- 1 Department of Pathology, University of Texas Medical Branch, Galveston, TX, USA
- 2 Sylvius Laboratory, Institute of Biology, Leiden University, Leiden, Netherlands
- 3 Microbiology and Immunology, University of Texas Medical Branch, Galveston, TX, USA
- 4 Center for Biodefense and Emerging Infectious Diseases, University of Texas Medical Branch, Galveston, TX, USA
- 5 Sealy Center for Vaccine Development, University of Texas Medical Branch, Galveston, TX, USA
- 6 Institute for Human Infections and Immunity, University of Texas Medical Branch, Galveston, TX, USA
Ehrlichia chaffeensis has type 1 and 4 secretion systems (T1SS and T4SS), but the substrates have not been identified. Potential substrates include secreted tandem repeat protein (TRP) 47, TRP120, and TRP32, and the ankyrin repeat protein, Ank200, that are involved in molecular host–pathogen interactions including DNA binding and a network of protein–protein interactions with host targets associated with signaling, transcriptional regulation, vesicle trafficking, and apoptosis. In this study we report that E. chaffeensis TRP47, TRP32, TRP120, and Ank200 were not secreted in the Agrobacterium tumefaciens Cre recombinase reporter assay routinely used to identify T4SS substrates. In contrast, all TRPs and the Ank200 proteins were secreted by the Escherichia coli complemented with the hemolysin secretion system (T1SS), and secretion was reduced in a T1SS mutant (ΔTolC), demonstrating that these proteins are T1SS substrates. Moreover, T1SS secretion signals were identified in the C-terminal domains of the TRPs and Ank200, and a detailed bioinformatic analysis of E. chaffeensis TRPs and Ank200 revealed features consistent with those described in the repeats-in-toxins (RTX) family of exoproteins, including glycine- and aspartate-rich tandem repeats, homology with ATP-transporters, a non-cleavable C-terminal T1SS signal, acidic pIs, and functions consistent with other T1SS substrates. Using a heterologous E. coli T1SS, this investigation has identified the first Ehrlichia T1SS substrates supporting the conclusion that the T1SS and corresponding substrates are involved in molecular host–pathogen interactions that contribute to Ehrlichia pathobiology. Further investigation of the relationship between Ehrlichia TRPs, Ank200, and the RTX exoprotein family may lead to a greater understanding of the importance of T1SS substrates and specific functions of T1SS in the pathobiology of obligately intracellular bacteria.
Introduction
Members of the family Anaplasmataceae consist of a group of Gram-negative obligately intracellular alphaproteobacteria belonging to the order Rickettsiales, and are responsible for various arthropod-borne diseases of mammalian hosts including ehrlichioses and anaplasmoses. Human monocytotropic the ehrlichiosis (HME) is an emerging life-threatening tick-borne zoonosis caused by Ehrlichia chaffeensis, which exhibits tropism for mononuclear phagocytes, and survives by evading the innate host defenses, most likely by secreting multiple effectors into the host cell (Barnewall et al., 1997; Lee and Rikihisa, 1998; Lin and Rikihisa, 2004). Genes encoding Sec-dependent and Sec-independent Tat, TRAP-T (tripartite ATP-independent periplasmic transporters), type 1 and 4 secretion systems have been identified in E. chaffeensis genome; however, genes representing components of other secretion systems (type 2, 3, 5, 6) are not present (Hotopp et al., 2006).
Recent studies have reported an increasing number of tyrosine phosphorylated bacterial effector proteins translocated into host cells by type 3 or type 4 secretion systems (T3SS or T4SS; Deibel et al., 1998; Stein et al., 2000; Clifton et al., 2004; Backert and Selbach, 2005; Selbach et al., 2009). Several important human pathogens such as Helicobacter pylori, Legionella pneumophila, Coxiella burnetii, Bordetella pertussis, Brucella melitensis, and Bartonella henselae utilize T4SS for the delivery of bacterial effector proteins into the cytoplasm of mammalian host cells in order to manipulate host cell functions (Christie et al., 2005; Backert and Meyer, 2006; Alvarez-Martinez and Christie, 2009). The T4SS consists of a substrate translocation channel that spans the periplasm and both membranes of Gram-negative bacteria. The archetypal Agrobacterium tumefaciens T4SS comprises 12 proteins named VirB1 through 11 (clustered in a single virB locus) and VirD4 (VirB/D4). T4SS gene clusters (generally clustered into two to five groups) have been identified in the members of family Anaplasmataceae (Ohashi et al., 2002; Collins et al., 2005; Hotopp et al., 2006; Mavromatis et al., 2006; Alvarez-Martinez and Christie, 2009), and expression of Ehrlichia T4SS genes is upregulated during infection (Cheng et al., 2008). Furthermore, using the Cre recombinase reporter assay for translocation (CRAfT) system developed in A. tumefaciens (Vergunst et al., 2000), translocation of an ankyrin repeat protein of Anaplasma phagocytophilum, AnkA, was recently reported (Lin et al., 2007).
The type 1 secretion system (T1SS) is widespread among Gram-negative bacteria and directs the translocation of a variety of proteins of various sizes and diverse functions from the cytoplasm to the extracellular medium (Delepelaire, 2004). The common substrates for T1SS include pore-forming hemolysins (HlyA), adenylate cyclases, lipases, proteases, surface layers, and hemophores (Delepelaire, 2004). T1SS consists of three proteins; an inner membrane protein with a cytoplasmic ATPase domain operating as an ATP-binding cassette (ABC) transporter (Escherichia coli HlyB), a periplasmic adaptor (also known as membrane fusion protein, MFP; E. coli HlyD), and an outer membrane channel protein of the TolC family (E. coli TolC). The interaction of the T1SS substrate with HlyB and HlyD triggers recruitment of TolC, thereby creating a continuous, but transient channel-tunnel from the cytosol directly into the extracellular medium (Thanabalu et al., 1998; Benabdelhak et al., 2003). Previous studies have shown that E. coli hemolysin transporter promotes secretion of heterologous T1SS substrates expressed in E. coli, including exotoxins Cya of B. pertussis (Sebo and Ladant, 1993), LtkA of Aggregatibacter actinomycetemcomitans (Lally et al., 1989), PaxA of Pasteurella aerogenes (Kuhnert et al., 2000), and FrpA of Neisseria meningitidis (Thompson and Sparling, 1993). Maintenance of genes encoding the type 1 and 4 secretion system components in the small Ehrlichia spp. genome that has evolved through reductive evolution is indicative of the importance of these secretion systems for survival, yet knowledge regarding Ehrlichia spp. secreted effectors and secretion mechanisms remain undefined.
Major immunoreactive proteins of E. chaffeensis have been molecularly characterized and many are members of a tandem repeat protein (TRP) family that includes TRP120 (ECH_0039), TRP47 (ECH_0166), TRP32 (ECH_0170), and one member of an ankyrin repeat protein (Ank) family, Ank200 (ECH_0684; Yu et al., 1996; Doyle et al., 2006; Luo et al., 2008, 2010). Microscopic, proteomic, and ultrastructural evidence indicates that these proteins are secreted and exposed on the surface of the bacterium and extracellularly associated with the morula matrix and membrane (Popov et al., 2000; Doyle et al., 2006; Luo et al., 2008; Wakeel et al., 2009, 2010a; Zhu et al., 2009). Several functions have been associated with TRPs in pathogenic bacteria, including immune evasion, adhesion, actin nucleation, and other host–pathogen interactions (Gaillard et al., 1991; Wren, 1991; Kling et al., 1997; Jordan et al., 2003; Clifton et al., 2005; Palmer and Brayton, 2007; Shak et al., 2009; Wang et al., 2009). Recent studies have demonstrated that E. chaffeensis TRP47 interacts with a network of host cell proteins involved in signaling, modulation of gene expression, and intracellular vesicle trafficking (Wakeel et al., 2009) and is tyrosine phosphorylated (Wakeel et al., 2010a). TRP120 is a novel DNA binding protein that binds a G + C-rich DNA motif and is involved in host gene transcriptional regulation (Zhu et al., 2011). E. chaffeensis TRP120 also interacts with a diverse group of host cell proteins associated with major biological processes including transcription and regulation, cell signaling, protein trafficking, and actin cytoskeleton organization (Luo et al., 2011). TRP47 and TRP120 are differentially expressed on the surface of E. chaffeensis (Popov et al., 2000; Doyle et al., 2006), and degradation of surface exposed TRP120 by endogenous serine protease HtrA been linked to reduced bacterial aggregation and internalization into THP-1 cells (Kumagai et al., 2010, 2011). E. chaffeensis Ank200 is translocated to the host cell nucleus and interacts with Alu-Sx element motifs located in promoters and introns of various host cell genes involved in transcription, apoptosis, ATPase activity, and structural proteins associated with the nucleus and membrane-bound organelles (Zhu et al., 2009). Similarly, AnkA (APH_0740), an ankyrin repeat-containing protein of A. phagocytophilum that is a T4SS substrate (Lin et al., 2007), is also translocated to the nucleus of infected neutrophils (Garcia-Garcia et al., 2009) where it appears to bind regulatory regions of the CYBB gene and interacts with gene regulatory regions containing high AT content (Garcia-Garcia et al., 2009). Others have shown that AnkA of A. phagocytophilum binds to Abi-1 and is tyrosine phosphorylated by Abl-1 host cell tyrosine kinase (Lin et al., 2007).
The objective of this study was to determine the secretion mechanism of E. chaffeensis tandem repeat (TR) and Ank proteins (effectors) using well established T4SS (Vergunst et al., 2000) and T1SS models (Masure et al., 1990; Thompson and Sparling, 1993). We determined that the TRP120, TRP47, TRP32, and Ank200 are not translocated by a VirB/D4-dependent T4SS. Conversely, we demonstrated that secretion of E. chaffeensis TR and Ank proteins is mediated by the hemolysin secretion apparatus in E. coli. We demonstrated that the C-terminus is essential and required for the secretion of E. chaffeensis TRPs and Ank200. Collectively, this work demonstrated that E. chaffeensis TRP120, TRP47, TRP32, and Ank200 are E. chaffeensis T1SS substrates, and like other bacterial effector proteins, the Ank200 protein is tyrosine phosphorylated by a host cell tyrosine kinase. These findings provide new insights into the E. chaffeensis TRPs and Ank200 secretion mechanisms, substrates, and demonstrate the importance of the T1SS in ehrlichial pathobiology.
Results
Examination of E. chaffeensis-Secreted TRP and Ank Proteins in T4SS
Expression of E. chaffeensis-secreted TRP and Ank proteins in A. tumefaciens
We have previously demonstrated that TRP120, TRP47, TRP32, and Ank200 are secreted in E. chaffeensis-infected cells (Popov et al., 2000; Doyle et al., 2006; Luo et al., 2008; Zhu et al., 2009). However, the secretion mechanism of TRP120, TRP47, TRP32, and Ank200 are still unknown. Interestingly, the C-terminal 20 amino acids of Ank200 contains a potential VirB/D4 T4SS recognition motif (AVSPSTSQGADVKKSSCQSK) that is positively charged (pI 9.2), and has a hydropathy profile similar to the consensus secretory motif R-X(7)-R-X-R-X-R of A. tumefaciens effectors, where replacement of the Arg residues by Lys has negligible effect on substrate translocation efficiency (Vergunst et al., 2005). To investigate whether E. chaffeensis TRP120, TRP47, TRP32, and Ank200 are T4SS substrates, we used the previously developed CRAfT system, a surrogate system that has been used successfully to identify or verify the translocation of several substrates including AnkA of A. phagocytophilum from A. tumefaciens into plant cells (Vergunst et al., 2000, 2005; Lin et al., 2007). To demonstrate E. chaffeensis protein transport in a VirB/D4-dependent manner, the C-terminal (320 amino acids) of Ank200, near full length TRP120 (99%), and full length TRP47 and TRP32 were translationally fused to the C-terminus of the Cre protein (Cre::Ank200-C, Cre::TRP120, Cre::TRP47, Cre::TRP32; Figure 1A; Tables A1 and A2 in Appendix). The expression of the fusion proteins was brought under the control of the vir induction system in A. tumefaciens and confirmed by Western blot analysis with anti-Cre antibody (Figure 1B). Visualization of the large Cre::TRP120 was difficult, which may be due inefficient transfer of this large size protein. But after long exposure of the film a faint band was visible at 175 kDa (Figure 1B, lane 4).
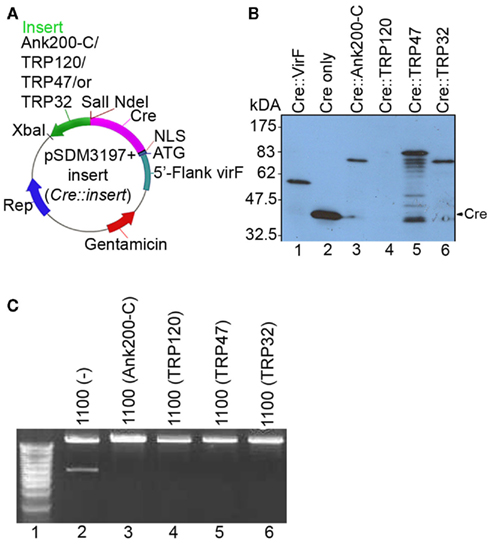
Figure 1. Cloning of Cre::Ehrlichia in-frame fusion constructs and their expression and Cre activity in A. tumefaciens. (A) Plasmids Cre::Ank200-C, Cre::TRP120, Cre::TRP47, and Cre::TRP32 harboring the fusion of Cre and C-terminal 320 amino acids of E. chaffeensis Ank200, TRP120, TRP47, and TRP32 were constructed from pSDM3197 (for details see Materials and Methods). (B) The expression of the fusion proteins was confirmed by western immunoblotting with anti-Cre antibody, lane 1, Cre::VirF (pSDM3155) 59.3 kDa; lane 2, Cre only (pSDM3197) 42.9 kDa; lane 3, Cre::Ank200-C (42.9 + 33.9 = 76.8 kDa; lane 4, Cre::TRP120 (42.9 + 60.8 = 103.7 kDa); lane 5, Cre::TRP47 (42.9 + 32.9 = 75.8 kDa); lane 6, Cre::TRP32 (42.9 + 22.5 = 65.4 kDa). (C) Plasmid pSDM3043 that contains a fragment with a BamHI restriction site between lox sites was introduced into A. tumefaciens strain LBA1100 harboring Cre::Ehrlichia fusion protein and grown overnight. The plasmid pSDM3043 was isolated and transformed into Escherichia coli strain DH5α. The plasmid pSDM3043 isolated from E. coli was digested with BamHI and then separated on agarose gel. Lane 1, DNA marker; lane 2, LBA1100 (wild-type) with pSDM3043; lane 3, LBA1100 (Cre::Ank200-C) with pSDM3043; lane 4, LBA1100 (Cre::TRP120) with pSDM3043; lane 5, LBA1100 (Cre::TRP47) with pSDM3043; lane 6, LBA1100 (Cre::TRP32) with pSDM3043. Cre activity causes excision of the blocking sequences (floxed DNA fragment).
Cre recombinase activity of Cre::Ehrlichia fusion proteins in A. tumefaciens
As the detection of protein translocation relies on Cre activity of the fusion proteins in the host cells we examined fusion protein Cre activity. A Cre recombinase activity assay was performed with Cre::Ehrlichia fusion proteins in A. tumefaciens strain LBA1100 containing the plasmid pSDM3043. Digestion of pSDM3043 by BamHI gives two fragments, but after deletion of a small floxed fragment by Cre recombination one of the BamHI sites is lost and only one fragment becomes visible after digestion with BamHI. The results showed that Cre is active in the Cre::Ehrlichia fusion proteins (Cre::Ank200-C, Cre::TRP120, Cre::TRP47, and Cre::TRP32) in A. tumefaciens strain LBA1100 as demonstrated by loss of the BamHI restriction site in the presence of these fusion proteins (Figure 1C, lanes 3, 4, 5, and 6). In contrast, two DNA fragments were detected in plasmid pSDM3043 isolated from A. tumefaciens strain LBA1100 lacking any Cre::Ehrlichia fusion protein, thus demonstrating the absence of Cre activity and served as a control (Figure 1C, lane 2).
Detection of protein translocation using CRAfT assay
Transformation of CB1 roots with A. tumefaciens strain LBA1100 with pSDM3155 expressing Cre–VirF fusion proteins (Cre::VirFΔ42N; A. tumefaciens fusion protein that serves as positive control) resulted in high numbers of CB1 cells expressing GFP 3 days after cocultivation (Figure 2B). Cocultivation with the negative control strain expressing Cre alone from the A. tumefaciens virF promoter, pSDM3197, rarely resulted in any GFP expression (Figure 2A). In contrast to the positive control, but similar to the negative control CB1 root explants cocultivated with A. tumefaciens strain LBA1100 transformed with the Cre::Ank200-C, Cre::TRP120, Cre::TRP47, and Cre::TRP32 fusion protein constructs, did not or only rarely result in any GFP expression (Figures 2C–F).
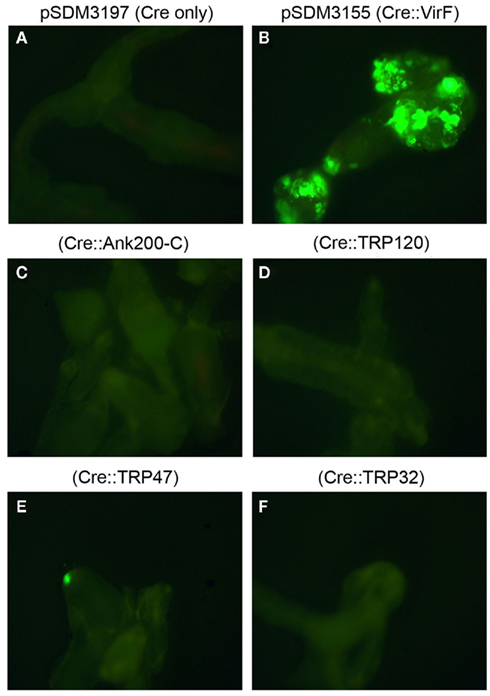
Figure 2. Visualization of protein translocation into host cells using CRAfT assay. Root explants of A. thaliana GFP reporter line CB1 4 days after cocultivation with A. tumefaciens wild-type strain LBA1100 containing (A) pSDM3197 plasmid (Cre only, serving as a negative control), (B) pSDM3155 (Cre:VirF, serving as a positive control), (C) Cre::Ank200-C, (D) Cre::TRP120, (E) Cre::TRP47, and (F) Cre::TRP32. Two Petri dishes, each containing at least 200 root explants were used per strain. Fluorescence microscopy was used to examine the GFP marker, which becomes active in CB1 cells after Cre-mediated excision of the blocking sequence, and thus indicates the successful translocation of Cre fusion protein into plant cells.
Ehrlichia VirD4 as coupling factor for translocation
The coupling factor VirD4 forms the interface between the translocated substrates and the VirB translocation channel. We hypothesized that in an effort to gain access to the VirB translocation channel Ehrlichia protein substrates may require their own cognate VirD4. To determine whether this is the case, E. chaffeensis virD4 with N-terminal c-Myc tag was cloned behind the virD promoter of A. tumefaciens into an incP plasmid (pSDM3668) so that protein transfer could be checked in the presence of E. chaffeensis VirD4. The expression of E. chaffeensis VirD4–c-Myc fusion protein in A. tumefaciens was confirmed by immunoblot using c-Myc specific antibodies (data not shown). Functional replacement of A. tumefaciens VirD4 by E. chaffeensis VirD4 was evaluated in a tumor assay on Nicotiana glauca. Strains with the wild-type A. tumefaciens Ti-plasmid, LBA1010 (octopine pTiB6; Beijersbergen et al., 1992), and strain LBA1010ΔVirD4 (LBA2586) complemented by A. tumefaciens VirD4 protein, caused similar levels of tumor formation. In contrast, strain LBA2586 complemented by the E. chaffeensis VirD4 protein induced no or much smaller overgrowths, hardly better than LBA2586 in N. glauca (Figure 3), corroborating that A. tumefaciens VirD4 is essential for virulence and that E. chaffeensis VirD4 cannot complement LBA2586 in the tumor assay on N. glauca. Thus, it is possible that the E. chaffeensis VirD4 cannot function as an intermediate in the transfer of the A. tumefaciens translocation substrates to the VirB channel. In the following step, protein translocation was tested in the CRAfT assay on A. thaliana CB1. In this assay, derivatives of the non-tumorigenic (oncogenic T-DNA lacking) helper strain LBA1100 and LBA2587, LBA1100 with the same virD4 deletion as in LBA2586, were used. A large number of CB1 cells expressing GFP were observed 3 days post cocultivation with A. tumefaciens strain LBA1100 [45] containing Cre::VirF (positive control), whereas no GFP expressing cells were seen after cocultivation with the virD4 mutant LBA2587 containing Cre::VirF (negative control). Complementation of the virD4 mutant by a plasmid containing A. tumefaciens virD4 restored its ability for Cre::VirF translocation, but introduction of the E. chaffeensis virD4 did not lead to translocation of the Cre::VirF protein. This further confirms that the E. chaffeensis VirD4 cannot mediate the translocation of the A. tumefaciens T4SS substrates to the VirB channel. In order to test whether E chaffeensis VirD4 could mediate translocation of the E. chaffeensis substrates, the above strains (LBA1100, 2587, 2587/3609, 2587/3668, 1100/3668) were tested for translocation of Cre::TRP32, Cre::TRP47, Cre::TRP120, and Cre::Ank200-C. However, also in the presence of E. chaffeensis VirD4 no or only rarely GFP expressing cells were seen in the CRAfT assays, indicating that even in the presence of E. chaffeensis VirD4 no clear indication for translocation of Cre::TRP32, Cre::TRP47, Cre::TRP120, and Cre::Ank200-C by the T4SS was obtained (Table A3 in Appendix). These findings demonstrate that E. chaffeensis TRP32, TRP47, TRP120, and Ank200 are not translocated to host cells by the T4SS and suggest that their translocation is mediated by another secretion system.

Figure 3. Determination of virD4-dependent virulence in tumor assay in N. glauca. Effects of virD4 deletion and/or replacement on A. tumefaciens virulence in N. glauca in a tumor assay. Tumor assay on N. glauca with A. tumefaciens strain (A) LBA1010 (wild-type), (B) LBA2586 (LBA1010ΔVirD4), (C) LBA2586 + pSDM3609 (A. tumefaciens wild-type VirD4), and (D) LBA2586 + pSDM3668 (E. chaffeensis wild-type VirD4).
E. chaffeensis Ank200 is a tyrosine phosphorylated effector protein
Ank200 is the largest immunoreactive protein identified in E. chaffeensis and is translocated to the nuclei of the infected monocytes, where it interacts with the mid-A-stretch of host promoter and intronic Alu elements (Zhu et al., 2009; Luo et al., 2010). It contains 11 potential tyrosine phosphorylation sites as predicted by NetPhos 2.0. In order to identify the E. chaffeensis tyrosine phosphorylated proteins we performed Western blotting analysis of uninfected and E. chaffeensis-infected THP-1 cell lysates with anti-pTyr monoclonal antibody (PY99). The Western blot analysis showed that E. chaffeensis infection of THP-1 cells led to a major tyrosine phosphorylated protein at ∼200 kDa (Figure 4A). To confirm the protein identity, an Ank200 specific antibody was used (Figure 4B). This 200 kDa protein was further detected by Western blot analysis using anti-Ank200 antibody in lysates of E. chaffeensis-infected THP-1 cells immunoprecipitated with anti-pTyr antibody and not in lysates of E. chaffeensis-infected THP-1 cells immunoprecipitated with normal mouse IgG confirming that the 200-kDa protein is tyrosine phosphorylated Ank200 (Figure 4C).
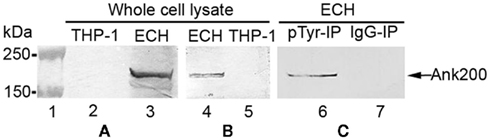
Figure 4. E. chaffeensis Ank200 protein was tyrosine phosphorylated in infected THP-1 cells. Whole cell lysates from normal (THP-1) and E. chaffeensis-infected THP-1 cells (ECH) were prepared and probed with (A) anti-pTyr antibody (lanes 2 and 3), (B) anti-Ank200 (lanes 4 and 5). (C) ECH whole cell lysates immunoprecipitated with mouse anti-pTyr antibody (pTyr-IP, lane 6) or normal mouse IgG (IgG-IP, lane 7) and detected with Ank200 antibody.
Comparative biophysical and domain analysis of tyrosine phosphorylated Ank proteins
The E. chaffeensis Ank200 and A. phagocytophilum AnkA proteins have recently been the focus of the several studies (McBride et al., 2003; Park et al., 2004; IJdo et al., 2007; Lin et al., 2007; Thomas and Fikrig, 2007; Garcia-Garcia et al., 2009; Zhu et al., 2009; Luo et al., 2010). The E. chaffeensis Ank200 and A. phagocytophilum AnkA proteins both contain Ank repeats and both are tyrosine phosphorylated (this study, IJdo et al., 2007; Lin et al., 2007). Some functional similarities have been reported between E. chaffeensis Ank200 and A. phagocytophilum AnkA, including translocation to the host cell nucleus and DNA interactions (Park et al., 2004; Garcia-Garcia et al., 2009; Zhu et al., 2009). Using the Cre recombinase reporter assay of A. tumefaciens a recent study reported that AnkA is translocated by the VirB/D4-dependent T4SS into the host cells (Lin et al., 2007). However, using the same Cre recombinase reporter assay, we found that Ank200 was not translocated by the VirB/D4-dependent T4SS, suggesting that Ank200 is translocated by another mechanism.
Although Ank200 and AnkA appear functionally similar, they have no significant sequence homology as demonstrated by their sequence alignment (BLASTN), and also have different biophysical properties, and thus, appear to be different in nature (Figure A1 in Appendix; Altschul et al., 1997). However, a search of E. chaffeensis Ank200 orthologs in the Integrated Microbial Genomes database identified A. phagocytophilum AnkA as an ortholog of Ank200, but with a limited (22%) sequence similarity that is primarily located in the Ank domain-containing regions of both the proteins. Ank200 (1463 amino acids) is more acidic (pI 4.9) with the majority of Ank motifs localized to the central region, while the tyrosine kinase, Src homology 2 (SH2), and Src homology 3 (SH3) domains are located in the N-terminus of the protein, which is more hydrophilic (Figure A1A in Appendix). In contrast, AnkA (1232 amino acids) is less acidic (pI 6.1), the Ank domains are localized to two distinct domains (N-terminus and central region) while the majority of tyrosine kinase, SH2, and SH3 domains were in the hydrophilic C-terminus of the protein (Figure A1B in Appendix). Moreover, the AnkA C-terminal 20 amino acids (SQPEAPQSEGPKSVKGGRGR) are more hydrophilic (grand average of hydropathy, −1.68, Expasy Proteomic Server) and in agreement with the requirements of the C-terminal T4SS signal [R-X(7)-R-X-R-X-R] (Vergunst et al., 2005) while the Ank200-C-terminal 20 amino acids (AVSPSTS QGADVKKSSCQSK) are less hydrophilic (grand average of hydropathy, −0.76) and do not have a prototypical T4SS signal (Figure A1C in Appendix).
Examination of E. chaffeensis-Secreted TRPs and Ank Proteins in T1SS
E. chaffeensis TRP47 TRP120, TRP32, and Ank200 amino acid composition and characteristics
The E. chaffeensis genome (NCBI accession number NC_007799) encodes T1SS genes (Hotopp et al., 2006). The E. coli hemolysin secretion system considered to be the prototype T1SS and is composed of the HlyB and HlyD proteins encoded by genes normally cotranscribed with hlyC and hlyA, while the outer membrane protein is encoded outside of the hly operon on the chromosome (Welch and Pellett, 1988; Wandersman and Delepelaire, 1990). We performed a BLASTP search for E. chaffeensis T1SS component genes (ECH_0383, ECH_0970, ECH_1020), and BLASTP identified a closest match for E. coli hlyB (YP_308793.1), hlyD (ZP_08360101.1), and tolC (EGB61997.1) genes with 27% (P = 5 × 10−56), 28% (P = 10−42), and 26% (P = 10−26) identity, respectively (Altschul et al., 1997). Although the similarity was low, the BLASTP results indicated that E. coli-like T1SS components exist in E. chaffeensis. Previous complementation studies have shown that the gene products of hlyB, hlyD, and tolC are required for the secretion of E. coli hemolysin (Mackman et al., 1985a,b; Wandersman and Delepelaire, 1990). The last 27 amino acids of the C-terminal region of hemolysin contain a specific signal sequence required for secretion (Nicaud et al., 1986; Mackman et al., 1987; Koronakis et al., 1989). The examination of the last 27 amino acids of the C-terminal region of the E. chaffeensis TRP47 and TRP120 proteins in a blast (BLASTP) search identified homology to several type 1 secretion substrates including ABC superfamily ABC transporter binding protein (Achromobacter piechaudii), ABC transporter periplasmic-binding protein (Bordetella petrii), and hemolysin (Sphingobacterium spiritivorum), and hemolysin A (S. spiritivorum; Table 1). A BLASTP search of the Ank200-C-terminal (last 27 amino acids) identified 69 and 89% homology to putative ABC transporter permease protein (Streptomyces cattleya) and nitrate/sulfonate/bicarbonate ABC transporter periplasmic protein (Starkeya novella), respectively (Table 1). Moreover, the E. chaffeensis TRP47 seven 19-mer TRs (ASVSEGDAVVNAVSQETPA, each repeat) covering a major part of the C-terminal region (42% of the full length protein) is glycine- and aspartate-rich and exhibits homology to adhesin (Staphylococcus epidermidis, SdrE), which is consistent with the common attributes of T1SS substrates (Delepelaire, 2004). A TRP32 C-terminal BLASTP search determined no substantial homology to other type 1 substrates; however, it identified homology to a Cyclic Nucleotide Gated Channel family member (Caenorhabditis elegans), an ion transport protein related to voltage and ligand gated potassium channel.
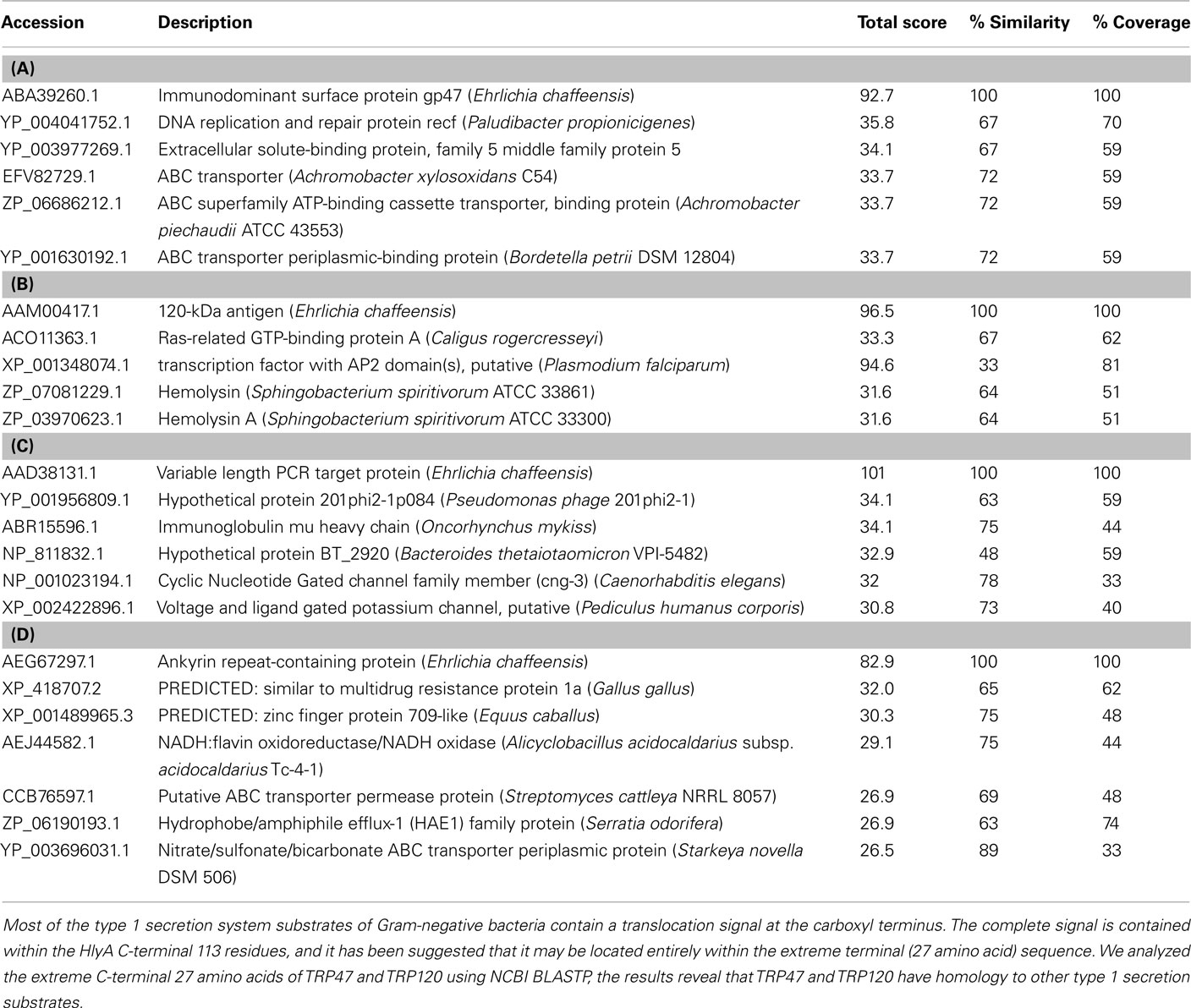
Table 1. NCBI BLASTP analysis result of C-terminal 27 amino acids of (A) TRP47, (B) TRP120, (C) TRP32, and (D) Ank200 identified homology to type 1 secretion substrates.
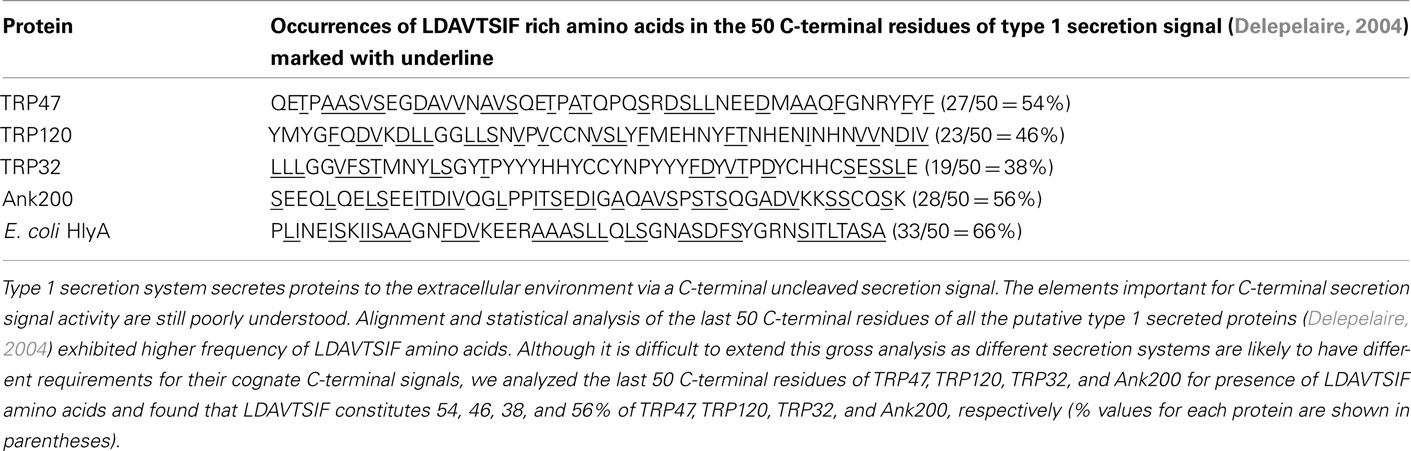
The T1SS translocates proteins to the extracellular environment via a C-terminal uncleaved secretion signal. We analyzed the last 50 C-terminal residues of TRP47, TRP120, TRP32, and Ank200 for the presence of LDAVTSIF amino acids and found that LDAVTSIF constitutes 54, 46, 38, and 56% of TRP47, TRP120, TRP32, and Ank200, respectively (Table 2). A previous study based on alignment and statistical analysis of the last 50 C-terminal residues of putative type 1 secreted proteins identified LDAVTSIF-enriched and KHPMWC-poor amino acids (Delepelaire, 2004).
Almost all the T1SS secreted proteins that have been characterized, including HlyA, LktA, CyaA, share a common domain structure and a secretion signal in the C-terminal domain of the protein (Delepelaire, 2004; Holland et al., 2005; Linhartova et al., 2010). E. chaffeensis TRPs and Ank200 exhibited a domain structure similar to repeats-in-toxin (RTX) exoprotein family such as HlyA, LktA, and CyaA (Figures 5A–C). Although the TRP47 19 amino acid TR sequence (ASVSEGDAVVNAVSQETPA) was not identical to RTX consensus sequence, it exhibited 69% similarity to S-layer protein in Methanotorris igneus (YP_004485351.1), 56% similarity to hemagglutinin in Stenotrophomonas sp. (ZP_05134659.1), 55% similarity to ABC transporter ATP-binding protein in Alteromonas sp. (YP_004469594.1) and 100% similarity to ABC superfamily ABC transporter, ABC protein in Kingella denitrificans (ZP_08132666.1), and metalloprotease, hemolysin-type calcium-binding region in Cupriavidus taiwanensis (YP_002008092.1). Furthermore, BLASTP identified amino acid sequence GDAVVN in each of the seven 19 amino acids TR sequences, which showed 100% similarity to ABC transporter ATP-binding protein in Gluconacetobacter hansenii (ZP_06834421.1) and Acetobacter pasteurianus (YP_003188074.1). An identical consensus sequence (GDAXXN) predicted to bind calcium ions has been identified in RTX proteins (Linhartova et al., 2010; Figure 5D).
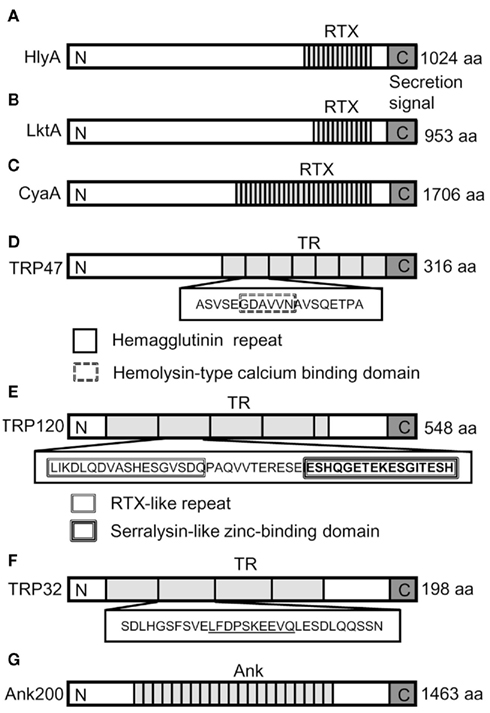
Figure 5. Schematic domain structures of the RTX toxins and E. chaffeensis TRPs and Ank200. RTX, Repeats-in-toxin in (A) E. coli HlyA, (B) P. haemolytica LktA, and (C) B. pertussis CyaA. (D) The putative hemagglutinin repeat and hemolysin-type calcium (Ca2+)-binding repeat in TRP47 TR are shown as white box with solid boundary and white box with double broken line boundary, respectively. (E) In TRP120, the RTX-like repeat of the aspartic acid and glycine-rich region and serralysin-like zinc-binding domain of histidine and glycine-rich region that are similar but not identical to RTX repeats and serralysin motif are indicated by double lined white box and triple lined white box, respectively. (F) TRP32 TR amino acids sequences are shown in white box with solid boundary and the amino acids sequences exhibiting greater than 75% sequence identity to ABC transporter permease and zinc metallopeptidase proteins are underlined. (G) Ank200 with centrally located ankyrin repeats (Ank). Overall, the boxed and underlined amino acid sequences represented in the figure indicated similarity to T1SS secreted proteins. The domain labeling is as follows: RTX, repeats-in-toxin domain; TR, tandem repeats domain; Ank, ankyrin repeats domain (map not to scale). The T1SS protein secretion signal is shown at the extreme C-terminal end of the proteins (gray colored box marked with C). The tandem repeat regions which vary in number and size of the repeat are shown as gray boxes. N and C represent the N and C-terminus of the protein, respectively.
Although the consensus sequence of RTX toxin (L/I/F-X-G-G-X-G-N/D-X, where X represents any amino acid) was not identified in the TRPs, but an RTX-like sequence (L/I/K-D-L-Q-D-VASHESGVSDQ) was found three times within the 80 amino acids long TRP120 TRs that exhibited 45% similarity with the ABC transporter, ATP-binding protein in Bacteroides clarus (ZP_08297392.1; Figure 5E). A part of the RTX-like sequence VASHESGVSDQ exhibited 64% similarity with putative ABC transporter ATP-binding protein in Marine actinobacterium (ZP_01129295.1) and ABC transporter ATP-binding protein in B. vulgates (YP_001297542.1), B. fluxus (ZP_08301787.1), and B. clarus (ZP_08297392.1). In addition, a unique TRP120 amino acid sequence (SEPFVAESEVSKVE) found within the TRs was similar to type 1 secretion membrane fusion protein, HlyD in Pectobacterium wasabiae and Pseudomonas mendocina, indicating that these regions may be required for TRP120 extracellular secretion by T1SS. Another unique glutamic acid- and histidine-rich amino acid sequence (ESHQGETEKESGITESH) was detected within the TRP120 TRs that exhibited similarity to zinc finger protein in Ailuropoda melanoleuca and Canis familiaris reminiscent of zinc-binding motif (HEXXHXXGXXH) analogous to that of the serralysin motif reported in P. pneumotropica RTX toxin PnxIIA (Sasaki et al., 2009; Figure 5E). Interestingly, the TRP32 TR showed homology to ATPase in Archaeoglobus profundus (YP_003400909.1) and beta-lactamase in Bacteroides vulgatus (ZP_06741900.1). Beta-lactamases were previously identified and predicted among the computationally detected RTX proteins (Linhartova et al., 2010). Moreover, TRP32 TR amino acid sequence (LFDPSKEEVQ) showed 80% identity to putative ABC transporter permease protein in Desulfovibrio magneticus (YP_002953007.1) and 75% identity to zinc metallopeptidase in Segniliparus rotundus (YP_003658757.1; Figure 5F). Although, we did not observe any homology of Ank200 to RTX proteins, a search for the RTX repeat structure GGXGXD using PATTINPROT software program set to find regions with 50 and 75% identity to the consensus RTX sequence identified a total of 27 and 4 repeat domains in Ank200. Moreover, the histidine-rich ankyrin repeat domain in Ank200 showed homology to zinc finger proteins that are involved in protein–protein and protein–DNA interactions (Figure 5G).
Secretion of E. chaffeensis TRPs and Ank200 by E. coli expressing HlyB and HlyD
Although, many previous studies using biochemical and molecular cellular imaging such as immunoconfocal and immunoelectron microscopy have clearly provided evidence of extracellular secretion of E. chaffeensis TRPs and Ank200 in infected mammalian cells, the secretion mechanism is unknown (Popov et al., 2000; Doyle et al., 2006; Luo et al., 2008; Zhu et al., 2009). TRP domain homology to RTX toxins and recent reviews of RTX toxins (Delepelaire, 2004; Linhartova et al., 2010) were supportive of E. chaffeensis TRPs as T1SS substrates. Thus, we investigated the ability of the E. coli HlyB and HlyD proteins to directly secrete E. chaffeensis TRPs and Ank200 into the extracellular medium. To this end, E. coli K-12 strain BW25113 that contains tolC, but does not contain the hlyCABD genes required for secretion of hemolysin was complemented with a dual vector, where vector pK184-HlyBD encodes inner membrane components HlyB and HlyD under the control of a lacZ promoter reconstituting the type 1 secretion apparatus and another vector pTRP/Ank200 encodes either E. chaffeensis TRP47, TRP120, TRP32, Ank200C4, or pHlyAc was used in the secretion assay.
In the type 1 secretion assay, large amounts of TRP47, TRP120, and TRP32 were secreted into the extracellular medium only in the presence of vector pK184-HlyBD compared to E. coli strain WM25113 harboring the single vector pTRP (Figures 6A,B). Although the expression levels of the TRPs were similar in E. coli lysates (data not shown), a higher concentration of E. chaffeensis TRP120 was detected in the supernatant compared to TRP47 and TRP32, and similar to that of HlyAc. Secretion of 23 kDa HlyAc into the medium was observed in the presence of the dual vector, pK184-HlyBD and pHlyAc, indicating that the HlyBD transport components were functional as previously demonstrated (Bakkes et al., 2010) and served as a positive control (Figure 6A). The size of the secreted TRP47, TRP120, and TRP32 was consistent with the sizes of the native proteins which migrate at larger than the actual molecular masses in SDS-PAGE (Wakeel et al., 2010a) as was demonstrated by Coomassie stain and confirmed by western immunoblotting using TRP47, TRP120, and TRP32 specific antibodies (Figures 6A,B). Ank200-C-terminus contains a type 1 secretion-like signal sequence as indicated by its similarity to ABC transporter permease and ABC transporter periplasmic proteins (Table 1). The Ank200C4 (Ank200-C-terminal 112 amino acids) secretion was detected in the extracellular medium only in the presence of HlyBD demonstrating that Ank200C4 is secreted by the functional T1SS system (Figures 6A,B).
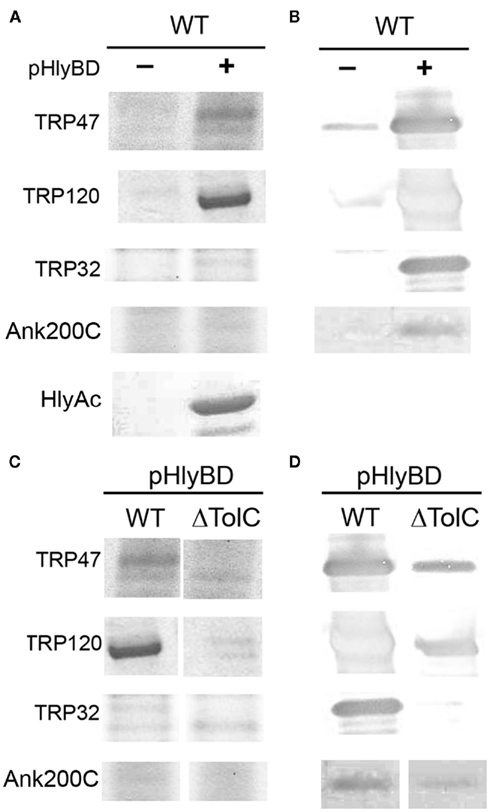
Figure 6. Extracellular secretion of E. chaffeensis TRP47, TRP120, TRP32, and Ank200 from E. coli by HlyB and HlyD. (A,B) E. coli BW25113 cells transfected with pK184-HlyBD (+) and a plasmid encoding TRP47, TRP120, TRP32, Ank200-C (C-terminal 112 amino acids; C4), or HlyAc as indicated were grown in LB medium supplemented with 1.5 mM IPTG to induce hlyBD coexpression. At OD660 = 0.8, the production of the TRP47, TRP120, TRP32, Ank200, or HlyAc proteins was induced by the addition of arabinose to a final concentration of 10 mM arabinose. Five hours after induction, protein in the culture supernatants was TCA-precipitated and analyzed by SDS-PAGE with Coomassie staining [(A), top left panel] or immunoblotting using TRP47, TRP120, TRP32, and Ank200 (C-terminal)-specific polyclonal antibodies [(B), top right panel]. E. coli BW25113 cells containing only a plasmid encoding TRP47, TRP120, TRP32, Ank200-C, or HlyAc protein but not containing pK184-HlyBD (indicated with −) were cultured, and proteins expressed and purified as described above and analyzed by SDS-PAGE with Coomassie staining [(A), top left panel] or immunoblotting using TRP47, TRP120, TRP32, and Ank200 (C-terminal)-specific polyclonal antibodies [(B), top right panel]. (C,D) E. coli BW25113 (WT) and CAG12184 (ΔTolC) cells containing pK184-HlyBD (+) and a plasmid encoding TRP47, TRP120, TRP32, or Ank200-C as indicated were cultured and proteins expressed and purified as described above. Coomassie staining, [(C) bottom left panel] or immunoblotting using TRP47, TRP120, TRP32, and Ank200 (C-terminal)-specific polyclonal antibodies [(D), bottom right panel]. “−” indicates in the absence presence of pK184-HlyBD and “+” indicates in the presence of pK184-HlyBD.
The structures at the C termini of RTX toxins that serve as secretion signals as well as the proteins required for their secretion are conserved among the bacteria secreting RTX toxins. This conservation is demonstrated by the ability of some of the transport proteins to mediate secretion of heterologous RTX toxins (Chang et al., 1989; Masure et al., 1990). In order to further define the domain necessary for secretion, we selected TRP47 as a model E. chaffeensis TRP and performed the secretion assay as described above for full length TRP47 using a dual vector system where type 1 secretion components HlyB and HlyD expressed by one vector and the substrate expressed by another vector in E. coli exhibited an increased level of secretion of C-terminal and full length GST–TRP47 fusion proteins in the presence of pHlyBD (Figures 7A,B). The N-terminal region of TRP47 was not secreted by itself to the extracellular medium. These results are consistent with the previous reports emphasizing the importance of the C-terminal domain of hemolysin which contains a secretion signal sequence that is required for secretion of the protein (Nicaud et al., 1986; Mackman et al., 1987; Koronakis et al., 1989) and demonstrating that secretion of TRP47 into the extracellular medium is dependent on type 1 secretion components similar to hemolysin.
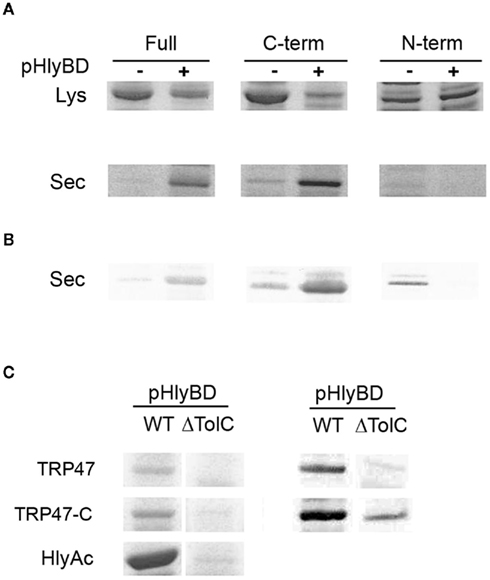
Figure 7. Extracellular secretion of E. chaffeensis full length, C-terminal, and N-terminal TRP47 fragment from E. coli. (A,B) E. coli BW25113 cells containing pK184-HlyBD (+) or not containing pK184-HlyBD (−) and a plasmid encoding GST–TRP47 full length (Full), GST–TRP47 C-terminal (C-term), or GST–TRP47 N-terminal (N-term) fusion protein as indicated were grown in LB medium supplemented with 1.5 mM IPTG to induce hlyBD coexpression and the production of the GST–TRP47 full length (Full), GST–TRP47 C-terminal (C-term), or GST–TRP47 N-terminal (N-term) fusion protein. Five hours after induction, protein in total cell extract [(A), Lys] or in the TCA-precipitated culture supernatants [(A), Sec] was analyzed by SDS-PAGE with Coomassie staining (A) or immunoblotting using anti-GST polyclonal antibodies [(B), Sec]. (C) E. coli BW25113 (WT) and CAG12184 (ΔTolC) cells containing pK184-HlyBD and a plasmid encoding GST–TRP47 full length (TRP47), GST–TRP47 C-terminal (TRP47C), or HlyAc protein as indicated were cultured and protein expressed and purified as described above. For E. coli WT and ΔTolC cells containing plasmid encoding HlyAc at OD660 = 0.8, the production of HlyAc protein was induced by the addition of arabinose to a final concentration of 10 mM arabinose. Five hours after induction, protein in the culture supernatants was TCA-precipitated and analyzed by SDS-PAGE with Coomassie staining [(C), left panel] or immunoblotting using anti-GST polyclonal antibodies [(C), right panel]. (Lys, indicates whole cell lysate; Sec, indicates secreted into the extracellular medium).
Extracellular secretion of Ehrlichia chaffeensis TRPs and Ank200 is reduced in the absence of Escherichia coli TolC protein
The type 1 secretion apparatus always includes a specific outer membrane protein, and in case of E. coli hemolysin secretion, this protein is TolC (Wandersman and Delepelaire, 1990). The TolC protein is an essential E. coli outer membrane protein that is required for hemolysin secretion (Wandersman and Delepelaire, 1990). In this study, we used a tolC210::Tn10, an insertional mutant derivative of E. coli K-12 strain CAG12184 (Singer et al., 1989). We cotransformed tolC210 with vector pK184-HlyBD and vector pTRP/Ank200C4 or pHlyAc to examine the extracellular secretion of E chaffeensis TRPs, Ank200C4, and HlyAc. This tolC mutant strain containing pK184-HlyBD exhibited a reduced level of E. chaffeensis TRP47, TRP120, TRP32, Ank200C4, and HlyAc secretion into the extracellular medium compared to wild-type E. coli (Figures 6C,D and 7C). Moreover, secretion of full length and C-terminal of GST–TRP47 fusion proteins was reduced in the tolC mutant compared to wild-type E. coli (Figure 7C). A small amount of protein (TRP47, TRP120, Ank200) was detected in supernatants of tolC mutant by western immunoblot, but no extracellular protein was detected for TRP32, which may be due to minimal lysis from overexpression or inefficient secretion due to the fact that HlyBD are expressed and functional (through complementation; Figures 6D and 7C). These results demonstrate that the outer membrane component, TolC, is important for translocation of the E. chaffeensis proteins from E. coli.
Discussion
In bacteria, secretion is essential for virulence and survival, and it is well established that TRPs and Ank200 proteins of Ehrlichia spp. are secreted and are involved in complex protein–protein and protein–DNA interactions with a diverse group of host cell targets and genes and are protective primary targets of the host humoral immune response (Yu et al., 1997; Sumner et al., 1999; McBride et al., 2003, 2007; Doyle et al., 2006; Nethery et al., 2007; Luo et al., 2009, 2010). E. chaffeensis, an obligately intracellular bacterium, resides and proliferates inside mononuclear phagocytes by manipulating host cell processes that affect cell signaling, transcription, and vesicle trafficking through specific interactions of its surface-expressed and secreted effector proteins (Popov et al., 2000; Doyle et al., 2006; Luo et al., 2008, 2009, 2011; Wakeel et al., 2010b; Zhu et al., 2011). Immunoelectron microscopy has identified TRP47 and TRP120 as differentially expressed proteins on the surface of dense-cored (DC) ehrlichiae, in addition to a non-differentially expressed TRP32, all of which are extracellularly associated with morular fibrillar matrix and the morula membrane, indicating that these proteins are secreted (Popov et al., 2000; Doyle et al., 2006; Luo et al., 2008). We have recently demonstrated that TRP47 interacts with multiple host proteins associated with cell signaling, transcriptional regulation, and vesicle trafficking and that TRP120 binds a G + C-rich motif in host cell DNA and exhibits eukaryotic transcriptional activator function and interacts with a diverse array of host proteins involved in transcription, signaling, and cytoskeleton organization similar to TRP47 (Wakeel et al., 2009; Luo et al., 2011; Zhu et al., 2011). Ank200 is translocated to the host cell nucleus where it binds with a specific adenine-rich motif of host promoter and intronic Alu elements (Zhu et al., 2009).
In general T1SS substrates are acidic proteins that contain TRs and a C-terminal secretion signal that is not cleaved during secretion. Protein BLAST (BLASTP) search of C-terminal amino acid sequence of TRP47, TRP120, TRP32, and Ank200 identified homology with type 1 secretion substrates (Altschul et al., 1997). Moreover, E. chaffeensis TRPs are acidic (pI ∼ 4) similar to type 1 substrates of other Gram-negative pathogens. A consensus T4SS substrate signal [R-X(7)-R-X-R-X-R] (Vergunst et al., 2005) is not present in TRPs. However, Ank200 contains a putative T4SS substrate motif, which is not similar to the prototypical T4SS signal. Although, previous studies have suggested secretion of the TRPs and Ank200 to be Sec-independent as they lack a classical signal peptide (SecretomeP 2.0), the secretion mechanisms of these E. chaffeensis effectors have remained undetermined. In this study we examined secretion of E. chaffeensis TRPs and Ank200 in T1SS and T4SS models and determined that TRPs and Ank200 are secreted into to the extracellular medium by T1SS similar to E. coli hemolysin and consistent with other RTX family exoproteins.
Recently, the use of a surrogate host enabled the identification of secretion substrates of a T4SS functioning in the obligate intracellular pathogen C. burnetii, which phylogenetically closely related to L. pneumophila. Both contain a Dot/Icm-like T4SS (Voth and Heinzen, 2009). Eleven C. burnetii Ank proteins expressed in L. pneumophila were found to be translocated through the L. pneumophila Dot/Icm system (Voth and Heinzen, 2009; Voth et al., 2009). In order to identify the substrates of the E. chaffeensis T4SS machinery, we investigated the secretion of E. chaffeensis Ank200, TRP32, TRP47, and TRP120 by using a previously developed CRAfT assay, which was used for the identification of T4SS translocation substrates from A. tumefaciens (Vergunst et al., 2000, 2005). The data obtained from the CRAfT assays demonstrated that translocation of Cre:: Ehrlichia Ank200, TRP32, TRP47, and TRP120 fusion proteins to A. thaliana CB1 plant cells by the T4SS does not occur. Although, the use of this heterologous T4SS system has provided insights into the translocation of many effector proteins, this study indicated that the E. chaffeensis TRPs and Ank200 were not translocated by the T4SS, underscoring the likelihood that another secretion mechanism may be involved in their secretion from E. chaffeensis into infected host cell (Doyle et al., 2006; Hotopp et al., 2006; Luo et al., 2008; Wakeel et al., 2009; Zhu et al., 2009).
Although the T4SS has been reported to be responsible for substrate translocation by Anaplasmataceae, only two T4SS substrates have been identified so far, one (AnkA) by the CRAfT assay and another (Ats-1) by using the bacterial two-hybrid assay (Lin et al., 2007; Niu et al., 2010; Rikihisa and Lin, 2010). Contrary to A. tumefaciens, in the E. chaffeensis genome the T4SS genes are spread over five groups, and several virB genes are duplicated (Hotopp et al., 2006; Cheng et al., 2008; Alvarez-Martinez and Christie, 2009). Although, trp120 is in the opposite orientation relative to the virB8-virD4 cluster (Yu et al., 1997), the close proximity of these genes is suggestive of a coordinated expression and function among T4SS and surface constituents (Alvarez-Martinez and Christie, 2009). Interestingly, although TRP120, which is located downstream of virD4 (ribA-virB8-virB9-virB10-virB11-virD4-trp120), it is not a T4SS substrate in contrast to other Gram-negative bacteria (Schulein et al., 2005; Hotopp et al., 2006; Alvarez-Martinez and Christie, 2009).
The results of this study are particularly important in the light of a previous report (Lin et al., 2007) and highlight our conclusion that Ank200 of E. chaffeensis is distinct from A. phagocytophilum AnkA in many respects. For instance, they have dissimilar nucleic acid sequences and exhibit a minimal (22%) amino acid identity limited to conserved Ank repeats. In Ank200 there are centralized Ank domains, and a majority of motifs including tyrosine kinase motif are localized in the N-terminus compared to AnkA where the Ank domains are spread over two major loci in the N-terminus and the central region, respectively, and the majority of motifs are in the C-terminus of the protein. However, most importantly, the C-terminal 20 amino acids of Ank200 and AnkA are clearly different, whereby the C-terminus of AnkA has more amino acids sequence similarity to the T4SS substrate signal [R-X(7)-R-X-R-X-R] (Vergunst et al., 2005) than that of Ank200, and thus AnkA, but not Ank200 is secreted by the T4SS machinery. Similarity of Ank200 domain structure and homology to TRPs and other T1SS substrates suggested that Ank200 is a T1SS substrate. Indeed, in this study, we demonstrated that Ank200-C-terminal (112 amino acids) peptide is secreted by T1SS.
Several previous studies reported that infection with Ehrlichia or Anaplasma induces tyrosine phosphorylation which is required for bacterial entry and proliferation (Zhang and Rikihisa, 1997; Lin et al., 2002, 2007; IJdo et al., 2007; Thomas and Fikrig, 2007). Tyrosine phosphorylation of the effector AnkA of A. phagocytophilum was reported recently (IJdo et al., 2007; Lin et al., 2007). However, no tyrosine phosphorylated effectors of E. chaffeensis were known until recently (Wakeel et al., 2010a; McBride et al., 2011). In this present study, we demonstrated that the strongly tyrosine phosphorylated 200 kDa protein in the E. chaffeensis-infected cell, is DNA binding protein Ank200, the largest major immunoreactive protein identified thus far in E. chaffeensis and E. canis (Nethery et al., 2007; Luo et al., 2010). Using mass spectrometry and immunoprecipitation, we have previously reported that E. chaffeensis TRP47, TRP75, and E. canis TRP95 are tyrosine phosphorylated (Wakeel et al., 2010a; McBride et al., 2011). Recent studies have shown that AnkA of A. phagocytophilum is tyrosine phosphorylated by host Abl-1 and Src tyrosine kinases and plays an important role in bacterial infection (IJdo et al., 2007; Lin et al., 2007). The E. chaffeensis effectors TRP47 (Wakeel et al., 2010a) and Ank200 (this study) are tyrosine phosphorylated; however, the host tyrosine kinases involved have not been identified. A recent study suggests that TRP47 physically interacts with Src family tyrosine kinase, Fyn, a key component of the αβTCR-coupled signaling pathway, and thus may be involved in tyrosine phosphorylation of TRP47 (Wakeel et al., 2009). The tyrosine kinase involved in Ank200 phosphorylation is unknown; however, Motif Scan prediction suggests that Abl and Lck tyrosine kinases may be involved.
T1SS in Gram-negative bacteria is dependent upon an ABC transporter but is Sec-independent, bypasses the periplasm and allows secretion of proteins of diverse sizes (19–800 kDa) and functions (proteases, adhesins or S-layer proteins, hemophores, hydrolases, lipases, toxins, or hemolytic enzymes) from the cytoplasm into the extracellular medium in a single step via a C-terminal uncleaved secretion signal (Delepelaire, 2004; Holland et al., 2005; Linhartova et al., 2010). Several unique features identified using bioinformatics in E. chaffeensis TRPs including glycine and aspartic acid-rich RTX-like repeats that specifically bind calcium ions in RTX proteins, are very acidic (pI ∼ 4), and a non-cleavable C-terminal secretion signal and exhibit homology with adhesins, are hallmarks of the T1SS substrates (Delepelaire, 2004; Linhartova et al., 2010). Alpha hemolysin (HlyA) of some uropathogenic E. coli isolates, leukotoxin (LktA) of Mannheimia haemolytica, bifunctional adenylate cyclase hemolysin (CyaA) of B. pertussis, metalloprotease PrtA and PrtB of Erwinia chrysanthemi, hemophore (HasA) and lipase (LipA) of Serratia marcescens, and FrpA and FrpC of N. meningitidis are some of the well characterized T1SS secreted proteins (Thompson and Sparling, 1993; Delepelaire, 2004; Linhartova et al., 2010). Although normally associated with the secretion of toxins or hydrolytic enzymes, the T1SS is essentially promiscuous and efficiently secretes a wide range of proteins carrying a type 1 secretion signal (Delepelaire, 2004; Linhartova et al., 2010).
The E. chaffeensis T1SS apparatus exhibits close similarity to the protease secretion apparatus in other bacteria. E. chaffeensis T1SS ATPase (ECH_0383) predicted to code for the T1SS ABC protein exhibited similarity to S. proteamaculans, E. amylovora, P. fluorescens, and Photorhabdus luminescens T1SS ABC transporter of the PrtD family. The type 1 secretion membrane fusion protein of the HlyD family is encoded by ECH_0970 showed homology with the HlyD family secretion proteins in Rhodospirillum centum, Marinomonas sp., and Pseudomonas syringae. The third component of the T1SS, the outer membrane protein TolC encoded by E. chaffeensis tolC (ECH_1020), exhibited similarity to type 1 secretion outer membrane protein, TolC in R. centenum and Parvibaculum lavamentivorans. E. coli hemolysin secretion system is considered to be the prototype T1SS and is composed of the HlyB and HlyD proteins encoded by genes normally cotranscribed with hlyC and hlyA, whereas the outer membrane protein TolC is encoded outside of the hly operon on the chromosome (Welch and Pellett, 1988; Wandersman and Delepelaire, 1990). Although the E. coli hemolysin secretion apparatus showed low homology to the E. chaffeensis T1SS apparatus, E. coli complemented with hlyB and hlyD efficiently secreted E. chaffeensis TRPs and Ank200 similar to FrpA of N. meningitidis (Thompson and Sparling, 1993). In contrast to the cistronic organization of the secretion genes, the E. chaffeensis genome encodes genes with similarity to E. coli hlyB and hlyD in two non-contiguous sequences like N. meningitidis where scattered genes encode a functional T1SS required for the secretion of meningococcal RTX proteins (Thompson and Sparling, 1993; Wooldridge et al., 2005).
The fact that the last 50 C-terminal residues of TRP47, TRP120, TRP32, and Ank200 contain a higher percentage of LDAVTSIF residues similar as previously reported to be present in T1SS secretion signals. This observation is consistent with and supports the concept that E. chaffeensis TRPs and Ank200 are typical T1SS substrates (Delepelaire, 2004). It is interesting to note that a LDAVTSIF residues-rich C-terminal secretion signal has been recently reported in A. phagocytophilum APH_0032 and APH_7378, which are proposed to be secreted by T1SS (Huang et al., 2010). Substantial similarity of seven E. chaffeensis TRP47 TRs to S-layer protein in M. igneus, hemagglutinin in Stenotrophomonas sp., ABC transporter ATP-binding protein in Alteromonas sp., and K. denitrificans, and metalloprotease, hemolysin-type calcium-binding region in C. taiwanensis is not only indicative of TRP47 being a T1SS secreted protein, but also points to its role as an E. chaffeensis effector. Furthermore, the presence of a consensus sequence (GDAXXN) seven times within TRP47 TRs predicted to bind calcium ions in RTX proteins and its similarity to ABC transporter ATP-binding protein in G. hansenii and A. pasteurianus provide additional evidence of the similarity of TRP47 to other T1SS substrates (Linhartova et al., 2010). Although the significance of a domain in E. chaffeensis TRP47 similar to hemagglutinin and hemolysin-type calcium-binding repeat domain is unknown, a recent study identified these repeat domains in RTX PnxIIIA of P. pneumotropica localized on the bacterial surface and associated with bacterial adherence and invasion of the host cell (Sasaki et al., 2011). The presence of multiple copies of RTX-like sequence (L/I/K-D-L-Q-D-VASHESGVSDQ) in TRP120 TRs showing similarity with the ABC transporter, ATP-binding protein in B. clarus, putative ABC transporter ATP-binding protein in Marine actinobacterium, ABC transporter ATP-binding protein in B. vulgates, B. fluxus, and B. clarus provides strong evidence that TRP120 is an RTX-like secreted protein. Furthermore, a glutamic acid and histidine-rich TRP120 amino acid sequence (ESHQGETEKESGITESH) exhibiting similarity to zinc finger protein in A. melanoleuca and C. familiaris and zinc-binding motif (HEXXHXXGXXH) reported in the major zinc-dependent metalloprotease secreted by S. marcescens serralysin and PnxIIA in P. pneumotropica supporting the conclusion that TRP120 is also secreted by T1SS (Sasaki et al., 2009).
Thus, overall the putative domains and repeat sequence in the primary structure of E. chaffeensis TRPs and Ank200 appear to be similar to those previously reported in E. coli HlyA, P. haemolytica LktA, B. pertussis CyaA and recently identified P. pneumotropica PnxIA, PnxIIA, and PnxIIIA RTX toxins (Sasaki et al., 2009, 2011; Linhartova et al., 2010). In addition, a unique TRP120 TR amino acid sequence (SEPFVAESEVSKVE) that showed similarity to type 1 secretion membrane fusion protein, HlyD in P. wasabiae and P. mendocina, indicates that these regions may be essential for TRP120 extracellular secretion by T1SS. HlyD forms a substrate-specific complex with inner membrane protein HlyB and subsequently recognizes the C-terminal signal peptide of HlyA. Upon binding of HlyA, the HlyD trimer interacts with the trimeric TolC protein inducing conformational change and extracellular export of HlyA (Andersen et al., 2001). Others have reported ABC transporter and MFP association only after substrate binding in S. marcescens hemophore HasA and E. chrysanthemi metalloprotease B and C (Letoffe et al., 1996).
The paradigm of type 1 secretion pathway is based on the analysis of the mechanism of secretion of the E. coli hemolysin. Complementation studies have shown that the gene products of hlyB, hlyD, and tolC are required for the secretion of hemolysin, the gene product hlyA of E. coli. The E. coli hemolysin secretion apparatus was shown promote secretion of heterologous RTX proteins expressed in E. coli, including the LktA of M. haemolytica, Cya of B. pertussis, HlyIA of Actinobacillus pleuropneumoniae, and FrpA of N. meningitides (Chang et al., 1989; Gygi et al., 1990; Masure et al., 1990; Thompson and Sparling, 1993). In this study we demonstrated that E. chaffeensis TRP47, TRP120, TRP32, and Ank200 possess features of T1SS substrates and are secreted by the E. coli T1SS secretion apparatus (HlyB, HlyD, and TolC). Secretion was exclusively dependent on E. coli HlyB and HlyD proteins, as secretion was significantly reduced in their absence. In the present work we also report that the TolC protein is required for E. chaffeensis TRP47, TRP120, TRP32, and Ank200 secretion. Previous studies have demonstrated the specific requirement of TolC for hemolysin and FrpC secretion, and that TolC is able to form an active T1SS transporter system in the presence of HlyB, HlyD to transport CyaA of B. pertussis, LtkA of A. actinomycetemcomitans, Pax of P. aerogenes, and FrpA and FrpC of N. meningitidis (Lally et al., 1989; Masure et al., 1990; Wandersman and Delepelaire, 1990; Thompson and Sparling, 1993; Kuhnert et al., 2000; Vakharia et al., 2001; Kamal et al., 2007). The small amount of extracellular TRPs and Ank200 detected in the supernatant of the TolC mutant in this study is consistent with similar TolC experiments reported previously (Thomas et al., 1992; Vakharia et al., 2001). Thus, E. chaffeensis TRPs and Ank200 are secreted only in the presence of a functional hemolysin secretion apparatus suggesting that TolC has a direct effect on TRPs and Ank200 secretion as reported for hemolysin (Wandersman and Delepelaire, 1990).
Using the heterologous type 1 secretion apparatus of E. coli, we demonstrated in this study that E. chaffeensis TR and Ank200 proteins/fragments are secreted into the extracellular medium by the T1SS dependent on HlyB, HlyD, and TolC indicating that these proteins may be transferred to the host cell by TISS. This study increases our knowledge of an E. chaffeensis secretion mechanism for the effectors to host cells. Our present study demonstrated for the first time the E. chaffeensis TRPs and Ank200 secretion by an ABC transporter dependent T1SS similar to RTX proteins. This knowledge may be useful for targeting and inhibiting Ehrlichia proliferation in future.
Materials and Methods
Propagation and Purification of E. chaffeensis
Propagation of uninfected and E. chaffeensis-infected THP-1 cells was performed as previously described (Wakeel et al., 2009).
Preparation of Whole Cell Lysates
Whole cell lysates were prepared as described previously (Wakeel et al., 2009) with some modifications. Briefly, 107 of uninfected and E. chaffeensis-infected (3 days post-infection) THP-1 cells were collected (500 × g, 5 min), washed twice in ice-cold phosphate buffered saline (PBS), resuspended in 1 ml of ice-cold RIPA lysis buffer (Pierce, Rockford, IL, USA) that contained complete Mini protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN, USA), phosphatase inhibitors cocktail (Pierce), 5 mM EDTA, and 1 mM of phenylmethylsulfonyl fluoride, sodium fluoride, sodium orthovanadate, and incubated for 20 min on ice. Cell lysates were prepared by sonication of cells for 1 min on ice. Lysates were collected by centrifugation at 12,000 × g for 10 min at 4°C.
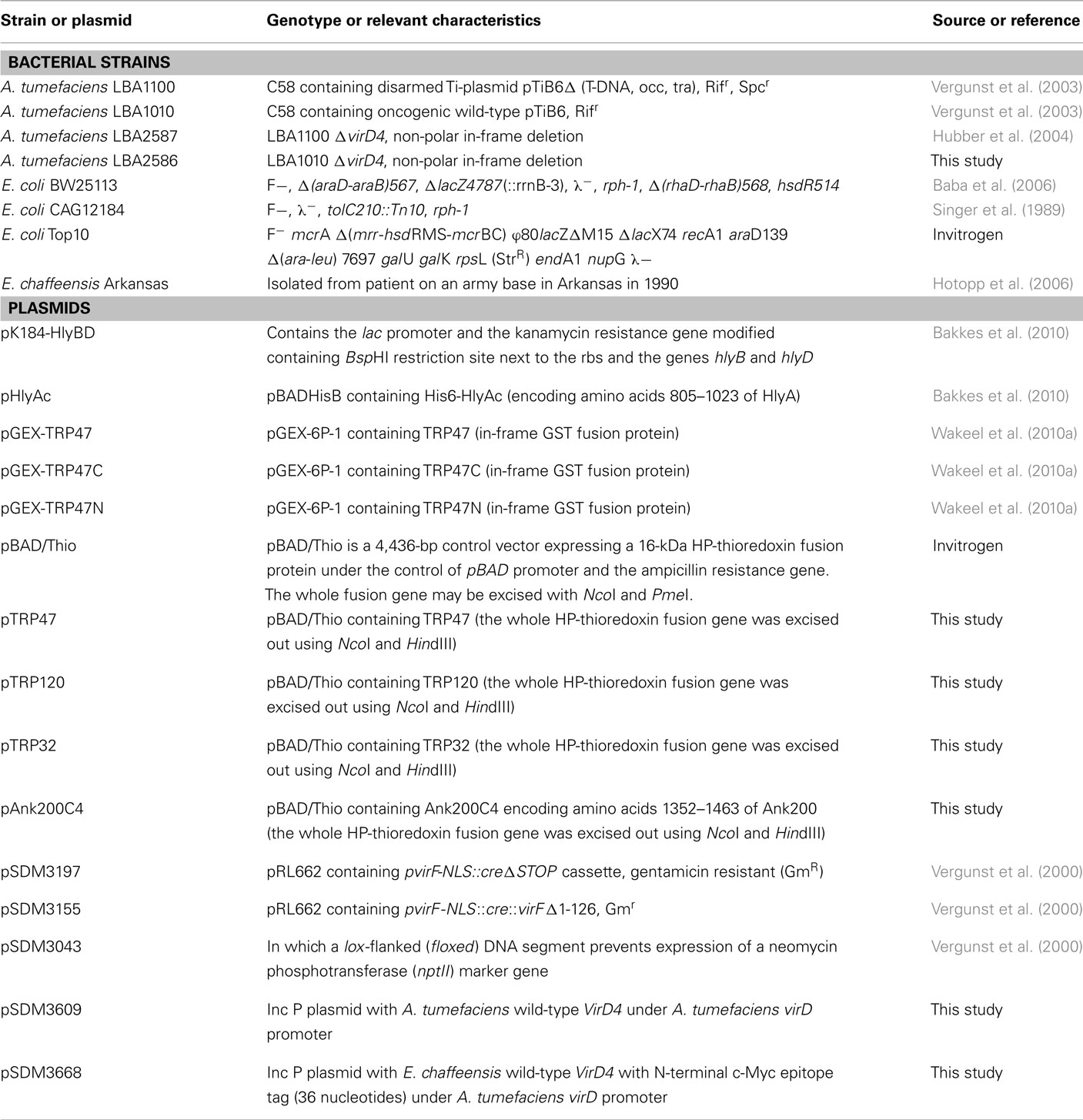
Cloning and Expression of Recombinant E. chaffeensis Ank200-C, TRP120, TRP47, and TRP32
For protein translocation study using T4SS model, in-frame fusions between the 3′ region of ank200 encoding the C-terminal 320 amino acids (Ank2003429–4392), nearly full length trp120 (trp12017-1647), trp47 (trp472-951), trp32 (trp322-597) and the cre coding region resulting in Cre::Ank200-C, Cre::TRP120, Cre::TRP47, Cre::TRP32 fusion proteins were generated by PCR, amplifying the corresponding coding regions from E. chaffeensis Arkansas strain genomic DNA using custom synthesized oligonucleotide primers (Table A1 in Appendix) in plasmid pSDM3197 (Schrammeijer et al., 2003). SalI/XbaI or SalI/NdeI-digested PCR product was translationally fused to cre via SalI/XbaI or SalI/NdeI-digested plasmid pSDM3197 (Schrammeijer et al., 2003). All cre control and cre-vir genes used in this study were expressed from the A. tumefaciens virF promoter sequence, and the chimeric proteins contained an N-terminally located simian virus 40 nuclear localization signal sequence to ensure nuclear targeting after Vir-mediated translocation into host cells. All plasmids were introduced into A. tumefaciens by electroporation (den Dulk-Ras and Hooykaas, 1995), and expression was confirmed by Western blot analysis as described (Vergunst et al., 2003). Briefly, the transformed A. tumefaciens strains including the control lines LBA1100 with pSDM3197 (Cre only) and pSDM3155 (Cre::VirFΔ42N of A. tumefaciens expressing Cre–VirF fusion proteins; Vergunst et al., 2000; Schrammeijer et al., 2003) were induced overnight with acetosyringone (Sigma). The pellets of the induced culture were boiled for 10 min and separated on SDS-PAGE gel prior to Western blot analysis using anti-Cre antibody.
For T1SS assay, the coding regions of the E. chaffeensis TRPs were amplified by PCR from E. chaffeensis genomic DNA using a forward primer that included a 5′ NcoI site and reverse primer with a 5′ HindIII site and stop codon, and ligated into the complementary sites of pBAD/Thio plasmid resulting in in-frame cloning of E. chaffeensis TRPs without thioredoxin fusion under the control of arabinose promoter and generation of plasmids pTRP47, pTRP120, pTRP32, and pAnk200C4 (see Tables A1 and A2 in Appendix for details). E. coli Top 10 (Invitrogen) was used for cloning procedures. E. coli K-12 strain BW25113 (wild-type) and tolC::Tn10 insertional mutant in E. coli K-12 strain CAG12184 (tolC mutant; Singer et al., 1989; Baba et al., 2006) were obtained from the Coli Genetic Resource Center (CGSC, E. coli Genetic resources at Yale University), and these cells were used for expression and secretion analysis in this study. The cloning and expression of the recombinant GST–TRP47 (full length, GST–TRP47; N-terminal, GST–NterTRP47; and C-terminal, GST–CterTRP47) fusion proteins have been described previously (Wakeel et al., 2010a). The plasmids pTRP47, pTRP120, pTRP32, pAnk200C4, pGEX-TRP47 (full length), pGEX-TRP47C-term (C-terminal), pGEX-TRP47N-term (N-terminal), and pHlyAC compatible with plasmid pK184-HlyBD were transformed into E. coli strains BW25113, CAG12184 (Tables 1 and 2) and selected on LB media containing appropriate antibiotics. The fusion proteins were expressed from an arabinose-inducible promoter (pBAD-Thio derivative) and isopropyl 1-thio-β-D-galactopyranoside (IPTG)-inducible promoter (pK184- and pGEX-derivative). E. coli (strains BW25113) cells harboring both compatible plasmids (pBAD-derived and pK184-HlyBD) were grown in the presence of ampicillin (100 μg/ml) and kanamycin (30 μg/ml). Secretion experiments in the absence of TolC were performed with E. coli CAG12184 tolC210::Tn10 (tetracycline resistant). Cells harboring both compatible plasmids (pBAD-derived and pK184-HlyBD) were grown in the presence of ampicillin (100 μg/ml), kanamycin (30 μg/ml), and tetracycline (10 μg/ml).
Expression and Secretion of Recombinant E. chaffeensis TRP and Ank Proteins by Wild-Type and tol C Mutant (tol C210::Tn10) E. coli Strains
Secretion experiments were performed as previously described with some modifications (Bakkes et al., 2010). Briefly, overnight cultures of E. coli strains (wild-type and tolC mutant) harboring the appropriate recombinant plasmids were diluted 1:20 into fresh LB supplemented with antibiotics. Cells were grown in LB medium containing isopropyl 1-thio-β-D-galactopyranoside at a final concentration of 1.5 mM for the production of the inner membrane proteins HlyB and HlyD with agitation at 30°C to an optical density at 600 nm (OD600) of 0.8, and then production of the TRP47, TRP120, TRP32, Ank200C4, and HlyAc fusion proteins was induced for 5 h by the addition of arabinose to a final concentration of 10 mM. Total cell extracts or culture supernatants were collected by centrifugation (12,000 × g, 10 min, 4°C). Proteins in the supernatants were concentrated by precipitation with 10% (v/v) trichloroacetic acid for 1 h at 4°C. The precipitated proteins were collected by centrifugation (16,000 × g, 30 min, 4°C) and washed in 80% acetone. Total cell extracts and precipitated proteins were resuspended in 1x sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, and total cell extract from 0.025 ml of culture pellet or protein precipitated from 1.0 ml of culture supernatant was analyzed by SDS-PAGE followed by staining with Coomassie brilliant blue (CBB) or probing by immunoblotting using anti-GST or anti-TRP47, -TRP120, -TRP32, and Ank200 specific antibodies.
Detection of Protein Translocation by CRAfT Assay
Translocation of Ank200, TRP120, TRP47, and TRP32 was performed as described previously by CRAfT assay (Vergunst et al., 2005). This system uses the site-specific recombinase Cre translationally fused to transport signals of the effector proteins (T4SS substrates). In brief, the seedlings from A. thaliana CB1 were grown for 10 days. Roots were collected and precultured for 3 days, followed by a 3-day cocultivation period with A. tumefaciens. Two Petri dishes, each containing at least 200 root explants, were used per strain. The GFP marker, which becomes active in CB1 cells only after Cre-mediated excision of the blocking sequence [lox-flanked (floxed) DNA sequence], allowed assaying for translocation directly after cocultivation by fluorescence microscopy (Leica MZ FLIII microscope and a Sony 3CCD color video camera).
Cre Recombinase Activity Assay of Cre::Ehrlichia Fusion Proteins in A. tumefaciens
To assay Cre activity in Cre::Ehrlichia fusion proteins, Cre::Ank200-C, Cre::TRP120, Cre::TRP47, and Cre::TRP32 containing A. tumefaciens strain LBA1100 further transformed with pSDM3043 (Vergunst and Hooykaas, 1998; Vergunst et al., 2000) and grown overnight. The plasmid pSDM3043 that contains a single BamHI restriction site between the floxed DNA fragments was rescued and transformed into E. coli strain DH5α, which was then grown overnight before isolation of plasmid pSDM3043. The plasmid pSDM3043 isolated from E. coli was digested with BamHI and then separated on an agarose gel.
Antibodies
Rabbit anti-E. chaffeensis Ank200 antiserum was generated against synthetic keyhole limpet hemocyanin-conjugated 25-mer C-terminus Ank200 peptide (1439-DIGAQAVSPSTSQGADVKKSSCQSK-1463) by a commercial vendor (Bio-Synthesis, Lewisville, TX, USA). Normal mouse IgG and other antibodies used in this study were mouse monoclonal anti-pTyr (PY99; Santa Cruz Biotechnology, Santa Cruz, CA, USA), goat anti-GST (GE Healthcare, Bio-Sciences Corp., Piscataway, NJ, USA) and anti-Cre (Eurogentec, Seraing, Belgium), and anti-TRP47, anti-TRP120, and anti-TRP32 described previously (Wakeel et al., 2010a).
Coimmunoprecipitation
Immunoprecipitation was performed as described (Wakeel et al., 2010a) except the nitrocellulose membranes were incubated with horseradish peroxidase-conjugated anti-pTyr or rabbit anti-Ank200 antibody.
Western Immunoblotting
Following electrophoresis, proteins were transferred to nitrocellulose membrane and detected with antibodies specific for TRP47, TRP120, TRP32, Ank200, E. chaffeensis, Cre, GST, or phosphotyrosine (pTyr; PY99; Santa Cruz Biotech) as previously described (Vergunst et al., 2003; Wakeel et al., 2010a).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge Lutz Schmitt and Patrick Bakkes for kindly providing pHlyBD and pHlyAc plasmids and CGSC Yale for E. coli strains BW25113 and CAG12184 used in this study. We are grateful to Tian Luo and Xiaofeng Zhang for constructing some plasmids used in this study. We thank Jason Carlyon for many invaluable discussions and David Walker and Xue-jie Yu for reviewing the manuscript and providing helpful suggestions. The Hooykaas lab was supported by a TOP grant from Chemical Sciences of the Netherlands Organization for Scientific Research (NWO). This work was supported by National Institute of Allergy and Infectious Diseases (AI 071145).
Abbreviations
Ank, ankyrin repeat protein; CRAfT, Cre recombinase reporter assay for translocation; HME, human monocytotropic ehrlichiosis; RTX, repeats-in-toxins; T1SS, type 1 secretion system; T3SS, type 3 secretion system; T4SS, type 4 secretion system; TRs, tandem repeats; TRP, tandem repeat protein.
References
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402.
Alvarez-Martinez, C. E., and Christie, P. J. (2009). Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 73, 775–808.
Andersen, C., Hughes, C., and Koronakis, V. (2001). Protein export and drug efflux through bacterial channel-tunnels. Curr. Opin. Cell Biol. 13, 412–416.
Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., Datsenko, K. A., Tomita, M., Wanner, B. L., and Mori, H. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008. doi: 10.1038/msb4100050
Backert, S., and Meyer, T. F. (2006). Type IV secretion systems and their effectors in bacterial pathogenesis. Curr. Opin. Microbiol. 9, 207–217.
Backert, S., and Selbach, M. (2005). Tyrosine-phosphorylated bacterial effector proteins: the enemies within. Trends Microbiol. 13, 476–484.
Bakkes, P. J., Jenewein, S., Smits, S. H., Holland, I. B., and Schmitt, L. (2010). The rate of folding dictates substrate secretion by the Escherichia coli hemolysin type 1 secretion system. J. Biol. Chem. 285, 40573–40580.
Barnewall, R. E., Rikihisa, Y., and Lee, E. H. (1997). Ehrlichia chaffeensis inclusions are early endosomes which selectively accumulate transferrin receptor. Infect. Immun. 65, 1455–1461.
Beijersbergen, A., Dulk-Ras, A. D., Schilperoort, R. A., and Hooykaas, P. J. (1992). Conjugative transfer by the virulence system of Agrobacterium tumefaciens. Science 256, 1324–1327.
Benabdelhak, H., Kiontke, S., Horn, C., Ernst, R., Blight, M. A., Holland, I. B., and Schmitt, L. (2003). A specific interaction between the NBD of the ABC-transporter HlyB and a C-terminal fragment of its transport substrate haemolysin A. J. Mol. Biol. 327, 1169–1179.
Chang, Y. F., Young, R., Moulds, T. L., and Struck, D. K. (1989). Secretion of the Pasteurella leukotoxin by Escherichia coli. FEMS Microbiol. Lett. 60, 169–173.
Cheng, Z., Wang, X., and Rikihisa, Y. (2008). Regulation of type IV secretion apparatus genes during Ehrlichia chaffeensis intracellular development by a previously unidentified protein. J. Bacteriol. 190, 2096–2105.
Christie, P. J., Atmakuri, K., Krishnamoorthy, V., Jakubowski, S., and Cascales, E. (2005). Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59, 451–485.
Clifton, D. R., Dooley, C. A., Grieshaber, S. S., Carabeo, R. A., Fields, K. A., and Hackstadt, T. (2005). Tyrosine phosphorylation of the chlamydial effector protein Tarp is species specific and not required for recruitment of actin. Infect. Immun. 73, 3860–3868.
Clifton, D. R., Fields, K. A., Grieshaber, S. S., Dooley, C. A., Fischer, E. R., Mead, D. J., Carabeo, R. A., and Hackstadt, T. (2004). A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc. Natl. Acad. Sci. U.S.A. 101, 10166–10171.
Collins, N. E., Liebenberg, J., de Villiers, E. P., Brayton, K. A., Louw, E., Pretorius, A., Faber, F. E., van, H. H., Josemans, A., van, K. M., Steyn, H. C., van Strijp, M. F., Zweygarth, E., Jongejan, F., Maillard, J. C., Berthier, D., Botha, M., Joubert, F., Corton, C. H., Thomson, N. R., Allsopp, M. T., and Allsopp, B. A. (2005). The genome of the heartwater agent Ehrlichia ruminantium contains multiple tandem repeats of actively variable copy number. Proc. Natl. Acad. Sci. U.S.A. 102, 838–843.
Deibel, C., Kramer, S., Chakraborty, T., and Ebel, F. (1998). EspE, a novel secreted protein of attaching and effacing bacteria, is directly translocated into infected host cells, where it appears as a tyrosine-phosphorylated 90 kDa protein. Mol. Microbiol. 28, 463–474.
Delepelaire, P. (2004). Type I secretion in gram-negative bacteria. Biochim. Biophys. Acta 1694, 149–161.
den Dulk-Ras, A., and Hooykaas, P. J. (1995). Electroporation of Agrobacterium tumefaciens. Methods Mol. Biol. 55, 63–72.
Doyle, C. K., Nethery, K. A., Popov, V. L., and McBride, J. W. (2006). Differentially expressed and secreted major immunoreactive protein orthologs of Ehrlichia canis and E. chaffeensis elicit early antibody responses to epitopes on glycosylated tandem repeats. Infect. Immun. 74, 711–720.
Gaillard, J. L., Berche, P., Frehel, C., Gouin, E., and Cossart, P. (1991). Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell 65, 1127–1141.
Garcia-Garcia, J. C., Rennoll-Bankert, K. E., Pelly, S., Milstone, A. M., and Dumler, J. S. (2009). Silencing of host cell CYBB gene expression by the nuclear effector AnkA of the intracellular pathogen Anaplasma phagocytophilum. Infect. Immun. 77, 2385–2391.
Gygi, D., Nicolet, J., Frey, J., Cross, M., Koronakis, V., and Hughes, C. (1990). Isolation of the Actinobacillus pleuropneumoniae haemolysin gene and the activation and secretion of the prohaemolysin by the HlyC, HlyB and HlyD proteins of Escherichia coli. Mol. Microbiol. 4, 123–128.
Holland, I. B., Schmitt, L., and Young, J. (2005). Type 1 protein secretion in bacteria, the ABC-transporter dependent pathway (review). Mol. Membr. Biol. 22, 29–39.
Hotopp, J. C., Lin, M., Madupu, R., Crabtree, J., Angiuoli, S. V., Eisen, J. A., Seshadri, R., Ren, Q., Wu, M., Utterback, T. R., Smith, S., Lewis, M., Khouri, H., Zhang, C., Niu, H., Lin, Q., Ohashi, N., Zhi, N., Nelson, W., Brinkac, L. M., Dodson, R. J., Rosovitz, M. J., Sundaram, J., Daugherty, S. C., Davidsen, T., Durkin, A. S., Gwinn, M., Haft, D. H., Selengut, J. D., Sullivan, S. A., Zafar, N., Zhou, L., Benahmed, F., Forberger, H., Halpin, R., Mulligan, S., Robinson, J., White, O., Rikihisa, Y., and Tettelin, H. (2006). Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2, e21. doi:10.1371/journal.pgen.0020021
Huang, B., Troese, M. J., Howe, D., Ye, S., Sims, J. T., Heinzen, R. A., Borjesson, D. L., and Carlyon, J. A. (2010). Anaplasma phagocytophilum APH_0032 is expressed late during infection and localizes to the pathogen-occupied vacuolar membrane. Microb. Pathog. 49, 273–284.
Hubber, A., Vergunst, A. C., Sullivan, J. T., Hooykaas, P. J., and Ronson, C. W. (2004). Symbiotic phenotypes and translocated effector proteins of the Mesorhizobium loti strain R7A VirB/D4 type IV secretion system. Mol. Microbiol. 54, 561–574.
IJdo, J. W., Carlson, A. C., and Kennedy, E. L. (2007). Anaplasma phagocytophilum AnkA is tyrosine-phosphorylated at EPIYA motifs and recruits SHP-1 during early infection. Cell. Microbiol. 9, 1284–1296.
Jordan, P., Snyder, L. A., and Saunders, N. J. (2003). Diversity in coding tandem repeats in related Neisseria spp. BMC Microbiol. 3, 23. doi:10.1186/1471-2180-3-23
Kamal, N., Rouquette-Loughlin, C., and Shafer, W. M. (2007). The TolC-like protein of Neisseria meningitidis is required for extracellular production of the repeats-in-toxin toxin FrpC but not for resistance to antimicrobials recognized by the Mtr efflux pump system. Infect. Immun. 75, 6008–6012.
Kling, D. E., Gravekamp, C., Madoff, L. C., and Michel, J. L. (1997). Characterization of two distinct opsonic and protective epitopes within the alpha C protein of the group B Streptococcus. Infect. Immun. 65, 1462–1467.
Koronakis, V., Koronakis, E., and Hughes, C. (1989). Isolation and analysis of the C-terminal signal directing export of Escherichia coli hemolysin protein across both bacterial membranes. EMBO J. 8, 595–605.
Kuhnert, P., Heyberger-Meyer, B., Nicolet, J., and Frey, J. (2000). Characterization of PaxA and its operon: a cohemolytic RTX toxin determinant from pathogenic Pasteurella aerogenes. Infect. Immun. 68, 6–12.
Kumagai, Y., Matsuo, J., Cheng, Z., Hayakawa, Y., and Rikihisa, Y. (2011). c-di-GMP signaling regulates intracellular aggregation, sessility, and growth of Ehrlichia chaffeensis. Infect. Immun. 79, 3905–3912.
Kumagai, Y., Matsuo, J., Hayakawa, Y., and Rikihisa, Y. (2010). Cyclic di-GMP signaling regulates invasion by Ehrlichia chaffeensis of human monocytes. J. Bacteriol. 192, 4122–4133.
Lally, E. T., Golub, E. E., Kieba, I. R., Taichman, N. S., Rosenbloom, J., Rosenbloom, J. C., Gibson, C. W., and Demuth, D. R. (1989). Analysis of the Actinobacillus actinomycetemcomitans leukotoxin gene. Delineation of unique features and comparison to homologous toxins. J. Biol. Chem. 264, 15451–15456.
Lee, E. H., and Rikihisa, Y. (1998). Protein kinase A-mediated inhibition of gamma interferon-induced tyrosine phosphorylation of Janus kinases and latent cytoplasmic transcription factors in human monocytes by Ehrlichia chaffeensis. Infect. Immun. 66, 2514–2520.
Letoffe, S., Delepelaire, P., and Wandersman, C. (1996). Protein secretion in gram-negative bacteria: assembly of the three components of ABC protein-mediated exporters is ordered and promoted by substrate binding. EMBO J. 15, 5804–5811.
Lin, M., den Dulk-Ras, A., Hooykaas, P. J., and Rikihisa, Y. (2007). Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell. Microbiol. 9, 2644–2657.
Lin, M., and Rikihisa, Y. (2004). Ehrlichia chaffeensis downregulates surface Toll-like receptors 2/4, CD14 and transcription factors PU.1 and inhibits lipopolysaccharide activation of NF-kappa B, ERK 1/2 and p38 MAPK in host monocytes. Cell. Microbiol. 6, 175–186.
Lin, M., Zhu, M. X., and Rikihisa, Y. (2002). Rapid activation of protein tyrosine kinase and phospholipase C-gamma2 and increase in cytosolic free calcium are required by Ehrlichia chaffeensis for internalization and growth in THP-1 cells. Infect. Immun. 70, 889–898.
Linhartova, I., Bumba, L., Masin, J., Basler, M., Osicka, R., Kamanova, J., Prochazkova, K., Adkins, I., Hejnova-Holubova, J., Sadilkova, L., Morova, J., and Sebo, P. (2010). RTX proteins: a highly diverse family secreted by a common mechanism. FEMS Microbiol. Rev. 34, 1076–1112.
Luo, T., Kuriakose, J. A., Zhu, B., Wakeel, A., and McBride, J. W. (2011). Ehrlichia chaffeensis TRP120 interacts with a diverse array of eukaryotic proteins involved in transcription, signaling and cytoskeleton organization. Infect. Immun. 79, 4382–4391.
Luo, T., Zhang, X., and McBride, J. W. (2009). Major species-specific antibody epitopes of the Ehrlichia chaffeensis p120 and E. canis p140 orthologs in surface-exposed tandem repeat regions. Clin. Vaccine Immunol. 16, 982–990.
Luo, T., Zhang, X., Nicholson, W. L., Zhu, B., and McBride, J. W. (2010). Molecular characterization of antibody epitopes of Ehrlichia chaffeensis ankyrin protein 200 and tandem repeat protein 47 and evaluation of synthetic immunodeterminants for serodiagnosis of human monocytotropic ehrlichiosis. Clin. Vaccine Immunol. 17, 87–97.
Luo, T., Zhang, X., Wakeel, A., Popov, V. L., and McBride, J. W. (2008). A variable-length PCR target protein of Ehrlichia chaffeensis contains major species-specific antibody epitopes in acidic serine-rich tandem repeats. Infect. Immun. 76, 1572–1580.
Mackman, N., Baker, K., Gray, L., Haigh, R., Nicaud, J. M., and Holland, I. B. (1987). Release of a chimeric protein into the medium from Escherichia coli using the C-terminal secretion signal of haemolysin. EMBO J. 6, 2835–2841.
Mackman, N., Nicaud, J. M., Gray, L., and Holland, I. B. (1985a). Genetical and functional organisation of the Escherichia coli haemolysin determinant 2001. Mol. Gen. Genet. 201, 282–288.
Mackman, N., Nicaud, J. M., Gray, L., and Holland, I. B. (1985b). Identification of polypeptides required for the export of haemolysin 2001 from E. coli. Mol. Gen. Genet. 201, 529–536.
Masure, H. R., Au, D. C., Gross, M. K., Donovan, M. G., and Storm, D. R. (1990). Secretion of the Bordetella pertussis adenylate cyclase from Escherichia coli containing the hemolysin operon. Biochemistry 29, 140–145.
Mavromatis, K., Doyle, C. K., Lykidis, A., Ivanova, N., Francino, M. P., Chain, P., Shin, M., Malfatti, S., Larimer, F., Copeland, A., Detter, J. C., Land, M., Richardson, P. M., Yu, X. J., Walker, D. H., McBride, J. W., and Kyrpides, N. C. (2006). The genome of the obligately intracellular bacterium Ehrlichia canis reveals themes of complex membrane structure and immune evasion strategies. J. Bacteriol. 188, 4015–4023.
McBride, J. W., Comer, J. E., and Walker, D. H. (2003). Novel immunoreactive glycoprotein orthologs of Ehrlichia spp. Ann. N. Y. Acad. Sci. 990, 678–684.
McBride, J. W., Doyle, C. K., Zhang, X., Cardenas, A. M., Popov, V. L., Nethery, K. A., and Woods, M. E. (2007). Identification of a glycosylated Ehrlichia canis 19-kilodalton major immunoreactive protein with a species-specific serine-rich glycopeptide epitope. Infect. Immun. 75, 74–82.
McBride, J. W., Zhang, X., Wakeel, A., and Kuriakose, J. A. (2011). Tyrosine-phosphorylated Ehrlichia chaffeensis and Ehrlichia canis tandem repeat orthologs contain a major continuous cross-reactive antibody epitope in lysine-rich repeats. Infect. Immun. 79, 3178–3187.
Nethery, K. A., Doyle, C. K., Zhang, X., and McBride, J. W. (2007). Ehrlichia canis gp200 contains dominant species-specific antibody epitopes in terminal acidic domains. Infect. Immun. 75, 4900–4908.
Nicaud, J. M., Mackman, N., Gray, L., and Holland, I. B. (1986). The C-terminal, 23 kDa peptide of E. coli haemolysin 2001 contains all the information necessary for its secretion by the haemolysin (Hly) export machinery. FEBS Lett. 204, 331–335.
Niu, H., Kozjak-Pavlovic, V., Rudel, T., and Rikihisa, Y. (2010). Anaplasma phagocytophilum Ats-1 is imported into host cell mitochondria and interferes with apoptosis induction. PLoS Pathog. 6, e1000774. doi:10.1371/journal.ppat.1000774
Ohashi, N., Zhi, N., Lin, Q., and Rikihisa, Y. (2002). Characterization and transcriptional analysis of gene clusters for a type IV secretion machinery in human granulocytic and monocytic ehrlichiosis agents. Infect. Immun. 70, 2128–2138.
Palmer, G. H., and Brayton, K. A. (2007). Gene conversion is a convergent strategy for pathogen antigenic variation. Trends Parasitol. 23, 408–413.
Park, J., Kim, K. J., Choi, K. S., Grab, D. J., and Dumler, J. S. (2004). Anaplasma phagocytophilum AnkA binds to granulocyte DNA and nuclear proteins. Cell. Microbiol. 6, 743–751.
Popov, V. L., Yu, X., and Walker, D. H. (2000). The 120 kDa outer membrane protein of Ehrlichia chaffeensis: preferential expression on dense-core cells and gene expression in Escherichia coli associated with attachment and entry. Microb. Pathog. 28, 71–80.
Rikihisa, Y., and Lin, M. (2010). Anaplasma phagocytophilum and Ehrlichia chaffeens is type IV secretion and Ank proteins. Curr. Opin. Microbiol. 13, 59–66.
Sasaki, H., Ishikawa, H., Sato, T., Sekiguchi, S., Amao, H., Kawamoto, E., Matsumoto, T., and Shirama, K. (2011). Molecular and virulence characteristics of an outer membrane-associated RTX exoprotein in Pasteurella pneumotropica. BMC Microbiol. 11, 55. doi:10.1186/1471-2180-11-55
Sasaki, H., Kawamoto, E., Tanaka, Y., Sawada, T., Kunita, S., and Yagami, K. (2009). Identification and characterization of hemolysin-like proteins similar to RTX toxin in Pasteurella pneumotropica. J. Bacteriol. 191, 3698–3705.
Schrammeijer, B., den Dulk-Ras, A., Vergunst, A. C., Jurado, J. E., and Hooykaas, P. J. (2003). Analysis of Vir protein translocation from Agrobacterium tumefaciens using Saccharomyces cerevisiae as a model: evidence for transport of a novel effector protein VirE3. Nucleic Acids Res. 31, 860–868.
Schulein, R., Guye, P., Rhomberg, T. A., Schmid, M. C., Schroder, G., Vergunst, A. C., Carena, I., and Dehio, C. (2005). A bipartite signal mediates the transfer of type IV secretion substrates of Bartonella henselae into human cells. Proc. Natl. Acad. Sci. U.S.A. 102, 856–861.
Sebo, P., and Ladant, D. (1993). Repeat sequences in the Bordetella pertussis adenylate cyclase toxin can be recognized as alternative carboxy-proximal secretion signals by the Escherichia coli alpha-haemolysin translocator. Mol. Microbiol. 9, 999–1009.
Selbach, M., Paul, F. E., Brandt, S., Guye, P., Daumke, O., Backert, S., Dehio, C., and Mann, M. (2009). Host cell interactome of tyrosine-phosphorylated bacterial proteins. Cell Host Microbe 5, 397–403.
Shak, J. R., Dick, J. J., Meinersmann, R. J., Perez-Perez, G. I., and Blaser, M. J. (2009). Repeat-associated plasticity in the Helicobacter pylori RD gene family. J. Bacteriol. 191, 6900–6910.
Singer, M., Baker, T. A., Schnitzler, G., Deischel, S. M., Goel, M., Dove, W., Jaacks, K. J., Grossman, A. D., Erickson, J. W., and Gross, C. A. (1989). A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53, 1–24.
Stein, M., Rappuoli, R., and Covacci, A. (2000). Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc. Natl. Acad. Sci. U.S.A. 97, 1263–1268.
Sumner, J. W., Childs, J. E., and Paddock, C. D. (1999). Molecular cloning and characterization of the Ehrlichia chaffeensis variable-length PCR target: an antigen-expressing gene that exhibits interstrain variation. J. Clin. Microbiol. 37, 1447–1453.
Thanabalu, T., Koronakis, E., Hughes, C., and Koronakis, V. (1998). Substrate-induced assembly of a contiguous channel for protein export from E. coli: reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J. 17, 6487–6496.
Thomas, V., and Fikrig, E. (2007). Anaplasma phagocytophilum specifically induces tyrosine phosphorylation of ROCK1 during infection. Cell. Microbiol. 9, 1730–1737.
Thomas, W. D. Jr., Wagner, S. P., and Welch, R. A. (1992). A heterologous membrane protein domain fused to the C-terminal ATP-binding domain of HlyB can export Escherichia coli hemolysin. J. Bacteriol. 174, 6771–6779.
Thompson, S. A., and Sparling, P. F. (1993). The RTX cytotoxin-related FrpA protein of Neisseria meningitidis is secreted extracellularly by meningococci and by HlyBD+ Escherichia coli. Infect. Immun. 61, 2906–2911.
Vakharia, H., German, G. J., and Misra, R. (2001). Isolation and characterization of Escherichia coli tolC mutants defective in secreting enzymatically active alpha-hemolysin. J. Bacteriol. 183, 6908–6916.
Vergunst, A. C., and Hooykaas, P. J. (1998). Cre/lox-mediated site-specific integration of Agrobacterium T-DNA in Arabidopsis thaliana by transient expression of cre. Plant Mol. Biol. 38, 393–406.
Vergunst, A. C., Schrammeijer, B., den Dulk-Ras, A., de Vlaam, C. M., Regensburg-Tuink, T. J., and Hooykaas, P. J. (2000). VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290, 979–982.
Vergunst, A. C., van Lier, M. C., den Dulk-Ras, A., and Hooykaas, P. J. (2003). Recognition of the Agrobacterium tumefaciens VirE2 translocation signal by the VirB/D4 transport system does not require VirE1. Plant Physiol. 133, 978–988.
Vergunst, A. C., van Lier, M. C., den Dulk-Ras, A., Stuve, T. A., Ouwehand, A., and Hooykaas, P. J. (2005). Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc. Natl. Acad. Sci. U.S.A. 102, 832–837.
Voth, D. E., and Heinzen, R. A. (2009). Coxiella type IV secretion and cellular microbiology. Curr. Opin. Microbiol. 12, 74–80.
Voth, D. E., Howe, D., Beare, P. A., Vogel, J. P., Unsworth, N., Samuel, J. E., and Heinzen, R. A. (2009). The Coxiella burnetii ankyrin repeat domain-containing protein family is heterogeneous, with C-terminal truncations that influence Dot/Icm-mediated secretion. J. Bacteriol. 191, 4232–4242.
Wakeel, A., Kuriakose, J. A., and McBride, J. W. (2009). An Ehrlichia chaffeensis tandem repeat protein interacts with multiple host targets involved in cell signaling, transcriptional regulation, and vesicle trafficking. Infect. Immun. 77, 1734–1745.
Wakeel, A., Zhang, X., and McBride, J. W. (2010a). Mass spectrometric analysis of Ehrlichia chaffeensis tandem repeat proteins reveals evidence of phosphorylation and absence of glycosylation. PLoS ONE. 5, e9552. doi:10.1371/journal.pone.0009552
Wakeel, A., Zhu, B., Yu, X. J., and McBride, J. W. (2010b). New insights into molecular Ehrlichia chaffeensis-host interactions. Microbes Infect. 12, 337–345.
Wandersman, C., and Delepelaire, P. (1990). TolC, an Escherichia coli outer membrane protein required for hemolysin secretion. Proc. Natl. Acad. Sci. U.S.A. 87, 4776–4780.
Wang, J., Chen, L., Chen, F., Zhang, X., Zhang, Y., Baseman, J., Perdue, S., Yeh, I. T., Shain, R., Holland, M., Bailey, R., Mabey, D., Yu, P., and Zhong, G. (2009). A chlamydial type III-secreted effector protein (Tarp) is predominantly recognized by antibodies from humans infected with Chlamydia trachomatis and induces protective immunity against upper genital tract pathologies in mice. Vaccine 27, 2967–2980.
Welch, R. A., and Pellett, S. (1988). Transcriptional organization of the Escherichia coli hemolysin genes. J. Bacteriol. 170, 1622–1630.
Wooldridge, K. G., Kizil, M., Wells, D. B., and Ala’Aldeen, D. A. (2005). Unusual genetic organization of a functional type I protein secretion system in Neisseria meningitidis. Infect. Immun. 73, 5554–5567.
Wren, B. W. (1991). A family of clostridial and streptococcal ligand-binding proteins with conserved C-terminal repeat sequences. Mol. Microbiol. 5, 797–803.
Yu, X. J., Crocquet-Valdes, P., Cullman, L. C., and Walker, D. H. (1996). The recombinant 120-kilodalton protein of Ehrlichia chaffeensis, a potential diagnostic tool. J. Clin. Microbiol. 34, 2853–2855.
Yu, X. J., Crocquet-Valdes, P., and Walker, D. H. (1997). Cloning and sequencing of the gene for a 120-kDa immunodominant protein of Ehrlichia chaffeensis. Gene 184, 149–154.
Zhang, Y., and Rikihisa, Y. (1997). Tyrosine phosphorylation is required for ehrlichial internalization and replication in P388D1 cells. Infect. Immun. 65, 2959–2964.
Zhu, B., Kuriakose, J. A., Luo, T., Ballesteros, E., Gupta, S., Fofanov, Y., and McBride, J. W. (2011). Ehrlichia chaffeensis TRP120 binds a G+C-rich motif in host cell DNA and exhibits eukaryotic transciptional activator function. Infect. Immun. 79, 4370–4381.
Zhu, B., Nethery, K. A., Kuriakose, J. A., Wakeel, A., Zhang, X., and McBride, J. W. (2009). Nuclear translocated Ehrlichia chaffeensis ankyrin protein interacts with a specific adenine-rich motif of host promoter and intronic Alu elements. Infect. Immun. 77, 4243–4255.
Appendix
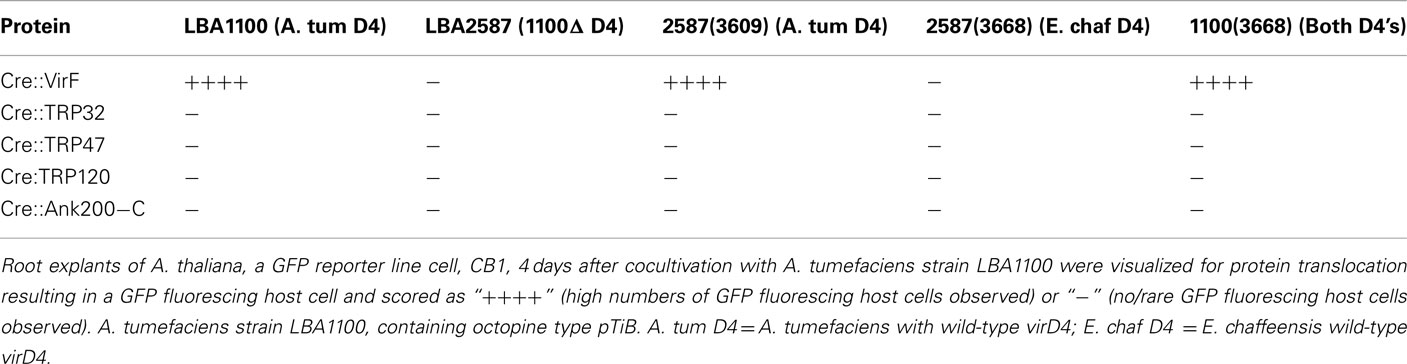
Table A3. Cre::Vir and Cre::Ehrlichia protein translocation from A. tumefacien s strain LBA1100 into A. thaliana (CB1), a reporter plant line that allows expression of GFP as a reporter for translocation upon Cre activity.
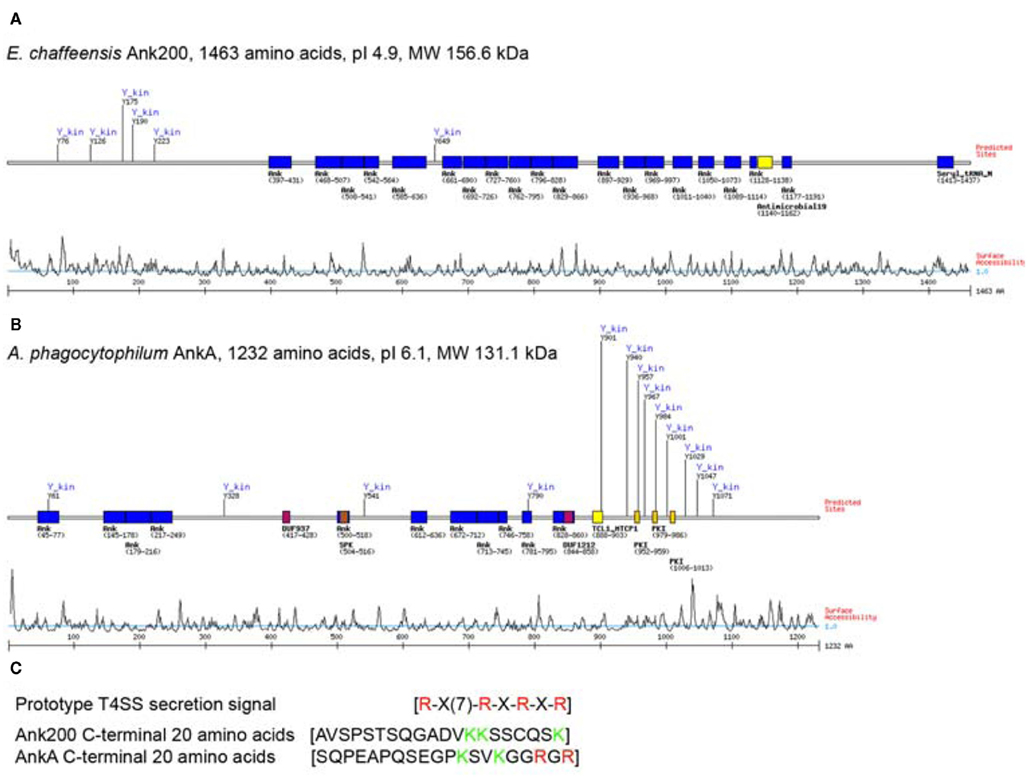
Figure A1. Comparison of biophysical characteristics and major motifs present on E. chaffeensis Ank200 and A. phagocytophilum AnkA. Major motifs and their relative positions in terms of amino acid sequences as predicted by Motif Scan (http://scansite.mit.edu) for (A) Ank200 and (B) AnkA. (C) A comparison of the C-terminal amino acid sequences of Ank200 and AnkA with prototype T4SS signal sequence.
Keywords: Ehrlichia, tandem repeat protein, ankyrin repeat protein, type 1 and 4 secretion systems, RTX family, tyrosine phosphorylation, exoproteins
Citation: Wakeel A, den Dulk-Ras A, Hooykaas PJJ and McBride JW (2011) Ehrlichia chaffeensis tandem repeat proteins and Ank200 are type 1 secretion system substrates related to the repeats-in-toxin exoprotein family. Front. Cell. Inf. Microbio. 1:22. doi: 10.3389/fcimb.2011.00022
Received: 22 September 2011;
Accepted: 14 December 2011;
Published online: 30 December 2011.
Edited by:
Robert Heinzen, NIH/NIAID-RML, USAReviewed by:
Michael Konkel, Washington State University, USAJon Audia, University of South Alabama School of Medicine, USA
Copyright: © 2011 Wakeel, den Dulk-Ras, Hooykaas and McBride. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Jere W. McBride, Department of Pathology, Center for Biodefense and Emerging Infectious Diseases, University of Texas Medical Branch, Galveston, TX 77555-0609, USA. e-mail:amVtY2JyaWRAdXRtYi5lZHU=

 Amke den Dulk-Ras2
Amke den Dulk-Ras2