- 1State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology, Beijing, China
- 2Clinical Laboratory, The Second Affiliated Hospital of Soochow University, Suzhou, China
- 3New Jersey Medical School, Public Health Research Institute Tuberculosis Center, Rutgers University, Newark, NJ, USA
Methicillin-resistant Staphylococcus aureus (MRSA), is one of the most prevalent clinical pathogens isolated from hospital settings, and has increasingly identified in community settings. In China, the SCCmecIII-ST239 strains are disseminated in different geographic regions, accounting for >75% of all MRSA isolates in some national studies. Here we characterized 150 non-duplicate MRSA isolates collected from February 2012 to May 2013 in a tertiary hospital in Suzhou, Eastern China, to explore the molecular epidemiology. All isolates were characterized by spa typing, SCCmec typing, and detection of genes encoding Panton-Valentine leukocidin (PVL) and toxic shock syndrome toxin (TSST-1). Representative genotypes were also subjected to multilocus sequence typing (MLST). Antibiotic susceptibility testing was performed using BD Phoenix™ Automated Microbiology System. Molecular typing identified 11 clonal complex (CC) and 28 spa types, with the CC5-spa t002 (29.3%) and CC239-spa t037 (14.7%) being the most prevalent. SCCmec types II, III, IV, and V were identified in 33.3, 21.3, 23.3, and 21.3% of all isolates, respectively. PVL genes (lukF/S-PV) were detected in 11.3% of all isolates and from 6 CCs (5, 8, 59, 88, 239, and 398). The TSST-1 gene (tst) was detected in 18.0% of the all isolates, predominantly in CC5 (96.3%). All the tst-1-positve CC5 isolates were spa t002. Eighteen patients died within 30 days of hospitalization, and the in-hospital 30-day mortality was 12.0%. Multivariable analysis showed that 60 years old (odds ratio [OR] = 7.2, P = 0.026), cancer diagnosis (OR = 9.6, P = 0.022), and MRSA isolate carriage of tst-1 (OR = 62.5, P < 0.001) were independent factors associated with 30-day mortality. Our study revealed unique MRSA dissemination patterns in our hospital in comparison to those of other regions in China. The finding that tst-1-positive CC5 strains were associated with higher mortality highlights the need for strict infection control measures in order to prevent further spread of these strains in our hospital, as well as others.
Introduction
Staphylococcus aureus (S. aureus) is one of the most prevalent clinical pathogens isolated from hospital settings, and has recently become widespread in community settings as well. S. aureus causes a broad variety of diseases including skin and soft-tissue infections, bacteremia, osteomyelitis, infective endocarditis, and necrotizing pneumonia (Lowy, 1998; Nadig et al., 2010; Tokajian et al., 2010; Song et al., 2013). Following its emergence in the early 1960s, methicillin-resistant Staphylococcus aureus (MRSA) has become highly epidemic in many hospitals and health care settings worldwide. Moreover, in recent years distinct community-acquired MRSA (CA-MRSA) strains have also emerged as a cause of invasive and life-threatening infections among young, healthy patients with no significant healthcare exposure (Deleo et al., 2010; Tokajian et al., 2010; Alon et al., 2011; Mediavilla et al., 2012; El-Mahdy et al., 2013).
Molecular typing techniques are routinely used to explore the evolution and epidemiology of MRSA, the most commonly-used being staphylococcal protein A (spa) typing (Shopsin et al., 1999; Harmsen et al., 2003), staphylococcal cassette chromosome (SCC) mec typing (International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements, 2009), multilocus sequence typing (MLST) (Enright et al., 2000), pulsed-field gel electrophoresis (PFGE) (Tenover et al., 1995), and multiple-locus variable-number tandem repeat analysis (MLVA) (Sabat et al., 2003). Basedon molecular typing, MRSA strains can be divided into various clones, usually denoted by their MLST sequence type (ST) or clonal complex (CC), followed by the SCCmec type. Some of these, for example CC8-MRSA-IV (USA300 clone) and CC30-MRSA-IV (Southwest Pacific clone) strains, appear to be pandemic, and have been found on nearly every continent, while others, such as ST59-MRSA-V (Taiwan clone) and ST80-MRSA-IV (European clone), appear to be only regionally disseminated (Mediavilla et al., 2012). In addition, virulence genes such as Panton-Valentine leukocid in (PVL) gene lukF-PV and toxic shock syndrome toxin gene tst-1, are often associated with certain S. aureus clones, and can be used as additional genetic markers with which to characterize MRSA strains. For example, lukF-PV has been frequently found in isolates causing community infections, and has traditionally been used as a surrogate maker for CA-MRSA. By contrast, tst-1 appears to be limited to a handful of clonal lineages, and is most frequently associated with methicillin-susceptible S. aureus (MSSA) strains belonging to CC30, while more recently, tst-positive CC5 and CC22 MRSA strains have also been documented (Dauwalder et al., 2008; Al Laham et al., 2015).
In China, a multi-drug resistant MRSA clone, defined as ST239 by MLST, is now widely disseminated in different geographic regions, accounting for >75% of all MRSA isolates in two national studies (Liu et al., 2009; Chen et al., 2014). MRSA ST239 strains usually harbor SCCmec type III elements, and largely correspond to two spa types: t037 and t030. Interestingly, recent studies demonstrated that spa t030 has displaced t037 and has thereby become the most frequently-isolated MRSA spa type in China (Chen et al., 2010). Second in prevalence following ST239-MRSA-III is the ST5-MRSA-II clone, of which t002 is the most commonly reported spa type (19). Other MRSA clones, such as ST398-MRSA-V (live stock-associated) and ST59-MRSA-IV (community-associated), have also been identified (Chen et al., 2010). Previous studies have shown that virulence genes lukF-PV and tst-1 are rarely identified in ST239-MRSA-III and ST5-MRSA-II strains in China. In this study, we report the spread of tst-1-harboring ST5-MRSA-II isolates in a tertiary hospital in Suzhou, Eastern China, with multivariate analysis further demonstrating that presence of tst-1 is an independent risk factor for 30-day mortality.
Materials and Methods
Bacterial Isolates
A total of 150 MRSA isolates collected from February 2012 to May 2013 in a tertiary hospital in Suzhou, China, were included. Identification of S. aureus isolates was performed using standard microbiologic methods and the Phoenix System-100 BD Automated Microbiology system (BD Diagnostics, USA). Presence of the mecA or mecC gene was determined by PCR as described previously (Murakami et al., 1991; Stegger et al., 2012).
Antimicrobial Susceptibility
Isolates of S. aureus were inoculated onto the Phoenix panel according to the manufacturer's instructions, following which species identification and antimicrobial susceptibility were determined using the Phoenix System-100 BD Automated Microbiology system (BD Diagnostics, USA). Results of Minimum Inhibitory Concentrations (MICs) were recorded according to Clinical and Laboratory Standards Institute recommendations (CLSI, 2012). S. aureus ATCC 29213 was used as a quality control strain for antimicrobial susceptibility testing.
Molecular Typing
All 150 S. aureus isolates were characterized by staphylococcal protein A (spa) typing (Shopsin et al., 1999), and spa types were assigned using eGenomics software (Shopsin et al., 1999; Mathema et al., 2008), with Ridom assignments made using the Spa Server website (http://spa.ridom.de/). In order to avoid confusion, eGenomics spa types were shown by the spa motif repeats (e.g., spa type 2, TJMBMDMGMK), and Ridom spa types shown as numbers (e.g., spa t030). All MRSA isolates were subjected to SCCmec typing using multiplex real-time PCR (Chen et al., 2009). MLST was performed as described previously (Enright et al., 2000) on a representative subset of 30 isolates, with clonal complexes inferred via eBURST analysis (Feil et al., 2004); all other clonal complexes were inferred from spa typing data as described previously (Mathema et al., 2008), using both the Ridom Spa Server website and the eGenomics database. Clonal complex sub-groups with distinct genotypic signatures were classified as individual CCs (Mediavilla et al., 2012), e.g., ST239 strains were classified as CC239 rather than CC8. PFGE and MLVA were performed as described previously (Tenover et al., 1995; Sabat et al., 2003).
Detection of PVL and TSST-1 Genes
All isolates were tested for the presence of the genes encoding Panton-Valentine leukocid in (PVL) and toxic shock syndrome toxin (TSST-1). The genes coding for PVL were detected by PCR amplification of lukS-PV and lukF-PV (Said-Salim et al., 2005), while the gene coding for TSST-1 was detected using a novel real-time PCR assay reported elsewhere (Al Laham et al., 2015).
Clinical Information
For each patient diagnosed with MRSA infection, we recorded demographics, comorbidities, patient location at isolation, specimen source, antimicrobial therapies, vancomycin treatment history, and 30-day mortality. Hospital-onset cases were defined as positive culture occurring ≥3 days after hospitalization. The study was approved by the Institutional Review Boards of the Second Affiliated Hospital of Soochow University.
Statistical Analysis
Characteristics of patients in different groups were compared using Chi-square or Fisher's exact tests for categorical variables, and Wilcoxon rank-sum test for continuous variables. P ≤ 0.05 (two-tailed) were considered statistically significant. A multivariable logistic regression model was constructed to identify baseline factors independently associated with 30-day mortality. All variables with P ≤ 0.1 in univariate analysis were entered into the multivariable model and a backward stepwise selection process was applied. SPSS, version 22.0 (IBM SPSS, IBM Corporation, Somers, NY) was used for all statistical analyses.
Ethical Approval
The Medical Ethics Committee of Second Affliated Hospital of Soochow University approved this study and all isolates were collected with the patients' written informed consent.
Results
Bacterial Isolates and Patient Data
A total of 150 unique MRSA isolates from different specimens, collected from February 2012 to May 2013, were included in this study. Among these, 71 were collected from internal medicine wards (47.3%), 48 from surgical wards (32.0%), and 31 from intensive care unit (ICU) wards (20.7%). All isolates were obtained from inpatients. 60% of patients were more than 60 years old, with an average age of 63, and 74.0% (n = 111) are male. A total of 87.3% (n = 131) of the cases were hospital-onset. The majority of the isolates were from sputum (n = 90, 60.0%), followed by drainage (n = 17, 11.3%) and wounds (n = 12, 8.0%); 4 isolates were from blood, and 5 were from urine, while the rest of the 22 isolates were from other sites. The Medical Ethics Committee of Second Affiliated Hospital of Soochow University approved this study and all isolates were collected with patient consent in this study.
Antimicrobial Susceptibility
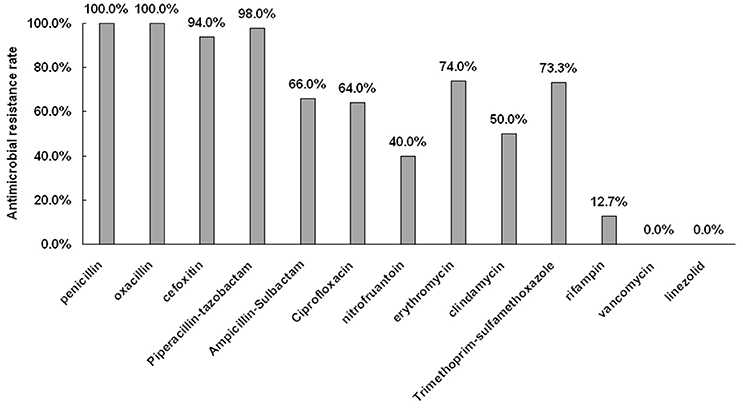
The antimicrobial resistance profiles for all 150 MRSA isolates are shown in Figure 1. All isolates were susceptible to vancomycin and linezolid. The percentage of resistance to both penicillin and oxacillin were 100.0% (150/150), while resistant tocefoxitin, piperacillin-tazobactam, ampicillin-sulbactam, ciprofloxacin, nitrofuantoin, erythromycin, clindamycin, trimethoprim-sulfame thoxazole, and rifampin were 94.0% (141/150), 98.0% (147/150), 66.0% (99/150), 64.0% (96/150), 60.0% (90/150), 74.0% (111/150), 50.0% (75/150), 72.0% (108/150), and 12.7% (19/150), respectively.
Molecular Characteristics of MRSA
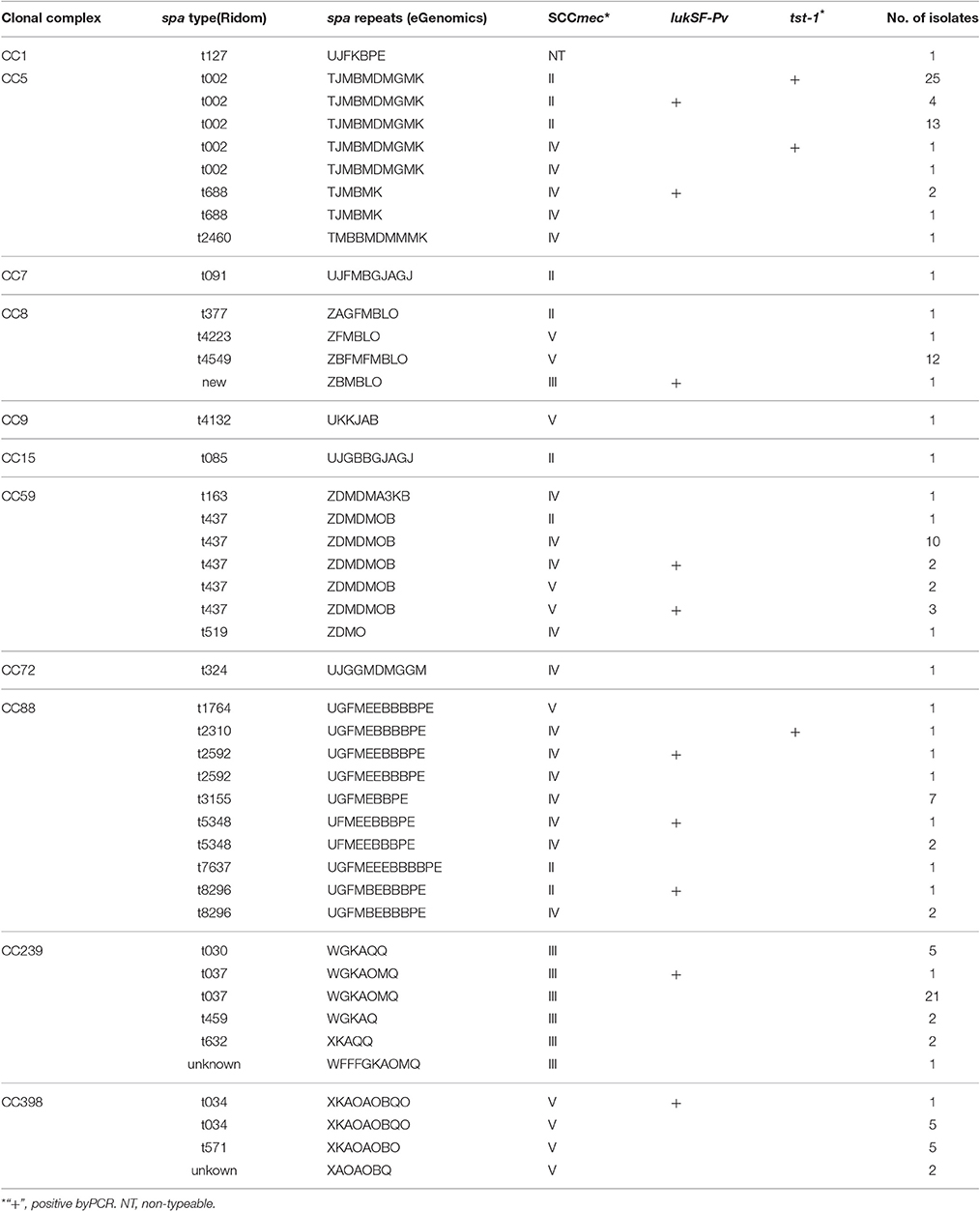
Among the 150 MRSA isolates, four SCCmec types were identified, including types II (50, 33.3%), III (32, 21.3%), IV (35, 23.3%), and V (32, 21.3%), while1 isolate (0.7%) was non-typeable (NT) (Table 1). A total of 28 spa types were identified among the 150 MRSA isolates, belonging to 11 CCs according to eBURST analysis. CC5 isolates were the most common, accounting for nearly one-third (48, 32.0%) of total isolates, followed by CC239 (32, 21.3%), CC59 (19, 12.7%), CC88 (18, 12.0%), CC8 (15, 10.0%), and CC398 (13, 8.7%). One isolate each of CC1, CC7, CC9, CC15, and CC72 was also identified.
The most common spa type was t002(29.3%, 44/150), followed by t037(14.7%, 22/150), t437(11.3%, 17/150), t4549(8.0%, 12/150), t3155(4.7%, 7/150), t034(4.0%, 6/150), and t571(3.3%, 5/150), while spa t030 was only found in 5 isolates (3.3%). The two predominant genotypes were (a) CC5, spa type t002, and SCCmec II (n = 42, 28.0%), and (b) CC239, spa t037, and SCCmec III (n = 22, 14.7%).
Prevalence of pvl and tst-1
Among the 150 MRSA isolates, 11.3% (17/150) tested positive for the presence of the genes coding for PVL (lukF-PV, lukS-PV). PVL-positive isolates were identified in six different clonal complexes (Table 1), including CC5, 8, 59, 88, 239, and 398. The two most common spa types among PVL-positive isolates were t437 (5/17, CC59) and t002 (4/17, CC5). Among these, 6 (35.3%), 5 (29.4%), and 4 (23.5%) isolates were classified as SCCmec type IV, II, and V, respectively.
Similarly, 18.0% of the MRSA isolates (27/150) tested positive for the presence of the TSST-1 gene(tst-1), which was detected predominantly in CC5 (n = 26, 96.3%); the other tst-1-positve isolate belonged to CC88. All the tst-1-positve CC5 isolates were spa t002. Among the 26 tst-1-positive CC5-t002 isolates, 25 were SCCmec typeII and 1 isolate was type IV. PFGE and MLVA both showed that these tst-positive CC5-t002 isolates have indistinguishable pulsotypes and MLVA patterns (data not shown), suggesting clonal spread.
Outcome and Risk Factors
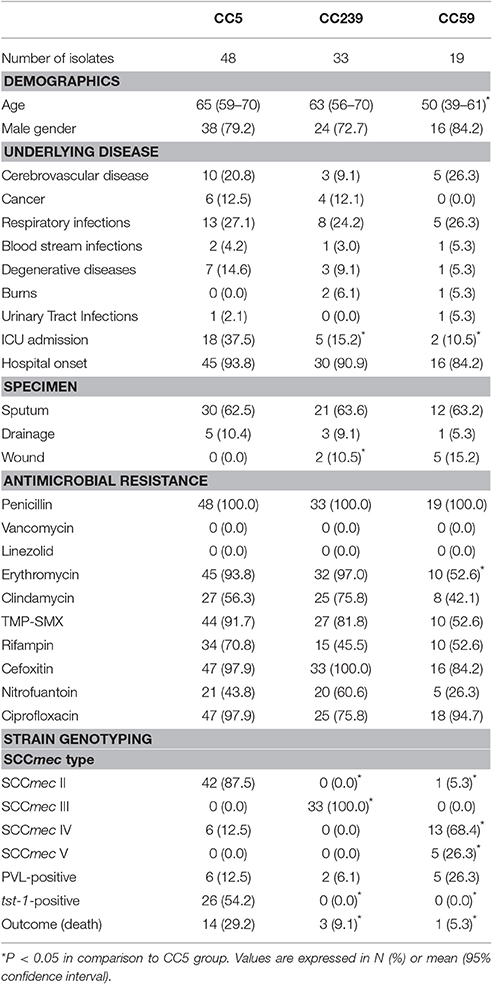
In this study, 18 patients died within 30 days of hospitalization, and the in-hospital 30-day mortality was 12.0%. We then compared the outcomes, clinical characteristics, and MRSA isolate genotypes within three major CCs (CC5, CC239, and CC59) (Table 2). In comparison to cases infected with CC239 or CC59 strains, patients with CC5 infections had significantly higher 30-day mortality (P < 0.05). In addition, CC5 strains had a higher frequency of SCCmec type II and tst-1(P < 0.01), and were more likely to be isolated from patients admitted to an ICU (P < 0.05) (Table 2).
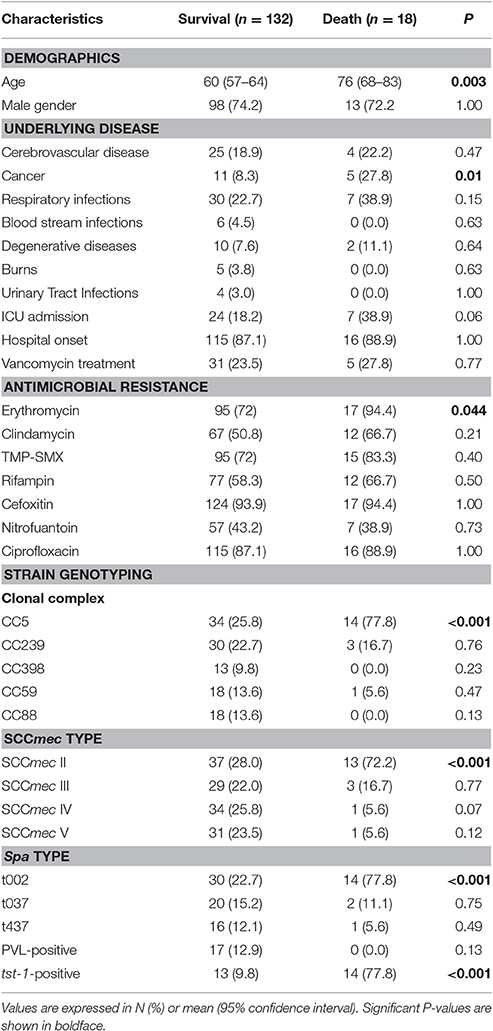
We then compared the clinical and molecular characteristics between patients with different clinical outcomes (survival vs. death) (Table 3). The following factors were associated with 30-day mortality in univariate analysis: age, cancer, erythromycin resistance, belonging to CC5, harboring SCCmec II, spa t002, and presence of tst-1 (P < 0.05). In addition, ICU admission displayed border line significance (P = 0.06). Notably, observed 30-day mortality in patients with tst-1-positive MRSA was significantly higher than that inpatient with tst-1-negative MRSA (51.9 vs. 3.3%, P < 0.001). By contrast, no significant difference in 30-day mortality was observed between PVL-positive vs.-negative groups (0.0 vs. 13.5%, P = 0.13). In multivariable analysis, factors independently associated with 30-day mortality included age greater than 60 years old (odds ratio [OR] = 7.2, 95% confidence interval [95% CI] = 1.26–41.6, P = 0.026), cancer diagnosis (OR = 9.6, 95% CI = 1.4–65.7, P = 0.02), and MRSA isolate carriage of tst-1 (OR = 62.5, 95% CI = 12.0–325.2, P < 0.001).
Discussion
MRSA is a major nosocomial pathogen worldwide. Infection due to MRSA imposes a high and increasing burden on health care resources, as well as increasing morbidity and mortality. Several major MRSA clones are spreading globally, and they often harbor virulence factors including PVL and TSST-1. Here we characterized 150 non-duplicated MRSA clinical isolates collected from a tertiary hospital in Suzhou, Eastern China. The overall 30-day mortality was as high as 12%. Risk factors for mortality were examined and we found that presence of tst-1, age greater than 60 years, and underlying cancer co-morbidity were independent risk factors associated with mortality.
This study revealed some interesting findings. Firstly, CC5 isolates were the main CC in our study, accounting for nearly one third (48, 32.0%) of the total isolates, with the major genotype identified consisting of CC5, spa type t002, and SCCmec type II (42/150, 28.0%). By contrast, several studies have demonstrated that CC239 is the predominant CC in China, associated with SCCmec type III and mainly comprised of spa types t037 and t030. A large survey screening a total of 702 MRSA isolates from 18 teaching hospitals in 14 Chinese cities between 2005 and 2006 showed that that the CC239 spa type t030 comprised 52.0% of the total isolates, whilet037 accounted for 25.5% (Liu et al., 2009). In addition, the distribution of spa types varied among different regions, with t002 (CC5) the most common in northern cities, t037 (CC239) predominant in eastern cities (e.g., Shanghai), and t030 (CC239) the most common in other cities (Liu et al., 2009). Another investigation highlighted a clonal shift from spa t037 (from 1994 to 2000) to spa t030 (since2000) as the major clone in a Beijing hospital. Similarly, a recent study involving seven hospitals in China showed an increasing prevalence of spa type t030, with 80.1% of all MRSA isolates belonging to t030 (Chen et al., 2014). The above studies suggest that ST239 spa t030 has replaced t037, which represents the ancestral CC239 spa type, as the most frequent MRSA spa type in China (Liu et al., 2009; Chen et al., 2010, 2014). By contrast, CC5 spa t002 emerged initially in 2002, but has subsequently exhibited a low prevalence rate (Chen et al., 2010). However, our study suggests that this may not be the case in all institutions.
The current study was conducted in Suzhou, which is the largest city near Shanghai, located ~60 miles away. Interestingly, our study showed that t002 was the most common spa type in our hospital, whereas over 70% isolates in Shanghai were typed as t037 (Liu et al., 2009). Although located in a similar geographic region, the epidemic pattern of CC and spa types in our city was different from that of Shanghai. In addition, our results showed that t037 (16.1%) was more common than t030 (3.6%), which appears to be inconsistent with the pattern of replacement of spa t037 by t030 in other studies (Liu et al., 2009; Chen et al., 2010, 2014). Our results therefore suggest differences in geographic distribution of MRSA clones within Chinese hospitals. Specifically, the predominance of CC5 isolates in our hospital is largely due to the high frequency of tst-1-positive CC5-MRSA-II, spa t002 strains (Table 1).
The CC5 strains in this study have been largely associated with tst-1, a gene encoding the toxic shock syndrome toxin TSST-1. In this study, we found that 18.0% (n = 27) of all MRSA isolates were tst-1-positive, with all but one belonging to CC5. However, tst-1 is rarely identified in CC5 MRSA strains in China. In a previous multicenter study from China, tst-1 was detected in 31.4% of isolates tested (including both MSSA and MRSA), but mostly in CC398, CC15, and CC188 (He et al., 2013). Another study from a city in central China has found tst-1 is mostly associated with CC398, CC59, and CC8, but not CC5 (Liu et al., 2015). Moreover, a recent study from another hospital in Suzhou, the same city as in our study, also reported CC5 as the predominant MRSA genotype, accounting for 50% of all MRSA isolates; however, none of the strains possessed tst-1 (Li et al., 2015). It appears therefore that tst-1-harboring CC5 strains are an emerging clone in our hospital. Alarmingly, our study showed that tst-1 is an independent risk factor associated with 30-day mortality. The mortality of patients infected by tst-1-positive MRSA strains was much greater than that of patients infected by tst-negative strains (51.8 vs. 3.5%, P < 0.01). Our findings therefore suggest that, in our hospital, tst-1-positive CC5-MRSA-II- isolates may be more virulent. Conversely, the presence of another virulence factor, PVL, was not associated with increased mortality. In this study, none of the 17 patients infected by PVL-positive strains died. PVL is commonly found in CA-MRSA strains, and has been associated with severe hospital-acquired pneumonia with increased mortality (Vandenesch et al., 2003; Zhang et al., 2016). However, our results are similar to those of other recent studies of MRSA which did not observe an association between the presence of PVL genes and increased mortality (Peyrani et al., 2011; Haque et al., 2012; Tadros et al., 2013).
Our study had several limitations. Firstly, this study is limited due to the relatively small sample size death cases, which is reflected by the wide range of some confidence intervals in multivariable analysis. Secondly, most of the isolates (60%) were collected from sputum, which is over-represented in the current study. Thirdly, we only tested for the presence of two virulence factors (PVL and TSST-1) while evaluating their associations with outcomes. Therefore, it is possible that other virulence factors co-harbored along with tst-1 may contribute to the increased mortality observed in the current study. Lastly, this study lacks detailed information about the underlying cause of death, partially due to the nature of retrospective data collection, therefore we were not able to determine the disease specific mortality but total mortality. Nevertheless, our study revealed unique MRSA dissemination patterns in our hospital. The finding that tst-1-positive CC5 strains were associated with higher total mortality highlights the need for strict infection control measures in order to prevent further spread of these strains in our hospital, as well as others. Further genome-based analysis of tst-1-positive CC5 strains should also be performed to identify additional virulence factors contributing to the high mortality observed in this study.
Author Contributions
MW, work, data analysis and manuscript preparation; YZ, work; JM, manuscript preparation; LC, study design, data analysis and manuscript preparation; BK, manuscript preparation; YS, manuscript preparation; RY, manuscript preparation; HD, study design, work, data analysis and manuscript preparation.
Funding
This study was supported by the National Natural Science Foundation of China (81572032, 81401636), Six talent peaks project in Jiangsu Province (2016-WSN-112), Gusu key health talent of Suzhou, the Natural Science Foundation for Colleges and Universities in Jiangsu Province (16KJB320006), the Science and Technology Program of Suzhou (SS201638), Pre-research Foundation of Young Workers (SDFEYQN1612), Jiangsu youth medical talents program (QN-867).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Al Laham, N., Mediavilla, J. R., Chen, L., Abdelateef, N., Elamreen, F. A., Ginocchio, C. C., et al. (2015). MRSA clonal complex 22 strains harboring toxic shock syndrome toxin (TSST-1) are endemic in the primary hospital in Gaza, Palestine. PLoS ONE 10:e0120008. doi: 10.1371/journal.pone.0120008
Alon, D., Abd-Elkadir, F., Chowers, M., and Paitan, Y. (2011). MRSA SCCmec epidemiology in Israel: development and implementation of an MRSA SCCmec typing strategy. Eur. J. Clin. Microbiol. Infect. Dis. 30, 1443–1452. doi: 10.1007/s10096-011-1243-9
Chen, H., Liu, Y., Jiang, X., Chen, M., and Wang, H. (2010). Rapid change of methicillin-resistant Staphylococcus aureus clones in a Chinese tertiary care hospital over a 15-year period. Antimicrob. Agents Chemother. 54, 1842–1847. doi: 10.1128/AAC.01563-09
Chen, L., Mediavilla, J. R., Oliveira, D. C., Willey, B. M., de Lencastre, H., and Kreiswirth, B. N. (2009). Multiplex real-time PCR for rapid Staphylococcal cassette chromosome mec typing. J. Clin. Microbiol. 47, 3692–3706. doi: 10.1128/JCM.00766-09
Chen, Y., Liu, Z., Duo, L., Xiong, J., Gong, Y., Yang, J., et al. (2014). Characterization of Staphylococcus aureus from distinct geographic locations in China: an increasing prevalence of spa-t030 and SCCmec type III. PLoS ONE 9:e96255. doi: 10.1371/journal.pone.0096255
CLSI, (2012). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement, M100-S22. Wayne, PA: CLSI.
Dauwalder, O., Lina, G., Durand, G., Bes, M., Meugnier, H., Jarlier, V., et al. (2008). Epidemiology of invasive methicillin-resistant Staphylococcus aureus clones collected in France in 2006 and 2007. J. Clin. Microbiol. 46, 3454–3458. doi: 10.1128/JCM.01050-08
Deleo, F. R., Otto, M., Kreiswirth, B. N., and Chambers, H. F. (2010). Community-associated meticillin-resistant Staphylococcus aureus. Lancet 375, 1557–1568. doi: 10.1016/S0140-6736(09)61999-1
El-Mahdy, T. S., El-Ahmady, M., and Goering, R. V. (2013). Molecular characterization of methicillin-resistant Staphylococcus aureus isolated over a 2-year period in a Qatari hospital from multinational patients. Clin. Microbiol. Infect. 20, 169–173. doi: 10.1111/1469-0691.12240
Enright, M. C., Day, N. P., Davies, C. E., Peacock, S. J., and Spratt, B. G. (2000). Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38, 1008–1015.
Feil, E. J., Li, B. C., Aanensen, D. M., Hanage, W. P., and Spratt, B. G. (2004). eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186, 1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004
Haque, N. Z., Arshad, S., Peyrani, P., Ford, K. D., Perri, M. B., Jacobsen, G., et al. (2012). Analysis of pathogen and host factors related to clinical outcomes in patients with hospital-acquired pneumonia due to methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 50, 1640–1644. doi: 10.1128/JCM.06701-11
Harmsen, D., Claus, H., Witte, W., Rothgänger, J., Claus, H., Turnwald, D., et al. (2003). Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41, 5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003
He, W., Chen, H., Zhao, C., Zhang, F., Li, H., Wang, Q., et al. (2013). Population structure and characterisation of Staphylococcus aureus from bacteraemia at multiple hospitals in China: association between antimicrobial resistance, toxin genes and genotypes. Int. J. Antimicrob. Agents 42, 211–219. doi: 10.1016/j.ijantimicag.2013.04.031
International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (2009). Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 53, 4961–4967. doi: 10.1128/AAC.00579-09
Li, Y., Zhao, R., Zhang, X., Han, Q., Qian, X., Gu, G., et al. (2015). Prevalence of enterotoxin genes and spa genotypes of methicillin-resistant Staphylococcus aureus from a tertiary care hospital in China. J. Clin. Diagn. Res. 9, Dc11–14. doi: 10.7860/jcdr/2015/12090.5990
Liu, C., Chen, Z. J., Sun, Z., Feng, X., Zou, M., Cao, W., et al. (2015). Molecular characteristics and virulence factors in methicillin-susceptible, resistant, and heterogeneous vancomycin-intermediate Staphylococcus aureus from central-southern China. J. Microbiol. Immunol. Infect. 48, 490–496. doi: 10.1016/j.jmii.2014.03.003
Liu, Y., Wang, H., Du, N., Shen, E., Chen, H., Niu, J., et al. (2009). Molecular evidence for spread of two major methicillin-resistant Staphylococcus aureus clones with a unique geographic distribution in Chinese hospitals. Antimicrob. Agents Chemother. 53, 512–518. doi: 10.1128/AAC.00804-08
Lowy, F. D. (1998). Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532. doi: 10.1056/NEJM199808203390806
Mathema, B., Mediavilla, J., and Kreiswirth, B. N. (2008). Sequence analysis of the variable number tandem repeat in Staphylococcus aureus protein a gene: spa typing. Methods Mol. Biol. 431, 285–305. doi: 10.1007/978-1-60327-032-8_22
Mediavilla, J. R., Chen, L., Mathema, B., and Kreiswirth, B. N. (2012). Global epidemiology of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA). Curr. Opin. Microbiol. 15, 588–595. doi: 10.1016/j.mib.2012.08.003
Murakami, K., Minamide, W., Wada, K., Nakamura, E., Teraoka, H., and Watanabe, S. (1991). Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J. Clin. Microbiol. 29, 2240–2244.
Nadig, S., Ramachandra Raju, S., and Arakere, G. (2010). Epidemic meticillin-resistant Staphylococcus aureus (EMRSA-15) variants detected in healthy and diseased individuals in India. J. Med. Microbiol. 59, 815–821. doi: 10.1099/jmm.0.017632-0
Peyrani, P., Allen, M., Wiemken, T. L., Haque, N. Z., Zervos, M. J., Ford, K. D., et al. (2011). Severity of disease and clinical outcomes in patients with hospital-acquired pneumonia due to methicillin-resistant Staphylococcus aureus strains not influenced by the presence of the Panton-Valentine leukocidin gene. Clin. Infect. Dis. 53, 766–771. doi: 10.1093/cid/cir541
Sabat, A., Krzyszton-Russjan, J., Strzalka, W., Filipek, R., Kosowska, K., Hryniewicz, W., et al. (2003). New method for typing Staphylococcus aureus strains: multiple-locus variable-number tandem repeat analysis of polymorphism and genetic relationships of clinical isolates. J. Clin. Microbiol. 41, 1801–1804. doi: 10.1128/JCM.41.4.1801-1804.2003
Saïd-Salim, B., Mathema, B., Braughton, K., Davis, S., Sinsimer, D., Eisner, W., et al. (2005). Differential distribution and expression of Panton-Valentine leucocidin among community-acquired methicillin-resistant Staphylococcus aureus strains. J. Clin. Microbiol. 43, 3373–3379. doi: 10.1128/JCM.43.7.3373-3379.2005
Shopsin, B., Gomez, M., Montgomery, S. O., Smith, D. H., Waddington, M., Dodge, D. E., et al. (1999). Evaluation of protein a gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37, 3556–3563.
Song, Y., Du, X., Li, T., Zhu, Y., and Li, M. (2013). Phenotypic and molecular characterization of Staphylococcusaureus recovered from different clinical specimens of inpatients at a teaching hospital in Shanghai between 2005 and 2010. J. Med. Microbiol. 62, 274–282. doi: 10.1099/jmm.0.050971-0
Stegger, M., Andersen, P. S., Kearns, A., Pichon, B., Holmes, M. A., Edwards, G., et al. (2012). Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecA (LGA251). Clin. Microbiol. Infect. 18, 395–400. doi: 10.1111/j.1469-0691.2011.03715.x
Tadros, M., Williams, V., Coleman, B. L., McGeer, A. J., Haider, S., Lee, C., et al. (2013). Epidemiology and outcome of pneumonia caused by methicillin-resistant Staphylococcus aureus (MRSA) in Canadian hospitals. PLoS ONE 8:e75171. doi: 10.1371/journal.pone.0075171
Tenover, F. C., Arbeit, R. D., Goering, R. V., Mickelsen, P. A., Murray, B. E., Persing, D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33, 2233–2239.
Tokajian, S. T., Khalil, P. A., Jabbour, D., Rizk, M., Farah, M. J., Hashwa, F. A., et al. (2010). Molecular characterization of Staphylococcus aureus in Lebanon. Epidemiol. Infect. 138, 707–712. doi: 10.1017/S0950268810000440
Vandenesch, F., Naimi, T., Enright, M. C., Lina, G., Nimmo, G. R., Heffernan, H., et al. (2003). Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerging Infect. Dis. 9, 978–984. doi: 10.3201/eid0908.030089
Zhang, C., Guo, L., Chu, X., Shen, L., Guo, Y., Dong, H., et al. (2016). Presence of the panton-valentine leukocidin genes in methicillin-resistant Staphylococcus aureus is associated with severity and clinical outcome of hospital-acquired pneumonia in a single center study in China. PLoS ONE 11:e0156704. doi: 10.1371/journal.pone.0156704
Keywords: MRSA, spa typing, SCCmec typing, tst, CC5, in-hospital 30-day mortality, multivariable analysis
Citation: Wang M, Zheng Y, Mediavilla JR, Chen L, Kreiswirth BN, Song Y, Yang R and Du H (2017) Hospital Dissemination of tst-1-Positive Clonal Complex 5 (CC5) Methicillin-Resistant Staphylococcus aureus. Front. Cell. Infect. Microbiol. 7:101. doi: 10.3389/fcimb.2017.00101
Received: 14 January 2017; Accepted: 14 March 2017;
Published: 31 March 2017.
Edited by:
Fabio Bagnoli, GlaxoSmithKline, ItalyReviewed by:
Giuseppe Lofano, Ragon Institute of MGH, MIT and Harvard, USASarah Maddocks, Cardiff Metropolitan University, UK
Copyright © 2017 Wang, Zheng, Mediavilla, Chen, Kreiswirth, Song, Yang and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Du, aG9uZ19kdUAxMjYuY29t
 Min Wang
Min Wang Yi Zheng
Yi Zheng Jose. R. Mediavilla3
Jose. R. Mediavilla3 Liang Chen
Liang Chen Hong Du
Hong Du