- 1Université de Caen Basse-Normandie, Normandie Université, Biology of Aquatic Organisms and Ecosystems (BOREA), Caen, France
- 2Muséum National d’Histoire Naturelle, Sorbonne Universités, BOREA, Paris, France
- 3Université Pierre et Marie Curie, BOREA, Paris, France
- 4UMR 7208 Centre National de la Recherche Scientifique, BOREA, Paris, France
- 5IRD 207, L’Institut de recherche pour le développement, BOREA, Paris, France
Since the initial characterization of the cardioexcitatory peptide FMRFamide in the bivalve mollusk Macrocallista nimbosa, a great number of FMRFamide-like peptides (FLPs) have been identified in mollusks. FLPs were initially isolated and molecularly characterized in model mollusks using biochemical methods. The development of recombinant technologies and, more recently, of genomics has boosted knowledge on their diversity in various mollusk classes. Today, mollusk FLPs represent approximately 75 distinct RFamide peptides that appear to result from the expression of only five genes: the FMRFamide-related peptide gene, the LFRFamide gene, the luqin gene, the neuropeptide F gene, and the cholecystokinin/sulfakinin gene. FLPs display a complex spatiotemporal pattern of expression in the central and peripheral nervous system. Working as neurotransmitters, neuromodulators, or neurohormones, FLPs are involved in the control of a great variety of biological and physiological processes including cardiovascular regulation, osmoregulation, reproduction, digestion, and feeding behavior. From an evolutionary viewpoint, the major challenge will then logically concern the elucidation of the FLP repertoire of orphan mollusk classes and the way they are functionally related. In this respect, deciphering FLP signaling pathways by characterizing the specific receptors these peptides bind remains another exciting objective.
Introduction
Mollusks exhibit great morphological diversity. They have adapted to marine, freshwater, and terrestrial habitats. They have distinct reproductive strategies; some species are gonochoric, simultaneous hermaphrodite, or alternative hermaphrodite. The phylum Mollusca is divided into eight classes (1): Monoplacophora (Neopilina), Polyplacophora (chitons), Bivalvia (clams, oysters, mussels), Gastropoda (snails, slugs), Aplacophora (worm-like mollusks), Cephalopoda (squid, cuttlefish, octopus), and Scaphopoda (tusk shells). Neuropeptides play a crucial neurotransmitter, neuromodulator, or neurohormone role in the elaboration of adapted physiological and behavioral responses to environmental constraints. As a result, phylogenetic distances, together with body plan and physiological behavior diversity probably reflect differences in the composition of mollusks’ neuropeptide record and their pattern of expression. The relatively simple central nervous system (CNS) of the gastropod mollusks Aplysia californica and Lymnaea stagnalis, with large identified neurons, has made them the most widely studied species for deciphering the role of neuropeptides in the control of physiological processes and behaviors (2–4). In mollusks, among the multiplicity of neuropeptides, the cardioexcitatory neuropeptide FMRFamide was first isolated in the clam Macrocallista nimbosa (5). So far, FMRFamide along with its structurally related neuropeptides or FMRFamide-like peptides (FLPs) that display varying sizes but harbor the common C-terminal RFamide sequence probably represent the best investigated family of neuropeptides in mollusks. Initial studies essentially concerned immunocytochemical screening of mollusk tissues with antibodies raised against the tetrapeptide FMRFamide. Then, with the refinement of peptide purification and sequencing methodologies, many new related peptides were discovered. By rendering the structure of the precursors encoding this family of peptides accessible, the development of recombinant technology has progressively extended knowledge about the diversity of FLPs. Knowledge includes FMRFamide-related peptides (FaRPs) and other RFamide peptides, especially in mollusks where purification was difficult for anatomical reasons (Figure 1).
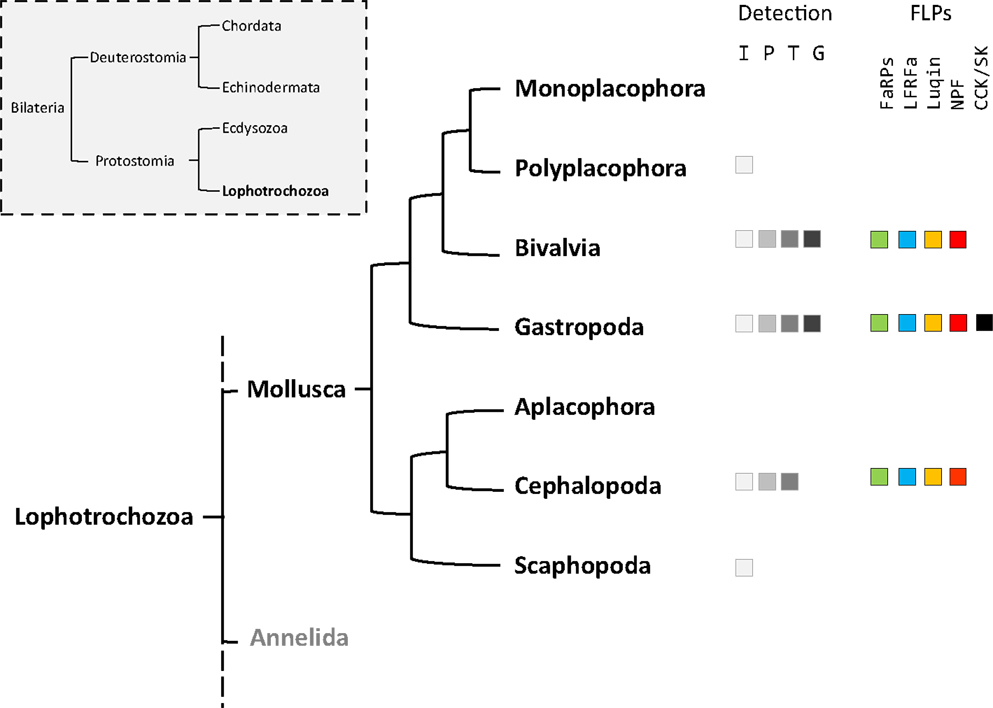
Figure 1. Phylogenic tree of mollusks based on Stöger et al. (1). The occurrence of the different FMRFamide-like peptides (FLPs) in each class is detailed. Fa, FMRFa-related peptides or FaRPs; Lr, LFRFamide peptides; Lu, Luqin; Np, NPF; Cck, cholecystokinin-/sulfakinin-like peptides. The methodologies used for FLPs detection are indicated: I, immunohistochemistry; P, peptide identification; T, transcript characterization; G, genome.
In addition to mollusks (6), the FLP family has been extensively described in all major animal phyla, from cnidarians to mammals (7, 8), and are involved in the regulation of a great variety of physiological processes (9–12). With the advent of the genomics era, bioinformatics analyses of transcriptome and genome databases now clearly suggest that the high diversity of RFamide peptides in mollusks results from the expression of only five distinct genes: the FMRF gene, the LFRFamide gene, the luqin gene, the neuropeptide F (NPF) gene, and the cholecystokinin/sulfakinin (CCK/SK)-related gene (Figure 1).
The FMRF Gene Codes for a Diversity of FMRFamide-Related Peptides
Since the discovery of the FMRFamide, a cardioactive neuropeptide in the clam M. nimbosa (5), an increasing number of studies has demonstrated the presence of this peptide and related peptides (FaRPs) in other mollusks (Table 1) and other protostomes [for reviews, see Ref. (6, 8)].
Immunohistochemistry assays using an anti-FMRFamide serum have evidenced the expression of a diversity of RFamide-like peptides in the five classes of mollusks through a variety of nervous structures and peripheral tissues. After the initial studies demonstrating the widespread distribution of FMRF-amide immunoreactivity in perikarya and nerve fibers in the central and peripheral nervous system of the pond snail L. stagnalis (29, 30), presence of FMRFamide immunoreactivity was investigated in a significant collection of mollusks. In the bivalve Placopecten magellanicus, immunoreactivity was not only concentrated in the cerebral, pedal, and parietovisceral ganglia but was also equally localized in the peripheral organs, including the gut and gills of juveniles and adults (31). In Dreissena polymorpha, immunoreactivity was observed in all nervous ganglia and in the neuromuscular system associated to the siphon and mantle (32). In the gastropod Helix aspersa, immunoreactivity revealed a highly developed peripheral nervous system where nerves containing FMRFamide were associated to the muscular fibers of the reproductive or digestive tracts (33, 34). In the cephalopod Sepia officinalis, immunoreactive neurons were detected in the CNS at the olfactory and basal-dorsal lobe of the supra-esophageal brain mass as well as in the vicinity of the optic gland (35). In this species, FMRFamide-containing motoneurons innervating the fin chromatophore muscles are localized to the posterior chromatophore and fin lobes in the posterior subesophageal mass of the brain (36). The occurrence of FLPs during mollusk development was also observed in the sensory system of chiton larvae (polyplacophores) (37) or during the development of the cerebral system, the visceral loop, and the buccal nerve cords in scaphodod larvae (38). However, the use of FMRFamide antisera recognizing the antigenic major determinant RFamide likely covers the detection of different sets of peptides.
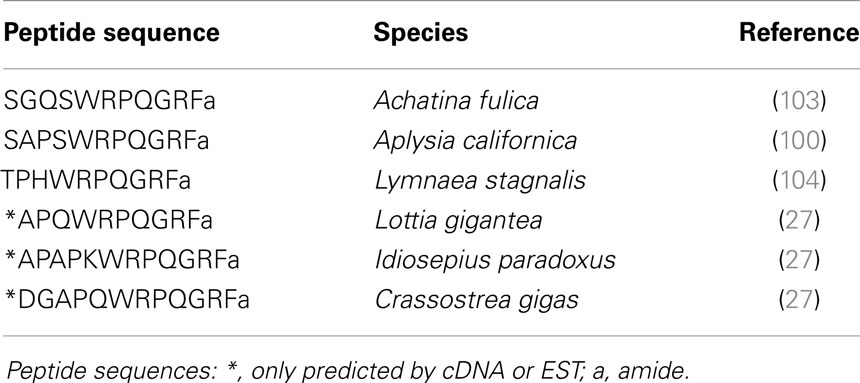
Different FLPs including FaRPs have been characterized by HPLC separation and amino acid sequencing by Edman degradation. Besides the tetrapeptide identified in several gastropod or cephalopod species, other peptides defined as FaRPs have been described; FaRPs include all the peptides with the C-terminal consensus sequence X1×2RFamide where X1 is an aromatic amino acid Phenylalanine (F), Tryptophane (W), or Tyrosine (Y) and X2 a hybrophobic amino acid Phenylalanine (F), Methionine (M), Isoleucine (I), or Leucine (L) (39). Thus, the pentapeptides AFLRFamide and TFLRFamide were identified from immunoreactive fractions of Octopus vulgaris vena cava (14). The heptapeptides SDPFLRFamide and GDPFLRFamide were identified from immunoreactive fractions in L. stagnalis (40) but also in H. aspersa where other heptapeptides (pQDPFLRFamide, NDPYLRFamide, and SEPYLRFamide) were isolated (13, 16). The first decapeptide (AdLAGDHFFRFamide) related to FaRPs was purified from Mytilus edulis based on its excitatory effect on the anterior byssus retractor muscle (ABRM) (20). This decapeptide has a d-leucine in position 2 that does not appear essential for biological activity of the peptide on ABRM (20). Surprisingly, in the venom of Conus spurius, two singular peptides inducing hyperactivity syndrome in mouse have been identified. The first one (GPMGWVPVFYRFamide) belongs to the FaRPs family (26), and the second one exhibits two gamma-carboxyglutamates and an IIRIa C-terminal sequence (GPMγDPLγIIRIa, with γ = gamma-carboxyglutamate) (41).
With the characterization of cDNAs, the number of FaRPs identified in mollusks increased and gave a more rational basis for this diversity. The first mRNA encoding FaRP precursors was characterized in A. californica (42, 43) and revealed the occurrence of 28 copies of the FMRFamide sequence associated with a single copy of FLRFamide. Thereafter, several transcripts coding for multiple copies of the FMRFamide sequence were identified in other gastropods like L. stagnalis (17) and H. aspersa (19), in the bivalve M. edulis (25) and in cephalopods such as S. officinalis (24). In cuttlefish and squid, FMRFamide transcripts encode four identical FaRPs: three tetrapeptides (FMRFamide, FLRFamide, FIRFamide) and one decapeptide (ALSGDAFLRFamide) (44). In O. vulgaris, partial cDNA predicts the presence of the same ALSGDAFLRFamide decapeptide associated to an uncommon FMKFamide tetrapeptide (45). A common model of FMRFamide precursor can be defined based on the analysis of all of these tetrapeptide encoding transcripts (Figure 2; Table 2). It contains a unique tetrabasic site corresponding to a furin-processing site (RXK/RR) that typically separates the precursor into two domains: the N-terminal region encoding the FL/IRFamide peptides and a decapeptide, while the C-terminal domain harbors the FMRFamides. As a result, peptides from the two separate domains may presumably be sorted differentially into distinct secretory vesicles, as suggested for L. stagnalis FMRF gene products (46) and demonstrated for egg-laying prohormone gene products in A. californica (47) and L. stagnalis (48).
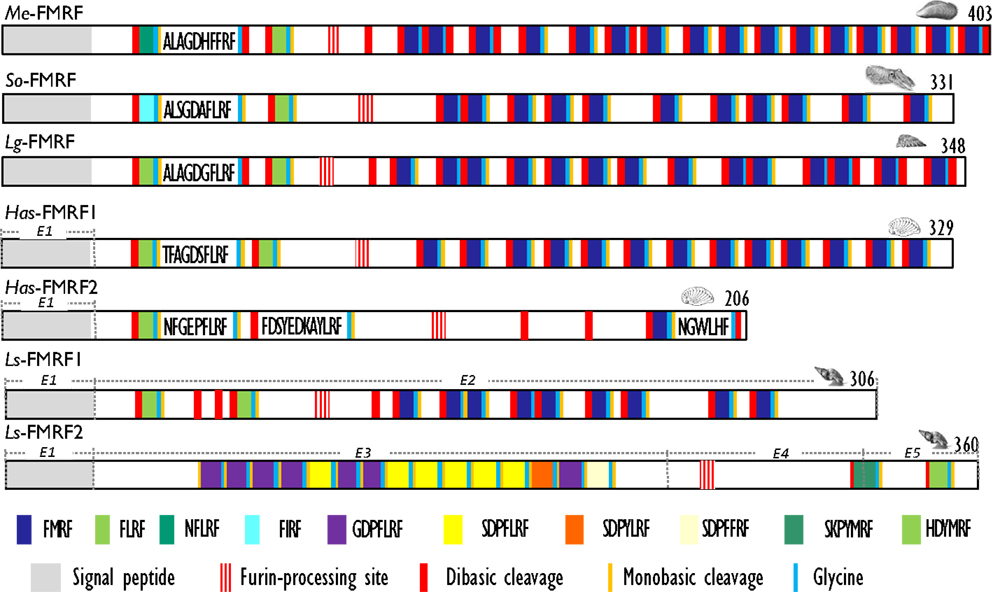
Figure 2. Comparison of the linear schematic organization of FMRFa precursors in mollusks: Me-FMRF identified in Mytilus edulis (25), So-FMRF in Sepia officinalis (24), Lg-FMRF in Lottia gigantea (27), Has-FMRF1 and Has-FMRF2 in Haliotis asinina (28), and Ls-FMRF1 and Ls-FMRF2 in Lymnaea stagnalis (18). Signal peptides, proteolytic processing sites, and C-terminal glycines for amidation are indicated by distinct labels. For Ls-FMRF1 and Ls-FMRF2 precursors, the respective encoding exons are indicated as E1, E2, E3, E4, and E5.
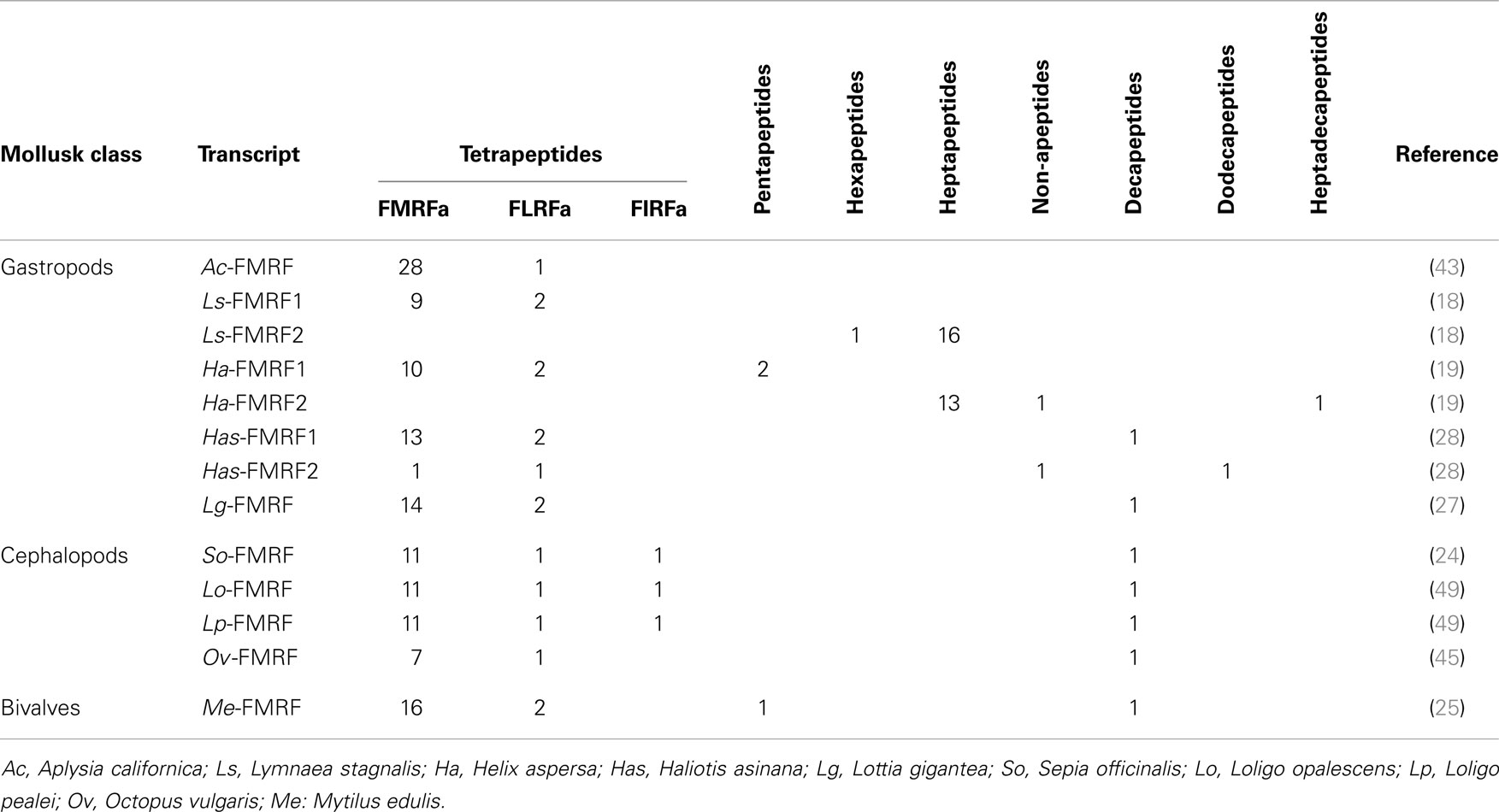
Table 2. FaRP precursors from different species of mollusks with indication of copy numbers for each peptide category.
There exists a second transcript coding for FaRPs in some gastropods. In the pulmonate mollusks L. stagnalis (17) and H. aspersa (19), this second mRNA mostly codes for heptapeptides (Table 2) and represents an alternatively spliced transcript that only shares the 5′exon with the tetrapeptide type transcript. In L. stagnalis (18), Ls-FMRF1 mRNA includes exons E1 and E2 and Ls-FMRF2 mRNA exons E1, E3, E4, and E5 (Figure 2). Singularly, two mRNAs were also characterized in the gastropod Haliotis asinana. These mRNAs share the first exon coding for the signal peptide and appear to be splice variants of a single gene, but in contrast to Lymnaea alternative transcripts they both encode copies of FMRFamide and FLRFamide along with a small number of new peptides. These two FaRP transcripts are differentially expressed in the gastropod nervous system. In adult H. aspersa snails, tetraFaRP mRNA (Ha-FMRF1) is located primarily in the cerebral ganglia, whereas heptaFaRP mRNA (Ha-FMRF2) is expressed almost exclusively in the parietal ganglia (50). In L. stagnalis, the two alternative mRNA transcripts are expressed in the CNS in a mutually exclusive manner at the single cell level; as a result, the distinct sets of FaRPs that they encode are differentially distributed in defined neuronal networks (51). FaRPs are expressed early during embryo development and larval development in mollusks with indirect development. In Lymnaea embryos, the differential localization of FaRPs could be established with specific antibodies against EFLRIa expressed by tetraFaRP mRNA (Ls-FMRF1) or against the acidic spacer peptide expressed by heptaFaRPs mRNA (Ls-FMRF2). Thus, the first neurons in Lymnaea embryos co-express the two transcript products, but neurons differentiate rapidly during ganglion development by expressing peptides of only one type of transcript. Otherwise, abundant EFLRIa-immunoreactive cells (corresponding to Ls-FMRF1) have been observed in the lip, mantle, and foot of larvae, but no peripheral cells immunoreactive to antibodies raised against the acidic peptide (corresponding to an Ls-FMRF2 cleaved product) have been found (52). In H. asinina, the two transcripts are always co-expressed in the larval ganglia but as in Lymnaea larvae, Has-FMRF1 is expressed alone in the periphery of putative sensory cells of the foot (28). In the cephalopod S. officinalis, expression of FaRP-encoding mRNAs was also observed during embryogenesis in the nervous system from the early steps of organogenesis (stage 16). Wider FaRP expression was observed concomitantly with brain differentiation (around stage 22) (53).
In mollusks, as in other invertebrates, FaRPS are involved in a variety of physiological processes including regulation of circulation, neuronal activity, feeding, digestion, reproduction, and osmoregulation. After the initial discovery of the excitatory effect of FMRFamide on the heart of the clam M. nimbosa (5), the cardioexcitatory or the cardioinhibitory effects of FaRPs on the cardiac muscle were later confirmed in other bivalves (54). In gastropods, FaRPs induced contractions of the ventricles by triggering calcium release from internal pools (55). Besides their direct activity on the heart, FaRPs also play a role in the circulatory system by acting on vessels, as shown in A. californica where the FMRFamide inhibited aorta contractions in synergy with serotonin (56). FaRPs are also involved in the control of the feeding behavior in mollusks, as observed in other invertebrates and for the other FLPs in vertebrates [for a review, see Ref. (57)]. The FMRFamide inhibited motoneurons B15 and B16 but excited interneurons B4 involved in the control of the feeding behavior in A. californica (58). FMRFamide affects the feeding system in other gastropods like Lymnaea and Helisoma where bath application of FMRFamide slowed down fictive feeding (59, 60). In Lymnaea, FMRFamide rapidly attenuated the rhythmic firing of buccal and cerebral ganglion neurons responsible for initiating and maintaining the repetitive motor outputs required for feeding (61). In addition, the tetrapeptide acts directly on muscles as described in Aplysia. FMRFamide modulate the activity of radula opener muscles in association with serotonin and myomodulins (62), and FaRPs play a role in digestion by modulating spontaneous gut activity or inducing contractions of the gizzard in Aplysia (63). In the gastropod Helisoma, the FMRFamide suppresses the activation of the salivary glands by directly acting on gland cells and on the effector neuron (neuron 4) (64). The effect on secretion activity was confirmed in scallop (Pecten maximus), where the FMRFamide exhibited a secretagogue effect on α-amylase secretion from the stomach-digestive gland complex (65). The FaRPs widely expressed in the CNS modulate neuronal activity. FMRFamide has a powerful inhibitory influence on bag cell neurons in A. californica by altering the properties of ion currents involved in the generation of action potentials and in the control of the resting potential (66). As bag cell neurons control egg-laying behavior, this effect on neurons can modulate a main physiological function like reproduction. Similarly, in the mollusk L. stagnalis, FMRFamide activates K+ currents that induce the inhibition of the caudodorsal neurons involved in the release of CDCH, the egg-laying hormone (ELH) (67). In cephalopods, FaRPs also appear to be involved in the control of reproduction regulation processes. In Octopus, the FMRFamide inhibits the activity of the optic gland involved in sexual maturity (45), whereas in cuttlefish, FaRPs control egg laying by directly regulating oocyte transport through the oviduct; the tetrapeptides FMRFamide and FLRFamide stimulate oviduct contractions, whereas FIRFamide and ALSGDAFLRFamide lower the tonus, the frequency, and the amplitude of the contractions and thus modulate them (68). FaRPs are also involved in specific functions in cephalopods. FMRFamide induces the contraction of chromatophore muscles involved in the control of body coloration patterning (69). The establishment of neural networks at the origin of this control was monitored during cuttlefish embryogenesis. FaRP staining throughout CNS development evidenced the implementation of neuronal networks involved in the control of coloration patterns. This suggests that the involvement of FaRPs in the chromatophore control pathway takes place early during embryonic development (53). In Helisoma trivolvis, the level of FaRPs in kidney, detected by radioimmunoassay, appeared to be lower in snails kept under hypoosmotic stress than in snails kept under isosmotic conditions, suggesting an involvement in gastropod osmoregulation (70).
FMRFamide-related peptides act synergistically with other mollusk-specific neuropeptides like APGWamide-related peptides. In H. aspersa, some cells contain both the APGWamide and FMRFamide; these cells may have dual projections in both the penial nerve and the nervus cutaneous pedalis primus dexter (71). In Lymnaea, different FaRPs and APGWamide have distinct actions on the penis retractor muscle; this demonstrates a complex peptidergic regulation of the male copulation behavior (72, 73). Together, these two tetrapeptides also modulate the feeding behavior in the gastropod mollusks H. trivolvis and L. stagnalis in association with other factors like biogenic amines (serotonin, dopamine, octopamine) (74). In the bivalve M. edulis, they act in synergy to modulate the contractile activity of the ABRM (75).
In mollusks, FaRPs appear to act via FMRFamide-gated sodium channels (FaNaC) and via G protein-coupled receptors. The first FaNaC was molecularly characterized in H. aspersa (76). Only gastropod FaNaCs have been characterized so far (77–79). FaNaC is a neuronal Na+-selective channel that is directly gated by micromolar concentrations of FMRFamide and related tetrapeptides inducing a fast and partially desensitizing response, and it is thought to participate in peptidergic neurotransmission. The sensitivity of L. stagnalis FaNaC to acids suggests common ancestry with mammalian acid-sensing ion channels (ASICs) (80), strengthened by the study of Askwith and collaborators concerning the direct modulation of rat ASICs by FMRFamide (81). Among the variety of candidate G-protein-coupled receptors (GPCRs) encoded by mollusk genomes, no FaRP-specific GPCR has been functionally characterized yet. Receptor-binding assays in H. aspersa suggest the presence of two receptors, one for tetrapeptides and another for heptapeptides localized in the heart, the brain, the reproductive, and digestive systems (82, 83). In a structure–activity relationship study of FMRFmide analogs in Lymnaea, the occurrence of a single receptor was proposed and demonstrated that activation of this receptor requires the C-terminal RFamide sequence, whereas the N-terminal amino acids are involved in binding. This unique receptor mediates the transient hyperpolarizing response and the long-lasting depression of excitability of the neurosecretory caudo-dorsal cells (CDCs) (84). In the squid, Loligo pealei, a radio receptor assay stressed the critical role of the RFamide moiety for binding (85).
The LFRFamide/Short Neuropeptide F-Related Gene
LFRFamide peptides (Table 3) have been identified later. The first heptapeptides GSLFRFamide and SSLFRFamide were discovered from their inhibitory activity on F2 neurons of the prosobranch gastropod Fusinus ferrugineus (23). In parallel, in the opisthobranch A. californica, GSLFRFamide peptides were characterized, along with two other LFRFamide peptides with the STLFRFamide and GGALFRFamide amino acid sequences, from their activities on the accessory radula closer (ARC) neuromuscular system (86).
For a long time, LFRFamide peptides were found only in gastropods, yet recent studies in cephalopods and oysters have evidenced this peptide family in other mollusk classes. In the cephalopod S. officinalis, two LFRFamide peptides (GNLFRFamide and TIFRFamide) were clearly characterized from LC-MS/MS analysis, with an experimental strategy based on the presence of the RFamide moiety (90). In oyster (Crassostrea gigas), the LFRF precursor was retrieved from the GigasDatabase [CU994925] (92). The first LFRFamide precursor was identified in the pulmonate snail L. stagnalis from its overexpression following parasitization by Trichobilharzia ocellata (93). The precursor contains one of several types of LFRFamide peptides in L. stagnalis (88) and Lottia gigantea (27) (Figure 3). Sometimes, several copies of the same peptide are present as in the case of the LFRF precursor (Ac-LFRF) isolated from A. californica. In the Ac-LFRF precursor, two copies of the GGALFRFamide are associated with one copy of the GSLFRFamide, TLFRFamide, STLFRFamide, and GAGTLFRFamide sequences (89) (Figure 3).
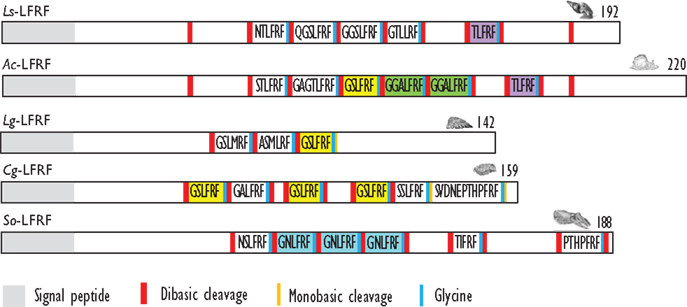
Figure 3. Comparison of the linear schematic organization of LFRFa precursors in mollusks: Ls-LFRF identified in Lymnaea stagnalis (88), Ac-LFRF in Aplysia californica (86), Lg-LFRF in Lottia gigantea (27), Cg-LFRF in Crassostrea gigas (87), and So-LFRF in Sepia officinalis (91). Signal peptides, proteolytic processing sites, and C-terminal glycines for amidation are indicated by distinct labels.
The characterization of the S. officinalis So-LFRFamide precursor revealed the presence of four different LFRF peptides with three copies of the GNLFRFamide associated to one copy of the TIRFamide, NSLRFamide, and PHTPFRFamide sequences (91) (Figure 3). The recent identification of LFRFamide peptides in the oyster C. gigas (87) confirms the widespread distribution of this peptide family in the mollusk phylum, with two of the bivalve peptides (GSLFRFamide and SSLFRFamide) previously identified in gastropods, particularly in F. ferrugineus. Similar to the So-LFRF precursor, the Cg precursor contains three copies of the GSLFRFamide peptide associated to one copy of the SSLFRFamide, GAFLRFamide, and SVDNEPTHPFRFamide sequences (Figure 3). That last sequence is located in the C-terminal moiety of the Cg-LFRFamide precursor and shares a common sequence with the last peptide (THPFRFamide) of the So-LFRFamide precursor.
Similar to FaRPs, LFRFamide peptides are involved in the regulation of main physiological functions. Studies on the localization of LFRFamide transcripts in L. stagnalis during parasitization show an expression in neurons in the buccal ganglia and in the cerebral ganglia and suggest an involvement in feeding and reproduction (88). Schistosome parasites adjust the physiology and behavior of their intermediate molluscan hosts to their own benefit by altering the expression of regulatory neuropeptides. The very close presence of LFRFamide-expressing neurons to Lymnaea CDCs and light green cells (LGCs), two cell clusters that regulate female reproduction, and growth and metabolism, respectively, together with the inhibitory activity of FLRFamide peptides on these cells suggest that the parasite induces LFRFamide gene expression to suppress host metabolism and reproduction (88). In cuttlefish, the myotropic activity of the GNLFRFamide peptide differs from that of FaRPs. Only the contractions of the rectum were modulated by this heptapeptide, but those of the oviduct were not, suggesting a role only in digestion-associated processes but not in reproduction (90). In situ hybridization (ISH) carried out in all parts of the cuttlefish CNS confirmed the involvement of LFRFamide peptides in feeding but also in learning and memory and in the control of body patterning (94, 95).
A recent study aimed at the functional characterization of a C. gigas short NPF like receptor (Cg-sNPFR-like) identified oyster LFRFamide family peptides as specific ligands (87). The Cg-sNPFR-like receptor was more abundantly expressed in the ganglia of females compared to males, and upregulated in starved oysters suggesting a role in reproduction and feeding. In the gonad area, highest receptor gene expression was observed at the start of gametogenesis, when glycogen storage activity is maximal. Oyster LFRFamide peptides are thought to play a role in the coordination of nutrition, energy storage, and metabolism through the Cg-sNPFR-like receptor by promoting storage at the expense of reproduction. The involvement of LFRFa in feeding and learning is very similar to sNPF in insects (96–98), further suggesting ancestral relationship The authors propose that mollusk FLRFamide peptides could represent functional orthologs of insect sNPFs (87).
The Luqin Gene
Using a differential screening strategy devoted to characterizing the neuropeptide-encoding transcripts specifically expressed in A. californica RF-amide immunoreactive L5 (left abdominal hemiganglion) neuron, a first precursor was characterized. L5 peptide precursor sequence predicts an RFamide decapeptide generated by signal peptide cleavage and by proteolytic cleavages at the Lys-Arg sequence. The C-terminal glycine of the resulting peptide is subsequently enzymatically converted into an amide group (99). A biochemical investigation showed that the processing of this precursor generates Luqin, a mature amidated decapeptide, together with a C-terminal proline-rich mature peptide (PRMP) of 76 amino acids (100). That latter peptide is subsequently proteolytically processed to generate two products, Luqin-B and Luqin-C (101) (Figure 4A). Alternative splicing by exon skipping of the gene encoding this precursor results in the production of a shorter PRMP with a distinct C-terminal sequence due to a reading frame shift (102). The A. californica decapeptide was named Luqin because it is differentially expressed in LUQ (left upper quadrant) neurons. Luqin displays a high degree of identity with the formerly identified cardioexcitatory undecapeptide ACEP1 isolated from atria of the African giant snail Achatina fulica (103) and with LyCEP, a decapeptide from L. stagnalis CNS isolated from its ability to activate an orphan GPCR (104). In addition to sequences from gastropod mollusks such as L. gigantea, expressed sequence tags (ESTs) encoding Luqin precursors were also identified from bivalve and cephalopod mollusks that potentially yield structurally highly conserved mature luqin peptides with only the first three amino acids exhibiting conservative substitutions (27) (Figure 4A). Interestingly, annelids also express Luqin precursors with an analogous organization and a predicted mature peptide of very similar structure (105) (Table 4). Recent studies on the evolution of neuropeptide-signaling components in animals suggest the ancestral presence of Luqin in bilaterians. Luqin has been proposed to have evolutionary links with ecdyzoan RYamide and ambulacrian RWamide precursors, as they all harbor a proline-rich C-terminal peptide displaying a conserved domain including two cysteine residues (106, 107).
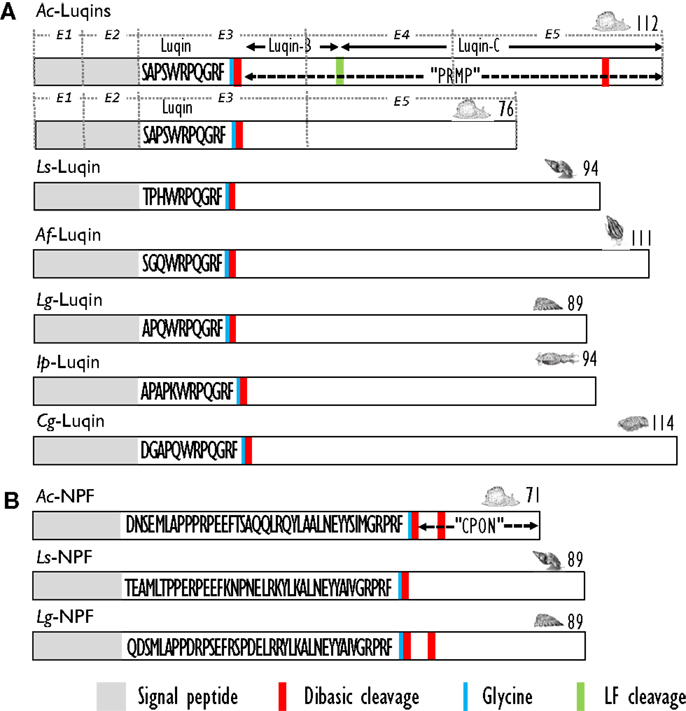
Figure 4. (A) Comparison of the linear schematic organization of Luqin precursors in mollusks: Ac-Luqin identified in Aplysia californica (101), Ls-Luqin (LyCEP: Lymnaea CardioExcitatory Peptide) in Lymnaea stagnalis (104), Af-Luqin (ACEP: Achatina CardioExcitatory Peptide) in Achatina fulica (108), Lg-Luqin in Lottia gigantea, Ip-Luqin in Idiosepius paradoxus, and Cg-Luqin in Crassostrea gigas (27). (B) Comparison of the linear schematic organization of NPF precursors in gastropods: Ac-NPF in A. californica (109), Ls-NPF in L. stagnalis (93), and Lg-NPF in Lottia gigantea (27). Signal peptides, proteolytic processing sites, and C-terminal glycines for amidation are indicated by distinct labels. CPON, carboxy-terminal peptide of NPF; PRMP, proline-rich mature peptide. For Ac-Luqin precursors, the respective encoding exons are indicated as E1, E2, E3, E4, and E5.
Luqin was initially known to enhance the amplitude of the tetanic contraction of the heart ventricle, induce the tetanic contraction of the penis retractor muscle in response to electrical stimulation in A. fulica, or enhance the phasic contraction of the ABRM produced by repetitive electrical stimulation in Mytilus (103). Besides its activity as myoactive or cardioactive peptide, Luqin functional involvement was also inferred from the pattern of expression of peptide precursors or receptor transcripts by ISH or by immunocytochemical detection (ICC) of mature Luqin peptides using specific antibodies.
Luqin-encoding transcripts or mature Luqin peptides appear to be distributed in each of the central ganglia in around 20 neurons in A. fulica (108) and approximately 100 neurons including LUQ cells in A. californica (110). In L. stagnalis, prominent immunoreactivity was mainly found in neurons in the pedal ganglia (104). In this species, the caudodorsal cells that produce ELHs are the predominant site of Luqin- (LyCEP-) specific receptor gene expression and appear to be innervated by LyCEP-containing fibers involved in their inhibition (104). Thus, Luqin seems to inhibit the egg-laying system in that snail. In L. stagnalis peripheral tissues, Luqin immunoreactivity was detected in fibers ending blindly at the pericardial lumen (104). In A. californica, Luqin distribution was consistent with each LUQ cell sending its major axon through the pericardial nerve to innervate the kidney and the renal pore, a sphincter that controls urine efflux (111). Immunoreactive fibers were detected in specific regions of the circulatory system, such as the auricle, the ventricle, the cristae, and the anterior aorta, in the reproductive system (especially in the genital ganglion), the large and small hermaphrodite ducts, and the ovotestis (110). Intestine and kidney also display immunoreactivity in widely ramifying varicose fibers (110). In kidney, both Luqin transcripts and gene products were detected in neurites located in a large nerve associated with muscles inside the renal pore (112). Besides the conventional transcript, an alternatively spliced Luqin transcript appears to be specifically transcribed in peripheral neurons located in or in the vicinity of the kidney. Its gene products may locally play a physiological role by complementing or antagonizing the action of Luqin/PRMP peptides (102). Altogether, these data suggest a role of Luqin in the control of reproduction. Given the involvement of the target organs in fluid mobilization, absorption, and secretion, Luqin is also probably involved in the regulation of water and/or salt balance in mollusks.
The Neuropeptide F (NPF) Gene
The first biochemical evidence for mollusk neuropeptide Y (NPY) homologs was provided by the amino acid sequence of a peptide isolated by means of a radioimmunoassay using an antibody raised against the C-terminal hexapeptide amide (LTRPRYamide) of mammalian pancreatic polypeptide (PP), in extracts of circumesophageal ganglia of the garden snail, H. aspersa (113). The 39 amino acid Helix peptide displayed significant homology with the vertebrate NPY/PP superfamily and with the first characterized invertebrate NPY homolog identified from the tapeworm Monieza expansa (114). Similar to its flatworm relative, the Helix C-terminal tyrosine-amide peptide (Ya) is substituted for a phenylalanine-amide peptide (Fa), so it was called NPF. Soon after the initial characterization of Helix NPF, A. californica NPF, a 40 amino acid long peptide, was isolated from the abdominal ganglia by using its distinctive, prolonged inhibitory effect on neurons L3 and L6. Next, the cDNA encoding its precursor protein was cloned and sequenced (115). A. californica NPF prohormone revealed an organization similar to vertebrate NPY precursor proteins; it had a single copy of NPF immediately following the signal peptide, and separated from the carboxy-terminal peptide of NPF (CPON) by a GKR sequence serving as combined amidation and prohormone convertase proteolytic signals (Figure 4B). At the same time, a nonapeptide with high homology with the C-terminal end of the other molluscan NPFs was purified from brain extracts of squid (Loligo vulgaris) (116). This peptide may represent a processed form of an authentic NPF peptide whose N-terminally truncated forms were also later characterized in other animal phyla [for a review, see Ref. (117)]. In L. stagnalis, a partial cDNA encoding an NPF prohormone was cloned by differential screening of a CNS-specific cDNA library of snails parasitized with T. ocellata (93) (Figure 4B). Then, the mature NPF peptide was structurally characterized after its purification to homogeneity using a reverse endocrinology approach. This method is based on the ability of fractionated brain extracts to induce changes in cAMP levels following the activation of an L. stagnalis orphan GPCR (GRL105) expressed in CHO cells (118). This receptor shares structural and functional similarity with vertebrate NPY-receptors and currently represents the only functionally characterized NPF receptor in mollusks. That a typical NPF could be isolated from the venom of the cone snail Conus betulinus (119) was somewhat intriguing though neuroendocrine-like peptides used for prey catching or defense have already been described (120). Completion of mollusk genome and transcriptome sequencing projects (91, 92) has now paved the way toward the identification of novel NPFs and their putative receptors. NPF members from owl limpet L. gigantea (27), pygmy squid Idiosepius paradoxus (121), and Pacific oyster C. gigas (87) have been identified (Table 5). In C. gigas, bioinformatics investigation of genome and transcriptome databases yielded two additional NPY sequences with a C-terminal tyrosine-amide instead of phenylalanine-amide (personal communication). All mollusk NPFs display a high degree of identity, especially among the last 20 C-terminal residues. They also have a conserved N-terminal pattern of proline residues probably involved in the formation of a polyproline helix proved essential for the three-dimensional structure of vertebrate NPYs (122).
Expression of NPF in these different species was investigated by ISH, mass spectrometry, and ICC using antibodies specific to each mollusk NPF or antibodies raised against vertebrate NPY/PP peptides, though in this later case, a cross-reaction with other FLPs cannot be discarded.
In L. stagnalis, ISH and whole-mount ICC studies revealed neurons expressing the NPF gene and peptide in all ganglia of the CNS, except the two buccal ganglia that innervate the buccal mass. In the visceral ganglion, one NPY-positive neuron projects its axon into the nervus intestinalis, which also innervates the reproductive tract including the accessory sex glands. In the CNS, NPF-positive axons run as a circular band through all ganglia passing the commissures connecting the ganglia. A close association was also found between NPF-positive axons and axons from ovulation hormone-producing neurons and molluscan insulin-like peptide-producing neurons involved in the regulation of growth (123). A subset of anterior lobe neurons expressing the gene encoding the APGWamide peptide was also shown to co-express NPF mRNA and peptide (124). These cells project via the penis nerve to the penial complex and play an important role in the control of male copulating behavior. NPF immunoreactivity was also detected in axons innervating the penis retractor muscle (124).
Aplysia californica NPF-encoding mRNAs are also widely expressed in the CNS, very abundantly in the abdominal ganglion, to a lesser extent in the pleural-pedal ganglia, and at much lower levels in the cerebral ganglion. In the abdominal ganglion, A. californica NPF coexists with ELH and other peptides processed from its precursor in bag cell neurons involved in the control of egg-laying behavior (115). In a study investigating the role of NPF in the control of feeding, whole-mount ISH and immunocytochemistry localized NPF mRNAs and peptides in several cerebral somata and in only one buccal neuron in the A. californica feeding system (109). NPF was present in fibers in all major connectives and especially in buccal ganglion afferent fibers originating in the gut-innervating esophageal nerve, a nerve involved in satiation and in the generation of egestive programs (109).
The widespread distribution of NPF immunoreactivity in the CNS and in a variety of target organs of H. aspersa was also demonstrated by ICC using antibodies specific to Helix NPF or vertebrate NPY (125). In O. vulgaris, ICC studies evidenced NPF immunoreactivity in the optic lobe and peduncle complex. In the optic gland, an endocrine organ that controls gonad maturation (126), NPF was detected in varicose fibers, suggesting a role in optic gland regulation (127).
The widespread and high expression of NPF in the CNS and the variety of target cells and organs innervated by NPF-containing fibers in mollusks emphasize the involvement of NPF signaling in the control of a number of physiological processes. In mollusks, the biological function of NPF has mainly been investigated in the two model species L. stagnalis and A. californica.
In L. stagnalis, the gene encoding NPF is upregulated in T. ocellata-parasitized animals (93). A role of NPF in the control of the energy flow was thus assumed. Indeed, parasites selectively interfere with neuroendocrine mechanisms that regulate the main determinants of the energy budget in the host’s reproduction and growth processes (128). NPF injection or implantation of slow-release peptide pellets in non-parasitized snails resulted in suppressed reproductive activity and reduced growth in a dose- and time-dependent manner (123). This activity appeared reminiscent of the well-known role of NPY in the regulation of the energy flow in vertebrates (129) but singularly differed by not affecting food intake (130). By contrast, NPF injection produced satiation-like effects in A. californica, i.e., a reduced meal-size and slower ingestion. NPF released from the esophageal nerve affects the feeding behavior; it modifies the feeding central pattern generator, a neuron network in the buccal ganglion, by eliciting egestive responses (109). Altogether, the distribution and the activity of NPF in mollusks point to a role in the coordination of the energy balance via the control of energy-consuming processes like growth and reproduction or possibly via the control of food intake. An NPF-related receptor-encoding mRNA was recently found significantly upregulated in oyster (C. gigas) lines with reduced reproductive investment and increased potential for energy production (131). Although care should be paid until the actual ligand of this receptor is identified, this result also suggests the control of NPF signaling in bivalve mollusk reproduction and energy management.
The Cholecystokinin/Sulfakinin-Related Gene
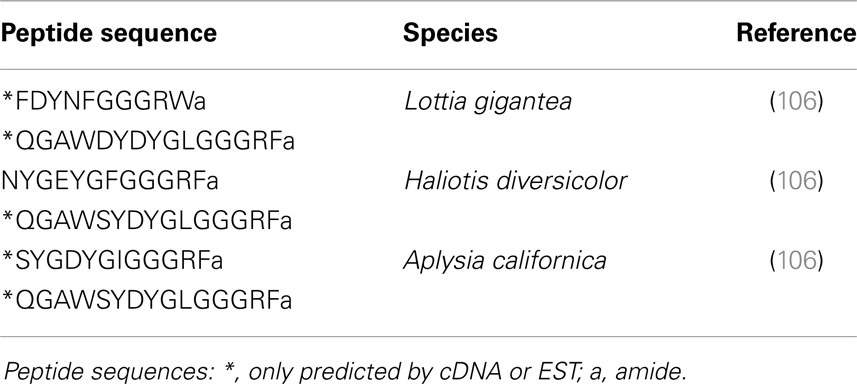
A recent extended comparative survey of peptide signaling systems in bilaterians demonstrated the early evolutionary origin of the CCK/SK signaling system (106). In this study, in silico data-mining of Haliotis diversicolor, A. californica, and L. gigantea sequence resources revealed the existence of peptides with the following consensus sequence: X(1-6)(D/E)Y(G/N)(L/F/I)GGGR(F/W)-amide. These mollusk CCK/SK peptides exhibit the C-terminal RF(W)amide sequence common to insect SKs and the DY motif shared by both insect SKs and vertebrate CCKs (Table 6). Whether mollusk peptides are sulfated remains to be investigated. As vertebrate CCKs and insect SKs reveal similar biological functions with respect to digestive enzyme secretion, satiety (food intake), and smooth muscle contraction (132), we could expect their mollusk counterparts to have retained these basic biological activities. In this respect, it is interesting to note that FMRFamide and FLPs, including insect SKs, elicit potent stimulation of the release of the digestive enzyme α-amylase from cell suspensions of the stomach-digestive gland complex of scallop (P. maximus) (65, 133, 134).
Conclusion
FMRFamide-like peptides exert pleiotropic activities and mediate a variety of physiological and behavioral processes in mollusks (Table 7). Interestingly, mollusks with diverse and well-defined habitats, life styles, and behaviors do not seem to exhibit significantly marked complexity levels in their FLP repertoires. This probably means that behavioral variety and adaptation to environmental cues result not only from the structural diversity of neuropeptide components but also from the intricacy of the spatiotemporal and stimulus-dependent patterns of expression of their encoding genes. Most of the functional assignments of this family of neuropeptides were obtained from the key model species A. californica and L. stagnalis because they have a CNS with large and reproducibly identified neurons, readily accessible and amenable to in vivo or in vitro handling. Since the activity of these neurons can also be correlated with physiological states or behaviors, establishing functional circuitries has become feasible in these species (61, 109, 135). As a result, a specific neuropeptide distribution within the CNS strongly infers its physiological involvement (Figure 5). Although sequence information is still lacking about so far completely understudied mollusk classes, such as monoplacophora, placophora, aplacophora, and scaphopoda, substantial genomic data are now becoming available in economically important classes of mollusks such as bivalves (91, 92) and cephalopods (136, 137). Such data offer the opportunity to investigate new mollusk models. These model species often have a compact and tiny CNS, so the methodologies developed for A. californica and L. stagnalis do not apply. Therefore, access to the neuropeptide-encoding gene repertoire of these non-conventional model species will necessarily require the improvement of currently operating post-genomic functional tools such as RNA interference (138) to succeed in deciphering their biological function.
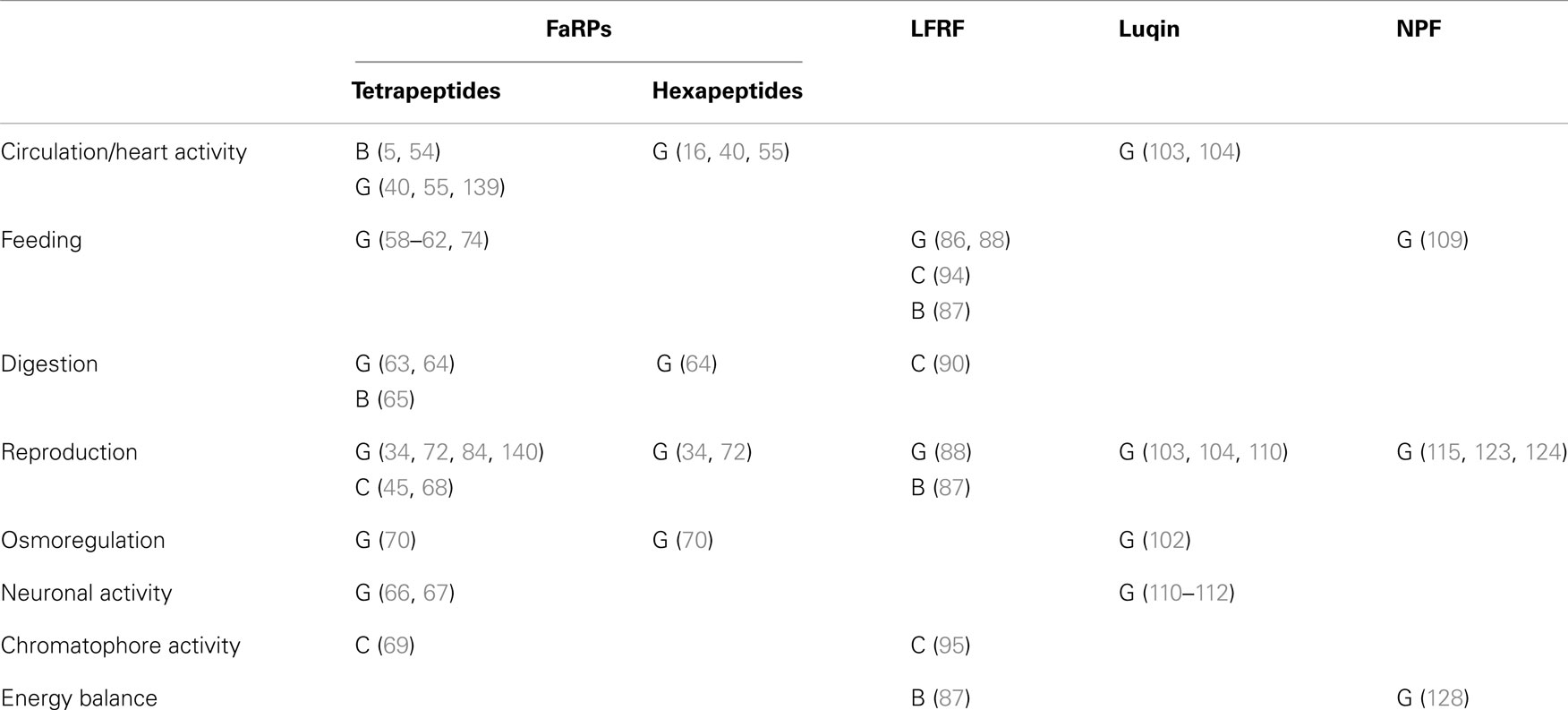
Table 7. Functional involvement of RFamide-like peptides in the three main mollusk classes: B, Bivalves; C, Cephalopods; G, Gastropods.
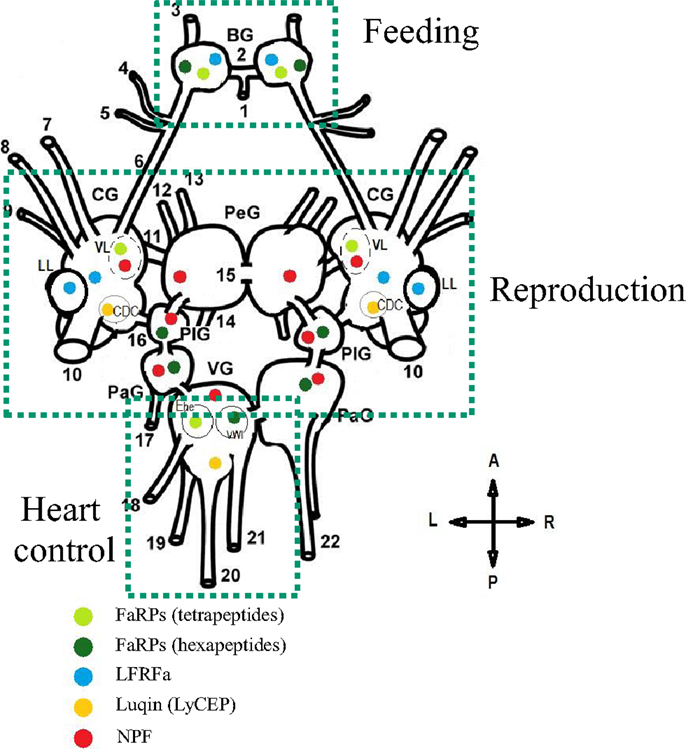
Figure 5. Schematic representation of the distribution and the role of distinct RFamide-like peptides (FLPs) in the central nervous system (dorsal view) of Lymnaea stagnalis. BG, buccal ganglion; CG, cerebral ganglion; PeG, pedal ganglion; PlG, pleural ganglion; PaG, parietal ganglion; VG, visceral ganglion; VL, ventral lobe; LL, lateral lobe; CDC, caudo-dorsal cell. Nerves: 1, postbuccal; 2, buccal commissure; 3, dorsobuccal; 4, laterobuccal; 5, ventrobuccal; 6, cerebro-buccal connective; 7, superior lip; 8, median lip; 9, tentacular; 10, cerebral commissure; 11, cerebro-pedal connective; 12, superior medial; 13, median pedal; 14, inferior pedal; 15, pedal commissure; 16, cerebro-pleural connective; 17, left parietal; 18, cutaneous; 19, anal; 20, intestinal; 21, genital; 22, right parietal. Ehe neuron and the visceral white interneuron (VWI) [Adapted from Ref. (72, 104, 130, 141, 142)].
In the field of neuroendocrinology, sequence data will become crucial for unraveling the complexity of signaling systems in an evolutionary context; genome-wide analysis gives access to the complexity of the repertoire of GPCRs, which constitute the main molecular targets of neuropeptides. In parallel, exhaustive interaction between bioinformatics and mass spectrometry analyses makes the molecular identification of most mature neuropeptides produced in a given species accessible (143). In the context of FLPs, gaining insight into the structure of the mature peptides appears crucial. Indeed, some peptides like CCK/SKs might be subjected to post-translational sulfatation. Others, like NPFs, are presumed to produce distinct truncated forms, with potentially distinct biological properties that derive from the non-conventional processing of a precursor protein (116, 144). The grouping of RFamide peptides within a same family might be somewhat misleading and restrictive, especially when considering the evolutionary links between the deuterostome and protostome NPY/F signaling systems, or between the Luqin and insect RYamide signaling systems (106, 107). Phenylalanine and tyrosine display very close physicochemical properties and the shift from one residue to the other only results from a single nucleotide mutation. We suggest that mollusk neuropeptides with the C-terminal RYamide might be included. These include the newly LFRYamide peptide family identified in Lottia (27) or neuropeptide KY (NKY) with the C-terminal RYamide initially identified in Aplysia (145) and also via the identification of ESTs encoding NKY precursors in a variety of mollusks (27). In other respects, sharing a common C-terminal RFamide moiety does not entail that the peptides are functionally or evolutionarily related. Thus, the major challenge will be to get further insight into the evolution and the functional assignment of these various FLPs, especially by focusing on poorly studied mollusk classes. One way of reaching this goal will rely on the characterization of ligand/receptor pairs thanks to the development of methodologies proven efficient in mollusks (87, 104, 118), and in other animal phyla (146–149). Such an approach was efficient to demonstrate the ancestral functional relationship between mollusk LFRFamide and insect sNPF signaling pathways (87). This indicates that sequence homology or disparity alone is not sufficient to ascribe evolutionary relatedness. Within a wide comparative context, the functional characterization of FLP/receptor couples in a variety of bilaterian or eumetozoan species will obviously help define the orthologies between the genes encoding the FLP signaling components. Then, functional and synteny analyses of these orthologous genes might generate unique data to propose convincing scenarios on the evolution of this complex family of neuropeptides in Eumetazoa.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The Guest Associate Editor Karine Rousseau declares that, despite being affiliated with the same institution as authors Céline Zatylny-Gaudin and Pascal Favrel, the review process was handled objectively and no conflict of interest exists.
Acknowledgments
This work was supported by grants from ANR (“Gametogenes” ANR-08-GENM-041) and EU FP7-KBBE-2009 (REPROSEED grant no. 245119).
References
1. Stöger I, Sigwart JD, Kano Y, Knebelsberger T, Marshall BA, Schwabe E, et al. The continuing debate on deep molluscan phylogeny: evidence for Serialia (Mollusca, Monoplacophora + Polyplacophora). Biomed Res Int (2013) 2013:407072. doi: 10.1155/2013/407072
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
2. Scheller RH, Kaldany RR, Kreiner T, Mahon AC, Nambu JR, Schaefer M, et al. Neuropeptides: mediators of behavior in Aplysia. Science (1984) 225(4668):1300–8. doi:10.1126/science.6474178
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
3. Jiménez R, Spijker S, de Schipper S, Lodder JC, Janse CK, Geraerts WP, et al. Peptidomics of a single identified neuron reveals diversity of multiple neuropeptides with convergent actions on cellular excitability. J Neurosci (2006) 26(2):518–29. doi:10.1523/JNEUROSCI.2566-05.2006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
4. El Filali Z, Van Minnen J, Liu WK, Smit AB, Li KW. Peptidomics analysis of neuropeptides involved in copulatory behavior of the mollusk Lymnaea stagnalis. J Proteome Res (2006) 5(7):1611–7. doi:10.1021/pr060014p
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
5. Price DA, Greenberg MJ. Structure of a molluscan cardioexcitatory neuropeptide. Science (1977) 197(4304):670–1. doi:10.1126/science.877582
6. López-Vera E, Aguilar MB, Heimer de la Cotera EP. FMRFamide and related peptides in the phylum Mollusca. Peptides (2008) 29(2):310–7. doi:10.1016/j.peptides.2007.09.025
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
7. Walker RJ, Papaioannou S, Holden-Dye L. A review of FMRFamide- and RFamide-like peptides in metazoa. Invert Neurosci (2009) 9(3–4):111–53. doi:10.1007/s10158-010-0097-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
8. Krajniak KG. Invertebrate FMRFamide related peptides. Protein Pept Lett (2013) 20(6):647–70. doi:10.2174/0929866511320060005
9. Mercier AJ, Friedrich R, Boldt M. Physiological functions of FMRFamide-like peptides (FLPs) in crustaceans. Microsc Res Tech (2003) 60(3):313–24. doi:10.1002/jemt.10270
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
10. Kriegsfeld LJ. Driving reproduction: RFamide peptides behind the wheel. Horm Behav (2006) 50(5):655–66. doi:10.1016/j.yhbeh.2006.06.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
11. Bechtold DA, Luckman SM. The role of RFamide peptides in feeding. J Endocrinol (2007) 192(1):3–15. doi:10.1677/JOE-06-0069
12. Mousley A, Novozhilova E, Kimber MJ, Day TA. Neuropeptide physiology in helminths. Adv Exp Med Biol (2010) 692:78–97. doi:10.1007/978-1-4419-6902-6_5
13. Price DA, Cottrell GA, Doble KE, Greenberg MJ, Jorenby W, Lehman HK, et al. A novel FMRFamide-related peptide in Helix: pQDPFLRFamide. Biol Bull (1985) 169(1):256. doi:10.2307/1541402
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
14. Martin R, Voigt KH. The neurosecretory system of the Octopus vena cava: a neurohemal organ. Experientia (1987) 43(5):537–43. doi:10.1007/BF02143582
15. Linacre A, Kellett E, Saunders S, Bright K, Benjamin PR, Burke JF. ardioactive neuropeptide Phe-Met-Arg-Phe-NH2 (FMRFamide) and novel related peptides are encoded in multiple copies by a single gene in the snail Lymnaea stagnalis. J Neurosci (1990) 10(2):412–9.
16. Price DA, Lesser W, Lee TD, Doble KE, Greenberg MJ. Seven FMRFamide-related and two SCP-related cardioactive peptides from Helix. J Exp Biol (1990) 154:421–37.
17. Saunders SE, Bright K, Kellett E, Benjamin PR, Burke JF. Neuropeptides Gly-Asp-Pro-Phe-Leu-Arg-Phe-Amide (GDPFLRFamide) and Ser-Asp-Pro-Phe-Leu-Arg-Phe-Amide (SDPFLRFamide) are encoded by an Exon 3’ to Phe-Met-Arg-Phe-NH2 (FMRFamide) in the snail Lymnaea stagnalis. J Neurosci (1991) 11(3):740–5.
18. Kellett E, Saunders SE, Li KW, Staddon JW, Benjamin PR, Burke JF. Genomic organization of the FMRFamide gene in Lymnaea: multiple exons encoding novel neuropeptides. J Neurosci (1994) 14(11 Pt 1):6564–70.
19. Lutz EM, MacDonald M, Hettle S, Price DA, Cottrell GA, Sommerville J. Structure of cDNA clones and genomic DNA FMRFamide-related peptides (FaRPs) in Helix. Mol Cell Neurosci (1992) 3(5):373–82. doi:10.1016/1044-7431(92)90049-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
20. Fujisawa Y, Ikeda T, Nomoto K, Yasuda-Kamatani Y, Minakata H, Kenny PT, et al. The FMRFamide-related decapeptide of Mytilus contains a d-amino acid residue. Comp Biochem Physiol C (1992) 102(1):91–5. doi:10.1016/0742-8413(92)90049-D
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
21. Minakata H, Ikeda T, Fujita T, Kiss T, Hiripi L, Muneoka Y, et al. Neuropeptides Isolated from Helix pomatia part 2. FMR Famide-related peptides, S-lamide peptides, FR peptides and others. In: Yanaihara N, editor. Peptide Chemistry – Japan Symposium-Escom. Leiden: ESCOM (1992). p. 579–82.
22. de With ND, van der Schors RC. SKPYMRFamide, a novel FMRFamide-related peptide in the snail Lymnaea stagnalis. Neuroreport (1992) 3(7):612–4. doi:10.1097/00001756-199207000-00017
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
23. Kuroki Y, Kanda T, Kubota I, Ikeda T, Fujisawa Y, Minakata H, et al. FMRFamide-related peptides isolated from the prosobranch mollusc Fusinus ferrugineus. Acta Biol Hung (1993) 44(1):41–4.
24. Loi PK, Tublitz N. Molecular analysis of FMRFamide- and FMRFamide-related peptides (FaRPS) in the cuttlefish Sepia officinalis. J Exp Biol (1997) 200(Pt 10):1483–9.
25. Favrel P, Lelong C, Mathieu M. Structure of the cDNA encoding the precursor for the neuropeptide FMRFamide in the bivalve mollusc Mytilus edulis. Neuroreport (1998) 9(13):2961–5. doi:10.1097/00001756-199809140-00008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
26. Maillo M, Aguilar MB, Lopéz-Vera E, Craig AG, Bulaj G, Olivera BM, et al. Conorfamide, a Conus venom peptide belonging to the RFamide family of neuropeptides. Toxicon (2002) 40(4):401–7. doi:10.1016/S0041-0101(01)00176-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
27. Veenstra JA. Neurohormones and neuropeptides encoded by the genome of Lottia gigantea, with reference to other mollusks and insects. Gen Comp Endocrinol (2010) 167(1):86–103. doi:10.1016/j.ygcen.2010.02.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
28. Cummins SF, Tollenaere A, Degnan BM, Croll RP. Molecular analysis of two FMRFamide-encoding transcripts expressed during the development of the tropical abalone Haliotis asinina. J Comp Neurol (2011) 519(10):2043–59. doi:10.1002/cne.22621
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
29. Boer HH, Schot LP, Veenstra JA, Reichelt D. Immunocytochemical identification of neural elements in the central nervous systems of a snail, some insects, a fish, and a mammal with an antiserum to the molluscan cardio-excitatory tetrapeptide FMRF-amide. Cell Tissue Res (1980) 213(1):21–7. doi:10.1007/BF00236917
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
30. Schot LP, Boer HH. Immunocytochemical demonstration of peptidergic cells in the pond snail Lymnaea stagnalis with an antiserum to the molluscan cardioactive tetrapeptide FMRF-amide. Cell Tissue Res (1982) 225(2):347–54. doi:10.1007/BF00214687
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
31. Too CK, Croll RP. Detection of FMRFamide-like immunoreactivities in the sea scallop placopecten magellanicus by immunohistochemistry and western blot analysis. Cell Tissue Res (1995) 281(2):295–304. doi:10.1007/BF00583398
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
32. Ram JL, Baidoun F, Ram ML, Croll RP. Cholinergic and peptidergic regulation of siphon/mantle function in the zebra mussel, Dreissena polymorpha. Comp Biochem Physiol C Pharmacol Toxicol Endocrinol (1997) 117(3):275–82. doi:10.1016/S0742-8413(97)00006-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
33. Cardot J, Fellman D. Immunofluorescent evidence of an FMRFamide-like peptide in the peripheral nervous system of the gastropod mollusc Helix aspersa. Neurosci Lett (1983) 43(2–3):167–72. doi:10.1016/0304-3940(83)90182-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
34. Lehman HK, Price DA. Localization of FMRFamide-like peptides in the snail Helix aspersa. J Exp Biol (1987) 131:37–53.
35. Le Gall S, Féral C, Van Minnen J, Marchand CR. Evidence for peptidergic innervation of the endocrine optic gland in Sepia by neurons showing FMRFamide-like immunoreactivity. Brain Res (1988) 462(1):83–8. doi:10.1016/0006-8993(88)90588-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
36. Loi P, Saunders R, Young D, Tublitz N. Peptidergic regulation of chromatophore function in the European cuttlefish Sepia officinalis. J Exp Biol (1996) 199(Pt 5):1177–87.
37. Haszprunar G, Friedrich S, Wanninger A, Ruthensteiner B. Fine structure and immunocytochemistry of a new chemosensory system in the Chiton larva (Mollusca: Polyplacophora). J Morphol (2002) 251(2):210–8. doi:10.1002/jmor.1084
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
38. Wanninger A, Haszprunar G. Muscle development in Antalis entalis (Mollusca, Scaphopoda) and its significance for scaphopod relationships. J Morphol (2002) 254(1):53–64. doi:10.1002/jmor.10004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
39. Espinoza E, Carrigan M, Thomas SG, Shaw G, Edison AS. A statistical view of FMRFamide neuropeptide diversity. Mol Neurobiol (2000) 21(1–2):35–56. doi:10.1385/MN:21:1-2:035
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
40. Ebberink RH, Price DA, van Loenhout H, Doble KE, Riehm JP, Geraerts WP, et al. The brain of Lymnaea contains a family of FMRFamide-like peptides. Peptides (1987) 8(3):515–22. doi:10.1016/0196-9781(87)90018-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
41. Aguilar MB, Luna-Ramírez KS, Echeverría D, Falcón A, Olivera BM, Heimer de la Cotera EP, et al. Conorfamide-Sr2, a gamma-carboxyglutamate-containing FMRFamide-related peptide from the venom of Conus spurius with activity in mice and mollusks. Peptides (2008) 29(2):186–95. doi:10.1016/j.peptides.2007.09.022
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
42. Schaefer M, Picciotto MR, Kreiner T, Taussig R, Scheller RH. Aplysia neurons express a gene encoding multiple FMRFamide neuropeptides late the cardiovascular. Cell (1984) 1985:457–67. doi:10.1016/S0092-8674(85)80019-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
43. Taussig R, Scheller RH. The Aplysia FMRFamide gene encodes sequences related to mammalian brain peptides. DNA (1986) 5(6):453–61. doi:10.1089/dna.1.1986.5.453
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
44. Sweedler JV, Li L, Floyd P, Gilly W. Mass spectrometric survey of peptides in cephalopods with an emphasis on the FMRFamide-related peptides. J Exp Biol (2000) 203(Pt 23):3565–73.
45. Di Cristo C, Delli Bovi P, Di Cosmo A. Role of FMRFamide in the reproduction of Octopus vulgaris: molecular analysis and effect on visual input. Peptides (2003) 24(10):1525–32. doi:10.1016/j.peptides.2003.07.018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
46. Santama N, Li KW, Geraerts WP, Benjamin PR, Burke JF. Post-translational processing of the alternative neuropeptide precursor encoded by the FMRFamide gene in the pulmonate snail Lymnaea stagnalis. Eur J Neurosci (1996) 8(5):968–77. doi:10.1111/j.1460-9568.1996.tb01584.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
47. Sossin WS, Fisher JM, Scheller RH. Cellular and molecular biology of neuropeptide processing and packaging. Neuron (1989) 2(5):1407–17. doi:10.1016/0896-6273(89)90186-4
48. Li KW, Jiménez CR, Van Veelen PA, Geraerts WP. Processing and targeting of a molluscan egg-laying peptide prohormone as revealed by mass spectrometric peptide fingerprinting and peptide sequencing. Endocrinology (1994) 134(4):1812–9. doi:10.1210/endo.134.4.8137747
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
49. Burbach JPH, Grant P, Hellemons AJCGM, Degiorgis JA, Li KW, Pant HC. Differential expression of the FMRF gene in adult and hatchling stellate ganglia of the squid Loligo pealei. Biol Open (2014) 3(1):50–8. doi:10.1242/bio.20136890
50. MacDonald M, Lutz EM, Lesser W, Cottrell GA, Sommerville J. Expression of mRNA encoding FMRFamide-related peptides (FaRPs) in the nervous system of Helix aspersa. Mol Cell Neurosci (1994) 5(1):23–34. doi:10.1006/mcne.1994.1003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
51. Santama N, Benjamin PR. Gene expression and function of FMRFamide-related neuropeptides in the snail Lymnaea. Microsc Res Tech (2000) 49(6):547–56. doi:10.1002/1097-0029(20000615)49:6<547::AID-JEMT5>3.0.CO;2-Y
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
52. Voronezhskaya EE, Elekes K. Expression of FMRFamide gene encoded peptides by identified neurons in embryos and juveniles of the pulmonate snail Lymnaea stagnalis. Cell Tissue Res (2003) 314(2):297–313. doi:10.1007/s00441-003-0800-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
53. Aroua S, Andouche A, Martin M, Baratte S, Bonnaud L. FaRP cell distribution in the developing CNS suggests the involvement of FaRPs in all parts of the chromatophore control pathway in Sepia officinalis (Cephalopoda). Zoology (Jena) (2011) 114(2):113–22. doi:10.1016/j.zool.2010.11.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
54. Painter SD, Greenberg MJ. A survey of the responses of bivalve hearts to the molluscan neuropeptide FMRFamide and to 5-hydroxytryptamine. Biol Bull (1982) 162(3):311. doi:10.2307/1540986
55. Moulis A. RFamide neuropeptide actions on the molluscan heart. Acta Biol Hung (2004) 55(1–4):335–41. doi:10.1556/ABiol.55.2004.1-4.39
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
56. Alevizos A, Bailey CH, Chen M, Koester J. Innervation of vascular and cardiac muscle of Aplysia by multimodal motoneuron L7. J Neurophysiol (1989) 61(5):1053–63.
57. Dockray GJ. The expanding family of RFamide peptides and their effects on feeding behaviour. Exp Physiol (2004) 89(3):229–35. doi:10.1113/expphysiol.2004.027169
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
58. Sossin WS, Kirk MD, Scheller RH. Peptidergic modulation of neuronal circuitry controlling feeding in Aplysia. J Neurosci (1987) 7(3):671–81.
59. Kyriakides MA, McCrohan CR. Effect of putative neuromodulators on rhythmic buccal motor output in Lymnaea stagnalis. J Neurobiol (1989) 20(7):635–50. doi:10.1002/neu.480200704
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
60. Murphy AD. An identified pleural ganglion interneuron inhibits patterned motor activity in the buccal ganglia of the snail, Helisoma. Brain Res (1990) 525(2):300–3. doi:10.1016/0006-8993(90)90878-F
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
61. Alania M, Sakharov DA, Elliott CJH. Multilevel Inhibition of feeding by a peptidergic pleural interneuron in the mollusc Lymnaea stagnalis. J Comp Physiol A Neuroethol Sens Neural Behav Physiol (2004) 190(5):379–90. doi:10.1007/s00359-004-0503-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
62. Evans CG, Vilim FS, Harish O, Kupfermann I, Weiss KR, Cropper EC. Modulation of radula opener muscles in Aplysia. J Neurophysiol (1999) 82(3):1339–51.
63. Austin T, Weiss S, Lukowiak K. FMRFamide effects on spontaneous and induced contractions of the anterior gizzard in Aplysia. Can J Physiol Pharmacol (1983) 61(8):949–53. doi:10.1139/y83-142
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
64. Bulloch AG, Price DA, Murphy AD, Lee TD, Bowes HN. FMRFamide peptides in Helisoma: identification and physiological actions at a peripheral synapse. J Neurosci (1988) 8(9):3459–69.
65. Favrel P, Giard W, Benlimane N, Boucaud-Camou E, Henry M. A new biological activity for the neuropeptide FMRFamide: experimental evidence for a secretagogue effect on amylase secretion in the scallop Pecten maximus. Experientia (1994) 50(11–12):1106–10. doi:10.1007/BF01923468
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
66. Fisher T, Lin CH, Kaczmarek LK. The peptide FMRFa terminates a discharge in Aplysia bag cell neurons by modulating calcium, potassium, and chloride conductances. J Neurophysiol (1993) 69(6):2164–73.
67. Kits KS, Lodder JC, Veerman MJ. Phe-Met-Arg-Phe-amide activates a novel voltage-dependent K+ current through a lipoxygenase pathway in molluscan neurones. J Gen Physiol (1997) 110(5):611–28. doi:10.1085/jgp.110.5.611
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
68. Henry J, Zatylny C, Boucaud-Camou E. Peptidergic control of egg-laying in the cephalopod Sepia officinalis: involvement of FMRFamide and FMRFamide-related peptides. Peptides (1999) 20(9):1061–70. doi:10.1016/S0196-9781(99)00102-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
69. Loi PK, Tublitz NJ. Roles of glutamate and FMRFamide-related peptides at the chromatophore neuromuscular junction in the cuttlefish, Sepia officinalis. J Comp Neurol (2000) 420(4):499–511. doi:10.1002/(SICI)1096-9861(20000515)420:4<499::AID-CNE7>3.0.CO;2-E
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
70. Madrid KP, Price DA, Greenberg MJ, Khan HR, Saleuddin AS. FMRFamide-related peptides from the kidney of the snail, Helisoma trivolvis. Peptides (1994) 15(1):31–6. doi:10.1016/0196-9781(94)90166-X
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
71. Li G, Chase R. Correlation of axon projections and peptide immunoreactivity in mesocerebral neurons of the snail Helix aspersa. J Comp Neurol (1995) 353(1):9–17. doi:10.1002/cne.903530103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
72. van Golen FA, Li KW, de Lange RP, Jespersen S, Geraerts WP. Mutually exclusive neuronal expression of peptides encoded by the FMRFa gene underlies a differential control of copulation in Lymnaea. J Biol Chem (1995) 270(47):28487–93. doi:10.1074/jbc.270.47.28487
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
73. Li KW, Smit AB, Geraerts WP. Structural and functional characterization of neuropeptides involved in the control of male mating behavior of Lymnaea stagnalis. Peptides (1992) 13(4):633–8. doi:10.1016/0196-9781(92)90166-Z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
74. Elliott CJ, Vehovszky A. Comparative pharmacology of feeding in molluscs. Acta Biol Hung (2000) 51(2–4):153–63.
75. Henry J, Zatylny C, Favrel P. HPLC and electrospray ionization mass spectrometry as tools for the identification of APGWamide-related peptides in gastropod and bivalve mollusks: comparative activities on Mytilus muscles. Brain Res (2000) 862(1–2):162–70. doi:10.1016/S0006-8993(00)02108-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
76. Lingueglia E, Champigny G, Lazdunski M, Barbry P. Cloning of the amiloride-sensitive FMRFamide peptide-gated sodium channel. Nature (1995) 378(6558):730–3. doi:10.1038/378730a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
77. Jeziorski MC, Green KA, Sommerville J, Cottrell GA. Cloning and expression of a FMRFamide-gated Na(+) channel from Helisoma trivolvis and comparison with the native neuronal channel. J Physiol (2000) 526(Pt 1):13–25. doi:10.1111/j.1469-7793.2000.00013.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
78. Perry SJ, Straub VA, Schofield MG, Burke JF, Benjamin PR. Neuronal expression of an FMRFamide-gated Na+ channel and its modulation by acid pH. J Neurosci (2001) 21(15):5559–67.
79. Furukawa Y, Miyawaki Y, Abe G. Molecular cloning and functional characterization of the Aplysia FMRFamide-gated Na+ channel. Pflügers Arch (2006) 451(5):646–56. doi:10.1007/s00424-005-1498-z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
80. Lingueglia E, Deval E, Lazdunski M. FMRFamide-gated sodium channel and ASIC channels: a new class of ionotropic receptors for FMRFamide and related peptides. Peptides (2006) 27(5):1138–52. doi:10.1016/j.peptides.2005.06.037
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
81. Askwith CC, Cheng C, Ikuma M, Benson C, Price MP, Welsh MJ. Neuropeptide FF and FMRFamide potentiate acid-evoked currents from sensory neurons and proton-gated DEG/ENaC channels. Neuron (2000) 26(1):133–41. doi:10.1016/S0896-6273(00)81144-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
82. Payza K. FMRFamide receptors in Helix aspersa. Peptides (1987) 8(6):1065–74. doi:10.1016/0196-9781(87)90138-0
83. Payza K, Greenberg MJ, Price DA. Further characterization of Helix FMRFamide receptors: kinetics, tissue distribution, and interactions with the endogenous heptapeptides. Peptides (1989) 10(3):657–61. doi:10.1016/0196-9781(89)90156-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
84. Brussaard AB, Kits KS, Ter Maat A. One receptor type mediates two independent effects of FMRFa on neurosecretory cells of Lymnaea. Peptides (1989) 10(2):289–97. doi:10.1016/0196-9781(89)90032-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
85. Chin GJ, Payza K, Price DA, Greenberg MJ, Doble KE. Characterization and solubilization of the FMRFamide receptor of squid. Biol Bull (1994) 187(2):185–99. doi:10.2307/1542241
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
86. Cropper EC, Brezina V, Vilim FS, Harish O, Price DA, Rosen S, et al. FRF peptides in the ARC neuromuscular system of Aplysia: purification and physiological actions. J Neurophysiol (1994) 72(5):2181–95.
87. Bigot L, Beets I, Dubos M-P, Boudry P, Schoofs L, Favrel P. Functional characterization of a short neuropeptide F-related receptor in a lophotrochozoan, the mollusk Crassostrea gigas. J Exp Biol (2014) 217(Pt 16):2974–82. doi:10.1242/jeb.104067
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
88. Hoek RM, Li KW, van Minnen J, Lodder JC, de Jong-Brink M, Smit AB, et al. LFRFamides: a novel family of parasitation-induced -RFamide neuropeptides that inhibit the activity of neuroendocrine cells in Lymnaea stagnalis. J Neurochem (2005) 92(5):1073–80. doi:10.1111/j.1471-4159.2004.02927.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
89. Vilim FS, Sasaki K, Rybak J, Alexeeva V, Cropper EC, Jing J, et al. Distinct mechanisms produce functionally complementary actions of neuropeptides that are structurally related but derived from different precursors. J Neurosci (2010) 30(1):131–47. doi:10.1523/JNEUROSCI.3282-09.2010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
90. Zatylny-Gaudin C, Bernay B, Zanuttini B, Leprince J, Vaudry H, Henry J. Characterization of a novel LFRFamide neuropeptide in the cephalopod Sepia officinalis. Peptides (2010) 31(2):207–14. doi:10.1016/j.peptides.2009.11.021
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
91. Zhang G, Fang X, Guo X, Li L, Luo R, Xu F, et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature (2012) 490(7418):49–54. doi:10.1038/nature11413
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
92. Fleury E, Huvet A, Lelong C, de Lorgeril J, Boulo V, Gueguen Y, et al. Generation and analysis of a 29,745 unique expressed sequence tags from the Pacific oyster (Crassostrea gigas) assembled into a publicly accessible database: the GigasDatabase. BMC Genomics (2009) 10:341. doi:10.1186/1471-2164-10-341
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
93. Hoek RM, van Kesteren RE, Smit AB, de Jong-Brink M, Geraerts WP. Altered gene expression in the host brain caused by a trematode parasite: neuropeptide genes are preferentially affected during parasitosis. Proc Natl Acad Sci U S A (1997) 94(25):14072–6. doi:10.1073/pnas.94.25.14072
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
94. Zhang Z, Tublitz NJ. Expression of the SOFaRP2 gene in the central nervous system of the adult cuttlefish Sepia officinalis. Neuropeptides (2013) 47(3):149–55. doi:10.1016/j.npep.2013.01.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
95. Zhang Z, Goodwin E, Loi PK, Tublitz NJ. Molecular analysis of a novel FMRFamide-related peptide gene (SOFaRP(2)) and its expression pattern in the brain of the European cuttlefish Sepia officinalis. Peptides (2012) 34(1):114–9. doi:10.1016/j.peptides.2011.07.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
96. Lee K-S, You KH, Choo J-K, Han Y-M, Yu K. Drosophila short neuropeptide F regulates food intake and body size. J Biol Chem (2004) 279(49):50781–9. doi:10.1074/jbc.M407842200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
97. Kapan N, Lushchak OV, Luo J, Nässel DR. Identified peptidergic neurons in the Drosophila brain regulate insulin-producing cells, stress responses and metabolism by coexpressed short neuropeptide F and corazonin. Cell Mol Life Sci (2012) 2012:4051–66. doi:10.1007/s00018-012-1097-z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
98. Knapek S, Kahsai L, Winther AM, Tanimoto H, Nässel DR. Short neuropeptide F acts as a functional neuromodulator for olfactory memory in Kenyon cells of Drosophila mushroom bodies. J Neurosci (2013) 33(12):5340–5. doi:10.1523/JNEUROSCI.2287-12.2013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
99. Shyamala M, Fisher JM, Scheller RH. A neuropeptide precursor expressed in Aplysia neuron L5. DNA (1986) 5(3):203–8. doi:10.1089/dna.1986.5.203
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
100. Aloyz RS, DesGroseillers L. Processing of the L5-67 precursor peptide and characterization of LUQIN in the LUQ neurons of Aplysia californica. Peptides (1995) 16(2):331–8. doi:10.1016/0196-9781(94)00140-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
101. Li L, Moroz TP, Garden RW, Floyd PD, Weiss KR, Sweedler JV. Mass spectrometric survey of interganglionically transported peptides in Aplysia. Peptides (1998) 19(8):1425–33. doi:10.1016/S0196-9781(98)00094-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
102. Angers A, DesGroseillers L. Alternative splicing and genomic organization of the L5-67 gene of Aplysia californica. Gene (1998) 208(2):271–7. doi:10.1016/S0378-1119(98)00009-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
103. Fujimoto K, Ohta N, Yoshida M, Kubota I, Muneoka Y, Kobayashi M. A novel cardio-excitatory peptide isolated from the atria of the African giant snail, Achatina fulica. Biochem Biophys Res Commun (1990) 167(2):777–83. doi:10.1016/0006-291X(90)92093-F
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
104. Tensen C, Cox K, Smit A, van der Schors R, Meyerhof W, Richter D, et al. The Lymnaea cardioexcitatory peptide (LyCEP) receptor: a G-protein-coupled receptor for a novel member of the RFamide neuropeptide family. J Neurosci (1998) 18(23):9812–21.
105. Veenstra JA. Neuropeptide evolution: neurohormones and neuropeptides predicted from the genomes of Capitella teleta and Helobdella robusta. Gen Comp Endocrinol (2011) 171(2):160–75. doi:10.1016/j.ygcen.2011.01.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
106. Mirabeau O, Joly J-S. Molecular evolution of peptidergic signaling systems in bilaterians. Proc Natl Acad Sci U S A (2013) 110(22):E2028–37. doi:10.1073/pnas.1219956110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
107. Jékely G. Global view of the evolution and diversity of metazoan neuropeptide signaling. Proc Natl Acad Sci U S A (2013) 110(21):8702–7. doi:10.1073/pnas.1221833110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
108. Satake H, Takuwa K, Minakata H. Characterization of cDNA and expression of mRNA encoding an Achatina cardioexcitatory RFamide peptide. Peptides (1999) 20(11):1295–302. doi:10.1016/S0196-9781(99)00134-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
109. Jing J, Vilim FS, Horn CC, Alexeeva V, Hatcher NG, Sasaki K, et al. From hunger to satiety: reconfiguration of a feeding network by Aplysia neuropeptide Y. J Neurosci (2007) 27(13):3490–502. doi:10.1523/JNEUROSCI.0334-07.2007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
110. Giardino ND, Aloyz RS, Zollinger M, Miller MW, DesGroseillers L. L5-67 and LUQ-1 peptide precursors of Aplysia californica: distribution and localization of immunoreactivity in the central nervous system and in peripheral tissues. J Comp Neurol (1996) 374(2):230–45. doi:10.1002/(SICI)1096-9861(19961014)374:2<230::AID-CNE6>3.3.CO;2-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
111. Koester J, Alevizos A. Innervation of the kidney of Aplysia by L10, the LUQ cells, and an identified peripheral motoneuron. J Neurosci (1989) 9(11):4078–88.
112. Angers A, Zappulla JP, Zollinger M, DesGroseillers L. Gene products from LUQ neurons in the abdominal ganglion are present at the renal pore of Aplysia californica. Comp Biochem Physiol B Biochem Mol Biol (2000) 126(3):435–43. doi:10.1016/S0305-0491(00)00217-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
113. Leung PS, Shaw C, Maule AG, Thim L, Johnston CF, Irvine GB. The primary structure of neuropeptide F (NPF) from the garden snail, Helix aspersa. Regul Pept (1992) 41(1):71–81. doi:10.1016/0167-0115(92)90515-V
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
114. Maule AG, Shaw C, Halton DW, Thim L, Johnston CF, Fairweather I, et al. Neuropeptide F: a novel parasitic flatworm regulatory peptide from moniezia expansa (Cestoda: Cyclophyllidea). Parasitology (1991) 102(02):309. doi:10.1017/S0031182000062648
115. Rajpara SM, Garcia PD, Roberts R, Eliassen JC, Owens DF, Maltby D, et al. Identification and molecular cloning of a neuropeptide Y homolog that produces prolonged inhibition in Aplysia Neurons. Neuron (1992) 9(3):505–13. doi:10.1016/0896-6273(92)90188-J
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
116. Smart D, Shaw C, Johnston C, Thim L, Halton D, Buchanan K. Peptide tyrosine phenylalanine: a novel neuropeptide F-related nonapeptide from the brain of the squid, Loligo vulgaris. Biochem Biophys Res Commun (1992) 186(3):1616–23. doi:10.1016/S0006-291X(05)81593-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
117. Nässel DR, Wegener C. A comparative review of short and long neuropeptide F signaling in invertebrates: any similarities to vertebrate neuropeptide Y signaling? Peptides (2011) 32(6):1335–55. doi:10.1016/j.peptides.2011.03.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
118. Tensen CP, Cox KJ, Burke JF, Leurs R, van der Schors RC, Geraerts WP, et al. Molecular cloning and characterization of an invertebrate homologue of a neuropeptide Y receptor. Eur J Neurosci (1998) 10(11):3409–16. doi:10.1046/j.1460-9568.1998.00350.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
119. Wu X, Shao X, Guo Z-Y, Chi C-W. Identification of neuropeptide Y-like conopeptides from the venom of Conus betulinus. Chin J Biochem Biophys (2010) 42(7):502–5. doi:10.1093/abbs/gmq042
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
120. Nielsen DB, Dykert J, Rivier JE, McIntosh JM. Isolation of Lys-conopressin-G from the venom of the worm-hunting snail, Conus imperialis. Toxicon (1994) 32(7):845–8. doi:10.1016/0041-0101(94)90009-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
121. Nuss AB, Forschler BT, Crim JW, TeBrugge V, Pohl J, Brown MR. Molecular characterization of neuropeptide f from the eastern subterranean termite Reticulitermes flavipes (Kollar) (Isoptera: Rhinotermitidae). Peptides (2010) 31(3):419–28. doi:10.1016/j.peptides.2009.09.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
122. Allen J, Novotný J, Martin J, Heinrich G. Molecular structure of mammalian neuropeptide Y: analysis by molecular cloning and computer-aided comparison with crystal structure of avian homologue. Proc Natl Acad Sci U S A (1987) 84(8):2532–6. doi:10.1073/pnas.84.8.2532
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
123. De Jong-Brink M, Reid CN, Tensen CP, Ter Maat A. Parasites flicking the NPY gene on the host’s switchboard: why NPY? FASEB J (1999) 13(14):1972–84.
124. De Lange RP, van Golen FA, van Minnen J. Diversity in cell specific co-expression of four neuropeptide genes involved in control of male copulation behaviour in Lymnaea stagnalis. Neuroscience (1997) 78(1):289–99. doi:10.1016/S0306-4522(96)00576-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
125. Leung PS, Shaw C, Johnston CF, Irvine GB. Immunocytochemical distribution of neuropeptide F (NPF) in the gastropod mollusc, Helix aspersa, and in several other invertebrates. Cell Tissue Res (1994) 275(2):383–93. doi:10.1007/BF00319438
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
127. Suzuki H, Muraoka T, Yamamoto T. Localization of corticotropin-releasing factor-immunoreactive nervous tissue and colocalization with neuropeptide Y-like substance in the optic lobe and peduncle complex of the Octopus (Octopus vulgaris). Cell Tissue Res (2003) 313(1):129–38. doi:10.1007/s00441-003-0734-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
128. De Jong-Brink M. How schistosomes profit from the stress responses they elicit in their hosts. Adv Parasitol (1995) 35:177–256. doi:10.1016/S0065-308X(08)60072-X
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
129. Frankish HM, Dryden S, Hopkins D, Wang Q, Williams G. Neuropeptide Y, the hypothalamus, and diabetes: insights into the central control of metabolism. Peptides (1995) 16(4):757–71. doi:10.1016/0196-9781(94)00200-P
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
130. De Jong-Brink M, Ter Maat A, Tensen CP. NPY in invertebrates: molecular answers to altered functions during evolution. Peptides (2001) 22(3):309–15. doi:10.1016/S0196-9781(01)00332-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
131. Fleury E, Moal J, Boulo V, Daniel J-Y, Mazurais D, Hénaut A, et al. Microarray-based identification of gonad transcripts differentially expressed between lines of pacific oyster selected to be resistant or susceptible to summer mortality. Mar Biotechnol (NY) (2010) 12(3):326–39. doi:10.1007/s10126-009-9227-9
132. Schoofs L, Nachman RJ, Sulfakinins. In: Kastin A, editor. Handbook of Biological Active Peptides. New York, NY: Elsevier (2006). p. 183–87.
133. Nachman RJ, Giard W, Favrel P, Suresh T, Sreekumar S. Insect myosuppressins and sulfakinins stimulate release of the digestive enzyme α-amylase in two invertebrates: the scallop Pecten maximus and insect rynchophorus ferrugineus. Ann N Y Acad Sci (1997) 814:335–8. doi:10.1111/j.1749-6632.1997.tb46178.x
134. Nachman RJ, Giard W, Lange A, Favrel P. Stimulation of alpha-amylase release in the scallop Pecten maximus by the myosuppressins. Structure-activity relationships. Ann N Y Acad Sci (1999) 897:273–81. doi:10.1111/j.1749-6632.1999.tb07898.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
135. Alania M, Vorontsov DD, Sakharov DA. Higher-order control of the feeding network in Lymnaea. Acta Biol Hung (2008) 59(Suppl):23–8. doi:10.1556/ABiol.59.2008.Suppl.3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
136. Bassaglia Y, Bekel T, Da Silva C, Poulain J, Andouche A, Navet S, et al. ESTs library from embryonic stages reveals tubulin and reflectin diversity in Sepia officinalis (Mollusca – Cephalopoda). Gene (2012) 498(2):203–11. doi:10.1016/j.gene.2012.01.100
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
137. Zhang X, Mao Y, Huang Z, Qu M, Chen J, Ding S, et al. Transcriptome analysis of the Octopus vulgaris central nervous system. PLoS One (2012) 7(6):e40320. doi:10.1371/journal.pone.0040320
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
138. Fabioux C, Corporeau C, Quillien V, Favrel P, Huvet A. In vivo RNA interference in oyster – vasa silencing inhibits germ cell development. FEBS J (2009) 276(9):2566–73. doi:10.1111/j.1742-4658.2009.06982.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
139. Moulis A, Huddart H. RFamide neuropeptide actions on the molluscan ventricle: interactions with primary neurotransmitters. Comp Biochem Physiol C Toxicol Pharmacol (2004) 142(1–2):95–102. doi:10.1016/j.cbpc.2005.10.018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
140. Brussaard AB, Kits KS, Ter Maat A, Van Minnen J, Moed PJ. Dual inhibitory action of FMRFamide on neurosecretory cells controlling egg laying behavior in the pond snail. Brain Res (1988) 447(1):35–51. doi:10.1016/0006-8993(88)90963-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
141. Worster BM, Yeoman MS, Benjamin PR. Matrix-assisted laser desorption/ionization time of flight mass spectrometric analysis of the pattern of peptide expression in single neurons resulting from alternative mRNA splicing of the FMRFamide gene. Eur J Neurosci (1998) 10(11):3498–507. doi:10.1046/j.1460-9568.1998.00361.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
142. Koene JM. Neuro-endocrine control of reproduction in hermaphroditic freshwater snails: mechanisms and evolution. Front Behav Neurosci (2010) 4:167. doi:10.3389/fnbeh.2010.00167
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
143. Boerjan B, Cardoen D, Bogaerts A, Landuyt B, Schoofs L, Verleyen P. Mass spectrometric profiling of (neuro)-peptides in the worker honeybee, Apis mellifera. Neuropharmacology (2010) 58(1):248–58. doi:10.1016/j.neuropharm.2009.06.026
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
144. Clynen E, Husson SJ, Schoofs L. Identification of new members of the (short) neuropeptide F family in locusts and Caenorhabditis elegans. Ann N Y Acad Sci (2009) 1163:60–74. doi:10.1111/j.1749-6632.2008.03624.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
145. Wickham L, Desgroseillers L. A bradykinin-like neuropeptide precursor gene is expressed in neuron L5 of Aplysia californica. DNA Cell Biol (1991) 10(4):249–58. doi:10.1089/dna.1991.10.249
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
146. Staubli F, Jorgensen TJ, Cazzamali G, Williamson M, Lenz C, Sondergaard L, et al. Molecular identification of the insect adipokinetic hormone receptors. Proc Natl Acad Sci U S A (2002) 99(6):3446–51. doi:10.1073/pnas.052556499
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
147. Janssen T, Meelkop E, Nachman RJ, Schoofs L. Evolutionary conservation of the cholecystokinin/gastrin signaling system in nematodes. Ann N Y Acad Sci (2009) 1163:428–32. doi:10.1111/j.1749-6632.2008.03649.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
148. Lindemans M, Janssen T, Beets I, Temmerman L, Meelkop E, Schoofs L. Gonadotropin-releasing hormone and adipokinetic hormone signaling systems share a common evolutionary origin. Front Endocrinol (2011) 2:16. doi:10.3389/fendo.2011.00016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
149. Beets I, Janssen T, Meelkop E, Temmerman L, Suetens N, Rademakers S, et al. Vasopressin/oxytocin-related signaling regulates gustatory associative learning in C. elegans. Science (2012) 338(6106):543–5. doi:10.1126/science.1226860
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: FaRPs, LFRFamide, luqin, NPF, CCK/SK, mollusks
Citation: Zatylny-Gaudin C and Favrel P (2014) Diversity of the RFamide peptide family in mollusks. Front. Endocrinol. 5:178. doi: 10.3389/fendo.2014.00178
Received: 15 July 2014; Accepted: 06 October 2014;
Published online: 24 October 2014.
Edited by:
Karine Rousseau, Muséum National d’Histoire Naturelle, FranceReviewed by:
Dick R. Nässel, Stockholm University, SwedenJan Adrianus Veenstra, Université de Bordeaux, France
Copyright: © 2014 Zatylny-Gaudin and Favrel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pascal Favrel, Université de Caen Basse-Normandie, Esplanade de la Paix, CS 14032, Caen Cedex 5 14032, France e-mail:cGFzY2FsLmZhdnJlbEB1bmljYWVuLmZy
 Celine Zatylny-Gaudin
Celine Zatylny-Gaudin Pascal Favrel
Pascal Favrel