- 1Anhui Medical University, Hefei, China
- 2State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology, Beijing, China
- 3Department of Clinical Laboratory, The 307th Hospital of the People's Liberation Army, Beijing, China
- 4Department of Clinical Laboratory, The First Affiliated Hospital of Anhui Medical University, Hefei, China
- 5College of Food Science and Project Engineering, Bohai University, Jinzhou, China
- 6Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing, China
This work presents the complete nucleotide sequences of p0801-IMP from Klebsiella pneumoniae, p7121-IMP from K. oxytoca, and p17285-IMP from Citrobacter freundii, which are recovered from three different cases of nosocomial infection. These three plasmids represent the first fully sequenced blaIMP-carrying IncN2 plasmids. Further comparative genomics analysis of all the five integron-carrying IncN2 plasmids p0801-IMP, p7121-IMP, p17285-IMP, pJIE137, and p34983-59.134kb indicates that they possess conserved IncN2 backbones with limited genetic variations with respect to gene content and organization. Four class 1 integrons (blaIMP-1-carrying In1223 in p0801-IMP/p7121-IMP, blaIMP-8-carrying In655 in p17285-IMP, In27 in pJIE137, and In1130 in p34983-59.134kb), two insertion sequence-based transposition units (ISEcp1-orfRA1-14 in p17285-IMP, and ISEcp1-blaCTX-M-62-Δorf477-orfRA1-14 in pJIE137), and a novel Tn1696-related transposon Tn6325 carrying In1130 in p34983-59.134kb are indentified in the plasmid accessory regions. In1223 and In655 represent ancestral Tn402-associated integrons, while In27 and In1130 belong to complex class 1 integrons. The relatively small IncN2 backbones are able to integrate different mobile elements which carry various resistance markers, promoting the accumulation and spread of antimicrobial resistance genes among enterobacterial species.
Introduction
The Ambler B metallo-β-lactamases IMPs are capable of hydrolyzing almost all β-lactams including carbapenems and, to date, 52 IMP-variant enzymes have been reported in at least 26 species of clinically important Gram-negative organisms such as Pseudomonas, Acinetobacter and Enterobacteriaceae species all over the world (Zhao and Hu, 2015). IMP producers often employ additional mechanisms (e.g., membrane permeability defects) and have gained significant attention due to their high-level resistance to carbapenems.
Class 1 integrons commonly carry a 5′-conserved segment [5′-CS], which is composed of the integrase gene intI1, a specific recombination site attI1 located next to intI1 and recognized by intI1, and a promoter Pc driving the transcription of cassette-borne genes and lying within intI1 (Partridge et al., 2009; Domingues et al., 2012; Gillings, 2014). The blaIMP genes are often found together with other resistance genes in the variable gene cassette arrays of class 1 integrons, and these integrons are further associated with mobile elements such as transposons and plasmids, leading to the easily mobilization of cassette-borne resistance genes across various bacterial species (Gillings et al., 2008).
Plasmids belonging to the IncN incompatibility group are the important mobile genetic platforms for dissemination of clinically important resistance genes among enterobacterial species (Poirel et al., 2011; Chen et al., 2012; Partridge et al., 2012; Netikul et al., 2014; Sun et al., 2015; Tijet et al., 2016). The IncN group can be further divided into three subgroups IncN1 to IncN3. These three subgroups have very similar backbone gene organization but with limited nucleotide sequence homology over the backbones. There is still no report of blaIMP-carrying IncN2 or IncN3 plasmid.
This work present the complete nucleotide sequences of three novel IncN2 plasmids, p0801-IMP from Klebsiella pneumoniae, p7121-IMP from K. oxytoca, and p17285-IMP from Citrobacter freundii. p0801-IMP and p17285-IMP harbor the class 1 integrons In1223 and In655 carrying the cassette arrays blaIMP-1-gcu162-aacA4- aadA6 and blaIMP-8-aacA4, respectively. Further comparative genomics assay of all the fully sequenced integron-carrying IncN2 plasmids indicates that different mobile elements including integrons, transposons and insertion sequence-based transposition units can be inserted through transposition at different sites of the relatively small IncN2 backbones. Data presented here would promote us to gain insights into genetic variation and evolutionary history of IncN2 plasmids.
Materials and Methods
Bacterial Isolation and Identification
The use of human specimens and all related experimental protocols were approved by the Committee on Human Research of the 307th Hospital of the People's Liberation Army and that of the First Affiliated Hospital of Anhui Medicial University, and carried out in accordance with the approved guidelines. The research involving biohazards and all related procedures were approved by the Biosafety Committee of the Beijing Institute of Microbiology and Epidemiology. Bacterial species was identified by 16S rRNA gene sequencing (Frank et al., 2008). The major plasmid-borne carbapenemase genes were screened for by PCR (Chen et al., 2015), followed by amplicon sequencing on ABI 3730 Sequencer.
Plasmid Conjugal Transfer
Plasmid conjugal transfer experiments were carried out with the rifampin-resistant Escherichia coli EC600 (LacZ−, NalR, RifR) being used as recipient and strain 0801 or 7121 or 17285 or as donor (Chen et al., 2015). 3 ml of overnight culture of each of donor and recipient bacteria were mixed together, harvested and resuspended in 80 μl of Brain Heart Infusion (BHI) broth (BD Biosciences). The mixture was spotted on a 1 cm2 filter membrane that was placed on BHI agar (BD Biosciences) plate, and then incubated for mating at 37°C for 12–18 h. Bacteria were washed from filter membrane and spotted on Muller-Hinton (MH) agar (BD Biosciences) plate containing 1,000 μg/ml rifampin and 2 μg/ml imipenem for selection of blaIMP-positive E. coli transconjugants.
Detection of Carbapenemase Activity
Activity of class A/B/D carbapenemases in bacterial cell extracts was determined via a modified CarbaNP test (Chen et al., 2015). Overnight bacterial cell culture in MH broth was diluted 1:100 into 3 ml of fresh MH broth, and bacteria were allowed to grow at 37°C with shaking at 200 rpm to reach an OD600 of 1.0 to 1.4. If required, ampicillin was used at 200 μg/ml. Bacterial cells were harvested from 2 ml of the above culture, and washed twice with 20 mM Tris-HCl (pH 7.8). Cell pellets were resuspended in 500 μl of 20 mM Tris-HCl (pH 7.8), and lysed by soniation, followed by centrifugation at 10,000 × g at 4°C for 5 min. 50 μl of the supernatant (the enzymatic bacterial suspension) were mixed with 50 μl of substrate I to V, respectively, followed by incubation at 37°C for a maximum of 2 h. Substrate I: 0.054% phenol red plus 0.1 mM ZnSO4 (pH7.8). Substrate II: 0.054% phenol red plus 0.1 mM ZnSO4 (pH7.8), and 0.6 mg/μl imipenem. Substrate III: 0.054% phenol red plus 0.1 mM ZnSO4 (pH7.8), 0.6 mg/μl mg imipenem, and 0.8 mg/μl tazobactam. Substrate IV: 0.054% phenol red plus 0.1 mM ZnSO4 (pH7.8), 0.6 mg/μl mg imipenem, and 3 mM EDTA (pH7.8). Substrate V: 0.054% phenol red plus 0.1 mM ZnSO4 (pH7.8), 0.6 mg/μl mg imipenem, 0.8 mg/μl tazobactam, and 3 mM EDTA (pH7.8).
Bacterial Antimicrobial Susceptibility Test
Bacterial antimicrobial susceptibility was tested by BioMérieux VITEK 2 and interpreted as per Clinical and Laboratory Standards Institute (CLSI) guidelines (ClSI, 2015).
Plasmid Sequencing and Sequence Assembly
Plasmid DNA was isolated from E. coli transconjugant using a Qiagen Large Construct Kit, and then sequenced with a paired-end library with an average insert size of 500 bp and a mate-pair library with average insert size of 5,000 bp, using an Illumina MiSeq sequencer (Illumina). Reads from each sample were trimmed to remove poor quality sequences, and then the contigs were assembled with Newbler 2.6 (Nederbragt, 2014).
Sequence Annotation and Genome Comparison
Open reading frames and pseudogenes were predicted using RAST 2.0 (Brettin et al., 2015) combined with BLASTP/BLASTN searches (Boratyn et al., 2013) against the UniProtKB/Swiss-Prot (Boutet et al., 2016) and RefSeq (O'leary et al., 2016) databases. Annotation of resistance genes, mobile elements, and other features was carried out using the online databases including CARD (Jia et al., 2016), ResFinder (Zankari et al., 2012), BacMet (Pal et al., 2014), ISfinder (Siguier et al., 2006), INTEGRALL (Moura et al., 2009), and the Tn Number Registry (Roberts et al., 2008). Multiple and pairwise sequence comparisons were performed using MUSCLE 3.8.31 (Edgar, 2004) and BLASTN, respectively. Gene organization diagrams were drawn in Inkscape 0.48.1.
Nucleotide Sequence Accession Numbers
The complete sequences of p0801-IMP, p7121-IMP, and p17285-IMP were submitted to GenBank under accession numbers KT345947, KX784502, and KX784503, respectively.
Results
Case Reports
K. pneumoniae 0801, K. oxytoca 7121, and C. freundii 17285 were isolated from three different inpatients designated Patient 1 to Patient 3, respectively, with nosocomial infections. Patient 1 was a 35-year-old woman admitted to Hospital 1 in May 2013 and diagnosed to have acute lymphoblastic leukemia, and she received chemotherapy for 1 week. Pulmonary infection, septicemia and recurrent fever occurred during chemotherapy, and she received empirical intravenous administration with moxifloxacin. K. pneumoniae 0801was isolated from the blood specimens on the next day after chemotherapy. The patient was discharged 3 days later upon request from her family members.
Patient 2 was a 43-year-old woman admitted to Hospital 1 in January 2014 and diagnosed to have acute myeloid leukemia, and she received hematopoietic stem cell transplantation. Pulmonary infection occurred in the convalescent period, and K. oxytoca 7121 was then isolated from the sputum specimens. The patient received intravenous administration with flucloxacillin empirically at first, which was switched into levofloxacin based on antimicrobial susceptibility test results. Her symptoms associated with pulmonary infections progressively improved. The patient was discharged at 2 weeks after transplantation.
Patient 3 was a 66-year-old woman admitted to Hospital 2 in July 2013 and diagnosed to have adult onset Still's disease, and she received antianaphylactic treatment. Urinary tract infection occurred at 1 week after hospitalization, and C. freundii 17285 was then isolated from the voided midstream urine specimens. The patient received intravenous administration with teicoplanin. Her symptoms associated with infection and adult onset Still's disease progressively improved. The patient was discharged at 2 weeks after hospitalization.
General Features of Resistant Strains
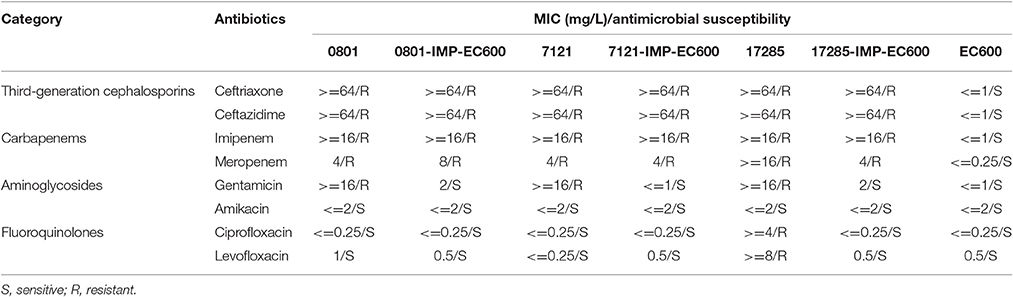
PCR screening assay indicated the presence of blaIMP but not any of the other carbapenemase genes tested in strains 0801, 7121, and 17285, and the blaIMP-carrying plasmids were designated p0801-IMP, p7121-IMP, and p17285-IMP, respectively. Each of these plasmids could be transferred into strain EC600 through conjugation, generating E. coli transconjugants 0801-IMP-EC600, 7121-IMP-EC600, and 17285-IMP-EC600, respectively, indicating that all these three plasmids are conjugative. All the above wild-type and transconjugant strains have the class B carbapenemase activity (data not shown), and are resistant to ceftriaxone, ceftazidime, imipenem and meropenem (Table 1). Strains 0801, 7121, and 17285 are resistant to gentamicin but the other strains remain susceptible to this drug, and all of them are susceptible to amikacin.
Overview of Sequenced Plasmids
Genome sequencing shows that p0801-IMP, p7121-IMP, and p17285-IMP have 42,580-bp, 42,461-bp and 43797-bp circularly closed DNA sequences, respectively, all of which carry 54 predicted open reading frames in total (Figure S1). These three plasmids belong to the IncN2 group because each of them contains an IncN2-type repA (plasmid replication initiation) gene. Further comparative genomics analysis is applied to all the five integron-carrying IncN2 plasmids p0801-IMP, p7121-IMP and p17285-IMP (this study), pJIE137 (Partridge et al., 2012), and p34983-59.134kb (accession number CP010378), together with the IncN2 reference plasmid pYNKP001-NDM (Sun et al., 2015), and the modular structure of each plasmid is divided into the IncN2 backbone as well as one or more accessory modules inserted at different sites of the backbone (Figure S1, Figure 1). Although p271A is the first fully sequenced IncN2 plasmid, pYNKP001-NDM is more appropriate as the IncN2 reference, because a 5.2-kb backbone region within the CUP-controlled regulon is absent from p271A relative to pYNKP001-NDM (Poirel et al., 2011; Sun et al., 2015).
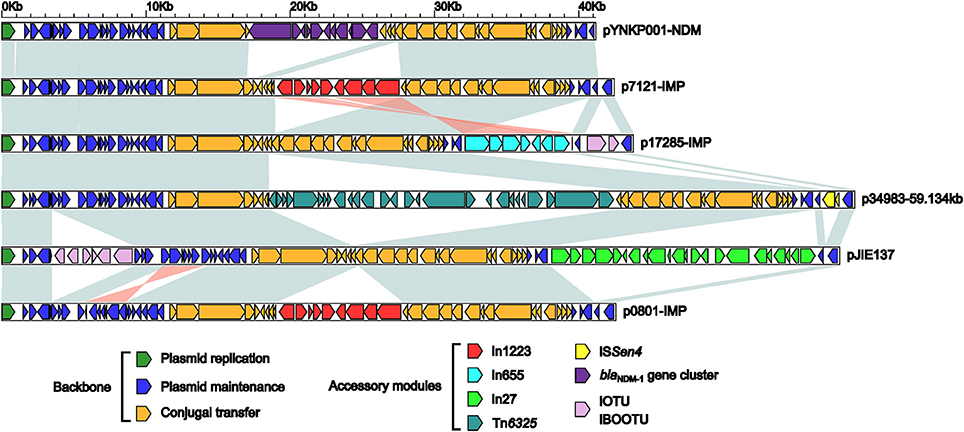
Figure 1. Linear comparison of sequenced plasmids. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on function classification. Shading denotes regions of homology (>95% nucleotide identity).
Backbones of Integron-Carrying IncN2 Plasmids
The six plasmids involved in genomic comparison possess conserved IncN2 backbones, each of which can be further divided into the regions responsible for plasmid replication (repA and its iterons), maintenance [the CUP (conserved upstream repeat)-controlled regulon, the stbABC-orfD operon, and resD), and conjugal transfer (tra and kikA-korB) (Figure S1, Figure 1). There are two major differences among the backbones of these six plasmids: (i) a region between repA and its iterons from pYNKP001-NDM differs from all the other counterparts, and (ii) insertion, deletion, and rearrangement occur within the CUP-controlled regulons.
Gene organization and function of the IncN1 CUP-controlled regulon (Delver and Belogurov, 1997) have been described in the IncN1 reference plasmid R46 (accession number AY046276). Similarly, four putative operons (i.e., the ardK operon, the CUP-4 operon, the CUP-3 operon, the CUP-2 operon and the CUP-1 operon) arranged in the same orientation are annotated within the IncN2 CUP-controlled regulon, and CUP-4, CUP-3, CUP-2 and CUP-1 are located at the 5′-ends of the last four operons, respectively (Figure 2). Each of these operons contains a putative ArdK-binding site and a promoter, accounting for ArdK-dependent expression of operon-borne genes. A 40-bp deletion is found within CUP-3 of p7121-IMP. An ISEcp1-blaCTX-M-62-Δorf477-orfRA1-14 transposition unit is inserted between CUP-4 and ardB in pJIE137 (Partridge et al., 2012), which would impair the gene expression of the CUP-4 operon. In p0801-IMP, homologous recombination medicated by CUP-3 and CUP-1 likely leads to an inversion of the orf792 to ros region as well as the disruption of CUP-3 and CUP-1.
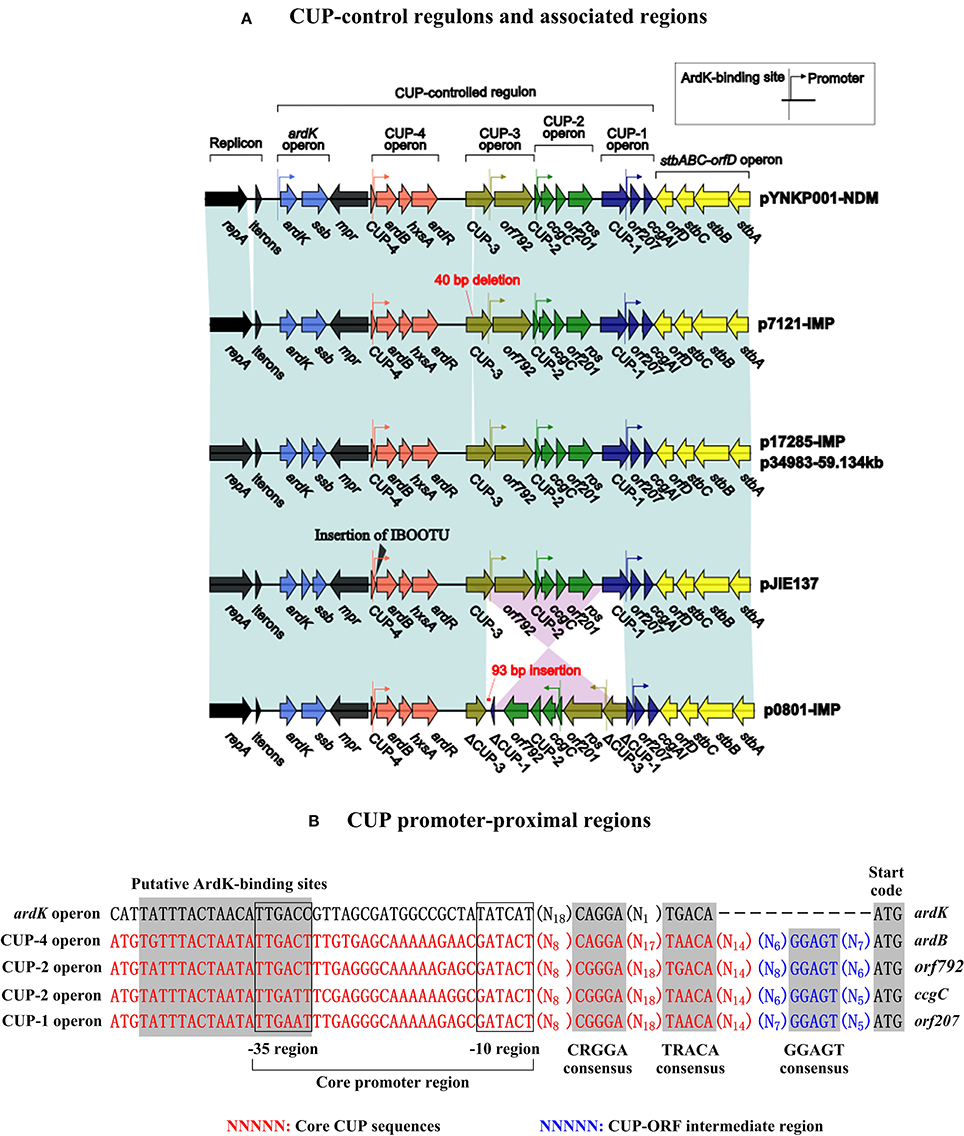
Figure 2. CUP-related sequences. (A) CUP-control regulons and associated regions. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on function classification. Shading denotes regions of homology (>95% nucleotide identity). (B) CUP promoter-proximal regions. Shown are putative ArdK-binding sites, core promoter −10 and −35 regions, and 3 different consensus sequences within CUP promoter-proximal regions.
Accessory Regions of Integron-Carrying IncN2 Plasmids
p0801-IMP and p7121-IMP carry a novel class 1 integron In1223, containing 5′-CS, the cassette array blaIMP-1-gcu162-aacA4-aadA6, and the complete Tn402 tni module (tniABQ-res-tniR), which is bordered by IRi (inverted repeat at the integrase end) and IRt (inverted repeat at the tni end) (Figure 3A). blaIMP-1 and aacA4/aadA6 account for resistance to carbapenems and aminoglycosides, respectively, while gcu162 is a novel gene cassette of unknown function.
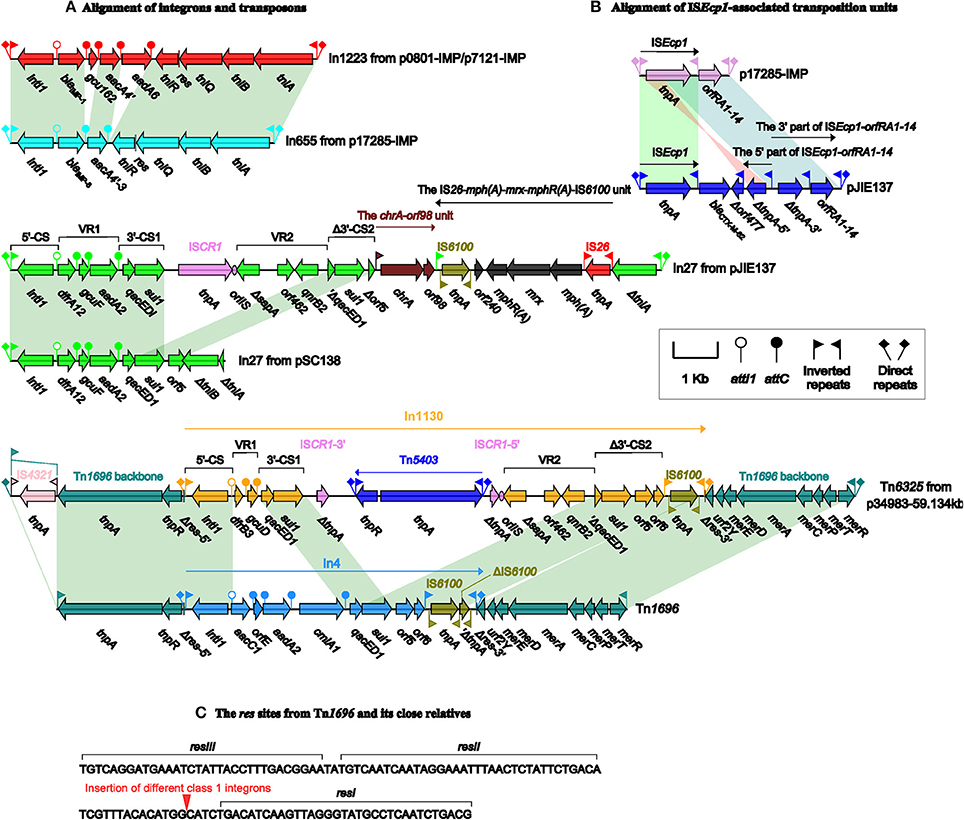
Figure 3. Plasmid accessory modules. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on function classification. Shading denotes regions of homology (>95% nucleotide identity). Shown are the alignment of intergrons and transpsons (A) and ISEcp1-associated transposition units (B), and also the organization of the res sites from Tn1696 and its derivatives (C).
There are two accessory modules in each of pJIE137 (Partridge et al., 2012) and p17285-IMP. As shown in Figures 3A,B, p17285-IMP contains In655 (inserted into resD) and a 2554-bp ISEcp1-orfRA1-14 transposition unit (inserted between orf333 and orf648), while pJIE137 harbors ISEcp1-blaCTX-M-62-Δorf477-orfRA1-14 (inserted within the CUP-4 operon) and In27 (inserted between resD and orf333). In655 differs from In1223 by presence of a distinct cassette array blaIMP-8-aacA4 and, notably, its Tn402-family tni module is maximally only 95% identical to the others at nucleotide level. ISEcp1 captures and arranges orfRA1-14 and blaCTX-M-62-Δorf477 at its immediately downstream, which generates the transposition units ISEcp1-orfRA1-14 and ISEcp1-blaCTX-M-62-Δorf477, respectively, bordered by terminal inverted repeats IRLISEcp1 and IRRISEcp1. Similar ISEcp1-blaCTX-M-Δorf477 structures (containing different variants of the blaCTX-M-1 group) are found on plasmids from various bacterial hosts, while the ISEcp1-orfRA1-14 elements are found on plasmids from only enterobacterial species. ISEcp1-blaCTX-M-62-Δorf477-orfRA1-14 is a hybrid of ISEcp1-blaCTX-M-62-Δorf477 and an ISEcp1-orfRA1-14-related element that originates from splitting of ISEcp1-orfRA1-14 into the 5′- and 3′-parts, followed by inversion of the 5′-part (Zong et al., 2010).
Tn6325 (Figure 3A) from p34983-59.134kb is a novel derivative of Tn1696 belonging to the Tn21 subgroup of the Tn3 transposon family. Tn1696, located in the IncP1 plasmid R1033 from clinical P. aeruginosa, is generated from insertion of In4 within the resolution (res) site of a transposon backbone structure IRL-tnpA (transposase)-tnpR (resolvase)-res-mer (mercury resistance locus)-IRR, interrupting res into two separate parts (Partridge et al., 2001). Close Tn1696 relatives, which contain different In4-type integrons inserted at exactly the same position as In4, have been found on plasmids such as pHCM1, pSRC125 and pSRC26 (Cain et al., 2010). Tn6325 differs from Tn1696 by (i) insertion a distinct integron In1130 at the same position as In4 within res (Figure 3C), and (ii) disruption of IRLTn6325 into two parts by IS4321 (Figure 3A) that is a hunter of terminal inverted repeats of Tn21 subgroup transposons (Partridge and Hall, 2003).
The modular structure of a typical complex class 1 integron is organized sequentially as IRi, 5′-CS, variable region 1 (VR1), the first copy of 3′-CS [3′-CS1: qacED1 (quaternary ammonium compound resistance)-sul1 (sulfonamide resistance)], the common region ISCR1, VR2, the second copy of 3′-CS (3′-CS2; qacED1-sul1-orf5-orf6), tni, and IRt (Partridge et al., 2009). In27 from pJIE137 and In1130 from p34983-59.134kb (Figure 3A) belong to complex class 1 integrons because they contain all of the above core components with modifications of 3′-CS2. For both In27 and In1130, the connection of VR2 [ΔsapA-orf462-qnrB2 (quinolone resistance)] with 3′-CS2 leads to the truncation of qacED1 at the 5′-terminus of 3′-CS2. In addition, the tni module within the 3′-CS2 has been replaced by an IS6100 element in In1130, while the 3′-CS2 of In27 is interrupted into two separate parts ΔqacED1-sul1-Δorf5 and ΔtniA due to the insertion of a 6.8-kb region [composed of the chromate-resistance unit chrA-orf98 and the macrolide-resistance unit IS26-mph(A)-mrx-mphR(A)-IS6100], which is highly similar to the 3′-region of In37 from p112298-KPC (Feng et al., 2015). In In1130, ISCR1 is interrupted by the insertion of a cryptic Tn3-family unit transposon Tn5403.
The insertion of each of In1223, In655, ISEcp1-orfRA1-14, In27, ISEcp1-blaCTX-M-62- Δorf477-orfRA1-14, In1130 and Tn6325 into the relevant plasmids leaves target site duplication signals of transposition, manifesting as various types of 5-bp direct repeat at the sites of insertion.
Discussion
A collection of fully sequenced plasmids including p0801-IMP, p7121-IMP, p17285-IMP, p271A, pYNKP001-NDM, pNDM-ECS01, pTR3, p34983-59.134kb, pJIE137, pKPC-SMH, and p34998-53.129kb (Poirel et al., 2011; Chen et al., 2012; Partridge et al., 2012; Netikul et al., 2014; Sun et al., 2015; Tijet et al., 2016) carry the IncN2 replicon and very similar backbones, which dramatically differ from IncN1 and IncN3, and thereby they are assigned into the IncN2 subgroup (Figure 4).
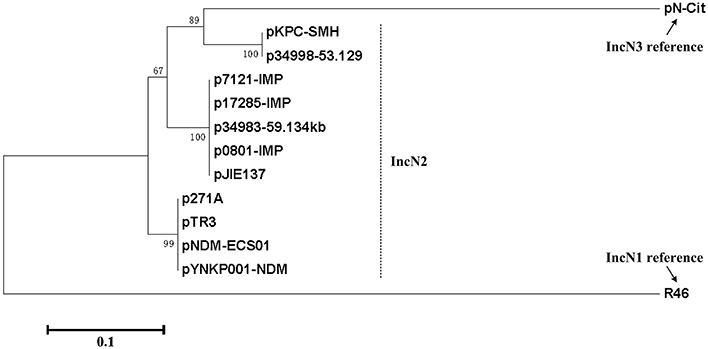
Figure 4. Phylogenetic tree of repA sequences. The nucleotide sequences of the repA coding regions from all the fully sequenced IncN2 plasmids together with R46 and pN-Cit (Villa et al., 2013) as the IncN1 and IncN3 reference, respectively, are aligned with MUSCLE 3.5 (Edgar, 2004). An unrooted neighbor-joining tree is inferred from the aligned sequences by using MEGA7 (Kumar et al., 2016) with calculation of evolutionary distances by the Maximum Composite Likelihood method. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches.
Each of the four class 1 integrons including In1223 from p0801-IMP/p7121-IMP, In655 from p17285-IMP, In27 from pJIE137, and In1130 from p34983-59.134kb has a complete set of IRi/IRt, intI1, and attI1. In1223/In27, In655, and In1130 have the promoters PcWTGN-10 (Strong) (Nesvera et al., 1998), PcS (Strong) (Collis and Hall, 1995), and PcW (weak) combined with P2 (strong) (Wei et al., 2011), respectively, which would drive the high-level expression of cassette-borne genes.
Tn402 acts as a primary carrier of class 1 integrons, and the evolution of Tn402-associated class 1 integrons involves at least three stages as summarized previously: stage I, insertion of ancestral integron sequence (containing captured gene cassettes but lacking 3'-CS) into Tn402 (harboring the tni module) to generate a hybrid structure, thereby combining the ability of integron to capture gene cassettes with the mobility of Tn402, which occurs prior to or concomitant with capture of qacE; stage II, capture of sul1-orf5-orf6 and then formation of 3′-CS (qacED1-sul1-orf5-orf6-tni) after deletions between qacE and sul1; and stage III, deletions within the tni region, impairing the tni-mediated mobility (Chen et al., 2014). In1223 and In655 represent ancestral Tn402-associated integrons at stage I, while In27 from pSC138 (Chiu et al., 2005) and In4 from Tn1696 are at stage III (Figure 3A). In27 from pJIE137 and In1130 from p34983-59.134kb have evolved into complex class 1 integron with integration of one or more additional regions containing several resistance markers, which might involve complex homologous recombination events involving IS6100 and IS26(Feng et al., 2015).
The 7 mobile elements including In1223 from p0801-IMP/p7121-IMP, In655 and ISEcp1-orfRA1-14 from p17285-IMP, In27 and ISEcp1-blaCTX-M-62-Δorf477-orfRA1-14 from pJIE137, and In1130 and Tn6325 from p34983-59.134 kb are inserted at different sites and their mobilization into relevant plasmids leaves targeting signals of transposition, indicating that they are simple insertions due to transposition without adjacent deletions and rearrangements. The relatively small IncN2 backbones are able to integrate different mobile elements such as integrons, transposons and insertion sequence-based transposition units, which carry different resistance markers, thereby promoting accumulation and spread of antimicrobial resistance among bacterial species. Comparative genomics analysis of a larger collection of fully sequenced IncN1, IncN2, and IncN3 plasmids would promote us to gain deeper understanding of the horizontal transfer of antimicrobial resistance genes through mobile genetic elements as well as the molecular evolution mechanisms of diversification of IncN plasmid scaffolds. The combination of additional molecular epidemiological investigation will gain the highlights into not only the ability of plasmids to transmit among bacterial species and genera but also the underlying mechanisms of antibiotic resistance spread associated with hospitalized patients.
Author Contributions
DSZ, SC, and JW conceived the study and designed experimental procedures. XJ, ZY, XY, and HF, performed the experiments. XJ, DSZ, QS, and DFZ analyzed the data. XJ, YT, YX, JF, WC, and YS contributed reagents and materials. DSZ, SC, XJ, and JW wrote this manuscript.
Consent Statement
Written informed consent was obtained from all participants.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Basic Research Program of China (2015CB554202), the National Natural Science Foundation of China (81302608 and 81501716), and the Foundation of State Key Laboratory of Pathogen and Biosecurity of China (SKLPBS1402).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fcimb.2017.00102/full#supplementary-material
Figure S1. Schematic maps of sequenced plasmids. Genes are denoted by arrows, and the backbone and accessory module regions are highlighted in black and color, respectively. The innermost circle presents GC-skew [(G−C)/(G+C)], with a window size of 500 bp and a step size of 20 bp. The next-to-innermost circle presents GC content.
References
Boratyn, G. M., Camacho, C., Cooper, P. S., Coulouris, G., Fong, A., Ma, N., et al. (2013). BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41, W29–W33. doi: 10.1093/nar/gkt282
Boutet, E., Lieberherr, D., Tognolli, M., Schneider, M., Bansal, P., Bridge, A. J., et al. (2016). UniProtKB/Swiss-Prot, the manually annotated section of the UniProt KnowledgeBase: how to use the entry view. Methods Mol. Biol. 1374, 23–54. doi: 10.1007/978-1-4939-3167-5_2
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Cain, A. K., Liu, X., Djordjevic, S. P., and Hall, R. M. (2010). Transposons related to Tn1696 in IncHI2 plasmids in multiply antibiotic resistant Salmonella enterica serovar Typhimurium from Australian animals. Microb. Drug Resist. 16, 197–202. doi: 10.1089/mdr.2010.0042
Chen, Y. T., Lin, A. C., Siu, L. K., and Koh, T. H. (2012). Sequence of closely related plasmids encoding blaNDM−1 in two unrelated Klebsiella pneumoniae isolates in Singapore. PLoS ONE 7:e48737. doi: 10.1371/journal.pone.0048737
Chen, Z., Fang, H., Wang, L., Sun, F., Wang, Y., Yin, Z., et al. (2014). IMP-1 encoded by a novel Tn402-like class 1 integron in clinical Achromobacter xylosoxidans, China. Sci. Rep. 4:7212. doi: 10.1038/srep07212
Chen, Z., Li, H., Feng, J., Li, Y., Chen, X., Guo, X., et al. (2015). NDM-1 encoded by a pNDM-BJ01-like plasmid p3SP-NDM in clinical Enterobacter aerogenes. Front. Microbiol. 6:294. doi: 10.3389/fmicb.2015.00294
Chiu, C. H., Tang, P., Chu, C., Hu, S., Bao, Q., Yu, J., et al. (2005). The genome sequence of Salmonella enterica serovar Choleraesuis, a highly invasive and resistant zoonotic pathogen. Nucleic Acids Res. 33, 1690–1698. doi: 10.1093/nar/gki297
CLSI. (2015). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fifth Informational Supplement M100–S25. Wayne, PA: CLSI.
Collis, C. M., and Hall, R. M. (1995). Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemother. 39, 155–162. doi: 10.1128/AAC.39.1.155
Delver, E. P., and Belogurov, A. A. (1997). Organization of the leading region of IncN plasmid pKM101 (R46): a regulation controlled by CUP sequence elements. J. Mol. Biol. 271, 13–30. doi: 10.1006/jmbi.1997.1124
Domingues, S., Da Silva, G. J., and Nielsen, K. M. (2012). Integrons: vehicles and pathways for horizontal dissemination in bacteria. Mob. Genet. Elem. 2, 211–223. doi: 10.4161/mge.22967
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Feng, J., Qiu, Y., Yin, Z., Chen, W., Yang, H., Yang, W., et al. (2015). Coexistence of a novel KPC-2-encoding MDR plasmid and an NDM-1-encoding pNDM-HN380-like plasmid in a clinical isolate of Citrobacter freundii. J. Antimicrob. Chemother. 70, 2987–2991. doi: 10.1093/jac/dkv232
Frank, J. A., Reich, C. I., Sharma, S., Weisbaum, J. S., Wilson, B. A., and Olsen, G. J. (2008). Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 74, 2461–2470. doi: 10.1128/AEM.02272-07
Gillings, M., Boucher, Y., Labbate, M., Holmes, A., Krishnan, S., Holley, M., et al. (2008). The evolution of class 1 integrons and the rise of antibiotic resistance. J. Bacteriol. 190, 5095–5100. doi: 10.1128/JB.00152-08
Gillings, M. R. (2014). Integrons: past, present, and future. Microbiol. Mol. Biol. Rev. 78, 257–277. doi: 10.1128/MMBR.00056-13
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2016). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Moura, A., Soares, M., Pereira, C., Leitao, N., Henriques, I., and Correia, A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–1098. doi: 10.1093/bioinformatics/btp105
Nederbragt, A. J. (2014). On the middle ground between open source and commercial software - the case of the Newbler program. Genome Biol. 15:113. doi: 10.1186/gb4173
Nesvera, J., Hochmannova, J., and Patek, M. (1998). An integron of class 1 is present on the plasmid pCG4 from gram-positive bacterium Corynebacterium glutamicum. FEMS Microbiol. Lett. 169, 391–395. doi: 10.1111/j.1574-6968.1998.tb13345.x
Netikul, T., Sidjabat, H. E., Paterson, D. L., Kamolvit, W., Tantisiriwat, W., Steen, J. A., et al. (2014). Characterization of an IncN2-type blaNDM-1-carrying plasmid in Escherichia coli ST131 and Klebsiella pneumoniae ST11 and ST15 isolates in Thailand. J. Antimicrob. Chemother. 69, 3161–3163. doi: 10.1093/jac/dku275
O'leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., Mcveigh, R., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745. doi: 10.1093/nar/gkv1189
Pal, C., Bengtsson-Palme, J., Rensing, C., Kristiansson, E., and Larsson, D. G. (2014). BacMet: antibacterial biocide and metal resistance genes database. Nucleic Acids Res. 42, D737–D743. doi: 10.1093/nar/gkt1252
Partridge, S. R., Brown, H. J., Stokes, H. W., and Hall, R. M. (2001). Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45, 1263–1270. doi: 10.1128/AAC.45.4.1263-1270.2001
Partridge, S. R., and Hall, R. M. (2003). The IS1111 family members IS4321 and IS5075 have subterminal inverted repeats and target the terminal inverted repeats of Tn21 family transposons. J. Bacteriol. 185, 6371–6384. doi: 10.1128/JB.185.21.6371-6384.2003
Partridge, S. R., Paulsen, I. T., and Iredell, J. R. (2012). pJIE137 carrying blaCTX−M−62 is closely related to p271A carrying blaNDM−1. Antimicrob. Agents Chemother. 56, 2166–2168. doi: 10.1128/AAC.05796-11
Partridge, S. R., Tsafnat, G., Coiera, E., and Iredell, J. R. (2009). Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol. Rev. 33, 757–784. doi: 10.1111/j.1574-6976.2009.00175.x
Poirel, L., Bonnin, R. A., and Nordmann, P. (2011). Analysis of the resistome of a multidrug-resistant NDM-1-producing Escherichia coli strain by high-throughput genome sequencing. Antimicrob. Agents Chemother. 55, 4224–4229. doi: 10.1128/AAC.00165-11
Roberts, A. P., Chandler, M., Courvalin, P., Guedon, G., Mullany, P., Pembroke, T., et al. (2008). Revised nomenclature for transposable genetic elements. Plasmid 60, 167–173. doi: 10.1016/j.plasmid.2008.08.001
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Sun, F., Yin, Z., Feng, J., Qiu, Y., Zhang, D., Luo, W., et al. (2015). Production of plasmid-encoding NDM-1 in clinical Raoultella ornithinolytica and Leclercia adecarboxylata from China. Front. Microbiol. 6:458. doi: 10.3389/fmicb.2015.00458
Tijet, N., Muller, M. P., Matukas, L. M., Khan, A., Patel, S. N., and Melano, R. G. (2016). Lateral dissemination and inter-patient transmission of blaKPC−3: role of a conjugative plasmid in spreading carbapenem resistance. J. Antimicrob. Chemother. 71, 344–347. doi: 10.1093/jac/dkv356
Villa, L., Carattoli, A., Nordmann, P., Carta, C., and Poirel, L. (2013). Complete sequence of the IncT-type plasmid pT-OXA-181 carrying the blaOXA−181 carbapenemase gene from Citrobacter freundii. Antimicrob. Agents Chemother. 57, 1965–1967. doi: 10.1128/aac.01297-12
Wei, Q., Jiang, X., Li, M., Chen, X., Li, G., Li, R., et al. (2011). Transcription of integron-harboured gene cassette impacts integration efficiency in class 1 integron. Mol. Microbiol. 80, 1326–1336. doi: 10.1111/j.1365-2958.2011.07648.x
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Zhao, W. H., and Hu, Z. Q. (2015). Acquired metallo-beta-lactamases and their genetic association with class 1 integrons and ISCR elements in Gram-negative bacteria. Future Microbiol. 10, 873–887. doi: 10.2217/fmb.15.18
Keywords: IncN2 plasmids, class 1 integron, transposon, blaIMP, antimicrobial resistance
Citation: Jiang X, Yin Z, Yin X, Fang H, Sun Q, Tong Y, Xu Y, Zhang D, Feng J, Chen W, Song Y, Wang J, Chen S and Zhou D (2017) Sequencing of blaIMP-Carrying IncN2 Plasmids, and Comparative Genomics of IncN2 Plasmids Harboring Class 1 Integrons. Front. Cell. Infect. Microbiol. 7:102. doi: 10.3389/fcimb.2017.00102
Received: 16 December 2016; Accepted: 15 March 2017;
Published: 30 March 2017.
Edited by:
Brian Ahmer, Ohio State University at Columbus, USAReviewed by:
Baolin Sun, University of Science and Technology of China, ChinaMax Maurin, Université Grenoble Alpes, France
Copyright © 2017 Jiang, Yin, Yin, Fang, Sun, Tong, Xu, Zhang, Feng, Chen, Song, Wang, Chen and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinglin Wang, d2psd2psMDgwMUBzaW5hLmNvbQ==
Shuiping Chen, c2hwY2hlbkBob3RtYWlsLmNvbQ==
Dongsheng Zhou, ZG9uZ3NoZW5nemhvdTE5NzdAZ21haWwuY29t
†These authors have contributed equally to this work.
 Xiaoyuan Jiang1†
Xiaoyuan Jiang1† Zhe Yin
Zhe Yin Haihong Fang
Haihong Fang Yigang Tong
Yigang Tong Yajun Song
Yajun Song Shuiping Chen
Shuiping Chen Dongsheng Zhou
Dongsheng Zhou