- 1Department of Medical Instrumental Analysis, Zunyi Medical University, Zunyi, China
- 2School of Stomatology, Zunyi Medical University, Zunyi, China
- 3Department of Immunology, Zunyi Medical University, Zunyi, China
Periodontitis, a prevalent inflammatory oral disease, is intricately linked to disruptions in the oral microbiome, a state known as microbial dysbiosis. This review explores the pivotal roles of key pathogens, including Porphyromonas gingivalis and Tannerella forsythia, in driving periodontitis and examines the underlying molecular mechanisms that disrupt microbial homeostasis. We discuss how interactions among bacterial species affect the oral ecosystem’s balance and how microbial metabolites influence the host immune responses, contributing to disease progression. Leveraging these insights, we propose cutting-edge therapeutic approaches aimed at restoring microbial equilibrium. These include personalized pharmacological interventions tailored to individual microbiome profiles and innovative microbiome-targeted strategies such as probiotic formulations and bacteriophage therapy. By precisely modulating microbial communities, these strategies hold promise for enhancing treatment efficacy, preventing disease recurrence, and mitigating issues like antimicrobial resistance. Overall, this review paves the way for novel prevention and management techniques in periodontitis, offering significant improvements in oral health outcomes for patients.
1 Introduction
The oral cavity constitutes a highly complex microbial ecosystem, hosting a diverse array of microorganisms. These microbes play a pivotal role in maintaining oral health and in the transition to disease states. The oral microbiome is the second most diverse after the gut, and this complex microbial community is crucial in balancing health and disease, providing essential metabolic functions and immune support for the host (Caselli et al., 2020). In a healthy state, a balanced relationship exists among these microbes, with mutual regulation maintaining oral homeostasis (Hussein et al., 2022; Santonocito et al., 2022). However, when this balance is disrupted, certain pathogenic microbes begin to overgrow, leading to a loss of biodiversity, which is a key factor in the development of oral diseases such as periodontal disease (Mensi et al., 2023).
Periodontitis is an extremely common chronic oral disorder that significantly impacts patients’ oral health. It is caused by a complex polymicrobial community within dental plaque and an imbalance in the oral microbial ecology, resulting from uncontrolled gingivitis (Hajishengallis et al., 2012). Affecting approximately 20-50% of the global population, it is characterized by the loss of periodontal tissue support, alveolar bone resorption, and gingival inflammation (Nazir et al., 2020). The formation of pathogenic biofilm around the teeth is a key factor in the occurrence of periodontitis. In the early stages of the disease, multiple bacteria synergistically act on the tooth surfaces, forming a complex biofilm structure (Tomasi et al., 2000). The bacteria embed and proliferate within the biofilm matrix, leading to plaque formation, which interacts with the host immune system to trigger an inflammatory response (Johnston et al., 2021). If left untreated, the persistent biofilm can result in tooth loss as the gingiva and alveolar bone recede from the tooth surface, leading to the formation of periodontal pockets as a consequence of periodontal ligament destruction (Sanz et al., 2017). The periodontal pocket provides an ideal hiding place for bacteria, making them more difficult to eliminate, thereby promoting the persistence and exacerbation of the inflammatory response.
Current conventional treatments for periodontitis primarily rely on mechanical debridement and antimicrobial agents (Sanz et al., 2020; Herrera et al., 2022). However, conventional treatments have limited efficacy and unclear long-term effects (Khattri et al., 2020). Thus, active treatment should be followed by supportive care to maintain periodontal health (Kwon et al., 2021). Accordingly, advancing our knowledge of the pathogenic mechanisms involved in periodontitis and developing new effective treatment strategies is critically necessary. This review summarizes the mechanisms of how microbial dysbiosis leads to periodontitis and the treatment approaches based on restoring microbial balance. The novelty of this review is manifested not only through an in-depth exploration of how main periodontal pathobionts contribute to the process of periodontitis but, more significantly, in the proposition of innovative therapeutic strategies aimed at restoring microbial equilibrium. By integrating targeted host microbiome interventions with conventional treatment modalities, this review aspires to offer a more comprehensive and enduring approach to the management of periodontitis. Furthermore, this review aims to enhance our understanding of the etiology of periodontitis, laying a solid foundation for preventive health management and the development of innovative treatment methodologies with wide-ranging clinical implications.
2 The microbial code of periodontitis
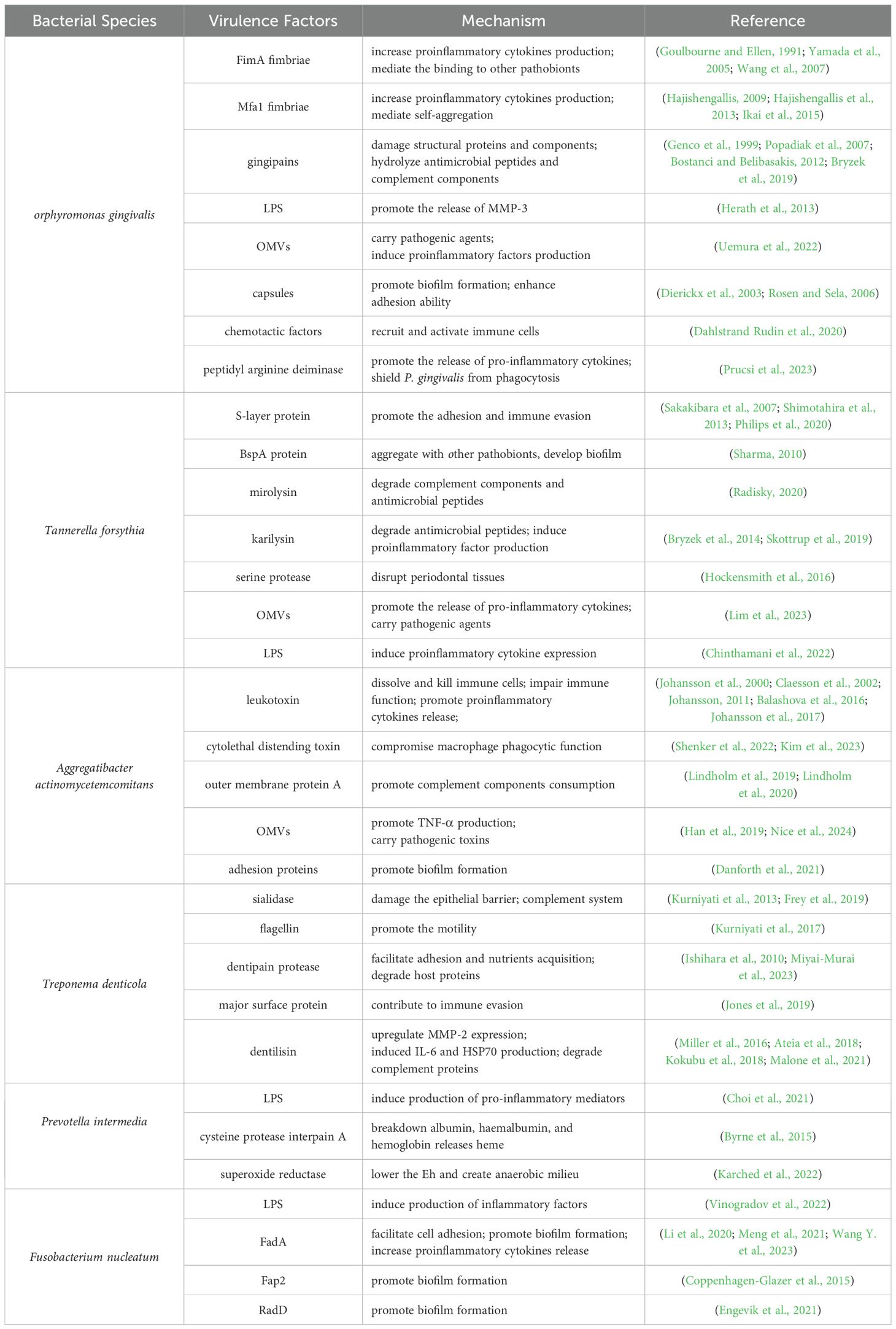
Under normal conditions, the oral cavity hosts a balanced symbiotic relationship among various microbial populations (Bacali et al., 2022). However, when the proportion of certain pathobionts (such as anaerobic Gram-negative bacteria) increases significantly, it leads to an imbalance in the oral microbial ecology (Li et al., 2022; Zhu et al., 2023) These pathobionts possess strong virulence and can trigger inflammatory responses. The major pathobionts identified include Porphyromonas gingivalis (P. gingivalis), Tannerella forsythia (T. forsythia), Aggregatibacter actinomycetemcomitan (A. actinomycetemcomitan), Treponema denticola (T. denticola), Prevotella intermedia (P. intermedia) and Fusobacterium nucleatum (F. nucleatum). These organisms are associated with the development of periodontitis under dysbiotic conditions.
2.1 P. gingivalis
P. gingivalis is an anaerobic, Gram-negative, short-rod bacterium and is recognized as the primary etiological agent of periodontitis (Torrungruang et al., 2015). P. gingivalis exerts its pathogenicity through various virulence factors (Figure 1), which play crucial roles in the initiation and progression of periodontal disease.
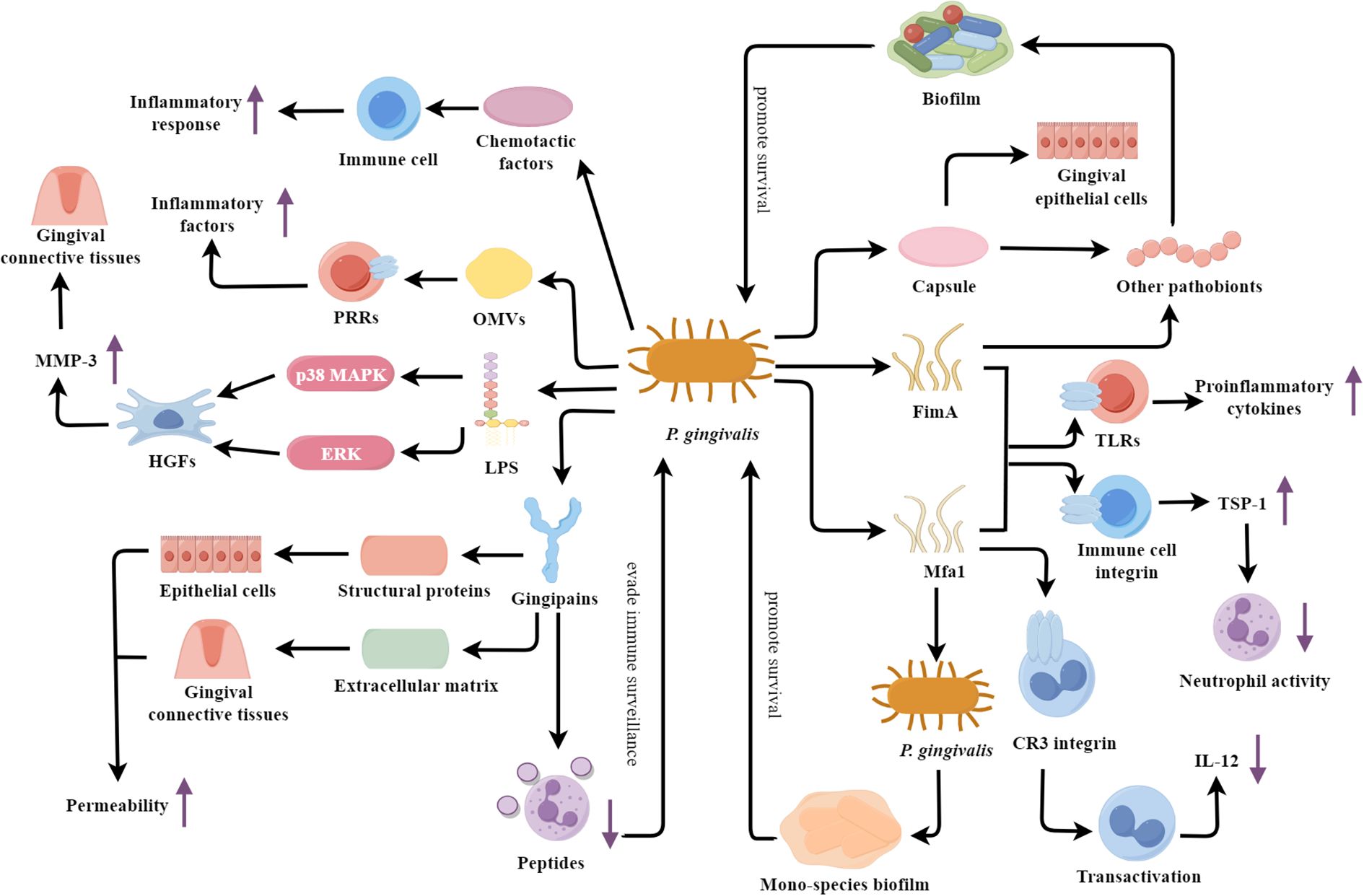
Figure 1. P. gingivalis: a veritable arsenal of virulence factors fueling periodontal destruction. TLRs, Toll-like receptors; LPS, Lipolyaccharide; OMVs, Outer membrane vesicles; HGFs, Human gingival fibroblasts; MMP-3, Matrix metalloproteinase-3.
One of the key virulence factors is the bacterial fimbriae, including FimA and Mfa1. These fimbrial structures mediate the adhesion and invasion of P. gingivalis into host epithelial and immune cells (Hasegawa and Nagano, 2021). Recent studies have elucidated the structural basis of FimA and Mfa1 interaction with host cell receptors, revealing a sophisticated mechanism of host manipulation and immune evasion. FimA and Mfa1 fimbriae can induce immune responses via interaction with Toll-like receptors (TLRs), resulting in proinflammatory cytokine production (Hajishengallis et al., 2006; Hasegawa and Nagano, 2021). They can also engage with host immune cell integrins, initiating a signaling pathway that culminates in TSP-1 secretion (Angabo et al., 2024). TSP-1 then inhibits neutrophil activity by decreasing ROS levels, ultimately improving bacterial survival (Angabo et al., 2024). In addition, FimA fimbriae can mediate the binding of P. gingivalis to other pathobionts, such as Actinomyces and Prevotella spp., then promoting biofilm formation (Goulbourne and Ellen, 1991; Yamada et al., 2005). Meanwhile, Mfa1 fimbriae participate in the self-aggregation of P. gingivalis and regulate the formation of mono-species biofilms (Ikai et al., 2015). This self-aggregation is now understood to be a critical step in biofilm maturation, significantly enhancing resistance to antimicrobial agents. Mfa1 can bind to the complement receptor 3 (CR3) integrin on the surface of macrophages, inducing their transactivation and suppressing the production of interleukin-12 (IL-12) (Hajishengallis, 2009; Hajishengallis et al., 2013). FimA can promote the entry of P. gingivalis into macrophages and inhibit the production of the key immune factor IL-12p70 through its interaction with host TLR2 and CR3 receptors (Wang et al., 2007).
Another important virulence factor is the secreted gingipains, which can disrupt the host cell-cell junctions and extracellular matrix. The gingipains of P. gingivalis mainly include arginine-specific gingipains (RgpA, RgpB) and lysine-specific gingipains (Kgp) (Kadowaki, 2021). These enzymes are capable of hydrolyzing the structural proteins of the host, compromising the tight junctions between epithelial cells and increasing the permeability of the epithelial barrier, thus providing an easier route for P. gingivalis to invade the host tissues (Genco et al., 1999; Bostanci and Belibasakis, 2012). Furthermore, the gingipains can also degrade components of the extracellular matrix, such as cell adhesion proteins and collagen, disrupting the integrity of the gingival connective tissues, amplifying the inflammatory response, and promoting the deeper invasion of P. gingivalis into the host (Genco et al., 1999; Bostanci and Belibasakis, 2012). Additionally, the gingipains can hydrolyze the antimicrobial peptides of neutrophils, suppressing the microbicidal functions of these immune cells and enabling P. gingivalis to evade host immune surveillance (Bryzek et al., 2019). These gingipains can also cleave and inactivate key complement components C3 and C5, thereby blocking the complement cascade reaction and achieving immune evasion (Popadiak et al., 2007). In addition, RgpA specifically binds to the complement inhibitory protein C4b-binding protein through its unique hemagglutinin-adhesin domain, further preventing the deposition of the membrane attack complex on the surface of P. gingivalis, protecting the bacteria from direct complement attacks (Potempa et al., 2008).
The lipopolysaccharide (LPS) of P. gingivalis is also a key contributor to the pathogenesis of periodontitis. The penta-acylated P. gingivalis LPS (LPS1690) significantly upregulates MMP-3 expression in HGFs through the activation of p38 mitogen-activated protein kinase (MAPK) and extracellular signal-regulated kinases (ERK) signaling pathways (Herath et al., 2013). The dysregulated expression of MMP-3 may disrupt the homeostatic balance of the gingival connective tissues and facilitate the chronic inflammatory response (Razavi et al., 2023). Additionally, the outer membrane vesicles (OMVs) secreted by P. gingivalis can carry multiple virulence factors and bind to the pattern recognition receptors (PRRs) of host cells, activating immune cells and inducing the release of large amounts of inflammatory factors (Uemura et al., 2022). P. gingivalis also possesses another crucial virulence factor—capsule. The capsule structure not only facilitates the binding of P. gingivalis with other pathobionts (such as F. nucleatum) to form biofilms, but also enhances the adhesion of P. gingivalis to gingival epithelial cells. These properties contribute to the persistent infection of P. gingivalis (Dierickx et al., 2003; Rosen and Sela, 2006). Chemotactic factors released by P. gingivalis significantly influence the progression of periodontitis. For instance, research by Rudin et al (Dahlstrand Rudin et al., 2020). has identified various water-soluble chemotactic factors, including short-chain fatty acids, which effectively recruit and activate neutrophils into the periodontal pocket via the FFAR2 receptor-mediated signaling pathway, thereby enhancing local inflammatory responses. The peptidylarginine deiminase (PPAD) secreted by P. gingivalis also plays a significant role in the progression of periodontitis. PPAD not only stimulates neutrophils to secrete abnormal levels of IL-6 and TNF-α but also affects the generation of ROS and phagocytic activity in neutrophils, thereby enhancing the survival of P. gingivalis (Prucsi et al., 2023).
2.2 T. forsythia
T. forsythia is a Gram-negative, anaerobic, spindle-shaped bacterium that is considered a key pathobiont responsible for periodontitis in humans (Kirst et al., 2015; Hottmann et al., 2021). T. forsythia is highly prevalent in subgingival biofilms of patients with advanced periodontal disease and its abundance is strongly correlated with disease severity (Bloch et al., 2017). It employs a multifaceted arsenal of virulence factors to subvert host defenses and drive the pathogenesis of periodontitis (Figure 2).
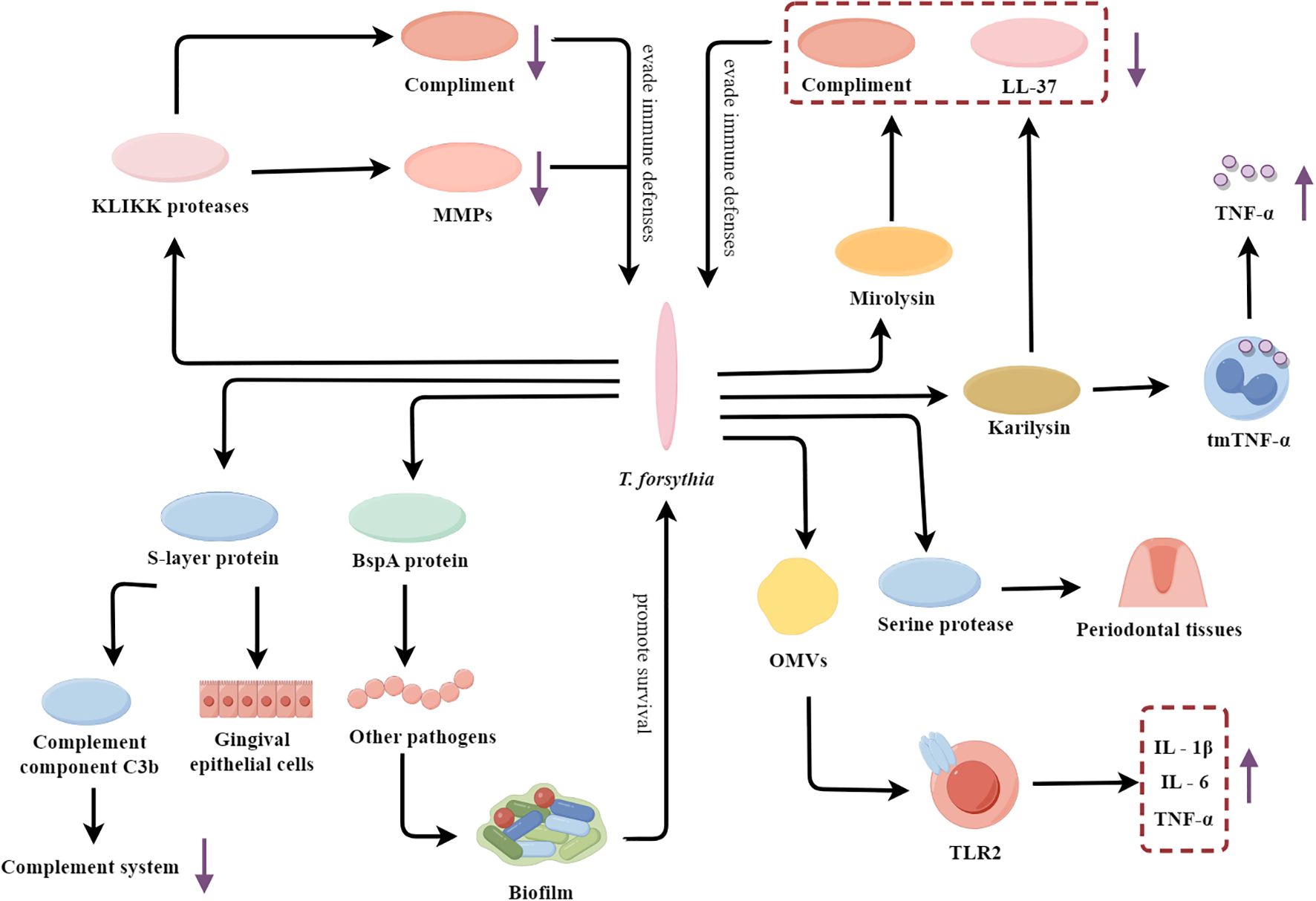
Figure 2. T. forsythia’s multifaceted arsenal: fueling the flames of periodontal inflammation. OMVs, Outer membrane vesicles; TLR2, Toll-like receptor 2; tmTNF-α, The membrane-bound form of TNF-α; MMPs, Matrix metalloproteinases.
T. forsythia contains a variety of virulence factors such as surface-layer (S-layer) protein, Bacteroides surface protein A (BspA), and KLIKK protease, which enhance its virulence and adaptability to the host environment. The S-layer protein can mediate the specific adhesion of the bacteria to the human gingival epithelial cells, enhancing the affinity of the bacteria to the human gingival epithelial cells (Sakakibara et al., 2007; Philips et al., 2020). Furthermore, the S-layer protein plays a crucial role in the immune evasion strategies of T. forsythia. It has been shown to inhibit the deposition of complement component C3b onto the surface of T. forsythia cells, thereby suppressing the complement activation pathway to resist complement-mediated bactericidal action (Shimotahira et al., 2013). A key virulence strategy of T. forsythia involves the evasion and interference of host immune defenses. The BspA protein promotes the formation of more pathogenic multispecies biofilms through mediating interspecies interactions (Sharma, 2010). The KLIKK proteases secreted by T. forsythia not only cleave and inactivate complement proteins to evade complement-mediated killing, but also specifically inhibit MMPs, which are critical components of the host’s antimicrobial arsenal (Koziel et al., 2010; Książek et al., 2023). Among the KLIKK proteases, mirolysin and karilysin are the primary enzymes that play significant pathological roles. Mirolysin and karilysin can cleave complement components and degrade the antimicrobial peptide LL-37 (Skottrup et al., 2019; Radisky, 2020). And karilysin has been demonstrated that it can cleave the membrane-bound form of tumor necrosis factor-α (TNF-α) on the surface of macrophages, leading to the release of the pro-inflammatory cytokine TNF-α and triggering an inflammatory response (Bryzek et al., 2014). This cleavage amplifies inflammation and contributes to tissue destruction and bone resorption, leading to chronic periodontitis. T. forsythia can also secrete another protease which is serine protease, an enzyme that directly cleaves gelatin and type I collagen, thus disrupting the structural integrity of periodontal tissues (Hockensmith et al., 2016). In addition, T. forsythia also actively induces a dysregulated inflammatory response by releasing OMVs containing many virulence factors. These OMVs activate TLR2 signaling in host cells and then induce host cells to produce excessive inflammatory mediators (IL‐1β, IL‐6, and TNF-α) by activating the TLR2 pathway (Lim et al., 2023). The LPS of T. forsythia is also capable of inducing the release of pro-inflammatory factors. For instance, its LPS significantly induces the release of the chemokine IP-10 in macrophages through the activation of the TLR4 and STAT1 signaling pathways (Chinthamani et al., 2022). This uncontrolled inflammatory response eventually leads to severe damage and degradation of periodontal tissue.
2.3 A. actinomycetemcomitans
A. actinomycetemcomitans is a key etiological agent of periodontitis in adolescents, and its pathogenicity has been extensively studied (Fine et al., 2007). This Gram-negative, facultative anaerobic coccobacillus utilizes a multifaceted arsenal of virulence factors to subvert host immunity and exacerbate periodontal tissue destruction (Figure 3).
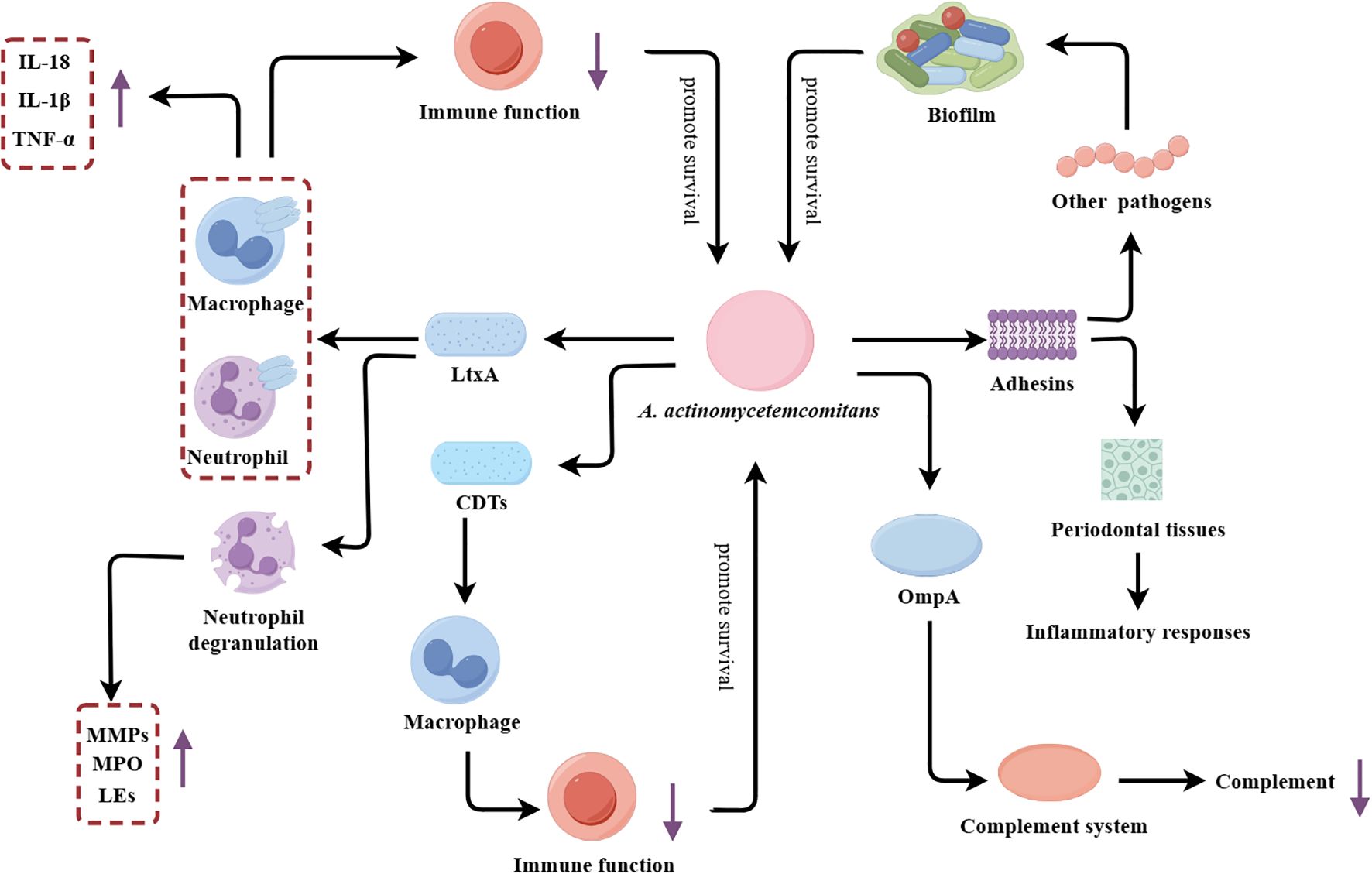
Figure 3. A. actinomycetemcomitans’s multifaceted virulence factors driving periodontal pathogenesis. LtxA, Leukotoxin A; CDTs, Cytotoxic distending toxins; MMPs, Matrix metalloproteinases; MPO, Myeloperoxidase; Les, Lysosomal enzymes; OmpA, Outer membrane protein A.
The principal virulence determinant of A. actinomycetemcomitans is repeats in toxin (RTX) toxin. The RTX toxin of A. actinomycetemcomitans is leukotoxin (LtxA), which plays a pivotal role in the pathogenesis of this organism. LtxA is a potent leukotoxin that selectively disrupts host immune cells, inducing both humoral and cellular immune responses, and ultimately amplifying tissue damage (Johansson, 2011; Johansson et al., 2017). Mechanistically, LtxA facilitates the degradation of type I collagen in periodontal tissues by inducing degranulation and dissolution of neutrophils, thereby promoting the release and activation of MMP-8 (Claesson et al., 2002). In addition to MMP-8, LtxA has been demonstrated to facilitate the release of other substances by inducing neutrophil degranulation, including lactoferrin and elastase, which contribute to the destruction of periodontal tissue (Johansson et al., 2000). LtxA also impairs host immune function by recognizing and binding to cells expressing human lymphocyte function-associated antigen 1 (CD11a/CD18), inducing lysosomal damage and causing cytoplasmic acidification (Balashova et al., 2016). Moreover, LtxA can activate inflammatory signaling pathways in immune cells, leading to the production of inflammatory cytokines. For instance, previous research has demonstrated that LtxA can induce human macrophages to release a significant amount of the pro-inflammatory cytokine IL-1β through the activation of caspase-1 (Kelk et al., 2005).
Beyond the RTX toxin, A. actinomycetemcomitans possesses additional virulence factors that amplify its pathogenic effects. Similar to other pathobionts, A. actinomycetemcomitans enhances its adaptability by forming biofilms. The diverse array of adhesins such as EmaA and Aae expressed on the bacterial surface promote adherence to other microbes, further facilitating biofilm formation (Danforth et al., 2021). In addition to LtxA, A. actinomycetemcomitans can also secrete another toxin, namely cytolethal-distending toxins (CDTs). CDTs can impair macrophage function by disrupting phagosome maturation and phagolysosome formation, thereby inhibiting the clearance of A. actinomycetemcomitans (Shenker et al., 2022; Kim et al., 2023). Additionally, the secreted outer membrane protein OmpA, which activates the complement system and consumes complement components, allowing the bacterium to evade host humoral immunity and enhance antibiotic tolerance (Lindholm et al., 2019; Lindholm et al., 2020).
2.4 T. denticola
T. denticola is a Gram-negative, anaerobic spirochete that is widely recognized as a key etiological agent of periodontitis (Ng et al., 2019; Goetting-Minesky et al., 2021). T. denticola employs a sophisticated arsenal of virulence factors to subvert host defenses and promote the development and progression of destructive periodontal disease (Figure 4).
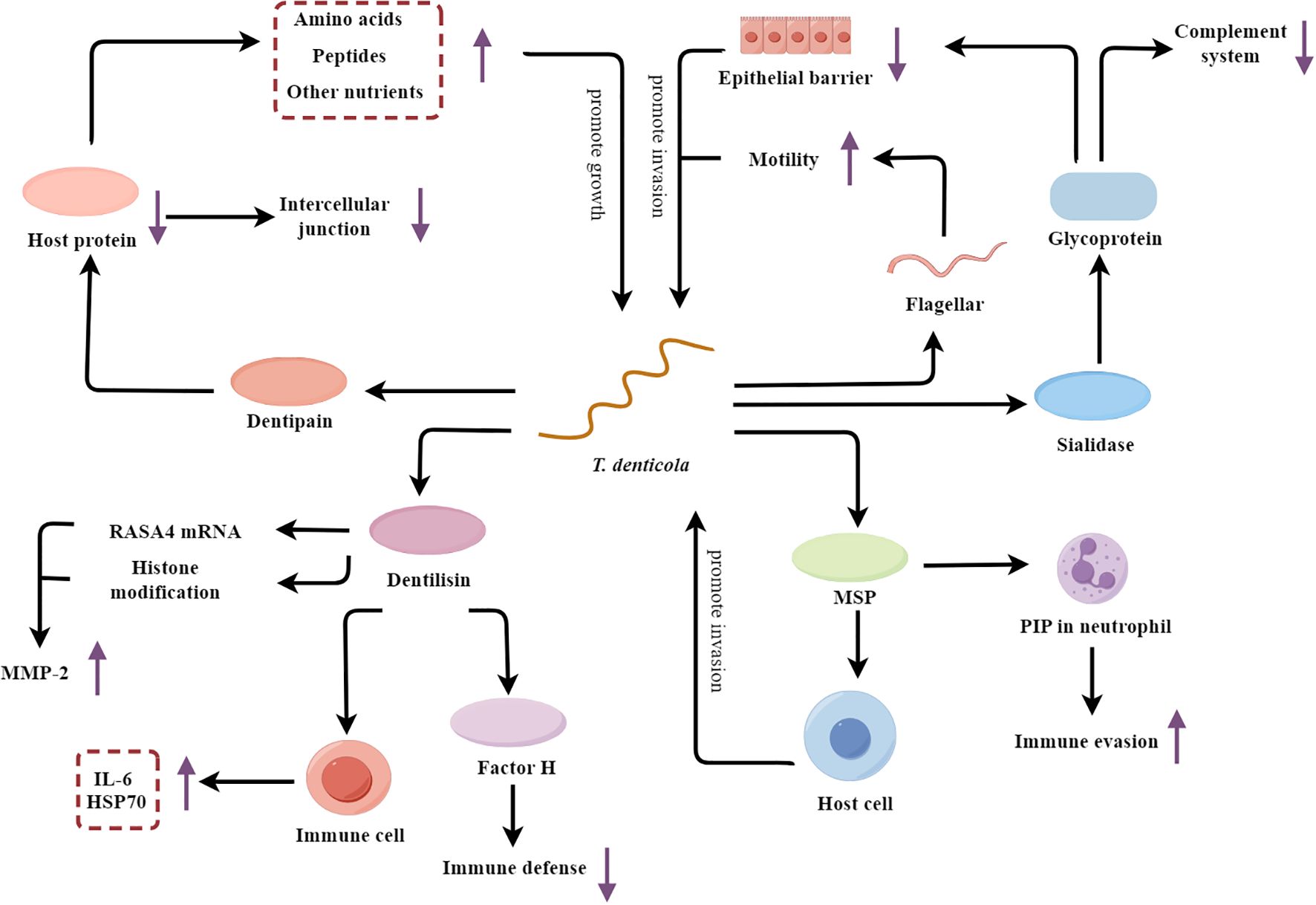
Figure 4. T. denticola’s multifaceted mechanisms fueling periodontal pathogenesis. MSP, major surface protein; PIP, Phosphoinositide processing; MMP-2, Metalloproteinase-2; HSP70, Heat shock protein 70.
T. denticola secretes sialidase, which hydrolyzes sialic acid from host glycoproteins at the tight junctions between epithelial cells, thereby compromising the integrity of the epithelial barrier and facilitating the invasion of host tissues (Frey et al., 2019). The hydrolysis of sialic acid not only disrupts host tissues, but also releases hydrolyzed sialic acid as a nutritional source for T. denticola’s growth (Kurniyati et al., 2013). Furthermore, T. denticola alters the glycosylation patterns on the host cell surfaces by hydrolyzing sialic acid, thereby interfering with the function of several key complement factors, such as C1q and Factor H (Kurniyati et al., 2013). This interference weakens the activity of the complement system, enhancing the bacterium’s survival rate. Except for sialidase, the flagellar motility of T. denticola enhances its ability to penetrate the epithelium and access the underlying periodontal tissues (Kurniyati et al., 2017). Glycosylation at specific sites on flagellin (such as at the S116 position) may interfere with the interaction between flagellin and TLR5 in the host immune system, facilitating bacterial evasion of the host immune response (Kurniyati et al., 2017).
In addition to breaching the epithelial barrier, T. denticola also actively adheres to and invades the host cells. The dentipain protease secreted by the bacterium promotes its adhesion and facilitates the acquisition of essential nutrients (Miyai-Murai et al., 2023). Dentipain is a cysteine protease that can degrade a variety of host proteins, including those involved in cell-cell junctions and the extracellular matrix (Ishihara et al., 2010; Miyai-Murai et al., 2023). By degrading these host proteins, dentipain allows T. denticola to more easily adhere to and invade the host epithelial and connective tissues. Additionally, the breakdown of host proteins provides T. denticola with a source of amino acids, peptides, and other nutrients that the bacterium can utilize for its growth and proliferation within the periodontal environment. Besides the dentipain, the major surface protein (MSP) of T. denticola also synergistically enhances the bacterium’s adherence and invasive capabilities (Godovikova et al., 2019; Jones et al., 2019). Interestingly, MSP also contributes to immune evasion by blocking phosphoinositide processing (PIP) in neutrophils, thereby impairing their phagocytic and microbicidal functions (Jones et al., 2019). Furthermore, T. denticola secretes the protease dentilisin, which upregulates the expression of MMP-2 through mechanisms involving RAS p21 protein activator 4 (RASA4) mRNA expression or histone modification (Ateia et al., 2018; Malone et al., 2021). This leads to the degradation of the extracellular matrix and the destruction of periodontal tissues. The dentilisin also stimulates host immune cells, causing these cells to produce more inflammatory mediators IL-6 and heat shock protein 70 (HSP70), thus exacerbating the inflammatory response (Kokubu et al., 2018). Dentilisin also degrades complement proteins such as factor H, thereby diminishing the effectiveness of innate immune defenses (Miller et al., 2016).
2.5 P. intermedia
P. intermedia is an anaerobic, Gram-negative short-rod bacterium that is strongly associated with the development and progression of chronic periodontitis (Petrović et al., 2018). It contributes to the initiation and progression of periodontitis through various mechanisms (Figure 5).
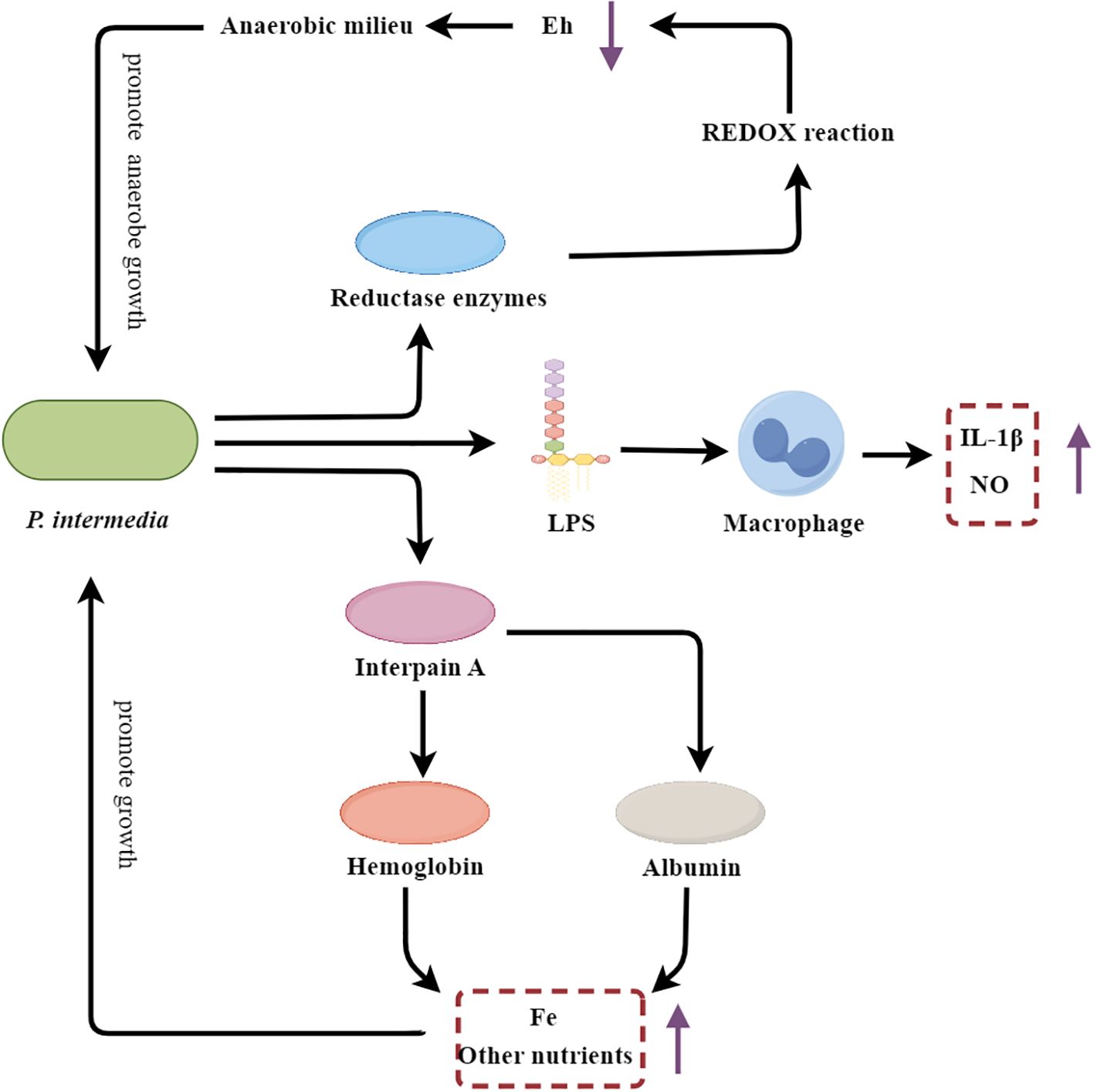
Figure 5. P. intermedia’s multifarious strategies fueling periodontal inflammation and destruction. Eh, REDOX potential; LPS, Lipopolysaccharide.
P. intermedia contributes to the initiation of the inflammatory cascade through the activation of host immune cells. The LPS component of the P. intermedia cell wall stimulates macrophages to produce pro-inflammatory mediators, such as NO and IL-1β, thereby amplifying the inflammatory response within the periodontal tissues (Choe et al., 2021). Additionally, P. intermedia secretes a cysteine protease known as interpain A, which degrades host proteins, including albumin and hemoglobin (Byrne et al., 2015). The resulting albumin fragments can serve as a nutrient source, particularly for iron, which is essential for the growth and proliferation of P. intermedia and other oral pathobionts (Byrne et al., 2015). Moreover, interpain A-mediated degradation of hemoglobin releases heme, a critical iron source that supports the growth of the anaerobic microbial community within the periodontal pocket (Byrne et al., 2015). Beyond its direct proinflammatory and nutrient-scavenging capabilities, P. intermedia also plays a crucial role in shaping the local periodontal environment to favor the establishment and persistence of anaerobic pathobionts. This bacterium secretes various reductase enzymes, including superoxide reductase, which lower the redox potential (Eh) within the periodontal pocket, creating a more anaerobic milieu that selectively promotes the growth of anaerobic species (Karched et al., 2022). Furthermore, P. intermedia has been shown to suppress the phagocytic activity of host neutrophils through the inhibition of phagosome maturation and the reduction of ROS production, thereby diminishing the production of oxygen radicals and further contributing to the development of an anaerobic environment (Doke et al., 2017).
2.6 F. nucleatum
F. nucleatum is an anaerobic, Gram-negative, fusiform-shaped bacterium that is widely recognized as a major etiological agent of periodontal disease (Song et al., 2023). F. nucleatum employs a multifaceted approach to promote the initiation and progression of destructive periodontitis (Figure 6).
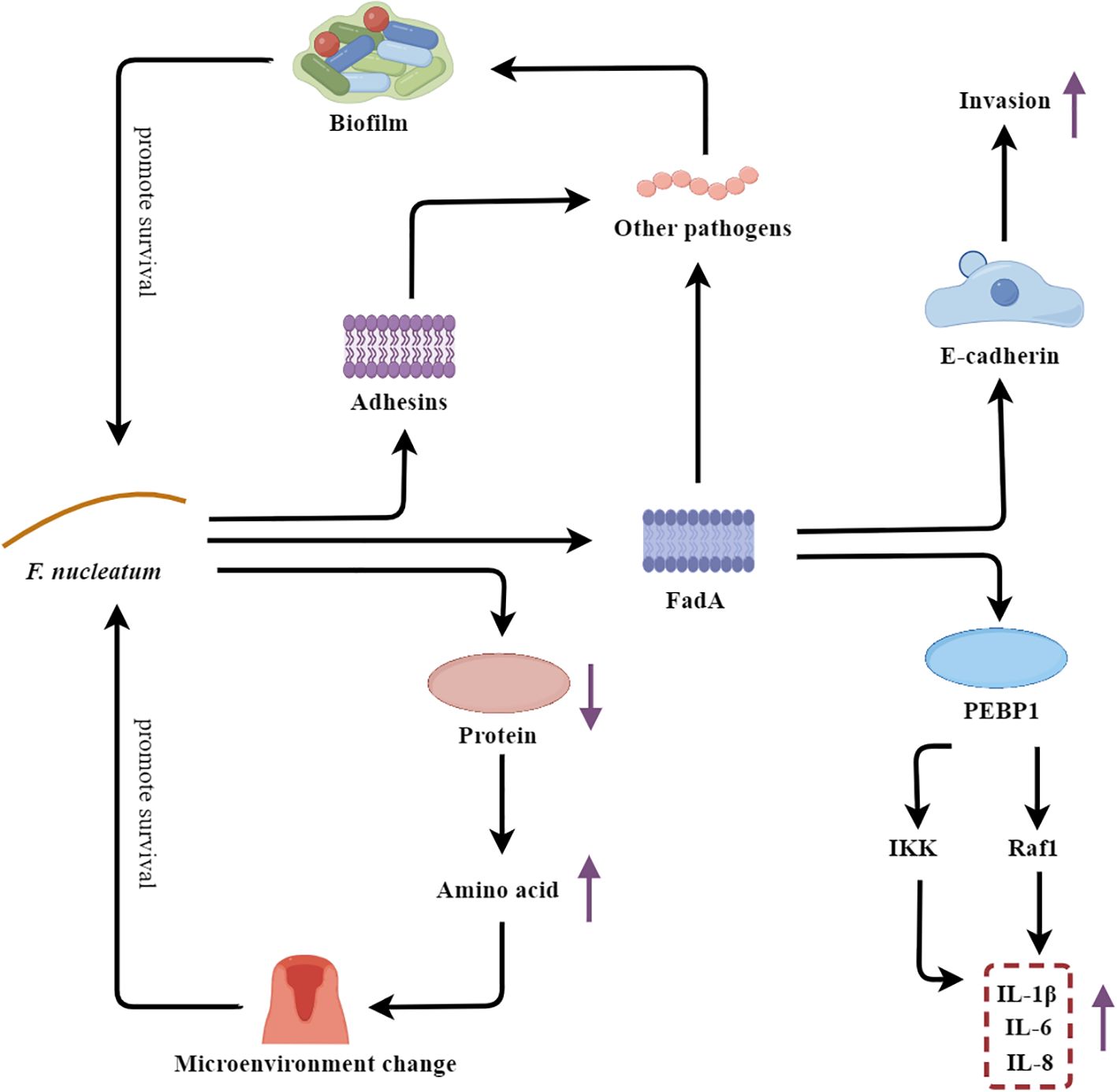
Figure 6. F. nucleatum’s multifaceted strategies fueling the vicious cycle of periodontal disease. PEBP1, Cytoplasmic protein phosphatidylethanolamine-binding protein 1.
As an adhesive bacterium, F. nucleatum plays a crucial role in maintaining the integrity of the polymicrobial biofilms associated with periodontal disease. One of the key virulence factors of F. nucleatum is the adhesin FadA, which facilitates the bacterium’s colonization and invasion of the host. FadA binds to E-cadherin on the surface of oral epithelial cells, enabling F. nucleatum to adhere to and penetrate the epithelial barrier (Meng et al., 2021). Moreover, FadA also promotes the adhesion of F. nucleatum to other pathobionts, contributing to the formation of a stable and cohesive biofilm structure within the dental plaque (Li et al., 2020; Meng et al., 2021). In addition to FadA, F. nucleatum also expresses other adhesins, such as RadD and Fap2, which further enhance the bacterium’s ability to interact with and integrate into the polymicrobial biofilm community (Coppenhagen-Glazer et al., 2015; Shokeen et al., 2020). The formation of this complex biofilm matrix confers increased resistance to host defenses and antimicrobial agents, allowing F. nucleatum to thrive in the adverse periodontal environment. Additionally, the FadA adhesin of F. nucleatum also functions as a potent virulence factor by triggering a robust host immune response. FadA binds to the cytoplasmic protein phosphatidylethanolamine-binding protein 1 (PEBP1), leading to the activation of the Raf1 and IKK signaling pathways, which in turn activate the ERK-JNK-p38 MAPK and NF-κB-p65 cascades. This signaling culminates in the production of a range of proinflammatory mediators (IL-1β, IL-6, and IL-8), thereby amplifying the inflammatory response within the periodontal tissues (Wang Y. et al., 2023). Furthermore, F. nucleatum is capable of hydrolyzing proteins and generating an abundance of amino acid metabolites, which can alter the local oral microenvironment, such as increasing the pH and redox potential. These changes in the periodontal milieu create conditions that favor the growth and proliferation of other pathogenic microorganisms, thereby contributing to the dysbiosis and progression of periodontal disease (Wang M. et al., 2023; Hara et al., 2024).
3 Dysbiosis and periodontitis
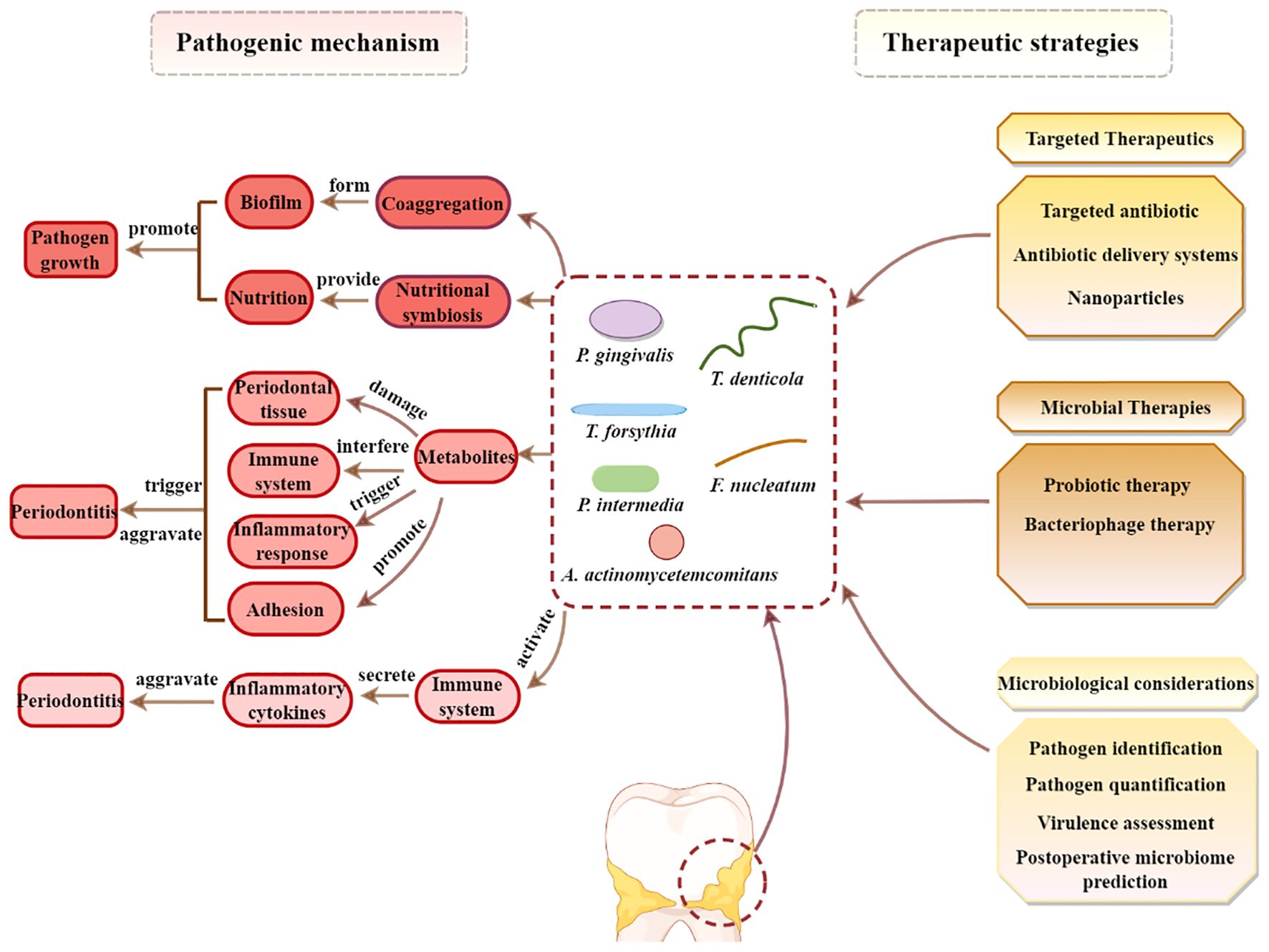
Dysbiosis, an imbalance in the microbial community, is a key factor in the development of periodontitis. Periodontopathic bacteria initiate and perpetuate periodontitis through various mechanisms (Figure 7). These mechanisms involve bacterial interactions, host immune responses, and the production of bacterial virulence factors, which interact and form a vicious cycle, ultimately triggering and aggravating periodontitis, leading to the progressive destruction of periodontal tissues (Santos et al., 2020; Xu et al., 2020; Teles et al., 2022).
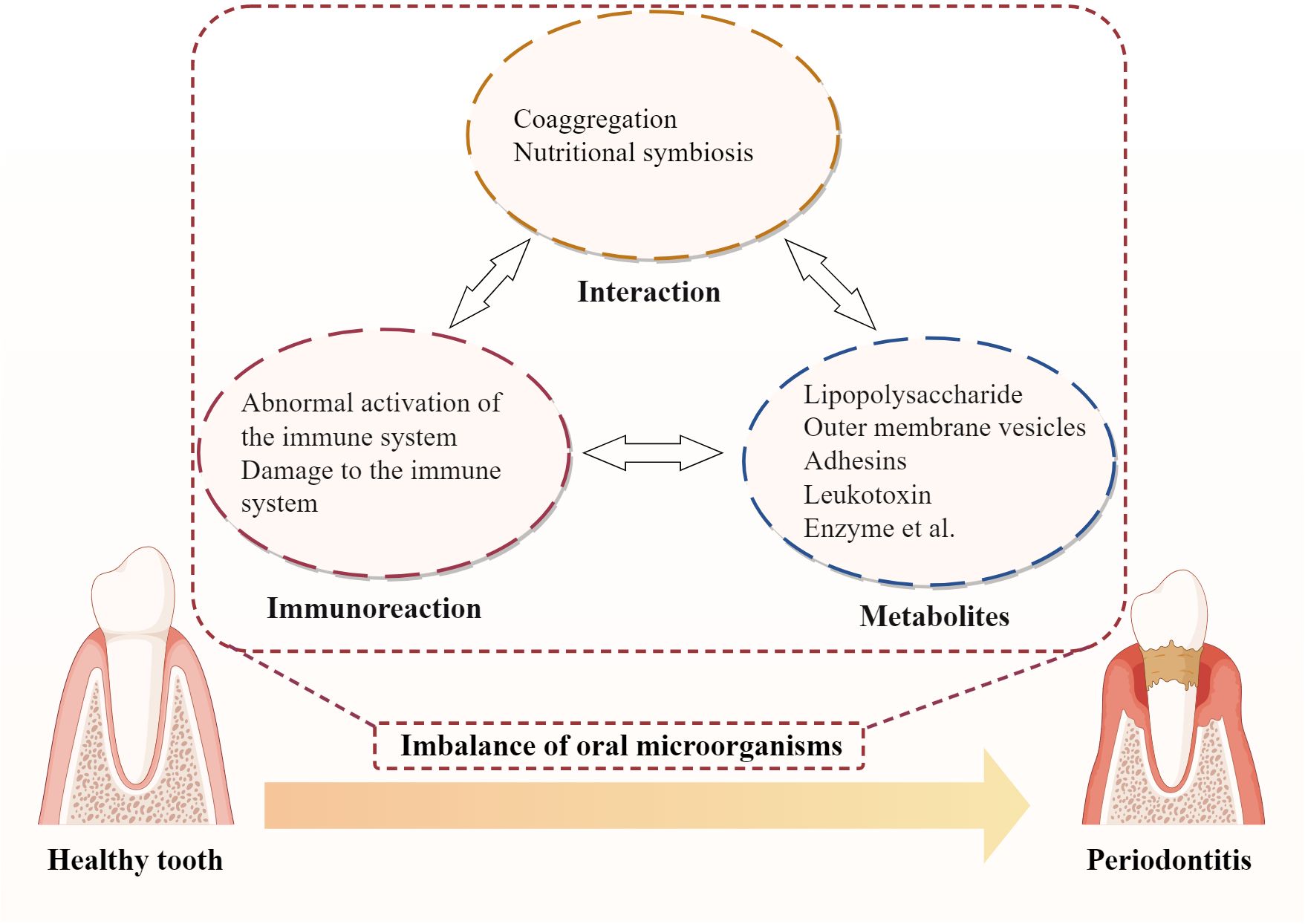
Figure 7. Vicious cycles of periodontal pathogenesis: microbial interaction, host immunity and bacterial virulence.
3.1 Microbial interactions in Periodontitis
The oral cavity is a complex microbial ecosystem, harboring a diverse range of microorganisms. In a healthy state, these microbes coexist in a balanced equilibrium, maintaining oral health. However, when this balance is disrupted, an overgrowth of certain pathobionts can lead to the development of periodontitis (Łasica et al., 2024).
Microbial interactions play a significant role in the progression of periodontitis. One such interaction is coaggregation, where certain bacteria secrete signaling molecules that promote the aggregation and growth of other pathobionts, collectively forming a resilient dental plaque biofilm (Valm, 2019; Jakubovics et al., 2021; Qiao et al., 2023). For instance, P. gingivalis is capable of establishing biofilms in conjunction with pathobionts such as Actinomyces and Prevotella spp., facilitated by its fimbriae (Goulbourne and Ellen, 1991; Yamada et al., 2005). These biofilms provide a protective environment, shielding both P. gingivalis and other pathobionts from the host immune system and antibiotic treatments. A critical determinant in the successful colonization of oral pathobionts is their capacity to form stable biofilm structures in conjunction with other microbial communities present in the oral cavity. A successful establishment of P. gingivalis in the oral environment relies on its ability to interact with primary colonizers within the dental biofilm, such as Streptococcus oralis and Streptococcus gordonii (Lamont et al., 1993; Kuboniwa et al., 2009). Similarly, the ability of T. forsythia to successfully colonize the oral environment is predicated upon its synergistic interactions with other pathobionts within the red complex, such as P. gingivalis and T. denticola (Jiang et al., 2021). Additionally, quorum sensing mechanisms allow bacteria to communicate and coordinate their activities, enhancing biofilm robustness. Research conducted by Su et al (Su et al., 2023). has demonstrated that quorum-sensing signaling molecules promote the aggregation and information exchange among key microbial communities such as Streptococcus and the Prevotella-Fusobacteria complex, which can enhance the structural stability and resilience of biofilms. Moreover, QS-mediated metabolic cooperation and environmental adaptability enable pathogens to establish an ecological niche conducive to their survival and proliferation.
Another type of microbial interaction is nutritional symbiosis, where bacteria exchange metabolic products provide essential nutrients for bacteria’s growth. For example, amino acids constitute a significant source of energy for F. nucleatum’s growth. F. nucleatum is capable of engaging in nutritional complementation and metabolic product exchange with bacteria possessing proteolytic capabilities, such as P. gingivalis, Streptococcus gordonii, and Veillonella parvula (Sakanaka et al., 2022). This interaction facilitates the effective acquisition of the requisite amino acids. P. gingivalis degrades hemoglobin to produce heme, which is utilized by Actinomyces for growth (Zhang L. et al., 2020; Śmiga et al., 2024). Veillonella utilizes lactic acid produced by Streptococcus and Actinomyces for its metabolism, while Streptococcus and Actinomyces use acetic acid produced by Veillonella as a carbon source (Scheiman et al., 2019; Zmysłowska-Polakowska et al., 2023). This metabolic exchange not only supports bacterial proliferation but also creates a suitable environment conducive to pathogenicity. F. nucleatum is able to enhance the availability of ornithine in co-culture with Streptococcus gordonii, thereby facilitating putrescine production (Sakanaka et al., 2022). This metabolic augmentation not only creates a putrescine-rich microenvironment on the surface of the F. nucleatum biofilm but also alters the biofilm phenotype of P. gingivalis, accelerating its transition from a planktonic state to a mature biofilm (Sakanaka et al., 2022). Additionally, P. intermedia secretes a variety of reductases, including superoxide reductase, which reduce the Eh within the periodontal pocket, thereby creating a more anaerobic environment (Karched et al., 2022). Such an environment not only selectively promotes the growth of anaerobes such as P. gingivalis, but also creates favorable conditions for the development of pathogenicity.
3.2 Microbial metabolites and pathological changes in periodontal tissues
Microbial metabolites are closely associated with pathological changes in periodontal tissues. Pathogenic bacteria in the oral cavity produce various toxic substances, such as enzymes and endotoxins, through their metabolic activities. These substances can directly compromise the integrity of periodontal tissues and indirectly cause damage via the induction of the host’s immune response. Table 1 summarizes the major pathobionts and the mechanisms of action of their virulence factors. The metabolic products of these microorganisms have potent biological activities that directly stimulate gingival tissue, initiating a local inflammatory response.
Various substances associated with oral pathobionts, such as the cell wall component LPS and metabolic products like short-chain fatty acids, exhibit potent biological activity (Huang and Guan, 2023; Park et al., 2023). Table 1 summarizes the major pathobionts and the mechanisms of action of their virulence factors. As shown in Table 1, these metabolites can directly stimulate gingival tissues, triggering local inflammatory responses. For instance, LPS from P. gingivalis and Escherichia coli can directly stimulate gingival fibroblasts and epithelial cells, initiating an inflammatory response (Yang et al., 2021; Yang et al., 2024). Moreover, these metabolites can suppress the defensive functions of host cells, exacerbating periodontal damage. Butyrate, produced by P. gingivalis, inhibits the clearance of inflammatory factors by fibroblasts and promotes their apoptosis, compromising the periodontal barrier (Shirasugi et al., 2017; Tsukahara et al., 2020; Nakagawa et al., 2021). Furthermore, these microorganisms and their products can activate the host’s specific immune response, inducing the production of a plethora of inflammatory mediators, such as cytokines and matrix metalloproteinases. For example, LPS from P. gingivalis can activate the NF-κB signaling pathway, leading to the activation of M1 macrophages. M1 macrophages secrete cytokines like IL-1β and TNF-α, which activate osteoclasts and release matrix metalloproteinases, causing the destruction of periodontal tissues (Huang et al., 2019; Xu et al., 2023; Li et al., 2024). The OMVs secreted by P. gingivalis can penetrate host tissues, promoting neutrophil infiltration and inducing macrophages to produce pro-inflammatory mediators, thereby exacerbating periodontitis (Fleetwood et al., 2017; De Souza et al., 2022; Kim et al., 2022; Wielento et al., 2022).
3.3 Dysbiosis and the host immune response
The local periodontal microenvironment in periodontitis patients exhibits immunological differences compared to healthy individuals. In a healthy state, the host immune system can recognize and eliminate oral pathobionts, maintaining oral health. This involves a balanced interaction between the innate and adaptive immune responses, with antimicrobial peptides and immune surveillance mechanisms effectively controlling microbial balance. When dysbiosis occurs, it triggers the host immune response, leading to inflammation to clear the pathobionts (Shahbeik et al., 2021). However, persistent dysbiosis can overwhelm these regulatory mechanisms, leading to chronic inflammatory response and persistent microbial dysbiosis.
The inflammatory response involves the recruitment and activation of immune cells, which have both pro-inflammatory and anti-inflammatory functions. Immune cells, such as macrophages, neutrophils, and T cells, are not only defensive cells in the progression of periodontitis but can also contribute to the development of inflammation (Marinho et al., 2015; Mishra et al., 2023). On the one hand, the pro-inflammatory factors they secrete, such as IL-1β and IL-6, attract more immune cells to the lesion, prolonging the inflammation (Marinho et al., 2015; Mishra et al., 2023). Additionally, these cytokines can activate osteoclasts, leading to the resorption of alveolar bone and furthering tissue destruction (Alippe et al., 2017). The prolonged inflammatory response can lead to excessive activation and damage to the host immune system, resulting in tissue destruction and pain (Zhang J. et al., 2020). Specifically, the activity of these cells can also affect the balance of the host immune system, promoting detrimental immune regulatory mechanisms. For instance, IL-6 and IL-23 secreted by M1 macrophages can promote the expansion of pathogenic Th17 cells, exacerbating periodontitis (Dutzan et al., 2018). Moreover, the polarization of T cells toward a Th17 phenotype can inhibit regulatory T cells, which are crucial for maintaining immune tolerance and preventing excessive inflammation. Pro-inflammatory cytokines such as IL-1β, IL-6, and IL-8 in the environment of periodontitis stimulate neutrophils to transition into a more aggressive and pro-inflammatory state (Williams et al., 2021). This shift toward a pro-inflammatory phenotype disrupts the balance between effector and regulatory immune responses, leading to the excessive production of inflammatory mediators (Williams et al., 2021). These altered neutrophils are also capable of releasing reactive oxygen species and proteases, which not only exacerbate tissue destruction but also further recruit additional immune cells, amplifying the inflammatory cascade (Williams et al., 2021).
4 Therapeutic strategies for periodontitis based on microbial dysbiosis
Recent research on the microbiological mechanisms of periodontitis has demonstrated that its development is not only associated with the presence of specific pathobionts but also closely related to the overall balance of the oral microbiome (Albuquerque-Souza and Sahingur, 2022; Di Stefano et al., 2022; Liu et al., 2023). Based on this concept, novel therapeutic strategies have been developed that aim to prevent and treat periodontitis by regulating and managing the composition of the microbial community.
4.1 Targeted therapeutics for microbial dysbiosis
Nonsurgical treatment of periodontitis primarily involves the removal of dental plaque and calculus through scaling and root planning (SRP), often supplemented with local or systemic antibiotic therapy (Khattri et al., 2020). However, the escalating risk of bacterial resistance, coupled with the potential disruption of the beneficial oral microbiota balance due to antibiotic use, presents significant challenges. Additionally, pathobionts can form biofilm structures that protect them from antibiotic attacks, further diminishing the efficacy of treatment. Consequently, strategies for eliminating pathobionts are continually being updated and optimized. Research on therapeutic drugs has focused on developing novel antibiotics, antibiotic delivery systems, and nanoparticles.
SRP stands as the primary treatment for periodontitis, adept at efficiently eliminating plaque from both teeth and periodontal pockets. However, it’s worth noting that certain pathobionts, particularly A. actinomycetemcomitans, may continue to persist despite treatment (Takamatsu et al., 1999). Therefore, antimicrobial therapy is often combined with SRP for the treatment of periodontitis in clinical practice. Studies have shown that adjunctive systemic antimicrobial therapy can improve the outcomes of periodontitis treatment (Lu et al., 2022). Table 2 summarizes the currently used antibiotic therapies for pathobionts. In addition to directly inhibiting pathobionts, these antibiotics can be utilized in various other aspects of periodontitis treatment. For instance, Zhang et al (Zhang et al., 2021). developed a minocycline hydrochloride-loaded microsphere/sucrose acetate isobutyrate hybrid depot, which ensures the sustained release and maintenance of minocycline at appropriate concentrations. At these optimal concentrations, minocycline not only exhibits antimicrobial activity but also promotes osteoblast growth, demonstrating significant potential in the treatment of periodontitis. Similarly, doxycycline, beyond its antibacterial properties, can inhibit the pathological upregulation of MMPs involved in tissue degradation during periodontitis (Mittal et al., 2022). This inhibition helps to mitigate the tissue destruction caused by periodontitis. Furthermore, the combination of antibiotics with natural antimicrobial agents can enhance the efficacy of antibiotics, effectively addressing the rising issue of antibiotic resistance. Ibrahim et al. demonstrated that the combination of metronidazole with extracts from Symphytum officinale and Panax ginseng significantly enhances the inhibitory effect of metronidazole on P. gingivalis biofilms (Ibrahim et al., 2023). Additionally, this combination suppresses the production of acylated homoserine lactones, thereby impairing the bacterium’s quorum-sensing capabilities and ultimately reducing its pathogenicity (Bacali et al., 2022).
Antibiotic delivery systems are devices or materials that deliver antibiotics precisely to periodontal pockets, such as polymer nanoparticles and nanogels. These systems are capable of effectively encapsulating antibiotics, offering the advantage of precise drug delivery to targeted sites. Additionally, they facilitate the sustained release of the drug, ensuring that an effective concentration of the medication is maintained over an extended period (Brun et al., 2020). For instance, Lin et al (Lin et al., 2018). developed pH-responsive PLGA/chitosan nanoparticles encapsulating metronidazole, enabling targeted and controlled drug release in acidic environments. This design minimizes the drug’s impact on healthy tissues and prevents initial burst release, enhancing treatment sustainability and safety. The nanoparticles significantly reduce inflammatory responses and alveolar bone loss, and facilitate tissue repair. Additionally, this delivery system effectively preserves beneficial oral microbiota, offering a more efficient and comprehensive solution for periodontitis management.
With the problem of antibiotic resistance caused by antibiotic overuse, new antimicrobial materials are receiving attention. Nanotechnology provides new antimicrobial materials for the oral field, including metal nanoparticles and polymer nanoparticles. Silver nanoparticles (AgNPs), exemplifying metal nanoparticles, can significantly inhibit bacterial growth in oral biofilms associated with periodontitis by disrupting bacterial cell walls and preventing biofilm formation (Hernández-Venegas et al., 2023). In Hernández-Venegas et al’s study, AgNPs were evaluated in vitro using biofilms collected from patients with and without periodontal disease, demonstrating that their antimicrobial efficacy increases as particle size decreases (Hernández-Venegas et al., 2023). AgNPs, in particular, exhibit excellent dispersibility and a large surface area, which enhances their contact and binding with bacteria while effectively preventing particle aggregation (Hernández-Venegas et al., 2023). This combination of properties significantly boosts their bactericidal capabilities, making silver nanoparticles a promising antimicrobial material for treating periodontitis. Polymer nanoparticles, such as curcumin nanoparticles, have been shown to exhibit antimicrobial efficacy comparable to 1% chlorhexidine gel in a clinical study involving patients with chronic periodontitis (Guru et al., 2020). In this randomized controlled trial, curcumin nanoparticles effectively improve clinical parameters and reduce the levels of P. gingivalis, T. forsythia, and A. actinomycetemcomitans compared to SRP solely (Guru et al., 2020).
4.2 Bacteriophage therapy in periodontitis
Phage therapy is a promising and emerging approach for the treatment of periodontitis. Phage therapy encompasses not only the use of a single bacteriophage but also phage-derived enzymes, phage-antibiotic combinations, synthetic phages, and phage cocktails (Guo et al., 2024). This diversification of phage applications offers more options and possibilities for clinical use.
Compared with conventional antibiotic therapy, phage therapy offers several advantages. Firstly, considering that bacteriophages generally possess a narrow host range, typically exhibiting specificity toward only a limited subset of strains within a single species. Phages are capable of specifically targeting and eliminating pathobionts without causing significant harm to the beneficial oral microbiota, thereby mitigating the risk of microbial dysbiosis. For example, the five novel lytic bacteriophages (JD-Fnp1 ~ JD-Fnp5) isolated by Wang et al (Wang et al., 2022). exhibit specificity in targeting and lysing F. nucleatum, resulting in a significant reduction of its abundance within the periodontal microbiome. Furthermore, these bacteriophages do not cause substantial damage to beneficial oral microbial communities, thereby enabling a more effective maintenance and restoration of the healthy balance within the periodontal microecosystem. In past studies, phage therapy has been commonly used to treat biofilm-embedded pathobionts. For example, the biofilm of Streptococcus mutans demonstrated a reduction in cell viability by 5 log10upon treatment with bacteriophage ϕAPCM01 in an in vitro biofilm assay, highlighting its potent anti-biofilm activity (Dalmasso et al., 2015). Tinoco et al (Tinoco et al., 2016). have demonstrated that the engineered bacteriophage ϕEf11/ϕFL1C(Δ36) PnisA can effectively disrupt Enterococcus faecalis biofilms, including antibiotic-resistant strains in a vitro study. Additionally, depolymerases encoded by bacteriophages, including enzymes such as sialidases, levanases, and xylosidases, degrade the polysaccharide matrix within the biofilm (Pires et al., 2016). This enzymatic degradation significantly enhances the phage’s ability to penetrate bacterial biofilms. In addition, bacteria are less likely to develop phage resistance compared with antibiotics, further enhancing the clinical application prospects of phage therapy. However, this does not imply that bacteria are incapable of developing resistance to bacteriophages. Bacteria can acquire phage resistance through mechanisms such as receptor gene mutations, phase variation, and epigenetic modifications (Burmeister et al., 2020). To mitigate the potential emergence of phage resistance, the utilization of phage cocktails is primarily recommended to decrease the likelihood of resistance development. Furthermore, the isolation of novel phage strains specifically designed to target resistant bacterial populations serves as an alternative strategy (Khalifa et al., 2015).
4.3 Probiotic therapy in periodontitis
Oral probiotics play an essential role in maintaining the stability of oral microbiota. Common oral probiotics include Lactobacillus, Bifidobacterium, Streptococcus, and Bacillus (Homayouni Rad et al., 2023). These beneficial microorganisms can suppress the growth of pathobionts through various mechanisms, thereby preserving the dynamic balance of the oral microbial community. Specifically, probiotics can directly inhibit the growth and colonization of pathobionts by secreting antimicrobial substances and competing for nutrients. For instance, Lactobacillus fermentum (L. fermentum) ALAL020 can inhibit the growth of pathobionts such as P. gingivalis by producing a cyclic dipeptide (Kawai et al., 2022). Furthermore, probiotics can create an acidic environment and disrupt harmful bacteria’s cell membranes, enzyme activities, and metabolic processes, thereby further suppressing their growth (Argandoña Valdez et al., 2021). In addition to regulating the balance of the oral microbiome, probiotics can also stimulate the activity of oral immune cells, enhancing the local immune defense against pathogenic microorganisms. L. acidophilus La5 downregulates the release of CXCL-8, GM-CSF, and IL-1β by A. actinomycetemcomitans-stimulated gingival epithelial cells, modulating local immune responses and inflammation in the gingiva (Bueno et al., 2022). In oral diseases such as periodontitis, the oral beneficial bacteria can become disrupted due to host or environmental factors, leading to the overgrowth of pathobionts and persistent inflammatory responses. Therefore, re-establishing the equilibrium of oral beneficial bacteria is a crucial aspect of periodontitis treatment. Moreover, probiotic-based therapies are gaining attention as an alternative approach in the context of increasing antibiotic resistance.
Unlike conventional antibiotic treatments, probiotic therapy offers the advantage of selectively targeting pathobionts without disrupting the commensal oral microbiota, thereby avoiding microbiome dysbiosis. This selective action and ability to modulate the oral ecosystem make probiotic therapy a valuable adjunct to other innovative treatment modalities, such as phage therapy, in periodontal disease management. Studies have demonstrated that certain probiotic strains possess antimicrobial, anti-inflammatory, and immunomodulatory properties, effectively inhibiting the growth and virulence of pathobionts. For instance, L. fermentum ALAL020 exerts growth-inhibitory effects against P. gingivalis and P. intermedia by producing a cyclic dipeptide (Kawai et al., 2022). L. acidophilus La5 and L. rhamnosus Lr32 significantly reduce the adhesion/invasion of A. actinomycetemcomitans to gingival epithelial cells OBA-9 and decrease the transcription of virulence factors associated with host defense (Ishikawa et al., 2021; Bueno et al., 2022). Furthermore, probiotics can suppress pathobiont growth by counteracting biofilm formation. For example, L. plantarum 14917 significantly inhibits the growth of Streptococcus mutans and Candida albicans within a multispecies biofilm model (Wu et al., 2015). Concurrently, L. plantarum 14917 further disrupts the biofilm structure of the pathogens by lowering the medium’s pH and reducing the biofilm’s dry weight and total protein content (Wu et al., 2015). Probiotics such as L. acidophilus JCM 1021 and L. casei NBRC 15883 have also been found to inhibit A. actinomycetemcomitans biofilm formation (Jaffar et al., 2016). Topical application of probiotic supplements, such as probiotic mouthwashes or toothpaste, has also shown promising clinical efficacy (Parčina Amižić et al., 2017; Seminario-Amez et al., 2017). Additionally, probiotics exhibit anti-inflammatory properties by modulating the host’s immune response. For instance, the experiments conducted by Invernici et al (Invernici et al., 2018). have successfully demonstrated that Bifidobacterium animalis subsp. lactis HN019 can modulate the inflammatory response in periodontal disease treatment by downregulating pro-inflammatory cytokines IL-1β and IL-8, while upregulating anti-inflammatory factors IL-10 and TGF-β. Furthermore, Bifidobacterium animalis subsp. lactis HN019 has been reported in another study to enhance the expression of β-defensin 3 and TLR4 in gingival tissues, thereby boosting the activity of CD-4+ T cells (Invernici et al., 2020), and then contributing to the reduction of excessive pro-inflammatory cytokine production. This anti-inflammatory effect helps maintain the balance of the oral microenvironment by inhibiting the growth of pathogenic bacteria under inflammatory conditions.
Although probiotic therapy has demonstrated potential in the treatment of periodontitis, current research predominantly focuses on the short-term effects of such treatments, leaving the sustainability of probiotic therapy’s efficacy largely unexplored. The Meta-Analysis conducted by Mendonça et al (Mendonça et al., 2024). revealed that probiotic treatments extending beyond one month did not exhibit sustained benefits, suggesting that providing probiotic therapy for more than one month yields no significant difference. Therefore, future studies should consider extending the follow-up period to assess the long-term effects and potential sustainability of probiotic therapy.
4.4 Microbiological considerations in treating periodontitis
Microbiological considerations for periodontitis therapy involve identifying and quantifying pathobionts, assessing pathobiont virulence factors, and predicting post-treatment microbial changes. Dental plaque samples can be collected from periodontal pockets for microbial analysis. Given that a significant portion of the oral microbiome cannot be cultured, techniques such as polymerase chain reaction (PCR) and fluorescence in situ hybridization (FISH) can be employed to achieve a more comprehensive understanding of the microbial landscape associated with periodontal disease (Gu J et al., 2022; Murugaiyan et al., 2024). Furthermore, sequence-based culture-independent approaches, such as 16S rRNA gene sequencing and metagenomic sequencing, offer a more detailed analysis of the identification and quantification of unculturable microorganisms (Jünemann S et al., 2012; Shi C et al., 2021). In addition to identifying pathobionts, understanding their virulence factors is crucial. These factors dictate the severity and invasiveness of the disease caused by the pathobionts. Relevant virulence factors include biofilm formation, toxin production, and protease activity, as detailed in Section 2. Because the composition of the periodontal microbiota changes following periodontal therapy, it is important to predict post-treatment outcomes. Typically, there is a decrease in the abundance of pathobionts and an increase in the abundance of beneficial bacteria. However, in some cases, pathobionts may persist or recolonize, leading to treatment failure. Therefore, prediction of post-treatment outcomes is paramount.
These microbiological considerations are essential for treatment plans for periodontitis. Based on pathobionts identification, antibiotics and other antibacterial substances that target specific pathobionts are selected to achieve the goal of precision treatment. Adjunctive use of biofilm disruptors, such as chlorhexidine (Merchant et al., 2022), polysaccharides (Alsalhi, 2024), and hydrogen peroxide (Brookes et al., 2023), can enhance the efficacy of antibacterial substances. Due to the periodontal tissue being reduced during the process of periodontitis, regenerative techniques such as guided tissue regeneration can be used to promote bone and periodontal tissue regeneration (Janjić et al., 2020). Accurate identification and quantification of pathobionts, understanding their virulence factors, and predicting post-treatment microbial changes aid in developing personalized treatment plans, thereby improving treatment outcomes.
5 Conclusions and perspectives
5.1 Challenges
While we have come to understand that dysbiosis of the oral microbiome is a key factor in the pathogenesis of periodontitis, there are still challenges in its practical application. Firstly, the oral microbiome is highly complex and dynamic (Peng et al., 2022). The oral cavity harbors a multitude of bacteria that interact in intricate ways, making it difficult to comprehensively monitor and modulate the microbiome. Additionally, the microbiome is subject to dynamic changes influenced by factors such as age, diet, and lifestyle, adding to the difficulty of precise modulation (Li et al., 2021). Concurrently, the composition of the microbiome varies among individuals (Nearing et al., 2020). These differences are not only reflected in the types and abundances of microorganisms but also may influence responses to antimicrobial treatments. This leads to variability in therapeutic outcomes, yet concurrently highlights the advantages of precise treatment. Current periodontitis treatment modalities, such as mechanical debridement and antimicrobial agents, often overlook the role of the oral microbiome and fail to fully restore microbial homeostasis (Kwon et al., 2021). Thus, there is a need for the development of more targeted and effective therapeutic strategies. The emergence of antimicrobial resistance due to the prolonged use of antimicrobial agents poses a significant challenge to treatment. Therefore, the overuse of antimicrobial agents should be curtailed (Yuan et al., 2023).
For probiotic therapy, the critical hurdles include identifying the optimal probiotic strains that can effectively colonize the oral cavity and compete with pathogenic bacteria (Seminario-Amez et al., 2017; Bustamante et al., 2020). It is also essential to ensure the viability and targeted delivery of these probiotics to the periodontal tissues, as well as maintaining the long-term colonization of the beneficial microbes (Zidar et al., 2021). In the case of phage therapy, the challenges lie in isolating phages with the appropriate specificity to target the pathogenic bacterial species involved in periodontitis and successfully delivering and maintaining therapeutic concentrations of the phages at the site of infection (Shlezinger et al., 2017; Szafrański et al., 2017; Chen et al., 2021). Furthermore, addressing the unique regulatory and safety considerations for the clinical implementation of phage therapy is a critical step before widespread adoption. A pivotal aspect inherent in strategies targeting oral microbial therapeutics lies in the intricate relationship between the oral microbiome and the microbiomes of other bodily sites, such as the gut. Treatments aimed at modulating the oral microbiome may inadvertently affect the microbial communities in the gut, potentially leading to unintended health consequences. Consequently, understanding these interconnections is crucial for developing holistic and safe therapeutic approaches.
5.2 Perspectives
Future research and therapeutic strategies should center on personalized and precision medicine approaches. The standard clinical treatment for periodontitis involves the combination of SRP with systemic antibiotics. However, the highly complex nature of the oral microbiome makes comprehensive monitoring and regulation of microbial communities particularly challenging. Consequently, there is a pressing need to develop personalized microbiome profiling and precision treatment plans. Given that the majority of oral microorganisms are difficult to culture, strain identification typically relies on advanced techniques such as PCR and metagenomics. Nevertheless, the high costs associated with these methods remain a significant barrier to their widespread adoption. Future research should aim to reduce the costs of microbiome analysis while simultaneously enhancing its accuracy and efficiency. Additionally, in-depth investigations into the inter-individual variations in microbiomes and their influencing factors will provide a scientific foundation for devising more precise therapeutic strategies.
Moreover, emerging therapeutic strategies, including probiotic therapy and phage therapy, hold promise as alternative treatments. However, translating these approaches into clinical practice presents numerous challenges. Regarding probiotic therapy, existing studies are predominantly short-term, and the long-term efficacy and safety of probiotics remain unclear (Mendonça et al., 2024). Therefore, future efforts should focus on optimizing the screening and delivery technologies for probiotics to ensure their effective colonization and long-term maintenance within the oral environment, thereby enabling the assessment of their sustained therapeutic benefits. Phage therapy has demonstrated effectiveness against various microbial infections, but its application in the treatment of oral diseases has been scarcely explored (Łasica et al., 2024). Additionally, there is a limited availability of phages that specifically target oral pathobionts. Consequently, future research should prioritize the discovery of phages with specificity toward periodontitis-associated pathobionts and address challenges related to phage screening, delivery, and clinical safety. Furthermore, establishing comprehensive regulatory frameworks and conducting large-scale clinical trials are essential steps to facilitate the clinical application of phage therapy.
5.3 Conclusion
This review presents a robust and detailed analysis of the role of microbial dysbiosis in periodontitis and explores innovative therapeutic strategies to address this condition. However, the imitations of this review mainly include challenges in translating innovative therapies from the lab to clinical practice, insufficient depth in exploring host-microbiome interactions and immune responses, and a lack of consideration for geographical and demographic variations that could affect microbial dysbiosis and treatment outcomes. Despite these limitations, the elucidation of microbial dysbiosis as a central mechanism in periodontitis paves the way for transformative therapeutic strategies that transcend conventional treatments. By harnessing the power of personalized medicine and microbiome engineering, it is possible to develop precise interventions that not only treat but also prevent periodontitis. Continued research and interdisciplinary collaboration are essential to translate these innovative approaches from the laboratory to clinical practice, ultimately improving oral health outcomes and enhancing the quality of life for patients.
Author contributions
YL: Data curation, Formal Analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. XH: Data curation, Formal Analysis, Investigation, Methodology, Resources, Writing – original draft. GL: Methodology, Resources, Software, Validation, Writing – original draft. JZ: Formal Analysis, Investigation, Resources, Supervision, Writing – review & editing. GB: Data curation, Funding acquisition, Project administration, Software, Writing – review & editing. DX: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. We acknowledge financial support from the National Natural Science Foundation of China (82160181, 32260089, 82360018); the Science and Technology Department Foundation of Guizhou Province, China (QKHJC-MS[2025]371); Zunyi Oral Disease Immune Prevention and Medical Biomaterials Research and Development Innovation Talent Team (Zunyi Science Talent 2022 No.1); the “12345 Future Talent Training Plan” of Zunyi Medical University (XJ2023-JX-01-06, ZYSE-2022-04); the Undergraduate Education and Teaching Reform Project of Zunyi Medical University (XJKCSZ2023-9, XJJG2022-22, XJJG2024-09); and the first batch of Class Advisor Studios at Zunyi Medical University (2024BZR-01).
Acknowledgments
We greatly appreciate the assistance of Figdraw in creating the figures.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Śmiga, M., Ślęzak, P., Olczak, T. (2024). Comparative analysis of Porphyromonas gingivalis A7436 and ATCC 33277 strains reveals differences in the expression of heme acquisition systems. Microbiol Spectr. 12, e0286523. doi: 10.1128/spectrum.02865-23
Albuquerque-Souza, E., Sahingur, S. E. (2022). Periodontitis, chronic liver diseases, and the emerging oral-gut-liver axis. Periodontol 2000 89, 125–141. doi: 10.1111/prd.12427
Alippe, Y., Wang, C., Ricci, B., Xiao, J., Qu, C., Zou, W., et al. (2017). Bone matrix components activate the NLRP3 inflammasome and promote osteoclast differentiation. Sci. Rep. 7, 6630. doi: 10.1038/s41598-017-07014-0
Alsalhi, A. (2024). Applications of selected polysaccharides and proteins in dentistry: A review. Int. J. Biol. Macromol 260, 129215. doi: 10.1016/j.ijbiomac.2024.129215
Angabo, S., Pandi, K., David, K., Steinmetz, O., Makkawi, H., Farhat, M., et al. (2024). CD47 and thrombospondin-1 contribute to immune evasion by Porphyromonas gingivalis. Proc. Natl. Acad. Sci. U S A 121, e2405534121. doi: 10.1073/pnas.2405534121
Argandoña Valdez, R. M., Ximenez-Fyvie, L. A., Caiaffa, K. S. (2021). Antagonist effect of probiotic bifidobacteria on biofilms of pathogens associated with periodontal disease. Microb Pathog 150, 104657. doi: 10.1016/j.micpath.2020.104657
Ateia, I. M., Sutthiboonyapan, P., Kamarajan, P., Jin, T., Godovikova, V., Kapila, Y. L., et al. (2018). Treponema denticola increases MMP-2 expression and activation in the periodontium via reversible DNA and histone modifications. Cell Microbiol 20, 10. doi: 10.1111/cmi.12815
Bacali, C., Vulturar, R., Buduru, S., Cozma, A., Fodor, A., Chiș, A., et al. (2022). Oral microbiome: Getting to know and befriend neighbors, a biological approach. Biomedicines 10, 671. doi: 10.3390/biomedicines10030671
Balashova, N., Dhingra, A., Boesze-Battaglia, K., Lally, E. T. (2016). Aggregatibacter actinomycetemcomitans leukotoxin induces cytosol acidification in LFA-1 expressing immune cells. Mol. Microbiol 31, 106–114. doi: 10.1111/omi.2016.31.issue-1
Bloch, S., Thurnheer, T., Murakami, Y., Belibasakis, G. N., Schäffer, C. (2017). Behavior of two Tannerella forsythia strains and their cell surface mutants in multispecies oral biofilms. Mol. Microbiol 32, 404–418. doi: 10.1111/omi.2017.32.issue-5
Bostanci, N., Belibasakis, G. N. (2012). Porphyromonas gingivalis: an invasive and evasive opportunistic oral pathogen. FEMS Microbiol Lett. 333, 1–9. doi: 10.1111/j.1574-6968.2012.02579.x
Brookes, Z., Mcgrath, C., Mccullough, M. (2023). Antimicrobial mouthwashes: an overview of mechanisms-what do we still need to know? Int. Dent. J. 73, S64–Ss8. doi: 10.1016/j.identj.2023.08.009
Brun, A., Moignot, N., Colombier, M. L., Dursun, E. (2020). Emerging nanotechnology in non-surgical periodontal therapy in animal models: A systematic review. Nanomater (Basel) 10, 1414. doi: 10.3390/nano10071414
Bryzek, D., Ciaston, I., Dobosz, E., Gasiorek, A., Makarska, A., Sarna, M., et al. (2019). Triggering NETosis via protease-activated receptor (PAR)-2 signaling as a mechanism of hijacking neutrophils function for pathogen benefits. PloS Pathog 15, e1007773. doi: 10.1371/journal.ppat.1007773
Bryzek, D., Ksiazek, M., Bielecka, E., Karim, A. Y., Potempa, B., Staniec, D., et al. (2014). A pathogenic trace of Tannerella forsythia - shedding of soluble fully active tumor necrosis factor α from the macrophage surface by karilysin. Mol. Microbiol 29, 294–306. doi: 10.1111/mom-omi.2014.29.issue-6
Bueno, M. R., Ishikawa, K. H., Almeida-Santos, G., Ando-Suguimoto, E S., Shimabukuro, N., Kawamoto, D., et al. (2022). Lactobacilli attenuate the effect of aggregatibacter actinomycetemcomitans infection in gingival epithelial cells. Front. Microbiol 13, 846192. doi: 10.3389/fmicb.2022.846192
Burmeister, A. R., Fortier, A., Roush, C., Lessing, A. J., Bender, R. G., Barahman, R., et al. (2020). Pleiotropy complicates a trade-off between phage resistance and antibiotic resistance. Proc. Natl. Acad. Sci. U S A 117, 11207–11216. doi: 10.1073/pnas.1919888117
Bustamante, M., Oomah, B. D., Mosi-Roa, Y., Rubilar, M., Burgos-Díaz, C. (2020). Probiotics as an adjunct therapy for the treatment of halitosis, dental caries and periodontitis. Probiotics Antimicrob Proteins 12, 325–334. doi: 10.1007/s12602-019-9521-4
Byrne, D. P., Manandhar, S. P., Potempa, J., Smalley, J. W. (2015). Breakdown of albumin and haemalbumin by the cysteine protease interpain A, an albuminase of Prevotella intermedia. BMC Microbiol 15, 185. doi: 10.1186/s12866-015-0516-3
Caselli, E., Fabbri, C., D’accolti, M., Soffritti, I., Bassi, C., Mazzacane, S., et al. (2020). Defining the oral microbiome by whole-genome sequencing and resistome analysis: the complexity of the healthy picture. BMC Microbiol 20, 120. doi: 10.1186/s12866-020-01801-y
Chen, Z., Guo, Z., Lin, H., Tian, Y., Zhang, P., Chen, H., et al. (2021). The feasibility of phage therapy for periodontitis. Future Microbiol 16, 649–656. doi: 10.2217/fmb-2020-0161
Chinthamani, S., Settem, R. P., Honma, K., Stafford, G. P., Sharma, A. (2022). Tannerella forsythia strains differentially induce interferon gamma-induced protein 10 (IP-10) expression in macrophages due to lipopolysaccharide heterogeneity. Pathog Dis. 80, ftac008. doi: 10.1093/femspd/ftac008
Choe, S. H., Choi, E. Y., Hyeon, J. Y., Keum, B. R., Choi, I. S., Kim, S. J., et al. (2021). Effect of nifedipine, a calcium channel blocker, on the generation of nitric oxide and interleukin-1β by murine macrophages activated by lipopolysaccharide from Prevotella intermedia. Naunyn Schmiedebergs Arch. Pharmacol. 394, 59–71. doi: 10.1007/s00210-020-01958-3
Choi, E. Y., Keum, B. R., Choe, S. H., Hyeon, J. Y., Choi, I. S., Kim, S. J., et al. (2021). Tricarbonyldichlororuthenium(II) dimer, the lipid-soluble carbon monoxide-releasing molecule, attenuates Prevotella intermedia lipopolysaccharide-induced production of nitric oxide and interleukin-1β in murine macrophages. Int. Immunopharmacol 90, 107190. doi: 10.1016/j.intimp.2020.107190
Claesson, R., Johansson, A., Belibasakis, G., Hänström, L., Kalfas, S.. (2002). Release and activation of matrix metalloproteinase 8 from human neutrophils triggered by the leukotoxin of Actinobacillus actinomycetemcomitans. J. Perio Res. 37, 353–359. doi: 10.1034/j.1600-0765.2002.00365.x
Coppenhagen-Glazer, S., Sol, A., Abed, J., Naor, R., Zhang, X., Han, Y. W., et al. (2015). Fap2 of Fusobacterium nucleatum is a galactose-inhibitable adhesin involved in coaggregation, cell adhesion, and preterm birth. Infect. Immun. 83, 1104–1113. doi: 10.1128/IAI.02838-14
Dahlstrand Rudin, A., Khamzeh, A., Venkatakrishnan, V., Persson, T., Gabl, M., Savolainen, O., et al. (2020). Porphyromonas gingivalis produce neutrophil specific chemoattractants including short chain fatty acids. Front. Cell Infect. Microbiol 10, 620681. doi: 10.3389/fcimb.2020.620681
Dalmasso, M., De Haas, E., Neve, H., Strain, R., Cousin, F. J., Stockdale, S. R., et al. (2015). Isolation of a Novel Phage with Activity against Streptococcus mutans Biofilms. PloS One 10, e0138651. doi: 10.1371/journal.pone.0138651
Danforth, D. R., Melloni, M., Tristano, J., Mintz, K. P. (2021). Contribution of adhesion proteins to Aggregatibacter actinomycetemcomitans biofilm formation. Mol. Microbiol 36, 243–253. doi: 10.1111/omi.12346
De Souza, P. P. C., Henning, P., Lerner, U. H. (2022). Stimulation of osteoclast formation by oncostatin M and the role of WNT16 as a negative feedback regulator. Int. J. Mol. Sci. 23, 3287. doi: 10.3390/ijms23063287
Dierickx, K., Pauwels, M., Laine, M. L., Van Eldere, J., Cassiman, J. J., van Winkelhoff, A. J., et al. (2003). Adhesion of Porphyromonas gingivalis serotypes to pocket epithelium. J. Periodontol 74, 844–848. doi: 10.1902/jop.2003.74.6.844
Di Stefano, M., Polizzi, A., Santonocito, S., Romano, A., Lombardi, T., Isola, G., et al. (2022). Impact of oral microbiome in periodontal health and periodontitis: A critical review on prevention and treatment. Int. J. Mol. Sci. 23, 5142. doi: 10.3390/ijms23095142
Doke, M., Fukamachi, H., Morisaki, H., Arimoto, T., Kataoka, H., Kuwata, H., et al. (2017). Nucleases from Prevotella intermedia can degrade neutrophil extracellular traps. Mol. Microbiol 32, 288–300. doi: 10.1111/omi.2017.32.issue-4
Dutzan, N., Kajikawa, T., Abusleme, L., Greenwell-Wild, T., Zuazo, C. E., Ikeuchi, T., et al. (2018). A dysbiotic microbiome triggers T(H)17 cells to mediate oral mucosal immunopathology in mice and humans. Sci. Transl. Med. 10, EAAT0797. doi: 10.1126/scitranslmed.aat0797
Engevik, M. A., Danhof, H. A., Auchtung, J., Endres, B. T., Ruan, W., Bassères, E., et al. (2021). Fusobacteriumnucleatum Adheres to Clostridioides difficile via the RadD Adhesin to Enhance Biofilm Formation in Intestinal Mucus. Gastroenterology 160, 1301–14.e8. doi: 10.1053/j.gastro.2020.11.034
Fine, D. H., Markowitz, K., Furgang, D., Fairlie, K., Ferrandiz, J., Nasri, C., et al. (2007). Aggregatibacter actinomycetemcomitans and its relationship to initiation of localized aggressive periodontitis: Longitudinal cohort study of initially healthy adolescents. J. Clin. Microbiol 45, 3859–3869. doi: 10.1128/JCM.00653-07
Fleetwood, A. J., Lee, M. K. S., Singleton, W., Achuthan, A., Lee, M. C., O'Brien-Simpson, N. M., et al. (2017). Metabolic remodeling, inflammasome activation, and pyroptosis in macrophages stimulated by porphyromonas gingivalis and its outer membrane vesicles. Front. Cell Infect. Microbiol 7, 351. doi: 10.3389/fcimb.2017.00351
Frey, A. M., Satur, M. J., Phansopa, C., Honma, K., Urbanowicz, P. A., Spencer, D. I. R., et al. (2019). Characterization of Porphyromonas gingivalis sialidase and disruption of its role in host-pathogen interactions. Microbiol. (Reading) 165, 1181–1197. doi: 10.1099/mic.0.000851
Genco, C. A., Potempa, J., Mikolajczyk-Pawlinska, J., Travis, J. (1999). Role of gingipains R in the pathogenesis of Porphyromonas gingivalis-mediated periodontal disease. Clin. Infect. Dis. 28, 456–465. doi: 10.1086/cid.1999.28.issue-3
Godovikova, V., Goetting-Minesky, M. P., Timm, J. C., Fenno, J. C. (2019). Immunotopological analysis of the treponema denticola major surface protein (Msp). J. Bacteriol 201, e00528–e00518. doi: 10.1128/jb.00528-18
Goetting-Minesky, M. P., Godovikova, V., Fenno, J. C. (2021). Approaches to understanding mechanisms of dentilisin protease complex expression in treponema denticola. Front. Cell Infect. Microbiol 11, 668287. doi: 10.3389/fcimb.2021.668287
Goulbourne, P. A., Ellen, R. P. (1991). Evidence that Porphyromonas (Bacteroides) gingivalis fimbriae function in adhesion to Actinomyces viscosus. J. Bacteriol 173, 5266–5274. doi: 10.1128/jb.173.17.5266-5274.1991
Gu, J., Wang, H., Zhang, M., Xiong, Y., Yang, L., Ren, B., et al. (2022). Application of fluorescence in situ hybridization (FISH) in oral microbial detection. Pathogens 11, 1450. doi: 10.3390/pathogens11121450
Guo, X., Wang, X., Shi, J., Ren, J., Zeng, J., Li, J., et al. (2024). A review and new perspective on oral bacteriophages: Manifestations in the ecology of oral diseases. J. Microbiol 16, 2344272. doi: 10.1080/20002297.2024.2344272
Guru, S. R., Reddy, K. A., Rao, R. J., Padmanabhan, S., Guru, R., Srinivasa, T. S., et al. (2020). Comparative evaluation of 2% turmeric extract with nanocarrier and 1% chlorhexidine gel as an adjunct to scaling and root planing in patients with chronic periodontitis: A pilot randomized controlled clinical trial. J. Indian Soc. Periodontol 24, 244–252. doi: 10.4103/jisp.jisp_207_19
Hajishengallis, G. (2009). Porphyromonas gingivalis-host interactions: Open war or intelligent guerilla tactics? Microbes Infect. 11, 637–645. doi: 10.1016/j.micinf.2009.03.009
Hajishengallis, G., Darveau, R. P., Curtis, M. A. (2012). The keystone-pathogen hypothesis. Nat. Rev. Microbiol 10, 717–725. doi: 10.1038/nrmicro2873
Hajishengallis, G., Mcintosh, M. L., Nishiyama, S. I., Yoshimura, F. (2013). Mechanism and implications of CXCR4-mediated integrin activation by Porphyromonas gingivalis. Mol. Microbiol 28, 239–249. doi: 10.1111/omi.12021
Hajishengallis, G., Tapping, R. I., Harokopakis, E., Nishiyama, S., Ratti, P., Schifferle, R. E., et al. (2006). Differential interactions of fimbriae and lipopolysaccharide from Porphyromonas gingivalis with the Toll-like receptor 2-centred pattern recognition apparatus. Cell Microbiol 8, 1557–1570. doi: 10.1111/j.1462-5822.2006.00730.x
Han, E. C., Choi, S. Y., Lee, Y., Park, J. W., Hong, S. H., Lee, H. J., et al. (2019). Extracellular RNAs in periodontopathogenic outer membrane vesicles promote TNF-α production in human macrophages and cross the blood-brain barrier in mice. FASEB J. 33, 13412–13422. doi: 10.1096/fj.201901575R
Hara, T., Sakanaka, A., Lamont, R. J., Amano, A., Kuboniwa, M. (2024). Interspecies metabolite transfer fuels the methionine metabolism of Fusobacterium nucleatum to stimulate volatile methyl mercaptan production. mSystems 9, e0076423. doi: 10.1128/msystems.00764-23
Hasegawa, Y., Nagano, K. (2021). Porphyromonas gingivalis FimA and Mfa1 fimbriae: Current insights on localization, function, biogenesis, and genotype. Jpn Dent. Sci. Rev. 57, 190–200. doi: 10.1016/j.jdsr.2021.09.003
Herath, T. D., Wang, Y., Seneviratne, C. J., Darveau, R. P., Wang, C. Y., Jin, L., et al. (2013). The expression and regulation of matrix metalloproteinase-3 is critically modulated by Porphyromonas gingivalis lipopolysaccharide with heterogeneous lipid A structures in human gingival fibroblasts. BMC Microbiol 13, 73. doi: 10.1186/1471-2180-13-73
Hernández-Venegas, P. A., Martínez-Martínez, R. E., Zaragoza-Contreras, E. A., Domínguez-Pérez, R. A., Reyes-López, S. Y., Donohue-Cornejo, A., et al. (2023). Bactericidal activity of silver nanoparticles on oral biofilms related to patients with and without periodontal disease. J. Funct. Biomater 14, 311. doi: 10.3390/jfb14060311
Herrera, D., Sanz, M., Kebschull, M., Jepsen, S., Sculean, A., Berglundh, T., et al. (2022). Treatment of stage IV periodontitis: The EFP S3 level clinical practice guideline. J. Clin. Periodontol 49, 4–71. doi: 10.1111/jcpe.v49.S24
Hockensmith, K., Dillard, K., Sanders, B., Harville, B. A. (2016). Identification and characterization of a chymotrypsin-like serine protease from periodontal pathogen, Tannerella forsythia. Microb Pathog 100, 37–42. doi: 10.1016/j.micpath.2016.08.041
Homayouni Rad, A., Pourjafar, H., Mirzakhani, E. (2023). A comprehensive review of the application of probiotics and postbiotics in oral health. Front. Cell Infect. Microbiol 13, 1120995. doi: 10.3389/fcimb.2023.1120995
Hottmann, I., Borisova, M., Schäffer, C., Mayer, C. (2021). Peptidoglycan Salvage Enables the Periodontal Pathogen Tannerella forsythia to Survive within the Oral Microbial Community. Microb Physiol. 31, 123–134. doi: 10.1159/000516751
Huang, X. Y., Guan, W. Q. (2023). CTHRC1 expressed in periodontitis and human periodontal fibroblasts exposed to inflammatory stimuli. Dis. 29, 1738–1746. doi: 10.1111/odi.14151
Huang, Y., Tian, C., Li, Q., Xu, Q. (2019). TET1 Knockdown Inhibits Porphyromonas gingivalis LPS/IFN-γ-Induced M1 Macrophage Polarization through the NF-κB Pathway in THP-1 Cells. Int. J. Mol. Sci. 20, 2023. doi: 10.3390/ijms20082023
Hussein, N. A., Soliman, Z. S., Edrees, M. F. (2022). Oral microbiota associated with gingiva of healthy, gingivitis and periodontitis cases. Microb Pathog 171, 105724. doi: 10.1016/j.micpath.2022.105724
Ibrahim, S. M., Al-Mizraqchi, A. S., Haider, J. (2023). Metronidazole Potentiation by Panax Ginseng and Symphytum officinale: A New Strategy for P. gingivalis Infection Control. ATB (Basel) 12, 1288. doi: 10.3390/antibiotics12081288
Ikai, R., Hasegawa, Y., Izumigawa, M., Nagano, K., Yoshida, Y., Kitai, N., et al. (2015). Mfa4, an accessory protein of mfa1 fimbriae, modulates fimbrial biogenesis, cell auto-aggregation, and biofilm formation in porphyromonas gingivalis. PloS One 10, e0139454. doi: 10.1371/journal.pone.0139454
Invernici, M. M., Furlaneto, F. A. C., Salvador, S. L., Ouwehand, A. C., Salminen, S., Mantziari, A., et al. (2020). Bifidobacterium animalis subsp lactis HN019 presents antimicrobial potential against periodontopathogens and modulates the immunological response of oral mucosa in periodontitis patients. PloS One 15, e0238425. doi: 10.1371/journal.pone.0238425
Invernici, M. M., Salvador, S. L., Silva, P. H. F., Soares, M. S. M., Casarin, R., Palioto, D. B., et al. (2018). Effects of Bifidobacterium probiotic on the treatment of chronic periodontitis: A randomized clinical trial. J. Clin. Periodontol 45, 1198–1210. doi: 10.1111/jcpe.2018.45.issue-10
Ishihara, K., Wawrzonek, K., Shaw, L. N., Inagaki, S., Miyamoto, M., Potempa, J., et al. (2010). Dentipain, a Streptococcus pyogenes IdeS protease homolog, is a novel virulence factor of Treponema denticola. Biol. Chem. 391, 1047–1055. doi: 10.1515/bc.2010.113
Ishikawa, K. H., Bueno, M. R., Kawamoto, D., Simionato, M. R. L., Mayer, M. P. A. (2021). Lactobacilli postbiotics reduce biofilm formation and alter transcription of virulence genes of Aggregatibacter actinomycetemcomitans. Mol. Microbiol 36, 92–102. doi: 10.1111/omi.12330
Jaffar, N., Ishikawa, Y., Mizuno, K., Okinaga, T., Maeda, T. (2016). Mature Biofilm Degradation by Potential Probiotics: Aggregatibacter actinomycetemcomitans versus Lactobacillus spp. PloS One 11, e0159466. doi: 10.1371/journal.pone.0159466
Jakubovics, N. S., Goodman, S. D., Mashburn-Warren, L., Stafford, G. P., Cieplik, F. (2021). The dental plaque biofilm matrix. Periodontol 2000 86, 32–56. doi: 10.1111/prd.12361
Janjić, K., Schädl, B., Andrukhov, O., Agis, H. (2020). The response of gingiva monolayer, spheroid, and ex vivo tissue cultures to collagen membranes and bone substitute. J. Tissue Eng Regener. Med. 14, 1307–1317. doi: 10.1002/term.3102
Jiang, Y., Song, B., Brandt, B. W., Cheng, L., Zhou, X., Exterkate, R. A. M., et al. (2021). Comparison of red-complex bacteria between saliva and subgingival plaque of periodontitis patients: A systematic review and meta-analysis. Front. Cell Infect. Microbiol 11, 727732. doi: 10.3389/fcimb.2021.727732
Johansson, A. (2011). Aggregatibacter actinomycetemcomitans leukotoxin: a powerful tool with capacity to cause imbalance in the host inflammatory response. Toxins (Basel) 3, 242–259. doi: 10.3390/toxins3030242
Johansson, A., Buhlin, K., Sorsa, T., Pussinen, P. J. (2017). Systemic aggregatibacter actinomycetemcomitans leukotoxin-neutralizing antibodies in periodontitis. J. Periodontol 88, 122–129. doi: 10.1902/jop.2016.160193
Johansson, A., Claesson, R., Hänström, L., Sandström, G., Kalfas, S. (2000). Polymorphonuclear leukocyte degranulation induced by leukotoxin from Actinobacillus actinomycetemcomitans. J. Perio Res. 35, 85–92. doi: 10.1034/j.1600-0765.2000.035002085.x
Johnston, W., Rosier, B. T., Artacho, A., Paterson, M., Piela, K., Delaney, C., et al. (2021). Mechanical biofilm disruption causes microbial and immunological shifts in periodontitis patients. Sci. Rep. 11, 9796. doi: 10.1038/s41598-021-89002-z
Jones, M. M., Vanyo, S. T., Visser, M. B. (2019). The msp protein of treponema denticola interrupts activity of phosphoinositide processing in neutrophils. Infect. Immun. 87, e00553–e00519. doi: 10.1128/IAI.00553-19
Jünemann, S., Prior, K., Szczepanowski, R., Harks, I., Ehmke, B., Goesmann, A., et al. (2012). Bacterial community shift in treated periodontitis patients revealed by ion torrent 16S rRNA gene amplicon sequencing. PloS One 7, e41606. doi: 10.1371/journal.pone.0041606
Kadowaki, T. (2021). Enzymatic characteristics and activities of gingipains from porphyromonas gingivalis. Methods Mol. Biol. 2210, 97–112. doi: 10.1007/978-1-0716-0939-2_10
Karched, M., Bhardwaj, R. G., Qudeimat, M., Al-Khabbaz, A., Ellepola, A. (2022). Proteomic analysis of the periodontal pathogen Prevotella intermedia secretomes in biofilm and planktonic lifestyles. Sci. Rep. 12, 5636. doi: 10.1038/s41598-022-09085-0
Kawai, T., Ohshima, T., Tanaka, T., Ikawa, S., Tani, A., Inazumi, N., et al. (2022). Limosilactobacillus (Lactobacillus) fermentum ALAL020, a Probiotic Candidate Bacterium, Produces a Cyclic Dipeptide That Suppresses the Periodontal Pathogens Porphyromonas gingivalis and Prevotella intermedia. Front. Cell Infect. Microbiol 12, 804334. doi: 10.3389/fcimb.2022.804334
Kelk, P., Claesson, R., Hänström, L., Lerner, U. H., Kalfas, S., Johansson, A., et al. (2005). Abundant secretion of bioactive interleukin-1beta by human macrophages induced by Actinobacillus actinomycetemcomitans leukotoxin. Infect. Immun. 73, 453–458. doi: 10.1128/IAI.73.1.453-458.2005
Khalifa, L., Coppenhagen-Glazer, S., Shlezinger, M., Kott-Gutkowski, M., Adini, O., Beyth, N., et al. (2015). Complete genome sequence of enterococcus bacteriophage EFLK1. Genome Announc 3, e01308–e01315. doi: 10.1128/genomeA.01308-15
Khattri, S., Kumbargere Nagraj, S., Arora, A., Eachempati, P., Kusum, C. K., Bhat, K. G., et al. (2020). Adjunctive systemic antimicrobials for the non-surgical treatment of periodontitis. Cochrane Database Syst. Rev. 11, Cd012568. doi: 10.1002/14651858.CD012568.pub2
Kim, T. J., Shenker, B. J., Macelory, A. S., Spradlin, S., Walker, L. P., Boesze-Battaglia, K., et al. (2023). Aggregatibacter actinomycetemcomitans cytolethal distending toxin modulates host phagocytic function. Front. Cell Infect. Microbiol 13, 1220089. 10.3389/fcimb.2023.1220089
Kim, H. Y., Song, M. K., Lim, Y., Jang, J. S., An, S. J., Kim, H. H., et al. (2022). Effects of extracellular vesicles derived from oral bacteria on osteoclast differentiation and activation. Sci. Rep. 12, 14239. doi: 10.1038/s41598-022-18412-4
Kirst, M. E., Li, E. C., Alfant, B., Chi, Y. Y., Walker, C., Magnusson, I., et al. (2015). Dysbiosis and alterations in predicted functions of the subgingival microbiome in chronic periodontitis. Appl. Environ. Microbiol 81, 783–793. doi: 10.1128/AEM.02712-14
Kokubu, E., Inoue, T., Ishihara, K. (2018). Response of epithelial cells infected by Treponema denticola. Dis. 24, 14–18. doi: 10.1111/odi.12794
Koziel, J., Karim, A. Y., Przybyszewska, K., Ksiazek, M., Rapala-Kozik, M., Nguyen, K. A., et al. (2010). Proteolytic inactivation of LL-37 by karilysin, a novel virulence mechanism of Tannerella forsythia. J. Innate Immun. 2, 288–293. doi: 10.1159/000281881
Książek, M., Goulas, T., Mizgalska, D., Rodríguez-Banqueri, A., Eckhard, U., Veillard, F., et al. (2023). A unique network of attack, defence and competence on the outer membrane of the periodontitis pathogen Tannerella forsythia. Chem. Sci. 14, 869–888. doi: 10.1039/D2SC04166A
Kuboniwa, M., Amano, A., Hashino, E., Yamamoto, Y., Inaba, H., Hamada, N., et al. (2009). Distinct roles of long/short fimbriae and gingipains in homotypic biofilm development by Porphyromonas gingivalis. BMC Microbiol 9, 105. doi: 10.1186/1471-2180-9-105
Kurniyati, K., Kelly, J. F., Vinogradov, E., Robotham, A., Tu, Y., Wang, J., et al. (2017). A novel glycan modifies the flagellar filament proteins of the oral bacterium Treponema denticola. Mol. Microbiol 103, 67–85. doi: 10.1111/mmi.2017.103.issue-1
Kurniyati, K., Zhang, W., Zhang, K., Li, C. (2013). A surface-exposed neuraminidase affects complement resistance and virulence of the oral spirochaete Treponema denticola. Mol. Microbiol 89, 842–856. doi: 10.1111/mmi.2013.89.issue-5
Kwon, T., Lamster, I. B., Levin, L. (2021). Current concepts in the management of periodontitis. Int. Dent. J. 71, 462–476. doi: 10.1111/idj.12630
Lamont, R. J., Bevan, C. A., Gil, S., Persson, R. E., Rosan, B. (1993). Involvement of Porphyromonas gingivalis fimbriae in adherence to Streptococcus gordonii. Microbiol Immunol. 8, 272–276. doi: 10.1111/j.1399-302X.1993.tb00573.x
Łasica, A., Golec, P., Laskus, A., Zalewska, M., Gędaj, M., Popowska, M., et al. (2024). Periodontitis: Etiology, conventional treatments, and emerging bacteriophage and predatory bacteria therapies. Front. Microbiol 15, 1469414. doi: 10.3389/fmicb.2024.1469414
Li, Y., Cui, J., Liu, Y., Chen, K., Huang, L., Liu, Y., et al. (2021). Oral, tongue-coating microbiota, and metabolic disorders: A novel area of interactive research. Front. Cardiovasc. Med. 8, 730203. doi: 10.3389/fcvm.2021.730203
Li, Y., Li, X., Guo, D., Meng, L., Feng, X., Zhang, Y., et al. (2024). Immune dysregulation and macrophage polarization in peri-implantitis. Front. Bioeng Biotechnol. 12, 1291880. doi: 10.3389/fbioe.2024.1291880
Li, X., Liu, Y., Yang, X., Li, C., Song, Z. (2022). The oral microbiota: Community composition, influencing factors, pathogenesis, and interventions. Front. Microbiol 13, 895537. doi: 10.3389/fmicb.2022.895537
Li, Q., Tan, L., Wang, H., Kou, Y., Shi, X., Zhang, S., et al. (2020). Fusobacterium nucleatum Interaction with Pseudomonas aeruginosa Induces Biofilm-Associated Antibiotic Tolerance via Fusobacterium Adhesin A. ACS Infect. Dis. 6, 1686–1696. doi: 10.1021/acsinfecdis.9b00402
Lim, Y., Kim, H. Y., Han, D., Choi, B. K. (2023). Proteome and immune responses of extracellular vesicles derived from macrophages infected with the periodontal pathogen Tannerella forsythia. J. Extracell Vesicles 12, e12381. doi: 10.1002/jev2.v12.12
Lin, J. H., Feng, F., Yu, M. C., Wang, C. H., Chang, P. C. (2018). Modulation of periodontitis progression using pH-responsive nanosphere encapsulating metronidazole or N-phenacylthialzolium bromide. J. Perio Res. 53, 22–28. doi: 10.1111/jre.2018.53.issue-1
Lindholm, M., Metsäniitty, M., Granström, E., Oscarsson, J. (2020). Outer membrane vesicle-mediated serum protection in Aggregatibacter actinomycetemcomitans. J. Microbiol 12, 1747857. doi: 10.1080/20002297.2020.1747857
Lindholm, M., Min Aung, K., Nyunt Wai, S., Oscarsson, J. (2019). Role of OmpA1 and OmpA2 in Aggregatibacter actinomycetemcomitans and Aggregatibacter aphrophilus serum resistance. J. Microbiol 11, 1536192. doi: 10.1080/20002297.2018.1536192
Liu, S., Xie, G., Chen, M., He, Y., Yu, W., Chen, X., et al. (2023). Oral microbial dysbiosis in patients with periodontitis and chronic obstructive pulmonary disease. Front. Cell Infect. Microbiol 13, 1121399. doi: 10.3389/fcimb.2023.1121399
Lu, H., He, L., Jin, D., Zhu, Y., Meng, H. (2022). Effect of adjunctive systemic antibiotics on microbial populations compared with scaling and root planing alone for the treatment of periodontitis: A pilot randomized clinical trial. J. Periodontol 93, 570–583. doi: 10.1002/JPER.20-0764
Luchian, I., Goriuc, A., Martu, M. A., Covasa, M. (2021). Clindamycin as an alternative option in optimizing periodontal therapy. ATB (Basel) 10, 814. doi: 10.3390/antibiotics10070814
Malone, E. T., Ganther, S., Mena, N., Nascimento, G. G., Gomes, B. P. (2021). Treponema denticola-induced RASA4 upregulation mediates cytoskeletal dysfunction and MMP-2 activity in periodontal fibroblasts. Front. Cell Infect. Microbiol 11, 671968. doi: 10.3389/fcimb.2021.671968
Marinho, A. C., Martinho, F. C., Leite, F. R., Radaic, A., Shariati, K., Kindberg, A., et al. (2015). Proinflammatory Activity of Primarily Infected Endodontic Content against Macrophages after Different Phases of the Root Canal Therapy. J. Endod. 41, 817–823. doi: 10.1016/j.joen.2015.01.017
Mendonça, C. D., Mata, A., Azevedo, L. F. R., Marques, J. F., Silveira, J. M. L., Marques, D., et al. (2024). Probiotics in the non-surgical treatment of periodontitis: a systematic review and network meta-analysis. BMC Health 24, 1224. doi: 10.1186/s12903-024-05027-6
Meng, Q., Gao, Q., Mehrazarin, S., Tangwanichgapong, K., Wang, Y., Huang, Y., et al. (2021). Fusobacterium nucleatum secretes amyloid-like FadA to enhance pathogenicity. EMBO Rep. 22, e52891. doi: 10.15252/embr.202152891
Mensi, M., Caselli, E., D’accolti, M., Soffritti, I., Farina, R., Scotti, E., et al. (2023). Efficacy of the additional use of subgingival air-polishing with erythritol powder in the treatment of periodontitis patients: a randomized controlled clinical trial. Part II: Effect on sub-gingival microbiome. Clin. Invest. 27, 2547–2563. doi: 10.1007/s00784-022-04811-4
Merchant, A. T., Gupta, R. D., Akonde, M., Reynolds, M., Smith-Warner, S., Liu, J., et al. (2022). Association of chlorhexidine use and scaling and root planing with birth outcomes in pregnant individuals with periodontitis: A systematic review and meta-analysis. JAMA Netw Open 5, e2247632. doi: 10.1001/jamanetworkopen.2022.47632
Miller, D. P., Oliver, L. D., Jr., Tegels, B. K., Reed, L. A., O'Bier, N. S., Kurniyati, K., et al. (2016). The treponema denticola fhbB protein is a dominant early antigen that elicits fhbB variant-specific antibodies that block factor H binding and cleavage by dentilisin. Infect. Immun. 84, 2051–2058. doi: 10.1128/iai.01542-15
Mishra, S., Johnson, L., Gazala, M. P., Dahiya, S., Rahman, W., Sreeraj, V. S., et al. (2023). Systemic immune-inflammation index in patients with generalized stage III grade C periodontitis. Dis. 29, 3599–3609. doi: 10.1111/odi.14328
Mittal, P., Gokhale, S. T., Manjunath, S., Al-Qahtani, S. M., Magbol, M. A., Nagate, R. R., et al. (2022). Comparative evaluation of locally administered 2% Gel fabricated from lemongrass polymer and 10% Doxycycline hyclate gel as an adjunct to scaling and root planing in the treatment of chronic periodontitis-A randomized controlled trial. Polym (Basel) 14, 2766. doi: 10.3390/polym14142766
Miyai-Murai, Y., Okamoto-Shibayama, K., Sato, T., Kikuchi, Y., Kokubu, E., Potempa, J., et al. (2023). Localization and pathogenic role of the cysteine protease dentipain in Treponema denticola. Mol. Microbiol 38, 212–223. doi: 10.1111/omi.12406
Murugaiyan, V., Utreja, S., Hovey, K. M., Sun, Y., LaMonte, M. J., Wactawski-Wende, J., et al. (2024). Defining Porphyromonas gingivalis strains associated with periodontal disease. Sci. Rep. 14, 6222. doi: 10.1038/s41598-024-56849-x
Nakagawa, M., Shirasugi, M., Yamamoto, T., Nakaya, T., Kanamura, N. (2021). Long-term exposure to butyric acid induces excessive production of matrix metalloproteases in human gingival fibroblasts. Arch. Biol. 123, 105035. doi: 10.1016/j.archoralbio.2020.105035
Nazir, M., Al-Ansari, A., Al-Khalifa, K., Alhareky, M., Gaffar, B., Almas, K., et al. (2020). Global prevalence of periodontal disease and lack of its surveillance. ScientificWorldJournal 2020, 2146160. doi: 10.1155/2020/2146160
Nearing, J. T., Declercq, V., Van Limbergen, J., Langille, M. G. I. (2020). Assessing the variation within the oral microbiome of healthy adults. mSphere 5, e00451–e00420. doi: 10.1128/mSphere.00451-20
Ng, H. M., Slakeski, N., Butler, C. A., Veith, P. D., Chen, Y. Y., Liu, S. W., et al. (2019). The Role of Treponema denticola Motility in Synergistic Biofilm Formation With Porphyromonas gingivalis. Front. Cell Infect. Microbiol 9, 432. doi: 10.3389/fcimb.2019.00432
Nice, J. B., Collins, S. M., Agro, S. M. J., Sinani, A., Moros, S. D., Pasch, L. M., et al. (2024). Heterogeneity of size and toxin distribution in aggregatibacter actinomycetemcomitans outer membrane vesicles. Toxins (Basel) 16, 138. doi: 10.3390/toxins16030138
Parčina Amižić, I., Cigić, L., Gavić, L., Radić, M., Biočina Lukenda, D., Tonkić, M., et al. (2017). Antimicrobial efficacy of probiotic-containing toothpastes: an in vitro evaluation. Med. Glas (Zenica) 14, 139–144. doi: 10.17392/870-16
Park, O. J., Ha, Y. E., Sim, J. R., Lee, D., Lee, E. H., Kim, S. Y., et al. (2023). Butyrate potentiates Enterococcus faecalis lipoteichoic acid-induced inflammasome activation via histone deacetylase inhibition. Cell Death Discov. 9, 107. doi: 10.1038/s41420-023-01404-2
Peng, X., Cheng, L., You, Y., Tang, C., Ren, B., Li, Y., et al. (2022). Oral microbiota in human systematic diseases. Int. J. Sci. 14, 14. doi: 10.1038/s41368-022-00163-7
Petrović, M. S., Kannosh, I. Y., Milašin, J. M., Mihailović, D. S., Obradović, R. R., Bubanj, S. R., et al. (2018). Clinical, microbiological and cytomorphometric evaluation of low-level laser therapy as an adjunct to periodontal therapy in patients with chronic periodontitis. Int. J. Dent. Hyg 16, e120–e1e7. doi: 10.1111/idh.12328
Philips, A., Stolarek, I., Handschuh, L., Nowis, K., Juras, A., Trzciński, D., et al. (2020). Analysis of oral microbiome from fossil human remains revealed the significant differences in virulence factors of modern and ancient Tannerella forsythia. BMC Genomics 21, 402. doi: 10.1186/s12864-020-06810-9
Pires, D. P., Oliveira, H., Melo, L. D., Sillankorva, S., Azeredo, J. (2016). Bacteriophage-encoded depolymerases: their diversity and biotechnological applications. Appl. Microbiol Biotechnol. 100, 2141–2151. doi: 10.1007/s00253-015-7247-0
Popadiak, K., Potempa, J., Riesbeck, K., Blom, A. M. (2007). Biphasic effect of gingipains from Porphyromonas gingivalis on the human complement system. J. Immunol. 178, 7242–7250. doi: 10.4049/jimmunol.178.11.7242
Potempa, M., Potempa, J., Okroj, M., Popadiak, K., Eick, S., Nguyen, K. A., et al. (2008). Binding of complement inhibitor C4b-binding protein contributes to serum resistance of Porphyromonas gingivalis. J. Immunol. 181, 5537–5544. doi: 10.4049/jimmunol.181.8.5537
Prucsi, Z., Zimny, A., Płonczyńska, A., Zubrzycka, N., Potempa, J., Sochalska, M., et al. (2023). Porphyromonas gingivalis peptidyl arginine deiminase (PPAD) in the context of the feed-forward loop of inflammation in periodontitis. Int. J. Mol. Sci. 24, 12922. doi: 10.3390/ijms241612922
Qiao, X., Tang, J., Dou, L., Yang, S., Sun, Y., Mao, H., et al. (2023). Dental pulp stem cell-derived exosomes regulate anti-inflammatory and osteogenesis in periodontal ligament stem cells and promote the repair of experimental periodontitis in rats. Int. J. Nanomed 18, 4683–4703. doi: 10.2147/IJN.S420967
Radisky, E. S. (2020). Mirolysin structures open a window on gum disease. IUCrJ 7, 3–4. doi: 10.1107/S2052252519016968
Razavi, P., Rezaee, S. A., Akhondian, S., Asgari, N., Fatemi, K., Mohajertehran, F., et al. (2023). Matrix metalloproteinase-3 but not matrix metalloproteinase-9, implicated in the manifestation of chronic periodontitis. Rep. Biochem. Mol. Biol. 11, 656–662. doi: 10.52547/rbmb.11.4.656
Rosen, G., Sela, M. N. (2006). Coaggregation of Porphyromonas gingivalis and Fusobacterium nucleatum PK 1594 is mediated by capsular polysaccharide and lipopolysaccharide. FEMS Microbiol Lett. 256, 304–310. doi: 10.1111/j.1574-6968.2006.00131.x
Sakakibara, J., Nagano, K., Murakami, Y., Higuchi, N., Nakamura, H., Shimozato, K., et al. (2007). Loss of adherence ability to human gingival epithelial cells in S-layer protein-deficient mutants of Tannerella forsythensis. Microbiol. (Reading) 153, 866–876. doi: 10.1099/mic.0.29275-0
Sakanaka, A., Kuboniwa, M., Shimma, S., Alghamdi, S. A., Mayumi, S., Lamont, R. J., et al. (2022). Fusobacterium nucleatum metabolically integrates commensals and pathogens in oral biofilms. mSystems 7, e0017022. doi: 10.1128/msystems.00170-22
Santonocito, S., Giudice, A., Polizzi, A., Troiano, G., Merlo, E. M., Sclafani, R., et al. (2022). A cross-talk between diet and the oral microbiome: Balance of nutrition on inflammation and immune system’s response during periodontitis. Nutrients 14, 2426. doi: 10.3390/nu14122426
Santos, T. A., Scorzoni, L., Correia, R., Junqueira, J. C., Anbinder, A. L. (2020). Interaction between Lactobacillus reuteri and periodontopathogenic bacteria using in vitro and in vivo (G. mellonella) approaches. Pathog Dis. 78, FTAA044. doi: 10.1093/femspd/ftaa044
Sanz, M., Beighton, D., Curtis, M. A., Cury, J. A., Dige, I., Dommisch, H., et al. (2017). Role of microbial biofilms in the maintenance of oral health and in the development of dental caries and periodontal diseases. Consensus report of group 1 of the Joint EFP/ORCA workshop on the boundaries between caries and periodontal disease. J. Clin. Periodontol 44, S5–s11. doi: 10.1111/jcpe.12682
Sanz, M., Herrera, D., Kebschull, M., Chapple, I., Jepsen, S., Beglundh, T., et al. (2020). Treatment of stage I-III periodontitis-The EFP S3 level clinical practice guideline. J. Clin. Periodontol 47, 4–60. doi: 10.1111/jcpe.v47.s22
Scheiman, J., Luber, J. M., Chavkin, T. A., MacDonald, T., Tung, A., Pham, L. D., et al. (2019). Meta-omics analysis of elite athletes identifies a performance-enhancing microbe that functions via lactate metabolism. Nat. Med. 25, 1104–1109. doi: 10.1038/s41591-019-0485-4
Seminario-Amez, M., López-López, J., Estrugo-Devesa, A., Ayuso-Montero, R., Jané-Salas, E. (2017). Probiotics and oral health: A systematic review. Med. Patol Cir Bucal 22, e282–e2e8. doi: 10.4317/medoral.21494
Senarat, S., Rojviriya, C., Puyathorn, N., Lertsuphotvanit, N., Phaechamud, T. (2023a). Levofloxacin HCl-incorporated zein-based solvent removal phase inversion in situ forming gel for periodontitis treatment. Pharmaceutics 15, 1199. doi: 10.3390/pharmaceutics15041199
Senarat, S., Rojviriya, C., Sarunyakasitrin, K., Charoentreeraboon, J., Pichayakorn, W., Phaechamud, T., et al. (2023b). Moxifloxacin HCl-incorporated aqueous-induced nitrocellulose-based in situ gel for periodontal pocket delivery. Gels 9, 572. doi: 10.3390/gels9070572
Shahbeik, S., Taleghani, F., Sattari, M., Mahvash Mohammadi, M., Moravej, M. (2021). Evaluation of NLRP3 and IL-18 levels after periodontal therapy. Iran J. Allergy Asthma Immunol. 20, 764–770. doi: 10.18502/ijaai.v20i6.8028
Sharma, A. (2010). Virulence mechanisms of Tannerella forsythia. Periodontol 2000 54, 106–116. doi: 10.1111/j.1600-0757.2009.00332.x
Shenker, B. J., Walker, L. P., Zekavat, A., Korostoff, J., Boesze-Battaglia, K. (2022). Aggregatibacter actinomycetemcomitans cytolethal distending toxin-induces cell cycle arrest in a glycogen synthase kinase (GSK)-3-dependent manner in oral keratinocytes. Int. J. Mol. Sci. 23, 11831. doi: 10.3390/ijms231911831
Shi, C., Cai, L., Xun, Z., Zheng, S., Shao, F., Wang, B., et al. (2021). Metagenomic analysis of the salivary microbiota in patients with caries, periodontitis and comorbid diseases. J. Dent. Sci. 16, 1264–1273. doi: 10.1016/j.jds.2020.12.002
Shimotahira, N., Oogai, Y., Kawada-Matsuo, M., Yamada, S., Fukutsuji, K., Nagano, K., et al (2013). The surface layer of Tannerella forsythia contributes to serum resistance and oral bacterial coaggregation. Infect. Immun. 81, 1198–1206. doi: 10.1128/IAI.00983-12
Shirasugi, M., Nishioka, K., Yamamoto, T., Nakaya, T., Kanamura, N. (2017). Normal human gingival fibroblasts undergo cytostasis and apoptosis after long-term exposure to butyric acid. Biochem. Biophys. Res. Commun. 482, 1122–1128. doi: 10.1016/j.bbrc.2016.11.168
Shlezinger, M., Khalifa, L., Houri-Haddad, Y., Coppenhagen-Glazer, S., Resch, G., Que, Y. A., et al. (2017). Phage therapy: A new horizon in the antibacterial treatment of oral pathogens. Curr. Top. Med. Chem. 17, 1199–1211. doi: 10.2174/1568026616666160930145649
Shokeen, B., Park, J., Duong, E., Rambhia, S., Paul, M., Weinberg, A., et al. (2020). Role of FAD-I in fusobacterial interspecies interaction and biofilm formation. Microorganisms 8, 70. doi: 10.3390/microorganisms8010070
Skottrup, P. D., López, R., Ksiazek, M., Højrup, P., Baelum, V., Potempa, J., et al. (2019). An IgY-based immunoassay to evaluate the biomarker potential of the Tannerella forsythia virulence factor karilysin in human saliva. J. Immunol. Methods 469, 26–32. doi: 10.1016/j.jim.2019.03.003
Song, B., Xian, W., Sun, Y., Gou, L., Guo, Q., Zhou, X., et al. (2023). Akkermansia muciniphila inhibited the periodontitis caused by Fusobacterium nucleatum. NPJ Biofilms Microbiomes 9, 49. doi: 10.1038/s41522-023-00417-0
Su, Y., Xu, M. Y., Cui, Y., Chen, R. Z., Xie, L. X., Zhang, J. X., et al. (2023). Bacterial quorum sensing orchestrates longitudinal interactions to shape microbiota assembly. Microbiome 11, 241. doi: 10.1186/s40168-023-01699-4
Szafrański, S. P., Winkel, A., Stiesch, M. (2017). The use of bacteriophages to biocontrol oral biofilms. J. Biotechnol. 250, 29–44. doi: 10.1016/j.jbiotec.2017.01.002
Takamatsu, N., Yano, K., He, T., Umeda, M., Ishikawa, I. (1999). Effect of initial periodontal therapy on the frequency of detecting Bacteroides forsythus, Porphyromonas gingivalis, and Actinobacillus actinomycetemcomitans. J. Periodontol 70, 574–580. doi: 10.1902/jop.1999.70.6.574
Teles, F., Collman, R. G., Mominkhan, D., Wang, Y. (2022). Viruses, periodontitis, and comorbidities. Periodontol 2000 89, 190–206. doi: 10.1111/prd.12435
Tinoco, J. M., Buttaro, B., Zhang, H., Liss, N., Sassone, L., Stevens, R., et al. (2016). Effect of a genetically engineered bacteriophage on Enterococcus faecalis biofilms. Arch. Biol. 71, 80–86. doi: 10.1016/j.archoralbio.2016.07.001
Tomasi, C., Abrahamsson, K. H., Apatzidou, D. (2000). Subgingival instrumentation. Periodontol 2000. doi: 10.1111/prd.12485
Torrungruang, K., Jitpakdeebordin, S., Charatkulangkun, O., Gleebbua, Y. (2015). Porphyromonas gingivalis, Aggregatibacter actinomycetemcomitans, and Treponema denticola/Prevotella intermedia Co-Infection Are Associated with Severe Periodontitis in a Thai Population. PloS One 10, e0136646. doi: 10.1371/journal.pone.0136646
Tsukahara, T., Toyoda, A., Kawase, T., Nakamura, S. I., Ochiai, K. (2020). Consecutive intra-gingival injections of lipopolysaccharide and butyric acid to mice induce abnormal behavior and changes in cytokine concentrations. J. NI 17, 331. doi: 10.1186/s12974-020-02008-8
Uemura, Y., Hiroshima, Y., Tada, A., Murakami, K., Yoshida, K., Inagaki, Y., et al. (2022). Porphyromonas gingivalis Outer Membrane Vesicles Stimulate Gingival Epithelial Cells to Induce Pro-Inflammatory Cytokines via the MAPK and STING Pathways. Biomedicines 10, 2643. doi: 10.3390/biomedicines10102643
Valm, A. M. (2019). The structure of dental plaque microbial communities in the transition from health to dental caries and periodontal disease. J. Mol. Biol. 431, 2957–2969. doi: 10.1016/j.jmb.2019.05.016
Vinogradov, E., Michael, F. S., Cairns, C., Cox, A. D. (2022). Structure of the lipopolysaccharide O-antigens from Fusobacterium nucleatum strains SB-106CP and HM-992 and immunological comparison to the O-antigen of strain 12230. Carbohydr Res. 517, 108576. doi: 10.1016/j.carres.2022.108576
Wang, Y., Liu, Z., Chen, Q., Yi, L., Xu, Z., Cai, M., et al. (2022). Isolation and characterization of novel Fusobacterium nucleatum bacteriophages. Front. Microbiol 13, 945315. doi: 10.3389/fmicb.2022.945315
Wang, M., Shakhatreh, M. A., James, D., Liang, S., Nishiyama, S., Yoshimura, F., et al. (2007). Fimbrial proteins of porphyromonas gingivalis mediate in vivo virulence and exploit TLR2 and complement receptor 3 to persist in macrophages. J. Immunol. 179, 2349–2358. doi: 10.4049/jimmunol.179.4.2349
Wang, M., Wang, Z., Lessing, D. J., Guo, M., Chu, W. (2023). Fusobacterium nucleatum and its metabolite hydrogen sulfide alter gut microbiota composition and autophagy process and promote colorectal cancer progression. Microbiol Spectr. 11, e0229223. doi: 10.1128/spectrum.02292-23
Wang, Y., Wang, L., Sun, T., Shen, S., Li, Z., Ma, X., et al. (2023). Study of the inflammatory activating process in the early stage of Fusobacterium nucleatum infected PDLSCs. Int. J. Sci. 15, 8. doi: 10.1038/s41368-022-00213-0
Wielento, A., Bereta, G. P., Łagosz-Ćwik, K. B., Eick, S., Lamont, R. J., Grabiec, A. M., et al. (2022). TLR2 activation by porphyromonas gingivalis requires both PPAD activity and fimbriae. Front. Immunol. 13, 823685. doi: 10.3389/fimmu.2022.823685
Williams, D. W., Greenwell-Wild, T., Brenchley, L., Dutzan, N., Overmiller, A., Sawaya, A. P., et al. (2021). Human oral mucosa cell atlas reveals a stromal-neutrophil axis regulating tissue immunity. Cell 184, 4090–104.e15. doi: 10.1016/j.cell.2021.05.013
Wu, C. C., Lin, C. T., Wu, C. Y., Peng, W. S., Lee, M. J., Tsai, Y. C., et al. (2015). Inhibitory effect of Lactobacillus salivarius on Streptococcus mutans biofilm formation. Mol. Microbiol 30, 16–26. doi: 10.1111/mom-omi.2015.30.issue-1
Xu, Y., Luo, Y., Weng, Z., Xu, H., Zhang, W., Li, Q., et al. (2023). Microenvironment-responsive metal-phenolic nanozyme release platform with antibacterial, ROS scavenging, and osteogenesis for periodontitis. ACS Nano 17, 18732–18746. doi: 10.1021/acsnano.3c01940
Xu, W., Zhou, W., Wang, H., et al. (2020). Roles of Porphyromonas gingivalis and its virulence factors in periodontitis. Adv. Protein Chem. Struct. Biol. 120, 45–84. doi: 10.1016/bs.apcsb.2019.12.001
Yamada, M., Ikegami, A., Kuramitsu, H. K. (2005). Synergistic biofilm formation by Treponema denticola and Porphyromonas gingivalis. FEMS Microbiol Lett. 250, 271–277. doi: 10.1016/j.femsle.2005.07.019
Yang, X., Cai, X., Lin, J., Zheng, Y., Liao, Z., Lin, W., et al. (2024). E. Coli LPS-induced calcium signaling regulates the expression of hypoxia-inducible factor 1α in periodontal ligament fibroblasts in a non-hypoxia-dependent manner. Int. Immunopharmacol 128, 111418. doi: 10.1016/j.intimp.2023.111418
Yang, K., Xu, S., Zhao, H., Liu, L., Lv, X., Hu, F., et al. (2021). Hypoxia and Porphyromonas gingivalis-lipopolysaccharide synergistically induce NLRP3 inflammasome activation in human gingival fibroblasts. Int. Immunopharmacol 94, 107456. doi: 10.1016/j.intimp.2021.107456
Yuan, X., Zhou, F., Wang, H., Xu, X., Xu, S., Zhang, C., et al. (2023). Systemic antibiotics increase microbiota pathogenicity and oral bone loss. Int. J. Sci. 15, 4. doi: 10.1038/s41368-022-00212-1
Zhang, L., Hendrickson, R. C., Meikle, V., Lefkowitz, E. J., Ioerger, T. R., Niederweis, M., et al. (2020). Comprehensive analysis of iron utilization by Mycobacterium tuberculosis. PloS Pathog 16, e1008337. doi: 10.1371/journal.ppat.1008337
Zhang, J., Liu, X., Wan, C., Liu, Y., Wang, Y., Meng, C., et al. (2020). NLRP3 inflammasome mediates M1 macrophage polarization and IL-1β production in inflammatory root resorption. J. Clin. Periodontol 47, 451–460. doi: 10.1111/jcpe.13258
Zhang, T., Qiu, Y., Song, J., Zhou, P., Liao, H., Cheng, Y., et al. (2021). Electrosprayed minocycline hydrochloride-loaded microsphere/SAIB hybrid depot for periodontitis treatment. Drug Delivery 28, 620–633. doi: 10.1080/10717544.2021.1902020
Zhu, Y., Wang, Y., Zhang, S., Li, J., Li, X., Ying, Y., et al. (2023). Association of polymicrobial interactions with dental caries development and prevention. Front. Microbiol 14, 1162380. doi: 10.3389/fmicb.2023.1162380
Zidar, A., Kristl, J., Kocbek, P., Zupančič, Š. (2021). Treatment challenges and delivery systems in immunomodulation and probiotic therapies for periodontitis. Expert Opin. Drug Delivery 18, 1229–1244. doi: 10.1080/17425247.2021.1908260
Keywords: oral microbiome, microbial dysbiosis, periodontitis, mechanistic insights, therapeutic strategies inflammatory response
Citation: Li Y, He X, Luo G, Zhao J, Bai G and Xu D (2025) Innovative strategies targeting oral microbial dysbiosis: unraveling mechanisms and advancing therapies for periodontitis. Front. Cell. Infect. Microbiol. 15:1556688. doi: 10.3389/fcimb.2025.1556688
Received: 07 January 2025; Accepted: 08 April 2025;
Published: 30 April 2025.
Edited by:
Bastiaan P. Krom, VU Amsterdam, NetherlandsReviewed by:
Marcia Pinto Alves Mayer, University of São Paulo, BrazilRoberta Gasparro, University of Naples Federico II, Italy
Copyright © 2025 Li, He, Luo, Zhao, Bai and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Delin Xu, eHVkZWxpbjIwMDBAMTYzLmNvbQ==; Juanjuan Zhao, amouei4yMDA4QDE2My5jb20=; Guohui Bai, YmFpZ3VvaHVpMTIyOEAxMjYuY29t
†These authors share first authorship
‡ORCID: Juanjuan Zhao, orcid.org/0000-0001-9253-808
Delin Xu, orcid.org/0000-0003-3695-2997
 Yang Li
Yang Li Xinyu He
Xinyu He Guocheng Luo1
Guocheng Luo1 Juanjuan Zhao
Juanjuan Zhao Guohui Bai
Guohui Bai Delin Xu
Delin Xu