- 1Sorbonne Paris Cité, Bâtiment Leriche, Université Paris Descartes, Paris, France
- 2Institut National de la Santé et de la Recherche Médicale, Institut Necker-Enfants Malades, INSERM U1151 -Team 11, Pathogenesis of Systemic Infections, Paris, France
- 3Centre National de la Recherche Scientifique, UMR8253, Paris, France
Francisella tularensis is able to invade, survive and replicate inside a variety of cell types. However, in vivo F. tularensis preferentially enters host macrophages where it rapidly escapes to the cytosol to avoid phagosomal stresses and to multiply to high numbers. We previously showed that human monocyte infection by F. tularensis LVS triggered deglycosylation of the glutamine transporter SLC1A5. However, this deglycosylation, specifically induced by Francisella infection, was not restricted to SLC1A5, suggesting that host protein deglycosylation processes in general might contribute to intracellular bacterial adaptation. Indeed, we later found that Francisella infection modulated the transcription of numerous glycosidase and glycosyltransferase genes in human macrophages and analysis of cell extracts revealed an important increase of N and O-protein glycosylation. In eukaryotic cells, glycosylation has significant effects on protein folding, conformation, distribution, stability, and activity and dysfunction of protein glycosylation may lead to development of diseases like cancer and pathogenesis of infectious diseases. Pathogenic bacteria have also evolved dedicated glycosylation machineries and have notably been shown to use these glycoconjugates as ligands to specifically interact with the host. In this review, we will focus on Francisella and summarize our current understanding of the importance of these post-translational modifications on its intracellular niche adaptation.
Introduction
Protein glycosylation is one of the most common post-translational modifications (PTM) of proteins, as present in all kingdoms of life. It consists in the covalent attachment of glycans onto amino acid side chains, this reaction being catalyzed by an enzyme. In eukaryotic cells, glycosylation has significant effects on protein folding, conformation, distribution, stability, and activity. Particularly, the sugar chains of glycoproteins are essential for maintaining the order of intercellular interactions among all differentiated cells in multicellular organisms. Therefore, alterations in the sugar chains may range from being essentially undetectable to a complete loss in particular functions (Varki, 1993). Indeed, dysfunction of protein glycosylation may lead to development of diseases like cancer and pathogenesis of infectious diseases (Moran et al., 2011). In the innate immune system, which is the major actor for protection against microbial pathogens, several host glycoproteins have been shown to function as pattern recognition receptors (PRRs), involved in pathogen binding (Di Gioia and Zanoni, 2015). Cell-surface glycoproteins facing the extracellular environment are ideally located to facilitate this host–pathogen interaction. The receptors of the innate immune response i.e., Toll-like receptors (TLRs) and nucleotide oligomerization domain (NOD-like) receptors (NLRs) are glycoproteins. In the adaptative immune response, the major components, which include class I and class II major histocompatibility complex proteins, chemokine and cytokine receptors, and essentially all cytokines and chemokines are glycosylated (Opdenakker et al., 2016).
Bacterial pathogens have also evolved dedicated glycosylation machineries. When compared to higher organisms, bacteria are capable of producing an extraordinary amount of unique and diverse glycans, which are principally attached to the cell surface, and secreted molecules. Bacteria are able to use these glycoconjugates as a range of unique and specific ligands, which specifically interact with the host (Tytgat and de Vos, 2016). Bacteria are covered with various types of carbohydrate moieties. These surface-exposed bacterial structures are often called pathogen-associated molecular patterns (or PAMPS).
Oligosaccharides may either mediate “specific recognition” events or provide “modulation” of biological processes. For example, they may allow interaction of bacterial proteins with host-derived proteins or they may modulate bacteria- and/or host-related events (Bastos et al., 2017). All these events may be essential for bacterial colonization, its survival and the subsequent infection. Therefore, host immunization may be dependent on these PTM, whether mediated by the pathogen or by the host.
Francisella tularensis is a Gram-negative bacterium causing the zoonotic disease tularemia in a number of mammalian species, including humans (Sjöstedt, 2011). F. tularensis invades, survives and replicates inside a variety of cell types, including phagocytic and non-phagocytic cells of various species (Meibom and Charbit, 2010), as well as arthropod-derived cells (Santic et al., 2010). “In vivo,” F. tularensis preferentially enters host macrophages (Clemens et al., 2005), rapidly escapes to the cytosol where it actively multiplies (Case et al., 2014). While the cytoplasm was initially considered as a safe nutrient-replete haven (Ray et al., 2009), it is now clearly established that the host cytosol may be a harsh environment by depriving nutrients against invading bacteria (Abu Kwaik and Bumann, 2013; Zhang and Rubin, 2013). Conversely, invading intracellular pathogens may also “steal” nutrients of the host cell that, in turn, needs to adapt its metabolism to control its cytosolic content (Barel et al., 2015). Indeed, upon addition of gluconeogenic substrates, such as oaxaloacetate and pyruvate, to the cell culture medium increased intracellular multiplication of F. tularensis LVS was observed, suggesting that these nutrients served as sources of glucose to feed multiplying bacteria.
We will herein summarize what is known about the glycosylation-deglycosylation processes occurring during Francisella infection, as observed from either the host or the pathogen.
Host Point of View
Francisella infection modifies numerous “glyco-genes” involved in glycosylation pathways in human macrophages. Indeed, using a glycan processing gene microarray (Chacko et al., 2011), we observed significant changes in the level of glycosyltransferase and glycosidase gene expression profiles in human THP-1 monocytes, infected for 24 h with F. tularensis LVS (Barel et al., 2016). Expression of eight genes, encoding four glycosyltransferases and four glycosidases, was down-regulated upon infection. These four glycosidase belonged to the EDEM family, which is involved in ER-associated degradation (ERAD). The expression of six genes was up-regulated upon infection, corresponding to five glycosyltransferases and one glycosidase. The up-regulated glycosyltransferases were involved either in N-glycosylation or in O-glycosylation of glycoproteins. The glycosidase gene whose expression was up-regulated, encoded the glycosidase HEXA, which is involved in the Hexosamine Biosynthetic Pathway (HBP) (Vaidyanathan et al., 2014).
Glycosylation occurred as soon as 1 h after entry of the bacteria into the cells. Only three proteins were found and characterized as carrying potential N-glycosylation residues, while nine proteins contained potential O-glycosylation residues. Among them, we characterized BiP/GRP78/HSPA5 protein, a member of the HSP70 heat shock protein family. BiP expression was increased both at transcription and translation level, by F. tularensis LVS infection immediately after binding to the cells. BiP glycosylation was also induced at early stage of infection. BiP being a key regulator of the UPR (Ni et al., 2009; Pfaffenbach and Lee, 2011), we hypothesized that the glycosylation-deglycosylation processes could be modified by Francisella. This could result in direct triggering of the UPR (including BiP) in infected cells with a decrease of the load of newly synthesized “abnormal” proteins. In addition, among the nine proteins containing potential O-glycosylation residues and being glycosylated by Francisella infection, we also found PRKCSH, the beta-subunit of glucosidase 2. This enzyme is acting upstream BiP, in the calnexin pathway, which is also involved in correcting misfolded proteins (Hetz et al., 2011).
Infection of human monocytes by F. tularensis LVS also triggered the deglycosylation of the glycosylated amino acid transporter SLC1A5 and other glycoproteins (Barel et al., 2012). Deglycosylation induced by F. tularensis LVS was maximum at 24 h when intracellular multiplication occurred and depended on the capacity of the bacteria to escape from the phagosomes (Barel et al., 2012). It was not an inhibition of glycosylation since tunicamycine had no inhibiting effect on this deglycosylation.
The enzymes involved in these glycosylation-deglycosylation mechanisms are still not characterized.
We tried to summarize the cascade of events triggered upon infection of macrophages by Francisella in the hypothetical model depicted in Figure 1. The transporter SLC1A5 was chosen as a prototypic glycosylated membrane protein. After its synthesis and translocation into the ER, the protein is transported to the Golgi where it is first glycosylated ➀ and, from there, addressed to the membrane via secretory vesicles. In the plasma membrane, SLC1A5 is present only as a glycosylated protein ➁ (Console et al., 2015). Upon re-entry into the cytoplasm via endocytosis, glycosylated SLC1A5 becomes available to glucosidases ➂ such as HEXA (whose expression is induced upon Francisella infection). The deglycosylated form of SLC1A5 has been indeed localized only in the cytoplasm (Console et al., 2015). This deglycosylated form of the protein (possibly misfolded) could trigger increase of BiP expression and its glycosylation ➃.
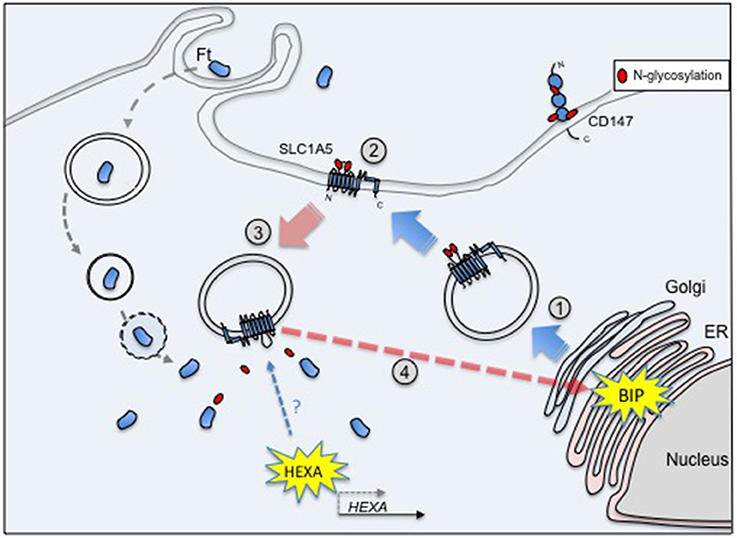
Figure 1. SLC1A5 is glycosylated after passage into the Golgi ① and is exported to the membrane ②. Its endocytosis into the cytoplasm renders it available to glucosidases ➂, e.g., HEXA, whose transcription level is increased upon Francisella infection. The deglycosylated form of SLC1A5 has been indeed localized only in the cytoplasm. In turn, these deglycosylated (and possibly misfolded) proteins could trigger the increase of BiP expression and its glycosylation ➃.
It is tempting to suggest that the intracellular survival of Francisella would be favored both by the control exerted on the UPR response of the host and by the availability of free oligosaccharides resulting from deglycosylation processes, that could serve as nutrients.
Pathogen Point of View
A large number of bacterial proteins have been found to be glycosylated (Tan et al., 2015). They show a surprising degree of diversity, both within and between bacterial species. Protein glycosylation can be classified according to the glycosidic linkage. Attachment to the amide nitrogen of asparagine (Asn) is known as N-glycosylation, with that of serine or threonine (Ser/Thr) to the hydroxyl oxygen being known as O-glycosylation. N- and O-linked glycosylation may occur either through the action of an oligosaccharyltransferase (OST) or via the action of glycosyltransferases (GTs). OSTs substrates are lipid-linked oligosaccharides while the GTs substrates are usually nucleotide-activated sugars. It was only very recently (Dankova et al., 2016) that the glycosylation machinery of Francisella was found to involve a variety of sugar biogenesis enzymes, glycosyltransferases, a flippase, and a protein-targeting oligosaccharyltransferase. As both type A and type B strains of F. tularensis subspecies expressed an O-linked protein glycosylation system, which utilizes core biosynthetic and assembly pathways, O-linked protein glycosylation may be a feature common to members of the Francisella genus (Egge-Jacobsen et al., 2011).
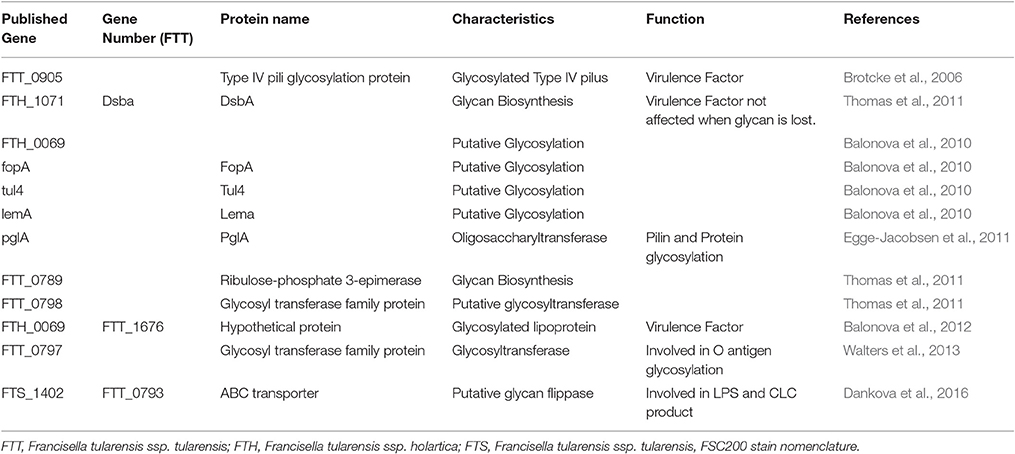
The initial attempts to elucidate the glycan repertoire of Francisella and their structures had failed because of the enzymatic and chemical release techniques used. Some proteins were found after transcriptional profiling of mutants. Indeed, FTT_0905 was characterized as a glycosylated Type IV pili protein, which is transcriptionally regulated by MglA. As MglA controls the expression of the Francisella pathogenicity island, FTT_0905 was considered as a new virulence factor (Brotcke et al., 2006). However, by mapping the glycoproteome of the FSC200 strain of F. tularensis subsp. holarctica, several candidate proteins were found that could be target for glycosylation as DsbA (FTH_1071), an uncharacterized protein FTH_0069, FopA, Tul4, and LemA (Balonova et al., 2010). In contrast, the PglA protein was identified as a targeting oligosaccharyltransferase because it is necessary for PilA glycosylation in F. tularensis (Egge-Jacobsen et al., 2011). Indeed, this protein undergoes multisite O-linked glycosylation, with a pentasaccharide of the structure HexNac-Hex-Hex-HexNac-HexNac. PglA is highly conserved in Francisella genus, supporting the general feature of O-glycosylation. Then, the detailed characterization of the DsbA glycan and the putative role of the FTT0789–FTT0800 gene cluster in glycan biosynthesis were reported (Thomas et al., 2011). Indeed, these authors observed that the essential virulence factor DsbA migrated as multiple protein spots on two-dimensional electrophoresis gels. The protein was modified with a 1,156-Da glycan moiety in O-linkage. The glycan is a hexasaccharide, comprised of N-acetylhexosamines, hexoses, and an unknown monosaccharide. Loss of DsbA glycan modification was obtained by disruption of two genes within the FTT0789–FTT0800 putative polysaccharide locus, including a galE homolog (FTT0791) and one gene encoding a putative glycosyltransferase (FTT0798). As the mutants remained virulent in the murine model of subcutaneous tularemia, it indicated that glycosylation of DsbA does not play a major role in virulence under these conditions (Thomas et al., 2011). When defining the previously uncharacterized FTH_0069 protein as a novel glycosylated lipoprotein required for virulence, Balonova et al. (2012) also showed that the glycan structure modifying its two C-terminal peptides was identical to that of DsbA glycoprotein, as well as to one of the multiple glycan structures modifying the type IV pilin PilA. They therefore suggested a common biosynthetic pathway for the protein modification and a relationship between synthesis of the O-antigen and the glycan in the early steps of their biosynthetic pathways. Indeed, the pglA gene, encoding pilin oligosaccharyl transferase PglA, was involved in both pilin and general F. tularensis protein glycosylation.
In another study on activation of pulmonary inflammation after F. tularensis Schu S4 exposure (Walters et al., 2013), altered expression level of bacteria-specific mRNA transcripts was found. Among these transcripts, a hypothetical protein FTT_0797 was characterized which shared homology with a glycosyl transferase. This protein is part of a gene cluster, which is thought to encode a polysaccharide additional to the lipopolysaccharide O antigen. Another protein, encoded by FTS_1402, was found to be involved in glycoprotein synthesis and to also contribute in part to LPS/capsule and/or Capsule Like Complex (CLC) production (Dankova et al., 2016). The resulting FTS_1402 mutant presented more sensitivity to serum complement.
All these proteins are summarized in Table 1.
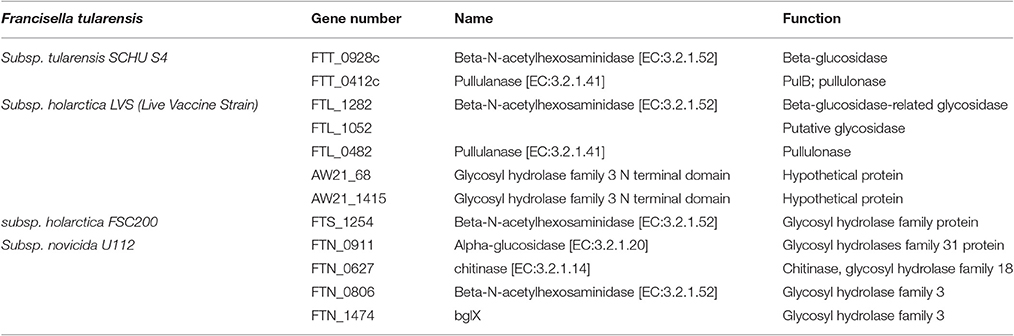
Concerning enzymes involved in degradation pathways, analysis of F. tularensis genomes showed a difference in the number of genes coding for proteins with such enzymatic activity (Table 2). Five genes were found in LVS, while only two genes were found in SchuS4 strain and only one gene in FSC200 strain. None of them was characterized.
Role of Post-Translational Modifications (PTM) on Bacteria/Host Cell Proteins
While two-third of all eukaryotic proteins are estimated to be glycosylated, the number of prokaryotic glycoproteins is still way behind understanding. This is mainly due to the enormous variability of their glycan structures and variations in the underlying glycosylation processes. In 2016, Schäffer and and Messner (2016) combined glycan structural information with bioinformatic, genetic, biochemical and enzymatic data for in-depth analyses of glycosylation processes in prokaryotes. This study included the major classes of prokaryotic (i.e., bacterial and archaeal) glycoconjugates without any example on Francisella. Furthermore, in a very recent publication (Bastos et al., 2017), while F. tularensis was shown to exhibit the largest number of glycoproteins in common with M. tuberculosis (Mtb), by sharing 16% of its glycoproteome, none of the glycosylated proteins of Francisella, as well as none of the enzymes involved in glycosylation pathway, have been found to play a specific role in pathogenesis. At the opposite, in M. tuberculosis, glycosylation of HbN, a truncated hemoglobin protein, was demonstrated to be necessary for its maintainance at the bacterial membrane and wall (Arya et al., 2013). Mutation in its mannose glycan linkage disrupted the facilitation of Mtb and M. smegmatis entry within the macrophages. These data suggested that glycosylation processes allowed Mtb survival within the hazardous environment of macrophages and the establishment of long term persistent infection in the host (Dey and Bishai, 2014).
Of note, Francisella did not belong to the list of prokaryotes that catalyzed glycosylation of host cell proteins (Bastos et al., 2017). In contrast, Legionella was cited as targeting eEF1A through effect of the glucosyl transferase Lgt1, with as result, the killing of eukaryotic cells (Belyi et al., 2008).
Conclusion
While 146 examples of protein glycosylation were cited for Francisella and only 111 for Helicobacter pylori (Bastos et al., 2017), the importance of these PTM, observed in Francisella and those induced in the host, is still largely unknown, notably on the outcome of the infectious cycle. Indeed, a large correlation between glycosylation and bacterial pathogenicity has already been proven for various species e.g., Campylobacter jejuni, Legionella and enteropathogenic Escherichia coli (EPEC) (Lu et al., 2015).
Francisella infection modifies the unfolded protein response (UPR) (Barel et al., 2016) and manipulates autophagy (Miller and Celli, 2016). Both processes are involved in maintaining cellular homeostasis and helping destroy invading microorganisms. Glycosylation and deglycosylation could be involved in molecular mimicry of common host cell glycans therefore helping the bacteria to avoid immune recognition. At this stage, we have all the reasons to believe that the glycosylation-deglycosylation processes observed in THP-1 cells were originated from eukaryotic enzymes. However, we cannot formerly exclude that Francisella enzymes might also be involved. Glycans and glycan-binding receptors influence all stages of infection, starting from initial colonization of host epithelial surfaces to spreading in tissue and inducing inflammation or host-cell injury, which may results in clinical symptoms (Nizet and Esko, 2009). Therefore, knowledge of glycosylation pathways involved during Francisella infection remains fundamental for prevention and treatment strategies.
Author Contributions
MB and AC wrote the review.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
INSERM, CNRS, and Université Paris Descartes Paris Cité Sorbonne supported these studies. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
Abu Kwaik, Y., and Bumann, D. (2013). Microbial quest for food in vivo: “Nutritional virulence” as an emerging paradigm. Cell. Microbiol. 15, 882–890. doi: 10.1111/cmi.12138
Arya, S., Sethi, D., Singh, S., Hade, M. D., Singh, V., Raju, P., et al. (2013). Truncated hemoglobin, hbn, is post-translationally modified in Mycobacterium tuberculosis and modulates host-pathogen interactions during intracellular infection. J. Biol. Chem. 288, 29987–29999. doi: 10.1074/jbc.M113.507301
Balonova, L., Hernychova, L., Mann, B. F., Link, M., Bilkova, Z., Novotny, M. V., et al. (2010). A multimethodological approach to identification of glycoproteins from the proteome of Francisella tularensis, an intracellular microorganism. J. Proteome Res. 9, 1995–2005. doi: 10.1021/pr9011602
Balonova, L., Mann, B. F., Cerveny, L., Alley, W. R., Chovancova, E., Forslund, A.-L., et al. (2012). Characterization of protein glycosylation in Francisella tularensis subsp. holarctica: identification of a novel glycosylated lipoprotein required for virulence. Mol. Cell Proteomics 11:M111.015016. doi: 10.1074/mcp.M111.015016
Barel, M., Grall, N., and Charbit, A. (2015). Pathogenesis of Francisella tularensis in Humans. Hoboken, NJ: John Wiley & Sons, Inc.
Barel, M., Harduin-Lepers, A., Portier, L., Slomianny, M.-C., and Charbit, A. (2016). Host glycosylation pathways and the unfolded protein response contribute to the infection by Francisella. Cell. Microbiol. 18, 1763–1781. doi: 10.1111/cmi.12614
Barel, M., Meibom, K., Dubail, I., Botella, J., and Charbit, A. (2012). Francisella tularensis regulates the expression of the amino acid transporter SLC1A5 in infected THP-1 human monocytes. Cell. Microbiol. 14, 1769–1783. doi: 10.1111/j.1462-5822.2012.01837.x
Bastos, P. A. D., da Costa, J. P., and Vitorino, R. (2017). A glimpse into the modulation of post-translational modifications of human-colonizing bacteria. J. Proteomics 152, 254–275. doi: 10.1016/j.jprot.2016.11.005
Belyi, Y., Tabakova, I., Stahl, M., and Aktories, K. (2008). Lgt: a family of cytotoxic glucosyltransferases produced by Legionella pneumophila. J. Bacteriol. 190, 3026–3035. doi: 10.1128/JB.01798-07
Brotcke, A., Weiss, D. S., Kim, C. C., Chain, P., Malfatti, S., Garcia, E., et al. (2006). Identification of Mgla-regulated genes reveals novel virulence factors in Francisella tularensis. Infect. Immun. 74, 6642–6655. doi: 10.1128/IAI.01250-06
Case, E. D. R., Chong, A., Wehrly, T. D., Hansen, B., Child, R., Hwang, S., et al. (2014). The Francisella O-antigen mediates survival in the macrophage cytosol via autophagy avoidance. Cell. Microbiol. 16, 862–877. doi: 10.1111/cmi.12246
Chacko, B. K., Scott, D. W., Chandler, R. T., and Patel, R. P. (2011). Endothelial surface N-glycans mediate monocyte adhesion and are targets for anti-inflammatory effects of peroxisome proliferator-activated receptor γ ligands. J. Biol. Chem. 286, 38738–38747. doi: 10.1074/jbc.M111.247981
Clemens, D. L., Lee, B. Y., and Horwitz, M. A. (2005). Francisella tularensis enters macrophages via a novel process involving pseudopod loops. Infect. Immun. 73, 5892–5902. doi: 10.1128/IAI.73.9.5892-5902.2005
Console, L., Scalise, M., Tarmakova, Z., Coe, I. R., and Indiveri, C. (2015). N-linked glycosylation of human SLC1A5 (ASCT2) transporter is critical for trafficking to membrane. Biochim. Biophys. Acta 1853, 1636–1645. doi: 10.1016/j.bbamcr.2015.03.017
Dankova, V., Balonova, L., Link, M., Straskova, A., Sheshko, V., and Stulik, J. (2016). Inactivation of Francisella tularensis gene encoding putative ABC transporter has a pleiotropic effect upon production of various glycoconjugates. J. Proteome Res. 15, 510–524. doi: 10.1021/acs.jproteome.5b00864
Dey, B., and Bishai, W. R. (2014). Crosstalk between Mycobacterium tuberculosis and the host cell. Semin. Immunol. 26, 486–496. doi: 10.1016/j.smim.2014.09.002
Di Gioia, M., and Zanoni, I. (2015). Toll-like receptor co-receptors as master regulators of the immune response. Mol. Immunol. 63, 143–152. doi: 10.1016/j.molimm.2014.05.008
Egge-Jacobsen, W., Salomonsson, E. N., Aas, F. E., Forslund, A.-L., Winther-Larsen, H. C., Maier, J., et al. (2011). O-linked glycosylation of the pila pilin protein of Francisella tularensis: identification of the endogenous protein-targeting oligosaccharyltransferase and characterization of the native oligosaccharide. J. Bacteriol. 193, 5487–5497. doi: 10.1128/JB.00383-11
Hetz, C., Martinon, F., Rodriguez, D., and Glimcher, L. H. (2011). The unfolded protein response: integrating stress signals through the stress sensor IRE1α. Physiol. Rev. 91, 1219–1243. doi: 10.1152/physrev.00001.2011
Lu, Q., Li, S., and Shao, F. (2015). Sweet talk: protein glycosylation in bacterial interaction with the host. Trends Microbiol. 23, 630–641. doi: 10.1016/j.tim.2015.07.003
Meibom, K. L., and Charbit, A. (2010). The unraveling panoply of Francisella tularensis virulence attributes. Curr. Opin. Microbiol. 13, 11–17. doi: 10.1016/j.mib.2009.11.007
Miller, C., and Celli, J. (2016). Avoidance and subversion of eukaryotic homeostatic autophagy mechanisms by bacterial pathogens. J. Mol. Biol. 428, 3387–3398. doi: 10.1016/j.jmb.2016.07.007
Moran, A. P., Gupta, A., and Joshi, L. (2011). Sweet-talk: role of host glycosylation in bacterial pathogenesis of the gastrointestinal tract. Gut 60, 1412–1425. doi: 10.1136/gut.2010.212704
Ni, M., Zhou, H., Wey, S., Baumeister, P., and Lee, A. Y. (2009). Regulation of PERK signaling and leukemic cell survival by a novel cytosolic isoform of the UPR regulator GRP78/BiP. PLoS ONE 4:e6868. doi: 10.1371/journal.pone.0006868
Nizet, V., and Esko, J. (2009). “Chapter 39: Bacterial and viral infections,” in Essentials of Glycobiology, 2nd Edn., eds R. D. Cummings, A. Varki, J. D. Esko H. H. Freeze, P. Stanley, C. R. Bertozzi, G. W. Hart, and M. E. Etzler (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), 1–16.
Opdenakker, G., Proost, P., and Van Damme, J. (2016). Microbiomic and posttranslational modifications as preludes to autoimmune diseases. Trends Mol. Med. 22, 746–757. doi: 10.1016/j.molmed.2016.07.002
Pfaffenbach, K. T., and Lee, A. S. (2011). The critical role of of GRP78 in physiologic and pathologic stress. Curr. Opin. Cell Biol. 23, 150–156. doi: 10.1016/j.ceb.2010.09.007
Ray, K., Marteyn, B., Sansonetti, P. J., and Tang, C. M. (2009). Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat. Rev. Microbiol. 7, 333–340. doi: 10.1038/nrmicro2112
Santic, M., Al Khodor, S., and Abu Kwaik, Y. (2010). Cell biology and molecular ecology of Francisella tularensis. Cell. Microbiol. 12, 129–139. doi: 10.1111/j.1462-5822.2009.01400.x
Schäffer, C., and Messner, P. (2016). Emerging facets of prokaryotic glycosylation. FEMS Microbiol. Rev. 41, 49–91. doi: 10.1093/femsre/fuw036
Sjöstedt, A. (2011). Special topic on Francisella tularensis and tularemia. Front. Cell. Infect. Microbiol. 2:86. doi: 10.3389/fmicb.2011.00086
Tan, F. Y. Y., Tang, C. M., and Exley, R. M. (2015). Sugar coating: bacterial protein glycosylation and host–microbe interactions. Trends Biochem. Sci. 40, 342–350. doi: 10.1016/j.tibs.2015.03.016
Thomas, R. M., Twine, S. M., Fulton, K. M., Tessier, L., Kilmury, S. L. N., Ding, W., et al. (2011). Glycosylation of DsbA in francisella tularensis subsp. tularensis. J Bacteriol 193, 5498–5509. doi: 10.1128/JB.00438-11
Tytgat, H. L. P., and de Vos, W. M. (2016). Sugar coating the envelope: glycoconjugates for microbe–host crosstalk. Trends Microbiol. 24, 853–861. doi: 10.1016/j.tim.2016.06.004
Vaidyanathan, K., Durning, S., and Wells, L. (2014). Functional O-GlcNac modifications: implications in molecular regulation and pathophysiology. Crit. Rev. Biochem. Mol. Biol. 49, 140–163. doi: 10.3109/10409238.2014.884535
Varki, A. (1993). Biological roles of oligosaccharides: all of the theories are correct. Glycobiology 3, 97–130. doi: 10.1093/glycob/3.2.97
Walters, K.-A., Olsufka, R., Kuestner, R. E., Cho, J. H., Li, H., Zornetzer, G. A., et al. (2013). Francisella tularensis subsp. tularensis induces a unique pulmonary inflammatory response: role of bacterial gene expression in temporal regulation of host defense responses. PLoS ONE 8:e62412. doi: 10.1371/journal.pone.0062412
Keywords: glycosylation, host-pathogen interaction
Citation: Barel M and Charbit A (2017) Role of Glycosylation/Deglycolysation Processes in Francisella tularensis Pathogenesis. Front. Cell. Infect. Microbiol. 7:71. doi: 10.3389/fcimb.2017.00071
Received: 13 January 2017; Accepted: 27 February 2017;
Published: 21 March 2017.
Edited by:
Marina Santic', University of Rijeka, CroatiaReviewed by:
Jiri Stulik, University of Defence, CzechiaPaul Edward Carlson, Food and Drug Administration, USA
Copyright © 2017 Barel and Charbit. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Monique Barel, TW9uaXF1ZS5iYXJlbEBpbnNlcm0uZnI=
 Monique Barel
Monique Barel Alain Charbit
Alain Charbit