- 1Center for Clinical Epidemiology and Population Health, Marshfield Clinic Research Institute, Marshfield, WI, United States
- 2Cancer Care and Research Center, Marshfield Clinic Research Institute, Marshfield, WI, United States
- 3Security Health Plan, Marshfield Clinic Health System, Marshfield, WI, United States
- 4Berkeley Bioinformatics Open-source Projects, Lawrence Berkeley National Laboratory, Berkeley, CA, United States
- 5Department of Oral Biology, School of Dental Medicine, University at Buffalo, Buffalo, NY, United States
- 6Advocate Aurora Research Institute, Advocate Aurora Health, Chicago, IL, United States
Background: The objective of this study was to build models that define variables contributing to pneumonia risk by applying supervised Machine Learning (ML) to medical and oral disease data to define key risk variables contributing to pneumonia emergence for any pneumonia/pneumonia subtypes.
Methods: Retrospective medical and dental data were retrieved from the Marshfield Clinic Health System's data warehouse and the integrated electronic medical-dental health records (iEHR). Retrieved data were preprocessed prior to conducting analyses and included matching of cases to controls by (a) race/ethnicity and (b) 1:1 Case: Control ratio. Variables with >30% missing data were excluded from analysis. Datasets were divided into four subsets: (1) All Pneumonia (all cases and controls); (2) community (CAP)/healthcare-associated (HCAP) pneumonias; (3) ventilator-associated (VAP)/hospital-acquired (HAP) pneumonias; and (4) aspiration pneumonia (AP). Performance of five algorithms was compared across the four subsets: Naïve Bayes, Logistic Regression, Support Vector Machine (SVM), Multi Layer Perceptron (MLP), and Random Forests. Feature (input variables) selection and 10-fold cross validation was performed on all the datasets. An evaluation set (10%) was extracted from the subsets for further validation. Model performance was evaluated in terms of total accuracy, sensitivity, specificity, F-measure, Mathews-correlation-coefficient, and area under receiver operating characteristic curve (AUC).
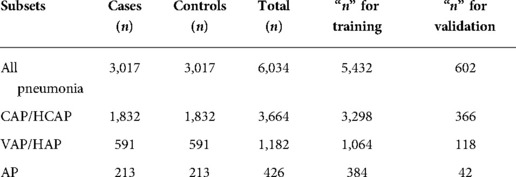
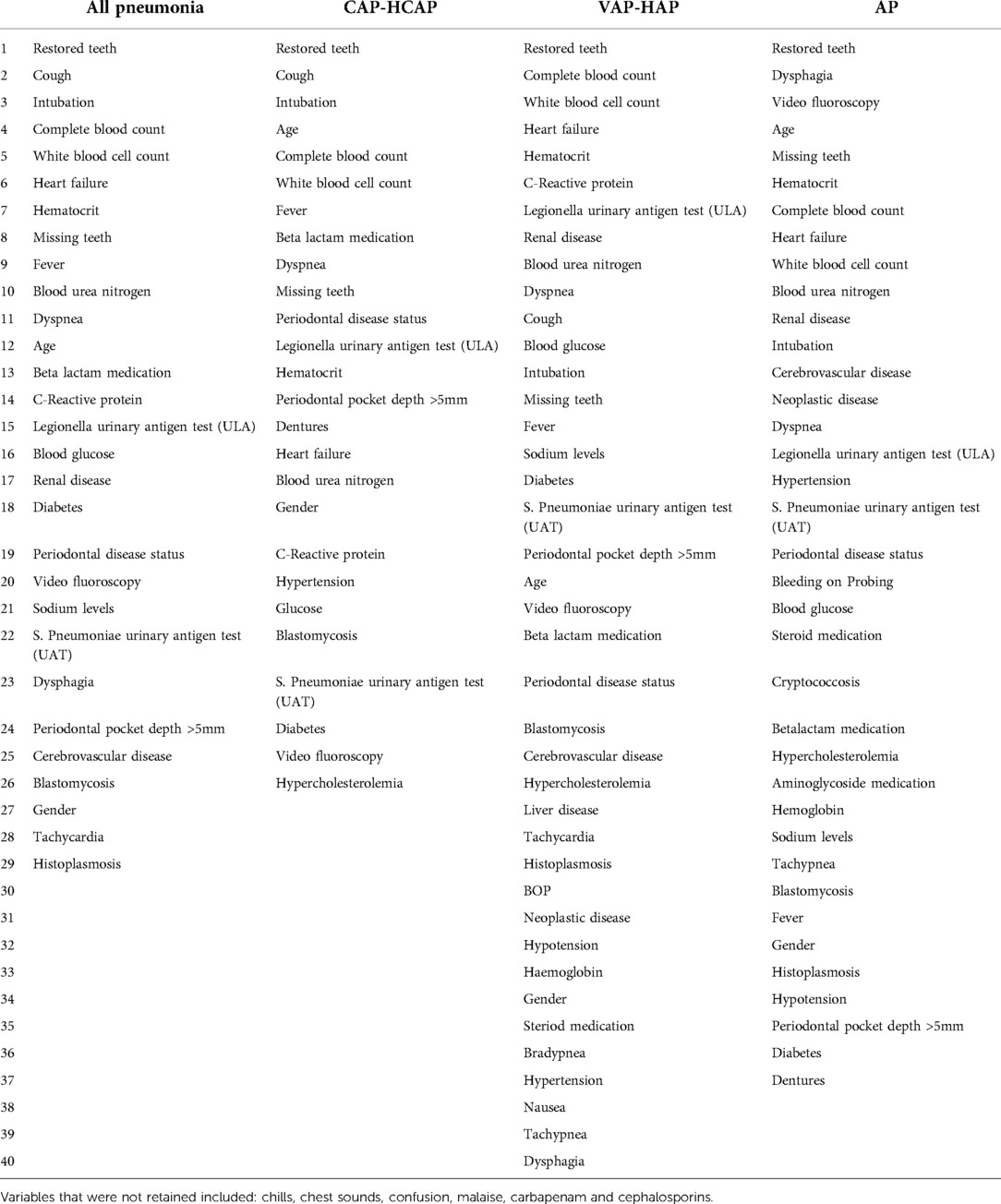
Results: In total, 6,034 records (cases and controls) met eligibility for inclusion in the main dataset. After feature selection, the variables retained in the subsets were: All Pneumonia (n = 29 variables), CAP-HCAP (n = 26 variables), VAP-HAP (n = 40 variables), and AP (n = 37 variables). Variables retained (n = 22) were common across all four pneumonia subsets. Of these, the number of missing teeth, periodontal status, periodontal pocket depth more than 5 mm, and number of restored teeth contributed to all the subsets and were retained in the model. MLP outperformed other predictive models for All Pneumonia, CAP-HCAP, and AP subsets, while SVM outperformed other models in VAP-HAP subset.
Conclusion: This study validates previously described associations between poor oral health and pneumonia. Benefits of an integrated medical-dental record and care delivery environment for modeling pneumonia risk are highlighted. Based on findings, risk score development could inform referrals and follow-up in integrated healthcare delivery environments and coordinated patient management.
Introduction
Pneumonia continues to represent a significant medical condition associated with substantially increased morbidity, mortality, and healthcare cost, especially with advancing age. The American Lung Association defines pneumonia as a common infection of the lung caused by bacteria, fungi, and/or viruses (1). Treatment and management vary depending on the cause of pneumonia and symptom severity. In 2017, the National Hospital Ambulatory Medical Care Survey reported approximately 1.3 million visits to emergency departments with a primary diagnosis of pneumonia (2) and the CDC reported 49,157 deaths due to pneumonia in the same year (3). Pneumonia has five subtypes: aspiration pneumonia (AP), community-acquired pneumonia (CAP), hospital-acquired pneumonia (HAP), health care-acquired pneumonia (HCAP), and ventilator-associated pneumonia (VAP) (4, 5). In the timespan between 2010 and 2014, Corrado et al. reported that CAP is the most frequent subtype (54.3%) while VAP (1.6%) is the least frequent subtype (6).
The current evidence base supports poor oral health as a risk factor for VAP and AP. In contrast with the now well-studied association between oral health and HAP, CAP remains under-explored by population-level studies (7). Notably, a 2017 systematic review of risk factors for adult CAP identified poor oral/dental health including periodontal disease (PD) as potential risk factors (8, 9). However, a large population-based study did not find PD to be a risk factor but did find dental caries and tooth loss to be associated with pneumonia (7). Notably, difficulty in defining the causal organism(s) in over 60% of pneumonia cases may be partially attributable to pathogenic emergence of normal oral flora (10), including anaerobes (11), consequential to environmental perturbation, indicating that some pneumonia may originate from oral dysbiosis (11). Viral infection may also represent a potential cause of infection unrelated to oral health status (12, 13). Moreover, poor oral health leads to a more anaerobic environment, which may contribute organisms that colonize both lungs and the oral cavity (14). Growing evidence supports the potential role of oral flora in the etiology of pneumonia (15–18). Comparison of pulmonary microbiota of patients admitted to the ICU with CAP, VAP, and other HAP, in the context of VAP (15), demonstrates clear overlaps across both microbiomes. A systematic review supported a 40% reduction in HAP incidence following improvement of oral hygiene in the hospital setting (19). By contrast, a clinical trial implementing improved oral hygiene in a nursing home setting was terminated early due to futility (20), so further study is warranted.
Currently, trends to transform care delivery across the siloed medical-dental domains include development of integrated patient-centric care delivery models. This has been supported by application of Artificial Intelligence (AI) in healthcare to develop translational e-Health approaches to facilitate implementation of precision care delivery (21–23). For example, machine learning (ML), a subdomain of AI, involves development of algorithms and makes decisions or predictions relative to future data based on iterative modeling of historic patient data (21). Algorithms developed by these models can be translated at point of care in the form of clinical decision support tools or risk prediction models (24–26).
Secondary use of electronically collected medical and dental data for elucidating associations between oral-systemic health conditions may expand insights into potential risk factors that contribute to pneumonia emergence in the context of the various pneumonia subtypes. Such characterization will contribute to development of eHealth approaches in emerging integrated medical and dental care delivery models. Because poor oral health is a modifiable risk factor, targeting oral disease prevention and treatment in the general population and high-risk subpopulations could help reduce pneumonia risk (27, 28). The objective of this study was to build models that define variables contributing to pneumonia risk by applying supervised Machine Learning (ML) to medical and oral disease data to define key risk variables contributing to pneumonia emergence for any pneumonia/pneumonia subtypes.
Methods
Study setting
This study was conducted at Marshfield Clinic Health System (MCHS) (28), a large multispecialty healthcare practice with an expansive service area spanning largely rural tracts of central, western, and northern Wisconsin. MCHS has partnered with the Family Health Center of Marshfield (FHC-M), whose service area largely overlaps that of MCHS (30). Medical and oral healthcare are delivered across the service area via a network of over 50 clinics and hospitals supported by an integrated medical-dental electronic health record (iEHR) that captures healthcare encounter data in real time. Data are backed up daily in the MCHS enterprise data warehouse (EDW), making this repository and iEHR among the largest combined medical and dental record systems in the country.
This study was reviewed and approved by the Institutional Review Board (IRB) of Marshfield Clinic Research Institute.
Definition of the subject eligibility criteria
From a cohort of patients with medical and dental data, the following inclusion/exclusion criteria were applied:
a. Patients more than 21 years of age.
b. With at least one oral examination at the FHC dental center between 2007 and 2019.
c. With at least two ambulatory visits within 3 years of their latest medical visit.
d. Recurrent pneumonia episodes were excluded. Pneumonia recurrence was identified by documenting (additional ICD9/10 CM pneumonia associated codes) less than 90 days of the index pneumonia diagnosis.
Eligible patients (n = 6,034) were assigned to one of the two cohorts, based on their alignment with the following inclusion criteria:
Cases were defined as patients who had documented evidence of ICD9 CM (480.0–497.0) or ICD10 CM (J12-J18.9) and a pneumonia encounter between 01/01/2007 and 12/30/2019 as previously defined (31):
a. Rule of one: pneumonia encounter documented by ICD9/10CM codes and associated antibiotics prescription and/or a chest radiograph collected within ±30 days of the index pneumonia encounter.
b. Rule of two: two pneumonia encounters in patients documented by ICD9/10 CM codes during a pneumonia episode.
There was no overlap between “a” and “b”. Patients who did not meet the above definitions were excluded from further analyses.
Controls: Patients with no ICD9/10 CM code for pneumonia were included in the dataset.
Data retrieval
To achieve the objective of the study, retrospective data from 2007 to 2019 were extracted from the MCHS EDW. A comprehensive list of potential data features were shortlisted following comprehensive review of prior studies that defined risk variables associated with definition of pneumonia and its various subtypes. Candidate potential risk factors targeted for further analysis are catalogued in Supplementary Table S1. The goal of predicting Pneumonia subtype risk was treated as a classification problem, stratifying patients who had a pneumonia diagnosis (cases) as “high risk” and those with no pneumonia diagnosis (controls) as “low risk”.
Data preparation
Retrieved data were preprocessed prior to conducting analyses and included matching of cases to controls by (a) race/ethnicity and (b) 1:1 Case: Control ratio (24). Variables with >30% missing data were excluded from analysis. Datasets were divided into four subsets: (1) All Pneumonia (all cases and controls); (2) community (CAP) and healthcare-associated (HCAP) pneumonias; (3) ventilator-associated (VAP) or hospital-acquired (HAP) pneumonias; and (4) aspiration pneumonia (AP). An evaluation set (10%) was extracted from the subsets for further validation. There was no overlap among subtypes based on subclassification process. The data were exhaustively classified using a rule-based algorithm that examined each case for carefully defined sub-type specific features that would allow for unequivocal assignment to a specific subtype (31). Moreover, only validated pneumonia applying NLP to radiographic notes was included in the dataset (32).
Feature selection
To identify a representative subset of attributes, a univariate filter, i.e., information gain with the ranker method was employed (33). Feature selection was conducted on all the four datasets using WEKA® (34). This allowed for evaluating the contribution of each variable by measuring the information gain with respect to the class, using the following formula:
class="mb15">where E stands for entropy, which is defined as:
class="mb15">We set a cut-off value of ∼3*10−3 as low information gain and variables with less than this value were further excluded from the analyses.
Machine learning algorithms
Performances of five algorithms were compared across the four subsets: Naïve Bayes (NB), Logistic Regression (LR), Support Vector Machine (SVM), Multi Layer Perceptron (MLP), and Random Forests (RF). Feature (input variables) selection and 10-fold cross validation were performed on all the datasets. Model performance was evaluated in terms of total accuracy, sensitivity, specificity, F-measure, Mathews-correlation-coefficient, and area under receiver operating characteristic curve (AUC). Performance estimation was conducted using a stratified 10-fold cross validation approach. Table 1 summarizes the list of all data features included in the prediction model. The study utilized the implementation of these ML algorithms available in the Waikato Environment for Knowledge Analyses (WEKA) open source tool (34). ML was applied to rigorously processed datasets that included only validated pneumonia cases subtyped using a rule-based algorithm to prevent misclassification error.
Performance measures
To assess the prediction model performance of different algorithms, the study compared ML algorithms using the following performance measures.
(1) The area under the ROC curve (AUC) as defined by Hand and Till for binary classification (35).
class="mb15">where n0 and n1 are the numbers of “pneumonia cases” and “controls”, respectively, and , where ri is the rank of the ith “pneumonia cases” in the ranked list.
(2) Sensitivity, also termed recall, is the ratio of the number of correctly classified “pneumonia cases” instances to the total number of “controls” instances.
class="mb15">where TP = true positive and FN = false negative.
(3) Precision is the ratio of the number of correctly classified “pneumonia cases” instances to the total number of instances that are classified as “controls”.
class="mb15">where FP = false positive.
(4) Specificity is the ratio of the number of correctly classified “pneumonia cases” instances to the total number of instances that are classified as “controls”.
class="mb15">where TN = true negative.
(5) Accuracy is the ratio of the number of correctly classified instances to the total number of instances.
style="margin-top:0em;margin-bottom:0em;margin-left:1em;text-indent:-1em;text-align:left">(6) F-measure is the harmonic mean of precision and recall.
style="margin-top:0em;margin-bottom:0em;margin-left:1em;text-indent:-1em;text-align:left">(7) Matthew’s Correlation Coefficient (MCC) considers the accuracy and error rates and is calculated by the following equation:
Results
In total, 6,034 records (equal number of cases and controls) met eligibility for inclusion in the main dataset. Table 1 shows the distribution of the datasets. A total of 52 variables which included demographic (n = 2), oral health (n = 6), and medical/environmental/behavioral (n = 44) were identified and retrieved from the data warehouse. Preprocessing of data resulted in deletion of three features (Arterial blood gas, Blood oxygen saturation levels, and Pro-calcitonin levels) based on a high proportion of missing data. Among these (n = 49), a total of 43 variables (22 common and 21 unique variables) showed association with pneumonia. After performing feature selection on all 4 datasets, variables were excluded due to low information gains from the subsets respectively, thus bringing the variable countdown to: All Pneumonia (29 variables), CAP/HCAP (26 variables), VAP/HAP (40 variables), and AP (37 variables). Table 2 shows the variables retained in the different subsets. Variables retained (n = 22) were common across all four pneumonia subsets. The most significant feature in terms of information gain for dental variables included “restored teeth” (∼0.3). Restored teeth was consistently the highest ranked variable across all pneumonia and pneumonia subtypes. While other dental variables such as missing teeth, periodontal pocket depth ≥5 mm, periodontal disease status, and dentures were retained in all pneumonia models, they were differentially ranked with respect to level of contribution across the various pneumonia subtypes (see Table 2). Bleeding on probing was only retained in the AP model.
Machine learning
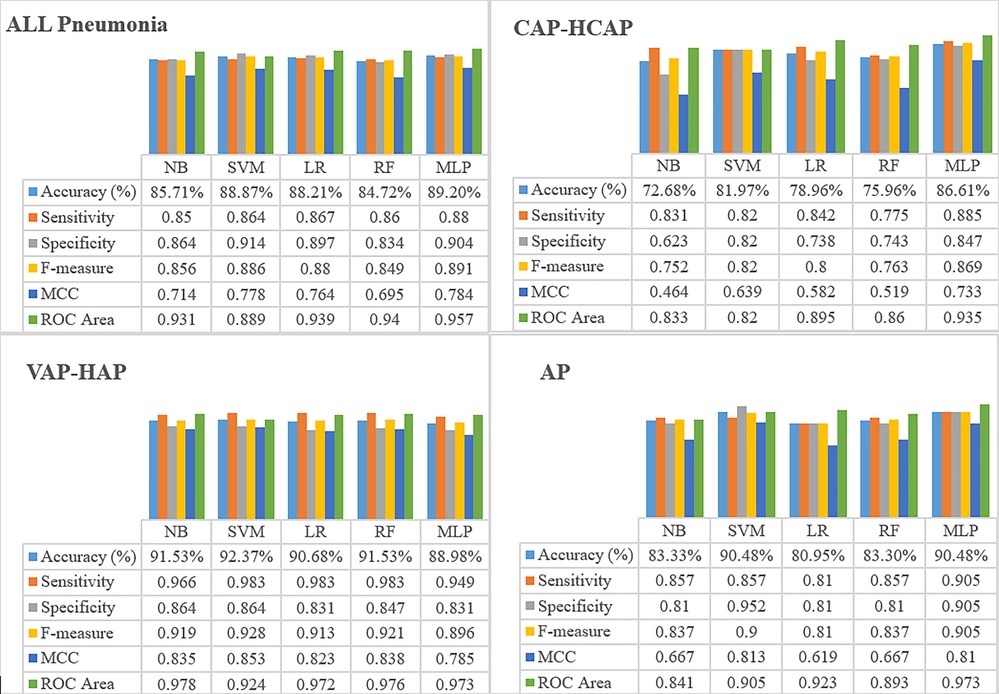
Results of the performance estimated through 10-fold cross validation are shown in Figure 1. MLP demonstrated higher accuracy in classifying the patients with Pneumonia risk as compared to NB, LR, and SVM in all Pneumonia and CAP-HCAP subsets. In terms of sensitivity and specificity of the resultant models for all Pneumonia and CAP-HCAP subsets, the MLP algorithm demonstrated all Pneumonia (sensitivity: 88% and specificity: 90%) and CAP-HCAP (sensitivity: 88% and specificity: 84%). By comparison, the sensitivity and specificity in VAP and AP subsets were: VAP [SVM, (sensitivity: 98%, specificity: 86%)], AP [SVM, (sensitivity: 85%, specificity: 95%)], and [MLP, (sensitivity: 90.5%, specificity: 90.5%)].
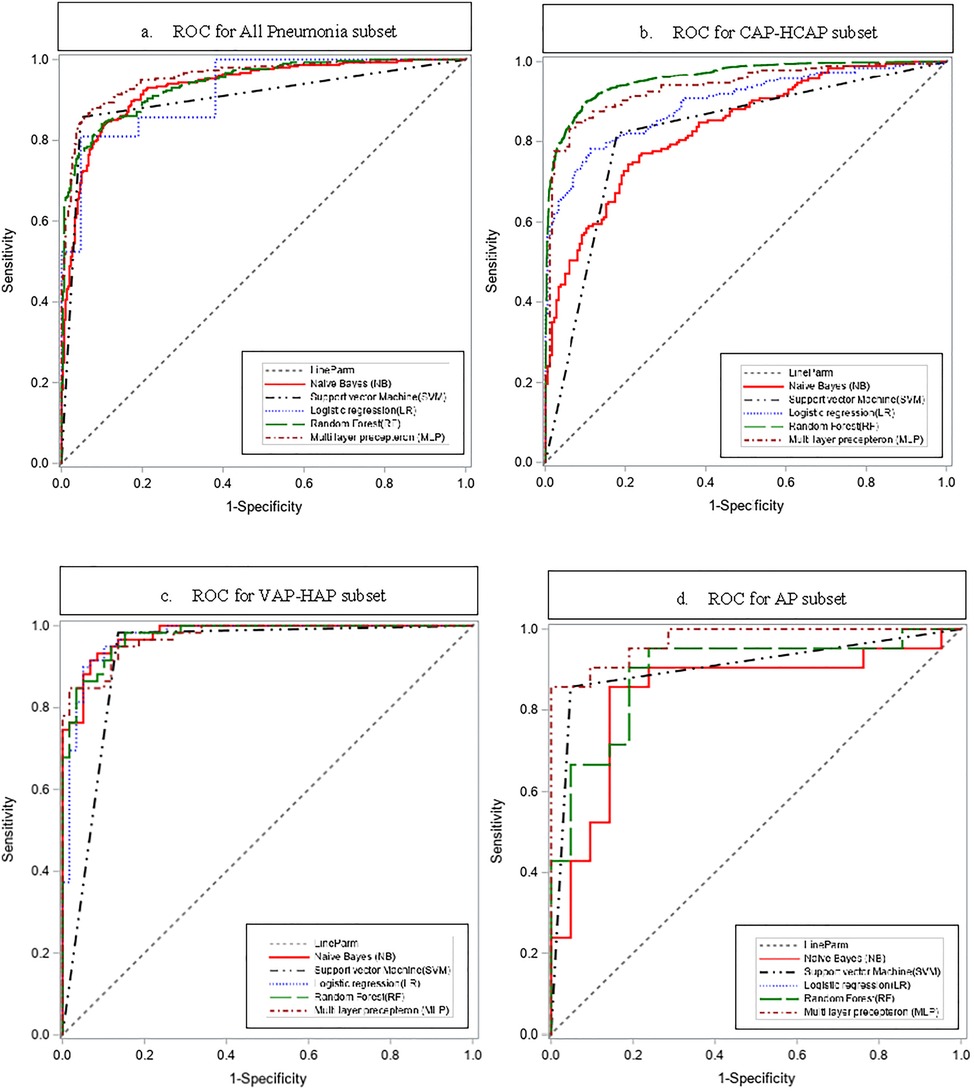
The ROC curves are shown in Figure 2.
MLP outperformed other predictive models for All Pneumonia, CAP-HCAP, and AP subsets, while SVM outperformed other models in VAP-HAP subset.
Discussion
This study capitalized on the availability of rich, clinical real-world, population-based, “big data” from an integrated medical-dental record and care delivery environment for modeling pneumonia risk through application of ML. Validated ML analytical approaches were applied to population-level data to vet association of established pneumonia risk factors including oral/dental variables with incidence of any pneumonia, as well as pneumonia subtypes including “CAP-HCAP”, “VAP-HAP”, and “AP”. Among the ML algorithms used in this study, MLP yielded the best AUC (0.9) in “ALL pneumonia”, “CAP-HCAP”, and “AP” subsets. Although predictive modeling using ML approaches has been used for various health conditions, use of ML approaches to define variables most predictive of pneumonia risk has been limited.
Notably, oral health-related variables defined in the current study that contributed most significantly to pneumonia risk are consistent with outcomes of oral diseases associated with infectious/inflammatory etiologies. Increasingly, a growing body of evidence supports plausibility of microbial pathogenesis as an important contributory factor underlying the association between pneumonia and oral diseases. Distinct pulmonary microbiomes in the upper and lower airways have recently been reported (11, 14). Notably, resident airway flora reflects microbiota found in the oral cavity, likely due to close proximity and interconnections between the lung and oral cavity (11). Further, shifts in microbial representation associated with disease processes such as cariogenesis and PD may also cause shifts in relative representation of oral microbiota in the pulmonary microbiomes (14). Such shifts in the relative representation of microbial species in the oral cavity in the context of infectious/inflammatory processes elicited by periodontal or cariogenic pathogens can lead to dysbiosis. Dysbiosis is associated with perturbation of the microbial content and environment giving rise to conditions unfavorable for normal flora which normally maintain microbial balance and the microbial environment. Subsequently, shifts in microbial representation may favor oral pathogens and establishment of conditions favorable to colonization by potential pulmonary pathogens. This positions these organisms to become opportunistic pathogens when conditions become favorable, especially in immunocompromised hosts. Moreover, direct transfer of oral bacteria from the oral cavity to the lungs may occur in the context of aspiration and VAP subtypes. Causality of VAP in conjunction with microbial transfer from the oral cavity during intubation has been definitively established by demonstrating genetic identity between the isolated pneumonia pathogen and bacterial isolates from dental plaque of the affected patient (16).
This study builds on two previous studies that applied informatics approaches to achieve pneumonia sub-classification into CAP, HCAP, VAP, HAP, and ASP pneumonia subtypes in the same population analyzed in the current study (31, 32). We performed pneumonia case validation by using Natural Language Processing (NLP) on the observations recorded by radiologists on chest radiographs (31). The rules followed were based on the study published by Dublin et al. (36). A NLP-based software was developed which enforced the rule-set prescribed by Dublin et al. and used to classify radiological records to have “positive”, “negative”, or “unknown” mentions of pneumonia. This validated the presence of the pneumonia diagnoses in patients through unstructured data in addition to “Rule of one” and “Rule of two”. The validated pneumonia episodes after case validation were then classified into six cohorts. After extensive literature review, key medical features that were identified to classify pneumonia episodes and rules were set in place to develop a rule-based algorithm which allowed for classification of pneumonia episodes (32). Application of ML to datasets from our population-based pneumonia cohort that had previously undergone algorithm-driven subclassification and validation of pneumonia status ensured that all pneumonia subclassification assigned in the current study was accurate and represented true, validated pneumonia cases. Following propensity score adjustment for potential confounding by other established pneumonia risk factors, our group also conducted time to event analysis and statistical modeling in the same dataset. This alternative approach to exploring association between oral health status preceding pneumonia events similarly identified “missing teeth” and “periodontal status assigned by a dental professional” as two variables that were retained in statistical models as significant independent risk factors for pneumonia emergence.
Historically, similar studies developed predictive models to predict pneumonia risk in patients with specific systemic conditions including schizophrenia (37), liver transplantation (38), and traumatic brain injury (39). Another study modeled risk factors for 30-day hospital readmission following incidence of pneumonia (40). A recent study (41) developed ML VAP risk prediction models using EHR data from adult ICU encounters (n = 524 positive VAP patients) during the patients’ hospital stay (41). The authors reported logistic regression as the best performing model followed by MLP. The AUC (ROC) reported by the investigators was 0.8 after reviewing 48 h of data (41). The performance for VAP/HAP subset in our study was 0.9 and is likely attributable to volume of data used (591 positive VAP patients), the number of variables used (n = 40) [vs. 10 variables modeled in the study (41)], and use of 10% vs. 20% evaluation set in their study. Similarly, Xu et al. built models to predict adverse outcomes in patients with CAP using nine ML algorithms and reported AUC of 0.8 using MLP for prediction of death in pneumonia patients (42). The study reported using variables including fever, cough, tachypnea, dyspnea, hypertension, hematocrit, hemoglobin, WBC, creatinine, BUN, glucose, heart disease, immunosuppression, malignancy, cerebrovascular disease, renal disease, and liver disease, which were also retained in our models using feature selection (42).
Our study focused on improving overall predictive accuracy by including medical and dental variables to develop a risk model for assessing patient risk for pneumonia. This study demonstrated that dental variables, such as restored teeth, missing teeth, periodontal pocket depth ≥5 mm, periodontal disease status assigned by dental provider, and presence of dentures, displayed high predictive performance. Selection of dental data variables also led to a novel observation: tooth restoration history and missing teeth play a significant role in pneumonia risk. Increased numbers of restorations and/or missing teeth may be related to higher risk of aspiration and perhaps dysphagia, well-known risk factors for pneumonia (43). These findings are similar to a recent population-based study conducted by Son et al. who showed that the risk of pneumonia significantly increased in patients with a higher number of dental caries and missing teeth. In the present study, PPD ≥ 5 mm and periodontal status assigned by a dental provider were identified as significant factors contributing to pneumonia risk in all subsets. This observation further reinforces the association between periodontal disease and pneumonia risk as shown in other studies (8, 10). Further in our study, “restored teeth” was the dental variable contributing to highest pneumonia risk. Two additional studies that applied ML to evaluate potential risk variables associated with HAP (37) and VAP (39) identified WBC count and serum sodium levels as additional crucial risk factors in their respective studies. WBC count was also identified as a significant variable in the current study. A recent study by Zhao et el. applied ML approaches to datasets of pneumonia and COVID patients and similarly showed that clinical indicators including WBC count may be a significant factor to predict the disease progression and outcomes in patients with pneumonia and COVID-19 (44).
The study acknowledges some limitation. The study data used were collected from a single healthcare system. This may raise potential for selection bias within the healthcare system. Generalizability of the predictive models developed in this study will require further testing and validation in other healthcare systems. Some variables such as clinical attachment loss were not included due to incomplete and missing data. The addition of these variables may be necessary to further improve the pneumonia risk predictability in healthcare settings.
Conclusion
The results of this study show that ML approaches that include medical and dental data show an association of oral health variables with pneumonia subtypes. Thus, consideration of oral health outcomes in the integrated healthcare environment would improve patient care through early detection of pneumonia risk and further help in clinical decision support to undertake preventive approaches.
To the best of our knowledge, this study is the first to develop predictive models using ML techniques for identifying risk factors associated with emergence of different pneumonia subtypes based on modeling of medical and dental data. Risk scores could be developed to inform patient referral and follow-up in integrated medical-dental care delivery settings and coordination of oral health and pneumonia management. Future studies would test portability and translation of e-Health approaches into clinical care delivery in diverse healthcare settings.
Data availability statement
The datasets presented in this article are not readily available because of private clinical data, are owned by the Marshfield Clinic and are not available for public sharing. Requests to access the datasets should be directed to amit.acharya@aah.org.
Ethics statement
The studies involving human participants were reviewed and approved by the Marshfield Clinic Research Institute. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author contributions
NS: is a biomedical informatician and a dental surgeon, Co-I, performed machine learning analyses, contributed to analytical interpretation, involved in drafting the article, revising it critically for important intellectual content, and the final approval of the version. IG: microbiologist/immunologist; participated in grant development, contributed to analyses and data interpretation, and involved in manuscript drafting, final editing, and final approval of the version. AP: data analyst and dentist, contributed to dataset development and quality assurance, and participated in analytical discussion, manuscript review, and final approval. HH: programmer and lead architect, contributed to data interpretation, and was involved in manuscript review and final approval of the version. FS: dentist (periodontist), dental domain and pneumonia expert and contributed to grant development, analytical discussion, and interpretation, and critical manuscript editing. AA: bioinformatition, dental surgeon, grant PI, conceptualized the study and led grant development. AA procured project funding and provided project oversight, led analytical planning and interpretation discussions, and participated in critical review and final manuscript editing. AA will serve as the corresponding author on the study. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a grant from the National Institute of Dental and Craniofacial Research branch of the National Institutes of Health, Award Number 1R03DE027020 and funding from Marshfield Clinic Research Institute. Funding sources had no involvement in: study design; collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Acknowledgments
The study acknowledges the efforts of Brooke Delgoffe and Steffani Roush, senior research programmer analyst in the Office of Research Computation and Analytics of the Marshfield Clinic Research Institute for assisting with clinical medical and dental data retrieval from the electronic health records and enterprise data warehouse of Marshfield Clinic Health System to support data set development.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fdmed.2022.1005140/full#supplementary-material.
References
1. American Lung Association. Pneumococcal pneumonia. American Lung Association (2017). Available at: https://www.lung.org/lung-health-diseases/lung-disease-lookup/pneumonia/pneumococcal?gclid=EAIaIQobChMI9__Z_Pzi-QIVrsmUCR2R-gJbEAAYASAAEgKTofD_BwE (Accessed August 25, 2022).
2. National Center for Health Statistics. National Hospital Ambulatory Medical Care Survey: 2017 emergency department summary tables (2017). Available at: https://www.cdc.gov/nchs/data/nhamcs/web_tables/2017_ed_web_tables-508.pdf (Accessed August 25, 2022).
3. National Center for Health Statistics. FastStats - pneumonia. CDC (2014). Available at: http://medbox.iiab.me/modules/en-cdc/www.cdc.gov/nchs/fastats/pneumonia.htm (Accessed August 25, 2022).
4. Burnham JP, Kollef MH. CAP, HCAP, HAP, VAP: the diachronic linguistics of pneumonia. Chest. (2017) 152:909–10. doi: 10.1016/j.chest.2017.05.002
5. Mandell LA, Niederman MS. Aspiration pneumonia. N Engl J Med. (2019) 380:651–63. doi: 10.1056/NEJMra1714562
6. Corrado RE, Lee D, Lucero DE, Varma JK, Vora NM. Burden of adult community-acquired, health-care-associated, hospital-acquired, and ventilator-associated pneumonia: New York city, 2010 to 2014. Chest. (2017) 152:930–42. doi: 10.1016/j.chest.2017.04.162
7. Son M, Jo S, Lee D, Lucero DE, Varma JK, Vora NM. Association between oral health and incidence of pneumonia: a population-based cohort study from Korea. Sci Rep. (2020) 10:9576. doi: 10.1038/S41598-020-66312-2.32533077
8. Scannapieco FA. Role of oral bacteria in respiratory infection. J Periodontol. (1999) 70:793–802. doi: 10.1902/jop.1999.70.7.793
9. Almirall J, Serra-Prat M, Bolibar I, Balassi V. Risk factors for community-acquired pneumonia in adults: a systematic review of observational studies. Respiration. (2017) 94:299–311. doi: 10.1159/000479089
10. Mammen MJ, Scannapieco FA, Sethi S. Oral-lung microbiome interactions in lung diseases. Periodontol 2000. (2020) 83:234–41. doi: 10.1111/prd.12301
11. Huffnagle GB, Dickson RP, Lukacs NW. The respiratory tract microbiome and lung inflammation: a two-way street. Mucosal Immunol. (2017) 10:299–306. doi: 10.1038/mi.2016.108
12. Shima K, Coopmeiners J, Graspeunter S, Dalhoff K, Rupp J. Impact of micro-environmental changes on respiratory tract infections with intracellular bacteria. FEBS Lett. (2016) 590:3887–904. doi: 10.1002/1873-3468.12353
13. Jain S, Self WH, Wunderink RG, et al. Community-Acquired pneumonia requiring hospitalization. N Engl J Med. (2015) 373:415–27. doi: 10.1056/NEJMoa1500245
14. Wu BG, Segal LN. The lung microbiome and its role in pneumonia. Clin Chest Med. (2018) 39:677–89. doi: 10.1016/j.ccm.2018.07.003
15. Bousbia S, Papazian L, Saux P, Forel JM, Auffray J-P, Martin C, et al. Repertoire of intensive care unit pneumonia Microbiota. PLoS One. (2012) 7:e32486. doi: 10.1371/journal.pone.0032486
16. Heo SM, Haase EM, Lesse AJ, Gill SR, Scannapieco FA. Genetic relationships between respiratory pathogens isolated from dental plaque and bronchoalveolar lavage fluid from patients in the intensive care unit undergoing mechanical ventilation. Clin Infect Dis. (2008) 47:1562–70. doi: 10.1086/593193
17. Sands KM, Twigg JA, Lewis MAO, Wise MP, Marchesi JR, Smith A, et al. Microbial profiling of dental plaque from mechanically ventilated patients. J Med Microbiol. (2016) 65:147. doi: 10.1099/jmm.0.000212
18. Kageyama S, Takeshita T, Furuta M, Tomioka M, Asakawa M, Suma S, et al. Relationships of variations in the tongue Microbiota and pneumonia mortality in nursing home residents. J Gerontol A Biol Sci Med Sci. (2018) 73:1097–102. doi: 10.1093/gerona/glx205
19. Silvestri L, Weir I, Gregori D, Taylor N, Zandstra D, Van Saene JJ, et al. Effectiveness of oral chlorhexidine on nosocomial pneumonia, causative micro-organisms and mortality in critically ill patients: a systematic review and meta-analysis. Minerva Anestesiol. (2013) 80:805–20. https://pubmed.ncbi.nlm.nih.gov/24257147/24257147
20. Juthani-Mehta M, Ness PH, McGloin J, Argraves S, Chen S, Charpentier P. A cluster-randomized controlled trial of a multicomponent intervention protocol for pneumonia prevention among nursing home elders. Clin Infect Dis. (2015) 60:849–57. doi: 10.1093/cid/ciu935
21. Aİ T. Artificial intelligence in healthcare: past, present and future. Anatol J Cardiol. (2019) 22:8–9. doi: 10.14744/AnatolJCardiol.2019.28661
22. Millet L. Artificial intelligence in healthcare and the transformation of healthcare professions. Soins. (2019) 64:51–2. doi: 10.1016/j.soin.2019.06.012
23. Kohli M, Prevedello LM, Filice RW, Geis JR. Implementing machine learning in radiology practice and research. Am J Roentgenol. (2017) 208:754–60. doi: 10.2214/AJR.16.17224
24. Hegde H, Shimpi N, Panny A, Glurich I, Christie P, Acharya A. Development of non-invasive diabetes risk prediction models as decision support tools designed for application in the dental clinical environment. Informatics Med Unlocked. (2019) 17:100254. doi: 10.1016/J.IMU.2019.100254.
25. Shimpi N, Glurich I, Rostami R, Hegde H, Olson B, Acharya A. Development and validation of a non-invasive, chairside oral cavity cancer risk assessment prototype using machine learning approach. J Pers Med. (2022) 12:614. doi: 10.3390/jpm12040614
26. Shimpi N, McRoy S, Zhao H, Wu M, Acharya A. Development of a periodontitis risk assessment model for primary care providers in an interdisciplinary setting. Technol Health Care. (2020) 28:143–54. doi: 10.3233/THC-191642
27. Wise MP, Williams DW, Lewis MA, Thomas JG, Frost PJ. Impact of poor dental health on pneumonia. Eur Respir J. (2008) 32:1123–4. doi: 10.1183/09031936.00096808
28. Glurich I, Shimpi N, Scannapieco F, et al. Interdisciplinary care model: pneumonia and oral health. In: Acharya A, Powell V, Torres-Urquidy MH, et al., editors. Integration of medical and dental care and patient data. Cham, Switzerland: Springer Nature Switzerland AG, Springer International Publishing (2019). p. 123–39.
29. Shimpi N, Glurich I, Acharya A, et al. Integrated care case study: marshfield clinic health system. In: Acharya A, Powell V, Torres-Urquidy MH, et al., editors. Integration of medical and dental care and patient data. Cham, Switzerland: Springer Nature Switzerland AG, Springer International Publishing (2019). p. 315–26.
30. Nycz G, Shimpi N, Glurich I, Ryan M, Sova G, Weiner S, et al. Positioning operations in the dental safety net to enhance value-based care delivery in an integrated health-care setting. J Public Health Dent. (2020) 80(Suppl 2):S71–6. doi: 10.1111/jphd.12392
31. Panny A, Hegde H, Glurich I, Scannapieco FA, Vedre JG, VanWormer JJ. A methodological approach to validate pneumonia encounters from radiology reports using natural language processing (NLP). Methods Inf Med. (2022) 61(1-2):38–45. doi: 10.1055/A-1817-7008
32. Hegde H, Glurich I, Panny A, Vedre JG, VanWormer JJ. Identifying pneumonia sub-types from electronic health records using rule-based algorithms. Methods Inf Med. (2022) 61(1-2):29–37. doi: 10.1055/a-1801-2718
33. Wilcoxon F. Individual comparisons by ranking methods. Available at: http://www.jstor.org/about/terms.html (Accessed June 27, 2020).
34. Witten IH, Frank E, Hall MA. Data mining: practical machine learning tools and techniques. In: The Morgan Kaufmann Series in Data Management Systems, 3rd Edition. Burlington, MA: Morgan Kaufmann Publishers (2011).
35. Hand DJ. Measuring classifier performance: a coherent alternative to the area under the ROC curve. Mach Learn. (2009) 77:103–23. doi: 10.1007/s10994-009-5119-5
36. Dublin S, Baldwin E, Walker RL, Christensen LM, Haug PJ, Jackson ML. Natural language processing to identify pneumonia from radiology reports. Pharmacoepidemiol Drug Saf. (2013) 22:834–41. doi: 10.1002/pds.3418
37. Kuo KM, talley PC, Huang CH, Cheng LC. Predicting hospital-acquired pneumonia among schizophrenic patients: a machine learning approach. BMC Med Inform Decis Mak. (2019) 19:1–8. doi: 10.1186/s12911-018-0723-6
38. Chen C, Yang D, Gao S, Zhnag Y, Chen L, Wang B, et al. Development and performance assessment of novel machine learning models to predict pneumonia after liver transplantation. Respir Res. (2021) 22:1–12. doi: 10.1186/s12931-020-01578-8
39. Abujaber A, Fadlalla A, Gammoh D, Al-Thani H, El-Menyar A. Machine learning model to predict ventilator associated pneumonia in patients with traumatic brain injury: the C.5 decision tree approach. Brain Inj. (2021) 35:1095–102. doi: 10.1080/0269905220211959060
40. Caruana R, Lou Y, Gehrke J, Koch P, Sturm M, Elhadad N. Intelligible models for healthcare: predicting pneumonia risk and hospital 30-day readmission. Proceedings of the ACM SIGKDD international conference on knowledge discovery and data mining. New York, NY: Association for Computing Machinery (2015). p. 1721–30.
41. Giang C, Calvert J, Rahmani K, Barnes G, Siefkas A, Green-Saxena A, et al. Predicting ventilator-associated pneumonia with machine learning. Medicine (Baltimore). (2021) 100:e26246. doi: 10.1097/MD.0000000000026246
42. Xu Z, Guo K, Chu W, Lou J, Chen C. Performance of machine learning algorithms for predicting adverse outcomes in community-acquired pneumonia. Front Bioeng Biotechnol. (2022) 10:903426. doi: 10.3389/FBIOE.2022.903426
43. Terpenning M. Geriatric oral health and pneumonia risk. Clin Infect Dis. (2005) 40:1807–10. doi: 10.1086/430603
Keywords: data mining, decision support tools, eHealth, health information system (HIS), electronic health records (EHR), information storage and retrieval, pneumonia, medical-dental integration
Citation: Shimpi N, Glurich I, Panny A, Hegde H, Scannapieco FA and Acharya A (2022) Identifying oral disease variables associated with pneumonia emergence by application of machine learning to integrated medical and dental big data to inform eHealth approaches. Front. Dent. Med 3: 1005140. doi: 10.3389/fdmed.2022.1005140
Received: 28 July 2022; Accepted: 7 September 2022;
Published: 22 September 2022.
Edited by:
Elsbeth Kalenderian, University of California, San Francisco, United StatesReviewed by:
Hasan Tahir Abbas, University of Glasgow, United KingdomAbhiram Maddi, Medical University of South Carolina, United States
© 2022 Shimpi, Glurich, Panny, Hegde, Scannapieco and Acharya. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amit Acharya, YW1pdC5hY2hhcnlhQGFhaC5vcmc=
Specialty Section: This article was submitted to Systems Integration, a section of the journal Frontiers in Dental Medicine
 Neel Shimpi
Neel Shimpi Ingrid Glurich2
Ingrid Glurich2 Frank A. Scannapieco
Frank A. Scannapieco Amit Acharya
Amit Acharya