- 1School of Life Sciences, Shandong University of Technology, Zibo, China
- 2UMR BIPAR, INRA, ANSES, École Nationale Vétérinaire d'Alfort, Université Paris-Est Créteil Val-de-Marne, Maisons-Alfort, France
Bartonellosis is an infectious disease caused by Bartonella species that are distributed worldwide with animal and public health impact varying according to Bartonella species, infection phase, immunological characteristics, and geographical region. Bartonella is widely present in various mammals including cats, rodents, ruminants, and humans. At least 13 Bartonella species or subspecies are zoonotic. Each species has few reservoir animals in which it is often asymptomatic. Bartonella infection may lead to various clinical symptoms in humans. As described in the B.tribocorum-rat model, when Bartonella was seeded into the blood stream, they could escape immunity, adhered to and invaded host erythrocytes. They then replicated and persisted in the infected erythrocytes for several weeks. This review summarizes the current knowledge of how Bartonella prevent phagocytosis and complement activation, what pathogenesis factors are involved in erythrocyte adhesion and invasion, and how Bartonella could replicate and persist in mammalian erythrocytes. Current advances in research will help us to decipher molecular mechanisms of interactions between Bartonella and mammalian erythrocytes and may help in the development of biological strategies for the prevention and control of bartonellosis.
Introduction
Bartonella species are fastidious, Gram-negative hemotropic organisms. Bartonella have been isolated from a range of species; from diverse animals, such as canids, rodents, ruminants, and felids. They are mainly transmitted via direct contact (animal scratches and bites) or by numerous arthropods such as sand flies, fleas, lice, biting flies, and ticks (Deng et al., 2012).
Until now, at least 40 species or subspecies of Bartonella have been found (Mullins et al., 2017). Each species can establish a lasting intraerythrocytic bacteraemia in its reservoir host, but typically not with obvious detriments (Vayssier-Taussat et al., 2009; Deng et al., 2012). In contrast, when Bartonella accidentally infects the incidental hosts, which means that absolutely no erythrocytes are involved during the acute phase of a zoonotic infection, the acute clinical manifestations can be provoked (Raoult, 2007; Mosepele et al., 2012).
B. henselae is the most prevalent zoonotic Bartonella species (Yuan et al., 2011). B.henselae infection is typically asymptomatic in the reservoir cats, in spite of up to 108 CFU/ml blood. However, various clinical symptoms can be caused in humans, such as cat scratch disease and bacillary peliosis in immunocompetent and immunocompromised individuals, respectively (Dehio, 1999, 2001; Chomel et al., 2009; Pulliainen and Dehio, 2012; Deng et al., 2016). The distribution of Bartonella in animal and public health varies with Bartonella species, infection phase, immunological characteristics, and geographical region.
Bartonella spp., Plasmodium spp., Babesia spp., Theileria spp., Mycoplasma suis, and Anaplasma marginale are important intracellular pathogens which can infect mammalian erythrocytes (Barbour and Restrepo, 2000; Schülein et al., 2001; Groebel et al., 2009). In contrast to other pathogens, all Bartonella species could survive within the infected erythrocytes for several weeks with only subtle changes of the erythrocyte membrane, except the deadly B. bacilliformis (Dehio, 2005; Harms and Dehio, 2012).
The infection course of Bartonella has been described in natural and experimental animal models, such as the B. birtlesii-mouse, B.tribocorum-rat, and B. henselae-cat models (Guptill et al., 1997; Boulouis et al., 2001; Seubert et al., 2002; Birtles, 2005; Marignac et al., 2010). All of them show similar results, which suggest a universal infection course of the different species in their respective mammalian reservoir hosts. Following initial inoculation, Bartonella could be rapidly cleared from the blood, which was considered due to Bartonella infection of the so called primary niche outside of circulating blood, potentially endothelial cells, erythrocytic precursors, liver, and possibly other cell types or organs (Dehio et al., 1997; Dehio, 1999, 2001; Mändle et al., 2005; Deng et al., 2012b). Bartonella was released into the blood stream between 2 and 5 days post-infection. Followed by erythrocyte adhesion and invasion. They then replicated in the infected erythrocytes until eight daughter cells were reproduced. The infected erythrocytes could persist for many weeks (Schülein et al., 2001; Guptill, 2010; Harms and Dehio, 2012). This review will discuss the current understanding of Bartonella and erythrocyte interactions, especially focusing on the required factors involved in virulence of Bartonella in their reservoir hosts (Figure 1).
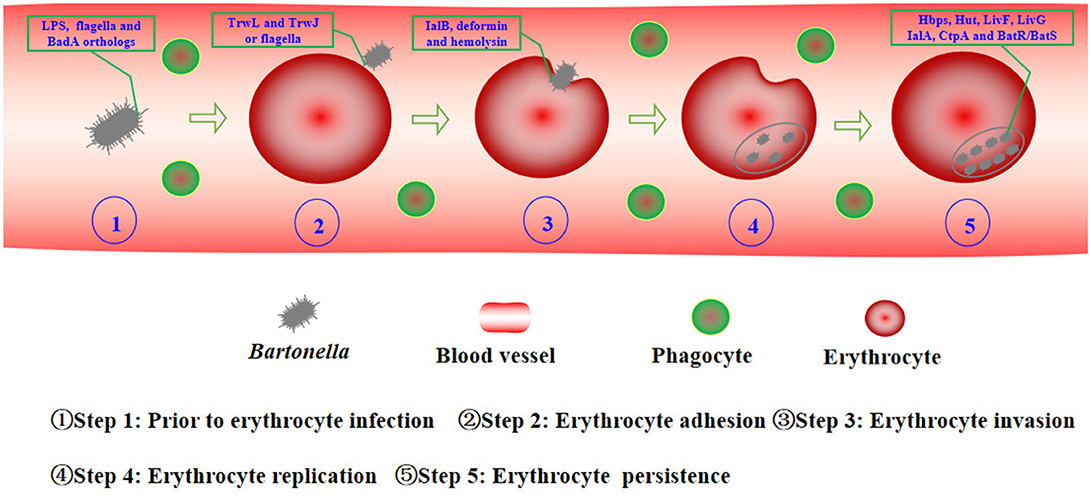
Figure 1. Holistic view of Bartonella interactions with erythrocytes. Prior to erythrocyte infection, Bartonella must escape the host immune responses to facilitate their extracellular longevity to approach and infect erythrocytes efficiently. Bartonella use LPS, flagella, and BadA orthologs against phagocytes and complement activation (Step 1). Erythrocyte adhesion is mediated by multiple copies of TrwL and TrwJ variants of pilus components or flagella (Step 2). IalB, deformin, and hemolysin cause some subtle changes of erythrocyte membrane and erythrocyte invasion by Bartonella species (Step 3). Once inside, the bacterium replicates in a membrane-bound compartment, until the number of intracellular bacteria reaches static levels for the remaining lifespan of the infected erythrocytes, the distinguishable changes will disappear (Step 4). Within erythrocytes, Bartonella uses Hbps, Hut, LivF, and LivG to get nutrients, and IalA, CtpA, and BatR/BatS to cope with stressors (Step 5).
Step 1: Prior to Erythrocyte Infection
As mentioned above, prior to mammalian erythrocytes infection, Bartonella could infect the primary niche and reappear in the bloodstream. Bartonella must escape the host immune responses to facilitate their extracellular survival to approach and infect erythrocytes efficiently in this step (Arvand et al., 2001; Kabeya et al., 2003; Resto-Ruiz et al., 2003; Ben-Tekaya et al., 2013; Dehio and Tsolis, 2017; Scherler et al., 2017).
The First Strategy Is Replication of Large Numbers of Bartonella
Following intravenous inoculation, the bacteria were unable to infect the erythrocytes. Instead, they were disappeared from circulation and maintained undetectable during ~4 days (Schülein et al., 2001). During this time, the primary niche may support Bartonella replication and allow them to gain competency for erythrocyte interactions (Dehio, 2005; Harms and Dehio, 2012). On day 5 post-inoculation, numerous Bartonella are seeded into the bloodstream and cause autoagglutination (Kaiser et al., 2008; Schmidgen et al., 2014). This is one of the first steps of biofilm formation (Okaro et al., 2017; Tu et al., 2017). The bacterial factors that are responsible for replication are presently unknown.
The Second Strategy Is Against Phagocytes
On day 5 post-inoculation, Bartonella are extracellular, thus they are completely exposed to the immune system. Phagocytes such as macrophages are the first line of immune defense against the infection (Dornand et al., 2002; Weiss and Schaible, 2015). Pattern recognition receptors (PRRs) such as Toll-like receptors (TLRs) on the phagocytes are considered to recognize Bartonella spp. (Kloch et al., 2018). Generally, LPS and particularly its lipid A part is mainly recognized by TLR4 and causes pro-inflammatory cytokines secretion to induce various inflammatory cells to move to the infection site (Malgorzata-Miller et al., 2016). It was reported that LPS of B.henselae and B. bacilliformis has a deep-rough structure, and B.henselae LPS contains an unusual lipid A with a long chain fatty acid and without an O-chain polysaccharide (Gorczynski et al., 2004; Focà et al., 2012). The unusual features of Bartonella LPS were weakly recognized by TLR4 and did not evoke TLR4 activation (Minnick, 1994; Raetz and Whitfield, 2002; Focà et al., 2012). As B. henselae LPS was 1,000-10,000-fold less active than Salmonella LPS in activating TLR4 signaling, B. quintana LPS could not induce pro-inflammatory cytokines production (Zähringer et al., 2004; Popa et al., 2007). And B. quintana LPS could be a TLR4 activation antagonist to inhibit release of cytokines mediated by Escherichia coli LPS, such as interleukin-1β, interleukin-6 and tumor necrosis factor α (Boonjakuakul et al., 2007; Popa et al., 2007; Matera et al., 2008). Moreover, it could also block TLR4 signaling transduction in rheumatoid arthritis (Abdollahi-Roodsaz et al., 2007). Compared with Salmonella flagellin, the flagellin of Bartonella species which possess flagella such as B. bacilliformis, B. bovis, B. capreoli, B. chomelii, B. clarridgeiae, and B. schoenbuchensis contains amino acid differences in the site of TLR5 recognition. Which does not cause flagellin-mediated TLR5-dependent NF-κB activation and might escape TLR5 recognition (Andersen-Nissen et al., 2005; Deng et al., 2012; Kloch et al., 2018). The unusual structures of LPS and flagellin are important for Bartonella spp. to escape the TLR4 and TLR5 recognition by phagocytes, respectively.
Bartonella adhesion A (BadA) is an outer membrane protein which is homologous to Yersinia adhesin A (YadA), Haemophilus surface fibrils (Hsf), Moraxella surface protein A (UspA), and Haemophilus adhesin (Hia) (Lafontaine et al., 2000; St Geme and Cutter, 2000; Biedzka-Sarek et al., 2008). BadA belongs to the trimeric autotransporter adhesion (TAA) family, which all share similar modular architectures, consisting of a head, neck/stalk repeats, and C-membrane anchor domains (Hoiczyk et al., 2000; Wollmann et al., 2006). The number of neck/stalk repeats are variable in different Bartonella species (Kaiser et al., 2012). BadA could cause bacterial autoaggregation and encode antigenic variation of repetitive tandem stalk domains to prevent phagocytosis (Riess et al., 2004). Bartonella could also temporarily enter macrophages in a unique Bartonella-containing vacuole (BCV) and delay lysosomal targeting and destruction (Kyme et al., 2005).
The Third Strategy Is Preventing Complement Activation
The complement system has the function of microbial infection control, either directly by membrane-attack complex (MAC) formation or via phagocyte opsonization. It was considered that the absence of O-side chain of Bartonella LPS could decrease complement fixation and increase serum resistance (Zähringer et al., 2004). Recent observations suggested that BadA was involved in preventing complement activation, since mouse serum could kill B. birtlesii badA-knockout (ΔbadA) mutants, while not the wild type B. birtlesii. Moreover, anti-BadA antibodies could neutralized this killing activity and ΔbadA was resistant to heat-inactivated serum (Deng et al., 2012).
Since Bartonella LPS, flagellin, and BadA could inhibit the function of the immune system, such as the complement and phagocytic cells, the inflammatory response decreased resulting in reduced phagocytes migration, antigen presenting, and B cells activation.
Step 2: Erythrocyte Adhesion
Intracellular pathogens must bind to host cells to successfully initiate infection (Barnett et al., 2015). Bacteria use various components to adhere to host cells, ranging from complex substances, such as fimbriae or pili, to proteins, such as Brucella suis BmaC, BtaE, and BatF adhesins (Ruiz-Ranwez et al., 2013; Wu et al., 2014). Recognition of host molecules by adhesins is the first step of bacterial infection (Coutte et al., 2003; Caswell et al., 2010; Ruiz-Ranwez et al., 2013). Exploitation of erythrocytes by Bartonella spp. is a complex progression through a series of different infection stages, beginning with erythrocyte adhesion. Although some factors have been shown to be essential for this step, the knowledge about erythrocyte adhesion factors of Bartonella is nominal. It is difficult to perform genetic studies, since no liquid medium can support rapid growth of Bartonella spp. and suitable animal models for study on pathogenicity of this bacteria are limited.
The First Factor Is the Trw System
The Trw system is the third type 4 secretion system (T4SS) found in certain Bartonella spp. and has a short-path of evolution (Frank et al., 2005). It shares high homology with plasmid R388 which is a broad-host-range conjugation system of the IncW group that confers resistance to sulfonamide and trimethoprim and produces constitutively rigid conjugative pili called W pili (Bolland et al., 1990). Both encode an identical and interchangeable transcription regulatory circuit KorA/KorB repressor which could negatively regulate T4SS expression by binding to kor box sequences (Figure 2).
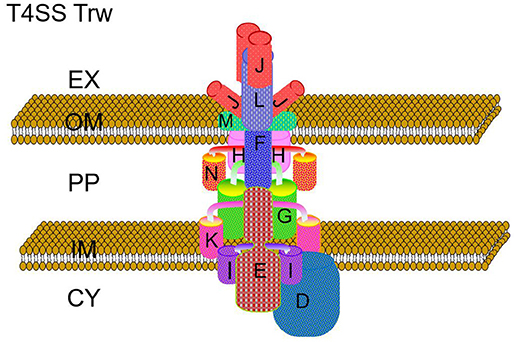
Figure 2. Hypothetical model of the architecture of the Trw system of Bartonella species. The Trw system is a multi-component protein complex spanning the inner and outer membranes of Bartonella species. The multiple copies of TrwL and TrwJ represent the major and minor pilus components, while the other duplicated proteins, TrwI and TrwH are involved in pilus elongation and for pilus anchorage to the outer membrane respectively. EX, extracellular matrix; OM, outer membrane; PP, periplasm; IM, inner membrane; CY, cytoplasm. Letters denote individual Trw subunits.
Although the Trw system shares homology with plasmid R388, this system lacks a TrwB which is the key protein required for transfering effectors. This suggests that the Trw system is not a secretion system (Seubert et al., 2003; Dehio, 2004; Vayssier-Taussat et al., 2010; Larrea et al., 2013). In fact, to date no substrates translocated by this system have been characterized. Upregulation of the Trw system during endothelial cell infection would decrease the time in the hostile environment of the bloodstream. Bartonella could then adhere to and invade erythrocytes quickly (Dehio, 2004; Pulliainen and Dehio, 2012). The Trw system required for intraerythrocytic infection was identified by large-scale signature tagged mutagenesis (STM) screens in the B. birtlesii-mouse and B. tribocorum-rat models in vivo (Saenz et al., 2007; Vayssier-Taussat et al., 2010).
Recently, the function of Trw in erythrocyte infection was identified by an in vitro model for erythrocyte adhesion and invasion (Vayssier-Taussat et al., 2010). In the study, we identified nine mutants that could not invade erythrocytes in vitro. The nine mutants included an invasion-associated locus (ialA/B) mutant, a leucine-isoleucine-valine (livG) mutant, and seven mutants for genes encoding Trw components. When we horizontally transferred the trw locus of rat-specific B. tribocorum into human-specific B. quintana and cat-specific B. henselae, they were able to interact with rat erythrocytes, suggesting that the Trw sytstem is a key factor of erythrocyte infection and host specificity.
The trw genes of Bartonella species are collinear with their homologous genes of plasmid R388, except for the gene duplications of trwJ-I-H (the virB5, virB6, and virB7 homologs) and trwL (the virB2 homolog) (Figure 3). There are variable copy numbers of the duplicated genes in the different species, which is evidence of gene conversion and rapid evolution (Schröder and Dehio, 2005; Schulein et al., 2005; O'Rourke et al., 2011). For example, the trans and inner membrane regions of TrwL are almost identical, but the outer membrane regions are different across Bartonella species (Figure 4). The duplicated copies of trwJ and trwL encode variant forms of pilus components, while trwH and trwI are involved in pilus elongation and anchorage, respectively (Dehio, 2004; Deng et al., 2012).
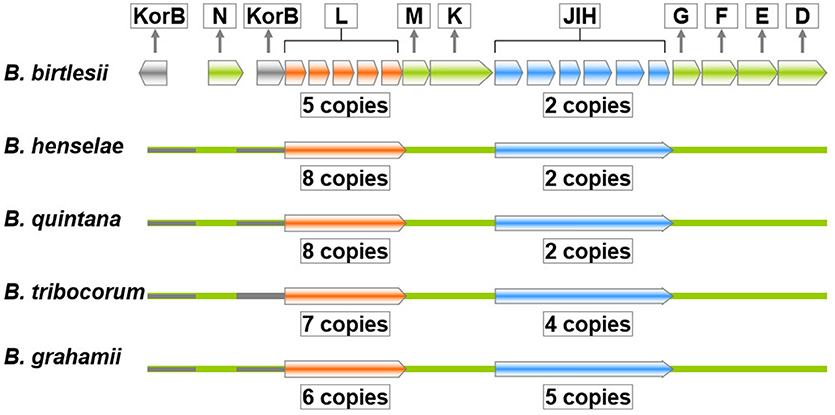
Figure 3. Gene order structure of the trw locus of Bartonella species. The trw genes of Bartonella species are collinear with the respective genes of plasmid R388, except for the multiple tandem gene duplications of trwL and trwJ-I-H, which are present in variable copy numbers in the different species. The copy number of amplified genes or segments of the trw locus of five Bartonella species is indicated within the boxes. The copy number trwJ and trwL displays a large degree of sequence variation.
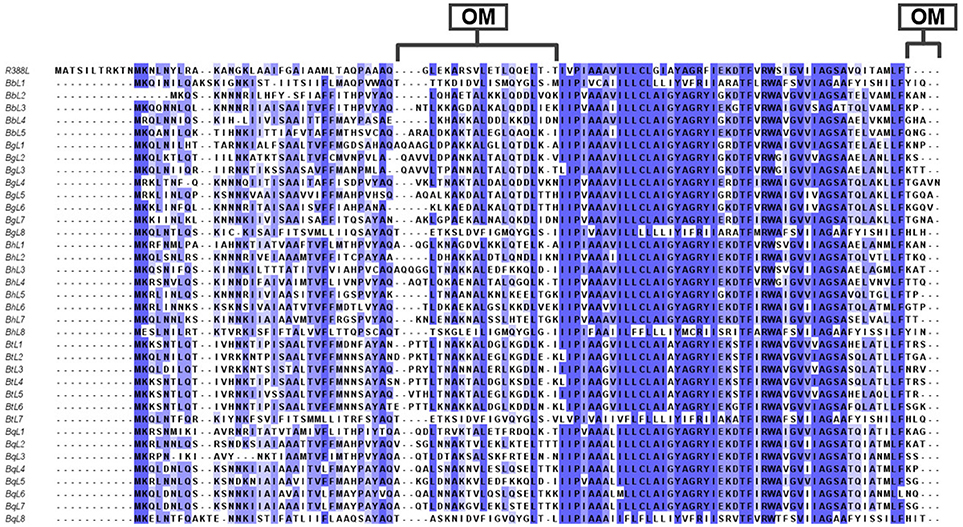
Figure 4. Multiple amino acid sequence alignments of TrwL of Bartonella species. Multiple amino acid sequences of TrwL from different Bartonella species of B. birtlesii (BbL1 to BbL5), B. grahamii (BgL1 to BbL7), B. henselae (BhL1 to BhL8), B. quintana (BqL1 to BhL8), B. tribocorum (BqL1 to BhL7), and R388 TrwL (R388L) were aligned by Clustal Omega. Conserved amino acids are shaded and each shade represented a degree of conservation (Blue, 100%). The major outer membrane proteins were calculated by TMHMM 2.0. All the sequences were taken from the NCBI GenBank. OM, Outer membrane proteins.
However, the direct function of Trw system has only recently been obtained by using different technologies (Deng et al., 2012,a). In the study, both TrwJ1 and TrwJ2 were found at B. birtlesii surface and could bind to band 3 of mouse erythrocytes. It was considered that TrwL might also bind to the surface of erythrocytes, and the outer membrane parts of TrwL proteins might be responsible for this ability. Further studies are required to identify the erythrocytic receptors of TrwL. Bacteria usually use some virulence factors to bind to host cells more intimately after initial adherence. We considered that the specific and stable interactions between Bartonella and host erythrocytes were mediated by both TrwJ and TrwL. Moreover, TrwJ might have the capability to interact with TrwL, which represents the major and minor pilus (Figure 5). TrwJ and TrwL might be involved in initial or intimate adhesion during infection of erythrocytes. The interactions of TrwJ and TrwL with host erythrocytes were associated with Bartonella invasion, although direct evidence is lacking to confirm this theory.
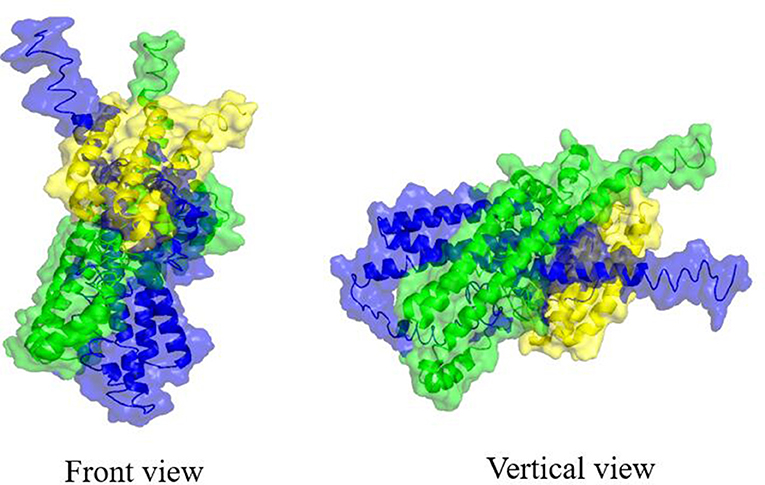
Figure 5. Molecular docking between B. birtlesii TrwJ1, TrwJ2, and TrwL. Three-dimensional models of B. birtlesii TrwJ1, TrwJ2, and the outer membrane parts of TrwL were modeled by the I-TASSER server based on the amino acid sequences (Yang et al., 2015). The best identified structural analogs of both TrwJ1 (C-score of −1.27 and TM-score of 0.74) and TrwJ2 (C-score of −1.45 and TM-score of 0.69) in protein data bank (PDB) are 1R8I, and the outer membrane parts of TrwL (C-score of −3.17 and TM-score of 0.70) in PDB is 4TQL. Molecular docking between B. birtlesii TrwJ1, TrwJ2 and the outer membrane parts of TrwL was analyzed using the ClusPro 2.0 server and visualized by the PyMOL program (Kozakov et al., 2017). The front and vertical view of the docked conformations were shown as B. birtlesii TrwJ1 (green cartoon with semitransparent surface), TrwJ2 (blue cartoon with semitransparent surface), and the outer membrane parts of TrwL (yellow cartoon with semitransparent surface).
The multiple gene copies have many advantages. First, they can increase expression of the dosage of pilus proteins and the length or the number of pili for rapid adhesion to erythrocytes in the bloodstream of the mammalian host (Gillespie et al., 2015). Second, multiple Trw pilus variants encoded by those gene copies might bind to various surface components of different blood groups in different reservoir populations (Dehio, 2008). Third, they provide variability within the genome for antigenic variation (Lindroos et al., 2006). Fourth, they might increase potential for new functions.
The Second Factor Is Flagella
The role of Trw evolved to replace the flagella, since the expression of Trw and flagella is mutually exclusive among the Bartonella species (Dehio, 2008; Harms and Dehio, 2012). The multiple flagella which let B.bacilliformis with highly motile could be important for the high rate of erythrocyte infection in Oroya fever (Scherer et al., 1993; Dehio, 2001). It has been reported that B. bacilliformis flagellin site-directed mutants bind poorly to erythrocytes, and this phenomenon can be partially rescued by trans-complementation with nature flagellin (Battisti and Minnick, 1999; Sander et al., 2000). The flagellin subunit antibodies could partially inhibit the adhesion between B. bacilliformis and erythrocytes (Scherer et al., 1993; Sander et al., 2000). Early work indicated that B. bacilliformis could interact with many human erythrocyte membrane proteins, including glycophorins A and B (Buckles and McGinnis Hill, 2000). Those observations correspond with the former views that the flagella of Bartonella may serve as an adhesin, although it remains unknown whether flagella can directly bind to host erythrocytes (Walker and Winkler, 1981; Benson et al., 1986).
Step 3: Erythrocyte Invasion
After erythrocyte adhesion, Bartonella invaded mature erythrocytes within 2 days, which has been demonstrated in the B. tribocorum-rat infection model (Seubert et al., 2002). The unusual structure and physiology of erythrocytes could allow Bartonella to escape antigen presentation and immune surveillance. We have little knowledge about how Bartonella enter host erythrocytes, but some factors have been shown to be essential for this step.
The First Factor Is IalB
As described above, IalA/B was identified by STM screens in the B.birtlesii-mouse and B. tribocorum-rat models in vivo and by an in vitro model for erythrocyte adhesion and invasion.
IalB which is a 19.9 kDa protein with putative signal peptides (Figure 6), shares high homology with the Yersinia enterocolitica protein Ail, that plays a major role in cell invasion (Kirjavainen et al., 2008; Deng et al., 2016). Early work demonstrated that E. coli could invade erythrocytes when it was transformed with B. bacilliformis ialB, and deletion of ialB decreased the erythrocyte infection of B.birtlesii and B.tribocorum in vivo (Mitchell and Minnick, 1995; Saenz et al., 2007; Vayssier-Taussat et al., 2010). Moreover, the B. birtlesii IalB mutant caused a 10-fold decrease in erythrocyte invasion, but it has no significant effect on erythrocyte adhesion in vitro (Vayssier-Taussat et al., 2010). The B. bacilliformis mutant can be restored to erythrocyte invasiveness when trans-complemented with wild-type IalB locus (Coleman and Minnick, 2001). Our recent study showed that IalB was immunogenic and anti-IalB antibodies could inhibit mouse erythrocyte invasion by B. birtlesii (Deng et al., 2016).
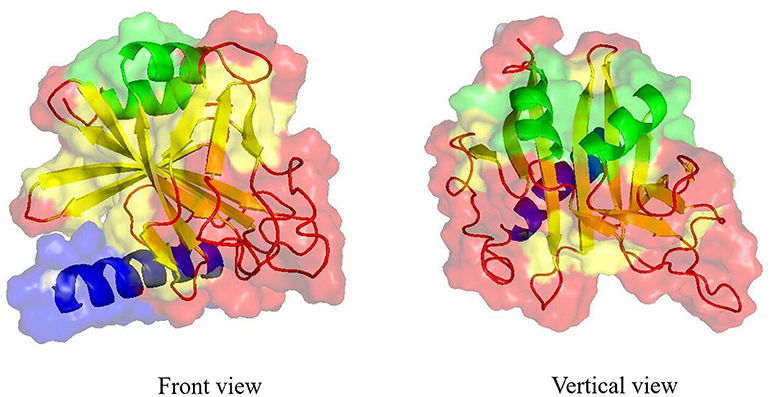
Figure 6. Three-dimensional structure model of B. birtlesii IalB. Three-dimensional structure model of B. birtlesii IalB was built using the I-TASSER server based on the amino acid sequence (Yang et al., 2015). The front and vertical view of the crystal structure of IalB (C-score of −1.15 and TM-score of 0.77) was shown as the cartoon model with semitransparent surface. The best identified structural analogs in PDB is 3DTD. The structure that contained 1 signal peptide domain (blue bands), 2 α-helices (green bands), 11 β-strands (yellow bands), and 14 coils (red bands) in the secondary structure constituted a stable region.
There was confusion about the location of IalB in Bartonella. B. bacilliformis IalB was an inner membrane protein, while B. henselae IalB was an outer membrane protein (Mitchell and Minnick, 1995; Coleman and Minnick, 2001; Chenoweth et al., 2004). In our recent study, a small quantity of IalB was detected on B. birtlesii surface, while most of IalB was expressed in Bartonella lysate supernatants of different species (Deng et al., 2016). So, we hypothesized that most of the B. birtlesii IalB might be secreted proteins that mediated erythrocyte invasion by unknown mechanisms.
The Second Factor Is Deformin
B. bacilliformis could cause production of trenches, pits, conical invaginations, and internal vacuoles in the erythrocyte membrane (Benson et al., 1986; Xu et al., 1995). This phenomenon is mediated by deformin, which has been found in the culture supernatants of B. henselae and B. bacilliformis, suggesting that this mechanism might be present in several Bartonella species (Iwaki-Egawa and Ihler, 1997).
There was confusion about the identity of deformin in Bartonella. Early work demonstrated that deformin was a protease- and heat- resistant, water-soluble, and albumin binding molecule with a molecular weight of ~1.4 kDa (Derrick and Ihler, 2001). More recent work has indicated that deformin is several proteins present in the supernatant of B. bacilliformis with a molecular weight of ~36 kDa (Hendrix and Kiss, 2003). The 36 kDa proteins appear to either necessary for deformin secretion or directly deforming human erythrocytes. The nature of deformin and the molecular mechanisms of erythrocyte deformation require further studies.
The Third Factor Is Hemolysin
Two types of Bartonellla hemolytic factors have been found including a contact-dependent hemolysin of B. bacilliformis and an autotransporter cohemolysin of B. henselae (Hendrix, 2000; Litwin and Johnson, 2005). B. bacilliformis contact-dependent hemolysin is maximally expressed during exponential growth phase, and might be used to escape from the vacuoles or erythrocytes during intracellular parasitism (Hendrix, 2000; Litwin and Johnson, 2005). B. henselae cohemolysin which is a 180 kDa autotransporter protein, has homologs in B. quintana and causes lysis of erythrocytes (Litwin and Johnson, 2005; Minnick and Battisti, 2009).
Step 4: Erythrocyte Replication and Persistence
Bartonella spp. attach, invade and replicate within a vacuole of erythrocytes in the B. tribocorum-rat infection model. After several days, bacterial replication stops until an approximately eight daughter cells are reproduced. There are some subtle changes in the physiology of erythrocytes during erythrocyte invasion and replication. B. tribocorum-infected erythrocytes are removed more rapidly than uninfected erythrocytes from circulation. However, once the number of intraerythrocytic Bartonella reaches static levels, the distinguishable changes and the rapid clearance rates will disappear (Schülein et al., 2001). Within an erythrocyte, Bartonella must not only get nutrients, but also cope with stressors.
The First Strategy Is Nutrient Uptake
Bartonella species use two gene families of heme binding proteins (Hbps) and the heme utilization locus (Hut) to sequester heme (Carroll et al., 2000; Minnick et al., 2003; Zimmermann et al., 2003; Parrow et al., 2009). Hbps are required for intraerythrocytic bacteraemia and have been identified by STM screens in the B. birtlesii-mouse and B. tribocorum-rat models in vivo (Saenz et al., 2007; Vayssier-Taussat et al., 2010). B. quintana HbpA is a 29.3 kDa protein and part of a hbpA-E gene family (Carroll et al., 2000). Compared with parental strains, an HbpA mutant of B. quintana showed an enhanced heme binding phenotype (Minnick et al., 2003). It was also reported that anti-HbpA antibodies could inhibit the hemin binding in a dose-dependent manner (Carroll et al., 2000).
LivF and LivG, which are highly conserved among the Bartonella species, are required for intraerythrocytic bacteraemia and have been identified by STM screens in the B. birtlesii-mouse and B. tribocorum-rat models in vivo. Moreover, the B. birtlesii LivG mutant provoked a dramatic decrease in bacterial entry into erythrocytes in vitro (Vayssier-Taussat et al., 2010). LivF and LivG which are ATPase components of ABC transporters are required for amino acid nutrient uptake during Bartonella inside erythrocytes (Saenz et al., 2007).
The Second Strategy Is Against Stressors
In order to adapt to the intraerythrocytic environment, Bartonella must cope with a variety of stressors, including reactive oxygen species, fluctuations in osmolarity, changes in pH, and misfolded proteins.
IalA which is a 20.1 kDa protein, has homologs in other invasive bacteria and has been demonstrated as a (de)nucleoside polyphosphate hydrolase of the MutT motif family (Mitchell and Minnick, 1995, 1997; Cartwright et al., 1999; Conyers and Bessman, 1999). IalA hydrolysates including ATP and inorganic phosphate could he recycled. IalA and its homologs are believed to regulate the level of stress-induced nucleotides and their derivatives during invasion. The carboxy-terminal protease (CtpA), which is encoded upstream of the ialA gene, could degrade misfolded or aberrant proteins from stress or anomalous processing (Mitchell and Minnick, 1997; Cartwright et al., 1999).
It has been reported that BatR/BatS which is an important two-component regulator/sensor is probably used by Bartonella to regulate the expression of some pathogenic genes such as the T4SS, BadA, and Hbps, and respond to environmental cues in the mammalian circulatory system (Quebatte et al., 2010; Harms and Dehio, 2012).
None of the molecular factors of mechanisms allowing for Bartonella spp. replication and persistence in the infected erythrocytes have been identified to date.
Conclusion
Bartonella species are intraerythrocytic pathogens. They are mainly transmitted by animal contact and arthropods. For example, B. henselae is transmitted between cats by cat fleas (Ctenocephalides felis) and transmitted from cats to humans by cat scratches or bites (Chomel et al., 1996). In order to prevent the spread of the disease, it is important for scientists to explore the mechanisms of Bartonella infection.
Despite significant amounts of effort and advances to understand the molecular mechanisms of how Bartonella infects host erythrocytes, many uncertain aspects need further studies. The functions of the above-mentioned strategies and virulence determinants are still not fully elucidated and many other virulence factors have yet to be found. Moreover, the gene expression, regulation, and signal transduction pathways of those factors are still elusive. We also know little about the physiological changes and recognition receptors of erythrocytes during their infection.
In summary, with so many exciting and important questions yet to be answered, future studies would not only better clarify the functions of the factors, but also increase our understanding of the network between the factors and erythrocytes at a molecular level.
Author Contributions
HD, QP, and BZ wrote the initial draft of the paper. HD and MV-T organized and proofread the paper. BZ helped to draft the figure. HD approved the version to be published. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 31302125), Natural Science Foundation of Shandong Province of China (Grant No. ZR2014DM015), the Research Fund of Shandong University of Technology (Grant No. 413016 and No. 114023).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mr. Chad Risch for linguistic assistance and critical reading of the manuscript.
References
Abdollahi-Roodsaz, S., Joosten, L. A., Roelofs, M. F., Radstake, T. R., Matera, G., Popa, C., et al. (2007). Inhibition of Toll-like receptor 4 breaks the inflammatory loop in autoimmune destructive arthritis. Arthritis Rheum. 56, 2957–2967. doi: 10.1002/art.22848
Andersen-Nissen, E., Smith, K. D., Strobe, K. L., Barrett, S. L., Cookson, B. T., Logan, S. M., et al. (2005). Evasion of Toll-like receptor 5 by flagellated bacteria. Proc. Natl. Acad. Sci. U.S.A. 102, 9247–9252. doi: 10.1073/pnas.0502040102
Arvand, M., Ignatius, R., Regnath, T., Hahn, H., and Mielke, M. E. (2001). Bartonella henselae-specific cell-mediated immune responses display a predominantly Th1 phenotype in experimentally infected C57BL/6 mice. Infect. Immun. 69, 6427–6433. doi: 10.1128/IAI.69.10.6427-6433.2001
Barbour, A. G., and Restrepo, B. I. (2000). Antigenic variation in vector-borne pathogens. Emerg. Infect. Dis. 6, 449–457. doi: 10.3201/eid0605.000502
Barnett, T. C., Lim, J. Y., Soderholm, A. T., Rivera-Hernandez, T., West, N. P., and Walker, M. J. (2015). Host-pathogen interaction during bacterial vaccination. Curr. Opin. Immunol. 36, 1–7. doi: 10.1016/j.coi.2015.04.002
Battisti, J. M., and Minnick, M. F. (1999). Development of a system for genetic manipulation of Bartonella bacilliformis. Appl. Environ. Microbiol. 65, 3441–3448.
Benson, L. A., Kar, S., McLaughlin, G., and Ihler, G. M. (1986). Entry of Bartonella bacilliformis into erythrocytes. Infect. Immun. 54, 347–353.
Ben-Tekaya, H., Gorvel, J. P., and Dehio, C. (2013). Bartonella and Brucella–weapons and strategies for stealth attack. Cold Spring Harb. Perspect. Med. 3:a010231. doi: 10.1101/cshperspect.a010231
Biedzka-Sarek, M., Salmenlinna, S., Gruber, M., Lupas, A. N., Meri, S., and Skurnik, M. (2008). Functional mapping of YadA- and Ail-mediated binding of human factor H to Yersinia enterocolitica serotype O:3. Infect. Immun. 76, 5016–5027. doi: 10.1128/IAI.00314-08
Birtles, R. J. (2005). Bartonellae as elegant hemotropic parasites. Ann. N. Y. Acad. Sci. 1063, 270–279. doi: 10.1196/annals.1355.044
Bolland, S., Llosa, M., Avila, P., and de la Cruz, F. (1990). General organization of the conjugal transfer genes of the IncW plasmid R388 and interactions between R388 and IncN and IncP plasmids. J. Bacteriol. 172, 5795–5802. doi: 10.1128/jb.172.10.5795-5802.1990
Boonjakuakul, J. K., Gerns, H. L., Chen, Y. T., Hicks, L. D., Minnick, M. F., Dixon, S. E., et al. (2007). Proteomic and immunoblot analyses of Bartonella quintana total membrane proteins identify antigens recognized by sera from infected patients. Infect. Immun. 75, 2548–2561. doi: 10.1128/IAI.01974-06
Boulouis, H. J., Barrat, F., Bermond, D., Bernex, F., Thibault, D., Heller, R., et al. (2001). Kinetics of Bartonella birtlesii infection in experimentally infected mice and pathogenic effect on reproductive functions. Infect. Immun. 69, 5313–5317. doi: 10.1128/IAI.69.9.5313-5317.2001
Buckles, E. L., and McGinnis Hill, E. (2000). Interaction of Bartonella bacilliformis with human erythrocyte membrane proteins. Microb. Pathog. 29, 165–174. doi: 10.1006/mpat.2000.0381
Carroll, J. A., Coleman, S. A., Smitherman, L. S., and Minnick, M. F. (2000). Hemin-binding surface protein from Bartonella quintana. Infect. Immun. 68, 6750–6757. doi: 10.1128/IAI.68.12.6750-6757.2000
Cartwright, J. L., Britton, P., Minnick, M. F., and McLennan, A. G. (1999). The IalA invasion gene of Bartonella bacilliformis encodes a (de)nucleoside polyphosphate hydrolase of the MutT motif family and has homologs in other invasive bacteria. Biochem. Biophys. Res. Commun. 256, 474–479. doi: 10.1006/bbrc.1999.0354
Caswell, C. C., Oliver-Kozup, H., Han, R., Lukomska, E., and Lukomski, S. (2010). Scl1, the multifunctional adhesin of group A Streptococcus, selectively binds cellular fibronectin and laminin, and mediates pathogen internalization by human cells. FEMS Microbiol. Lett. 303, 61–68. doi: 10.1111/j.1574-6968.2009.01864.x
Chenoweth, M. R., Greene, C. E., Krause, D. C., and Gherardini, F. C. (2004). Predominant outer membrane antigens of Bartonella henselae. Infect. Immun. 72, 3097–3105. doi: 10.1128/IAI.72.6.3097-3105.2004
Chomel, B. B., Boulouis, H. J., Breitschwerdt, E. B., Kasten, R. W., Vayssier-Taussat, M., Birtles, R. J., et al. (2009). Ecological fitness and strategies of adaptation of Bartonella species to their hosts and vectors. Vet. Res. 40:29. doi: 10.1051/vetres/2009011
Chomel, B. B., Kasten, R. W., Floyd-Hawkins, K., Chi, B., Yamamoto, K., Roberts-Wilson, J., et al. (1996). Experimental transmission of Bartonella henselae by the cat flea. J. Clin. Microbiol. 34, 1952–1956.
Coleman, S. A., and Minnick, M. F. (2001). Establishing a direct role for the Bartonella bacilliformis invasion-associated locus B (IalB) protein in human erythrocyte parasitism. Infect. Immun. 69, 4373–4381. doi: 10.1128/IAI.69.7.4373-4381.2001
Conyers, G. B., and Bessman, M. J. (1999). The gene, ialA, associated with the invasion of human erythrocytes by Bartonella bacilliformis, designates a nudix hydrolase active on dinucleoside 5'-polyphosphates. J. Biol. Chem. 274, 1203–1206. doi: 10.1074/jbc.274.3.1203
Coutte, L., Alonso, S., Reveneau, N., Willery, E., Quatannens, B., Locht, C., et al. (2003). Role of adhesin release for mucosal colonization by a bacterial pathogen. J. Exp. Med. 197, 735–742. doi: 10.1084/jem.20021153
Dehio, C. (1999). Interactions of Bartonella henselae with vascular endothelial cells. Curr. Opin. Microbiol. 2, 78–82. doi: 10.1016/S1369-5274(99)80013-7
Dehio, C. (2001). Bartonella interactions with endothelial cells and erythrocytes. Trends Microbiol. 9, 279–285. doi: 10.1016/S0966-842X(01)02047-9
Dehio, C. (2004). Molecular and cellular basis of Bartonella pathogenesis. Annu. Rev. Microbiol. 58, 365–390. doi: 10.1146/annurev.micro.58.030603.123700
Dehio, C. (2005). Bartonella-host-cell interactions and vascular tumour formation. Nat. Rev. Microbiol. 3, 621–631. doi: 10.1038/nrmicro1209
Dehio, C. (2008). Infection-associated type IV secretion systems of Bartonella and their diverse roles in host cell interaction. Cell. Microbiol. 10, 1591–1598. doi: 10.1111/j.1462-5822.2008.01171.x
Dehio, C., Meyer, M., Berger, J., Schwarz, H., and Lanz, C. (1997). Interaction of Bartonella henselae with endothelial cells results in bacterial aggregation on the cell surface and the subsequent engulfment and internalisation of the bacterial aggregate by a unique structure, the invasome. J. Cell Sci. 18, 2141–2154.
Dehio, C., and Tsolis, R. M. (2017). Type IV effector secretion and subversion of host functions by Bartonella and Brucella species. Curr. Top. Microbiol. Immunol. 413, 269–295. doi: 10.1007/978-3-319-75241-9_11
Deng, H., Le Rhun, D., Buffet, J. P., Cotte, V., Read, A., Birtles, R. J., et al. (2012). Strategies of exploitation of mammalian reservoirs by Bartonella species. Vet. Res. 43:15. doi: 10.1186/1297-9716-43-15
Deng, H., Pang, Q., Xia, H., Le Rhun, D., Le Naour, E., Yang, C., et al. (2016). Identification and functional analysis of invasion associated locus B (IalB) in Bartonella species. Microb. Pathog. 98, 171–177. doi: 10.1016/j.micpath.2016.05.007
Deng, H. K., Le Rhun, D., Le Naour, E., Bonnet, S., and Vayssier-Taussat, M. (2012a). Identification of Bartonella Trw host-specific receptor on erythrocytes. PLoS ONE 7:e41447. doi: 10.1371/journal.pone.0041447
Deng, H. K., Le Rhun, D., Lecuelle, B., Le Naour, E., and Vayssier-Taussat, M. (2012b). Role of the spleen in Bartonella spp. FEMS Immunol. Med. Microbiol. 64, 143–145. doi: 10.1111/j.1574-695X.2011.00908.x
Derrick, S. C., and Ihler, G. M. (2001). Deformin, a substance found in Bartonella bacilliformis culture supernatants, is a small, hydrophobic molecule with an affinity for albumin. Blood Cells Mol. Dis. 27, 1013–1019. doi: 10.1006/bcmd.2001.0475
Dornand, J., Gross, A., Lafont, V., Liautard, J., Oliaro, J., and Liautard, J. P. (2002). The innate immune response against Brucella in humans. Vet. Microbiol. 90, 383–394. doi: 10.1016/S0378-1135(02)00223-7
Focà, A., Liberto, M. C., Quirino, A., and Matera, G. (2012). Lipopolysaccharides: from erinyes to charites. Mediators Inflamm. 2012:684274. doi: 10.1155/2012/684274
Frank, A. C., Alsmark, C. M., Thollesson, M., and Andersson, S. G. (2005). Functional divergence and horizontal transfer of type IV secretion systems. Mol. Biol. Evol. 22, 1325–1336. doi: 10.1093/molbev/msi124
Gillespie, J. J., Phan, I. Q., Scheib, H., Subramanian, S., Edwards, T. E., Lehman, S. S., et al. (2015). Structural insight into how bacteria prevent interference between multiple divergent type IV secretion systems. MBio 6, e01867–e01815. doi: 10.1128/mBio.01867-15
Gorczynski, R. M., Alexander, C., Bessler, W., Fournier, K., Hoffmann, P., Mach, J. P., et al. (2004). Characterization of an interaction between fetal hemoglobin and lipid A of LPS resulting in augmented induction of cytokine production in vivo and in vitro. Int. Immunopharmacol. 4, 1859–1872. doi: 10.1016/j.intimp.2004.08.006
Groebel, K., Hoelzle, K., Wittenbrink, M. M., Ziegler, U., and Hoelzle, L. E. (2009). Mycoplasma suis invades porcine erythrocytes. Infect. Immun. 77, 576–584. doi: 10.1128/IAI.00773-08
Guptill, L. (2010). Feline bartonellosis. Vet. Clin. North Am. Small Anim. Pract. 40, 1073–1090. doi: 10.1016/j.cvsm.2010.07.009
Guptill, L., Slater, L., Wu, C. C., Lin, T. L., Glickman, L. T., Welch, D. F., et al. (1997). Experimental infection of young specific pathogen-free cats with Bartonella henselae. J. Infect. Dis. 176, 206–216. doi: 10.1086/514026
Harms, A., and Dehio, C. (2012). Intruders below the radar: molecular pathogenesis of Bartonella spp. Clin. Microbiol. Rev. 25, 42–78. doi: 10.1128/CMR.05009-11
Hendrix, L. R. (2000). Contact-dependent hemolytic activity distinct from deforming activity of Bartonella bacilliformis. FEMS Microbiol. Lett. 182, 119–124. doi: 10.1111/j.1574-6968.2000.tb08884.x
Hendrix, L. R., and Kiss, K. (2003). Studies on the identification of deforming factor from Bartonella bacilliformis. Ann. N. Y. Acad. Sci. 990, 596–604. doi: 10.1111/j.1749-6632.2003.tb07433.x
Hoiczyk, E., Roggenkamp, A., Reichenbecher, M., Lupas, A., and Heesemann, J. (2000). Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 19, 5989–5999. doi: 10.1093/emboj/19.22.5989
Iwaki-Egawa, S., and Ihler, G. M. (1997). Comparison of the abilities of proteins from Bartonella bacilliformis and Bartonella henselae to deform red cell membranes and to bind to red cell ghost proteins. FEMS Microbiol. Lett. 157, 207–217. doi: 10.1111/j.1574-6968.1997.tb12775.x
Kabeya, H., Tsunoda, E., Maruyama, S., and Mikami, T. (2003). Immune responses of immunocompetent and immunocompromised mice experimentally infected with Bartonella henselae. J. Vet. Med. Sci. 65, 479–484. doi: 10.1292/jvms.65.479
Kaiser, P. O., Linke, D., Schwarz, H., Leo, J. C., and Kempf, V. A. (2012). Analysis of the BadA stalk from Bartonella henselae reveals domain-specific and domain-overlapping functions in the host cell infection process. Cell. Microbiol. 14, 198–209. doi: 10.1111/j.1462-5822.2011.01711.x
Kaiser, P. O., Riess, T., Wagner, C. L., Linke, D., Lupas, A. N., Schwarz, H., et al. (2008). The head of Bartonella adhesin A is crucial for host cell interaction of Bartonella henselae. Cell. Microbiol. 10, 2223–2234. doi: 10.1111/j.1462-5822.2008.01201.x
Kirjavainen, V., Jarva, H., Biedzka-Sarek, M., Blom, A. M., Skurnik, M., and Meri, S. (2008). Yersinia enterocolitica serum resistance proteins YadA and ail bind the complement regulator C4b-binding protein. PLoS Pathog. 4:e1000140. doi: 10.1371/journal.ppat.1000140
Kloch, A., Wenzel, M. A., Laetsch, D. R., Michalski, O., Welc-Faleciak, R., and Piertney, S. B. (2018). Signatures of balancing selection in toll-like receptor (TLRs) genes - novel insights from a free-living rodent. Sci. Rep. 8:8361. doi: 10.1038/s41598-018-26672-2
Kozakov, D., Hall, D. R., Xia, B., Porter, K. A., Padhorny, D., Yueh, C., et al. (2017). The ClusPro web server for protein-protein docking. Nat. Protoc. 12, 255–278. doi: 10.1038/nprot.2016.169
Kyme, P. A., Haas, A., Schaller, M., Peschel, A., Iredell, J., and Kempf, V. A. (2005). Unusual trafficking pattern of Bartonella henselae-containing vacuoles in macrophages and endothelial cells. Cell. Microbiol. 7, 1019–1034. doi: 10.1111/j.1462-5822.2005.00531.x
Lafontaine, E. R., Cope, L. D., Aebi, C., Latimer, J. L., McCracken, G. H. Jr., and Hansen, E. J. (2000). The UspA1 protein and a second type of UspA2 protein mediate adherence of Moraxella catarrhalis to human epithelial cells in vitro. J. Bacteriol. 182, 1364–1373. doi: 10.1128/JB.182.5.1364-1373.2000
Larrea, D., de Paz, H. D., Arechaga, I., de la Cruz, F., and Llosa, M. (2013). Structural independence of conjugative coupling protein TrwB from its Type IV secretion machinery. Plasmid 70, 146–153. doi: 10.1016/j.plasmid.2013.03.006
Lindroos, H., Vinnere, O., Mira, A., Repsilber, D., Naslund, K., and Andersson, S. G. (2006). Genome rearrangements, deletions, and amplifications in the natural population of Bartonella henselae. J. Bacteriol. 188, 7426–7439. doi: 10.1128/JB.00472-06
Litwin, C. M., and Johnson, J. M. (2005). Identification, cloning, and expression of the CAMP-like factor autotransporter gene (cfa) of Bartonella henselae. Infect. Immun. 73, 4205–4213. doi: 10.1128/IAI.73.7.4205-4213.2005
Malgorzata-Miller, G., Heinbockel, L., Brandenburg, K., van der Meer, J. W., Netea, M. G., and Joosten, L. A. (2016). Bartonella quintana lipopolysaccharide (LPS): structure and characteristics of a potent TLR4 antagonist for in-vitro and in-vivo applications. Sci. Rep. 6:34221. doi: 10.1038/srep34221
Mändle, T., Einsele, H., Schaller, M., Neumann, D., Vogel, W., Autenrieth, I. B., et al. (2005). Infection of human CD34+ progenitor cells with Bartonella henselae results in intraerythrocytic presence of B. henselae. Blood 106, 1215–1222. doi: 10.1182/blood-2004-12-4670
Marignac, G., Barrat, F., Chomel, B., Vayssier-Taussat, M., Gandoin, C., Bouillin, C., et al. (2010). Murine model for Bartonella birtlesii infection: new aspects. Comp. Immunol. Microbiol. Infect. Dis. 33, 95–107. doi: 10.1016/j.cimid.2008.07.011
Matera, G., Liberto, M. C., Joosten, L. A., Vinci, M., Quirino, A., Pulicari, M. C., et al. (2008). The Janus face of Bartonella quintana recognition by Toll-like receptors (TLRs): a review. Eur. Cytokine Netw. 19, 113–118. doi: 10.1684/ecn.2008.0128
Minnick, M. F. (1994). Identification of outer membrane proteins of Bartonella bacilliformis. Infect. Immun. 62, 2644–2648.
Minnick, M. F., and Battisti, J. M. (2009). Pestilence, persistence and pathogenicity: infection strategies of Bartonella. Future Microbiol. 4, 743–758. doi: 10.2217/fmb.09.41
Minnick, M. F., Sappington, K. N., Smitherman, L. S., Andersson, S. G., Karlberg, O., and Carroll, J. A. (2003). Five-member gene family of Bartonella quintana. Infect. Immun. 71, 814–821. doi: 10.1128/IAI.71.2.814-821.2003
Mitchell, S. J., and Minnick, M. F. (1995). Characterization of a two-gene locus from Bartonella bacilliformis associated with the ability to invade human erythrocytes. Infect. Immun. 63, 1552–1562.
Mitchell, S. J., and Minnick, M. F. (1997). A carboxy-terminal processing protease gene is located immediately upstream of the invasion-associated locus from Bartonella bacilliformis. Microbiology 143 (Pt 4), 1221–1233. doi: 10.1099/00221287-143-4-1221
Mosepele, M., Mazo, D., and Cohn, J. (2012). Bartonella infection in immunocompromised hosts: immunology of vascular infection and vasoproliferation. Clin. Dev. Immunol. 2012:612809. doi: 10.1155/2012/612809
Mullins, K. E., Hang, J., Clifford, R. J., Onmus-Leone, F., Yang, Y., Jiang, J., et al. (2017). Whole-genome analysis of Bartonella ancashensis, a novel pathogen causing verruga peruana, rural ancash region, Peru. Emerg. Infect. Dis. 23, 430–438. doi: 10.3201/eid2303.161476
Okaro, U., Addisu, A., Casanas, B., and Anderson, B. (2017). Bartonella species, an emerging cause of blood-culture-negative endocarditis. Clin. Microbiol. Rev. 30, 709–746. doi: 10.1128/CMR.00013-17
O'Rourke, F., Schmidgen, T., Kaiser, P. O., Linke, D., and Kempf, V. A. (2011). Adhesins of Bartonella spp. Adv. Exp. Med. Biol. 715, 51–70. doi: 10.1007/978-94-007-0940-9_4
Parrow, N. L., Abbott, J., Lockwood, A. R., Battisti, J. M., and Minnick, M. F. (2009). Function, regulation, and transcriptional organization of the hemin utilization locus of Bartonella quintana. Infect. Immun. 77, 307–316. doi: 10.1128/IAI.01194-08
Popa, C., Abdollahi-Roodsaz, S., Joosten, L. A., Takahashi, N., Sprong, T., Matera, G., et al. (2007). Bartonella quintana lipopolysaccharide is a natural antagonist of Toll-like receptor 4. Infect. Immun. 75, 4831–4837. doi: 10.1128/IAI.00237-07
Pulliainen, A. T., and Dehio, C. (2012). Persistence of Bartonella spp. stealth pathogens: from subclinical infections to vasoproliferative tumor formation. FEMS Microbiol. Rev. 36, 563–599. doi: 10.1111/j.1574-6976.2012.00324.x
Quebatte, M., Dehio, M., Tropel, D., Basler, A., Toller, I., Raddatz, G., et al. (2010). The BatR/BatS two-component regulatory system controls the adaptive response of Bartonella henselae during human endothelial cell infection. J. Bacteriol. 192, 3352–3367. doi: 10.1128/JB.01676-09
Raetz, C. R., and Whitfield, C. (2002). Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700. doi: 10.1146/annurev.biochem.71.110601.135414
Raoult, D. (2007). From cat scratch disease to Bartonella henselae infection. Clin. Infect. Dis. 45, 1541–1542. doi: 10.1086/523716
Resto-Ruiz, S., Burgess, A., and Anderson, B. E. (2003). The role of the host immune response in pathogenesis of Bartonella henselae. DNA Cell Biol. 22, 431–440. doi: 10.1089/104454903767650694
Riess, T., Andersson, S. G., Lupas, A., Schaller, M., Schafer, A., Kyme, P., et al. (2004). Bartonella adhesin a mediates a proangiogenic host cell response. J. Exp. Med. 200, 1267–1278. doi: 10.1084/jem.20040500
Ruiz-Ranwez, V., Posadas, D. M., Estein, S. M., Abdian, P. L., Martin, F. A., and Zorreguieta, A. (2013). The BtaF trimeric autotransporter of Brucella suis is involved in attachment to various surfaces, resistance to serum and virulence. PLoS ONE 8:e79770. doi: 10.1371/journal.pone.0079770
Saenz, H. L., Engel, P., Stoeckli, M. C., Lanz, C., Raddatz, G., Vayssier-Taussat, M., et al. (2007). Genomic analysis of Bartonella identifies type IV secretion systems as host adaptability factors. Nat. Genet. 39, 1469–1476. doi: 10.1038/ng.2007.38
Sander, A., Zagrosek, A., Bredt, W., Schiltz, E., Piemont, Y., Lanz, C., et al. (2000). Characterization of Bartonella clarridgeiae flagellin (FlaA) and detection of antiflagellin antibodies in patients with lymphadenopathy. J. Clin. Microbiol. 38, 2943–2948.
Scherer, D. C., DeBuron-Connors, I., and Minnick, M. F. (1993). Characterization of Bartonella bacilliformis flagella and effect of antiflagellin antibodies on invasion of human erythrocytes. Infect. Immun. 61, 4962–4971.
Scherler, A., Jacquier, N., and Greub, G. (2017). Chlamydiales, Anaplasma and Bartonella: persistence and immune escape of intracellular bacteria. Microbes Infect. 20, 416–423. doi: 10.1016/j.micinf.2017.11.002
Schmidgen, T., Kaiser, P. O., Ballhorn, W., Franz, B., Gottig, S., Linke, D., et al. (2014). Heterologous expression of Bartonella adhesin A in Escherichia coli by exchange of trimeric autotransporter adhesin domains results in enhanced adhesion properties and a pathogenic phenotype. J. Bacteriol. 196, 2155–2165. doi: 10.1128/JB.01461-13
Schröder, G., and Dehio, C. (2005). Virulence-associated type IV secretion systems of Bartonella. Trends Microbiol. 13, 336–342. doi: 10.1016/j.tim.2005.05.008
Schulein, R., Guye, P., Rhomberg, T. A., Schmid, M. C., Schroder, G., Vergunst, A. C., et al. (2005). A bipartite signal mediates the transfer of type IV secretion substrates of Bartonella henselae into human cells. Proc. Natl. Acad. Sci. U.S.A. 102, 856–861. doi: 10.1073/pnas.0406796102
Schülein, R., Seubert, A., Gille, C., Lanz, C., Hansmann, Y., Piemont, Y., et al. (2001). Invasion and persistent intracellular colonization of erythrocytes. J. Exp. Med. 193, 1077–1086. doi: 10.1084/jem.193.9.1077
Seubert, A., Hiestand, R., de la Cruz, F., and Dehio, C. (2003). A bacterial conjugation machinery recruited for pathogenesis. Mol. Microbiol. 49, 1253–1266. doi: 10.1046/j.1365-2958.2003.03650.x
Seubert, A., Schulein, R., and Dehio, C. (2002). Bacterial persistence within erythrocytes: a unique pathogenic strategy of Bartonella spp. Int. J. Med. Microbiol. 291, 555–560. doi: 10.1078/1438-4221-00167
St Geme, J. W. III., and Cutter, D. (2000). The Haemophilus influenzae Hia adhesin is an autotransporter protein that remains uncleaved at the C terminus and fully cell associated. J. Bacteriol. 182, 6005–6013. doi: 10.1128/JB.182.21.6005-6013.2000
Tu, N., Carroll, R. K., Weiss, A., Shaw, L. N., Nicolas, G., Thomas, S., et al. (2017). A family of genus-specific RNAs in tandem with DNA-binding proteins control expression of the badA major virulence factor gene in Bartonella henselae. Microbiologyopen 6:e00420. doi: 10.1002/mbo3.420
Vayssier-Taussat, M., Le Rhun, D., Bonnet, S., and Cotte, V. (2009). Insights in Bartonella host specificity. Ann. N. Y. Acad. Sci. 1166, 127–132. doi: 10.1111/j.1749-6632.2009.04531.x
Vayssier-Taussat, M., Le Rhun, D., Deng, H. K., Biville, F., Cescau, S., Danchin, A., et al. (2010). The Trw type IV secretion system of Bartonella mediates host-specific adhesion to erythrocytes. PLoS Pathog. 6:e1000946. doi: 10.1371/journal.ppat.1000946
Walker, T. S., and Winkler, H. H. (1981). Bartonella bacilliformis: colonial types and erythrocyte adherence. Infect. Immun. 31, 480–486.
Weiss, G., and Schaible, U. E. (2015). Macrophage defense mechanisms against intracellular bacteria. Immunol. Rev. 264, 182–203. doi: 10.1111/imr.12266
Wollmann, P., Zeth, K., Lupas, A. N., and Linke, D. (2006). Purification of the YadA membrane anchor for secondary structure analysis and crystallization. Int. J. Biol. Macromol. 39, 3–9. doi: 10.1016/j.ijbiomac.2005.11.009
Wu, H. Y., Chen, C. Y., and Lai, E. M. (2014). Expression and functional characterization of the Agrobacterium VirB2 amino acid substitution variants in T-pilus biogenesis, virulence, and transient transformation efficiency. PLoS ONE 9:e101142. doi: 10.1371/journal.pone.0101142
Xu, Y. H., Lu, Z. Y., and Ihler, G. M. (1995). Purification of deformin, an extracellular protein synthesized by Bartonella bacilliformis which causes deformation of erythrocyte membranes. Biochim. Biophys. Acta 1234, 173–183. doi: 10.1016/0005-2736(94)00271-P
Yang, J., Yan, R., Roy, A., Xu, D., Poisson, J., and Zhang, Y. (2015). The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8. doi: 10.1038/nmeth.3213
Yuan, C., Zhu, C., Wu, Y., Pan, X., and Hua, X. (2011). Bacteriological and molecular identification of Bartonella species in cats from different regions of China. PLoS Negl. Trop. Dis. 5:e1301. doi: 10.1371/journal.pntd.0001301
Zähringer, U., Lindner, B., Knirel, Y. A., van den Akker, W. M., Hiestand, R., Heine, H., et al. (2004). Structure and biological activity of the short-chain lipopolysaccharide from Bartonella henselae ATCC 49882T. J. Biol. Chem. 279, 21046–21054. doi: 10.1074/jbc.M313370200
Keywords: bartonellosis, erythrocyte interactions, adhesion and invasion, replicate and persist, pathogenesis factors
Citation: Deng H, Pang Q, Zhao B and Vayssier-Taussat M (2018) Molecular Mechanisms of Bartonella and Mammalian Erythrocyte Interactions: A Review. Front. Cell. Infect. Microbiol. 8:431. doi: 10.3389/fcimb.2018.00431
Received: 12 September 2018; Accepted: 30 November 2018;
Published: 12 December 2018.
Edited by:
Eric Ghigo, IHU Mediterranee Infection, FranceReviewed by:
Michael F. Minnick, University of Montana, United StatesVictoria A. Blaho, Sanford Burnham Prebys Medical Discovery Institute (SBP), United States
Copyright © 2018 Deng, Pang, Zhao and Vayssier-Taussat. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongkuan Deng, aG9uZ2t1YW5kZW5nQGdtYWlsLmNvbQ==
 Hongkuan Deng
Hongkuan Deng Qiuxiang Pang1
Qiuxiang Pang1 Muriel Vayssier-Taussat
Muriel Vayssier-Taussat