- 1College of Veterinary Medicine, Mississippi State University, Mississippi State, MS, United States
- 2Institute for Genomics, Biocomputing and Biotechnology, Mississippi State University, Mississippi State, MS, United States
- 3Bioinformatics & Systems Biology, Justus-Liebig-University Giessen, Giessen, Germany
Aeromonas veronii is a Gram-negative species ubiquitous in different aquatic environments and capable of causing a variety of diseases to a broad host range. Aeromonas species have the capability to carry and acquire antimicrobial resistance (AMR) elements, and currently multi-drug resistant (MDR) Aeromonas isolates are commonly found across the world. A. veronii strain MS-17-88 is a MDR strain isolated from catfish in the southeastern United States. The present study was undertaken to uncover the mechanism of resistance in MDR A. veronii strain MS-17-88 through the detection of genomic features. To achieve this, genomic DNA was extracted, sequenced, and assembled. The A. veronii strain MS-17-88 genome comprised 5,178,226-bp with 58.6% G+C, and it encoded several AMR elements, including imiS, ampS, mcr-7.1, mcr-3, catB2, catB7, catB1, floR, vat(F), tet(34), tet(35), tet(E), dfrA3, and tetR. The phylogeny and resistance profile of a large collection of A. veronii strains, including MS-17-88, were evaluated. Phylogenetic analysis showed a close relationship between MS-17-88 and strain Ae5 isolated from fish in China and ARB3 strain isolated from pond water in Japan, indicating a common ancestor of these strains. Analysis of phage elements revealed 58 intact, 63 incomplete, and 15 questionable phage elements among the 53 A. veronii genomes. The average phage element number is 2.56 per genome, and strain MS-17-88 is one of two strains having the maximum number of identified prophage elements (6 elements each). The profile of resistance against various antibiotics across the 53 A. veronii genomes revealed the presence of tet(34), mcr-7.1, mcr-3, and dfrA3 in all genomes (100%). By comparison, sul1 and sul2 were detected in 7.5% and 1.8% of A. veronii genomes. Nearly 77% of strains carried tet(E), and 7.5% of strains carried floR. This result suggested a low abundance and prevalence of sulfonamide and florfenicol resistance genes compared with tetracycline resistance among A. veronii strains. Overall, the present study provides insights into the resistance patterns among 53 A. veronii genomes, which can inform therapeutic options for fish affected by A. veronii.
Introduction
Aeromonas species are Gram-negative rods in the family Aeromonadaceae. They are among the most common bacteria in aquatic environments and have been isolated from virtually all water source types including freshwater, estuarine environments, drinking waters, wastewaters, and sewage (Janda and Abbott, 2010). The disease caused by Aeromonas species affects a broad host range, including freshwater fish, amphibians, reptiles, and birds (Barony et al., 2015). Mesophilic Aeromonas species such as A. hydrophila, A. caviae, and A. veronii, are associated with several kinds of human infections, including gastroenteritis, wound infections, septicemia, and respiratory infections (Figueras, 2005; Igbinosa et al., 2012). A. veronii is one member of the genus Aeromonas, which is known for causing hemorrhagic septicemia in both wild and farmed fish such as channel catfish (Ictalurus punctatus) (Liu et al., 2016; Yang et al., 2017), snakehead (Ophiocephalus argus), whitefish (Coregonus clupeaformis) (Loch and Faisal, 2010), obscure puffer (Takifugu obscurus), Nile tilapia (Oreochromis niloticus) (Hassan et al., 2017), and common carp (Cyprinus carpio) (Sun et al., 2016). In recent years, an increasing number of Aeromonas species have become associated with disease of predominantly freshwater fish in most countries (Goni-Urriza et al., 2000), which results in causing severe outbreaks in different important aquaculture industries.
Sulfonamides potentiated with trimethoprim or ormethoprim, oxytetracycline, florfenicol, and erythromycin are the most commonly used antimicrobial (AM) agents for treatment of Aeromonas-related diseases in global aquaculture (Serrano, 2005). Although the judicious use of AM-medicated feeds is important for treatment purposes when faced with outbreaks of bacterial infections (Okocha et al., 2018), a substantial number of reports suggest that indiscriminate use of AMs can foster selection pressure and enable development of multi-drug resistant (MDR) bacteria in aquatic environments (Arslan and Küçüksari, 2015). MDR in bacterial pathogens can result in therapeutic challenges for control of bacterial diseases (Marshall and Levy, 2011).
Over the past two decades, A. veronii has gained epidemiological and ecological importance by several research groups due to its potential as an opportunistic and primary pathogen for fish and the prevalence of MDR strains (Sanchez-Cespedes et al., 2008). Previous studies have reported the isolation of MDR A. veronii strains from different regions of the world such as Sri Lanka (Jagoda et al., 2017), China (Yang et al., 2017), and United States (Abdelhamed et al., 2019; Tekedar et al., 2019). The resistance elements in Aeromonas species are often harbored in mobile genetic elements such as class 1 integrons, plasmids, IS elements, transposons, and genomic islands (Piotrowska and Popowska, 2015). These mobile elements can facilitate the spread of resistance among bacteria via transduction and conjugation (Sanchez-Cespedes et al., 2008; Hossain et al., 2013; Piotrowska and Popowska, 2015). Therefore, it is important to investigate the pattern of resistance, genetic relatedness, and mobile elements in Aeromonas species. High-throughput sequencing provides an opportunity to detect MDR bacteria, discover potential resistance mechanisms, and explore the mechanisms underlying resistance gene transfer (Liu et al., 2012).
The purpose of the current work was to uncover mechanisms of resistance in A. veronii strain MS-17-88, which was isolated from catfish in the southeastern U.S., and to assess the diversity, resistance profiles, and mobile elements in a large collection of A. veronii strains. Here we present the draft genome of A. veronii and results from a comparative analysis with 52 publicly available A. veronii genomes with a special emphasis on patterns of AMR genes and prophage elements distribution. To the best of our knowledge, no study has been published reporting a core-genome based phylogenetic relationship of sequenced A. veronii genomes, AMR profiles, and their mobilomes. Overall, our work provides a basis to understand the AMR profile for A. veronii in aquatic environments, which is an important step toward curtailing AMR spread and informing a treatment of disease caused by A. veronii.
Materials and Methods
Bacterial Strains and Data Source for Comparative Genome Analysis
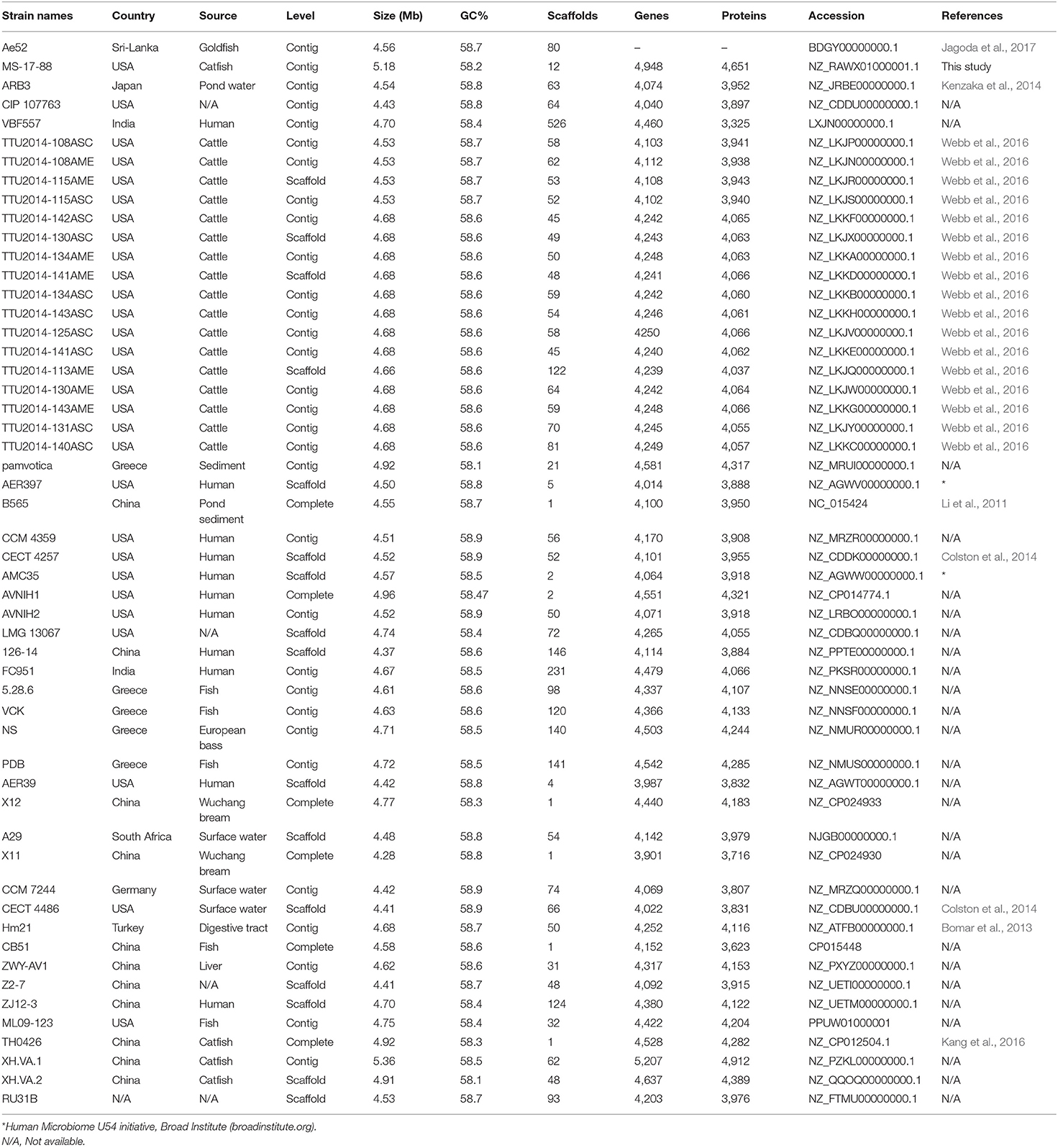
A. veronii strain MS-17-88 was recovered from a diseased channel catfish in 2017 from the Aquatic Diagnostic Laboratory at the College of Veterinary Medicine, Mississippi State University. The isolate was confirmed phenotypically as A. veronii. A 20% glycerol stock culture was stored at −80°C. A. veronii strain MS-17-88 was cultured in brain heart infusion (BHI) agar or broth (Difco) and incubated at 30°C. Fifty-two A. veronii genomes were retrieved from the National Center for Biotechnology Information (NCBI) genomes database on October 9, 2018, including five complete genome sequences and forty-seven draft genome sequences (Table 1).
Antibiotic Resistance Phenotypes of A. veronii Strain MS-17-88
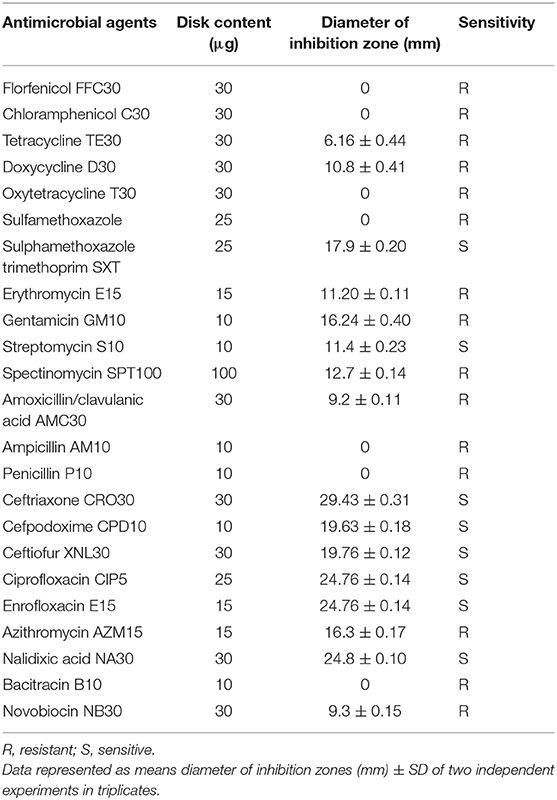
The AM susceptibility of A. veronii strain MS-17-88 was determined by the Kirby-Bauer disk diffusion method (Bauer et al., 1966). Strain MS-17-88 was streaked on Mueller-Hinton agar plates, and the AM disks were applied on the streaked cultures with a Dispens-O-Disc dispenser. The AM agents tested were florfenicol (30 μg), chloramphenicol (30 μg), tetracycline (30 μg), doxycycline (30 μg), oxytetracycline (30 μg), sulfamethoxazole-trimethoprim (23:75; 1.25 μg), sulfamethoxazole (25 μg), erythromycin (15 μg), gentamicin (10 μg), streptomycin (10 μg), spectinomycin (100 μg), amoxicillin/clavulanic acid (30 μg), ampicillin (30 μg), penicillin (10 μg), ceftriaxone (30 μg), cefpodoxime (10 μg), ceftiofur (30 μg), ciprofloxacin (5 μg), enrofloxacin (15 μg), azithromycin (15 μg), nalidixic acid (30 μg), bacitracin (10 μg), and novobiocin (30 μg). These AM agents were selected based on the World Health Organization's list of the most common classes of antimicrobials (aminoglycosides, tetracyclines, macrolides, beta-lactam, phenicols, quinolones, and sulfonamides) that are regularly used in agriculture and aquaculture and linked to human medicine (Done et al., 2015). After 24 h of incubation at 30°C, the zones of inhibition diameter were measured and compared to the criteria of the National Committee for Clinical Laboratory Standards. The assay was performed in triplicate and repeated as two independent experiments.
DNA Extraction, Whole-Genome Sequencing, Assembly, and Annotation
Genomic DNA of A. veronii strain MS-17-88 was extracted using the DNeasy Blood & Tissue Kit (Qiagen., USA) according to the manufacturer's instructions. Genome sequencing was conducted using HiSeq X Ten (Illumina, San Diego, CA, USA) and MinION (Oxford Nanopore Technologies, Oxford, UK), producing approximately 848.86X and 229.35X genome coverages, respectively. Together, the genome coverage is 1077X. Trimmomatic (Bolger et al., 2014) was used to trim Illumina reads, Nanopore reads were corrected with Canu (version 1.6) (Koren et al., 2017), and contig errors were corrected using Pilon (version 1.21) (Walker et al., 2014). Assembly of the Illumina and Nanopore reads into contigs was done using MaSuRCA (version 3.2.4 (Zimin et al., 2013). Average nucleotide identity (ANI) was calculated based on whole genome sequencing using BLAST alignments (Richter and Rossello-Mora, 2009).
Phylogenetic Tree
A phylogenetic tree was constructed based on the complete core genome of A. veronii strain MS-17-88 and 52 A. veronii genomes to evaluate taxonomic positions. All publicly available A. veronii genome sequences (52 genomes) were downloaded from NCBI. Gene sets of the core genome were aligned using MUSCLE (Edgar, 2004) and concatenated. Concatenated alignment files were used as an input to compute a Kimura distance matrix, which was followed by using the concatenated files for the Neighbor-Joining algorithm as implemented in PHYLP (Felsenstein, 1989).
Subsystem Coverages and Genomes Structure Variation
A. veronii strain MS-17-88 and 52 A. veronii genomes were submitted to Rapid Annotations using Subsystems Technology (RAST) for annotation, subsystem categorization, and comparison purposes (Aziz et al., 2008). The following criteria were used for annotation pipeline: classic RAST for annotation, RAST gene caller for open reading frame (ORF) identification, and Figfam (version release70 with automatic fix errors and fix frameshifts options). A. veronii strain MS-17-88 genome was compared against 52 A. veronii using BRIG (BLAST Ring Image Generator) (Alikhan et al., 2011).
Prophages
The presence of prophages in the 53 A. veronii genomes was determined using PHASTER (PHAge Search Tool Enhanced Release) (Arndt et al., 2016, 2019). Nucleotide sequences from 53 genomes were concatenated using Sequencher 5.4.5 to serve as an input file prior to submission to the PHASTER server. Results from PHASTER were arranged into three categories: score > 90 was considered intact phage element; a score between 70 and 90 was deemed questionable; and score <70 was considered incomplete phage region (Arndt et al., 2019).
Comparative Analysis of Putative AMR Elements
The potential AMR genes and related elements for each genome were identified using ResFinder 3.1 (Zankari et al., 2012). ResFinder database was downloaded and used in CLC Workbench version 11.0.1 (CLC Bio) for the BLAST search. The contig files for each genome were concatenated, and concatenated nucleotide files were uploaded to CLC Workbench. A BLAST search was run with the following settings: 40% minimum identity and 40% minimum matching length.
Results and Discussion
General Genome Features of A. veronii Strain MS-17-88
The present study reported the draft genome of A. veronii strain MS-17-88 isolated from diseased catfish in the southeastern U.S. The draft genome of A. veronii strain MS-17-88 consisted of 5,178,226 bp with 58.6% G+C content and encoded 4,944 predicted coding sequences (CDSs). A total of 181 RNA genes were predicted in the genome including 139 tRNAs, 4 ncRNAs, and 38 rRNAs (12, 13, 13 for 5, 16, and 23 s, respectively). The final assembly contained 13 contigs. The largest contig assembled was 1,457,362-bp length, and the smallest contig was 7,082-bp. The genome has been deposited in GenBank (accession number NZ_RAWX01000000). A. veronii contains two biovars (A. veronii biovar veronii and A. veronii biovar sobria) (Janda and Abbott, 2010). ANI and phylogenetic tree calculation confirmed that strain MS-17-88 belongs to A. veronii biovar veronii (ANI score higher than 95%).
Genotypic and Phenotypic Characterization of A. veronii Strain MS-17-88
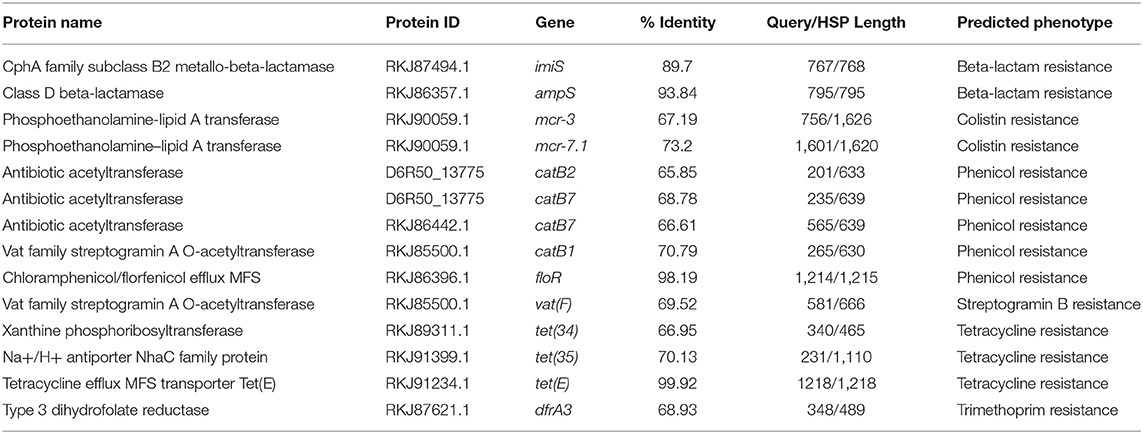
The Rasfinder and CARD analysis revealed 14 resistance elements in the A. veronii strain MS-17-88 genome (Table 2), including beta-lactamase resistance genes (imiS and ampS), chloramphenicol and florfenicol resistance gene (floR), macrolide resistance genes (mcr-3 and mcr-7.1), streptogramin B resistance vat(F), tetracycline resistance genes [tet(34), tet(35), and tet(E)], and acetyltransferase genes conferring resistance to phenicol compounds. The disk diffusion results (Table 3) demonstrated that A. veronii strain MS-17-88 strain is resistant to phenicol class (florfenicol and chloramphenicol), tetracyclines (tetracycline, doxycycline, and oxytetracycline), macrolides (erythromycin, azithromycin,), aminoglycoside (gentamicin), sulfamethoxazole, beta-lactam class (amoxicillin/clavulanic acid, ampicillin, and penicillin), spectinomycin, bacitracin, and novobiocin.
Resistance to β-lactam antibiotics in Aeromonas species is primarily mediated by β-lactamases, whose mode of action involves hydrolyzing the β-lactam ring. Many different β-lactamases have been detected in Aeromonas species, such as TEM-, SHV-, OXA-, CMY-, and CTX-M-type β-lactamases. In the present study, ampS and imiS were detected in A. veronii stain 17-88. AmpS is a class 2d penicillinase and ImiS is a class 3 metallo-β-lactamase. The imiS gene has been detected in clinical isolates of A. veronii biovar sobria (Wu et al., 2012). MS-17-88 also harbors floR, which encodes a major facilitator superfamily efflux pump that exports florfenicol (Schwarz et al., 2004). Florfenicol is one among the three approved AMs for use in catfish aquaculture in the U.S (Bowker et al., 2010). Most fish pathogenic bacteria mediate florfenicol resistance through FloR (Dang et al., 2007; Gordon et al., 2008). Dissemination of florfenicol resistance among bacterial pathogens isolated from aquaculture can limit the efficacy of this agent as an important treatment option.
Phylogenetic Tree
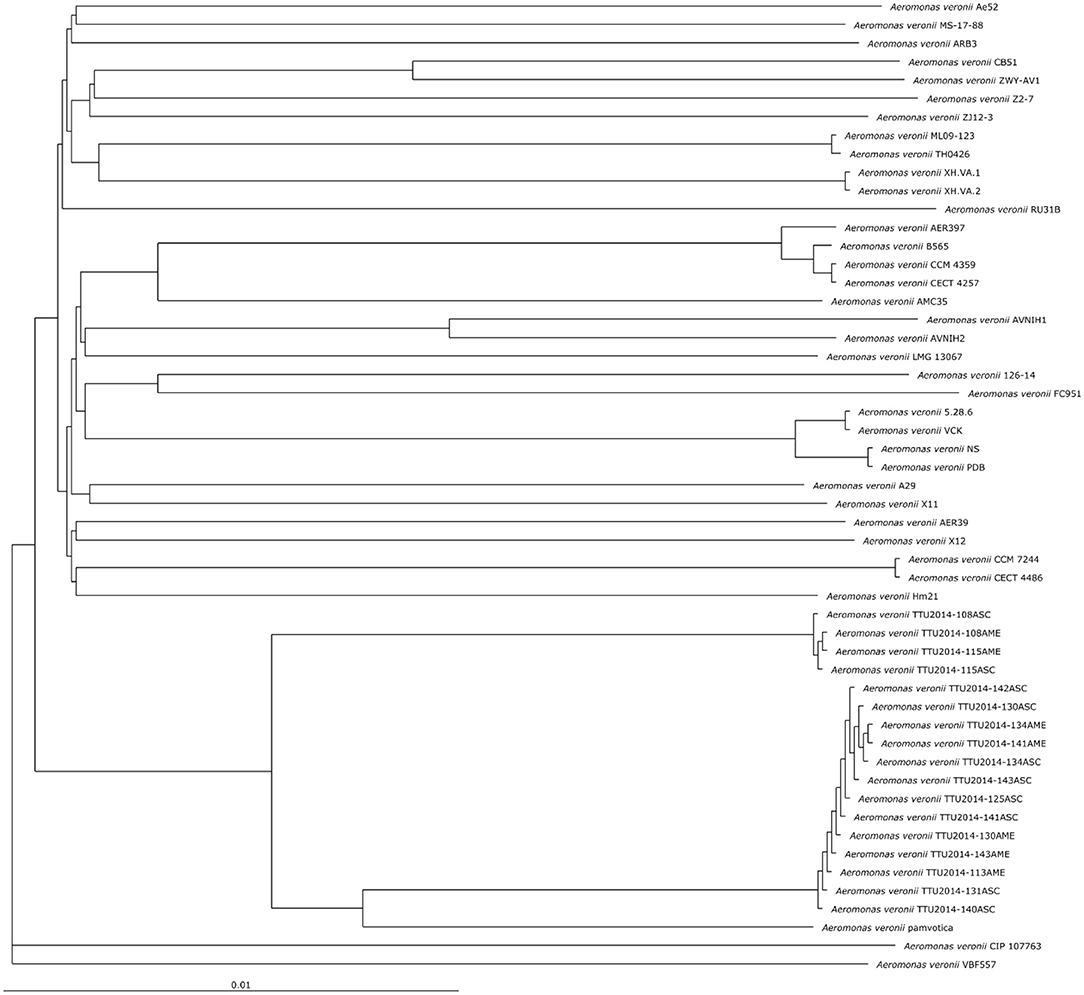
The phylogenetic relationship between the A. veronii MS-17-88 genome and 52 other A. veronii genomes was assessed. The strains used in this analysis are from different countries and hosts, and they had distinct resistance profiles (Table 1). The phylogenetic tree for the 53 A. veronii genomes was built from a core genome of 2,563 genes per genome (135,839 genes in total). The core has 2,538,377 bp per genome (134,533,981 bp in total).
The phylogenetic tree showed that there are multiple highly conserved branches that are separated from the other A. veronii genomes. MDR A. veronii strain MS-17-88 from U.S. channel catfish, goldfish (Carassius auratus) MDR strain Ae5 from China, and A. veronii ARB3 from pond water in Japan were clustered together, which may indicate a common origin. Similarly, U.S. channel catfish isolate ML09-123 and China catfish isolate TH0426 were closely related, which also may suggest derivation from the same monophyletic origin despite their geographic disparity. Moreover, dairy cattle isolates and Greece surface sediment isolates (strain pamvotica) formed another closely related group. U.S. human isolates (strains AER 397, CECT 4257, and CCM 4359) and China pond sediment isolate B565 formed another clade. Lastly, U.S. surface water and Germany surface water isolates were closely related, and Greece fish isolates (strains 5.28.6, VCK, NS, and PDB) were clustered together. These data clearly suggest that the ecological niche is a more important factor contributing to relatedness among A. veronii isolates than geographical location. However, two isolates (CIP 107763 and VBF557) showed a distinct genetic relationship to the other isolates.
It has been postulated that a fish pathogenic Aeromonas hydrophila clonal group was transferred to the U.S. channel catfish aquaculture industry from China (Hossain et al., 2014). In the current study, we observed two different clonal groups of A. veronii that contain isolates from both U.S. and China. One clade has MDR A. veronii strain MS-17-88 from U.S. catfish aquaculture and MDR isolate Ae5 from goldfish in China, and another clade has U.S. channel catfish isolate ML09-123 and China catfish strain TH0426 (Figure 1).
Subsystems Coverage
The subsystems categorization based on RAST annotation is shown in Figure 2. SEED subsystem categorization analysis predicted 26 different categories for the evaluated A. veronii genomes. The most abundant systems are “amino acid and derivatives,” followed by “carbohydrates” and “protein metabolism.” These subsystems are essential for bacteria to perform basic cellular processes and may indicate the potential ability of A. veronii to utilize different kinds of sugars and amino acids available in the environment (Liang et al., 2019). On the other hand, A. veronii genomes show remarkably low numbers of mobile genetic elements including phages, prophages, transposable elements, and plasmids. These mobile elements can mediate alteration of genotypes, and these findings may suggest that horizontal gene exchange may not contribute to A. veronii genomic variation as much as other species. Interestingly, U.S. catfish isolate strain MS-17-88 carries the most abundant subsystems (63 elements) associated with phages, prophages, transposable elements, and plasmids. Chinese catfish isolate strain TH0426 has the second largest number (52 elements) of phage and transposable elements. Therefore, these two strains (MS-17-88 and TH0426) may have acquired significant genome structure changes by gene acquisitions from mobile elements. It is notable that both strains were isolated from aquatic environments, and we speculate that the aquatic environment may be favorable for genetic exchange and horizontal gene acquisition.
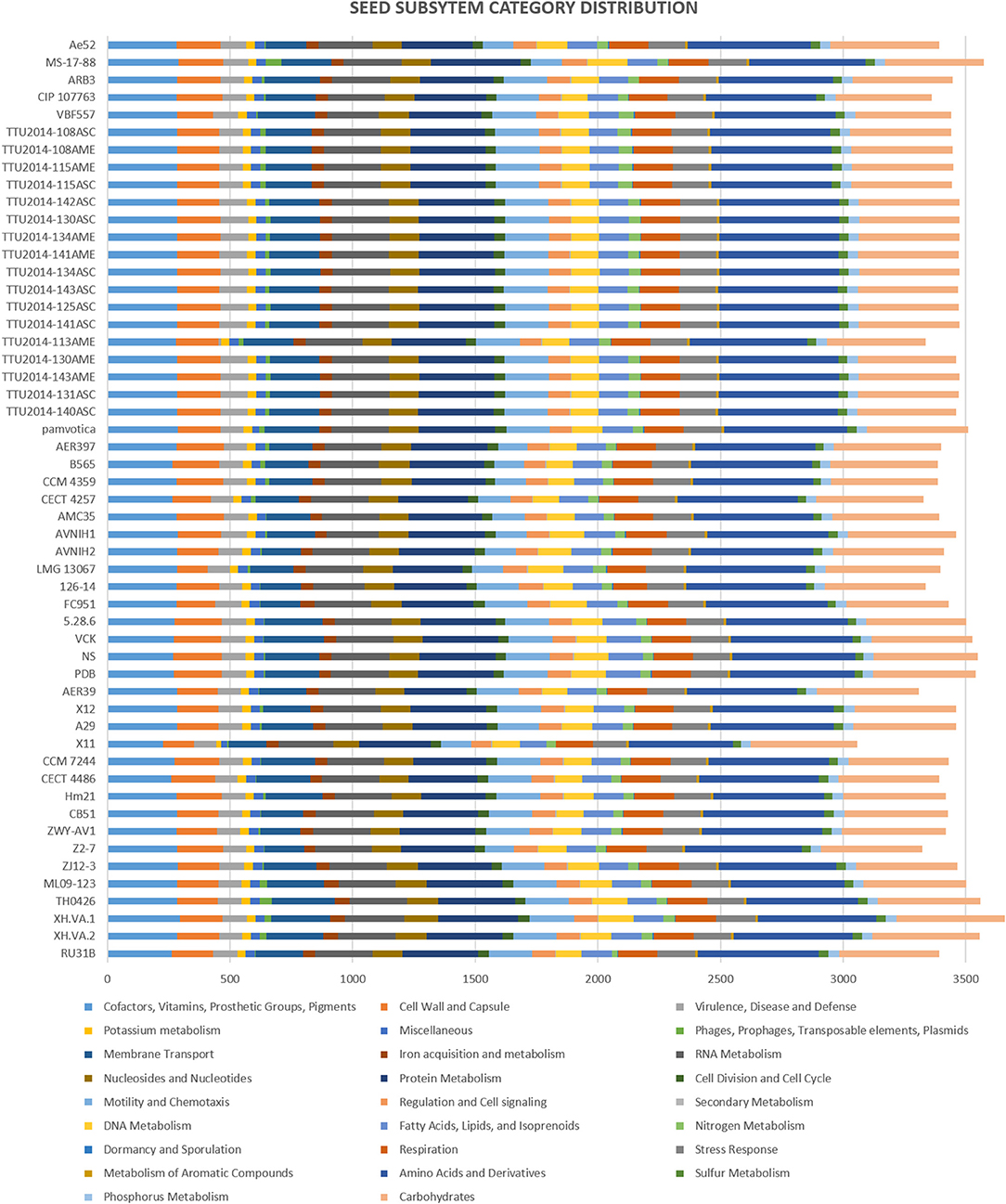
Figure 2. Comparison of functional categories in 53 A. veronii genomes based on SEED. Functional categorization is based on roles of annotated and assigned genes. Each colored bar represents the number of genes assigned to each category.
Genome Structure Variation
Visualization of the alignment between A. veronii MS-17-88 and 52 other A. veronii genomes revealed that phage elements, transposons, and genomic islands comprise many of the MS-17-88-specific regions (Figure 3). This suggests that mobile elements, especially phages and transposons, play an important role in A. veronii genome variation. These mobile elements can result in genomic rearrangements and evolution through acquisition of novel virulence or antibiotic resistance genes which may result in emergence of new phenotypes (Brown-Jaque et al., 2015).
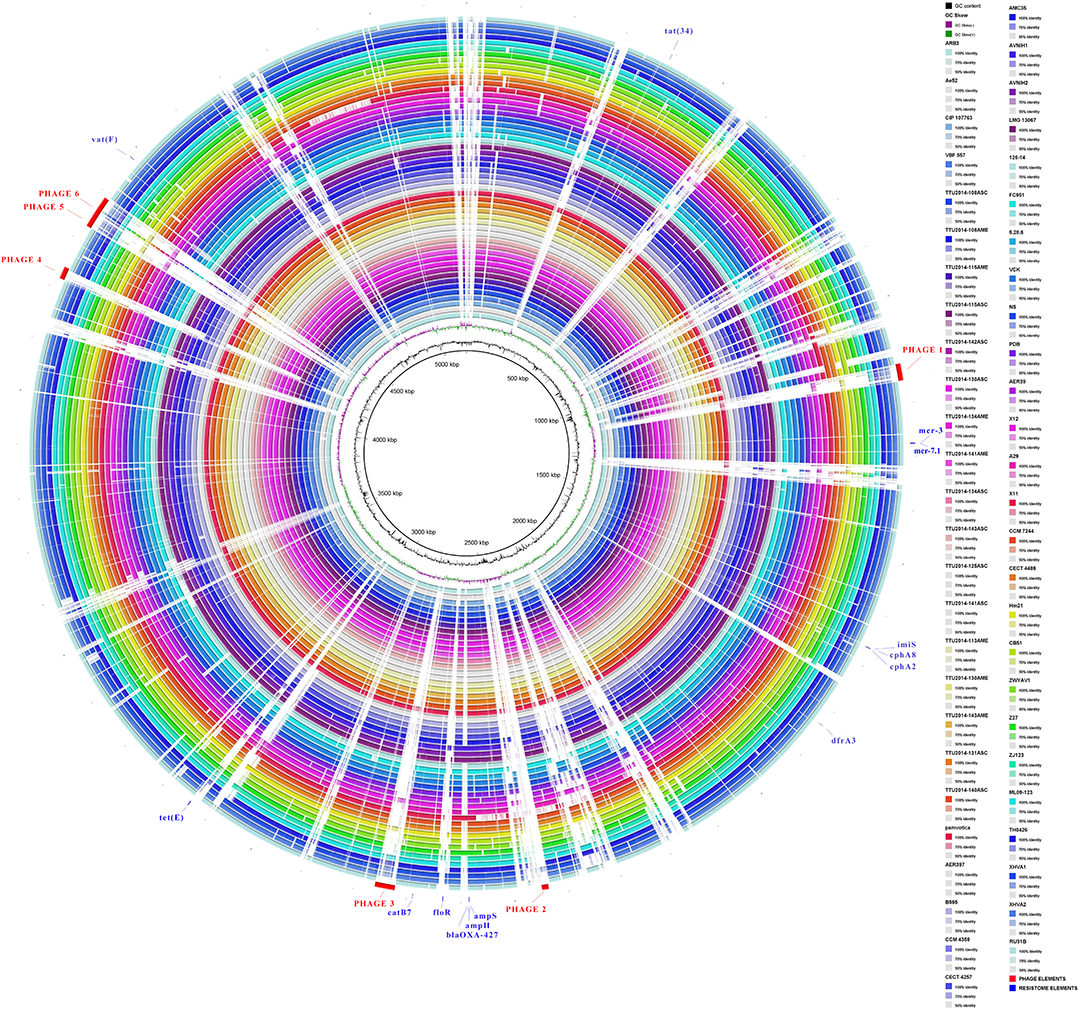
Figure 3. Comparative circular map of the A. veronii MS-17-88 genome. Phage regions are highlighted with red color: phage region-1 encodes 41 proteins (31.8 Kb), phage region-2 encodes 12 proteins (12.6 Kb), phage region-3 encodes 39 proteins (37.7 Kb), phage region-4 encodes 31 proteins (22.9 Kb), phage region-5 encodes 45 proteins (48.6 Kb), and phage region-6 encodes 30 proteins (24.1 Kb).
Prophages
Bacteriophages are responsible for loci rearrangements and deletions and are recognized as an important element in bacterial evolution (Tinsley et al., 2006). The vast majority of aquatic bacteria (about 70%) are infected with prophages (Chen et al., 2006). Of particular interest, bacteriophages can mediate horizontal gene transfer, including genes encoding virulence factors and antibiotic resistance (Colomer-Lluch et al., 2011). A higher number of phages may represent a concern because they can expand the pathogenicity of a bacterial strain or convert an avirulent strain into a virulent one (Canchaya et al., 2004). Even though phage elements may not be the main spreading factors of resistance elements, recent studies indicate that they can infrequently contribute to the dissemination of these elements (Allen et al., 2011; Enault et al., 2017). In Aeromonas, the transfer of resistance gene by phage elements has never been observed previously (Piotrowska and Popowska, 2015).
In the present study, we identified 58 intact, 63 incomplete, and 15 questionable phage elements among the 53 A. veronii genomes (Figure 4). The average phage element number is 2.56 per genome (136 identified phage elements/53 A. veronii strains). In a previous study, A. hydrophila genomes were found to harbor an average of 2.91 phage elements per genome (143 identified phage elements/49 A. hydrophila strains) (Awan et al., 2018). Almost all the strains carry one or more prophage elements, with the exceptions being strains FC951, CECT4486, and CCM7244, which do not carry any type of prophage. Strains MS-17-88 and TH0426 had the maximum number of identified prophage elements (6 elements per strain). The maximum number of complete prophage elements (5 complete phages) was present in strain MS-17-88 along with one incomplete phage element (Figure 4). The maximum number of incomplete prophage elements (4 incomplete phages) was present in strains TTU2014-108ASC and TTU2014-115AME. Strain MS-17-88 carries three different types of phage elements: vB_AbaM_ME3, phi018P, and RSA1. Interestingly, two of these phage elements (vB_AbaM_ME3 and RSA1) are not carried by any other evaluated A. veronii genomes, suggesting that this strain has been exposed to different environments and acquired unique phage elements. Phage vB_AbaM_ME3 was previously isolated from wastewater effluent using the propagating host Acinetobacter baumannii DSM 30007 (Kropinski et al., 2009). A. baumannii is a known nosocomial pathogen that causes pneumonia, urinary tract infection, and septicemia (Buttimer et al., 2016). Phage vB_AbaM_ME3 of A. baumannii has a size of 234,900 bp and 326 ORFs (Buttimer et al., 2016). The Myovirus-type phage RSA1 is relatively small (39 to 40 kb) and has lytic activity. It has restricted host range, mainly Ralstonia solanacearum, a soil-borne species pathogenic to many important crops (Yamada et al., 2007; Addy et al., 2019).
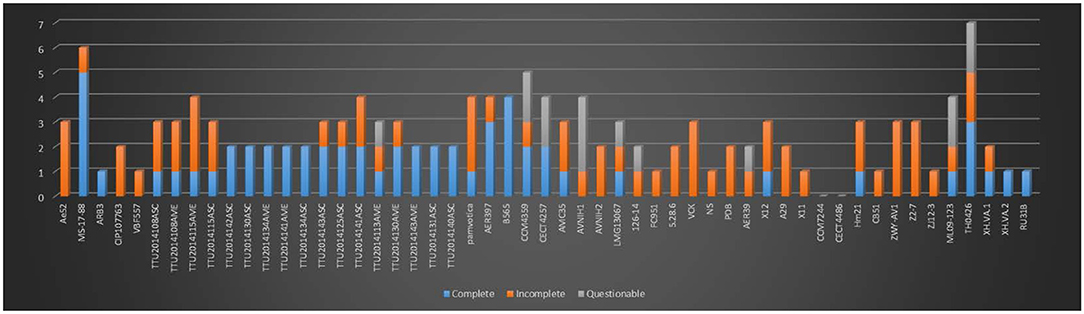
Figure 4. Number of prophages with their completeness profiles in A. veronii genomes. Strains FC91, CCM7244, and CECT4486 did not have any prophage elements.
Interestingly, the MS-17-88 genome has four prophages sharing structural similarities with temperate Aeromo_phiO18P elements found in Aeromonas media isolated from a pond in Germany (Beilstein and Dreiseikelmann, 2008). The phiO18P phage type belongs to the Myoviridae phage family and consists of 33 kb. The phiO18P phage elements typically have 46 ORFs encoding proteins responsible for integration and regulation, replication, packaging, head and tail, and lysis (Beilstein and Dreiseikelmann, 2008). Unlike other prophage elements, Aeromo_phiO18P does not have a lytic phase; it replicates lysogenically by integrating its genome into the bacterial chromosome (Vincent et al., 2017). In some instances, the phage-encoded genes are advantageous to the host bacteria (Dziewit and Radlinska, 2016). Aeromo_phiO18P shows significant similarity to the P2 phage family in Aeromonas salmonicida and Vibrio cholerae K139 genomes (Beilstein and Dreiseikelmann, 2008).
The present study documented 40 different types of phage elements across the 53 A. veronii genomes (Figure 5). Among these 40 phage elements, phage type “Aeromo_phiO18P” is the most abundant type in all the evaluated A. veronii genomes as well as the most abundant in strain MS-17-88. Strains CCM 4359 and XH.VA.1 carry five different phage elements. Incomplete Staphy_SPbeta_like phage element was detected in two strains (MS-17-88 and AVNIH1). In our initial analysis of the MS-17-88 genome using PHASTER, we identified that the strain carries the floR gene inside incomplete phage element PHAGE_Staphy_SPbeta_like_NC_029119 (genome position 2631302-2644102), but later analysis with PHASTER showed that this may not be a true phage element. Further investigation of this region is warranted to determine whether a phage element mediated dissemination of the floR resistance gene (Garriss et al., 2009).
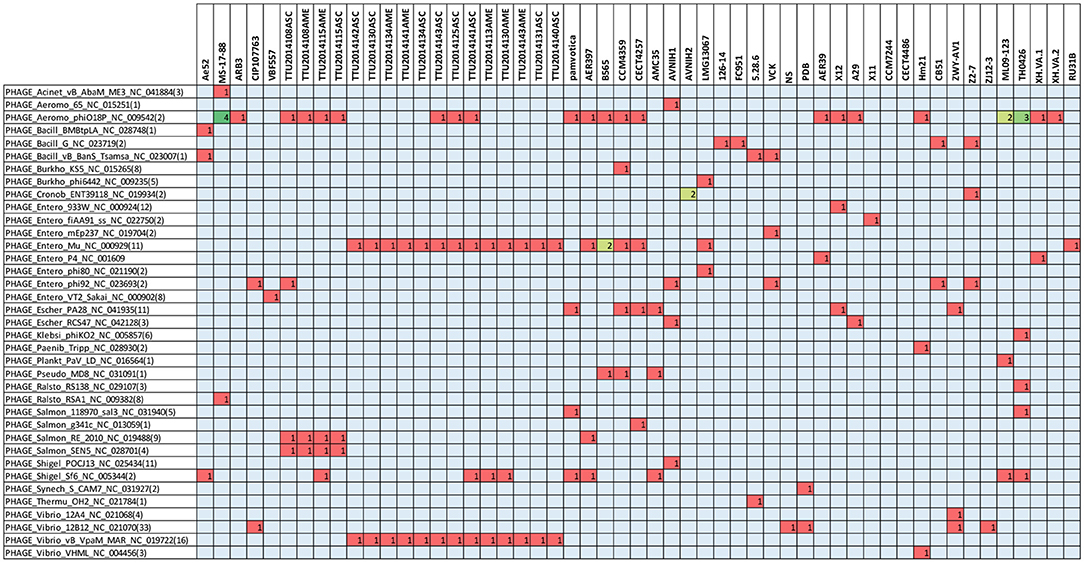
Figure 5. Type of prophage elements present in the A. veronii genomes. Red color represents presence of the gene.
Comparative Analysis of Antibiotic Resistance Determinants
A comparative analysis of 87 AMR determinants and components was conducted to determine their distribution among the 53 A. veronii strains. A. veronii MS-17-88 shared a common AMR gene composition with the other A. veronii genomes. Figure 6 shows the distribution of antibiotic resistance genes in each strain. All the A. veronii genomes carry tetracycline [tet(34) and tet(E)], and trimethoprim (dfrA3) resistance genes. Oxytetracycline, tetracycline, and trimethoprim/sulfamethoxazole have been extensively used in human clinical, veterinary, and agricultural sectors for decades. The linkage between AM use and resistance has been demonstrated for other bacteria in aquaculture ecosystems and other animal husbandry facilities (Verner-Jeffreys et al., 2009; Lagana et al., 2011; Tamminen et al., 2011). In addition to tet(34), tet(E), and dfrA3, A. veronii MS-17-88 genome carries colistin resistance genes (mcr-7.1 and mcr-3). There have been an increasing number of reports on the identification of mcr genes in many bacterial species globally (Stoesser et al., 2016; Elbediwi et al., 2019). A recent study reported that mcr-3 variants are more common in Aeromonas than in other bacterial species but aeromonads do not inherently carry the mcr-3 gene (Shen et al., 2018). However, it is speculated that Aeromonas isolates from the aquatic environment may be the major reservoir for the dissemination of mcr-3 genes to other bacteria (Ling et al., 2017; Eichhorn et al., 2018). Furthermore, A. veronii MS-17-88 genome harbors a macrolide resistance gene vat(F) that encodes an acetyltransferase that acetylates class A streptogramins (Seoane and García Lobo, 2000). Six genes encoding resistance to β-lactamases were identified in A. veronii MS-17-88 genome including ampH and blaOXA-427 belong to class D beta-lactamase, ampS encoding a class 2d penicillinase and hydrolyzing mainly penicillins (Walsh et al., 1995), and cphA2, cphA8, and imiS encoding a class 3 metallo-β-lactamase and active mainly against carbapenems (Walsh et al., 1998).
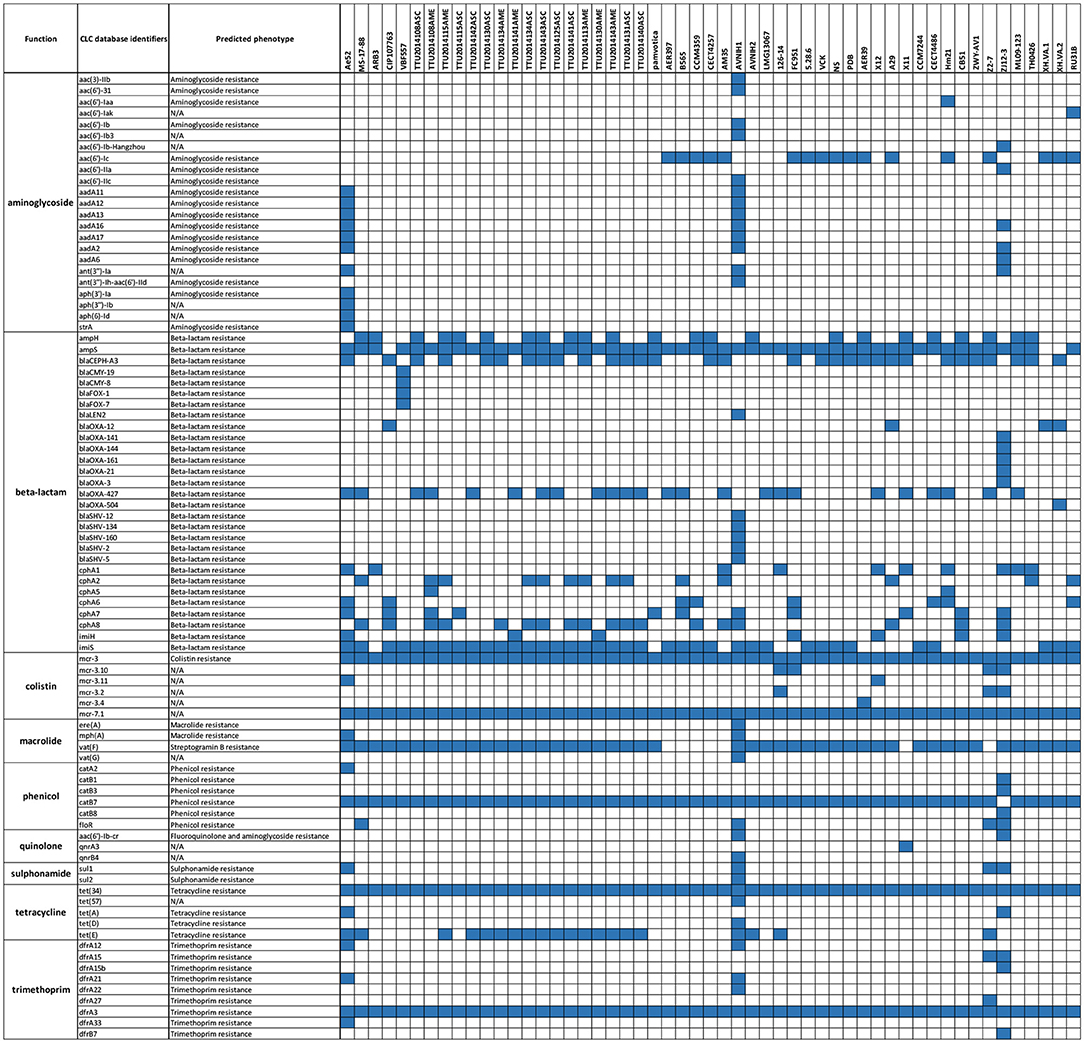
Figure 6. AMR genes distribution across the 53 A. veronii genomes. Blue color represents presence of the gene.
Among the 53 A. veronii strains, AVNIH1 strain had resistance genes to almost all the antibiotic classes. This clinical strain was isolated from human stool, and the genome exhibits clear evidence of horizontal gene transfer (Hughes et al., 2016). In our analyses, we did not observe any pattern of antimicrobial resistance in specific bacterial host or sources. However, there was one exception: the highest number of AMR elements were observed in two human isolates (strain AVNIH from U.S.A; (Hughes et al., 2016) and ZJ12-3 from China; Shen et al., 2018), and one isolate from septicemic goldfish (strain Ae52 from Sri-Lanka; Jagoda et al., 2017). The use of antimicrobial agents in human medicine may be associated with these nosocomial trends (Hughes et al., 2016).
In regard to sulphonamide resistance, sul1 was detected in four strains (Ae52, ANIH1, Z2-7, and ZJ12-3) representing 7.5% of the A. veronii strains, and sul2 was present in one strain (Z2-7) representing 1.8%. This suggests that sul1 is the most frequent gene encoding sulfonamides resistance in A. veronii. Nearly all strains (100%) carried tet(34), and a significant proportion of strains carried tet(E) (20 strains; 37%). In contrast, tet(57) and tet(D) were detected in only one strain (ANIH1) (1.8%), and tet(A) was detected in two strains (Ae52 and ZJ12-3) (3.8%). floR gene conferring resistance to florfenicol and chloramphenicol was present in only four strains (MS-17-88, AVNIH1, Z2-7, and ZJ12-3) (7.5%). The prevalence of beta-lactam resistance genes in the 53 A. veronii genomes was as follows: ampH was detected in 41.5% of the strains, ampS was detected in 92.4% of the strains, imiH was detected in 13.2% of the strains, imiS was detected in 71.7% of the strains, blaCEPH-A3 was detected in 56.6% of the strains, blaOXA-427 was detected in 45.3% of the strains, cphA1 was detected in 17% of the strains, cphA2 was detected in 26.41% of the strains, cphA6 was detected in 13.2% of the strains, cphA7 was detected in 20.7% of the strains, and cphA8 was detected in 44% of the strains. Of the aminoglycoside resistance genes, aac(6′)-Ic was the most prevalent (32%). Interestingly, A. veronii AVNIH1 and Ae52 strains carried multiple aminoglycoside-resistance genes. Several studies reported that the majority of Aeromonas species exhibit only a single aminoglycoside modifying gene (Dahanayake et al., 2019). However, Pseudomonas aeruginosa isolated from hospitals from Iran was reported to carry up to four aminoglycoside resistance genes (Perez-Vazquez et al., 2009).
In conclusion, we used genome sequencing to investigate genetic variation and AMR gene distribution in 53 A. veronii genomes. We found significant genetic differences and a high degree of genomic plasticity in the evaluated A. veronii genomes. Overall, the AMR gene frequency against sulfamethoxazole and florfenicol is low, while AMR genes against tetracycline are very high. Among tetracycline-resistant isolates, tet(34) and tet(E) were the most frequent AMR genes. Taken together, our results show that AMR genes are common and are distributed among A. veronii genomes; however, the frequency of most AMR genes in individual strains is still low. Identified phage elements may be useful for future development of an efficient and effective bio-treatment method to control bacterial diseases in aquaculture. The knowledge generated from this study can benefit our understanding of A. veronii evolution and provide insight into how A. veronii isolates are intrinsically resistant to multiple antimicrobials. In addition, A. veronii species are important considerations as potential sources for resistance determinants in the environment. Therefore, it is important to continue surveillance of resistance and genetic mechanisms of resistance in this species.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article.
Author Contributions
HT, ML, and HA designed and conceived the analysis and experiments. HT, MA, C-YH, AT, JB, and HA performed the experiments and analyzed the data. HT, ML, and HA wrote the manuscript. All authors read and approved the final manuscript.
Funding
Salary support to HA was provided by the Center for Biomedical Research Excellence in Pathogen–Host Interactions, National Institute of General Medical Sciences, and National Institutes of Health awarded grant number P20GM103646-07. This work was supported by College of Veterinary Medicine and by USDA-ARS SCA no. 58-6066-7081 titled MS Center for Food Safety and Post-Harvest Technology, MS Agricultural and Forestry Experiment Station.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the Aquatic Diagnostic Laboratory at the College of Veterinary Medicine for providing Aeromonas veronii strain MS 17-88.
References
Abdelhamed, H., Lawrence, M. L., and Waldbieser, G. (2019). Complete genome sequence data of multidrug-resistant Aeromonas veronii strain MS-18-37. Data Brief 23:103689. doi: 10.1016/j.dib.2019.01.037
Addy, H. S., Ahmad, A. A., and Huang, Q. (2019). Molecular and biological characterization of ralstonia phage RsoM1USA, a new species of P2virus, isolated in the United States. Front. Microbiol. 10:267. doi: 10.3389/fmicb.2019.00267
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Allen, H. K., Looft, T., Bayles, D. O., Humphrey, S., Levine, U. Y., Alt, D., et al. (2011). Antibiotics in feed induce prophages in swine fecal microbiomes. MBio 2. doi: 10.1128/mBio.00260-11
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Arndt, D., Marcu, A., Liang, Y., and Wishart, D. S. (2019). PHAST, PHASTER and PHASTEST: tools for finding prophage in bacterial genomes. Brief. Bioinform. 20, 1560–1567. doi: 10.1093/bib/bbx121
Arslan, S., and Küçüksari, R. (2015). Phenotypic and genotypic virulence factors and antimicrobial resistance of motile Aeromonas spp. from Fish and Ground Beef. J. Food Saf. 35, 551–559. doi: 10.1111/jfs.12205
Awan, F., Dong, Y., Liu, J., Wang, N., Mushtaq, M. H., Lu, C., et al. (2018). Comparative genome analysis provides deep insights into Aeromonas hydrophila taxonomy and virulence-related factors. BMC Genomics 19:712. doi: 10.1186/s12864-018-5100-4
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Barony, G. M., Tavares, G. C., Assis, G. B., Luz, R. K., Figueiredo, H. C., and Leal, C. A. (2015). New hosts and genetic diversity of Flavobacterium columnare isolated from Brazilian native species and Nile tilapia. Dis. Aquat. Org. 117, 1–11. doi: 10.3354/dao02931
Bauer, A. W., Kirby, W. M., Sherris, J. C., and Turck, M. (1966). Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45, 493–496. doi: 10.1093/ajcp/45.4_ts.493
Beilstein, F., and Dreiseikelmann, B. (2008). Temperate bacteriophage ΦO18P from an Aeromonas media isolate: characterization and complete genome sequence. Virology 373, 25–29. doi: 10.1016/j.virol.2007.11.016
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bomar, L., Stephens, W. Z., Nelson, M. C., Velle, K., Guillemin, K., and Graf, J. (2013). Draft genome sequence of Aeromonas veronii Hm21, a symbiotic isolate from the medicinal leech digestive tract. Genome Announc. 1:e00800-13. doi: 10.1128/genomeA.00800-13
Bowker, J. D., Ostland, V. E., Carty, D., and Bowman, M. P. (2010). Effectiveness of aquaflor (50% florfenicol) to control mortality associated with Streptococcus iniae in freshwater-reared subadult sunshine bass. J. Aquat. Anim. Health 22, 254–265. doi: 10.1577/H09-010.1
Brown-Jaque, M., Calero-Caceres, W., and Muniesa, M. (2015). Transfer of antibiotic-resistance genes via phage-related mobile elements. Plasmid 79, 1–7. doi: 10.1016/j.plasmid.2015.01.001
Buttimer, C., O'Sullivan, L., Elbreki, M., Neve, H., McAuliffe, O., Ross, R. P., et al. (2016). Genome sequence of jumbo phage vB_AbaM_ME3 of Acinetobacter baumanni. Genome Announc. 4:e00431-16. doi: 10.1128/genomeA.00431-16
Canchaya, C., Fournous, G., and Brussow, H. (2004). The impact of prophages on bacterial chromosomes. Mol. Microbiol. 53, 9–18. doi: 10.1111/j.1365-2958.2004.04113.x
Chen, F., Wang, K., Stewart, J., and Belas, R. (2006). Induction of multiple prophages from a marine bacterium: a genomic approach. Appl. Environ. Microbiol. 72, 4995–5001. doi: 10.1128/AEM.00056-06
Colomer-Lluch, M., Imamovic, L., Jofre, J., and Muniesa, M. (2011). Bacteriophages carrying antibiotic resistance genes in fecal waste from cattle, pigs, and poultry. Antimicrob. Agents Chemother. 55, 4908–4911. doi: 10.1128/AAC.00535-11
Colston, S. M., Fullmer, M. S., Beka, L., Lamy, B., Gogarten, J. P., and Graf, J. (2014). Bioinformatic genome comparisons for taxonomic and phylogenetic assignments using Aeromonas as a test case. MBio 5:e02136. doi: 10.1128/mBio.02136-14
Dahanayake, P. S., Hossain, S., Wickramanayake, M., and Heo, G. J. (2019). Antibiotic and heavy metal resistance genes in Aeromonas spp. isolated from marketed Manila Clam (Ruditapes philippinarum) in Korea. J. Appl. Microbiol. 127, 941–952. doi: 10.1111/jam.14355
Dang, H., Zhang, X., Song, L., Chang, Y., and Yang, G. (2007). Molecular determination of oxytetracycline-resistant bacteria and their resistance genes from mariculture environments of China. J. Appl. Microbiol. 103, 2580–2592. doi: 10.1111/j.1365-2672.2007.03494.x
Done, H. Y., Venkatesan, A. K., and Halden, R. U. (2015). Does the recent growth of aquaculture create antibiotic resistance threats different from those associated with land animal production in agriculture? AAPS J. 17, 513–524. doi: 10.1208/s12248-015-9722-z
Dziewit, L., and Radlinska, M. (2016). Two novel temperate bacteriophages co-existing in Aeromonas sp. ARM81 - characterization of their genomes, proteomes and DNA methyltransferases. J. Gen. Virol. 97, 2008–2022. doi: 10.1099/jgv.0.000504
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Eichhorn, I., Feudi, C., Wang, Y., Kaspar, H., Fessler, A. T., Lubke-Becker, A., et al. (2018). Identification of novel variants of the colistin resistance gene mcr-3 in Aeromonas spp. from the national resistance monitoring programme GERM-Vet and from diagnostic submissions. J. Antimicrob. Chemother. 73, 1217–1221. doi: 10.1093/jac/dkx538
Elbediwi, M., Li, Y., Paudyal, N., Pan, H., Li, X., Xie, S., et al. (2019). Global burden of colistin-resistant bacteria: mobilized colistin resistance genes study (1980-2018). Microorganisms 7:461. doi: 10.3390/microorganisms7100461
Enault, F., Briet, A., Bouteille, L., Roux, S., Sullivan, M. B., and Petit, M. A. (2017). Phages rarely encode antibiotic resistance genes: a cautionary tale for virome analyses. ISME J. 11, 237–247. doi: 10.1038/ismej.2016.90
Figueras, M. J. (2005). Clinical relevance of Aeromonas sM503. Rev. Med. Microbiol. 16, 145–153. doi: 10.1097/01.revmedmi.0000184410.98677.8a
Garriss, G., Waldor, M. K., and Burrus, V. (2009). Mobile antibiotic resistance encoding elements promote their own diversity. PLoS Genet. 5:e1000775. doi: 10.1371/journal.pgen.1000775
Goni-Urriza, M., Pineau, L., Capdepuy, M., Roques, C., Caumette, P., and Quentin, C. (2000). Antimicrobial resistance of mesophilic Aeromonas spp. isolated from two European rivers. J. Antimicrob. Chemother. 46, 297–301. doi: 10.1093/jac/46.2.297
Gordon, L., Cloeckaert, A., Doublet, B., Schwarz, S., Bouju-Albert, A., Ganiere, J. P., et al. (2008). Complete sequence of the floR-carrying multiresistance plasmid pAB5S9 from freshwater Aeromonas bestiarum. J. Antimicrob. Chemother. 62, 65–71. doi: 10.1093/jac/dkn166
Hassan, M. A., Noureldin, E. A., Mahmoud, M. A., and Fita, N. A. (2017). Molecular identification and epizootiology of Aeromonas veronii infection among farmed Oreochromis niloticus in Eastern Province, KSA. Egyptian J. Aquat. Res. 43, 161–167. doi: 10.1016/j.ejar.2017.06.001
Hossain, M. J., Sun, D., McGarey, D. J., Wrenn, S., Alexander, L. M., Martino, M. E., et al. (2014). An Asian origin of virulent Aeromonas hydrophila responsible for disease epidemics in United States-farmed catfish. MBio 5:e00848-14. doi: 10.1128/mBio.00848-14
Hossain, M. J., Waldbieser, G. C., Sun, D., Capps, N. K., Hemstreet, W. B., Carlisle, K., et al. (2013). Implication of lateral genetic transfer in the emergence of Aeromonas hydrophila isolates of epidemic outbreaks in channel catfish. PLoS ONE 8:e80943. doi: 10.1371/journal.pone.0080943
Hughes, H. Y., Conlan, S. P., Lau, A. F., Dekker, J. P., Michelin, A. V., Youn, J.-H., et al. (2016). Detection and whole-genome sequencing of carbapenemase-producing Aeromonas hydrophila isolates from routine perirectal surveillance culture. J. Clin. Microbiol. 54, 1167–1170. doi: 10.1128/JCM.03229-15
Igbinosa, I. H., Igumbor, E. U., Aghdasi, F., Tom, M., and Okoh, A. I. (2012). Emerging Aeromonas species infections and their significance in public health. Sci. World J. 2012:625023. doi: 10.1100/2012/625023
Jagoda, S. S. D. S., Honein, K., Arulkanthan, A., Ushio, H., and Asakawa, S. (2017). Genome sequencing and annotation of Aeromonas veronii strain Ae52, a multidrug-resistant isolate from septicaemic gold fish (Carassius auratus) in Sri Lanka. Genom Data 11, 46–48. doi: 10.1016/j.gdata.2016.11.011
Janda, J. M., and Abbott, S. L. (2010). The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23, 35–73. doi: 10.1128/CMR.00039-09
Kang, Y., Pan, X., Xu, Y., Siddiqui, S. A., Wang, C., Shan, X., et al. (2016). Complete genome sequence of the fish pathogen Aeromonas veronii TH0426 with potential application in biosynthesis of pullulanase and chitinase. J. Biotechnol. 227, 81–82. doi: 10.1016/j.jbiotec.2016.04.009
Kenzaka, T., Nakahara, M., Higuchi, S., Maeda, K., and Tani, K. (2014). Draft genome sequences of amoeba-resistant Aeromonas spp. isolated from aquatic environments. Genome Announc. 2:e01115-14. doi: 10.1128/genomeA.01115-14
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., and Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Kropinski, A. M., Prangishvili, D., and Lavigne, R. (2009). Position paper: the creation of a rational scheme for the nomenclature of viruses of Bacteria and Archaea. Environ. Microbiol. 11, 2775–2777. doi: 10.1111/j.1462-2920.2009.01970.x
Lagana, P., Caruso, G., Minutoli, E., Zaccone, R., and Santi, D. (2011). Susceptibility to antibiotics of Vibrio spp. and Photobacterium damsela ssp. piscicida strains isolated from Italian aquaculture farms. New Microbiol. 34, 53–63.
Li, Y., Liu, Y., Zhou, Z., Huang, H., Ren, Y., Zhang, Y., et al. (2011). Complete genome sequence of Aeromonas veronii strain B565. J. Bacteriol. 193, 3389–3390. doi: 10.1128/JB.00347-11
Liang, C. Y., Yang, C. H., Lai, C. H., Huang, Y. H., and Lin, J. N. (2019). Genomic features, comparative genomic analysis, and antimicrobial susceptibility patterns of Chryseobacterium arthrosphaerae strain ED882-96 isolated in Taiwan. Genes 10:309. doi: 10.3390/genes10040309
Ling, Z., Yin, W., Li, H., Zhang, Q., Wang, X., Wang, Z., et al. (2017). Chromosome-mediated mcr-3 variants in Aeromonas veronii from chicken meat. Antimicrob. Agents Chemother. 61:e01272-17. doi: 10.1128/AAC.01272-17
Liu, D., Geng, Y., Wang, K., Chen, D., Huang, X. L., Ou, Y., et al. (2016). Aeromonas veronii infection in cultured channel catfish, ictalurus punctatus, in Southwest China. Israeli J. Aquacult. 68.
Liu, W., Yang, M., Xu, Z., Zheng, H., Liang, W., Zhou, R., et al. (2012). Complete genome sequence of Pasteurella multocida HN06, a toxigenic strain of serogroup D. J. Bacteriol. 194, 3292–3293. doi: 10.1128/JB.00215-12
Loch, T. P., and Faisal, M. (2010). Infection of lake whitefish (Coregonus clupeaformis) with motile Aeromonas spp. in the Laurentian great lakes. J. Great Lakes Res. 36, 6–12. doi: 10.1016/j.jglr.2009.07.001
Marshall, B. M., and Levy, S. B. (2011). Food animals and antimicrobials: impacts on human health. Clin. Microbiol. Rev. 24, 718–733. doi: 10.1128/CMR.00002-11
Okocha, R. C., Olatoye, I. O., and Adedeji, O. B. (2018). Food safety impacts of antimicrobial use and their residues in aquaculture. Public Health Rev. 39:21. doi: 10.1186/s40985-018-0099-2
Perez-Vazquez, M., Vindel, A., Marcos, C., Oteo, J., Cuevas, O., Trincado, P., et al. (2009). Spread of invasive Spanish Staphylococcus aureus spa-type t067 associated with a high prevalence of the aminoglycoside-modifying enzyme gene ant(4')-Ia and the efflux pump genes msrA/msrB. J. Antimicrob. Chemother. 63, 21–31. doi: 10.1093/jac/dkn430
Piotrowska, M., and Popowska, M. (2015). Insight into the mobilome of Aeromonas strains. Front. Microbiol. 6:494. doi: 10.3389/fmicb.2015.00494
Richter, M., and Rossello-Mora, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Sanchez-Cespedes, J., Blasco, M. D., Marti, S., Alba, V., Alcalde, E., Esteve, C., et al. (2008). Plasmid-mediated QnrS2 determinant from a clinical Aeromonas veronii isolate. Antimicrob. Agents Chemother. 52, 2990–2991. doi: 10.1128/AAC.00287-08
Schwarz, S., Kehrenberg, C., Doublet, B., and Cloeckaert, A. (2004). Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 28, 519–542. doi: 10.1016/j.femsre.2004.04.001
Seoane, A., and García Lobo, J. M. (2000). Identification of a streptogramin A acetyltransferase gene in the chromosome of Yersinia enterocolitica. Antimicrobial Agents Chemother. 44, 905–909. doi: 10.1128/AAC.44.4.905-909.2000
Serrano, P. H. (2005). Responsible Use of Antibiotics in Aquaculture. Rome: Food and Agriculture Organization.
Shen, Y., Xu, C., Sun, Q., Schwarz, S., Ou, Y., Yang, L., et al. (2018). Prevalence and genetic analysis of mcr-3-positive Aeromonas species from humans, retail meat, and environmental water samples. Antimicrob. Agents Chemother. 62:e00404-18. doi: 10.1128/AAC.00404-18
Stoesser, N., Mathers, A. J., Moore, C. E., Day, N. P., and Crook, D. W. (2016). Colistin resistance gene mcr-1 and pHNSHP45 plasmid in human isolates of Escherichia coli and Klebsiella pneumoniae. Lancet Infect. Dis. 16, 285–286. doi: 10.1016/S1473-3099(16)00010-4
Sun, J., Zhang, X., Gao, X., Jiang, Q., Wen, Y., and Lin, L. (2016). Characterization of virulence properties of Aeromonas veronii isolated from diseased gibel carp (Carassius gibelio). Int. J. Mol. Sci. 17:496. doi: 10.3390/ijms17040496
Tamminen, M., Karkman, A., Lõhmus, A., Muziasari, W. I., Takasu, H., Wada, S., et al. (2011). Tetracycline resistance genes persist at aquaculture farms in the absence of selection pressure. Environ. Sci. Technol. 45, 386–391. doi: 10.1021/es102725n
Tekedar, H. C., Kumru, S., Blom, J., Perkins, A. D., Griffin, M. J., Abdelhamed, H., et al. (2019). Comparative genomics of Aeromonas veronii: identification of a pathotype impacting aquaculture globally. PLoS ONE 14:e0221018. doi: 10.1371/journal.pone.0221018
Tinsley, C. R., Bille, E., and Nassif, X. (2006). Bacteriophages and pathogenicity: more than just providing a toxin? Microbes Infect. 8, 1365–1371. doi: 10.1016/j.micinf.2005.12.013
Verner-Jeffreys, D. W., Welch, T. J., Schwarz, T., Pond, M. J., Woodward, M. J., Haig, S. J., et al. (2009). High prevalence of multidrug-tolerant bacteria and associated antimicrobial resistance genes isolated from ornamental fish and their carriage water. PLoS ONE 4:e8388. doi: 10.1371/journal.pone.0008388
Vincent, A. T., Paquet, V. E., Bernatchez, A., Tremblay, D. M., Moineau, S., and Charette, S. J. (2017). Characterization and diversity of phages infecting Aeromonas salmonicida subsp. salmonicida. Sci Rep 7:7054. doi: 10.1038/s41598-017-07401-7
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9:e112963. doi: 10.1371/journal.pone.0112963
Walsh, T. R., Hall, L., MacGowan, A. P., and Bennett, P. M. (1995). Sequence analysis of two chromosomally mediated inducible beta-lactamases from Aeromonas sobria, strain 163a, one a class D penicillinase, the other an AmpC cephalosporinase. J. Antimicrob. Chemother. 36, 41–52. doi: 10.1093/jac/36.1.41
Walsh, T. R., Neville, W. A., Haran, M. H., Tolson, D., Payne, D. J., Bateson, J. H., et al. (1998). Nucleotide and amino acid sequences of the metallo-beta-lactamase, ImiS, from Aeromonas veronii bv. sobria. Antimicrob. Agents Chemother. 42, 436–439. doi: 10.1128/AAC.42.2.436
Webb, H. E., Bugarel, M., den Bakker, H. C., Nightingale, K. K., Granier, S. A., Scott, H. M., et al. (2016). Carbapenem-resistant bacteria recovered from faeces of dairy cattle in the high plains region of the USA. PLoS ONE 11:e0147363. doi: 10.1371/journal.pone.0147363
Wu, C-J., Chen, P.-L., Wu, J.-J., Yan, J.-J., Lee, C.-C., Lee, H.-C., et al. (2012). Distribution and phenotypic and genotypic detection of a metallo-β-lactamase, CphA, among bacteraemic aeromonas isolates. J Med Microbiol. 61, 712–719. doi: 10.1099/jmm.0.038323-0
Yamada, T., Kawasaki, T., Nagata, S., Fujiwara, A., Usami, S., and Fujie, M. (2007). New bacteriophages that infect the phytopathogen Ralstonia solanacearum. Microbiology 153, 2630–2639. doi: 10.1099/mic.0.2006/001453-0
Yang, Q., Zhao, M., Wang, K. Y., Wang, J., He, Y., Wang, E. L., et al. (2017). Multidrug-Resistant Aeromonas veronii recovered from channel catfish (Ictalurus punctatus) in China: prevalence and mechanisms of fluoroquinolone resistance. Microbial Drug Resist. 23, 473–479. doi: 10.1089/mdr.2015.0296
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Keywords: Aeromonas veronii, antibiotic resistant, phage elements, comparative genomics, phylogenetic tree
Citation: Tekedar HC, Arick MA II, Hsu C-Y, Thrash A, Blom J, Lawrence ML and Abdelhamed H (2020) Identification of Antimicrobial Resistance Determinants in Aeromonas veronii Strain MS-17-88 Recovered From Channel Catfish (Ictalurus punctatus). Front. Cell. Infect. Microbiol. 10:348. doi: 10.3389/fcimb.2020.00348
Received: 31 March 2020; Accepted: 08 June 2020;
Published: 17 July 2020.
Edited by:
Xiangmin Lin, Fujian Agriculture and Forestry University, ChinaReviewed by:
Shengkang Li, Shantou University, ChinaXiaofeng Shan, Jilin Agricultural University, China
Copyright © 2020 Tekedar, Arick, Hsu, Thrash, Blom, Lawrence and Abdelhamed. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hossam Abdelhamed, YWJkZWxoYW1lZEBjdm0ubXNzdGF0ZS5lZHU=
 Hasan C. Tekedar
Hasan C. Tekedar Mark A. Arick II2
Mark A. Arick II2 Jochen Blom
Jochen Blom Mark L. Lawrence
Mark L. Lawrence Hossam Abdelhamed
Hossam Abdelhamed