- State Key Laboratory of Veterinary Etiological Biology, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Lanzhou, China
Clostridium perfringens is an important human and animal pathogen that is the primary causative agent of necrotizing enteritis and enterotoxemia in many types of animals; it causes traumatic gas gangrene in humans and animals and is associated with cases of food poisoning in humans. C. perfringens produces a variety of toxins as well as many enzymes, including three sialidases, NanH, NanI, and NanJ. Sialidases could be important virulence factors that promote the pathogenesis of C. perfringens. Among them, NanI promotes the colonization of C. perfringens in the intestinal tract and enhances the cytotoxic activity and association of several major C. perfringens toxins with host cells. In recent years, studies on the structure and functions of sialidases have yielded interesting results, and the functions of sialic acid and sialidases in bacterial pathogenesis have become a hot research topic. An in-depth understanding and additional studies of sialidases will further elucidate mechanisms of C. perfringens pathogenesis and could promote the development and clinical applications of sialidase inhibitors. This article reviews the structural characteristics, expression regulation, roles of sialidases in C. perfringens pathogenesis, and effects of their inhibitors.
Introduction
Clostridium perfringens is a gram-positive anaerobic bacillus that is widely present in nature, especially in the soil and intestines of human and animals. C. perfringens can infect humans and various animals (primarily cows and sheep), causing food poisoning in humans, necrotizing enteritis, and enterotoxemia in animals and traumatic gas gangrene in both humans and animals, as well as other diseases. These diseases not only seriously threaten the health of humans and animals but also cause enormous economic losses (Parent et al., 2017; Silva et al., 2018).
At present, more than 20 C. perfringens toxins have been identified, and there may be additional toxins that have yet to be identified (Hatheway, 1990; Petit et al., 1999; Amimoto et al., 2007; Keyburn et al., 2008; Yonogi et al., 2014; Irikura et al., 2015; Mehdizadeh Gohari et al., 2016). Based on the primary toxins produced by C. perfringens and differences in pathogenesis among strains, C. perfringens strains are divided into seven types according to a recent revision (Type A–G, Table 1; Rood et al., 2018). The α-toxin gene is the most prevalent toxin gene carried among C. perfringens and is encoded on chromosome. The genes encoding the beta, epsilon, iota, and NetB toxins are plasmid-borne, whereas CPE can be encoded either on the chromosome or on a plasmid (Hassan et al., 2015). Of the seven types of C. perfringens, the type F strains are considered the most common pathogen in humans (Rood et al., 2018).
So far many studies have shown that sialidases play important roles in the pathogenesis of pathogenic bacteria, providing nutrition for bacteria and promoting bacterial colonization, bacterial adhesion, bacterial internalization, biofilm formation, and the binding of toxins to host cells (Traving and Schauer, 1998; Vimr et al., 2004; Tong et al., 2005; Pastoriza Gallego and Hulen, 2006; Soong et al., 2006; Severi et al., 2007; Parker et al., 2009; Thompson et al., 2009; Trappetti et al., 2009; Banerjee et al., 2010; King, 2010; Honma et al., 2011; Li et al., 2011; Brittan et al., 2012; Lewis and Lewis, 2012; Awad et al., 2016; Blanchette et al., 2016). Recently, a study found that NanA, NanB, and NanC increased the interaction of S. pneumoniae with human airway epithelial cells (Janesch et al., 2018). Some pathogens also use sialic acid to coat their cell surface, flagella, capsule polysaccharides, or lipopolysaccharides, concealing themselves to evade the host immune system (Severi et al., 2007). Free sialic acid also participates in capsule formation in Neisseria meningitidis, Escherichia coli, and Porphyromonas gingivalis and defends cells against the immune responses of the host (Vimr et al., 2004; Allen et al., 2005), although the mechanism associated with this activity is unclear. Meningococcal capsule can block the killing effect of human serum, which may be due to sialic acids concealing the membrane attack complex on the bacterial cell membrane (Vimr and Lichtensteiger, 2002). In addition, a Vibrio cholerae sialidase can hydrolyze ganglioside on the surfaces of intestinal mucosal epithelial cells, and ganglioside GM1 binds the enterotoxin of V. cholerae to disrupt the normal function of cellular ion channels, leading to dehydration and other symptoms in the human body (Vimr and Lichtensteiger, 2002). In a recent study, sialidases from microorganisms in the cervix and vagina were observed to modify gonococci and enhance the successful transmission of the pathogen to men (Ketterer et al., 2016). Therefore, sialidases play significant roles in the survival and pathogenesis of bacteria. Furthermore, recent studies have shown that a sialidase deficiency in P. gingivalis can weaken the activation of CR3 in macrophages, reduce the inhibition of lncRNA GAS5 by CR3, and induce less miR-21 and more IL-12 production in macrophages. These results suggest that the inhibition of sialidase activity in P. gingivalis renders the bacteria easier to be cleared by macrophages (Yang et al., 2018), and this discovery will open up a new direction for the prevention and treatment of chronic periodontitis. Sialidases are also a marker of some diseases (Liu et al., 2018). Similarly, sialidases can also contribute to important steps in the pathogenesis of C. perfringens, and studies on this activity have made substantial progress. This article reviews the structural characteristics, expression regulation, and roles of sialidases in the pathogenesis of C. perfringens.
Sialidases
Sialidases, also known as neuraminidases (E.C.3.2.1.18), hydrolyze the α-glycoside bond of the terminal sialic acid in glycoproteins and glycolipids to produce free sialic acid (Traving and Schauer, 1998; Vimr et al., 2004; Severi et al., 2007; Lewis and Lewis, 2012). Hydrolytic-sialidases usually have wide substrate specificity and cleave α2-3-, α2-6-, and α2-8-linked terminal sialic acids (Juge et al., 2016). Sialidases are the key enzymes responsible for the catabolism of oligosaccharides containing sialic acid (Lewis and Lewis, 2012).
Sialidases participate in cell metabolism, adhesion, proliferation, immune functions, and infectious processes under various pathological and physiological conditions (Imai and Kawaoka, 2012; Varki and Gagneux, 2012; Walther et al., 2013; de Graaf and Fouchier, 2014; Arabyan et al., 2017; Miyagi et al., 2018). Sialidases are known to be produced by a variety of viruses and bacterial pathogens, including influenza viruses, V. cholerae, S. pneumoniae, E. coli, Staphylococcus aureus, and C. perfringens (Traving and Schauer, 1998; Vimr et al., 2004; Severi et al., 2007; Nishiyama et al., 2018). Therefore, sialidases are virulence factors involved in the pathogenesis of infections (Rohmer et al., 2011; Lewis and Lewis, 2012; Li et al., 2016).
Molecular and Structural Characteristics of Sialidases From C. perfringens
C. perfringens produces three sialidases, NanH, NanI, and NanJ (Li and McClane, 2014a), all of which are encoded by genes located on different regions of the chromosome (Shimizu et al., 2002; Myers et al., 2006). They are classified in the GH family 33 (GH33) of the CAZy classification (Lombard et al., 2014). Sequencing analysis of C. perfringens sialidase ORFs showed that the similarity of nanJ, nanI, and nanH gene sequences from different C. perfringens strains ranges from 96 to 100, 98 to 100, and 93 to 100%, respectively (Shimizu et al., 2002; Myers et al., 2006; Li et al., 2011). The nanI and nanJ genes have 2–3 hypothetical transcription initiation sites (Therit et al., 2015), which are located within 500 bp of the start codon of these genes. These diverse promoters allow for sialidase expression to be regulated by a variety of different regulatory factors (Therit et al., 2015). The nanI and nanJ promoter regions and the hypothetical promoter region of nanH in strain SM101 contain a conserved 14-bp sequence (a consensus sequence of 5′-GAAAAATATTTTC-3′). These conserved repetitive sequences are located in the transcription initiation region of the promoters and may function as recognition sequences of transcription factors that regulate the production of C. perfringens sialidases.
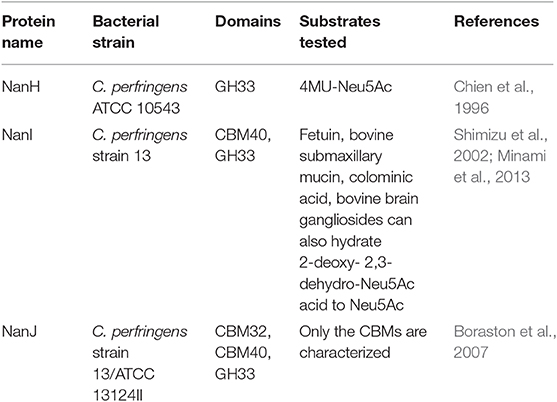
The small protein NanH (43 kDa) lacks a signal peptide and is located in the cytoplasm during logarithmic growth (Li et al., 2011; Li and McClane, 2014a). In contrast, the NanI (77 kDa) and NanJ (129 kDa) are secreted into the extracellular matrix (Li and McClane, 2014a). These three sialidases have similarities and differences in structure (Figure 1). The amino acid sequences of the catalytic domains of these sialidases are conserved among sialidases (Boraston et al., 2007). NanH contains only the catalytic domain, while NanI has a carbohydrate-binding domain (CBM40) in addition to the catalytic domain (Boraston et al., 2007). The structure of NanJ is complex, consisting of a catalytic domain, two carbohydrate binding domains (CBM32 and CBM40), and three additional auxiliary regions (Boraston et al., 2007). These carbohydrate-binding regions increase the binding affinity of NanI and NanJ toward their polyvalent substrates. The characteristics of sialidases from C. perfringens are summarized in Table 2. The structure of the catalytic domain (residues 243–694) of NanI has been studied (Newstead et al., 2004, 2008), showing that NanI folds into two distinct domains: a regular six-blade β-propeller catalytic domain formed by residues 243–359 and 429–691 and a small β-barrel domain formed by residues 360–428 (Figure 2) (Newstead et al., 2008). The carbohydrate binding domains CBM32 and CBM40 of NanJ recognize Gal and Neu5Ac, respectively (Boraston et al., 2007).
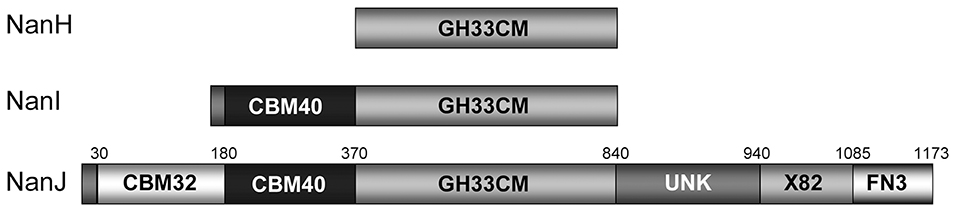
Figure 1. Modular organization of the clostridial sialidases (Boraston et al., 2007) Copyright © 2007, with permission from American Chemical Society. Amino acid numbers corresponding to the module boundaries are shown above the schematic of NanJ (Boraston et al., 2007).
Figure 2. Overall structure of the NanI sialidase. Reprinted from Newstead et al. (2008) Copyright © 2008, with permission from the American Society for Biochemistry and Molecular Biology, Inc. (A,B) Represent orthogonal views of a comparison of the fold of NanI (yellow) with the leech sialidase (blue). Sialic acid is drawn as spheres to locate the active site. (C,D) Show a surface representation of NanI, in the same orientations as above, colored according to electrostatic potential from −7 to +7 kT/e, calculated using APBS.
The Enzymatic Characteristics of Three C. perfringens Sialidases
Most C. perfringens strains produce three sialidases, NanH, NanI, and NanJ, while some produce only one or two of these sialidases. The activity of NanI typically accounts for 70% of the total extracellular sialidase activity in strains that produce the three types of sialidase (Chiarezza et al., 2009; Li and McClane, 2014a). To study the properties of the three enzymes, those sialidase genes were inactivated in C. perfringens type D strain CN3718 to construct a series of isogenic mutants (Li and McClane, 2014a). The results showed that NanJ and NanH were most active at 37 and 43°C, respectively. These two sialidases showed low activity at 48°C, while the activity of NanI increased steadily as the temperature increased to 48°C. At 25°C, the activity of the three sialidases was relatively low. After incubating at 60°C for 5 min in the same buffer, the loss of enzyme activity of NanI in the culture supernatant was ~50%, while that of NanJ and NanH exceeded 80%. Thus, compared to NanJ and NanH, NanI exhibits higher heat resistance (Li and McClane, 2014a).
The activity of the three C. perfringens sialidases was the highest at pH 5, and each enzyme exhibits different sensitivity to various metal ions (Li and McClane, 2014a). Fe2+, Mn2+, and Mg2+ were found to enhance NanI enzyme activity, while Fe3+ and Zn2+ decreased NanI enzyme activity. More metal ions (Fe2+, Mn2+, Co2+, Mg2+, Ni2+, and Zn2+) increased NanJ activity, only Fe3+ decreased NanJ activity. In contrast, for NanH activity, all metal ions mentioned above, except Mg2+, were found to decrease its activity. Furthermore, unlike sialidases from Streptococcus, those of C. perfringens are sensitive to p-chloromethylbenzoate, which reacts with the sulfhydryl groups of proteins. Among the three enzymes, NanJ is the least sensitive to p-chloromethylbenzoate, and 10 mM p-chloromethylbenzoate can inhibit the activity of all three sialidases. EDTA has a minor inhibitory effect on NanI, while NanH and NanJ are not affected (Li and McClane, 2014a). These results further confirm differences between the activities of the C. perfringens sialidases.
The three C. perfringens sialidases exhibit different substrate preferences. NanI showed preferential activity in the order of α-2,3 > α-2,6 > α-2,8 linkages. NanJ activities showed a preference for α-2,6 > α-2,8 > α-2,3 sialic acid linkages. NanH activities showed a preference for α-2,8 > α-2,3 > α-2,6 linkages (Li and McClane, 2014a). The diversity of these preferences suggests that these three sialidases in combination can hydrolyze and release free sialic acid from complex substrates (Li and McClane, 2014a).
Regulation of C. perfringens Sialidase Expression
Previous studies have reported that the addition of free sialic acid to culture medium could induce extracellular sialidase activity in C. perfringens (Nees and Schauer, 1974), suggesting the presence of a specific regulatory system that responds to sialic acid and regulates the transcription of sialidase-encoding genes. Subsequent studies showed that multiple proteins are involved in regulating sialic acid-related gene expression (Figure 3). It has been demonstrated that VirS/VirR, ReeS/ReeR, RevS/RevR, CodY, and NanR directly or indirectly affect the production of sialidases (Ohtani et al., 2010; Hiscox et al., 2011, 2013; Li et al., 2013; Therit et al., 2015). The VirS/VirR bicomponent signal transduction system indirectly upregulates the expression of nanI and nanJ (Ohtani et al., 2010), where VirS first upregulates the expression of the vrr gene (encoding an RNA (VR-RNA) regulated by VirR), and then this regulatory RNA regulates the expression of sialidase-encoding genes (Ohtani et al., 2010). The RevS/RevR system directly or indirectly positively regulates nanJ expression and negatively regulates that of nanI. However, since more than 100 genes are differentially expressed in revR mutants, the regulation of sialidase genes by this system is likely indirect (Hiscox et al., 2011).
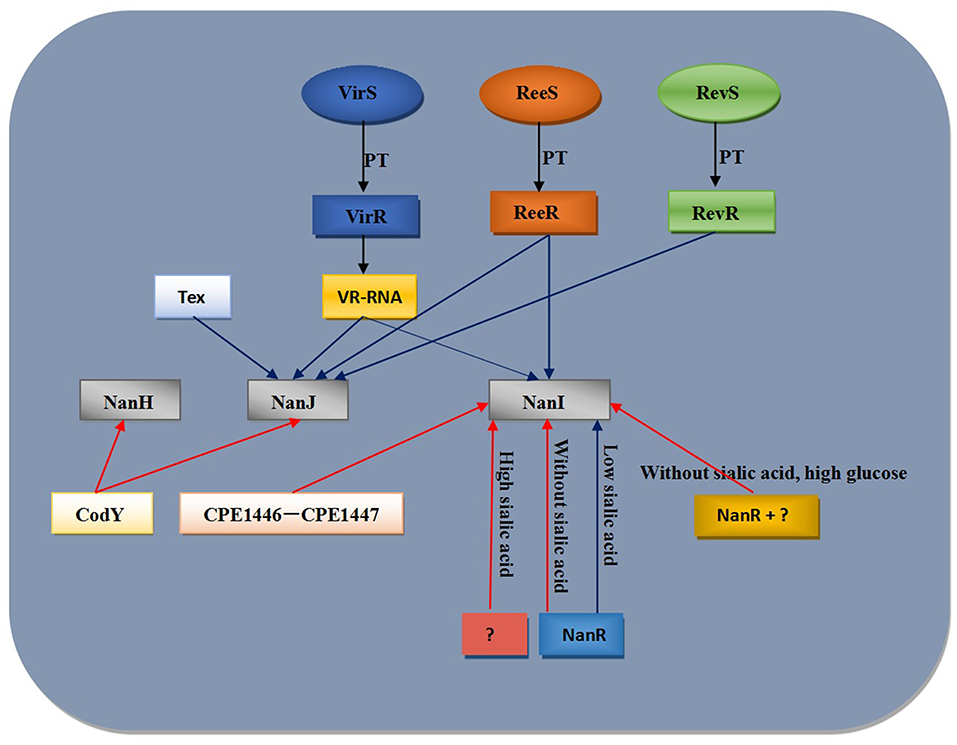
Figure 3. Model for regulation of C. perfringens sialidase expression. Blue lines indicate positive regulation of sialidase expression while red lines indicate negative regulation regulation of sialidase expression. PT, phosphotransfer.
Abe et al. (2010) observed that an RNA binding protein called Tex (CPE2168) affected nanJ mRNA levels in a non-sialic acid dependent manner. However, it is important to note that the results obtained with tex mutant have not been confirmed by complementation. And another study reported that the expression of nanI gene was up-regulated in both a CPE1446 mutant and a CPE1447-CPE1446 deletion mutant. Complementation of CPE1446 and CPE1447 revealed that a heterologous complex of the proteins CPE1446 and CPE1447 negatively regulated nanI expression in a non-sialic acid-dependent manner (Obana and Nakamura, 2011). In the absence of sialic acid in the growth medium, the ReeS/ReeR system positively regulates the transcription of nanI and nanJ (Hiscox et al., 2013). Therefore, there are additional sialic acid specific transcription regulatory factors that have yet to be identified in C. perfringens.
NanR is a member of the RpiR transcription factor family. NanR finely regulates the generation and use of sialic acids to support sporulation and CPE production (Mi et al., 2018). There are six NanR binding sites in the nanI promoter region (Therit et al., 2015). When C. perfringens is grown in the absence of sialic acid, NanR binds to some or all of the binding sites in the nanI promoter. NanR represses the production of NanI and the expression of sialic acid metabolism-related proteins (Therit et al., 2015). However, because there are similar NanR binding sites near the nan operon, NanR may reduce the expression of this operon in the absence of sialic acid (Mi et al., 2018). In the presence of free sialic acid, sialic acid from glycoconjugates is metabolized by C. perfringens to produce ManNAc-6P, which binds to NanR to relieve the inhibition of nanI expression (or possibly that of the nan operon). Another unknown repressor that may reduce nanI expression in the presence of high concentration of sialic acid. Moreover, the expression of nanI decreases in the presence of high glucose concentrations, suggesting that in addition to NanR, there is an additional repressor that represses nanI expression. Whether this repressor is the same as that repressing the expression of nanI in the presence of a high concentration of sialic acid remains to be determined (Li et al., 2017). Since NanI can affect the expression of ccpA and codY (Li et al., 2015), the CcpA and CodY regulatory proteins may be candidates for the aforementioned nanI repressors, but that remains to be verified. Furthermore, an investigation using a codY null mutant of the type D C. perfringens strain CN3718 showed that CodY repressed the production of NanJ and NanH, while failing to affect the production of NanI (Li et al., 2013). Thus, sialidases are regulated by multiple systems, and the relationship between these systems awaits further elucidation.
Potential Role of Sialidases in C. Perfringens Pathogenesis
Effects of Sialidase on C. perfringens Growth and Survival
The ability of bacteria to grow in the gut requires tenacity, as the competition for nutrition among intestinal microbes is extremely intense (Tailford et al., 2015). Particularly during intestinal diseases, the damaging effect of diarrhea can also reduce the concentration of enteral nutrients. One of the coping strategies bacteria use to respond to limited enteral nutrients is the production of sialidases (Severi et al., 2007), which enable sialic acid to be obtained from the host as a source of carbon, nitrogen, amino acids, and energy. Sialic acid is typically transported into bacterial cells by a sialic acid transporter (NanT), sialic acid-specific subfamily of TRAP transporters (SiaPQM), or an ATP binding transporter (SatABCD), after which it is metabolized to fructose-6-P (Vimr et al., 2004).
C. perfringens was one of the first bacteria to have been shown to be able to use sialic acid as a carbon source (Nees and Schauer, 1974). NanI and NanJ have been shown to cause Caco-2 cells (human intestinal-like cells) to release sialic acid, while NanH does not have this activity (Li and McClane, 2014b). A recent study (Li and McClane, 2018) has shown that C. perfringens strain F4969, a non-food-borne human intestinal infectious strain, can use mucin or Caco-2 cells to support its growth and survival. In addition, natural levels of NanI can enhance the growth and survival of bacteria in the gut, in part due to the ability of NanI to release free sialic acid from mucin or sialic acid-modified macromolecules from host cells to support bacterial survival. Furthermore, NanI exposes the underlying carbohydrates and amino acids by removing the terminal sialic acids from macromolecules, allowing other glycoside hydrolases or proteases to hydrolyze and release nutrients for bacterial growth and utilization. However, NanI is not a major contributor to the early logarithmic growth of this strain. When bacteria enter the late logarithmic growth period and encounter more restricted nutrient levels, they seek alternative nutrients by increasing the production of sialidases, especially NanI. Moreover, free sialic acids produced by NanI allow other enzymes to produce carbohydrates and amino acids for bacterial growth. Thus, other factors may act synergistically with NanI to enhance the growth of C. perfringens (Li and McClane, 2018).
Previous in vitro studies have shown that NanI promotes the growth and survival of bacteria, facilitating bacterial colonization in the gut (Li and McClane, 2014b). A recent in vivo study showed that the type F NFD strain F4969 could survive for at least 4 days in the small intestine, caecum, and colon of mice (Navarro et al., 2018). However, when the mice were infected with the nanI mutant, the number of nanI mutant recovered from each intestinal segment was significantly lower than that observed for the wild-type strain. In addition, the mutants were completely cleared from the small intestine on the 4th day, while the complementation of the nanI mutation restored the colonization ability of this strain. When mice were inoculated simultaneously with the same nanI null mutant and a nanI-producing strain, the number of mutant bacteria in any of the assayed intestinal segments returned to the level of the wild-type strain F4969. These results showed for the first time that NanI is an important contributor for the colonization of NFD strains in the intestinal tract, clearly demonstrating that sialidases produced by bacterial pathogens can enhance the colonization of bacteria in the intestinal tract and are an important contributor to chronic intestinal infections (Navarro et al., 2018). This result also provides a reasonable explanation for the presence of the NanI gene in C. perfringens strains that persist and colonize the intestinal tract as well as the absence of the nanI gene in C. perfringens type F strains carrying a chromosomal cpe gene, which cause acute infections.
Although some roles for sialidases in intestinal infections caused by C. perfringens have been elucidated, the effect of sialidase on the growth and colonization of C. perfringens in parenteral infections is unclear. A study showed that a nanJ and nanI double null mutant of C. perfringens strain 13 remained virulent in a murine myonecrosis model (Chiarezza et al., 2009). This result indicates that Neu5Ac metabolism is not necessary for the growth or colonization of C. perfringens in muscle tissue (Chiarezza et al., 2009). However, this result could not exclude the possibility of some subtle effects of sialidases on the colonization or growth of C. perfringens because the mouse myonecrosis model requires a large number of inoculum, which may mask subtle effects (Chiarezza et al., 2009). Thus, the role of sialidases in parenteral infections caused by C. perfringens awaits further study.
Effects of Sialidase on C. perfringens Adhesion
The type D C. perfringens strain CN3718 adheres weakly to fibroblasts and renal cells, whereas this strain can adhere to cultured Caco-2 cells. Compared with a triple mutant strain that does not produce any sialidases, the wild-type strain CN3718 exhibits significantly increased adhesion to Caco-2 cells. Complementation studies showed that, of the three sialidases made by CN3718, restoring NanI production to the triple sialidase mutant yielded the greatest enhancement of adherence. Thus, NanI can significantly promote the specific adhesion of the CN3718 strain to Caco-2 cells and plays a major role in enhancing the adhesion of this strain to Caco-2 cells among the three sialidases (Li et al., 2011).
The process of C. perfringens adhesion to host cells can be divided into two steps. First, secreted NanI specifically modifies the surface of host cells, after which an unknown adhesin on the surface of C. perfringens binds to an unknown receptor on the surface of intestinal epithelial cells. The affinity of the C. perfringens CN3718 adhesin to some mammalian cells (such as Caco-2 and HT-29 intestinal cells) is higher than that observed to non-intestinal cell lines (Li et al., 2011). Thus, NanI can modify the surface of host cells to specifically increase the adhesion of CN3718.
Sialidases also non-specifically enhance the adhesion of C. perfringens to host cells, primarily due to the negative charges of sialic acids at the distal end of carbohydrate chains (Severi et al., 2007; Lewis and Lewis, 2012). Moreover, terminal sialic acids can destroy the integrity of the endothelial barrier. The treatment of monolayers of epithelial cells with C. perfringens sialidases results in barrier damage, which promotes the ability of C. perfringens to approach and adhere epithelial cells (Cioffi et al., 2012). The non-specific interaction between secreted NanI and electrical charges associated with the an intact epithelial barrier contributes to the enhancement of toxin binding and the colonization of C. perfringens.
Studies on the mechanisms of the adherence of C. perfringens to host cells and tissues are very limited (Jost et al., 2006; Hitsumoto et al., 2014; Katayama et al., 2015). Although NanI can significantly promote the specific adhesion of CN3718 to Caco-2 cells, NanI itself is not the major adhesin (Li et al., 2013). Thus, the adhesin used by C. perfringens in vivo remains unclear.
Effects of Sialidases on the Cytotoxicity of C. perfringens Toxins
In addition to affecting bacterial colonization, NanI can enhance the activity of α-toxin that causes gas gangrene (Flores-Díaz et al., 2005; Chiarezza et al., 2009), while also increasing the cytotoxicity of β- and ε-toxins and NetF and CPE toxin associated with gastrointestinal diseases (Li et al., 2011; Mehdizadeh Gohari et al., 2018; Theoret et al., 2018). Although many C. perfringens pathogenic strains only produce a small amount of CPB, CPE, or ETX (Collie et al., 1998; Sayeed et al., 2005; Fisher et al., 2006; Fernandez-Miyakawa et al., 2007; Ma et al., 2014), and the enhancement effect of natural levels of NanI on CPB, CPE and ETX is relatively moderate (1.5–2-fold), this activity may be important for the pathogenesis of C. perfringens during enteritis or enterotoxemia. The primary mechanism by which NanI enhances the cytotoxicity of CPB, CPE, and ETX is to enhance the binding of toxins to host cells. Because CPE, CPB, and ETX do not share the same receptors, the enhancement of the binding of toxins to host cells induced by NanI is essentially non-specific. This enhancement may involve one or more mechanisms, one of which is that most of the charges on mammalian cell surfaces are provided by sialic acids, and NanI reduces electrostatic repulsion and enhances toxin binding by removing surface sialic acid residues. Alternatively, NanI may facilitate the accessibility of the toxins to their receptors by removing sialic acids from the toxin receptors. A third possibility is that NanI may enhance toxin binding by trimming glycoproteins or glycolipids adjacent to toxin receptors, removing some of the sialic acids that block the toxin receptors. In addition, sialidases can increase paracellular permeability (Cioffi et al., 2012). Finally, NanI makes toxins more accessible to receptors on the basolateral surface of host cells, leading to increased toxin binding. The mechanism(s) by which NanI enhances the cytotoxicity of CPB, CPE, and ETX requires additional investigations (Theoret et al., 2018).
Hydrolysis and Activation of Sialidases
When intestinal diseases occur, the proteins such as ETX and CPE toxins secreted by C. perfringens contact host proteases (such as trypsin) in the intestinal cavity and become hydrolyzed and activated (Hanna et al., 1992; Freedman et al., 2014). NanI can also be hydrolyzed and activated by trypsin, whereas NanJ can't be activated. The activation of NanI by trypsin may promote its activity (Li et al., 2011; Li and McClane, 2014a). The activation of trypsin by NanI promotes intestinal diseases caused by C. perfringens (Theoret et al., 2018). Recently, purified chymotrypsin has been shown to activate NanI, and it is worth mentioning that small intestine fluid can activate NanI in vitro. During the course of intestinal diseases, NanI is produced in the gastrointestinal tract and is activated through contact with fluid in the small intestine that contains a mixture of intestinal proteases (including trypsin and chymotrypsin). The hydrolysis and activation of NanI can further enhance the effects of CPE, ETX, and CPB, promoting the occurrence of disease (Theoret et al., 2018). Thus, the hydrolysis and activation of NanI may contribute to the pathophysiology of C. perfringens (Theoret et al., 2018). Therefore, NanI is a potential auxiliary virulence factor promoting enteritis and enterotoxemia caused by C. perfringens.
Sialidase Inhibitors
Sialidases have become a drug target for the treatment of viral and bacterial infections (Soong et al., 2006; Memoli et al., 2008). Some studies have shown that a number of sialidase inhibitors may become effective drugs for the treatment of sialic acid toxicosis, cancers, infections, immune diseases, atherosclerosis, and many other diseases (Karagodin et al., 2018). Recently, researchers have reported encouraging findings. One study showed that sialidase inhibitors could attenuate pulmonary fibrosis in mice, suggesting that sialidase inhibitors may be helpful in the treatment of fibrosis (Karhadkar et al., 2017). Another study indicated that C9-BA-DANA inhibits endogenous and ectopically expressed sialidase activity and established NEU1-mediated bioactivities in human airway epithelia, lung microvascular endothelia, and fibroblasts in vitro and murine lungs in vivo (Hyun et al., 2016).
Two classical inhibitors of C. perfringens sialidases, Siastatin B (SB), and N-acetyl-2,3-dehydro-2-deoxyneuraminic acid (NADNA), have been shown to inhibit C. perfringens sialidase activity (Li and McClane, 2014a,b; Li et al., 2015). SB or NADNA can reduce the adhesion of C. perfringens AAD strain F4969 to Caco-2 cells (Li and McClane, 2014b) and effectively inhibit the activity of sialidases in cell-free supernatants collected from C. perfringens D strain CN3718 cultures. Moreover, SB can reduce the production of ETX in strain CN3718 (Li et al., 2015). A recent study showed that SB reduced the growth and survival rate of strain F4969 in the presence of Caco-2 cells (Li and McClane, 2018). Diseases caused by C. perfringens are challenging to treat with antibiotics since toxins that have already been synthesized continue to work after the administration of antibiotics. Similarly, because some strains produce many different toxins, it is often difficult to treat or prevent C. perfringens infections with vaccines or neutralizing antibodies (Li et al., 2015). Although NanI can enhance C. perfringens adherence and toxin binding, it is worth mentioning that NanI would not be needed for nutritional purposes or to enhance bacteria adherence or colonization during the acute food poisoning (Li and McClane, 2014b). So sialidase inhibitors may be a potential candidate for the development of drugs against human non-foodborne gastrointestinal disease such as antibiotic-associated diarrhea and sporadic diarrhea, and the development of highly effective sialidase inhibitors will be an important direction in generating anti-C. perfringens infection drugs in the near future.
The development of targeted sialidase inhibitors is based on random screening of synthetic compounds or substrate (sialic acid) mimics. In recent years, researchers have studied natural non-substrate mimic sialidase inhibitors, such as flavonoids. Natural flavonoids have potential in promoting resistance to trypanosomiasis (Arioka et al., 2010). Research has shown that these compounds also have a significant competitive inhibitory effect on C. perfringens sialidases (Cp-NanI) (Lee et al., 2014).
Biochemical and structural data on sialidases can provide valuable information for the design of new selective antimicrobial or antiviral drugs. A recent study has identified the crystal structure of the Cp-NanI catalytic domain (residues 243-694) complexed with diplacone and elucidated their interaction in detail. The interactions between the inhibitor and the enzyme are primarily hydrogen bonds and hydrophobic interactions. The 3′,4′,5′,7′-hydroxy-4-oxo group and O atom at the 1-position of diplacone forms direct and indirect hydrogen bonds with the tri-Arg cluster (Arg266, Arg555, and Arg615), Arg285, Asp328, Phe353, Tyr485, Gln493, Thr538, and Tyr655 residues on the side chain and the Ala292 residue on the primary chain, as well as with three water molecules (Figure 4). The flavanone backbone of diplacone and the Ile327, Phe347, Phe460, Tyr485, and Tyr655 residues correctly position the conformation of the stable inhibitor complex through hydrophobic interactions (Lee et al., 2014).
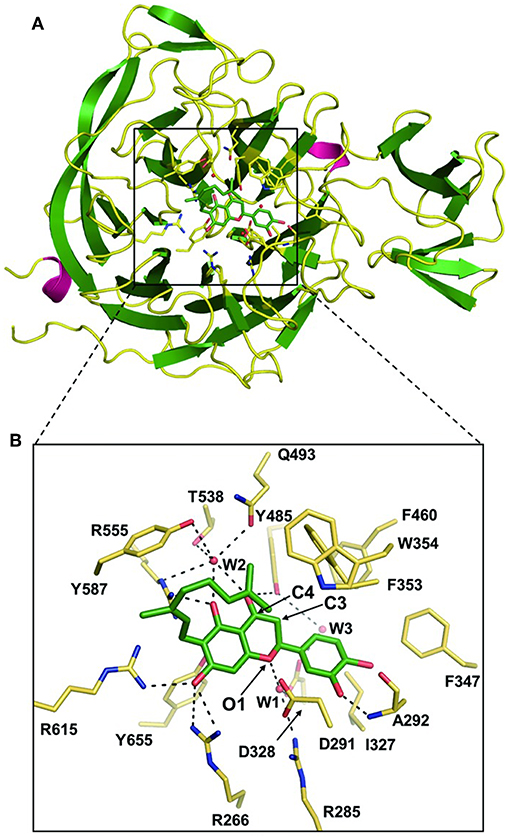
Figure 4. The Cp-NanI catalytic site with diplacone. Reprinted from Lee et al. (2014), Copyright © (Lee et al., 2014). (A) Overall structure of the Cp-NanI catalytic domain bound to diplacone. (B) Details of the mode of binding. Diplacone and three water molecules (W1, W2, and W3) are shown as green sticks and red spheres, respectively. Hydrogen bonds are displayed as dashed lines.
Microorganism drug resistance remains a difficult problem. But what's exciting is that studies found various natural flavonoids contain flavanone backbones, which have an inhibitory effect on viral sialidases (Grienke et al., 2012). This finding may aid in solving the problem of sialidase inhibitor resistance, because regardless of the I223R or H275Y mutation, diplacone appears to have anti-sialidase activity. As with the wild-type protein, the C4 hydroxyl group of diplacone can interact with the Glu276 of the H275Y mutant, suggesting that diplacone may become a potential lead compound for the development of inhibitors for viral sialidase drug resistant mutants (Lee et al., 2014). Therefore, the elucidation of the structural information of the Cp-NanI-diplacone complex is extremely helpful in the development of antibacterial and antiviral sialidase inhibitors.
At present, the development of new natural antibacterial sialidase inhibitors has become a novel direction of research. Kim et al. (2018) focused on the natural product turmeric and observed that a curcumin derivative compound, 7-(3,4-dihydroxyphenyl)-5-hydroxy-1-(3-hydroxy-4-methoxyphenyl) hepta-1,4,6-trien-3-one, inhibits S. pneumoniae NanA and sialidases from V. cholerae and C. perfringens. The methoxy or hydroxyl group, heptadienyl, and α, β unsaturated ketone groups play important roles in the activity of this inhibitor. Although the positions of the methoxy and hydroxyl groups do not affect the inhibitory effects on the enzymes, the inhibition increases with the increase in the number of hydroxyl groups substituted in benzene rings. The activity of the C. perfringens sialidase inhibitor is not affected by an electronic effect, and the catechol moiety group in this compound plays an important role in enhancing its inhibitory effect.
Conclusion and Prospection
Sialidases, especially NanI, are emerging as potential pathogenic factors during C. perfringens intestinal infections. These enzymes can function by upregulating the production of toxins associated with intestinal infections, enhancing the cytotoxicity of toxins, increasing the adhesion of C. perfringens to host cells, and producing substrates used for bacterial growth and metabolism (Li and McClane, 2014a; Li et al., 2015). In the past, studies of C. perfringens virulence were focused on toxins. Although studies on the functions of sialidases in the pathogenesis of C. perfringens have yielded interesting results in recent years, there are still many questions to be answered. Among the questions to be answered is whether NanI directly contributes to disease. Future studies on C. perfringens sialidases may also involve investigating the following aspects: (1) the relationship between various systems involved in the regulation of sialidases; (2) the roles of sialidase NanH and NanJ in intestinal infections caused by C. perfringens and whether NanH, NanI, and NanJ have synergistic effects; (3) the differences in the specific sialidases produced by epidemiologically different strains of C. perfringens. And whereas the catalytic mechanism of the S. pneumoniae sialidase NanC has been reported (Xiong et al., 2018), those of the C. perfringens sialidases have not been reported. To facilitate the design and development of sialidase inhibitors in the future, studies on the catalytic mechanism of sialidases are required. In addition, the inhibition of sialidase activity in P. gingivalis renders the bacteria easier to be cleared by macrophages (Yang et al., 2018). Studies on this aspect of the C. perfringens sialidase have not been reported and should therefore be a major focus of future studies. Thus, the continued study of sialidases will promote new breakthroughs in the prevention and treatment of patients suffering infections by C. perfringens in the near future.
Author Contributions
Y-HW collected the references and wrote the paper.
Funding
The author thank Science and Technology Innovation Project of China Academy of Agricultural Sciences (CAAS-ASTIP-IVFCAAS) for the financial support.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abe, K., Obana, N., and Nakamura, K. (2010). Effects of depletion of RNA-binding protein Tex on the expression of toxin genes in Clostridium perfringens. Biosci. Biotechnol. Biochem. 74, 1564–1571. doi: 10.1271/bbb.100135
Allen, S., Zaleski, A., Johnston, J. W., Gibson, B. W., and Apicella, M. A. (2005). Novel sialic acid transporter of Haemophilus influenzae. Infect. Immun. 73, 5291–5300. doi: 10.1128/IAI.73.9.5291-5300.2005
Amimoto, K., Noro, T., Oishi, E., and Shimizu, M. (2007). A novel toxin homologous to large clostridial cytotoxins found in culture supernatant of Clostridium perfringens type C. Microbiology 153(Pt 4), 1198–1206. doi: 10.1099/mic.0.2006/002287-0
Arabyan, N., Weis, A. M., Huang, B. C., and Weimer, B. C. (2017). Implication of sialidases in Salmonella infection: genome release of sialidase knockout strains from Salmonella enterica serovar typhimurium LT2. Genome Announc. 5:e00341–17. doi: 10.1128/genomeA.00341-17
Arioka, S., Sakagami, M., Uematsu, R., Yamaguchi, H., Togame, H., Takemoto, H., et al. (2010). Potent inhibitor scaffold against Trypanosoma cruzi trans-sialidase. Bioorg. Med. Chem. 18, 1633–1640. doi: 10.1016/j.bmc.2009.12.062
Awad, M. M., Singleton, J., and Lyras, D. (2016). The sialidase NanS enhances non-TcsL mediated cytotoxicity of Clostridium sordellii. Toxins 8:189. doi: 10.3390/toxins8060189
Banerjee, A., Van Sorge, N. M., Sheen, T. R., Uchiyama, S., Mitchell, T. J., and Doran, K. S. (2010). Activation of brain endothelium by pneumococcal neuraminidase NanA promotes bacterial internalization. Cell. Microbiol. 12, 1576–1588. doi: 10.1111/j.1462-5822.2010.01490.x
Blanchette, K. A., Shenoy, A. T., Milner, J., Gilley, R. P., McClure, E., Hinojosa, C. A., et al. (2016). Neuraminidase A-exposed galactose promotes Streptococcus pneumoniae biofilm formation during colonization. Infect. Immun. 84, 2922–2932. doi: 10.1128/IAI.00277-16
Boraston, A. B., Ficko-Blean, E., and Healey, M. (2007). Carbohydrate recognition by a large sialidase toxin from Clostridium perfringens. Biochemistry 46, 11352–11360. doi: 10.1021/bi701317g
Brittan, J. L., Buckeridge, T. J., Finn, A., Kadioglu, A., and Jenkinson, H. F. (2012). Pneumococcal neuraminidase A: an essential upper airway colonization factor for Streptococcus pneumoniae. Mol. Oral Microbiol. 27, 270–283. doi: 10.1111/j.2041-1014.2012.00658.x
Chiarezza, M., Lyras, D., Pidot, S. J., Flores-Diaz, M., Awad, M. M., Kennedy, C. L., et al. (2009). The NanI and NanJ sialidases of Clostridium perfringens are not essential for virulence. Infect. Immun. 77, 4421–4428. doi: 10.1128/IAI.00548-09
Chien, C. H., Shann, Y. J., and Sheu, S. Y. (1996). Site-directed mutations of the catalytic and conserved amino acids of the neuraminidase gene, nanH, of Clostridium perfringens ATCC 10543. Enzyme Microb. Technol. 19, 267–276. doi: 10.1016/0141-0229(95)00245-6
Cioffi, D. L., Pandey, S., Alvarez, D. F., and Cioffi, E. A. (2012). Terminal sialic acids are an important determinant of pulmonary endothelial barrier integrity. Am. J. Physiol. Lung Cell. Mol. Physiol. 302, L1067–L1077. doi: 10.1152/ajplung.00190.2011
Collie, R. E., Kokai-Kun, J. F., and McClane, B. A. (1998). Phenotypic characterization of enterotoxigenic Clostridium perfringens isolates from non-foodborne human gastrointestinal diseases. Anaerobe 4, 69–79. doi: 10.1006/anae.1998.0152
de Graaf, M., and Fouchier, R. A. (2014). Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J. 33, 823–841. doi: 10.1002/embj.201387442
Fernandez-Miyakawa, M. E., Fisher, D. J., Poon, R., Sayeed, S., Adams, V., Rood, J. I., et al. (2007). Both epsilon-toxin and beta-toxin are important for the lethal properties of Clostridium perfringens type B isolates in the mouse intravenous injection model. Infect. Immun. 75, 1443–1452. doi: 10.1128/IAI.01672-06
Fisher, D. J., Fernandez-Miyakawa, M. E., Sayeed, S., Poon, R., Adams, V., Rood, J. I., et al. (2006). Dissecting the contributions of Clostridium perfringens type C toxins to lethality in the mouse intravenous injection model. Infect. Immun. 74, 5200–5210. doi: 10.1128/IAI.00534-06
Flores-Díaz, M. A., Alape-Girón, G., Clark, B., Catimel, Y., Hirabayashi, E., Nice, J. M., et al. (2005). A cellular deficiency of gangliosides causes hypersensitivity to Clostridium perfringens phospholipase C. J. Biol. Chem. 280, 26680–26689. doi: 10.1074/jbc.M500278200
Freedman, J. C., Li, J., Uzal, F. A., and McClane, B. A. (2014). Proteolytic processing and activation of Clostridium perfringens epsilon toxin by caprine small intestinal contents. MBio 5, e01994–e01914. doi: 10.1128/mBio.01994-14
Grienke, U., Schmidtke, M., von Grafenstein, S., Kirchmair, J., Liedl, K. R., and Rollinger, J. M. (2012). Influenza neuraminidase: a druggable target for natural products. Nat. Prod. Rep. 29, 11–36. doi: 10.1039/C1NP00053E
Hanna, P. C., Wieckowski, E. U., Mietzner, T. A., and McClane, B. A. (1992). Mapping functional regions of Clostridium perfringens type A enterotoxin. Infect. Immun. 60, 2110–2114.
Hassan, K. A., Elbourne, L. D., Tetu, S. G., Melville, S. B., Rood, J. I., and Paulsen, I. T. (2015). Genomic analyses of Clostridium perfringens isolates from five toxinotypes. Res. Microbiol. 166, 255–263. doi: 10.1016/j.resmic.2014.10.003
Hiscox, T. J., Chakravorty, A., Choo, J. M., Ohtani, K., Shimizu, T., Cheung, J. K., et al. (2011). Regulation of virulence by the RevR response regulator in Clostridium perfringens. Infect. Immun. 79, 2145–2153. doi: 10.1128/IAI.00060-11
Hiscox, T. J., Harrison, P. F., Chakravorty, A., Choo, J. M., Ohtani, K., Shimizu, T., et al. (2013). Regulation of sialidase production in Clostridium perfringens by the orphan sensor histidine kinase ReeS. PLoS ONE 8:e73525. doi: 10.1371/journal.pone.0073525
Hitsumoto, Y., Morita, N., Yamazoe, R., Tagomori, M., Yamasaki, T., and Katayama, S. (2014). Adhesive properties of Clostridium perfringens to extracellular matrix proteins collagens and fibronectin. Anaerobe 25, 67–71. doi: 10.1016/j.anaerobe.2013.11.002
Honma, K., Mishima, E., and Sharma, A. (2011). Role of Tannerella forsythia NanH sialidase in epithelial cell attachment. Infect. Immun. 79, 393–401. doi: 10.1128/IAI.00629-10
Hyun, S. W., Liu, A., Liu, Z., Cross, A. S., Verceles, A. C., Magesh, S., et al. (2016). The NEU1-selective sialidase inhibitor, C9-butyl-amide-DANA, blocks sialidase activity and NEU1-mediated bioactivities in human lung in vitro and murine lung in vivo. Glycobiology 26, 834–849. doi: 10.1093/glycob/cww060
Imai, M., and Kawaoka, Y. (2012). The role of receptor binding specificity in interspecies transmission of influenza viruses. Curr. Opin. Virol. 2, 160–167. doi: 10.1016/j.coviro.2012.03.003
Irikura, D., Monma, C., Suzuki, Y., Nakama, A., Kai, A., Fukui-Miyazaki, A., et al. (2015). Identification and characterization of a new enterotoxin produced by Clostridium perfringens isolated from food poisoning outbreaks. PLoS ONE 10:e0138183. doi: 10.1371/journal.pone.0138183
Janesch, P., Rouha, H., Badarau, A., Stulik, L., Mirkina, I., Caccamo, M., et al. (2018). Assessing the function of pneumococcal neuraminidases NanA, NanB and NanC in in vitro and in vivo lung infection models using monoclonal antibodies. Virulence 9, 1521–1538. doi: 10.1080/21505594.2018.1520545
Jost, B. H., Billington, S. J., Trinh, H. T., and Songer, J. G. (2006). Association of genes encoding beta2 toxin and a collagen binding protein in Clostridium perfringens isolates of porcine origin. Vet. Microbiol. 115, 173–182. doi: 10.1016/j.vetmic.2006.01.012
Juge, N., Tailford, L., and Owen, C. D. (2016). Sialidases from gut bacteria: a mini-review. Biochem. Soc. Trans. 44, 166–175. doi: 10.1042/BST20150226
Karagodin, V. P., Sukhorukov, V. N., Myasoedova, V. A., Grechko, A. V., and Orekhov, A. N. (2018). Diagnostics and therapy of human diseases - focus on sialidases. Curr. Pharm. Des. 24, 2870–2875. doi: 10.2174/1381612824666180910125051
Karhadkar, T. R., Pilling, D., Cox, N., and Gomer, R. H. (2017). Sialidase inhibitors attenuate pulmonary fibrosis in a mouse model. Sci. Rep. 7:15069. doi: 10.1038/s41598-017-15198-8
Katayama, S., Tagomori, M., Morita, N., Yamasaki, T., Nariya, H., Okada, M., et al. (2015). Determination of the Clostridium perfringens-binding site on fibronectin. Anaerobe 34, 174–181. doi: 10.1016/j.anaerobe.2014.11.007
Ketterer, M. R., Rice, P. A., Gulati, S., Kiel, S., Byerly, L., Fortenberry, J. D., et al. (2016). Desialylation of neisseria gonorrhoeae lipooligosaccharide by cervicovaginal microbiome sialidases: the potential for enhancing infectivity in men. J. Infect. Dis. 214, 1621–1628. doi: 10.1093/infdis/jiw329
Keyburn, A. L., Boyce, J. D., Vaz, P., Bannam, T. L., Ford, M. E., Parker, D., et al. (2008). NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathog. 4:e26. doi: 10.1371/journal.ppat.0040026
Kim, B. R., Park, J. Y., Jeong, H. J., Kwon, H. J., Park, S. J., Lee, I. C., et al. (2018). Design, synthesis, and evaluation of curcumin analogues as potential inhibitors of bacterial sialidase. J. Enzyme Inhib. Med. Chem. 33, 1256–1265. doi: 10.1080/14756366.2018.1488695
King, S. J. (2010). Pneumococcal modification of host sugars: a major contributor to colonization of the human airway? Mol. Oral Microbiol. 25, 15–24. doi: 10.1111/j.2041-1014.2009.00564.x
Lee, Y., Ryu, Y. B., Youn, H. S., Cho, J. K., Kim, Y. M., Park, J. Y., et al. (2014). Structural basis of sialidase in complex with geranylated flavonoids as potent natural inhibitors. Acta Crystallogr. D Biol. Crystallogr. 70(Pt 5), 1357–1365. doi: 10.1107/S1399004714002971
Lewis, A. L., and Lewis, W. G. (2012). Host sialoglycans and bacterial sialidases: a mucosal perspective. Cell. Microbiol. 14, 1174–1182. doi: 10.1111/j.1462-5822.2012.01807.x
Li, J., Evans, D. R., Freedman, J. C., and McClane, B. A. (2017). NanR regulates nanI sialidase expression by Clostridium perfringens F4969, a human enteropathogenic strain. Infect. Immun. 85:e00241–17. doi: 10.1128/IAI.00241-17
Li, J., Freedman, J. C., and McClane, B. A. (2015). NanI sialidase, CcpA, and CodY work together to regulate epsilon toxin production by Clostridium perfringens type D strain CN3718. J. Bacteriol. 197, 3339–3353. doi: 10.1128/JB.00349-15
Li, J., Ma, M., Sarker, M. R., and McClane, B. A. (2013). CodY is a global regulator of virulence-associated properties for type D strain CN3718. MBio 4, e00770–e00713. doi: 10.1128/mBio.00770-13
Li, J., and McClane, B. A. (2014a). The sialidases of Clostridium perfringens type D strain CN3718 differ in their properties and sensitivities to inhibitors. Appl. Environ. Microbiol. 80, 1701–1709. doi: 10.1128/AEM.03440-13
Li, J., and McClane, B. A. (2014b). Contributions of NanI sialidase to Caco-2 cell adherence by Clostridium perfringens type A and C strains causing human intestinal disease. Infect. Immun. 82, 4620–4630. doi: 10.1128/IAI.02322-14
Li, J., and McClane, B. A. (2018). NanI sialidase can support the growth and survival of Clostridium perfringens strain F4969 in the presence of sialyated host macromolecules (mucin) or Caco-2 cells. Infect. Immun. 86:e00547–17. doi: 10.1128/IAI.00547-17
Li, J., Sayeed, S., Robertson, S., Chen, J., and McClane, B. A. (2011). Sialidases affect the host cell adherence and epsilon toxin-induced cytotoxicity of Clostridium perfringens type D strain CN3718. PLoS Pathog. 7:e1002429. doi: 10.1371/journal.ppat.1002429
Li, J., Uzal, F. A., and McClane, B. A. (2016). Clostridium perfringens sialidases: potential contributors to intestinal pathogenesis and therapeutic targets. Toxins 8:E341. doi: 10.3390/toxins8110341
Liu, G. J., Wang, B., Zhang, Y., Xing, G. W., Yang, X., and Wang, S. (2018). A tetravalent sialic acid-coated tetraphenylethene luminogen with aggregation-induced emission characteristics: design, synthesis and application for sialidase activity assay, high-throughput screening of sialidase inhibitors and diagnosis of bacterial vaginosis. Chem. Commun. 54, 10691–10694. doi: 10.1039/C8CC06300A
Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M., and Henrissat, B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495. doi: 10.1093/nar/gkt1178
Ma, M., Gurjar, A., Theoret, J. R., Garcia, J. P., Beingesser, J., Freedman, J. C., et al. (2014). Synergistic effects of Clostridium perfringens enterotoxin and beta toxin in rabbit small intestinal loops. Infect. Immun. 82, 2958–2970. doi: 10.1128/IAI.01848-14
Mehdizadeh Gohari, I., Brefo-Mensah, E. K., Palmer, M., Boerlin, P., and Prescott, J. F. (2018). Sialic acid facilitates binding and cytotoxic activity of the pore-forming Clostridium perfringens NetF toxin to host cells. PLoS ONE 13:e0206815. doi: 10.1371/journal.pone.0206815
Mehdizadeh Gohari, I., Parreira, V. R., Timoney, J. F., Fallon, L., Slovis, N., and Prescott, J. F. (2016). NetF-positive Clostridium perfringens in neonatal foal necrotising enteritis in Kentucky. Vet. Rec. 178:216. doi: 10.1136/vr.103606
Memoli, M. J., Morens, D. M., and Taubenberger, J. K. (2008). Pandemic and seasonal influenza: therapeutic challenges. Drug Discov. Today 13, 590–595. doi: 10.1016/j.drudis.2008.03.024
Mi, E., Li, J., and McClane, B. A. (2018). NanR regulates sporulation and enterotoxin production by Clostridium perfringens type F strain F4969. Infect. Immun. 86:e00416–18. doi: 10.1128/IAI.00416-18
Minami, A., Ishibashi, S., Ikeda, K., Ishitsubo, E., Hori, T., Tokiwa, H., et al. (2013). Catalytic preference of Salmonella typhimurium LT2 sialidase for N-acetylneuraminic acid residues over N-glycolylneuraminic acid residues. FEBS Open Bio. 3, 231–236. doi: 10.1016/j.fob.2013.05.002
Miyagi, T., Takahashi, K., Yamamoto, K., Shiozaki, K., and Yamaguchi, K. (2018). Biological and pathological roles of ganglioside sialidases. Prog. Mol. Biol. Transl. Sci. 156, 121–150. doi: 10.1016/bs.pmbts.2017.12.005
Myers, G. S., Rasko, D. A., Cheung, J. K., Ravel, J., Seshadri, R., DeBoy, R. T., et al. (2006). Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 16, 1031–1040. doi: 10.1101/gr.5238106
Navarro, M. A., Li, J., McClane, B. A., Morrell, E., Beingesser, J., and Uzal, F. A. (2018). NanI sialidase is an important contributor to Clostridium perfringens type F strain F4969 intestinal colonization in mice. Infect. Immun. 86:e00462–18. doi: 10.1128/IAI.00462-18
Nees, S., and Schauer, R. (1974). Induction of neuraminidase from Clostridium perfringens and the correlation of this enzyme with acylneuraminate pyruvate-lyase. Behring Inst. Mitt. 55, 68–78.
Newstead, S., Chien, C. H., Taylor, M., and Taylor, G. (2004). Crystallization and atomic resolution X-ray diffraction of the catalytic domain of the large sialidase, NanI, from Clostridium perfringens. Acta Crystallogr. D Biol. Crystallogr. 60 (Pt 11), 2063–2066. doi: 10.1107/S090744490402181X
Newstead, S. L., Potter, J. A., Wilson, J. C., Xu, G., Chien, C. H., Watts, A. G., et al. (2008). The structure of Clostridium perfringens NanI sialidase and its catalytic intermediates. J. Biol. Chem. 283, 9080–9088. doi: 10.1074/jbc.M710247200
Nishiyama, K., Nagai, A., Uribayashi, K., Yamamoto, Y., Mukai, T., and Okada, N. (2018). Two extracellular sialidases from Bifidobacterium bifidum promote the degradation of sialyl-oligosaccharides and support the growth of Bifidobacterium breve. Anaerobe 52, 22–28. doi: 10.1016/j.anaerobe.2018.05.007
Obana, N., and Nakamura, K. (2011). A novel toxin regulator, the CPE1446-CPE1447 protein heteromeric complex, controls toxin genes in Clostridium perfringens. J. Bacteriol. 193, 4417–4424. doi: 10.1128/JB.00262-11
Ohtani, K., Hirakawa, H., Tashiro, K., Yoshizawa, S., Kuhara, S., and Shimizu, T. (2010). Identification of a two-component VirR/VirS regulon in Clostridium perfringens. Anaerobe 16, 258–264. doi: 10.1016/j.anaerobe.2009.10.003
Parent, E., Archambault, M., Charlebois, A., Bernier-Lachance, J., and Boulianne, M. (2017). A chicken intestinal ligated loop model to study the virulence of Clostridium perfringens isolates recovered from antibiotic-free chicken flocks. Avian Pathol. 46, 138–149. doi: 10.1080/03079457.2016.1228825
Parker, D., Soong, G., Planet, P., Brower, J., Ratner, A. J., and Prince, A. (2009). The NanA neuraminidase of Streptococcus pneumoniae is involved in biofilm formation. Infect. Immun. 77, 3722–3730. doi: 10.1128/IAI.00228-09
Pastoriza Gallego, M., and Hulen, C. (2006). Influence of sialic acid and bacterial sialidase on differential adhesion of Pseudomonas aeruginosa to epithelial cells. Colloids Surf. B Biointerfaces 52, 154–156. doi: 10.1016/j.colsurfb.2006.04.013
Petit, L., Gibert, M., and Popoff, M. R. (1999). Clostridium perfringens: toxinotype and genotype. Trends Microbiol. 7, 104–110. doi: 10.1016/S0966-842X(98)01430-9
Rohmer, L., Hocquet, D., and Miller, S. I. (2011). Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol. 19, 341–348. doi: 10.1016/j.tim.2011.04.003
Rood, J. I., Adams, V., Lacey, J., Lyras, D., McClane, B. A., Melville, S. B., et al. (2018). Expansion of the Clostridium perfringens toxin-based typing scheme. Anaerobe 53, 5–10. doi: 10.1016/j.anaerobe.2018.04.011
Sayeed, S., Fernandez-Miyakawa, M. E., Fisher, D. J., Adams, V., Poon, R., Rood, J. I., et al. (2005). Epsilon-toxin is required for most Clostridium perfringens type D vegetative culture supernatants to cause lethality in the mouse intravenous injection model. Infect. Immun. 73, 7413–7421. doi: 10.1128/IAI.73.11.7413-7421.2005
Severi, E., Hood, D. W., and Thomas, G. H. (2007). Sialic acid utilization by bacterial pathogens. Microbiology 153 (Pt 9), 2817–2822. doi: 10.1099/mic.0.2007/009480-0
Shimizu, T., Ohtani, K., Hirakawa, H., Ohshima, K., Yamashita, A., Shiba, T., et al. (2002). Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. U.S.A. 99, 996–1001. doi: 10.1073/pnas.022493799
Silva, R. O. S., Duarte, M. C., Oliveira Junior, C. A., de Assis, R. A., Lana, A. M. Q., and Lobato, F. C. F. (2018). Comparison of humoral neutralizing antibody response in rabbits, guinea pigs, and cattle vaccinated with epsilon and beta toxoids from Clostridium perfringens and C. botulinum types C and D toxoids. Anaerobe 54, 19–22. doi: 10.1016/j.anaerobe.2018.07.014
Soong, G., Muir, A., Gomez, M. I., Waks, J., Reddy, B., Planet, P., et al. (2006). Bacterial neuraminidase facilitates mucosal infection by participating in biofilm production. J. Clin. Invest. 116, 2297–2305. doi: 10.1172/JCI27920
Tailford, L. E., Crost, E. H., Kavanaugh, D., and Juge, N. (2015). Mucin glycan foraging in the human gut microbiome. Front. Genet. 6:81. doi: 10.3389/fgene.2015.00081
Theoret, J. R., Li, J., Navarro, M. A., Garcia, J. P., Uzal, F. A., and McClane, B. A. (2018). Native or proteolytically activated nani sialidase enhances the binding and cytotoxic activity of Clostridium perfringens enterotoxin and beta toxin. Infect. Immun. 86:e00730–17. doi: 10.1128/IAI.00730-17
Therit, B., Cheung, J. K., Rood, J. I., and Melville, S. B. (2015). NanR, a transcriptional regulator that binds to the promoters of genes involved in sialic acid metabolism in the anaerobic pathogen Clostridium perfringens. PLoS ONE 10:e0133217. doi: 10.1371/journal.pone.0133217
Thompson, H., Homer, K. A., Rao, S., Booth, V., and Hosie, A. H. (2009). An orthologue of Bacteroides fragilis NanH is the principal sialidase in Tannerella forsythia. J. Bacteriol. 191, 3623–3628. doi: 10.1128/JB.01618-08
Tong, H. H., Li, D., Chen, S., Long, J. P., and DeMaria, T. F. (2005). Immunization with recombinant Streptococcus pneumoniae neuraminidase NanA protects chinchillas against nasopharyngeal colonization. Infect. Immun. 73, 7775–7778. doi: 10.1128/IAI.73.11.7775-7778.2005
Trappetti, C., Kadioglu, A., Carter, M., Hayre, J., Iannelli, F., Pozzi, G., et al. (2009). Sialic acid: a preventable signal for pneumococcal biofilm formation, colonization, and invasion of the host. J. Infect. Dis. 199, 1497–1505. doi: 10.1086/598483
Traving, C., and Schauer, R. (1998). Structure, function and metabolism of sialic acids. Cell. Mol. Life Sci. 54, 1330–1349. doi: 10.1007/s000180050258
Varki, A., and Gagneux, P. (2012). Multifarious roles of sialic acids in immunity. Ann. N. Y. Acad. Sci. 1253, 16–36. doi: 10.1111/j.1749-6632.2012.06517.x
Vimr, E., and Lichtensteiger, C. (2002). To sialylate, or not to sialylate: that is the question. Trends Microbiol. 10, 254–257. doi: 10.1016/S0966-842X(02)02361-2
Vimr, E. R., Kalivoda, K. A., Deszo, E. L., and Steenbergen, S. M. (2004). Diversity of microbial sialic acid metabolism. Microbiol. Mol. Biol. Rev. 68, 132–153. doi: 10.1128/MMBR.68.1.132-153.2004
Walther, T., Karamanska, R., Chan, R. W., Chan, M. C., Jia, N., Air, G., et al. (2013). Glycomic analysis of human respiratory tract tissues and correlation with influenza virus infection. PLoS Pathog. 9:e1003223. doi: 10.1371/journal.ppat.1003223
Xiong, J., Zhang, C., and Xu, D. (2018). Catalytic mechanism of type C sialidase from Streptococcus pneumoniae: from covalent intermediate to final product. J. Mol. Model. 24:297. doi: 10.1007/s00894-018-3822-5
Yang, X., Pan, Y., Xu, X., Tong, T., Yu, S., Zhao, Y., et al. (2018). Sialidase deficiency in porphyromonas gingivalis increases IL-12 secretion in stimulated macrophages through regulation of CR3, IncRNA GAS5 and miR-21. Front. Cell. Infect. Microbiol. 8:100. doi: 10.3389/fcimb.2018.00100
Keywords: C. perfringens, sialidase, molecular properties, regulatory mechanism, pathogenesis, sialidase inhibitor
Citation: Wang Y (2020) Sialidases From Clostridium perfringens and Their Inhibitors. Front. Cell. Infect. Microbiol. 9:462. doi: 10.3389/fcimb.2019.00462
Received: 23 August 2019; Accepted: 16 December 2019;
Published: 10 January 2020.
Edited by:
Jorge Eugenio Vidal, University of Mississippi Medical Center, United StatesReviewed by:
Bruce A. McClane, University of Pittsburgh, United StatesAlberto Alape-Girón, University of Costa Rica, Costa Rica
Copyright © 2020 Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan-hua Wang, d2FuZ3lhbmh1YUBjYWFzLmNu
 Yan-hua Wang
Yan-hua Wang