- Division of Molecular and Cellular Function, School of Biological Sciences, Faculty of Biology, Medicine and Health, The University of Manchester, Manchester, United Kingdom
The formation of a functional nervous system during development and its maintenance in adulthood rely on precise regulation of neural stem cell (NSC) proliferation and differentiation. During neurogenesis, progenitor cells use various cellular and molecular mechanisms to balance these processes. Among these, dynamic signal encoding, specifically ultradian oscillations, which are regular protein fluctuations occurring over a few hours, has emerged as a key mechanism underlying NSC fate decisions. In adults, reactivation of quiescent NSCs, proliferation, and differentiation are also controlled by ultradian oscillations. Furthermore, these ultradian dynamics signals are modulated by microRNAs and are considered critical for the ability of neural progenitors to transition between different states. Altogether, these findings may have potential significance for our understanding of NSC reactivation and differentiation in the context of injury or neurodegeneration. The mammalian spinal cord harbours endogenous multipotent NSCs that respond to injury but mostly generate astrocytes and do not undergo neurogenesis. By contrast, many anamniotes regenerate spinal cord neurons from endogenous progenitors, despite the same molecular signalling pathways being activated, suggesting that subtle differences in how these pathways are regulated may result in different regenerative outcomes. Whether oscillatory dynamics could influence the reactivation and differentiation of NSCs upon spinal cord injury remains to be determined. This review explores the role of transcription factor ultradian oscillations in neurogenesis and how microRNAs modulate them. Additionally, we examine evidence for the role of ultradian dynamics in the reactivation of quiescent NSCs and their potential significance for regenerative neurogenesis in the context of spinal cord injury.
1 Introduction
The embryonic development of the central nervous system (CNS) is a complex multi-step process that is characterised by the formation of the neural tube, neurogenesis, and gliogenesis, which are critical for establishing the structure and function of the CNS. This process begins with the symmetric division of neuroepithelial cells, the first type of neural stem cell (NSC), to expand the NSC population. These cells then undergo a morphological transformation into radial glial cells (RGCs). Subsequently, RGCs begin to divide asymmetrically, initially producing neurons and later generating glial cell types, including oligodendrocytes, ependymal cells, and astrocytes (Kageyama et al., 2015; Paridaen and Huttner, 2014; Imayoshi and Kageyama, 2014). While NSCs persist in the adult brain and support ongoing neurogenesis in localised regions of the CNS (Alvarez-Buylla et al., 2001), adult mammalian NSCs normally stay in a non-proliferative and undifferentiated state known as quiescence, infrequently transitioning to an active state to produce new neurons, thus having a limited overall cellular turnover (Kaise and Kageyama, 2024; Reynolds and Weiss, 1992; Doetsch et al., 1997; Zhou et al., 2022). Adult NSCs can be activated under pathological conditions, such as spinal cord injury. In mammals, which have limited regenerative capacity, they primarily differentiate into astrocytes. By contrast, in regenerative species such as zebrafish (Reimer et al., 2008) and salamanders (Tazaki et al., 2017), NSCs differentiate into neurons upon injury. In these species, radial glial cells within the spinal ependymal zone can undergo de novo neurogenesis to repair the spinal cord. Equivalent cell types exist in the mammalian spinal cord (Becker et al., 2018) and show latent differentiation potential that can be activated in the right conditions (Llorens-Bobadilla et al., 2020; Stenudd et al., 2022). Therefore, studying the mechanisms that activate regenerative neurogenesis in fish or amphibians can inform potential therapeutic approaches to human spinal cord injury.
The generation of neurons and glia during development and adulthood is tightly controlled by factors that regulate whether NSCs remain in an undifferentiated state, enter quiescence, or differentiate. In addition to asymmetric inheritance of neuronal fate determinants (Casas Gimeno and Paridaen, 2022), other cell mechanisms control the rate of differentiation to ensure the maintenance of a pool of undifferentiated progenitors over time. For example, cell-cell signalling that represses or promotes neurogenesis can act to delineate tissue domains with different rates of differentiation (Bally-Cuif and Hammerschmidt, 2003; Janesick et al., 2015; Kicheva et al., 2014), while juxtracrine signalling (e.g., Notch signalling and downstream transcription factors) can balance the rates of differentiation over time in NSC populations (Simpson, 1990); reviewed in Henrique and Schweisguth, 2019). Additionally, microRNAs that target transcription factors and other cell fate determinants regulate both cell fate and the timing of differentiation during neurogenesis and gliogenesis (reviewed in Rajman and Schratt, 2017).
Neural progenitor cell (NPC) differentiation is regulated by transcriptional regulatory networks that integrate signals from transcription factors that either promote or inhibit differentiation. Among these, members of the basic helix-loop-helix (bHLH) transcription factor family have diverse roles: some, like the HES/Her transcription factors, act as negative regulators of differentiation by repressing proneural genes (Hu and Zou, 2022). Proneural genes, including the Neurogenin family, NeuroD/NeuroM, and Ascl1, promote cell cycle exit and neuronal differentiation (Bertrand et al., 2002; Imayoshi and Kageyama, 2014; Dennis et al., 2019). Additionally, some bHLH factors, e.g., Neurog1, suppress gliogenesis (Sun et al., 2001) or, like the Olig gene family, contribute to both neuronal and glial differentiation (Lu et al., 2001; Sugimori et al., 2008). The specific outcomes of NPC fate decisions often depend on the combinations of bHLH proteins expressed, which can either promote or inhibit distinct lineages within the nervous system (Sugimori et al., 2007). HES/Her proteins are modulated by miRNAs, short non-coding RNAs which mediate sequence-specific repression of mRNAs by binding to the 3′ untranslated regions (UTRs) of their targets (Bartel, 2018). They are predicted to be key modulators of dynamical behaviour generated by interactions within gene regulatory networks (Lai et al., 2016) and are involved in various cellular activities including CNS development and homeostasis (Saliminejad et al., 2019).
Recent evidence has revealed changes in the expression dynamics of bHLH factors as cells differentiate and points to ways in which these dynamics can enable cell fate decisions. Here, we highlight the importance of the dynamic expression of transcription factors, the role of oscillatory gene expression, and how external factors such as microRNAs fine-tune oscillations during CNS development. Additionally, we discuss the significance of these dynamic processes for the activation of quiescent NSCs. We will attempt to address this complex dynamic interaction in the context of reactivation of neurogenesis during spinal cord repair and discuss possible reasons why mammals fail to regenerate neurons.
2 Neurogenesis in development: the role of oscillatory gene expression
Oscillatory dynamics can encode more information than only levels, including duration (how long a signal is on), frequency, fold change, and number of oscillations (Figure 1A; reviewed in Sonnen and Aulehla, 2014), thus diversifying the potential number of responses to the same signal. The ubiquity of oscillations in biological control systems has been attributed to the fact that signalling based on detection of frequency or fold change of a signal is relatively robust to noise (Rapp, 1987; Rapp et al., 1981; Sonnen and Aulehla, 2014). Oscillatory signalling can span several orders of magnitude and can be classified according to the periodicity of the oscillations. Widely known types are infradian and circadian oscillations, with periods longer than or around 24 h, respectively (Uriu, 2016; Laje et al., 2018). In this review, we will focus on a type of oscillation known as ultradian, with periodicities of a few hours (Isomura and Kageyama, 2014; Soto et al., 2020) as they have been implicated in the control of neurogenesis during development (Kageyama et al., 2020).
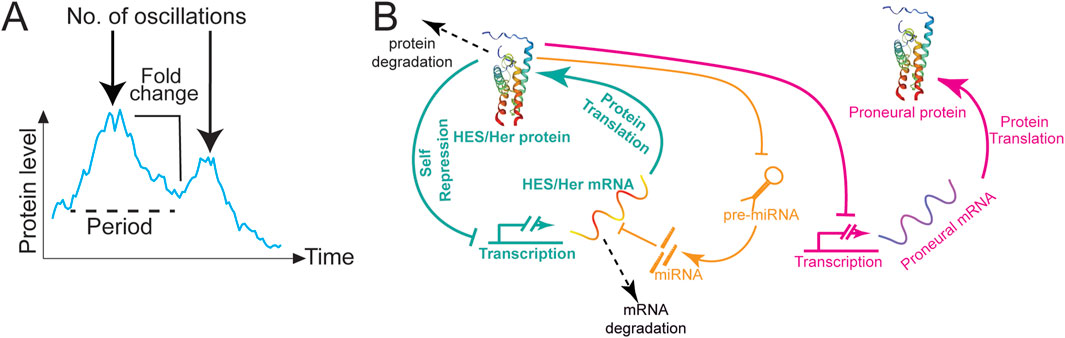
Figure 1. HES/Her oscillations in neural development. (A) Types of information encoded by oscillations. Dynamics of Her6:Venus protein expression modified from Doostdar et al. (2024). The graph depicts the intensity mean of a single cell over time (cyan line). The following dynamic parameters are highlighted: period between peaks or frequency (dashed black line); number of oscillations (black arrow); and fold-change (black line). (B) Schematic representation of the HES/Her autorepression network (green) including miRNA regulation (orange). HES/Her protein represses the transcription of HES/Her mRNA, leading to the periodic fluctuations in protein concentration shown in Figure 2A. miRNAs repress HES/Her mRNAs, stabilising oscillations or dampening them depending on miRNA levels. HES/Her proteins repress proneural gene expression (magenta), driving their oscillatory expression, shown in Figure 2A. Arrow lines: protein translation. T-bars: transcriptional repression. Dashed arrows: protein or mRNA degradation.
Recent evidence has highlighted ultradian oscillations as a powerful mechanism that facilitates cell state transitions. This relies heavily on their ability to encode more information than just protein levels. For example, ultradian oscillators allow the cell to control the period that a regulator is on, rather than controlling the concentration of active regulator (Albeck et al., 2013) and can also provide a more robust mechanism for implementing cell-autonomous developmental timers to determine when cells differentiate (Levine et al., 2012). They can also harness stochastic processes to control the timing of differentiation events (Miller et al., 2025; Soto et al., 2020) and may be important for the coordination of signalling pathways that drive cell fate changes (Kageyama et al., 2019; Purvis et al., 2012). Lastly, different protein expression dynamics can drive different outcomes during cell state transitions (Imayoshi et al., 2013; Maeda et al., 2023; Manning et al., 2019; Marinopoulou et al., 2021; Sueda et al., 2019), which will be reviewed in more detail below.
The mechanisms behind ultradian oscillations typically involve a negative feedback loop (Goodwin, 1965; Novak and Tyson, 2008) coupled with biological time delays (Lewis, 2003; Luo et al., 2023). These delays are intrinsically linked to various cellular processes, such as transcription and translation, as well as the stability of mRNA and proteins. For ultradian oscillations to be generated by negative feedback, relatively unstable mRNA and proteins are required (Bonev et al., 2012; Hirata et al., 2002; Kiparissides et al., 2011; Tan et al., 2012), and longer mRNA half-lives may result in longer oscillation periods (Kobayashi et al., 2009). In addition, mathematical models of transcription factor oscillators predict that a sufficiently long delay in mRNA processing is required for sustained oscillations to occur (Lewis, 2003; Hirata et al., 2004; Monk, 2003b; Jensen et al., 2003). In agreement with the predictions, experimentally altering intron length of oscillatory genes involved in Notch signalling, such as Hes7 (Harima et al., 2013), Dll1 (Shimojo et al., 2016) and Hes1 (Ochi et al., 2020), has been shown to influence the duration of time delays in a transcription-translation feedback loop, thus modulating the overall frequency and stability of oscillations (Ochi et al., 2020; Shimojo et al., 2016).
Among the most well-characterized ultradian oscillators expressed in NSCs and NPCs are the downstream targets of Notch signalling, Hairy and Enhancer of Split (HES/Her) transcriptional inhibitors, which are key members of the bHLH protein family expressed in neural progenitors. HES/Her proteins bind E-box DNA sequences as homo- or heterodimers and recruit co-repressors to repress target gene expression (Hu and Zou, 2022). The fact that HES/Her factors generally act as transcriptional repressors and bind their own promoter (Hirata et al., 2002) means that they can potentially drive oscillatory expression of themselves and their targets through negative autoregulation Figure 1B; (Goodwin, 1965; Novak and Tyson, 2008). Indeed, Hes1 was shown to give rise to oscillations via feedback inhibition in mice (Hirata et al., 2002; Bessho and Kageyama, 2003; Bonev et al., 2012), and this is true for related genes in other vertebrates, including zebrafish (Lewis, 2003; Giudicelli et al., 2007; Brend and Holley, 2009). The dynamic oscillation of HES/Her proteins relies heavily on the interplay between transcriptional feedback and the inherent delays that occur during mRNA processing, translation, and protein accumulation (Monk, 2003a; Jensen et al., 2003). The HES/Her negative feedback loop likely drives oscillatory expression of the proneural genes Ngn2 and Ascl1 Figure 1B; (Imayoshi et al., 2013; Shimojo et al., 2008; Sueda et al., 2019) and the Notch ligand Dll1 (Shimojo et al., 2008), whereas other transcription factors, such as Olig2, may show HES/Her-independent oscillations (Imayoshi et al., 2013). Other ultradian oscillators are likely to be expressed in neural progenitors. However, in contrast to the presomitic mesoderm, where up to 100 cyclically expressed genes may be present in oscillatory gene networks (Krol et al., 2011), the asynchronous nature of oscillations in NSCs has precluded direct transcriptome-wide characterisation of oscillatory gene expression.
2.1 HES/Her ultradian oscillations in neural development
In vertebrates, HES/Her protein oscillations play crucial roles during neural development by coordinating the timing of cellular events necessary for proper tissue formation. HES/Her oscillations were initially described in somitogenesis (Aulehla and Pourquie, 2008; Cooke and Zeeman, 1976; Palmeirim et al., 1997) and more recently discovered in neurogenesis (Shimojo et al., 2008). This later discovery was because, unlike in somitogenesis, where oscillations are synchronous within the presomitic mesoderm, they tend to be asynchronous in NPCs and so detecting them requires appropriate reporters and single cell imaging (Imayoshi et al., 2015).
The first hints of the presence of oscillations in neurogenesis came from experiments where NPCs were sorted into subpopulations of high or low HES1 expression. These cells then re-established heterogeneous protein expression levels, suggesting that high or low levels of transcription factors represented different phases of oscillations, rather than stably different expression levels in subpopulations of cells, This was supported by studies using live imaging reporters for Dll1 (a Notch ligand that promotes HES/Her protein expression) and HES1, revealing that both mRNA expression and protein fusion oscillates in the mouse embryonic neuroepithelium in a Notch-dependent manner (Figure 2A) (Shimojo et al., 2016; Shimojo et al., 2008). Furthermore, the proneural gene Neurogenin2 (NGN2) is also expressed in an oscillatory manner in neural progenitors. Inhibition of Notch signalling, a condition known to promote neuronal differentiation, leads to downregulation of HES1 and a sustained upregulation of NGN2 and Dll1. This suggests that oscillatory expression of HES1 regulates the oscillations of NGN2 and Dll1, which in turn contribute to the maintenance of neural progenitors through the mutual activation of Notch signalling (Shimojo et al., 2008). This cycling through states of high and low Notch component expression prompted researchers to ask whether gene expression oscillations have any significance for cell state transitions.
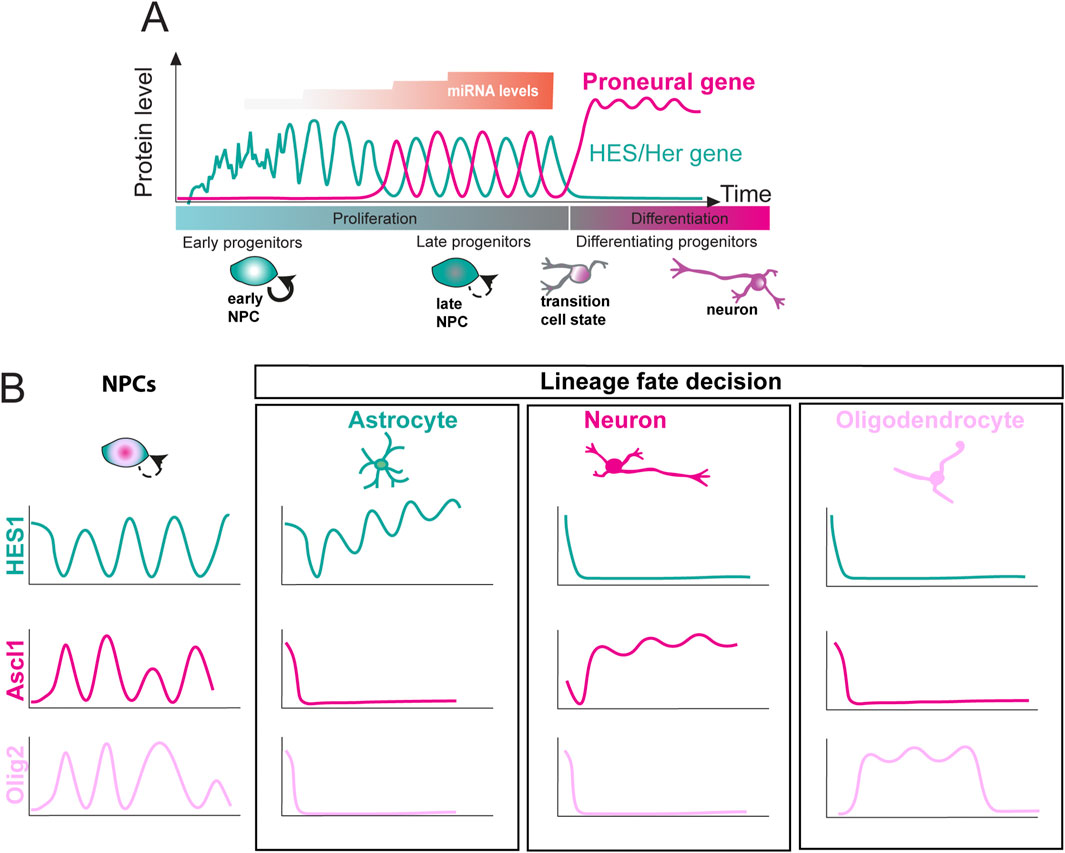
Figure 2. Expression dynamics of basic helix-loop-helix (bHLH) factors during neural and glial differentiation. (A) HES/Her and proneural protein oscillations over time during development (green and magenta lines, respectively). In early progenitors, HES/Her expression shows noisy fluctuations. miRNA levels increase over time in a stepwise manner (orange), stabilising HES/Her oscillations and reducing noise. This allows proneural gene expression, which oscillates out of phase with HES/Her genes in late progenitors. Upon differentiation, HES/Her proteins are downregulated, whereas proneural genes are upregulated with sustained dynamics. (B) Multiple bHLH factor cell fate determinants are co-expressed, and show oscillatory expression, in undifferentiated neural progenitor cells (NPCs). By contrast, upon commitment to a neural or glial fate, the expression of a single fate determinant becomes sustained, whereas the others are repressed. Ascl1 (magenta), Olig2 (pink), or Hes1 (green) becomes sustained in differentiating neurons (magenta), oligodendrocytes (pink), and astrocytes (green), respectively (after Kageyama et al., 2019).
2.2 Ultradian oscillations and neural cell fate choice
Some cues revealing the importance of oscillatory gene expression for cell fate choice could be gleaned from observing gene expression dynamics upon neuronal or glial differentiation. Multiple cell fate determinants, such as HES1, Ascl1s and Olig2, are co-expressed in undifferentiated neural progenitors and show oscillatory expression in the undifferentiated state (Figure 2B) (Imayoshi et al., 2013; Shimojo et al., 2008). By contrast, upon differentiation into neurons, oligodendrocytes, or astrocytes, a single fate determinant is typically expressed at high levels in a sustained, non-oscillatory manner, (Ascl1, Olig2 and HES1, respectively) which precedes the upregulation of downstream factors that drive differentiation (Figure 2B) (Imayoshi et al., 2013; Shimojo et al., 2008). This suggests that different (oscillatory vs. sustained) dynamic expression of transcription factors may mediate the choice between proliferation and differentiation, with oscillatory expression of proneural genes (i.e., Ascl1) promoting proliferation, and sustained expression priming progenitors for differentiation along one of the neural or glial lineages (Sueda and Kageyama, 2020).
There is compelling functional evidence to support this theory. For example, oscillatory induction of the proneural gene Ascl1 using a light-inducible gene expression construct promotes proliferation but does not rescue neurogenesis in Ascl1-deficient progenitors, whereas sustained expression results in neuronal differentiation. Furthermore, increasing the amplitude of the Ascl1 oscillations leads to more proliferation but no differentiation, arguing that the choice between proliferation and differentiation is determined by Ascl1 expression dynamics but not absolute protein levels (Imayoshi et al., 2013). Furthermore, manipulating gene expression delays by artificially lengthening or shortening the length of Dll1 or Hes1 (a progenitor marker) genes leads to the abrogation of Hes1 oscillations in neural progenitors. Mice carrying the lengthened or shortened alleles exhibit a reduction in brain size, likely due to accelerated neural differentiation and consequent depletion of the progenitor pool (Ochi et al., 2020; Shimojo et al., 2016), arguing that oscillatory Hes1 expression is important for normal progression of neural progenitor differentiation.
Why are oscillatory dynamics important for NPC state transitions? One possibility is that the oscillatory expression of factors repressing and promoting neurogenesis allows differentiation to take place. Indeed, two recent studies examining HES/Her transcription factor oscillations in the tissue context reported that differentiation of neural progenitors is preceded by a transition from noisy, aperiodic fluctuations in gene expression levels to stable, oscillatory dynamics Figure 2A; (Manning et al., 2019; Soto et al., 2020). For example, in the zebrafish hindbrain, a transition from noisy aperiodic expression to oscillatory dynamics can be observed in single neural progenitors expressing HES1/Her6 and correlates with the peak of neurogenesis as development progresses. Furthermore, preventing this transition by specifically abolishing the regulation of her6 by miR-9 results in a delay in neurogenesis, with cells typically being stuck in an intermediate stage of differentiation, suggesting that oscillatory transcription factor dynamics endorse the progression of cell fate decisions, and that oscillator noise is an important feature of the decoding of oscillatory expression (Soto et al., 2020).
2.3 miRNAs: fine-tuning ultradian oscillations during development
miRNAs can regulate their target mRNAs by inhibiting translation and/or promoting deadenylation and mRNA decay (Bagga et al., 2005; Jonas and Izaurralde, 2015; Olsen and Ambros, 1999; Standart and Jackson, 2007; Wu et al., 2006). The predominant mechanism depends on the context, with mRNA decay contributing more towards target gene repression at steady state and in post-embryonic cells (Baek et al., 2006; Hendrickson et al., 2009; Guo et al., 2010; Bazzini et al., 2012; Djuranovic et al., 2012; Eichhorn et al., 2014). Since both modes of regulation decrease protein output, regulation by miRNAs can influence the dynamics of genetic oscillators based on delayed negative feedback, including HES/Her gene oscillations (Hirata et al., 2002; Jensen et al., 2003; Lewis, 2003; Monk, 2003a).
Mathematical modelling has demonstrated that when translation inhibition and mRNA degradation are combined into a single decay parameter within a model of delayed negative feedback (autorepression), the system can produce sustained oscillations in gene expression. These oscillations occur only within a specific range of mRNA half-lives: if the mRNA is degraded too quickly or too slowly, the oscillations are dampened and eventually disappear (Xie et al., 2007), suggesting that miRNAs can influence oscillatory behaviour by modulating mRNA degradation rates.
Such regulation can cause a system to switch between oscillatory and non-oscillatory states, as demonstrated in several studies (Nandi et al., 2009; Zhou et al., 2012; Cai et al., 2013; Goodfellow et al., 2014; Li S. et al., 2016; Gao et al., 2020). For example, in nervous system development, miRNA-9 has been shown to regulate the expression of progenitor genes, including HES/Her transcription factors (Darnell et al., 2006; Leucht et al., 2008; Shibata et al., 2008; Shibata et al., 2011; Bonev et al., 2011; Coolen et al., 2012; Tan et al., 2012), as well as proneural genes (Coolen et al., 2012). Depleting or overexpressing miRNA-9 results in dampened HES1 oscillations (Bonev et al., 2012; Tan et al., 2012), supporting the notion of a “window” of miRNA concentrations that can sustain transcription factor oscillations, predicted by (Xie et al., 2007). Reciprocally, HES1 also suppresses miRNA-9 precursor expression, forming a mutual repression loop (Bonev et al., 2012). Although this can generate out-of-phase oscillations between HES1 and the miRNA-9 precursor, the mature miRNA-9 is stable and accumulates in progenitors supporting a model where rising miRNA-9 levels initially sustain HES1 oscillations, but eventually suppress them, triggering a shift to a stable HES1-low, differentiated state.
Incorporating the mutual repression motif into mathematical models introduces bistability, enabling cells to adopt either HES1-low (differentiated) or HES1-high (quiescent) states (Goodfellow et al., 2014; Castella et al., 2000; Baek et al., 2006; Sang et al., 2008; Shimojo et al., 2008; Sueda et al., 2019). Notably, while quiescent cells can resume oscillations, differentiated cells are resistant to reactivation. Thus, miRNA-mediated control of HES1 dynamics may be key to balancing progenitor maintenance, quiescence, and differentiation. Moreover, in vivo studies using zebrafish embryos have shown that miRNA-9 fine-tunes HES1/Her6 oscillatory dynamics by optimizing noise characteristics, enabling progenitor cells to effectively transition into differentiated neural states. This reveals a novel mechanism by which miRNAs regulate temporal gene expression patterns during development (Soto et al., 2020; Figures 1B, 2A).
In summary, ultradian oscillations are important for the progression of neural progenitors through different stages of differentiation during development and are fine-tuned by post-translational factors such as miRNAs. However, it is not clear if these tuneable dynamic processes play a role in the context of nervous system dysfunction, such as neurodegeneration or injury to the CNS. There, differentiation of resident stem or progenitor cells, or the lack thereof, can contribute to recovery outcomes. The following section will explore the significance of ultradian oscillations in regulating NSC quiescence.
3 Gene expression dynamics and reactivation of quiescent progenitor cells: implications for neural repair
Notch signalling is known to regulate the transitioning of neural progenitors between quiescent and proliferative states. For example, high and sustained levels of HES1 expression is associated with areas of low proliferation, such as the midbrain-hindbrain boundary (Baek et al., 2006; Hirata et al., 2001) and HES1 overexpression prolongs G-phase and attenuates proliferation (Baek et al., 2006; Maeda et al., 2023), suggesting that Notch positively regulates quiescence in mouse embryos and NSCs. On the other hand, loss of the Notch1 receptor (Ables et al., 2010) or the Notch effector Rbpj (Ehm et al., 2010; Imayoshi et al., 2010) leads to premature differentiation and consequent depletion of NSC, while deletion of Dll1 decreases the number of quiescent stem cells and increases the number of activated and differentiated NPCs in the adult mouse brain (Kawaguchi et al., 2013) suggesting that Notch is key for maintaining a reservoir of quiescent stem cells in the adult mouse. Other Notch effectors and modulators, such as non-oscillatory expression of the Notch effector Hey1, and ID proteins, have been proposed to maintain neural stem cell quiescence (Harada et al., 2021; Tzeng et al., 2001). ID proteins can dimerise with HES1, disrupting its negative autoregulatory feedback loop while preserving its ability to regulate downstream target genes, leading to suppression of neurogenesis (Bai et al., 2007).
More recently, dynamic patterns of gene expression have been implicated in regulating NPC quiescence. Oscillatory and sustained HES1 dynamics have opposite effects on cell cycle progression: oscillatory HES1 expression supports proliferation and drives oscillations of p21, a negative regulator of the G1/S transition. Sustained expression upregulates p21 expression through inhibition of ERK signalling, thus suppressing proliferation (Maeda et al., 2023). Hes1 oscillates in both quiescent and activated cells but the expression levels of oscillatory HES1 are higher in the quiescent state. By contrast, Ascl1 is not observed in quiescent cells, but shows oscillatory expression in the activated state, and inducing oscillatory Ascl1 expression leads to stem cell activation in the adult mouse brain (Sueda et al., 2019). This suggests that oscillatory Ascl1 dynamics activate quiescent neural stem cells, and high levels of HES1 inhibit reactivation by suppressing Ascl1 (Figures 3A,B).
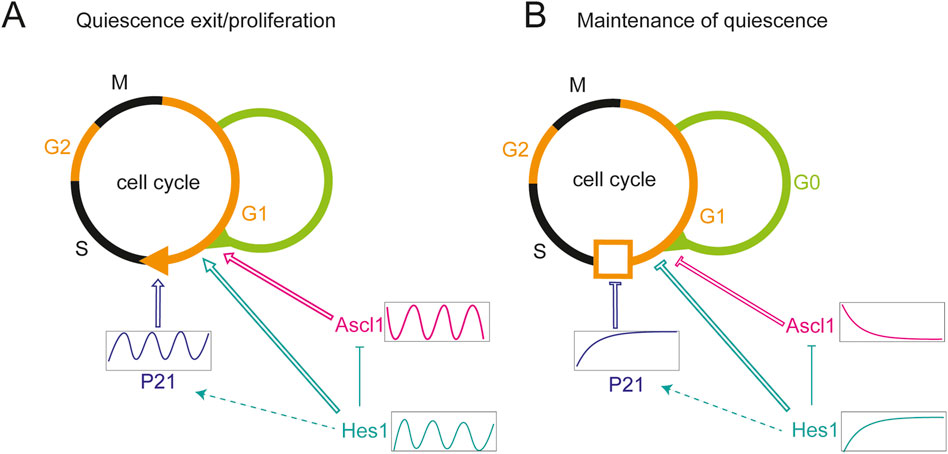
Figure 3. Regulation of proliferation and quiescence by HES/Her dynamics. (A) HES1 oscillations positively regulate proliferation by inducing oscillatory expression of P21, a negative regulator of the G1/S transition. Hes1 oscillations are permissive to quiescence exit. Oscillatory expression of Ascl1 promotes exit from quiescence, proliferation and differentiation of neural stem cells. Ascl1 oscillations may depend on Hes1 oscillations via transcriptional repression. (B) Sustained, high level expression of Hes1 results in high level expression of P21, inhibiting the G1/S transition. Sustained Hes1 likewise inhibits Ascl1 expression and exit from quiescence. Arrows and T-bars with open lines: activation and repression, respectively, on the level of molecular or cellular processes. Filled T-shape lines: direct repression on the molecular level. Dashed arrow: indirect activation on the molecular level. Hes1 (cyan), Ascl1 (magenta), p21 (blue).
Whilst Ascl1 oscillations can drive neural stem cell exit from quiescence, this likely depends on HES1 dynamics or protein levels. In a NSC culture model of quiescence, HES1 maintains its oscillatory expression across proliferative, quiescent, and reactivated states, indicating that oscillations are compatible with all three conditions. However, sustained HES1 expression, while supporting both proliferation and quiescence entry, prevents cells from exiting quiescence, suggesting that HES1 oscillations are required for quiescent cells to exit the dormant state (Marinopoulou et al., 2021; Figure 3B).
Emerging evidence suggests that oscillations in quiescent and proliferative NSCs also play a significant role in disease contexts. For instance, HES1 has been shown to oscillate in a proliferative breast cancer model (Sabherwal et al., 2021), Moreover, recent studies have demonstrated that HES1 dynamics changes during the dormancy and reactivation of breast cancer cells, and disruption of these oscillations prevents cell cycle re-entry and induces cell death (Cottrell et al., 2025). Interestingly, oscillatory expression of SOX2 has been observed in quiescent glioblastoma cells (Fu et al., 2024), reminiscent of HES1 oscillations in quiescent NSCs. Given that HES1 oscillations are known to facilitate the transition out of quiescence (Marinopoulou et al., 2021; Sueda et al., 2019), it is plausible to hypothesise that sustained, non-oscillatory SOX2 expression could prevent quiescent glioblastoma cells from reactivating, thereby impairing tumour reactivation and progression. Other studies have demonstrated that aged dormant NSCs, which have lost their proliferative and neurogenic potential, can regain juvenile like properties and resume functional neurogenesis following the delivery of the lentiviral vector iPaD (Kaise et al., 2022). Extraordinarily, this reprogramming induces oscillatory expression of Ascl1, a hallmark feature of active NSCs (Imayoshi et al., 2013; Sueda et al., 2019), underscoring the importance of studying of oscillation-based reactivation strategies in regenerative medicine (Kaise et al., 2022). Collectively, these findings highlight an expanding repertoire of dynamically regulated transcription factors orchestrating transitions between quiescence and proliferation in both disease and regeneration.
Another neurological condition relevant to address in the context of reactivation of neurogenesis is spinal cord injury (SCI). In mammals, it results in complete or partial paralysis due to the limited ability of the central nervous system (CNS) to repair. This is due to the lack of intrinsic capacity for axon regeneration and neurogenesis, impeding functional recovery after injury (see Table 1, for a comparison of processes occurring in non-regenerative and regenerative species) (Grossman et al., 2001). Interestingly, Notch signalling and its downstream HES/Her family transcription factors are reactivated in neural stem cells (NSCs) following SCI in both regenerative and non-regenerative species (Dias et al., 2012; Yamamoto et al., 2001). Additionally, ID genes, known positive regulators of quiescence, are triggered by injury (Tzeng et al., 2001) suggesting that ID proteins may modulate HES/Her dynamics in response to injury, thereby limiting neurogenesis and promoting the maintenance of a quiescent NSC pool. Another factor known to modulate HES/Her dynamics, named miR-9, has been implicated in the regrowth of injured axons in the larva, through repression of Her6 protein expression (Shen Y. et al., 2024). However, it remains unclear if dynamic gene expression is involved. Altogether, these findings hint that HES/Her expression dynamics (e.g., oscillatory versus non-oscillatory) may be relevant upon injury as it may influence the differentiation of resident NSCs or the lack thereof, and therefore might impact recovery outcomes. In the section below, we discuss the concept of ultradian oscillations in the context of quiescent NSC reactivation and neurogenesis during spinal cord repair.
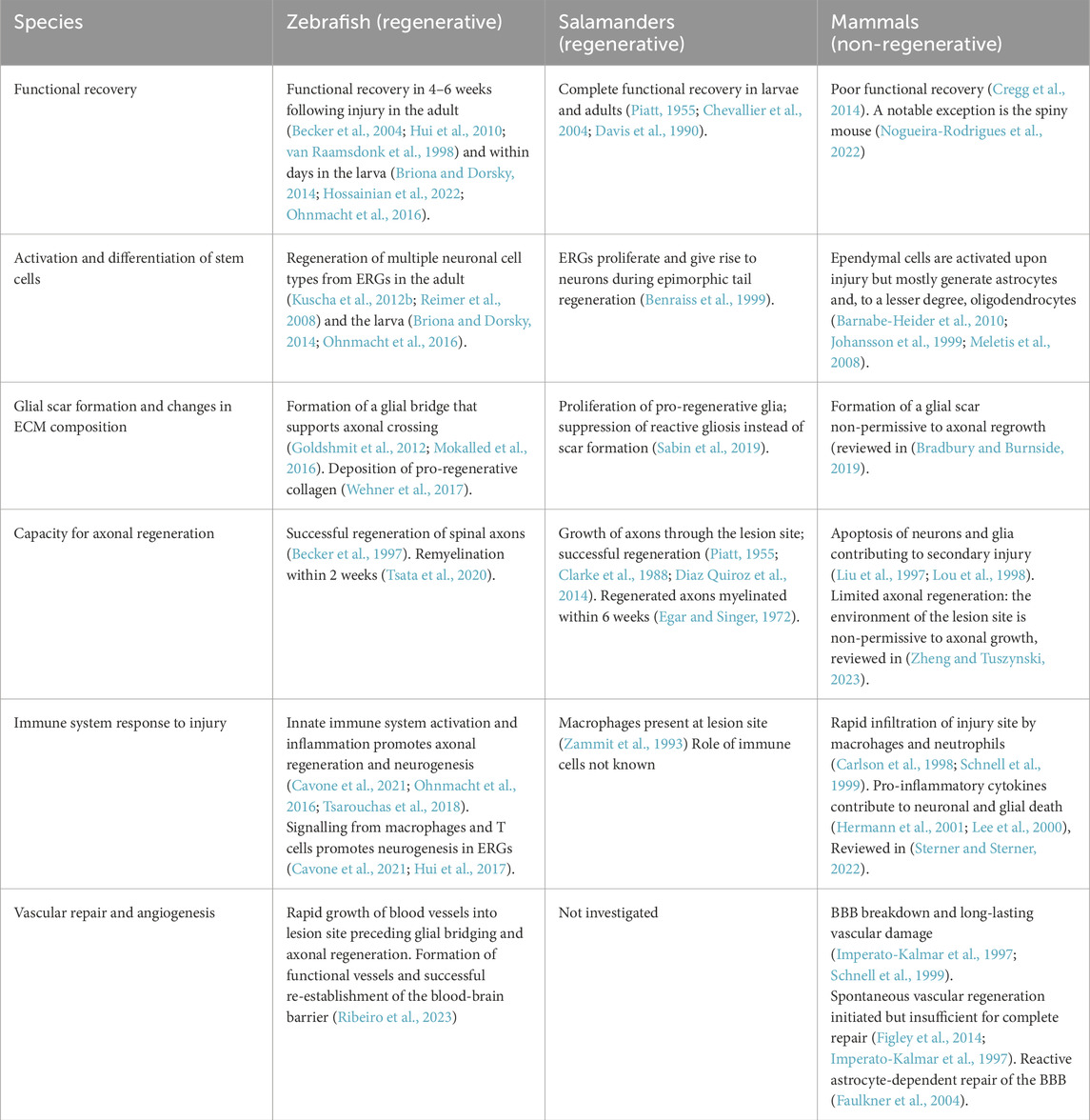
Table 1. Differences between regenerative responses in regenerative (zebrafish, salamanders) and non-regenerative (mammals) species.
4 Discussion
4.1 Reactivation of quiescent NSCs for spinal cord repair
In all vertebrate species studied so far, the adult CNS harbours NSCs that sustain constitutive neurogenesis, as well as latent NPCs that can be reactivated in response to lesions (Alunni and Bally-Cuif, 2016; Zhao et al., 2008). Nevertheless, in mammals, most of the CNS, except for NSCs of the ventricular zone (SVZ) of the lateral ventricles and the dentate gyrus of the hippocampus (Reynolds and Weiss, 1992), does not support the addition of new neurons in adulthood, and overall cellular turnover remains limited. However, quiescent cells with NSC-like properties can be isolated from various regions of the adult CNS, including the spinal cord. While these cells normally generate astrocytes and a small number of oligodendrocytes in the lesioned spinal cord, they have the potency to generate new neurons in vitro (Barnabe-Heider and Frisen, 2008; Barnabe-Heider et al., 2010; Deleyrolle et al., 2006; Doetsch et al., 1997; Johansson et al., 1999; Shihabuddin et al., 1997; Stenudd et al., 2022; Verma et al., 2008; Weiss et al., 1996). Their activation following injury suggests that the endogenous stem/progenitor cell niche may play a role in the limited regeneration observed after SCI (Ghosh and Hui, 2018; Vajn et al., 2013). Understanding the intrinsic properties and regenerative capacity of these endogenous glial progenitors and NSCs could significantly inform the development of SCI therapies.
To begin understanding the intrinsic mechanisms that promote neurogenesis after spinal cord injury, it is valuable to study regenerative species such as zebrafish and Salamanders (amniotes), and Xenopus larvae (Cardozo et al., 2017; Diaz Quiroz and Echeverri, 2013). Among these, zebrafish are particularly notable for their exceptional regenerative capacity, displaying full recovery of locomotor function even after complete spinal cord transection (Becker et al., 2004; Briona and Dorsky, 2014; Hossainian et al., 2022; Hui et al., 2010; Ohnmacht et al., 2016). This recovery is attributed to several key processes: the regeneration of lost neurons, generation of a permissive environment, remyelination of axons, and the re-establishment of functional neural circuits (Figure 4), further illustrated in a comparative analysis of regenerative and non-regenerative species (Table 1) (Becker et al., 2004; Becker and Becker, 2022; Becker et al., 1997; Briona and Dorsky, 2014; Cigliola et al., 2020; Mokalled et al., 2016; Tsata and Wehner, 2021; Vasudevan et al., 2021; Wehner et al., 2017).
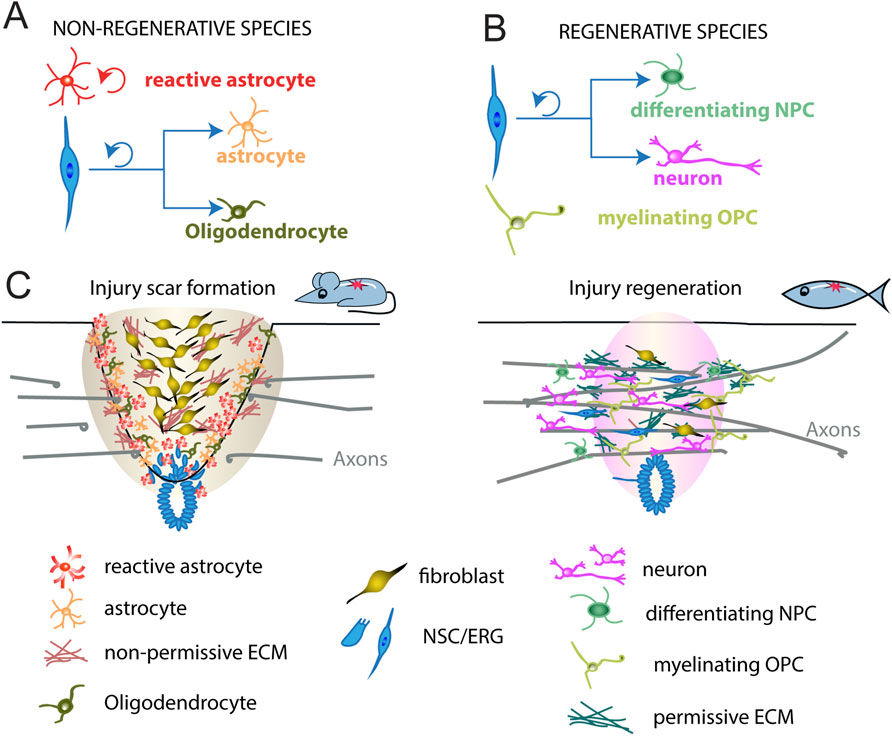
Figure 4. Different spinal cord repair in non-regenerative and regenerative species. (A) In non-regenerative animals, ependymal neural stem cells (blue) divide, leave the central canal region, and generate astrocytes (orange) and some oligodendrocytes (dark green) after spinal cord injury. Injury reactivates astrocytes to proliferate (red). (B) In regenerative animals, ependymo-radial cells (blue) divide, leave the central canal region, and generate new neurons (magenta) and neural progenitor cells undergoing differentiation (differentiating NPC, green). Injury reactivates OPCs to myelinate the axons undergoing repair (myelinating OPCs, light green). (C) In mammals, such as mouse, an injury scar is formed by invading immune cells and fibroblasts, as well as reactive astrocytes and oligodendrocytes, constituting a hostile environment to axon growth. In zebrafish, cells similarly respond to injury but, instead, generate new neurons, differentiating NPCs and myelinating OPCs to promote axonal regrowth (bridging) across the lesion site. Shown are major cell types underlying the different modes of repair in either species. NSC, neural stem cells; ERG, ependymo-radial glia. OPC, oligodendrocyte progenitor cell; NPC, neural progenitor cells; ECM, extracellular matrix.
In adult zebrafish, SCI triggers the reactivation and proliferation of progenitor cells known as ependymo-radial glia (ERGs; (Becker and Becker, 2015; Goldshmit et al., 2012; Hui et al., 2010; Reimer et al., 2009; Reimer et al., 2008). These cells possess the capacity to regenerate multiple neuronal subtypes, including motor neurons, glutamatergic interneurons, and GABAergic interneurons, within the dorso-ventral domains equivalent to those present during spinal cord development (Jessell, 2000; Becker and Becker, 2015; Becker et al., 2018; Kuscha et al., 2012b). Under normal conditions, ERGs divide slowly and primarily give rise to oligodendrocytes (Ohnmacht et al., 2016; Reimer et al., 2013; Park et al., 2007). However, following injury, they can revert to a neurogenic state (Figures 4B,C; reviewed in Becker and Becker, 2022). Similar ependymal cells are present in mammals and are triggered to proliferate after injury, but predominantly undergo gliogenesis, contributing to the formation of scar tissue (Figures 4A,C; Gregoire et al., 2015; Stenudd et al., 2015). Interestingly, unlike adults, neonatal mice exhibit enhanced spinal cord regeneration after injury, characterized by an increase in ependymal cell-like populations (Ikeda-Yorifuji et al., 2022). Similarly, juvenile mice display upregulated neuronal markers in glial cells following SCI, suggesting that regenerative neurogenesis diminishes with age and is largely lost by adulthood (Zhao et al., 2025). This divergence in injury response between species highlights the value of studying how neural stem cells (or ERGs) can be reactivated and directed toward neuronal fates following injury.
Functional studies in species that successfully regenerate spinal cord tissue after injury have identified several signalling pathways and downstream transcription factors that regulate the response of ependymal cells to injury (Table 2). This includes the reactivation of developmental signalling pathways that promote ependymal cell proliferation and neurogenesis, such as Hedgehog (Reimer et al., 2009), as well as pathways that negatively regulate both proliferation and differentiation, such as Notch (Dias et al., 2012). Presumably, the balance of these signals is important to ensure the regeneration of neurons whilst avoiding the depletion of the ependymal stem cell pool. Likewise, the transcription factor Sox2, a regulator of neural stem cell behaviour in development, is implicated in the proliferation of ependymal cells in response to injury (Fei et al., 2014; Ogai et al., 2014). Furthermore, regenerative neurogenesis in the spinal cord involves the reactivation of more than just embryonic developmental programs. For instance, macrophages recruited to the lesion site directly signal to ependymal progenitors via the cytokine Tnf-α to promote neurogenesis (Cavone et al., 2021) and neurotransmitter release following injury also regulates neurogenesis (Barreiro-Iglesias et al., 2015; Chang et al., 2021; Reimer et al., 2013). How these diverse signals converge to regulate proneural gene expression, and thereby control the rate and extent of neurogenesis, underscores the complexity of the regenerative processes making it an important area of investigation. Elucidating the dynamic gene regulatory networks that orchestrate the reactivation of neurogenesis will form the basis for uncovering the mechanisms that underlie successful regenerative outcomes.
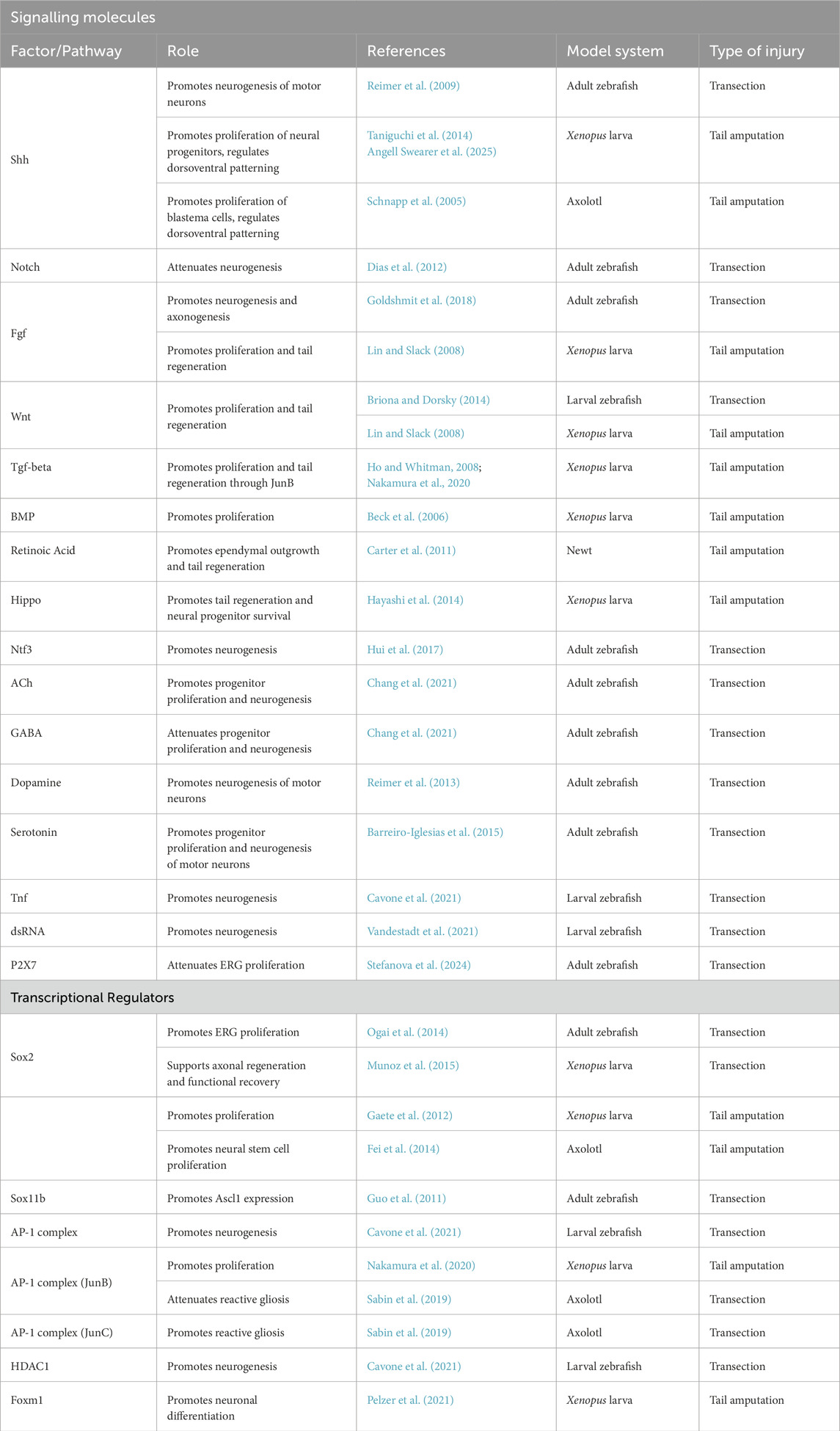
Table 2. Signalling pathways and transcription factors implicated in regenerative neurogenesis. The following criteria were considered: (1) the availability of functional data in a regenerative species; (2) Pathway/factor is involved in regulation of neural progenitor behaviour during spinal cord regeneration (tissue regeneration or epimorphic regeneration).
Ultradian oscillations of HES/Her family transcription factor expression has been demonstrated to pace neurogenesis during neural development in the spinal cord (Biga et al., 2021; Manning et al., 2019). Therefore, it is plausible that precise temporal control of these oscillatory patterns is also required for effective regeneration of spinal neurons from progenitors. Notably, the decline in NSC function with age, characterized by reduced proliferative and neurogenic capacity, can be reversed, leading to the reactivation of functional neurogenesis. The underlying mechanism involves the reactivation of oscillatory Ascl1 expression, a well-established downstream target of Hes1 and a defining feature of activated NSCs (Imayoshi et al., 2013; Sueda et al., 2019), suggesting the possibility that NSC reactivation in neuronal regeneration could be driven by transcription factor oscillations (Kaise et al., 2022). Since ultradian oscillations have only recently emerged as key mechanisms involved in quiescence exit, cell fate decisions, and pathological contexts, investigating gene expression dynamics following SCI in both regenerative and non-regenerative systems could provide valuable insights. A deeper understanding of these temporal patterns may inform the development of novel therapeutic strategies aimed at enhancing spinal cord regeneration.
4.2 Notch signalling dynamics and the regulation of NSC quiescence: implications for spinal cord repair
Adult quiescent NSCs isolated from the mouse spinal cord retain the potential to reactivate and generate multiple cell types in vitro (Weiss et al., 1996). However, this potential appears to be restricted in vivo, and following spinal cord injury is mostly limited to the generation of astrocytes, likely due to the activation of Notch signalling in the NSC niche (Yamamoto et al., 2001). Paradoxically, Notch signalling is also upregulated in adult zebrafish following SCI (Dias et al., 2012), yet in this context, it supports successful reactivation of neurogenesis and regeneration of multiple neuronal subtypes (Kuscha et al., 2012a; Kuscha et al., 2012b; Reimer et al., 2009; Reimer et al., 2008). Therefore, details in how the Notch signalling pathway is regulated may be important for whether regenerative neurogenesis takes place. Given the established role of Notch signalling oscillations in regulating progenitor maintenance, neural cell fate decisions and NSC quiescence, it would be pertinent to investigate whether the downstream bHLH transcription factors dynamics may be facilitators of regenerative neurogenesis in species that successfully repair their spinal cords. Understanding the context dependent role of these dynamics could thus provide critical insight into why regenerative capacity is lost in mammals and how it might be therapeutically restored.
During development, the Notch target Ascl1 is required for the specification of both neurons and oligodendrocytes (Parras et al., 2004; Sugimori et al., 2008) and its expression during oligodendrogenesis is regulated by HES5 (Kondo and Raff, 2000; Liu et al., 2006), which exhibits oscillatory expression in the embryonic mouse spinal cord (Manning et al., 2019). Notably, Ascl1 itself oscillates in oligodendrocyte precursor cells, albeit at lower amplitude than in neuronal precursors. Sustained Ascl1 expression biases cell fate toward neurons at the expense of oligodendrocytes (Sueda and Kageyama, 2021). These observations suggest that Ascl1 expression dynamics, driven by HES/her dynamics, may act as a critical regulatory mechanism in determining the balance between neuronal and glial fates.
In adult mammalian NSCs, Ascl1 is required for the reactivation of quiescent cells (Andersen et al., 2014), and its oscillatory expression drives the transition from quiescence to neuronal differentiation in mice neurogenic zones (Sueda et al., 2019). This process is likely governed by HES1 oscillations (Chen et al., 1997; Imayoshi et al., 2013; Sueda et al., 2019). For example, Marinopoulou et al. (2021) showed that NSCs exhibiting sustained HES1 expression failed to exit quiescence, potentially due to the inability to reinitiate Ascl1 oscillations (Figure 3). Ascl1a expression is detected in differentiating neurons after SCI in adult zebrafish (Guo et al., 2011) while in adult rats, Ascl1 is expressed in a small number of oligodendrocyte progenitors and its overexpression stimulates the production of oligodendrocytes instead of neurons (Ohori et al., 2006). Whether differences in the expression dynamics of Ascl1 could influence differentiation outcomes upon SCI, for example by inducing a fate switch between choice of neurons or oligodendrocytes, remains to be investigated.
4.3 miRNAs in spinal cord repair: regulators of gene expression dynamics?
miRNAs have been implicated in the pathogenesis of spinal cord injuries (Bhalala et al., 2013), showing dysregulated expression following SCI (Li P. et al., 2016; Liu et al., 2009; Shen W. et al., 2024; Strickland et al., 2011; Tang et al., 2014; Yunta et al., 2012). Thus, manipulation of miRNAs has received considerable interest as a strategy for treating SCI (Li P. et al., 2016). There is some evidence that modulation of miRNA expression can enable the creation of a regeneration-permissive environment (Diaz Quiroz et al., 2014). For example, miRNA-9 has been implicated in the regulation of her6 to facilitate regrowth of injured axons in the zebrafish larva (Shen Y. et al., 2024). Furthermore, it has been shown that activation of miRNA-9 leads to enhanced neuronal survival and reduced apoptosis in rodents (He et al., 2022; Wang et al., 2021) and miRNA-219, through HES5, promotes oligodendrocyte differentiation (Wang et al., 2017). Further studies have provided evidence that miRNAs regulate proneural genes. For example, in rodents, miRNA-20a and miRNA-486 are upregulated following SCI (Jee et al., 2012a; Jee et al., 2012b; Liu et al., 2009). miRNA-20a directly targets Neurogenin 1 (Ngn1), while miRNA-486 suppresses NeuroD6 leading to impaired neuronal survival and thus suggesting that their suppression would improve recovery of SCI. Since transcription factor dynamics coupled with miRNA feedback has been suggested to endow neural progenitors with the ability to enter reversible quiescence and progress to neuronal differentiation (Bonev et al., 2012; Goodfellow et al., 2014; Soto et al., 2020), it will be interesting to learn how miRNAs regulate the underlying cellular processes contributing to spinal cord repair as well as ascertain whether miRNA-regulated dynamics could influence cell fate decisions upon SCI. Some miRNAs whose function has been tested in vivo in animal models of SCI are listed in Table 3.
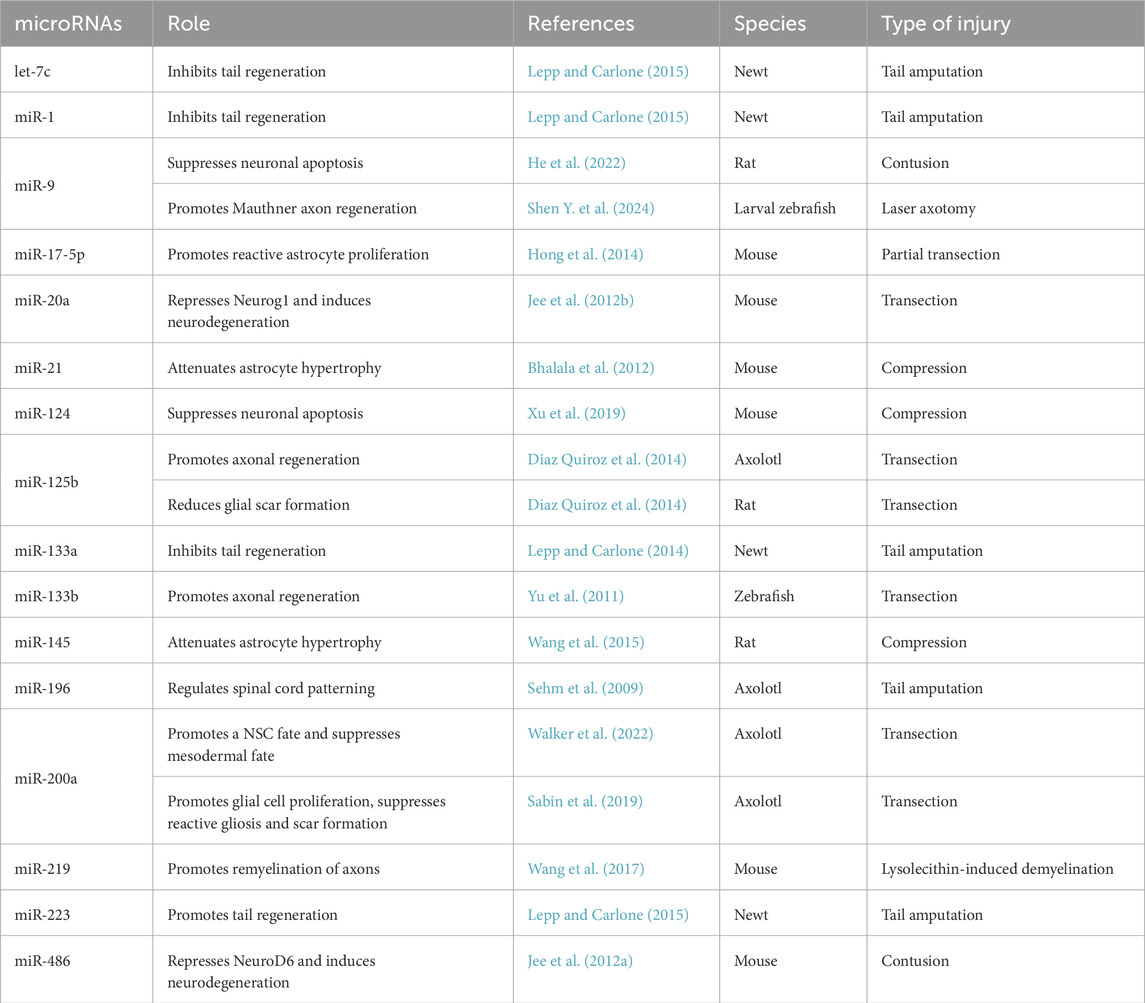
Table 3. microRNAs implicated in spinal cord repair. The following criteria were considered: (1) functional experiments carried out in a regenerative or non-regenerative species; (2) miRNA depletion or overexpression leads to a phenotype indicative of impaired or improved spinal cord repair.
For miRNAs to be able to tune the dynamics of target gene expression, their levels must be precisely controlled. For example, too much or too little miR-9 can lead to dampening of HES1 oscillations (Bonev et al., 2012; Goodfellow et al., 2014). In hindbrain development, the levels of miR-9 increase over time in a sharp, stepwise manner, controlled spatiotemporally by the differential onset of expression of the different miR-9 precursors (Soto et al., 2022). However, the mechanisms that regulate the transcriptional expression of miRNA precursors in response to SCI remain unknown. In adult vertebrates, miRNA can also be delivered to the injury site through cerebrospinal fluid encapsulated in exosomes. This exosomal delivery of miRNAs has been extensively studied as a potential therapeutic strategy for promoting recovery after SCI. Yet, the levels and timing of delivery for optimal therapeutic effect needs to be addressed (Pan et al., 2021; Feng et al., 2021).
In summary, factors that modulate bHLH transcriptional oscillations are implicated in the regulation of NSC quiescence and are also expressed in the context of central nervous system (CNS) injury. These observations provide hints of the potential involvement of dynamic gene expression in neural regeneration. Based on recent studies demonstrating a role for oscillatory gene expression dynamics in the reactivation of stem cells (Cottrell et al., 2025; Kaise et al., 2022; Marinopoulou et al., 2021), we hypothesise that, during spinal cord repair, dynamic gene expression is necessary for cells to make cell fate decisions precisely orchestrated in time and space. Further, we hypothesise that these dynamics are fine-tuned by microRNAs, which may influence cellular differentiation outcomes following spinal cord injury. So far, the role of HES/Her oscillatory dynamics in spinal cord repair remains poorly understood, primarily due to the lack of experimental approaches for intravital imaging of dynamic gene expression in adult animals.
In the following section, we will explore current strategies for detecting and manipulating gene expression dynamics in both regenerative and non-regenerative contexts.
4.4 Potential approaches to the study of gene expression dynamics during spinal cord repair
Understanding ultradian oscillations of transcription factors has gained increasing attention over the past 2 decades, closely paralleling the development of techniques for visualizing gene expression dynamics in living organisms. Despite these advances, visualizing ultradian oscillations in the CNS in whole organisms remains a significant challenge. The development of tools to label endogenous proteins has become more accessible with the advent of CRISPR-Cas technology and the generation of knock-in models. However, live imaging remains a major hurdle. To date, most studies have been conducted in isolated NSCs, NPCs, ex vivo mouse tissue sections, or in vivo in zebrafish embryos and larvae. Due to the complex tissue context of nervous system regeneration, cell culture or tissue slice models are not appropriate for addressing gene expression dynamics in these conditions. Therefore, moving the field forward, particularly in the context of spinal cord injury, will require further innovation to enable such studies in adult organisms.
Recent advances in imaging techniques offer promising directions. For example, in adult zebrafish, the application of multiphoton microscopy, specifically intravital three-photon microscopy (3PM), has allowed deep-tissue imaging up to 300 µm with subcellular resolution in the adult brain of albino zebrafish (Choe et al., 2022; Hontani et al., 2022). In mammals, however, reduced tissue transparency presents an additional challenge. Because mammalian tissues are relatively transparent to near-infrared (NIR) light, the use of NIR fluorescent proteins (NIR-FPs) in combination with 3PM could enable non-invasive imaging of tissues at depths of up to 3 mm (Shcherbakova et al., 2018) which has already been demonstrated using two-photon microscopy (2PM) (Tong et al., 2021).
Moreover, the use of implanted glass windows in combination with advanced imaging could revolutionize the ability to monitor ultradian oscillations in the CNS (Fenrich et al., 2013). Nevertheless, a major obstacle remains: ultradian oscillations occur on timescales of minutes to hours, necessitating high-frequency, long-duration intravital imaging. To address this, artificial intelligence (AI) and mathematical modelling can be employed to infer dynamic gene expression between imaging time points. For example, the OscoNet framework can infer transcriptional oscillators from single-cell RNA sequencing data (Cutillo et al., 2020). Finally, to not only observe but also manipulate gene expression dynamics in adult animals, optogenetic approaches are rapidly advancing and may offer powerful tools to induce controlled oscillations in vivo (Riefolo et al., 2019; Sanchez-Sanchez et al., 2025).
In summary, miRNA-modulated ultradian oscillations in transcription factor expression are essential for normal development of the nervous system, allowing transitions between quiescence, proliferating and differentiating states. Notch signalling and the ultradian dynamics of its downstream effectors have emerged as key regulators of stem cell quiescence in the adult CNS. However, the significance of these dynamics for nervous system repair in response to injury remains unclear, and addressing this question will require live imaging of dynamics in the native tissue context. While intravital imaging of ultradian oscillations remains a formidable challenge, emerging technologies, including high-resolution microscopy, photo-switchable proteins, and AI-driven analysis, are rapidly evolving and promise to significantly advance our understanding of spinal cord repair and dynamic gene regulation.
Author contributions
SL: Writing – original draft, Writing – review and editing. XS: Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This review was supported by UKRI Medical Research Council, MR/X020754/1.
Acknowledgments
We are grateful to Prof. Martin Lowe, Dr Clare Buckley and Dr Veronica Biga for advice and discussions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Correction note
This article has been corrected with minor changes. These changes do not impact the scientific content of the article.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ables, J. L., Decarolis, N. A., Johnson, M. A., Rivera, P. D., Gao, Z., Cooper, D. C., et al. (2010). Notch1 is required for maintenance of the reservoir of adult hippocampal stem cells. J. Neurosci. 30, 10484–10492. doi:10.1523/JNEUROSCI.4721-09.2010
Albeck, J. G., Mills, G. B., and Brugge, J. S. (2013). Frequency-modulated pulses of ERK activity transmit quantitative proliferation signals. Mol. Cell 49, 249–261. doi:10.1016/j.molcel.2012.11.002
Alunni, A., and Bally-Cuif, L. (2016). A comparative view of regenerative neurogenesis in vertebrates. Development 143, 741–753. doi:10.1242/dev.122796
Alvarez-Buylla, A., Garcia-Verdugo, J. M., and Tramontin, A. D. (2001). A unified hypothesis on the lineage of neural stem cells. Nat. Rev. Neurosci. 2, 287–293. doi:10.1038/35067582
Andersen, J., Urban, N., Achimastou, A., Ito, A., Simic, M., Ullom, K., et al. (2014). A transcriptional mechanism integrating inputs from extracellular signals to activate hippocampal stem cells. Neuron 83, 1085–1097. doi:10.1016/j.neuron.2014.08.004
Angell Swearer, A., Perkowski, S., and Wills, A. (2025). Shh signaling directs dorsal ventral patterning in the regenerating X. tropicalis spinal cord. Dev. Biol. 520, 191–199. doi:10.1016/j.ydbio.2025.01.015
Aulehla, A., and Pourquie, O. (2008). Oscillating signaling pathways during embryonic development. Curr. Opin. Cell Biol. 20, 632–637. doi:10.1016/j.ceb.2008.09.002
Baek, J. H., Hatakeyama, J., Sakamoto, S., Ohtsuka, T., and Kageyama, R. (2006). Persistent and high levels of Hes1 expression regulate boundary formation in the developing central nervous system. Development 133, 2467–2476. doi:10.1242/dev.02403
Bagga, S., Bracht, J., Hunter, S., Massirer, K., Holtz, J., Eachus, R., et al. (2005). Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122, 553–563. doi:10.1016/j.cell.2005.07.031
Bai, G., Sheng, N., Xie, Z., Bian, W., Yokota, Y., Benezra, R., et al. (2007). Id sustains Hes1 expression to inhibit precocious neurogenesis by releasing negative autoregulation of Hes1. Dev. Cell 13, 283–297. doi:10.1016/j.devcel.2007.05.014
Bally-Cuif, L., and Hammerschmidt, M. (2003). Induction and patterning of neuronal development, and its connection to cell cycle control. Curr. Opin. Neurobiol. 13, 16–25. doi:10.1016/s0959-4388(03)00015-1
Barnabe-Heider, F., and Frisen, J. (2008). Stem cells for spinal cord repair. Cell Stem Cell 3, 16–24. doi:10.1016/j.stem.2008.06.011
Barnabe-Heider, F., Goritz, C., Sabelstrom, H., Takebayashi, H., Pfrieger, F. W., Meletis, K., et al. (2010). Origin of new glial cells in intact and injured adult spinal cord. Cell Stem Cell 7, 470–482. doi:10.1016/j.stem.2010.07.014
Barreiro-Iglesias, A., Mysiak, K. S., Scott, A. L., Reimer, M. M., Yang, Y., Becker, C. G., et al. (2015). Serotonin promotes development and regeneration of spinal motor neurons in zebrafish. Cell Rep. 13, 924–932. doi:10.1016/j.celrep.2015.09.050
Bazzini, A. A., Lee, M. T., and Giraldez, A. J. (2012). Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336, 233–237. doi:10.1126/science.1215704
Beck, C. W., Christen, B., Barker, D., and Slack, J. M. (2006). Temporal requirement for bone morphogenetic proteins in regeneration of the tail and limb of Xenopus tadpoles. Mech. Dev. 123, 674–688. doi:10.1016/j.mod.2006.07.001
Becker, C. G., and Becker, T. (2015). Neuronal regeneration from ependymo-radial glial cells: cook, little pot, cook. Dev. Cell 32, 516–527. doi:10.1016/j.devcel.2015.01.001
Becker, T., and Becker, C. G. (2022). Regenerative neurogenesis: the integration of developmental, physiological and immune signals. Development 149, dev199907. doi:10.1242/dev.199907
Becker, T., Wullimann, M. F., Becker, C. G., Bernhardt, R. R., and Schachner, M. (1997). Axonal regrowth after spinal cord transection in adult zebrafish. J. Comp. Neurol. 377, 577–595. doi:10.1002/(sici)1096-9861(19970127)377:4<577::aid-cne8>3.0.co;2-#
Becker, C. G., Lieberoth, B. C., Morellini, F., Feldner, J., Becker, T., and Schachner, M. (2004). L1.1 is involved in spinal cord regeneration in adult zebrafish. J. Neurosci. 24, 7837–7842. doi:10.1523/JNEUROSCI.2420-04.2004
Becker, C. G., Becker, T., and Hugnot, J. P. (2018). The spinal ependymal zone as a source of endogenous repair cells across vertebrates. Prog. Neurobiol. 170, 67–80. doi:10.1016/j.pneurobio.2018.04.002
Benraiss, A., Arsanto, J. P., Coulon, J., and Thouveny, Y. (1999). Neurogenesis during caudal spinal cord regeneration in adult newts. Dev. Genes Evol. 209, 363–369. doi:10.1007/s004270050265
Bertrand, N., Castro, D. S., and Guillemot, F. (2002). Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 3, 517–530. doi:10.1038/nrn874
Bessho, Y., and Kageyama, R. (2003). Oscillations, clocks and segmentation. Curr. Opin. Genet. Dev. 13, 379–384. doi:10.1016/s0959-437x(03)00083-2
Bhalala, O. G., Pan, L., Sahni, V., McGuire, T. L., Gruner, K., Tourtellotte, W. G., et al. (2012). microRNA-21 regulates astrocytic response following spinal cord injury. J. Neurosci. 32, 17935–17947. doi:10.1523/JNEUROSCI.3860-12.2012
Bhalala, O. G., Srikanth, M., and Kessler, J. A. (2013). The emerging roles of microRNAs in CNS injuries. Nat. Rev. Neurol. 9, 328–339. doi:10.1038/nrneurol.2013.67
Biga, V., Hawley, J., Soto, X., Johns, E., Han, D., Bennett, H., et al. (2021). A dynamic, spatially periodic, micro-pattern of HES5 underlies neurogenesis in the mouse spinal cord. Mol. Syst. Biol. 17, e9902. doi:10.15252/msb.20209902
Bonev, B., Pisco, A., and Papalopulu, N. (2011). MicroRNA-9 reveals regional diversity of neural progenitors along the anterior-posterior axis. Dev. Cell 20, 19–32. doi:10.1016/j.devcel.2010.11.018
Bonev, B., Stanley, P., and Papalopulu, N. (2012). MicroRNA-9 modulates Hes1 ultradian oscillations by forming a double-negative feedback loop. Cell Rep. 2, 10–18. doi:10.1016/j.celrep.2012.05.017
Bradbury, E. J., and Burnside, E. R. (2019). Moving beyond the glial scar for spinal cord repair. Nat. Commun. 10, 3879. doi:10.1038/s41467-019-11707-7
Brend, T., and Holley, S. A. (2009). Expression of the oscillating gene her1 is directly regulated by hairy/enhancer of split, T-box, and suppressor of hairless proteins in the zebrafish segmentation clock. Dev. Dyn. 238, 2745–2759. doi:10.1002/dvdy.22100
Briona, L. K., and Dorsky, R. I. (2014). Radial glial progenitors repair the zebrafish spinal cord following transection. Exp. Neurol. 256, 81–92. doi:10.1016/j.expneurol.2014.03.017
Cai, S., Zhou, P., and Liu, Z. (2013). Functional characteristics of a double negative feedback loop mediated by microRNAs. Cogn. Neurodyn 7, 417–429. doi:10.1007/s11571-012-9236-7
Cardozo, M. J., Mysiak, K. S., Becker, T., and Becker, C. G. (2017). Reduce, reuse, recycle - developmental signals in spinal cord regeneration. Dev. Biol. 432, 53–62. doi:10.1016/j.ydbio.2017.05.011
Carlson, S. L., Parrish, M. E., Springer, J. E., Doty, K., and Dossett, L. (1998). Acute inflammatory response in spinal cord following impact injury. Exp. Neurol. 151, 77–88. doi:10.1006/exnr.1998.6785
Carter, C., Clark, A., Spencer, G., and Carlone, R. (2011). Cloning and expression of a retinoic acid receptor β2 subtype from the adult newt: evidence for an early role in tail and caudal spinal cord regeneration. Dev. Dyn. 240, 2613–2625. doi:10.1002/dvdy.22769
Casas Gimeno, G., and Paridaen, J. (2022). The symmetry of neural stem cell and progenitor divisions in the vertebrate brain. Front. Cell Dev. Biol. 10, 885269. doi:10.3389/fcell.2022.885269
Castella, P., Sawai, S., Nakao, K., Wagner, J. A., and Caudy, M. (2000). HES-1 repression of differentiation and proliferation in PC12 cells: role for the helix 3-helix 4 domain in transcription repression. Mol. Cell Biol. 20, 6170–6183. doi:10.1128/MCB.20.16.6170-6183.2000
Cavone, L., McCann, T., Drake, L. K., Aguzzi, E. A., Oprisoreanu, A. M., Pedersen, E., et al. (2021). A unique macrophage subpopulation signals directly to progenitor cells to promote regenerative neurogenesis in the zebrafish spinal cord. Dev. Cell 56, 1617–1630.e6. doi:10.1016/j.devcel.2021.04.031
Chang, W., Pedroni, A., Bertuzzi, M., Kizil, C., Simon, A., and Ampatzis, K. (2021). Locomotion dependent neuron-glia interactions control neurogenesis and regeneration in the adult zebrafish spinal cord. Nat. Commun. 12, 4857. doi:10.1038/s41467-021-25052-1
Chen, H., Thiagalingam, A., Chopra, H., Borges, M. W., Feder, J. N., Nelkin, B. D., et al. (1997). Conservation of the Drosophila lateral inhibition pathway in human lung cancer: a hairy-related protein (HES-1) directly represses achaete-scute homolog-1 expression. Proc. Natl. Acad. Sci. U S A. 94, 5355–5360. doi:10.1073/pnas.94.10.5355
Chevallier, S., Landry, M., Nagy, F., and Cabelguen, J. M. (2004). Recovery of bimodal locomotion in the spinal-transected salamander, Pleurodeles waltlii. Eur. J. Neurosci. 20, 1995–2007. doi:10.1111/j.1460-9568.2004.03671.x
Choe, K., Hontani, Y., Wang, T., Hebert, E., Ouzounov, D. G., Lai, K., et al. (2022). Intravital three-photon microscopy allows visualization over the entire depth of mouse lymph nodes. Nat. Immunol. 23, 330–340. doi:10.1038/s41590-021-01101-1
Cigliola, V., Becker, C. J., and Poss, K. D. (2020). Building bridges, not walls: spinal cord regeneration in zebrafish. Dis. Model Mech. 13, dmm044131. doi:10.1242/dmm.044131
Clarke, J. D., Alexander, R., and Holder, N. (1988). Regeneration of descending axons in the spinal cord of the axolotl. Neurosci. Lett. 89, 1–6. doi:10.1016/0304-3940(88)90471-5
Cooke, J., and Zeeman, E. C. (1976). A clock and wavefront model for control of the number of repeated structures during animal morphogenesis. J. Theor. Biol. 58, 455–476. doi:10.1016/s0022-5193(76)80131-2
Coolen, M., Thieffry, D., Drivenes, O., Becker, T. S., and Bally-Cuif, L. (2012). miR-9 controls the timing of neurogenesis through the direct inhibition of antagonistic factors. Dev. Cell 22, 1052–1064. doi:10.1016/j.devcel.2012.03.003
Cottrell, O. R. A., Noble, B., Clarke, R. B., and Papalopulu, N. (2025). HES1 oscillations are required for cell cycle re-entry in oestrogen receptor positive breast cancer cells. BioRxiv. doi:10.1101/2025.08.04.668440
Cregg, J. M., DePaul, M. A., Filous, A. R., Lang, B. T., Tran, A., and Silver, J. (2014). Functional regeneration beyond the glial scar. Exp. Neurol. 253, 197–207. doi:10.1016/j.expneurol.2013.12.024
Cutillo, L., Boukouvalas, A., Marinopoulou, E., Papalopulu, N., and Rattray, M. (2020). OscoNet: inferring oscillatory gene networks. BMC Bioinforma. 21, 351. doi:10.1186/s12859-020-03561-y
Darnell, D. K., Kaur, S., Stanislaw, S., Konieczka, J. H., Yatskievych, T. A., and Antin, P. B. (2006). MicroRNA expression during chick embryo development. Dev. Dyn. 235, 3156–3165. doi:10.1002/dvdy.20956
Davis, B. M., Ayers, J. L., Koran, L., Carlson, J., Anderson, M. C., and Simpson, S. B. Jr. (1990). Time course of salamander spinal cord regeneration and recovery of swimming: HRP retrograde pathway tracing and kinematic analysis. Exp. Neurol. 108, 198–213. doi:10.1016/0014-4886(90)90124-b
Deleyrolle, L., Marchal-Victorion, S., Dromard, C., Fritz, V., Saunier, M., Sabourin, J. C., et al. (2006). Exogenous and fibroblast growth factor 2/epidermal growth factor-regulated endogenous cytokines regulate neural precursor cell growth and differentiation. Stem Cells 24, 748–762. doi:10.1634/stemcells.2005-0138
Dennis, D. J., Han, S., and Schuurmans, C. (2019). bHLH transcription factors in neural development, disease, and reprogramming. Brain Res. 1705, 48–65. doi:10.1016/j.brainres.2018.03.013
Dias, T. B., Yang, Y. J., Ogai, K., Becker, T., and Becker, C. G. (2012). Notch signaling controls generation of motor neurons in the lesioned spinal cord of adult zebrafish. J. Neurosci. 32, 3245–3252. doi:10.1523/JNEUROSCI.6398-11.2012
Diaz Quiroz, J. F., and Echeverri, K. (2013). Spinal cord regeneration: where fish, frogs and salamanders lead the way, can we follow? Biochem. J. 451, 353–364. doi:10.1042/BJ20121807
Diaz Quiroz, J. F., Tsai, E., Coyle, M., Sehm, T., and Echeverri, K. (2014). Precise control of miR-125b levels is required to create a regeneration-permissive environment after spinal cord injury: a cross-species comparison between salamander and rat. Dis. Model Mech. 7, 601–611. doi:10.1242/dmm.014837
Djuranovic, S., Nahvi, A., and Green, R. (2012). miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336, 237–240. doi:10.1126/science.1215691
Doetsch, F., Garcia-Verdugo, J. M., and Alvarez-Buylla, A. (1997). Cellular composition and three-dimensional organization of the subventricular germinal zone in the adult mammalian brain. J. Neurosci. 17, 5046–5061. doi:10.1523/JNEUROSCI.17-13-05046.1997
Doostdar, P., Hawley, J., Chopra, K., Marinopoulou, E., Lea, R., Arashvand, K., et al. (2024). Cell coupling compensates for changes in single-cell Her6 dynamics and provides phenotypic robustness. Development 151, dev202640. doi:10.1242/dev.202640
Egar, M., and Singer, M. (1972). The role of ependyma in spinal cord regeneration in the urodele, Triturus. Exp. Neurol. 37, 422–430. doi:10.1016/0014-4886(72)90085-4
Ehm, O., Goritz, C., Covic, M., Schaffner, I., Schwarz, T. J., Karaca, E., et al. (2010). RBPJkappa-dependent signaling is essential for long-term maintenance of neural stem cells in the adult hippocampus. J. Neurosci. 30, 13794–13807. doi:10.1523/JNEUROSCI.1567-10.2010
Eichhorn, S. W., Guo, H., McGeary, S. E., Rodriguez-Mias, R. A., Shin, C., Baek, D., et al. (2014). mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol. Cell 56, 104–115. doi:10.1016/j.molcel.2014.08.028
Faulkner, J. R., Herrmann, J. E., Woo, M. J., Tansey, K. E., Doan, N. B., and Sofroniew, M. V. (2004). Reactive astrocytes protect tissue and preserve function after spinal cord injury. J. Neurosci. 24, 2143–2155. doi:10.1523/JNEUROSCI.3547-03.2004
Fei, J. F., Schuez, M., Tazaki, A., Taniguchi, Y., Roensch, K., and Tanaka, E. M. (2014). CRISPR-mediated genomic deletion of Sox2 in the axolotl shows a requirement in spinal cord neural stem cell amplification during tail regeneration. Stem Cell Rep. 3, 444–459. doi:10.1016/j.stemcr.2014.06.018
Feng, J., Zhang, Y., Zhu, Z., Gu, C., Waqas, A., and Chen, L. (2021). Emerging exosomes and exosomal MiRNAs in spinal cord injury. Front. Cell Dev. Biol. 9, 703989. doi:10.3389/fcell.2021.703989
Fenrich, K. K., Weber, P., Rougon, G., and Debarbieux, F. (2013). Implanting glass spinal cord windows in adult mice with experimental autoimmune encephalomyelitis. J. Vis. Exp., e50826. doi:10.3791/50826
Figley, S. A., Khosravi, R., Legasto, J. M., Tseng, Y. F., and Fehlings, M. G. (2014). Characterization of vascular disruption and blood-spinal cord barrier permeability following traumatic spinal cord injury. J. Neurotrauma 31, 541–552. doi:10.1089/neu.2013.3034
Fu, R. Z., Cottrell, O., Cutillo, L., Rowntree, A., Zador, Z., Wurdak, H., et al. (2024). Identification of genes with oscillatory expression in glioblastoma: the paradigm of SOX2. Sci. Rep. 14, 2123. doi:10.1038/s41598-024-51340-z
Gaete, M., Munoz, R., Sanchez, N., Tampe, R., Moreno, M., Contreras, E. G., et al. (2012). Spinal cord regeneration in Xenopus tadpoles proceeds through activation of Sox2-positive cells. Neural Dev. 7, 13. doi:10.1186/1749-8104-7-13
Gao, C., Liu, H., and Yan, F. (2020). Dynamic behavior of p53 driven by delay and a Microrna-34a-Mediated feedback loop. Int. J. Mol. Sci. 21, 1271. doi:10.3390/ijms21041271
Ghosh, S., and Hui, S. P. (2018). Axonal regeneration in zebrafish spinal cord. Regen. (Oxf) 5, 43–60. doi:10.1002/reg2.99
Giudicelli, F., Ozbudak, E. M., Wright, G. J., and Lewis, J. (2007). Setting the tempo in development: an investigation of the zebrafish somite clock mechanism. PLoS Biol. 5, e150. doi:10.1371/journal.pbio.0050150
Goldshmit, Y., Sztal, T. E., Jusuf, P. R., Hall, T. E., Nguyen-Chi, M., and Currie, P. D. (2012). Fgf-dependent glial cell bridges facilitate spinal cord regeneration in zebrafish. J. Neurosci. 32, 7477–7492. doi:10.1523/JNEUROSCI.0758-12.2012
Goldshmit, Y., Tang, J., Siegel, A. L., Nguyen, P. D., Kaslin, J., Currie, P. D., et al. (2018). Different Fgfs have distinct roles in regulating neurogenesis after spinal cord injury in zebrafish. Neural Dev. 13, 24. doi:10.1186/s13064-018-0122-9
Goodfellow, M., Phillips, N. E., Manning, C., Galla, T., and Papalopulu, N. (2014). microRNA input into a neural ultradian oscillator controls emergence and timing of alternative cell states. Nat. Commun. 5, 3399. doi:10.1038/ncomms4399
Goodwin, B. C. (1965). Oscillatory behavior in enzymatic control processes. Adv. Enzyme Regul. 3, 425–438. doi:10.1016/0065-2571(65)90067-1
Gregoire, C. A., Goldenstein, B. L., Floriddia, E. M., Barnabe-Heider, F., and Fernandes, K. J. (2015). Endogenous neural stem cell responses to stroke and spinal cord injury. Glia 63, 1469–1482. doi:10.1002/glia.22851
Grossman, S. D., Rosenberg, L. J., and Wrathall, J. R. (2001). Temporal-spatial pattern of acute neuronal and glial loss after spinal cord contusion. Exp. Neurol. 168, 273–282. doi:10.1006/exnr.2001.7628
Guo, H., Ingolia, N. T., Weissman, J. S., and Bartel, D. P. (2010). mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840. doi:10.1038/nature09267
Guo, Y., Ma, L., Cristofanilli, M., Hart, R. P., Hao, A., and Schachner, M. (2011). Transcription factor Sox11b is involved in spinal cord regeneration in adult zebrafish. Neuroscience 172, 329–341. doi:10.1016/j.neuroscience.2010.10.026
Harada, Y., Yamada, M., Imayoshi, I., Kageyama, R., Suzuki, Y., Kuniya, T., et al. (2021). Cell cycle arrest determines adult neural stem cell ontogeny by an embryonic Notch-nonoscillatory Hey1 module. Nat. Commun. 12, 6562. doi:10.1038/s41467-021-26605-0
Harima, Y., Takashima, Y., Ueda, Y., Ohtsuka, T., and Kageyama, R. (2013). Accelerating the tempo of the segmentation clock by reducing the number of introns in the Hes7 gene. Cell Rep. 3, 1–7. doi:10.1016/j.celrep.2012.11.012
Hayashi, S., Ochi, H., Ogino, H., Kawasumi, A., Kamei, Y., Tamura, K., et al. (2014). Transcriptional regulators in the Hippo signaling pathway control organ growth in Xenopus tadpole tail regeneration. Dev. Biol. 396, 31–41. doi:10.1016/j.ydbio.2014.09.018
He, X., Zhang, J., Guo, Y., Yang, X., Huang, Y., and Hao, D. (2022). Exosomal miR-9-5p derived from BMSCs alleviates apoptosis, inflammation and endoplasmic reticulum stress in spinal cord injury by regulating the HDAC5/FGF2 axis. Mol. Immunol. 145, 97–108. doi:10.1016/j.molimm.2022.03.007
Hendrickson, D. G., Hogan, D. J., McCullough, H. L., Myers, J. W., Herschlag, D., Ferrell, J. E., et al. (2009). Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 7, e1000238. doi:10.1371/journal.pbio.1000238
Henrique, D., and Schweisguth, F. (2019). Mechanisms of notch signaling: a simple logic deployed in time and space. Development 146, dev172148. doi:10.1242/dev.172148
Hermann, G. E., Rogers, R. C., Bresnahan, J. C., and Beattie, M. S. (2001). Tumor necrosis factor-alpha induces cFOS and strongly potentiates glutamate-mediated cell death in the rat spinal cord. Neurobiol. Dis. 8, 590–599. doi:10.1006/nbdi.2001.0414
Hirata, H., Tomita, K., Bessho, Y., and Kageyama, R. (2001). Hes1 and Hes3 regulate maintenance of the isthmic organizer and development of the mid/hindbrain. EMBO J. 20, 4454–4466. doi:10.1093/emboj/20.16.4454
Hirata, H., Yoshiura, S., Ohtsuka, T., Bessho, Y., Harada, T., Yoshikawa, K., et al. (2002). Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science 298, 840–843. doi:10.1126/science.1074560
Hirata, H., Bessho, Y., Kokubu, H., Masamizu, Y., Yamada, S., Lewis, J., et al. (2004). Instability of Hes7 protein is crucial for the somite segmentation clock. Nat. Genet. 36, 750–754. doi:10.1038/ng1372
Ho, D. M., and Whitman, M. (2008). TGF-beta signaling is required for multiple processes during Xenopus tail regeneration. Dev. Biol. 315, 203–216. doi:10.1016/j.ydbio.2007.12.031
Hong, P., Jiang, M., and Li, H. (2014). Functional requirement of dicer1 and miR-17-5p in reactive astrocyte proliferation after spinal cord injury in the mouse. Glia 62, 2044–2060. doi:10.1002/glia.22725
Hontani, Y., Akbari, N., Kolkman, K. E., Wu, C., Xia, F., Choe, K., et al. (2022). Deep-tissue three-photon fluorescence microscopy in intact mouse and zebrafish brain. J. Vis. Exp. doi:10.3791/63213
Hossainian, D., Shao, E., Jiao, B., Ilin, V. A., Parris, R. S., Zhou, Y., et al. (2022). Quantification of functional recovery in a larval zebrafish model of spinal cord injury. J. Neurosci. Res. 100, 2044–2054. doi:10.1002/jnr.25118
Hu, N., and Zou, L. (2022). Multiple functions of hes genes in the proliferation and differentiation of neural stem cells. Ann. Anat. 239, 151848. doi:10.1016/j.aanat.2021.151848
Hui, S. P., Dutta, A., and Ghosh, S. (2010). Cellular response after crush injury in adult zebrafish spinal cord. Dev. Dyn. 239, 2962–2979. doi:10.1002/dvdy.22438
Hui, S. P., Sheng, D. Z., Sugimoto, K., Gonzalez-Rajal, A., Nakagawa, S., Hesselson, D., et al. (2017). Zebrafish regulatory T cells mediate organ-specific regenerative programs. Dev. Cell 43, 659–672.e5. doi:10.1016/j.devcel.2017.11.010
Ikeda-Yorifuji, I., Tsujioka, H., Sakata, Y., and Yamashita, T. (2022). Single-nucleus RNA sequencing identified cells with ependymal cell-like features enriched in neonatal mice after spinal cord injury. Neurosci. Res. 181, 22–38. doi:10.1016/j.neures.2022.04.006
Imayoshi, I., and Kageyama, R. (2014). Oscillatory control of bHLH factors in neural progenitors. Trends Neurosci. 37, 531–538. doi:10.1016/j.tins.2014.07.006
Imayoshi, I., Sakamoto, M., Yamaguchi, M., Mori, K., and Kageyama, R. (2010). Essential roles of notch signaling in maintenance of neural stem cells in developing and adult brains. J. Neurosci. 30, 3489–3498. doi:10.1523/JNEUROSCI.4987-09.2010
Imayoshi, I., Isomura, A., Harima, Y., Kawaguchi, K., Kori, H., Miyachi, H., et al. (2013). Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science 342, 1203–1208. doi:10.1126/science.1242366
Imayoshi, I., Ishidate, F., and Kageyama, R. (2015). Real-time imaging of bHLH transcription factors reveals their dynamic control in the multipotency and fate choice of neural stem cells. Front. Cell Neurosci. 9, 288. doi:10.3389/fncel.2015.00288
Imperato-Kalmar, E. L., McKinney, R. A., Schnell, L., Rubin, B. P., and Schwab, M. E. (1997). Local changes in vascular architecture following partial spinal cord lesion in the rat. Exp. Neurol. 145, 322–328. doi:10.1006/exnr.1997.6449
Isomura, A., and Kageyama, R. (2014). Ultradian oscillations and pulses: coordinating cellular responses and cell fate decisions. Development 141, 3627–3636. doi:10.1242/dev.104497
Janesick, A., Wu, S. C., and Blumberg, B. (2015). Retinoic acid signaling and neuronal differentiation. Cell Mol. Life Sci. 72, 1559–1576. doi:10.1007/s00018-014-1815-9
Jee, M. K., Jung, J. S., Choi, J. I., Jang, J. A., Kang, K. S., Im, Y. B., et al. (2012a). MicroRNA 486 is a potentially novel target for the treatment of spinal cord injury. Brain 135, 1237–1252. doi:10.1093/brain/aws047
Jee, M. K., Jung, J. S., Im, Y. B., Jung, S. J., and Kang, S. K. (2012b). Silencing of miR20a is crucial for Ngn1-mediated neuroprotection in injured spinal cord. Hum. Gene Ther. 23, 508–520. doi:10.1089/hum.2011.121
Jensen, M. H., Sneppen, K., and Tiana, G. (2003). Sustained oscillations and time delays in gene expression of protein Hes1. FEBS Lett. 541, 176–177. doi:10.1016/s0014-5793(03)00279-5
Jessell, T. M. (2000). Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 1, 20–29. doi:10.1038/35049541
Johansson, C. B., Momma, S., Clarke, D. L., Risling, M., Lendahl, U., and Frisen, J. (1999). Identification of a neural stem cell in the adult mammalian central nervous system. Cell 96, 25–34. doi:10.1016/s0092-8674(00)80956-3
Jonas, S., and Izaurralde, E. (2015). Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 16, 421–433. doi:10.1038/nrg3965
Kageyama, R., Shimojo, H., and Imayoshi, I. (2015). Dynamic expression and roles of hes factors in neural development. Cell Tissue Res. 359, 125–133. doi:10.1007/s00441-014-1888-7
Kageyama, R., Shimojo, H., and Ohtsuka, T. (2019). Dynamic control of neural stem cells by bHLH factors. Neurosci. Res. 138, 12–18. doi:10.1016/j.neures.2018.09.005
Kageyama, R., Ochi, S., Sueda, R., and Shimojo, H. (2020). The significance of gene expression dynamics in neural stem cell regulation. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 96, 351–363. doi:10.2183/pjab.96.026
Kaise, T., and Kageyama, R. (2024). Transcriptional control of neural stem cell activity. Biochem. Soc. Trans. 52, 617–626. doi:10.1042/BST20230439
Kaise, T., Fukui, M., Sueda, R., Piao, W., Yamada, M., Kobayashi, T., et al. (2022). Functional rejuvenation of aged neural stem cells by Plagl2 and anti-Dyrk1a activity. Genes Dev. 36, 23–37. doi:10.1101/gad.349000.121
Kawaguchi, D., Furutachi, S., Kawai, H., Hozumi, K., and Gotoh, Y. (2013). Dll1 maintains quiescence of adult neural stem cells and segregates asymmetrically during mitosis. Nat. Commun. 4, 1880. doi:10.1038/ncomms2895
Kicheva, A., Bollenbach, T., Ribeiro, A., Valle, H. P., Lovell-Badge, R., Episkopou, V., et al. (2014). Coordination of progenitor specification and growth in mouse and chick spinal cord. Science 345, 1254927. doi:10.1126/science.1254927
Kiparissides, A., Koutinas, M., Moss, T., Newman, J., Pistikopoulos, E. N., and Mantalaris, A. (2011). Modelling the Delta1/Notch1 pathway: in search of the mediator(s) of neural stem cell differentiation. PLoS One 6, e14668. doi:10.1371/journal.pone.0014668
Kobayashi, T., Mizuno, H., Imayoshi, I., Furusawa, C., Shirahige, K., and Kageyama, R. (2009). The cyclic gene Hes1 contributes to diverse differentiation responses of embryonic stem cells. Genes Dev. 23, 1870–1875. doi:10.1101/gad.1823109
Kondo, T., and Raff, M. (2000). Basic helix-loop-helix proteins and the timing of oligodendrocyte differentiation. Development 127, 2989–2998. doi:10.1242/dev.127.14.2989
Krol, A. J., Roellig, D., Dequeant, M. L., Tassy, O., Glynn, E., Hattem, G., et al. (2011). Evolutionary plasticity of segmentation clock networks. Development 138, 2783–2792. doi:10.1242/dev.063834
Kuscha, V., Barreiro-Iglesias, A., Becker, C. G., and Becker, T. (2012a). Plasticity of tyrosine hydroxylase and serotonergic systems in the regenerating spinal cord of adult zebrafish. J. Comp. Neurol. 520, 933–951. doi:10.1002/cne.22739
Kuscha, V., Frazer, S. L., Dias, T. B., Hibi, M., Becker, T., and Becker, C. G. (2012b). Lesion-induced generation of interneuron cell types in specific dorsoventral domains in the spinal cord of adult zebrafish. J. Comp. Neurol. 520, 3604–3616. doi:10.1002/cne.23115
Lai, X., Wolkenhauer, O., and Vera, J. (2016). Understanding microRNA-mediated gene regulatory networks through mathematical modelling. Nucleic Acids Res. 44, 6019–6035. doi:10.1093/nar/gkw550
Laje, R., Agostino, P. V., and Golombek, D. A. (2018). The times of our lives: interaction among different biological periodicities. Front. Integr. Neurosci. 12, 10. doi:10.3389/fnint.2018.00010
Lee, Y. B., Yune, T. Y., Baik, S. Y., Shin, Y. H., Du, S., Rhim, H., et al. (2000). Role of tumor necrosis factor-alpha in neuronal and glial apoptosis after spinal cord injury. Exp. Neurol. 166, 190–195. doi:10.1006/exnr.2000.7494
Lepp, A. C., and Carlone, R. L. (2014). RARβ2 expression is induced by the down-regulation of microRNA 133a during caudal spinal cord regeneration in the adult newt. Dev. Dyn. 243, 1581–1590. doi:10.1002/dvdy.24210
Lepp, A. C., and Carlone, R. L. (2015). MicroRNA dysregulation in response to RARβ2 inhibition reveals a negative feedback loop between MicroRNAs 1, 133a, and RARβ2 during tail and spinal cord regeneration in the adult newt. Dev. Dyn. 244, 1519–1537. doi:10.1002/dvdy.24342
Leucht, C., Stigloher, C., Wizenmann, A., Klafke, R., Folchert, A., and Bally-Cuif, L. (2008). MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat. Neurosci. 11, 641–648. doi:10.1038/nn.2115
Levine, J. H., Fontes, M. E., Dworkin, J., and Elowitz, M. B. (2012). Pulsed feedback defers cellular differentiation. PLoS Biol. 10, e1001252. doi:10.1371/journal.pbio.1001252
Lewis, J. (2003). Autoinhibition with transcriptional delay: a simple mechanism for the zebrafish somitogenesis oscillator. Curr. Biol. 13, 1398–1408. doi:10.1016/s0960-9822(03)00534-7
Li, P., Teng, Z. Q., and Liu, C. M. (2016). Extrinsic and intrinsic regulation of axon regeneration by MicroRNAs after Spinal cord injury. Neural Plast. 2016, 1279051. doi:10.1155/2016/1279051
Li S., S., Liu, Y., Liu, Z., and Wang, R. (2016). Neural fate decisions mediated by combinatorial regulation of Hes1 and miR-9. J. Biol. Phys. 42, 53–68. doi:10.1007/s10867-015-9391-2
Lin, G., and Slack, J. M. (2008). Requirement for Wnt and FGF signaling in Xenopus tadpole tail regeneration. Dev. Biol. 316, 323–335. doi:10.1016/j.ydbio.2008.01.032
Liu, X. Z., Xu, X. M., Hu, R., Du, C., Zhang, S. X., McDonald, J. W., et al. (1997). Neuronal and glial apoptosis after traumatic spinal cord injury. J. Neurosci. 17, 5395–5406. doi:10.1523/JNEUROSCI.17-14-05395.1997
Liu, A., Li, J., Marin-Husstege, M., Kageyama, R., Fan, Y., Gelinas, C., et al. (2006). A molecular insight of Hes5-dependent inhibition of myelin gene expression: old partners and new players. EMBO J. 25, 4833–4842. doi:10.1038/sj.emboj.7601352
Liu, N. K., Wang, X. F., Lu, Q. B., and Xu, X. M. (2009). Altered microRNA expression following traumatic spinal cord injury. Exp. Neurol. 219, 424–429. doi:10.1016/j.expneurol.2009.06.015
Llorens-Bobadilla, E., Chell, J. M., Le Merre, P., Wu, Y., Zamboni, M., Bergenstrahle, J., et al. (2020). A latent lineage potential in resident neural stem cells enables spinal cord repair. Science 370, eabb8795. doi:10.1126/science.abb8795
Lou, J., Lenke, L. G., Ludwig, F. J., and O'Brien, M. F. (1998). Apoptosis as a mechanism of neuronal cell death following acute experimental spinal cord injury. Spinal Cord. 36, 683–690. doi:10.1038/sj.sc.3100632
Lu, Q. R., Cai, L., Rowitch, D., Cepko, C. L., and Stiles, C. D. (2001). Ectopic expression of Olig1 promotes oligodendrocyte formation and reduces neuronal survival in developing mouse cortex. Nat. Neurosci. 4, 973–974. doi:10.1038/nn718
Luo, L., Liu, H., and Yan, F. (2023). Dynamic behavior of P53-Mdm2-Wip1 gene regulatory network under the influence of time delay and noise. Math. Biosci. Eng. 20, 2321–2347. doi:10.3934/mbe.2023109
Maeda, Y., Isomura, A., Masaki, T., and Kageyama, R. (2023). Differential cell-cycle control by oscillatory versus sustained Hes1 expression via p21. Cell Rep. 42, 112520. doi:10.1016/j.celrep.2023.112520
Manning, C. S., Biga, V., Boyd, J., Kursawe, J., Ymisson, B., Spiller, D. G., et al. (2019). Quantitative single-cell live imaging links HES5 dynamics with cell-state and fate in murine neurogenesis. Nat. Commun. 10, 2835. doi:10.1038/s41467-019-10734-8
Marinopoulou, E., Biga, V., Sabherwal, N., Miller, A., Desai, J., Adamson, A. D., et al. (2021). HES1 protein oscillations are necessary for neural stem cells to exit from quiescence. iScience 24, 103198. doi:10.1016/j.isci.2021.103198
Meletis, K., Barnabe-Heider, F., Carlen, M., Evergren, E., Tomilin, N., Shupliakov, O., et al. (2008). Spinal cord injury reveals multilineage differentiation of ependymal cells. PLoS Biol. 6, e182. doi:10.1371/journal.pbio.0060182
Miller, A., Biga, V., Rowntree, A., Chhatriwala, M., Woods, F., Noble, B., et al. (2025). NGN3 oscillatory expression controls the timing of human pancreatic endocrine differentiation. Dev. Cell 60, 2518–2532.e9. doi:10.1016/j.devcel.2025.06.004
Mokalled, M. H., Patra, C., Dickson, A. L., Endo, T., Stainier, D. Y., and Poss, K. D. (2016). Injury-induced ctgfa directs glial bridging and spinal cord regeneration in zebrafish. Science 354, 630–634. doi:10.1126/science.aaf2679
Monk, N. A. (2003a). Oscillatory expression of Hes1, p53, and NF-kappaB driven by transcriptional time delays. Curr. Biol. 13, 1409–1413. doi:10.1016/s0960-9822(03)00494-9
Monk, N. A. (2003b). Unravelling Nature's networks. Biochem. Soc. Trans. 31, 1457–1461. doi:10.1042/bst0311457
Munoz, R., Edwards-Faret, G., Moreno, M., Zuniga, N., Cline, H., and Larrain, J. (2015). Regeneration of Xenopus laevis spinal cord requires Sox2/3 expressing cells. Dev. Biol. 408, 229–243. doi:10.1016/j.ydbio.2015.03.009
Nakamura, M., Yoshida, H., Takahashi, E., Wlizla, M., Takebayashi-Suzuki, K., Horb, M. E., et al. (2020). The AP-1 transcription factor JunB functions in Xenopus tail regeneration by positively regulating cell proliferation. Biochem. Biophys. Res. Commun. 522, 990–995. doi:10.1016/j.bbrc.2019.11.060
Nandi, A., Vaz, C., Bhattacharya, A., and Ramaswamy, R. (2009). miRNA-regulated dynamics in circadian oscillator models. BMC Syst. Biol. 3, 45. doi:10.1186/1752-0509-3-45
Nogueira-Rodrigues, J., Leite, S. C., Pinto-Costa, R., Sousa, S. C., Luz, L. L., Sintra, M. A., et al. (2022). Rewired glycosylation activity promotes scarless regeneration and functional recovery in spiny mice after complete spinal cord transection. Dev. Cell 57, 440–450.e7. doi:10.1016/j.devcel.2021.12.008
Novak, B., and Tyson, J. J. (2008). Design principles of biochemical oscillators. Nat. Rev. Mol. Cell Biol. 9, 981–991. doi:10.1038/nrm2530
Ochi, S., Imaizumi, Y., Shimojo, H., Miyachi, H., and Kageyama, R. (2020). Oscillatory expression of Hes1 regulates cell proliferation and neuronal differentiation in the embryonic brain. Development 147, dev182204. doi:10.1242/dev.182204
Ogai, K., Nakatani, K., Hisano, S., Sugitani, K., Koriyama, Y., and Kato, S. (2014). Function of Sox2 in ependymal cells of lesioned spinal cords in adult zebrafish. Neurosci. Res. 88, 84–87. doi:10.1016/j.neures.2014.07.010
Ohnmacht, J., Yang, Y., Maurer, G. W., Barreiro-Iglesias, A., Tsarouchas, T. M., Wehner, D., et al. (2016). Spinal motor neurons are regenerated after mechanical lesion and genetic ablation in larval zebrafish. Development 143, 1464–1474. doi:10.1242/dev.129155
Ohori, Y., Yamamoto, S., Nagao, M., Sugimori, M., Yamamoto, N., Nakamura, K., et al. (2006). Growth factor treatment and genetic manipulation stimulate neurogenesis and oligodendrogenesis by endogenous neural progenitors in the injured adult spinal cord. J. Neurosci. 26, 11948–11960. doi:10.1523/JNEUROSCI.3127-06.2006
Olsen, P. H., and Ambros, V. (1999). The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216, 671–680. doi:10.1006/dbio.1999.9523
Palmeirim, I., Henrique, D., Ish-Horowicz, D., and Pourquie, O. (1997). Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell 91, 639–648. doi:10.1016/s0092-8674(00)80451-1
Pan, D., Liu, W., Zhu, S., Fan, B., Yu, N., Ning, G., et al. (2021). Potential of different cells-derived exosomal microRNA cargos for treating spinal cord injury. J. Orthop. Transl. 31, 33–40. doi:10.1016/j.jot.2021.09.008
Paridaen, J. T., and Huttner, W. B. (2014). Neurogenesis during development of the vertebrate central nervous system. EMBO Rep. 15, 351–364. doi:10.1002/embr.201438447
Park, H. C., Shin, J., Roberts, R. K., and Appel, B. (2007). An olig2 reporter gene marks oligodendrocyte precursors in the postembryonic spinal cord of zebrafish. Dev. Dyn. 236, 3402–3407. doi:10.1002/dvdy.21365
Parras, C. M., Galli, R., Britz, O., Soares, S., Galichet, C., Battiste, J., et al. (2004). Mash1 specifies neurons and oligodendrocytes in the postnatal brain. EMBO J. 23, 4495–4505. doi:10.1038/sj.emboj.7600447
Pelzer, D., Phipps, L. S., Thuret, R., Gallardo-Dodd, C. J., Baker, S. M., and Dorey, K. (2021). Foxm1 regulates neural progenitor fate during spinal cord regeneration. EMBO Rep. 22, e50932. doi:10.15252/embr.202050932
Piatt, J. (1955). Regeneration of the spinal cord in the salamander. J. Exp. Zool. 129, 177–207. doi:10.1002/jez.1401290109
Purvis, J. E., Karhohs, K. W., Mock, C., Batchelor, E., Loewer, A., and Lahav, G. (2012). p53 dynamics control cell fate. Science 336, 1440–1444. doi:10.1126/science.1218351
Rajman, M., and Schratt, G. (2017). MicroRNAs in neural development: from master regulators to fine-tuners. Development 144, 2310–2322. doi:10.1242/dev.144337
Rapp, P. E. (1987). Why are so many biological systems periodic? Prog. Neurobiol. 29, 261–273. doi:10.1016/0301-0082(87)90023-2
Rapp, P. E., Mees, A. I., and Sparrow, C. T. (1981). Frequency encoded biochemical regulation is more accurate than amplitude dependent control. J. Theor. Biol. 90, 531–544. doi:10.1016/0022-5193(81)90304-0
Reimer, M. M., Sorensen, I., Kuscha, V., Frank, R. E., Liu, C., Becker, C. G., et al. (2008). Motor neuron regeneration in adult zebrafish. J. Neurosci. 28, 8510–8516. doi:10.1523/JNEUROSCI.1189-08.2008
Reimer, M. M., Kuscha, V., Wyatt, C., Sorensen, I., Frank, R. E., Knuwer, M., et al. (2009). Sonic hedgehog is a polarized signal for motor neuron regeneration in adult zebrafish. J. Neurosci. 29, 15073–15082. doi:10.1523/JNEUROSCI.4748-09.2009
Reimer, M. M., Norris, A., Ohnmacht, J., Patani, R., Zhong, Z., Dias, T. B., et al. (2013). Dopamine from the brain promotes spinal motor neuron generation during development and adult regeneration. Dev. Cell 25, 478–491. doi:10.1016/j.devcel.2013.04.012
Reynolds, B. A., and Weiss, S. (1992). Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 255, 1707–1710. doi:10.1126/science.1553558
Ribeiro, A., Rebocho da Costa, M., de Sena-Tomas, C., Rodrigues, E. C., Quiteria, R., Macarico, T., et al. (2023). Development and repair of blood vessels in the zebrafish spinal cord. Open Biol. 13, 230103. doi:10.1098/rsob.230103
Riefolo, F., Matera, C., Garrido-Charles, A., Gomila, A. M. J., Sortino, R., Agnetta, L., et al. (2019). Optical control of cardiac function with a photoswitchable muscarinic agonist. J. Am. Chem. Soc. 141, 7628–7636. doi:10.1021/jacs.9b03505
Sabherwal, N., Rowntree, A., Marinopoulou, E., Pettini, T., Hourihane, S., Thomas, R., et al. (2021). Differential phase register of Hes1 oscillations with mitoses underlies cell-cycle heterogeneity in ER(+) breast cancer cells. Proc. Natl. Acad. Sci. U S A. 118, e2113527118. doi:10.1073/pnas.2113527118
Sabin, K. Z., Jiang, P., Gearhart, M. D., Stewart, R., and Echeverri, K. (2019). AP-1(cFos/JunB)/miR-200a regulate the pro-regenerative glial cell response during axolotl spinal cord regeneration. Commun. Biol. 2, 91. doi:10.1038/s42003-019-0335-4
Saliminejad, K., Khorram Khorshid, H. R., Soleymani Fard, S., and Ghaffari, S. H. (2019). An overview of microRNAs: biology, functions, therapeutics, and analysis methods. J. Cell Physiol. 234, 5451–5465. doi:10.1002/jcp.27486
Sanchez-Sanchez, J. M., Riefolo, F., Barbero-Castillo, A., Sortino, R., Agnetta, L., Manasanch, A., et al. (2025). Control of cortical slow oscillations and epileptiform discharges with photoswitchable type 1 muscarinic ligands. PNAS Nexus 4, pgaf009. doi:10.1093/pnasnexus/pgaf009
Sang, L., Coller, H. A., and Roberts, J. M. (2008). Control of the reversibility of cellular quiescence by the transcriptional repressor HES1. Science 321, 1095–1100. doi:10.1126/science.1155998
Schnapp, E., Kragl, M., Rubin, L., and Tanaka, E. M. (2005). Hedgehog signaling controls dorsoventral patterning, blastema cell proliferation and cartilage induction during axolotl tail regeneration. Development 132, 3243–3253. doi:10.1242/dev.01906
Schnell, L., Fearn, S., Klassen, H., Schwab, M. E., and Perry, V. H. (1999). Acute inflammatory responses to mechanical lesions in the CNS: differences between brain and spinal cord. Eur. J. Neurosci. 11, 3648–3658. doi:10.1046/j.1460-9568.1999.00792.x
Sehm, T., Sachse, C., Frenzel, C., and Echeverri, K. (2009). miR-196 is an essential early-stage regulator of tail regeneration, upstream of key spinal cord patterning events. Dev. Biol. 334, 468–480. doi:10.1016/j.ydbio.2009.08.008
Shcherbakova, D. M., Stepanenko, O. V., Turoverov, K. K., and Verkhusha, V. V. (2018). Near-infrared fluorescent proteins: multiplexing and optogenetics across scales. Trends Biotechnol. 36, 1230–1243. doi:10.1016/j.tibtech.2018.06.011
Shen, W., Cai, J., Li, J., Li, W., Shi, P., Zhao, X., et al. (2024). Regulation of MicroRNAs after spinal cord injury in adult zebrafish. J. Mol. Neurosci. 74, 66. doi:10.1007/s12031-024-02242-2
Shen, Y., Chen, X., Song, Z., Yao, H., Han, A., Zhang, Y., et al. (2024). MicroRNA-9 promotes axon regeneration of mauthner-cell in zebrafish via her6/calcium activity pathway. Cell Mol. Life Sci. 81, 104. doi:10.1007/s00018-024-05117-2
Shibata, M., Kurokawa, D., Nakao, H., Ohmura, T., and Aizawa, S. (2008). MicroRNA-9 modulates Cajal-Retzius cell differentiation by suppressing Foxg1 expression in mouse medial pallium. J. Neurosci. 28, 10415–10421. doi:10.1523/JNEUROSCI.3219-08.2008
Shibata, M., Nakao, H., Kiyonari, H., Abe, T., and Aizawa, S. (2011). MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J. Neurosci. 31, 3407–3422. doi:10.1523/JNEUROSCI.5085-10.2011
Shihabuddin, L. S., Ray, J., and Gage, F. H. (1997). FGF-2 is sufficient to isolate progenitors found in the adult mammalian spinal cord. Exp. Neurol. 148, 577–586. doi:10.1006/exnr.1997.6697
Shimojo, H., Ohtsuka, T., and Kageyama, R. (2008). Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron 58, 52–64. doi:10.1016/j.neuron.2008.02.014
Shimojo, H., Isomura, A., Ohtsuka, T., Kori, H., Miyachi, H., and Kageyama, R. (2016). Oscillatory control of Delta-like1 in cell interactions regulates dynamic gene expression and tissue morphogenesis. Genes Dev. 30, 102–116. doi:10.1101/gad.270785.115
Simpson, P. (1990). Lateral inhibition and the development of the sensory bristles of the adult peripheral nervous system of Drosophila. Development 109, 509–519. doi:10.1242/dev.109.3.509
Sonnen, K. F., and Aulehla, A. (2014). Dynamic signal encoding--from cells to organisms. Semin. Cell Dev. Biol. 34, 91–98. doi:10.1016/j.semcdb.2014.06.019
Soto, X., Biga, V., Kursawe, J., Lea, R., Doostdar, P., Thomas, R., et al. (2020). Dynamic properties of noise and Her6 levels are optimized by miR-9, allowing the decoding of the Her6 oscillator. EMBO J. 39, e103558. doi:10.15252/embj.2019103558
Soto, X., Burton, J., Manning, C. S., Minchington, T., Lea, R., Lee, J., et al. (2022). Sequential and additive expression of miR-9 precursors control timing of neurogenesis. Development 149, dev200474. doi:10.1242/dev.200474
Standart, N., and Jackson, R. J. (2007). MicroRNAs repress translation of m7Gppp-capped target mRNAs in vitro by inhibiting initiation and promoting deadenylation. Genes Dev. 21, 1975–1982. doi:10.1101/gad.1591507
Stefanova, E. E., Dychiao, J. V. T., Chinn, M. C., Borhani, M., and Scott, A. L. (2024). P2X7 regulates ependymo-radial glial cell proliferation in adult Danio rerio following spinal cord injury. Biol. Open 13, bio060270. doi:10.1242/bio.060270
Stenudd, M., Sabelstrom, H., and Frisen, J. (2015). Role of endogenous neural stem cells in spinal cord injury and repair. JAMA Neurol. 72, 235–237. doi:10.1001/jamaneurol.2014.2927
Stenudd, M., Sabelstrom, H., Llorens-Bobadilla, E., Zamboni, M., Blom, H., Brismar, H., et al. (2022). Identification of a discrete subpopulation of spinal cord ependymal cells with neural stem cell properties. Cell Rep. 38, 110440. doi:10.1016/j.celrep.2022.110440
Sterner, R. C., and Sterner, R. M. (2022). Immune response following traumatic spinal cord injury: pathophysiology and therapies. Front. Immunol. 13, 1084101. doi:10.3389/fimmu.2022.1084101
Strickland, E. R., Hook, M. A., Balaraman, S., Huie, J. R., Grau, J. W., and Miranda, R. C. (2011). MicroRNA dysregulation following spinal cord contusion: implications for neural plasticity and repair. Neuroscience 186, 146–160. doi:10.1016/j.neuroscience.2011.03.063
Sueda, R., and Kageyama, R. (2020). Regulation of active and quiescent somatic stem cells by notch signaling. Dev. Growth Differ. 62, 59–66. doi:10.1111/dgd.12626
Sueda, R., and Kageyama, R. (2021). Oscillatory expression of Ascl1 in oligodendrogenesis. Gene Expr. Patterns 41, 119198. doi:10.1016/j.gep.2021.119198
Sueda, R., Imayoshi, I., Harima, Y., and Kageyama, R. (2019). High Hes1 expression and resultant Ascl1 suppression regulate quiescent vs. active neural stem cells in the adult mouse brain. Genes Dev. 33, 511–523. doi:10.1101/gad.323196.118
Sugimori, M., Nagao, M., Bertrand, N., Parras, C. M., Guillemot, F., and Nakafuku, M. (2007). Combinatorial actions of patterning and HLH transcription factors in the spatiotemporal control of neurogenesis and gliogenesis in the developing spinal cord. Development 134, 1617–1629. doi:10.1242/dev.001255
Sugimori, M., Nagao, M., Parras, C. M., Nakatani, H., Lebel, M., Guillemot, F., et al. (2008). Ascl1 is required for oligodendrocyte development in the spinal cord. Development 135, 1271–1281. doi:10.1242/dev.015370
Sun, Y., Nadal-Vicens, M., Misono, S., Lin, M. Z., Zubiaga, A., Hua, X., et al. (2001). Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell 104, 365–376. doi:10.1016/s0092-8674(01)00224-0
Tan, S. L., Ohtsuka, T., Gonzalez, A., and Kageyama, R. (2012). MicroRNA9 regulates neural stem cell differentiation by controlling Hes1 expression dynamics in the developing brain. Genes cells. 17, 952–961. doi:10.1111/gtc.12009
Tang, Y., Ling, Z. M., Fu, R., Li, Y. Q., Cheng, X., Song, F. H., et al. (2014). Time-specific microRNA changes during spinal motoneuron degeneration in adult rats following unilateral brachial plexus root avulsion: ipsilateral vs. contralateral changes. BMC Neurosci. 15, 92. doi:10.1186/1471-2202-15-92
Taniguchi, Y., Watanabe, K., and Mochii, M. (2014). Notochord-derived hedgehog is essential for tail regeneration in Xenopus tadpole. BMC Dev. Biol. 14, 27. doi:10.1186/1471-213X-14-27
Tazaki, A., Tanaka, E. M., and Fei, J. F. (2017). Salamander spinal cord regeneration: the ultimate positive control in vertebrate spinal cord regeneration. Dev. Biol. 432, 63–71. doi:10.1016/j.ydbio.2017.09.034
Tong, L., Hill, R. A., Damisah, E. C., Murray, K. N., Yuan, P., Bordey, A., et al. (2021). Imaging and optogenetic modulation of vascular mural cells in the live brain. Nat. Protoc. 16, 472–496. doi:10.1038/s41596-020-00425-w
Tsarouchas, T. M., Wehner, D., Cavone, L., Munir, T., Keatinge, M., Lambertus, M., et al. (2018). Dynamic control of proinflammatory cytokines Il-1β and Tnf-α by macrophages in zebrafish spinal cord regeneration. Nat. Commun. 9, 4670. doi:10.1038/s41467-018-07036-w
Tsata, V., and Wehner, D. (2021). Know how to regrow-axon regeneration in the zebrafish spinal cord. Cells 10, 1404. doi:10.3390/cells10061404
Tsata, V., Kroehne, V., Wehner, D., Rost, F., Lange, C., Hoppe, C., et al. (2020). Reactive oligodendrocyte progenitor cells (Re-) myelinate the regenerating zebrafish spinal cord. Development 147, dev193946. doi:10.1242/dev.193946
Tzeng, S. F., Bresnahan, J. C., Beattie, M. S., and de Vellis, J. (2001). Upregulation of the HLH Id gene family in neural progenitors and glial cells of the rat spinal cord following contusion injury. J. Neurosci. Res. 66, 1161–1172. doi:10.1002/jnr.10089
Uriu, K. (2016). Genetic oscillators in development. Dev. Growth Differ. 58, 16–30. doi:10.1111/dgd.12262
Vajn, K., Plunkett, J. A., Tapanes-Castillo, A., and Oudega, M. (2013). Axonal regeneration after spinal cord injury in zebrafish and mammals: differences, similarities, translation. Neurosci. Bull. 29, 402–410. doi:10.1007/s12264-013-1361-8
van Raamsdonk, W., Maslam, S., de Jong, D. H., Smit-Onel, M. J., and Velzing, E. (1998). Long term effects of spinal cord transection in zebrafish: swimming performances, and metabolic properties of the neuromuscular system. Acta histochem. 100, 117–131. doi:10.1016/S0065-1281(98)80021-4
Vandestadt, C., Vanwalleghem, G. C., Khabooshan, M. A., Douek, A. M., Castillo, H. A., Li, M., et al. (2021). RNA-induced inflammation and migration of precursor neurons initiates neuronal circuit regeneration in zebrafish. Dev. Cell 56, 2364–2380.e8. doi:10.1016/j.devcel.2021.07.021
Vasudevan, D., Liu, Y. C., Barrios, J. P., Wheeler, M. K., Douglass, A. D., and Dorsky, R. I. (2021). Regenerated interneurons integrate into locomotor circuitry following spinal cord injury. Exp. Neurol. 342, 113737. doi:10.1016/j.expneurol.2021.113737
Verma, P., Garcia-Alias, G., and Fawcett, J. W. (2008). Spinal cord repair: bridging the divide. Neurorehabil Neural Repair 22, 429–437. doi:10.1177/1545968307313500
Walker, S. E., Sabin, K. Z., Gearhart, M. D., Yamamoto, K., and Echeverri, K. (2022). Regulation of stem cell identity by miR-200a during spinal cord regeneration. Development 149, dev200033. doi:10.1242/dev.200033
Wang, C. Y., Yang, S. H., and Tzeng, S. F. (2015). MicroRNA-145 as one negative regulator of astrogliosis. Glia 63, 194–205. doi:10.1002/glia.22743
Wang, H., Moyano, A. L., Ma, Z., Deng, Y., Lin, Y., Zhao, C., et al. (2017). miR-219 cooperates with miR-338 in myelination and promotes Myelin repair in the CNS. Dev. Cell 40, 566–582.e5. doi:10.1016/j.devcel.2017.03.001
Wang, F., Chang, S., Li, J., Wang, D., Li, H., and He, X. (2021). Lithium alleviated spinal cord injury (SCI)-induced apoptosis and inflammation in rats via BDNF-AS/miR-9-5p axis. Cell Tissue Res. 384, 301–312. doi:10.1007/s00441-020-03298-3
Wehner, D., Tsarouchas, T. M., Michael, A., Haase, C., Weidinger, G., Reimer, M. M., et al. (2017). Wnt signaling controls pro-regenerative Collagen XII in functional spinal cord regeneration in zebrafish. Nat. Commun. 8, 126. doi:10.1038/s41467-017-00143-0
Weiss, S., Dunne, C., Hewson, J., Wohl, C., Wheatley, M., Peterson, A. C., et al. (1996). Multipotent CNS stem cells are present in the adult mammalian spinal cord and ventricular neuroaxis. J. Neurosci. 16, 7599–7609. doi:10.1523/JNEUROSCI.16-23-07599.1996
Wu, L., Fan, J., and Belasco, J. G. (2006). MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. U S A. 103, 4034–4039. doi:10.1073/pnas.0510928103
Xie, Z. R., Yang, H. T., Liu, W. C., and Hwang, M. J. (2007). The role of microRNA in the delayed negative feedback regulation of gene expression. Biochem. Biophys. Res. Commun. 358, 722–726. doi:10.1016/j.bbrc.2007.04.207
Xu, Z., Zhang, K., Wang, Q., and Zheng, Y. (2019). MicroRNA-124 improves functional recovery and suppresses Bax-dependent apoptosis in rats following spinal cord injury. Mol. Med. Rep. 19, 2551–2560. doi:10.3892/mmr.2019.9904
Yamamoto, S., Nagao, M., Sugimori, M., Kosako, H., Nakatomi, H., Yamamoto, N., et al. (2001). Transcription factor expression and notch-dependent regulation of neural progenitors in the adult rat spinal cord. J. Neurosci. 21, 9814–9823. doi:10.1523/JNEUROSCI.21-24-09814.2001
Yu, Y. M., Gibbs, K. M., Davila, J., Campbell, N., Sung, S., Todorova, T. I., et al. (2011). MicroRNA miR-133b is essential for functional recovery after spinal cord injury in adult zebrafish. Eur. J. Neurosci. 33, 1587–1597. doi:10.1111/j.1460-9568.2011.07643.x
Yunta, M., Nieto-Diaz, M., Esteban, F. J., Caballero-Lopez, M., Navarro-Ruiz, R., Reigada, D., et al. (2012). MicroRNA dysregulation in the spinal cord following traumatic injury. PLoS One 7, e34534. doi:10.1371/journal.pone.0034534
Zammit, P. S., Clarke, J. D., Golding, J. P., Goodbrand, I. A., and Tonge, D. A. (1993). Macrophage response during axonal regeneration in the axolotl central and peripheral nervous system. Neuroscience 54, 781–789. doi:10.1016/0306-4522(93)90247-d
Zhao, C., Deng, W., and Gage, F. H. (2008). Mechanisms and functional implications of adult neurogenesis. Cell 132, 645–660. doi:10.1016/j.cell.2008.01.033
Zhao, Q., Fan, P., Gu, Q., Yang, H., Xie, Y., Xia, S., et al. (2025). NG2 glia promote injured spinal cord repair through neuronal transdifferentiation and keratins expression in juvenile mice. Spine J. doi:10.1016/j.spinee.2025.07.028
Zheng, B., and Tuszynski, M. H. (2023). Regulation of axonal regeneration after mammalian spinal cord injury. Nat. Rev. Mol. Cell Biol. 24, 396–413. doi:10.1038/s41580-022-00562-y
Zhou, P., Cai, S., Liu, Z., and Wang, R. (2012). Mechanisms generating bistability and oscillations in microRNA-mediated motifs. Phys. Rev. E Stat. Nonlin Soft Matter Phys. 85, 041916. doi:10.1103/PhysRevE.85.041916
Keywords: oscillations, neurogenesis, regeneration, development, microRNA, BHLH factors
Citation: Leino SA and Soto X (2025) Importance of ultradian oscillations in neurogenesis during development and its implications for spinal cord regeneration. Front. Cell Dev. Biol. 13:1680322. doi: 10.3389/fcell.2025.1680322
Received: 05 August 2025; Accepted: 25 September 2025;
Published: 08 October 2025; Corrected: 09 October 2025.
Edited by:
Samir Vaid, University Hospital Carl Gustav Carus, GermanyReviewed by:
Sayako Katada, Kyushu University, JapanAdam Matthew Elkin, University of Plymouth, United Kingdom
Copyright © 2025 Leino and Soto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ximena Soto, eGltZW5hLnNvdG9AbWFuY2hlc3Rlci5hYy51aw==
 Sami A. Leino
Sami A. Leino Ximena Soto
Ximena Soto