- 1Medical College, Yanbian University, Yanji, China
- 2Institute of Military Veterinary Medicine, Academy of Military Medical Science, Changchun, China
- 3Changchun University of Traditional Chinese Medicine, Changchun, China
- 4Institute of Virology, Wenzhou University Town, Wenzhou, China
- 5Department of Head and Neck Surgery, Tumor Hospital of Jilin Province, Changchun, China
An attenuated vaccinia virus-MVTTEAB-was constructed by deletion of non-essential gene segments related to the immunomodulatory and virulence functions of the vaccinia virus Tiantan strain (VVTT). The shuttle plasmids pTC-EGFP, pTE-EGFP, pTA35-EGFP, pTB-EGFP, and pTA66-EGFP were constructed and combined with the early and late strong promoter pE/L and EGFP as an exogenous selectable marker. Then, through the homologous recombination technology and Cre/loxP system, the following gene fragments were gradually knocked out one by one: TC7L-TK2L, TE3L, TA35R, TB13R, and TA66R. Ultimately, the five-segment-deleted attenuated strain MVTTEAB was obtained. Knockout of these segments and genetic stability of MVTTEAB were confirmed, and it was also shown that knockout of these segments did not affect the replication ability of the virus. Further, a series of in vivo and in vitro experiments demonstrated that the virulence of MVTTEAB was attenuated significantly, but at same time, high immunogenicity was maintained. These results indicate that MVTTEAB has potential for clinical use as a safe viral vector or vaccine with good attenuation and immunogenicity.
Introduction
The vaccinia virus (VV), which is used as a smallpox vaccine, belongs to the family Poxviridae and genus Orthopoxvirus. It is a complex double-stranded DNA virus that replicates and forms peculiar viral particles in host cells. The VV genome size is 185–200 kb, and it can encode about 200 different proteins (Qin et al., 2015). The highly conserved central part of the genome, which comprises the majority of the VV genome, contains the essential genes that play a key role in virus replication, such as transcription, DNA replication, and viral particle assembly. In contrast, the genes present at both ends of the genome are commonly used to identify species or host specificities and to encode proteins that regulate the host immune system and virulence factors (Liu and McFadden, 2015). Genome analysis of VV has led to new breakthroughs in the phylogeny and evolution of VV, and has shown that the VV proteins are more analogous to eukaryotic proteins than bacterial proteins. Research findings have indicated that the genes in this virus are likely to have come from their eukaryotic host genes via horizontal gene transfer, and that these slow and sustained processes have contributed to the evolution of VV. Many laboratories have initiated research on the use of naturally evolved strains for vaccination, especially research on improving the safety of VV and other poxviruses. In particular, many studies have shown that VV is useful for the study of vaccine vectors and exogenous gene expression systems (Garcia-Arriaza et al., 2013; Noisumdaeng et al., 2013; Adelfinger et al., 2014; Remy-Ziller et al., 2014).
VV is widely used in the field of gene engineering vaccine vectors and exogenous gene expression systems (Garcia-Arriaza et al., 2013; Adelfinger et al., 2014). Furthermore, live genetically engineered vector vaccines using VV as the vectors have been developed, and to be directed against more than 30 species of virus including herpes simplex virus, hepatitis A virus, hepatitis B virus, human immunodeficiency virus, and others, such as the genetically engineered vaccine of rabies virus glycoprotein (G) expressed using VV Copenhagen strain as the vector (Pastoret and Brochier, 1996), that of Newcastle Disease Virus fusion protein (Fusion protein, F) expressed using VV Elstree strain as the vector (Meulemans et al., 1988), that of vesicular stomatitis virus G protein expressed using VV vaccine Western Reserve strain as the vector (Mackett et al., 1985)and etc. Although research on the use of VV as vectors has made remarkable progress, there are still some limitations, such as difficulty in the screening of recombinant viruses and the insertion of exogenous selection markers, and the complexity of vector construction procedures (MacNeil et al., 2009). Currently, reducing the side effects of VV vaccines, improving the efficiency of vector vaccines, and simplifying the preparation procedures are hot topics in research on VV vectors.
In this study, the Cre-loxP recombination system was used to delete the following non-essential gene segments of the VVTT strain one by one to ultimately to obtain the attenuated strain MVTTEAB: TC7L-TK2L (15,262–25,450: TC, TC, TC, TC, TC, TC, TC, TN, TN, TM, TM, TK1L, and TK), TE3L (47,348–47,921), TA35R (138,881–139,570), TB13R (173,213–174,206), and TA66R (161,870–162,817). The knockout fragments included a variety of virulence-related genes, host-related genes and immunomodulatory genes. The TC7L-TK2L fragment is involved in the regulation of pathogenicity, virulence, and host range of the gene. NYVAC, as one of the most successful gene-knockout attenuated VV vectors, was obtained by phenotypic attenuation after knockout of the C-K1L fragment (12 ORFs) (Tartaglia et al., 1992). E3L is a virulence and immunomodulatory gene that encodes a protein which inhibits the activation of interferon-induced pathways, thereby inhibiting the antiviral response of host cells (Guerra et al., 2011). Several previous reports have shown that knockout of E3L in the Copenhagen, WR and NYCBH strains of VV can result in highly attenuated virus (Vijaysri et al., 2008). A35R is a virulence gene that modulates the adaptive immune response. A study has shown that knockout of A35R can lead to a decrease in viral replication capacity and reduce viral virulence (Brennan et al., 2015). B13R is a non-essential immunomodulatory gene with anti-apoptotic and anti-inflammatory effects and has sequence homology with serpins (Legrand et al., 2005). The TA66R gene encodes a viral hemagglutinin that has similar function to A56R of the VACV WR strain, which is inhibition of cell fusion. The total size of the deleted sequences was about 20 kb, which accounted for 10.6% of the sequence of the VVTT genome. Enhanced green fluorescent protein (EGFP) was used as the exogenous screening marker, and the Cre/loxP system was introduced into the shuttle vector plasmid for knockout of exogenous selection markers. In subsequent in vitro and in vivo experiments, MVTTEAB was found to have good attenuation and good immunogenicity as a vaccine. Thus, the recombinant VVTT strain MVTTEAB with the five gene segment deletion that was constructed in this study may have a wide range application prospects as a live vector vaccine and exogenous gene expression vector. Further, the number of cycles of recombination for constructing recombinant VVs could be significantly reduced and the efficiency of screening could be significantly improved by using the construction and screening strategies used in the present study. Thus, these construction and screening methods for the recombinant virus could present optimized solutions for studying new recombinant VV vector vaccines.
Materials and Methods
Cells, Viruses, and Animals
BHK-21, HeLa, PK-15, MDCK, and Vero cells were purchased from the China Center for Type Culture Collection. All cells were cultured in Dulbecco's modified Eagle medium (DMEM) (Invitrogen, Beijing, China) supplemented with 10% fetal bovine serum (FBS) (Hyclone, Beijing, China), 1% streptomycin (10 mg/mL), and 1% penicillin (10,000 U/mL). The VVTT strain (GenBank accession no. AF095689) was obtained from the Institute of Virology at the Chinese Center for Disease Control and Prevention.
Female New Zealand white rabbits and female BALB/c mice (aged 3–8 weeks) were obtained from the Experimental Animal Center of the Academy of Military Medical Sciences of China.
The animal experimental protocols were approved by the Institutional Animal Care and Use Committee of the Chinese Academy of Military Medical Science, Changchun, China (10ZDGG007). All surgical procedures were performed under sodium pentobarbital-induced anesthesia, and all efforts were made to minimize suffering.
Construction of VVTT Transfer Vectors
We constructed the pSK-TC-EGFP, pSK-TE-EGFP, pSK-TA35-EGFP, pSK-TB-EGFP, and pSK-TA66-EGFP vectors with standard gene synthesis techniques (Kan et al., 2012). The pSK-TC-EGFP vector has a DNA fragment containing TCL-loxP-PE/L-EGFP-loxP-TCR sites, and both ends of the EGFP fragment had EcoRI and PstI restriction enzyme sites. The other four vectors were constructed using the same strategy. These five vectors were constructed by Shanghai Generay Biotech Co. Ltd. The five transfer vectors were identified by digestion with EcoRI and PstI.
Construction of MVTTEAB
The TC, TE, TA35, TB, and TA66 genes were replaced with the EGFP gene to generate the recombinant virus MVTTEAB (Figure 1D). The BHK-21 cells were prepared in six-well plates into which VVTT was added at an MOI of 0.1. The plates were cultured in DMEM containing FBS, streptomycin and penicillin (as mentioned previously) for 2 h before transfection with the mixture of the shuttle plasmid pSK-TC-EGFP and Lipofectamine 2000 (Invitrogen). After culture for 72 h, the recombinant virus was released by three freeze-thaw cycles and used for further infection. Under an inverted fluorescence microscope, green fluorescent plaques were picked out, and this purification process was repeated six times until the monoclonal fluorescent plaque (i.e., the recombinant virus rVVTT-C−EGFP+) was purified. rVVTT-C−EGFP+ and the plasmid pVAX1-Cre were co-infected/transfected in BHK-21 cells and cultured for 72 h. Then, the non-green fluorescent plaques (i.e., the non-EGFP-expressing virus) were picked out and purified; this step was repeated six times. Deletion of the TC and EGFP genes in the recombinant virus rVVTT-C− was confirmed by PCR. This process was used to successively delete the other four segments—TE, TA35, TB, and TA66—from the recombinant virus rVVTT-C−. Ultimately, an attenuated strain of VVTT with five deleted gene segments was obtained MVTTEAB.
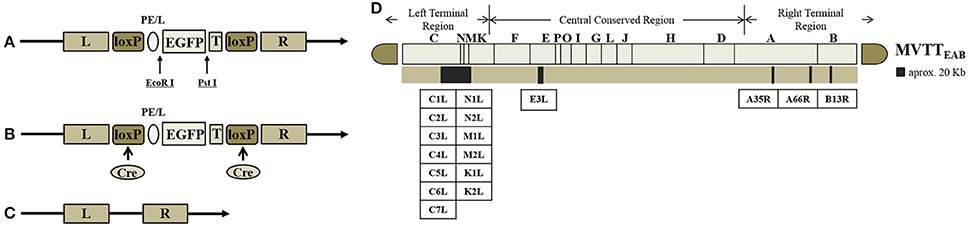
Figure 1. Construction of MVTTEAB–an attenuated vaccinia virus with five deleted gene segments. Structure of the plasmid vector (A), recombinant vaccinia virus (B), and gene-deleted virus (C). EGFP as a screening marker was inserted at the TC7L-TK2L gene site of VVTT (A) and driven by the synthetic early/late promoter (pE/L) to obtain the recombinant virus with deletion of the TC-TK2L gene but containing the EGFP gene (B). The recombinant virus VVTTC− was generated through Cre/loxP site-specific recombination to remove the EGFP gene (C). The same method was used to successively knock out the other four segments: TE3L, TA35R, TB13R, and TA66R. Ultimately, the five-segment-deleted attenuated strain MVTTEAB was obtained. T, termination signal of vaccinia virus; loxP, loxP site; Cre, Cre recombinase. (D) The TC7L-TK2L, TE3L, TA35R, TB13R, and TA66R genomic deletions were found in the viral genome.
MVTTEAB was identified by PCR as follows. The MVTTEAB genome was extracted and used as the template for PCR amplification of the partial fragments TC, TE, TA35, TB, and TA66. The amplification protocol was as follows: 95°C for 5 min; 35 cycles of 95°C for 30 s, 57°C for 30 s, and 72°C for 30 s; and a final extension at 72°C for 10 min. The VVTT genome was used as a control to identify the recombinant virus MVTTEAB. The identity of the products was confirmed by nucleotide sequencing. The sequences of the primers used for identification are shown in Table 1.
Genetic Stability of MVTTEAB
BHK-21 cells were infected with MVTTEAB and serially passaged 20 times to detect any reverse mutations of the deleted fragments (Guirakhoo et al., 1999). The amplification protocol consisted of 95°C for 5 min; 35 cycles of 95°C for 30 s, 57°C for 30 s, and 72°C for 30 s; and a final extension at 72°C for 10 min. The VVTT genome was used as a control to determine the genetic stability of MVTTEAB. The sequences of the primers used for identification are shown in Table 1.
Growth Curve
The BHK-21, HeLa, PK-15, MDCK, and Vero cells were cultured at a density of 5 × 105 cells/well in six-well plates and then infected with MVTTEAB or VVTT at 0.5 MOI. The cells were collected at 3, 12, 24, and 48 h after infection, and the viral titer in the BHK-21 cells was determined after three freeze-thaw cycles (Embry et al., 2011). The number of plaque-forming units (PFU) contained in 1 mL of viral fluid was calculated as follows: PFU/mL = (number of viral plaques × dilution ratio)/inoculation volume.
MTS Assay
The BHK-21, HeLa, PK-15, MDCK, and Vero cells were cultured at a density of 1 × 104 cells/well in 96-well plates for 24 h at 37°C in a 5% CO2 atmosphere. Then the cells were infected with MVTTEAB or VVTT at 0.5 MOI/well, and the infected cells were cultured at 37°C with 5% CO2. At 24, 48, 72, and 96 h, 20 mL of MTS solution (Promega) was added to each of the 96-well plate, which was cultured for 1 h at 37°C in a 5% CO2 atmosphere. Subsequently, we measured the absorption values at a wavelength of 490 nm using a microplate reader, which indirectly reflected the number of viable cells. Cell viability was calculated according to the following formula: 100 × (absorbance of culture in the treated wells/absorbance of culture in the control wells) (Mosmann, 1983; Li et al., 2006).
Weight Changes in Mice after Infection with MVTTEAB
Five-week-old BALB/c mice were inoculated intranasally with 1 × 105 PFU/20 μL PBS, 1 × 106 PFU/20 μL PBS, or 1 × 107 PFU/20 μL PBS of MVTTEAB or VVTT; a control group of mice was inoculated with PBS. There were 10 mice in each of the seven groups. The body weight of each mouse was recorded daily for 25 days after the inoculation (Vijaysri et al., 2008).
Skin Pathogenicity of MVTTEAB in Rabbits
In the rabbit skin pathogenicity assay, 1 × 106 PFU/0.1 mL PBS, 1 × 107 PFU/0.1 mL PBS, or 1 × 108 PFU/0.1 mL PBS of MVTTEAB or VVTT was injected intradermally into the backs of New Zealand white rabbits after their hair was shaved. Each concentration was injected into three rabbits. After the inoculation, skin lesions were formed on the back of the rabbits, and the lesions were measured with a vernier caliper and observed for 18 consecutive days.
Detection of Neurotoxicity in Mice after Infection with MVTTEAB
Three-week-old BALB/C mice (n = 10) were inoculated intracranially with 10 μL of MVTTEAB or VVTT diluted with sterile PBS at doses of 1 × 105 PFU/10 μL PBS, 1 × 106 PFU/10 μL PBS, or 1 × 107 PFU/10 μL PBS; the control group of mice was inoculated with PBS. Deaths were observed and recorded for 14 days after the inoculation. The intracranial 50% lethal infectious dose (ICLD50) was calculated using the Reed and Muench method (Reed and Muench, 1938).
In Vivo Immunogenicity Assay
Six-week-old BALB/c mice (n = 10) were inoculated intramuscularly with 0.1 mL of MVTTEAB or VVTT diluted with sterile PBS at a dose of 1 × 106 PFU/0.1 mL PBS, and the control group of mice was inoculated with PBS. The first immunization was performed after collection of the first blood sample, and blood samples were collected every week after the first immunization. Three weeks later, booster immunization via the same route and of the same dose was performed. At the end of the fifth week, the mice were euthanized after the blood samples were collected. All serum samples were separated from the mouse blood samples, and the serum level of IL-2, IL-4, IL-10, and IFN-γ was detected using ELISA kits (GBD). And neutralization assay was performed as described previously (Kan et al., 2012). The results were calculated using the method of Reed and Muench (Reed and Muench, 1938).
Statistical Analysis
Statistical analysis was conducted using data from at least three independent experiments. SPSS or SigmaStat 3.5 (Systat Software) was used for the analysis. P < 0.05 was considered to indicate statistical significance. Data are presented as the mean ± standard deviation (SD) values.
Results
Construction and Screening of the Recombinant Virus MVTTEAB
The shuttle plasmids were identified by double-restriction enzyme digestion with EcoRI and PstI. The EGFP fragment (720 bp) was obtained by double digestion of the shuttle vectors pTC-EGFP, pTE-EGFP, pTA35-EGFP, pTB-EGFP, and pTA66-EGFP. These results indicated that the five shuttle plasmids were constructed successfully (Figures 1A–C).
The recombinant shuttle plasmid pTC-EGFP was co-infected/transfected into BHK-21 cells with VVTT, and the recombinant VV rVVTT-C−EGFP+ was obtained by 10 rounds of fluorescence plaque screening. The EGFP gene of rVVTT-C-EGFP+ was knocked out with the Cre-loxP system, and then the recombinant vaccinia virus rVVTT-C− that did not contain the TC7L–TK2L genes was obtained by 10 rounds of fluorescence plaque screening (Figures 1, 2). The same screening method was used to knock out the other deletion fragments and construct a multi-gene-deleted attenuated strain of VVTT (MVTTEAB) in which five gene fragments and an exogenous selectable marker gene were deleted (Figure 2).
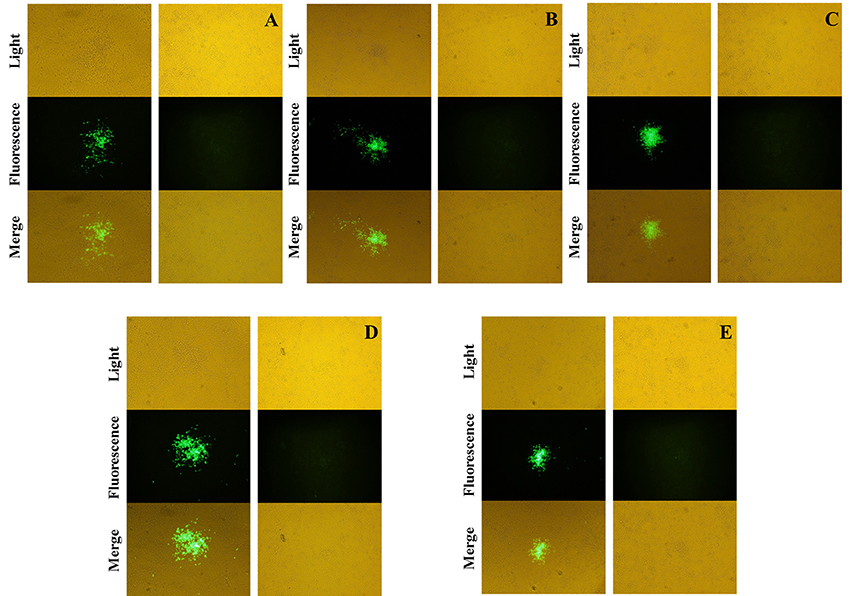
Figure 2. Screening and identification of mutants. The fused EGFP was expressed by the mutants rVVTT-C−EGFP+ (A), rVVTT-C−E−EGFP+ (B), rVVTT-C−E−A35−EGFP+ (C), rVVTT-C−E−A35−B−EGFP+ (D), and rVVTT-C−E−A35−B−A66−EGFP+ (MVTTEAB-EGFP+) (E) in BHK-21 cells. The virus-infected cells were identified by visualization of isolated fluorescent plaques in the same visual fields. Non-fluorescent plaques were observed for rVVTT-C− (A), rVVTT-C−E− (B), rVVTT-C−E−A35− (C), rVVTT-C−E−A35−B− (D), and rVVTT-C−E−A35−B−A66− (MVTTEAB) (E) mutants in BHK-21 cells (magnification 200×, A–E).
With the VVTT genome as the template, we obtained products of five different sizes by PCR amplification: TC, 322 bp; TE, 359 bp; TA35, 307 bp; TB, 406 bp; TA66, 352 bp (Figure 3). With the MVTTEAB genome as the template, PCR amplification under the same conditions did not produce similar bands. Then, the virus was identified by sequencing, and the results showed that the five target gene fragments were knocked out from the VVTT genome: TC-TK, TE, TA35R, TB13R, and TA66R.
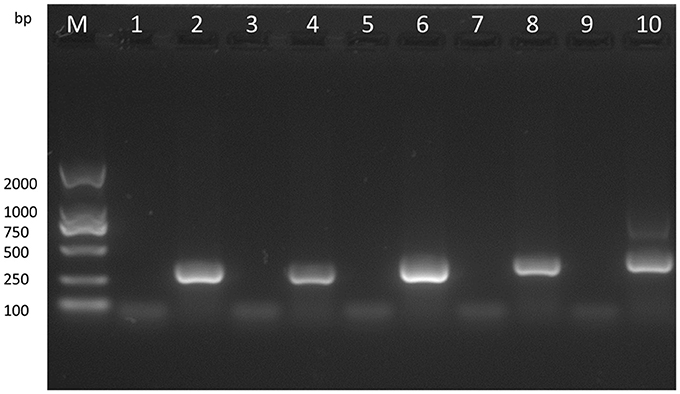
Figure 3. Analysis of the recombinant virus MVTTEAB by PCR. PCR was performed to identify the final mutant MVTTEAB. Lane 2: positive control containing the TC7L-TK2L gene (322 bp), lane 4: positive control containing the TE3L gene (359 bp), lane 6: positive control containing the TA35L gene (307 bp), lane 8: positive control containing the TB13R gene (406 bp), lane 10: positive control containing the TA66R gene (352 bp), and lanes 1, 3, 5, 7, and 9: PCR products of the MVTTEAB genome.
Genetic Stability of MVTTEAB
The five products of different sizes that were obtained by PCR amplification of the VVTT genome corresponded to the deleted fragments in the recombinant virus MVTTEAB: TC, 322 bp; TE, 359 bp; TA35, 307 bp; TB, 406 bp; TA66, 352 bp (Figure 4). However, PCR amplification of the 5, 10, 15, and 20th generation of the MVTTEAB genome under the same conditions did not produce the corresponding bands. These results showed that MVTTEAB had good genetic stability.
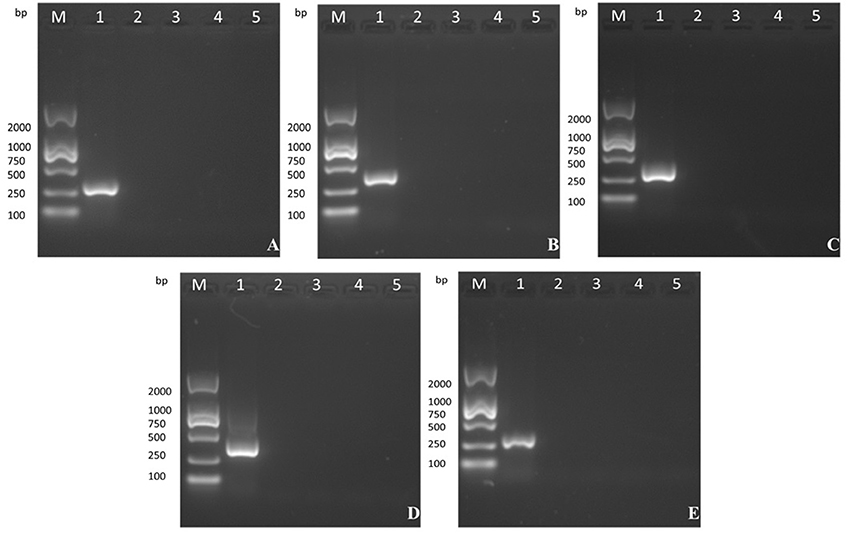
Figure 4. Evaluation of the genetic stability of the recombinant virus in BHK-21 cells after 5, 10, 15, and 20 passages. (A) TC7L-TK2L gene of MVTTEAB (lanes 2–5), (B) TE3L gene of MVTTEAB (lanes 2–5), (C) TA35L gene of MVTTEAB (lane 2–5), (D) TB13R gene of MVTTEAB (lanes 2–5), and (E) TA66R gene of MVTTEAB (lanes 2–5). PCR of all gene-deleted mutants produced negative results, compared to the corresponding positive PCR controls (lane 1).
Replication of MVTTEAB in Cells
As shown in Figure 5, MVTTEAB and VVTT showed similar growth trends in the same cell lines, but the viral titer decreased. The results indicated that the deletion of the gene segments in MVTTEAB did not influence normal replication of the virus in the cells.
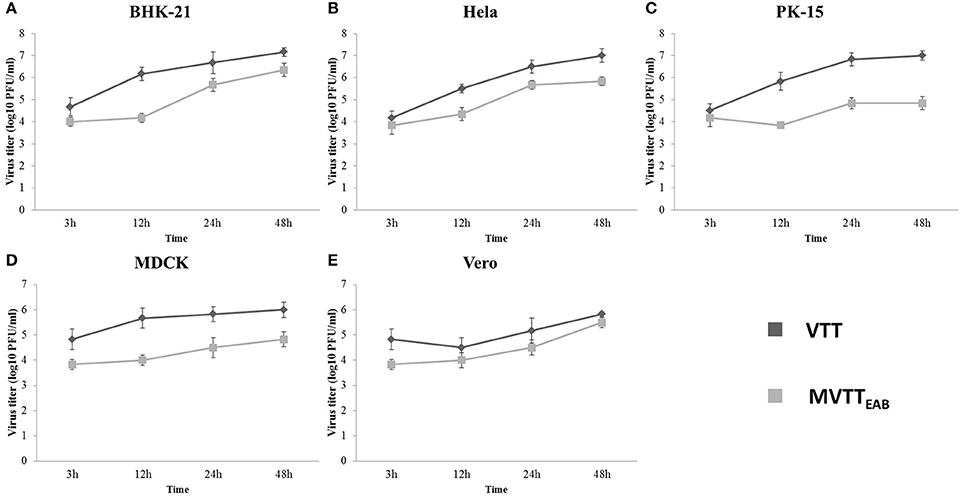
Figure 5. In vitro replication of MVTTEAB and VVTT. Growth curve of MVTTEAB and VVTT in BHK-21 (A), HeLa (B), PK-15 (C), MDCK (D), and Vero (E) cells. Cells were infected with MVTTEAB and VVTT at an MOI of 0.5, and the virus titers were measured at 3, 12, 24, and 48 h after infection.
Cell Virulence of MVTTEAB
BHK-21, HeLa, PK-15, MDCK, and Vero cells are sensitive to VVTT, which easily replicates in these cells. Here, the cytopathic effects of the recombinant virus MVTTEAB were detected and analyzed through MVTTEAB and VVTT infection of the five cell lines (Figures 6A–E). As shown in Figure 6, the five cell types that were infected with VVTT exhibited varying levels of cytocidal effects. The number of living cells decreased with time, and in the BHK-21, PK-15, and Vero cells, the cell viability rate was 30, 60, and 50%, respectively, at 96 h. The cytopathic effect of MVTTEAB on the growth of cells was lower than that of VVTT. From the data in Figure 6, it can be seen that in the BHK, PK-15, and Vero cells, the cell survival rate of the VVTT-infected cells was significantly lower than that of the MVTT-infected cells at 48 and 72 h (P < 0.01). At 96 h, the cell survival rate of the VVTT-infected cells (of all five cell types) was significantly lower than that of the MVTT-infected cells at 48 and 72 h. The difference was more significant in the BHK cells than in the other cell types (P < 0.001). The results indicated that the absence of the removed gene segments resulted in a decrease in the virulence of MVTTEAB. This demonstrated that the recombinant virus would be a safer vector or vaccine for vaccination than VVTT.
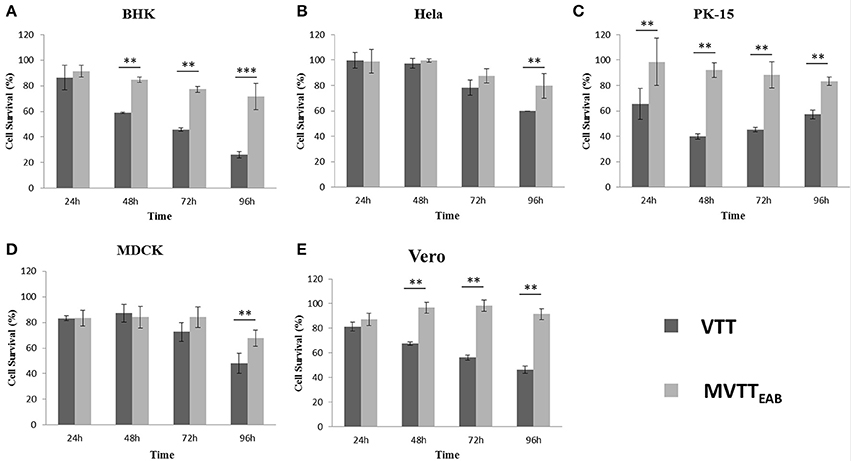
Figure 6. In vitro cytotoxicity of MVTTEAB and VVTT. BHK-21 (A), HeLa (B), PK-15 (C), MDCK (D), and Vero (E) cells were seeded in 96-well plates (1 × 104 cells/well) and infected with MVTTEAB and VVTT at an MOI of 0.5. Cell viability was measured every 24 h over a 96-h period. All measurements were performed in triplicate. Data are presented as the mean ± standard deviation (SD) values. When P < 0.01 (**) or P < 0.001 (***), the difference was considered to be significant.
Lesion Formation in Rabbits Infected Intradermally with MVTTEAB
MVTTEAB and wild-type VVTT were inoculated intradermally into the back of the rabbits, and the size of the pock lesions formed was measured daily for 18 days. The pock lesions induced by VVTT were extremely obvious, while the lesions induced by MVTTEAB were much smaller. Furthermore, the higher viral titers of both VVTT and MVTTEAB resulted in the formation of larger pock lesions. In addition, the trends in the size of the pock lesions formed by the two virus infections over time are similar: that is, the size of the pock lesions first increased and then decreased. As shown in Figure 7A, the pock lesions induced by VVTT became red and swollen from the next day of the inoculation. The size of the VVTT-induced lesions peaked on day 2, after which they gradually started festering. This was followed by scab formation, and on the tenth day, an obvious scab could be observed. At the end of the experiment, the size of the pock lesions induced by VVTT had been reduced, but they had not all disappeared. In contrast to the VVTT-induced lesions, the pock lesions produced by MVTTEAB infection only appeared as a slight swelling on day 2, which subsided on day 4 for the lowest MVTTEAB titer. From the data in Figure 7B, it can be seen that the size of pock lesions formed by MVTTEAB infection at the lowest and medium viral titer were significantly smaller than that of lesions formed by VVTT infection at the lowest, medium and highest titer (P < 0.05). No significant difference was observed between the size of the pock lesions formed by MVTTEAB infection at the highest viral titer and the MVTT infection at the lowest titer, but the former did not result in the formation of a fester or scab (P > 0.05). Thus, the deletion of the five genes in MVTTEAB significantly reduced its virulence compared to wild-type VVTT. The dose-dependent pattern observed was comparable to that of VVTT and WR previously reported in mice (Brandt and Jacobs, 2001; Fang et al., 2005).
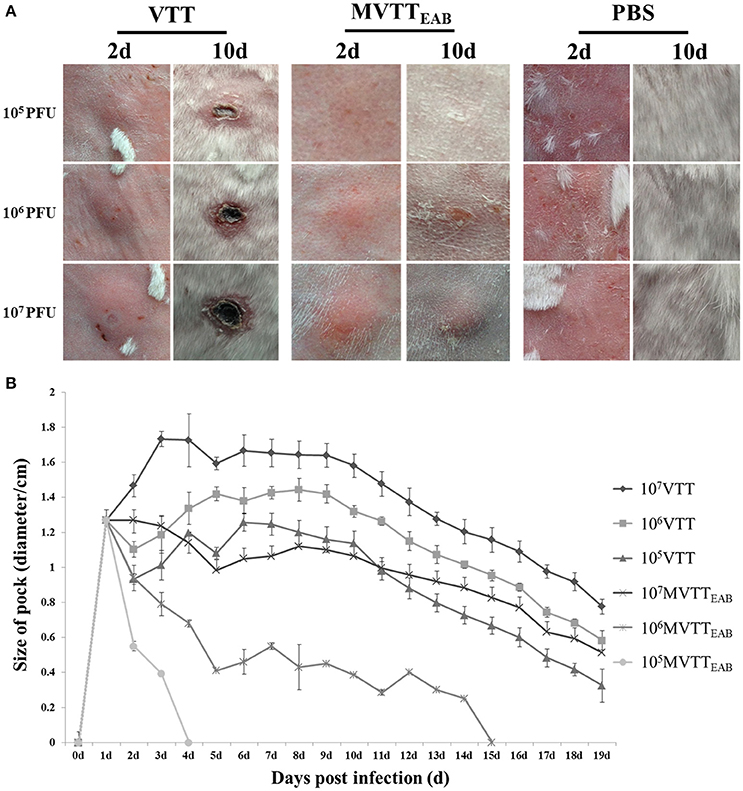
Figure 7. Virulence of MVTTEAB or VVTT in rabbits. Rabbits were infected with 105, 106, or 107 PFU of MVTTEAB or VVTT (as a positive control) or PBS (as a negative control) by intradermal injection. Infection with 105, 106, and 107 PFU of MVTTEAB resulted in mild swelling by day 2 that subsided by day 4, and no visible scars were observed by day 10 at the sites of infection (A). By contrast, infection with VVTT resulted in the formation of relatively severe lesions at the infection site with ulcerations; the size of the lesions increased until they peaked on day 2, after which they gradually decreased but left scars (A). The diameter of the lesions was associated with the VVTT and MVTTEAB dose (B).
MVTTEAB Virulence in BALB/c Mice
The recombinant virus MVTTEAB and wild-type VVTT were intranasally inoculated into 5-week-old female BALB/c mice, and their body weights were observed for 25 days. As shown in Figure 8A, the weight of the mice infected with the highest dose of VVTT decreased from the fourth day after inoculation and reached the lowest level on the ninth day, with an average reduction of 22.8%. The weight started to gradually increase on the tenth day, but until the end of the experiment, the mean body weight of the VVTT group was significantly lower than that of the MVTTEAB and PBS control groups (P < 0.05). The change in body weight in the low- and middle-dose VVTT group was similar to that in the high-dose VVTT group, but the degree of weight loss and period over which the weight were significantly different (P < 0.05). The weight of the mice in all the three MVTTEAB groups did not decrease, and the trend in body weight changes was similar to that of the PBS control group. Further, the low-dose MVTTEAB group showed more obvious weight gain than the other two groups with higher MVTTEAB doses (P < 0.05). The results showed that the loss of body weight was positively correlated with the viral titer of the inoculated mice, and that the virulence of MVTTEAB was lower than that of VVTT. These findings indicate that the deletion of the five genes in MVTTEAB significantly reduced the in vivo virulence of VVTT.
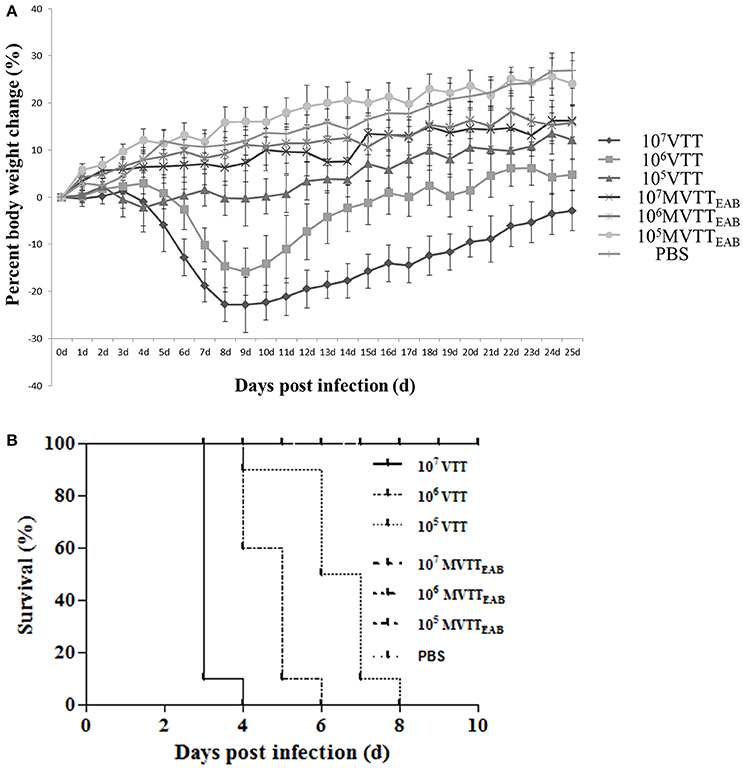
Figure 8. Virulence of VVTT and MVTTEAB in mice after intranasal and intracranial infection. (A) Body weight changes were monitored in mice that were intranasally infected with 105, 106, or 107 PFU of MVTTEAB or VVTT (as a positive control) or PBS (as a negative control). Error bars indicate the standard error of the mean, and differences between groups were determined by two-way repeated measures analysis of variance. Mice inoculated with VVTT showed significant signs of illness, and a clear positive correlation was found between the viral dose and weight loss (P < 0.05). No distinct weight changes or signs of illness were observed in the animals inoculated with MVTTEAB or PBS. (B) Mice were inoculated intracranially with 105, 106, or 107 PFU of MVTTEAB or VVTT (PBS in the negative control group), and the survival rates were observed for 14 days. All the mice inoculated with MVTTEAB survived, while all the mice infected with VVTT died.
The recombinant VV MVTTEAB and wild-type VVTT were intracranially inoculated in mice for 14 days. Mice infected with VVTT exhibited neurological symptoms such as scruffy fur, lassitude, convulsions, and stiffness; the mice in the VVTT groups finally died. By comparison, mice infected with the three doses of MVTTEAB did not exhibit such symptoms and were alive at the end of the observation period. Survival curves drawn for the mice (Figure 8B) showed that mice inoculated with the three different doses of MVTTEAB showed 100% survival until the end of the experiment (Figure 8B), which was significantly different from the survival rate of the VVTT-infected mice (P < 0.05). On the fourth day after inoculation with 1.0 × 107 PFU of VVTT, the survival rate was 0%, and on the sixth and eighth day after inoculation with 1.0 × 106 PFU and 1.0 × 105 PFU of VVTT, the survival rate was 0%. The ICLD50 of VVTT-inoculated mice was 1.16 × 104 PFU. The results showed that the deletion of the five genes in MVTTEAB significantly reduced the neurotoxicity of the virus in mice. In short, the safety of the recombinant vaccinia virus in mice was improved by knocking out multiple genes, which improved its potential as a vaccine vector or vaccine for MVTTEAB.
Humoral and Cellular Immune Response to Infection with MVTTEAB
The serum levels of IL-2, IL-4, IL-10, and IFN-γ were measured in all the mouse groups with mouse serum cytokine assay kits at the third and fifth weeks after immunization. The results indicated that the levels of IL-2, IL-4, IL-10, and IFN-γ in mouse serum in the VVTT- and MVTTEAB-infected groups were significantly higher than those in the PBS control group after the first immunization and the booster immunization (Figures 9A–D) (P < 0.01). In contrast, no significant differences were observed between the MVTTEAB-infected groups and VVTT-infected groups (P > 0.05). The results indicated that despite the deletion of the five genes in MVTTEAB, the virus could still induce high levels of these cytokines and maintain its immunogenicity as a vaccine.
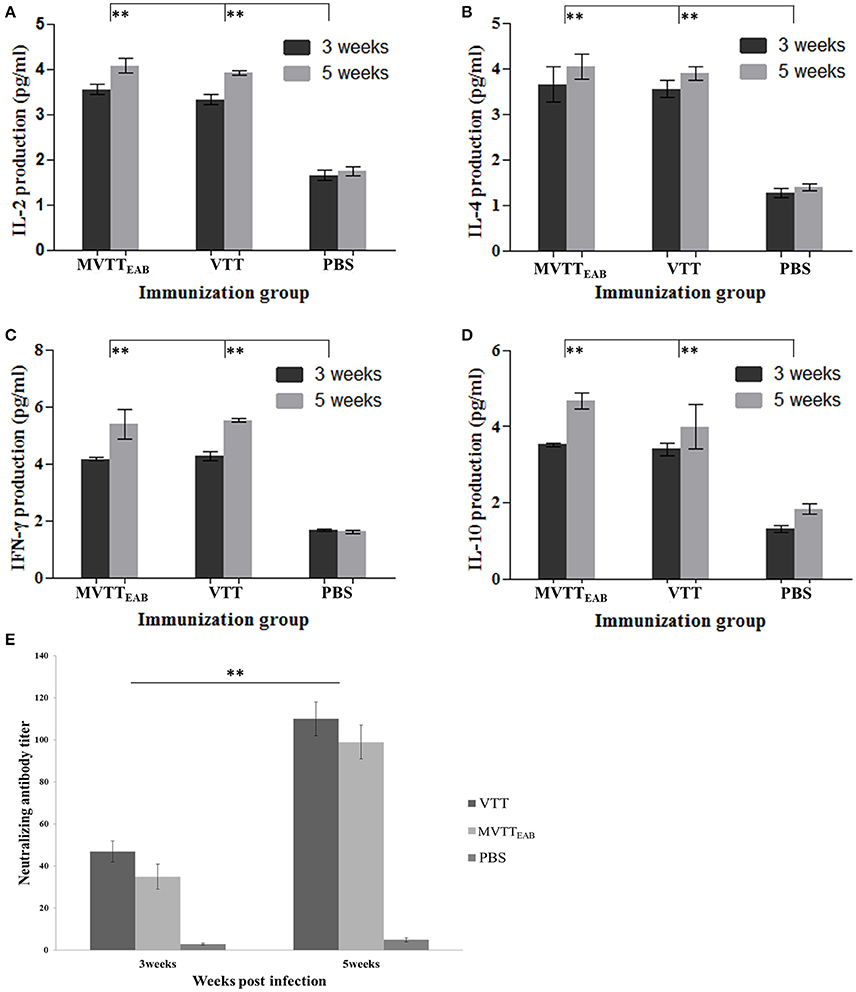
Figure 9. Immune responses to MVTTEAB or VVTT in vaccinated mice. BABL/c mice were inoculated intramuscularly with 106 PFU MVTTEAB or VVTT, and given a booster immunization of the same dose in 0.1 ml PBS 3 weeks later. The serum samples were harvested at weeks 3 and 5 after immunization. The IL-2 (A), IL-4 (B), IFN-γ (C), and IL-10 (D) levels in the two vaccinated groups were measured by mouse ELISA kits. All measurements were performed in triplicate. Data are presented as the mean ± SD values. The neutralization antibody titer was calculated by determining the highest serum dilution required to generate a 50% viral plaque reduction (E). All measurements were performed in triplicate. Data are presented as the mean ± SD values. When P < 0.01 (**), the difference was considered to be significant.
The mice were immunized with MVTTEAB and VVTT at a dose of 1 × 106 PFU to detect neutralizing antibodies in the serum. As shown in Figure 9E, the serum neutralizing antibody titer of the MVTTEAB and VVTT groups was not significantly different after immunization (P > 0.05). After the booster immunization, the serum neutralizing antibody titers of the MVTTEAB and VVTT groups increased by about five times and reached about 100. Thus, the recombinant MVTTEAB virus retains its immunogenicity as a vaccine despite the deletion of five genes and can stimulate a strong systemic immune response.
Discussion
In this study, in order to construct a safer and effective attenuated VVTT strain, the genome of this strain was modified on the basis of analysis of the whole genome of several VV strains that are widely used. The non-essential gene fragments of the viral genome were deleted using the homologous recombination technique and the Cre/loxP system. The deleted gene segments were TC7L-TK2L, TE3L, TA35R, TB13R, and TA66R. TC7L-TK2L is not only a host-range gene but also a host defense regulatory gene (McFadden, 2005; Zhu et al., 2007; Yu et al., 2010). TE3L is a virulence-associated gene that inhibits the antiviral activity of interferon, and it is also associated with host range determinants, host defense regulation factors, and cell apoptosis regulatory factors (Wang et al., 2012). TA35R is a vaccinia A-type inclusion body protein gene fragment that is homologous with A26L of Vaccinia virus Copenhagen strain-encoded A-type envelope proteins (Rehm and Roper, 2011). TB13R is an immunoregulatory gene that encodes serine protein inhibitor and is also involved in the Fas-mediated death receptor pathway and lipoxygenase pathway. TA66R is a gene encoding viral hemagglutinin, which has the same function as the A56R gene of the Vaccinia virus WR strain and plays a role in inhibiting cell fusion (Buller et al., 1985). The absence of these genes in the newly constructed MVTTEAB strain was confirmed by PCR. Additionally, the genetic stability of the newly constructed MVTTEAB strain was confirmed, and it was also shown that the deletion of these segments did not affect the replication ability of the virus.
In the present study, a significant decline in the virulence of MVTTEAB was verified in both in vitro and in vivo experiments. Similar to our findings, other studies have also reported that the deletion of certain non-essential gene fragments led to a decrease in virulence in VV strains. For example, it has been shown that the virulence of VV is reduced after deletion of A35R but does not affect the size of the plaque formed; thus, A35R is a non-essential gene for viral replication that is associated with the virulence of VV (Roper, 2006). Further, deletion of C, C2L, and N1L has also been found to result in a significant reduction in the virulence of VV (Legrand et al., 2004). Another study showed that the virulence decreased significantly in the absence of B13L in the WR strain in nude mice and normal mice, while humoral immunity and cellular immune response were still high (Legrand et al., 2004). Knockout of E3L has also been shown to inhibit interferon activation and antiviral response (Langland and Jacobs, 2002), which means that deletion of this gene attenuates viral virulence (Wang et al., 2012). Altogether, the present findings as well the findings of previous studies show that the deletion of these specific non-essential genes that are not associated with viral replication can attenuate the virulence of VV and therefore improve its prospects as a vaccine.
In our in vitro experiments, the MTS assay was used to detect the cytotoxicity of the recombinant virus MVTTEAB that was constructed and wild-type VVTT in five different cell lines. From 48 h after infection, the survival rate of the infected cells began to significantly differ between the VVTT- and MVTTEAB-infected groups, with the survival rate of the cells infected with MVTTEAB being significantly higher than that of the cells infected with VVTT. Thus, the cytotoxicity of VVTT was significantly weakened as a result of deletion of the gene fragments. The recombinant virus MVTTEAB therefore seems to have better safety than the wild-type VVTT, which means that it may be safer for use as a viral vector or vaccine for disease prevention/treatment.
In the skin lesion formation experiment in rabbits, the lesion size peaked on the third day after inoculation with VVTT, and then decreased slowly. On the contrary, only mild swelling was observed in the MVTTEAB groups, and the swelling subsided in the low-dose group on the fourth day. Thus, deletion of the five gene fragments seems to have significantly reduced the skin damage caused by the wild-type VV in rabbits. Similarly, it has been shown that after inoculation of MVA, a recombinant VVTT strain, in rabbits, the lesions formed recovered at 12 days (Melamed et al., 2013).
In our in vivo virulence experiments in mice, the body weight of the mice was the lowest on the ninth day in the high-dose VVTT group, with an average reduction of 22.8%. In contrast, the weight trend of mice in the MVTTEAB group was similar to that in the PBS control group. Thus, there was a significant reduction in the virulence of VV after deletion of the five gene fragments. Similarly, it has been reported that inoculation of MVA did not result in a decrease in the weight of the inoculated mice compared to mice inoculated with the wild-type virus (Melamed et al., 2013). With regard to its neurotoxicity, MVTTEAB was found to be highly safe in BALB/c mice, as all the mice that were intracranially inoculated with MVTTEAB survived with no neurological symptoms. This was in contrast to the observations in the VVTT-inoculated group, in which all the mice died on the eighth day after inoculation. The findings for MVTTEAB are similar to those reported for MVA and NYVAC, which are the most commonly used attenuated VV strains in which multiple host-range genes, virulence genes and other non-essential genes are deleted (Tartaglia et al., 1992; Melamed et al., 2013).
Cytokines play an important role in the body's immune response, such as antiviral and mediated inflammatory responses. Research has shown that IL-4, IL-10, and IFN-γ play an important role in the immune response of mice to VV infection (van Den Broek et al., 2000). Most studies on the deletion of immunoregulatory genes in VV have shown that the absence of several VV genes can reduce the toxicity of the virus (Smith et al., 2013), but the effect on the immunogenicity of the virus is variable. For example, deletion of these immunomodulatory genes from different strains (mainly WR and MVA) was found to increase the immunogenicity of this virus: E3L, B15R/B16R, A41L, B22R, C12L, and C6L. However, the absence of immunomodulatory genes, such as B8R, was found to have no effect on virulence or pathogenicity, while the deletion of genes such as N2L and C16L was found to have no effect on immunogenicity (Alcami and Smith, 1995; Fahy et al., 2008; Ferguson et al., 2013). In addition, deletion of C12L, A44L, A46R, or B7R in MVA did not significantly affect VACV-specific CD8 + T cell immunogenicity in BALB/c mice (Cottingham et al., 2008). Based on the studies described above, we deleted five gene fragments, including TC7L-TK2L, to detect and analyze changes in the immunogenicity of MVTTEAB. The results of this study show that the degree of immune response in mice infected with MVTTEAB is almost equal to that of VTT infection, and there is no significant difference (P > 0.05). Further, MVTTEAB seems to have attenuated virulence compared to VVTT, which makes it safer for clinical application. In summary, MVTTEAB has demonstrated excellent safety and immunogenicity than the wild-type virus and has potential as a new virus vector and vaccine for the prevention or treatment of diseases caused by different pathogens.
Author Contributions
Conceived and designed the experiments: YL, XL, LS, and NJ. Performed the experiments: YL, YZ, SC, WL, XY, SL, and NJ. Analyzed the data: YL, XL, LS, and NJ. Contributed reagents/materials/analysis tools: YZ, SC, WL, XY, SL, PX, and JH. Wrote the paper: YL and NJ. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer MF and handling Editor declared their shared affiliation.
Acknowledgments
This work was supported in part by the National Science and Technology Major Project (Major New Drugs Innovation and Development) [grant number 2014ZX09304314-002]; the Key Technologies R&D Program of Jilin Province, China [grant numbers 20140309006YY, 20150201002YY]; the National Key R&D Project [grant number 2016YFC1200901]; and the Major Technological Program of Changchun City, China [grant number 16ss11].
References
Adelfinger, M., Gentschev, I., Grimm de Guibert, J., Weibel, S., Langbein-Laugwitz, J., Härtl, B., et al. (2014). Evaluation of a new recombinant oncolytic vaccinia virus strain GLV-5b451 for feline mammary carcinoma therapy. PLoS ONE 9:e104337. doi: 10.1371/journal.pone.0104337
Alcamí, A., and Smith, G. L. (1995). Cytokine receptors encoded by poxviruses: a lesson in cytokine biology. Immunol. Today 16, 474–478. doi: 10.1016/0167-5699(95)80030-1
Brandt, T. A., and Jacobs, B. L. (2001). Both carboxy- and amino-terminal domains of the vaccinia virus interferon resistance gene, E3L, are required for pathogenesis in a mouse model. J. Virol. 75, 850–856. doi: 10.1128/JVI.75.2.850-856.2001
Brennan, G., Kitzman, J. O., Shendure, J., and Geballe, A. P. (2015). Experimental evolution identifies vaccinia virus mutations in A24R and A35R that antagonize the protein kinase r pathway and accompany collapse of an extragenic gene amplification. J. Virol. 89, 9986–9997. doi: 10.1128/JVI.01233-15
Buller, R. M., Smith, G. L., Cremer, K., Notkins, A. L., and Moss, B. (1985). Decreased virulence of recombinant vaccinia virus expression vectors is associated with a thymidine kinase-negative phenotype. Nature 317, 813–815. doi: 10.1038/317813a0
Cottingham, M. G., Andersen, R. F., Spencer, A. J., Saurya, S., Furze, J., Hill, A. V., et al. (2008). Recombination-mediated genetic engineering of a bacterial artificial chromosome clone of modified vaccinia virus Ankara (MVA). PLoS ONE 3:e1638. doi: 10.1371/journal.pone.0001638
Embry, A., Meng, X., Cantwell, A., Dube, P. H., and Xiang, Y. (2011). Enhancement of immune response to an antigen delivered by vaccinia virus by displaying the antigen on the surface of intracellular mature virion. Vaccine 29, 5331–5339. doi: 10.1016/j.vaccine.2011.05.088
Fahy, A. S., Clark, R. H., Glyde, E. F., and Smith, G. L. (2008). Vaccinia virus protein C16 acts intracellularly to modulate the host response and promote virulence. J. Gen. Virol. 89, 2377–2387. doi: 10.1099/vir.0.2008/004895-0
Fang, Q., Yang, L., Zhu, W., Liu, L., Wang, H., Yu, W., et al. (2005). Host range, growth property, and virulence of the smallpox vaccine: vaccinia virus Tian Tan strain. Virology 335, 242–251. doi: 10.1016/j.virol.2005.02.014
Ferguson, B. J., Benfield, C. T., Ren, H., Lee, V. H., Frazer, G. L., Strnadova, P., et al. (2013). Vaccinia virus protein N2 is a nuclear IRF3 inhibitor that promotes virulence. J. Gen. Virol. 94, 2070–2081. doi: 10.1099/vir.0.054114-0
García-Arriaza, J., Arnáez, P., Gómez, C. E., Sorzano, C. Ó., and Esteban, M. (2013). Improving adaptive and memory immune responses of an HIV/AIDS vaccine candidate MVA-B by deletion of vaccinia virus genes (C6L and K7R) blocking interferon signaling pathways. PLoS ONE 8:e66894. doi: 10.1371/journal.pone.0066894
Guerra, S., Abaitua, F., Martinez-Sobrido, L., Esteban, M., Garcia-Sastre, A., and Rodriguez, D. (2011). Host-range restriction of vaccinia virus E3L deletion mutant can be overcome in vitro, but not in vivo, by expression of the influenza virus NS1 protein. PLoS ONE 6:e28677. doi: 10.1371/journal.pone.0028677
Guirakhoo, F., Zhang, Z. X., Chambers, T. J., Delagrave, S., Arroyo, J., Barrett, A. D., et al. (1999). Immunogenicity, genetic stability, and protective efficacy of a recombinant, chimeric yellow fever-Japanese encephalitis virus (ChimeriVax-JE) as a live, attenuated vaccine candidate against Japanese encephalitis. Virology 257, 363–372. doi: 10.1006/viro.1999.9695
Kan, S., Wang, Y., Sun, L., Jia, P., Qi, Y., Su, J., et al. (2012). Attenuation of vaccinia Tian Tan strain by removal of viral TC7L-TK2L and TA35R genes. PLoS ONE 7:e31979. doi: 10.1371/journal.pone.0031979
Langland, J. O., and Jacobs, B. L. (2002). The role of the PKR-inhibitory genes, E3L and K3L, in determining vaccinia virus host range. Virology 299, 133–141. doi: 10.1006/viro.2002.1479
Legrand, F. A., Verardi, P. H., Chan, K. S., Peng, Y., Jones, L. A., and Yilma, T. D. (2005). Vaccinia viruses with a serpin gene deletion and expressing IFN-gamma induce potent immune responses without detectable replication in vivo. Proc. Nat. Acad. Sci. U.S.A. 102, 2940–2945. doi: 10.1073/pnas.0409846102
Legrand, F. A., Verardi, P. H., Jones, L. A., Chan, K. S., Peng, Y., and Yilma, T. D. (2004). Induction of potent humoral and cell-mediated immune responses by attenuated vaccinia virus vectors with deleted serpin genes. J. Virol. 78, 2770–2779. doi: 10.1128/JVI.78.6.2770-2779.2004
Li, X., Jin, N., Mi, Z., Lian, H., Sun, L., Li, X., et al. (2006). Antitumor effects of a recombinant fowlpox virus expressing Apoptin in vivo and in vitro. Int. J. Cancer 119, 2948–2957. doi: 10.1002/ijc.22215
Liu, J., and McFadden, G. (2015). SAMD9 is an innate antiviral host factor with stress response properties that can be antagonized by poxviruses. J. Virol. 89, 1925–1931. doi: 10.1128/JVI.02262-14
Mackett, M., Yilma, T., Rose, J. K., and Moss, B. (1985). Vaccinia virus recombinants: expression of VSV genes and protective immunization of mice and cattle. Science 227, 433–435. doi: 10.1126/science.2981435
MacNeil, A., Reynolds, M. G., and Damon, I. K. (2009). Risks associated with vaccinia virus in the laboratory. Virology 385, 1–4. doi: 10.1016/j.virol.2008.11.045
Melamed, S., Wyatt, L. S., Kastenmayer, R. J., and Moss, B. (2013). Attenuation and immunogenicity of host-range extended modified vaccinia virus Ankara recombinants. Vaccine 31, 4569–4577. doi: 10.1016/j.vaccine.2013.07.057
Meulemans, G., Letellier, C., Gonze, M., Carlier, M. C., and Burny, A. (1988). Newcastle disease virus F glycoprotein expressed from a recombinant vaccinia virus vector protects chickens against live-virus challenge. Avian Pathol. 17, 821–827. doi: 10.1080/03079458808436504
Mosmann, T. (1983). Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 65, 55–63. doi: 10.1016/0022-1759(83)90303-4
Noisumdaeng, P., Pooruk, P., Kongchanagul, A., Assanasen, S., Kitphati, R., Auewarakul, P., et al. (2013). Biological properties of H5 hemagglutinin expressed by vaccinia virus vector and its immunological reactivity with human sera. Viral Immunol. 26, 49–59. doi: 10.1089/vim.2012.0055
Pastoret, P. P., and Brochier, B. (1996). The development and use of a vaccinia-rabies recombinant oral vaccine for the control of wildlife rabies; a link between Jenner and Pasteur. Epidemiol. Infect. 116, 235–240. doi: 10.1017/S0950268800052535
Qin, L., Favis, N., Famulski, J., and Evans, D. H. (2015). Evolution of and evolutionary relationships between extant vaccinia virus strains. J. Virol. 89, 1809–1824. doi: 10.1128/JVI.02797-14
Reed, L. J., and Muench, H. (1938). A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27, 493–497.
Rehm, K. E., and Roper, R. L. (2011). Deletion of the A35 gene from Modified Vaccinia Virus Ankara increases immunogenicity and isotype switching. Vaccine 29, 3276–3283. doi: 10.1016/j.vaccine.2011.02.023
Remy-Ziller, C., Germain, C., Spindler, A., Hoffmann, C., Silvestre, N., Rooke, R., et al. (2014). Immunological characterization of a modified vaccinia virus Ankara vector expressing the human papillomavirus 16 E1 protein. Clin. Vaccine Immunol. 21, 147–155. doi: 10.1128/CVI.00678-13
Roper, R. L. (2006). Characterization of the vaccinia virus A35R protein and its role in virulence. J. Virol. 80, 306–313. doi: 10.1128/JVI.80.1.306-313.2006
Smith, G. L., Benfield, C. T., Maluquer de Motes, C., Mazzon, M., Ember, S. W., Ferguson, B. J., et al. (2013). Vaccinia virus immune evasion: mechanisms, virulence and immunogenicity. J. Gen. Virol. 94, 2367–2392. doi: 10.1099/vir.0.055921-0
Tartaglia, J., Perkus, M. E., Taylor, J., Norton, E. K., Audonnet, J. C., Cox, W. I., et al. (1992). NYVAC: a highly attenuated strain of vaccinia virus. Virology 188, 217–232. doi: 10.1016/0042-6822(92)90752-B
van Den Broek, M., Bachmann, M. F., Köhler, G., Barner, M., Escher, R., Zinkernagel, R., et al. (2000). IL-4 and IL-10 antagonize IL-12-mediated protection against acute vaccinia virus infection with a limited role of IFN-gamma and nitric oxide synthetase 2. J. Immunol. 164, 371–378. doi: 10.4049/jimmunol.164.1.371
Vijaysri, S., Jentarra, G., Heck, M. C., Mercer, A. A., McInnes, C. J., and Jacobs, B. L. (2008). Vaccinia viruses with mutations in the E3L gene as potential replication-competent, attenuated vaccines: intra-nasal vaccination. Vaccine 26, 664–676. doi: 10.1016/j.vaccine.2007.11.045
Wang, Y., Kan, S., Du, S., Qi, Y., Wang, J., Liu, L., et al. (2012). Characterization of an attenuated TE3L-deficient vaccinia virus Tian Tan strain. Antiviral Res. 96, 324–332. doi: 10.1016/j.antiviral.2012.10.002
Yu, W., Fang, Q., Zhu, W., Wang, H., Tien, P., Zhang, L., et al. (2010). One time intranasal vaccination with a modified vaccinia Tiantan strain MVTT(ZCI) protects animals against pathogenic viral challenge. Vaccine 28, 2088–2096. doi: 10.1016/j.vaccine.2009.12.038
Keywords: attenuated vaccinia virus vector, homologous recombination, vaccinia Tiantan strain virus, virulence, immunogenicity
Citation: Li Y, Zhu Y, Chen S, Li W, Yin X, Li S, Xiao P, Han J, Li X, Sun L and Jin N (2017) Generation of an Attenuated Tiantan Vaccinia Virus Strain by Deletion of Multiple Genes. Front. Cell. Infect. Microbiol. 7:462. doi: 10.3389/fcimb.2017.00462
Received: 27 June 2017; Accepted: 18 October 2017;
Published: 31 October 2017.
Edited by:
Wenjun Liu, Institute of Microbiology (CAS), ChinaReviewed by:
Min Fang, Institute of Microbiology (CAS), ChinaGuoqiang Zhu, Yangzhou University, China
Copyright © 2017 Li, Zhu, Chen, Li, Yin, Li, Xiao, Han, Li, Sun and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao Li, bGl4aWFvMDZAbWFpbHMuamx1LmVkdS5jbg==
Lili Sun, bGluamlheGlhb3lhQDE2My5jb20=
Ningyi Jin, c2t5bGVlNjIyNkAxNjMuY29t
 Yiquan Li
Yiquan Li Yilong Zhu2,3
Yilong Zhu2,3 Jicheng Han
Jicheng Han