- 1Juntendo University Graduate School of Medicine, Bunkyo, Japan
- 2Department of Animal Sciences, Graduate School of Agriculture and Life Sciences, The University of Tokyo, Bunkyo, Japan
- 3Department of Applied Biological Chemistry, Graduate School of Agriculture and Life Sciences, The University of Tokyo, Bunkyo, Japan
- 4Faculty of Natural System, Institute of Science and Engineering, Kanazawa University, Kanazawa, Japan
- 5Department of Oncology and Pathology, Cancer Centre Karolinska, Karolinska Institutet, Stockholm, Sweden
- 6Department of Veterinary Medical Sciences, Graduate School of Agriculture and Life Science, The University of Tokyo, Bunkyo, Japan
The short-stature homeobox-containing gene (SHOX) was originally discovered as one of genes responsible for idiopathic short-stature syndromes in humans. Previous studies in animal models have shown the evolutionarily conserved link between this gene and skeletal formation in early embryogenesis. Here, we characterized developmental roles of shox/SHOX in zebrafish embryos and human mesenchymal stem cells (hMSCs) using loss-of-function approaches. Morpholino oligo-mediated knockdown of zebrafish shox markedly hindered cell proliferation in the anterior region of the pharyngula embryos, which was accompanied by reduction in the dlx2 expression at mesenchymal core sites for future pharyngeal bones. In addition, the impaired shox expression transiently increased expression levels of skeletal differentiation genes in early larval stage. In cell culture studies, we found that hMSCs expressed SHOX; the siRNA-mediated blockade of SHOX expression significantly blunted cell proliferation in undifferentiated hMSCs but the loss of SHOX expression did augment the expressions of subsets of early osteogenic genes during early osteoblast differentiation. These data suggest that shox/SHOX maintains the population of embryonic bone progenitor cells by keeping its proliferative status and by repressing the onset of early osteogenic gene expression. The current study for the first time shows cellular and developmental responses caused by shox/SHOX deficiency in zebrafish embryos and hMSCs, and it expands our understanding of the role of this gene in early stages of skeletal growth.
Introduction
Leri–Weill dyschondrosteosis (LWD) is an inherited skeletal deformity accompanied by short stature, mesomelia, and Madelung wrist deformity (1). The LWD gene is linked to abnormalities in the pseudoautosomal region (PAR1) on the sex chromosomes, and to the short-stature homeobox-containing gene (SHOX). The SHOX gene resides within the PAR1 and was first discovered as a gene strongly linked with the LWD (2–4). The phenotypes in LWD are complex, but the major mark of the skeletal deformity is shortening of the zeugopods (5). The proliferative chondrocytes in the growth plates of the LWD patients’ fore-/hind-limb zeugopods are highly disrupted, and it is noteworthy that the zeugopodial SHOX expression in embryonic human limb buds is spatially corresponding to the defect in LWD patients (6–11). Since the most prominent feature of LWD is the defect in cartilages and bones, the primary action of the SHOX is likely exerted in chondrocytes, osteocytes, and potentially in their progenitor cells.
Bone formation is initiated during the embryonic/fetal period, and it continues until the time of adolescence. The chondrocytes and osteocytes play central roles in bone formation, and it is also well accepted that the mesenchymal stem cells (MSCs) are major skeletal progenitor cells of both these types of cells (12–14). The SHOX gene is indeed expressed in the embryonic limb bud, mesenchymal core of early pharyngeal tissues, and in hypertrophic chondrocytes of adolescent bone, suggesting crucial developmental role(s) of this gene in these cells (15, 16). Given the prominent expression of SHOX gene in early embryonic limb bud, it is plausible that SHOX is expressed in skeletal progenitor cells and controlling their cell fate thus governs bone development and skeletal growth. Despite the overt involvement of SHOX in early embryonic skeletal formation, to date, we have limited information about the developmental role of this gene. In addition, the SHOX gene does not exist in major rodent model species (such as mice and rat) (5, 17–19), which largely restricts in vivo studies on the developmental role of SHOX using rodent models.
Zebrafish is an ideal model species to study the role of SHOX gene in early embryogenesis due to its fast development and growth, easy manipulation of embryos and genes using well-established methods (20–23). Most importantly, shox presents in zebrafish genome and conserved between fish and human (24). In the previous study, we characterized the shox gene in zebrafish embryos and found that the impaired expression of shox resulted in hindered embryonic growth even before the bone mineralization became evident (25). These data suggested crucial roles of shox in early embryonic skeletal progenitor cells, but the molecular and cellular changes caused by shox deficiency were unclear. In addition, as a cell culture model, the human MSCs (hMSCs) with their established methodologies for differentiation provide an ideal system to analyze the role of genes in skeletal formation (12–14); however, the influence of SHOX deficiency in hMSCs has yet to be definitively determined. The major scope of the current study is to increase our understanding of the developmental roles of shox/SHOX. Toward this goal, we characterized the cellular and molecular changes induced by the shox/SHOX deficiency in the developing zebrafish embryo and hMSC models. Our data suggest that shox/SHOX regulates both mitogenic and differentiation events in skeletal progenitor cells in developing animals.
Materials and Methods
Materials
Chemicals and reagents were purchased from Wako (Tokyo, Japan) and Nacalai Tesque (Kyoto, Japan) unless noted otherwise. DNA ligase and restriction endonuclease were purchased from Promega (Madison, WI, USA). For reverse-transcription (RT)-PCR, Trizol reagent, M-MLV reverse transcriptase and oligonucleotide primers were purchased from Invitrogen Life Technologies (Invitrogen, Carlsbad, CA, USA). The translation block antisense-morpholino oligo (MO) was purchased from Gene Tools, LLC (Philomath, OR, USA). A human osteosarcoma cell line, U2OS cells, was obtained from the American Type Culture Collection (ATCC; HTB-96) (ATCC, Manassas, VA, USA).
Zebrafish Husbandry
Wild-type zebrafish (Danio rerio) were maintained at 28°C on a 14-h:10-h (light:dark) cycle as previously described. Embryos were obtained from natural crossings and the fertilized embryos were grown in E3 embryo rearing solution (20) at 28.5°C according to the standard method (21). For the sampling, embryos were anesthetized in tricainemesylate (ethyl 3-aminobenzoate methanesulfonate; Sigma-Aldrich Japan, Tokyo). This study was carried out in accordance with the recommended guidelines of the committee of the Life Science Research Ethics and Safety in the Graduate School of Agriculture and Life Sciences at The University of Tokyo, and the Guide for the Care and Use of Laboratory Animals prepared by Kanazawa University.
Microinjection Experiments
The translational block antisense MO against zebrafish shox mRNA and the standard control MO (ctr MO) were prepared by Gene Tools, LLC (Philomath, OR, USA) as previously reported (25), and specificity and efficiency of the shox MO were verified in that previous report. The MO-injected embryos were raised in E3 medium and were sampled at the determined time points.
Whole-Mount Immunostaining and In Situ Hybridization Analyses
For the whole-mount staining analyses, specimens were fixed by transferring to 4% paraformaldehyde for a couple of hours to overnight. The whole-mount immunostaining was conducted using anti-phospho-Histone H3 (3377, Cell Signaling Technology Japan, Tokyo, Japan), anti-active-caspase-3 (559565, BD Pharmingen™, BD Biosciences Japan, Tokyo, Japan) and anti-myosin heavy chain (05-716, Millipore, Darmstadt, Germany) according to the instruction manuals. The anti-phospho-Histone H3 and anti-active-caspase-3 antibodies were further labeled with anti-Rabbit IgG conjugated with Alexa-546 or -596 as second antibody. The signals corresponding to the myosin heavy chain (MyHC) antibody were visualized by using anti-mouse IgG conjugated with Alexa488. The specimen was counterstained with Hoechst33342. To detect specific mRNA, the fixed specimens were subjected to the whole-mount in situ hybridization using specific cRNA probes for dlx2 mRNA, pax2a mRNA, and ntl mRNA. The digoxigenin-labeled cRNA probes were prepared as previously described (26). Pictures were taken by Canon iVIS HF M52 (Canon, Tokyo, Japan) mounted on a stereomicroscope or by BZ9000 fluorescence microscope (Keyence, Tokyo, Japan).
RT-PCR and Quantitative Real-time (qRT)-PCR Analyses
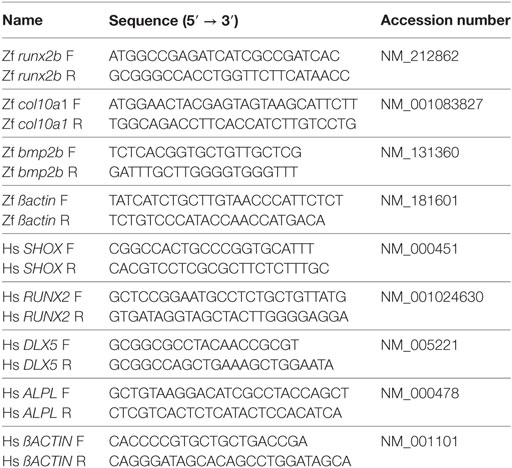
Total RNA was isolated from 10–25 zebrafish embryos or from more than 0.5 × 105 culture cells by using TRIzol reagent (Invitrogen). Total RNA (1–2 µg) was reverse-transcribed to generate the first-strand cDNA using SuperScript II reverse transcriptase according to the manufacturer’s instructions. The qPCR analysis was performed using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA, USA) or SYBR® Premix Ex Taq™ II (Tli RNaseH Plus, TaKaRa, Shiga, Japan) according to the standard method. Standard curves were constructed from serial dilution of standard samples to analyzed relative amount of the target transcript in each specimen. The amount of particular gene transcript was calculated based on the standard curve and normalized to the β-actin mRNA level. The specificity of the PCR was verified by denaturing curve analysis. Primer sets used in RT-PCR and qRT-PCR are described in the Table 1.
Reporter DNA Constructs and Dual Luciferase Assays
To construct a luciferase reporter plasmid pLuc-NPPB1940, the previously reported 1,940 bp-long promoter sequences of the NPPB gene, which contains human SHOX binding sequences, were amplified by PCR using a specific primer set (5′-AAAGTCGACAAGCTTGCTTTTTGTAGAAA-3′, 5′-AAACCATGGGTCTCTGGAGGGACTGGG-3′) (27). The amplified DNA fragment was then subcloned into the 5′-upstream cloning sites of the firefly luciferase reporter gene in the pGL3-basic vector (Promega, Madison, WI, USA) using SalI and NcoI sites. For testing the transfection efficiency, we used pRL-TK (Promega), which constitutively expresses the renilla lucierase under control of the thymidine kinase promoter. The hMSCs were transfected with 0.3 µg of pLuc-NPPB1940 in combination with 0.1 µg of pRL-TK by a polyethyleneimine-mediated lipofection method. The luciferase assay was carried out using the dual luciferase assay kit (Promega), and the firefly luciferase activities were normalized to the renilla luciferase activities.
Cell Culture and siRNA Transfection
The cells were cultured at 37.0°C under the 5% CO2 condition. For the culture of U2OS cells, Dulbecco’s modified Eagle medium supplemented with 3.5 g/L glucose, penicillin 0.25 µg/mL, streptomycin 5 µg/mL, kanamycin 10 µg/mL, amphotericin-B, and 10% fetal bovine serum was used. Human mesenchymal stem cells (hMSCs) were obtained from a commercial dealer (Poietics™, Lonza Japan, Tokyo, Japan), and the cells were cultured at 37.0°C under the 5% CO2 condition. The hMSCs were cultured in MSC growth medium (MSCGM™, Lonza Japan) supplemented with MCGS (mesenchymal cell growth supplement cell culture tested, Lonza Japan), l-glutamine, and GA-1000 (Gentamicin sulfate, amphotericin-B, Lonza Japan). For the induction of osteogenic differentiation of hMSCs, the cells were kept in Differentiation medium BulletKits™—osteogenic (osteogenic basal medium supplemented with dexamethasone, l-glutamine, ascorbate, penicillin/streptomycin MCGS, β-glycerophosphate, Lonza Japan). The siRNAs targeting human SHOX mRNA (Sense: 5′-GUGGCACCCUACGUCAACAUG-3′; Antisense: 5′-UGUUGACGUAGGGUGCCACUC-3′) and the control siRNA (Sense: 5′-GUACCGCACGUCAUUCGUAUC-3′; Antisense: 5′-UACGAAUGACGUGCGGUACGU-3′) was synthesized by RNAi Inc. (Tokyo, Japan) and introduced to the hMSCs by standard protocol using RNAiMAX Reverse Transfections Lipofectamine® reagent (Invitrogen).
Immunoprecipitation (IP) and Immunoblotting (IB)
Immunoprecipitation was performed using 0.3 mg of protein from total cell lysate by conventional method using Protein G Sepharose 4 Fast Flow (GE Healthcare UK Ltd., England) according to the instruction manual. Immunoblot analyses were conducted as previously reported (25). The antibodies for SHOX (SAB2102136, Sigma, St. Louis, MO, USA), RUNX2 (#12556 D1L7F Rabbit mAb, Cell Signaling Technology Japan, Tokyo, Japan), DLX5 (ab109737, Abcam Japan, Tokyo, Japan), and Tubulin (#2148 α/β-Tubulin Antibody, Cell Signaling Technology Japan, Tokyo, Japan) were purchased from commercial dealers and used at indicated dilutions as manufacturer’s instruction. Total protein concentration was measured by Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA), and equal amount of proteins were subjected to SDS-PAGE followed by IB analysis.
DNA Synthesis Assay
To assess DNA synthesis of siRNA-transfected cells, EdU assay was performed using Click-iT® Plus EdU Alexa Fluor® 488 Imaging Kit(Thermo Fisher Scientific)according to the instruction manual. At the 3 day post transfection of the siRNA, the hMSCs were incubated with 10 µM of EdU (5-ethyl-2′-deoxyuridine, a nucleoside analog of thymidine) under GM conditions for 10 h for labeling the S-phase cells. After the EdU-detection reaction, cells were counterstained with Hoechst33342 for counting. More than 1.0 × 103 cells were counted to calculate the rate of EdU-positive cell emergence in each experimental group.
Cell Counting of hMSCs
To compare the cellular growth in the siRNA-transfected hMSCs, the cells were suspended in BD Trucount™ Tubes (BD Biosciences, Tokyo, Japan) and cell numbers were counted using a flow cytometer FACS Verse (BD Biosciences). The relative cell numbers were calculated as the cell numbers of day-0 group of siCTR introduced group is 1.0.
Statistics
Student’s t-test was used to determine the statistical significance between two groups. Difference among multiple groups were determined by one-way analysis of variance, followed by the Tukey’s multiple comparison tests. Statistical calculation was performed using GraphPad Prism 6.0 software (GraphPad Software Inc., San Diego, CA, USA), and significance was accepted at P < 0.05.
Results
Blunted shox Expression Reduced Cell Proliferation of Developing Embryos
In a previous study, we showed the basic spatiotemporal developmental expression of shox assessed by quantitative PCR, in situ hybridization, and IB analyses, and, importantly, we also found that the loss of shox expression induced slightly reduced embryonic growth in early pharyngula stages (25). To test if the cell proliferation and cell death are controlled by early developmental shox, we analyzed the levels of phospho-Histone H3 as markers of cell proliferation and active caspase-3 as a cell death marker in shox MO or ctr MO-injected zebrafish embryos. As a result, the phospho-Histone H3 signals in shox MO-injected embryos were significantly reduced in anterior regions of the embryos compared to those in ctr MO embryos (Figure 1A, phospho-Histone H3, anterior). The effect of shox knockdown was not prominent in the posterior part of the body (Figure 1A, phospho-Histone H3, posterior). We then also analyzed active caspase-3 signals in both shox MO-injected embryos and ctr MO-injected embryos. Unlike the data for phospho-Histone H3, there were no obvious differences in active caspase-3 signals between the two groups (Figure 1A, active caspase-3). These data indicate that the shox participates in the cell cycle progression rather than the regulation of cell death in zebrafish embryo model.
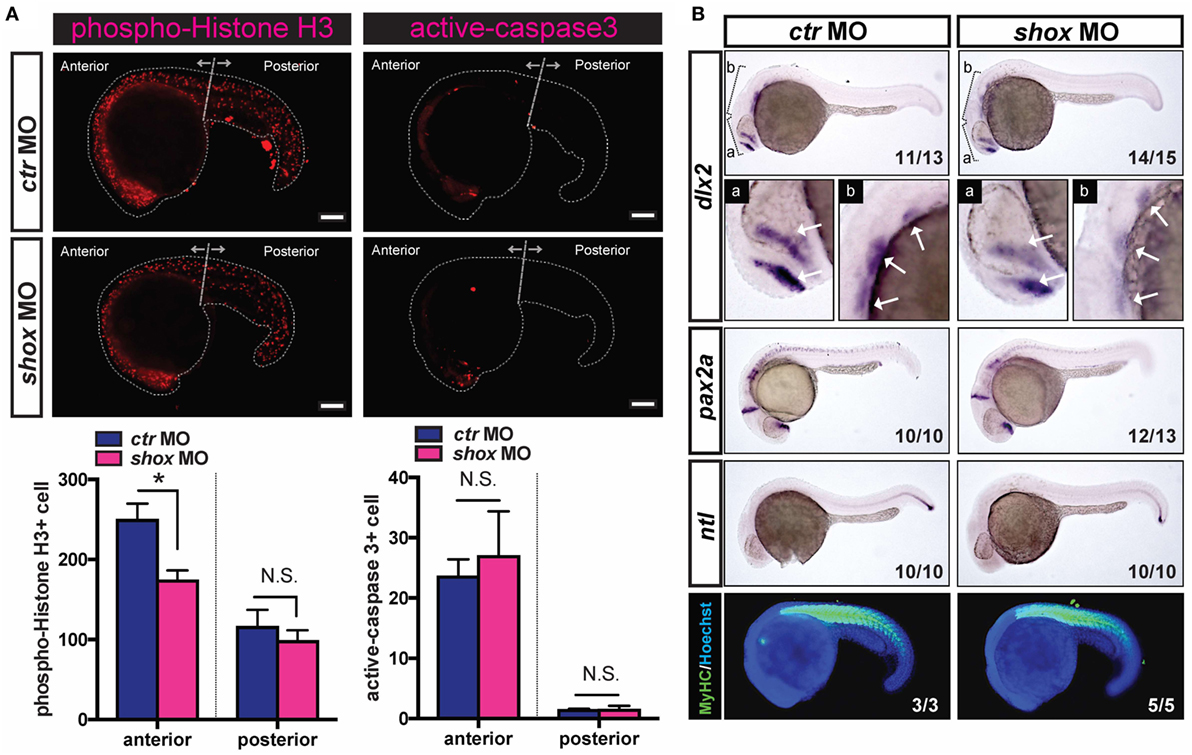
Figure 1. Translational blockade of zebrafish shox reduced cell proliferation and specific gene expression of skeletal progenitors in developing embryos. (A) Analyses of cell proliferation and cell death of shox morpholino oligo (MO) or control MO (ctr MO)-injected 22 hpf embryos. Bars, 100 µm. Representative results of phospho-Histone H3-staining and active-caspase-3-staining. Quantification data of phospho-Histone H3-staining of ctr MO-injected embryos (n = 5) and shox MO-injected embryos (n = 9). *P < 0.05. Quantification data of active caspase-3-staining of ctr MO-injected embryos (n = 3) and shox MO-injected embryos (n = 9). (B) Whole-mount in situ hybridization analysis and whole-mount immunostaining of 24 hpf embryos. The cRNA probe either for dlx2 mRNA, pax2a, or ntl mRNA was used. The arrows indicate domains with dlx2 positive signals. Whole-mount immunostaining analysis of myosin heavy chain (MyHC) (green) and Hoechst33342 (blue) was conducted. Representative results are displayed and the penetrance is shown in the lower right corner of each panel.
Impaired shox Expression Reduced dlx2 Expression in Pharyngula Stage Embryos
To investigate the effects of the blocked shox expression in embryonic development, the expression levels of several developmental marker genes were examined by whole-mount in situ hybridization. The dlx2 gene is expressed in cells at the pharyngeal mesenchymal core, which contains the progenitor cells of the craniofacial skeletal bone. The dlx2 mRNA signal levels were clearly reduced in shox MO-injected embryos (Figure 1B, dlx2). The majority of embryos had reduced dlx2 mRNA level in the shox MO-injected group as indicated by its high phenotype penetrance (14/15). On the other hand, the expressions of pax2a (labeling subsets of neurons) and ntl (labeling posterior notochord) were also analyzed. Unlike the result for dlx2, in embryos with reduced shox expression neither pax2a nor ntl expression were dramatically changed (Figure 1B, pax2a and ntl). Furthermore, the effect of shox MO on muscle tissue development was investigated by staining the MyHC in developing embryos. As a result, the MyHC signals were not altered when shox expression was reduced (Figure 1B, MyHC), suggesting that the shox is not intimately involved in skeletal muscle development at early pharyngula stage.
Knockdown of shox Transiently Increased Expression of Bone Differentiation Markers
Given the fact that the shox expression is increased in accordance with the developmental stage advances and that the loss of shox expression resulted in clearly reduced Shox protein level with defective bone ossification in our previous study (25), we have analyzed several bone differentiation marker genes in shox MO-injected embryos at 1, 3, 5, 7, and 10 days post fertilization. In ctr MO-injected embryos, the expression of runx2b, an osteogenic marker and col10a1, a chondrogenic marker, increased with embryonic growth. Expression levels of both genes peaked at 5 dpf, but in the case of col10a1, it declined thereafter (Figure 2, runx2b and col10a1, ctr MO). On the contrary, shox MO-injected embryos exhibited the increased runx2b and col10a1 expression at earlier time points: the highest expression levels of these genes were marked at 3 dpf in shox MO-injected embryos (Figure 2, runx2b and col10a1, shox MO). The expression level of bmp2b, another bone differentiation-related gene, was not significantly changed at any sampling time point examined (Figure 2, bmp2b, ctr MO, shox MO). These data indicate that shox controls expression timing of a subset of genes involved in skeletal development during early larval period.
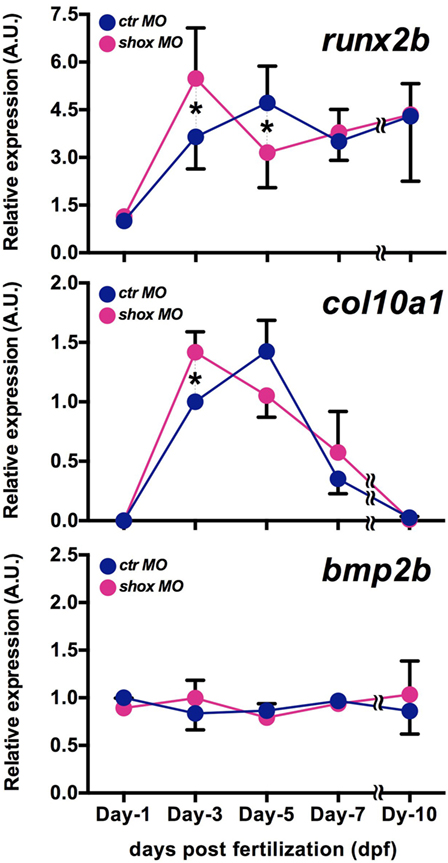
Figure 2. Translational blockade of zebrafish shox resulted in precocious expression of bone differentiation markers in developing embryos. Quantitative real-time-PCR analysis of bone differentiation markers (runx2b, col10a1, bmp2b) in morpholino oligo (MO)-injected embryos. Embryos were sampled at 1, 3, 5, 7, and 10 dpf. Transcript levels were normalized to the β-actin level, and the value is shown as relative abundance compared to that of control MO (ctr MO) at 1 dpf (runx2b, bmp2b) or at 3 dpf (col10a1). Data are mean ± SEM, 4–6 independent experiments. *P < 0.05.
SHOX Is Expressed in Human MSCs, and Its Expression Was Increased During Osteogenic Differentiation
Since the MSCs are indispensable for developing skeletal organs including cartilage and bone, and since our data showed a possibility that the expression of osteogenic differentiation marker genes were altered in shox knocked down zebrafish embryos, we focused on the role of this gene in hMSC. To begin with, expression levels of SHOX in hMSCs were measured by the qRT-PCR analysis. The U2OS cells whose SHOX expression level is known to be very low were used for comparison. Results demonstrated that the hMSCs had much higher SHOX gene expression (approximately 500 times higher) than that of U2OS cells (Figure 3A). This supports the appropriateness of the use of hMSCs for loss-of-function studies rather than the gain-of-function approach as usually applied in U2OS cells. We also examined the changes in SHOX gene expression level during osteogenic differentiation in a specific differentiation medium (osteogenic differentiation medium: O-DM). The SHOX expression was significantly increased by the 7-day long incubation in O-DM (Figure 3B). The change in endogenous SHOX activity was also tested by luciferase assay using the NPPB reporter construct that contains human SHOX binding sequences of NPPB gene promoter region as previously reported (27). The endogenous transcriptional activity of SHOX tended to be decreased within a few days post osteogenic differentiation compared to that in undifferentiated hMSCs (Figure 3C).
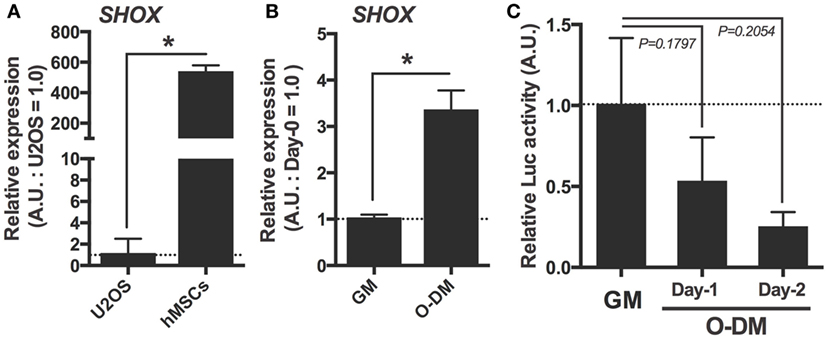
Figure 3. Expression and transcriptional activation activity of SHOX in human mesenchymal stem cells (hMSCs) under undifferentiated and under osteogenic differentiation conditions. (A) Quantitative real-time (qRT)-PCR analysis of SHOX gene in U2OS and undifferentiated hMSCs. Data are means of duplicated assays. Data are mean ± SEM, three independent experiments. *P < 0.05. (B) qRT-PCR analysis of SHOX gene in hMSCs under undifferentiated condition for 2 days or osteogenic differentiation condition for 7 days. Data are mean ± SEM, three independent experiments. *P < 0.05. GM: growth medium; O-DM: osteogenic differentiation medium (C) Relative luciferase activities of differentiating hMSCs transfected with luciferase reporter plasmid. At 12 h after transfection, the cells were collected and the luciferase activity was determined. Data are expressed as fold increase over the control group (cells not transfected pLuc-NPPB1940) in each experimental group. Data are mean ± SEM, 2–8 independent experiments.
siRNA-Mediated Knockdown of SHOX Expression Caused Reduced Cell Growth
The hMSCs were transfected with siRNA against human SHOX mRNA and the expression levels of SHOX in hMSCs were evaluated by both qRT-PCR analysis and IB following the IP using SHOX antibody. As shown in Figure 4A, loss of the majority of SHOX transcripts in hMSCs was confirmed. Since we did not find clear SHOX protein signals in a simple IB analysis using a total cell lysates, we performed IP to concentrate the SHOX protein in the sample. After the IP–IB analysis, in agreement with the RNA levels, we found that the SHOX protein level in siSHOX group was clearly reduced from the one in siCTR group (Figure 4B). These data validate that the siRNA-mediated knockdown of SHOX worked efficiently in hMSCs. Since the shox MO-injected fish embryos showed reduced cell proliferation, we next tested the effect of impaired SHOX on cell proliferation under undifferentiated conditions (culture in growth medium: GM). First, the DNA synthesis was tested by the thymidine analog incorporation assay (EdU assay). In our assay, 15.0–24.6% of siCTR cells showed EdU-positive signals in 10 h in growth medium, but only 3.0–5.1% of siSHOX-treated cells showed EdU-positive signals (Figure 4C). Furthermore, we directly measure the cell number by using the flow cytometry-based cell counting method. In analogy with the results in EdU assay, the cell number of siCTR group was clearly increased by 7 days post transfection (2.4 times as many as initially), while the cells transfected with siRNA against SHOX (siSHOX) showed blunted cell growth (nearly no increase from initial) (Figure 4D). Since the loss of shox reduced embryonic cell proliferation and mesenchymal core gene expression (dlx2) and since hMSCs reduced its proliferation due to loss of SHOX, it is suggested that the shox/SHOX maintains skeletal mass via regulating proliferation of bone progenitor cells such as MSCs in a cell-autonomous manner.
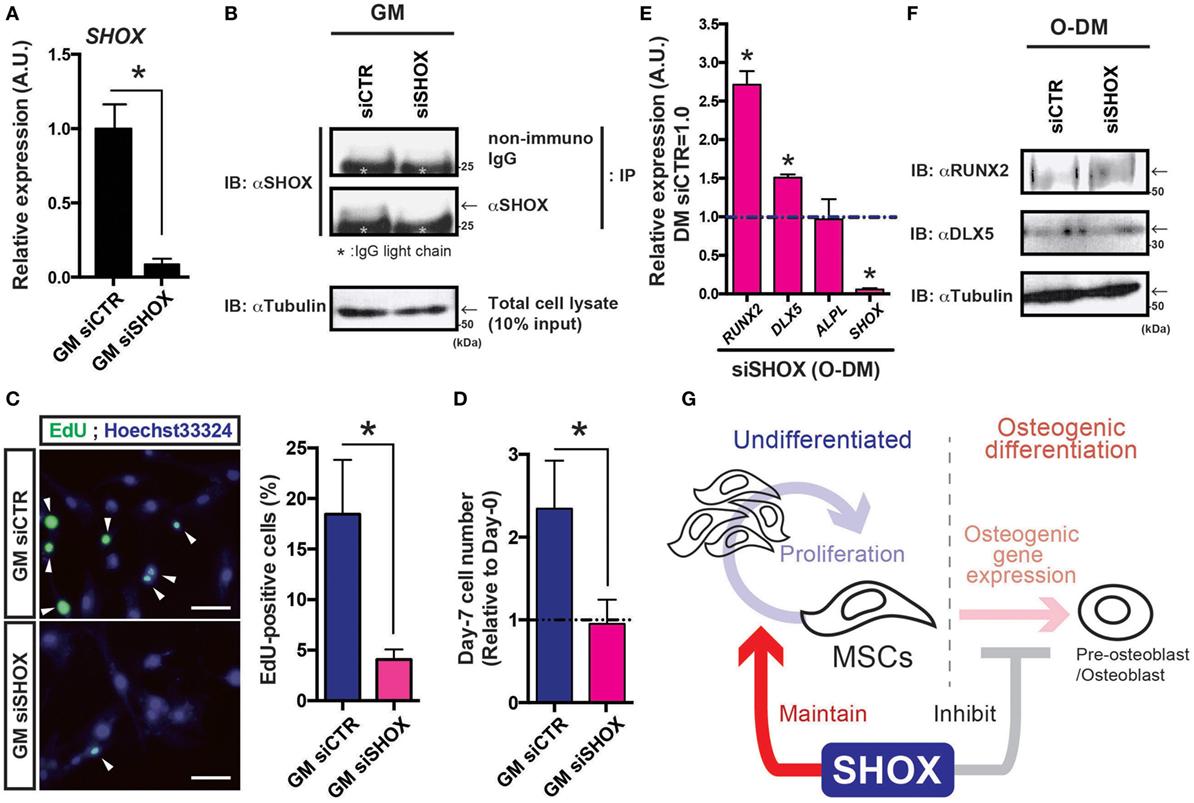
Figure 4. Reduced expression of SHOX hampered cell proliferation in human mesenchymal stem cells (hMSCs) under undifferentiated condition and disrupts osteogenic gene expression under differentiation-induction condition. (A) Changes in SHOX gene expression by siRNA in hMSCs. At the 4 days post siRNA transfection, total RNA was extracted from the cells and the levels of SHOX gene expression were examined by quantitative real-time (qRT)-PCR analysis. Data are mean ± SEM, three independent experiments. *P < 0.05. GM: growth medium. (B) Cell lysates were prepared at the 3 days post transfection of each siRNA and the changes in SHOX protein level are assessed by immunoprecipitation (IP) followed by immunoblotting (IB) using indicated antibodies/IgG. The IgG light chain is marked with asterisk. Arrows indicate the expected SHOX signal just above the IgG light chain. Tubulin was detected as an internal control. (C) The EdU assay was performed to test DNA synthesis of siRNA-treated cells under GM condition. Representative pictures and quantification data are shown. Arrowheads indicate EdU signals. Data are mean ± SEM, three independent experiments. *P < 0.05. (D) Changes in cell proliferation in undifferentiated hMSCs. The relative cell numbers are expressed by showing the value in initial (day-0) as 1.0, and the value in day-7 was compared. Data are mean ± SEM, three independent experiments. *P < 0.05. GM: growth medium (E) qRT-PCR analysis of SHOX, ALPL, DLX5, RUNX2 genes in hMSCs under osteogenic differentiation condition. Data are mean ± SEM, three independent experiments. *P < 0.05. Undiffer., undifferentiated hMSCs; Osteo., hMSCs under osteogenic differentiation at day-4. (F) Cell lysates were prepared at the 4 days post transfection of each siRNA and the changes in protein levels of RUNX2, DLX5, and tubulin were assessed by IB using indicated antibodies. Arrows indicate the expected signal. (G) Schematic illustration of working hypothesis. The shox/SHOX expressed in mesenchymal stem cells (MSCs) maintains cell proliferation in early developmental period. On the other hand, during the osteogenic differentiation, shox/SHOX may coordinate the proper timing of the specific gene expression(s) required for differentiation process. Through these actions in MSCs, shox/SHOX regulates normal skeletal growth and development.
Impaired SHOX Expression Promoted the Osteogenic Differentiation
Furthermore, the results showing advanced profile of osteogenic gene expression in shox MO-injected zebrafish led us to analyze the role of SHOX during the osteogenic differentiation of hMSCs. The hMSCs treated with each siRNA were incubated in O-DM and the osteogenic genes expressions were tested at 4-day post O-DM incubation. The SHOX transcript at day-4 post transfection of the siRNA against SHOX significantly reduced its expression level (<1/20 of siCTR) in O-DM culture (Figure 4C, SHOX). Knockdown of SHOX resulted in significant gains in RUNX2 and DLX5 expressions, both of which are early osteogenic lineage marker genes (Figure 4E, RUNX2, DLX5). Expression levels of the other osteogenic gene such as ALPL were less affected by knockdown of SHOX under osteogenic differentiation condition (Figure 4E, ALPL). The protein levels of RUNX2 and DLX5 were also measured by IB experiment (Figure 4F). In agreement with the results from RNA quantification assay, the RUNX2 protein level was clearly increased in siSHOX group (1.45-fold increase from siCTR group), and the change in DLX5 protein level was 1.19-fold increase from siCTR group, by the data of two independent experiments. The increased RUNX2 expression in siSHOX-treated hMSCs was consistent with zebrafish results, and all these results suggest that shox/SHOX cell autonomously inhibits early onset of osteogenic differentiation of growing skeleton.
Discussion
Since the SHOX was discovered, their molecular and cellular functions have been studied in various models using human osteosarcoma U2OS cells, primary human oral fibroblasts, primary human chondrocytes, chicken micromass culture, and so on (8, 18, 19, 27–31). In this study, we examined roles of shox/SHOX in zebrafish embryos and in a human bone progenitor cell (hMSCs). We found that the blockade of shox expression caused blunted cell proliferation in pharyngula embryos with reduced expression of anterior bone progenitor marker gene such as dlx2 (Figure 1). In addition, the reduced shox expression upregulated expression of bone differentiation marker genes in relatively earlier stages (Figure 2). The cell culture study demonstrated that the SHOX was expressed in undifferentiated hMSCs, and its transactivation activity tended to be reduced under osteogenic differentiation (Figure 3). The loss of SHOX expression clearly reduced both DNA synthesis and cell proliferation in undifferentiated hMSCs and it also augmented early osteogenic gene expression (Figure 4). All the data suggest that SHOX positively regulates the proliferation in the undifferentiated skeletal bone progenitor cells and negatively regulates early-stage osteogenic differentiation (Figure 4G). To our knowledge, it is the first time to examine the cellular and developmental changes caused by shox/SHOX deficiency in both early stage of developing animal and cultured MSCs. Our data would provide an insight for the deeper understanding of the role of shox/SHOX in skeletal growth and development.
In zebrafish experiments, the blunted shox expression reduced the cell proliferation in anterior region of pharyngula embryos, in conjunction with hindered expression of dlx2. Meanwhile, we failed to see obvious cell death in this stage of fish embryos induced by the loss of shox expression. Since the dlx2 expressing cells are one of the major populations of future anterior skeleton (32), our current data imply that shox is required for maintaining skeletal progenitor cells. Indeed, current cell culture study using hMSCs showed that the hMSCs expressed SHOX and the siRNA-mediated SHOX knockdown affected the cell proliferation. Thus, data in both zebrafish embryo and hMSC models support the notion that shox/SHOX facilitates cell proliferation in embryonic skeletal progenitor cells. On the other hand, previous studies in U2OS osteosarcoma cells, human primary oral fibroblasts, and chondrocytes showed that overexpression of SHOX induced cell cycle arrest at G2/M phase and the activation of apoptotic pathway (8, 33). We presume the seemingly contradicting results between these previous studies and our current study would be due to the differences in experimental systems: previous studies took the gain-of-function approach in differentiated cell line, and the current one carried out the loss-of-function experiments of endogenously expressed shox/SHOX genes in embryonic cells/undifferentiated hMSCs. It is noteworthy that, in the case of the closely related SHOX paralog genes (Shox2 in mice; SHOX2 in human), overexpression and knockdown of these genes often did not reach the same conclusion: the knockout or knockdown of Shox2 impaired cell proliferation but the overexpression of the gene never upregulated cell proliferation (19, 34). Alternatively, it is possible that shox/SHOX is required for cell proliferation in undifferentiated skeletal progenitor cells but not in the cells under differentiation or under fully differentiated status. Further study is needed to explain this issue, but it is plausible that the levels of SHOX expression and the cellular context cooperatively define its role in cell proliferation. Since the expression levels of shox increase during embryonic growth and development in zebrafish model, understanding the relationship between shox/SHOX expression level and its role in cell proliferation seems a very important issue to interpret the biological function of SHOX in skeletal formation.
Gene expression analysis showed the transient increase of both osteogenic and chondrogenic marker expressions in shox MO-injected embryos. Recent studies have demonstrated that the loss of mouse Shox2 resulted in accelerated chondrogenic differentiation of prechondrocyte and MSCs (30). These data encouraged us to analyze the role of shox/SHOX in skeletal progenitor cells. As we found in zebrafish model, the loss of SHOX expression in hMSCs also increased the expression of differentiation-related genes such as RUNX2 in early osteogenic differentiation. The mouse model of early onset of osteogenesis induced by precocious RUNX2 expression resulted in hindered skeletal growth likely due to the inappropriate calcification and abnormal chondrocyte differentiation (35). The early onset of chondrocyte differentiation is also linked with premature ossification of developing bone that restricts skeletal growth (36). Thus, the earlier elevation of differentiation genes (such as runx2b/RUNX2) would not be beneficial for the future skeletal growth in shox/SHOX deficient animals.
Previous studies showed that the fully differentiated osteoblast did not express prominent levels of SHOX and it suggests a less important role of SHOX in osteoblasts (5, 8). In our data, SHOX expressed in undifferentiated hMSCs and the loss of SHOX expression in hMSCs increased expression of early osteogenic marker genes (such as RUNX2 and DLX5) under osteogenic differentiation condition. Given its increased expression and decreased transcription activity during early osteogenesis (as our results in Figure 3C), SHOX would act as a transcriptional repressor in differentiating osteogenic progenitor cells. Indeed, SHOX protein is known to behave not only as a transcriptional activator but also as a transcriptional repressor (19, 27, 29, 37). Currently, it is uncertain whether or not these osteogenic marker genes are the direct targets of SHOX. Investigations of its specific target genes in undifferentiated and differentiating hMSCs would be needed for delineating the developmental roles of this gene.
The SHOX family proteins are homeobox transcription factors, which often act as heterodimers to control target gene expressions (5). It might be supposed that the interacting partners of SHOX proteins are diverse depending on the cell type, cellular differentiation status, or environmental conditions, thus we see various, sometime opposite, functions of this protein in various experimental models. It is of great interest to identify binding partners of the SHOX protein in undifferentiated and/or differentiating skeletal progenitor cells such as MSCs. In addition to the bone and cartilage development, evidence from a recent study implies the potential link between the SHOX2 expression and the specification of adipocytes, another important lineage originated from the MSCs (38). Furthermore, as well as its binding partners, we have been gradually developing greater understanding of the novel regulators of the SHOX family genes including microRNA (34). Since shox/SHOX is expressed in diverse types of cells and tissues (11, 25), it is likely that Shox/SHOX proteins have pleiotropic actions in many types of cells. Future studies on how the shox/SHOX expression and its action are regulated by other factors are doubtlessly required for comprehensive understanding of the role of this gene in animal growth and development.
Ethics Statement
This study was carried out in accordance with the recommended guidelines of the committee of the Life Science Research Ethics and Safety in the Graduate School of Agriculture and Life Sciences at The University of Tokyo, and the Guide for the Care and Use of Laboratory Animals prepared by Kanazawa University.
Author Contributions
TY, HK, TaS, DY, RS-Y, FH, S-IT, and ToS conceived and designed these experiments. TY, HK, TaS, DY, RS-Y, and FH performed the experiments. TY, HK, TaS, DY, RS-Y, and FH analyzed the data. HK, DY, FH, S-IT, and ToS contributed reagents and materials/analysis tools. TY, HK, TaS, FH, and S-IT wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Michihiro Shibata at BD Biosciences for invaluable discussions. We also acknowledge the help with proofreading the manuscript of Dr. Susan Hall (The University of North Carolina at Chapel Hill).
Funding
This work was partially supported by Grant-in-Aid [(A) (2)#16208028, (A) #22248030, (A) #25252047, (S) #25221204], Challenging Exploratory Research (#20658065, #25660210, #15K14840), core-to-core program from the Japan Society for the Promotion of Science (JSPS) and Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry (#25006A) to S-IT and by Grant-in-Aid [(B) #243801521] to FH. This work was also partially supported by Grant-in-Aid for Young Scientists [(Start-up) #23880008 (B) #25850220, (B) #15K18799] to HK from JSPS. HK has been supported by National Institute of Genetics, Cooperative Research Program Grant (2012-B11, 2013-A28, 2014-A26).
Abbreviations
ISS, idiopathic short stature; LWD, Leri–Weill dyschondrosteosis; PAR1, the pseudo autosomal region 1; DIG, digoxigenin; hpf, hours post fertilization; dpf, days post fertilization.
References
1. Bernasconi S, Mariani S, Falcinelli C, Milioli S, Iughetti L, Forabosco A. SHOX gene in Leri-Weill syndrome and in idiopathic short stature. J Endocrinol Invest (2001) 24:737–41. doi:10.1007/BF03343919
2. Rao E, Weiss B, Fukami M, Rump A, Niesler B, Mertz A, et al. Pseudoautosomal deletions encompassing a novel homeobox gene cause growth failure in idiopathic short stature and Turner syndrome. Nat Genet (1997) 16:54–63. doi:10.1038/ng0597-54
3. Belin V, Cusin V, Viot G, Girlich D, Toutain A, Moncla A, et al. SHOX mutations in dyschondrosteosis (Leri-Weill syndrome). Nat Genet (1998) 19:67–9. doi:10.1038/ng0198-67
4. Shears DJ, Vassal HJ, Goodman FR, Palmer RW, Reardon W, Superti-Furga A, et al. Mutation and deletion of the pseudoautosomal gene SHOX cause Leri-Weill dyschondrosteosis. Nat Genet (1998) 19:70–3. doi:10.1038/ng0198-70
5. Marchini A, Ogata T, Rappold GA. A track record on SHOX: from basic research to complex models and therapy. Endocr Rev (2016) 37:417–48. doi:10.1210/er.2016-1036
6. Rappold G. SHOX: a geneticist’s view. J Clin Endocrinol Metab (2002) 87:1911–2; author reply 1912. doi:10.1210/jc.87.4.1911-a
7. Stuppia L, Calabrese G, Gatta V, Pintor S, Morizio E, Fantasia D, et al. SHOX mutations detected by FISH and direct sequencing in patients with short stature. J Med Genet (2003) 40:E11. doi:10.1136/jmg.40.2.e11
8. Marchini A, Marttila T, Winter A, Caldeira S, Malanchi I, Blaschke RJ, et al. The short stature homeodomain protein SHOX induces cellular growth arrest and apoptosis and is expressed in human growth plate chondrocytes. J Biol Chem (2004) 279:37103–14. doi:10.1074/jbc.M307006200
9. Munns CJ, Haase HR, Crowther LM, Hayes MT, Blaschke R, Rappold G, et al. Expression of SHOX in human fetal and childhood growth plate. J Clin Endocrinol Metab (2004) 89:4130–5. doi:10.1210/jc.2003-032230
10. Thomas NS, Maloney V, Bass P, Mulik V, Wellesley D, Castle B. SHOX mutations in a family and a fetus with Langer mesomelic dwarfism. Am J Med Genet A (2004) 128A:179–84. doi:10.1002/ajmg.a.30095
11. Durand C, Roeth R, Dweep H, Vlatkovic I, Decker E, Schneider KU, et al. Alternative splicing and nonsense-mediated RNA decay contribute to the regulation of SHOX expression. PLoS One (2011) 6:e18115. doi:10.1371/journal.pone.0018115
12. Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, et al. Multilineage potential of adult human mesenchymal stem cells. Science (1999) 284:143–7. doi:10.1126/science.284.5411.143
13. Uccelli A, Moretta L, Pistoia V. Mesenchymal stem cells in health and disease. Nat Rev Immunol (2008) 8:726–36. doi:10.1038/nri2395
14. Giuliani N, Lisignoli G, Magnani M, Racano C, Bolzoni M, Dalla Palma B, et al. New insights into osteogenic and chondrogenic differentiation of human bone marrow mesenchymal stem cells and their potential clinical applications for bone regeneration in pediatric orthopaedics. Stem Cells Int (2013) 2013:312501. doi:10.1155/2013/312501
15. Clement-Jones M, Schiller S, Rao E, Blaschke RJ, Zuniga A, Zeller R, et al. The short stature homeobox gene SHOX is involved in skeletal abnormalities in Turner syndrome. Hum Mol Genet (2000) 9:695–702. doi:10.1093/hmg/9.5.695
16. Durand C, Rappold GA. Height matters-from monogenic disorders to normal variation. Nat Rev Endocrinol (2013) 9:171–7. doi:10.1038/nrendo.2012.251
17. Yu L, Gu S, Alappat S, Song Y, Yan M, Zhang X, et al. Shox2-deficient mice exhibit a rare type of incomplete clefting of the secondary palate. Development (2005) 132:4397–406. doi:10.1242/dev.02013
18. Cobb J, Dierich A, Huss-Garcia Y, Duboule D. A mouse model for human short-stature syndromes identifies Shox2 as an upstream regulator of Runx2 during long-bone development. Proc Natl Acad Sci U S A (2006) 103:4511–5. doi:10.1073/pnas.0510544103
19. Yu L, Liu H, Yan M, Yang J, Long F, Muneoka K, et al. Shox2 is required for chondrocyte proliferation and maturation in proximal limb skeleton. Dev Biol (2007) 306:549–59. doi:10.1016/j.ydbio.2007.03.518
20. Mullins MC, Hammerschmidt M, Haffter P, Nusslein-Volhard C. Large-scale mutagenesis in the zebrafish: in search of genes controlling development in a vertebrate. Curr Biol (1994) 4:189–202. doi:10.1016/S0960-9822(00)00048-8
21. Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn (1995) 203:253–310. doi:10.1002/aja.1002030302
22. Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell (2004) 7:133–44. doi:10.1016/j.devcel.2004.06.005
23. Walker MB, Kimmel CB. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech Histochem (2007) 82:23–8. doi:10.1080/10520290701333558
24. Kenyon EJ, Mcewen GK, Callaway H, Elgar G. Functional analysis of conserved non-coding regions around the short stature hox gene (shox) in whole zebrafish embryos. PLoS One (2011) 6:e21498. doi:10.1371/journal.pone.0021498
25. Sawada R, Kamei H, Hakuno F, Takahashi S, Shimizu T. In vivo loss of function study reveals the short stature homeobox-containing (shox) gene plays indispensable roles in early embryonic growth and bone formation in zebrafish. Dev Dyn (2015) 244:146–56. doi:10.1002/dvdy.24239
26. Kamei H, Lu L, Jiao S, Li Y, Gyrup C, Laursen LS, et al. Duplication and diversification of the hypoxia-inducible IGFBP-1 gene in zebrafish. PLoS One (2008) 3:e3091. doi:10.1371/journal.pone.0003091
27. Marchini A, Hacker B, Marttila T, Hesse V, Emons J, Weiss B, et al. BNP is a transcriptional target of the short stature homeobox gene SHOX. Hum Mol Genet (2007) 16:3081–7. doi:10.1093/hmg/ddm266
28. Aza-Carmona M, Shears DJ, Yuste-Checa P, Barca-Tierno V, Hisado-Oliva A, Belinchon A, et al. SHOX interacts with the chondrogenic transcription factors SOX5 and SOX6 to activate the aggrecan enhancer. Hum Mol Genet (2011) 20:1547–59. doi:10.1093/hmg/ddr032
29. Decker E, Durand C, Bender S, Rodelsperger C, Glaser A, Hecht J, et al. FGFR3 is a target of the homeobox transcription factor SHOX in limb development. Hum Mol Genet (2011) 20:1524–35. doi:10.1093/hmg/ddr030
30. Bobick BE, Cobb J. Shox2 regulates progression through chondrogenesis in the mouse proximal limb. J Cell Sci (2012) 125:6071–83. doi:10.1242/jcs.111997
31. Aza-Carmona M, Barca-Tierno V, Hisado-Oliva A, Belinchon A, Gorbenko-Del Blanco D, Rodriguez JI, et al. NPPB and ACAN, two novel SHOX2 transcription targets implicated in skeletal development. PLoS One (2014) 9:e83104. doi:10.1371/journal.pone.0083104
32. Sperber SM, Saxena V, Hatch G, Ekker M. Zebrafish dlx2a contributes to hindbrain neural crest survival, is necessary for differentiation of sensory ganglia and functions with dlx1a in maturation of the arch cartilage elements. Dev Biol (2008) 314:59–70. doi:10.1016/j.ydbio.2007.11.005
33. Hristov G, Marttila T, Durand C, Niesler B, Rappold GA, Marchini A. SHOX triggers the lysosomal pathway of apoptosis via oxidative stress. Hum Mol Genet (2014) 23:1619–30. doi:10.1093/hmg/ddt552
34. Hong S, Noh H, Teng Y, Shao J, Rehmani H, Ding HF, et al. SHOX2 is a direct miR-375 target and a novel epithelial-to-mesenchymal transition inducer in breast cancer cells. Neoplasia (2014) 16(279–290):e271–5. doi:10.1016/j.neo.2014.03.010
35. Maeno T, Moriishi T, Yoshida CA, Komori H, Kanatani N, Izumi S, et al. Early onset of Runx2 expression caused craniosynostosis, ectopic bone formation, and limb defects. Bone (2011) 49:673–82. doi:10.1016/j.bone.2011.07.023
36. Vega RB, Matsuda K, Oh J, Barbosa AC, Yang X, Meadows E, et al. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell (2004) 119:555–66. doi:10.1016/j.cell.2004.10.024
37. Rao E, Blaschke RJ, Marchini A, Niesler B, Burnett M, Rappold GA. The Leri-Weill and Turner syndrome homeobox gene SHOX encodes a cell-type specific transcriptional activator. Hum Mol Genet (2001) 10:3083–91. doi:10.1093/hmg/10.26.3083
Keywords: SHOX, zebrafish embryo, human mesenchymal stem cells, proliferation, osteogenic differentiation
Citation: Yokokura T, Kamei H, Shibano T, Yamanaka D, Sawada-Yamaguchi R, Hakuno F, Takahashi S-I and Shimizu T (2017) The Short-Stature Homeobox-Containing Gene (shox/SHOX) Is Required for the Regulation of Cell Proliferation and Bone Differentiation in Zebrafish Embryo and Human Mesenchymal Stem Cells. Front. Endocrinol. 8:125. doi: 10.3389/fendo.2017.00125
Received: 23 January 2017; Accepted: 22 May 2017;
Published: 08 June 2017
Edited by:
Yong Zhu, East Carolina University, United StatesReviewed by:
Julang Li, University of Guelph, CanadaToshinobu Tokumoto, Shizuoka University, Japan
Chun Peng, York University, Canada
Copyright: © 2017 Yokokura, Kamei, Shibano, Yamanaka, Sawada-Yamaguchi, Hakuno, Takahashi and Shimizu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hiroyasu Kamei, aGthbWVpQHNlLmthbmF6YXdhLXUuYWMuanA=;
Shin-Ichiro Takahashi, YXRrc2hpbkBtYWlsLmVjYy51LXRva3lvLmFjLmpw
†These authors have contributed equally to this work.
 Tomoaki Yokokura
Tomoaki Yokokura Hiroyasu Kamei
Hiroyasu Kamei Takashi Shibano
Takashi Shibano Daisuke Yamanaka
Daisuke Yamanaka Rie Sawada-Yamaguchi1,2,3
Rie Sawada-Yamaguchi1,2,3 Fumihiko Hakuno
Fumihiko Hakuno Shin-Ichiro Takahashi
Shin-Ichiro Takahashi