- 1Center of Clinical Laboratory, Zhongshan Hospital, School of Medicine, Xiamen University, Xiamen, China
- 2Institute of Infectious Disease, School of Medicine, Xiamen University, Xiamen, China
In total, 49 clinical samples were analyzed using two typing schemes, Enhanced Centers for Disease Control and Prevention (ECDC) and multilocus sequence typing (MLST), to describe the molecular characteristics of circulating Treponema pallidum isolates in Xiamen between 2016 and 2017. In addition, genetic mutations potentially related to antibiotic resistance of T. pallidum were also analyzed. Forty five samples were fully typed by ECDC, and 14 different subtypes were detected. The most common subtype was 16d/f (24.4%), followed by 14d/f (20.0%). All forty nine samples were successfully typed by MLST, while only four allelic profiles were identified, including three SS14-like profiles and one Nichols-like profile. Among them, the major allelic profile was 1.1.8 (85.7%). Interestingly, the allelic profile 1.3.1 widespread in Europe and North America was not detected in this region. Additionally, A2058G mutation in 23S rRNA was found in all detectable samples (38/38), and no mutation in 16S rRNA was observed (36/36). Four non-synonymous single-nucleotide polymorphisms in penicillin-binding protein genes were found in the 35 samples eligible for Sanger sequencing. Among them, the variant in tp0500 (P564I) can only be found in the SS14-like isolates. Homoplastic changes in tp0760 (I415F/I415M) and tp0705 (A506V/A506T) were found. Moreover, the variant tp0705 A506V and the variant tp0705 A506T separately appeared in the SS14-like isolates and Nichols-like isolates, respectively. This study showed that the genotypes of T. pallidum isolates in Xiamen between 2016 and 2017 were different from those in other geographic areas. The resistance-related variants of T. pallidum isolates identified in this study could provide awareness for clinicians in the treatment of syphilis.
Introduction
Syphilis, caused by Treponema pallidum subsp. pallidum, is a chronic sexually transmitted disease. Recently, the number of cases of syphilis has dramatically increased in many countries, including China (Shen et al., 2018; Liu et al., 2019). Because of the difficulty in culture of T. pallidum in vitro, T. pallidum was cultured through in vivo rabbit testicular models in the past decades, which greatly hindered the understanding of T. pallidum biology (Ho and Lukehart, 2011). To date, molecular characterization has been an important tool to understand T. pallidum biology. Strain typing, as a powerful tool for determining the diversity of circulating T. pallidum isolates and the dynamic transmission of the infection, is widely applied in many countries (Peng et al., 2011; Dai et al., 2012; Gallo Vaulet et al., 2017; Giacani et al., 2018).
In 1998, the Centers for Disease Control and Prevention (CDC) firstly introduced a CDC typing scheme based on determining the number of 60-bp repeats in the arp gene and sequence differences by restriction fragment length polymorphism analysis (RFLP) in the tpr genes to distinguish among subtypes of T. pallidum (Pillay et al., 1998). Then, the scheme was supplemented by additional sequencing of a portion of the tp0548 gene and designated as the enhanced CDC (ECDC) typing system (Marra et al., 2010). The two typing schemes were commonly used with over 3,000 clinical T. pallidum isolates worldwide, and they enabled identification of several associations between the subtypes and specific clinical status, including the 14d/f type with neurosyphilis and the 14i/a with the serofast status (Marra et al., 2010; Zhang et al., 2017). However, the typing scheme has weaknesses, including the low amplification efficiency of the tpr genes and possible instability of the arp and tpr loci (Mikalová et al., 2013). Therefore, sequencing-based molecular typing based on sequencing of the tp0136, tp0548, and 23S rRNA genes has been introduced in parallel (Flasarova et al., 2012). With the prevalence of SS14-like strains worldwide, one of the two main T. pallidum clades, sequencing-based molecular typing was supplemented with additional sequencing of the tp0705 gene, known as the multilocus sequence typing (MLST) system, to greatly increase the genotype resolution power for SS14-like isolates (Grillova et al., 2018). The typing scheme based on direct sequencing of typing loci effectively improved the success rate of fully typed isolates and ensured stability. Moreover, it enabled the construction of phylogenetic trees to trace infections and distinguish T. pallidum from other treponemal subspecies (Grillova et al., 2018; Grillova et al., 2019a).
Additionally, there is no effective vaccine to prevent syphilis infection, antibiotic treatment remains an important strategy for syphilis control. However, the emergence of antibiotic-resistant T. pallidum isolates, similar to other bacteria that increase antimicrobial resistance due to the use of antibiotics, has always been a concern. Recently, a high prevalence of azithromycin resistance to T. pallidum isolates in geographically different areas in China and treatment failure cases have been reported (Zhou et al., 2010; Chen et al., 2013). It has been demonstrated that there is a strong association between the A2058G and A2059G mutations in 23S rRNA and macrolide resistance (Lukehart et al., 2004; Molini et al., 2016). Mutations in 16S rRNA was indicated to associate with tetracycline resistance in T. pallidum isolates (Wu et al., 2014). Moreover, previous analysis of T. pallidum genomes have shown amino acid changes in penicillin-binding proteins, although the clinical relevance of these changes is unclear so far (Pinto et al., 2016; Sun et al., 2016). This fact reminds us of the necessity to detect mutations in relevant genes and investigate the prevalence of resistances in a region.
Hence, we applied the two commonly used typing schemes, ECDC and MLST, to describe the circulating genotypes of T. pallidum isolates in Xiamen and comprehensively understand the epidemic change in T. pallidum between 2016 and 2017. In addition, we explored the 23S rRNA gene, 16S rRNA gene and three penicillin-binding protein (pbp) genes (tp0500, tp0760, and tp0705) for potential mutations associated with antibiotic resistance in the T. pallidum isolates.
Materials and Methods
Collection of Clinical Samples
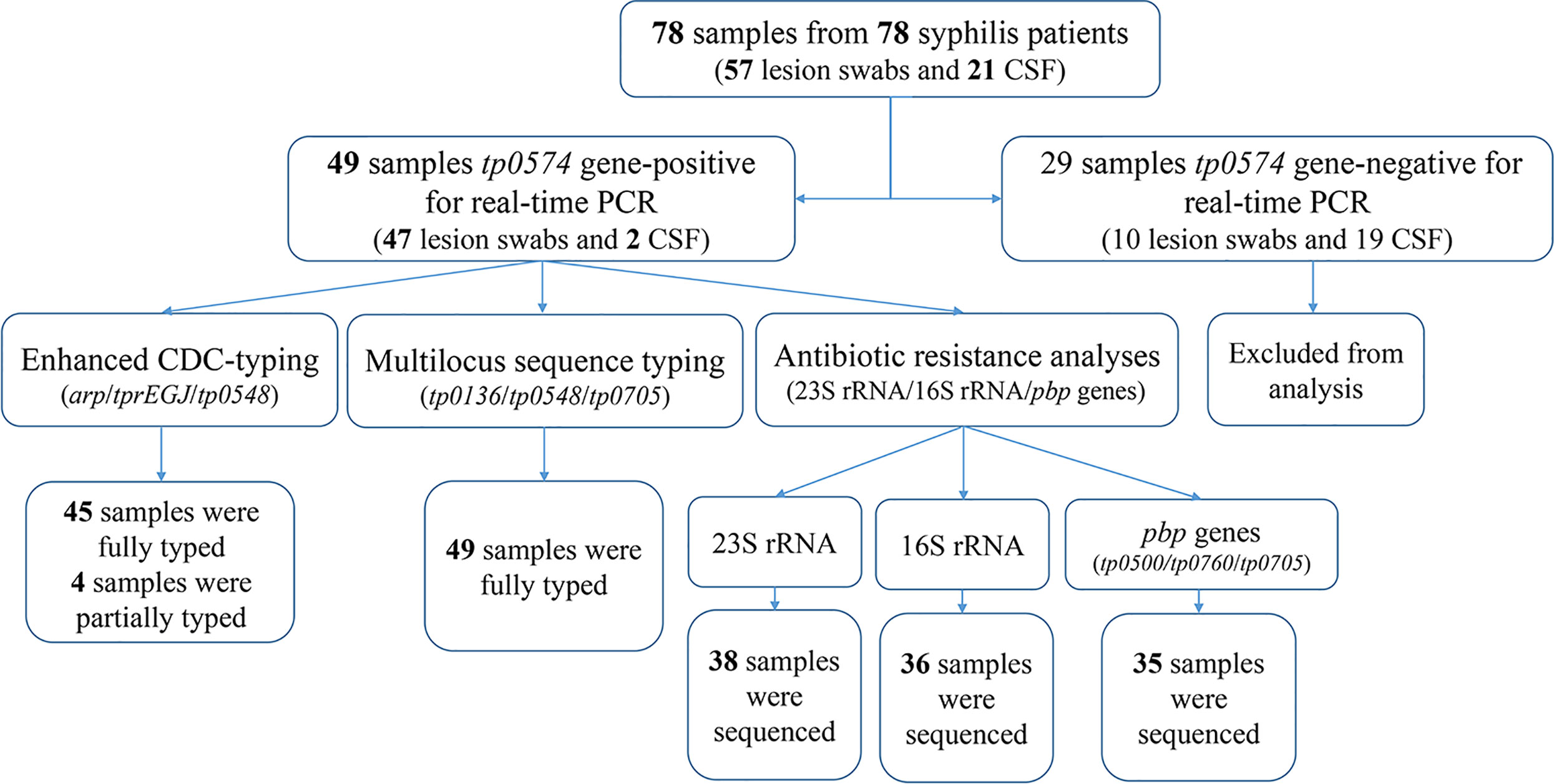
Between 2016 and 2017, 78 clinical samples were collected from 78 patients with syphilis in Xiamen, China, including 57 lesion swabs and 21 cerebrospinal fluid (CSF) samples. According to the US Centers for Disease Control and Prevention (CDC) and the European CDC (ECDC) guidelines (Janier et al., 2014; Workowski and Bolan, 2015), syphilis patients were clinically diagnosed based on clinical findings with laboratory tests including serological tests and dark-filed microscopy. Serology tests were performed using the rapid plasma regain (RPR) test (InTec, Xiamen, China) and Treponema pallidum particle agglutination (TPPA) test (Fujirebio, Tokyo, Japan). Patients were considered to have syphilis when one of the tests (serology or/and dark-field microscopy) was positive. Clinical data of the enrolled patients were collected.
Isolation and Detection of Treponemal DNA
DNA was isolated from the patient’s lesion swabs or CSF using a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) in accordance with the manufacturer’s protocol (Tong et al., 2017; Zhang et al., 2020). Real-time PCR targeting tp0574 was performed to determine the copies of treponemal DNA. The DNA samples that tested positive were stored at -20°C for the next procedure.
Molecular Typing of Treponema pallidum Isolates
The ECDC typing scheme, including the determination of the number of 60-bp repetitions in the arp genes, difference analysis in RFLP of the tprE, G, and J genes, and sequencing of an 83-bp region in the tp0548 gene, were performed as described previously (Marra et al., 2010). MLST was performed as described previously (Grillova et al., 2018). Briefly, three loci, tp0136, tp0548, and tp0705, were amplified, and the purified amplicons were sequenced. A high-fidelity PCR polymerase, KOD FX Neo polymerase (TOYOBO, Osaka, Japan), was used for amplification of the two typing schemes. DNA sequencing was performed by Sangon Biotech Company (Shanghai, China).
Detection of Mutations Potentially Associated With Antibiotic Resistance
The 23S rDNA and 16S rDNA genes were amplified and sequenced following previously described methods (Xiao et al., 2016). Three pbp genes, tp0500, tp0760, and tp0705, were amplified and sequenced to identify the reported mutations and the development of new mutations (Sun et al., 2016).
Phylogenetic Analyses
Sequence Matrix 1.8 software was used for sequence concatenation. Phylogenetic trees were generated with MEGA 5 using the Tamura 3-parameter model and 1,000 pseudorandom bootstrap replicates.
Results
Clinical Characteristics of Syphilis Patients
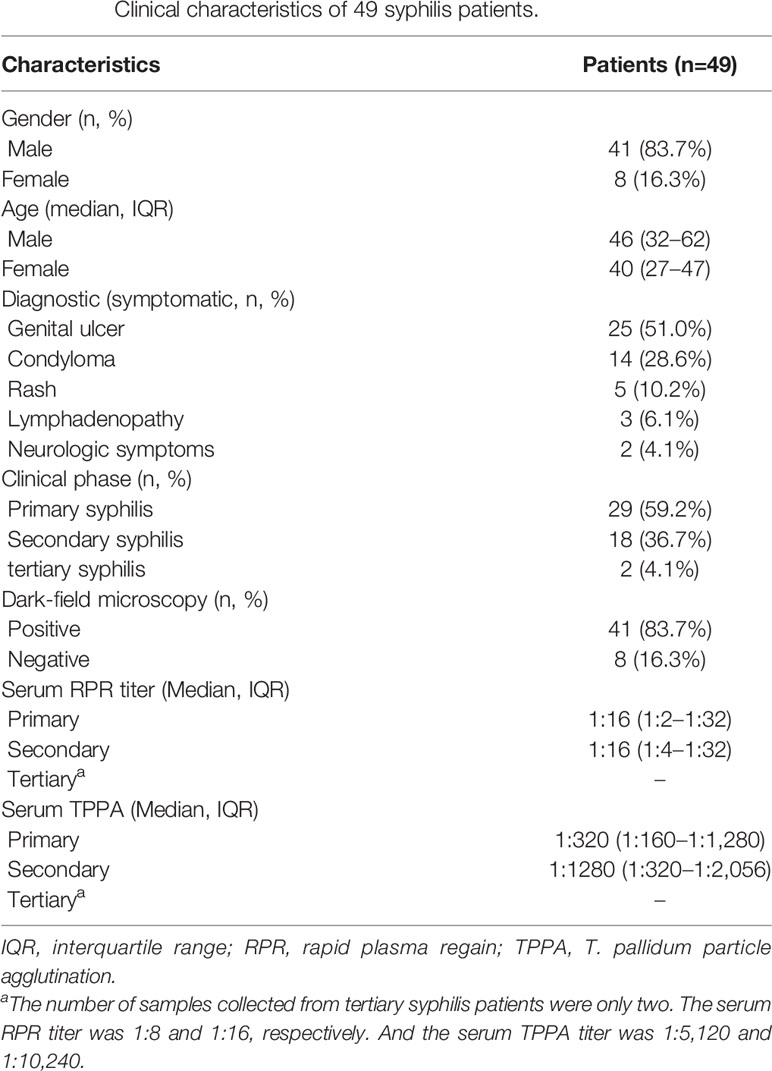
Treponemal DNA was detected positively in 49 syphilis patients, including 29 primary syphilis patients and 18 secondary syphilis patients (lesion swabs collected) and two tertiary syphilis patients (CSF collected) (Figure 1). The samples from the 49 syphilis patients were further analyzed using ECDC and MLST. The clinical information for the 49 syphilis patients is listed in Table 1. Most of the patients included in this study were male (83.7%), with a mean age of 46 years. Regarding diagnostics and clinical status, the majority of recruited patients (95.9%) were at an early syphilis stage, and 59.2% were patients with primary syphilis. Out of 49 samples, for which dark field microscopy was performed, 83.7% were positive, showing an appreciable microscopy-positive rate. Except for three primary syphilis patients with negative rapid plasma regain (RPR), all other patients were positive for serological tests, including RPR and T. pallidum particle agglutination (TPPA).
Typing of Clinical Samples Based on ECDC Typing Scheme
Forty-five samples (91.8%) were fully typeable with the ECDC typing system (including the arp, tprE/G/J, and tp0548 loci), and four samples were partially typed. The plurality of the fully typeable specimens belonged to strain type 16d/f, accounting for 24.4% (11/45). Strain type 14d/f was found to be the second most common strain, accounting for 20.0% (9/45), followed by 15d/f (13.3%), 18d/f (8.9%), 11d/f (6.7%), 17d/f (6.7%), and 22d/c (4.4%). All other subtypes, including 11d/c, 13d/ao, 13d/f, 14b/f, 19d/f, 21d/c, and 23d/c, were detected only once (Figure 2). In terms of the three typing loci, a total of 11 arp types (11, 13, 14, 15, 16, 17, 18, 19, 21, 22, and 23) were identified in these specimens, of which type 16 was the most commonly detected (11/45, 24.4%), followed by type 14, accounting for 22.2% (10/45). Two tpr RFLP patterns (b and d) were found, the majority of which belonged to type d. For the tp0548 locus, three sequence-types (c, f, and ao), including a new type ao, were identified.
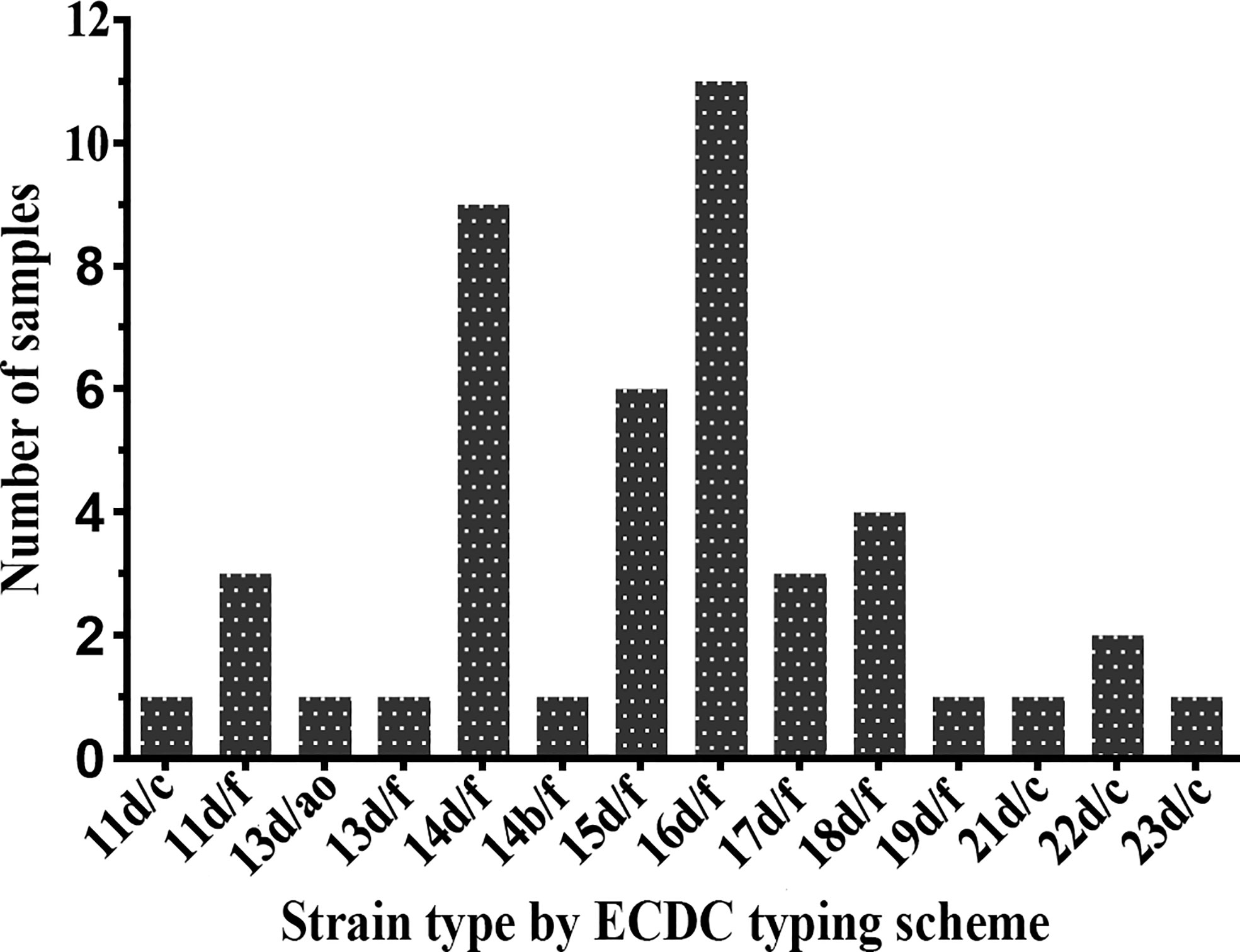
Figure 2 Distribution of Treponema pallidum subtypes by Enhanced Centers for Disease Control and Prevention (ECDC) typing scheme between 2016 and 2017.
Typing of Clinical Samples Based on MLST Typing Scheme
The 49 samples were fully typed at the tp0136, tp0548, and tp0705 loci by the MLST typing system. In contrast to the ECDC typing system results, only four different allelic profiles (including 1.1.8, 1.56.8, 1.57.8, and 3.2.3) were observed for T. pallidum isolates circulating in Xiamen (Figure 3). Among them, the 1.1.8 allelic profile (the SS14-like profile) was the most common (85.7%, 42/49). Two additional new allelic profiles (1.56.8 and 1.57.8) belonging to the SS14-like group contained novel variants in the tp0548 locus (Figure 4A). In addition, we found one profile that belonged to the Nichols-like group (3.2.3). Notably, the tp0136 locus of the strains belonging to Nichols-like group were all contained, apart from one single-nucleotide variant (SNV) (G1205A), a 6-nucleotide deletion (TTCTTC) related to TPANIC_0136 (GenBank accession number CP004010.2) (Figure 4B). Then, we compared the discrimination ability of the two typing schemes for these 49 samples and found that the common allelic profile 1.1.8 was further divided into nine subtypes by ECDC. However, the most prevalent subtypes, 16d/f and 14d/f, were not further divided by MLST (Table 2).

Figure 3 Phylogeny of allelic profiles obtained from all typed samples from 49 syphilis patients by multilocus sequence typing (MLST) typing scheme. Treponema pallidum Nichols (CP004010.1) and T. pallidum SS14 (CP04011.2) concatenated sequences of typing loci were used as reference.
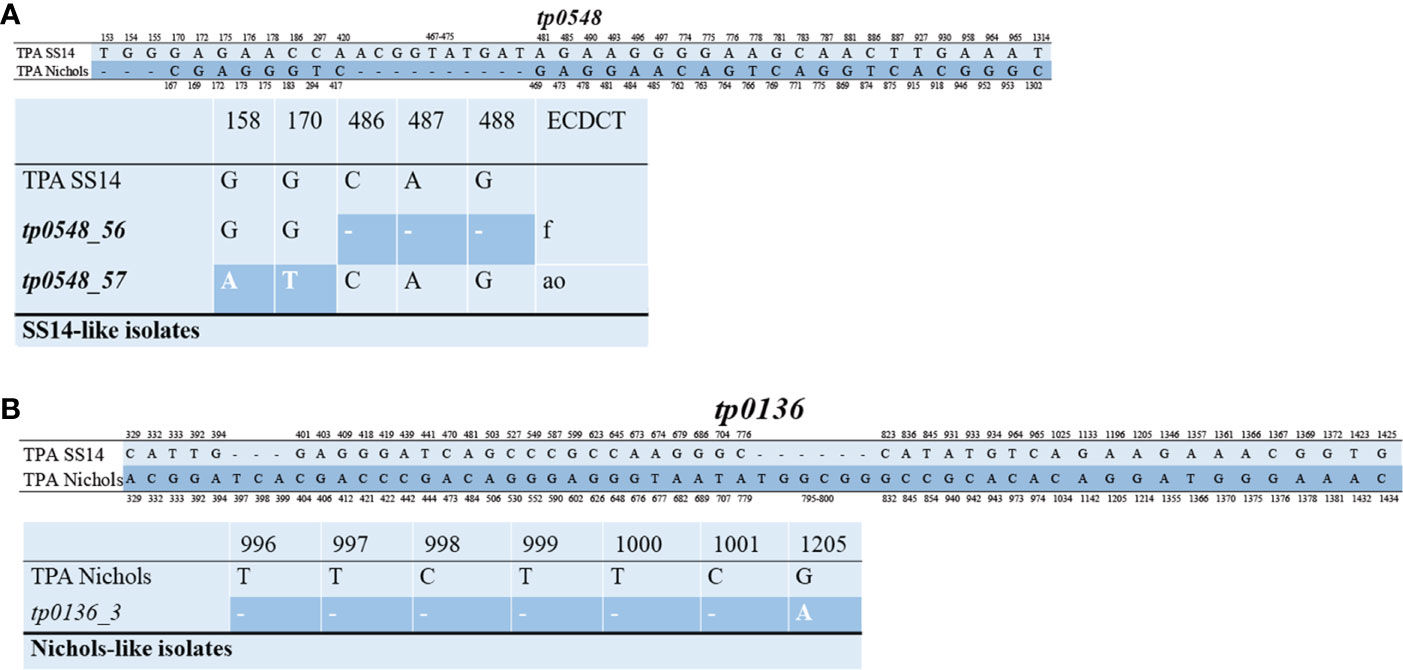
Figure 4 Alignment of the sequences in the tp0548 and tp0136 locus. Only position containing nucleotide variants are shown. Deletions are shown with dashes. (A) Upper part represents nucleotide difference between reference TPA SS14 (CP004011.1) and TPA Nichols (CP004010.2) in the tp0548 locus. Lower patter represents the novel sequence identified in SS14-like isolates in the study using MLST (coordinates correspond to TPASS_0548) and the translation to ECDCT subtypes is shown in the last column. (B) Upper part represents nucleotide difference between reference TPA SS14 (CP004011.1) and TPA Nichols (CP004010.2) in the tp0136 locus. Lower patter represents the sequence identified in Nichols-like isolates in the study by MLST (coordinates correspond to TPANIC_0136).
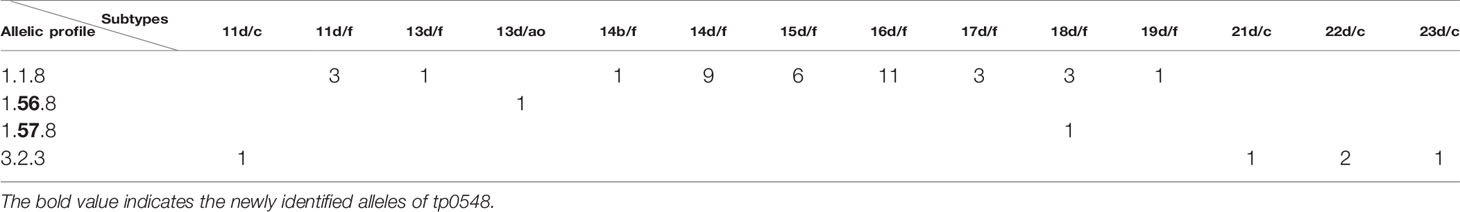
Table 2 Comparison of fully typed samples using Enhanced Centers for Disease Control and Prevention typing (ECDCT) and multilocus sequence typing (MLST). Only samples with fully typed in both typing schemes are shown.
Prevalence of Potential Mutations Associated With Antibiotic Resistance in the Treponema pallidum Isolates
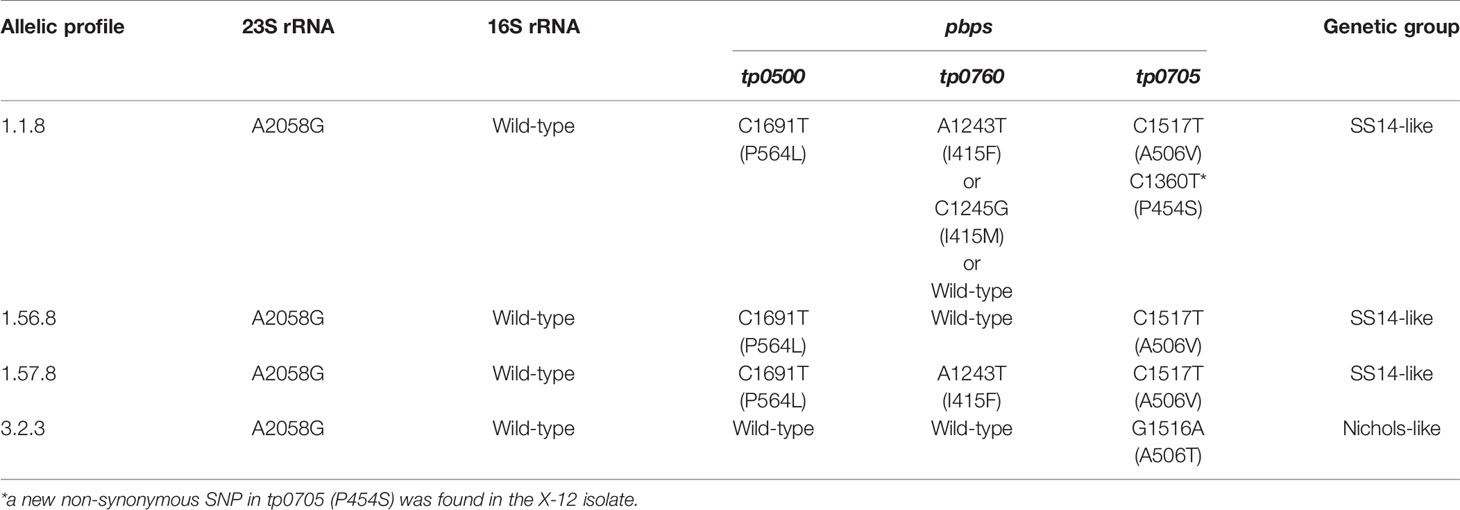
The 23S rRNA gene locus was amplified in 38 samples (out of 49) and revealed by Sanger sequencing. No isolates were found to harbor the A2059G mutation; instead, all of them harbored the A2058G mutation, which meant that the isolates belonging to the SS14-like group or Nichols-like group were potentially resistant to macrolides (Table 3). Additionally, the sequences of the 16S rRNA gene locus, potentially associated with tetracycline resistance, from 36 samples were obtained, and no mutations were found. Additionally, we detected mutations in the genes encoding penicillin-binding protein of T. pallidum (tp0500, tp0760, and tp0705). A total of 35 samples were fully identified, and four non-synonymous single-nucleotide polymorphisms (SNPs) were found. A variant in tp0500 (P564I) was exclusively present in the isolates belonging to the SS14-like group. And two SNPs affecting the same amino acid position in tp0760 (I415F/I415M) and tp0705 (A506V/A506T) were identified. In tp0760, the variant tp0760 I415F was more frequently detected. In tp0705, the variant tp0705 A506V only appeared in the SS14-like isolates and that the variant tp0705 A506T only appeared in the Nichols-like isolates. In addition, we found a new non-synonymous SNP in the position 1,360 (C->T) of tp0705 (P454S) in the X-12 isolate, which is beyond the sequence used for the MLST analysis (Table 3).
Discussion
In this study, we firstly explored the characteristics of T. pallidum isolates circulating in Xiamen, China. Two typing schemes (ECDC and MLST) were applied to determine the diversity of the T. pallidum isolates. In addition, we surveilled 23S rRNA mutations and 16S rRNA mutations to track the distribution of resistant strains in Xiamen. And we detected the genes encoding penicillin-binding proteins to monitor the reported mutations and identify the development of new mutations, although phenotypic or clinical penicillin resistance has never been documented in more than 60 years of clinical treatment for syphilis.
The study included 49 samples from 49 syphilis patients, of which 45 samples were fully characterized using the ECDC typing scheme. The results showed that the most common genotype in Xiamen was 16d/f (24.4%), while the prevalent genotype (14d/f) in Shanghai, Jiangsu, and Hunan (Dai et al., 2012; Xiao et al., 2016; Zhang et al., 2017) was the secondary, although the proportion of the two genotypes was basically equal (24.4 vs. 20.0%). This result suggested that there was a difference in the geographic distribution of syphilis-causing isolates. Moreover, the strain distribution evolved over time (Giacani et al., 2018), as previously described, with the introduction and loss of some strains in Seattle from 1999 to 2008 and the replacement of the prevalent subtype (14d/g replacing 14d/f) (Marra et al., 2010). Continuous tracking of the distribution of strains in Xiamen should be encouraged to determine whether the subtype 16d/f completely replace the 14d/f or whether a new strain type will appear and become prevalent.
In this study, all the samples were fully typed using a recently proposed typing scheme, MLST, which effectively avoids the difficulty in amplifying and analyzing tandem repeats in the arp and ambiguous restriction patterns in the tprE/G/J genes. As revealed in this study, only four allelic profiles were found in Xiamen, demonstrating less genetic diversity in comparison with that in other countries where MLST has been performed (Grillova et al., 2018; Pospisilova et al., 2018; Grillova et al., 2019b; Marangoni et al., 2019). Moreover, the most common allelic profile (1.1.8) identified in Xiamen between 2016 and 2017 was less common in other countries, and the most common allelic profile (1.3.1) reported in other areas was not found in this region. This result might indicate that the most prevalent allelic profile across the globe has not yet been introduced into China and that the strains tend to remain restricted to specific geographic areas. Notably, our study only provided a snapshot of T. pallidum isolates circulating in one area of China (Xiamen), and more research using MLST is needed to reveal the diversity of T. pallidum isolates in different areas of China to track the transmission of syphilis. In summary, we applied two typing schemes to investigate the diversity of T. pallidum isolates in Xiamen. The common allelic profile 1.1.8 was further divided by ECDC; however, the most prevalent subtype by ECDC was not further divided by MLST, which might be a consequence of less genetic diversity in the typing loci. It also indicated that the ECDC typing scheme might be better than MLST to investigate the genetic diversity of T. pallidum isolates in Xiamen. Of course, this study also demonstrated that the two typing schemes were independent and would be useful in different aspects to explore the epidemiology and evolution of T. pallidum isolates. In addition, MLST based on sequencing data yielded phylogenetic trees to better understand the epidemiology of the two genetically distinct groups of T. pallidum isolates. As described in previous studies (Beale et al., 2019), most of the sequence-characterized clinical T. pallidum isolates belonged to SS14-like groups, and a limited number of clinical isolates belonged to Nichols-like groups, which was further verified in our study. Only five Nichols-like strains were identified, accounting for 10.2% of all the studied T. pallidum isolates, and the remaining 89.8% were SS14-like strains.
In recent years, there have been several cases of clinical syphilis treatment failure associated with macrolide resistance that was conferred by the A2058G or A2059G mutations in the 23S rRNA (Zhou et al., 2010; Molini et al., 2016). Based on previous molecular analysis, multiple locations in China showed a high prevalence rate (up to over 90%) of macrolide-resistant T. pallidum isolates (Martin et al., 2009; Chen et al., 2013; Lu et al., 2015). Similarly, in Xiamen, we detected 38 T. pallidum isolates (35 SS14-like isolates and 3 Nichols-like isolates) and found all of them harbored the A2058G mutation, suggesting that macrolide-resistant T. pallidum isolates might have spread throughout the country. Recent research using direct whole-genome sequencing combined with phylogenetic analyses revealed that both SS14-like and Nichols-like lineages with genotypic macrolide resistance were simultaneously circulating, which was corroborated by our findings (Beale et al., 2019). Moreover, the study showed that the widespread macrolide-resistant SS14-like isolates in the population were due to macrolide selective pressure rather than expansion of a single fitness advantage resistant SS14-like isolate. In fact, although macrolide antibiotics are an attractive regimen for syphilis treatment, they are not frequently in the list of physicians’ prescriptions for syphilis (Lu et al., 2015). Instead, in China, macrolide antibiotics are widely used in other indications, such as upper respiratory tract infections and other sexually transmitted infections. The use of antibiotics in China was approximately 10-fold higher than that in other nations (Chen et al., 2013). The widespread macrolide-resistant T. pallidum isolates may be an indication of broader issues of antimicrobial resistance.
In contrast, all T. pallidum isolates in this study had no mutations in the 16S rRNA that could potentially confer tetracycline resistance, consistent with other reports (Xiao et al., 2016; Grillova et al., 2018). In addition, previous studies identified a number of variants affecting penicillin-binding proteins (Sun et al., 2016; Beale et al., 2019). Our study further confirmed the characteristics of homoplastic changes in tp0760 (I415F/I415M) and tp0705 (A506V/A506T). Interestingly, variants of tp0760 were exclusively reported in isolates from China and were not found in isolates from other countries (Beale et al., 2019). Moreover, Mathew et al. identified the variant tp0705 A506T both in Nichols-like isolates and in SS14-like isolates, but our study showed that the variant tp0705 A506T only appeared in Nichols-like isolates and that the variant tp0705 A506V only appeared in SS14-like isolates. The inconsistent results might reflect different evolutionary selection in T. pallidum isolates from different geographic areas. However, none of the variants had a significant functional effect on pbp genes, according to analysis by SIFT software. Despite this result, we still cannot ignore the possibility of evolutionary pressure causing resistance in an era of uncontrolled antibiotic use, and we need to keep monitoring mutations in pbp genes that are putatively involved in a decrease in susceptibility to penicillin (Beale et al., 2019).
Finally, the limitations of this study are discussed. First, the clinical and epidemiological data were not sufficient to further analyze the clinical relevance of the genotypes. Second, the data of syphilis patients regarding the history of antibiotic use or treatment was lacking, and thus, this study could not provide direct evidence about the relationship among treatment failure, macrolide resistance, and genetic mutations in T. pallidum.
This study made use of traditional typing scheme (ECDC) and a recently refined typing scheme (MLST) to reveal the genetic diversity of circulating T. pallidum isolates in Xiamen between 2016 and 2017. The most common genotype by ECDC was 16d/f and the prevalent allelic profile by MLST was 1.1.8, demonstrating that the group of T. pallidum isolates in Xiamen has geographic differences. The findings from this study offer valuable information on syphilis epidemiology as well as syphilis evolution. Moreover, this study for the first time identified genetic mutations associated with antibiotic resistance in Xiamen. It could be a indicator for clinicians in the treatment of syphilis patients.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repositories is PubMLST database of Treponema pallidum subsp. pallidum (https://pubmlst.org/tpallidum/).
Ethics Statement
The study was approved by the Ethics Committee of Zhongshan Hospital, Xiamen University, and complied with the Declaration of Helsinki. All patients provided written consent in accordance with the institutional guidelines.
Author Contributions
J-JN and T-CY conceived the study. DL, S-MH, and T-CY designed the study. DL, L-RL, and L-LL performed the wet lab experiments. DL and J-JN performed bioinformatics analyses. S-MH and X-ZZ performed research and analyzed data. L-RL performed sample collection. DL, S-MH, and T-CY wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China [grant numbers 81971147, 81972028, 81973104, 81772260, 81771312, 82001292], the Key Projects for Province Science and Technology Program of Fujian Province [grant number 2018D0014, 2019D008]. The funders played no role in the study design, data collection, or analyses, the decision to publish, or manuscript preparation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Beale M. A., Marks M., Sahi S. K., Tantalo L. C., Nori A. V., French P., et al. (2019). Genomic epidemiology of syphilis reveals independent emergence of macrolide resistance across multiple circulating lineages. Nat. Commun. 10 (1), 3255. doi: 10.1038/s41467-019-11216-7
Chen X. S., Yin Y. P., Wei W. H., Wang H. C., Peng R. R., Zheng H. P., et al. (2013). High prevalence of azithromycin resistance to Treponema pallidum in geographically different areas in China. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 19 (10), 975–979. doi: 10.1111/1469-0691.12098
Dai T., Li K., Lu H., Gu X., Wang Q., Zhou P. (2012). Molecular typing of Treponema pallidum: a 5-year surveillance in Shanghai, China. J. Clin. Microbiol. 50 (11), 3674–3677. doi: 10.1128/JCM.01195-12
Flasarova M., Pospisilova P., Mikalova L., Valisova Z., Dastychova E., Strnadel R., et al. (2012). Sequencing-based molecular typing of treponema pallidum strains in the Czech Republic: all identified genotypes are related to the sequence of the SS14 strain. Acta Derm. Venereol. 92 (6), 669–674. doi: 10.2340/00015555-1335
Gallo Vaulet L., Grillova L., Mikalova L., Casco R., Rodriguez Fermepin M., Pando M. A., et al. (2017). Molecular typing of Treponema pallidum isolates from Buenos Aires, Argentina: Frequent Nichols-like isolates and low levels of macrolide resistance. PloS One 12 (2), e0172905. doi: 10.1371/journal.pone.0172905
Giacani L., Ciccarese G., Puga-Salazar C., Dal Conte I., Colli L., Cusini M., et al. (2018). Enhanced Molecular Typing of Treponema pallidum subspecies pallidum Strains From 4 Italian Hospitals Shows Geographical Differences in Strain Type Heterogeneity, Widespread Resistance to Macrolides, and Lack of Mutations Associated With Doxycycline Resistance. Sex. Transm. Dis. 45 (4), 237–242. doi: 10.1097/olq.0000000000000741
Grillova L., Bawa T., Mikalova L., Gayetageron A., Nieselt K., Strouhal M., et al. (2018). Molecular characterization of Treponema pallidum subsp. pallidum in Switzerland and France with a new multilocus sequence typing scheme. PloS One 13 (7), e0200773. doi: 10.1371/journal.pone.0200773
Grillova L., Jolley K., Smajs D., Picardeau M. (2019a). A public database for the new MLST scheme for Treponema pallidum subsp. pallidum: surveillance and epidemiology of the causative agent of syphilis. PeerJ 6, e6182. doi: 10.7717/peerj.6182
Grillova L., Noda A. A., Lienhard R., Blanco O., Rodriguez I., Smajs D. (2019b). Multilocus Sequence Typing of Treponema pallidum subsp. pallidum in Cuba From 2012 to 2017. J. Infect. Dis. 219 (7), 1138–1145. doi: 10.1093/infdis/jiy604
Ho E. L., Lukehart S. A. (2011). Syphilis: using modern approaches to understand an old disease. J. Clin. Invest. 121 (12), 4584–4592.
Janier M., Hegyi V., Dupin N., Unemo M., Tiplica G. S., Potočnik M., et al. (2014). European guideline on the management of syphilis. J. Eur. Acad. Dermatol. Venereol. 2014) 28 (12), 1581–1593. doi: 10.1111/jdv.12734
Liu L. L., Lin Y., Zhuang J. C., Ren J., Jiang X. Y., Chen M. H., et al. (2019). Analysis of serum metabolite profiles in syphilis patients by untargeted metabolomics. J. Eur. Acad. Dermatol. Venereol. 33 (7), 1378–1385. doi: 10.1111/jdv.15530
Lu H., Li K., Gong W., Yan L., Gu X., Chai Z., et al. (2015). High frequency of the 23S rRNA A2058G mutation of Treponema pallidum in Shanghai is associated with a current strategy for the treatment of syphilis. Emerg. Microbes Infect. 4 (2), e10. doi: 10.1038/emi.2015.10
Lukehart S. A., Godornes C., Molini B. J., Sonnett P., Hopkins S., Mulcahy F., et al. (2004). Macrolide resistance in Treponema pallidum in the United States and Ireland. New Engl. J. Med. 351 (2), 154–158.
Marangoni A., Vrbová E., Grillová L., Mikalová L., Pospíšilová P., Strnadel R., et al. (2019). MLST typing of Treponema pallidum subsp. pallidum in the Czech Republic during 2004-2017: Clinical isolates belonged to 25 allelic profiles and harbored 8 novel allelic variants. PloS One 14 (5), e0217611. doi: 10.1371/journal.pone.0217611
Marra C., Sahi S., Tantalo L., Godornes C., Reid T., Behets F., et al. (2010). Enhanced molecular typing of treponema pallidum: geographical distribution of strain types and association with neurosyphilis. J. Infect. Dis. 202 (9), 1380–1388. doi: 10.1086/656533
Martin I. E., Gu W., Yang Y., Tsang R. S. (2009). Macrolide resistance and molecular types of Treponema pallidum causing primary syphilis in Shanghai, China. Clin. Infect. Dis. an Off. Publ. Infect. Dis. Soc. America 49 (4), 515–521. doi: 10.1086/600878
Mikalová L., Pospíšilová P., Woznicová V., Kuklová I., Zákoucká H., Smajs D. (2013). Comparison of CDC and sequence-based molecular typing of syphilis treponemes: tpr and arp loci are variable in multiple samples from the same patient. BMC Microbiol. 13:178. doi: 10.1186/1471-2180-13-178
Molini B. J., Tantalo L. C., Sahi S. K., Rodriguez V. I., Brandt S. L., Fernandez M. C., et al. (2016). Macrolide Resistance in Treponema pallidum Correlates With 23S rDNA Mutations in Recently Isolated Clinical Strains. Sex. Transm. Dis. 43 (9), 579–583. doi: 10.1097/olq.0000000000000486
Peng R. R., Wang A. L., Li J., Tucker J. D., Yin Y. P., Chen X. S. (2011). Molecular typing of Treponema pallidum: a systematic review and meta-analysis. PloS Negl. Trop. Dis. 5 (11), e1273. doi: 10.1371/journal.pntd.0001273
Pillay A., Liu H., Chen C. Y., Holloway B., Sturm A. W., Steiner B., et al. (1998). Molecular subtyping of Treponema pallidum subspecies pallidum. Sex. Transm. Dis. 25 (8), 408–414. doi: 10.1097/00007435-199809000-00004
Pinto M., Borges V., Antelo M., Pinheiro M., Nunes A., Azevedo J., et al. (2016). Genome-scale analysis of the non-cultivable Treponema pallidum reveals extensive within-patient genetic variation. Nat. Microbiol. 2, 16190. doi: 10.1038/nmicrobiol.2016.190
Pospisilova P., Grange P. A., Grillova L., Mikalova L., Martinet P., Janier M., et al. (2018). Multi-locus sequence typing of Treponema pallidum subsp. pallidum present in clinical samples from France: Infecting treponemes are genetically diverse and belong to 18 allelic profiles. PloS One 13 (7), e0201068. doi: 10.1371/journal.pone.0201068
Shen X., Liu D., Lin Y., Zhu X. Z., Lin L. R., Tong M. L., et al. (2018). The characteristics of beta 2-glycoprotein I-dependent anticardiolipin antibody and blood coagulation status in subjects with classical biological false-positive syphilis reactions. Int. Immunopharmacol. 62, 132–138. doi: 10.1016/j.intimp.2018.05.033
Sun J., Meng Z. F., Wu K. Q., Liu B. A., Zhang S. F., Liu Y. D., et al. (2016). Tracing the origin of Treponema pallidum in China using nextgeneration sequencing. Oncotarget 7 (28), 42904–42918.
Tong M.-L., Zhao Q., Liu L.-L., Zhu X.-Z., Gao K., Zhang H.-L., et al. (2017). Whole genome sequence of the Treponema pallidum subsp. pallidum strain Amoy: An Asian isolate highly similar to SS14. PloS One 12 (8), e0182768. doi: 10.1371/journal.pone.0182768
Workowski K. A., Bolan G. A. (2015). Sexually Transmitted Diseases Treatment Guidelines. 2015 MMWR Recomm. Rep. Morb. Mortal. Wkly. Rep. Recomm. Rep. 64 (RR-03), 1–137.
Wu B. R., Liu W. C., Wu P. Y., Su Y. C., Yang S. P., Hung C. C., et al. (2014). Surveillance study of Treponema pallidum harbouring tetracycline resistance mutations in patients with syphilis. Int. J. Antimicrob. Agents 44 (4), 370–372. doi: 10.1016/j.ijantimicag.2014.06.009
Xiao Y., Liu S., Liu Z., Xie Y., Jiang C., Xu M., et al. (2016). Molecular Subtyping and Surveillance of Resistance Genes In Treponema pallidum DNA From Patients With Secondary and Latent Syphilis in Hunan, China. Sex. Transm. Dis. 43 (5), 310–316. doi: 10.1097/OLQ.0000000000000445
Zhang R. L., Wang Q. Q., Zhang J. P., Yang L. J. (2017). Molecular subtyping of Treponema pallidum and associated factors of serofast status in early syphilis patients: Identified novel genotype and cytokine marker. PloS One 12 (4), e0175477. doi: 10.1371/journal.pone.0175477
Zhang S., Yang M.-J., Peng B., Peng X.-x., Li H. (2020). Reduced ROS-mediated antibiotic resistance and its reverting by glucose in Vibrio alginolyticus. Environ. Microbiol. 22 (10), 4367–4380. doi: 10.1111/1462-2920.15085
Keywords: syphilis, Treponema pallidum isolates, strain typing, ECDC, MLST, antibiotic resistance
Citation: Liu D, He S-M, Zhu X-Z, Liu L-L, Lin L-R, Niu J-J and Yang T-C (2021) Molecular Characterization Based on MLST and ECDC Typing Schemes and Antibiotic Resistance Analyses of Treponema pallidum subsp. pallidum in Xiamen, China. Front. Cell. Infect. Microbiol. 10:618747. doi: 10.3389/fcimb.2020.618747
Received: 29 October 2020; Accepted: 31 December 2020;
Published: 19 February 2021.
Edited by:
Yang Zhang, University of Pennsylvania, United StatesReviewed by:
Hao Zhang, University of Pennsylvania, United StatesLinda Grillova, Institut Pasteur, France
Copyright © 2020 Liu, He, Zhu, Liu, Lin, Niu and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Jun Niu, bml1amlhbmp1bjIxMUB4bXUuZWR1LmNu; Tian-Ci Yang, eWFuZ3RpYW5jaUB4bXUuZWR1LmNu
†These authors have contributed equally to this work
 Dan Liu1,2†
Dan Liu1,2† Li-Li Liu
Li-Li Liu Li-Rong Lin
Li-Rong Lin Jian-Jun Niu
Jian-Jun Niu Tian-Ci Yang
Tian-Ci Yang