- Faculty of Medicine and Dentistry, University of Alberta, Edmonton, AB, Canada
Objective: Various health-related fields have applied Machine learning (ML) techniques such as text mining, topic modeling (TM), and artificial neural networks (ANN) to automate tasks otherwise completed by humans to enhance patient care. However, research in dentistry on the integration of these techniques into the clinic arena has yet to exist. Thus, the purpose of this study was to: introduce a method of automating the reviewing patient chart information using ML, provide a step-by-step description of how it was conducted, and demonstrate this method's potential to identify predictive relationships between patient chart information and important oral health-related contributors.
Methods: A secondary data analysis was conducted to demonstrate the approach on a set of anonymized patient charts collected from a dental clinic. Two ML applications for patient chart review were demonstrated: (1) text mining and Latent Dirichlet Allocation (LDA) were used to preprocess, model, and cluster data in a narrative format and extract common topics for further analysis, (2) Ordinal logistic regression (OLR) and ANN were used to determine predictive relationships between the extracted patient chart data topics and oral health-related contributors. All analysis was conducted in R and SPSS (IBM, SPSS, statistics 22).
Results: Data from 785 patient charts were analyzed. Preprocessing of raw data (data cleaning and categorizing) identified 66 variables, of which 45 were included for analysis. Using LDA, 10 radiographic findings topics and 8 treatment planning topics were extracted from the data. OLR showed that caries risk, occlusal risk, biomechanical risk, gingival recession, periodontitis, gingivitis, assisted mouth opening, and muscle tenderness were highly predictable using the extracted radiographic and treatment planning topics and chart information. Using the statistically significant predictors obtained from OLR, ANN analysis showed that the model can correctly predict >72% of all variables except for bruxism and tooth crowding (63.1 and 68.9%, respectively).
Conclusion: Our study presents a novel approach to address the need for data-enabled innovations in the field of dentistry and creates new areas of research in dental analytics. Utilizing ML methods and its application in dental practice has the potential to improve clinicians' and patients' understanding of the major factors that contribute to oral health diseases/conditions.
Introduction
Dental records, known also as patient charts, are legal documents that provide detailed information of patients' oral health history and consist of significant amounts of data about the patient. They include: demographics, clinical, medical, and radiographic findings, corresponding diagnosis, and treatment planning (1–3). Accurate records and access to such information are necessary for dentists to adequately monitor patients' oral and systemic health, determine and provide appropriate and quality care interventions, and facilitate optimal communication between the treating dentist and patient as well as between the dentist and other health professionals for continuity of care (1, 4). For example, high-quality documentation of information on patients' oral health status and treatment course serves as a source of evaluation over time to determine and implement the best approach to address a patient's unique dental needs (1, 4).
As patient charts enable clinicians to deliver quality patient care and follow-up (1), it is essential that the information comprising these records is accurate, organized, clear, and easily accessible so pertinent data can be readily identified and utilized over time. Although many dentists have traditionally employed paper patient charts in their practice, an increasing number of dentists are utilizing electronic records to collect patient clinical data (4). While both formats provide a reasonable option to record and store patient information, electronic records are often cited as advantageous due to their storage capacity, accessibility, ease of information relay, time efficiency, and scalability (e.g., features such as fillable templates and typed notes enable dentists to spend more time with patients and completing procedures than writing or interpreting notes) (4, 5). However, it is important to note that human errors in the accuracy and relevance of information recorded can occur in either paper or electronic formations (e.g., illegible handwritten notes or auto-fillable templates may result in meaningless or incorrect entries) (5).
In addition to providing electronic options for creating and storing patient records, technological advances in computerized systems generate new opportunities to enhance patient care through the application of artificial intelligence (AI) to these records. AI refers to computer algorithms that simulate the process of human intelligence to solve problems (6). Machine learning (ML) is a subset of AI techniques, which enable computers to learn from and identify relationships within data in different ways based on the specific algorithm used (7). For example, supervised algorithms are trained to learn using predictive models based on labeled data (i.e., data that humans give meaning to and describe by using preexisting understanding and knowledge) (8). Supervised algorithms apply the information learned from labeled data sets (e.g., classifications, outcome predictions, etc.) to new, unlabeled datasets to identify similar information. In contrast, unsupervised algorithms aim to discover unknown patterns in unlabeled data (i.e., data with no assigned meaning) and do not require human intervention for learning as these algorithms are tasked to identify key information on their own (7, 9–11). Medical and dental clinicians can utilize ML algorithms to manage and analyze the immense data present in patients' health records to extract meaningful information that can be used to improve population health and wellbeing (12). Diagnostic medicine incorporates advanced AI technologies to predict or diagnose disease and assist in the clinical decision-making process (9–11), and several studies have shown that AI-based comprehensive care system has great potential to enhance patient care, spur innovative research, and facilitate information sharing (13–15).
In dentistry, AI methods are also being applied to diagnose patients and treatment planning (16), identify patients at risk of oral cancer and pre-cancer (17), provide oral radiographic differential diagnosis (ORAD) (18), pulpal diagnosis (19), lower third molar treatment planning decisions (20) and prognosis evaluation (21). However, despite the important role oral health-related factors (22) play in dentists' diagnosis and treatment planning process (23), the use of ML techniques as a method to review chart information on these contributors has not yet been studied to the authors' knowledge. Given the extensive and valuable information patient charts hold and the functions they serve (e.g., to enable clinicians to deliver quality patient care and follow-up), applying ML methods to these data has the potential to identify significant predictive relationships between pertinent patient chart information and important oral health-related contributors that dentists can use as a complementing source of information in their care process. Thus, the purpose of this study was to introduce an automated method of reviewing patient chart information using ML and demonstrate this method's ability to identify predictive relationships between patient chart information and important oral health-related outcomes. We first describe the proposed method, and then use it to complete a secondary data analysis of patients' charts to provide an example of its utility in real-life contexts.
Methods
Proposed Method
The proposed method includes three phases of ML algorithms to facilitate information extraction and analysis: text mining, topic modeling, and predictive modeling. The first two phases (text mining and topic modeling) aim to import and organize data and extract key information and topics for further analysis of predictive relationships between patient chart data and important oral health-related contributors. The last phase (predictive modeling) uses two ML components (ordinal logistic regression and Artificial Neural Network) to identify the presence or absence of such predictive relationships by analyzing the extracted information and topics.
Text Mining
Text mining is a ML technique for quick extraction of key information from vast quantities of textual data. It is a powerful research tool that can be used for information retrieval, information extraction, and text categorization (24). Text mining is appropriate for patient chart data as it is useful for analyzing large data sets to extract latent (unknown) patterns in the data to create comprehensible information (12). Further, as text mining extracts key information from textual data that can be used to make sense of large quantities of both relevant and irrelevant information, patient chart data is a suitable application as it consists of textual, unstructured data (i.e., information that either does not have a predefined data model or is not organized in a pre-defined manner) (12).
Text Preprocessing and Data Cleaning
For text mining to perform properly, raw textual data (i.e., data that has not been organized in a predefined manner) needs to be preprocessed or cleaned as this step prepares the data for subsequent analysis and enables meaningful analytic results. Data cleaning aims to standardize (normalize) the data and ensure consistency across the data set, eliminate unnecessary data features (those containing irrelevant information), remove duplicates, and manage any missing data (data of interest that is not available or recorded) (25). High-quality data is an essential part of developing effective ML models; thus, raw data should be cleaned before utilization since errors within raw data (e.g., duplicate data) might confuse the ML process and cause erroneous pattern detection (26).
Topic Modeling
Topic modeling (TM) is an unsupervised ML technique that utilizes statistical analysis methods to extract underlying information from a collection of unstructured documents (in TM, one patient chart would reflect one document). TM algorithms can summarize a large collection of text documents and identify co-occurring texts between documents (27). This co-occurrence can then be used to extrapolate semantic structures from a set of narrative passages. The extracted semantic structures are called topics and represent recurring patterns or clusters of co-occurring words in documents (27). Topics are extracted based on a probabilistic model that determines the most frequent co-occurring words over all documents (28). Key elements of TM are words or terms (a basic unit of discrete data), documents (a sequence of terms), corpus (a collection of documents), and document-term-matrix (DTM; a matrix that presents the frequency of each word in each document) (28). An example of a DTM is presented in Table 1 where each cell is a frequency of terms used (column) in each document (row).
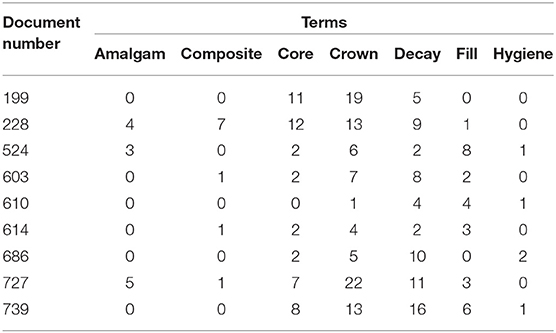
Table 1. A sample of DTM representing the frequency of terms (words) across some documents (patient charts).
Latent Dirichlet Allocation
Latent Dirichlet Allocation (LDA) is a common TM method and one of the most popular ML algorithms (28). LDA extracts previously unknown or latent information from an immense number of documents' original texts and unstructured data (28, 29). As patient charts are frequently found in the form of text files without any organized format, LDA is an appropriate method to extract salient information from these charts. Few TM alternatives are also available such as latent semantic analysis (LSA), probabilistic LSA (pLSA), or LDA. Given the information and the length of text in these charts, we selected LDA for use as it has greater accuracy and is easy to interpret the results as it provides a more efficient representation of results (30, 31).
Ordinal Logistic Regression
As our proposed model aimed to use the extracted LDA analysis data to identify the relationship between predictive variables (independent variables) and oral-health-related outcomes (dependent variables) in an ordinal scale (categorized by order), ordinal logistic regression (OLR) analysis was chosen. OLR is a supervised ML algorithm that uses a statistical approach to examine the relationship between ordinal outcomes and predictive variables (32). For example, OLR analysis could be used to find the relationship between independent variables (e.g., tooth brushing) and dependent variables (e.g., caries risk), which have an ordinal scale (e.g., low caries risk, moderate caries risk, high caries risk).
Artificial Neural Network
We included artificial neural networks (ANN) as an adjunct ML approach to OLR to find the predictability of independent variables. In contrast to OLR, ANN models do not require a predetermined assumption for either input or output variables (which may be ordinal or non-ordinal) and can engage in unsupervised learning to estimate complicated processes (33). This forward-feeding, multilayer perceptron approach toward supervised ML was important to include as it provides a prototypical framework for discovering and accounting for complex relationships between oral health outcomes and predictors. For example, oral health outcomes such as caries risk are dependent on a wide range of predictors including the patient's diet, oral hygiene, genetics, and socioeconomic factors (34, 35). Moreover, ANN allows for the prediction of multiple dependent variables. As OLR algorithms require pre-determined categories to identify category-related relationships and ANN algorithms do not, integrating this extra layer of analysis can help identify relationships outside of pre-determined categories that may be missed with OLR.
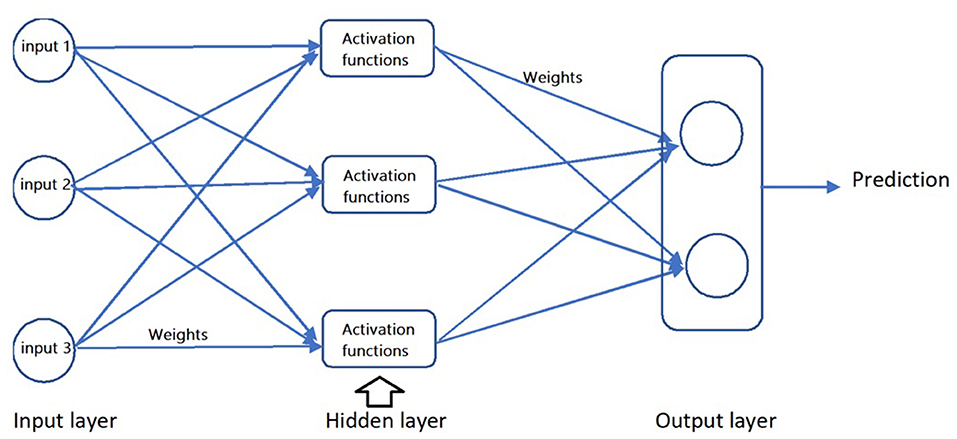
ANN simulates the function and structure of neural networks in the human brain and consists of three sets of neurons (see Figure 1): input (incoming information), hidden (analysis of incoming information), and output (outcome prediction) (36). In each hidden and output neuron, a mathematical function is used to produce an output (i.e., a value representing the prediction of the desired variable) from information in the preceding set of neurons (either the input or hidden neurons). ANN may involve a single-layer structure (having one input and output layer) or a multilayer structure (having at least one hidden layer in addition to input and output layers). Predictability of the input neuron and/or layer for the output neuron and/or layer (i.e., the predictive relationship between the variables) are determined by mathematical multipliers (weights) and constant numbers added to the input neuron (bias) that make connections between the neurons (37, 38). Training of these nodes involves a process named Gradient descent, a non-linear optimization approach that iteratively refines the mathematical function of each node to yield the best predictive outcome of the training data.
Application
To demonstrate how the proposed method can be used to facilitate the patient chart review process and predict for various oral health outcomes, we conducted a secondary data analysis on the intake patient charts referred to a private dental clinic from 2017 to 2020. The University Human Research Ethics Board approved this study (Pro00109728). Patient charts were collected in the initial visit and contain raw text files of patients' medical and dental history, clinician notes that indicated clinical findings including intra- and extra-oral findings, radiographic findings, and suggested treatment plans. As outlined in the previous section, we analyzed the anonymized chart data in three phases using the following steps.
Text Mining (Data Cleaning and Text Preprocessing)
1) A total of 785 patient charts were imported as individual text files. Chart data on radiographic findings and suggested treatment planning were separated for future TM purposes in later phases. The remaining components (patients' medical and dental history, clinical findings including intra- and extra-oral findings) were first compiled into R (software environment for statistical computing and graphics) using batch commands list.files(), and lapply(list.files, read.delim). Once all patient chart data was imported into the R environment, it was then transformed into one long narrative text (corpus) using the command Corpus().
2) After step 1, there was a corpus of raw data. Cleaning raw data is a critical step in text preprocessing. To complete data cleaning on the corpus, we removed stopwords (words and symbols that do not impact text meaning) using the command tm_map(docs,removeWords, stopwords(“english”)), since these words independently do not carry any relevant information for processing (39). In this command, the “docs” in parentheses was the name given to the corpus. This part of the command will vary based on what the corpus is named. We also removed white spaces and punctuations using the command tm_map from the package tm (40). Additionally, texts were transformed to lowercase letters to normalize the text using the command tm_map(docs,content_transformer(tolower). Similarly, the “docs” in this command also reflects the corpus name.
3) After step 2, the corpus consisted of cleaned text. To prepare the cleaned corpus for future analysis, we completed tokenization to break the corpus up into separate words called tokens. We used the tokenize_words command from the tokenizers package to split the corpus into tokens, which were individual words (41). These tokens reflected important information from the patient charts. For example, “caries,” “tenderness,” “bruxism,” and “wnl” were tokens. However, some of the tokens, such as “wnl” were not consistent across all data. For instance, some practitioners recorded “wnl” in the patient charts to reflect no significant findings, whereas others recoded “negative” to represent no significant findings. Therefore, to ensure consistency across all data, we recoded existing words such as “wnl” and “yes” with “negative” and “positive,” respectively, using the command str_replace_all from the stringi package in R for all variables.
4) After step 3, we were left with a collection of tokens that had been recoded for consistency. To prepare the tokenized data for analysis, we created a matrix of the tokens using the command matrix(). This command transforms the tokenized data into a tabular format to better visualize and export data in a well-organized format (42). To create the matrix, we first needed to determine the number of rows and columns. The rows were determined by the number of patient charts (785), whereas the columns were determined based on the information collected in patient charts (Appendix 1). Specifically, the research team found 66 main headings across all patient chart information (Appendix 2), which were used to determine the number of columns, as well as their name. After the number of rows (785) and columns [66] were determined, we used the command matrix() to transform the data with the appropriate dimensions and names. We used the command clnames to sort all patient chart information under the relevant heading in the respective columns.
5) Finally, the matrix was exported as a comma-separated values (CSV) format using the command write.xlsx from the xlsx package. CSV format was chosen as the format can be read easily by other software (43). The research team then reviewed the CSV file and used the extracted information from the matrix columns to select the outcome and predictive variables. We selected 10 column headings as outcome variables at random to demonstrate the modeling aspect of the study. Selected outcome variables were caries risk, occlusal risk, periodontitis, dental crowding, gingivitis, muscle tenderness, biomechanical risk, recession, bruxism, and assisted mouth opening. Then, we selected column headings as predictive variables based on their degree of predictability for the selected outcomes as described in the literature (e.g., allergies have been reported as predictive for caries risk) (44). As some outcome variables also held predictive qualities for other outcome variables (e.g., caries risk and gingivitis were predictive for each other), these were also included as predictive variables. In total, we selected 45 out of the 66 total columns as variables. The result was a CSV file with 785 rows (number of patient charts) and 45 columns (outcome and predictive variables).
6) We then changed the 10 selected outcomes to variables with an ordinal scale to prepare them for OLR analysis. For example, caries risk was one of the identified outcomes and was categorized as “low,” “moderate,” and “high” using CSV tools and “=IF” function.
7) The separated patient chart data on radiographic findings and suggested treatment planning were then used to repeat the first two steps identified above to create a corpus of cleaned data to use for TM (Code required to facilitate text mining as described in this phase is available in Appendix 3).
Topic Modeling AND Latent Dirichlet Allocation
We completed LDA on two cleaned texts from each patient: radiographic findings and suggested treatments. These texts were obtained during text mining to extract informative topics (cluster of words). We used the required TM packages in R programming (tm, LDAvis, topicmodels and ldatuning). To build the model, a DTM was created in which the terms and document numbers constructed the matrix dimensions.
A critical step in TM is determining the number of topics since having an excessive number of topics could lead to an extremely complex model that makes interpretation and validation difficult. Similarly, an inadequate number of topics could result in a coarse model that is not able to recognize accurate classifiers (45). Several approaches have been devised to apply various identification criteria. We compared approaches from Cao et al. (46), Aruen et al. (47), and Deveaud et al. (48) to find the most appropriate number of topics for LDA models, via the function FindTopicsNumber() from the package ldatuning (49). Cao et al.'s algorithm focuses on the number of topics that minimizes the average cosine between topics (therefore minimizing words used in multiple topics). An illustration of these algorithms for a given number of topics are presented in Figure 2. Arun et al.'s algorithm returns the value that minimizes the symmetric Kullback-Leibler (KL) divergence (47) between the matrices demonstrating the word-per-topic and topic-per-document distributions (therefore minimizes overlaps between words in topics and documents). Deveaud et al.'s algorithm selects the value that maximizes the sum of the divergences between topic pairs (therefore maximizing topic to topic). Typically, multiple metrics are applied for finding the best number of topics and are compared against Log Likelihood results as baseline (46–48).
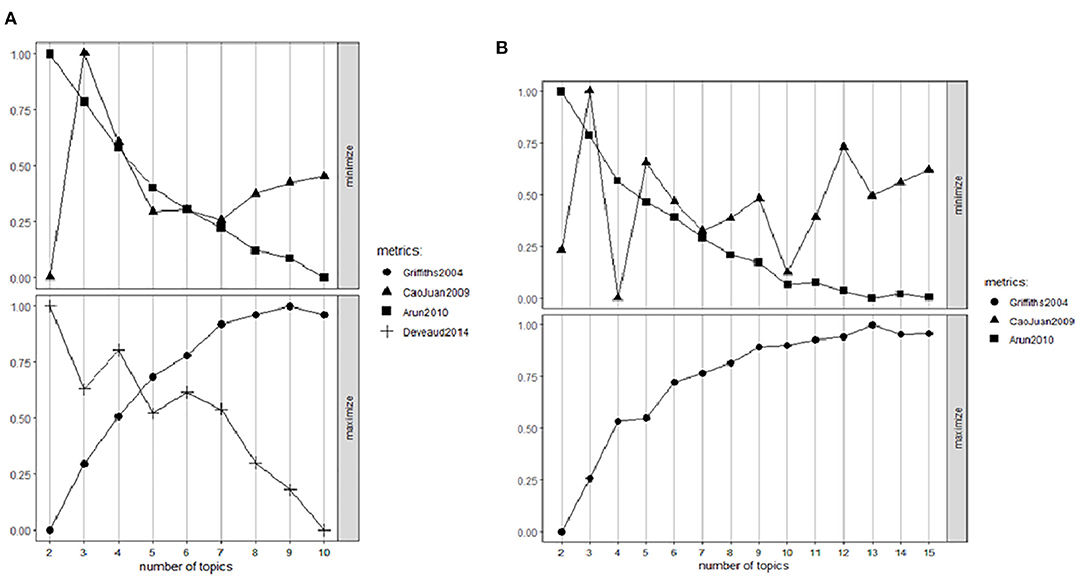
Figure 2. Metrics analysis to find the most appropriate number of topics for radiographic (A) and treatment planning (B) data.
After finding the appropriate number of topics, the determined number of topics were fit to LDA models using the LDA function from the package topic models (50) in conjunction with a Gibbs sampler to perform all future modeling activities (51).
Prediction (Ordinal Logistic Regression and Artificial Neural Network)
After TM was completed, both the radiographic and treatment topics (from TM) and 45 variables (from text mining) were used to find the predictors for various oral health outcomes (chosen at random through text mining) through OLR. First, OLR was used to identify significant predictors for each oral health outcome. Next, ANN analysis was completed to determine both the predictability of the model and the most important predictors among those significant variables.
Ordinal logistic regression was first computed to reduce dimensionality before ANN analysis. This was conducted prior to ANN to reduce irrelevant and unnecessary data and improve training outcomes and output comprehensibility (12, 52). Dimensionality reduction achieved these features through: (1) feature selection, which selects data pertaining to the most relevant information to solve a particular problem, and (2) feature extraction, which transforms the original features into a new, smaller set of more significant features (52). We used OLR to determine the most significant predictors, which were then utilized as the input data for our ANN model.
Both prediction modeling approaches (OLR and ANN) were conducted using SPSS (IBM SPSS Statistics version 26). For our ANN analysis the following procedures were used:
1) Multilayer-perceptron is the type of nodes selected to build the network. We selected the independent variables for each oral health outcome (dependent variable) based on the results obtained from OLR. For the independent variables, the categorical variables were put in the factors section while numerical variables were placed in the covariate sections.
2) Design of the network was specified next. The architecture of the network was set at one layer of hidden nodes for simplicity in interpreting the prediction results, with an automated number of nodes set. For the activation of nodes, the hyperbolic tangent function of the hidden nodes was chosen to minimize the number of training rounds, where a softmax activation function used as outcome variables was categorical in nature. The alternative, sigmoid function, would be used if outcomes were binary.
3) Training of the network required a partition of data into 70% training and 30% testing categories. The training was done in a batch sampling using gradient descent. The learning criteria were specified with an initial learning rate of 0.1 and a momentum of 0.5. Training stoppage criterion was set using a relative error ratio of 0.0001. The output was set to be reported as diagrams, synaptic weights, classification results and independent variable importance analysis.
Results
Text Mining
Text mining extracted various clinical features recorded in patients' charts which were exported as a CSV file for further analysis. A full list of the clinical features extracted from the charts and used for future prediction analysis is listed in Appendix 4.
Topic Modeling
Using the topic modeling thresholds as discussed, 10 topics were identified for radiographic findings and eight topics were identified for suggested treatment plans using the topic modeling packages in R. Topic modeling defines topics based on the most frequent words/co-occurring words. Thus, the extracted topics are defined as follows. Radiographic topics included: (1) recession, (2) structurally compromised teeth, core, crown, (3) composite, tooth chip, pit, (4) lingual, wire, anterior, (5) exo, wisdom teeth, (6) wear, occlusal, (7) fracture, filling, (8) root canal therapy, amalgam, (9) decay, recurrent, (10) bone. Suggested treatment planning topics included: (1) tooth wear and invisalign, (2) possible replacing the composite filling, (3) root canal therapy and mesio-occluso-distal (MOD) filling, (4) tooth fracture and amalgam filling, (5) impression, (6) post, core and crown, (7) wisdom teeth extraction and possible orthodontic treatment, (8) oral hygiene and possible sedation. Common topics with their five most frequent terms are presented in Figure 3.
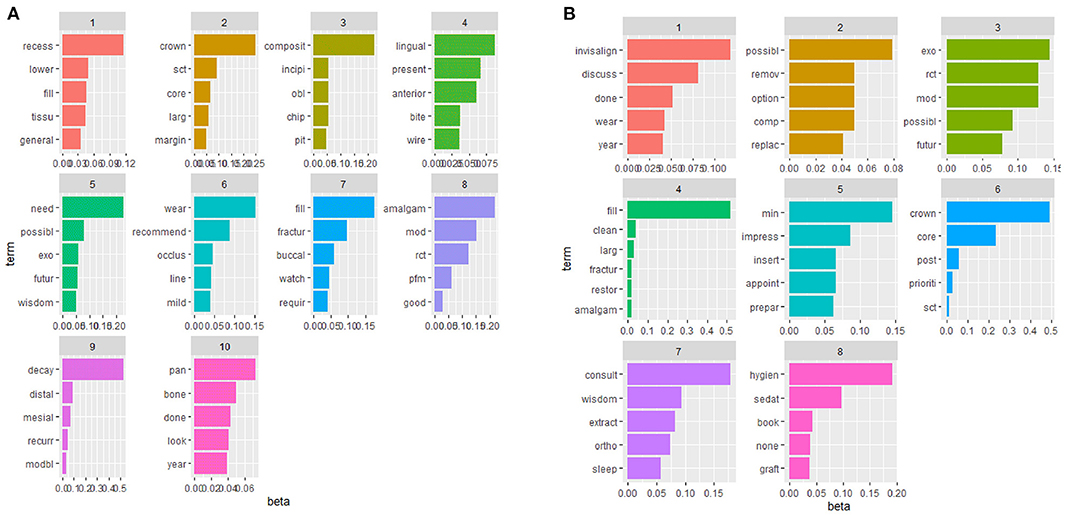
Figure 3. Extracted topics for radiographic findings and treatment plan using TM [x-axis denotes for beta (per-topic word distribution) and y-axis represents the five most frequent terms across each topic].
Ordinal Logistic Regression and Artificial Neural Network
OLR analysis found that all studied oral health outcomes were significantly predictable using various information recorded in patients' charts. For example, history of allergy (OR = 0.784, 95%CI, 0.235–1.332, p = 0.005), night guard use (OR = −0.670, 95% CI, −1.245 to −0.096, p = 0.022), presence of gingivitis (OR = −1.093, 95% CI, −1.513 to −0.672, p = 0.000), periodontitis (OR = −0.975, 95% CI, −1.507 to −0.407, p = 0.001) and mild occlusal risks (OR = −0.640, 95% CI, −1.097 to −0.182, p = 0.006) were significant predictors for caries risk after controlling for other variables. Similarly, caries risk (OR = −2.495, 95% CI, −3.942 to −1.047, p = 0.001), night guard use (OR = 0.624, 95% CI, 0.069 to 1.179, p = 0.027), mild gingivitis (OR = −0.456, 95% CI, −0.901 to −0.011, p = 0.045), normal salivary flow (OR = 1.616, 95% CI, 0.233 to 3.00, p = 0.022), structural integrity of the teeth (OR = −5.649, 95% CI, −6.520 to −4.777, p = 0.000) and left-side class II canine relationship (OR = −1.703, 95% CI, −3.393 to −0.013, p = 0.048) were significant predictors for occlusal risk.
Studied variables and their significant predictors (p < 0.05) after controlling for other variables are presented in Table 2. All extracted radiographic topics were significant predictors for muscle tenderness while all 8 treatment plan topics and topic 10 of radiographic findings (topic related to bone) were found to predict bruxism significantly. For the other eight evaluated oral health outcomes, the extracted topics were not significant predictors.
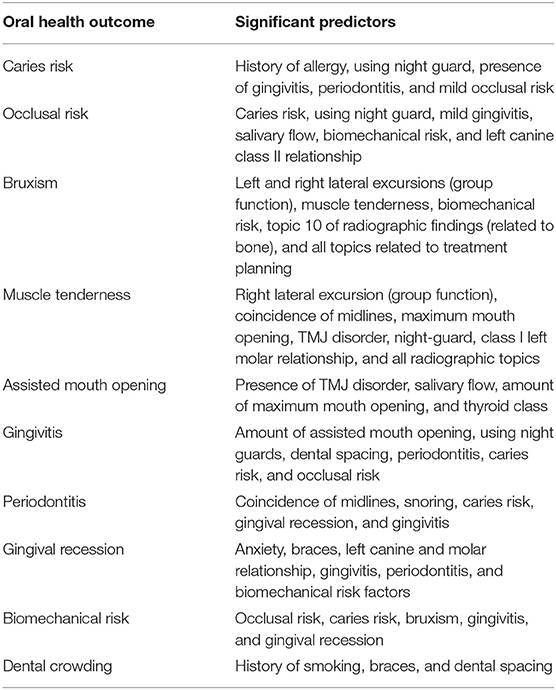
Table 2. Variables with significant predictability for different oral health outcomes according to ordinal logistic regression analysis results.
ANN analysis showed that the model can make >72% correct prediction for 8 out of 10 evaluated features (except for dental crowding and bruxism) using other recorded variables. Table 3 represents the predictability percentage, and the false positive rate of identifying different oral health outcomes with the most important predictor for each obtained from ANN analysis.
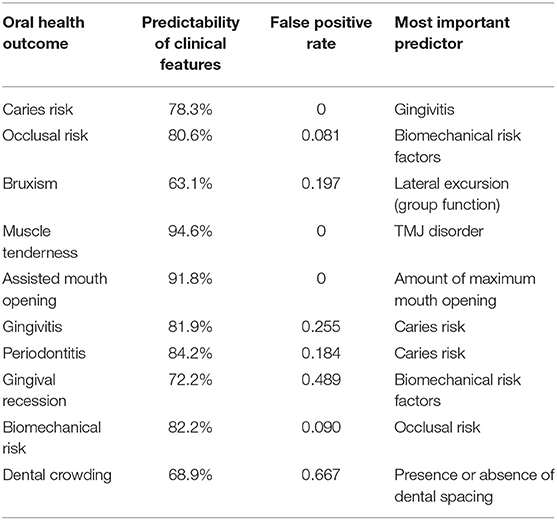
Table 3. Predictability of various clinical features and topics for oral health outcome and the most important predictors.
Table 4 represents the weights associated with the ANN analysis to calculate caries risk (output layer) as the dependent variable using gingivitis, periodontitis, occlusal risk, using night guard and history of allergy as independent variables (input layer). Caries risk was categorized into three levels: 0, indicating low caries risk, 1 indicating moderate caries risk, and 2 indicating high caries risk. The hidden layer consists of nine neurons, and the connection between all neurons in all three layers are shown by weights (blue and gray lines). The weight table represents a validation tool to verify how the values are calculated from the input layer.
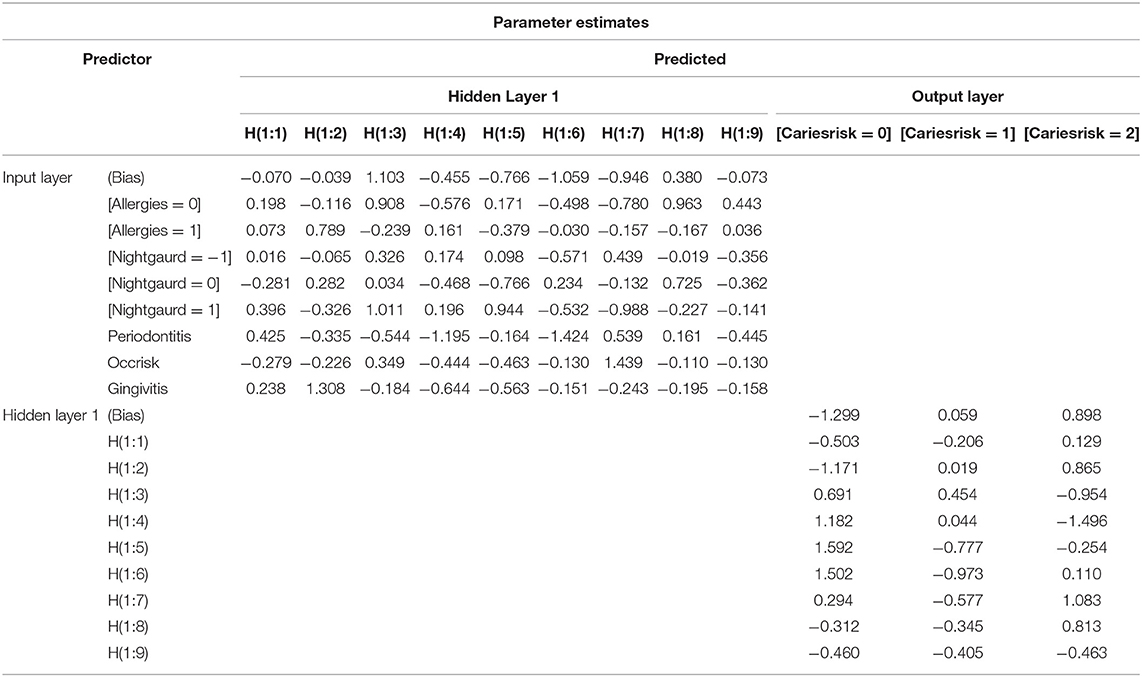
Table 4. Weights associated with the ANN analysis to calculate caries risk as an outcome using its significant predictors.
Discussion
Although the increasing popularity of digital dental records grants the opportunity to utilize AI-based technologies such as ML to streamline and enhance patient care (9), a recent review by Schwendicke et al. reported dentistry's clinical integration of such techniques has lagged. This study introduced a replicable, automated method of reviewing patient chart information using ML techniques and demonstrated its ability to identify predictive relationships between patient chart information and important oral health-related outcomes.
The proposed ML method was able to extract relevant topics and predict 8 out of 10 oral health outcomes with >72% predictability rate, which indicates that this model is well-suited to predict oral health-related outcomes using patient chart data. Similarly, other areas of dental research have used ML methods to successfully predict various health-related outcomes. For example, a study conducted by Miladinović et al. (53) evaluated indirect causes of tooth extraction using data mining methods and ML algorithms and found significant relationships between gender, age and patients' occupation and tooth extraction. Cooray et al. (54) also applied ML methods to explore predictors of tooth loss in older adults and found that demographic, baseline oral health and socioeconomic variables were important in predicting future tooth loss. As these studies' findings indicate, different oral health-related outcomes are intertwined with innumerable demographic, health, and socioeconomic factors. As the current study sought to examine several different oral health-related factors using dental patient data, ML methods were best suited to address the complexity and multi-dimensional nature of each (9) due to their documented ability to detect complex relationships and make accurate predictions (54). Compared to traditional statistical methods, ML models can consider a broader range of factors without strict, predetermined predictor and outcome parameters and make predictions with impressive accuracy (54). For example, a recent systematic review by Bichu et al. (55) found that ML predictive models have shown greater accuracy than statistic-based models in orthodontic applications. Notably, the majority of studies included in this review used ANNs as the predictive model, which parallels the current study's method and demonstrates the rationale behind integrating ML techniques. Similarly, another systematic review conducted by Adeoye et al. (32) reported that ML algorithms have satisfactory to excellent accuracy for predicting most oral cavity cancer outcomes including malignant transformation, nodal metastasis, and prognosis. However, the authors noted that the training approach required to use these models impedes their application to routine clinical practice. In contrast, the current study's model is well-suited for integration into routine clinical practice as it can be formatted into a readily usable program such as SPSS that requires minimal training. The only component of the proposed method that would require hands-on action from the clinician is the data cleaning process, which is easily replicable and outlined step-by-step above.
Sun et al. (56) emphasize the necessity of data cleaning and preprocessing in their review of text mining techniques on electronic medical records. Specifically, the authors underline the need to carefully clean and preprocess semi-structured or unstructured data (e.g., patient charts) to account for characteristics and content that can hamper analysis (e.g., grammatical/spelling errors). Ensuring sufficient preparation of such data is particularly important, as research has found that the application of text mining to medical records can expedite information extraction and enhance health-related outcomes. For example, Labrosse et al. (57) extracted information from 344 patients' electronic health records using both manual tracking and text mining techniques to identify the rare event of pregnancy following breast cancer (incidence rate of 0.01 pregnancy per person-year after breast cancer diagnosis). The authors found that text mining was more efficient than manual tracking (13 vs. 244 min, respectively) in identifying rare events in electronic health records. As digital dental records share the same unstructured texts as electronic medical records, these results demonstrate the utility of text mining in dentistry to enhance patient care by enabling precise and timely data retrieval and analysis.
In a review of the effectiveness of AI applications in endodontics, Boreak (58) reported that neural networks perform similarly to experienced professionals in terms of accuracy and precision. Interestingly, some of the studies included in the review indicated these models even outperformed the specialists. Considering these findings within the current study's context, the application of our method (which utilizes ANNs) may serve as a useful adjunct to dentists' care process by providing a second set of eyes to account for the possibility of human error. García-Pola et al. (59) reported that AI provides excellent opportunities for the automation of tasks in medical and dental practice through the detection of complex patterns. Using ML to automate patient chart review and data analysis will decrease dentists' time spent on routine work (e.g., retrieving and analyzing patients' records) and ultimately create greater opportunities for them to engage in humanizing care (i.e., the face-to-face time between dentists and their patients) and discuss their findings with patients (60). This additional time strengthens the opportunity to facilitate patients' understanding of their oral health status and capacity to self-monitor, specifically if the collection and review of the oral health information follow a continuous pattern, as is the case for periodontal disease (61). Similarly, as ML analysis results provide an easily accessible and comprehensible summary of patients' oral health status and contributing factors, they may serve as a helpful communication tool with other healthcare professionals to enhance continuity of care (62).
Study Limitations
This study's design and findings exhibit the potential utility and value of applying ML techniques to automate dental patient chart review. The application was conducted to demonstrate how the ML techniques can be applied in each step using broadly available software. However, these findings must be interpreted with caution given the preliminary nature of this research as it is a novel method applied on unstructured patient charts recruited from a private dental clinic, which might show different variabilities and qualities compared to other types of medical data. Therefore, the text mining and TM techniques used in the present study require validation to confirm their efficiency on different types of medical/dental records from other centers. Considering this, future studies should analyze larger and nationwide health records using these ML techniques to provide additional validation. However, while our method is novel, our findings are well-corroborated within the literature. Several studies have used established methods to successfully predict the same oral health outcomes we were able to identify using our proposed method (44, 63, 64), which suggests that our method was successful in finding and extracting the relevant information from patient charts.
Conclusion
Evolving technology has generated new opportunities and implications for health professions such as dentistry to adopt new advancements in computerized technology to enhance patient care, save time, enable follow-up and communication with patients and other health providers (1, 6). This study's proposed method can help meet the dental field's need for data-enabled innovations and create new areas of research in dental analytics. ML techniques such as text mining, TM and OLR and/or ANN are well-suited for application within a dental practice context and present substantial value through their ability to improve timely access to pertinent patient information and identify important predictors/contributors to oral health outcomes (12, 61). ML techniques can be also applied to other health-related fields that record patients' information as large amounts of unstructured texts, such as medicine (65). Integrating and using ML techniques to facilitate patient chart review will save time, reduce human error, provide a communication medium to use with patients and other providers, and enable practitioners and patients to locate and act upon the factors that play a major role in determining oral and general health status (7).
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by University of Alberta HREB Pro00109728. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
NA, HL, and AK was involved in the design of the study. AK provided the data for the study. NA, MH, and HL was involved in the analysis of the data and results. NA and MH was involved in the drafting of the manuscript. All authors were involved in the review and revision of the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fdmed.2022.833191/full#supplementary-material
References
1. Charangowda BK. Dental records: an overview. J Forensic Dent Sci. (2010) 2:5-−10. doi: 10.4103/0974-2948.71050
2. Arora KS, Bansal R. The use of dental records as a tool for the Unique Identification Authority of India in personal identification: a proposal. J Forens Dental Sci. (2018) 10:119-−22. doi: 10.4103/jfo.jfds_80_18
3. Gupta A, Mishra G, Bhutani H, Hoshing C, Bhalla A. Forensic revolution needs maintenance of dental records of patients by the dentists: a descriptive study. J Int Soc Prev Commun Dentistry. (2016). 6:316-−20. doi: 10.4103/2231-0762.186799
4. Starting Your Dental Practice: A Complete Guide. Chicago, IL: Council on Dental Practice, American Dental Association (2003).
5. Hadden AM The The FGDP(UK) Clinical Examination and Record-Keeping Working Group. Clinical examination and record-keeping: Part 1: Dental records. Br Dent J. (2017) 223:765-−8. doi: 10.1038/sj.bdj.2017.984
6. Bernauer SA, Zitzmann NU, Joda T. The use and performance of artificial intelligence in prosthodontics: a systematic review. Sensors. (2021) 21:6628. doi: 10.3390/s21196628
7. Ahmed N, Abbasi MS, Zuberi F, Qamar W, Halim M, Maqsood A, et al. Artificial intelligence techniques: analysis, application, and outcome in dentistry-a systematic review. BioMed Res Int. (2021) 2021:9751564. doi: 10.1155/2021/9751564
8. Jiang T, Gradus JL, Rosellini AJ. Supervised machine learning: a brief primer. Behav Ther. (2020) 51:675-−87. doi: 10.1016/j.beth.2020.05.002
9. Schwendicke F, Samek W, Krois J. Artificial Intelligence in Dentistry: Chances and Challenges. J Dent Res. (2020). 99:769-−74. doi: 10.1177/0022034520915714
10. Vougas K, Sakellaropoulos T, Kotsinas A, Foukas GP, Ntargaras A, Koinis F, et al. Machine learning and data mining frameworks for predicting drug response in cancer: an overview and a novel in silico screening process based on association rule mining. Pharmacol Ther. (2019) 203:107395. doi: 10.1016/j.pharmthera.2019.107395
11. Sultan AS, Elgharib MA, Tavares T, Jessri M, Basile JR. The use of artificial intelligence, machine learning and deep learning in oncologic histopathology. J Oral Pathol Med. (2020) 49:849-−56. doi: 10.1111/jop.13042
12. Al Turkestani N, Bianchi J, Deleat-Besson R, Le C, Tengfei L, Prieto JC, et al. Clinical decision support systems in orthodontics: a narrative review of data science approaches. Orthod Craniofac Res. (2021) 24(Suppl 2):26–36. doi: 10.1111/ocr.12492
13. Bahng J, Lee CH. Topic modeling for analyzing patients' perceptions and concerns of hearing loss on social Q&A sites: incorporating patients' perspective. Int J Environ Res Public Health. (2020) 17:6209. doi: 10.3390/ijerph17176209
14. Pirri S, Lorenzoni V, Andreozzi G, Mosca M, Turchetti G. Topic modeling and user network analysis on Twitter during world lupus awareness day. Int J Environ Res Public Health. (2020) 17:5440. doi: 10.3390/ijerph17155440
15. Chen YW, Stanley K, Att W. Artificial intelligence in dentistry: current applications and future perspectives. Quintessence Int. (2020) 51:248–57. doi: 10.3290/j.qi.a43952
16. Machoy ME, Szyszka-Sommerfeld L, Vegh A, Gedrange T, Wozniak K. The ways of using machine learning in dentistry. Adv Clin Exp Med. (2020) 29:375–84. doi: 10.17219/acem/115083
17. Speight PM, Elliott AE, Jullien JA, Downer MC, Zakrzewska JM. The use of artificial intelligence to identify people at risk of oral cancer and precancer. Br Dent J. (1995) 179:382–7. doi: 10.1038/sj.bdj.4808932
18. White SC. Computer-aided differential diagnosis of oral radiographic lesions. Dentomaxillofac Radiol. (1989) 18:53–9. doi: 10.1259/dmfr.18.2.2699592
19. Hyman JJ, Doblecki W. Computerized endodontic diagnosis. J Am Dent Assoc. (1983) 107:755–8. doi: 10.14219/jada.archive.1983.0349
20. Brickley MR, Shepherd JP. Performance of a neural network trained to make third-molar treatment-planning decisions. Med Decis Making. (1996) 16:153–60. doi: 10.1177/0272989X9601600207
21. Rennels GD, Shortliffe EH, Stockdale FE, Miller PL. A computational model of reasoning from the clinical literature. Comput Methods Programs Biomed. (1987) 24:139–49. doi: 10.1016/0169-2607(87)90025-3
22. American Dental Association. Health Policy Institute: Oral Health and Well-Being in the United States. Philadelphia, PA (2020).
23. Hackley DM, Jain S, Pagni SE, Finkelman M, Ntaganira J, Morgan JP. Oral health conditions and correlates: a National Oral Health Survey of Rwanda. Glob Health Action. (2021) 14:1904628. doi: 10.1080/16549716.2021.1904628
24. Przybyła P, Shardlow M, Aubin S, Bossy R, Eckart de Castilho R, Piperidis S, et al. Text mining resources for the life sciences. Database. (2016) 2016:baw145. doi: 10.1093/database/baw145
25. Moore R, Archer KR, Choi L. Statistical and machine learning models for classification of human wear and delivery days in accelerometry data. Sensors. (2021) 21:2726. doi: 10.3390/s21082726
26. Al-Jabery K, Obafemi-Ajayi T, Olbricht G, Wunsch D. Computational Learning Approaches to Data Analytics in Biomedical Applications. Basrah: Academic Press, An Imprint of Elsevier (2020).
27. Liu L, Tang L, Dong W, Yao S, Zhou W. An overview of topic modeling and its current applications in bioinformatics. Springerplus. (2016) 5:1608. doi: 10.1186/s40064-016-3252-8
28. Muchene L, Safari W. Two-stage topic modelling of scientific publications: a case study of University of Nairobi, Kenya. PLoS ONE. (2021) 16:e0243208. doi: 10.1371/journal.pone.0243208
30. Wahid JA, Shi L, Gao Y, Yang B, Tao Y, Wei L, et al. Topic2features: a novel framework to classify noisy and sparse textual data using LDA topic distributions. PeerJ Comput Sci. (2021) 7:e677. doi: 10.7717/peerj-cs.677
31. Jelodar H, Wang Y, Yuan C, Feng X, Jiang X, Li Y, et al. Latent Dirichlet allocation (LDA) and topic modelling: models, applications, a survey. Multimed Tools Appl. (2019). 78:15169–211. doi: 10.1007/s11042-018-6894-4
32. Adeoye J, Tan JY, Choi SW, Thomson P. Prediction models applying machine learning to oral cavity cancer outcomes: a systematic review. Int J Med Inform. (2021) 154:104557. doi: 10.1016/j.ijmedinf.2021.104557
33. Larasati A, DeYong C, Slevitch L. Comparing neural network and ordinal logistic regression to analyze attitude responses. Serv Sci. (2011) 3:304–12. doi: 10.1287/serv.3.4.304
34. André Kramer AC, Petzold M, Hakeberg M, Östberg AL. Multiple socioeconomic factors and dental caries in Swedish children and adolescents. Caries Res. (2018) 52:42–50. doi: 10.1159/000481411
35. Opal S, Grag S, Jian J, Walia I. Genetic factors affecting dental caries risk. Aust Dent J. (2015) 60:2–11. doi: 10.1111/adj.12262
36. Abiodun OI, Jantan A, Omolara AE, Dada KV, Mohamed NA, Arshad H. State-of-the-art in artificial neural network applications: a survey. Heliyon. (2018) 4:e00938. doi: 10.1016/j.heliyon.2018.e00938
38. Schmidhuber J. Deep learning in neural networks: an overview. Neural Netw. (2015) 61:85–117. doi: 10.1016/j.neunet.2014.09.003
39. Rashmi A., Vaishali A. Importance of text data preprocessing and implementation in rapidminer. In: The First International Conference on Information Technology and Knowledge Management. Delhi (2017). p. 71–5. doi: 10.15439/2018KM46
40. Feinerer I, Hornik K, Meyer D. Text Mining Packages. R Package Version 0.7-3. (2017). Available online at: https://CRAN.R-project.org/package=TM
41. Arnold T, Tilton L. Basic text processing in R. In: Programming Historian 6. Richmond: Editorial Board of the Programming Historian (2017). doi: 10.46430/phen0061
42. Pang H, Liu H, Vanderbei R. The fastclime package for linear programming and large-scale precision matrix estimation in R. J Mach Learn Res. (2014) 15:489–93. Avaiable online at: http://www.jmlr.org/
43. St Sauver JL, Carr AB, Yawn BP, Grossardt BR, Bock-Goodner CM, Klein LL, et al. Linking medical and dental health record data: a partnership with the Rochester Epidemiology Project. BMJ Open. (2017) 7:e012528. doi: 10.1136/bmjopen-2016-012528
44. Chuang CY, Sun HL, Ku MS. Allergic rhinitis, rather than asthma, is a risk factor for dental caries. Clin Otolaryngol. (2018) 43:131–6. doi: 10.1111/coa.12912
45. Zhao W, Chen JJ, Perkins R, Liu Z, Ge W, Ding Y, et al. A heuristic approach to determine an appropriate number of topics in topic modeling. BMC Bioinformatics. (2015) 16(Suppl 13):S8. doi: 10.1186/1471-2105-16-S13-S8
46. Cao J, Xia T, Li J, Zhang Y, Tang S. A density-based method for adaptive lda model selection. Neurocomputing. (2009) 72: 1775–81. doi: 10.1016/j.neucom.2008.06.011
47. Arun R, Suresh V, Veni Madhavan CE, Murthy MNN. On finding the natural number of topics with latent dirichlet allocation: some observations. In: Zaki MJ, Yu JX, Ravindran B, Pudi V, editors. Advances in Knowledge Discovery and Data Mining. Berlin; Heidelberg: Springer (2010). p. 391–402. doi: 10.1007/978-3-642-13657-3_43
48. Deveaud T, SanJuan E, Bellot P. Accurate and effective latent concept modeling for ad hoc information retrieval. Doc Num. (2014) 1:61–84. doi: 10.3166/dn.17.1.61-84
49. Murzintcev N. ldatuning: Tuning of the Latent Dirichlet Allocation Models Parameters. R package version 1.0.0. (2019). Available online at: https://CRAN.R-project.org/package=ldatuning (accessed May 12, 2019).
50. Grün B, Hornik K. topicmodels: an R package for fitting topic models. J Stat Softw. (2011) 40:1–30. doi: 10.18637/jss.v040.i13
51. Griffiths TL, Steyvers M. Finding scientific topics. Proc Natl Acad Sci USA. (2004) 101(Suppl):5228–35. doi: 10.1073/pnas.0307752101
52. Bianchi J, de Oliveira Ruellas AC, Gonçalves JR, Paniagua B, Prieto JC, Styner M, et al. Osteoarthritis of the temporomandibular joint can be diagnosed earlier using biomarkers and machine learning. Sci Rep. (2020) 10:8012. doi: 10.1038/s41598-020-64942-0
53. Miladinović M, Mihailović B, Janković A, Tošić G, Mladenović D, Živković D, et al. Reasons for extraction obtained by artificial intelligence. Acta Fac Med Naissensis. (2010). 27:143–58. Available online at: http://www.medfak.ni.ac.rs/Acta%20Facultatis/2010/3-2010/5%20rad.pdf
54. Cooray U, Watt RG, Tsakos G, Heilmann A, Hariyama M, Yamamoto T, et al. Importance of socioeconomic factors in predicting tooth loss among older adults in Japan: evidence from a machine learning analysis. Soc Sci Med. (2021) 291:114486. doi: 10.1016/j.socscimed.2021.114486
55. Bichu YM, Hansa I, Bichu AY, Premjani P, Flores-Mir C, Vaid NR. Applications of artificial intelligence and machine learning in orthodontics: a scoping review. Prog Orthod. (2021) 22:18. doi: 10.1186/s40510-021-00361-9
56. Sun W, Cai Z, Li Y, Liu F, Fang S, Wang G. Data processing and text mining technologies on electronic medical records: a review. J Healthc Eng. (2018) 2018:4302425. doi: 10.1155/2018/4302425
57. Labrosse J, Lam T, Sebbag C, Benque M, Abdennebi I, Merckelbagh H, et al. Text mining in electronic medical records enables quick and efficient identification of pregnancy cases occurring after breast cancer. JCO Clin Cancer Inform. (2019) 3:1–12. doi: 10.1200/CCI.19.00031
58. Boreak N. Effectiveness of artificial intelligence applications designed for endodontic diagnosis, decision-making, and prediction of prognosis: a systematic review. J Contemp Dent Pract. (2020) 21:926–34. doi: 10.5005/jp-journals-10024-2894
59. García-Pola M, Pons-Fuster E, Suárez-Fernández C, Seoane-Romero J, Romero-Méndez A, López-Jornet P. Role of artificial intelligence in the early diagnosis of oral cancer. A scoping review. Cancers. (2021) 13:4600. doi: 10.3390/cancers13184600
60. Israni ST, Verghese A. Humanizing artificial intelligence. JAMA. (2019) 321:29–30. doi: 10.1001/jama.2018.19398
61. -Topol E. Deep Medicine: How Artificial Intelligence Can Make Healthcare Human Again. New York, NY: Basic Books (2019).
62. Goecks J, Jalili V, Heiser LM, Gray JW. How machine learning will transform biomedicine. Cell. (2020) 181:92–101. doi: 10.1016/j.cell.2020.03.022
63. Sagtani RA, Thapa S, Sagtani A. Smoking, general and oral health related quality of life - a comparative study from Nepal. Health Qual Life Outcomes. (2020) 18:257. doi: 10.1186/s12955-020-01512-y
64. Du M, Haag D, Song Y, Lynch J, Mittinty M. Examining bias and reporting in oral health prediction modeling studies. J Dent Res. (2020) 99:374–87. doi: 10.1177/0022034520903725
Keywords: neural network, topic model (LDA), clinical data analysis, oral health, machine learning (ML)
Citation: Ameli N, Gibson MP, Khanna A, Howey M and Lai H (2022) An Application of Machine Learning Techniques to Analyze Patient Information to Improve Oral Health Outcomes. Front. Dent. Med. 3:833191. doi: 10.3389/fdmed.2022.833191
Received: 10 December 2021; Accepted: 31 January 2022;
Published: 04 March 2022.
Edited by:
Francisco Nociti, Universidade Estadual de Campinas, BrazilReviewed by:
Rashmi Mishra, University of Washington, United StatesVincenzo Grassia, University of Campania Luigi Vanvitelli, Italy
Copyright © 2022 Ameli, Gibson, Khanna, Howey and Lai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hollis Lai, aG9sbGlzMUB1YWxiZXJ0YS5jYQ==
 Nazila Ameli
Nazila Ameli Monica Prasad Gibson
Monica Prasad Gibson Amreesh Khanna
Amreesh Khanna Madison Howey
Madison Howey Hollis Lai
Hollis Lai