Abstract
Background:
Changes in circadian-related behaviors (e.g., the timing of food intake, sleep cycles) and the environment (e.g., light-dark cycles) increase the risk of numerous cardiometabolic diseases, including diabetes mellitus and cardiac disease. Recent studies indicate a close interrelationship between circadian clocks and the posttranslational modification, protein O-GlcNAcylation. The current study was designed to investigate whether a modest elevation of protein O-GlcNAcylation in the adult mouse heart, similar to levels observed during pathologic states, influenced circadian governance of the heart.
Methods and results:
Cardiomyocyte-specific expression of a dominant negative O-GlcNAcase (dnOGAh) for a 2-week period resulted in an approximate 1.5-fold increase in cardiac protein O-GlcNAcylation, impacting 70% of core circadian clock components in the heart at the mRNA level. Further interrogation of cardiac mRNA species in dnOGAh hearts at candidate (RT-PCR) and unbiased (RNAseq) levels revealed a 95% loss of circadian governance of the cardiac transcriptome. This was despite persistent/augmented 24 h oscillations of the core circadian clock proteins BMAL1, REVERBα, and PER2 in dnOGAh hearts. Direct comparison of dnOGAh hearts with cardiomyocyte-specific BMAL1 knockout (CBK) hearts underscored an apparent uncoupling of the core clock mechanism from clock control of downstream target genes in dnOGAh hearts, and highlighted that loss of circadian governance results in interstitial fibrosis.
Conclusions:
Sustained protein O-GlcNAcylation in the heart causes loss of circadian governance, likely downstream of the core circadian clock mechanism. Moreover, interstitial fibrosis appears to be a universal adverse outcome following impaired circadian governance.
Introduction
Reversible posttranslational modification (PTM) of cellular proteins is an established means by which biological processes are modulated in both acute and chronic manners. Several hundred different PTMs have been identified, although the function of many of these modifications are not known. Most extensively studied PTMs include phosphorylation, acetylation, palmitoylation, SUMOylation, and O-GlcNAcylation ( 1). The latter involves the reversible, catalytic O-linked addition of β-N-acetyl-glucosamine to serine and threonine residues of target proteins by the enzyme O-GlcNAc transferase (OGT) (2). This PTM affects protein stability, cellular localization, and functions such as protein-protein interactions and intrinsic activity (2). It has been estimated that >5,000 proteins can be O-GlcNAc modified; O-GlcNAcylated proteins mediate/regulate diverse biological processes, ranging from transcription, translation, and protein quality control, to signaling, electrophysiology, and metabolism (3). Given that the carbon required for this PTM is derived from glucose, fatty acids, and amino acids (via the hexosamine biosynthetic pathway), protein O-GlcNAcylation has classically been considered a nutrient sensing pathway (3). However, it has become increasingly clear that alterations in protein O-GlcNAcylation occur in response to a variety of nutrient-independent stimuli/stresses, in both acute and chronic settings (2, 4). This can be exemplified by studies in the heart, for which perturbations in protein O-GlcNAcylation have been causally linked to acute adaptations to physiologic stimuli (e.g., exercise) and chronic pathologic conditions (e.g., diabetes mellitus and cardiac hypertrophy) (2, 4–6). In the latter case, recently published studies indicate that selective augmentation of protein O-GlcNAcylation in the adult murine heart results in interstitial fibrosis and diastolic dysfunction, which is comparable to the pathologic cardiac remodeling observed during diabetes (7).
Consistent with the dynamic and reversible nature of protein O-GlcNAcylation, levels of this PTM fluctuate over the course of the day in multiple organs, including the heart, peaking during the awake period ( 8, 9). For the heart, these oscillations appear to be driven in large part by the cardiomyocyte circadian clock (8). This cell autonomous timekeeping molecular mechanism is comprised of a series of interconnected transcription-translation feedback loops, which have a free running periodicity of approximately 24 h (10). Numerous core circadian clock components are regulated by PTMs, which are critical for the temporal nature of this timekeeping mechanism (11). One such PTM is protein O-GlcNAcylation. For example, BMAL1 (brain and muscle ARNT-like 1), CLOCK (circadian locomotor output cycles kaput), and PER (period) proteins can be O-GlcNAc modified, which affects the nuclear-cytosolic localization, DNA binding, and/or stability of these core circadian clock components (8, 9, 12, 13). Consistent with these reports, acute (i.e., <24 h) pharmacologic augmentation of protein O-GlcNAcylation cause a rapid phase shift in the heart circadian clock (8).
Considerable overlap exists between the cellular processes regulated by circadian clocks and protein O-GlcNAcylation. Moreover, cardiomyocyte circadian clock disruption and chronic elevation of cardiomyocyte protein O-GlcNAcylation both result in cardiac fibrosis and contractile dysfunction ( 7, 14–16). Despite these observations, and the aforementioned interrelationship between the circadian clock and protein O-GlcNAcylation, it is unclear whether sustained elevation of O-GlcNAcylation (as might be observed during a disease state) contributes towards adverse cardiac remodeling through impairment of circadian governance (i.e., the ability of the circadian clock to temporally regulate biologic processes). To gain insight into this possibility, we compared the impact of sustained elevation of cardiomyocyte protein O-GlcNAcylation and cardiomyocyte BMAL1 deletion on circadian governance. These studies revealed that a modest (∼1.5-fold) increase in cardiac protein O-GlcNAcylation for 2 weeks is sufficient to severely attenuate the cardiac diurnal transcriptome, including circadian governance of genes involved in the extracellular matrix and collagen deposition.
Methods
Animals
The present study utilized inducible, cardiomyocyte-specific dominant negative O-GlcNAcase (dnOGAh) transgenic mice, as described previously ( 7). Briefly, to generate dnOGAh mice, TRE-EGFP-dnOGA (tetracycline-responsive element, enhanced green fluorescent protein, rat OGA splice variant) mice were bred with αMHC-rtTA (α-myosin heavy chain promoter driven codon optimized reverse tetracycline transactivator) mice. To induce the transgene, 10 week old male dnOGAh mice were injected with 100 μg doxycycline (DOX) (MilliporeSigma, Burlington, MA, USA) in 0.9% NaCl solution and immediately switched to 1 g/kg DOX-supplemented NIH31 diet (Envigo, Madison, WI, USA). αMHC-rtTA mice treated with DOX for the same duration served as controls (CON). This study also utilized male cardiomyocyte-specific BMAL1 knockout (CBK) mice; littermate floxed mice served as controls (CON), as described previously (14). All mice were housed at the Center for Comparative Medicine at the University of Alabama at Birmingham (UAB), under temperature-, humidity-, and light- controlled conditions. A strict 12 h light/12 h dark cycle regime was enforced [lights on at 6 a.m.; zeitgeber time (ZT) 0]; the light/dark cycle was maintained throughout these studies, facilitating investigation of diurnal variations (as opposed to circadian rhythms). Mice were housed either in standard micro-isolator cages or CLAMS (Comprehensive Laboratory Animal Monitoring System) cages, and received food and water in an ad libitum fashion. CLAMS cages allowed the non-invasive assessment of energy expenditure, food intake and physical activity in a continuous manner, as described previously (17). All mice were on the C57BL/6 background strain, were male, and were 12 weeks old at the time of assessments and/or tissue collection. All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Alabama at Birmingham.
Quantitative RT-PCR
RNA was extracted from biventricular samples using standard procedures. Candidate gene expression analysis was performed by quantitative RT-PCR, using previously described methods ( 18, 19). For quantitative RT-PCR, custom-designed Taqman assay were utilized; sequences for custom-designed assays have been reported previously (14, 20). All quantitative RT-PCR data are presented as fold change from an indicated control group.
RNA sequencing
Transcriptomic analysis was performed using RNA sequencing (RNA-seq) in the UAB Genomics Core facility. Following initial testing of RNA samples using an Agilent BioAnalyzer, RNA with RIN values greater than 7.0 were subsequently utilized for library preparation (after DNAse treatment). RNA-Sequencing libraries were next generated using the NEBNext Ultra II RNA kit (NEB, Ipswich MA); resulting libraries were sequenced on the Illumina NextSeq 500 (Illumina, Inc. San Diego CA) using paired end 75 bp sequencing reads, per standard methods. Sequencing data can be accessed in GEO.
Western blotting
Qualitative analysis of protein expression was performed via standard western blotting procedures, as described previously (8). Briefly, 15–30 μg protein lysate was separated on polyacrylamide gels and transferred to PVDF membranes. Membranes were probed for with anti- BMAL1 (Cell Signaling Tech #14020; 1:2000), REVERBα (Cell Signaling Tech #13418; 1:2000), PER2 (Alpha Diagnostics Inc #PER21-A; 1:2500), and E4BP4 (Cell Signaling Tech #14312; 1:2000) antibodies, or with the CTD110.6 antibody (UAB Epitope Recognition Core) for detection of O-GlcNAcylation. Rabbit and mouse HRP-conjugated secondary antibodies (Cell Signaling 7074 and 7076, respectively; 1:2000–5000) were used for chemiluminescent detection with Luminata Forte Western Blotting substrate (Millipore, WBLUF0100). All densitometry data were normalized to amido black staining. Importantly, to minimize the contribution that position on the gel might have on outcomes, samples were randomized on gels; all original gels are included in Supplemental files.
Histologic assessment
Cross sections from the middle region of the left ventricle were taken immediately upon removal of the heart, and were fixed in formalin for 24 h (followed by storage in 70% ethanol at 4°C prior to paraffin embedding and sectioning). Assessment of left ventricular interstitial fibrosis employed either Picrosirius Red staining (CBK hearts) or Masson Trichrome staining (dnOGAh hearts), followed by semi-quantitative analysis using Image-J software (NIH), as described previously (21). All original images are included in Supplemental files.
Statistical analysis
All data are presented as mean ± standard error of the mean (SEM). For non-omics data (i.e., not RNA-seq data), comparisons among groups with two variables were analyzed by two-way analysis of variance (ANOVA) through use of Prism statistical software to investigate main effects of genotype and time. Normality of data was assessed through use of the Shapiro–Wilks test, followed by either parametric (t-tests for only two experimental groups or Sidak's post-hoc test for multiple pairwise comparisons) or non-parametric (Mann–Whitney for only two experimental groups or Kruskal–Wallis with Dunn's correction for multiple pairwise comparisons). Cosinor analyses were performed to determine whether 24 h time series data significantly fit a cosine curve; if they did, then mesor (daily average value), amplitude (peak-to-mesor difference), and acrophase (timing of the peak) were calculated and compared between experimental groups, as described previously (22). For RNA-seq data, JTK cycle was performed using the online tool NiteCap. For the JTK cycle analysis, transcripts were considered to have a 24 h oscillation if they significantly fit a cosine curve (q < 0.05); amplitude and acrophase were calculated within NiteCap, which also determined significant differences between genotypes for these cosine parameters. NiteCap was further utilized for heat map generation and principal component analysis (PCA). In addition, the online tool PANTHER was utilized for pathway enrichment analysis. In all analyses, the null hypothesis of no model effects was rejected at p < 0.05.
Biological variables
The primary outcomes of this study included gene expression, protein expression, and fibrosis; one or more of these outcomes are influenced by time-of-day, sex, age, diet, and genetic background. Consistent with the stated purpose of the study, two main variables were investigated: time-of-day and genotype (i.e., CBK, dnOGAh, and littermate controls). This resulted in a total of 28 experimental groups: 16 experimental groups for the CBK model and 12 experimental groups for the dnOGAh model. To the extent that was possible based on the study design, other biological variables remained constant. Male 12 week old mice on the C57BL/6 background were chosen, allowing direct comparison of RNA-seq data with previously published transcriptomic data obtained from CBK and littermate control hearts ( 14). Our recent studies investigating male and female dnOGAh mice at this age revealed comparable phenotypes (7). One variable that was not identical between CBK and dnOGAh models was diet; dnOGAh mice were fed a doxycycline containing diet (to induce the transgene), whereas CBK mice were fed a normal rodent chow.
Results
Cardiomyocyte-specific BMAL1 deletion impacts both circadian clock gene expression and protein O-GlcNAcylation in the heart
To identify roles of the cardiomyocyte circadian clock, we previously generated a cardiomyocyte-specific BMAL1 knockout (CBK) mouse model ( 14). To confirm that the cardiac clock is disrupted in CBK mice, hearts were collected from CBK and littermate control mice at 3 h intervals over a 24 h period, followed by assessment of circadian clock components at mRNA and protein levels. At the transcript level, 10 core circadian clock components were investigated; bmal1, clock, npas2, reverbα, reverbβ, per1, per2, per3, cry1, and cry2 mRNA. A 2-way ANOVA revealed significant time main effects for 9 out of 10 core clock component mRNAs (Figure 1A;Supplementary Table 1); only cry2 mRNA did not exhibit a significant time main effect (p = 0.187). Significant genotype main effects were also observed for 9 out of 10 core clock component mRNAs (Figure 1A;Supplementary Table 1); per2 mRNA did not exhibit a significant genotype main effect (p = 0.183). Of those transcripts that exhibited significant genotype main effects, bmal1, reverbα, reverbβ, per1, and per3 were repressed, while clock, npas2, cry1, and cry2 were induced (Figure 1A;Supplementary Table 1). Cosinor analysis indicated that all 10 core clock component mRNAs exhibit 24hr oscillations in CON hearts; these oscillations were either abolished or significantly attenuated in CBK hearts for 8 out of 10 core clock component mRNAs (cry2 and per2 mRNA 24 h oscillations persisted in CBK hearts;Supplementary Table 2). At the protein level, 3 core circadian clock components were investigated; BMAL1, REVERBα, and PER2. A 2-way ANOVA revealed significant time and genotype main effects for all 3 core clock proteins; both BMAL1 and REVERBα protein levels were decreased in CBK hearts, while PER2 levels were increased (Figure 1B;Supplementary Table 1). Cosinor analysis indicated that all 3 core clock component proteins exhibit 24 h oscillations in CON hearts; REVERBα oscillations were significantly attenuated in CBK hearts (Supplementary Table 2). Consistent with the cardiomyocyte-specific nature of the murine model, CBK mice exhibited 24 h oscillations in energy expenditure, food intake, and physical activity that were comparable to littermate control mice (Supplementary Figure 1A, as well asSupplementary Tables 1, 2). Similarly, we have previously reported that distinct circadian clock components are not altered in livers of CBK mice (14). Next, we investigated protein O-GlcNAcylation levels in CON and CBK hearts isolated during the light phase (ZT9); protein O-GlcNAcylation levels were significantly increased in CBK hearts (2.26-fold;Figure 1C). Collectively, these data are consistent with the concept that cardiomyocyte BMAL1 deletion disrupts the clock in the heart, and this is associated with augmentation of protein O-GlcNAcylation.
Figure 1
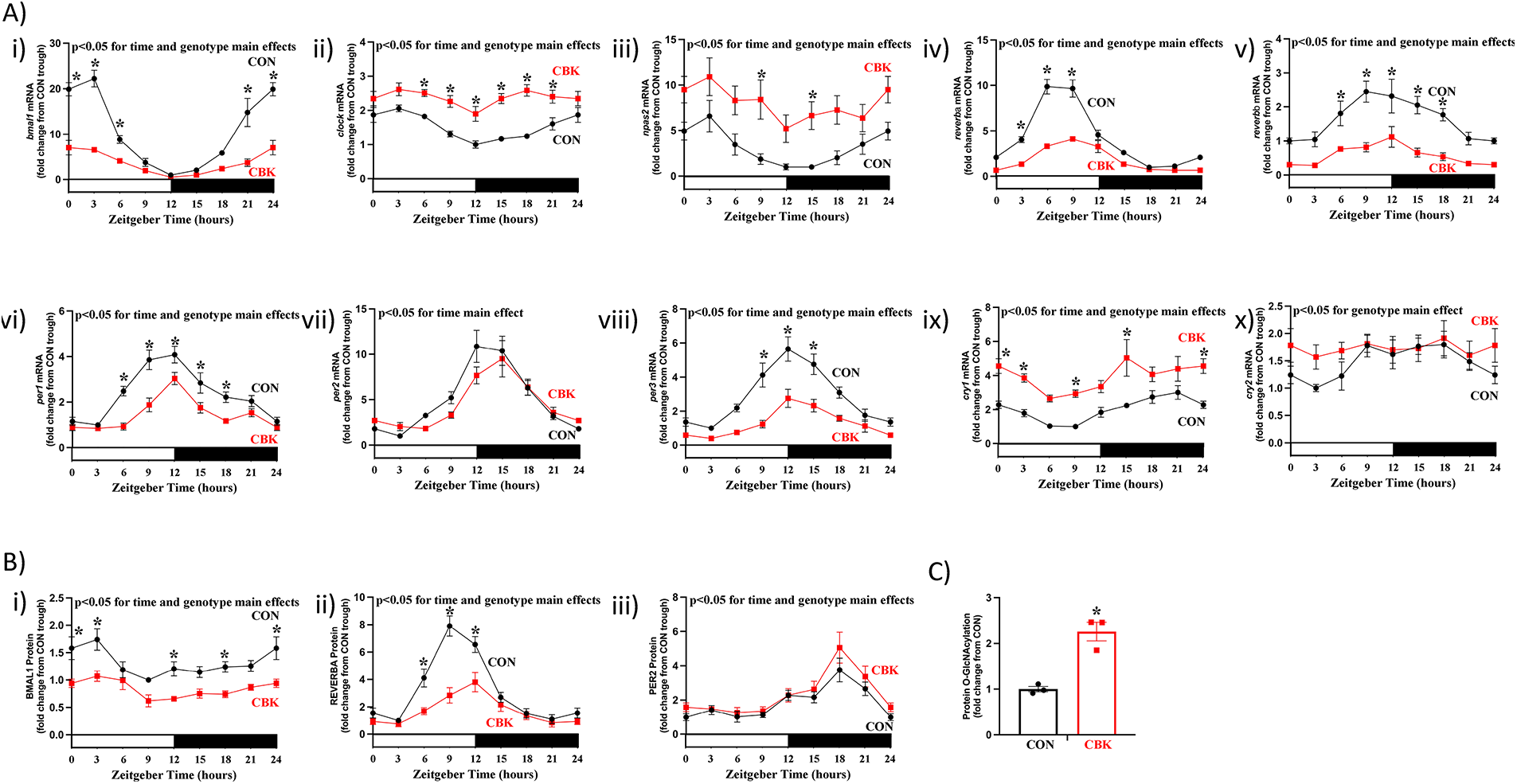
Impact of cardiomyocyte-specific BMAL1 deletion on circadian clock components and protein O-GlcNAcylation in the heart. Hearts were collected from CBK and littermate CON mice at 3 h intervals over a 24 h period, followed by RT-PCR and Western Blot analyses. (A) mRNA levels of bmal1 (i), clock (ii), npas2 (iii), reverbα (iv), reverbβ (v), per1 (vi), per2 (vii), per3 (viii), cry1 (ix), and cry2 (x) in CON and CBK hearts (n = 5−6). (B) Protein levels of BMAL1 (i), REVERBα (ii), and PER2 (iii) in CON and CBK hearts (n = 5−8). (C) Protein O-GlcNAcylation levels in CON and CBK hearts at ZT9 (n = 3). In panels A and B, ZT0 and ZT24 are identical (the data are double plotted purely for the sake of presentation). Please see Supplemental files for all original images. Data are presented as mean ± SEM, and have been normalized to the lowest (trough) value in CON hearts. Main effects of time and genotype are reported at the top of the figure panels. *, p < 0.05 for CON vs. CBK hearts (at the same time-of-day).
Impact of inducible dnOGA overexpression on core circadian clock components in the heart
We, and others, have reported that several circadian clock components are O-GlcNAc modified, impacting their cellular localization, DNA binding, and/or protein stability ( 8, 9, 12, 13). Moreover, disruption of the cardiomyocyte circadian clock impacts protein O-GlcNAcylation levels (Figure 1C), consistent with an interrelationship between the clock and O-GlcNAcylation (13). We therefore hypothesized that cardiomyocyte dnOGA induction would impact the cardiac circadian clock and protein O-GlcNAcylation in a manner similar to that observed in CBK hearts. To test this hypothesis, hearts were isolated from dnOGAh and littermate CON mice at 4 h intervals over the 24 h day, 2 weeks after doxycycline treatment initiation; this duration of transgene induction is not associated with cardiac dysfunction (7). Following RNA isolation, RT-PCR was performed for the same 10 core circadian clock components investigated in CBK hearts. A 2-way ANOVA revealed significant time main effects for all 10 core clock component mRNAs (Figure 2A;Supplementary Table 3). Significant genotype main effects were observed for 7 out of 10 core clock component mRNAs (Figure 2A;Supplementary Table 3); clock, reverbα, and per1 mRNA did not exhibit significant genotype main effects (p-values of 0.054, 0.821, and 0.217, respectively). Of those transcripts that exhibited significant genotype main effects, reverbβ, per3, cry1, and cry2 were repressed, while bmal1, npas2, and per2 were induced (Figure 2A;Supplementary Table 3). Cosinor analysis indicated that all 10 core clock component mRNAs exhibit 24 h oscillations in CON hearts; these oscillations were significantly altered in dnOGAh hearts for only npas2 mRNA (Supplementary Table 4). At the protein level, 3 core circadian clock components were investigated; BMAL1, REVERBα, and PER2. A 2-way ANOVA revealed significant time main effects for all 3 core clock proteins, and significant genotype main effects for BMAL1 and PER2 (Figure 2B;Supplementary Table 3); REVERBα did not exhibit a significant genotype main effect (p = 0.916). Both BMAL1 and PER2 protein levels were increased in dnOGAh hearts (Figure 2B;Supplementary Table 3). Cosinor analysis indicated that all 3 core clock component proteins exhibit 24 h oscillations in CON hearts; PER2 oscillations were significantly augmented in dnOGAh hearts (Supplementary Table 4). Consistent with the cardiomyocyte-specific nature of the murine model, dnOGA mice exhibited 24 h oscillations in energy expenditure, food intake, and physical activity that were comparable to littermate control mice (Supplementary Figure 1B, as well asSupplementary Tables 3, 4). Similarly, 24 h oscillations in bmal1, reverbαand dbp mRNA levels were not different between CON and dnOGAh livers (Supplementary Figure 2). Next, we investigated protein O-GlcNAcylation levels in CON and dnOGA hearts isolated during the light phase (ZT8); protein O-GlcNAcylation levels were significantly increased in dnOGAh hearts (Figure 2C). Collectively, these data are consistent with the concept that increasing protein O-GlcNAcylation in the heart impacts the circadian clock, in a manner that is distinct from CBK mice.
Figure 2
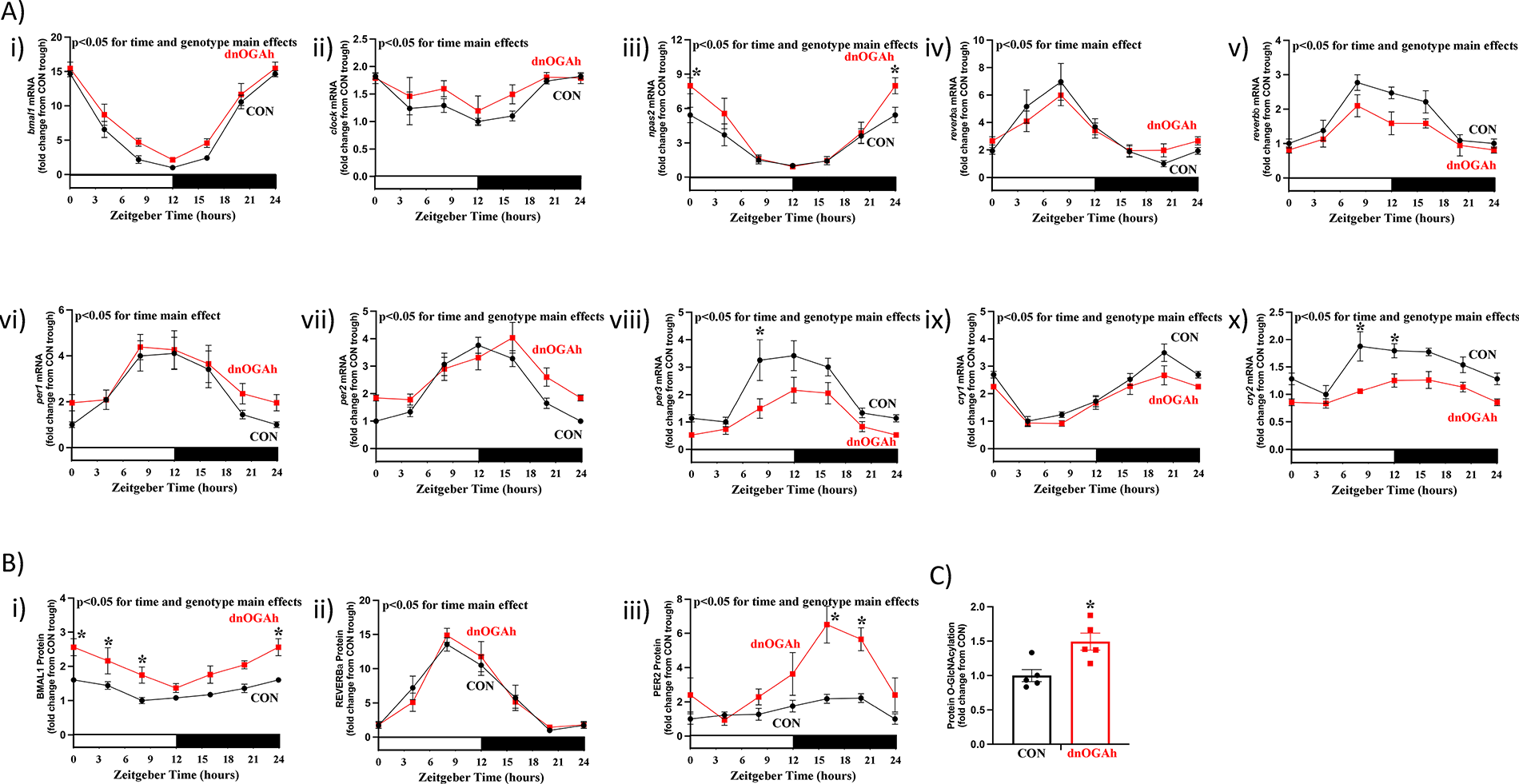
Impact of augmented protein O-GlcNAcylation on circadian clock components in the heart. Hearts were collected from dnOGAh and littermate CON mice at 4 h intervals over a 24 h period, 2 weeks after doxycycline treatment initiation, followed by RT-PCR and Western Blot analyses. (A) mRNA levels of bmal1 (i), clock (ii), npas2 (iii), reverbα (iv), reverbβ (v), per1 (vi), per2 (vii), per3 (viii), cry1 (ix), and cry2 (x) in CON and dnOGAh hearts (n = 5−9). (B) Protein levels of BMAL1 (i), REVERBα (ii), and PER2 (iii) in CON and dnOGAh hearts (n = 5). (C) Protein O-GlcNAcylation levels in CON and dnOGAh hearts at ZT8 (n = 5). In panels A and B, ZT0 and ZT24 are identical (the data are double plotted purely for the sake of presentation). Please see Supplemental files for all original images. Data are presented as mean ± SEM, and have been normalized to the lowest (trough) value in CON hearts. Main effects of time and genotype are reported at the top of the figure panels. *, p < 0.05 for CON vs. dnOGAh hearts (at the same time-of-day).
Attenuation of circadian clock output following dnOGA induction
Core circadian clock components directly regulate the expression of numerous genes that do not feedback on the clock mechanism, but instead impact cellular functions ( 10). These clock output genes include the PAS transcription factors, DBP and TEF, which are direct BMAL1/CLOCK targets; DNA binding activity of DBP and TEF is directly antagonized by the REV-ERBα/β target E4BP4 (23, 24). As such, dbp, tef, and e4bp4 mRNA levels were next assessed in both CBK and dnOGAh mouse models. A 2-way ANOVA revealed significant time and genotype main effects for all 3 clock output mRNAs in both CBK and dnOGAh hearts (Figures 3A,B, as well asSupplementary Tables 1, 3). Transcripts for dbp and tef were significantly decreased in CBK and dnOGAh hearts, while e4bp4 mRNA was significantly increased in hearts of both mouse models (Figures 3A,B, as well asSupplementary Tables 1, 3). Cosine analysis revealed that 24 h oscillations of these 3 output mRNAs were either abolished or significantly attenuated in CBK hearts (Supplementary Table 2). Similarly, 24 h oscillations were significantly attenuated for dbp mRNA in dnOGAh hearts (Supplementary Table 4). E4BP4 protein levels were next investigated in both mouse models. A 2-way ANOVA revealed significant time and genotype main effects for E4BP4 protein in both CBK and dnOGAh; E4BP4 protein was increased in both models (Figure 3C, as well asSupplementary Tables 1, 3). Although 24 h oscillations in E4BP4 protein were abolished in CBK hearts, these oscillations were not significantly altered in dnOGAh hearts (Supplementary Tables 2–4).
Figure 3
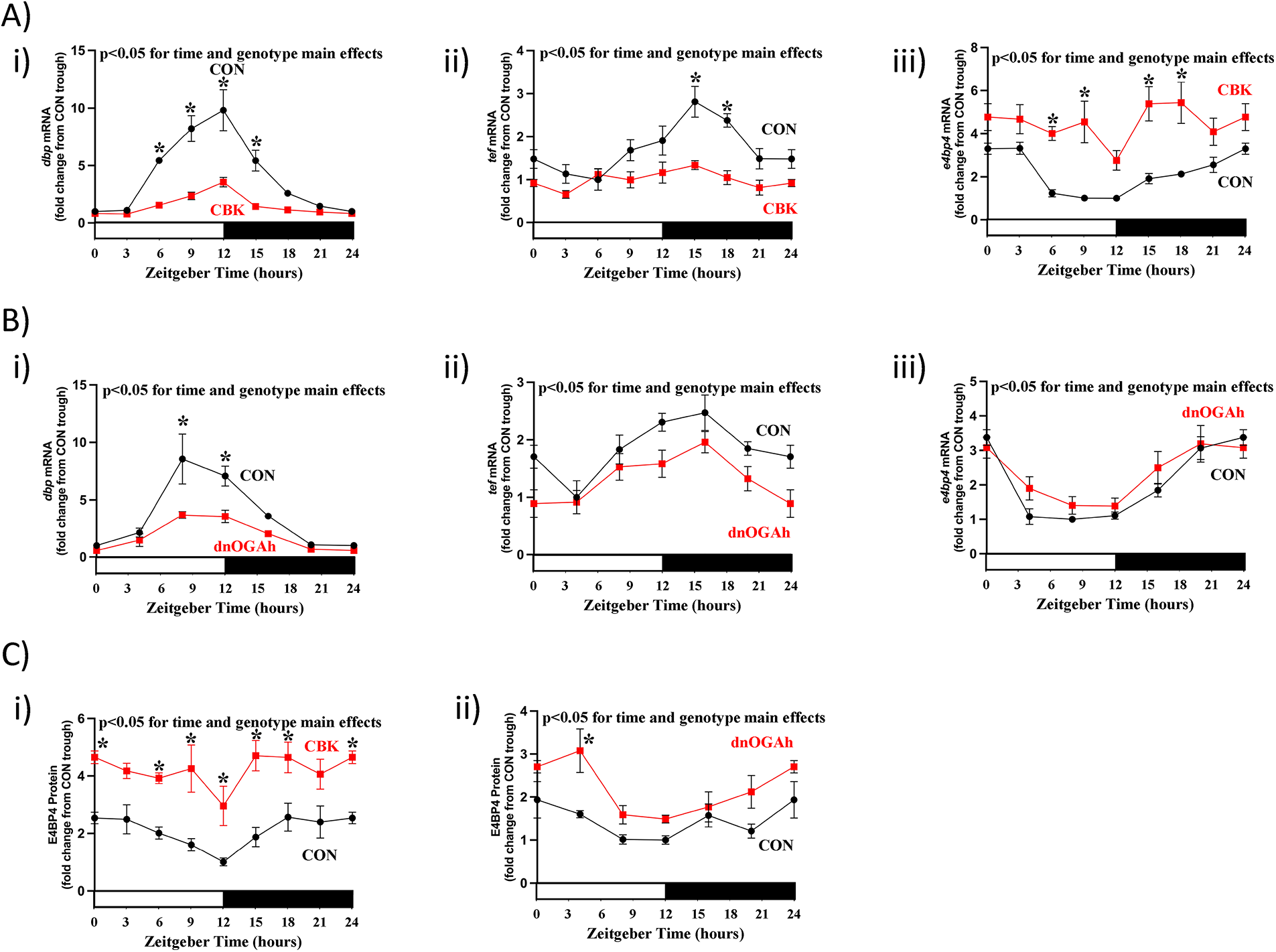
Diurnal variations in direct circadian clock-controlled genes/proteins in CBK and dnOGA hearts. Hearts were collected either from CBK and littermate CON mice at 3 h intervals over a 24 h period, or from dnOGAh and littermate CON mice at 4 h intervals over a 24 h period (2 weeks after doxycycline treatment initiation), followed by RT-PCR and Western Blot analyses. (A) mRNA levels of direct circadian clock-controlled transcription factors dbp (i), tef (ii), and e4bp4 (iii) in CON and CBK hearts (n = 5−6). (B) mRNA levels of direct circadian clock-controlled transcription factors dbp (i), tef (ii), and e4bp4 (iii) in CON and dnOGA hearts (n = 5−9). (C) E4BP4 protein levels in CBK (i) and dnOGAh (ii) hearts compared to littermate CON hearts (n = 5−6). In all panels, ZT0 and ZT24 are identical (the data are double plotted purely for the sake of presentation). Please see Supplemental files for all original images. Data are presented as mean ± SEM, and have been normalized to the lowest (trough) value in CON hearts. Main effects of time and genotype are reported at the top of the figure panels. *, p < 0.05 for CON vs. CBK/dnOGAh hearts (at the same time-of-day).
Given similarities between CBK and dnOGAh hearts, in terms of impact on established clock output genes (i.e., dbp, tef, e4bp4), we next investigated transcripts involved in metabolism (dgat2, nampt) and signaling (pik3r1, rhobtb1) that we have previously reported to be regulated by the cardiomyocyte circadian clock ( 14, 25). A 2-way ANOVA revealed significant time main effects for dgat2, pik3r1, and rhobtb1 (Figures 4A,B, as well asSupplementary Tables 1, 3). In addition, genotype main effects were found for all 4 clock-controlled mRNAs, in both CBK and dnOGAh hearts; these 4 transcripts were decreased in both models (Figures 4A,B, as well asSupplementary Tables 1, 3. Cosine analysis revealed significant 24 h oscillations of these 4 transcripts in CON hearts (except for CBK littermate CON hearts, for which 24 h oscillations did not achieve statistical significance;Supplementary Tables 2–4). 24 h oscillations in these transcripts were abolished in CBK and dnOGA hearts (except for pik3r1 in CBK hearts and rhobtb1 in dnOGAh hearts;Supplementary Table 2–4). Collectively, these data are consistent with attenuated circadian clock output in the heart following increased protein O-GlcNAcylation.
Figure 4
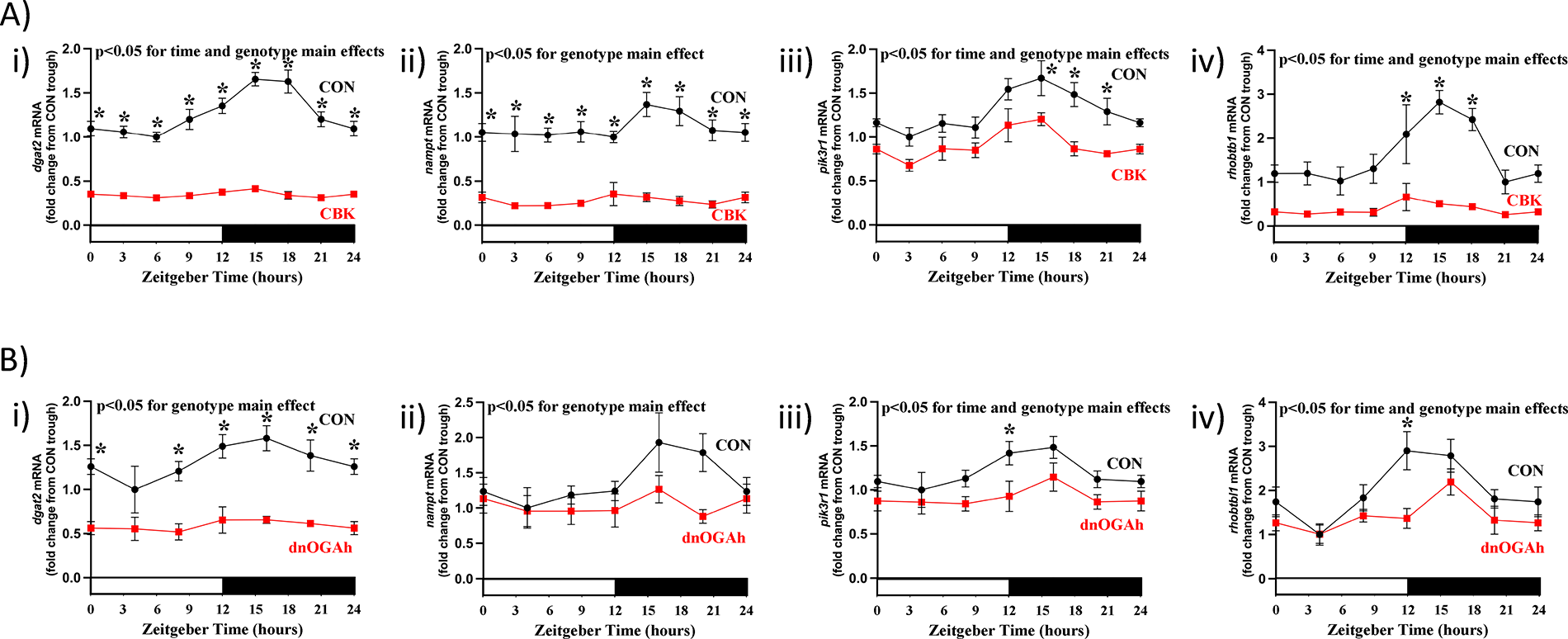
Diurnal variations in candidate clock-controlled genes in CBK and dnOGA hearts. Hearts were collected either from CBK and littermate CON mice at 3 h intervals over a 24 h period, or from dnOGAh and littermate CON mice at 4 h intervals over a 24 h period (2 weeks after doxycycline treatment initiation), followed by RT-PCR and Western Blot analyses. (A) mRNA levels of the clock-controlled genes dgat2 (i), nampt (ii), pik3r1 (iii), and rhobtb1 (iv) in CON and CBK hearts (n = 5−6). (B) mRNA levels of the clock-controlled genes dgat2 (i), nampt (ii), pik3r1 (iii), and rhobtb1 (iv) in CON and dnOGA hearts (n = 5−9). In all panels, ZT0 and ZT24 are identical (the data are double plotted purely for the sake of presentation). Data are presented as mean ± SEM, and have been normalized to the lowest (trough) value in CON hearts. Main effects of time and genotype are reported at the top of the figure panels. *, p < 0.05 for CON vs. CBK/dnOGAh hearts (at the same time-of-day).
The diurnal transcriptome of dnOGAh hearts is severely attenuated
Previous transcriptomic analyses indicate that the cardiomyocyte circadian clock orchestrates diurnal variations of approximately 5%–10% of the cardiac transcriptome ( 14, 26). Given that our candidate gene approach suggested that circadian clock output is attenuated in dnOGAh hearts (Figures 3B,4B), we next evaluated the diurnal transcriptome at a global level. More specifically, RNAseq was performed on RNA isolated from dnOGAh and littermate CON hearts collected at 4 h intervals over the 24 h day (2 weeks after doxycycline treatment initiation). JTK cycle identified 528 transcripts that significantly oscillate with a periodicity of 24 h in CON hearts; the number of oscillating transcripts was only 45 in dnOGA hearts (Supplementary Tables 5, 6, respectively). Further analysis revealed that 496 transcripts oscillate only in CON hearts, 13 transcripts oscillate only in dnOGA hearts, while 32 transcripts oscillate in both CON and dnOGA hearts (Supplementary Tables 7–9, respectively). Of the transcripts oscillating in both CON and dnOGA hearts, 5 had an amplitude that was significantly reduced in dnOGA hearts (Supplementary Table 9). As such, a total of 501 oscillating transcripts in CON hearts were either abolished or significantly attenuated in dnOGA hearts, representing 95% of all oscillating transcripts in CON hearts (Figure 5A). The impact of dnOGA induction on the diurnal transcriptome was further illustrated in a PCA plot; in addition to marked differential expression at the level of genotype, dnOGA hearts do not exhibit the same time-of-day-dependent pattern observed for CON hearts (Figure 5B). Comparing our previously published microarray analysis of CBK hearts (14) with the newly generated RNAseq analysis of dnOGAh hearts, we identified 125 transcripts exhibiting attenuated/abolished 24 h oscillations in both CBK and dnOGA hearts (Figure 5C;Supplementary Table 10). Pathway analysis of these 125 transcripts indicated notable enrichment in collagen and extracellular matrix (Figure 5D). Collectively, these data indicate that the diurnal transcriptome is markedly impaired in dnOGAh hearts.
Figure 5
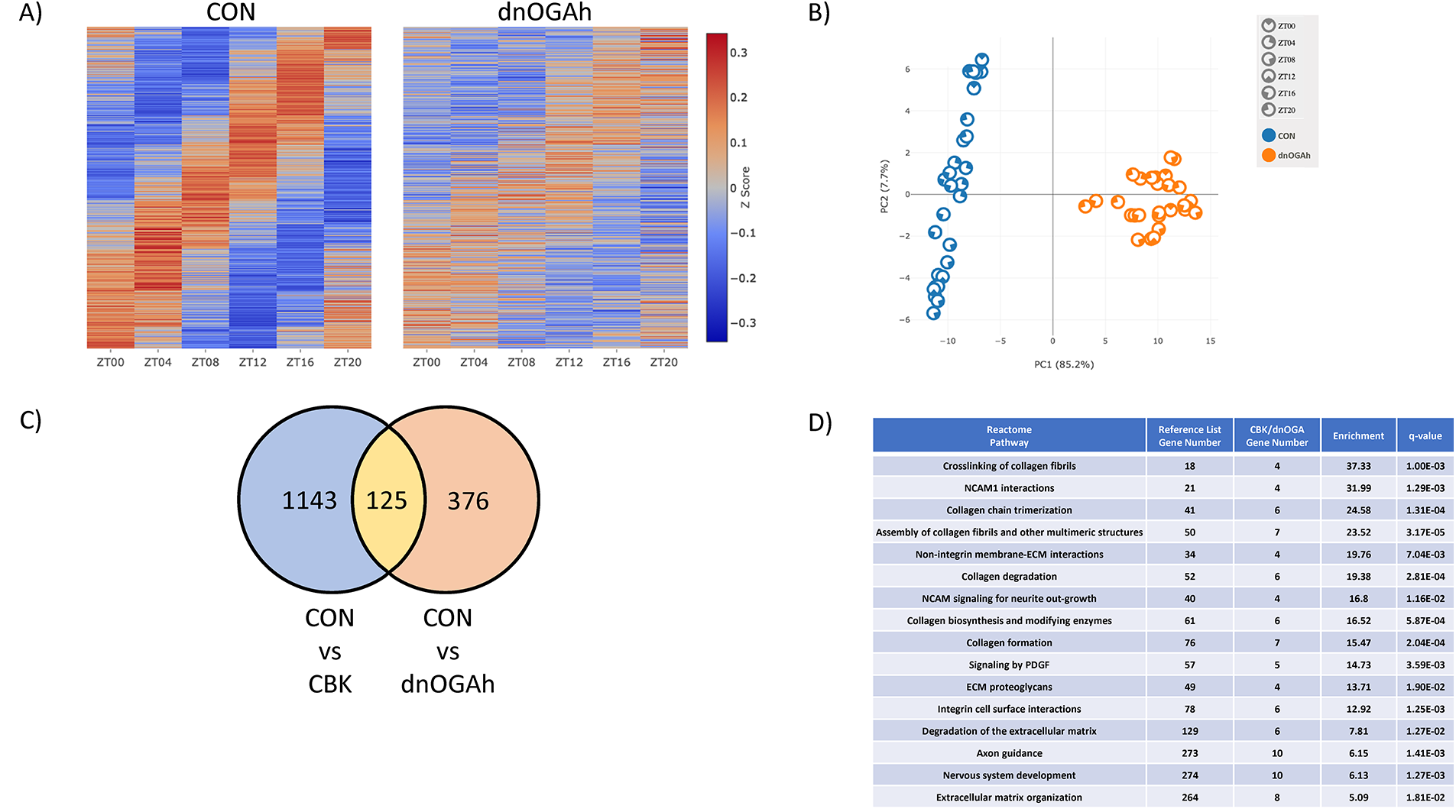
Impact of augmented protein O-GlcNAcylation on circadian governance of the cardiac transcriptome. Hearts were collected from dnOGAh and littermate CON mice at 4 h intervals over a 24 h period, 2 weeks after doxycycline treatment initiation, followed by RNAseq analysis (n = 5). (A) Heat map illustrating level of expression for genes exhibiting significant 24 h oscillations in CON hearts, that are either abolished or attenuated in dnOGAh hearts. (B) PCA plot for genes exhibiting significant 24 h oscillations in CON hearts, that are either abolished or attenuated in dnOGAh hearts. (C) Venn Diagram comparing differentially oscillating genes in CBK hearts vs. differentially oscillating genes in dnOGAh hearts. (D) Reactome pathway analysis results for oscillating genes that are commonly impacted in CBK and dnOGAh hearts.
Interstitial fibrosis following disruption of the cardiac diurnal transcriptome
As indicated in Figure 5D , multiple transcripts involved in the extracellular matrix exhibited attenuated/abolished 24hr oscillations in both CBK and dnOGAh hearts; these transcripts included several collagen isoforms ( Supplementary Table 10 ). RT-PCR was next performed to validate 24 h patterns of col1a1, col3a1, and col4a1 mRNA in CBK, dnOGAh, and littermate control hearts. A 2-way ANOVA revealed significant time main effects for col1a1, col3a1, and col4a1 mRNA ( Figure 6A ). In addition, genotype main effects were observed for all 3 collagen isoforms, in both CBK and dnOGAh hearts; these 3 transcripts were increased in both models ( Figures 6A,B ). Importantly, consistent with increased collagen isoform expression, interstitial fibrosis was augmented in both CBK and dnOGAh hearts ( Figure 6C ). These observations suggest that increased protein O-GlcNAcylation and attenuated circadian governance are associated with cardiac fibrosis.
Figure 6
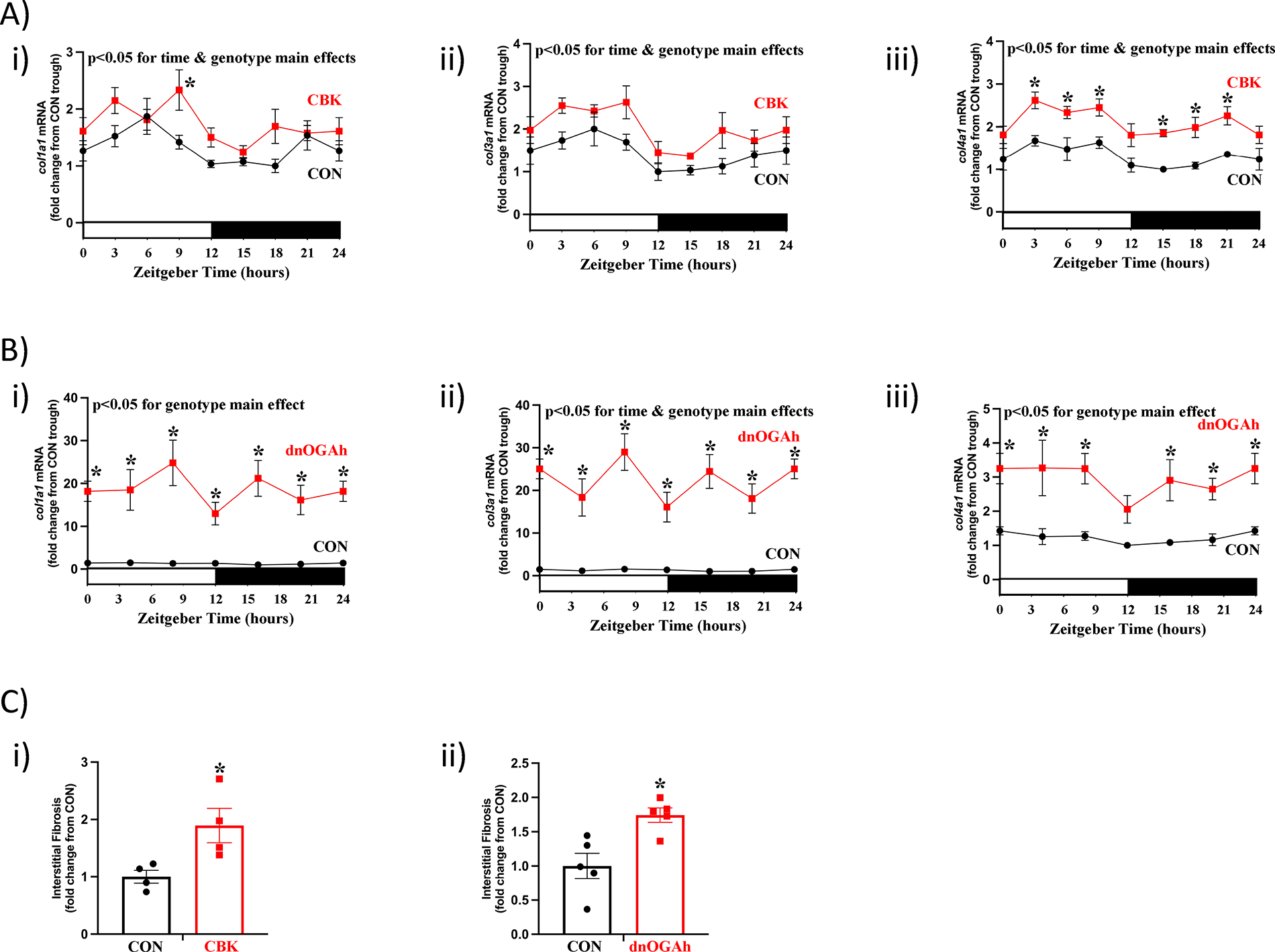
Increased interstitial fibrosis in CBK and dnOGA hearts. Hearts were collected either from CBK and littermate CON mice at 3 h intervals over a 24 h period, or from dnOGAh and littermate CON mice at 4 h intervals over a 24 h period (2 weeks after doxycycline treatment initiation), followed by RT-PCR and histologic analyses. (A) mRNA levels of collagen isoforms col1a1 (i), col3a1 (ii), and col4a1 (iii) in CON and CBK hearts (n = 5−6). (B) mRNA levels of collagen isoforms col1a1 (i), col3a1 (ii), and col4a1 (iii) in CON and dnOGA hearts (n = 5−9). (C) Interstitial fibrosis in CBK (i) and dnOGAh (ii) hearts compared to littermate CON hearts (n = 4−5). In panels A and B, ZT0 and ZT24 are identical (the data are double plotted purely for the sake of presentation). Please see Supplemental files for all original images. Data are presented as mean ± SEM, and have been normalized to the lowest (trough) value in CON hearts. Main effects of time and genotype are reported at the top of the figure panels. *, p < 0.05 for CON vs. CBK/dnOGAh hearts (at the same time-of-day).
Discussion
Aberrant protein O-GlcNAcylation is associated with cardiac pathology in both humans and animal models ( 2). Protein O-GlcNAcylation is elevated in the heart during diabetes mellitus, hypertrophy, and ischemia (6, 16, 27). Reducing this PTM within normal levels, through AAV-mediated OGA overexpression, normalized markers of adverse cardiac remodeling and diastolic dysfunction during diabetes (6). Similar observations have been reported in the setting of pressure overload; transgenic mice overexpressing OGA were resistant to transaortic constriction induced systolic dysfunction (16). Conversely, genetic augmentation of protein O-GlcNAcylation specifically in cardiomyocytes in vivo is sufficient to induce adverse cardiac remodeling and dysfunction, the severity of which is dependent on the manner by which this PTM is enhanced. More specifically, cardiac-specific OGT overexpression resulted in an approximate 6-fold increase in protein O-GlcNAcylation and heart failure (16), whereas dnOGAh mice exhibit more modest increase in O-GlcNAcylation (<2-fold) and diastolic dysfunction (without impact on systolic parameters) (7).
Despite appreciation that sustained augmentation of protein O-GlcNAcylation contributes towards the pathogenesis of cardiac disease, the mechanisms by which this occurs remain elusive. This likely reflects the large number of proteins that are O-GlcNAc modified, as well as technical limitations associated with both identification and manipulation of specific protein sites for this PTM. Interestingly, multiple similarities exist between aberrant protein O-GlcNAcylation and circadian clock dysfunction. The latter has been reported in multiple cardiac disease states (e.g., diabetes mellitus, pressure overload induced hypertrophy) ( 20, 28, 29), while genetic ablation of the cardiomyocyte circadian clock results in adverse cardiac remodeling and heart failure (14). Even in humans, perturbed circadian biology secondary to behaviors (e.g., shift work) and/or genetic polymorphisms in clock components, is associated with increased cardiovascular disease risk (30, 31). Much like protein O-GlcNAcylation, circadian clocks regulate a large number of cellular processes, making it difficult to define the precise mechanisms by which circadian disruption leads to cardiac disease.
Protein O-GlcNAcylation and circadian clocks are interconnected, a relationship that was first described in the heart. The cardiomyocyte circadian clock governs 24 h oscillations in cardiac glucose utilization, OGT expression, and protein O-GlcNAcylation levels, all of which peak during the middle of the active period ( 8). This includes augmented BMAL1 O-GlcNAcylation during the active period, which was later reported to enhance protein stability through inhibition of ubiquitination (8, 9). CLOCK and PER isoforms are also subjected to O-GlcNAcylation; this PTM increases CLOCK protein stability, and affects the subcellular localization of PER proteins (9, 12). Although REVERBαbinds with OGT, this circadian clock component does not appear to be O-GlcNAc modified. Instead, REVERBα affects OGT subcellular localization, thereby promoting O-GlcNAcylation of nuclear proteins during periods when REVERBα levels are elevated (32).
The majority of previously published studies investigating the impact of protein O-GlcNAcylation on circadian clock function have been acute in nature (i.e., <24 h), in vitro, and/or in extra-cardiac tissues. Moreover, the impact of sustained augmentation of protein O-GlcNAcylation on circadian governance of cellular processes is lacking. To gain greater insight, the purpose of the present study was to determine whether a modest elevation of protein O-GlcNAcylation in the adult heart for a prolonged period of time (2 weeks) influenced either the cardiac circadian clock or circadian governance of the cardiac transcriptome. Here, we report that an approximate 1.5-fold increase in protein O-GlcNAcylation in dnOGAh hearts impacted mRNA levels of 7 out of 10 circadian clock components ( Figure 2 ). Consistent with aforementioned previously published studies reporting the impact of O-GlcNAcylation on stability of clock component proteins, BMAL1 and PER2 protein levels were augmented in dnOGAh hearts (to an extent that was greater than changes in mRNA levels), while REVERBα protein levels were unaffected (Figure 2B; Supplementary Table 3). Importantly, circadian governance was severely attenuated, at both the candidate gene level (Figures 3B,4B) and whole transcriptome level (Figures 5A,B). In the latter case, 95% of oscillating transcripts in control hearts were either abolished or significantly attenuated in dnOGAh hearts. Such observations not only underscore the importance of protein O-GlcNAcylation in the operation of circadian clock mechanism, but also highlight the scale by which this PTM affects clock output and temporal control of the transcriptome. With regards to potential functional consequences of aberrant circadian governance, comparison of transcriptomic data from dnOGAh and CBK hearts indicated that the extracellular matrix was commonly impacted in both models (Figure 5D), which was confirmed through RT-PCR and histology (Figure 6).
The present study has led to a number of subsequent questions, including those related to identification of the mechanisms by which circadian governance over the cardiac transcriptome is so dramatically abolished by sustained protein O-GlcNAcylation. Of the 10 circadian clock components investigated at the mRNA level, only clock was non-rhythmic in dnOGA hearts ( Supplementary Table 4 ). At the protein level, 24 h oscillations in circadian clock components were either unaffected (i.e., REVERBα) or enhanced (BMAL1 and PER2; p = 0.11 and p < 0.05, respectively) in dnOGAh hearts (Figure 2B). This is in striking contrast to CBK hearts, for which 24 h oscillations in clock components were largely abolished/attenuated (Figures 1A,B). These observations suggest a potential uncoupling of the core clock mechanism from circadian governance of the transcriptome following a 2-week elevation of protein O-GlcNAcylation. Several possible explanations exist, including influences on direct clock output genes that serve as mechanistic links from the clock to cellular processes. The PAR transcription factors DBP, HLF, and TEF are excellent examples. At the mRNA, dbp and tef were reduced in dnOGAh hearts, while an antagonist to these transcription factors, E4BP4, was increased at both mRNA and protein levels (Figures 3B,C;Supplementary Table 3). Interestingly, E4BP4 protein levels were augmented in dnOGAh hearts to a greater extent compared with e4bp4 mRNA, suggesting potential posttranscriptional regulation (Figures 3B,C;Supplementary Table 3). Recent studies indicate that E4BP4 links the cardiomyocyte circadian clock with cardiac metabolism and electrophysiology (33). Moreover, chronic elevations in E4BP4 levels in CBK hearts play a causal role in adverse remodeling and cardiac dysfunction. More specifically, pharmacologic and genetic attenuation of E4BP4 in CBK hearts reduces cardiac fibrosis and heart failure development (33, 34). Whether elevated E4BP4 levels in dnOGAh hearts (Figure 3C) promotes fibrosis (Figure 6) and previously reported contractile dysfunction (7) requires further investigation.
It is noteworthy that we and others have previously reported that approximately 10% of the cardiac transcriptome oscillates with a periodicity of 24 h in wild-type control hearts ( 35). However, the number of oscillating transcripts identified for CON hearts in the current dnOGAh RNAseq analysis was considerably lower. This is potentially secondary to two experimental factors. First, dnOGAh and littermate CON mice were both treated with doxycycline (initially injection, followed by ingestion) for 2 weeks. Doxycycline has established effects on the microbiome, as well as biological processes in the host (including metabolism) (36, 37). Interestingly, previously published studies have reported that antibiotic-mediated perturbations in the microbiome impacts both the heart and circadian clocks (38). Second, a higher time resolution was utilized for our previously published microarray studies (3 h intervals; 8 time points) compared to the current RNAseq analysis in dnOGA and littermate CON hearts (4 h intervals; 6 time points) (14). An additional limitation of the bulk RNA approach employed by our microarray and RNAseq analyses is that they fail to consider the heterogeneity of the heart. The possibility exists that O-GlcNAcylation phase shifts the cardiomyocyte clock in dnOGAh hearts, whereas the clock remains relatively intact in non-cardiomyocytes. Such an intercellular dyssynchrony would be perceived as attenuated/abolished 24 h oscillations when RNA is pooled from all cell types in the heart (i.e., bulk methods). Finally, the current study investigated diurnal variations, as opposed to circadian rhythms. Future studies should be performed under circadian conditions (e.g., constant darkness), preventing any potential masking effects of light-dark cycles.
In summary, the present study has revealed that a modest increase in protein O-GlcNAcylation for a 2-week period in adult hearts is sufficient to severely attenuate circadian governance of the cardiac transcriptome, in a manner that appears to be distinct from circadian clock disruption. This abolishment of circadian governance in the heart was associated with increased interstitial fibrosis. Future studies are required to define the precise mechanisms by which protein O-GlcNAcylation impairs circadian governance, and whether this plays a role in the etiology of cardiac disease during diabetes mellitus, pressure overload, and/or ischemia.
Statements
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE301657.
Ethics statement
The animal study was approved by Animal Care and Use Committee of the University of Alabama at Birmingham. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
GS: Methodology, Investigation, Data curation, Writing – review & editing. SC: Investigation, Writing – review & editing. HC: Investigation, Writing – review & editing. CH: Writing – review & editing, Investigation. MK: Writing – review & editing, Investigation. LP: Investigation, Writing – review & editing. LX: Writing – review & editing, Investigation. LZ: Writing – review & editing, Investigation. JZ: Writing – review & editing, Conceptualization, Funding acquisition. JC: Conceptualization, Funding acquisition, Writing – review & editing. AW: Writing – review & editing, Conceptualization. MY: Conceptualization, Investigation, Validation, Funding acquisition, Supervision, Project administration, Methodology, Formal analysis, Data curation, Resources, Writing – review & editing, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Institutes of Health (HL142216, GM008361, HD071866, HL172687, HL133011, HL152354).
Acknowledgments
We would like to thank Maximiliano Grenett for technical assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2025.1601407/full#supplementary-material
Supplementary Figure 1Diurnal variations in whole-body energy balance in CBK (A) and dnOGA (B) mice. CBK, dnOGA, and littermate CON mice were placed in CLAMS cages, followed by continuous non-invasive assessment of energy expenditure, food intake, and physical activity. In all panels, ZT0 and ZT24 are identical (the data are double plotted purely for the sake of presentation). Data are presented as mean ± SEM, for 5–7 independent observations. Main effects of time are reported at the top of the figure panels.
Supplementary Figure 2Diurnal variations in hepatic gene expression in dnOGA mice. Livers were collected from dnOGAh and littermate CON mice at 4hr intervals over a 24 h period, 2 weeks after doxycycline treatment initiation, followed by RT-PCR analyses. mRNA levels of bmal1 (i), reverba (ii), and dbp (iii) in CON and dnOGAh livers (n = 5–9). ZT0 and ZT24 are identical (the data are double plotted purely for the sake of presentation). Data are presented as mean ± SEM, and have been normalized to the lowest (trough) value in CON hearts. Main effects of time are reported at the top of the figure panels.
References
1.
Lee JM Hammarén HM Savitski MM Baek SH . Control of protein stability by post-translational modifications. Nat Commun. (2023) 14:201. 10.1038/s41467-023-35795-8
2.
Chatham JC Zhang J Wende AR . Role of O-linked N-acetylglucosamine protein modification in cellular (patho)Physiology. Physiol Rev. (2021) 101:427–93. 10.1152/physrev.00043.2019
3.
Zachara N Akimoto Y Boyce M Hart G . The O-GlcNAc modification. In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, et al., editors.Essentials of Glycobiology. 4th ed. Chapter 19. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press (2022). 10.1101/glycobiology.4e
4.
Collins HE Chatham JC . Regulation of cardiac O-GlcNAcylation: more than just nutrient availability. Biochim Biophys Acta Mol Basis Dis. (2020) 1866:165712. 10.1016/j.bbadis.2020.165712
5.
Belke DD . Swim-exercised mice show a decreased level of protein O-GlcNAcylation and expression of O-GlcNAc transferase in heart. J Appl Physiol. (2011) 111:157–62. 10.1152/japplphysiol.00147.2011
6.
Prakoso D Lim SY Erickson JR Wallace RS Lees JG Tate M et al Fine-tuning the cardiac O-GlcNAcylation regulatory enzymes governs the functional and structural phenotype of the diabetic heart. Cardiovasc Res. (2022) 118:212–25. 10.1093/cvr/cvab043
7.
Ha CM Bakshi S Brahma MK Potter LA Chang SF Sun Z et al Sustained increases in cardiomyocyte protein O-linked N-acetylglucosamine levels lead to cardiac hypertrophy and reduced mitochondrial function without systolic contractile impairment. J Am Heart Assoc. (2023) 12:e029898. 10.1161/JAHA.123.029898
8.
Durgan DJ Pat BM Laczy B Bradley JA Tsai JY Grenett MH et al O-GlcNAcylation, novel post-translational modification linking myocardial metabolism and cardiomyocyte circadian clock. J Biol Chem. (2011) 286:44606–19. 10.1074/jbc.M111.278903
9.
Li MD Ruan HB Hughes ME Lee JS Singh JP Jones SP et al O-GlcNAc signaling entrains the circadian clock by inhibiting BMAL1/CLOCK ubiquitination. Cell Metab. (2013) 17:303–10. 10.1016/j.cmet.2012.12.015
10.
Takahashi JS Hong HK Ko CH McDearmon EL . The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. (2008) 9:764–75. 10.1038/nrg2430
11.
Hirano A Fu Y-H Ptáček LJ . The intricate dance of post-translational modifications in the rhythm of life. Nat Struct Mol Biol. (2016) 23:1053–60. 10.1038/nsmb.3326
12.
Li YH Liu X Vanselow JT Zheng H Schlosser A Chiu JC . O-GlcNAcylation of PERIOD regulates its interaction with CLOCK and timing of circadian transcriptional repression. PLoS Genet. (2019) 15:e1007953. 10.1371/journal.pgen.1007953
13.
Fu M Yang X . The sweet tooth of the circadian clock. Biochem Soc Trans. (2017) 45:871–84. 10.1042/bst20160183
14.
Young ME Brewer RA Peliciari-Garcia RA Collins HE He L Birky TL et al Cardiomyocyte-specific BMAL1 plays critical roles in metabolism, signaling, and maintenance of contractile function of the heart. J Biol Rhythms. (2014) 29:257–76. 10.1177/0748730414543141
15.
Ingle KA Kain V Goel M Prabhu SD Young ME Halade GV . Cardiomyocyte-specific Bmal1 deletion in mice triggers diastolic dysfunction, extracellular matrix response, and impaired resolution of inflammation. Am J Physiol Heart Circ Physiol. (2015) 309:H1827–36. 10.1152/ajpheart.00608.2015
16.
Umapathi P Mesubi OO Banerjee PS Abrol N Wang Q Luczak ED et al Excessive O-GlcNAcylation causes heart failure and sudden death. Circulation. (2021) 143:1687–703. 10.1161/circulationaha.120.051911
17.
Bray MS Ratcliffe WF Grenett MH Brewer RA Gamble KL Young ME . Quantitative analysis of light-phase restricted feeding reveals metabolic dyssynchrony in mice. Int J Obes. (2013) 37:843–52. 10.1038/ijo.2012.137
18.
Gibson UE Heid CA Williams PM . A novel method for real time quantitative RT-PCR. Genome Res. (1996) 6:995–1001. 10.1101/gr.6.10.995
19.
Heid CA Stevens J Livak KJ Williams PM . Real time quantitative PCR. Genome Res. (1996) 6:986–94. 10.1101/gr.6.10.986
20.
Young M Razeghi P Taegtmeyer H . Clock genes in the heart: characterization and attenuation with hypertrophy. Circ Res. (2001) 88:1142–50. 10.1161/hh1101.091190
21.
Durgan DJ Tsai JY Grenett MH Pat BM Ratcliffe WF Villegas-Montoya C et al Evidence suggesting that the cardiomyocyte circadian clock modulates responsiveness of the heart to hypertrophic stimuli in mice. Chronobiol Int. (2011) 28:187–203. 10.3109/07420528.2010.550406
22.
Peliciari-Garcia RA Bargi-Souza P Young ME Nunes MT . Repercussions of hypo and hyperthyroidism on the heart circadian clock. Chronobiol Int. (2018) 35:147–59. 10.1080/07420528.2017.1388253
23.
Gachon F Fonjallaz P Damiola F Gos P Kodama T Zakany J et al The loss of circadian PAR bZip transcription factors results in epilepsy. Genes Dev. (2004) 18:1397–412. 10.1101/gad.301404
24.
Mitsui S Yamaguchi S Matsuo T Ishida Y Okamura H . Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev. (2001) 15:995–1006. 10.1101/gad.873501
25.
Latimer MN Williams LJ Shanmugan G Carpenter BJ Lazar MA Dierickx P et al Cardiomyocyte-specific disruption of the circadian BMAL1–REV-ERBa/b regulatory network impacts distinct miRNA species in the murine heart. Comm Biol. (2023) 6(1):1149. 10.1038/s42003-023-05537-z
26.
Bray M Shaw C Moore M Garcia R Zanquetta M Durgan D et al Disruption of the circadian clock within the cardiomyocyte influences myocardial contractile function; metabolism; and gene expression. Am J Physiol Heart Circ Physiol. (2008) 294:H1036–47. 10.1152/ajpheart.01291.2007
27.
Fülöp N Zhang Z Marchase RB Chatham JC . Glucosamine cardioprotection in perfused rat hearts associated with increased O-linked N-acetylglucosamine protein modification and altered p38 activation. Am J Physiol Heart Circ Physiol. (2007) 292:H2227–36. 10.1152/ajpheart.01091.2006
28.
Young M Wilson C Razeghi P Guthrie P Taegtmeyer H . Alterations of the circadian clock in the heart by streptozotocin-induced diabetes. J Mol Cell Cardiol. (2002) 34:223–31. 10.1006/jmcc.2001.1504
29.
Mohri T Emoto N Nonaka H Fukuya H Yagita K Okamura H et al Alterations of circadian expressions of clock genes in dahl salt-sensitive rats fed a high-salt diet. Hypertension. (2003) 42:189–94. 10.1161/01.HYP.0000082766.63952.49
30.
Woon PY Kaisaki PJ Braganca J Bihoreau MT Levy JC Farrall M et al Aryl hydrocarbon receptor nuclear translocator-like (BMAL1) is associated with susceptibility to hypertension and type 2 diabetes. Proc Natl Acad Sci U S A. (2007) 104:14412–7. 10.1073/pnas.0703247104
31.
Smolensky MH Hermida RC Reinberg A Sackett-Lundeen L Portaluppi F . Circadian disruption: new clinical perspective of disease pathology and basis for chronotherapeutic intervention. Chronobiol Int. (2016) 33:1101–19. 10.1080/07420528.2016.1184678
32.
Berthier A Vinod M Porez G Steenackers A Alexandre J Yamakawa N et al Combinatorial regulation of hepatic cytoplasmic signaling and nuclear transcriptional events by the OGT/REV-ERBalpha complex. Proc Natl Acad Sci U S A. (2018) 115:E11033–42. 10.1073/pnas.1805397115
33.
Mia S Sonkar R Williams L Latimer M Rawnsley D Rana S et al Novel roles for the transcriptional repressor E4BP4 in both cardiac physiology and pathophysiology. J Am Coll Cardiol Basic Trans Sci. (2023) 8(9):1141–56. 10.1016/j.jacbts.2023.03.016
34.
Mia S Kane MS Latimer MN Reitz CJ Sonkar R Benavides GA et al Differential effects of REV-ERBalpha/beta agonism on cardiac gene expression, metabolism, and contractile function in a mouse model of circadian disruption. Am J Physiol Heart Circ Physiol. (2020) 318:H1487–508. 10.1152/ajpheart.00709.2019
35.
Martino TA Young ME . Influence of the cardiomyocyte circadian clock on cardiac physiology and pathophysiology. J Biol Rhythms. (2015) 30:183–205. 10.1177/0748730415575246
36.
Moullan N Mouchiroud L Wang X Ryu D Williams Evan G Mottis A et al Tetracyclines disturb mitochondrial function across eukaryotic models: a call for caution in biomedical research. Cell Rep. (2015) 10:1681–91. 10.1016/j.celrep.2015.02.034
37.
Cho I Yamanishi S Cox L Methé BA Zavadil J Li K et al Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. (2012) 488:621–6. 10.1038/nature11400
38.
Mistry P Reitz CJ Khatua TN Rasouli M Oliphant K Young ME et al Circadian influence on the microbiome improves heart failure outcomes. J Mol Cell Cardiol. (2020) 149:54–72. 10.1016/j.yjmcc.2020.09.006
Summary
Keywords
chronobiology, fibrosis, gene expression, post-translational modification, glycobiology
Citation
Shanmugam G, Chang SF, Collins HE, Ha C-M, Kane MS, Potter L, Xie L, Zou L, Zhang J, Chatham JC, Wende AR and Young ME (2025) Severe attenuation of circadian clock output in the heart following sustained augmentation of cardiomyocyte protein O-GlcNAcylation. Front. Cardiovasc. Med. 12:1601407. doi: 10.3389/fcvm.2025.1601407
Received
17 April 2025
Accepted
16 June 2025
Published
17 July 2025
Volume
12 - 2025
Edited by
Thomas Pulinilkunnil, Dalhousie University, Canada
Reviewed by
Beshay Zordoky, University of Minnesota Twin Cities, United States
Inna Rabinovich-Nikitin, University of Manitoba, Canada
Noemi Polgar, University of Hawaii at Manoa, United States
Updates
Copyright
© 2025 Shanmugam, Chang, Collins, Ha, Kane, Potter, Xie, Zou, Zhang, Chatham, Wende and Young.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
* Correspondence: Martin E. Young meyoung@uab.edu
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.