- Cognitive Systems Lab, Department for Mathematics and Computer Science, University of Bremen, Bremen, Germany
1 Background
In this digital age, human activity recognition (HAR) plays an increasingly important role in almost all aspects of life to improve people’s quality of life, such as auxiliary medical care, rehabilitation technology, and interactive entertainment. Besides external sensing, sensor-based internal sensing for HAR is also intensively studied. A large body of research involves recognizing various kinds of everyday human activities, including walking, standing, jumping, and performing gestures. HAR research relies on large amounts of data, which includes the collection of laboratory data that meet in-house research goals, as well as the usage of external and public databases to verify models and methods. Therefore, data collection is an essential part of our entire HAR research work, for which we will detail this extensive progress.
Many public HAR datasets are available online, providing various sorts of collected data, some of which have some similarities with our in-house data acquisition in terms of purpose, sensor selection, or protocol design. For instance, the Opportunity benchmark database (Chavarriaga et al., 2013) contains naturalistic daily living activities recorded with a large set of on-body sensors. The UniMiB SHAR dataset (Micucci et al., 2017) includes 11,771 samples of both human activities and falls divided into 17 fine-grained classes. The GaitAnalysisDataBase (Loose et al., 2020) contains 3D walking kinematics and muscle activity data from healthy adults walking at normal, slow or fast pace on the flat ground or at incremental speeds on a treadmill. The RealWorld dataset (Sztyler and Stuckenschmidt, 2016) covers acceleration, GPS, gyroscope, light, magnetic field, and sound level data of the activities climbing stairs down and up, jumping, lying, standing, sitting, running/jogging, and walking of 15 subjects. The FORTH-TRACE dataset (Karagiannaki et al., 2016) is collected from 15 participants wearing five Shimmer wearable sensor nodes on the left/right wrist, the torso, the right thigh, and the left ankle. The ENABL3S dataset (Hu et al., 2018) contains bilateral electromyography (EMG) and joint and limb kinematics recorded from wearable sensors for ten able-bodied individuals as they freely transitioned between sitting, standing, and five walking-related activities.
In this article, we disclose our in-house collected sensor-based dataset, CSL-SHARE (Cognitive Systems Lab Sensor-based Human Activity REcordings). Based on the improvement of the recording plan and organization through the experience gathered from the pilot datasets’ collection of CSL17 (one subject, seven activities of daily living, 15 minutes) and CSL18 (four subjects, 18 activities of daily living and sports, 90 minutes), the CSL-SHARE dataset covers 22 types of activities of daily living and sports from 20 subjects in a total time of 691 minutes, of which 363 minutes are segmented and annotated. In this dataset, we used two triaxial accelerometers, two triaxial gyroscopes, four surface electromyography (sEMG) sensors, one biaxial electrogoniometer, and one airborne microphone integrated into a knee bandage, bringing the total number of channels to 19, as these sensors can provide usable and reliable biosignals for HAR research, gait analysis, and health assessment according to existing studies, such as Whittle (1996), Rowe et al. (2000), Mathie et al. (2003), Kwapisz et al. (2010), Rebelo et al. (2013), and Teague et al. (2016). We also tried to use a piezoelectric microphone and a force sensor for sensing the acoustic and physical pressure signals from the knee during the acquisition. Nevertheless, in subsequent analysis and research, we did not have evidence to support their contribution to HAR research. Therefore, we removed these two channels of signal from the public dataset. In addition, although our two pilot datasets mentioned above, CSL17 and CSL18, are not publicly available due to the relatively smaller data volume, they can also be obtained from us for scientific research purposes.
2 Dataset Description
2.1 Devices, Sensors, and Sensor Placement
We chose the biosignalsplux Researcher Kit1 with the selected various types of sensors supplied together. One hub from the kit records biosignals from eight channels, each up to 16 bits, simultaneously. Since we needed to record over 20 channels, we connected 3 hubs via synchronization cables that connect the hubs and synchronize all channels automatically between the hubs at the beginning of each recording session, which ensured the synchronicity during the entire recording sessions.
The sensor positioning on the right-leg-worn knee bandage was decided in collaboration with kinesiologists of the Institute of Sport and Sports Science at Karlsruhe Institute of Technology based on their substantial research experience in knee kinematics (Stetter et al., 2018; Stetter et al., 2019) to capture ambulation activities. The CSL-SHARE sensor positions and their measured muscles/body parts are listed as follows:
• Triaxial accelerometer 1 (upper): thigh, proximal ventral
• Triaxial accelerometer 2 (lower): shank, distal ventral
• Triaxial gyroscope 1 (upper): thigh, proximal ventral
• Triaxial gyroscope 2 (lower): shank, distal ventral
• EMG 1 (upper-front): musculus vastus medialis
• EMG 2 (lower-front): musculus tibialis anterior
• EMG 3 (upper-back): musculus biceps femoris
• EMG 4 (lower-back): musculus gastrocnemius
• Biaxial electrogoniometer (lateral): knee of the right leg
• Airborne microphone (lateral): knee of the right leg.
EMG and microphone signals were recorded with a sampling rate of 1000 Hz and all other signals with 100 Hz. The low-sampled channels at 100 Hz were up-sampled to 1000 Hz to be synchronized and aligned with high-sampled channels. All channels have a quantization resolution of 16 Bits.
2.2 Software for Data Acquisition
We developed a software called Activity Signal Kit (ASK) with a Graphical User Interface (GUI) and multi-functionalities using the driver library provided by biosignalsplux, as introduced in (Liu and Schultz, 2018). ASK automatically connects and synchronizes several recording hubs, then collects up to 24-channel sensor data from all hubs simultaneously and continuously. All recorded data are archived automatically in HDF5 files with the filename of dates and timestamps for further research.
A protocol-for-pushbutton mechanism of segmentation and annotation has been implemented in the ASK software, which will be introduced in Section 2.3. Moreover, the baseline ASK software also provides the functionalities of digital signal processing, feature extraction, modeling, training, and recognition by applying our in-house developed HMM-based decoder BioKIT (Telaar et al., 2014).
2.3 Annotation and Segmentation
The task of segmentation in HAR research is to split a relatively long sequence of activities into several segments of single activity, while annotation is the process of labeling each segment, such as “walk,” “run,” “stand-to-sit,” among others. Segmentation, which can be performed manually (Rebelo et al., 2013), semi-supervised Barbič et al. (2004), or automatically (Guenterberg et al., 2009; Micucci et al., 2017), is undoubtedly a prerequisite for annotation, and its output will be input for digital signal processing and feature extraction. Annotation, which can be performed directly after each segmentation subtask, helps two follow-up operations: training and evaluation.
In our research, we applied the pushbutton of the biosignalsplux Research Kit in our proposed semi-automated segmentation and annotation solution. In subsequent research, the applicability of the semi-automated segmented data has been verified for our research purpose during numerous experiments (see Section 4), so we have been applying this mechanism to our successively acquired datasets.
The so-called protocol-for-pushbutton mechanism of segmentation and annotation has been implemented in the ASK software (Liu and Schultz, 2018). When the “segmentation and annotation” mode is switched on during the data acquisition, a predefined activity sequence protocol will be loaded into the software, which prompts the user to perform the activities one after the other. Each activity is displayed on the screen one-by-one while the user controls the activity recording by pushing, holding, and releasing the pushbutton. The user follows the instructions of the software step-by-step. For example, the prompted activity states “walk,” the user sees the instruction “Please hold the pushbutton and do: walk.” The user prepares for it, then pushes the button and starts to “walk.” She/he keeps holding the pushbutton while walking for a duration at will, then releases the pushbutton to finish this activity. With the release, the system displays the next activity instruction, e.g., “stand-to-sit,” the process continues until the predefined acquisition protocol is fully processed.
The ASK software records all timestamps/sample numbers of each button push and button release during the data recording. These data are archived in CSV files as segmentation and annotation results for each activity. Since we synchronized all data at 1,000 Hz, each sample represents data from 1 ms. For example, a line “sit, 3647, 6163” in a CSV file means that the activity segment labeled “sit” lasts 2,516 samples from the timestamp 3,647 to 6,162, which corresponds to 2.516 seconds. The corresponding 2,516 samples form one segment for training the activity model “sit,” or for the recognition results evaluation. The protocol-for-pushbutton mechanism was implemented to reduce the time and labor costs of manual annotation. The resulting segmentations are excellent and required little to no manual correction, and lay a good foundation for subsequent research. Nevertheless, this mechanism has some limitations:
• The mechanism can only be applied during acquisition and is incapable of segmenting archived data
• Clear activity start-/endpoints need to be defined, which is impossible in cases like field studies
• Activities requiring both hands are not possible due to participants holding the pushbutton
• The pushbutton operation may consciously or subconsciously affect the activity execution
• The participant forgetting to push or release the button results in subsequent segmentation errors.
None of these limitations, except forgetting to release the pushbutton, hold in a laboratory setting with clear instructions and protocols. Hence, misapplication of the pushbutton was addressed by real-time human monitoring of the incoming sensor signals, including the pushbutton channel, during acquisition. Additionally, a mobile phone video camera for post verification and adjustments was used (see Section 2.4).
2.4 Post Verification
Although the “segmentation and annotation” mode of the ASK software was switched on to segment and annotate the recorded data efficiently, a mobile phone video camera was used in addition to record the whole biosignal acquisition sessions to manually correct the misoperation of pushing/holding/releasing after the data recording.
After each recording event with one subject, the collected data and the automatically generated segments with annotation labels were examined thoroughly based on the video. Segments with minor human-factor errors were corrected by shifting the start-/endpoints forward/backward a short distance manually, while segments with problems that cannot be easily corrected were discarded, which is one of the reasons leading to the slight divergence among the activity occurrences in Table 1 (see Section 2.8 for another reason). A script to automatically detect the activity length outlier was also implemented to assist the post verification. After finishing the correction and verification, we deleted all recorded videos to preserve privacy.
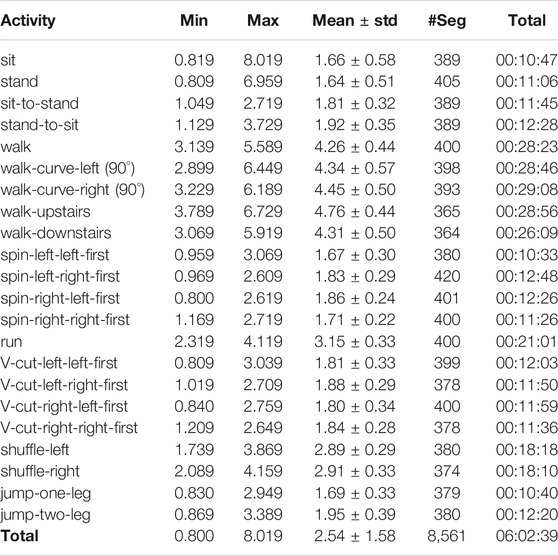
TABLE 1. Statistics of segmented corpus in the CSL-SHARE dataset. The minimum (Min), maximum (Max), mean, and standard deviation (std) values are in seconds. #Seg: number of segments.
2.5 Activities and Protocols
The CSL-SHARE dataset was recorded in a controlled laboratory environment at the Cognitive Systems Lab, University of Bremen, comprised of 22 daily living and sports-based activities. The acquisition protocols of CSL-SHARE recording events were strictly and normatively designed. The body steering angles and the number of steps related to the activity parameters are restricted. Most of the acquisition protocols contain only one activity. However, there are two protocols with two activities and one protocol with four activities because these activities can be practically and logically recorded one after another in a sequence, which also keeps the balance of the activity occurrences. To follow the logical sequence of the activities and the protocol-for-pushbutton segmentation and annotation mechanism (see Section 2.3), the order of the activities in these three multi-activity protocols must be observed during recording.
Figure 1 illustrates the diagrammatic sketch of all recording protocols, helping more intuitively understand the recording procedure and activity details. The 22 activities and the 17 acquisition protocols are described as follows:
•Protocol 1: walk (Figure 1A)
<push + hold> walk forward with three gait cycles, left foot starts, i.e., left-right-left-right-left-right <release> → (turn around in place) → …
20 repetitions (20 activities with 60 gait cycles) per subject.
Note: the “turn around in place” between two “walk”/“run” activities is due to the limited space in our laboratory.
•Protocol 2: walk-curve-left (90°) (Figure 1B)
<push + hold> turn left 90° with three gait cycles, left foot starts <release> → (turn left 90° in place) → …
20 repetitions (20 activities with 60 gait cycles) per subject.
•Protocol 3: spin-left-left-first (Figure 1C)
<push + hold> turn left 90° in one step, left foot starts <release> → …
20 repetitions (20 activities with 20 gait cycles) per subject.
•Protocol 4: spin-left-right-first (Figure 1C)
<push + hold> turn left 90° in one step, right foot starts <release> → …
20 repetitions (20 activities with 20 gait cycles) per subject.
•Protocol 5: walk-curve-right (90°) (Figure 1D)
<push + hold> turn right 90° with three gait cycles, left foot starts <release> → (turn right 90° in place) → …
20 repetitions (20 activities with 60 gait cycles) per subject.
•Protocol 6: spin-right-left-first (Figure 1E)
<push + hold> turn right 90° in one step, left foot starts <release> → …
20 repetitions (20 activities with 20 gait cycles) per subject.
•Protocol 7: spin-right-right-first (Figure 1E)
<push + hold> turn right 90° in one step, right foot starts <release> → …
20 repetitions (20 activities with 20 gait cycles) per subject.
•Protocol 8: run (Figure 1A)
<push + hold> go forward at fast speed with three gait cycles, left foot starts <release> → (turn around in place) → …
20 repetitions (20 activities with 60 gait cycles) per subject.
•Protocol 9: V-cut-left-left-first (30°) (Figure 1F)
<push + hold> turn 30° left forward in one step at jogging speed, left foot starts <release> → (return backward to the start point with three steps, left foot starts) → …
20 repetitions (20 activities with 20 gait cycles) per subject.
•Protocol 10: V-cut-left-right-first (30°) (Figure 1F)
<push + hold> turn 30° left forward in one step at jogging speed, right foot starts <release> → (return backward to the start point with three steps, left foot starts) → …
20 repetitions (20 activities with 20 gait cycles) per subject.
•Protocol 11: V-cut-right-left-first (30°) (Figure 1G)
<push + hold> turn 30° right forward in one step at jogging speed, left foot starts <release> → (return backward to the start point with three steps, left foot starts) → …
20 repetitions (20 activities with 20 gait cycles) per subject.
•Protocol 12: V-cut-right-right-first (30°) (Figure 1G)
<push + hold> turn 30° right forward in one step at jogging speed, right foot starts <release> → (return backward to the start point with three steps, left foot starts) → …
20 repetitions (20 activities with 20 gait cycles) per subject.
•Protocol 13: shuffle-left + shuffle-right (Figure 1H)
<push + hold> move leftward with three lateral gaits cycles, left foot starts, right foot follows <release> → <push + hold> move rightward with three lateral gaits cycles, right foot starts, left foot follows <release> → …
20 repetitions (40 activities with 120 gait cycles) per subject.
•Protocol 14: sit + sit-to-stand + stand + stand-to-sit (Figure 1I)
<push + hold> sit <release> → <push + hold> stand up <release> → <push + hold> stand <release> → <push + hold> sit down <release> → …
20 repetitions (80 activities) per subject.
•Protocol 15: jump-one-leg (Figure 1J)
<push + hold> squat, then jump upwards using the bandaged right leg, land in <release> → …
20 repetitions (20 activities) per subject.
•Protocol 16: jump-two-leg (Figure 1J)
<push + hold> squat, then jump upwards using both legs, land in <release> → …
20 repetitions (20 activities) per subject.
•Protocol 17: walk-upstairs + walk-downstairs (Figure 1K)
<push + hold> go up six stairs with three gait cycles, left foot starts <release> → (turn around in place) → <push + hold> go down six stairs with three gait cycles, left foot starts <release> → (turn around in place) → …
20 repetitions (40 activities with 120 gait cycles) per subject.
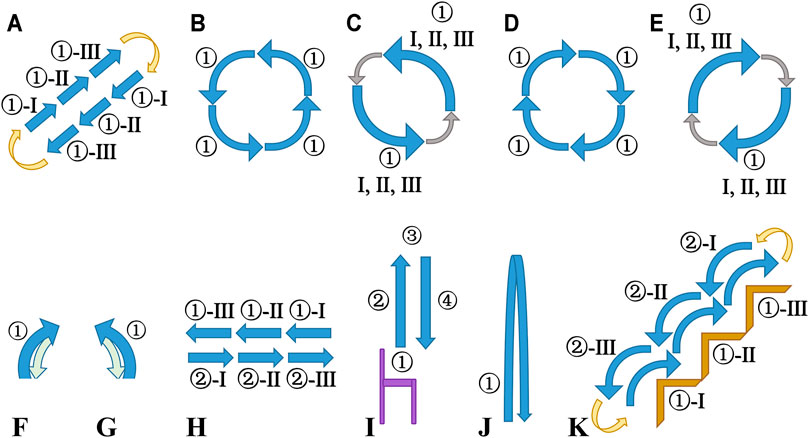
FIGURE 1. Diagrammatic sketch of all recording protocols. Sub-diagrams (A–G) are from the top view; Sub-diagram H is from the front view; Sub-diagrams (I–K) are from the side view. Blue arrows: activities or gaits; I, II, III: gait cycles; yellow arrows: turn around (180°) in place; gray arrows: turn left/right (90°) in place; green arrows: return backward to the start point. (A) Protocol 1 and Protocol 8: ① walk/run. (B) Protocol 2: ① walk-curve-left (90°). (C) Protocol 3 and Protocol 4: ① spin-left-left-first/spin-left-right-first. (D) Protocol 5: ① walk-curve-right (90°). (E) Protocol 6 and Protocol 7: ① spin-right-left-first/spin-right-right-first. (F) Protocol 9 and Protocol 10: ① V-cut-left-left-first/V-cut-left-right-first. (G) Protocol 11 and 12: ① V-cut-right-left-first/V-cut-right-right-first. (H) Protocol 13:① shuffle-left; ② shuffle-right. (I) Protocol 14: ① sit; ② sit-to-stand; ③ stand; ④ stand-to-sit; purple: a chair. (J) Protocol 15 and Protocol 16: ① jump-two-leg/jump-one-leg. (K) Protocol 17: ① walk-upstairs; ② walk-downstairs; brown: stairs.
The number of repetitions/to-record activities per each protocol is a pre-designed plan. In the post verification (see Section 2.4), a few non-conformity and erroneous segments were removed.
The meaning of most of the activities in the CSL-SHARE dataset can be self-explanatory from their names or the description in the protocols. The “spin-left/right” activity can be understood as the “left/right face” action in the army (but in daily situations, not so stressful as in military training). The “V-cut” activity is a step in which the body rotation (instead of the directional change) takes place, as shown in Figures 1F, G.
Some activities in the CSL-SHARE dataset are the subdivision of original activities. For example, “spin-left” is divided into “spin-left-left-first” and “spin-left-right-first,” denoting which foot should be moved first. Similarly, “spin-right,” “V-cut-left,” and “V-cut-right” are also divided into two activities in regard to the first-moved foot. The activities mentioned above are subdivided because they only involve one gait, and we only use the sensors placed on the right-leg-worn bandage. Therefore, the “left foot first” and “right foot first” of these activities will lead to very different signal patterns. On the contrary, for activities involving multiple (three) steps/gait cycles, such as “walk,” “walk-curve left/right,” “walk-upstairs,” “walk-downstairs,” “run,” and “shuffle-left/right,” we did not further subdivide them. Instead, we restricted in the protocols the number of gaits for each segment of these activities to three and defined the left foot as the start.
2.6 Subjects
Twenty subjects without any gait impairments, five female and fifteen male, aged between 23 and 43 (30.5 ± 5.8), participated in the data collection events, among which one subject had knee inflammation and could not perform certain activities. Each subject’s participation time is approximately 2 h, including announcement and precautions, questions and answers, equipment wearing and adjusting, software preparation and test-running, acquisition following all protocols, taking breaks, and equipment release.
2.7 Privacy Preservation and Data Security
All subjects signed a written informed consent form, and the study was conducted in accordance with Helsinki’s WMA (World Medical Association) Declaration (Association, 2013). According to the consent form, we only kept the wearable sensor data pseudonymized and did not leave any identification information of the participants. The to-share CSL-SHARE dataset is available in an anonymized form. As mentioned in Section 2.4, we used videos to verify the segmentation and annotation, and all videos have been deleted after the post verification to protect privacy.
In addition, the consent form stipulates that the use of the data is limited to non-commercial research purposes, and the data users guarantee not to attempt to identify the participating persons. Furthermore, the data users guarantee to pass on the data (or data derived from it) only to third parties who are bound by the same rules of use (for non-commercial research purposes, no identification attempts, restricted disclosure). Data users who violate the usage regulation mentioned above will bear the legal consequences themselves, where the dataset publisher takes no responsibility.
2.8 Data Format
We provide our CSL-SHARE dataset in an anonymized form in the following directory structure and file format: The root directory contains a total of 20 sub-directories with order numbers 1–20, representing the data of 20 subjects. In each subdirectory, there are 34 files. The seventeen.H5 files, named by the order number of protocols, use HDF5 format to save the raw recorded data of seventeen protocols, while the seventeen.CSV files are the corresponding annotation results.
Each row in the .H5 files is according to the following sensor order: EMG 1, EMG 2, EMG 3, EMG 4, airborne microphone, accelerometer upper X, accelerometer upper Y, accelerometer upper Z, electrogoniometer X, accelerometer lower X, accelerometer lower Y, accelerometer lower Z, electrogoniometer Y, gyroscope upper X, gyroscope upper Y, gyroscope upper Z, gyroscope lower X, gyroscope lower Y, gyroscope lower Z.
There are three sub-directories/sub-datasets with exceptions:
• Sub-directory 2: The 02.CSV and 05.CSV files are different from protocols 2 and 5: the labels are mixed with each other. Subject 2 performed wrong angles when turning the body between activities. We were not aware of it during the data collection process, and the problem was first discovered through the video in the post-verification. However, this mixture affects neither the integrity of the dataset nor the number of times each activity should occur
• Sub-directory 11: Protocol 13 is divided into two parts due to the device communication breaking
• Sub-directory 16: Not all activities were performed due to the subject’s knee inflammation, which is one of the reasons leading to the slight divergence among the activity occurrences in Table 1 (see Section 2.4 for another reason).
3 Statistical Analysis
The 22-activity CSL-SHARE dataset contains 11.52 hours of data (of which 6.03 hours have been segmented and annotated) from 20 subjects, 5 female and 15 male. Table 1 gives the number of activity segments, the total effective length over all segments, and the minimal/maximal/mean length of the 22 activities.
By analyzing the duration distribution of each activity of all subjects in histograms, we find that all activities’ duration over all segments approximately accords with the normal distribution. The distribution of the activities “sit” and “stand” deviates slightly, as they can last arbitrarily long.
4 Conclusion
We share our in-house collected dataset CSL-SHARE (Cognitive Systems Lab Sensor-based Human Activity REcordings) in this article and introduce its recording procedure and technical details. This 19-channel 22-activity 20-subject dataset applies two triaxial accelerometers, two triaxial gyroscopes, four EMG sensors, one biaxial electrogoniometer, and one airborne microphone with sampling rates up to 1000 Hz and uses a knee bandage as a novel wearable sensor carrier. Six-hour data of a totally 11.52-h recording are well segmented, annotated, and post-verified. The reliability and applicability of the CSL-SHARE dataset and its previous pilot data collection can be observed through literature in various research aspects here and there, such as HAR research pipeline (Liu and Schultz, 2018), real-time end-to-end HAR system (Liu and Schultz, 2019), visualized verification of multimodal feature extraction (Barandas et al., 2020), feature dimensionality study (Hartmann et al., 2020; Hartmann et al., 2021), human activity modeling (Liu et al., 2021), among others.
To the best of our knowledge, CSL-SHARE is the first publicly available dataset recorded with sensors integrated into a knee bandage and one of the most comprehensive HAR datasets with an ample number of sensors, activities, and subjects, as well as complete synchronization, segmentation, and annotation.
Standing on the dataset robustness, we publish the CSL-SHARE dataset as an open-source sensor-based biosignals dataset for HAR, hoping to contribute research materials to the researchers in the same or similar fields.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.uni-bremen.de/en/csl/research/human-activity-recognition.
Ethics Statement
Ethical approval was not provided for this study on human participants because the study was conducted in accordance with Helsinki's WMA (World Medical Association) Declaration. The participants provided their written informed consent to participate in this study. According to the consent form, we only kept the wearable sensor data pseudonymized and did not leave any identification information of the participants. The dataset is shared in an anonymized form.
Author Contributions
HL developed the software Activity Signal Kit (ASK), designed the protocols, organized the recording events, and processed the recorded data afterward. YH helped collect the pilot dataset CSL18 at the collaborative labor in Karlsruhe Institute of Technology and is responsible for disclosing and maintaining the data. HL and YL cooperatively researched human activity modeling, real-time recognition, feature dimensionality, and feature selection based on the published dataset and verify its completeness, correctness, and applicability. TS supervised and supported the work, advised the manuscript, and provided critical feedback. All authors read and approved the final version.
Funding
Open access was supported by the Open Access Initiative of the University of Bremen and the DFG via SuUB Bremen.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We gratefully acknowledge Bernd Stetter, Frieder Krafft, and Thorsten Stein for the cooperative work with the sensor placement design and laboratory data acquisition collaboration.
Footnotes
1https://biosignalsplux.com/products/kits/researcher.html, accessed August 10, 2021
References
Association, W. M. (2013). World Medical Association Declaration of Helsinki. Jama 310, 2191–2194. doi:10.1001/jama.2013.281053
Barandas, M., Folgado, D., Fernandes, L., Santos, S., Abreu, M., Bota, P., et al. (2020). TSFEL: Time Series Feature Extraction Library. SoftwareX 11, 100456. doi:10.1016/j.softx.2020.100456
Barbič, J., Safonova, A., Pan, J.-Y., Faloutsos, C., Hodgins, J. K., and Pollard, N. S. (2004). “Segmenting Motion Capture Data into Distinct Behaviors,” in Proceedings of Graphics Interface (Citeseer), 185–194.
Chavarriaga, R., Sagha, H., Calatroni, A., Digumarti, S. T., Tröster, G., Millán, J. d. R., et al. (2013). The Opportunity challenge: A Benchmark Database for On-Body Sensor-Based Activity Recognition. Pattern Recognition Lett. 34, 2033–2042. doi:10.1016/j.patrec.2012.12.014
Guenterberg, E., Ostadabbas, S., Ghasemzadeh, H., and Jafari, R. (2009). “An Automatic Segmentation Technique in Body Sensor Networks Based on Signal Energy,” in Proceedings of the Fourth International Conference on Body Area Networks (Institute for Computer Sciences (Brussels: Social-Informatics and Telecommunications Engineering). doi:10.4108/icst.bodynets2009.6036
Hartmann, Y., Liu, H., and Schultz, T. (2021). “Feature Space Reduction for Human Activity Recognition Based on Multi-Channel Biosignals,” in Proceedings of the 14th International Joint Conference on Biomedical Engineering Systems and Technologies (Setúbal: INSTICC (SciTePress)), 215–222. doi:10.5220/0010260802150222
Hartmann, Y., Liu, H., and Schultz, T. (2020). “Feature Space Reduction for Multimodal Human Activity Recognition,” in Proceedings of the 13th International Joint Conference on Biomedical Engineering Systems and Technologies - Volume 4 (Setúbal: BIOSIGNALS. INSTICC (SciTePress), 135–140. doi:10.5220/0008851401350140
Hu, B., Rouse, E., and Hargrove, L. (2018). Benchmark Datasets for Bilateral Lower-Limb Neuromechanical Signals from Wearable Sensors during Unassisted Locomotion in Able-Bodied Individuals. Front. Robot. AI 5, 14. doi:10.3389/frobt.2018.00014
Karagiannaki, K., Panousopoulou, A., and Tsakalides, P. (2016). The Forth-Trace Dataset for Human Activity Recognition of Simple Activities and Postural Transitions Using a Body Area Network.. https://zenodo.org/record/841301#.YVX4XLgzaUk. doi:10.5281/zenodo.841301
Kwapisz, J. R., Weiss, G. M., and Moore, S. A. (2010). “Activity Recognition Using Cell Phone Accelerometers,” in Proceedings of the 4th International Workshop on Knowledge Discovery from Sensor Data, 10–18.
Liu, H., Hartmann, Y., and Schultz, T. (2021). “Motion Units: Generalized Sequence Modeling of Human Activities for Sensor-Based Activity Recognition,” in EUSIPCO 2021 - 29th European Signal Processing Conference (IEEE) (New York: IEEE).
Liu, H., and Schultz, T. (2019). “A Wearable Real-Time Human Activity Recognition System Using Biosensors Integrated into a Knee Bandage,” in Proceedings of the 12th International Joint Conference on Biomedical Engineering Systems and Technologies - Volume 1 (Setúbal: BIODEVICES. INSTICC (SciTePress), 47–55. doi:10.5220/0007398800470055
Liu, H., and Schultz, T. (2018). “ASK: A Framework for Data Acquisition and Activity Recognition,” in Proceedings of the 11th International Joint Conference on Biomedical Engineering Systems and Technologies - Volume 1 (Setúbal: BIOSIGNALS. INSTICC (SciTePress), 262–268. doi:10.5220/0006732902620268
Loose, , Tetzlaff, H., Bolmgren, L., and Lindstr, J. (2020). A Public Dataset of Overground and Treadmill Walking in Healthy Individuals Captured by Wearable IMU and sEMG Sensors. BIOSIGNALS, 164–171.
Mathie, M. J., Coster, A. C., Lovell, N. H., and Celler, B. G. (2003). Detection of Daily Physical Activities Using a Triaxial Accelerometer. Med. Biol. Eng. Comput. 41 (3), 296–301. doi:10.1007/BF02348434
Micucci, D., Mobilio, M., and Napoletano, P. (2017). UniMiB SHAR: A Dataset for Human Activity Recognition Using Acceleration Data from Smartphones. Appl. Sci. 7, 1101. doi:10.3390/app7101101
Rebelo, D., Amma, C., Gamboa, H., and Schultz, T. (2013). “Human Activity Recognition for an Intelligent Knee Orthosis,” in BIOSIGNALS 2013 - 6th International Conference on Bio-inspired Systems and Signal Processing, 368–371. doi:10.5220/0004254903680371
Rowe, P. J., Myles, C. M., Walker, C., and Nutton, R. (2000). Knee Joint Kinematics in Gait and Other Functional Activities Measured Using Flexible Electrogoniometry: How Much Knee Motion Is Sufficient for normal Daily Life? Gait Posture 12, 143–155. doi:10.1016/s0966-6362(00)00060-6
Stetter, B. J., Krafft, F. C., Ringhof, S., Gruber, R., Sell, S., and Stein, T. (2018). “Estimation of the Knee Joint Load in Sport-specific Movements Using Wearable Sensors,” in SPINFORTEC 2018 - 12th Symposium der Sektion Sportinformatik und Sporttechnologie der Deutschen Vereinigung.
Stetter, B., Krafft, F., Ringhof, S., Sell, S., and Stein, T. (2019). “Assessing Knee Joint Forces Using Wearable Sensors and Machine Learning Techniques,” in Proceedings der DVS-Jahrestagung Biomechanik 2019 (Konstanz: Universität Konstanz), 55–56.
Sztyler, T., and Stuckenschmidt, H. (2016). “On-body Localization of Wearable Devices: An Investigation of Position-Aware Activity Recognition,” in 2016 IEEE International Conference on Pervasive Computing and Communications (PerCom) (IEEE Computer Society), 1–9. doi:10.1109/percom.2016.7456521
Teague, C. N., Hersek, S., Toreyin, H., Millard-Stafford, M. L., Jones, M. L., Kogler, G. F., et al. (2016). Novel Methods for Sensing Acoustical Emissions from the Knee for Wearable Joint Health Assessment. IEEE Trans. Biomed. Eng. 63, 1581–1590. doi:10.1109/tbme.2016.2543226
Telaar, D., Wand, M., Gehrig, D., Putze, F., Amma, C., Heger, D., et al. (2014). “Biokit—real-time Decoder for Biosignal Processing,” in INTERSPEECH 2014 - 15th Annual Conference of the International Speech Communication Association.
Keywords: human activity recognition, activity of daily living, EMG, inertial sensor, internal sensing, wearable sensing, accelerometer, gyroscope
Citation: Liu H, Hartmann Y and Schultz T (2021) CSL-SHARE: A Multimodal Wearable Sensor-Based Human Activity Dataset. Front. Comput. Sci. 3:759136. doi: 10.3389/fcomp.2021.759136
Received: 15 August 2021; Accepted: 20 September 2021;
Published: 15 October 2021.
Edited by:
Pekka Siirtola, University of Oulu, FinlandReviewed by:
Jun Shen, University of Wollongong, AustraliaJerry Chu-Wei Lin, Western Norway University of Applied Sciences, Norway
Copyright © 2021 Liu, Hartmann and Schultz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Liu, aHVpLmxpdUB1bmktYnJlbWVuLmRl
 Hui Liu
Hui Liu Yale Hartmann
Yale Hartmann Tanja Schultz
Tanja Schultz