- Departments of Microbiology and Internal Medicine, The Carver College of Medicine, The University of Iowa, Iowa City, IA, USA
Nontypeable Haemophilus influenzae (NTHi) is an exclusive human pathogen, which has evolved a number of unique mechanisms to survive within the human environment. An important part of this is the ability of the organism to take up and incorporate sialic acid into its surface structures. This protects the organism against host adaptive and innate immune factor as well as serving as a mechanism for sustaining itself within biofilms. Recent evidence suggests that this also may be the source of the evolution of human antibodies to non-human sialic acid structures, which can lead to inflammation in the host. In very rare instances, evolution of antibodies to sialylated lipooligosaccharide (LOS) mimics of human antigens can result in autoimmune disease.
Introduction
Nontypeable (non-encapsulated) Haemophilus influenzae (NTHi), a Gram-negative cocco-baccillus, is normally a human nasopharyngeal commensal organism, which can become an important human pathogen in both adults and children. In years past, some investigators suggested that NTHi were variant forms of encapsulated H. influenzae that had lost their capsule during passage in vitro. With the identification of the capsular biosynthesis genes and the advent of whole genome analysis, it has been established that strains of NTHi lack the genetic machinery involved in capsule biosynthesis. In addition, nontypeable H. influenzae affect different patient populations, cause different infections, present different surface antigens to the host, and are genetically different from the encapsulated strains.
In view of the fact that NTHi strains lack a capsular antigen, investigators have searched for virulence factors associated with this organism, which would confer protection against host defense mechanisms, particularly those associated with complement mediated bactericidal activity. Among other Gram-negative species, the lipopolysaccharide (LPS) is frequently a factor in conferring resistance to complement mediated lysis. These organisms generally have an LPS with repetitive O-antigen structures that act as a shield, which deters formation of the complement attack complex on the surface of the bacterium. NTHi also lacks the genes associated with O-antigen formation and forms a surface glycolipid structure, which has a lipid A–ketooxyoctanoate-heptose core region capped by no more than nine to ten sugars. This structure is embedded in the outer membrane of the organism. Given the short carbohydrate component, this glycolipid has been termed a “lipooligosaccharide (LOS)” to distinguish it from the O-antigen containing LPS structures found on Gram-negative bacteria present in the gastrointestinal tract or in the environment. In general, Gram-negative bacteria (Neisseria, Haemophilus, and Moraxella) found on the non-gastrointestinal mucosal surfaces of the airway and genital tract have LOS as their principle glycolipids rather than LPS. A number of these species are exclusive human colonizers and have evolved such that their LOS structures mimic human glycosphingolipid antigens including paragloboside, pK antigen and “i.” An important component of this mimicry is the addition of N-acetyl-5-neuraminic acid (Neu5Ac) to the terminus of the LOS structures. This enhances the immune evasion provided by the antigenic mimicry and adds a strong negative charge and hydrophilicity to the surface of the organism, which also acts to reduce the potential for hydrophobic interactions with host innate immune factors that would be detrimental to the organisms. Mandrell and co-workers first showed that the majority of NTHi strains expressed LOS structures which were sialylated and formed structures which mimicked human glycosphingolipid antigens (Mandrell et al., 1992). The addition of Neu5Ac to the LOS of NTHi increases the resistance of the organism to killing by normal human serum. In chinchilla models of otitis media, the ability of NTHi to incorporate Neu5Ac onto the LOS is crucial for development of the infectious process. This would indicate that the ability of the organism to cap its LOS structures with Neu5Ac is critical for the pathogenic potential of the organism.
The genome of NTHi is relatively small (∼2.1–2.2 Mb) and it has adapted well to the very narrow nasopharyngeal environmental niche in which it resides. This has resulted in the loss of synthetic capabilities found in other Gram-negative organisms with larger genomes. As an excellent example of environmental adaptability, NTHi has lost the ability to synthesize Neu5Ac. This might be surprising considering the importance of Neu5Ac for its survival as a pathogen in the human environment. However, NTHi lives in a sea of sialic acid (Neu5Ac) associated with the mucins (15% of the carbohydrate structure is Neu5Ac) found on the airway surfaces. The organism has adapted a number of mechanisms to acquire, control, and incorporate this important sugar into its membrane structures.
Sialic Acid
Sialic acids (NeuAc) are a diverse family of nine carbon acidic sugars, of which there are around 40 derivatives known to date (Schauer et al., 1995). The most common sialic acid is N-5-acetylneuraminic acid or Neu5Ac (Figure 1). This sugar is formed by the condensation of N-acetylmannosamine-6-phosphate and pyruvate (Figure 2). The most distinguishing features of the molecule are the carboxyl group at C1 and the amino group at C5. The carboxyl group at C1 gives the molecule a net negative charge, and is responsible for its acidic nature. The molecule's negative charge and relatively large size are two key features in how it plays a role in biology. NeuAc is usually the terminal sugar on carbohydrate structures (oligo- and polysaccharides), glycoproteins and glycolipids of mammals, birds, fungi, bacteria metazoans, and archaea.
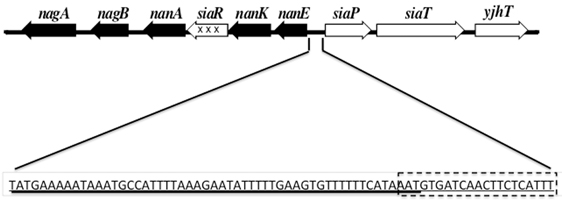
Figure 1. A diagrammatic representation of the catabolic (■), uptake genes (□), and regulatory gene ( ) of the two Sia operons in the H. influenzae genome are diagrammed in Figure 1. The intergenic region between the catabolic and uptake operons contains the binding sites for SiaR (solid Line) and Crp (dotted rectangle).
) of the two Sia operons in the H. influenzae genome are diagrammed in Figure 1. The intergenic region between the catabolic and uptake operons contains the binding sites for SiaR (solid Line) and Crp (dotted rectangle).
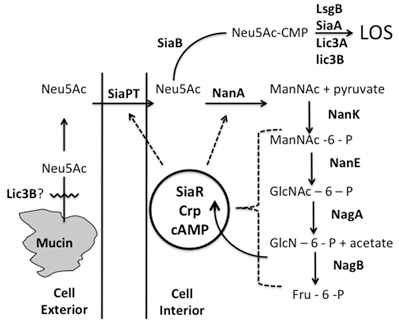
Figure 2. A composite model of Neu5Ac uptake, incorporation into LOS and catabolism into the Krebs cycle is shown. The different steps in the process is described in text.
NeuAc and its derivatives are found on cell membranes and body fluids in all mammals and many higher order animals, as well as pathogenic microorganisms. Typically only a few derivatives of Neu5Ac are seen in each individual species. In addition to this, the derivatives of NeuAc can be tissue or developmentally specific. The molecule is typically modified at positions C1, C4, C5, C7, C8, or C9. The most common NeuAc of mammalian cells are the N-acetylneuraminic acids (Neu5Ac) and N-glycolylneuraminic acid (Neu5Gc) (Varki, 2001). The addition of a hydroxyl group to the amino moiety at position C5 forms Neu5Gc. Neu5Gc is fairly common in animal species other than humans, but is found in humans only in association with certain forms of cancer (Narayanan, 1994; Sillanaukee et al., 1999). Normal humans are the exception, because of a mutation in CMP-sialic acid hydrolase. This enzyme is responsible for the addition of a single oxygen atom to Neu5Ac converting it to Neu5Gc (Brinkman-Van der Linden et al., 2000). In humans, this enzyme has no hydroxylase activity because of a large deletion caused by a deletion/frame shift mutation in the human gene. In contrast, the great apes including the chimpanzee, bonobo, gorilla, and orangutan, our closest animal relatives, have a functional CMP-Neu5Ac hydrolase, and can express Neu5Gc in large amounts. In some tissues, this is the predominate NeuAc.
Other common forms of modification are O-methylation and O-acetylation. These modifications can have a profound impact on the properties of sialic acid. For example, acetylation at C9 can make the sialic acid resistant to cleavage by sialidases, which catalyze the removal of sialic acid from glycoproteins and glycolipids.
The metabolism and nucleotide derivatization of Neu5Ac is well described (Figure 2). Entry of metabolites into the pathway can occur at multiple points. Fructose and D-glucosamine can be an entry point for the pathway. Neu5Ac and CMP-Neu5Ac can enter the pathway in bacteria from the external environment through dedicated cell membrane transporters (Martinez et al., 1995).
Synthesis and Incorporation of Sialic Acid by Microbes
Many pathogenic bacteria have evolved mechanisms to evade human defense mechanisms based on the surface exposure of Neu5Ac. A common theme in this mimicry is the incorporation of Neu5Ac as a component of either the capsular polysaccharide or the LOS or both. Based on an understanding of the biosynthesis of sialic acid and the evolving elucidation of the genomes of multiple microbes, Vimr and Lichtensteiger have described at least four mechanisms of microbial surface sialylation (Vimr and Lichtensteiger, 2002). These include de novo synthesis, donor scavenging, trans-sialylation, and precursor scavenging. Neisseria meningitidis serogroups B, C, Y, and W-135 can synthesize sialic acid for incorporation into capsular polysaccharides and LOS. Neisseria gonorrhoeae cannot synthesize or convert Neu5Ac to the nucleotide sugar form and must scavenge the donor, CMP-Neu5Ac, to sialylate its LOS structure. Trypanosoma cruzi does not synthesize or catabolize free sialic acid. It expresses a developmentally regulated sialidase, which it uses for surface sialylation by a trans-sialidase mechanism. The sialyltransferase activity of the sialidase is greater than its hydrolytic activity as long as the appropriate galactosyl acceptor is available.
The Neu5Ac Operons in NTHi
H. influenzae scavenges the precursor, Neu5Ac, to sialylate its LOS and biofilm. It must rely on scavenging of Neu5Ac from the environment because it cannot synthesize Neu5Ac. In addition, it has the means to use Neu5Ac as a source of carbon and nitrogen through a complex Neu5Ac catabolic system. The genes involved in the transport and metabolism of Neu5Ac are arranged in two adjacent tandem operox1ns, which are transcribed in opposite directions (Figure 1). The operon involved in Neu5Ac transport contains three genes encoding for a Neu5Ac binding protein (siaP), a transporter (siaT), and a protein which appears to be a homolog of Escherichia coli yjhT that is involved in the conversion of the β-anomer of Neu5Ac to the α-anomer (Severi et al., 2008). The binding protein (SiaP) and transporter (siaT) are homologous to a system of tripartite ATP-independent periplasmic transporters (TRAP) found in other bacterial species. These transporters differ from the better-characterized ABC-protein transporter family (Colli, 1993) in that they do not possess an ATP-binding cassette protein and are not driven by ATP hydrolysis but rather by an electrochemical ion gradient (Haft et al., 1996) (Figure 3). When the amino acid sequences of the NTHi binding protein and other members of this family of solute-binding proteins were aligned, the comparison allowed for the identification of three major groups based on sequence similarity and putative substrate: Neu5Ac, C4-dicarboxylates, and pentose sugars. Surface plasmon resonance studies in which the Neu5Ac was replaced with glucose, galactose, glucosamine, N-acetylglucoamine, or N-acetylmannosamine showed that SiaP did not bind any of these sugars. From the isothermal calorimetry data it was determined that the stoichiometry of binding of NeuAc to SiaP was 1:1. Severi et al. reported that the SiaP from H. influenzae RM118 had a stoichiometry of 1:1 and a Kd of 0.12 μM (Severi et al., 2005). The Kd values suggest that the SiaP/NeuAc binding interaction is a high affinity interaction. The failure of SiaP to bind KDO, an eight-carbon keto sugar, is indicative of the high specificity of this binding protein. The crystallographic structure and binding characteristics of this protein have been described (Severi et al., 2005; Johnston et al., 2008).
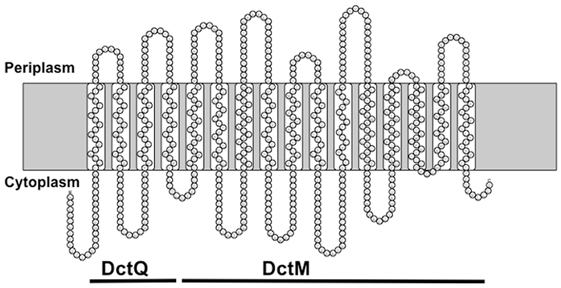
Figure 3. A diagram showing the transmembrane loops of SiaT based on a hydrophobicity plot. The regions corresponding to DctQ and DctM are shown. Model based on prediction program at http://proteinformatics.charite.de/rhythm/index.php?site=helix.
Evidence that SiaT was the Neu5Ac transporter was shown by the fact that NTHi siaT and siaTnanA mutants were both incapable of sialic acid uptake (Johnston et al., 2008). This indicated that the siaT gene product is required prior to activation of Neu5Ac by the CMP-sialic acid synthetase, thus implicating SiaT as the transporter in NTHi. Based on hydrophobicity plots, the Neu5Ac transporter appears to have 16 transmembrane domains. It is a fused version of the DctP/DctM proteins found in other TRAP transporter systems (Figure 2).
Sialic acids on glycoconjugates are sequestered as the α-anomer, while sialic acid in solution has been shown to slowly mutorotate from the α-anomer to the β-anomer. The E. coliyjhT homolog has been shown to convert this β-anomer of sialic acid back to the α-anomer (Severi et al., 2008). It is believed that the function of the NTHI gene in this uptake operon fulfills this function for NTHi.
The metabolic operon consists of five genes involved in metabolizing Neu5Ac and a gene involved in the regulation of the uptake and metabolic process (Figure 1). The five metabolic genes are nanE, nanK, nanA, nagB, and nagA, which encode N-acetyl-mannosamine-6-phosphate-2-epimerase, N-acetyl-mannosamine kinase, N-acetyl-neuraminate lyase, glucosamine-6-phosphate deaminase, and N-acetyl-glucosamine-6-phosphate deacetylase.
NTHi CMP-Neu5Ac Synthase
Prior to its transfer to glycolipids and glycoproteins, sialic acid has to be activated by the addition of a cytidine-5′-monophosphate (CMP) group (Figure 2). This activation of sialic acid is absolutely essential for sialylation to occur. A very unique feature of this activation is the presence of only one phosphate in the nucleotide/sugar linkage (Tullius et al., 1996), as opposed to two present in most other sugar nucleotides (Kean, 1991). A gene responsible for this activity has been identified in a number of bacterial organisms, including E. coli, Streptococcusagalactiae, N. meningitidis, H. ducreyi, and H. influenzae (Zapata et al., 1989; Ganguli et al., 1994; Haft et al., 1996; Tullius et al., 1996; Hood et al., 1999). The gene, called neuA, in E. coli, and N. meningitidis is part of their capsular loci. The neuA gene in H. ducreyi shares homology to other known CMP-NeuAc synthetases, and is transcriptionally coupled to its sialyltransferase gene (Tullius et al., 1996; Bozue et al., 1999). This same gene in H. influenzae (designated siaB) is not in close proximity with any other known LOS, capsule, or sialylation genes (Hood et al., 1999). In all cases, a deletion in the CMP-NeuAc synthetase results in no detectable sialic acid in the LOS, LPS, or capsules of the organisms.
NTHi Sialyltransferases
The sialyltransferases from bacteria catalyze the same basic reaction as the mammalian sialyltransferases. There are key differences in how they are isolated, their structures, and the specificity for acceptor structures, however. Bacteria contain sialylated structures in either their LOS/LPS or in their capsule. A great deal of work has been done on Neisserial sialyltransferases from both N. gonorrhoeae and N. meningitidis. To date, there have not been any bacterial sialyltransferases identified that have homology to mammalian sialyltransferases. Most notably, there is no homology to the “classic” sialylmotifs of these mammalian enzymes. For this reason, identification by PCR or Southern hybridization using mammalian sequences has not been possible.
Sialyltransferases from both N. meningitidis and N. gonorrhoeae LOS were first cloned and characterized by Gilbert et al. (1996). They were cloned by screening a DNA library made from a strain of N. meningitidis. Using a sensitive enzyme assay based on the transfer of sialic acid to synthetic fluorescent acceptors, this group was able to identify a sialyltransferase capable of transferring NeuAc with an α2,3-linkage to a N-acetyllactosamine acceptor. Strains of N. meningitidis capable of producing a sialic acid capsule contain a α2,8-polysialyltransferase in addition to the α2,3-sialyltransferase (Edwards et al., 1994).
Once the gene from N. meningitidis was cloned, probes were made to amplify a similar enzyme from N. gonorrhoeae. There was a high degree of homology between the two sialyltransferases, with only 16–17 amino acid differences sporadically located along the length of the 371 amino acid proteins (Gilbert et al., 1996). In addition to not containing any homology to the sialylmotifs of the mammalian sialyltransferases, these bacterial enzymes had more ambiguity in the acceptor structures they could recognize. Although there was a preference for the N-acetyllactosamine structure, sialic acid could also be transferred to lactose or galactose. In a more detailed study of the enzymatic properties of this enzyme, it was shown that NeuAc could be transferred to α-terminal or β-terminal galactose residues and β1,4-linked or β1,3-linked galactose (Gilbert et al., 1996). Ambiguity was also observed in the type of sialic acid able to be transferred, as N-propionylneuraminic acid and NeuGc could also be used with reduced efficiency.
A sialyltransferase has also been identified using a genetic approach in H. ducreyi. The LOS structure sialylated in H. ducreyi contains a terminal lactosamine structure (Melaugh et al., 1992, 1994). A gene was identified in this organism, designated lst, which was located downstream of neuA (a CMP-NeuAc synthetase) (Bozue et al., 1999). Mutational analysis of the two genes indicated that they are transcriptionally linked. The lst gene is thought to represent a novel class of bacterial sialyltransferases, because it does not contain strong homology to any known bacterial or mammalian sialyltransferases and contains only weak homology to the E. coli capsular polysialyltransferase (Vimr et al., 1992; Bozue et al., 1999).
Campylobacter jejuni is able to synthesize a number of sialylated LPS structures, some of which mimic human gangliosides (Moran et al., 1991; Aspinall et al., 1993; Prendergast et al., 1998; Gilbert et al., 2000; Guerry et al., 2000). C. jejuni strain OH4384 produces an LPS similar to the ganglioside GD1a (NeuAcα2–8NeuAcα2–3Galβ1–3GalNAcβ1–4Gal(NeuAcα2–3)β1,3Hep). Using the same strategy used to clone the Neisseria sialyltransferases, two genes were identified that contained sialyltransferase activity, designated cstI and cstII (Gilbert et al., 2000). There was a great deal of homology between the two genes, and it was hypothesized that cstI occurred through a duplication event and subsequent genetic drift. CstI transfers NeuAc α2,3-linked to lactose, while CstII was shown to be bi-functional, capable of transferring sialic acid α2,3-linked to either lactose or α2,8-linked to another NeuAc.
Another novel class of bacterial sialyltransferases was identified in the marine bacterium Photobacterium damsela (Yamamoto et al., 1998). The protein was purified using a detergent extract, which was also required for activity, indicating it was a membrane-associated protein. Further analysis of the cloned gene (bst) revealed a predicted α-helical structure in the C-terminus that could function in membrane insertion (Yamamoto et al., 1998). Deletion of this C-terminal sequence resulted in the detection of the soluble enzyme in the supernatant. Enzyme assay analysis showed that both lactose and N-acetyllactosamine were the preferred acceptor structures, however NeuAc could be transferred to either a galactose or a N-acetylgalactosamine (Yamamoto et al., 1996). The most surprising function of this enzyme was its linkage specificity, transferring NeuAc α2,6-linked to the terminal sugar. This was the first identified bacterial sialyltransferase to preferentially add sialic acid with this linkage. Wakarchuk et al. identified an α2–6-linked sialic acid attached to a Pk antigen structure (Galα1–4Galβ1–4Glc) on the LOS of N. meningitidis (Wakarchuk et al., 1998). It is not known if there is a second sialyltransferase involved in this linkage, or if the α2,3-sialyltransferase already identified in this organism can account for this activity.
The sequenced genome of H. influenzae strain Rd has greatly increased the ability to identify genes involved in LOS biosynthesis, including sialyltransferases (Fleischmann et al., 1995). There are at least four genes in the NTHi databases with homology to known sialyltransferases, these are lic3A (Hood et al., 2001), lsgB (Jones et al., 2002), siaA (Jones et al., 2002), and lic3B (Fox et al., 2006). The first gene encoding an NTHi sialyltransferase identified was lic3A. Lic3A has approximately 40% identity (at the protein level) to cstII from C. jejuni (Hood et al., 2001). Lic3A has been shown to sialylate a galactose (sialyl-α-(2−3)-lactosyl extension) as part of a lactosyl group coming from the distal heptose. As shown by Hood and co-workers, the activity is modulate by competition for the acceptor by LgtC (Hood et al., 2001). The second and third sialyltransferase found in H. influenzae were LsgB and SiaA (Jones et al., 2002). LsgB has 27% identity and 46% similarity to the N. meningitidis α2–3 sialyltransferases (Phillips et al., 2000; Jones et al., 2002; Fox et al., 2006). Sia has 44% identity and 60% similarity with Lst from H. ducreyi over the entire length of the proteins (Bozue et al., 1999; Jones et al., 2002). Lst has been shown to be an H. ducreyi a2–3 sialyltransferase (Bozue et al., 1999; Jones et al., 2002). Findings indicate that this protein adds sialic acid to a terminal N-acetyllactosamine structure on the LOS (Jones et al., 2002). Fox and co-workers have identified the most recent sialyltransferase in NTHi, Lic3B. This enzyme is a very close homology of Lic3A and is present in over 50% of NTHi strains tested (Fox et al., 2006). Like lic3A, this enzyme has been shown to be phase variable based on the tandem repeats tetranucleotide CAAT repeats just within the 5' end of the open reading frame. Lic3B also appears to be bifunctional similar the Cst1 found in C. jejuni. Lic3B has both α2–3 and α2–8 sialyltransferase activities as well as trans-sialidase activity (Fox et al., 2006).
A trans-sialidase is a bi-functional enzyme first found in the genus Trypanosoma. The enzyme catalyzes two reactions. The first is a trans-sialidase reaction, where sialic acid is cleaved from glycoconjugates free in solution or attached to a host membrane. The sialic acid can then either be hydrolyzed to form free sialic acid, or transferred to a mucin like glycoprotein found on the Trypanosoma cell surface (Schenkman et al., 1994). The enzyme has been identified in two pathogenic forms of Trypanosomes; T. brucei, the causative agent of African sleeping sickness, and T. cruzi, the causative agent of Chagas disease. The biological properties of trans-sialidases have been studied by a number of investigators (Colli, 1993; Cross and Takle, 1993; Schenkman et al., 1994). Trans-sialidases have a very high specificity for α2,3-linked sialic acid attached to a penultimate galactose residue on host glycoconjugates. The enzyme is not able to transfer either free Neu5Ac or CMP-Neu5Ac. The best acceptor structure is lactose, but maltose (Glcα1–4Glc) or cellobiose (Glcβ1–4Glc) can be used with decreased efficiency. The pH optimum for TS is around 7.0, which is different from the other sialidase enzymes, which have a pH optimum between 5.0 and 5.5.
While evidence for similar trans-sialidase activity in NTHi has not been conclusively shown, Lic3B could potentially have the capability of removing Neu5Ac from mucin if it can be secreted. Whether it would function extracellularly similar to the trans-sialidase in Trypanosomes has not been shown and there is no evidence for secretion or an export mechanism for the enzyme.
Role of Neu5Ac in NTHi Pathogenesis
Neu5Ac plays an important role in pathogenesis of NTHi infection in serum sensitivity, biofilm formation and the evolution of human antibodies to glycosyl structures containing N-glycol-5-neuraminic acid.
Hood and co-workers demonstrated that sialic acid incorporation into the LOS structure had the capability of protecting the organism against complement mediated antibody-dependent bactericidal activity (Hood et al., 1999). These investigators demonstrated that a CMP-NANA synthetase mutant (siaB) resulted in a sialylation deficient mutant, which was altered the serum resistance of the wildtype strain.
Greiner and co-workers (Greiner et al., 2004) and Swords and co-workers (Swords et al., 2004) demonstrated that Neu5Ac promoted biofilm formation. Confocal and electron microscopy demonstrated that the biofilm produced by the siaB mutant had a significantly reduced high and a smaller mass than that formed by the wildtype organism. After 24 h, the majority of the organisms in the biofilm formed by the mutant were not viable (Greiner et al., 2004). Of note, one feature on electron microscopy was the absence of patent water channels in the biofilm formed by the siaB mutant. Based on this it is interesting to postulate that the sialic acid on the LOS might be a factor in maintaining the patency of the water channels allowing flow of nutrients and water into the interstices of the biofilm.
Recent studies have demonstrated that the non-human sialic acid N-glycolneuraminic (Neu5Gc) can be metabolically incorporated into human tissue from certain mammalian-derived foods. Recent evidence indicates that this process can contribute to chronic inflammation in some individuals (Surolia et al., 2010). Studies have demonstrated that while Neu5Gc cannot be synthesize by bacterial species it can be taken up by NTHi and incorporated into its LOS surface structures. Appearance of antibodies in infants to Neu5Gc structures correlates with nasopharyngeal colonization with NTHi and it has been postulated that this may be the source of these antibodies (Taylor et al., 2010). Thus, xeno-autoantibody formation in humans may be a bystander reaction of sialic acid incorporation during NTHi colonization and infection.
Conclusions
NTHi is an exclusive human pathogen, which has evolved a number of unique mechanisms to survive within the human environment. An important part of this is the ability of the organism to take up and incorporate sialic acid into its surface structures. This protects the organism against host adaptive and innate immune factor as well as serving as a mechanism for sustaining itself within biofilms. Recent evidence suggests that this also may be the source of the evolution of human antibodies to non-human sialic acid structures which can lead to inflammation in the host.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aspinall, G. O., McDonald, A. G., Raju, T. S., Pang, H., Kurjanczyk, L. A., Penner, J. L., and Moran, A. P. (1993). Chemical structure of the core region of Campylobacter jejuni serotype O:2 lipopolysaccharide. Eur. J. Biochem. 213, 1029–1037.
Bozue, J. A., Tullius, M. V., Wang, J., Gibson, B. W., and Munson, R. S. Jr. (1999). Haemophilus ducreyi produces a novel sialyltransferase. Identification of the sialyltransferase gene and construction of mutants deficient in the production of the sialic acid-containing glycoform of the lipooligosaccharide. J. Biol. Chem. 274, 4106–4114.
Brinkman-Van der Linden, E. C., Sjoberg, E. R., Juneja, L. R., Crocker, P. R., Varki, N., and Varki, A. (2000). Loss of N-glycolylneuraminic acid in human evolution. Implications for sialic acid recognition by siglecs. J. Biol. Chem. 275, 8633–8640.
Colli, W. (1993). Trans-sialidase: a unique enzyme activity discovered in the protozoan Trypanosoma cruzi. FASEB J. 7, 1257–1264.
Cross, G. A., and Takle, G. B. (1993). The surface trans-sialidase family of Trypanosoma cruzi. Annu. Rev. Microbiol. 47, 385–411.
Edwards, U., Muller, A., Hammerschmidt, S., Gerardy-Schahn, R., and Frosch, M. (1994). Molecular analysis of the biosynthesis pathway of the alpha-2,8 polysialic acid capsule by Neisseria meningitidis serogroup B. Mol. Microbiol. 14, 141–149.
Fleischmann, R. D., Adams, M. D., White, O., Clayton, R. A., Kirkness, E. F., Kerlavage, A. R., Bult, C. J., Tomb, J. F., Dougherty, B. A., Merrick, J. M., McKenney, K., Sutton, G., FitzHugh, W., Fields, C., Gocayne, J. D., Scott, J., Shirley, R., Liu, L.-I., Glodek, A., Kelley, J. M., Weidner, J. F., Phillips, C. A., Spriggs, T., Hedblom, E., Cotton, M. D., Utterbach, T. R., Hanna, M. C., Nguyen, D. T., Saudek, D. M., Brandon, R. C., Fine, L. D., Fritchman, J. L., Fuhrmann, J. L., Geoghagen, N. S. M., Gnehm, C. L., McDonald, L. A., Small, K. V., Fraser, C. M., Smith, H. O., and Venter, J. C. (1995). Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269, 496–512.
Fox, K. L., Cox, A. D., Gilbert, M., Wakarchuk, W. W., Li, J., Makepeace, K., Richards, J. C., Moxon, E. R., and Hood, D. W. (2006). Identification of a bifunctional lipopolysaccharide sialyltransferase in Haemophilus influenzae: incorporation of disialic acid. J. Biol. Chem. 281, 40024–40032.
Ganguli, S., Zapata, G., Wallis, T., Reid, C., Boulnois, G., Vann, W. F., and Roberts, I. S. (1994). Molecular cloning and analysis of genes for sialic acid synthesis in Neisseria meningitidis group B and purification of the meningococcal CMP-NeuNAc synthetase enzyme. J. Bacteriol. 176, 4583–4589.
Gilbert, M., Brisson, J. R., Karwaski, M. F., Michniewicz, J., Cunningham, A. M., Wu, Y., Young, N. M., and Wakarchuk, W. W. (2000). Biosynthesis of ganglioside mimics in Campylobacter jejuni OH4384. Identification of the glycosyltransferase genes, enzymatic synthesis of model compounds, and characterization of nanomole amounts by 600-mhz (1)h and (13)c NMR analysis. J. Biol. Chem. 275, 3896–3906.
Gilbert, M., Watson, D., Cunningham, A.M., Jennings, M., Young, N., and Wakarchuk, W. (1996). Cloning of the Lipooligosaccharide alpha-2, 3-sialyltransferase from the bacterial pathogens Neisseria meningitidis and Neisseria gonorrhoeae. J. Biol. Chem. 273, 28271–28276.
Greiner, L. L., Watanabe, H., Phillips, N. J., Shao, J., Morgan, A., Zaleski, A., Gibson, B. W., and Apicella, M. A. (2004). Nontypeable Haemophilus influenzae strain 2019 produces a biofilm containing N-acetylneuraminic acid that may mimic sialylated O-linked glycans. Infect. Immun. 72, 4249–4260.
Guerry, P., Ewing, C. P., Hickey, T. E., Prendergast, M. M., and Moran, A. P. (2000). Sialylation of lipooligosaccharide cores affects immunogenicity and serum resistance of Campylobacter jejuni. Infect. Immun. 68, 6656–6662.
Haft, R. F., Wessels, M. R., Mebane, M. F., Conaty, N., and Rubens, C. E. (1996). Characterization of cpsF and its product CMP-N-acetylneuraminic acid synthetase, a group B streptococcal enzyme that can function in K1 capsular polysaccharide biosynthesis in Escherichia coli. Mol. Microbiol. 19, 555–563.
Hood, D. W., Cox, A. D., Gilbert, M., Makepeace, K., Walsh, S., Deadman, M. E., Cody, A., Martin, A., Mansson, M., Schweda, E. K., Brisson, J. R., Richards, J. C., Moxon, E. R., and Wakarchuk, W. W. (2001). Identification of a lipopolysaccharide alpha-2,3-sialyltransferase from Haemophilus influenzae. Mol. Microbiol. 39, 341–350.
Hood, D. W., Makepeace, K., Deadman, M. E., Rest, R. F., Thibault, P., Martin, A., Richards, J. C., and Moxon, E. R. (1999). Sialic acid in the lipopolysaccharide of Haemophilus influenzae: strain distribution, influence on serum resistance and structural characterization. Mol. Microbiol. 33, 679–692.
Johnston, J. W., Coussens, N. P., Allen, S., Houtman, J. C., Turner, K. H., Zaleski, A., Ramaswamy, S., Gibson, B. W., and Apicella, M. A. (2008). Characterization of the N-acetyl-5-neuraminic acid-binding site of the extracytoplasmic solute receptor (SiaP) of nontypeable Haemophilus influenzae strain 2019. J. Biol. Chem. 283, 855–865.
Jones, P. A., Samuels, N. M., Phillips, N. J., Munson, R. S. Jr., Bozue, J. A., Arseneau, J. A., Nichols, W. A., Zaleski, A., Gibson, B. W., and Apicella, M. A. (2002). Haemophilus influenzae type b strain A2 has multiple sialyltransferases involved in lipooligosaccharide sialylation. J. Biol. Chem. 277, 14598–14611.
Mandrell, R. E., McLaughlin, R., Aba Kwaik, Y., Lesse, A., Yamasaki, R., Gibson, B., Spinola, S. M., and Apicella, M. A. (1992). Lipooligosaccharides (LOS) of some Haemophilus species mimic human glycosphingolipids, and some LOS are sialylated. Infect. Immun. 60, 1322–1328.
Martinez, J., Steenbergen, S., and Vimr, E. (1995). Derived structure of the putative sialic acid transporter from Escherichia coli predicts a novel sugar permease domain. J. Bacteriol. 177, 6005–6010.
Melaugh, W., Phillips, N. J., Campagnari, A. A., Karalus, R., and Gibson, B. W. (1992). Partial characterization of the major lipooligosaccharide from a strain of Haemophilus ducreyi, the causative agent of chancroid, a genital ulcer disease. J. Biol. Chem. 267, 13434–13439.
Melaugh, W., Phillips, N. J., Campagnari, A. A., Tullius, M. V., and Gibson, B. W. (1994). Structure of the major oligosaccharide from the lipooligosaccharide of Haemophilus ducreyi strain 35000 and evidence for additional glycoforms. Biochemistry 33, 13070–13078.
Moran, A. P., Zahringer, U., Seydel, U., Scholz, D., Stutz, P., and Rietschel, E. T. (1991). Structural analysis of the lipid a component of Campylobacter jejuni CCUG 10936 (serotype O:2) lipopolysaccharide. Eur. J. Biochem. 198, 459–469.
Phillips, N. J., Miller, T. J., Engstrom, J. J., Melaugh, W., McLaughlin, R., Apicella, M. A., and Gibson, B. W. (2000). Characterization of chimeric lipopolysaccharides from Escherichia coli strain JM109 transformed with lipooligosaccharide synthesis genes (lsg) from Haemophilus influenzae. J. Biol. Chem. 275, 4747–4758.
Prendergast, M. M., Lastovica, A. J., and Moran, A. P. (1998). Lipopolysaccharides from Campylobacter jejuni O:41 strains associated with Guillain-Barre syndrome exhibit mimicry of GM1 ganglioside. Infect. Immun. 66, 3649–3655.
Schauer, R., Kelm, S., Reuter, G., Roggentin, P., Shaw, L.,and (1995). “Biochemistry and role of sialic acids,” in Biology of the Sialic Acids, ed A. Rosenberg (New York, NY: Plenum Press), 7–67.
Schenkman, S., Eichinger, D., Pereira, M. E., and Nussenzweig, V. (1994). Structural and functional properties of Trypanosoma trans-sialidase. Annu. Rev. Microbiol. 48, 499–523.
Severi, E., Muller, A., Potts, J. R., Leech, A., Williamson, D., Wilson, K. S., and Thomas, G. H. (2008). Sialic acid mutarotation is catalyzed by the Escherichia coli beta-propeller protein YjhT. J. Biol. Chem. 283, 4841–4849.
Severi, E., Randle, G., Kivlin, P., Whitfield, K., Young, R., Moxon, R., Kelly, D., Hood, D., and Thomas, G. H. (2005). Sialic acid transport in Haemophilus influenzae is essential for lipopolysaccharide sialylation and serum resistance and is dependent on a novel tripartite ATP-independent periplasmic transporter. Mol. Microbiol. 58, 1173–1185.
Sillanaukee, P., Ponnio, M., and Jaaskelainen, I. P. (1999). Occurrence of sialic acids in healthy humans and different disorders. Eur. J. CliN. Invest. 29, 413–425.
Surolia, I., Pirnie, S. P., Chellappa, V., Taylor, K. N., Cariappa, A., Moya, J., Liu, H., Bell, D. W., Driscoll, D. R., Diederichs, S., Haider, K., Netravali, I., Le, S., Elia, R., Dow, E., Lee, A., Freudenberg, J., De Jager, P. L., Chretien, Y., Varki, A., Macdonald, M. E., Gillis, T., Behrens, T. W., Bloch, D., Collier, D., Korzenik, J., Podolsky, D. K., Hafler, D., Murali, M., Sands, B., Stone, J. H., Gregersen, P. K., and Pillai, S. (2010). Functionally defective germline variants of sialic acid acetylesterase in autoimmunity. Nature 466, 243–247.
Swords, W. E., Moore, M. L., Godzicki, L., Bukofzer, G., Mitten, M. J., and VonCannon, J. (2004). Sialylation of lipooligosaccharides promotes biofilm formation by nontypeable Haemophilus influenzae. Infect. Immun. 72, 106–113.
Taylor, R. E., Gregg, C. J., Padler-Karavani, V., Ghaderi, D., Yu, H., Huang, S., Sorensen, R. U., Chen, X., Inostroza, J., Nizet, V., and Varki, A. (2010). Novel mechanism for the generation of human xeno-autoantibodies against the nonhuman sialic acid N-glycolylneuraminic acid. J. Exp. Med. 207, 1637–1646.
Tullius, M. V., Munson, R. S. Jr., Wang, J., and Gibson, B. W. (1996). Purification, cloning, and expression of a cytidine 5'-monophosphate N-acetylneuraminic acid synthetase from Haemophilus ducreyi. J. Biol. Chem. 271, 15373–15380.
Vimr, E., and Lichtensteiger, C. (2002). To sialylate, or not to sialylate: that is the question. Trends Microbiol. 10, 254–257.
Vimr, E. R., Bergstrom, R., Steenbergen, S. M., Boulnois, G., and Roberts, I. (1992). Homology among Escherichia coli K1 and K92 polysialytransferases. J. Bacteriol. 174, 5127–5131.
Wakarchuk, W. W., Gilbert, M., Martin, A., Wu, Y., Brisson, J. R., Thibault, P., and Richards, J. C. (1998). Structure of an alpha-2,6-sialylated lipooligosaccharide from Neisseria meningitidis immunotype L1. Eur. J. Biochem. 254, 626–633.
Yamamoto, T., Nakashizuka, M., Kodama, H., Kajihara, Y., and Terada, I. (1996). Purification and characterization of a marine bacterial beta-galactoside alpha 2,6-sialyltransferase from Photobacterium damsela JT0160. J. Biochem. 120, 104–110.
Yamamoto, T., Nakashizuka, M., and Terada, I. (1998). Cloning and expression of a marine bacterial beta-galactoside alpha2,6-sialyltransferase gene from Photobacterium damsela JT0160. J. Biochem. 123, 94–100.
Keywords: nontypeable Haemophilus influenzae, Neu5Ac, sialic acid, sialyltransferases
Citation: Apicella MA (2012) Nontypeable Haemophilus influenzae: the role of N-acetyl-5-neuraminic acid in biology. Front. Cell. Inf. Microbio. 2:19. doi: 10.3389/fcimb.2012.00019
Received: 20 December 2011; Paper pending published: 13 January 2012;
Accepted: 07 February 2012; Published online: 13 March 2012.
Edited by:
W. Edward Swords, Wake Forest University Health Sciences, USAReviewed by:
Robert J. C. McLean, TX State University, USAX. Frank Yang, Indiana University School of Medicine, USA
Copyright: © 2012 Apicella. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Michael A. Apicella, Departments of Microbiology and Internal Medicine, The Carver College of Medicine, The University of Iowa, 51 Newton Road, Iowa City, IA 52242, USA. e-mail:bWljaGFlbC1hcGljZWxsYUB1aW93YS5lZHU=
