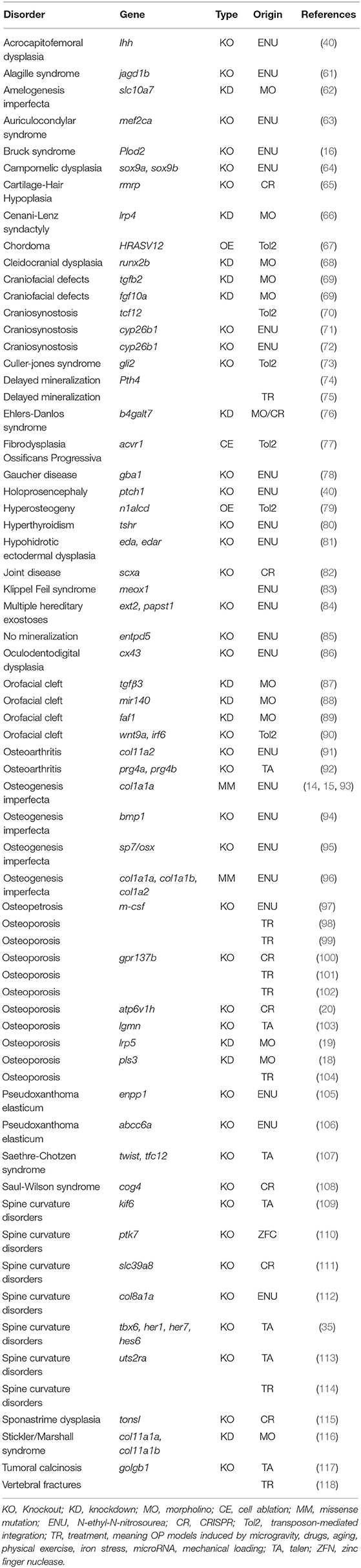
- 1Biochemistry Unit, Department of Molecular Medicine, University of Pavia, Pavia, Italy
- 2Department of Biomolecular Medicine, Center of Medical Genetics, Ghent University-University Hospital, Ghent, Belgium
- 3Bruker microCT, Kontich, Belgium
Animal models are essential tools for addressing fundamental scientific questions about skeletal diseases and for the development of new therapeutic approaches. Traditionally, mice have been the most common model organism in biomedical research, but their use is hampered by several limitations including complex generation, demanding investigation of early developmental stages, regulatory restrictions on breeding, and high maintenance cost. The zebrafish has been used as an efficient alternative vertebrate model for the study of human skeletal diseases, thanks to its easy genetic manipulation, high fecundity, external fertilization, transparency of rapidly developing embryos, and low maintenance cost. Furthermore, zebrafish share similar skeletal cells and ossification types with mammals. In the last decades, the use of both forward and new reverse genetics techniques has resulted in the generation of many mutant lines carrying skeletal phenotypes associated with human diseases. In addition, transgenic lines expressing fluorescent proteins under bone cell- or pathway- specific promoters enable in vivo imaging of differentiation and signaling at the cellular level. Despite the small size of the zebrafish, many traditional techniques for skeletal phenotyping, such as x-ray and microCT imaging and histological approaches, can be applied using the appropriate equipment and custom protocols. The ability of adult zebrafish to remodel skeletal tissues can be exploited as a unique tool to investigate bone formation and repair. Finally, the permeability of embryos to chemicals dissolved in water, together with the availability of large numbers of small-sized animals makes zebrafish a perfect model for high-throughput bone anabolic drug screening. This review aims to discuss the techniques that make zebrafish a powerful model to investigate the molecular and physiological basis of skeletal disorders.
Introduction
Preclinical animal models can be used to elucidate gene and protein function in ways often impossible in humans, by means of genome sequencing, advances in DNA manipulation and high resolution live-imaging (1). Mammals such as mice and non-human primates are traditionally the preferred models for biomedical research due to their close evolutionary relationship with humans. However, their use is costly and studies at early developmental stages raise ethical concerns. Furthermore, in most countries the adoption of the “Three R's” principles: Replacement, Reduction, and Refinement (2) for animal research is mandatory and encourages the use of alternative models, such as Danio rerio (zebrafish), Xenopus laevis/tropicalis (clawed toad), Drosophila melanogaster (fruit fly), and Caenorhabditis elegans (nematode). In these organisms in vivo techniques can be applied with the simplicity and versatility of in vitro assays and therefore they are frequently used in fundamental and biomedical research to quickly define gene functions and to develop novel therapeutic options (3). Zebrafish, the most frequently employed non-mammalian vertebrate animal model, is a freshwater bony fish, belonging to the Cyprinidae family and to the Teleostei infraclass of ray-finned fish which arose ~340 million years ago (4). This species was initially described by the Scottish physician and naturalist Hamilton (5) in a survey on South Asian flora and fauna. Starting from the pioneering research of George Streisinger in the 70s−80s, who was the first to clone a zebrafish and in this way demonstrated the easy genetic manipulation of this species (6), zebrafish became a powerful model organism for developmental studies, genetic research, drug and toxicology screenings and for understanding tissue regeneration and repair (7–9). In contrast to other vertebrate models such as mice, fertilization occurs externally, which together with transparency and rapid embryo to larval transition permits easy access and visualization of development (10) (Figure 1). Moreover, due to its rapid growth, a recognizable and complete vertebrate body plan is already in place by 1 day post fertilization (dpf) and embryogenesis is complete by 3 dpf (11). In contrast to other vertebrate models such as rodents, the small size and large number of offspring of zebrafish allow for increased sample numbers, thereby increasing the statistical power of experiments (3). Finally, the relatively low husbandry cost further contributed to the increasing popularity of the zebrafish as a model for research (11).
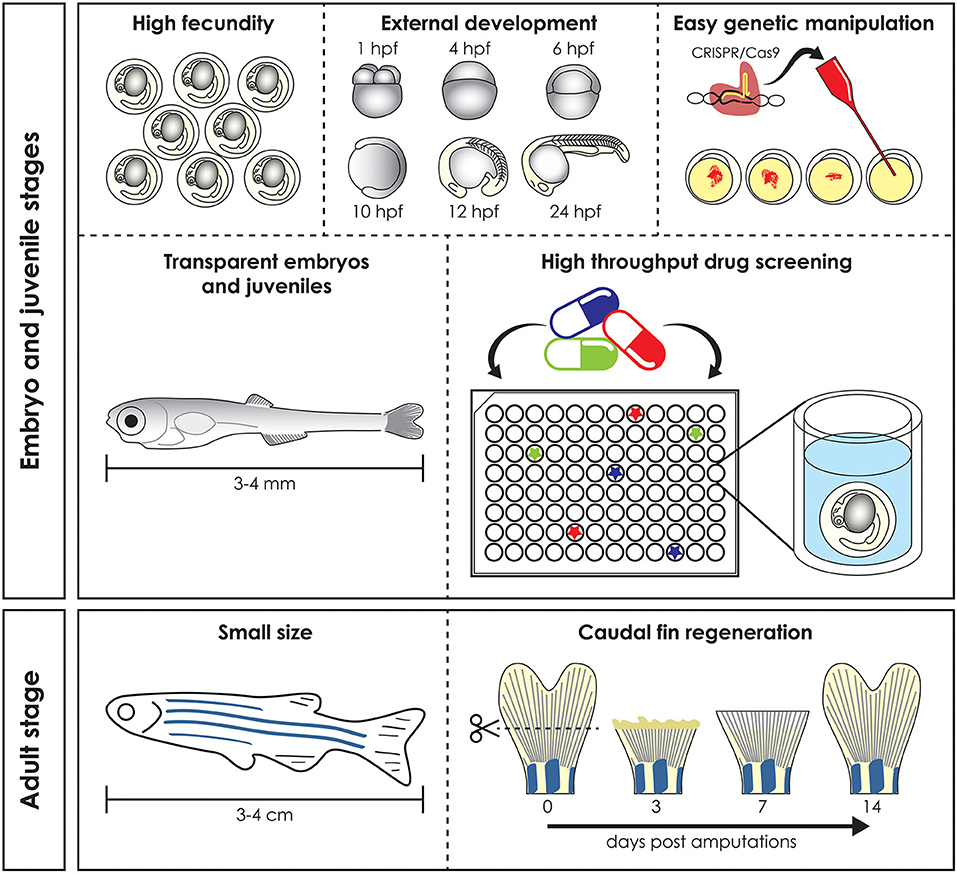
Figure 1. Advantages of the zebrafish model. Zebrafish has several advantages compared to mammal models. High fecundity and external fertilization and development allow easy genomic manipulation, transparent early life stages guarantee in vivo imaging and skin permeability makes them suitable for high throughput drug screening (top). Adult zebrafish reaches a maximum size of 3–4 cm and this make it easy and cheap to keep it in large numbers, reducing the husbandry cost (bottom left). Finally, zebrafish is used as a vertebrate model to study regeneration, due to its ability to regenerate different organs, such as the caudal fin, which is completely regenerated 14 days post amputation (bottom right). hpf, hours post fertilization.
Besides developmental studies, the zebrafish is an established research model in many other research fields. During the last 20 years, the zebrafish has proven itself as a useful model to study disease mechanisms (1). This is due to its physiological relevance and genetic tractability to model genetic variation in humans. Compared to mammalian model organisms, the zebrafish genome underwent an extra (third) whole duplication about 350 million years ago, with the result that for many genes in humans, there may be two copies (paralogues) in zebrafish. Despite this there is a relatively high level of genome conservation between zebrafish and humans with more than 70% of human protein-coding genes having at least one zebrafish ortholog. The haploid zebrafish genome has 25 chromosomes containing 1.7 billion base pairs (4). Various forward and reverse genetic approaches have been applied to generate mutant lines that mimic many different human diseases, including skeletal diseases ranging from secondary osteoporosis (OP) to rare disorders such as osteogenesis imperfecta (OI) (12–20). A major benefit of zebrafish is the simplicity of combining mutant and transgenic lines that express fluorescent reporter proteins under the control of responsive elements for signaling pathways or promoters of cell-type-specific markers. This in turn allows for in vivo investigation of the effect of a disease mutation on the spatio-temporal expression of specific genes, and on cell differentiation and signaling pathways.
Zebrafish larvae have been intensively used for pharmacological and toxicological screens, because of their small size (easy distribution in microtiter well plates), high abundance and their ability to absorb small compounds from the water through the skin and gills (21). Together with the availability of many different disease models, the zebrafish is a unique tool to develop novel targeted pharmacological approaches (Figure 1) (21).
Finally, their ability to regenerate some cells and tissues, such as fins and scales, makes the zebrafish a valuable model for understanding organ repair mechanisms during healthy and pathological conditions (Figure 1) (22).
This review, after providing a brief overview of zebrafish bone biology, will focus on the description and use of the various techniques and approaches which make Danio rerio a powerful model organism to investigate the molecular and physiological basis of skeletal disorders.
Zebrafish Bone Biology
The Skeleton
Skeletal development and gene expression and the general inventory of bone types are conserved between zebrafish and mammals, nevertheless few differences need to be considered when using this animal as model for skeletal study. Osteocytes are not present in all bones and/or at all developmental stages, endochondral ossification is rare in zebrafish and vertebral body do not build on a cartilaginous anlage (23, 24). The common perception of mammals being more complex than “lower” organisms, such as teleosts, is false, especially concerning the skeleton. Certain characteristics of the teleost skeleton are more advanced and elaborate compared to mammals, such as the zebrafish skull that contains at least twice the number of bones (24). At the tissue level, the mammalian skeleton mostly consists of cellular bone and hyaline cartilage. While other types of bone, such as hyperostotic and acellular bone and cartilage (i.e., fiber cartilage), can be present in mammalian skeletons, they are often associated with pathological processes. However, in teleosts many different bone and cartilage types with different cellularity and matrix composition exist in wild type conditions not related to disease (25). The zebrafish skeleton consists of a dermal skeleton and an endoskeleton. Scales, polarized structures of the exoskeleton, teeth, and fin rays are part of the dermal skeleton and are distinctive as skeletal structures in their ability to regenerate (25–27). In fish, teeth, scales, and fin rays can all be traced back in evolution to a single structure, called the odontode (28), and they arise at the epithelial-mesenchymal border (29, 30). It has been shown that the mesenchymal tissues that engender these skeletal elements have a neural crest origin (29, 31, 32).
The endoskeleton consists of cranial, axial, and appendicular skeletal elements (33). As in all vertebrates, the zebrafish cranial skeleton arises mostly from the cranial neural crest, while the appendicular skeleton develops from somite-derived paraxial mesoderm (31, 33). In contrast with tetrapods, in which vertebral centrum formation is controlled by somites patterned along the vertebral column, in teleosts the notochord has an instructive role in vertebral centrum patterning as the centra start out as mineralization foci in the notochord sheath (34, 35).
Skeletal Cells
The teleost and mammalian skeletal systems share similar cell types (Figure 2A). In cartilage there are (i) chondroblasts as the cartilage forming cells and (ii) chondrocytes maintaining the cartilage matrix. In bone there are (i) osteoblasts as the bone forming cells, (ii) osteocytes that act as the mechanosensors regulating osteoblast and osteoclast activity and (iii) osteoclasts which are the bone resorbing cells (24, 37). Similar to mammals, teleost skeletal histogenesis involves the differentiation of chondroblasts and osteoblasts, that secrete the collagen extracellular matrix, from mesenchymal stem cells (38, 39). Both in mammals and fish, skeletal cells are formed by a complex interplay of intracellular molecular pathways and secreted factors that regulate the timing, location, and pathway by which bone cells differentiate (40–42). Although not investigated in mammals before, in zebrafish osteoblasts are present in clusters at the end of growing bones and can be classified in two different groups (type I and type II) based on cell cluster size, location, and nuclei shape, although they have overlapping functions (36). Type I osteoblasts are located at the edges of growing flat bones, such as the dentary, maxillary, and frontal bone, in large clusters with more than 25 cells with a wide oval, round, or irregularly shaped nucleus. Laterally to these cells there is a zone of differentiating osteoblasts where cells are smaller and more elongated, assuming the typical spindle shape of osteoblast-like cells, which cover all zebrafish bones with a monolayer at the level of the perichondrium (36). Type II osteoblast clusters are smaller (4–12 cells) and are distributed throughout the skeleton. These osteoblasts have a reduced size, elongated nucleus and are present throughout the bony trabecular network of spongy bones. Type II osteoblast clusters can also be detected at the edges of cartilage break down zones and lateral to the epiphysial growth plate, at the outer surface of tubular bones (36).
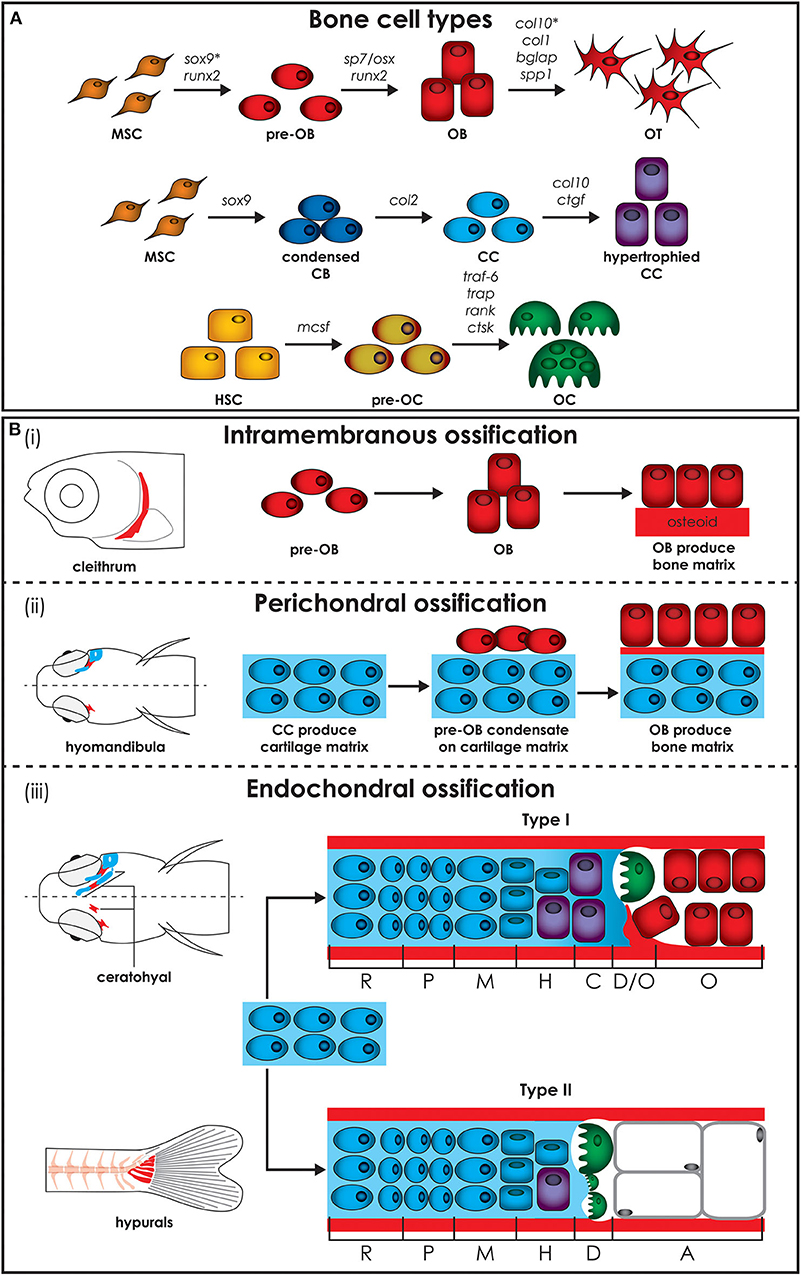
Figure 2. Zebrafish bone cells and ossification types. (A) Bone is formed by osteoblasts and osteocytes, while cartilage is formed by chondroblasts and chondrocytes, and both bone and cartilage are degraded by osteoclasts. All bone cell types develop from progenitors similar to the mammalian counterpart and share similar gene expression profiles (genes are indicated above arrows). Note however that HSCs in zebrafish are not present in the bone marrow but in the head kidney. In addition, the genes for collagen X, encoded by col10, and SRY-box transcription factor 9 (indicated by*), encoded by sox9, are expressed during osteoblasts differentiation in zebrafish, but not in humans. (B) Three types of ossification are present in zebrafish: (i) intramembranous ossification, (ii) perichondral ossification, present in teleosts but not in humans, and (iii) endochondral ossification. (i) During intramembranous ossification mesenchymal stem cells condensate and differentiate into pre-osteoblasts and finally into mature osteoblasts that deposit bone matrix (osteoid) that subsequently mineralizes. (ii) Perichondral ossification starts at the surface of a cartilaginous template where osteoblasts deposit bone matrix without replacing the cartilage. (iii) Endochondral ossification is the process by which growing cartilage is replaced by bone to allow the skeleton to grow. For ossification to start, matrix surrounding the chondrocytes must calcify so that osteoclasts can break down the cartilage. In teleost two types of endochondral ossification exist. Type I endochondral ossification, typical in the ceratohyal, resembles the mammalian endochondral ossification process. This is characterized by a hypertrophic zone, where the cartilage matrix calcifies, followed by a degradation zone where osteoclasts (also referred to as chondroclasts) degrade the cartilaginous matrix, and a bone formation zone. Type II ossification, in the hypurals, is characterized by a lack of the calcification and ossification zones, leading to tubular concave bones filled with adipose tissue. Schematics based on detail description in Weigele and Franz-Odendaal (36). A, adipose zone; C, calcification zone; CB, chondroblasts; CC, chondrocytes; D, degradation zone; H, hypertrophic zone; HSC, hematopoietic stem cell; M, maturation zone; MSC, mesenchymal stem cell; O, ossification zone; OB, osteoblasts; OC, osteoclasts; OT, osteocytes; P, proliferation zone; R, rest zone.
Most skeletal elements in the adult zebrafish skeleton contain osteocytes, but with a smaller volume and less canaliculi compared to mice and humans (36). The mechanosensing ability of osteocytes in zebrafish is not fully understood yet, but it was shown that osteocytes have a preferred orientation in adult zebrafish vertebrae (36). Acellular bone, without trapped osteocytes, can be found in many zebrafish cranial bones. Contrary to expectations, acellular bone does not appear to be stiffer due to the lack of osteocyte lacunae, making the role of acellular bone unclear (43). It is important to note that both cellular and acellular bone can occur within the same bony element. Osteon-like structures in zebrafish have been reported (for the lateral ethmoid bone) but these structures, composed of a central Haversian canal and bone lamella, do not have osteocytes (36).
In mammals, bone resorbing cells are multinucleated macrophages originating from the fusion and maturation of peripheral blood monocytes differentiated from hematopoietic bone marrow cells (44). Multinucleated osteoclasts can also be found in teleosts, especially in basal teleosts, such as salmonids and cyprinids (45). Nevertheless, in teleosts, smaller and mononucleated osteoclasts are predominant, but they retain the molecular regulators of mammalian osteoclast function (37). Examples include receptor activator of nuclear factor kappa-B (Rank) and Rank-ligand (Rankl) which are important for osteoclast maturation. Mature osteoclasts become tartrate-resistant acid phosphatase (Trap) and cathepsin K (CtsK) positive, which are both required for the cells to be able to degrade bone matrix components (37, 46). Zebrafish are characterized by an ontogenic change at 30 dpf when mononucleated osteoclasts evolve to multinucleated osteoclasts, which perform lacunar resorption and bone remodeling (37).
Each cell type achieves and performs its function by involving specific genes, acting as molecular fingerprints. All three bone cell types develop from similar progenitors as their mammalian counterpart and share similar profiles of gene expression (Figure 2A) (36). Gene expression of zebrafish collagen and transcription factor in skeletal cells of cartilage and bone are not completely conserved with mammals. Unlike mammals, zebrafish osteoblasts express collagen type X and various teleosts have been shown to have collagen type II in their bone matrix (47, 48). In addition, Sox9 expression, which is required for differentiation of chondrocytes, but not of osteoblasts in mammals, has been reported to be involved in bone development in teleosts (49). Unlike tetrapods, zebrafish type I collagen, the most abundant protein in bone, has three instead of two different α chains, namely α1, α3, and α2 encoded by col1a1a, col1a1b, and col1a2, respectively (50). Based on the amino acid sequence, the α3 chain is phylogenetically similar to α1, supporting the common origin of their coding genes, which derive from a genome duplication that occurred at an early stage in teleost evolution (51). Importantly, all amino acid residues involved in human/mouse collagen type I cross-links are conserved in zebrafish, suggesting the existence of similar extracellular assembly (50).
Bone Ossification
Bone formation starts in zebrafish around 4–5 dpf. The bony elements can have three modes of ossification: intramembranous, perichondral, or endochondral. Intramembranous ossification starts with mesenchymal cell condensation and differentiation into osteoblasts, without the need of a cartilage template (Figure 2Bi) (45). This type of ossification occurs in the skull, for example in the cranial roof and opercular bones, in the vertebral column, where most of the vertebral body is formed by this type of ossification, in scales and in the fin rays (45). In mammals, this ossification is mostly restricted to bones of the cranial vault and the dentary (52).
Perichondral ossification, characterized by bone formation in the perichondrium, is more common in the teleost compared to the mammalian skeleton, where it has been considered as a form of intramembranous ossification (45). In teleosts perichondral ossification is present in the hyomandibula and Meckel's cartilage, where osteoblasts aggregate on the surface of the cartilaginous template and deposit bone matrix into the perichondrium (Figure 2Bii).
Endochondral ossification, which is the main type of ossification in mammals, is uncommon in teleosts. In this type of ossification, mesenchymal cells condense and differentiate into chondroblasts and chondrocytes, which then secrete an extracellular cartilage matrix that functions as a template that is replaced by bone matrix (Figure 2Biii). In teleosts, two types of endochondral ossification exist. In a few bones, such as the ceratohyal and the radials in the pectoral fin, type I endochondral ossification takes place at the level of epiphysis and of the epiphysial growth plate resembling the mammalian endochondral ossification process. It is characterized by a resting zone, a proliferation zone with columnar cartilage, and a hypertrophic zone followed by a region in which cartilage matrix calcifies (36). Finally, chondroclasts degrade the cartilaginous matrix (degradation zone), allowing osteoblasts to lay down bone matrix (ossification zone). In the hyomandibula, branchial arches, ethmoid and hypuralia type II endochondral ossification takes place. Here, the calcification and ossification zones are absent and the cartilage template is replaced by adipose cells, leading to tubular concave bones filled with adipose tissue (36, 37).
Because the cranial skeleton is often too complex for screening by high throughput methods, the zebrafish vertebral body is the most investigated component of the skeleton both in early and adult life stages. Although the vertebrae in both mammals and teleosts consist of notochord and bone, there are a few key differences. First, the notochord is the de facto vertebral column in early teleost life stages and persists throughout life, while it only forms the intervertebral disc in mammals (53, 54). The notochord consists of a core of large and vacuolated chordocytes which is surrounded by an epithelial layer of chordoblasts that secrete the notochord sheath. This sheath is a stratified structure, composed of a thin external membrane containing elastin, covering a thicker layer of mainly collagen type II (54). Second, while the vertebrae in mammals have a cartilaginous precursor which endochondrally ossifies, zebrafish vertebrae form initially through direct mineralization of the notochord sheath, called chordacentra, in the absence of a cartilaginous precursor (55, 56). To this day, the exact cellular involvement of this notochord sheath mineralization remains unresolved. Third, the teleost vertebra is subsequently built via intramembranous ossification outside the notochord onto the chordacentrum, consisting of a compact autocentrum and trabecular arcocentrum, which forms the neural and haemal arches (56, 57). The osteoblasts produce collagen type I bone matrix and start to ossify the autocentrum at the level of the intervertebral disc, which acts as the growth center of the vertebral centrum (34).
Generation of Knock-Out and Knock-In Zebrafish Models
Forward Genetic Approach
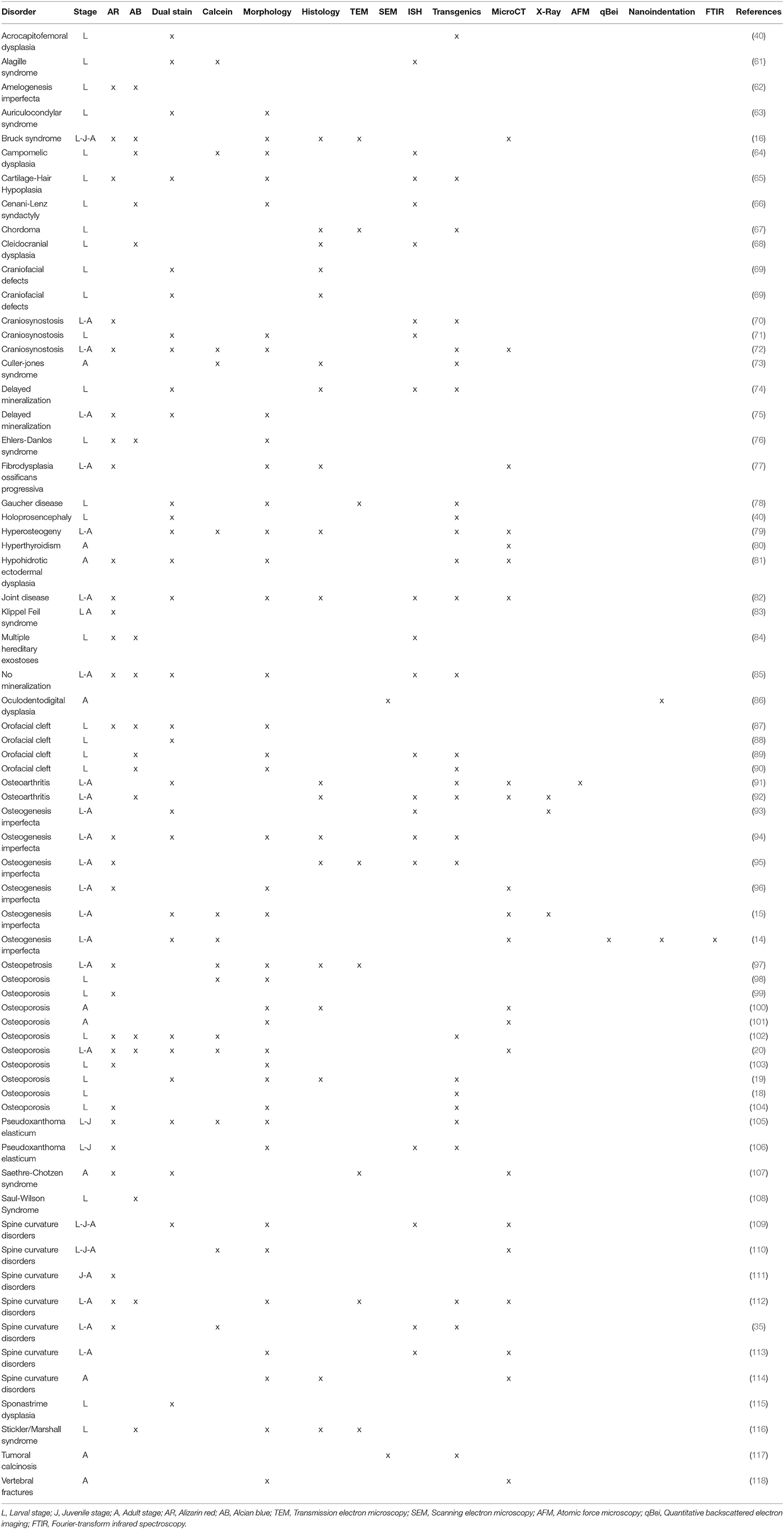
Different methods to generate zebrafish models of human disorders have been explored over the last decades. Initially, a number of large-scale forward genetic screens, based on random mutagenesis with radiation, chemicals, or insertional mutagenesis, revealed zebrafish mutants affecting different aspects of embryonic development and biological processes (58–60). This phenotype-driven approach was also applied to screen for genes involved in skeletal development and diseases (Table 1). Several mutants with defects in craniofacial cartilage elements and with mineralized tissue phenotypes (119), or with changes in the shape of the skeleton (96) were identified in large scale forward genetic screens. Mapping of the causative change established some of these mutants as models for human skeletal disorders. For instance, in a study by Gistelinck et al. (120), several type I collagen zebrafish mutants, previously discovered in a forward genetic screen (96), were established as representative models for the brittle bone disorder osteogenesis imperfecta.
Reverse Genetic Approach: Morpholino Knockdown and Gene Editing
Although forward genetics brought great progress to the field of disease modeling, still, for many causal human disease genes, this approach did not reveal corresponding zebrafish mutants, as there is incomplete genome coverage of mutagenesis. Consequently, the need to investigate the function of relevant candidate genes for specific diseases or developmental pathways, sparked the expansion of reverse genetic approaches in the zebrafish field.
The assessment of candidate gene function was initially enabled via knockdown through the use of antisense morpholinos (MO). Their ease of use made this approach increasingly popular for gene function analysis, and several early studies demonstrated that MO-mediated knockdown (“morphants”) recapitulated known mutant phenotypes (121, 122). Over the past years, MOs have also been used in zebrafish modeling of skeletal disorders (Table 1). An example includes the monogenetic form of X-linked osteoporosis, caused by loss-of-function variants in PLS3 encoding for plastin 3, a cytoskeletal protein involved in bone homeostasis. MO-mediated knockdown of pls3 in zebrafish (18) induced malformations of the developing craniofacial bone structure, which could be reversed by the administration of human PLS3 mRNA. Another example by Flores et al. (68) shows that depletion of runx2b by MO injection severely compromised craniofacial cartilage formation, phenocopying the human dominantly inherited disorder cleidocranial dysplasia, a condition characterized by impaired ossification and multiple skeletal abnormalities (68). Nevertheless, problems with the application of MOs in zebrafish emerged, such as the frequent occurrence of p53-dependent apoptosis (123–125), and off-target effects resulting in so-called “pseudophenotypes” (126, 127), but also MO-induced phenotypes that cannot be recapitulated in existing mutants (128). The latter issue has recently been studied in more detail leading to the insight that, at least for some genes, the phenotypic differences between morphants and mutants can be due to genetic compensation in the latter, but not in the former (129).
Definitive reverse genetic approaches in zebrafish recently became available in the form of site-specific nucleases enabling targeted gene modification. Initial work utilized zinc finger nucleases (ZFNs) (130, 131), and transcription activator-like effector nucleases (TALENs) (132). However, CRISPR/Cas9 genome editing is currently the most versatile and frequently employed reverse genetic technology for the creation of both knock-out and knock-in disease models. The CRISPR/Cas9 system induces a double-stranded DNA break (DSB), carried out by the Cas9 nuclease, at a specific target site, recognized by the binding of a single-guide RNA (sgRNA) molecule. Following DSB, different endogenous repair mechanisms can be initiated. On one hand, the error-prone non-homologous end joining (NHEJ) pathway can be activated, often leading to the introduction of indel mutations due to imprecise repair, resulting in gene knock-out. The generation of gene knock-outs in zebrafish is relatively straightforward and efficient. In a study by Zhang et al. (20) for instance, mutations in the ATP6V1H, coding for vacuolar ATPase, were identified in patients with short stature and osteoporosis. Loss-of-function mutants in atp6v1h were generated in zebrafish through CRISPR/Cas9-mediated gene knock-out (20). These mutants demonstrated loss of bone mass and increased expression of matrix metalloproteases mmp9 and mmp13. Indeed, pharmacological inhibition of mmp9 and mmp13 rescued the bone phenotype, suggesting that blockade of collagen degradation can be a valid therapeutic target. CRISPR/Cas9 gene editing has been recently used to generate knock-out zebrafish for crtap and p3h1, two genes that are part of a protein complex which is involved in prolyl 3-hydroxylation and proper folding of collagen type I. Loss-of-function mutations in the human ortholog genes cause recessive forms of OI. These zebrafish models faithfully mimic the human disease and support the defective chaperone role of the 3-hydroxylation complex as the primary cause of the skeletal phenotype (17).
In general, reverse genetic approaches are limited by the time required to generate mutant lines, where stable knock-out zebrafish are mostly obtained and analyzed from the F2 generation on. Therefore, an approach for rapid CRISPR-based reverse genetic screens was developed in which phenotyping is performed directly in F0 (mosaic) founders, which are called “crispants” (133, 134). This enables moderate to rapid throughput reverse genetic screens of candidate genes, contributing to skeletal disease. In a study by Watson et al. (133), the comparison between somatic, CRISPR-generated F0 mutants and homozygous germline mutants for plod2 and bmp1, two genes implicated in recessive OI, revealed phenotypic convergence, suggesting that CRISPR screens of F0 animals may faithfully recapitulate the phenotype of skeletal disease models (133).
As an alternative to NHEJ-mediated repair of CRISPR/Cas9-induced DSB, the homology-directed repair (HDR) pathway can be initiated, but only in the presence of a homologous repair template. In physiological circumstances, HDR occurs between sister chromatids during the G2 and S phase of the cell cycle. The knock-in modeling procedure exploits this mechanism by supplying the CRISPR/Cas9 system with an artificial repair template, homologous to the target sequence and containing a specific variant of interest. For the generation of knock-in models, mostly single-stranded oligodeoxynucleotide (ssODN) repair templates are used (135) mainly because the design and production of ssODNs is easier, cheaper and results in higher HDR efficiencies compared to double-stranded templates such as plasmids (136, 137). The need to complement knock-out models with these more precise knock-in disease models is growing, for various reasons. Firstly, specific point mutations may cause a highly divergent pathobiology compared to loss-of-function mutations modeled by knock-out models. More specifically, certain missense mutations may cause a gain-of-function rather than a loss-of-function, while missense mutations in genes encoding proteins included in protein complexes may exercise a dominant negative effect and change the function of the whole protein complex. For instance, in dominant types of OI caused by mutations in the genes encoding the type I collagen α chains, depending on the type of mutation, either the quantity or the structure of type I procollagen is altered (138). The “quantitative” mutations, mostly resulting in a null COL1A1 allele, typically cause mild forms of OI, while “qualitative or structural” defects, frequently associated with glycine substitutions, can be responsible for lethal, severe or moderate forms of the disease.
Also, missense mutations in vital developmental genes may be hypomorphic while their loss-of-function counterparts result in early lethality, as reported in the cdc6 zebrafish mutant for Meier-Gorlin syndrome. Hypomorphic mutations in the cdc6 gene recapitulate the patient's phenotype, while the knock-out mutants are embryonically lethal. In these cases, the introduction of such point mutations is a prerequisite to faithfully recapitulating human disease. Secondly, as mentioned before, several zebrafish knock-out models failed to generate a phenotype, which can be due to mRNA decay-induced genetic compensation (139), a phenomenon that is not expected to occur in knock-in models.
Nevertheless, several drawbacks mitigate the straightforward use of HDR knock-in zebrafish models. Firstly, HDR pathways have proved highly inefficient for genome editing (140) even despite proposed modifications, such as repair template modification (141, 142), cell cycle arrest (143) and chemical compound administration (144–151). Secondly, CRISPR/Cas9-mediated HDR mechanisms have been shown to be error-prone (152, 153). These issues hindered the development of knock-in zebrafish models and only a limited number have been reported, in contrast to numerous knock-outs. For instance, CRISPR/Cas9-mediated point mutation knock-ins have been generated for genetic variants implicated in inherited cardiac diseases (154–156), although to our knowledge none have been described so far for skeletal diseases. Different recently developed DSB-free alternatives for precise base pair substitution, such as programmable base editing (157–159) and prime editing (160) promise to be more efficient and versatile approaches, but more research is needed to further improve these methods for application to the zebrafish model system.
Transgenic Lines
Transgenic Zebrafish to Trace Bone Cells and Pathways
Despite the development of new approaches in large-scale and more recently single-cell transcriptomics, genomics, epigenomics, and proteomics (161), these techniques are time consuming, expensive and only available in specialized laboratories (162–164). Furthermore, retrospective -omic analyses exclude cells that do not survive to the point of cell harvest, a common and necessary event in growth and regeneration. Therefore, to be able to understand the dynamic nature of tissue development and regeneration, in vivo time-lapse imaging is essential.
The recent evolution of genetic engineering has allowed the generation of transgenic animal models, expressing fluorescent proteins under cell- or pathway- specific promoters, enabling in vivo imaging of differentiation and signaling (165). However, the generation of transgenic murine models remains technically demanding, time consuming and expensive (166). In addition, since mice develop in utero, it is almost impossible to investigate early developmental processes in real time and the visualization at cellular level usually requires post-mortem analyses (167).
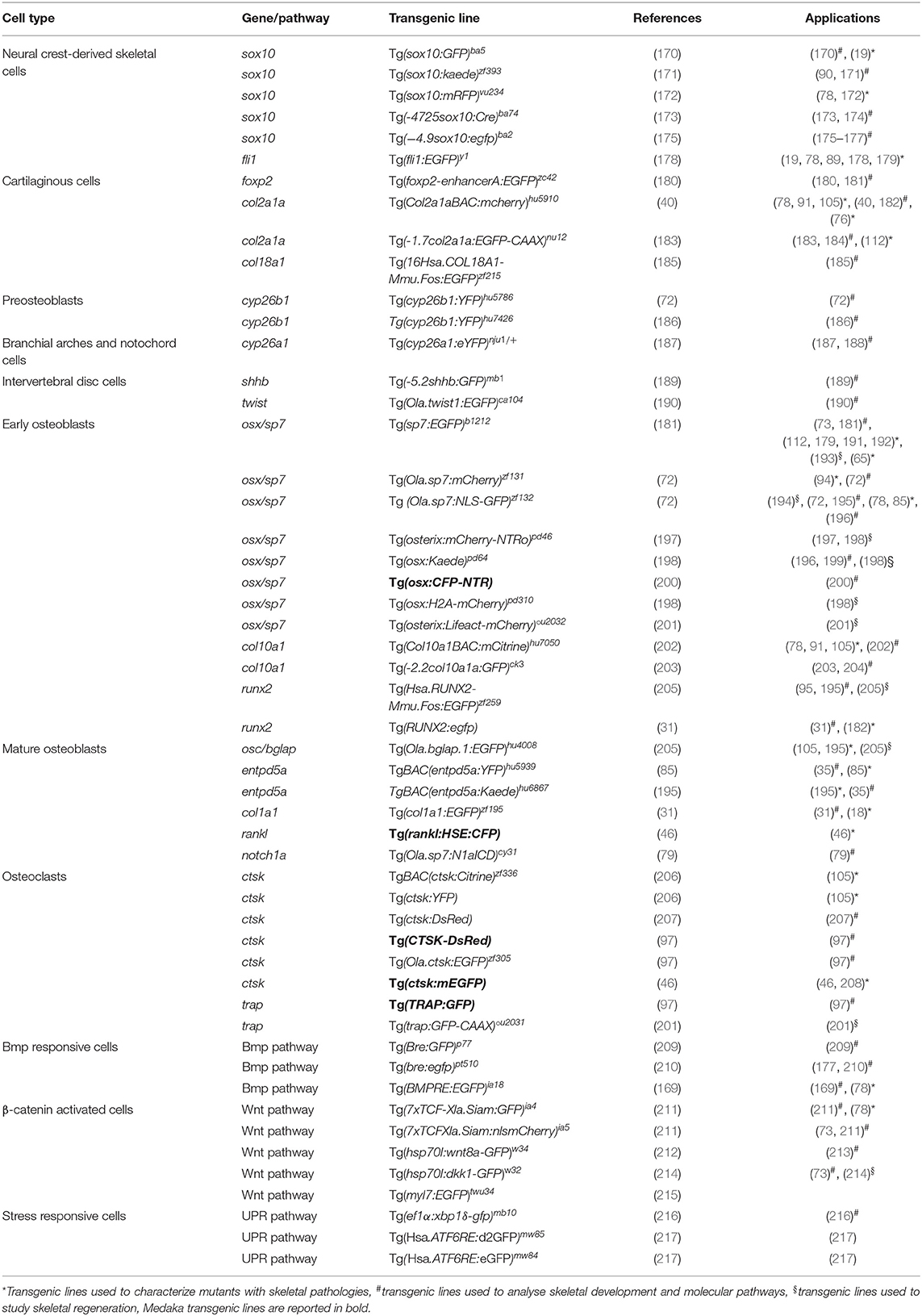
Zebrafish, with its fast external development, transparent early life stages and relative easy genetic manipulation, is rapidly becoming the model of choice for examining developmental processes via time-lapse microscopy. The introduction of reporter genes downstream of a specific promoter makes it possible to produce site-directed indicators in different organs, tissues or cells and permits real time imaging in developing embryos or post-hatch stages; or even in mature zebrafish by fluorescent microscopy on whole mount specimens (168, 169). A variety of transgenic reporter lines have been generated to mark skeletal cell lineages at different stages of differentiation and signal transduction pathways, by using the conserved regulators of skeletal development (Table 2). The availability of fluorescent reporter lines, together with the use of powerful techniques such as two or multi-photon or light sheet microscopy, has allowed imaging of tissues and organs at a cellular and subcellular level, especially by exploiting the transparency of early life stages (218).
Transgenic Lines to Trace Bone Cells
The most frequently used lines expressing fluorophores in chondrocytes include Tg(−4.9sox10:egfp)ba2 and Tg(Col2a1aBAC:mcherry)hu5910 (Table 2). The Tg(−4.9sox10:egfp)ba2 was employed to detect sox10 expression in head cartilage during embryo development and to follow migration of neural crest cells during cranium morphogenesis (175). The Tg(Col2a1aBAC:mcherry)hu5910 reporter line allowed impaired cartilage patterning and loss of chondrocyte organization to be shown in a zebrafish model of a recessive form of Ehlers-Danlos syndrome with partial loss of B4galt7, a transmembrane Golgi enzyme that plays a pivotal role in proteoglycan biosynthesis (76).
In order to trace the differentiation of bone forming cells, transgenic lines for both early and late osteoblast markers, expressing fluorophores under the osterix/sp7 and osteocalcin/bglap promoters, have been generated (Table 2). The Tg(sp7:EGFP)b1212 line allowed osteoblast behavior to be studied during both intramembranous and endochondral ossification. Moreover, this line was used to investigate the abnormal perichondral ossification in the RNA component of the mitochondrial RNA-processing endoribonuclease (rmrp) knock-out zebrafish model of cartilage hair hypoplasia (65). Tg(Ola.sp7:mCherry)zf131 was crossed with the OI type XIII zebrafish model frilly fins to elucidate the role of the bone morphogenic protein 1, encoded by bmp1a gene, in osteoblast differentiation and localization (94).
The Tg(Ola.bglap.1:EGFP)hu4008 line was used to understand the fundamental role of osteoblast dedifferentiation during bone healing in response to traumatic injury, and to show that adult zebrafish osteoblasts display an elevated cellular plasticity compared to their mammalian counterpart (195).
Despite the conservation of most of the osteoblastogenic markers, in zebrafish the expression of col10a is not limited to chondrocytes as in mammals, but is also present in osteoblasts (203). The transgenic line Tg(-2.2col10a1a:GFP)ck3, expressing GFP under col10a1 promoter, has therefore been used to investigate molecular events driving both chondrocyte and osteoblast development (203).
An interesting application of the transgenic reporter lines is their use in combination with a mineral stain, imaged at different fluorescent wavelengths, enabling the combined study of osteoblast dynamics and bone mineralization (196). For instance, alizarin red staining of the transgenic zebrafish Tg(Ola.sp7:NLS-GFP)zf132 localized osterix/sp7 positive osteoblasts in the mineralized bone and revealed the absence of osterix/sp7 expression in the anterior notochord region at 8 dpf (104). Similarly, mineral staining in combination with Tg(osx:Kaede)pd64 confirmed the osteoblast independent mineralisation of the notochord (196).
Most of the available osteoclast reporter lines express fluorophores under control of the promoter of cathepsin K (Ctsk), the osteoclast collagenase that mediates bone resorption (Table 2) (46). Chatani et al. (97) proved the absence of osteoclasts in the panther mutant, which lacks a functional receptor for the macrophage colony stimulator factor, taking advantage of the Tg(ctsk:mEGFP) transgenic line. A significantly reduced number of GFP-positive osteoclasts was found in the neural and haemal arches in panther larvae, indicating a crucial role of the protein in osteoclast proliferation and differentiation. Additionally, the medaka, another well-characterized teleost bony fish used for developmental and biomedical studies, was used to study osteoclasts by placing the gene encoding for the receptor activator of nuclear factor kappa-B ligand, rankl, a key osteoclast differentiation factor, under the control of a heat shock element (23). Increased osteoclast differentiation induced upon Rankl activation in this Tg(rankl:HSE:CFP) line resulted in an osteoporotic phenotype (46).
Transgenic Lines to Trace Signal Transduction Pathways
Zebrafish transgenic lines expressing in vivo reporter proteins under the control of signaling pathway responsive elements are a powerful tool to dissect dynamically the in vivo activation or repression of endogenous signaling pathways in real time (210, 219–221). Calcium, Bmp and Wnt pathways are crucial players during bone formation (222–224). Transgenic lines to further investigate these pathways have been generated (Table 2). The Tg(hsp70:bmp2b-GFP) line was used to analyze the role of the Bmp2 signaling pathway in an enteric disease, but the transgenic model could be employed to dissect BMP2b signaling in bone (225). To investigate Wnt pathway activation the Tg(7xTCF-Xla.Siam:GFP)ia4 and Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 transgenic lines, which contain multimerized tcf/lef binding sites for the transcription factor activated by β-catenin upstream to a siamois minimal promoter, were generated allowing the dynamics of neural crest-derived cell migration to be traced during development (211). Using the Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 transgenic line it was also possible to elucidate important regulatory steps in the osteogenic differentiation process of mesenchymal stem cells (73).
Finally, the unfolded protein response (UPR) was shown to play an important role in the modulation of the phenotype in rare skeletal diseases (226, 227). Interestingly, transgenic zebrafish lines allowing different branches of this pathway to be followed are already available (216, 217, 228, 229). For instance, the transgenic zebrafish model Tg(ef1α:xbp1δ-gfp)mb10 has been used to trace in vivo the splicing of xbp1, one of the terminal effectors of the UPR (216).
Live Imaging of Bone Regeneration
Tracing bone cells in vivo using transgenic lines in adult zebrafish is challenging due to tissue depth and complexity, but is possible in external structures such as fin rays or scales, which are easily accessible and suitable for regeneration studies (198, 230, 231). Indeed, the available panel of transgenic lines expressing fluorescent and photo-switchable reporter genes in bone cells is useful to trace regeneration in vivo (198). This strategy has clarified important biological aspects such as the cellular basis of integumentary bone regeneration. In vivo imaging of the Tg(sp7:EGFP)b1212 transgenic line during caudal fin regeneration showed the presence of GFP positive cells at the amputation plane starting from 2 days post amputation (dpa) and their association with the formation of newly mineralized matrix by 5 dpa (181). Osteoblast lineage tracing in the Tg(osx:Kaede)pd64 clarified migration and dedifferentiation of scleroblasts during fin regeneration (196).
However, the slow rates of regeneration require long-term live imaging to capture dynamic cellular events to improve the understanding of development, homeostasis, and regeneration by stem cell populations (232). Thus, to enable up to 24 h of continuous live imaging, specific protocols for long-term anesthesia of adult zebrafish have been optimized (198). Indeed, the transgenic line Tg(osx:H2A-mCherry)pd310 allowed spatio-temporally distinct cell division, motility, and death dynamic within a founder osteoblast pool to be imaged as bone regenerates (198).
Transgenic Lines as Tool for Drug Screening
Transgenesis is not only used to analyze bone development over time, to assess a mutant phenotype or track cell signaling, but also to evaluate drug screening effects (98, 104). Huang and colleagues employed the transgenic line Tg(Ola.sp7:NLS-GFP)zf132 to test anti-osteoporosis chemical drugs. This line, that expresses GFP under control of osterix/sp7, allowed for a faster in vivo evaluation of drug effects on bone mass and density compared to traditional staining methods. In another study, the osteocalcin/bglap reporter transgenic line Tg(Ola.Bglap:EGFP)hu4008 was employed to test chlorpropamide effects on the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB). The drug negatively regulated osteoblast-like cell dedifferentiation, thus helping to maintain bone forming cells in an active state promoting caudal fin ray regeneration (233).
Tips for Transgenic Lines Selection
For the proper selection of transgenic lines there are some aspects that require consideration. First, the choice of the reporter protein is influenced by differences such as color, brightness, toxicity, tissue penetration, subcellular localization, as well as the stability of the fluorescent protein. For instance, in order to study cell signaling dynamics or when performing prolonged cell lineage tracing, the use of long half-life fluorescent proteins is recommended. Furthermore, differences in signal pattern and intensity can be found among transgenic progeny possibly due to multiple insertions in the same founder, thus complicating the analysis (169). This aspect can be ameliorated by diluting the number of transgenic copies through subsequent generations.
Finally, in order to verify the localization of the reporter protein, the use of dual color analysis in the same transgenic line is recommended (196, 199) by for example complementary secondary techniques such as immunohistochemistry or in situ hybridization (169, 199).
X-Ray Imaging
One of the more frequently used techniques to visualize the human skeleton is x-ray imaging. Classic x-ray systems for human and veterinary purposes need to limit radiation exposure to the patient, and therefore have limited exposure settings, that is their range of tube accelerating voltage (kV), current (mA), and time of exposure. These parameters are set to optimize the image of the skeleton while keeping the radiation exposure to the patient as low as possible and cannot be easily changed. Consequently, these medical appliances are not appropriate to image the small zebrafish skeletons. Examples of x-ray sources that have a wide range of possible x-ray output settings are small manual units used to scan museum artifacts and fossils, a small animal radiation research platform (SARRP; Xstrahl, Surrey, UK) and the Faxitron© x-ray cabinets. Specifically, these sources can be set to low power but long exposure time parameters, and can be used in combination with high resolution technical film such as mammography film or x-ray film (e.g., AGFA D2) used in aerospace and petroleum factory applications. A Faxitron x-ray cabinet in combination with mammography film was used by Fisher et al. (93) to image the skeleton of WT and chihuahua mutant zebrafish to screen for skeletal abnormalities (Table 2).
With the revolution of digital sensors capturing the x-ray signal, it has become straightforward to take an x-ray image of a small or large part of the human skeleton. The use of digital x-ray sensors is however more challenging when using zebrafish (24, 234) as the resolution is too low in most cases to capture a quality image of the small zebrafish skeleton. A modern system such as a Faxitron Ultrafocus x-ray cabinet can provide digital x-ray images up to a 5 μm spatial resolution which can be geometrically magnified (Faxitron©) (Figure 3A). This technique was used to screen for deformed and fragile bones in chihuahua mutant zebrafish (15) and to assess the gross skeletal anatomy of prg4a−/−; prg4b−/− mutant zebrafish (92). Although these digital images may look clean and sharp, the thinner less mineralized bones may not be present in the image, which represents a loss of information about the zebrafish skeleton (234). In contrast, technical film such as AGFA D2 can theoretically capture extremely high-resolution images. Such technical film works well in combination with low energy settings needed for optimal imaging of the zebrafish skeleton. Moreover, this film is able to capture an image of smaller bones, which is not always possible when using a digital sensor.
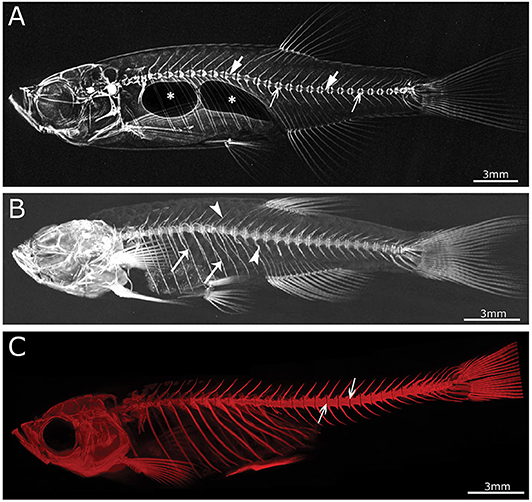
Figure 3. Imaging techniques in zebrafish. (A) Lateral x-ray image of a wild type zebrafish acquired with a Faxitron tabletop X-ray cabinet. Notice the outline of the major bones in the skull and vertebral column and the outline of the double chambered swim bladder (indicated by asterisks) in the abdominal cavity. The tissue inside the vertebrae (indicated by block arrows) and intervertebral spaces (indicated by line arrows), i.e., the notochord, can be easily assessed for the presence of mineral. (B) Lateral view of a 3D reconstructed microCT scanned adult zebrafish at 21 μm. More details are visible in the skull and especially the vertebral column compared to the x-ray image (neural and haemal arch are indicated by arrow heads and the ribs with a small arrows). (C) Lateral image in the fluorescent channel of a zebrafish whole mount cleared and stained with alizarin red for mineralized tissues. Compared to the images above, more details of the skeleton can be observed, especially in the vertebral column where all individual bones and their outlines can be noticed. The alizarin red image also allows to assess the presence of mineral in the intervertebral space (indicated by arrows). All images were taken of wild type zebrafish.
The main advantage of using x-rays to image the zebrafish skeleton is that it is a cheap and quick methodology. Furthermore, x-ray imaging can be repeated on live organisms and can be used as a preliminary diagnostic tool for skeletal imaging before applying a more specialized method such as micro computed tomography (microCT) or mineral staining (Figures 3B,C). For instance, x-ray imaging is frequently used in aquaculture related research where it is a first line tool to assess skeletal deformities (235, 236). Although x-ray imaging can be employed to assess skeletal deformities in adult zebrafish, its use for juvenile zebrafish, where the skeleton is too small to be captured on film or digitally, is not feasible. In addition, x-ray images of zebrafish are not suitable for quantification of tissue or bone mineral densities. MicroCT currently provides a better solution to estimate these bone parameters (80, 120).
Micro Computed Tomography
Computed tomography (CT) is a non-invasive technology based on x-ray analysis that allows detailed 3D reconstructions of large specimens. The generation of CT images involves the capturing and recording of x-rays that pass through the sample onto a detector. This process is repeated several times for multiple angles, followed by the virtual reconstruction into a 3D image (237). The required resolution for zebrafish imaging is beyond the capabilities of medical CT machines (≥70 μm), requiring higher resolutions, which can be obtained by microCT (Figure 3B) (237). The resolutions that can be achieved with modern microCT scanners vary from relatively low resolutions (≥20 μm), with quick scan times and large sample size, to higher resolutions (≤10 μm), with longer scanning durations and smaller sample size. It is important to note that the magnification, often described as the size of the voxels (3D pixels) is not identical to spatial resolution, which is roughly 2–3 times larger (238). MicroCT is less time consuming and provides excellent 3D resolution compared to optical microscopy/histology. Although mainly mineralized tissues are recorded, resulting in a loss of information on aspects such as cells and non-mineralized tissues, the use of contrast agents allows visualization of different tissues such as adipose or epithelial tissue and can even enhance the signal of poorly mineralized bone (239, 240). For example, scanning of juvenile stages can be performed by staining the samples with silver nitrate beforehand, allowing for visualization of early bone development where only low amounts of mineral are present (241). However, with this approach only relative mineralization densities can be determined, and not absolute hydroxyapatite levels, which is an important parameter when modeling skeletal disorders. The amount of hydroxyapatite present in samples can be determined by performing a calibration microCT scan of a reference object (phantom) with a known hydroxyapatite concentration. This approach was used in a study of the effect of aging on bone mineral density (BMD) in zebrafish, revealing progressively increased BMD with age, in contrast to humans (101). When interpreting skeletal phenotypes, it is important not to rely on a single method, because certain phenotypes can be better detected using other methods. For example, a mineralized notochord leading to completely solid centra is easier to assess using microCT compared to mineral staining (72). In addition to 3D renderings, microCT data allows the creation and viewing of individual slices throughout the sample, similar to histological sections. Histology of mineralized tissues is notoriously difficult and requires special protocols because samples cannot be demineralized for sectioning. As an example, a complementary approach of both histology and high resolution microCT (6 μm) was used in a zebrafish model for craniosynostosis revealing fusion of the coronal suture (107).
Although low resolution microCT (≥20 μm) does not allow the detection of subtle skeletal changes, such as fusions between adjacent bones, it is perfectly suitable for whole-body scanning and phenotyping of adult zebrafish with a moderate throughput (Figure 3B). Such a procedure was applied by Gistelink et al. (120), where individual vertebral bodies (neural/haemal arches and centrum) of different OI zebrafish models were manually segmented. Subsequently, tissue mineral density (TMD), vertebral length, bone volume, and thickness were determined for each component (80). Manual segmentation is a laborious process and possibly introduces human bias into the analysis, which can be overcome by semi-automated segmentation algorithms such as FishCut (80). FishCut enables the measuring of a large number of parameters in the vertebral column, and is supplemented by a statistical approach for analysis (80). Models for Bruck syndrome, osteogenesis imperfecta and hyperthyroidism have been successfully analyzed by this high-throughput pipeline, thereby standardizing zebrafish skeletal analyses (80, 120). High resolution microCT (≤10 μm) on the other hand, allows for more detailed analysis, but is very time consuming and limits the scanning to only small segments of the skeleton (Figure 4). MicroCT scans of a vertebral body at 1 μm voxel size revealed osteocyte lacunae, which is beyond the resolution range of whole body microCT scans (Figures 4B,D) (242). In a study by Newham et al. (118), high resolution scans of vertebral bodies before and after mechanical compression were analyzed via geometric morphometrics. The obtained measurements were successfully used to determine the deformation zones and subsequently used to predict the deformation and strain during loading (118).
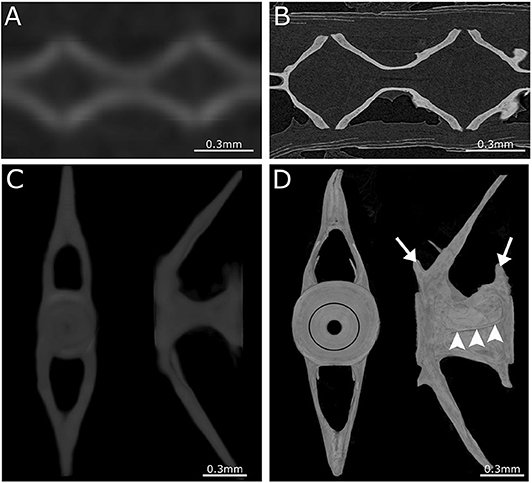
Figure 4. Comparison between low- and high-resolution microCT. (A) Image of parasagittal microCT plane at 21 μm. (B) Similar structure as in (A) but scanned at 0.75 μm. Comparison between low-resolution and high-resolution microCT clearly demonstrates the ability to distinguish separate vertebrae and compact bone only using high-resolution microCT. (C) Anterior and lateral view of a 3D maximal projection surface render of a vertebrae scanned at 21 μm. (D) Similar structure as in (C) but scanned at 0.75 μm. Notice the difference in detail where the growth rings (black circle) are visible in the vertebral endplate on the anterior view. The lateral view of high-resolution microCT shows the outline of the vertebra with the pre- and post-zygapophyses (white arrows), and an antero-posterior running medial vertebral trabecula (white arrowheads).
Bone Histology: From Whole Mount to Sections
Whole mount staining and high-resolution section analysis of the zebrafish skeleton represent complementary techniques, commonly used to describe bone development and structure at tissue and cellular levels.
Whole Mount Mineral and Cartilage Staining
In biomedical research, where the zebrafish is used as a model organism, whole mount staining is generally used to study the morphology of the skeleton (Table 3). The most commonly used techniques are staining of mineralized tissues with alizarin red S (ARS), staining of cartilage matrix with alcian blue (AB) or staining both tissues with a combination of both ARS and AB (Figure 5). These staining methods are based on well-established protocols, where a specimen is made translucent to transparent and cartilage matrix or mineralized tissues are stained with a dye. Images of whole mount cleared and stained animals, taken with a modern stereo microscope, have an even higher resolution than standard microCT images (Figures 3B,C). Therefore, the whole mount clearing and staining technique can be considered as the gold standard for observing the whole zebrafish skeleton in detail.
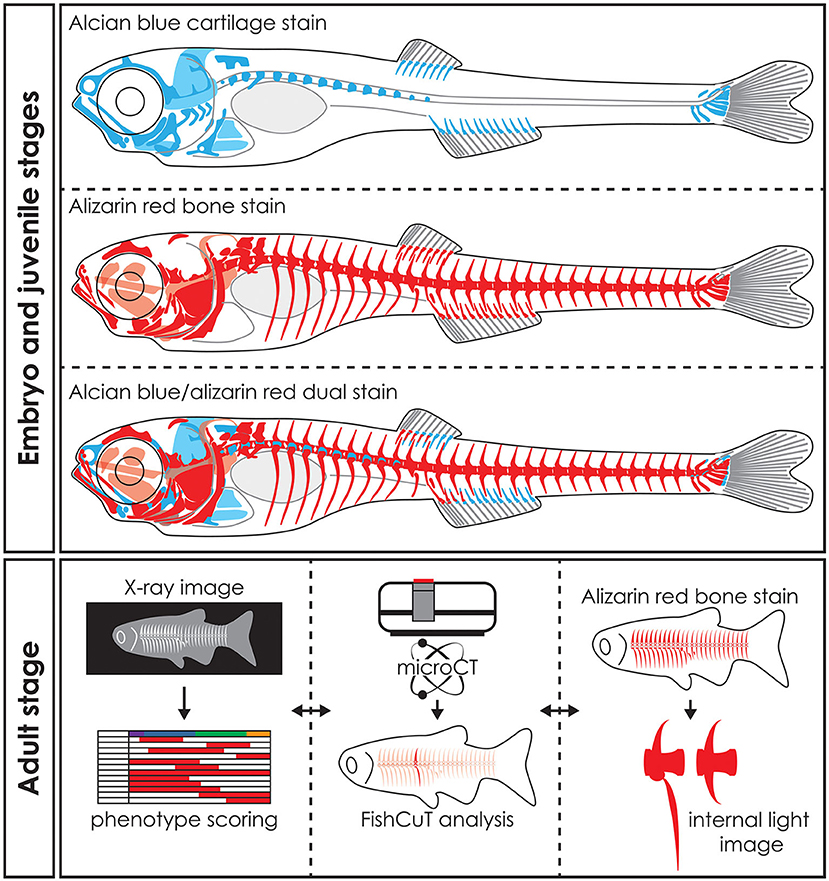
Figure 5. Whole mount staining in early stages and applications of visualization techniques in adult zebrafish. Schematic representation of whole mount cleared and stained early stage zebrafish for cartilage with alcian blue, mineralized tissues (bone) with alizarin red and dual stained for both cartilage and mineralized tissues. Notice that only part of the skull, the basiventrals [for definition see Gadow and Abbott (243)] of the abdominal vertebrae and the fins endoskeleton are pre-formed in cartilage. Many bones in the skull and especially in the vertebral column are formed by direct intramembranous ossification. Images of adult skeletons taken by x-ray can be used to score for skeletal abnormalities, while microCT data can be used in an analysis program such as FishCuT to obtain quantitative data of bone measurements such as size, volume, thickness, and bone mineral density (80, 120). Bright field images or fluorescent images of whole mount cleared and stained zebrafish for mineralized tissues with alizarin red can be used to study skeletal abnormalities in detail. The three techniques are mostly used on euthanized and fixed specimens and thus can be applied on the same specimen sequentially. Moreover, the data procured by these visualization techniques can be integrated into a large data matrix and allows detailed phenotypic descriptions of zebrafish disease models.
Alizarin Red S
Many different protocols exist for ARS staining of mineralized tissues, however the main steps are based on (i) removing the pigmentation of the tissue with a bleaching solution (basic pH), (ii) neutralization of depigmentation, (iii) staining the animal with ARS, and (iv) clearing the animal of excess stain (244). The ARS molecule is a dihydroxyanthraquinone, likely binding the Ca2+ on the hydroxyapatite surface to form either a salt or a chelate form (245), thus it specifically stains mineralized tissue. In disease models ARS will stain ectopic mineralization in soft tissues. For example, ectopic mineralization was shown surrounding the eye, in the wall of the bulbus arteriosus of the heart and in the ventral skin of the dragon fish (dgf−/−), a knock-out zebrafish model for the gene that encodes Enpp1, and modeled for generalized arterial calcification of infancy (GACI) and pseudoxanthoma elasticum (PXE) (105, 106). Bone collagen in teleosts can also be deposited without being mineralized, as was shown in salmon vertebral bone (246, 247) and in the dentine of replacement teeth of the African bichir (248). It is important to underline that the unmineralized collagen cannot be visualized with ARS, however, mineralization usually quickly follows collagen deposition. Finally, there is also one mineralized collagenous tissue that does not stain with alizarin red S, the hypermineralized enameloid of the tooth cusps (248, 249).
ARS staining for mineralized tissues is frequently used to assess the development of skeletal elements in the head, axial skeleton, and fins at early life stages (Figure 5). In addition, investigating the early skeletal phenotype can be focused on a delay or advance in the development or specifically on the mineralization status of early skeletal elements. Because ARS is autofluorescent in the rhodamine channel (red), it can be used in combination with skeletal transgenic zebrafish reporter lines in which the fluorescent signal of the skeletal cells is in a different light spectrum. Alternatively, a Kaede reporter line, where the spectrum of the fluorescent protein can be changed by exposing the specimen to UV-light, can be used in a more flexible way (196). While most studies using ARS for mineralized tissue examined fixed specimens, ARS can also be used as a live stain especially in early stages where pigmentation does not obscure the underlying skeleton yet [reviewed in (250)]. Staining with ARS can also be employed to assess the juvenile and adult skeleton (Figure 5) because mineralized bone is the main skeletal tissue present at these life stages and is easy to observe with this technique.
Alcian Blue
Staining cartilage whole mounts with AB 8GX, similar to ARS staining, is based on several basic steps including (i) removing the pigmentation of the tissue with a bleaching solution (basic pH), (ii) staining the specimens with AB (acid pH), (iii) rehydration and clearing the specimens of excess stain, and (iv) dehydration and storing the specimens. The AB molecule is part of the phthalocyanine dyes with most often copper (Cu2+) as the central metallic ion which results in a blue stain. AB has specifically four tetramethylisothiouronium solubility groups with S=C bonds that are easily broken to bind an insoluble AB molecule to the tissue (251). The stain binds as a salt to sulfated and carboxylated acid mucopolysaccharides and glycoproteins present in the cartilage matrix (251). Alcian blue is in most cases dissolved in a dehydrating ethanol/acetic acid solution and brought to a specific low pH. This low pH (1.5–2.5) causes AB to stain very specifically to the cartilage matrix (Figure 5).
Cartilage is the main skeletal tissue in early life stages of zebrafish, particularly in the skull (chondrocranium) and fins (252). Therefore, AB staining has been largely used in early life stages, i.e., 2–20 dpf, to study the morphology of the chondrocranium in different skeletal zebrafish models (62, 68) (Figure 5). Developing malformations are mainly defined as the irregular shape of skeletal elements, but can also be defined by the absence of skeletal elements or the incorrect morphogenesis of a single skeletal element (66, 84). Relative to the entire skeleton, not much cartilage is present in later life stages (late juveniles, adults) of zebrafish, yet AB staining can be used to assess for example cartilaginous joints (92).
Alcian Blue/Alizarin Red S Double Stain
Staining of cartilage and mineralized tissues can also be combined in a single specimen, as described in several papers by Kimmel et al. (253, 254). In this protocol tissues are stained first with AB followed by ARS staining (Figure 5). The dual staining for cartilage and mineralized tissues is similar to the single stain methods, except that AB can also be dissolved in a salt/ethanol solution, where the salts can be sodium acetate or the more commonly used magnesium chloride (244, 255).
The dual staining protocol is mostly used to assess development of malformations of the early skeleton but can also be used to investigate the normal development and developmental sequence of the skeleton (69). More specifically, dual staining has been used to assess ossification and mineralization status of cartilaginous bones (40, 87) and shape morphology of skeletal elements (61, 166).
The main advantage of this staining technique is the visualization of both cartilage and bone in an individual specimen, so that both connective tissues can be studied at the same time. However, this approach has also several disadvantages. First, when an acid/ethanol solution is used for AB staining, this acidic staining solution demineralizes the tissues that are subsequently visualized with ARS. This results in a reduced staining of mineralized tissues compromising the correct phenotypic assessment. This issue was reviewed by Witten et al. (24). Therefore, it is advisable to always use single staining protocols, either as an alternative or as a validation method in parallel to the double staining protocol. Second, dissolving AB in a non-acidic salt/ethanol solution is however challenging because pH higher then 6 decreases the specificity of the staining solution for mucopolysaccharides and glycoproteins (251).
ARS and AB Whole Mount Staining Advantages and Pitfalls
Considering the simplicity and above all the extensive use of the ARS and AB whole mount staining, a brief overview of its general advantages and disadvantages may be useful.
Both the single staining and double staining approaches are cheap and generally fast to use. Specimens that have not developed scales yet, can often be stained in a single day, with observations made the same day or the day after. In contrast, adult specimens can take up to 2 weeks to stain (244). Indeed, staining protocols need to be adapted to the size of the specimens. Therefore, a thorough description of the staining protocol is indispensable for the interpretation and reproducibility of results (251, 256).
Detailed observations of cartilaginous and mineralized connective tissues can be made owing to the high sensitivity and specificity of both the ARS and AB stains. In particular, small mineralized structures such as the initial mineralizations in early life stages and small intermuscular bones or tendons in adult life stages can be visualized by ARS with high fidelity (24, 234), especially when using fluorescent light which greatly enhances the visibility of these small structures (55, 250). Importantly, ARS stain disappears over time especially in small mineralized structures requiring immediate observation and imaging once the staining procedure is finished. In contrast, when specimens are stored correctly in 100% glycerol, AB staining will remain specific for a longer time (256).
Although AB stains cartilage matrix specifically when the correct pH is used, AB solutions with a pH that is too high or solutions that have a too high or too low salt concentration can result in non-specific staining of non-cartilaginous connective tissue, i.e., collagen type I bone matrix. Non-specific staining can lead to incorrect interpretations of results. Finally, careful interpretation is needed of single AB stained connective tissues in specimens of 15 dpf and older. During the perichondral ossification of cartilaginous bones in zebrafish (Figure 2Bii), when a collagenous sheath forms around cartilaginous bone, the AB solution fails to stain the cartilage, and therefore the cartilaginous connective tissue appears absent. The presence of cartilage beneath the collagen can however still be confirmed using oblique light settings.
Histological Stains
Bone histology is often necessary to complement other imaging techniques, such as whole mount imaging, and remains one of the methods of choice to investigate the skeletal phenotype and bone mineralization during developmental stages (Table 3). The small size of zebrafish has forced researchers to adapt existing, standard histological procedures performed on human and murine skeletal tissues. High quality histological preparations and extensive knowledge about the zebrafish skeletal anatomy and development are indispensable for a correct skeletal evaluation (36, 45). Since zebrafish share similar bone cell types and cellular markers with mammals, it is possible to apply the standard histological and histomorphometric staining protocols available for mammalian bone, although with some technical optimization. In zebrafish in particular, the cellular composition analysis requires high-magnification imaging because skeletal elements may consist of a very limited number of cells, that are smaller in comparison with mammalian cells (24).
Unlike humans and mice, histology on zebrafish can easily be performed on a whole specimen in different developmental stages. Skeletal development can be followed in early juvenile stages looking at the mineralization of the notochord sheath and of cranial bones, while in adult zebrafish histology is most often performed on the abdominal vertebra (the first 10 vertebrae articulated with ribs, although this number is variable), the scales and the caudal fin rays.
Histological Specimen Preparation
In general, the histological procedure for both whole adult zebrafish and dissected bone samples, involves fixation in 4% paraformaldehyde in phosphate buffer saline (PBS) pH 7.2 overnight at 4°C, decalcification in 10% EDTA pH 7.2 for 7 days at 4°C and dehydration according to standard histological protocols or in a gradient series of acetone solutions (199). Importantly, while no decalcification is required up to 20 dpf, for juvenile to adult life stages the time of decalcification varies and depends on the developmental stage and size. Juveniles from 21 dpf till adulthood are normally decalcified for 4 up to 7 days (257).
According to Oralova et al. (199), paraffin embedding does not provide high quality histological details of zebrafish embryos and of early juvenile stages. In these cases, epoxy, or methacrylate resin embedding media are recommended (258). From epoxy blocks, semi, and ultrathin sections can be obtained for light and transmission electron microscopy, respectively, while methacrylate is more suitable for histochemical reactions (24). When using transgenic zebrafish lines expressing fluorescent reporters, fluorescence is generally lost in paraffin embedded samples. Cryosections preserve fluorescence, but significantly decreases the quality of the morphological structure due to processing artifacts. For this reason, Orolova and colleagues developed a new protocol using glycol methacrylate (GMA) embedding, which preserves both fluorescent labeling, epitopes for immunostaining and morphology, making it a more suitable choice (199).
Staining of Skeletal Sections
Different stains can be applied to histological sections of the zebrafish skeleton. Masson's trichrome and toluidine blue are commonly used and generally allow visualization of collagen and particular aspects of bone. Masson's trichrome, which usually stains muscle fibers red, collagen and bone in blue/green, cytoplasm in light red/pink, and cell nuclei in dark brown to black, reveals much thinner layers of collagen fibrils in a mutant zebrafish model for type I collagenopathies, a heterogenous group of connective tissue disorders caused by genetic defects in type I collagen (120). Toluidine blue is often used to detect bone cells, but is also a powerful dye to visualize proteoglycans, elastin and, when using birefringent light—collagen type I and type II fiber organization. Toluidine blue was used to detect abnormalities in glycosaminoglycan pattern in the pharyngeal skeleton of a zebrafish model for a recessive OI knock-out of sec24C/sec24D, two components of the COPII vesicle complex required for collagen secretion (259). Moreover, sections stained with toluidine blue showed compressed and deformed vertebrae, and excessive bone formation and remodeling at the vertebral endplates in the Bruck syndrome plod2 mutant, characterized by the loss of type I collagen telopeptide lysyl hydroxylation (16).
The most widely used mineral staining assays include ARS, calcein and von Kossa staining, which specifically bind to calcium in the mineralized bone. In a study by Pasqualetti et al. (260), successive staining with ARS and calcein allowed evaluation of bone formation at the level of the circuli of growing scales in wild-type animals (260). In the panther fish, characterized by impaired osteoclast proliferation and differentiation, von Kossa staining enabled detection of altered mineralization of the neural arches (97).
Finally, collagen fiber maturation can be investigated by sirius red staining under polarized light, as performed to study the actinotrichia and lepidotrichia pattern in the chihuahua zebrafish, carrying a mutation in collagen type I α1 chain (15, 93, 261).
Transmission Electron Microscopy Analysis
Transmission electron microscopy (TEM) has also been used to investigate zebrafish bone. TEM represents a powerful method to analyze ultrastructural features of tissues since it provides much higher magnification and resolution compared to light microscopy, allowing visualization of cellular and matrix structures at a subnanometer scale. For instance, an altered distribution of bone collagen fiber diameter, a frequently described feature in various skeletal pathological conditions, was detected in the crtap and p3h1 knock-out models of OI type VII and VIII by TEM, revealing the crucial role of the collagen post translational modification complex in bone organization (17). TEM was also used to show enlarged endoplasmic reticulum cisterna in these models, reinforcing ER stress as a key element in the OI phenotype and a potential target for new therapeutic approaches (17, 226, 227).
Immunohistochemistry
Immunohistochemistry (IHC) on zebrafish sections is also possible but limited, compared to mammal specimens, given the reduced availability of specific zebrafish antibodies. Nonetheless, with IHC, the spatiotemporal pattern of distribution of several proteins, a key prerequisite for understanding development, have been elucidated in embryos both in physiological and pathological conditions (199). For example, a structural defect in the extracellular matrix (ECM) has been detected in the fndc3awue1/wue1 zebrafish where IHC of type II collagen showed a loss of mature actinotrichia in 52 h post fertilization (hpf) embryos and β-catenin staining revealed divergent ECM assembly in the regenerated adult fin (262).
Determining the exact spatial localization of the protein of interest in immunostained whole mount larvae is difficult, especially for more deeply located tissues. To overcome this limitation, it is possible to perform whole-mount IHC followed by GMA embedding and sectioning, as was shown by Oralova et al. (199). In this way, the distribution of labeled cells was mapped and quantified allowing for close investigation of the cellular behavior during tissue development, cell migration, and adhesion events, as well as growth and differentiation. As an example, the use of a pan cytokeratin antibody on Tg(sox17:egfp) embryos allowed the authors to localize the protein of interest, Sox17, and the epidermis in the same section (199).
Finally, alkaline phosphatase (Alp), expressed by osteoblasts and required for the mineralization of extracellular matrix, and Trap, expressed by osteoclasts, and important for bone resorption, can both be immunostained to detect active osteoblasts and osteoclasts, respectively, and have been used for example to follow cell differentiation in scales (260).
Histological Analysis of Tissue Regeneration
Zebrafish's ability to repair caudal fin rays and scales has led to the optimization of specific histological protocols for these tissues involving both tissue sectioning as well as whole organ analysis (263). The analysis of histological sections has made clear that during regeneration in the caudal fin rays, cells near the site of injury can dedifferentiate, proliferate and replace the damaged or missing cells (196, 264). Furthermore, histological studies have identified a population of Runx2/Sp7 positive chondrocytes involved in bone repair, and have helped to elucidate the ability of periosteal cells to generate cartilage in response to injury in indian hedgehog homolog a (Ihha) mutants (265).
To study mineralization and cellular compositions of caudal fin rays and scales, the tissue can also be isolated and directly stained without the need for dehydration and sectioning. For instance, by using ARS and calcein double staining and ALP immunohistochemistry, the specific mineralization pattern of bone forming cells in different areas of a scale was elucidated (260). Masson's trichrome staining of regenerating ray collagen proved that multiple amputations do not affect the regenerative bone capacity (266).
Is the Medaka an Alternative Tool in Skeletal Research?
Together with zebrafish, medaka (Oryzias latipes) is the other most frequently used small teleost in biomedical research. This species native to East Asia, belongs to the Adrianichthyidae family (order Beloniformes) and had an ancestor living in saltwater (267).
Evolutionarily, zebrafish and medaka are distantly related (268), with the last common ancestor dating back 110–200 million years ago (269). Being a small fish, medaka shares all the advantages already described for zebrafish, although it has a faster generation time, 2 vs. 3 months, shortening genetic experiments (23).
Similar to zebrafish, the medaka shares common skeletal developmental schemes as well as the presence of most of skeletal cells, chondrocytes, osteoblasts, and osteoclasts with tetrapods, but notably is missing osteocytes (23, 24).
The medaka genome, that underwent a whole duplication like that of the zebrafish, is available and easy to manipulate using the same techniques as in zebrafish research allowing easy generation of skeletal disease models and transgenic lines (46, 208, 270–273).
The almost completely conserved phenotypic features between zebrafish and medaka allow researchers to exploit the same imaging techniques to analyze skeletal components in both physiological and pathological conditions, either in terms of x-ray imaging or more specialized methods, such as microCT, whole mount or histological staining methods (23).
Limitations of the Zebrafish Model
To take full advantage of the zebrafish as a model of human diseases it is important to be aware of existing drawbacks. Due to the extra whole genome duplication compared to mammals, as mentioned above, about 20% of the zebrafish genes have two functional copies, complicating the generation of knock-out disease models (274). Furthermore, some of the duplicated genes have functionally diverged, thus limiting the use of zebrafish in accurately modeling human diseases (11, 24). Additionally, the limited availability of antibodies against zebrafish proteins and the difficulty in establishing tissue specific primary cell lines impairs zebrafish use in research. Finally, the generation of conditional knock-outs and knock-ins is still difficult in zebrafish. Although recently a method to integrate loxP sequences at specific sites in the zebrafish genome using the CRISPR/Cas9 technology has been developed, and conditional mutants of tbx20 and fleer have been generated employing Cre recombinase technology (275, 276).
Conclusions
In the last decade the zebrafish has emerged as a unique model to investigate common and rare human skeletal disorders. The advances in gene editing techniques, from the initial insertion of random genomic mutations by exposure to mutagenic substances, to the knockdown expression of specific genes by antisense morpholino oligonucleotides, to the change of the genome at a specific site by nuclease technologies and their simple use in zebrafish, have all allowed research groups to generate new bone disease models. In particular, the versatile and cheap CRISPR/Cas9 system has found a wide use in many laboratories and undergone a series of optimizations allowing an increasingly specific and error-free gene editing. Nevertheless, its use for knock-in mutations still requires further optimization. The combining of zebrafish skeletal disease models with already available or newly generated transgenic lines, has contributed tremendously to the advances made in in vivo analysis of bone cells. The advances in confocal microscopy and the emergence of light sheet microcopy allows for better visualization and characterization of larval phenotypes in skeletal disease models, taking advantage of larvae transparency. X-ray and microCT have been optimized for small adult zebrafish bones, allowing analysis of the whole skeleton or small elements at high resolution. On the other hand, traditional skeletal specific dyes, such as alizarin red and alcian blue remain a valuable tool to study bone in larvae and adults. Finally, biomedical research has an urgent need for high throughput drug screening platforms and zebrafish models of skeletal diseases represent a bridge from in vitro to in vivo approaches.
In conclusion, ongoing technological advances in analytical techniques are making the zebrafish emerge as a unique and powerful model for the investigation and understanding of human skeletal disorders, and additionally as an efficient platform for compound discovery.
Author Contributions
FT, JB, RB, AD, AW, and AF: writing—original draft. All authors: review and editing. LL, JB, and AD: figures.
Funding
This work was supported by the Italian Ministry of Education, University and Research (MIUR) [Dipartimenti di Eccellenza (2018–2022)] and the Brittle Bone Foundation Grant-2019 to AF. The study was supported by Ghent University (Methusalem Grant BOFMET2015000401).
Conflict of Interest
PS is a Bruker employee.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Gut P, Reischauer S, Stainier DYR, Arnaout R. Little fish, big data: zebrafish as a model for cardiovascular and metabolic disease. Physiol Rev. (2017) 97:889–938. doi: 10.1152/physrev.00038.2016
2. Russel WMS, Burch RL. The Principles of Humane Experimental Technique Wheathampstead: Universities Federation for Animal Welfare (1959).
3. Carnovali M, Banfi G, Mariotti M. Zebrafish models of human skeletal disorders: embryo and adult swimming together. Biomed Res Int. (2019) 2019:1253710. doi: 10.1155/2019/1253710
4. Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. (2013) 496:498–503. doi: 10.1038/nature12111
5. Hamilton F. An Account of the Fishes Found in the River Ganges and Its Branches. Edimburgh: Edinburgh Printed for Constable A, and Company (1822).
6. Streisinger G, Walker C, Dower N, Knauber D, Singer F. Production of clones of homozygous diploid zebra fish (Brachydanio rerio). Nature. (1981) 291:293–6. doi: 10.1038/291293a0
7. Kimmel CB. Genetics and early development of zebrafish. Trends Genet. (1989) 5:283–8. doi: 10.1016/0168-9525(89)90103-0
8. Poss KD, Keating MT, Nechiporuk A. Tales of regeneration in zebrafish. Dev Dyn. (2003) 226:202–10. doi: 10.1002/dvdy.10220
9. Zon LI, Peterson RT. In vivo drug discovery in the zebrafish. Nat Rev Drug Discov. (2005) 4:35–44. doi: 10.1038/nrd1606
10. Parichy DM, Elizondo MR, Mills MG, Gordon TN, Engeszer RE. Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Dev Dyn. (2009) 238:2975–3015. doi: 10.1002/dvdy.22113
12. Bergen DJM, Kague E, Hammond CL. Zebrafish as an emerging model for osteoporosis: a primary testing platform for screening new osteo-active compounds. Front Endocrinol. (2019) 10:6. doi: 10.3389/fendo.2019.00006
13. Besio R, Chow CW, Tonelli F, Marini JC, Forlino A. Bone biology: insights from osteogenesis imperfecta and related rare fragility syndromes. FEBS J. (2019) 286:3033–56. doi: 10.1111/febs.14963
14. Fiedler IAK, Schmidt FN, Wolfel EM, Plumeyer C, Milovanovic P, Gioia R, et al. Severely impaired bone material quality in chihuahua zebrafish resembles classical dominant human osteogenesis imperfecta. J Bone Min Res. (2018) 33:1489–99. doi: 10.1002/jbmr.3445
15. Gioia R, Tonelli F, Ceppi I, Biggiogera M, Leikin S, Fisher S, et al. The chaperone activity of 4PBA ameliorates the skeletal phenotype of chihuahua, a zebrafish model for dominant osteogenesis imperfecta. Hum Mol Genet. (2017) 26:2897–911. doi: 10.1093/hmg/ddx171
16. Gistelinck C, Witten PE, Huysseune A, Symoens S, Malfait F, Larionova D, et al. Loss of type I collagen telopeptide lysyl hydroxylation causes musculoskeletal abnormalities in a zebrafish model of bruck syndrome. J Bone Min Res. (2016) 31:1930–42. doi: 10.1002/jbmr.2977
17. Tonelli F, Cotti S, Leoni L, Besio R, Gioia R, Marchese L, et al. Crtap and p3h1 knock out zebrafish support defective collagen chaperoning as the cause of their osteogenesis imperfecta phenotype. Matrix Biol. (2020) 90:40–60. doi: 10.1016/j.matbio.2020.03.004
18. van Dijk FS, Zillikens MC, Micha D, Riessland M, Marcelis CL, de Die-Smulders CE, et al. PLS3 mutations in X-linked osteoporosis with fractures. N Engl J Med. (2013) 369:1529–36. doi: 10.1056/NEJMoa1308223
19. Willems B, Tao S, Yu T, Huysseune A, Witten PE, Winkler C. The Wnt co-receptor Lrp5 is required for cranial neural crest cell migration in zebrafish. PLoS ONE. (2015) 10:e0131768. doi: 10.1371/journal.pone.0131768
20. Zhang Y, Huang H, Zhao G, Yokoyama T, Vega H, Huang Y, et al. ATP6V1H deficiency impairs bone development through activation of MMP9 and MMP13. PLoS Genet. (2017) 13:e1006481. doi: 10.1371/journal.pgen.1006481
21. Lam PY, Peterson RT. Developing zebrafish disease models for in vivo small molecule screens. Curr Opin Chem Biol. (2019) 50:37–44. doi: 10.1016/j.cbpa.2019.02.005
22. Gemberling M, Bailey TJ, Hyde DR, Poss KD. The zebrafish as a model for complex tissue regeneration. Trends Genet. (2013) 29:611–20. doi: 10.1016/j.tig.2013.07.003
23. Lleras-Forero L, Winkler C, Schulte-Merker S. Zebrafish and medaka as models for biomedical research of bone diseases. Dev Biol. (2020) 457:191–205. doi: 10.1016/j.ydbio.2019.07.009
24. Witten PE, Harris MP, Huysseune A, Winkler C. Small teleost fish provide new insights into human skeletal diseases. Methods Cell Biol. (2017) 138:321–46. doi: 10.1016/bs.mcb.2016.09.001
25. Witten PE, Huysseune A, Hall BK. A practical approach for the identification of the many cartilaginous tissues in teleost fish. J Appl Ichthyol. (2010) 26:257–62. doi: 10.1111/j.1439-0426.2010.01416.x
26. Iwasaki M, Kuroda J, Kawakami K, Wada H. Epidermal regulation of bone morphogenesis through the development and regeneration of osteoblasts in the zebrafish scale. Dev Biol. (2018) 437:105–19. doi: 10.1016/j.ydbio.2018.03.005
27. Van der heyden C, Huysseune A. Dynamics of tooth formation and replacement in the zebrafish (Danio rerio) (Teleostei, Cyprinidae). Dev Dyn. (2000) 219:486–96. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1069>3.0.CO;2-Z
28. Reif WE. Evolution of Dermal Skeleton and Dentition in Vertebrates: The Odontode Regulation Theory Evolutionary Biology. Boston, MA: Springer (1982).
29. Huysseune A, Sire JY. Evolution of patterns and processes in teeth and tooth-related tissues in non-mammalian vertebrates. Eur J Oral Sci. (1998) 106(Suppl. 1):437–81. doi: 10.1111/j.1600-0722.1998.tb02211.x
30. Sire JY, Huysseune A. Formation of dermal skeletal and dental tissues in fish: a comparative and evolutionary approach. Biol Rev Camb Philos Soc. (2003) 78:219–49. doi: 10.1017/S1464793102006073
31. Kague E, Gallagher M, Burke S, Parsons M, Franz-Odendaal T, Fisher S. Skeletogenic fate of zebrafish cranial and trunk neural crest. PLoS ONE. (2012) 7:e47394. doi: 10.1371/journal.pone.0047394
32. Mongera A, Nusslein-Volhard C. Scales of fish arise from mesoderm. Curr Biol. (2013) 23:R338–9. doi: 10.1016/j.cub.2013.02.056
33. Bird NC, Mabee PM. Developmental morphology of the axial skeleton of the zebrafish, Danio rerio (Ostariophysi: Cyprinidae). Dev Dyn. (2003) 228:337–57. doi: 10.1002/dvdy.10387
34. Fleming A, Kishida MG, Kimmel CB, Keynes RJ. Building the backbone: the development and evolution of vertebral patterning. Development. (2015) 142:1733–44. doi: 10.1242/dev.118950
35. Lleras Forero L, Narayanan R, Huitema LF, VanBergen M, Apschner A, Peterson-Maduro J, et al. Segmentation of the zebrafish axial skeleton relies on notochord sheath cells and not on the segmentation clock. Elife. (2018) 7:e33843. doi: 10.7554/eLife.33843.043
36. Weigele J, Franz-Odendaal TA. Functional bone histology of zebrafish reveals two types of endochondral ossification, different types of osteoblast clusters and a new bone type. J Anat. (2016) 229:92–103. doi: 10.1111/joa.12480
37. Witten PE, Huysseune A. A comparative view on mechanisms and functions of skeletal remodelling in teleost fish, with special emphasis on osteoclasts and their function. Biol Rev Camb Philos Soc. (2009) 84:315–46. doi: 10.1111/j.1469-185X.2009.00077.x
38. Eames BF, Amores A, Yan YL, Postlethwait JH. Evolution of the osteoblast: skeletogenesis in gar and zebrafish. BMC Evol Biol. (2012) 12:27. doi: 10.1186/1471-2148-12-27
39. Schlombs K, Wagner T, Scheel J. Site-1 protease is required for cartilage development in zebrafish. Proc Natl Acad Sci USA. (2003) 100:14024–9. doi: 10.1073/pnas.2331794100
40. Hammond CL, Schulte-Merker S. Two populations of endochondral osteoblasts with differential sensitivity to Hedgehog signalling. Development. (2009) 136:3991–4000. doi: 10.1242/dev.042150
41. Mackie EJ, Ahmed YA, Tatarczuch L, Chen KS, Mirams M. Endochondral ossification: how cartilage is converted into bone in the developing skeleton. Int J Biochem Cell Biol. (2008) 40:46–62. doi: 10.1016/j.biocel.2007.06.009
42. Maes C, Kobayashi T, Selig MK, Torrekens S, Roth SI, Mackem S, et al. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev Cell. (2010) 19:329–44. doi: 10.1016/j.devcel.2010.07.010
43. Horton JM, Summers AP. The material properties of acellular bone in a teleost fish. J Exp Biol. (2009) 212(Pt 9):1413–20. doi: 10.1242/jeb.020636
44. Bar-Shavit Z. The osteoclast: a multinucleated, hematopoietic-origin, bone-resorbing osteoimmune cell. J Cell Biochem. (2007) 102:1130–9. doi: 10.1002/jcb.21553
45. Hall BK. Bones and Cartilage: Developmental and Evolutionary Skeletal Biology. 2nd ed. London: Elsevier/Academic Press (2015).
46. To TT, Witten PE, Renn J, Bhattacharya D, Huysseune A, Winkler C. Rankl-induced osteoclastogenesis leads to loss of mineralization in a medaka osteoporosis model. Development. (2012) 139:141–50. doi: 10.1242/dev.071035
47. Eames BF, Singer A, Smith GA, Wood ZA, Yan YL, He X, et al. UDP xylose synthase 1 is required for morphogenesis and histogenesis of the craniofacial skeleton. Dev Biol. (2010) 341:400–15. doi: 10.1016/j.ydbio.2010.02.035
48. Li N, Felber K, Elks P, Croucher P, Roehl HH. Tracking gene expression during zebrafish osteoblast differentiation. Dev Dyn. (2009) 238:459–66. doi: 10.1002/dvdy.21904
49. Yan YL, Willoughby J, Liu D, Crump JG, Wilson C, Miller CT, et al. A pair of sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development. (2005) 132:1069–83. doi: 10.1242/dev.01674
50. Gistelinck C, Gioia R, Gagliardi A, Tonelli F, Marchese L, Bianchi L, et al. Zebrafish collagen type I: molecular and biochemical characterization of the major structural protein in bone and skin. Sci Rep. (2016) 6:21540. doi: 10.1038/srep21540
51. Christoffels A, Koh EG, Chia JM, Brenner S, Aparicio S, Venkatesh B. Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. Mol Biol Evol. (2004) 21:1146–51. doi: 10.1093/molbev/msh114
52. Hirasawa T, Kuratani S. Evolution of the vertebrate skeleton: morphology, embryology, and development. Zoological Lett. (2015) 1:2. doi: 10.1186/s40851-014-0007-7
53. Corallo D, Schiavinato A, Trapani V, Moro E, Argenton F, Bonaldo P. Emilin3 is required for notochord sheath integrity and interacts with Scube2 to regulate notochord-derived Hedgehog signals. Development. (2013) 140:4594–601. doi: 10.1242/dev.094078
54. Stemple DL. Structure and function of the notochord: an essential organ for chordate development. Development. (2005) 132:2503–12. doi: 10.1242/dev.01812
55. Bensimon-Brito A, Cancela ML, Huysseune A, Witten PE. Vestiges, rudiments and fusion events: the zebrafish caudal fin endoskeleton in an evo-devo perspective. Evol Dev. (2012) 14:116–27. doi: 10.1111/j.1525-142X.2011.00526.x
56. Arratia G, Schultze HP. Reevaluation of the caudal skeleton of certain actinopterygian fishes: III. Salmonidae. Homologization of caudal skeletal structures. J Morphol. (1992) 214:187–249. doi: 10.1002/jmor.1052140209
57. Arratia G, Schultze HP, Casciotta J. Vertebral column and associated elements in dipnoans and comparison with other fishes: development and homology. J Morphol. (2001) 250:101–72. doi: 10.1002/jmor.1062
58. Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. (1996) 123:37–46.
59. Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. (1996) 123:1–36.
60. Lawson ND, Wolfe SA. Forward and reverse genetic approaches for the analysis of vertebrate development in the zebrafish. Dev Cell. (2011) 21:48–64. doi: 10.1016/j.devcel.2011.06.007
61. Zuniga E, Stellabotte F, Crump JG. Jagged-Notch signaling ensures dorsal skeletal identity in the vertebrate face. Development. (2010) 137:1843–52. doi: 10.1242/dev.049056
62. Ashikov A, Abu Bakar N, Wen XY, Niemeijer M, Osorio GRP, Brand-Arzamendi K, et al. Integrating glycomics and genomics uncovers SLC10A7 as essential factor for bone mineralization by regulating post-Golgi protein transport and glycosylation. Hum Mol Genet. (2018) 27:3029–45. doi: 10.1093/hmg/ddy213
63. Miller CT, Swartz ME, Khuu PA, Walker MB, Eberhart JK, Kimmel CB. mef2ca is required in cranial neural crest to effect endothelin1 signaling in zebrafish. Dev Biol. (2007) 308:144–57. doi: 10.1016/j.ydbio.2007.05.018
64. Yan YL, Miller CT, Nissen RM, Singer A, Liu D, Kirn A, et al. A zebrafish sox9 gene required for cartilage morphogenesis. Development. (2002) 129:5065–79.
65. Sun X, Zhang R, Liu M, Chen H, Chen L, Luo F, et al. Rmrp mutation disrupts chondrogenesis and bone ossification in zebrafish model of cartilage-hair hypoplasia via enhanced Wnt/beta-catenin signaling. J Bone Miner Res. (2019) 34:2101–16. doi: 10.1002/jbmr.3820
66. Tian J, Shao J, Liu C, Hou HY, Chou CW, Shboul M, et al. Deficiency of lrp4 in zebrafish and human LRP4 mutation induce aberrant activation of jagged-Notch signaling in fin and limb development. Cell Mol Life Sci. (2019) 76:163–78. doi: 10.1007/s00018-018-2928-3
67. Burger A, Vasilyev A, Tomar R, Selig MK, Nielsen GP, Peterson RT, et al. A zebrafish model of chordoma initiated by notochord-driven expression of HRASV12. Dis Models Mech. (2014) 7:907–13. doi: 10.1242/dmm.013128
68. Flores MV, Lam EY, Crosier P, Crosier K. A hierarchy of Runx transcription factors modulate the onset of chondrogenesis in craniofacial endochondral bones in zebrafish. Dev Dyn. (2006) 235:3166–76. doi: 10.1002/dvdy.20957
69. Swartz ME, Sheehan-Rooney K, Dixon MJ, Eberhart JK. Examination of a palatogenic gene program in zebrafish. Dev Dyn. (2011) 240:2204–20. doi: 10.1002/dvdy.22713
70. Blumel R, Zink M, Klopocki E, Liedtke D. On the traces of tcf12: investigation of the gene expression pattern during development and cranial suture patterning in zebrafish (Danio rerio). PLoS ONE. (2019) 14:e0218286. doi: 10.1371/journal.pone.0218286
71. Laue K, Janicke M, Plaster N, Sonntag C, Hammerschmidt M. Restriction of retinoic acid activity by Cyp26b1 is required for proper timing and patterning of osteogenesis during zebrafish development. Development. (2008) 135:3775–87. doi: 10.1242/dev.021238
72. Spoorendonk KM, Peterson-Maduro J, Renn J, Trowe T, Kranenbarg S, Winkler C, et al. Retinoic acid and Cyp26b1 are critical regulators of osteogenesis in the axial skeleton. Development. (2008) 135:3765–74. doi: 10.1242/dev.024034
73. Aman AJ, Fulbright AN, Parichy DM. Wnt/beta-catenin regulates an ancient signaling network during zebrafish scale development. Elife. (2018) 7:e37001. doi: 10.7554/eLife.37001.020
74. Suarez-Bregua P, Saxena A, Bronner ME, Rotllant J. Targeted Pth4-expressing cell ablation impairs skeletal mineralization in zebrafish. PLoS ONE. (2017) 12:e0186444. doi: 10.1371/journal.pone.0186444
75. Franz-Odendaal TA, Edsall SC. Long-term effects of simulated microgravity and vibration exposure on skeletal development in zebrafish. Stem Cells Dev. (2018) 27:1278–86. doi: 10.1089/scd.2017.0266
76. Delbaere S, Van Damme T, Syx D, Symoens S, Coucke P, Willaert A, et al. Hypomorphic zebrafish models mimic the musculoskeletal phenotype of beta4GalT7-deficient ehlers-danlos syndrome. Matrix Biol. (2019) 89:59–75. doi: 10.1016/j.matbio.2019.12.002
77. LaBonty M, Pray N, Yelick PC. A zebrafish model of human fibrodysplasia ossificans progressiva. Zebrafish. (2017) 14:293–304. doi: 10.1089/zeb.2016.1398
78. Zancan I, Bellesso S, Costa R, Salvalaio M, Stroppiano M, Hammond C, et al. Glucocerebrosidase deficiency in zebrafish affects primary bone ossification through increased oxidative stress and reduced Wnt/beta-catenin signaling. Hum Mol Genet. (2015) 24:1280–94. doi: 10.1093/hmg/ddu538
79. Liang ST, Chen JR, Tsai JJ, Lai YH, Hsiao CD. Overexpression of notch signaling induces hyperosteogeny in zebrafish. Int J Mol Sci. (2019) 20:3613. doi: 10.3390/ijms20153613
80. Hur M, Gistelinck CA, Huber P, Lee J, Thompson MH, Monstad-Rios AT, et al. MicroCT-based phenomics in the zebrafish skeleton reveals virtues of deep phenotyping in a distributed organ system. Zebrafish. (2018) 15:77–78. doi: 10.1089/zeb.2017.1540
81. Harris MP, Rohner N, Schwarz H, Perathoner S, Konstantinidis P, Nusslein-Volhard C. Zebrafish eda and edar mutants reveal conserved and ancestral roles of ectodysplasin signaling in vertebrates. PLoS Genet. (2008) 4:e1000206. doi: 10.1371/journal.pgen.1000206
82. Kague E, Hughes SM, Lawrence EA, Cross S, Martin-Silverstone E, Hammond CL, et al. Scleraxis genes are required for normal musculoskeletal development and for rib growth and mineralization in zebrafish. FASEB J. (2019) 33:9116–30. doi: 10.1096/fj.201802654RR
83. Dauer MVP, Currie PD, Berger J. Skeletal malformations of Meox1-deficient zebrafish resemble human Klippel-Feil syndrome. J Anat. (2018) 233:687–95. doi: 10.1111/joa.12890
84. Clement A, Wiweger M, von der Hardt S, Rusch MA, Selleck SB, Chien CB, et al. Regulation of zebrafish skeletogenesis by ext2/dackel and papst1/pinscher. PLoS Genet. (2008) 4:e1000136. doi: 10.1371/journal.pgen.1000136
85. Huitema LF, Apschner A, Logister I, Spoorendonk KM, Bussmann J, Hammond CL, et al. Entpd5 is essential for skeletal mineralization and regulates phosphate homeostasis in zebrafish. Proc Natl Acad Sci USA. (2012) 109:21372–7. doi: 10.1073/pnas.1214231110
86. Zhang Y, Cui FZ, Wang XM, Feng QL, Zhu XD. Mechanical properties of skeletal bone in gene-mutated stopsel(dtl28d) and wild-type zebrafish (Danio rerio) measured by atomic force microscopy-based nanoindentation. Bone. (2002) 30:541–6. doi: 10.1016/S8756-3282(02)00676-2
87. Cheah FS, Winkler C, Jabs EW, Chong SS. Tgfbeta3 regulation of chondrogenesis and osteogenesis in zebrafish is mediated through formation and survival of a subpopulation of the cranial neural crest. Mech Dev. (2010) 127:329–44. doi: 10.1016/j.mod.2010.04.003
88. Eberhart JK, He X, Swartz ME, Yan YL, Song H, Boling TC, et al. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat Genet. (2008) 40:290–8. doi: 10.1038/ng.82
89. Ghassibe-Sabbagh M, Desmyter L, Langenberg T, Claes F, Boute O, Bayet B, et al. FAF1, a gene that is disrupted in cleft palate and has conserved function in zebrafish. Am J Hum Genet. (2011) 88:150–61. doi: 10.1016/j.ajhg.2011.01.003
90. Dougherty M, Kamel G, Grimaldi M, Gfrerer L, Shubinets V, Ethier R, et al. Distinct requirements for wnt9a and irf6 in extension and integration mechanisms during zebrafish palate morphogenesis. Development. (2013) 140:76–81. doi: 10.1242/dev.080473
91. Lawrence EA, Kague E, Aggleton JA, Harniman RL, Roddy KA, Hammond CL. The mechanical impact of col11a2 loss on joints; col11a2 mutant zebrafish show changes to joint development and function, which leads to early-onset osteoarthritis. Philos Trans R Soc Lond B Biol Sci. (2018) 373:20170335. doi: 10.1098/rstb.2017.0335
92. Askary A, Smeeton J, Paul S, Schindler S, Braasch I, Ellis NA, et al. Ancient origin of lubricated joints in bony vertebrates. Elife. (2016) 5:e16415. doi: 10.7554/eLife.16415.016
93. Fisher S, Jagadeeswaran P, Halpern ME. Radiographic analysis of zebrafish skeletal defects. Dev Biol. (2003) 264:64–76. doi: 10.1016/S0012-1606(03)00399-3
94. Asharani PV, Keupp K, Semler O, Wang W, Li Y, Thiele H, et al. Attenuated BMP1 function compromises osteogenesis, leading to bone fragility in humans and zebrafish. Ame J Hum Genet. (2012) 90:661–74. doi: 10.1016/j.ajhg.2012.02.026
95. Kague E, Roy P, Asselin G, Hu G, Simonet J, Stanley A, et al. Osterix/Sp7 limits cranial bone initiation sites and is required for formation of sutures. Dev Biol. (2016) 413:160–72. doi: 10.1016/j.ydbio.2016.03.011
96. Henke K, Daane JM, Hawkins MB, Dooley CM, Busch-Nentwich EM, Stemple DL, et al. Genetic screen for postembryonic development in the zebrafish (Danio rerio): dominant mutations affecting adult form. Genetics. (2017) 207:609–23. doi: 10.1534/genetics.117.300187
97. Chatani M, Takano Y, Kudo A. Osteoclasts in bone modeling, as revealed by in vivo imaging, are essential for organogenesis in fish. Dev Biol. (2011) 360:96–109. doi: 10.1016/j.ydbio.2011.09.013
98. Chen JR, Lai YH, Tsai JJ, Hsiao CD. Live fluorescent staining platform for drug-screening and mechanism-analysis in zebrafish for bone mineralization. Molecules. (2017) 22:2068. doi: 10.3390/molecules22122068
99. Huo L, Wang L, Yang Z, Li P, Geng D, Xu Y. Prednisolone induces osteoporosis-like phenotypes via focal adhesion signaling pathway in zebrafish larvae. Biol Open. (2018) 7. doi: 10.1242/bio.029405
100. Urso K, Caetano-Lopes J, Lee PY, Yan J, Henke K, Sury M, et al. A role for G protein-coupled receptor 137b in bone remodeling in mouse and zebrafish. Bone. (2019) 127:104–13. doi: 10.1016/j.bone.2019.06.002
101. Monma Y, Shimada Y, Nakayama H, Zang L, Nishimura N, Tanaka T. Aging-associated microstructural deterioration of vertebra in zebrafish. Bone Rep. (2019) 11:100215. doi: 10.1016/j.bonr.2019.100215
102. Zhang W, Xu J, Qiu J, Xing C, Li X, Leng B, et al. Novel and rapid osteoporosis model established in zebrafish using high iron stress. Biochem Biophys Res Commun. (2018) 496:654–60. doi: 10.1016/j.bbrc.2017.12.172
103. Jafari A, Qanie D, Andersen TL, Zhang Y, Chen L, Postert B, et al. Legumain regulates differentiation fate of human bone marrow stromal cells and is altered in postmenopausal osteoporosis. Stem Cell Rep. (2017) 8:373–86. doi: 10.1016/j.stemcr.2017.01.003
104. Huang HX, Lin H, Lan F, Wu YF, Yang ZG, Zhang JJ. Application of bone transgenic zebrafish in anti-osteoporosis chemical screening. Anim Models Exp Med. (2018) 1:53–61. doi: 10.1002/ame2.12000
105. Apschner A, Huitema LF, Ponsioen B, Peterson-Maduro J, Schulte-Merker S. Zebrafish enpp1 mutants exhibit pathological mineralization, mimicking features of generalized arterial calcification of infancy (GACI) and pseudoxanthoma elasticum (PXE). Dis Models Mech. (2014) 7:811–22. doi: 10.1242/dmm.015693
106. Mackay EW, Apschner A, Schulte-Merker S. Vitamin K reduces hypermineralisation in zebrafish models of PXE and GACI. Development. (2015) 142:1095–101. doi: 10.1242/dev.113811
107. Teng CS, Ting MC, Farmer DT, Brockop M, Maxson RE, Crump JG. Altered bone growth dynamics prefigure craniosynostosis in a zebrafish model of Saethre-Chotzen syndrome. Elife. (2018) 7:e37024. doi: 10.7554/eLife.37024.041
108. Ferreira CR, Xia ZJ, Clement A, Parry DA, Davids M, Taylan F, et al. A recurrent de novo heterozygous COG4 substitution leads to saul-wilson syndrome, disrupted vesicular trafficking, and altered proteoglycan glycosylation. Am J Hum Genet. (2018) 103:553–67. doi: 10.1016/j.ajhg.2018.09.003
109. Buchan JG, Gray RS, Gansner JM, Alvarado DM, Burgert L, Gitlin JD, et al. Kinesin family member 6 (kif6) is necessary for spine development in zebrafish. Dev Dyn. (2014) 243:1646–57. doi: 10.1002/dvdy.24208
110. Grimes DT, Boswell CW, Morante NF, Henkelman RM, Burdine RD, Ciruna B. Zebrafish models of idiopathic scoliosis link cerebrospinal fluid flow defects to spine curvature. Science. (2016) 352:1341–4. doi: 10.1126/science.aaf6419
111. Haller G, McCall K, Jenkitkasemwong S, Sadler B, Antunes L, Nikolov M, et al. A missense variant in SLC39A8 is associated with severe idiopathic scoliosis. Nat Commun. (2018) 9:4171. doi: 10.1038/s41467-018-06705-0
112. Gray RS, Wilm TP, Smith J, Bagnat M, Dale RM, Topczewski J, et al. Loss of col8a1a function during zebrafish embryogenesis results in congenital vertebral malformations. Dev Biol. (2014) 386:72–85. doi: 10.1016/j.ydbio.2013.11.028
113. Zhang X, Jia S, Chen Z, Chong YL, Xie H, Feng D, et al. Cilia-driven cerebrospinal fluid flow directs expression of urotensin neuropeptides to straighten the vertebrate body axis. Nat Genet. (2018) 50:1666–73. doi: 10.1038/s41588-018-0260-3
114. Printzi A, Fragkoulis S, Dimitriadi A, Keklikoglou K, Arvanitidis C, Witten PE, et al. Exercise-induced lordosis in zebrafish Danio rerio (Hamilton, 1822). J Fish Biol. (2019). doi: 10.1111/jfb.14240
115. Burrage LC, Reynolds JJ, Baratang NV, Phillips JB, Wegner J, McFarquhar A, et al. Bi-allelic variants in TONSL Cause SPONASTRIME dysplasia and a spectrum of skeletal dysplasia phenotypes. Am J Hum Genet. (2019) 104:422–38. doi: 10.1016/j.ajhg.2019.01.007
116. Baas D, Malbouyres M, Haftek-Terreau Z, Le Guellec D, Ruggiero F. Craniofacial cartilage morphogenesis requires zebrafish col11a1 activity. Matrix Biol. (2009) 28:490–502. doi: 10.1016/j.matbio.2009.07.004
117. Bergen DJM, Stevenson NL, Skinner REH, Stephens DJ, Hammond CL. The golgi matrix protein giantin is required for normal cilia function in zebrafish. Biol Open. (2017) 6:1180–89. doi: 10.1242/bio.025502
118. Newham E, Kague E, Aggleton JA, Fernee C, Brown KR, Hammond CL. Finite element and deformation analyses predict pattern of bone failure in loaded zebrafish spines. J R Soc Interface. (2019) 16:20190430. doi: 10.1098/rsif.2019.0430
119. Andreeva V, Connolly MH, Stewart-Swift C, Fraher D, Burt J, Cardarelli J, et al. Identification of adult mineralized tissue zebrafish mutants. Genesis. (2011) 49:360–6. doi: 10.1002/dvg.20712
120. Gistelinck C, Kwon RY, Malfait F, Symoens S, Harris MP, Henke K, et al. Zebrafish type I collagen mutants faithfully recapitulate human type I collagenopathies. Proc Natl Acad Sci USA. (2018) 115:E8037–46. doi: 10.1073/pnas.1722200115
121. Draper BW, Morcos PA, Kimmel CB. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: a quantifiable method for gene knockdown. Genesis. (2001) 30:154–6. doi: 10.1002/gene.1053
122. Nasevicius A, Ekker SC. Effective targeted gene 'knockdown' in zebrafish. Nat Genet. (2000) 26:216–20. doi: 10.1038/79951
123. Ekker SC, Larson JD. Morphant technology in model developmental systems. Genesis. (2001) 30:89–93. doi: 10.1002/gene.1038
124. Pickart MA, Klee EW, Nielsen AL, Sivasubbu S, Mendenhall EM, Bill BR, et al. Genome-wide reverse genetics framework to identify novel functions of the vertebrate secretome. PLoS ONE. (2006) 1:e104. doi: 10.1371/journal.pone.0000104
125. Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, et al. p53 activation by knockdown technologies. PLoS Genet. (2007) 3:e78. doi: 10.1371/journal.pgen.0030078
126. Amoyel M, Cheng YC, Jiang YJ, Wilkinson DG. Wnt1 regulates neurogenesis and mediates lateral inhibition of boundary cell specification in the zebrafish hindbrain. Development. (2005) 132:775–85. doi: 10.1242/dev.01616
127. Gerety SS, Wilkinson DG. Morpholino artifacts provide pitfalls and reveal a novel role for pro-apoptotic genes in hindbrain boundary development. Dev Biol. (2011) 350:279–89. doi: 10.1016/j.ydbio.2010.11.030
128. Kok FO, Shin M, Ni CW, Gupta A, Grosse AS, van Impel A, et al. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell. (2015) 32:97–108. doi: 10.1016/j.devcel.2014.11.018
129. El-Brolosy MA, Stainier DYR. Genetic compensation: a phenomenon in search of mechanisms. PLoS Genet. (2017) 13:e1006780. doi: 10.1371/journal.pgen.1006780
130. Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. (2008) 26:702–8. doi: 10.1038/nbt1409
131. Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. (2008) 26:695–701. doi: 10.1038/nbt1398
132. Cade L, Reyon D, Hwang WY, Tsai SQ, Patel S, Khayter C, et al. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res. (2012) 40:8001–10. doi: 10.1093/nar/gks518
133. Watson CJ, Monstad-Rios AT, Bhimani RM, Gistelinck C, Willaert A, Coucke P, et al. Phenomics-based quantification of CRISPR-induced mosaicism in zebrafish. Cell Syst. (2020) 10:275–86.e5. doi: 10.1016/j.cels.2020.02.007
134. Wu RS, Lam, II, Clay H, Duong DN, Deo RC, Coughlin SR, et al. A rapid method for directed gene knockout for screening in G0 zebrafish. Dev Cell. (2018) 46:112–25.e4. doi: 10.1016/j.devcel.2018.06.003
135. Salsman J, Dellaire G. Precision genome editing in the CRISPR era. Biochem Cell Biol. (2017) 95:187–201. doi: 10.1139/bcb-2016-0137
136. Mikuni T, Nishiyama J, Sun Y, Kamasawa N, Yasuda R. High-throughput high-resolution mapping of protein localization in mammalian brain by in vivo genome editing. Cell. (2016) 165:1803–17. doi: 10.1016/j.cell.2016.04.044
137. Leonetti MD, Sekine S, Kamiyama D, Weissman JS, Huang B. A scalable strategy for high-throughput GFP tagging of endogenous human proteins. Proc Natl Acad Sci USA. (2016) 113:E3501–8. doi: 10.1073/pnas.1606731113
138. Forlino A, Marini JC. Osteogenesis imperfecta. Lancet. (2016) 387:1657–71. doi: 10.1016/S0140-6736(15)00728-X
139. El-Brolosy MA, Kontarakis Z, Rossi A, Kuenne C, Gunther S, Fukuda N, et al. Genetic compensation triggered by mutant mRNA degradation. Nature. (2019) 568:193–7. doi: 10.1038/s41586-019-1064-z
140. Coggins NB, Stultz J, O'Geen H, Carvajal-Carmona LG, Segal DJ. Methods for scarless, selection-free generation of human cells and allele-specific functional analysis of disease-associated SNPs and variants of uncertain significance. Sci Rep. (2017) 7:15044. doi: 10.1038/s41598-017-15407-4
141. Renaud JB, Boix C, Charpentier M, De Cian A, Cochennec J, Duvernois-Berthet E, et al. Improved genome editing efficiency and flexibility using modified oligonucleotides with TALEN and CRISPR-Cas9 nucleases. Cell Rep. (2016) 14:2263–72. doi: 10.1016/j.celrep.2016.02.018
142. Richardson CD, Kazane KR, Feng SJ, Zelin E, Bray NL, Schafer AJ, et al. CRISPR-Cas9 genome editing in human cells occurs via the fanconi anemia pathway. Nat Genet. (2018) 50:1132–9. doi: 10.1038/s41588-018-0174-0
143. Lin S, Staahl BT, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife. (2014) 3:e04766. doi: 10.7554/eLife.04766.010
144. Jayathilaka K, Sheridan SD, Bold TD, Bochenska K, Logan HL, Weichselbaum RR, et al. A chemical compound that stimulates the human homologous recombination protein RAD51. Proc Natl Acad Sci USA. (2008) 105:15848–53. doi: 10.1073/pnas.0808046105
145. Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. (2015) 33:538–42. doi: 10.1038/nbt.3190
146. Paquet D, Kwart D, Chen A, Sproul A, Jacob S, Teo S, et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. (2016) 533:125–9. doi: 10.1038/nature17664
147. Singh P, Schimenti JC, Bolcun-Filas E. A mouse geneticist's practical guide to CRISPR applications. Genetics. (2015) 199:1–15. doi: 10.1534/genetics.114.169771
148. Song J, Yang D, Xu J, Zhu T, Chen YE, Zhang J. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat Commun. (2016) 7:10548. doi: 10.1038/ncomms10548
149. Srivastava M, Nambiar M, Sharma S, Karki SS, Goldsmith G, Hegde M, et al. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell. (2012) 151:1474–87. doi: 10.1016/j.cell.2012.11.054
150. Xiong X, Chen M, Lim WA, Zhao D, Qi LS. CRISPR/Cas9 for human genome engineering and disease research. Annu Rev Genomics Hum Genet. (2016) 17:131–54. doi: 10.1146/annurev-genom-083115-022258
151. Yu C, Liu Y, Ma T, Liu K, Xu S, Zhang Y, et al. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. (2015) 16:142–7. doi: 10.1016/j.stem.2015.01.003
152. Boel A, De Saffel H, Steyaert W, Callewaert B, De Paepe A, Coucke PJ, et al. CRISPR/Cas9-mediated homology-directed repair by ssODNs in zebrafish induces complex mutational patterns resulting from genomic integration of repair-template fragments. Dis Model Mech. (2018) 11. doi: 10.1242/dmm.035352
153. Boel A, Steyaert W, De Rocker N, Menten B, Callewaert B, De Paepe A, et al. BATCH-GE: batch analysis of next-generation sequencing data for genome editing assessment. Sci Rep. (2016) 6:30330. doi: 10.1038/srep30330
154. Armstrong GA, Liao M, You Z, Lissouba A, Chen BE, Drapeau P. Homology directed knockin of point mutations in the zebrafish tardbp and fus genes in ALS using the CRISPR/Cas9 system. PLoS ONE. (2016) 11:e0150188. doi: 10.1371/journal.pone.0150188
155. Farr GH 3rd, Imani K, Pouv D, Maves L. Functional testing of a human PBX3 variant in zebrafish reveals a potential modifier role in congenital heart defects. Dis Model Mech. (2018) 11. doi: 10.1242/dmm.035972
156. Tessadori F, Roessler HI, Savelberg SMC, Chocron S, Kamel SM, Duran KJ, et al. Effective CRISPR/Cas9-based nucleotide editing in zebrafish to model human genetic cardiovascular disorders. Dis Model Mech. (2018) 11. doi: 10.1242/dmm.035469
157. Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature. (2017) 551:464–71. doi: 10.1038/nature24644
158. Qin W, Lu X, Lin S. Programmable base editing in zebrafish using a modified CRISPR-Cas9 system. Methods. (2018) 150:19–23. doi: 10.1016/j.ymeth.2018.07.010
159. Zhang Y, Qin W, Lu X, Xu J, Huang H, Bai H, et al. Programmable base editing of zebrafish genome using a modified CRISPR-Cas9 system. Nat Commun. (2017) 8:118. doi: 10.1038/s41467-017-00175-6
160. Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. (2019) 576:149–57. doi: 10.1038/s41586-019-1711-4
161. Stuart T, Satija R. Integrative single-cell analysis. Nat Rev Genet. (2019) 20:257–72. doi: 10.1038/s41576-019-0093-7
162. Farrell JA, Wang Y, Riesenfeld SJ, Shekhar K, Regev A, Schier AF. Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science. (2018) 360. doi: 10.1126/science.aar3131
163. Spanjaard B, Hu B, Mitic N, Olivares-Chauvet P, Janjuha S, Ninov N, et al. Simultaneous lineage tracing and cell-type identification using CRISPR-Cas9-induced genetic scars. Nat Biotechnol. (2018) 36:469–73. doi: 10.1038/nbt.4124
164. Wagner DE, Weinreb C, Collins ZM, Briggs JA, Megason SG, Klein AM. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science. (2018) 360:981–7. doi: 10.1126/science.aar4362
165. Long F. Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol. (2011) 13:27–38. doi: 10.1038/nrm3254
166. Miller RL. Transgenic mice: beyond the knockout. Am J Physiol Renal Physiol. (2011) 300:F291–300. doi: 10.1152/ajprenal.00082.2010
167. Tampieri A, Sprio S, Sandri M, Valentini F. Mimicking natural bio-mineralization processes: a new tool for osteochondral scaffold development. Trends Biotechnol. (2011) 29:526–35. doi: 10.1016/j.tibtech.2011.04.011
168. Gregg CL, Butcher JT. Quantitative in vivo imaging of embryonic development: opportunities and challenges. Differentiation. (2012) 84:149–62. doi: 10.1016/j.diff.2012.05.003
169. Moro E, Vettori A, Porazzi P, Schiavone M, Rampazzo E, Casari A, et al. Generation and application of signaling pathway reporter lines in zebrafish. Mol Genet Genomics. (2013) 288:231–42. doi: 10.1007/s00438-013-0750-z
170. Dutton JR, Antonellis A, Carney TJ, Rodrigues FS, Pavan WJ, Ward A, et al. An evolutionarily conserved intronic region controls the spatiotemporal expression of the transcription factor Sox10. BMC Dev Biol. (2008) 8:105. doi: 10.1186/1471-213X-8-105
171. Dougherty M, Kamel G, Shubinets V, Hickey G, Grimaldi M, Liao EC. Embryonic fate map of first pharyngeal arch structures in the sox10: kaede zebrafish transgenic model. J Craniofac Surg. (2012) 23:1333–7. doi: 10.1097/SCS.0b013e318260f20b
172. Kirby BB, Takada N, Latimer AJ, Shin J, Carney TJ, Kelsh RN, et al. In vivo time-lapse imaging shows dynamic oligodendrocyte progenitor behavior during zebrafish development. Nat Neurosci. (2006) 9:1506–11. doi: 10.1038/nn1803
173. Rodrigues FS, Doughton G, Yang B, Kelsh RN. A novel transgenic line using the Cre-lox system to allow permanent lineage-labeling of the zebrafish neural crest. Genesis. (2012) 50:750–7. doi: 10.1002/dvg.22033
174. Brunt LH, Begg K, Kague E, Cross S, Hammond CL. Wnt signalling controls the response to mechanical loading during zebrafish joint development. Development. (2017) 144:2798–09. doi: 10.1242/dev.153528
175. Wada N, Javidan Y, Nelson S, Carney TJ, Kelsh RN, Schilling TF. Hedgehog signaling is required for cranial neural crest morphogenesis and chondrogenesis at the midline in the zebrafish skull. Development. (2005) 132:3977–88. doi: 10.1242/dev.01943
176. Hochgreb-Hägelen T, Koo DES, Bronner M. Znf385C mediates a novel p53-dependent transcriptional switch to control timing of facial bone formation. Dev Biol. (2015) 400:23–32. doi: 10.1016/j.ydbio.2015.01.011
177. Li L, Mao A, Wang P, Ning G, Cao Y, Wang Q. Endodermal pouch-expressed dmrt2b is important for pharyngeal cartilage formation. Biol Open. (2018) 7. doi: 10.1242/bio.035444
178. Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. (2002) 248:307–18. doi: 10.1006/dbio.2002.0711
179. Flanagan-Steet H, Aarnio M, Kwan B, Guihard P, Petrey A, Haskins M, et al. Cathepsin-mediated alterations in TGFss-related signaling underlie disrupted cartilage and bone maturation associated with impaired lysosomal targeting. J Bone Miner Res. (2016) 31:535–48. doi: 10.1002/jbmr.2722
180. Bonkowsky JL, Wang X, Fujimoto E, Lee JE, Chien CB, Dorsky RI. Domain-specific regulation of foxP2 CNS expression by lef1. BMC Dev Biol. (2008) 8:103. doi: 10.1186/1471-213X-8-103
181. DeLaurier A, Eames BF, Blanco-Sanchez B, Peng G, He X, Swartz ME, et al. Zebrafish sp7:EGFP: a transgenic for studying otic vesicle formation, skeletogenesis, bone regeneration. Genesis. (2010) 48:505–11. doi: 10.1002/dvg.20639
182. Kague E, Witten PE, Soenens M, Campos CL, Lubiana T, Fisher S, et al. Zebrafish sp7 mutants show tooth cycling independent of attachment, eruption and poor differentiation of teeth. Dev Biol. (2018) 435:176–84. doi: 10.1016/j.ydbio.2018.01.021
183. Dale RM, Topczewski J. Identification of an evolutionarily conserved regulatory element of the zebrafish col2a1a gene. Dev Biol. (2011) 357:518–31. doi: 10.1016/j.ydbio.2011.06.020
184. Filipek-Górniok B, Carlsson P, Haitina T, Habicher J, Ledin J, Kjellén L. The NDST gene family in zebrafish: role of NDST1B in pharyngeal arch formation. PLoS ONE. (2015) 10:e0119040. doi: 10.1371/journal.pone.0119040
185. Kague E, Bessling SL, Lee J, Hu G, Passos-Bueno MR, Fisher S. Functionally conserved cis-regulatory elements of COL18A1 identified through zebrafish transgenesis. Dev Biol. (2010) 337:496–505. doi: 10.1016/j.ydbio.2009.10.028
186. Pogoda HM, Riedl-Quinkertz I, Lohr H, Waxman JS, Dale RM, Topczewski J, et al. Direct activation of chordoblasts by retinoic acid is required for segmented centra mineralization during zebrafish spine development. Development. (2018) 145. doi: 10.1242/dev.159418
187. Hu P, Tian M, Bao J, Xing G, Gu X, Gao X, et al. Retinoid regulation of the zebrafish cyp26a1 promoter. Dev Dyn. (2008) 237:3798–808. doi: 10.1002/dvdy.21801
188. Li J, Hu P, Li K, Zhao Q. Identification and characterization of a novel retinoic acid response element in zebrafish cyp26a1 promoter. Anat Rec (Hoboken). (2012) 295:268–77. doi: 10.1002/ar.21520
189. Haga Y, Dominique VJ 3rd, Du SJ. Analyzing notochord segmentation and intervertebral disc formation using the twhh:gfp transgenic zebrafish model. Transgenic Res. (2009) 18:669–83. doi: 10.1007/s11248-009-9259-y
190. Ma RC, Jacobs CT, Sharma P, Kocha KM, Huang P. Stereotypic generation of axial tenocytes from bipartite sclerotome domains in zebrafish. PLoS Genet. (2018) 14:e1007775. doi: 10.1371/journal.pgen.1007775
191. DeLaurier A, Huycke TR, Nichols JT, Swartz ME, Larsen A, Walker C, et al. Role of mef2ca in developmental buffering of the zebrafish larval hyoid dermal skeleton. Dev Biol. (2014) 385:189–99. doi: 10.1016/j.ydbio.2013.11.016
192. Eames BF, Yan YL, Swartz ME, Levic DS, Knapik EW, Postlethwait JH, et al. Mutations in fam20b and xylt1 reveal that cartilage matrix controls timing of endochondral ossification by inhibiting chondrocyte maturation. PLoS Genet. (2011) 7:e1002246. doi: 10.1371/journal.pgen.1002246
193. Stewart S, Gomez AW, Armstrong BE, Henner A, Stankunas K. Sequential and opposing activities of Wnt and BMP coordinate zebrafish bone regeneration. Cell Rep. (2014) 6:482–98. doi: 10.1016/j.celrep.2014.01.010
194. Blum N, Begemann G. Osteoblast de- and redifferentiation are controlled by a dynamic response to retinoic acid during zebrafish fin regeneration. Development. (2015) 142:2894–903. doi: 10.1242/dev.120204
195. Geurtzen K, Knopf F, Wehner D, Huitema LF, Schulte-Merker S, Weidinger G. Mature osteoblasts dedifferentiate in response to traumatic bone injury in the zebrafish fin and skull. Development. (2014) 141:2225–34. doi: 10.1242/dev.105817
196. Bek JW, De Clercq A, De Saffel H, Soenens M, Huysseune A, Witten PE, et al. Photoconvertible fluorescent proteins: a versatile tool in zebrafish skeletal imaging. J Fish Biol. (2020) doi: 10.1111/jfb.14335
197. Singh SP, Holdway JE, Poss KD. Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev Cell. (2012) 22:879–86. doi: 10.1016/j.devcel.2012.03.006
198. Cox BD, De Simone A, Tornini VA, Singh SP, Di Talia S, Poss KD. In toto imaging of dynamic osteoblast behaviors in regenerating skeletal bone. Curr Biol. (2018) 28:3937–47.e4. doi: 10.1016/j.cub.2018.10.052
199. Oralova V, Rosa JT, Soenens M, Bek JW, Willaert A, Witten PE, et al. Beyond the whole-mount phenotype: high-resolution imaging in fluorescence-based applications on zebrafish. Biol Open. (2019) 8. doi: 10.1242/bio.042374
200. Willems B, Buttner A, Huysseune A, Renn J, Witten PE, Winkler C. Conditional ablation of osteoblasts in medaka. Dev Biol. (2012) 364:128–37. doi: 10.1016/j.ydbio.2012.01.023
201. Kobayashi-Sun J, Yamamori S, Kondo M, Kuroda J, Ikegame M, Suzuki N, et al. Uptake of osteoblast-derived extracellular vesicles promotes the differentiation of osteoclasts in the zebrafish scale. Commun Biol. (2020) 3:190. doi: 10.1038/s42003-020-0925-1
202. Mitchell RE, Huitema LF, Skinner RE, Brunt LH, Severn C, Schulte-Merker S, et al. New tools for studying osteoarthritis genetics in zebrafish. Osteoarthritis Cartilage. (2013) 21:269–78. doi: 10.1016/j.joca.2012.11.004
203. Kim YI, Lee S, Jung SH, Kim HT, Choi JH, Lee MS, et al. Establishment of a bone-specific col10a1:GFP transgenic zebrafish. Mol Cells. (2013) 36:145–50. doi: 10.1007/s10059-013-0117-7
204. Hu Z, Chen B, Zhao Q. Hedgehog signaling regulates osteoblast differentiation in zebrafish larvae through modulation of autophagy. Biol Open. (2019) 8. doi: 10.1242/bio.040840
205. Knopf F, Hammond C, Chekuru A, Kurth T, Hans S, Weber CW, et al. Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev Cell. (2011) 20:713–24. doi: 10.1016/j.devcel.2011.04.014
206. Bussmann J, Schulte-Merker S. Rapid BAC selection for tol2-mediated transgenesis in zebrafish. Development. (2011) 138:4327–32. doi: 10.1242/dev.068080
207. Caetano-Lopes J, Henke K, Urso K, Duryea J, Charles JF, Warman ML, et al. Correction: unique and non-redundant function of csf1r paralogues in regulation and evolution of post-embryonic development of the zebrafish. Development. (2020) 147. doi: 10.1242/dev.192211
208. To TT, Witten PE, Huysseune A, Winkler C. An adult osteopetrosis model in medaka reveals the importance of osteoclast function for bone remodeling in teleost fish. Comp Biochem Physiol C Toxicol Pharmacol. (2015) 178:68–75. doi: 10.1016/j.cbpc.2015.08.007
209. Alexander C, Zuniga E, Blitz IL, Wada N, Le Pabic P, Javidan Y, et al. Combinatorial roles for BMPs and Endothelin 1 in patterning the dorsal-ventral axis of the craniofacial skeleton. Development. (2011) 138:5135–46. doi: 10.1242/dev.067801
210. Laux DW, Febbo JA, Roman BL. Dynamic analysis of BMP-responsive smad activity in live zebrafish embryos. Dev Dyn. (2011) 240:682–94. doi: 10.1002/dvdy.22558
211. Moro E, Ozhan-Kizil G, Mongera A, Beis D, Wierzbicki C, Young RM, et al. In vivo Wnt signaling tracing through a transgenic biosensor fish reveals novel activity domains. Dev Biol. (2012) 366:327–40. doi: 10.1016/j.ydbio.2012.03.023
212. Weidinger G, Thorpe CJ, Wuennenberg-Stapleton K, Ngai J, Moon RT. The Sp1-related transcription factors sp5 and sp5-like act downstream of Wnt/beta-catenin signaling in mesoderm and neuroectoderm patterning. Curr Biol. (2005) 15:489–500. doi: 10.1016/j.cub.2005.01.041
213. Felber K, Elks PM, Lecca M, Roehl HH. Expression of osterix is regulated by FGF and Wnt/beta-catenin signalling during osteoblast differentiation. PLoS ONE. (2015) 10:e0144982. doi: 10.1371/journal.pone.0144982
214. Stoick-Cooper CL, Weidinger G, Riehle KJ, Hubbert C, Major MB, Fausto N, et al. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. (2007) 134:479–89. doi: 10.1242/dev.001123
215. Huang CJ, Tu CT, Hsiao CD, Hsieh FJ, Tsai HJ. Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev Dyn. (2003) 228:30–40. doi: 10.1002/dvdy.10356
216. Li J, Chen Z, Gao LY, Colorni A, Ucko M, Fang S, et al. A transgenic zebrafish model for monitoring xbp1 splicing and endoplasmic reticulum stress in vivo. Mech Dev. (2015) 137:33–44. doi: 10.1016/j.mod.2015.04.001
217. Clark EM, Nonarath HJT, Bostrom JR, Link BA. Establishment and validation of an endoplasmic reticulum stress reporter to monitor zebrafish ATF6 activity in development and disease. Dis Model Mech. (2020) 13. doi: 10.1242/dmm.041426
218. Ding Y, Lee J, Hsu JJ, Chang CC, Baek KI, Ranjbarvaziri S, et al. Light-sheet imaging to elucidate cardiovascular injury and repair. Curr Cardiol Rep. (2018) 20:35. doi: 10.1007/s11886-018-0979-6
219. Collery RF, Link BA. Dynamic smad-mediated BMP signaling revealed through transgenic zebrafish. Dev Dyn. (2011) 240:712–22. doi: 10.1002/dvdy.22567
220. Dorsky RI, Itoh M, Moon RT, Chitnis A. Two tcf3 genes cooperate to pattern the zebrafish brain. Development. (2003) 130:1937–47. doi: 10.1242/dev.00402
221. Schwend T, Loucks EJ, Ahlgren SC. Visualization of gli activity in craniofacial tissues of hedgehog-pathway reporter transgenic zebrafish. PLoS ONE. (2010) 5:e14396. doi: 10.1371/journal.pone.0014396
222. Blair H, Schlesinger P, Huang C, Zaidi M. Calcium signalling and calcium transport in bone disease. Subcell Biochem. (2007) 45:539–62. doi: 10.1007/978-1-4020-6191-2_21
223. Cao X, Chen D. The BMP signaling and in vivo bone formation. Gene. (2005) 357:1–8. doi: 10.1016/j.gene.2005.06.017
224. Houschyar KS, Tapking C, Borrelli MR, Popp D, Duscher D, Maan ZN, et al. Wnt pathway in bone repair and regeneration - what do we know so far. Front Cell Dev Biol. (2019) 6:170. doi: 10.3389/fcell.2018.00170
225. Huang S, Wang Y, Luo L, Li X, Jin X, Li S, et al. BMP2 is related to hirschsprung's disease and required for enteric nervous system development. Front Cell Neurosci. (2019) 13:523. doi: 10.3389/fncel.2019.00523
226. Besio R, Garibaldi N, Leoni L, Cipolla L, Sabbioneda S, Biggiogera M, et al. Cellular stress due to impairment of collagen prolyl hydroxylation complex is rescued by the chaperone 4-phenylbutyrate. Dis Model Mech. (2019) 12. doi: 10.1242/dmm.038521
227. Besio R, Iula G, Garibaldi N, Cipolla L, Sabbioneda S, Biggiogera M, et al. 4-PBA ameliorates cellular homeostasis in fibroblasts from osteogenesis imperfecta patients by enhancing autophagy and stimulating protein secretion. Biochim Biophys Acta Mol Basis Dis. (2018) 1864(5 Pt A):1642–52. doi: 10.1016/j.bbadis.2018.02.002
228. Howarth DL, Lindtner C, Vacaru AM, Sachidanandam R, Tsedensodnom O, Vasilkova T, et al. Activating transcription factor 6 is necessary and sufficient for alcoholic fatty liver disease in zebrafish. PLoS Genet. (2014) 10:e1004335. doi: 10.1371/journal.pgen.1004335
229. Yeh KY, Lai CY, Lin CY, Hsu CC, Lo CP, Her GM. ATF4 overexpression induces early onset of hyperlipidaemia and hepatic steatosis and enhances adipogenesis in zebrafish. Sci Rep. (2017) 7:16362. doi: 10.1038/s41598-017-16587-9
230. Cardeira J, Gavaia PJ, Fernandez I, Cengiz IF, Moreira-Silva J, Oliveira JM, et al. Quantitative assessment of the regenerative and mineralogenic performances of the zebrafish caudal fin. Sci Rep. (2016) 6:39191. doi: 10.1038/srep39191
231. Chen CH, Puliafito A, Cox BD, Primo L, Fang Y, Di Talia S, et al. Multicolor cell barcoding technology for long-term surveillance of epithelial regeneration in zebrafish. Dev Cell. (2016) 36:668–80. doi: 10.1016/j.devcel.2016.02.017
232. Park S, Greco V, Cockburn K. Live imaging of stem cells: answering old questions and raising new ones. Curr Opin Cell Biol. (2016) 43:30–37. doi: 10.1016/j.ceb.2016.07.004
233. Mishra R, Sehring I, Cederlund M, Mulaw M, Weidinger G. NF-kappaB signaling negatively regulates osteoblast dedifferentiation during zebrafish bone regeneration. Dev Cell. (2020) 52:167–82.e7. doi: 10.1016/j.devcel.2019.11.016
234. Bruneel B, Witten PE. Power and challenges of using zebrafish as a model for skeletal tissue imaging. Connect Tissue Res. (2015) 56:161–73. doi: 10.3109/03008207.2015.1013193
235. Witten PE, Gil Martens L, Huysseune A, Takle H, Hjelde K. Towards a classification and an understanding of developmental relationships of vertebral body malformations in Atlantic salmon (Salmo salar Aquaculture L). Aquaculture. 295:6–14. doi: 10.1016/j.aquaculture.2009.06.037
236. Witten PE, Obach A, Huysseune A, Baeverfjord G. Vertebrae fusion in Atlantic salmon (Salmo salar): development, aggravation and pathways of containment. Aquaculture. (2006) 258:164–72. doi: 10.1016/j.aquaculture.2006.05.005
237. du Plessis A, Broeckhoven C, Guelpa A, le Roux SG. Laboratory x-ray micro-computed tomography: a user guideline for biological samples. Gigascience. (2017) 6:1–11. doi: 10.1093/gigascience/gix027
238. Rawson SD, Maksimcuka J, Withers PJ, Cartmell SH. X-ray computed tomography in life sciences. BMC Biol. (2020) 18:21. doi: 10.1186/s12915-020-0753-2
239. Babaei F, Hong TL, Yeung K, Cheng SH, Lam YW. Contrast-enhanced X-ray micro-computed tomography as a versatile method for anatomical studies of adult zebrafish. Zebrafish. (2016) 13:310–6. doi: 10.1089/zeb.2016.1245
240. Descamps E, Sochacka A, De Kegel B, Van Loo D, Van Hoorebeke L, Adriaens D. Soft tissue discrimination with contrast agents using micro-CT scanning. Belgian J Zool. (2014) 144:20–40. doi: 10.26496/bjz.2014.63
241. Charles JF, Sury M, Tsang K, Urso K, Henke K, Huang Y, et al. Utility of quantitative micro-computed tomographic analysis in zebrafish to define gene function during skeletogenesis. Bone. (2017) 101:162–71. doi: 10.1016/j.bone.2017.05.001
242. Suniaga S, Rolvien T, Vom Scheidt A, Fiedler IAK, Bale HA, Huysseune A, et al. Increased mechanical loading through controlled swimming exercise induces bone formation and mineralization in adult zebrafish. Sci Rep. (2018) 8:3646. doi: 10.1038/s41598-018-21776-1
243. Gadow H, Abbott EC. On the evolution of the vertebral coloumn of fishes. Philos Trans R Soc Lond. (1985) 186:163–221. doi: 10.1098/rstb.1895.0004
244. Sakata-Haga H, Uchishiba M, Shimada H, Tsukada T, Mitani M, Arikawa T, et al. A rapid and nondestructive protocol for whole-mount bone staining of small fish and Xenopus. Sci Rep. (2018) 8:7453–60. doi: 10.1038/s41598-018-25836-4
245. Moriguchi T, Yano K, Nakagawa S, Kaji F. Elucidation of adsorption mechanism of bone-staining agent alizarin red S on hydroxyapatite by FT-IR microspectroscopy. J Colloid Interface Sci. (2003) 260:19–25. doi: 10.1016/S0021-9797(02)00157-1
246. Fraser TWK, Witten PE, Albrektsen S, Breck O, Fontanillas R, Nankervis L, et al. Phosphorus nutrition in farmed Atlantic salmon (Salmo salar): life stage and temperature effects on bone pathologies. Aquaculture. (2019) 511:1–12. doi: 10.1016/j.aquaculture.2019.734246
247. Witten PE, Fjelldal PG, Huysseune A, McGurk C, Obach A, Owen MAG. Bone without minerals and its secondary mineralization in Atlantic salmon (Salmo salar): the recovery from phosphorus deficiency. J Exp Biol. (2019) 222:1–15. doi: 10.1242/jeb.188763
248. Vandenplas S, De Clercq A, Huysseune A. Tooth replacement without a dental lamina: the search for epithelial stem cells in Polypterus senegalus. J Exp Zool B Mol Dev Evol. (2014) 322:281–93. doi: 10.1002/jez.b.22577
249. Woltmann I, Shkil FN, De Clercq A, Huysseune A, Witten PE. Supernumerary teeth in the pharyngeal dentition of slow developing zebrafish. J Appl Ichthyol. (2018) 34:455–64. doi: 10.1111/jai.13668
250. Bensimon-Brito A, Cardeira J, Dionísio G, Huysseune A, Cancela ML, Witten PE. Revisiting in vivo staining with alizarin red S - a valuable approach to analyse zebrafish skeletal mineralization during development and regeneration. BMC Dev Biol. (2016) 16:1–9. doi: 10.1186/s12861-016-0102-4
251. Kiernan JA. Histological and Histochemical Methods: Theory and Practice. 5th ed. Banbury: Scion Publishing. (2015)
252. Cubbage CC, Mabee PM. Development of the cranium and paired fins in the zebrafish Danio rerio (Ostariophysi, Cyprinidae). J Morphol. (1996) 229:121–60. doi: 10.1002/(SICI)1097-4687(199608)229:2<121::AID-JMOR1>3.0.CO;2-4
253. Kimmel CB, Miller CT, Moens CB. Specification and morphogenesis of the zebrafish larval head skeleton. Dev Biol. (2001) 233:239–57. doi: 10.1006/dbio.2001.0201
254. Kimmel CB, Walker MB, Miller CT. Morphing the hyomandibular skeleton in development and evolution. J Exp Zool B Mol Dev Evol. (2007) 308:609–24. doi: 10.1002/jez.b.21155
255. Walker M, Kimmel C. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech Histochem. (2007) 82:23–28. doi: 10.1080/10520290701333558
256. Philip NS, Green DM. Recovery and enhancement of faded cleared and double stained specimens. Biotech Histochem. (2000) 75:193–6. doi: 10.3109/10520290009066500
257. Copper JE, Budgeon LR, Foutz CA, van Rossum DB, Vanselow DJ, Hubley MJ, et al. Comparative analysis of fixation and embedding techniques for optimized histological preparation of zebrafish. Comp Biochem Physiol C Toxicol Pharmacol. (2018) 208:38–46. doi: 10.1016/j.cbpc.2017.11.003
258. Witten PE. Enzyme histochemical characteristics of osteoblasts and mononucleated osteoclasts in a teleost fish with acellular bone (Oreochromis niloticus, Cichlidae). Cell Tissue Res. (1997) 287:591–9. doi: 10.1007/s004410050782
259. Sarmah S, Barrallo-Gimeno A, Melville DB, Topczewski J, Solnica-Krezel L, Knapik EW. Sec24D-dependent transport of extracellular matrix proteins is required for zebrafish skeletal morphogenesis. PLoS ONE. (2010) 5:e10367. doi: 10.1371/journal.pone.0010367
260. Pasqualetti S, Banfi G, Mariotti M. The zebrafish scale as model to study the bone mineralization process. J Mol Histol. (2012) 43:589–95. doi: 10.1007/s10735-012-9425-z
261. Duran I, Mari-Beffa M, Santamaria JA, Becerra J, Santos-Ruiz L. Actinotrichia collagens and their role in fin formation. Dev Biol. (2011) 354:160–72. doi: 10.1016/j.ydbio.2011.03.014
262. Liedtke D, Orth M, Meissler M, Geuer S, Knaup S, Koblitz I, et al. ECM alterations in Fndc3a (fibronectin domain containing protein 3A) deficient zebrafish cause temporal fin development and regeneration defects. Sci Rep. (2019) 9:13383. doi: 10.1038/s41598-019-50055-w
263. Sehring IM, Weidinger G. Recent advancements in understanding fin regeneration in zebrafish. Wiley Interdiscip Rev Dev Biol. (2019) 9:e367. doi: 10.1002/wdev.367
264. Stewart S, Stankunas K. Limited dedifferentiation provides replacement tissue during zebrafish fin regeneration. Dev Biol. (2012) 365:339–49. doi: 10.1016/j.ydbio.2012.02.031
265. Paul S, Schindler S, Giovannone D, de Millo Terrazzani A, Mariani FV, Crump JG. Ihha induces hybrid cartilage-bone cells during zebrafish jawbone regeneration. Development. (2016) 143:2066–76. doi: 10.1242/dev.131292
266. Azevedo AS, Grotek B, Jacinto A, Weidinger G, Saude L. The regenerative capacity of the zebrafish caudal fin is not affected by repeated amputations. PLoS ONE. (2011) 6:e22820. doi: 10.1371/journal.pone.0022820
267. Kinoshita M, Murata K, Naruse K, Tanaka M, Kamei Y. Medaka: Biology, Management, Experimental Protocols. Vol. 2. Wiley-Blackwell (2019).
268. Furutani-Seiki M, Wittbrodt J. Medaka and zebrafish, an evolutionary twin study. Mech Dev. (2004) 121:629–37. doi: 10.1016/j.mod.2004.05.010
269. Wittbrodt J, Shima A, Schartl M. Medaka–a model organism from the far East. Nat Rev Genet. (2002) 3:53–64. doi: 10.1038/nrg704
270. Azetsu Y, Inohaya K, Takano Y, Kinoshita M, Tasaki M, Kudo A. The sp7 gene is required for maturation of osteoblast-lineage cells in medaka (Oryzias latipes) vertebral column development. Dev Biol. (2017) 431:252–62. doi: 10.1016/j.ydbio.2017.09.010
271. Irie K, Kuroda Y, Mimori N, Hayashi S, Abe M, Tsuji N, et al. Histopathology of a wavy medaka. J Toxicol Pathol. (2016) 29:115–8. doi: 10.1293/tox.2015-0070
272. Mantoku A, Chatani M, Aono K, Inohaya K, Kudo A. Osteoblast and osteoclast behaviors in the turnover of attachment bones during medaka tooth replacement. Dev Biol. (2016) 409:370–81. doi: 10.1016/j.ydbio.2015.12.002
273. Yu T, Witten PE, Huysseune A, Buettner A, To TT, Winkler C. Live imaging of osteoclast inhibition by bisphosphonates in a medaka osteoporosis model. Dis Model Mech. (2016) 9:155–63. doi: 10.1242/dmm.019091
274. Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). Bioessays. (2005) 27:937–45. doi: 10.1002/bies.20293
275. Hadjantonakis AK, Pirity M, Nagy A. Cre recombinase mediated alterations of the mouse genome using embryonic stem cells. Methods Mol Biol. (2008) 461:111–32. doi: 10.1007/978-1-60327-483-8_8
Keywords: zebrafish, skeletal system, x-ray, microCT analyses, imaging techniques, skeletal diseases
Citation: Tonelli F, Bek JW, Besio R, De Clercq A, Leoni L, Salmon P, Coucke PJ, Willaert A and Forlino A (2020) Zebrafish: A Resourceful Vertebrate Model to Investigate Skeletal Disorders. Front. Endocrinol. 11:489. doi: 10.3389/fendo.2020.00489
Received: 25 April 2020; Accepted: 22 June 2020;
Published: 31 July 2020.
Edited by:
Wim Van Hul, University of Antwerp, BelgiumReviewed by:
Christoph Winkler, National University of Singapore, SingaporeMichaël R. Laurent, University Hospitals Leuven, Belgium
Copyright © 2020 Tonelli, Bek, Besio, De Clercq, Leoni, Salmon, Coucke, Willaert and Forlino. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Antonella Forlino, YWZvcmxpbm9AdW5pcHYuaXQ=
†These authors have contributed equally to this work
‡These authors share last authorship
 Francesca Tonelli
Francesca Tonelli Jan Willem Bek
Jan Willem Bek Roberta Besio
Roberta Besio Adelbert De Clercq
Adelbert De Clercq Laura Leoni
Laura Leoni Phil Salmon
Phil Salmon Paul J. Coucke
Paul J. Coucke Andy Willaert
Andy Willaert Antonella Forlino
Antonella Forlino