Class 10: Peripheral Retinal Degeneration (PRD); Class 11: Retinal Hole (RH); Class 12: Retinitis Pigmentosa (RP); Class 13: Normal; Class 14: Ocular Albinism Fundus (OAF); Class 15: Persistent Hyperplastic Primary Vitreous (PHPV).
Grad-CAM visualization
This study utilized the Grad-CAM technique to visually interpret the decision-making logic of the model. Its clinical diagnostic value has been strictly verified by ophthalmologists specializing in retinal diseases in tertiary hospitals. As shown in Figure 3, the visualization results exhibit a high degree of consistency with the typical pathological features of various retinal diseases. For retinal vascular diseases, the heat map of RVO highlighted the abnormally dilated venous vessels, whereas that of RAO emphasized the characteristic cherry red spot. In macular diseases, the significant regions of AMD were predominantly distributed in the lesion areas of the macula. The hotspots of DR exhibited prominent pathological features such as microaneurysms, hemorrhage, and hard exudates. The heat map of RD was prominently concentrated in the neuroepithelial layer detachment region. For HM, the feature activation regions were localized in the optic disc and choroidal atrophy lesions. The visualization results of peripheral retinal degeneration and holes accurately pinpointed the areas of retinal tissue defects. The heat distribution of uveitis was predominantly concentrated in the inflammatory exudation area. The significant mapping of VH precisely marked the suspended blood clots. The heat map of RP focused on the areas with dense pigment deposition, while the activation regions of CA were located in the thinned choroid. The area of interest for OAF corresponded to abnormal vessel regions, and the heat distribution of PHPV aligned accurately with the residual embryonic vitreous tissue. This high consistency with clinical pathological features substantiated the medical rationality of the model’s diagnostic decisions.
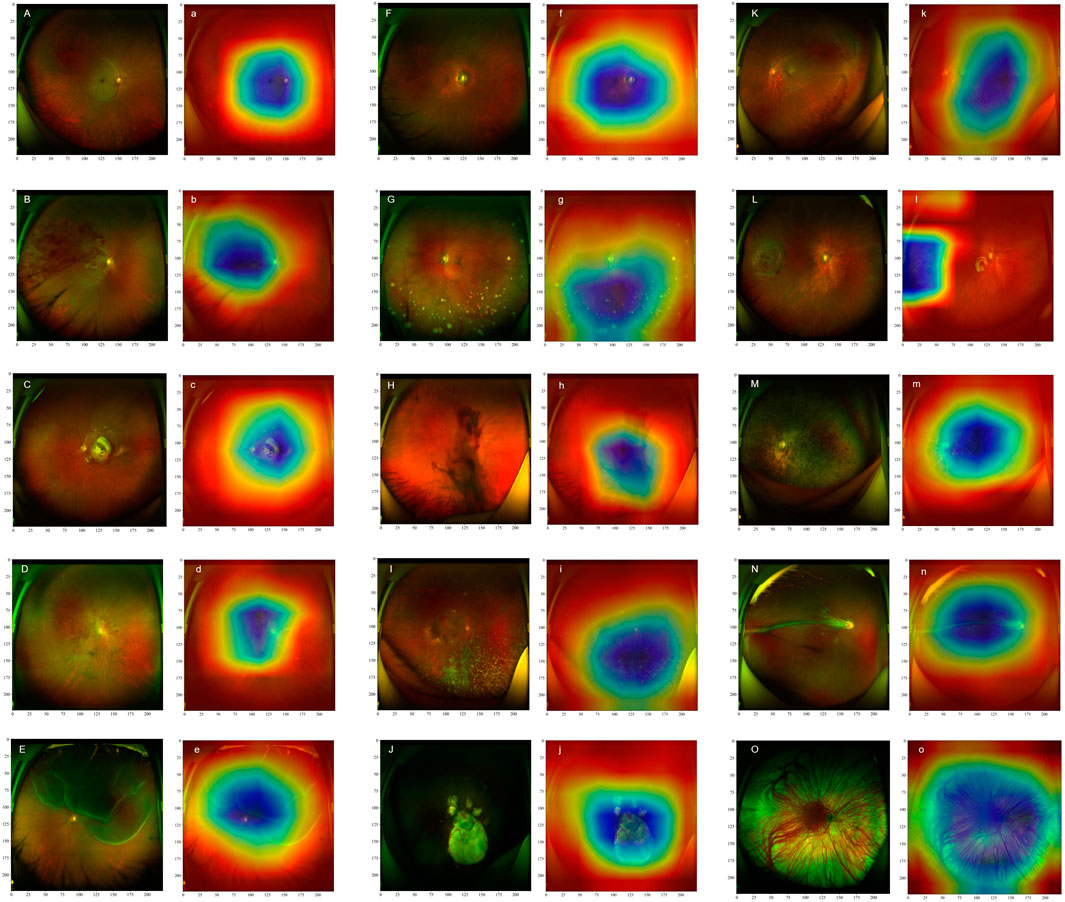
Figure 3. Ultra-Wide-Field (UWF) fundus images of different retinal diseases (marked with capital letters) and their corresponding heat maps (marked with lowercase letters) are presented. The heat maps illustrate the significance of image features for the DenseNet121 model, with different colors indicating the extent to which the model focuses on these features during classification. Specifically, blue areas highlight the regions that receive the greatest attention from the model during classification, whereas red areas correspond to regions with relatively lower attention. Retinal Artery Occlusion (A,a): Retinal Vein Occlusion (B,b); Age-related Macular Degeneration (C,c); Diabetic Retinopathy (D,d); Retinal Detachment (E,e); High Myopia (F,f); Uveitis (G,g); Vitreous Hemorrhage (H,h); Asteroid Hyalosis (I,i); Choroidal Atrophy (J,j); Peripheral Retinal Degeneration (K,k); Retinal Hole (L,l); Retinitis Pigmentosa (M,m); Ocular Albinism Fundus (N,n); Persistent Hyperplastic Primary Vitreous (O,o).
Comparison of diagnostic performance between ophthalmologists and AI models
We compared the artificial intelligence model with ophthalmologists of different levels. The results showed that the artificial intelligence model demonstrated higher accuracy in all disease diagnosis tasks compared to intermediate and junior ophthalmologists, approaching 1. The accuracy of intermediate ophthalmologists was slightly lower than that of the AI model but still maintained relatively high performance, with accuracy values all above 0.8. Junior ophthalmologists had the lowest accuracy among the three groups, ranging from 0.6 to 0.9. The results are shown in Figure 4.
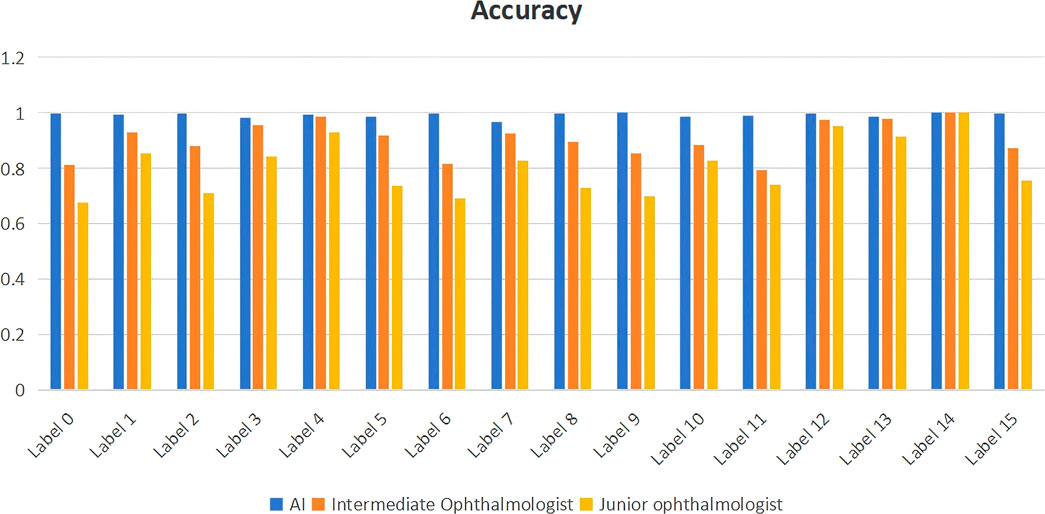
Figure 4. Comparison of diagnostic performance between ophthalmologists of different levels and artificial intelligence models. label 0: Retinal Artery Occlusion (RAO); label 1: Retinal Vein Occlusion (RVO); label 2: Age-related Macular Degeneration (AMD); label 3: Diabetic Retinopathy (DR); label 4: Retinal Detachment (RD); label 5: High Myopia (HM); label 6: Uveitis; label 7: Vitreous Hemorrhage (VH); label 8: Asteroid Hyalosis (AH); label 9: Choroidal Atrophy (CA); label 10: Peripheral Retinal Degeneration (PRD); label 11: Retinal Hole (RH); label 12: Retinitis Pigmentosa (RP); label 13: Normal; label 14: Ocular Albinism Fundus (OAF); label 15: Persistent Hyperplastic Primary Vitreous (PHPV).
Discussion
This study developed a deep learning model integrating DenseNet121 and XGBoost for diagnosing 16 types of retinal diseases in UWF fundus images. The results demonstrated that the model achieved exceptional diagnostic performance on both the validation set and the external test set. On the validation set, for common retinal diseases such as DR, AMD, and RVO, the model exhibited nearly perfect classification accuracy, with AUC values exceeding 0.975 and accuracy rates above 0.980. For rare retinal diseases such as PHPV, OAF, and RP, the model also demonstrated robust diagnostic capabilities, achieving AUC values above 0.970 and accuracy rates above 0.998. On the external test set, the model’s generalization ability was thoroughly validated. For common retinal diseases, AUC values exceeded 0.950 and accuracy rates were above 0.970; for rare retinal diseases, AUC values remained above 0.893 and accuracy rates were above 0.991. Consequently, the method proposed in this study holds significant clinical application potential in the field of intelligent diagnosis of retinal diseases.
It is important to highlight that the model’s superior performance in diagnosing both common and rare diseases does not signify overfitting. First, the complete independence between the training set and the external test set ensures no data leakage occurs (Chicco and Shiradkar, 2023; Gan et al., 2022). Second, the enhanced ability of the model to recognize rare diseases with limited samples can be attributed to the hybrid model architecture, which effectively alleviates the bias arising from class imbalance (Senan et al., 2022). Additionally, Grad-CAM visualization confirms that the regions the model focuses on align well with clinical lesions, thereby reinforcing the medical validity of its diagnostic outcomes (Long et al., 2025; Chen et al., 2021). Consequently, the high accuracy achieved by the model demonstrates its strong generalization capability rather than mere memorization of the training data.
Compared with the single-disease detection models of Ghasemi Falavarjani et al. (2017), Christ et al. (2024), this study, for the first time, realized comprehensive identification of 16 types of retinal diseases within a unified framework. It covered nine common retinal diseases, such as RVO, AMD, DR, RD, HM, Uveitis, VH, PRD, and RH, as well as six rare retinal diseases, including RAO, AH, CA, RP, OAF, and PHPV. The study abandoned the characteristic of traditional end-to-end deep learning models that rely on large-scale labeled data. Instead, it used transfer learning to extract high-level image features with the pre-trained DenseNet121 on ImageNet. Its dense connection structure enhanced the joint capture ability of subtle lesions and wide-field anatomical features through cross-layer feature reuse. Compared with Zhang et al. (2021) who directly fine-tuned large-scale networks such as seResNext50, this strategy reduced the computational load while ensuring the accuracy of feature extraction. The XGBoost classifier allocated weights dynamically to high-dimensional features through the Boosting algorithm, effectively solving the classification bias of the DenseNet pure convolutional network in unbalanced category scenarios (Ashwini and Bharathi, 2024; Seo et al., 2022). This ensured that rare diseases with a sample size of only 4% still achieved high classification performance.
Furthermore, Grad-CAM heat maps reveal that the model’s focus areas are highly consistent with clinical gold standards (e.g., in DR screening, it accurately highlights microaneurysms and hard exudates rather than irrelevant hemorrhages or fibroproliferative foci). Notably, for rare diseases such as PHPV, the activated regions precisely correspond to the vitreous proliferation foci. While Nagasawa et al. (2018) achieved high-precision detection of macular holes using traditional CNN-based processing of ultra-wide-angle fundus images, their model exhibits limitations in complex pathological scenarios. Specifically, an explanation analysis indicates that optical distortions in input data can cause activation regions to shift toward non-lesion areas such as the optic disc. In contrast, the deep learning framework proposed in this study demonstrates significant technical advantages and clinical applicability. This capability provides traceable pathological evidence for AI-assisted decision-making, thereby enhancing clinicians’ trust in diagnostic results.
The model demonstrated outstanding diagnostic performance on both the validation set and the test set, significantly outperforming junior and intermediate doctors. Its core clinical value lies in breaking the excessive reliance on experience in traditional manual reading of films and fundamentally solving the problem of diagnostic inconsistency caused by experience differences. Through advanced technical means, this model effectively compensates for the shortage of ophthalmic medical resources and provides strong support for improving the accessibility and equalization of medical services (Gan et al., 2025a; Zhu et al., 2022).
Certainly, this study has several limitations. While we have demonstrated the efficacy of the hybrid model, its real-time diagnostic performance and economic impact in actual clinical settings require further validation through additional prospective clinical studies. Specifically, its application value in long-term follow-up cohorts for rare diseases, such as PHPV, remains to be fully explored. Although the model exhibits clinically meaningful discrimination capabilities for rare diseases, the variability in its recall rate highlights the need for cautious evaluation of its standalone diagnostic reliability. This discrepancy primarily arises from insufficient feature learning due to the limited availability of rare disease samples (Zuo et al., 2022). Furthermore, the current model’s reliance on a single imaging modality represents a significant constraint. For complex retinal pathologies, the lack of OCT and other multimodal imaging inputs may compromise diagnostic accuracy (Gan et al., 2025b). Future research should focus on multi-center prospective clinical validation, exploration of cross-modal deep learning fusion models, and optimization of data augmentation strategies tailored for rare diseases (Yan et al., 2024).
Conclusion
In summary, the deep learning model developed in this study enables automatic identification and classification of multiple ophthalmic diseases on UWF devices, demonstrating robust performance. This model holds potential for ophthalmic disease screening and clinical diagnosis, assisting ophthalmologists in making more accurate and efficient diagnoses, thereby enhancing physician diagnostic efficiency and patient care quality.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by the case-control study received approval from the Medical Research Ethics Committee and the Institutional Review Board of the First People’s Hospital of Jiujiang City. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
M-MD: Supervision, Methodology, Validation, Conceptualization, Investigation, Funding acquisition, Resources, Writing – review and editing, Visualization, Software, Project administration, Writing – original draft. XT: Investigation, Software, Resources, Project administration, Writing – review and editing, Supervision, Visualization, Methodology, Formal Analysis, Writing – original draft, Validation.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
Some of our experiments were carried out on OnekeyAI platform. Thanks OnekeyAI and its developers’ help in this scientific research work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ashrafkhorasani, M., Habibi, A., Nittala, M. G., Corradetti, G., Emamverdi, M., and Sadda, S. R. (2024). Peripheral retinal lesions in diabetic retinopathy on ultra-widefield imaging. Saudi J. Ophthalmol. Off. J. Saudi Ophthalmol. Soc. 38 (2), 123–131. doi:10.4103/sjopt.sjopt_151_23
Ashwini, P., and Bharathi, S. H. (2024). Continuous feature learning representation to XGBoost classifier on the aggregation of discriminative features using DenseNet-121 architecture and ResNet 18 architectures towards apraxia recognition in the child speech therapy. Int. J. Speech Technol. 27 (1), 187–199. doi:10.1007/s10772-024-10089-6
Chen, T.-C., Lim, W. S., Wang, V. Y., Ko, M. L., Chiu, S. I., Huang, Y. S., et al. (2021). Artificial intelligence–assisted early detection of retinitis pigmentosa — the Most common inherited retinal degeneration. J. Digit. Imaging 34 (4), 948–958. doi:10.1007/s10278-021-00479-6
Chicco, D., and Shiradkar, R. (2023). Ten quick tips for computational analysis of medical images. PLOS Comput. Biol. 19 (1), e1010778. doi:10.1371/journal.pcbi.1010778
Christ, M., Habra, O., Monnin, K., Vallotton, K., Sznitman, R., Wolf, S., et al. (2024). Deep learning-based automated detection of retinal breaks and detachments on fundus photography. Transl. Vis. Sci. Technol. 13 (4), 1. doi:10.1167/tvst.13.4.1
Dong, H., Xu, X., Wang, L., and Pu, F. (2018). Gaofen-3 PolSAR image classification via XGBoost and polarimetric spatial information. Sensors 18 (2), 611. doi:10.3390/s18020611
Flaxman, S. R., Bourne, R. R. A., Resnikoff, S., Ackland, P., Braithwaite, T., Cicinelli, M. V., et al. (2017). Global causes of blindness and distance vision impairment 1990-2020: a systematic review and meta-analysis. Lancet Glob. Health 5 (12), e1221–e1234. doi:10.1016/S2214-109X(17)30393-5
Gan, F., Cao, J., Fan, H., Qin, W., Li, X., Wan, Q., et al. (2025b). Development and validation of the artificial intelligence-proliferative vitreoretinopathy (AI-PVR) insight system for deep learning-based diagnosis and postoperative risk prediction in proliferative vitreoretinopathy using multimodal fundus imaging. Quant. Imaging Med. Surg. 15 (4), 2774–2788. doi:10.21037/qims-24-1644
Gan, F., Chen, W.-Y., Liu, H., and Zhong, Y. L. (2022). Application of artificial intelligence models for detecting the pterygium that requires surgical treatment based on anterior segment images. Front. Neurosci. 16, 1084118. doi:10.3389/fnins.2022.1084118
Gan, F., Liu, H., Qin, W.-G., and Zhou, S. L. (2023b). Application of artificial intelligence for automatic cataract staging based on anterior segment images: comparing automatic segmentation approaches to manual segmentation. Front. Neurosci. 17, 1182388. doi:10.3389/fnins.2023.1182388
Gan, F., Long, X., Wu, X., Ji, W., Fan, H., et al. (2025a). Deep learning-enabled transformation of anterior segment images to corneal fluorescein staining images for enhanced corneal disease screening. Comput. Struct. Biotechnol. J. 28, 94–105. doi:10.1016/j.csbj.2025.02.039
Gan, F., Wu, F.-P., and Zhong, Y.-L. (2023a). Artificial intelligence method based on multi-feature fusion for automatic macular edema (ME) classification on spectral-domain optical coherence tomography (SD-OCT) images. Front. Neurosci. 17, 1097291. doi:10.3389/fnins.2023.1097291
Ghasemi Falavarjani, K., Tsui, I., and Sadda, S. R. (2017). Ultra-wide-field imaging in diabetic retinopathy. Vis. Res. 139, 187–190. doi:10.1016/j.visres.2017.02.009
Grassmann, F., Mengelkamp, J., Brandl, C., Harsch, S., Zimmermann, M. E., Linkohr, B., et al. (2018). A deep learning algorithm for prediction of age-related eye disease study severity scale for age-related macular degeneration from color fundus photography. Ophthalmology 125 (9), 1410–1420. doi:10.1016/j.ophtha.2018.02.037
Horie, S., and Ohno-Matsui, K. (2022). Progress of imaging in diabetic retinopathy-from the past to the present. Diagn. Basel Switz. 12 (7), 1684. doi:10.3390/diagnostics12071684
Li, B., Chen, H., Yu, W., et al. (2024). The performance of a deep learning system in assisting junior ophthalmologists in diagnosing 13 major fundus diseases: a prospective multi-center clinical trial: 1. Npj Digit. Med. 7 (1), 1–11. doi:10.1038/s41746-023-00991-9
Li, Z., Wang, L., Wu, X., Jiang, J., Qiang, W., Xie, H., et al. (2023). Artificial intelligence in ophthalmology: the path to the real-world clinic. Cell. Rep. Med. 4 (7), 101095. doi:10.1016/j.xcrm.2023.101095
Long, X., Gan, F., Fan, H., Qin, W., Li, X., Ma, R., et al. (2025). EfficientNetB0-Based end-to-end diagnostic system for diabetic retinopathy grading and macular edema detection. Diabetes Metab. Syndr. Obes. Targets Ther. 18, 1311–1321. doi:10.2147/DMSO.S506494
Masumoto, H., Tabuchi, H., Nakakura, S., Ohsugi, H., Enno, H., Ishitobi, N., et al. (2019). Accuracy of a deep convolutional neural network in detection of retinitis pigmentosa on ultrawide-field images. PeerJ 7, e6900. doi:10.7717/peerj.6900
Nagasawa, T., Tabuchi, H., Masumoto, H., Enno, H., Niki, M., Ohsugi, H., et al. (2018). Accuracy of deep learning, a machine learning technology, using ultra-wide-field fundus ophthalmoscopy for detecting idiopathic macular holes. PeerJ 6, e5696. doi:10.7717/peerj.5696
Nagiel, A., Lalane, R. A., Sadda, S. R., and Schwartz, S. D. (2016). Ultra-widefield fundus imaging: a review of clinical applications and future trends. Retina Phila. Pa 36 (4), 660–678. doi:10.1097/IAE.0000000000000937
Nguyen, T. D., Le, D.-T., Bum, J., Kim, S., Song, S. J., and Choo, H. (2024). Retinal disease diagnosis using deep learning on ultra-wide-field fundus images: 1. Diagnostics 14 (1), 105. doi:10.3390/diagnostics14010105
Ohsugi, H., Tabuchi, H., Enno, H., and Ishitobi, N. (2017). Accuracy of deep learning, a machine-learning technology, using ultra-wide-field fundus ophthalmoscopy for detecting rhegmatogenous retinal detachment. Sci. Rep. 7 (1), 9425. doi:10.1038/s41598-017-09891-x
Rajalakshmi, R., Subashini, R., Anjana, R. M., and Mohan, V. (2018). Automated diabetic retinopathy detection in smartphone-based fundus photography using artificial intelligence. Eye Lond. Engl. 32 (6), 1138–1144. doi:10.1038/s41433-018-0064-9
Selvaraju, R. R., Cogswell, M., Das, A., et al. (2024). “Grad-CAM: visual explanations from deep networks via gradient-based localization,” in 2017 IEEE int. conf. comput. vis. ICCV.
Senan, E. M., Jadhav, M. E., Rassem, T. H., Aljaloud, A. S., Mohammed, B. A., and Al-Mekhlafi, Z. G. (2022). Early diagnosis of brain tumour MRI images using hybrid techniques between deep and machine learning. Comput. Math. Methods Med. 2022, 8330833. doi:10.1155/2022/8330833
Seo, Y., Jang, H., and Lee, H. (2022). Potential applications of artificial intelligence in clinical trials for alzheimer’s disease. Life 12 (2), 275. doi:10.3390/life12020275
Shoughy, S. S., Arevalo, J. F., and Kozak, I. (2015). Update on wide- and ultra-widefield retinal imaging. Indian J. Ophthalmol. 63 (7), 575–581. doi:10.4103/0301-4738.167122
Silva, P. S., Cavallerano, J. D., Sun, J. K., Noble, J., Aiello, L. M., and Aiello, L. P. (2012). Nonmydriatic ultrawide field retinal imaging compared with dilated standard 7-field 35-mm photography and retinal specialist examination for evaluation of diabetic retinopathy. Am. J. Ophthalmol. 154 (3), 549–559. doi:10.1016/j.ajo.2012.03.019
Silva, P. S., Cavallerano, J. D., Sun, J. K., Soliman, A. Z., Aiello, L. M., and Aiello, L. P. (2013). Peripheral lesions identified by mydriatic ultrawide field imaging: distribution and potential impact on diabetic retinopathy severity. Ophthalmology 120 (12), 2587–2595. doi:10.1016/j.ophtha.2013.05.004
Tan, C. S., Li, K. Z., and Sadda, S. R. (2019). Wide-field angiography in retinal vein occlusions. Int. J. Retina Vitr. 5 (Suppl. 1), 18. doi:10.1186/s40942-019-0163-1
Wei, Q., Chi, L., Li, M., Qiu, Q., and Liu, Q. (2025). Practical applications of artificial intelligence diagnostic systems in fundus retinal disease screening. Int. J. Gen. Med. 18, 1173–1180. doi:10.2147/IJGM.S507100
Wei, W., Anantharanjit, R., Patel, R. P., and Cordeiro, M. F. (2023). Detection of macular atrophy in age-related macular degeneration aided by artificial intelligence. Expert Rev. Mol. diagn. 23 (6), 485–494. doi:10.1080/14737159.2023.2208751
Wong, W. L., Su, X., Li, X., Cheung, C. M. G., Klein, R., Cheng, C. Y., et al. (2014). Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob. Health 2 (2), e106–e116. doi:10.1016/S2214-109X(13)70145-1
Wu, M., Lu, Y., Hong, X., Zhang, J., Zheng, B., Zhu, S., et al. (2022). Classification of dry and wet macular degeneration based on the ConvNeXT model. Front. Comput. Neurosci. 16, 1079155. doi:10.3389/fncom.2022.1079155
Yan, C., Zhang, Z., Zhang, G., Liu, H., Zhang, R., Liu, G., et al. (2024). An ensemble deep learning diagnostic system for determining clinical activity scores in thyroid-associated ophthalmopathy: integrating multi-view multimodal images from anterior segment slit-lamp photographs and facial images. Front. Endocrinol. 15, 1365350. doi:10.3389/fendo.2024.1365350
Ye, X., Huang, Y., and Lu, Q. (2022). Automatic multichannel electrocardiogram record classification using XGBoost fusion model. Front. Physiol. 13, 840011. doi:10.3389/fphys.2022.840011
Zhang, C., He, F., Li, B., Wang, H., He, X., Li, X., et al. (2021). Development of a deep-learning system for detection of lattice degeneration, retinal breaks, and retinal detachment in tessellated eyes using ultra-wide-field fundus images: a pilot study. Graefes Arch. Clin. Exp. Ophthalmol. Albr. Von. Graefes Arch. Klin. Exp. Ophthalmol. 259 (8), 2225–2234. doi:10.1007/s00417-021-05105-3
Zhang, Y., Ning, C., and Yang, W. (2025). An automatic cervical cell classification model based on improved DenseNet121. Sci. Rep. 15 (1), 3240. doi:10.1038/s41598-025-87953-1
Zhu, S., Lu, B., Wang, C., Wu, M., Zheng, B., Jiang, Q., et al. (2022). Screening of common retinal diseases using six-category models based on EfficientNet. Front. Med. 9, 808402. doi:10.3389/fmed.2022.808402
Keywords: deep transfer learning, ultra-widefield fundus images, retinal diseases, DenseNet121, XGBoost, multiple fundus diseases
Citation: Duan M-M and Tu X (2025) Deep learning-based classification of multiple fundus diseases using ultra-widefield images. Front. Cell Dev. Biol. 13:1630667. doi: 10.3389/fcell.2025.1630667
Received: 18 May 2025; Accepted: 03 July 2025;
Published: 17 July 2025.
Edited by:
Weihua Yang, Southern Medical University, ChinaReviewed by:
Yu-Chen Chen, Nanjing Medical University, ChinaYan Tong, Renmin Hospital of Wuhan University, China
Karthik Karunakaran, Prediscan Medtech, India
Copyright © 2025 Duan and Tu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiang Tu, dHV4aWFuZ0BtYWlsLnN5c3UuZWR1LmNu
 Ming-Ming Duan1
Ming-Ming Duan1 Xiang Tu
Xiang Tu