- 1Laboratory of Epigenetics, Research Centre for Medical Genetics, Moscow, Russia
- 2I.M. Sechenov First Moscow State Medical University, Moscow, Russia
- 3Pirogov Russian National Research Medical University, Moscow, Russia
- 4Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Science, Moscow, Russia
- 5Privolzhsky Research Medical University, Nizhny Novgorod, Russia
- 6Morphological Department of Oncology, M.F. Vladimirsky Moscow Regional Research and Clinical Institute, Moscow, Russia
Insulinomatosis is characterized by monohormonality of multiple macro-tumors and micro-tumors that arise synchronously and metachronously in all regions of the pancreas, and often recurring hypoglycemia. One of the main characteristics of insulinomatosis is the presence of insulin-expressing monohormonal endocrine cell clusters that are exclusively composed of proliferating insulin-positive cells, are less than 1 mm in size, and show solid islet-like structure. It is presumed that insulinomatosis affects the entire population of β-cells. With regards to molecular genetics, this phenomenon is not related to mutation in MEN1 gene and is more similar to sporadic benign insulinomas, however, at the moment molecular genetics of this disease remains poorly investigated. NGS sequencing was performed with a panel of 409 cancer-related genes. Results of sequencing were analyzed by bioinformatic algorithms for detecting point mutations and copy number variations. DNA copy number variations were detected that harbor a large number of genes in insulinoma and fewer genes in micro-tumors. qPCR was used to confirm copy number variations at ATRX, FOXL2, IRS2 and CEBPA genes. Copy number alterations involving FOXL2, IRS2, CEBPA and ATRX genes were observed in insulinoma as well as in micro-tumors samples, suggesting that alterations of these genes may promote malignization in the β-cells population.
Introduction
Multiple insulinomas are most common in MEN1 syndrome, however, several studies show that they can also occur sporadically, albeit very rarely, and are referred to insulinomatosis (1–3). Term “insulinomatosis” was introduced and described by Anlauf et al. (1). It is characterized by monohormonality of multiple macrotumors and microtumors that arise synchronously and metachronously in all regions of the pancreas, rare of metastases, and often recurring hypoglycemia. In addition, one of the main characteristics of insulinomatosis is the presence of insulin-expressing monohormonal endocrine cell clusters that are exclusively composed of proliferating insulin-positive cells, are less than 1 mm in size, and show solid islet-like structure. Authors suggested that insulinomatosis affects the entire population of β-cells (1, 2). With regards to molecular genetics, this phenomenon is not related to mutation in MEN1 gene and is more similar to sporadic benign insulinomas, however, large-scale molecular genetic studies on this disease have not yet been carried out (1). Here we present a very rare case of insulinomatosis in a patient with hypoglycemia syndrome and without known hereditary syndromes.
Case Presentation
A male in his 60s with clinical signs of hypoglycemia was admitted to the surgical department of Sechenov University. The patient signed informed agreement to undergo diagnostic procedures and treatment, as well as to participate in the study, and for the presentation of clinical and molecular data in scientific and medical literature. This case report was approved by the local Ethics Committee at the Research Centre for Medical Genetics, Moscow, Russia.
The history of the disease is about 6 years with fasting serum glucose 2.7-3.7 mmol/l compensated by sugar intake. The 72-hour fasting test was negative. The past medical history is significant for combination treatment for laryngeal carcinoma T2N0M0, stage II. Currently there are no signs of recurrence. CT scan of the abdomen shows heterogeneous hypervascular soft tissue mass up to 26 mm suspicious for neuroendocrine tumor (NET), located within the tail of the pancreas, with a well-defined cystic component attached to the splenic artery and vein without invasion. Hypervascular lesions of up to 5 mm without clear borders are detected within the surrounding parenchyma of the pancreatic tail.
Treatment
Based on the obtained diagnostic data, the multidisciplinary board was held, implying the participation of surgical oncologist, medical oncologist, pathologist and radiation oncologist. Taking into account tumor size more than 2 cm and hypoglycemia syndrome radical pancreatic resection was planned. A distal spleen-preserving pancreatectomy was performed.
Outcome
Taking into account multiple microadenomas in the pancreatic parenchyma the recurrence rate of the hyperinsulinemic hypoglycemia is relatively high. Thus, patient is examined every 3-6 month. In our case there are no signs of hypoglycemia and no pancreatic tumors on imaging during 30 months of follow-up by CT and PET-CT scanning.
Microscopy and Immunohistochemistry
Microscopic examination showed well differentiated neuroendocrine tumor with 2 mitoses and no necrosis in 10 HPF. There are multiple large microadenomas in the surrounding pancreatic tissue. Tumor cells are positive for synaptophysin and chromogranin A, as well as for insulin (Figures 1A, B). There is no expression of glucagon, somatostatin and pancreatic polypeptide. Ki-67 is less than 1.5%, Grade 1. The cells of microadenomas express insulin and are negative for glucagon and somatostatin (Figures 1C, D).
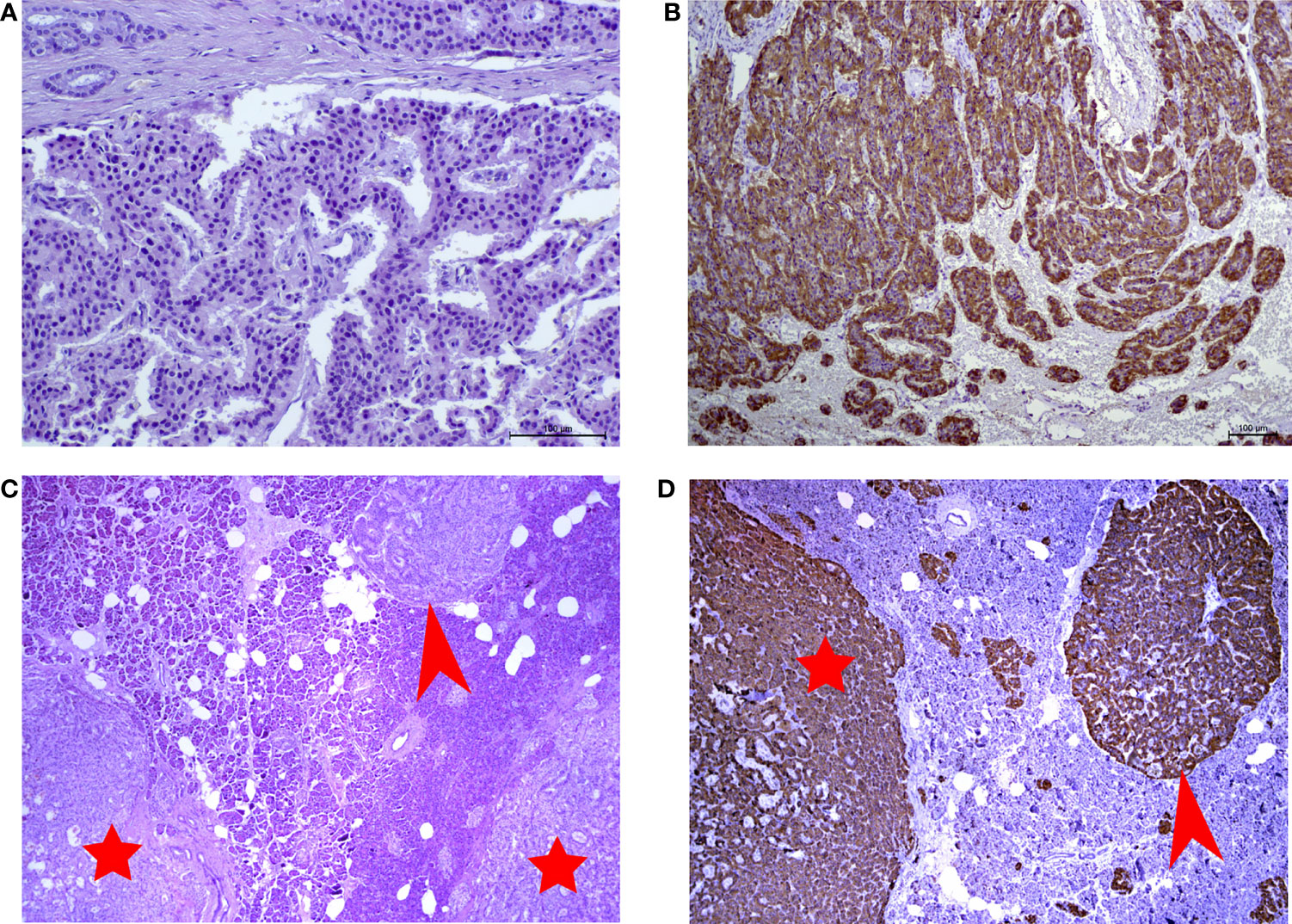
Figure 1 (A) Hematoxylin and eosin (H&E)-stained slide of insulinoma (x250 magnification). (B) Immunohistochemical reaction with antibodies to insulin in insulinoma (x125 magnification). (C) Hematoxylin and eosin (H&E)-stained slide of microtumors (x10 magnification). (D) Immunohistochemical reaction with antibodiesto insulin in microadenoma (x10 magnification). Red arrows point to insulin-producing microadenomas. Red stars indicate the microtumors.
Materials and Methods
DNA Extraction
Macrodissection of tumors was performed under guidance of pathologist. DNA was extracted from four formalin-fixed, paraffin-embedded (FFPE) tumor tissues by using GeneRead DNA FFPE kit (Qiagen, Germany), and from whole peripheral blood by using standard phenol-chloroform extraction protocol. One tumor sample was insulinoma, three others were small proliferative insulin-expressing monohormonal endocrine cell clusters (IMECCs).
NGS Sequencing
Next generation sequencing (NGS) was performed by using Ion AmpliSeq targeted amplification technology with AmpliSeq Comprehensive Cancer Panel (Thermo Fisher Scientific, United States) with exon coverage of 409 oncogenes and tumor suppressor genes.
Bioinformatic Analysis
The bioinformatic workflow for sequencing data analysis was based on Torrent Suite software (version 5.10.1). Annotation was performed by ANNOVAR (4). CNVs were called using CNVpanelizer R package (version 1.22.0) ran with default parameters (5). Putative CNV status is assigned when there is a significant difference between observed copy number and the one expected from the bootstrapped distribution. A reliable CNV is called when the upper bound of the copy number ratio is below the lower bound of background (noise) copy number for deletion or when the lower bound of the copy number ratio is above the upper bound of background (noise) copy number for amplification.
Sanger Sequencing
Sanger sequencing was performed for detection of the Trp372Arg mutation in YY1 gene. PCR was performed in 25 µl reactions. The reaction mixture for PCR consisted of the following reagents: 8% glycerol, 68 mM Tris-HCl with pH 8.3, 17 mM (NH4)2SO4, 0.01% Tween-20, 0.1 mg/ml BSA, 0.2 mM each dNTP, 1.5 units Taq polymerase, 0.12 pM each primer, 2.5 mM MgCl2. On top, 40-60 μl of mineral oil was layered. PCR reaction was performed with the following parameters: 95°C for 5 min, followed by 30 cycles of 95°C for 40 seconds, 61°C for 40 seconds and 72°C for 40 seconds. Final elongation was at 72°C for 5 minutes. Sanger sequencing was performed on ABI3500 according to the manufacturer’s protocol (Thermo Fisher, USA). Sequencing results were analyzed in Chromas software and compared to GenBank database using the BLAST algorithm. Primer sequences for Trp372Arg are listed in Table 1.
Quantitative PCR
Quantitative PCR (qPCR) is one of the methods that are widely used to detect copy number changes. This method is preferable due to its low consumable and instrumentation costs, high sensitivity and fast development time of assay (6–9). qPCR was performed to confirm the results of CNVPanelizer packages for CEBPA and FOXL2 gene, the first and the second exons of IRS2 gene and the first and the ninth exons of ATRX gene.
The qPCR mixes were prepared according to the GenPak PCR Core protocol (Isogene Lab ltd, Russia) in 20 µl reactions containing 1 ng genomic DNA and using SYBR green as a reporter and ROX as a reference dyes. All qPCR reactions were run in triplicate on a QuantStudio 5 Real-Time PCR System for Human Identification (Thermo Fisher Scientific, USA). The PCR conditions were as follows: 95°C for 5 min, followed by 40 cycles of 95°C for 20 seconds, 60°C for 20 seconds and 72°C for 20 seconds. Data were analyzed by the ΔΔCt method (10). The relative copy number was estimated by comparison with a normal blood control DNA sample. We used the B2M housekeeping gene as an endogenous control. Primer sequences for qPCR are shown in Table 1.
Results
Sanger Sequencing
Point mutation Trp372Arg (Chr14:100,743,807, hg19) in YY1 gene was not found in the tested samples.
Next Generation Sequencing
Median NGS read coverage was 486x, 461x, 471x, 454x for four tumor samples and 217x for a blood sample.
Single Nucleotide Variations
No pathogenic point mutations associated with any genetic syndrome including MEN1 were found in blood, neither pathogenic point mutations in tumor samples.
Copy Number Variations
CNVs in Tumors
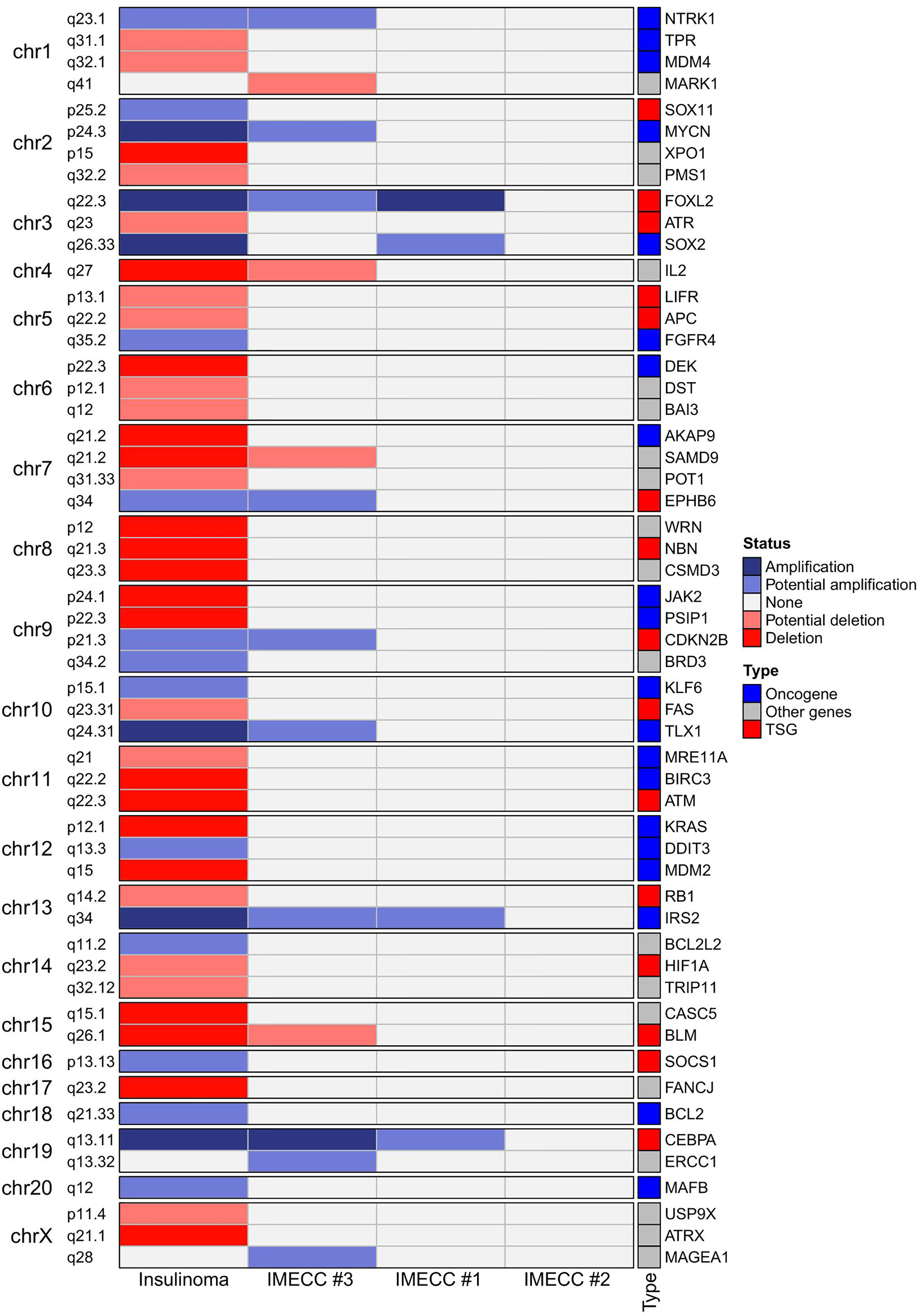
CNVPanelizer in total has detected 54 genes within CNV regions, where 51, 14 and 4 genes were found in insulinoma, IMECCs #1 and #3, respectively (Figure 2). By using ONgene (11) and TSGgene (12) databases that aggregate information about oncogenes and tumor suppressor genes respectively, we identified several oncogenes and tumor suppressor genes with different CNV status in our tumor samples.
In insulinoma sample, ten oncogenes (BCL2, DDIT3, FGFR4, IRS2, KLF6, MAFB, MYCN, NTRK1, SOX2 and TLX1) were with either putative or reliable status of gain, and nine tumor suppressor genes (APC, ATM, ATR, BLM, FAS, HIF1A, LIFR, NBN and RB1) were with putative or reliable status of loss.
With regards to IMECCs, in IMECC #1 sample two oncogenes, SOX2 and IRS2, were with putative status of gain. In IMECC #3 four oncogenes, IRS2, MYCN, NTRK1, TLX1, were with putative status of gain and one tumor suppressor gene, BLM, with putative status of loss. In IMECC #2 according to CNVPanelizer no CNVs were detected.
With regards to the most common genes that mutate in pancreatic neuroendocrine neoplasms (panNENs) (MEN1, ATRX and DAXX) only loss of ATRX was detected in insulinoma sample. Visualizing the results of CNVPanelizer on genome coordinates (hg19) we have noticed that in the insulinoma sample the data for the first exon of ATRX was higher (shown in red rectangle) than the reference values, whereas the downstream exons were lower (Figure 3).
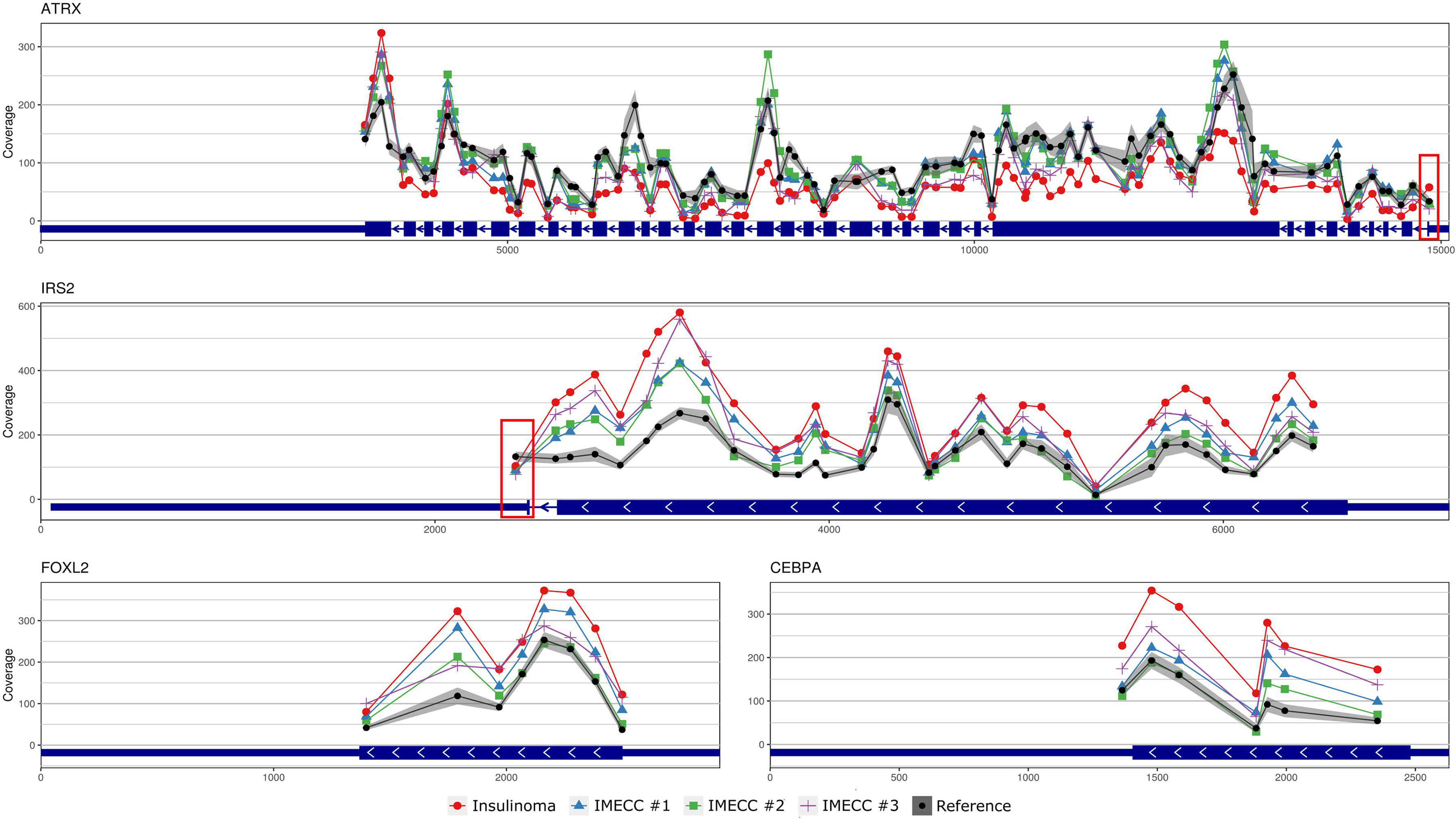
Figure 3 Visualization of CNVPanelizer results on genomic coordinates (genome assembly GRCh37/hg19) of the genes ATRX, IRS2, FOXL2 and CEBPA.
Recurrent CNVs
According to the results of CNVPanelizer, amplifications of FOXL2, IRS2 and CEBPA genes were found in all samples except IMECC #2, (Figure 2). However, visualization of CNVPanelizer results (Figure 3) showed that in IRS2 gene the second exon was lower in all tested samples including IMECC #2 compared to reference data, suggesting its loss.
Quantitative PCR
Gain status was confirmed in all samples for FOXL2 gene. For CEBPA, only in insulinoma and IMECC #1 gain status was confirmed (p-value <0.001 and <0.01 respectively).
Amplification of the first exon of the IRS2 gene was also confirmed in all samples (p-value <0.001 for all samples). However, the data on the second exon was various. Results qPCR with CNVPanelizer matched only in insulinoma sample (p-value <0.01).
Amplification of the first exon of ATRX in insulinoma sample that we noticed on CNVPanelizer was confirmed with a statistical significance of p-value < 0.01. The downstream loss of ATRX which we checked in the ninth exon was also confirmed with statistical significance of p-value < 0.05. These results show that CNVPanelizer can reliably detect CNVs in separate exons.
Discussion
In this study, we describe genetic alterations in tumors that refer to such rare phenomenon as insulinomatosis in a patient without known hereditary syndromes.
Pancreatic Neuroendocrine Neoplasms
Pancreatic neuroendocrine neoplasms (panNENs or pNENs) are rare tumors of the pancreas that account for up to 2% of all pancreatic neoplasms. However, based on autopsy studies, prevalence of panNENs has been reported to be up to 10% (13). The 2017 World Health Organization classification divided panNENs into two categories, well-differentiated pancreatic neuroendocrine tumors (panNETs or PNETs) and poorly differentiated pancreatic neuroendocrine carcinomas (panNECs) (14). In the vast majority PNETs occur sporadically (~90%), but up to 5-10% are associated with genetic syndromes like multiple endocrine neoplasia type 1, neurofibromatosis type I, Von Hippel–Lindau syndrome and tuberous sclerosis complex (15). The most common functioning PNETs are insulinomas with an incidence of 4-7 per 100,000 persons per year (16–18). Most of insulinomas, more than 90%, are benign. Insulinomas are composed of producing beta cells that are actively secreting a large amount of insulin that results in episodic hyperinsulinemia and is the most frequent cause of persistent hyperinsulinemic hypoglycemia (1). In addition to Ki-67, which differentiates malignant and benign nature, tumor size is critical for survival rate prognosis. Thus, insulinomas >2 cm in diameter have a 10-year survival rate nearer to 30%, whereas for those <2 cm, the survival rate is close to 100% (19).
Molecular Genetics of Tumors in Insulinomatosis
The molecular genetic of insulinomatosis is yet to be understood. This phenomenon is not related to mutation in MEN1 gene and is more similar to sporadic benign insulinomas (1). In insulinomas, mutations in MEN1, ATRX and DAXX that are often mutated in all PNETs occur in no more than 10 percent: 3%, 8% and 3%, respectively (20). In addition many other genes were seen to be mutated in insulinomas (20, 21) and several regions with amplification (7p, 3p, 5q and 13q) were identified as early events and may be involved in tumorogenesis (22). However, the most common mutation in insulinoma is a gain of function mutation Trp372Arg in YY1 gene that occurs in 30% in Asian population and in 13% in Caucasian (German) population (23, 24). Mutation Trp372Arg increases the activity of YY1 as transcription factor, which results in greater transcription of IDH3A, UCP2 and increases the expression of ADCY1 and CACNA2D2 which regulate the insulin secretion (22). It is assumed that mutations in YY1 gene are driver mutations for insulinomas (25). In our case of insulinoma, we did not find Trp372Arg mutation in the YY1 gene nor mutations in MEN1 or DAXX, however, loss of ATRX was found and confirmed by qPCR (Figures 2–4).
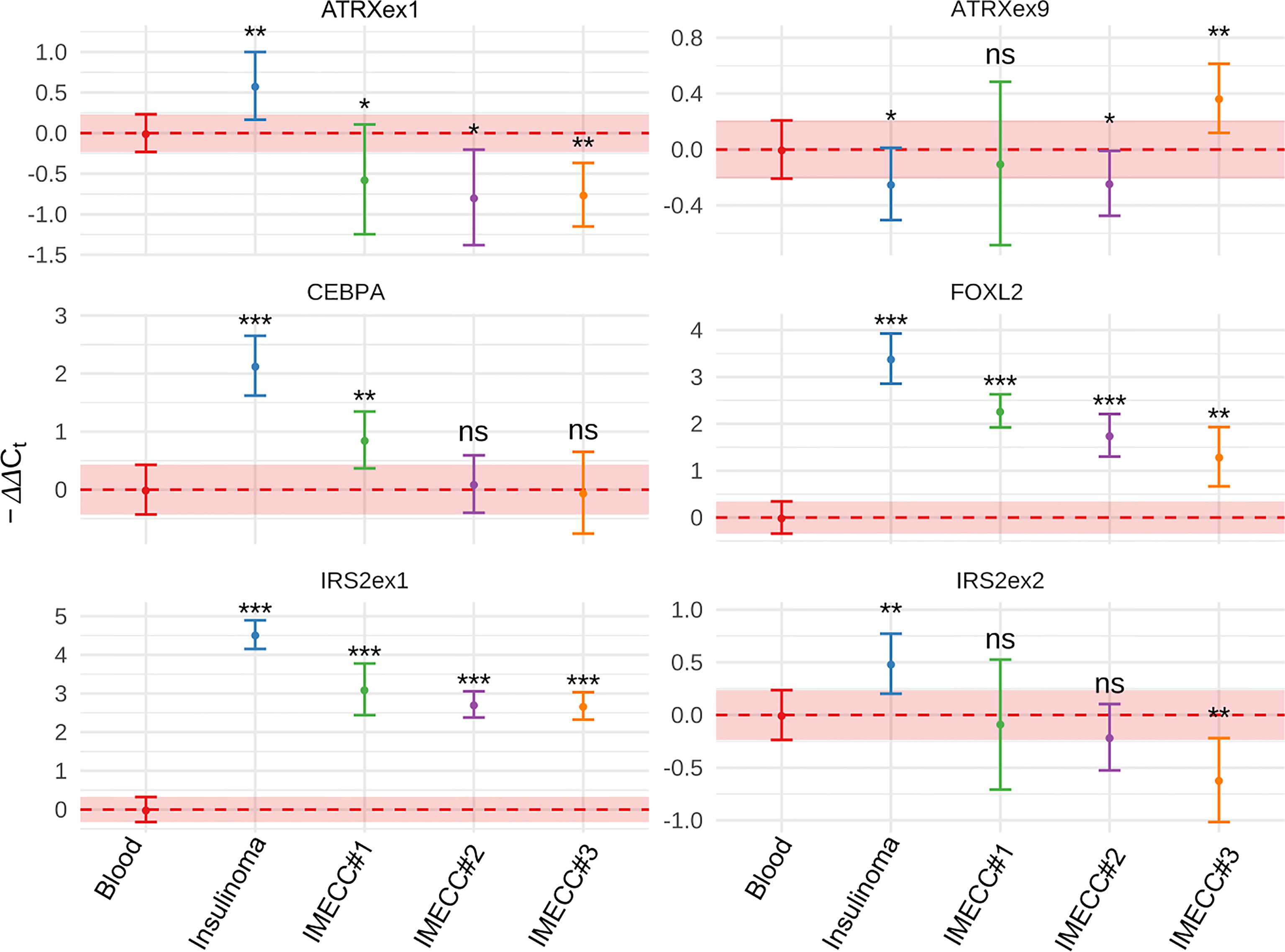
Figure 4 Results of qPCR in blood, insulinoma and IMECCs by using ΔΔCt method. *p-value < 0.05. **p-value < 0.01. ***p-value < 0.001. ns, not significant.
As for other CNVs, alterations were found in 13q34 (amplification of IRS2), 13q14.2 (loss of RB1) and 5q22.2 (loss of APC) (Figures 2, 3). With regards to the precursor lesions of PNETs, the information about their molecular genetic profile is very limited. Microadenomas are shown to harbor mutations in MEN1 (13, 14, 26, 27). Hadano et al. in their work showed that sporadic microadenomas have a significantly lower expression of ATRX and overexpression of cytokeratin-19 (CK19) compared to hyperplasia of pancreatic islet cells (13). In our studied samples of micro-tumors, we have not found alteration in MEN1, but gain status in IRS2, FOXL2 and CEBPA genes was found, suggesting that they can play a role in the development of micro- to macro-tumors, to wit insulinomas, and, accordingly, insulinomatosis.
Common Features and Filiation Between Micro- and Macro-Tumors
Besides the identity in monohormonality, our macro and micro-tumors samples share CNVs that harbor the same genes, CEBPA, FOXL2 and IRS2. CCAAT enhancer-binding protein alpha (CEBPA) and Forkhead Box L2 (FOXL2) are known for being able to arrest or suppress cell proliferation respectively (28–30). The IRS2 (Insulin receptor substrate 2) protein plays an important role in the response to stimuli for cytokines and growth factors, like insulin and insulin-like growth factor 1, and promotes proliferation and survival of normal and cancer cells by mediating signaling from INSR, IGF1R, EPOR, MPL, VEGFR, LEP, GH and IFNB1/IFNG proteins. The stimulation of insulin receptor results in IRS2 association with the p85 subunit of PI3K and GRB2, activating the PI3K/AKT/MTOR and MAPK pathways and leading to proliferation and differentiation (31, 32).
It was shown that increased expression of CEBPA is involved in apoptosis of pancreatic β-cells exposed to proinflammatory cytokines IL1β, IFNγ, and TNFα (33). Also it was shown that CEBPA induce the transdifferentiation of B cells into macrophages, and in co-expression with the transcription factors Oct4 (Pou5f1), Sox2, Klf4 and Myc enhances reprogramming into induced pluripotent stem cells (34).
Mohanty in his work showed that overexpression of IRS2 stimulates proliferation of β-cells and increases insulin secretion. It also protects β-cells from d-glucose-induced apoptosis. On the contrary, the repression of IRS2 in INS-1 cells leads to downregulation of proliferation (35).
As insulinomatosis seems to be a disease that affects the entire population of β-cells, we can assume that this triad of genes, CEBPA, FOXL2 and IRS2, participates in the disturbance of morphogenesis of β-cells, although of course, this statement requires more detailed investigations.
Conclusion
Despite the fact that detection of cancer associated CNVs has traditionally been performed by microarray techniques, NGS-based bioinformatic methods are actively developing and becoming increasingly popular due to cost-efficiency. However, the NGS-based bioinformatics results still need to be confirmed with other molecular approaches. On the other hand, molecular approaches such as qPCR are sensitive to the quality of material, which can be challenging when the samples are from FFPE tissues.
Here we described a rare case of insulinomatosis with novel CNVs that are seen in multiple micro-tumors and a macro-tumor and harbor CEBPA, FOXL2 and IRS2 genes that can be involved in pancreatic neuroendocrine tumor pathogenesis, specifically insulinomatosis, and can provide new insights into the disturbance of morphogenesis of β-cells.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA752875.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee at the Research Centre for Medical Genetics, Moscow, Russia. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
IV, AS, and AV performed the surgical operation. LG performed the microscopy and immunohistochemistry. KA performed the DNA extraction, NGS, bioinformatics analysis, Sanger sequencing, and literature review. KK performed bioinformatics analysis and data visualization. LK and KA performed qPCR analysis. MS performed statistical analysis of qPCR results and assisted with bioinformatic analysis of CNV data. AE, SK, and VS administered the project. All authors contributed to the article and approved the submitted version.
Funding
The research was carried out within the state assignment of Ministry of Science and Higher Education of the Russian Federation for RCMG. Statistical analysis in this work was supported by the Grant of the Government of the Russian Federation № 14.W03.31.0005.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the staff of the Shared Resource Centre “Genome” of FSBI RCMG for the help with sequencing. Also, we would like to thank Sandugash Tasmukanova for assistance with manuscript preparation (translation).
References
1. Anlauf M, Bauersfeld J, Raffel A, Koch CA, Henopp T, Alkatout I, et al. Insulinomatosis. Am J Surg Pathol (2009) 33(3):339–46. doi: 10.1097/PAS.0b013e3181874eca
2. Perren A, Anlauf M, Henopp T, Rudolph T, Schmitt A, Raffel A, et al. Multiple Endocrine Neoplasia Type 1 (MEN1): Loss of One MEN1 Allele in Tumors and Monohormonal Endocrine Cell Clusters But Not in Islet Hyperplasia of the Pancreas. J Clin Endocrinol Metab (2007) 92(3):1118–28. doi: 10.1210/jc.2006-1944
3. Babic B, Keutgen X, Nockel P, Miettinen M, Millo C, Herscovitch P, et al. Insulinoma Due to Multiple Pancreatic Microadenoma Localized by Multimodal Imaging. J Clin Endocrinol Metab (2016) 101(10):3559–63. doi: 10.1210/jc.2016-2717
4. Wang K, Li M, Hakonarson H. ANNOVAR: Functional Annotation of Genetic Variants From High-Throughput Sequencing Data. Nucleic Acids Res (2010) 38(16):e164–4. doi: 10.1093/nar/gkq603
5. Oliveira C, Wolf T. CNVPanelizer: Reliable CNV Detection in Targeted Sequencing Applications (2021).
6. D’haene B, Vandesompele J, Hellemans J. Accurate and Objective Copy Number Profiling Using Real-Time Quantitative PCR. Methods (2010) 50(4):262–70. doi: 10.1016/j.ymeth.2009.12.007
7. Daniell TJ, Davidson J, Alexander CJ, Caul S, Roberts DM. Improved Real-Time PCR Estimation of Gene Copy Number in Soil Extracts Using an Artificial Reference. J Microbiol Methods (2012) 91(1):38–44. doi: 10.1016/j.mimet.2012.07.010
8. Weaver S, Dube S, Mir A, Qin J, Sun G, Ramakrishnan R, et al. Taking qPCR to a Higher Level: Analysis of CNV Reveals the Power of High Throughput qPCR to Enhance Quantitative Resolution. Methods (2010) 50(4):271–6. doi: 10.1016/j.ymeth.2010.01.003
9. Ponchel F, Toomes C, Bransfield K, Leong FT, Douglas SH, Field SL, et al. Real-Time PCR Based on SYBR-Green I Fluorescence: An Alternative to the TaqMan Assay for a Relative Quantification of Gene Rearrangements, Gene Amplifications and Micro Gene Deletions. BMC Biotechnol (2003) 3(1):1–13. doi: 10.1186/1472-6750-3-18
10. Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2–ΔΔct Method. Methods (2001) 25(4):402–8. doi: 10.1006/meth.2001.1262
11. Liu Y, Sun J, Zhao M. ONGene: A Literature-Based Database for Human Oncogenes. J Genet Genomics (2017) 44(2):119–21. doi: 10.1016/j.jgg.2016.12.004
12. Zhao M, Kim P, Mitra R, Zhao J, Zhao Z. TSGene 2.0: An Updated Literature-Based Knowledgebase for Tumor Suppressor Genes. Nucleic Acids Res (2016) 44:1023–31. doi: 10.1093/nar/gkv1268
13. Hadano A, Hirabayashi K, Yamada M, Kawanishi A, Takanashi Y, Kawaguchi Y, et al. Molecular Alterations in Sporadic Pancreatic Neuroendocrine Microadenomas. Pancreatology (2016) 16(3):411–5. doi: 10.1016/j.pan.2016.01.011
14. Khanna L, Prasad SR, Sunnapwar A, Kondapaneni S, Dasyam A, Tammisetti VS, et al. Pancreatic Neuroendocrine Neoplasms: 2020 Update on Pathologic and Imaging Findings and Classification. RadioGraphics (2020) 40(5):1240–62. doi: 10.1148/rg.2020200025
15. Jeune F, Taibi A, Gaujoux S. Update on the Surgical Treatment of Pancreatic Neuroendocrine Tumors. Scand J Surg (2020) 109(1):42–52. doi: 10.1177/1457496919900417
16. AIRTUM Working Group, Busco S, Buzzoni C, Mallone S, Trama A, Castaing M, et al. Italian Cancer Figures–Report 2015: The Burden of Rare Cancers in Italy. Epidemiol Prev (2016) 40(1 Suppl 2):1–120. doi: 10.19191/EP16.1S2.P001.035
17. Dasari A, Shen C, Halperin D, Zhao B, Zhou S, Xu Y, et al. Trends in the Incidence, Prevalence, and Survival Outcomes in Patients With Neuroendocrine Tumors in the United States. JAMA Oncol (2017) 3(10):1335–42. doi: 10.1001/jamaoncol.2017.0589
18. Lawrence B, Gustafsson BI, Chan A, Svejda B, Kidd M, Modlin IM. The Epidemiology of Gastroenteropancreatic Neuroendocrine Tumors. Endocrinol Metab Clin N Am (2011) 40(1):1–18. doi: 10.1016/j.ecl.2010.12.005
19. Guilmette JM, Nosé V. Neoplasms of the Neuroendocrine Pancreas: An Update in the Classification, Definition, and Molecular Genetic Advances. Adv Anat Pathol (2019) 26(1):13–30. doi: 10.1097/PAP.0000000000000201
20. Stevenson M, Lines KE, Thakker RV. Molecular Genetic Studies of Pancreatic Neuroendocrine Tumors. Endocrinol Metab Clin N Am (2018) 47(3):525–48. doi: 10.1016/j.ecl.2018.04.007
21. Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res (2019) 47(D1):D941–7. doi: 10.1093/nar/gky1015
22. Hong X, Qiao S, Li F, Wang W, Jiang R, Wu H, et al. Whole-Genome Sequencing Reveals Distinct Genetic Bases for Insulinomas and Non-Functional Pancreatic Neuroendocrine Tumours: Leading to a New Classification System. Gut (2020) 69(5):877–87. doi: 10.1136/gutjnl-2018-317233
23. Cao Y, Gao Z, Li L, Jiang X, Shan A, Cai J, et al. Whole Exome Sequencing of Insulinoma Reveals Recurrent T372R Mutations in YY1. Nat Commun (2013) 4(1):2810. doi: 10.1038/ncomms3810
24. Lichtenauer UD, Di Dalmazi G, Slater EP, Wieland T, Kuebart A, Schmittfull A, et al. Frequency and Clinical Correlates of Somatic Ying Yang 1 Mutations in Sporadic Insulinomas. J Clin Endocrinol Metab (2015) 100(5):E776–82. doi: 10.1210/jc.2015-1100
25. Cromer MK, Choi M, Nelson-Williams C, Fonseca AL, Kunstman JW, Korah RM, et al. Neomorphic Effects of Recurrent Somatic Mutations in Yin Yang 1 in Insulin-Producing Adenomas. Proc Natl Acad Sci (2015) 112(13):4062–7. doi: 10.1073/pnas.1503696112
26. Anlauf M, Schlenger R, Perren A, Bauersfeld J, Koch CA, Dralle H, et al. Microadenomatosis of the Endocrine Pancreas in Patients With and Without the Multiple Endocrine Neoplasia Type 1 Syndrome. Am J Surg Pathol (2006) 30(5):560–74. doi: 10.1097/01.pas.0000194044.01104.25
27. Anlauf M, Perren A, Klöppel G. Endocrine Precursor Lesions and Microadenomas of the Duodenum and Pancreas With and Without MEN1: Criteria, Molecular Concepts and Clinical Significance. Pathobiology (2007) 74(5):279–84. doi: 10.1159/000105810
28. Timchenko NA, Harris TE, Wilde M, Bilyeu TA, Burgess-Beusse BL, Finegold MJ, et al. CCAAT/enhancer Binding Protein Alpha Regulates P21 Protein and Hepatocyte Proliferation in Newborn Mice. Mol Cell Biol (1997) 17(12):7353–61. doi: 10.1128/MCB.17.12.7353
29. Harris TE, Albrecht JH, Nakanishi M, Darlington GJ. CCAAT/Enhancer-Binding Protein-α Cooperates With P21 to Inhibit Cyclin-Dependent Kinase-2 Activity and Induces Growth Arrest Independent of DNA Binding. J Biol Chem (2001) 276(31):29200–9. doi: 10.1074/jbc.M011587200
30. Liu X-L, Meng Y-H, Wang J-L, Yang B-B, Zhang F, Tang S-J. FOXL2 Suppresses Proliferation, Invasion and Promotes Apoptosis of Cervical Cancer Cells. Int J Clin Exp Pathol (2014) 7(4):1534–43.
31. Patti M-E, Sun X-J, Bruening JC, Araki E, Lipes MA, White MF, et al. 4ps/Insulin Receptor Substrate (IRS)-2 Is the Alternative Substrate of the Insulin Receptor in IRS-1-Deficient Mice (∗). J Biol Chem (1995) 270(42):24670–3. doi: 10.1074/jbc.270.42.24670
32. Velloso LA, Folli F, Perego L, Saad MJA. The Multi-Faceted Cross-Talk Between the Insulin and Angiotensin II Signaling Systems. Diabetes Metab Res Rev (2006) 22(2):98–107. doi: 10.1002/dmrr.611
33. Barbagallo D, Giuseppe Condorelli A, Piro S, Parrinello N, Fløyel T, Ragusa M, et al. CEBPA Exerts a Specific and Biologically Important Proapoptotic Role in Pancreatic β Cells Through Its Downstream Network Targets. Mol Biol Cell (2014) 25:2333–41. doi: 10.1091/mbc.e14-02-0703
34. Di Stefano B, Sardina JL, van Oevelen C, Collombet S, Kallin EM, Vicent GP, et al. C/Ebpα Poises B Cells for Rapid Reprogramming Into Induced Pluripotent Stem Cells. Nature (2014) 506(7487):235–9. doi: 10.1038/nature12885
Keywords: neuroendocrine tumors, FOXL2, IRS2, CEBPA, copy number variation, insulinomatosis
Citation: Anoshkin K, Vasilyev I, Karandasheva K, Shugay M, Kudryavtseva V, Egorov A, Gurevich L, Mironova A, Serikov A, Kutsev S and Strelnikov V (2021) New Regions With Molecular Alterations in a Rare Case of Insulinomatosis: Case Report With Literature Review. Front. Endocrinol. 12:760154. doi: 10.3389/fendo.2021.760154
Received: 17 August 2021; Accepted: 28 September 2021;
Published: 19 October 2021.
Edited by:
Antongiulio Faggiano, Sapienza University of Rome, ItalyReviewed by:
Jean-Yves Scoazec, Institut Gustave Roussy, FranceGiovanni Vitale, University of Milan, Italy
Copyright © 2021 Anoshkin, Vasilyev, Karandasheva, Shugay, Kudryavtseva, Egorov, Gurevich, Mironova, Serikov, Kutsev and Strelnikov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kirill Anoshkin, YW5vc2hraW5raUBnbWFpbC5jb20=
 Kirill Anoshkin
Kirill Anoshkin Ivan Vasilyev2
Ivan Vasilyev2 Kristina Karandasheva
Kristina Karandasheva Mikhail Shugay
Mikhail Shugay Valeriya Kudryavtseva
Valeriya Kudryavtseva Vladimir Strelnikov
Vladimir Strelnikov