- Department of Biology, Ecology and Earth Sciences (DiBEST), University of Calabria, Rende, Italy
The plasticity of the individual epigenetic landscape that goes to countless rearrangements throughout life is closely the reflection of environmental factors such as chemical exposure, socio-economic status and nutrient intakes both early and late in life. The Mini Nutritional Assessment (MNA) is a well-validated tool for assessing malnutrition in old people. It includes 6 (MNA-SF) or 18 (MNA-LF) self-reported questions derived from general, anthropometric, dietary, and self- assessment. We evaluated the association between the nutritional status, as measured by MNA, and methylation biomarkers we previously demonstrated to be associated with chronological and biological age in human. We found that malnutrition is positively correlated with DNA methylation status at the global level, in line with our previous reports. On the contrary, most of the sites located within specific genes, which were previously reported to be correlated with chronological and biological aging, showed to be not affected by malnutrition, or even to have correlations with malnutrition opposite to those previously reported with frailty. These results may suggest that malnutrition is among the first effects of disability and other age- related problems and a generalized non-specific epigenetic remodeling may be the initial response of the organism. By contrast, the fine remodeling of specific genomic sites is scarcely affected by malnutrition and may respond to a more complex interaction of different factors. Therefore, although malnutrition in the elderly is certainly a risk factor for survival, this is partially independent of the aging process of the organism which leads to the methylation remodeling previously described to measure chronological and biological aging.
Introduction
The multi-causal phenotypic variability among elderly individuals has led researchers to evaluate changes in a wide variety of biological parameters in order to have a more comprehensive insight of health at old age and, possibly, predict both the quality of aging and the onset of age-related diseases (1, 2). In this context, in addition to non-directional changes in DNA methylation patterns, referred to as epigenetic drift, directional, and non-stochastic hyper- or hypo-methylation events, occurring over time at discrete CpG sites throughout the genome, turned out to be very useful, and also able to be predictive of both chronological and biological aging (3–7). These age-Differentially Methylated Regions (a-DMRs) have been found associated with survival chance, disability, frailty, multi-morbidity, thus determining the overall variation in life expectancy (8–12). What is more, the setup of a series of multi-tissue age estimator models, namely epigenetic clocks, is allowing the prediction of all-cause mortality independent of several risk factors (13–24).
Considered their function as substrates or cofactors for epigenetic enzymes, nutrients and their metabolites are the main environmental factors able to modify the epigenetic landscape as well as health and life expectancy throughout the lifetime (25–27). Dietary manipulation of methyl donors (either supplementation or deficiency) as well as calorie restriction (CR) have been recognized as responsible for global rearrangements of DNA methylation profiles (28–33). Furthermore, according to the “developmental origins of health and disease” hypothesis, already in utero, energy-rich, protein-deficient, micronutrient-deficient, and/or methyl donor-rich diets induce multi-tissue perturbations of methylation profile in mothers (34). These perturbations can be transmitted to next generation thus regulating in offspring long term metabolic processes and contributing to age phenotypes and age-related diseases (26, 29, 35–39). With aging, changes or loss in appetite, mainly due to the reduction of acuity in sense organs, in the secretion of hunger hormones, gastrointestinal motility as well as the inability in preparing food, depression, and dementia, lead to a significant reduction of food intake (40–42). Consequently, a loss in weight, muscle mass, strength, and physical function is generally observed and it is retained to be the main origin of the weakness and the decline in functional ability, including conditions such as sarcopenia and frailty (43–48). On the contrary, appropriate nutritional status was held responsible for the safeguard of healthy aging and linked to favorable outcomes in health, by retarding the detrimental consequences of aging and the prevention/treatment of a variety of diseases (44, 48–53).
Taking into account the relationship among nutritional status, DNA methylation patterns, and biological aging, we evaluated the association between nutritional status as measured by the Mini Nutritional Assessment (MNA), a validated assessment tool to measure nutrition status in elderly people, and previously identified age-related methylation biomarkers at both nuclear and mitochondrial level.
Materials and Methods
Study Population
Socio-demographic characteristics of the population sample analyzed in this study were previously reported (54). Briefly, the sample included 302 subjects living in Calabria (South Italy) subdivided into two groups: the first (S1) comprised 191 subjects younger than 85 years (101 women and 90 men, mean age 73.0 years), the second (S2), 111 subjects aged 85 or over (54 women and 57 men, mean age 97.1 years). Samples were collected within the framework of several recruitment campaigns carried out for monitoring the quality of aging in the whole Calabria region from 2002 onwards. Subjects older than 90 years were identified through the population registers and then contacted by specialized personnel and invited to join the study.
Younger subjects were contacted through general physicians. Finally, each subject was recruited after a complete multidimensional geriatric assessment with detailed clinical history, including anthropometric measures and a set of the most common tests to assess cognitive functioning, functional activity, physical performance, and depression. In addition, common clinical hematological tests were performed. White blood cells (WBC) from blood buffy coats were used as source of DNA.
Vital status at near 9 years from the baseline visit was traced for 189 subjects (98.95%) in S1 and for all the 111 subjects (100%) in S2 through the population registers of the municipalities where the respondents lived.
Mini Nutritional Assessment
The Mini Nutritional Assessment is a well-validated tool for assessing malnutrition in old people (55). It includes 18 self-reported questions derived from general, anthropometric, dietary, and self- assessment. In particular, the short form of the MNA (MNA-SF) is a screening tool consisting of six questions on food intake, weight loss, mobility, psychological stress, or acute disease, the presence of dementia or depression, and body mass index (BMI). The maximum score for this part is equal to 14. A score equal to or higher than 12 indicates that the subject under study has an acceptable nutritional status thus excluding malnutrition and/or malnutrition risk, meanwhile, a score ≤ 11 implicates to proceed with the complete version of the MNA (MNA-LF) (56). This version consists of 12 additional items and provides a maximum possible overall assessment of 30 scores: a score of fewer than 17 indicates malnutrition, a score of 17–23.5 indicates a risk for malnutrition and a score higher 23.5 indicates well-nourishment (57).
Epigenetic Biomarkers
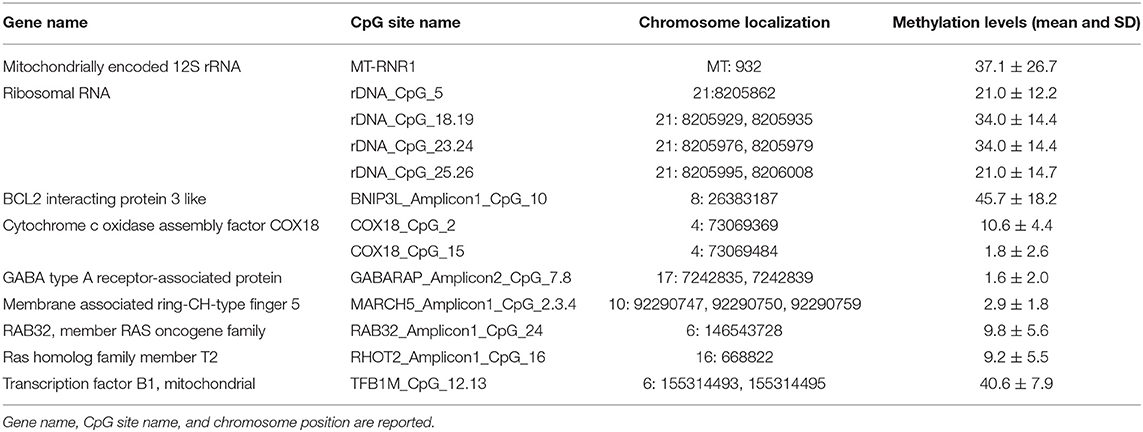
For the assessment of the correlation between MNA and the epigenetic status of biomarkers, we focused on DNA methylation of CpG sites, located within both nuclear and mitochondrial genes, we previously found associated with chronological and biological aging: one CpG site located within the human mitochondrial 12S ribosomal RNA (MT-RNR1) gene, 4 CpG units (CpG_5, CpG_18.19, CpG_23.24, CpG_25.26) falling into the promoter of the ribosomal RNA genes (rDNA), and 12 CpG sites (BNIP3L_Amplicon1_CpG_10, COX18_CpG_2, COX18_CpG_15, GABARAP_Amplicon2_CpG_7.8, MARCH5_Amplicon1_CpG_2.3.4, RAB32_Amplicon1_CpG_24, RHOT2_Amplicon1_CpG16, TFB1M_CpG_12.13) located within promoter regions of genes involved in mitochondrial quality control (10, 11, 58) (Table 1; Figure S1).
In addition, we had also considered the association of MNA and the overall degree of methylation of the human genome as measured by Global DNA Methylation Index (GDMI) (8).
Analytic Approach
We compared S1 and S2 groups with regard to study variables and covariates. We used the unpaired t-test for continuous variables and chi-square for categorical ones.
For each group, Kaplan-Meier survival curves were estimated for the MNA risk categories (MNA-SF < 12 and MNA-LF < 17 or MNA-LF < 23.5). In order to evaluate their predictive values with respect to mortality risk, the obtained survival curves were then compared by log-rank test. Subjects alive after the follow-up time were considered as censored, and this time was used as the censoring date in the survival analyses. In addition, Hazard ratios (HR) and 95% Confidence Intervals (95% CI) were estimated by using Cox proportional hazard models taking also into account possible confounder variables (age and gender).
A linear regression model was used to assess the association between the variability of epigenetic markers and MNA scores. Analyses were adjusted for age at the recruitment and gender.
All statistical analyses were performed using IBM SPSS statistics for Windows, V.25 (IBM Corp).
Results
MNA Assessment
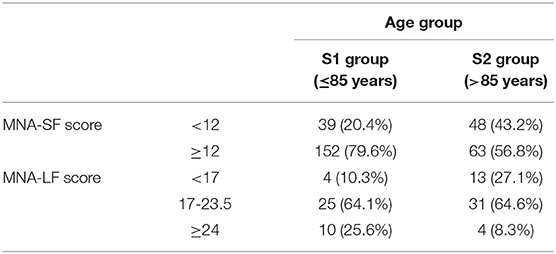
Subjects younger and older 85 years of age were included in the study. Table 2 shows the demographic characteristics of the analyzed sample and the nutritional status of the elderly as resulting by MNA assessment. Out of the 302 subjects we analyzed, 215 (71.2%) exhibited an adequate nutritional status scoring of 12 or more. In particular, this accounted for 79.6% of subjects younger than 85 years and 56.8% of older subjects. On the contrary, 87 subjects (28.8%) had a score lower than 12 and went through the complete assessment of MNA (MNA-LF). After the complete test, in S1 group, about 10% turned out to be malnourished, 64.1% at risk, while the remaining ones were well-nourished (25.6%). In S2 group we found a near-tripled proportion of malnourished subjects (27.1%) with respect to S1, a similar proportion of individuals at risk for malnutrition (about 65%), and a reduced number of well-nourished subjects (8.3%).
MNA Scores and Survival
Figure 1 shows the Kaplan–Meier estimates of the survival functions in people with malnutrition (MNA-SF < 12 or MNA-LF < 17) vs. those that did not show malnutrition in the S1 and S2 groups of the analyzed sample. We found that in S1 group people with malnutrition (MNA-SF < 12, MNA-LF < 17) lived shorter than people that did not show malnutrition, while in S2 group no association was found between malnutrition and mortality risk (Figures 1B,D). In fact, in S1 group after the follow-up period, 48.7% of those who were at risk for malnutrition (MNA-SF < 12) in S1 group had died (panel A), compared to 25.8% of those who were not a risk at baseline (P = 0.001). Among the subjects who obtained a screening score lower than 12 in S1 group, we found that after the follow-up period, all subjects with severe malnutrition (MNA-LF < 17) died (panel C), compared to 56.0% of those who were at risk for malnutrition (MNA-LF > 17 and MNA-LF < 23.5) and 10.0% of those who were not at risk (MNA-LF>23.5) (P = 0.002).
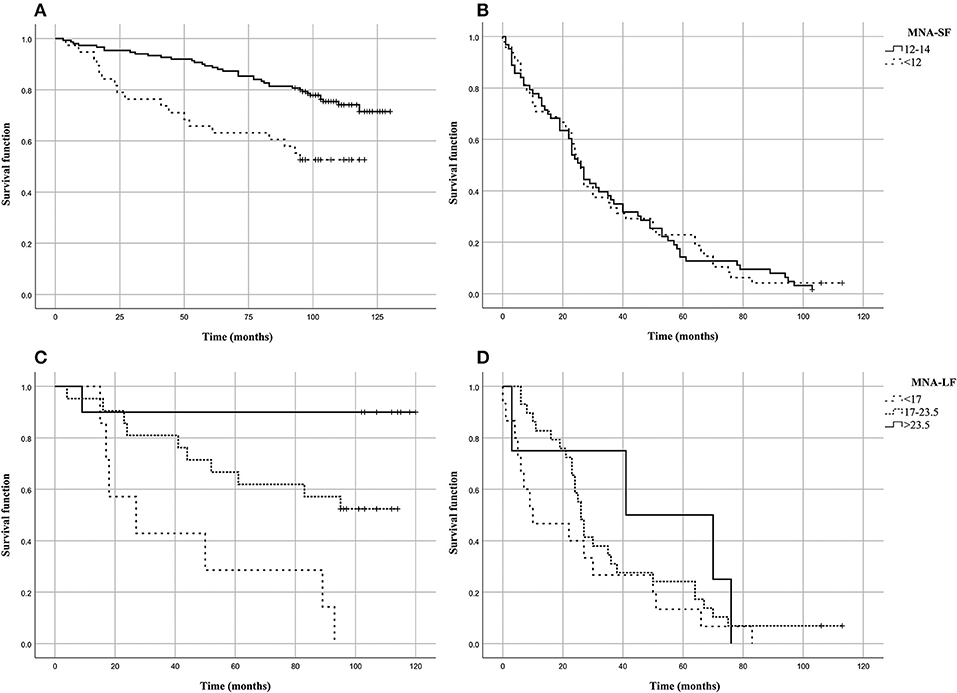
Figure 1. Kaplan–Meier estimates of the survival functions in people with malnutrition (MNA-SF<12 or MNA-LF<17) vs. those that did not show malnutrition in the analyzed sample. (A) MNA-SF in S1 group; (B) MNA-SF in S2 group; (C) MNA-LF in S1 group; (D) MNA-LF in S2 group.
To assess the independent predictive validity of malnutrition in terms of survival in S1 group, we evaluated its association with prospective mortality risk by Cox proportional hazard models. After adjusting for age at recruitment and gender the association between MNA-SF and mortality risk did not hold statistical significance (HR = 1.518, 95% CI: 0.828–2.785, P = 0.177). As it regards MNA-LF, we found that malnourished subjects (MNA-LF < 17) had a significantly increased risk of mortality with respect to subjects with a normal nutritional status (HR = 17.6, 95% CI = 1.583–195.588, P = 0.020) as well as those with a nutritional status at risk (HR = 5.854, 95% CI = 0.733–46.726, P = 0.095), also after adjusting for age and gender.
MNA Scores and Epigenetic Markers
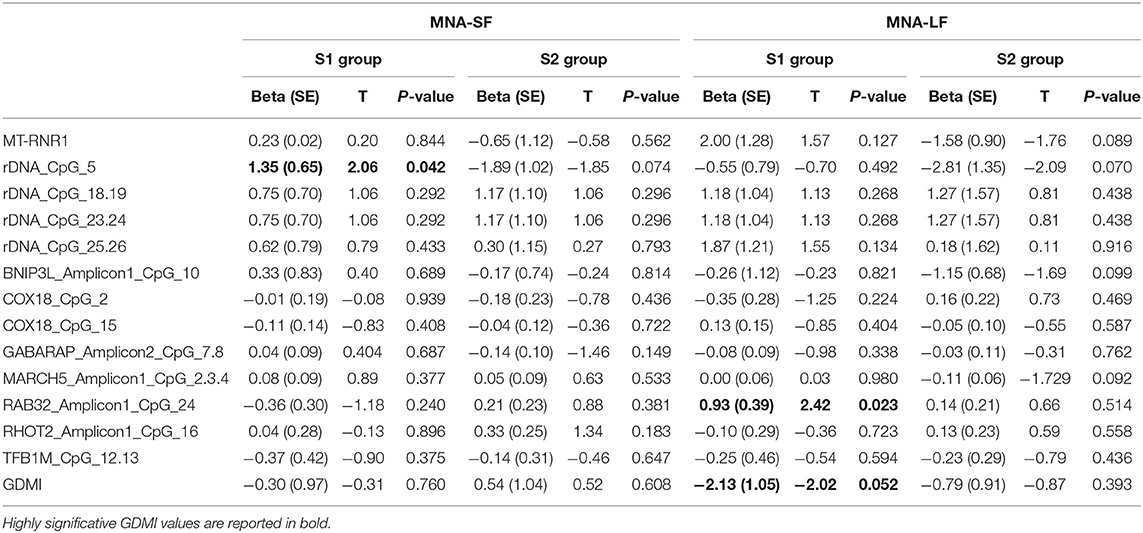
The analysis of the relationship between nutritional status and epigenetic markers revealed that after adjusting for age at recruitment and gender, GDMI values significantly decreased as MNA-LF scores increased (P = 0.052). No significant correlations between MNA-SF and epigenetics biomarkers were detected in S2 group (Table 3).
The analysis of specific sites showed that in most cases their methylation level was not associated with MNA. Among the few exceptions, rDNA_CpG_5 methylation levels which increased as MNA-SF scores also increased (P = 0.042). No significant correlations between MNA-SF and epigenetic biomarkers were detected in S2 group (Table 3).
Among the subjects who obtained a screening score lower than 12 in S1 group, we found that after adjusting for age at recruitment and gender, RAB32_Amplicon1_CpG_24 methylation levels were significantly correlated with the MNA-LF scores (P = 0.023).
After adjusting for multiple comparisons, none of the reported associations hold the statistical significance (Bonferroni corrected p-value 8.9 × 10−4). However, this correction seems more suitable when searching for associations without a priori hypotheses but too conservative when assessing a specific research question, as in our case, where a number of non-independent tests (due to the correlation between the analyzed markers) were also performed (59, 60).
Discussion
This study reports the relationship between nutritional status, assessed by the MNA tool, a method extensively used to identify the risk of malnutrition in the elderly, and epigenetic biomarkers we previously identified associated to chronological and biological aging in human.
According to MNA analysis, the majority of the participants to our study was found to be well-nourished and, thus, their analysis was limited to the short form of the MNA tool. Our results indicate that four subjects out of five in the younger group and about half of the subjects in the older group have a good nutritional status. Malnutrition significantly influenced mortality risk in particular among the subjects belonging to the youngest age group where its effect was independent of the age at recruitment and the sex of the participants. The presence of an adequate nutritional status in subjects of the aged Calabrian population is also confirmed by the evidence that most subjects of both groups exhibit MNA mean scores higher than 17 that is widely considered the threshold of malnutrition. In our population sample, high prevalence of subjects who were well-nourished was observed. Probably this is related to the fact that the subjects enrolled in our study originate from a population where the social-economic context does not promote malnutrition and, in addition, the adoption of the Mediterranean diet may have provided an adequate and balanced intake of food. What is more all subjects we analyzed were home-care elderly so that it is likely that the good nutritional status reflects great attention by caregivers, in most cases family members, as suggested by Soini et al. (61).
Results here reported suggest that malnutrition is correlated with global DNA methylation status since low global DNA methylation levels (high GDMI values) are associated with malnutrition. This adds new data in the already well-demonstrated relationship between DNA methylation and frailty (8). In fact, a number of literature reports suggest that health outcomes attributed to malnutrition seem to be associated with frailty. Nevertheless, as emphasized by Wei et al., the natural history of malnutrition and frailty with respect to each other is still unclear, and indeed our data on specific sites indicate that many aspects of this relationship need to be clarified (62). This history is even more complex if we introduce the state of methylation that affects both. It is plausible to retain that malnutrition through epigenetic changes at global levels exacerbates the reduction of muscle, fat and bone mass observed during aging. This reduction contributes to sarcopenia and, in more severe form, to the decline of physical performance that ultimately may decrease the survival rate.
On the contrary, most of the sites located within specific genes, which were previously reported to be correlated with chronological and biological aging, showed to be not affected by malnutrition. Moreover, the few methylated sites associated with MNA scores showed correlations not immediately in line with previous results. Indeed, we found a direct correlation between a good nutritional status and the methylation levels of rDNA_CpG_5 and RAB32_Amplicon1_CpG_24, while we have previously reported that high proportion of the methylation of these sites is correlated with impaired cognitive performance, decreased survival chance and disability (11, 58). These results may suggest that malnutrition is among the first effects of disability or of other age-related specific problems (such as socio-economic problems, depression, or other) and then the initial response is a generalized non-specific epigenetic remodeling. By contrast, the fine remodeling of specific genomic sites is scarcely affected by malnutrition and may respond to a more complex interaction of different factors, such as metabolites correlated to senescence or to oxidative stress. This confirms that these biomarkers are very reliable in gauging the organism aging and are not misled by malnutrition (63).
The above associations are restricted to the subjects of S1 group (<85), possibly because malnutrition may need some time to act on methylation and the higher mortality of the subjects older than 85 years (S2 group) may lead to death very soon after the start of malnutrition without the time to establish a methylation remodeling of these sites.
Finally, it is likely that genetic variants impairing nutrient intake and/or metabolism which could have a role in the interplay we observed and create an additional layer over epigenetics changes.
We are aware that our study has some weaknesses that should be addressed. A first limitation of the study is the reduced sample size. Its cross-sectional design does not allow us to assess the cause-effect relationship between the variability of epigenetic markers and the nutritional status of subjects under study. Another important limitation is the lack of proper correction for multiple testing. However, since this study was exploratory and hypothesis-driven, a Bonferroni correction would have eliminated potentially important findings if applied (59, 60). For these reasons, further explorations in additional study populations are needed before conclusions can be drawn.
The finding of the influence of nutrition on frailty through epigenetic modifications appears particularly relevant because of a balanced nutritional intervention could be an easy to use clinical approach for population-based strategies in aging to provide favorable functional and mortality outcomes and, at the same time, minimize the hospital assistance and long-term care, thus reducing the health care costs.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
PD'A, AM, GP, and DB designed the study. AM and VR performed the statistical analysis. DB and GP wrote the initial draft. AM, PD'A, VR, GP, and DB participated in critical revision and approved the final manuscript before submission.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The work has been made possible by the collaboration with the nursing homes of SADEL S.p.A (San Teodoro, San Raffaele, Villa del Rosario, A.G.I srl, SAVELLI HOSPITAL, Casa di Cura Madonna dello Scoglio) in the frame of the agreement SOLUZIONI INNOVATIVE PER L'INNALZAMENTO DELLA SALUTE E DELLA SICUREZZA DELLA POPOLAZIONE with the University of Calabria.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2019.00672/full#supplementary-material
References
1. Bürkle A, Moreno-Villanueva M, Bernhard J, Blasco M, Zondag G, Hoeijmakers JH, et al. MARK-AGE biomarkers of ageing. Mech Ageing Dev. (2015) 151:2–12. doi: 10.1016/j.mad.2015.03.006
2. Bacalini MG, D'Aquila P, Marasco E, Nardini C, Montesanto A, Franceschi C, et al. The methylation of nuclear and mitochondrial DNA in ageing phenotypes and longevity. Mech Ageing Dev. (2017) 165:156–61. doi: 10.1016/j.mad.2017.01.006
3. López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. (2013) 153:1194–217. doi: 10.1016/j.cell.2013.05.039
5. Li Y, Tollefsbol TO. Age-related epigenetic drift and phenotypic plasticity loss: implications in prevention of age-related human diseases. Epigenomics. (2016) 8:1637–51. doi: 10.2217/epi-2016-0078
6. Vaiserman A. Developmental tuning of epigenetic clock. Front Genet. (2018) 9:584. doi: 10.3389/fgene.2018.00584
7. Xiao FH, Wang HT, Kong QP. Dynamic DNA methylation during aging: a “Prophet” of age-related outcomes. Front Genet. (2019) 10:107. doi: 10.3389/fgene.2019.00107
8. Bellizzi D, D'Aquila P, Montesanto A, Corsonello A, Mari V, Mazzei B, et al. Global DNA methylation in old subjects is correlated with frailty. Age. (2012) 34:169–79. doi: 10.1007/s11357-011-9216-6
9. Heyn H, Li N, Ferreira HJ, Moran S, Pisano DG, Gomez A, et al. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci USA. (2012) 109:10522–7. doi: 10.1073/pnas.1120658109
10. D'Aquila P, Giordano M, Montesanto A, De Rango F, Passarino G, Bellizzi D. Age-and gender-related pattern of methylation in the MT-RNR1 gene. Epigenomics. (2015) 7:707–16. doi: 10.2217/epi.15.30
11. D'Aquila P, Montesanto A, Mandalà M, Garasto S, Mari V, Corsonello A, et al. Methylation of the ribosomal RNA gene promoter is associated with aging and age-related decline. Aging Cell. (2017) 16:966–75. doi: 10.1111/acel.12603
12. D'Aquila P, Bellizzi D, Passarino G. rRNA-gene methylation and biological aging. Aging. (2018) 10:7–8. doi: 10.18632/aging.101369
13. Bocklandt S, Lin W, Sehl ME, Sánchez FJ, Sinsheimer JS, Horvath S, et al. Epigenetic predictor of age. PLoS ONE. (2011) 6:e14821. doi: 10.1371/journal.pone.0014821
14. Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. (2013) 49:359–67. doi: 10.1016/j.molcel.2012.10.016
15. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. (2013) 14:R115. doi: 10.1186/gb-2013-14-10-r115
16. Marioni RE, Shah S, McRae AF, Chen BH, Colicino E, Harris SE, et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. (2015) 16:25. doi: 10.1186/s13059-015-0584-6
17. Chen BH, Marioni RE, Colicino E, Peters MJ, Ward-Caviness CK, et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging. (2016) 8:1844–65. doi: 10.18632/aging.101020
18. McCartney DL, Stevenson AJ, Walker RM, Gibson J, Morris SW, Campbell A, et al. Investigating the relationship between DNA methylation age acceleration and risk factors for Alzheimer's disease. Alzheimers Dement. (2018) 10:429–37. doi: 10.1016/j.dadm.2018.05.006
19. Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging. (2018) 10:573–91. doi: 10.18632/aging.101414
20. Guevara EE, Lawler RR. Epigenetic clocks. Evol Anthropol. (2018) 27:256–60. doi: 10.1002/evan.21745
21. Gale CR, Marioni RE, Harris SE, Starr JM, Deary IJ. DNA methylation and the epigenetic clock in relation to physical frailty in older people: the Lothian Birth Cohort 1936. Clin Epigenetics. (2018) 10:101. doi: 10.1186/s13148-018-0538-4
22. Lu AT, Quach A, Wilson JG, Reiner AP, Aviv A, Raj K, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging. (2019) 11:303–27. doi: 10.18632/aging.101684
23. Lund JB, Li S, Baumbach J, Svane AM, Hjelmborg J, Christiansen L, et al. DNA methylome profiling of all-cause mortality in comparison with age-associated methylation patterns. Clin Epigenetics. (2019) 11:23. doi: 10.1186/s13148-019-0622-4
24. White AJ, Kresovich JK, Xu Z, Sandler DP, Taylor JA. Shift work, DNA methylation and epigenetic age. Int J Epidemiol. (2019) doi: 10.1093/ije/dyz027. [Epub ahead of print].
25. D'Aquila P, Bellizzi D, Passarino G. Mitochondria in health, aging and diseases: the epigenetic perspective. Biogerontology. (2015) 16:569–85. doi: 10.1007/s10522-015-9562-3
26. Lillycrop KA, Burdge GC. Maternal diet as a modifier of offspring epigenetics. J Dev Orig Health Dis. (2015) 6:88–95. doi: 10.1017/S2040174415000124
27. Guarasci F, D'Aquila P, Mandalà M, Garasto S, Lattanzio F, Corsonello A, et al. Aging and nutrition induce tissue-specific changes on global DNA methylation status in rats. Mech Ageing Dev. (2018) 174:47–54. doi: 10.1016/j.mad.2018.02.001
28. Li Y, Saldanha SN, Tollefsbol TO. Impact of epigenetic dietary compounds on transgenerational prevention of human diseases. AAPS J. (2014) 16:27–36. doi: 10.1208/s12248-013-9538-7
29. Chango A, Pogribny IP. Considering maternal dietary modulators for epigenetic regulation and programming of the fetal epigenome. Nutrients. (2015) 7:2748–70. doi: 10.3390/nu7042748
30. Lee HS. Impact of maternal diet on the epigenome during in utero life and the developmental programming of diseases in childhood and adulthood. Nutrients. (2015) 7:9492–507. doi: 10.3390/nu7115467
31. Zhang N. Epigenetic modulation of DNA methylation by nutrition and its mechanisms in animals. Anim Nutr. (2015) 1:144–51. doi: 10.1016/j.aninu.2015.09.002
32. Hahn O, Grönke S, Stubbs TM, Ficz G, Hendrich O, Krueger F, et al. Dietary restriction protects from age-associated DNA methylation and induces epigenetic reprogramming of lipid metabolism. Genome Biol. (2017) 18:56. doi: 10.1186/s13059-017-1187-1
33. Maegawa S, Lu Y, Tahara T, Lee JT, Madzo J, Liang S, et al. Caloric restriction delays age-related methylation drift. Nat Commun. (2017) 8:539. doi: 10.1038/s41467-017-00607-3
34. Sinclair KD, Lea RG, Rees WD, Young LE. The developmental origins of health and disease: current theories and epigenetic mechanisms. Soc Reprod Fertil Suppl. (2007) 64:425–43. doi: 10.5661/RDR-VI-425
35. Vickers MH. Early life nutrition, epigenetics and programming of later life disease. Nutrients. (2014) 6:2165–78. doi: 10.3390/nu6062165
36. Geraghty AA, Lindsay KL, Alberdi G, McAuliffe FM, Gibney ER. Nutrition during pregnancy impacts offspring's epigenetic status-evidence from human and animal studies. Nutr Metab Insights. (2016) 8:41–7. doi: 10.4137/NMI.S29527
37. Geraghty AA, Sexton-Oates A, O'Brien EC, Alberdi G, Fransquet P, Saffery R, et al. A low glycaemic index diet in pregnancy induces DNA methylation variation in blood of newborns: results from the ROLO randomised controlled trial. Nutrients. (2018) 10:E455. doi: 10.3390/nu10040455
38. Pauwels S, Ghosh M, Duca RC, Bekaert B, Freson K, Huybrechts I, et al. Maternal intake of methyl-group donors affects DNA methylation of metabolic genes in infants. Clin Epigenetics. (2017) 9:16. doi: 10.1186/s13148-017-0321-y
39. Tao S, Zhou T, Saelao P, Wang Y, Zhu Y, Li T, et al. Intrauterine growth restriction alters the genome-wide DNA methylation profiles in small intestine, liver and longissimus dorsi muscle of newborn piglets. Curr Protein Pept Sci. (2019) 20:713–26. doi: 10.2174/1389203720666190124165243
40. Nieuwenhuizen WF, Weenen H, Rigby P, Hetherington MM. Older adults and patients in need of nutritional support: review of current treatment options and factors influencing nutritional intake. Clin Nutr. (2010) 29:160–9. doi: 10.1016/j.clnu.2009.09.003
41. Malafarina V, Uriz-Otano F, Gil-Guerrero L, Iniesta R. The anorexia of ageing: physiopathology, prevalence, associated comorbidity and mortality. A systematic review. Maturitas. (2013) 74:293–302. doi: 10.1016/j.maturitas.2013.01.016
42. Hedman S, Nydahl M, Faxén-Irving G. Individually prescribed diet is fundamental to optimize nutritional treatment in geriatric patients. Clin Nutr. (2016) 35:692–8. doi: 10.1016/j.clnu.2015.04.018
43. Cerri AP, Bellelli G, Mazzone A, Pittella F, Landi F, Zambon A, et al. Sarcopenia and malnutrition in acutely ill hospitalized elderly: Prevalence and outcomes. Clin Nutr. (2015) 34:745–51. doi: 10.1016/j.clnu.2014.08.015
44. Kelaiditi E, Guyonnet S, Cesari M. Is nutrition important to postpone frailty? Curr Opin Clin Nutr Metab Care. (2015) 18:37–42. doi: 10.1097/MCO.0000000000000129
45. Artaza-Artabe I, Sáez-López P, Sánchez-Hernández N, Fernández-Gutierrez N, Malafarina V. The relationship between nutrition and frailty: Effects of protein intake, nutritional supplementation, vitamin D and exercise on muscle metabolism in the elderly. A systematic review. Maturitas. (2016) 93:89–99. doi: 10.1016/j.maturitas.2016.04.009
46. Gabrovec B, Veninšek G, Samaniego LL, Carriazo AM, Antoniadou E, Jelenc M. The role of nutrition in ageing: a narrative review from the perspective of the European joint action on frailty–advantage JA. Eur J Intern Med. (2018) 56:26–32. doi: 10.1016/j.ejim.2018.07.021
47. Valentini A, Federici M, Cianfarani MA, Tarantino U, Bertoli A. Frailty and nutritional status in older people: the Mini Nutritional Assessment as a screening tool for the identification of frail subjects. Clin Interv Aging. (2018) 13:1237–44. doi: 10.2147/CIA.S164174
48. Veronese N, Stubbs B, Punzi L, Soysal P, Incalzi RA, Saller A, et al. Effect of nutritional supplementations on physical performance and muscle strength parameters in older people: a systematic review and meta-analysis. Ageing Res Rev. (2019) 51:48–54. doi: 10.1016/j.arr.2019.02.005
49. Haveman-Nies A, de Groot LC, van Staveren WA. Dietary quality, lifestyle factors and healthy ageing in Europe: the SENECA study. Age Ageing. (2003) 32:427–34. doi: 10.1093/ageing/32.4.427
50. Ohlhorst SD, Russell R, Bier D, Klurfeld DM, Li Z, Mein JR. Nutrition research to affect food and a healthy lifespan. Adv Nutr. (2013) 4:579–84. doi: 10.3945/an.113.004176
51. Passarino G, Rose G, Bellizzi D, De Luca M, Gonos ES. Aging and longevity between genetic background and lifestyle intervention. Biomed Res Int. (2014) 2014:516402. doi: 10.1155/2014/516402
52. Dato S, Bellizzi D, Rose G, Passarino G. The impact of nutrients on the aging rate: A complex interaction of demographic, environmental and genetic factors. Mech Ageing Dev. (2016) 154:49–61. doi: 10.1016/j.mad.2016.02.005
53. Bernstein M. Nutritional needs of the older adult. Phys Med Rehabil Clin N Am. (2017) 28:747–66. doi: 10.1016/j.pmr.2017.06.008
54. Montesanto A, Lagani V, Martino C, Dato S, De Rango F, Berardelli M, et al. A novel, population-specific approach to define frailty. Age. (2010) 32:385–95. doi: 10.1007/s11357-010-9136-x
55. Vellas B, Villars H, Abellan G, Soto ME, Rolland Y, Guigoz Y, et al. Overview of the MNA–its history and challenges. J Nutr Health Aging. (2006) 10:456–63.
56. Kaiser MJ, Bauer JM, Ramsch C, Uter W, Guigoz Y, Cederholm T, et al. Validation of the Mini Nutritional Assessment short-form (MNA-SF): a practical tool for identification of nutritional status. J Nutr Health Aging. (2009) 13:782–8. doi: 10.1007/s12603-009-0214-7
57. Guigoz Y, Vellas B, Garry PJ. Assessing the nutritional status of the elderly: the Mini Nutritional Assessment as part of the geriatric evaluation. Nutr Rev. (1996) 54:S59–65. doi: 10.1111/j.1753-4887.1996.tb03793.x
58. D'Aquila P, Montesanto A, De Rango F, Guarasci F, Passarino G, Bellizzi D. Epigenetic signature: implications for mitochondrial quality control in human aging. Aging. (2019) 11:1240–51. doi: 10.18632/aging.101832
59. Perneger TV. What's wrong with Bonferroni adjustments. BMJ. (1998) 316:1236–8. doi: 10.1136/bmj.316.7139.1236
60. Krzywinski M, Altman N. Points of significance: Comparing samples-part II. Nat Methods. (2014) 11:355–6. doi: 10.1038/nmeth.2900
61. Soini H, Routasalo P, Lagström H. Characteristics of the Mini-Nutritional Assessment in elderly home-care patients. Eur J Clin Nutr. (2004) 58:64–70. doi: 10.1038/sj.ejcn.1601748
62. Wei K, Nyunt MSZ, Gao Q, Wee SL, Ng TP. Frailty and malnutrition: related and distinct syndrome prevalence and association among community-dwelling older adults: singapore longitudinal ageing studies. J Am Med Dir Assoc. (2017) 18:1019–28. doi: 10.1016/j.jamda.2017.06.017
Keywords: Mini Nutritional Assessment, epigenetic, biomarkers, DNA methylation, aging, survival
Citation: Montesanto A, D'Aquila P, Rossano V, Passarino G and Bellizzi D (2019) Mini Nutritional Assessment Scores Indicate Higher Risk for Prospective Mortality and Contrasting Correlation With Age-Related Epigenetic Biomarkers. Front. Endocrinol. 10:672. doi: 10.3389/fendo.2019.00672
Received: 31 March 2019; Accepted: 16 September 2019;
Published: 01 October 2019.
Edited by:
Stefano Salvioli, University of Bologna, ItalyReviewed by:
Marian Beekman, Leiden University Medical Center, NetherlandsNicola Amodio, University of Catanzaro, Italy
Copyright © 2019 Montesanto, D'Aquila, Rossano, Passarino and Bellizzi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giuseppe Passarino, Z2l1c2VwcGUucGFzc2FyaW5vQHVuaWNhbC5pdA==; Dina Bellizzi, ZGluYS5iZWxsaXp6aUB1bmljYWwuaXQ=
†These authors have contributed equally to this work
 Alberto Montesanto
Alberto Montesanto Patrizia D'Aquila
Patrizia D'Aquila Veronica Rossano
Veronica Rossano Giuseppe Passarino
Giuseppe Passarino Dina Bellizzi
Dina Bellizzi