- Department of Endocrinology & Rheumatology, Shanghai University of Medicine & Health Sciences Affiliated Zhoupu Hospital, Shanghai, China
Objective: Epigenetic modifications in RNA are known to play critical roles in cell differentiation through regulating expressions of some key genes including members of the suppressor of cytokine signaling (SOCS) family. The present study aimed to unveil the relationship of SOCS mRNA methylation induced by methyltransferase like 3 (METTL3) with Graves’ disease (GD).
Methods: Differently expressed genes (DEG) in GD tissues were identified using microarray analysis and further validated using CD4+ T cell microarray of GD tissues and isolated peripheral blood mononuclear cells (PBMCs). Furthermore, expressions of METTL3 targeted genes were detected using METTL3 knock-down experiment in RAW264.7 cells.
Results: High throughput microarrays revealed that METTL3 and SOCS molecules were aberrantly expressed in thyroid tissues and CD4+T cells of GD compared to the controls. Bioinformatic analysis was undertaken by searching databases of found genes of the SOCS family that possessed many mRNA m6A modification loci. METTL3 knock-down experiment revealed that expressions of SOCS family members SOCS1, SOCS2, SOCS4, SOCS5, and SOCS6 were increased after METTL3 knock-down.
Conclusions: For the first time, the present study revealed the relationship between m6A modification and GD and indicated that METTL3 may be involved in the development of GD by inducing mRNA m6A methylation modification of SOCS family members.
Introduction
Graves’ Disease (GD) is the main clinical sub-type of autoimmune thyroid diseases (AITDs), leading to a decline in quality of life and increasing the risk of death, and thus threatening public health. It is the cause of 90% hyperthyroidism, annually affecting 20-50 in 100,000 people, mainly aged between 30-50 years old, among whom, 5% were females and only 0.5% were males (1, 2). In GD patients, thyrotropin-receptor antibody (TRAb) imitates the role of TSH and stimulates thyroid follicular epithelial cells to produce and secret hormones, thus inducing thyrotoxicosis. As of now, no therapy targets the etiology of GD, instead, all tend to focus on the treatment of basic symptoms (3). GD is an autoimmune disease caused by both genetic and environmental factors. Thus far, genetic-associated studies have identified mainly thyroglobulin (Tg), thyrotropin receptor (TSHR), PTPN22, IL-17, and IL-22 as the susceptible genes of GD (4–6). Similarly, immune dysfunction caused by the imbalance of immune cells was also found to play an important role in the pathogenesis of GD. Additionally, our previous studies, along with others, have also revealed that epigenetic factors play a pivotal part in the etiology of GD (7). Yet, despite these findings, the specific molecular mechanism underlying the above-mentioned multiple factors involved in the pathogenesis of GD has not been clarified.
N6-methyladenosine (m6A) is the most abundant and stable form of mRNA modification in most species and is also an important epigenetic marker involved in all aspects of the life process. mRNA m6A modification has been reported to influence mRNA alternative splicing, translation, and stability, thereby regulating the expression and function of targeted genes (8). Several studies have revealed that mRNA m6A modification is associated with the differentiation and immune response of immune cells (9–12). Furthermore, the recent identification of key enzymes for m6A modification including m6A methyltransferase (writer), m6A demethylase (eraser), and m6A RNA binding protein (reader) has dramatically deepened our understanding of RNA epigenesis (13, 14). Among these enzymes, methyltransferase like 3 (METTL3) is the key methylation modification enzyme. It has been evidenced that METTL3 is involved in multiple life processes by regulating mRNA level and the protein expression of some key molecules (15). Moreover, some studies found that polymorphisms of METTL3 and fat mass and obesity associated (FTO) genes are involved in the pathogenesis of some autoimmune diseases, such as rheumatoid arthritis (RA) (16) and latent autoimmune diabetes (LADA) (17). Our recent study also uncovered that METTL3 polymorphisms are involved in the pathogenesis of AITDs, including GD (18). However, the mechanism underlying the involvement of mRNA m6A modification in GD development is still incompletely understood.
mRNA m6A methylation alters mRNA levels and the subsequent protein expression of targeted genes by influencing mRNA stability and degradation rate. This mechanism is extremely important in regulating the expression of immediate-early genes (IEGs) (19). Among these IEGs, the suppressors of the cytokine signaling (SOCS) family, which mainly consist of one CIS and 7 SOCS proteins, are involved in many biological processes, such as cell proliferation, differentiation, and signal transduction. SOCS proteins also play a pivotal role in regulating the immune system and the development of immune-mediated diseases (20). Studies have revealed aberrant SOCS1 and SOCS3 expressions in diverse autoimmune diseases (21). However, whether METTL3 is involved in GD by inducing the methylation modification of the SOCS family has not been elucidated. Therefore, to explore the role of METTL3 in GD development and better understand the potential pathomechanism of GD, we analyzed our previous microarray data using bioinformatic analysis tools, validated changes in mRNA levels of METTL3 and SOCS family members using real-time quantitative polymerase chain reaction (qPCR) technology in clinical samples, and performed functional analyses in cultured cells by knocking-down METTL3.
Materials and Methods
Microarray and Bioinformatic Analyses
First, we screened the differentially expressed genes (DEGs) in our previously published GD microarray, which included 4 GD cases and 3 controls (22). In addition, to further explore the possible functions of these DEGs, by using the online functional annotation tools in the Database for Annotation, Visualization and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov/) (23, 24), we analyzed the functional pathways by the Gene Ontology (GO) functional enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway methods. Second, we searched the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) and found a genome-wide gene expression dataset, GSE9340, which includes 18 GD thyroid tissues. Adopting the same analysis, we screened the DEGs from GSE9340. We combined DEGs from our microarray data and GSE9340, and carried out the gene co-expression network bioinformatics analysis by using the WGCNA R software package as described previously (25). Finally, we further identified the potential functional pathways of genes in key co-expression modules using the DAVID database including GO and KEGG pathways.
Validation for Selected Genes
To further validate the expression of METTL3 and SOCS family members, we first explored Dataset GSE71956, which provides a genome-wide gene expression profile in CD4+ T cells from 15 GD patients and 10 controls. Furthermore, we isolated peripheral blood mononuclear cells (PBMCs) from whole blood samples of 26 newly recruited GD patients and 26 age- and sex-matched healthy volunteers and extracted total RNA from these PBMCs samples. The TRAb were positive for all the GD patients. For the healthy controls, we chose those who had normal thyroid function and negative TRAb, anti-thyroglobulin antibody (TgAb), and anti-thyroid peroxidase antibody (TPOAb). Those with thyroid diseases or other autoimmune diseases were excluded from the control group. After qualification and quantification by Nano Drop 2.0, we examined mRNA levels of METTL3, METTL14, FTO, and ALKBH5 using real-time qPCR with primers GTGATCGTAGCTGAGGTTCGT and GGGTTGCACATTGTGTGGTC for METTL3, TGGACCTTGGAAGAGTGTGTTT, and CATGAGGCAGTGTTCCTTTGTT for METTL14, TTGCCCGAACATTACCTGCT, and TGTGAGGTCAAAAAAGCGCAGAG for FTO, TCAAGCCTATTCGGGTGTCG, and AGCAGCATATCCACTGAGCA for ALKBH5, respectively.
Effects of the Knockdown of METTL3 on the Expression of SOCS Family Members
To explore the function of METTL3, we knocked down METTL3 in a mouse monocyte macrophage leukemic cell line RAW264.7 by transfecting METTL3 shRNA with METTL3 shRNA. The METTL3 shRNA sequences were designed as follows: upper-GATCCGGTTCGTTCCACCAGTCATAATTCAAGAGATTATGACTGGTGGAACGAACCTTTTTTG, lower-AATTCAAAAAAGGTTCGTTCCACCAGTCATAATCTCTTGAATTATGACTGGTGGAACGAACCG. Using the real-time PCR method, we identified that METTL3 gene expression was largely down-expressed after the interfering. Then we detected the gene expression of SOCS family members, such as SOCS1, SOCS2, SOCS3, SOCS4, SOCS5, SOCS6, and SOCS7, by real-time PCR. The primers of these genes are shown below:
SOCS1 former: GACGCCTGCGGCTTCTATT,
reverse: CAGCTCGAAAAGGCAGTCG;
SOCS2 former: ACGGAATGGGACTGTTCACC,
reverse: AAGGCAGTCCCCAGATCGTA;
SOCS3 former: TGGTCACCCACAGCAAGTTT,
reverse: TCGCTTTTGGAGCTGAAGGT;
SOCS4 former: CGGAGTCGAAGTGCTGACAG,
reverse: ACTCAATGGACGAACAGCTAAG;
SOCS5 former: TTCCCATGAGAACTTACAGCAAG,
reverse: TTTTGTGCTAAATCCGAGCCA;
SOCS6 former: AAGCAAAGACGAAACTGAGTTCA,
reverse: CAGCTCCCGAATAAAGAGTCATC;
SOCS7 former: GAAACCCAGGTTGACAAGAACT,
reverse: TCCACAAGCGATACTGTCTCA.
Statistical Analysis
SPSS 20.0 software (IBM, Chicago, USA) was used to carry out the statistical analysis. A T-test was performed to analyze the differences in gene expression between two groups. WGCNA R software package was used to find functional pathways including DEGs. p value smaller than 0.05 was considered statistically significant.
Results
mRNA Expressions Profile in GD
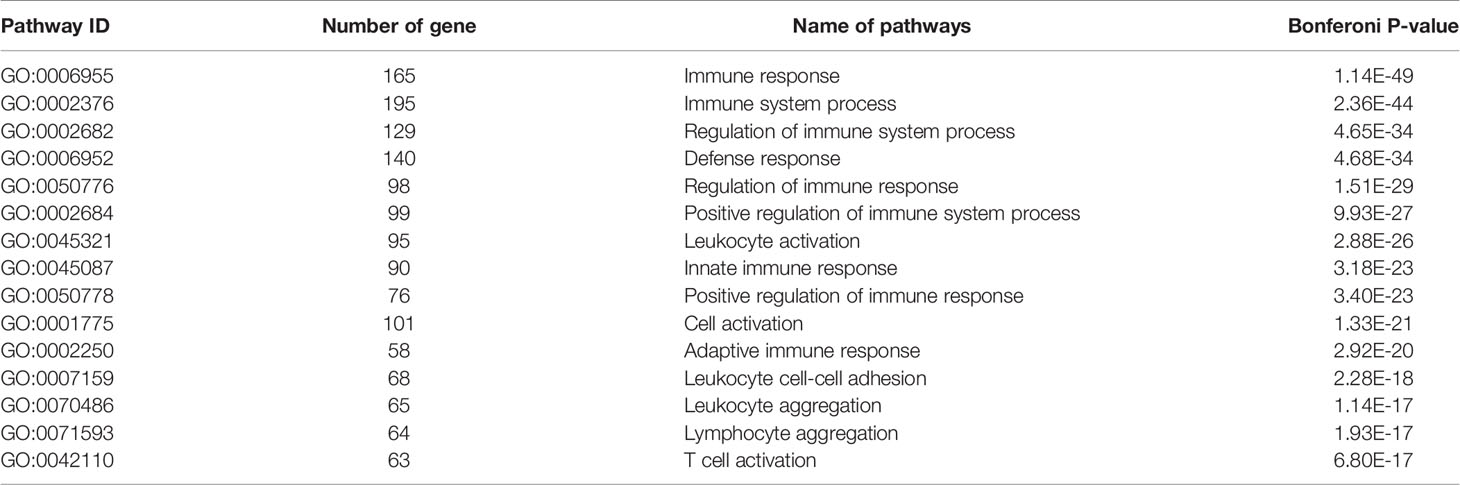
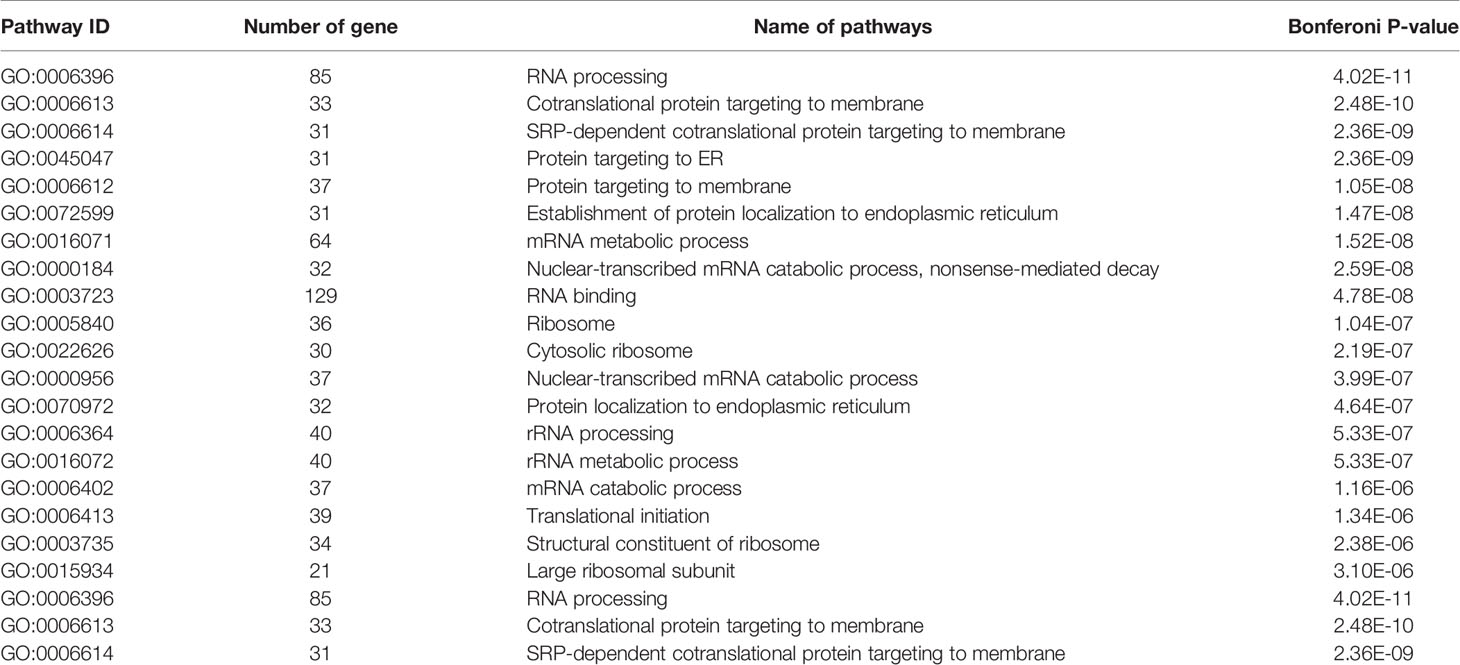
We have previously isolated 7 thyroid tissues from recruited 4 GD patients and 4 controls, screened the microRNA/mRNA profile by gene microarray, and found more than 2800 aberrant expressed mRNAs related to GD (22). Through GO and KEGG pathway analysis, we found that these aberrantly expressed mRNAs were enriched in the differentiation and regulation of immune cells (22). Among these genes, METTL3 and its target genes, SOCS family members, were all included. METTL3 mRNA level was significantly lower in GD thyroid tissues than in normal thyroid tissues (P=0.0026, Figure 1) while SOCS3 mRNA level was significantly higher in GD thyroid samples than in normal thyroid samples (P=0.04, Figure 2). Moreover, we combined DEGs in thyroid tissues from our precious microarray data and GSE9340 microarray data downloaded from the GEO database and created several GD related gene co-expression modules. Some of these modules were associated with the onset of GD (Figure 3), including a blue module composed of 553 genes and a green module composed of 903 genes. Among the 903 genes in the green module are key genes for m6A methylation, such as METTL3. GO function enrichment analysis indicated that the genes in the blue module are mainly involved in immune response pathways such as innate immune response (Bonferoni P = 3.18E-23) and adapted immune response (Bonferoni P = 2.92E-20) pathways (Table 1) and the genes in the green module are mainly correlated with RNA machinery and metabolism pathways (Table 2), implying that mRNA methylation plays vital roles in the etiology of GD.
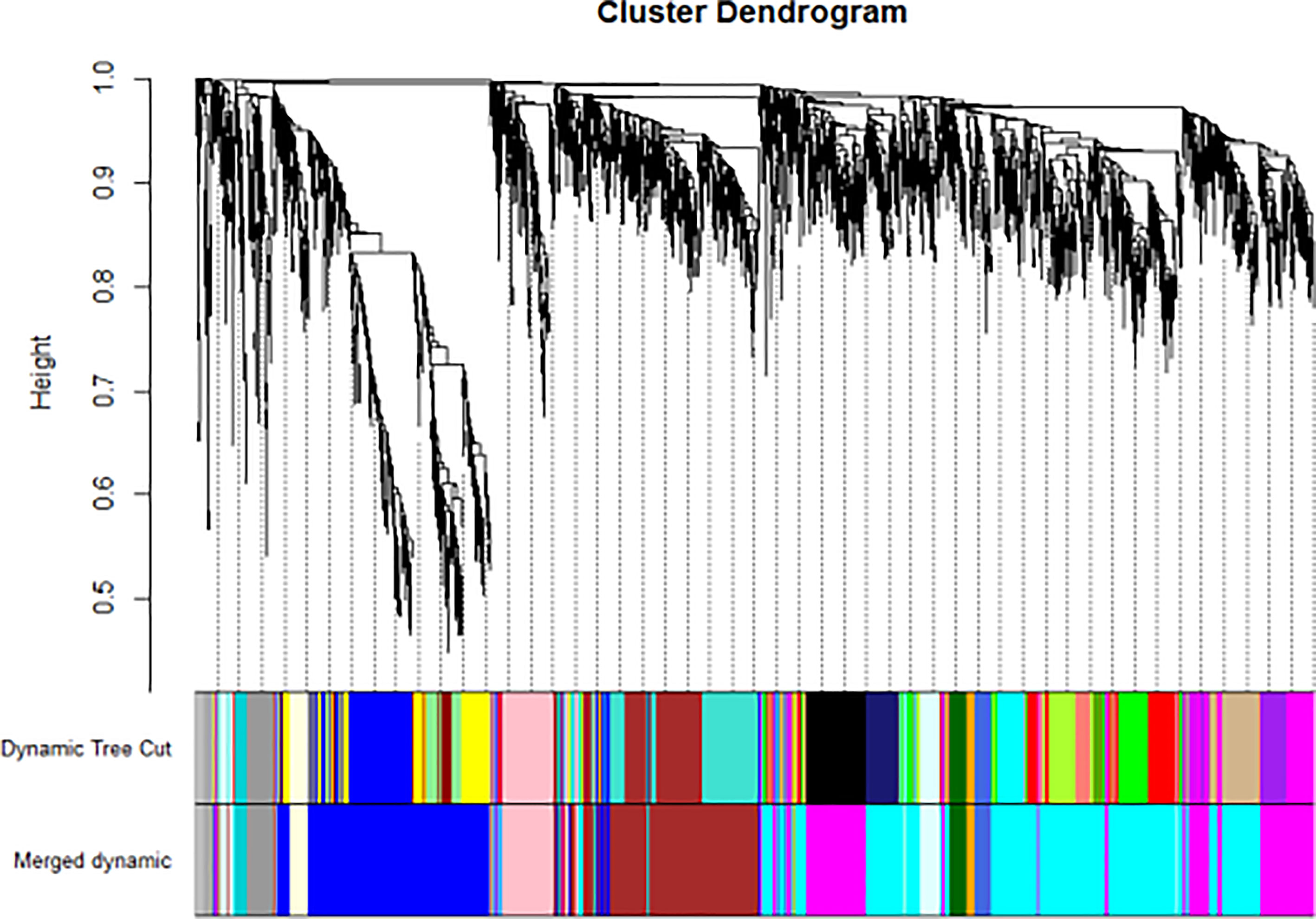
Figure 3 WGCNA found several GD related key gene expression modules (The most significant is the black module).
Validation of Aberrant Genes in GD
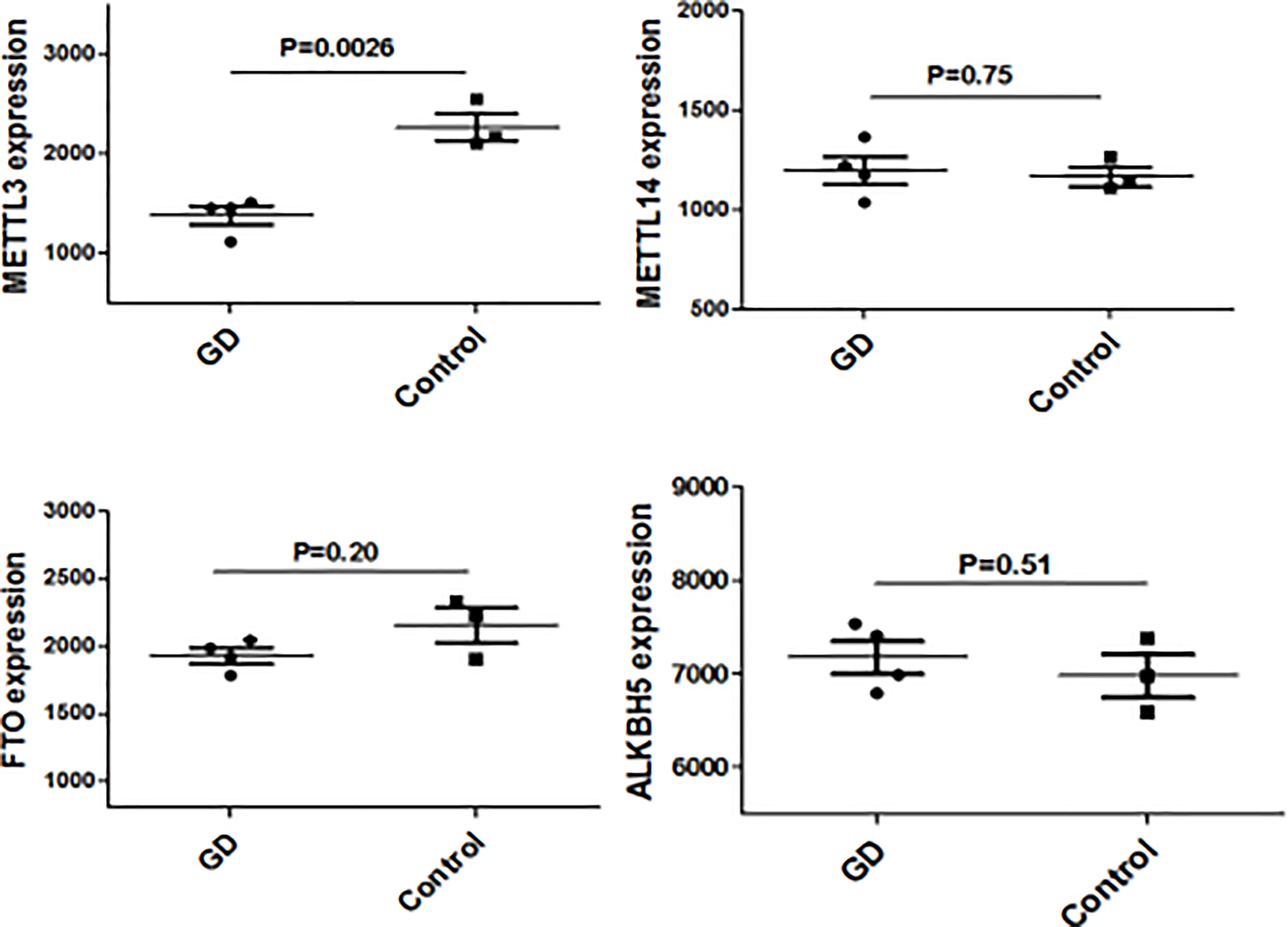
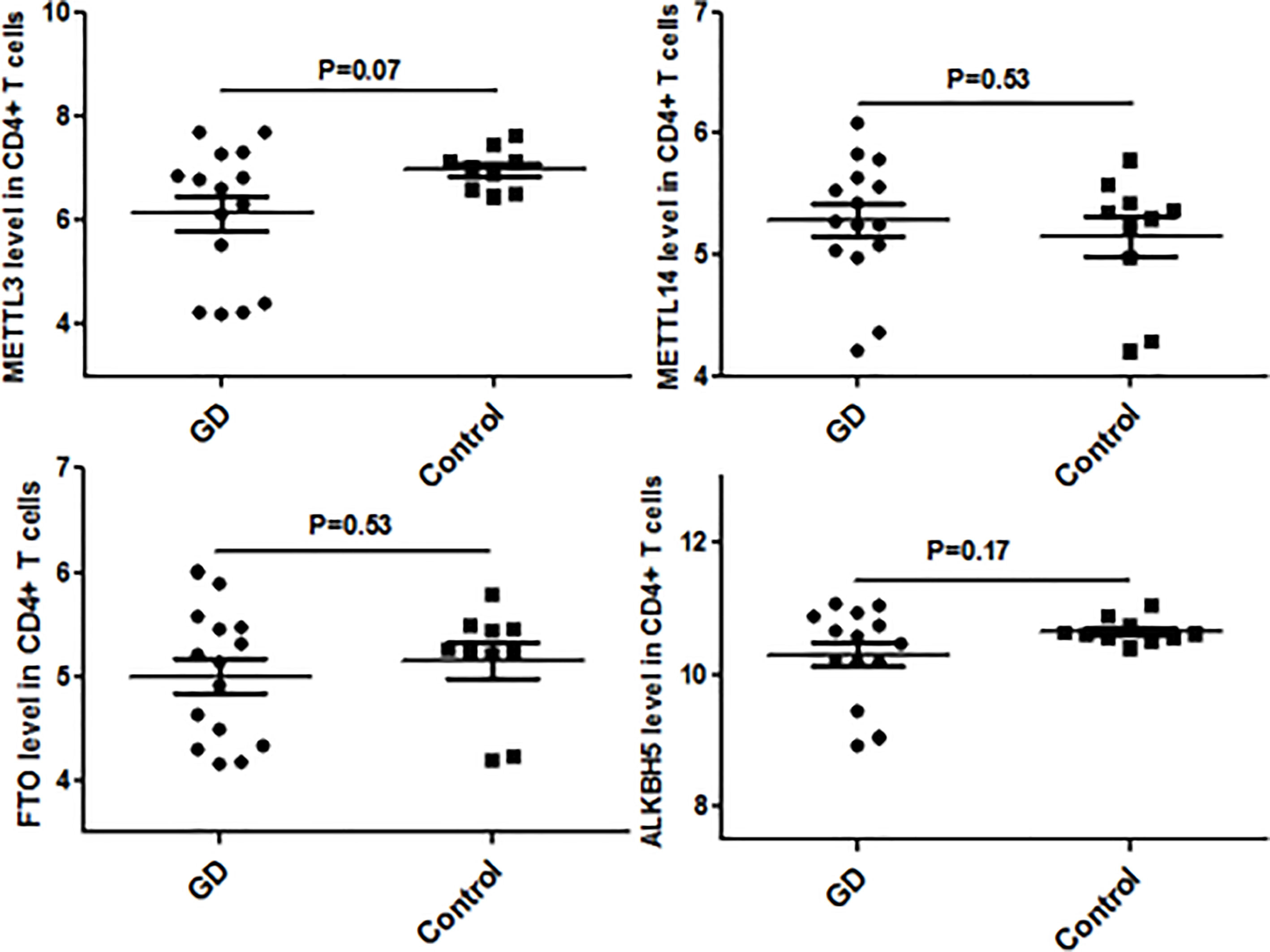
Since CD4+ T cells play important roles in GD, we then detected the expression of some genes that are the key enzymes of mRNA m6A methylation including METTL3 by using GSE71956, a CD4+ T cell gene expression profile microarray dataset. As shown in Figure 4, METTL3 expression in CD4+ T cells was lower in GD than in controls, although it was not statistically significant. A similar trend was also observed in other key genes involved in mRNA m6A modification, such as FTO and ALKBH5. It would be worthwhile to explore these trends in further depth in the future.
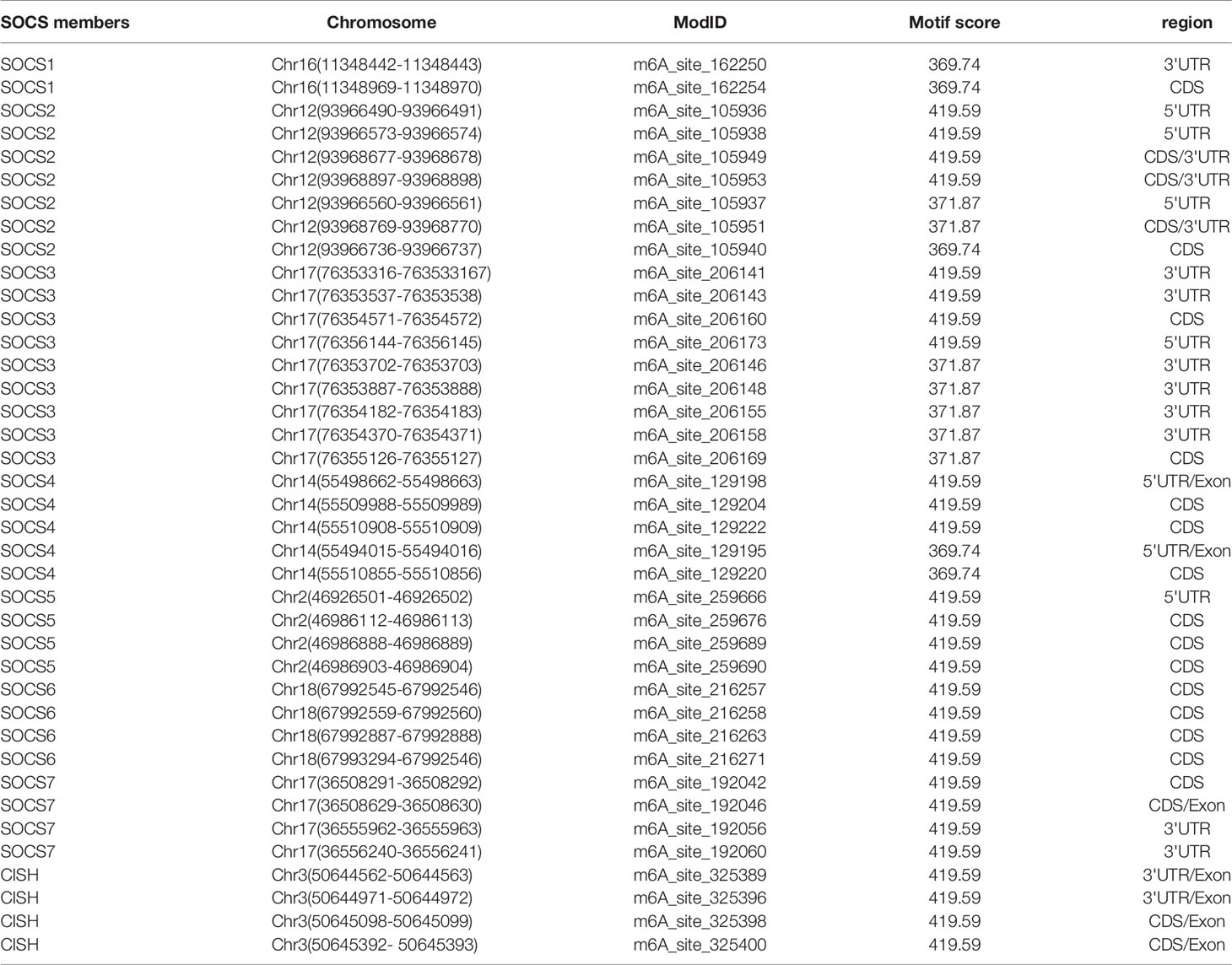
We used bioinformatic database RMBase v2.0 (http://rna.sysu.edu.cn/rmbase/index.php), which targets mRNA m6A modification to screen m6A modification loci. The results revealed numerous m6A modification loci on mRNA of SOCS family members. Table 3 displays those with motif score > 350 (motif score ranges from 0 to 500, the larger value represents higher accuracy).
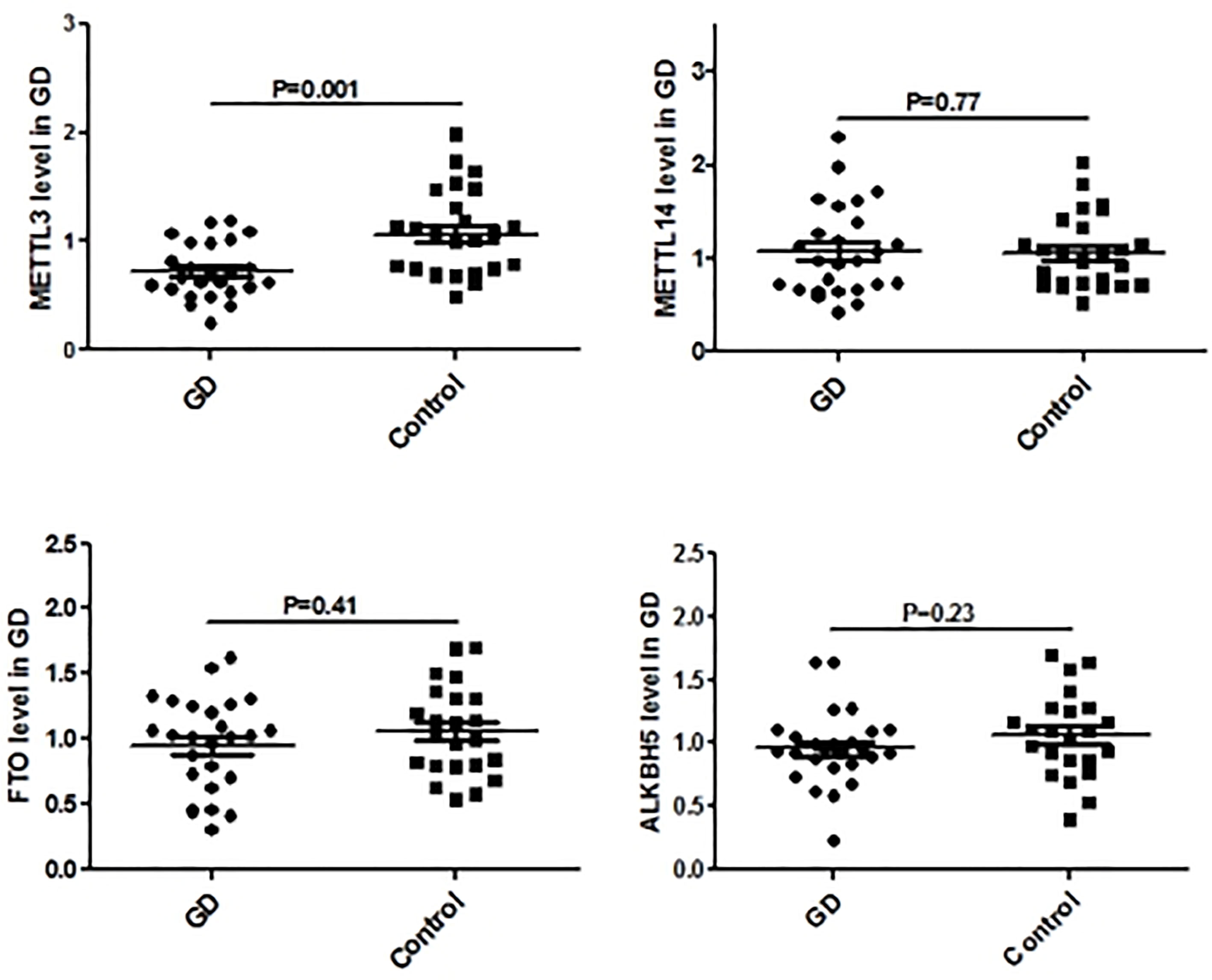
Figure 5 shows the real-time qPCR results on expressions of genes key to mRNA m6A methylation in PBMCs from clinical GD patients and healthy controls, indicating that METTL3 mRNA level was significantly lower in the PBMCs of GD patients than in healthy controls (P = 0.001). By contrast, mRNA levels of METTL14, FTO, and ALKBH5 were not significantly different among these two groups.
Influence of METTL3 Knockdown on the Expression of SOCS Family Members
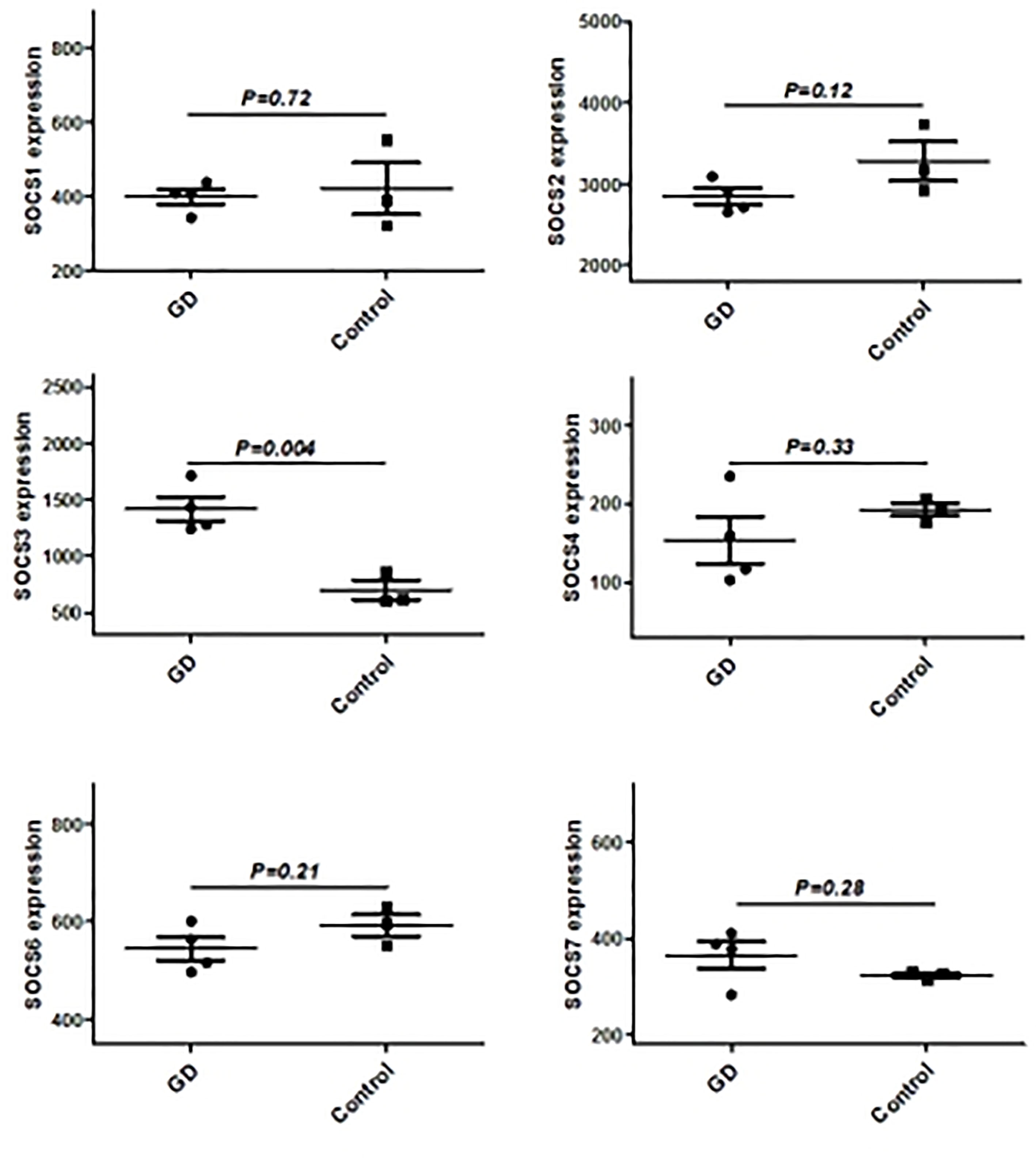
In vitro functional analysis found that the mRNA levels of SOCS1, SOCS2, SOCS4, SOCS5, and SOCS6 were significantly higher in METTL3 knockdown RAW264.7 cells (P<0.05), while SOCS3 mRNA level was not significantly changed after METTL3 knockdown (Figure 6).
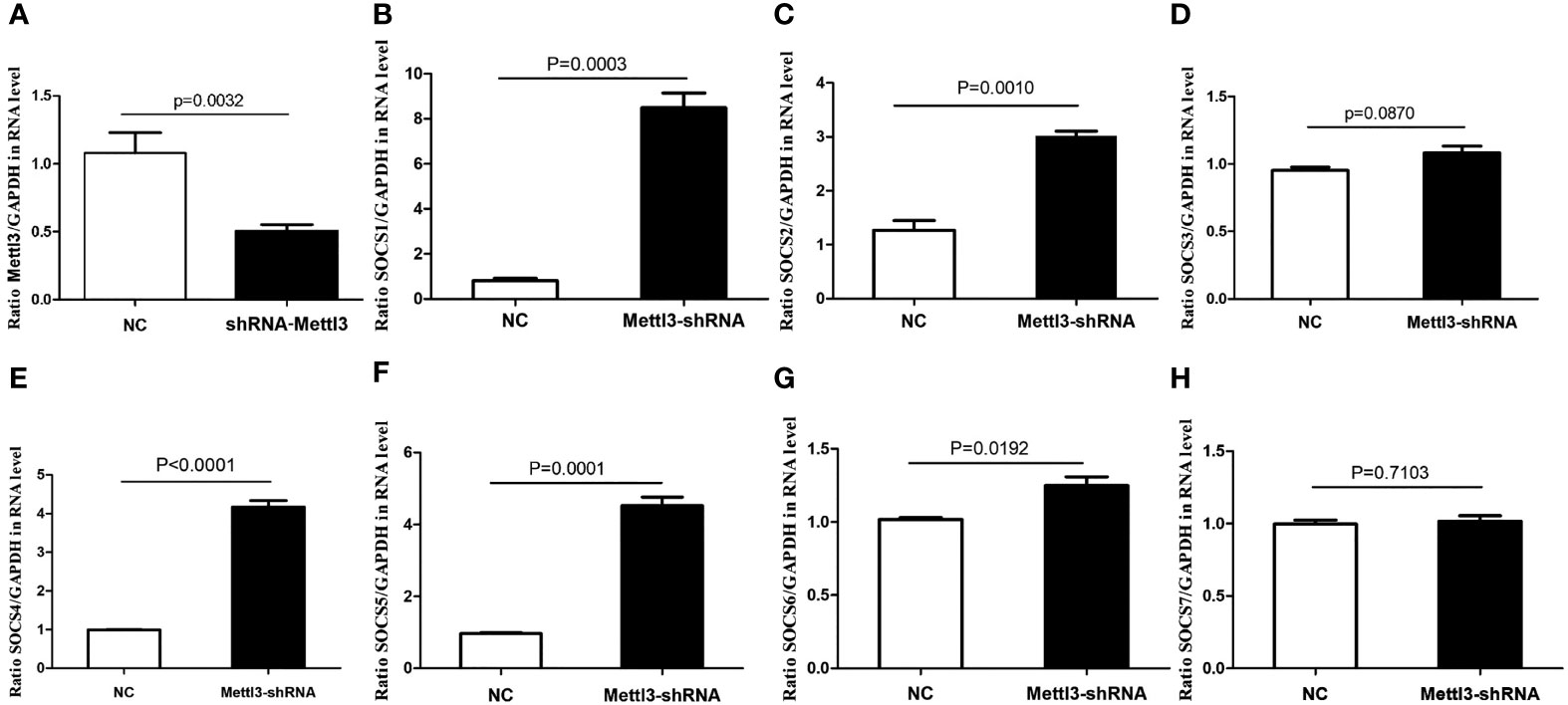
Figure 6 mRNA expressions of SOCS family members in METTL3 knock-down RAW 264.7 cells. (A) It showed the significant lower expression of METTL3 after METTL3 knock-down; (B) SOCS1 mRNA expression after METTL3 knock-down; (C) SOCS2 mRNA expression after METTL3 knock-down; (D) SOCS3 mRNA expression after METTL3 knock-down; (E) SOCS4 mRNA expression after METTL3 knock-down; (F) SOCS5 mRNA expression after METTL3 knock-down; (G) SOCS6 mRNA expression after METTL3 knock-down; (H) SOCS7 mRNA expression after METTL3 knock-down.
Discussion
GD is a common autoimmune disease that is susceptible to multiple factors. Among them, genetic, environmental, and immune factors all exert important roles (26). Additionally, epigenetics factors, which play vital roles in integrating environmental and genetic elements, are closely related to the predisposition of GD (7). Currently, mRNA m6A modification as a form of epigenetic modification has become a new research hotspot in various kinds of diseases including autoimmune diseases (13, 14). As a key RNA methylation modification enzyme, METTL3 is reported to regulate the mRNA and protein levels of several key molecules and plays a key role in diverse life processes (15). Although we have previously shown that METTL3 polymorphisms confer risk for the susceptibility of GD (18) and mRNA m6A modification is known to regulate immune cell differentiation and immune response (10–12) and is involved in the development of some autoimmune diseases (16, 17). The mechanisms underlying the effects of m6A modification on autoimmune diseases including GD are still incompletely understood and require more in-depth studies. In this study, we analyzed previous genome-wide expression microarray data on GD and found that METTL3, a key enzyme for mRNA m6A modification was aberrantly downregulated in GD thyroid tissues. Furthermore, METTL3 mRNA levels were also decreased in the PBMCs of GD patients. These data indicate that METTL3 induced mRNA m6A modification is linked to the pathomechanism of GD.
WGCNA is a widely used data mining method in a biological system to explore disease-related gene co-expression modules from high throughput data. The gene co-expression module involves a cluster of genes that are intensely related in expression and function levels and play key roles in disease development. In this study, WGCNA analysis was used to reveal gene co-expression modules that are intensely related to GD and enriched in the signaling pathway of RNA translation and metabolism. The results showed that METTL3 is a key component of these modules, which indicates that mRNA m6A modification by METTL3 may be involved in the development of GD.
It is well known that epigenetic modifications in RNA exert critical roles in regulating cell differentiation through changing the expressions of some key genes, like SOCS family members (19–21). Therefore we searched the bioinformatic database and found that there are many m6A modification loci in mRNA of SOCS family members, which implies that the key enzymes of m6A modification could regulate mRNA and protein expression of SOCS family members via methylation. SOCS1 and SOCS3 have been reported to be aberrantly expressed in some autoimmune diseases and involved in regulating the functions of diverse immune cells (21). Our microarray study also found that SOCS3 was upregulated in the thyroid tissues of GD, implying that SOCS3 and other SOCS family members are related to GD. Recent research showed that METTL3 is involved in liver cancer development by regulating the proliferation and differentiation of liver cells via inducing SOCS2 mRNA m6A modification (27). Another study found that METTL3 could regulate the differentiation of T cells by inducing SOCS1 and SOCS3 mRNA m6A modification (12). We found that METTL3 is downregulated in GD. Therefore, we hypothesized that METTL3 is intensively involved in the pathogenesis of GD by inducing an imbalance of key immune cells. To validate this hypothesis, we knocked down the METTL3 gene in in vitro cultured cells to study the influence of METTL3 on GD via inducing mRNA m6A modification of SOCS family members and its potential molecular mechanisms, which will hopefully provide a novel target for GD therapy. Indeed, we found that mRNA levels of SOCS1, SOCS2, SOCS4, SOCS5, and SOCS6 were increased in METTL3 knockdown cells.
In summary, in this study, we found that METTL3 mRNA declined in GD thyroid tissues using microarray data and functional pathway analysis and further validated the results in clinical PBMCs samples from GD patients. Subsequently, we searched several databases and found that there are multiple mRNA m6A modification loci in SOCS family members. At last, we knocked down METTL3 in in vitro cultured cells and found that the expression levels of some SOCS family members were enhanced in METTL3 knock-down cells. Overall, the study indicated that METTL3 is involved in the etiology of GD by regulating mRNA levels of SOCS family members including SOCS1, SOCS2, SOCS4, SOCS5, and SOCS6.
Conclusion
The present study was the first to explore the relationship between m6A modification and GD, and revealed that METTL3 may be involved in GD development via inducing mRNA m6A modification of SOCS family members.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethic Committee of Shanghai University of Medicine & Health Sciences Affiliated Zhoupu Hospital (reference number of 2018-C-027-E01). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
R-hS and J-aZ designed this study. C-qG, PD, and X-rL collected the samples. C-qG extracted RNA from samples. PD performed the real time qPCR. X-rL performed cell culture experiments. R-hS analyzed data and wrote the draft. J-aZ reviewed and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 81800696 and 81873636), Science and Technology Development Fund of Pudong New District Minsheng Scientific Research (Medical and Health) Project (No. PKJ2018-Y39), Talent Youth Cultivation Plan of Pudong New District (No. PWRq2020-11), Shanghai Medical Key Specialty (No. ZK2019C09) and “Top-100 Talent Cultivation Plan” of Shanghai University of Medicine and Health Sciences (No. B3-0200-20-311008-30).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Smith TJ, Hegedus L. Graves' Disease. N Engl J Med (2016) 375(16):1552–65. doi: 10.1056/NEJMra1510030
2. Zimmermann MB, Boelaert K. Iodine Deficiency and Thyroid Disorders. Lancet Diabetes Endocrinol (2015) 3(4):286–95. doi: 10.1016/S2213-8587(14)70225-6
3. Burch HB, Cooper DS. Management of Graves Disease: A Review. JAMA (2015) 314(23):2544–54. doi: 10.1001/jama.2015.16535
4. McLachlan SM, Rapoport B. Breaking Tolerance to Thyroid Antigens: Changing Concepts in Thyroid Autoimmunity. Endocr Rev (2014) 35(1):59–105. doi: 10.1210/er.2013-1055
5. Yan N, Yu YL, Yang J, Qin Q, Zhu YF, Wang X, et al. Association of Interleukin-17A and -17F Gene Single-Nucleotide Polymorphisms With Autoimmune Thyroid Diseases. Autoimmunity (2012) 45(7):533–9. doi: 10.3109/08916934.2012.702814
6. Song RH, Li Q, Wang W, Yao QM, Shao XQ, Zhang JA. Variants of Interleukin-22 Gene Confer Predisposition to Autoimmune Thyroid Disease. Int J Endocrinol (2017) 2017:3428236. doi: 10.1155/2017/3428236
7. Wang B, Shao X, Song R, Xu D, Zhang JA. The Emerging Role of Epigenetics in Autoimmune Thyroid Diseases. Front Immunol (2017) 8:396. doi: 10.3389/fimmu.2017.00396
8. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA Modifications in Gene Expression Regulation. Cell (2017) 169(7):1187–200. doi: 10.1016/j.cell.2017.05.045
9. Lichinchi G, Gao S, Saletore Y, Gonzalez GM, Bansal V, Wang YS, et al. Dynamics of the Human and Viral M(6)A RNA Methylomes During HIV-1 Infection of T Cells. Nat Microbiol (2016) 1:16011. doi: 10.1038/nmicrobiol.2016.11
10. Vu LP, Pickering BF, Cheng YM, Zaccara S, Nguyen D, Minuesa G, et al. The N(6)-Methyladenosine (M(6)A)-Forming Enzyme METTL3 Controls Myeloid Differentiation of Normal Hematopoietic and Leukemia Cells. Nat Med (2017) 23(11):1369–76. doi: 10.1038/nm.4416
11. Zhang C, Chen Y, Sun B, Wang L, Yang Y, Ma D, et al. M(6)A Modulates Haematopoietic Stem and Progenitor Cell Specification. Nature (2017) 549(7671):273–6. doi: 10.1038/nature23883
12. Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, et al. M(6)A mRNA Methylation Controls T Cell Homeostasis by Targeting the IL-7/STAT5/SOCS Pathways. Nature (2017) 548(7667):338–42. doi: 10.1038/nature23450
13. Zhao BS, Roundtree IA, He C. Post-Transcriptional Gene Regulation by mRNA Modifications. Nat Rev Mol Cell Biol (2017) 18(1):31–42. doi: 10.1038/nrm.2016.132
14. Gilbert WV, Bell TA, Schaening C. Messenger RNA Modifications: Form, Distribution, and Function. Science (2016) 352(6292):1408–12. doi: 10.1126/science.aad8711
15. Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, et al. Stem Cells. M6a mRNA Methylation Facilitates Resolution of Naive Pluripotency Toward Differentiation. Science (2015) 347(6225):1002–6. doi: 10.1126/science.1261417
16. Wang JH, Yan SS, Lu HY, Wang SF, Xu DH. METTL3 Attenuates LPS-Induced Inflammatory Response in Macrophages via NF-κ B Signaling Pathway. Mediators Inflamm (2019) 312–91. doi: 10.1155/2019/3120391
17. Elin P, Skorpen F, Kvaloy K, Midthjell K, Grill V. Genetic Heterogeneity in Latent Autoimmune Diabetes Is Linked to Various Degrees of Autoimmune Activity: Results From the Nord-Trøndelag Health Study. Diabetes (2010) 59(1):302–10. doi: 10.2337/db09-0923
18. Song RH, Liu XR, Gao CQ, Du P, Zhang JA. METTL3 Gene Polymorphisms Contribute to Susceptibility to Autoimmune Thyroid Disease. (2020) 72(2):495–504. doi: 10.1007/s12020-020-02503-1
19. Rabani M, Raychowdhury R, Jovanovic M, Rooney M, Stumpo DJ, Pauli A, et al. High-Resolution Sequencing and Modeling Identifies Distinct Dynamic RNA Regulatory Strategies. Cell (2014) 159(7):1698–710. doi: 10.1016/j.cell.2014.11.015
20. Renier N, Adams EL, Kirst C, Wu ZH, Azevedo R, Kohl J, et al. Mapping of Brain Activity by Automated Volume Analysis of Immediate Early Genes. Cell (2016) 165(7):1789–802. doi: 10.1016/j.cell.2016.05.007
21. Liang Y, Xu WD, Peng H, Pan HF, Ye DQ. SOCS Signaling in Autoimmune Diseases: Molecular Mechanisms and Therapeutic Implications. Eur J Immunol (2014) 44(5):1265–75. doi: 10.1002/eji.201344369
22. Qin Q, Wang X, Yan N, Song RH, Cai TT, Zhang W, et al. Aberrant Expression of miRNA and mRNAs in Lesioned Tissues of Graves' Disease. Cell Physiol Biochem (2015) 35(5):1934–42. doi: 10.1159/000374002
23. Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol (2003) 4(5):P3. doi: 10.1186/gb-2003-4-5-p3
24. Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for Integration and Interpretation of Large-Scale Molecular Data Sets. Nucleic Acids Res (2012) 40(Database issue):D109–14. doi: 10.1093/nar/gkr988
25. Langfelder P, Horvath S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform (2008) 9:559. doi: 10.1186/1471-2105-9-559
26. Lee HJ, Li CW, Hammerstad S, Stefan M, Tomer HJ. Immunogenetics of Autoimmune Thyroid Diseases: A Comprehensive Review. J Autoimmun (2015) 64:82–90. doi: 10.1016/j.jaut.2015.07.009
Keywords: Graves’ disease (GD), RNA modification, RNA methyltransferase, methyltransferase like 3 (METTL3), suppressor of cytokine signaling (SOCS)
Citation: Song R-h, Du P, Gao C-q, Liu X-r and Zhang J-a (2021) METTL3 Is Involved in the Development of Graves’ Disease by Inducing SOCS mRNA m6A Modification. Front. Endocrinol. 12:666393. doi: 10.3389/fendo.2021.666393
Received: 10 February 2021; Accepted: 30 June 2021;
Published: 20 September 2021.
Edited by:
Rauf Latif, Icahn School of Medicine at Mount Sinai, United StatesReviewed by:
Akira Sugawara, Tohoku University, JapanShengjun Wang, Jiangsu University Affiliated People’s Hospital, China
Copyright © 2021 Song, Du, Gao, Liu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin-an Zhang, emhhbmdqaW5hbkBob3RtYWlsLmNvbQ==
 Rong-hua Song
Rong-hua Song Peng Du
Peng Du Jin-an Zhang
Jin-an Zhang