- 1Wuxi Fisheries College, Nanjing Agricultural University, Wuxi, China
- 2Key Laboratory of Freshwater Fisheries and Germplasm Resources Utilization, Ministry of Agriculture, Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences, Wuxi, China
- 3East China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Shanghai, China
Molting and ovulation are the basic processes responsible for the growth and reproduction of Macrobrachium nipponense; however, the molecular mechanisms of molting and ovulation in M. nipponense are poorly understood. The present study aimed to use MnFtz-f1 as the starting point to study the molting and ovulation phenomena in M. nipponense at the molecular level. The full-length MnFtz-f1 cDNA sequence was 2,198 base pairs (bp) in length with an open reading frame of 1,899 bp encoding 632 amino acids. Quantitative real-time PCR analysis showed that MnFtz-f1 was highly expressed in the ovary at the cleavage stage and on the fifth day after hatching. In vivo administration of 20-hydroxyecdysone (20E) showed that 20E effectively inhibited the expression of the MnFtz-f1 gene, and the silencing of the MnFtz-f1 gene reduced the content of 20E in the ovary. In situ hybridization (ISH) analysis revealed the localization of MnFtz-f1 in the ovary. Silencing of MnFtz-f1 by RNA interference (RNAi) resulted in significant inhibition of the expression of the vitellogenin (Vg), Spook, and Phantom genes, thus confirming that MnFtz-f1 had a mutual regulatory relationship with Vg, Spook, and Phantom. After RNAi, the molting frequency and ovulation number of M. nipponense decreased significantly, which demonstrated that MnFtz-f1 played a pivotal role in the process of molting and ovulation.
Introduction
Molting is an important behavior in the growth and development of arthropods. A growing body of evidence shows that 20-hydroxyecdysone (20E) controls or triggers the molting process in arthropods, and the uncoordinated action of 20E is often fatal (1–5). The molting process of arthropods requires multilevel regulation, which involves some members of the nuclear receptor family of genes that perform important functions in the molting process (6). The synthesized 20E binds to the nuclear receptor genes to regulate downstream genes and jointly regulate molting (7). Thus, nuclear receptor-type transcription factors are essential for the molting process of arthropods (6).
Nuclear receptors are a family of transcription factors characterized by a central DNA binding region (8). The average insect has 21 genes encoding nuclear receptors (9). In-depth research has been conducted on the role of nuclear receptors in life activities of insects, such as oogenesis, embryonic development, and molting (9, 10). The nuclear receptor Ftz-f1, as the potential factor of molting response, plays a central role in coordinating different molting processes (11, 12). Ftz-f1 is induced after the level of 20E decreases (13–15). In Nilaparvata lugens, 20E was found to significantly inhibit the expression of Ftz-f1, indicating that Ftz-f1 was directly regulated by 20E (16). One isoform of Ftz-f1 has been detected in most insects such as Bombyx mori (17), Aedes aegypti (18), Manduca sexta (19), Blattella germanica (20), and Spodoptera litura (21); however, two isoforms of Ftz-f1, namely αFtz-f1 and βFtz-f1, have been detected in Drosophila (22) and Leptinotarsa decemlineata (23). Ftz-f1 is associated with molting in Tribolium castaneum (24) and acts as a competence factor for 20E in the vitellogenesis of mosquitoes (18). Ftz-f1 plays an essential role in embryogenesis, larval ecdysis, and pupation of Drosophila melanogaster (14, 15). In B. germanica, silencing of Ftz-f1 results in molting failure and larval death (20). In vertebrates, SF1 is the key factor that regulates steroid production, and SF1 is produced by Ftz-f1 (25). Previous studies have also shown that Ftz-f1 regulated the expression of genes related to ecdysone biosynthesis (26). The regulation of molting-related genes may be the original function of the Ftz-f1 protein (27, 28). In mammals, Ftz-f1 acts as a regulator of P450 steroid hydroxylase (29). In D. melanogaster, the loss of Ftz-f1 function leads to a significant decrease in the protein levels of the disembodied and phantom genes, which confirms that Ftz-f1 has a regulatory effect on these genes (26). Spook and Phantom are the upstream gene that catalyzes the synthesis of cholesterol into 20E, and MnFtz-f1 is the downstream gene of 20E (29). Therefore, MnFtz-f1, Spook and Phantom may have a synergistic effect between exercising the molting function.
Follicle maturation and ovulation are essential for successful reproduction in females. Studies have shown that Ftz-f1 regulates the occurrence of follicles through molting signals (30). In Drosophila, the disruption of Ftz-f1 expression leads to the failure of follicle cells to mature normally, eventually resulting in ovulation failure (31). Similarly, the knockdown of the Ftz-f1 gene severely hindered yolk formation and oogenesis in T. castaneum, and the reproductive ability of the insect was significantly inhibited (32). The Ftz-f1 gene also plays a role in the reproduction process of worker bees, and the size of their ovaries is regulated by Ftz-f1 (33). After the mosquitoes have a blood meal, under the effect of 20E, Ftz-f1 acts as a competence factor for the Vg gene (34). As noted above, Ftz-f1 performs basic functions in insects, but there are fewer reports of the role of Ftz-f1 in crustaceans. Presently, it is known that Ftz-f1 is involved in the regulation of Vg in Eriocheir sinensis (35) and Daphnia, and silencing of Ftz-f1 by interference results in molting failure. Previous studies have shown that both MnFtz-f1 and Vg are related to ovarian development and may have a regulatory relationship between them.
Crustaceans are very fragile due to the lack of a protective outer shell immediately after molting (36, 37). Because of a tendency to engage in combat and autophagy, crustaceans that have just molted are vulnerable to attack by their companions. In aquaculture, abnormal molting and damage to the new epidermis layer are important reasons for the high mortality of crustaceans (38). Macrobrachium nipponense is a decapod crustacean with an important economic value in China’s aquaculture industry (39, 40). The abnormal molting during the annual breeding period of M. nipponense causes a large number of deaths, which severely restricts the development of aquaculture for this crustacean (39, 40). In addition, although the relationship between gonadal development and molting is controversial in other species, the ovarian development of M. nipponense is closely related to molting during the breeding period (41). Molting and ovulation are very important processes for the growth and reproduction of M. nipponense; however, very few studies have been conducted on the molecular mechanisms underlying these processes. Therefore, it is important to study the molecular mechanisms of molting and ovulation in M. nipponense. Our previous studies have summarized in detail the entire process of the Halloween gene family that catalyzes the synthesis of 20E from cholesterol and showed that the Mn-Spook gene plays an indispensable role in the molting process of M. nipponense (41). To further understand the mechanism of molting and ovulation in M. nipponense, the present study continued to investigate the function of the nuclear receptor gene MnFtz-f1.
The current study identified the nuclear receptor gene MnFtz-f1 in M. nipponense. The expression of MnFtz-f1 in different tissues and developmental stages was analyzed by quantitative real-time PCR (qRT-PCR). The 20E was administered in vivo to detect its effect on the expression of MnFtz-f1. RNAi technology was used to knock-down the expression of MnFtz-f1 to study the regulation of MnFtz-f1 on the Mn-Spook, Phantom, and Vg genes. After silencing of MnFtz-f1, ISH was performed to localize MnFtz-f1 in the experimental and control groups, and the 20E content of M. nipponense was detected by ELISA. Finally, the role of MnFtz-f1 in the molting and ovulation of M. nipponense was studied by comparing the molting frequency and the number of ovulations completed in the experimental and control groups.
Results
Molecular Cloning and Structural Analysis of the MnFtz-f1 Gene
The full-length MnFtz-f1 cDNA sequence was 2,198 base pairs (bp); the 5ʹ and 3ʹ noncoding regions were 160 bp and 139 bp, respectively; and the open reading frame was 1,899 bp and encoded 632 amino acids (GenBank accession number: OK217288). The MnFtz-f1 cDNA included a polyadenylation signal (AATAAA) and a poly(A) tail in the 3ʹ-untranslated region (UTR), which indicated the integrity of the MnFtz-f1 gene sequence (Figure 1).

Figure 1 The nucleotide and amino acid sequences of the MnFtz-f1 gene in M. nipponense. The numbers on the left of the sequence indicate the positions of nucleotides and amino acids. The amino acids are presented as one-letter symbols and shown below their codons in each line. The starting codon (ATG) is underlined; the termination codon (TAA) is indicated by an asterisk (*); and the putative polyadenylation signal (AATAAA) is underlined. The DBD domain is marked with shadow.
The amino acid sequences of MnFtz-f1 were compared with those of other crustaceans by DNAMAN 6.0. The results showed that MnFtz-f1 had significant homology with Ftz-f1 of other crustaceans, and both had the DNA-binding domain (DBD) and activation factor-2 (AF-2) as conserved domains. MnFtz-f1 showed the highest amino acid identity (68.3%) with Ftz-f1 of Penaeus vannamei followed by Penaeus monodon (68.1%) and Homarus americanus (50.2%) (Figure 2).

Figure 2 Alignment of the deduced amino acid sequence of MnFtz-f1 with those of other species. The deduced amino acid sequence of MnFtz-f1 in M. nipponense (OK217288) was compared with that of Ftz-f1 from P. vannamei (QJI54417.1), P. monodon (XP_037803375.1), and H. americanus (KAG7156476.1) by the DNAMAN program.
A phylogenetic tree of insects and crustaceans was constructed by MEGA 5.1 software. The results showed that the amino acid sequence of H. americanus clustered with the amino acid sequence of MnFtz-f1. The phylogenetic tree was clearly divided into two major branches, i.e., insects and crustaceans (Figure 3).
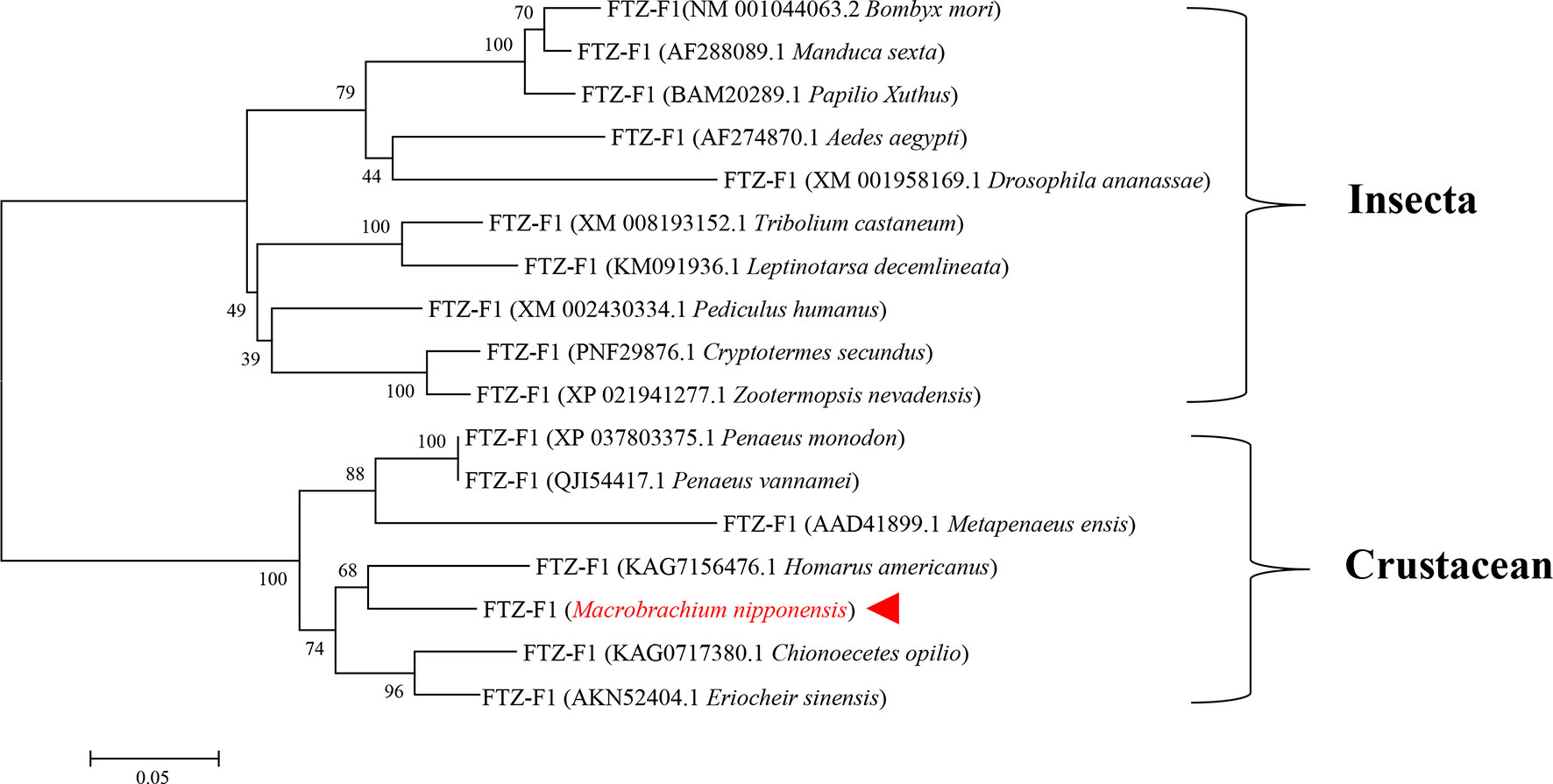
Figure 3 Phylogenetic tree of amino acid sequences of MnFtz-f1 from various species. GenBank accession numbers are shown in brackets. M. nipponense MnFtz-f1 is marked in red.
The iterative threading assembly refinement (I-TASSER) server (42, 43) was used to analyze and compare the Ftz-f1 amino acid sequences of M. nipponense and other crustaceans. The results of the three-dimensional (3D) atom model generated by I-TASSER showed that the Ftz-f1 amino acid sequences of M. nipponense, P. vannamei, and other crustaceans have the same DNA-binding domain (Figure 4).
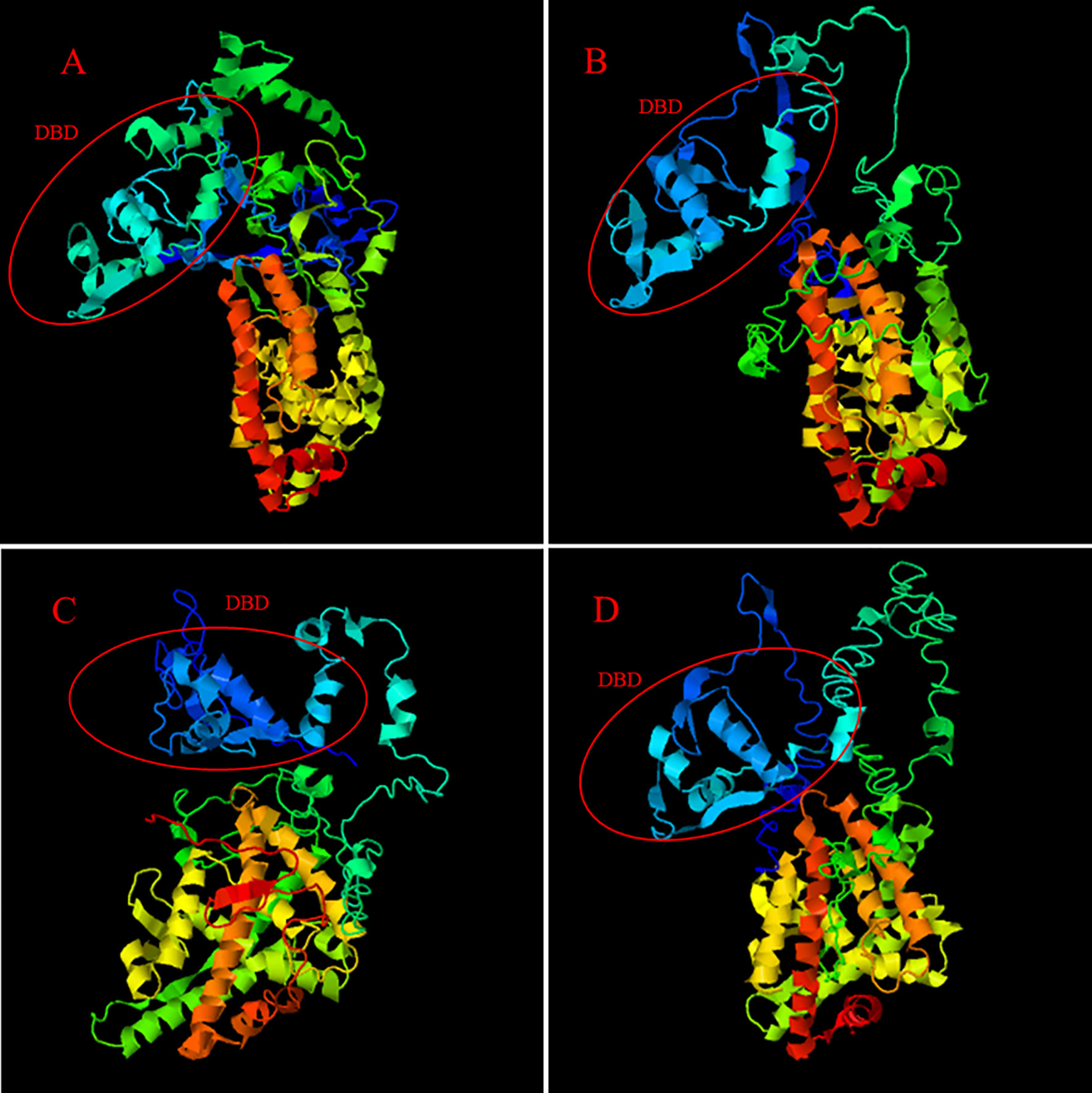
Figure 4 The 3D-structures of MnFtz-f1 and Ftz-f1 predicted by I-TASSER. (A) shows the 3D-structure of the MnFtz-f1 gene of M. nipponense. (B–D) show the 3D-structures of the Ftz-f1 gene of P. vannamei, H. americanus and P. monodon, respectively. The DNA-binding domain is marked with a red circle.
Expression of the MnFtz-f1M Gene in Different Tissues
The distribution of MnFtz-f1 gene expression in different tissues (ovary, heart, gill, eyestalk, hepatopancreas, and muscle) of M. nipponense was determined by qPCR. As shown in Figure 5, the highest mRNA expression was observed in the ovary, followed by that in the heart (P < 0.05). The expression levels of MnFtz-f1 in the ovary, heart and gill were 57.5-fold, 11.8-fold, and 6.2-fold higher than that in the muscle, respectively.
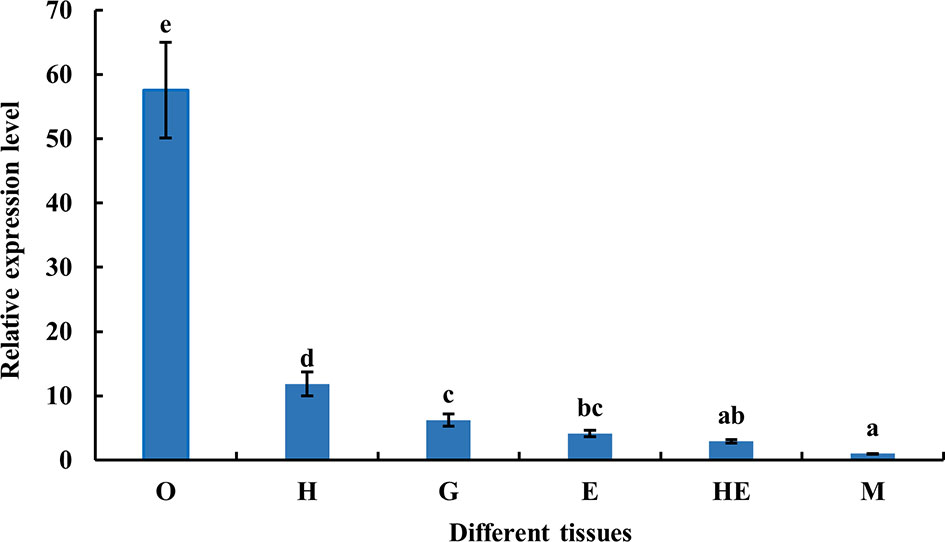
Figure 5 Expression of MnFtz-f1 mRNA in different tissues of M. nipponense. O, ovary; H, heart; G, gill; E, eyestalk; He, hepatopancreas; M, muscle. Statistical analyses were performed by one-way ANOVA. Data (mean ± SEM, n = 6) were expressed relative to the expression of the eukaryotic translation initiation factor 5A (EIF) gene. Bars with different letters indicate significant differences (P < 0.05).
Expression of the MnFtz-f1 Gene in Different Developmental Stages of the Ovaries
As shown in Figure 6, the expression level of MnFtz-f1 mRNA was the highest in the O2 stage and the lowest in the O3 stage (P < 0.05). There were no significant differences in the expression level of MnFtz-f1 mRNA between the other stages of ovarian development (P > 0.05).
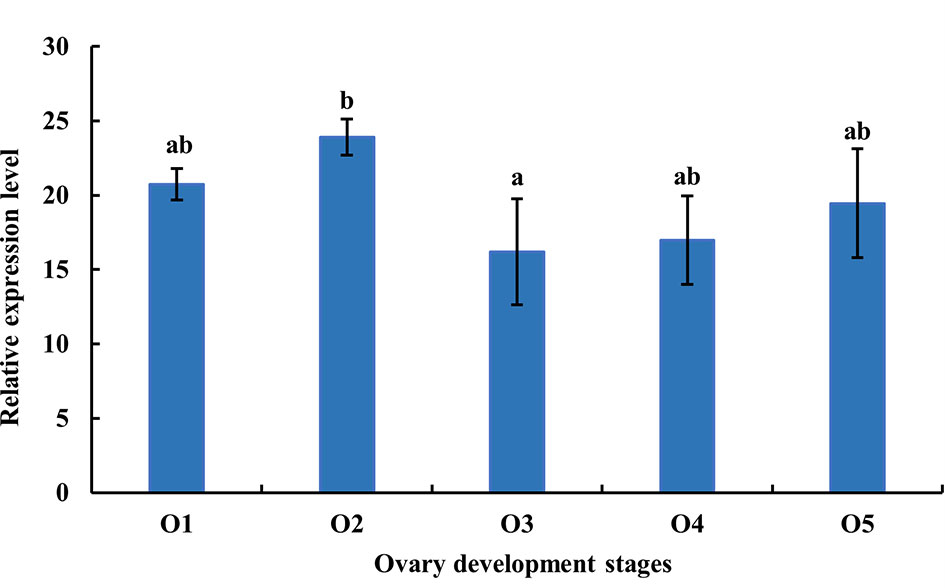
Figure 6 Expression of MnFtz-f1 mRNA in the developmental stages of the ovaries of M. nipponense. O1, undeveloped stage; O2, developing stage; O3, nearly ripe stage; O4, ripe stage; O5, spent stage. Statistical analyses were performed by one-way ANOVA. Data are expressed as mean ± SEM (n = 6). Bars with different letters indicate significant differences (P < 0.05).
Expression of the MnFtz-f1 Gene in Different Developmental Stages of Embryos and Individuals
The distribution of MnFtz-f1 gene expression in different developmental stages was investigated by qPCR (Figure 7). The MnFtz-f1 mRNA level was the highest in CS (P < 0.05), but no significant differences were observed between other embryonic developmental stages (BS, GS, NS, and ZS) (P > 0.05). The MnFtz-f1 mRNA level was reached the highest on the 5th day after hatching (L5), followed by that on the 5th day after larvae (PL5) and showed significant differences with those of other developmental stages (P < 0.05).
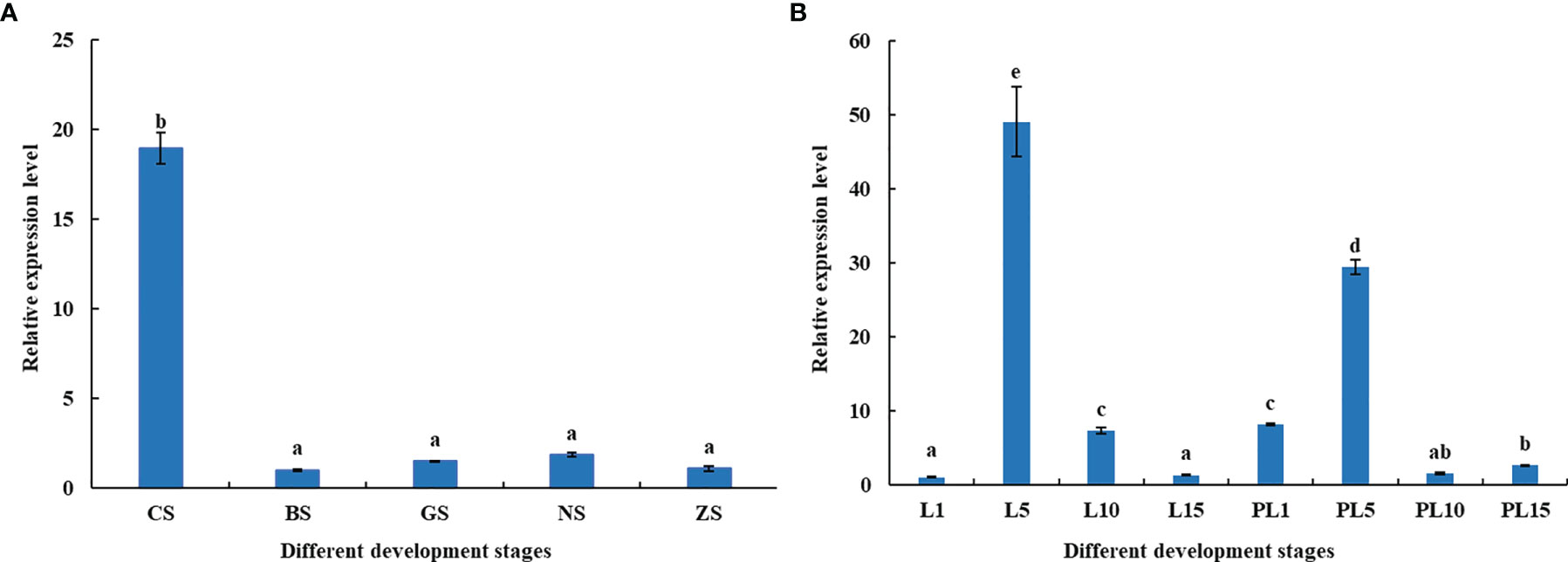
Figure 7 Expression of the MnFtz-f1 Gene in Different Developmental Stages of Embryos (A) and Individuals (B). CS, cleavage stage; BS, blastula stage; GS, gastrula stage; NS, nauplius stage; ZS, zoea stage; L1, the first day after hatching; PL1, the first day after larvae, and so on. Statistical analyses were performed by one-way ANOVA. Data are expressed as mean ± SEM (n = 6). Bars with different letters indicate significant differences (P < 0.05).
Effect of 20E on the Expression of MnFtz-f1
The expression level of MnFtz-f1 in the ovary under different concentrations of 20E was detected by qPCR (Figure 8). Compared to the control group, a low concentration of 20E (≤3 μg/g) had no significant effect on the expression of MnFtz-f1 (P > 0.05). When the concentration of 20E was ≥5 μg/g, the expression of MnFtz-f1 decreased significantly (P < 0.05). The expression of MnFtz-f1 was significantly inhibited under the action of a high concentration of 20E (20 μg/g) (P < 0.05). The expression level of MnFtz-f1 at different time points was detected at the same 20E concentration of 5 μg/g. The results showed that compared to the control group, the expression level of MnFtz-f1 was significantly decreased after 20E administration (P < 0.05). MnFtz-f1 expression decreased to the lowest level at 12 h and then increased gradually.
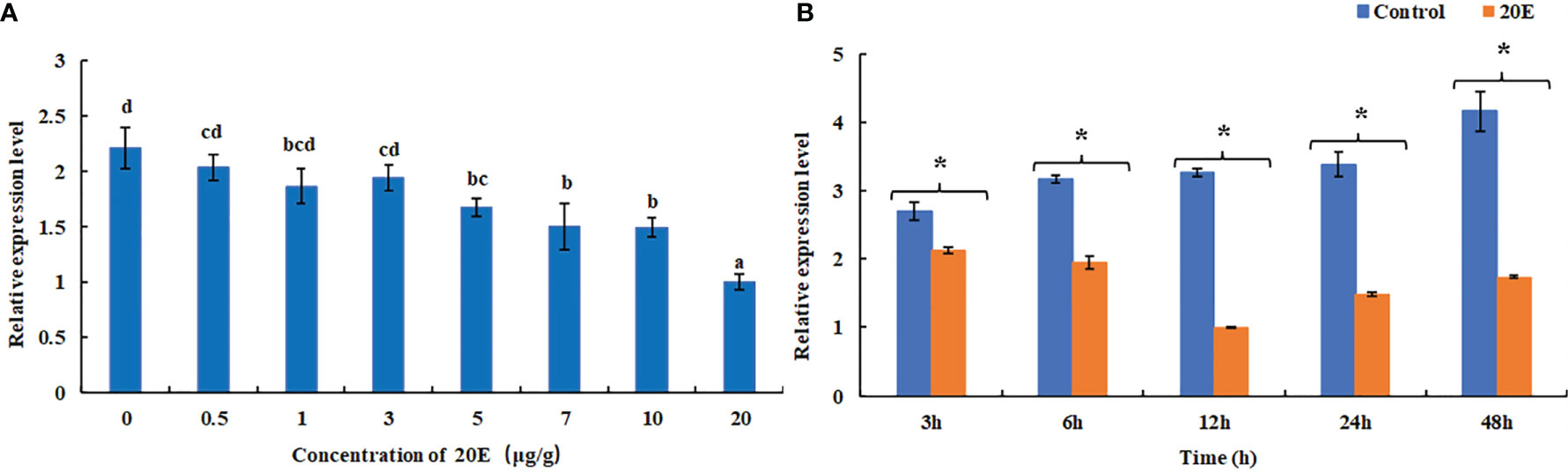
Figure 8 Expression of MnFtz-f1 mRNA under the influence of different concentrations of 20E (A). Effects of the same concentration of 20E (5 μg/g) on MnFTZ-F1 expression at different time points (B). Statistical analyses were performed by one-way ANOVA and Student’s t-test. Data are expressed as mean ± SEM (n = 6). Bars with different letters and (*) indicate significant differences (P < 0.05).
Effect of MnFtz-f1 Gene Knockdown on the Expression of MnFtz-f1, Vg, Mn-Spook, and Phantom in the Ovary
The function of MnFtz-f1 in M. nipponense and its regulatory relationship with other genes were studied by the RNAi method (Figure 9). Compared to the control group, the expression level of MnFtz-f1 did not decrease significantly within 24 h after dsMnFtz-f1 RNA administration (P > 0.05). The expression level of MnFtz-f1 at 48 and 96 h after the administration was significantly decreased by 97.12% and 86.09%, respectively, as compared to that of the control group (P < 0.05). After silencing of MnFtz-f1, the expression levels of Mn-Spook, Phantom, and Vg decreased significantly at 48 and 96 h after the administration, and the levels decreased by 51.42% and 66.06%, 56.16% and 69.82%, and 77.14% and 79.50%, respectively (P < 0.05).
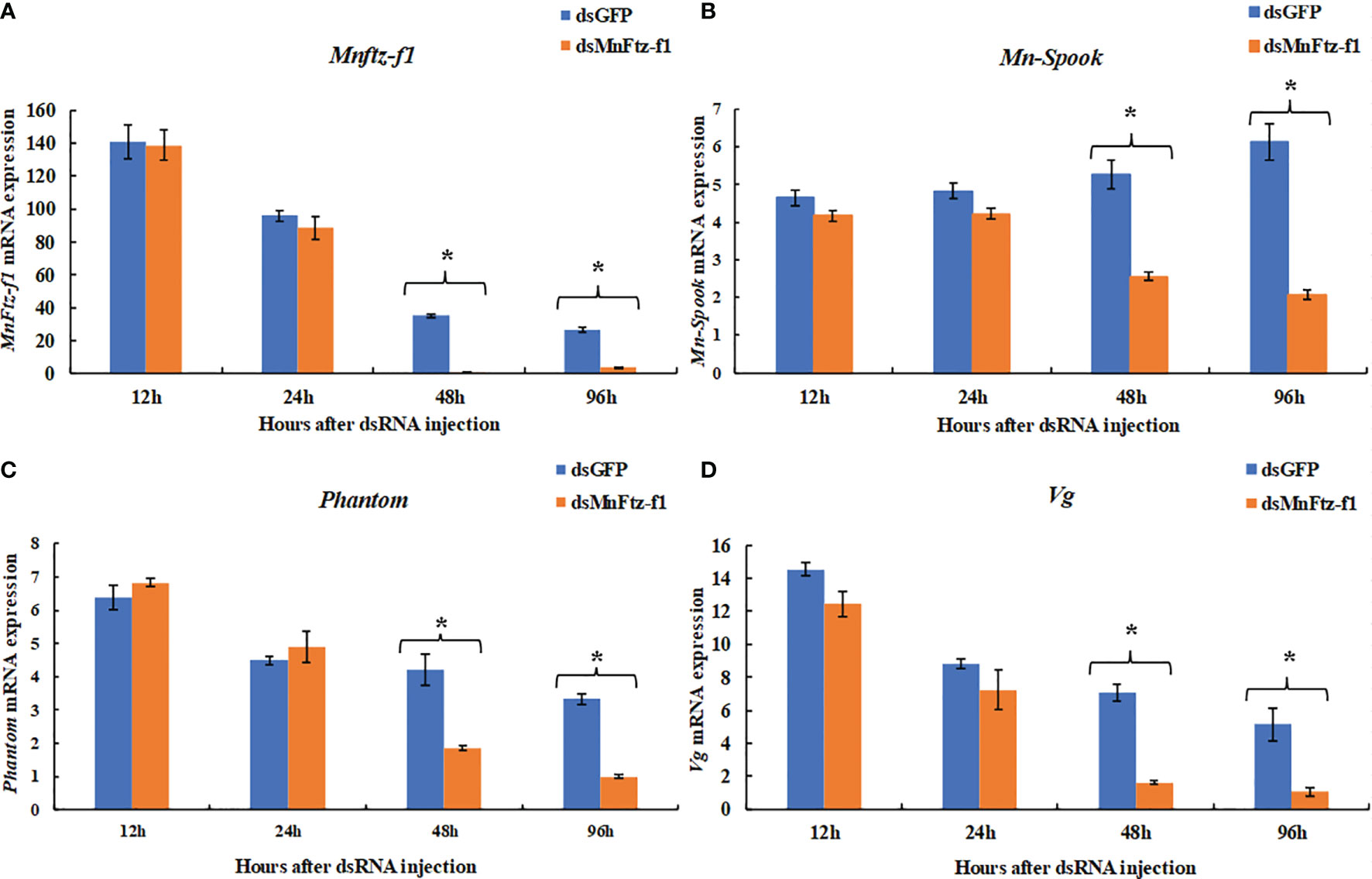
Figure 9 The expression levels of Mnftz-f1, Mn-Spook, Phantom and Vg after RNAi of Mnftz-f1. (A), MnFtz-f1; (B), Mn-Spook; (C), Phantom; (D), Vg. Data are expressed as mean ± SEM, and the differences were considered to be significant at P < 0.05 (*) by Student’s t-test (n = 6).
Effect of RNAi on the 20E Content of M. nipponense
The expression level of MnFtz-f1 on days 10 after the administration was significantly decreased by 54.70%, as compared to that of the control group (P < 0.05) (Figure 10A). The content of 20E in the ovaries of M. nipponense was measured by ELISA after the knockdown of Mnftz-f1 (Figure 10B). Compared to the control group (dsGFP administration), the 20E content did not decrease significantly on the first day after the administration of dsMnFtz-f1 RNA (P > 0.05). On the 10th day after RNAi, the content of 20E in the experimental group was significantly reduced and was 30.25% lower than that in the control group (P < 0.05).
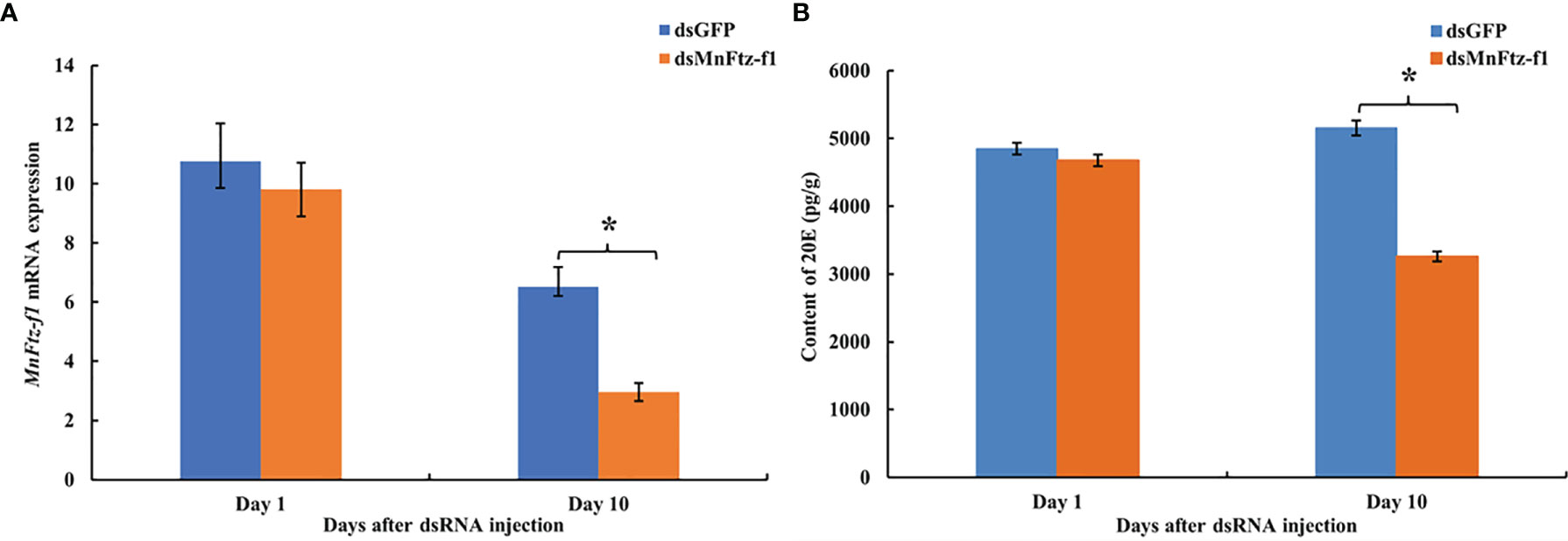
Figure 10 The expression level of Mnftz-f1 (A) and the content of 20E (B) in M. nipponense after RNAi of Mnftz-f1. Data are expressed as mean ± SEM, and the differences were considered to be significant at P < 0.05 (*) by Student’s t-test (n = 6).
Localization of the MnFtz-f1 Gene in the Ovaries
After the knockdown of the MnFtz-f1 gene, ISH was used to label the MnFtz-f1 mRNA in the experimental and control groups (Figure 11). MnFtz-f1 signals were detected in the cytoplasmic membrane and follicular cells. Compared to the control group, the MnFtz-f1 signals of the experimental group were weaker, and no signal was detected in the negative control.
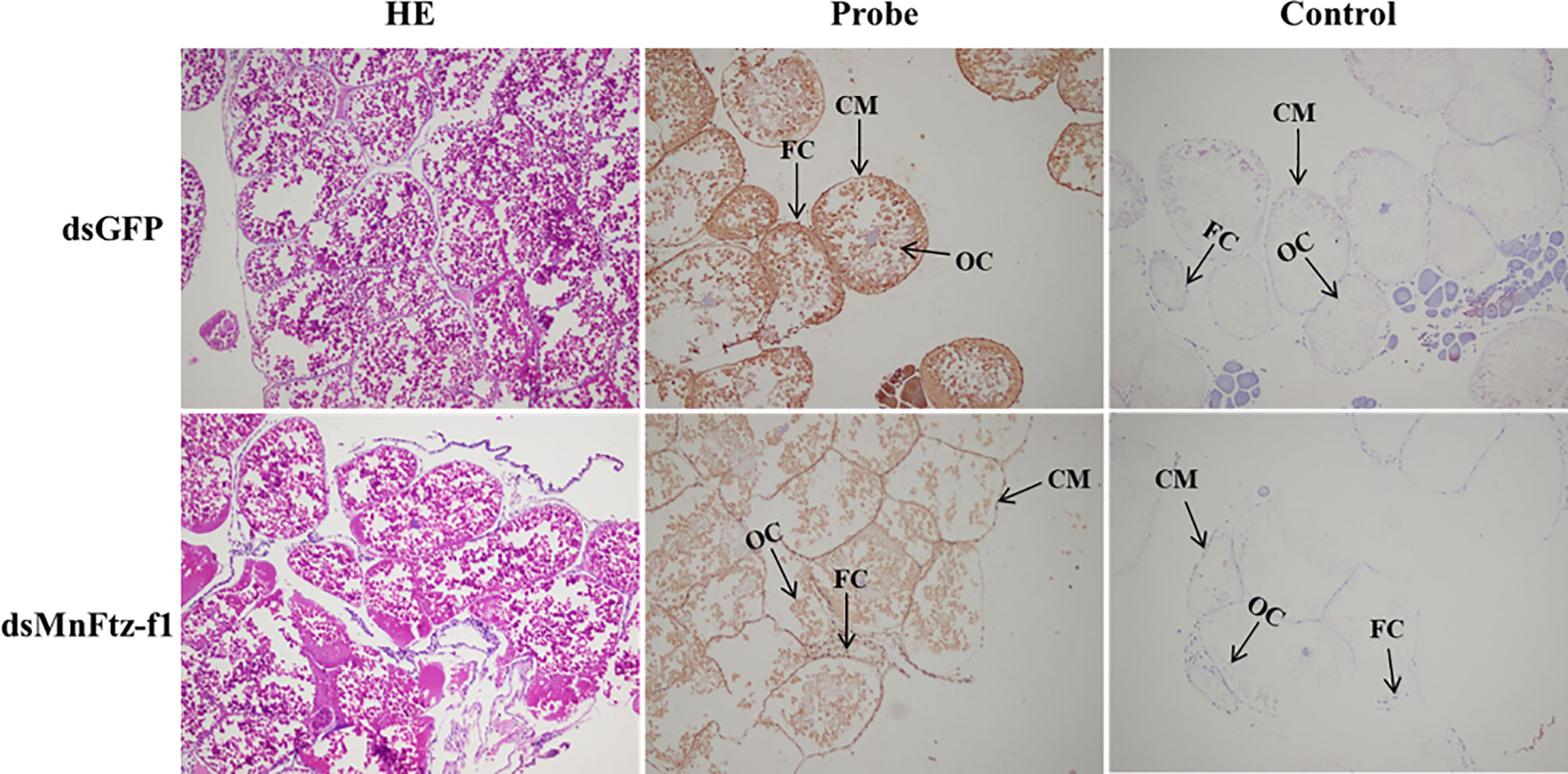
Figure 11 Histological sections of ovarian tissues of the experimental and control groups after RNAi. GFP was used as a control. OC, oocyte; CM, cytoplasmic membrane; FC, follicle cell; scale bar, 20 μm.
Effect of MnFtz-f1 Knockdown on the Molting Frequency and Ovulation of M. nipponense
Figure 12A shows the molting process of M. nipponense. After MnFtz-f1 knockdown, the molting frequency of M. nipponense was estimated (Figure 12B). The number of molting times was recorded by counting the procuticle of M. nipponense. M. nipponense began molting on the 3rd day. No significant differences were observed between the experimental and control groups on the 3rd and 4th days (P > 0.05). Starting from the 5th day, the molting frequency of the experimental group was significantly lower than that of the control group (P < 0.05).
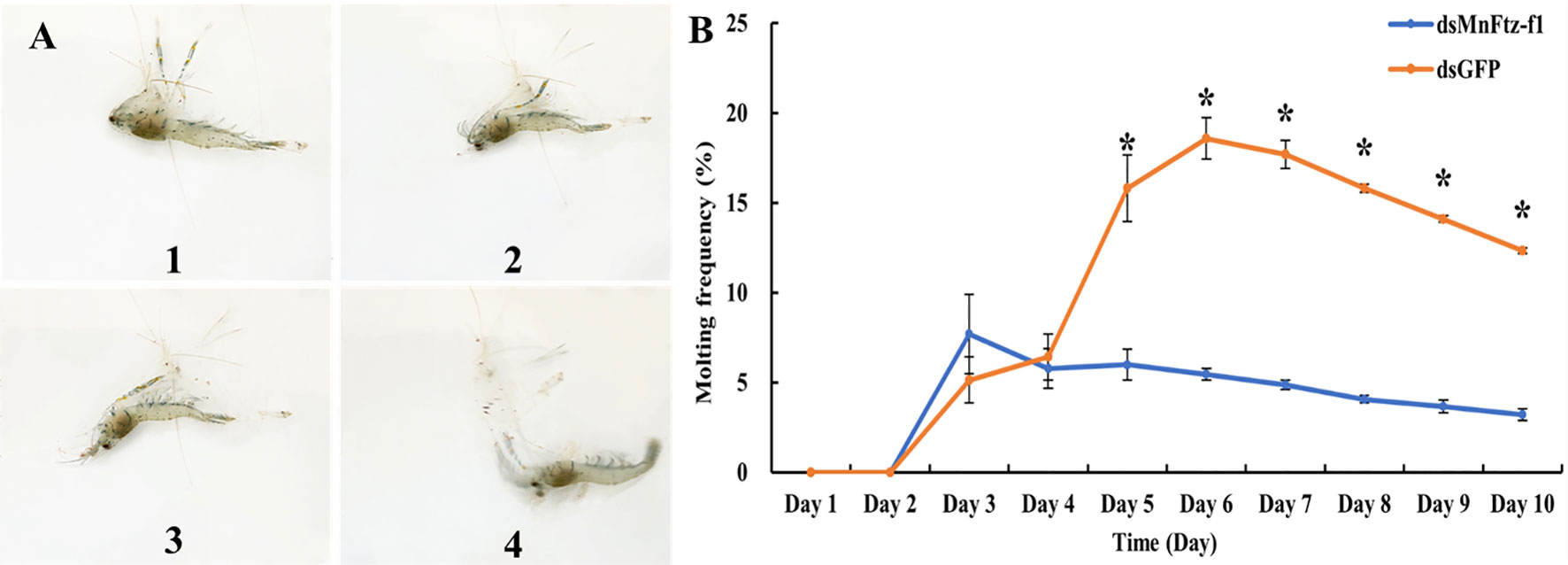
Figure 12 Molting frequency of M. nipponense in the experimental and control groups after RNAi (B). The molting order of prawn was 1- 4 (A). GFP was used as a control. Data are expressed as mean ± SEM, and the differences were considered to be significant at P < 0.05 (*) by Student’s t-test.
Figure 13A shows the comparison of ovulation and non-ovulation of M. nipponense. After RNAi, we counted the number of M. nipponense individuals that completed ovulation in the experimental and control groups (Figure 13B). M. nipponense started ovulation on the 3rd day after interference. On the 3rd day, no significant difference in ovulation was observed between the experimental group and the control group (P > 0.05). From the 4th day onwards, the ovulation frequency of the experimental group was significantly lower than that of the control group (P < 0.05).
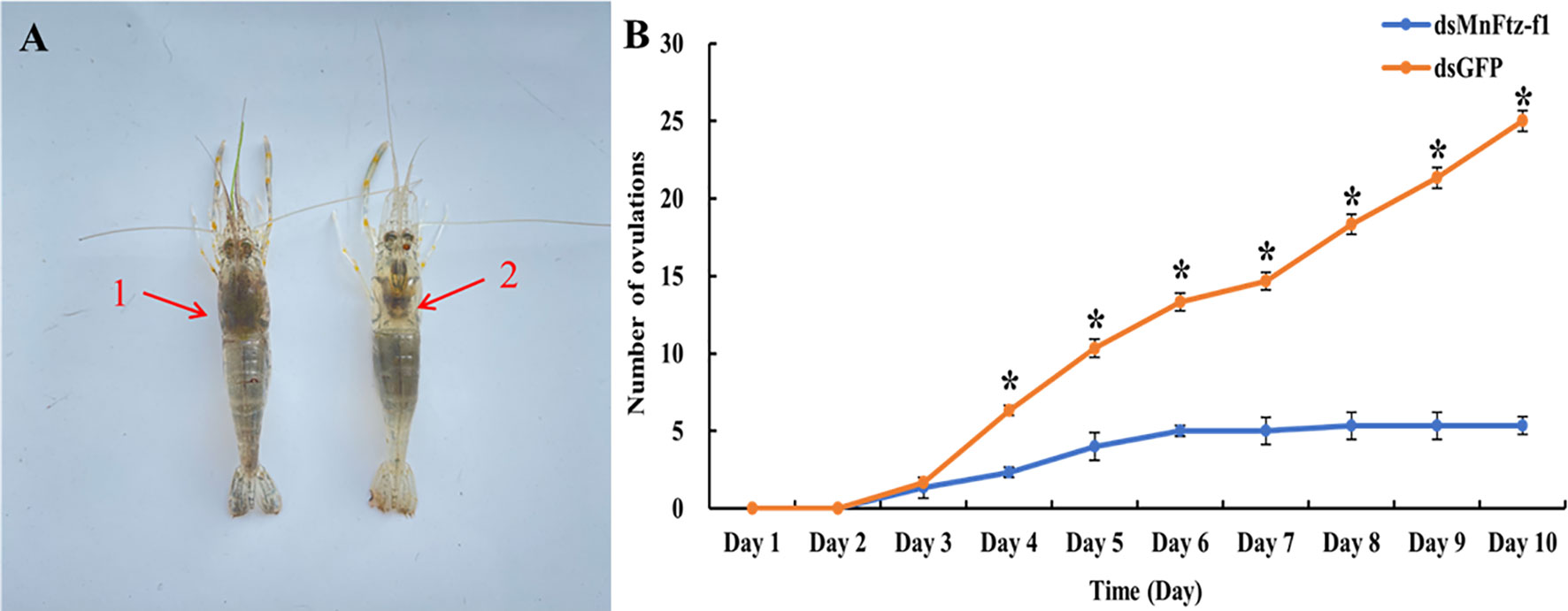
Figure 13 The number of ovulations of M. nipponense in the experimental and control groups after RNAi (B). GFP was used as a control. 1, non-ovulation, 2, ovulation (A). Data are expressed as mean ± SEM, and the differences were considered to be significant at P < 0.05 (*) by Student’s t-test.
Discussion
Nuclear receptor transcription factors are one of the most abundant transcription factors in metazoans, and they are involved in various developmental and physiological processes such as sex differentiation, ovarian and embryo development, and molting (44, 45). Ftz-f1 is one of the classical nuclear receptors (46). In the present study, we focused on the orphan receptor Ftz-f1 and successfully cloned the full-length MnFtz-f1 cDNA from M. nipponense (Figure 1). Multiple sequence alignments indicate that MnFtz-f1 has a nuclear receptor gene public DNA-binding domain (DBD) (10) (Figure 2). DBD has two Cys2-Cys2 zinc coordination modules, and subtle structural changes in DBD significantly affect transcriptional regulation (47). MnFtz-f1 is highly conserved, especially the DBD domain. The DBD domains of M. nipponense are identical to those of P. vannamei, H. americanus and P. monodon (Figure 2). Phylogenetic analysis showed that crustaceans and insects were clearly delimited and clustered together (Figure 3), indicating that Ftz-f1 was differentiated in crustaceans and insects and was more conserved in the same class.
In the current study, MnFtz-f1 was found to be expressed in different tissues of M. nipponense, among which the expression was highest in the ovary (Figure 5). Similar to previous results, Ftz-f1 has been shown to be involved in various developmental processes and is expressed in many different tissues (48). Ftz-f1 is essential for ovarian development in Drosophila (49) and is also essential for oogenesis in A. aegypti and T. castaneum (18, 32). The expression of MnFtz-f1 was highest in the ovary of M. nipponense, which was consistent with the finding that Ftz-f1 plays an important role in the reproductive process (50, 51). MnFtz-f1 expression in the different developmental stages of M. nipponense ovary did not show alterations with the development of the ovary; however, the expression level was the lowest in the O3 stage, and this level was significantly lower than that in the O2 stage (Figure 6). MnFtz-f1 expression in the O3 stage may be inhibited by 20E, which has been shown to significantly inhibit the expression of Ftz-f1 (16). When the concentration of 20E drops to a low level, the expression of Ftz-f1 initially inhibited by 20E begins to increase (48, 52–55). The embryonic stage is a special life stage with no food intake and no activity. Therefore, genes that are highly expressed at this stage are directly involved in embryonic development or in preparing for future physiological stages (56). The expression of MnFtz-f1 in the CS of M. nipponense was significantly higher than that in the other developmental stages (Figure 7); this showed that MnFtz-f1 might play an important role in the process of oocyte mitosis. A recent study in Drosophila revealed that Drosophila oocytes could not undergo normal mitosis in the absence of Ftz-f1, suggesting that Ftz-f1 was essential for oocyte division (57). In Drosophila, Ftz-f1 is divided into two subtypes: αFtz-f1 and βFtz-f1. The αFtz-f1 is mainly expressed in the early stage of embryogenesis, while βFtz-f1 is expressed in the late embryonic stage and pupal stage (58). In the current study, MnFtz-f1 was highly expressed in the early stage of major embryogenesis (CS), on the 5th day after hatching, and on the 5th day after larvae (Figure 7). MnFtz-f1 may have a similar function of αFtz-f1 and βFtz-f1 in the embryonic and hatching stages. Ftz-f1 is one of the 20E responsive genes, and the decrease in 20E level induces an increase in βFtz-f1 expression level (15, 17, 59). Consistent with previous research, in vivo administration of 20E significantly inhibited the expression level of MnFtz-f1 (Figure 8).
RNAi causes post-transcriptional gene silencing through double-stranded RNA (dsRNA) (60). In M. nipponense, RNAi has been widely used in gene function analysis (41, 61, 62). In the current study, the expression of MnFtz-f1 in M. nipponense ovaries was significantly reduced by the in vivo administration of dsRNA. To further study the mutual relationship of regulation between the genes, the expression levels of Mn-Spook, Phantom, and Vg were determined after MnFtz-f1 knockdown. Spook and Phantom are important members of the Halloween gene family and regulate molting by catalyzing the conversion of cholesterol to 20E (3). Mn-Spook plays a pivotal role in the molting of M. nipponense by participating in 20E production (41). In Schistocerca gregaria, silencing of Spook reduces ecdysteroid titer and leads to delayed nymphal development and failure to molt. Phantom is the enzyme required by the prothoracic glands of Bombyx and Drosophila to synthesize ecdysteroid (63). In crustaceans, Vg provides energy for ovarian development, and the maturation of ovaries depends on the synthesis and accumulation of Vg (64, 65). In general, Mn-Spook, Phantom, and Vg are closely related to the molting or ovarian development of crustaceans. Studying the regulatory relationship between MnFtz-f1 and these genes in M. nipponense is more conducive to our understanding of the molting and ovarian development processes of M. nipponense at the molecular level. In the current study, the expression levels of the Mn-Spook, Phantom, and Vg genes were also significantly reduced after silencing of MnFtz-f1 (Figure 9). Previous studies have shown that Ftz-f1 could regulate the expression of the Halloween genes and affect the ecdysone titer (26, 66). In the Drosophila ring gland, Ftz-f1 mutation caused a significant decrease in the expression level of Phantom, indicating that Ftz-f1 regulated the expression of Phantom (26). In T. castaneum, silencing the expression of Ftz-f1 results in a complete decrease in the expression of the Vg gene (32). Ftz-f1 plays a key role in the regulation of Vg in A. aegypti (30). In Apis mellifera, RNAi experiments showed that Ftz-f1 regulates the expression of Vg (51). In summary, our research confirmed that MnFtz-f1 regulated the expression of Mn-Spook, Phantom, and Vg. RNAi of MnFtz-f1 significantly reduced the content of 20E in M. nipponense (Figure 10). Similar to our results, Ftz-f1 plays a role in regulating ecdysone titer during the development of D. melanogaster (26, 67). Our results strongly confirmed that high concentrations of 20E inhibited the expression of MnFtz-f1, but knockdown MnFtz-f1 inhibited the expression of the Mn-spook and Phantom genes involved in the synthesis of 20E, thereby affecting the efficiency of 20E synthesis. Therefore, we speculated that MnFtz-f1 played a role of negative feedback regulation during the synthesis of 20E. The results of ISH showed that more MnFtz-f1 signals were detected in the oocyte plasma membrane and follicular cells, and more MnFtz-f1 signals were detected in the control group than in the experimental group (Figure 11). Similarly, Ftz-f1 was detected in the follicular cells of the ovary of D. melanogaster (68).
To determine whether MnFtz-f1 played a role in the molting and ovulation of M. nipponense, we estimated the molting frequency and ovulation number of M. nipponense after MnFtz-f1 knockdown. The results showed that the molting and ovulation of M. nipponense in the experimental group were significantly inhibited as compared to that in the control group (Figures 12 and 13). Similar studies in insects have shown that Ftz-f1 played a role in molting and ovarian development. In L. decemlineata, knockdown of Ftz-f1 causes surface defects in wings and legs and disrupts molting (23). Several studies have shown that silencing of Ftz-f1 could lead to failure of larvae to undergo pupation and molting (20, 24, 48, 69). Similar to our results, the role of Ftz-f1 in ovulation was also demonstrated in Drosophila. In Drosophila, Ftz-f1 promotes follicle maturation and ovulation. The interruption of Ftz-f1 expression prevents follicle maturation and causes ovulation failure (31). In B. germanica, Ftz-f1 knockdown leads to severe obstruction of ovulation (50), while Drosophila requires Ftz-f1 to promote ovulation in the final stage. Other studies have also shown that Ftz-f1 is essential for the oogenesis of A. aegypti (18) and T. castaneum (32).
In conclusion, we identified the nuclear receptor gene MnFtz-f1 in M. nipponense. The expression, distribution, and function of the MnFtz-f1 gene in M. nipponense were systematically analyzed by qRT-PCR, RNAi, ISH, ELISA, and other techniques. The results of the present study strongly confirmed that MnFtz-f1 played a pivotal role in the molting and ovulation processes of M. nipponense. This study enriched the molecular mechanisms of molting and ovulation during the reproduction period of M. nipponense and provided new insights for studying the relationship between molting and ovarian development in crustaceans.
Materials and Methods
Ethics Statement
All experimental animals (M. nipponense) in this study were handled according to the guidelines of the Institutional Animal Care and Use Ethics Committee of the Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences (Wuxi, China).
Animals
Healthy adult female prawns (2.19 ± 0.66 g) were obtained from the Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences (120◦13′44′′E, 31◦28′22′′N). The prawns were cultured in circulating water (26°C ± 1°C), and snails were fed twice a day. The experiment was conducted after 1 week of acclimatization.
RNA Isolation and cDNA Synthesis From Tissue
According to the manufacturer’s protocols, the RNAiso Plus kit (TaKaRa, Japan) was used to extract total RNA from the whole tissues of prawns (n=6). The quality of RNA was determined by 1.2% agarose gel. NanoDrop ND2000 (NanoDrop Technologies, Wilmington, DE, USA) was used to determine the concentration and purity of RNA, and the ratio of A260/A280 was estimated to determine the integrity of RNA. DNase I (Sangon, Shanghai, China) was used to process RNA samples to eliminate possible DNA contamination. The first-strand cDNA was synthesized using the reverse transcriptase M-MLV kit (TaKaRa). The synthesized cDNA was stored at -80°C for further experiments.
Cloning and Bioinformatics Analysis of MnFtz-f1
The cDNA fragment of the target gene MnFtz-f1 was obtained from the M. nipponense transcriptome cDNA library (ID: PRJNA533885) in our laboratory. The 3′-full RACE Core Set Ver. 2.0 kit and the 5′-full RACE kit (TaKaRa) were used to clone 3′-cDNA and 5′-cDNA according to the manufacturer’s protocols, respectively. Based on the known cDNA fragments, specific primers for MnFtz-f1 were designed for full-length cloning of the MnFtz-f1 cDNA. An automated DNA sequencer (ABI Biosystems, USA) was used to verify the nucleotide sequence of the cloned cDNA. All primers were synthesized by Shanghai Sangon Biotech Company (Shanghai, China) (Table 1). DNAMAN 6.0 was used to assemble the full length of the MnFtz-f1 cDNA. The MnFtz-f1 gene sequence was analyzed using GenBank BLASTX and BLASTN programs (http://www.ncbi.nlm.nih.gov/BLAST/). The online program ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) was used to analyze the open reading frame of the MnFtz-f1 gene. Phylogenetic trees based on the amino acid sequences were generated by the neighbor joining method with Molecular Evolutionary Genetics Analysis (MEGA5.0) software, and the bootstrapping replications were 1,000 (70, 71). Multiple sequence alignment of MnFtz-f1 amino acids was performed using DNAMAN 6.0 software. The spatial structure was predicted by I-TASSER (https://zhanglab.ccmb.med.umich.edu/I-TASSER/). The amino acid sequences of other arthropods investigated in this study were downloaded from the GenBank database (http://www.ncbi.nlm.nih.gov/).
The qRT-PCR Analysis
The Bio-Rad iCycler iQ5 Real-Time PCR System (Bio-Rad, Carlsbad, CA, USA) was used to perform the SYBR Green qRT-PCR assay. The reaction system and procedures of qRT-PCR were consistent with our previous study (41). MnEIF was used as the internal control gene (72). All primers used for qRT-PCR are listed in Table 1. The expression level of all genes in this experiment was calculated by the 2-ΔΔCt method (73). The ovarian development cycle was classified into different stages according to previous studies (74) as follows: O1 (undeveloped stage, transparent), O2 (developing stage, yellow), O3 (nearly ripe stage, light green), O4 (ripe stage, dark green), and O5 (spent stage, gray). All experiments were performed in triplicate for each group, with at least five samples in each group.
ISH
The localization of MnFtz-f1 mRNA was determined by ISH, and the detailed steps are described in Li et al. (75). According to the MnFtz-f1 cDNA sequence, the probe was designed with Primer5 software (http://www.premierbiosoft.com/primerdesign/). ISH experiments were performed in triplicate for each tissue, and the results were evaluated under a light microscope.
Effect of 20E on MnFtz-f1
On the basis of previous reports (76–78), 20E (Sigma-Aldrich, USA) with different concentration gradients (0.5, 1, 3, 5, 7, 10, and 20 µg/g) was administered through injection into prawns, and tissues were collected after 3 h to detect the expression level of MnFtz-f1. The same volume of ethanol was administered to the control group (0 µg/g). A fixed concentration based on the results of the 20E concentration experiment was selected and administered into M. nipponense to test its effect on the expression of MnFtz-f1 at different time points (3, 6, 12, 24, and 48 h). Six prawn tissues were collected in each group in triplicate. The collected tissues were rapidly frozen in liquid nitrogen and stored in a refrigerator at -80°C until mRNA extraction.
RNA Interfering
MnFtz-f1 primers and the Green Fluorescent Protein (GFP) gene were designed for RNAi using Snap Dragon tools (https://www.flyrnai.org/cgi-bin/RNAi_find_primers.pl). GFP was used as a control. The dsRNA was synthesized by the AidTMT7 High Yield Transcription Kit (Fermentas Inc., Waltham, MA, USA) according to the manufacturer’s instructions. The integrity and purity of dsRNA were detected by 1.2% agarose gel electrophoresis. A total of 300 healthy female prawns (2.19 ± 0.66 g) were randomly divided into the experimental group and the control group in triplicate (n=50). According to the previous 20E injection concentration, the experimental group was administered with MnFtz-f1 dsRNA, and the control group was administered with GFP (79) (4 µg/g of body weight). To prolong the interference efficiency of RNAi, dsRNA was administered every 5 days. Six prawns were randomly collected from each group at 12, 24, 48, and 96 h after injection, rapidly frozen with liquid nitrogen, and stored in a refrigerator at -80°C until mRNA extraction (n = 6). By silencing the MnFtz-f1 gene, we calculated the molting frequency (MF) and ovulation of M. nipponense. In addition, 180 prawns (O4) were divided into the experimental and control groups in triplicate to observe the number of molting and ovulation (n = 30). MF = (Nm/Ns)/D, where Nm is total molting times; Ns is the number of prawns in aquarium; and D is experimental days (80).
ELISA
After silencing the MnFtz-f1 gene, the ovaries of the experimental and control groups were collected on the 1st and 10th day to detect the content of 20E. As reported earlier (41), the Shrimp EH ELISA Kit (Lot number: E20210925-98502B; Meibo, Shanghai, China) was used to detect the content of 20E in the ovaries.
Statistical Analysis
All quantitative data conformed to homogeneity of variance and normal distribution and are expressed as mean ± standard error of the mean (SEM). Statistical analyses were performed using SPSS 20.0 software (IBM, New York, NY, USA). One-way ANOVA was used to analyze the differences in tissue distribution and different developmental stages. A two-sided t-test was used to compare the expression levels in the RNAi analysis. P < 0.05 was considered to be statistically significant.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Care and Use Ethics Committee of the Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences (Wuxi, China).
Author Contributions
HQ and HF: designed the study. HY: carried out the experiments and wrote the original draft. WZ and YF: provided technical assistance. HY and SZ: participated in methodology and data curation. YG, SJ, and YX: compiled resources. YW: performed software analysis. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by grants from the National Key R&D Program of China (2018YFD0901303); Central Public-interest Scientific Institution Basal Research Fund CAFS (2020TD36); Jiangsu Agricultural Industry Technology System; the New cultivar breeding Major Project of Jiangsu province (PZCZ201745); the China Agriculture Research System-48 (CARS-48).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
M. nipponense, Macrobrachium nipponense; MnFtz-f1, Macrobrachium nipponense Ftz-f1; 20E, 20-hydroxyecdysone; RNAi, RNA interference; ELISA, Enzyme Linked Immunosorbent Assay; qPCR, quantitative real-time reverse transcription PCR; ORF, open reading frame; O, ovary; H, heart; G, gill; E, eyestalk; HE, hepatopancreas; M, muscle; O1, undeveloped stage; O2, developing stage; O3, nearly-ripe stage; O4, ripe stage; O5, spent stage; CS, cleavage stage; BS, blastula stage; GS, gastrula stage; NS, nauplius stage; ZS, zoea stage; L1, the first day after hatching; PL1, the first day after larvae; OC, oocyte; CM, cytoplasmic membrane; FC, follicle cell; cDNA, complementary DNA; ISH, in situ, hybridization; Nm, total molting times; Ns, number of prawns in water aquarium; D, experimental days.
References
1. Nakagawa Y, Henrich VC. Arthropod Nuclear Receptors and Their Role in Molting. FEBS J (2009) 276(21):6128–57. doi: 10.1111/j.1742-4658.2009.07347.x
2. Thummel CS. Molecular Mechanisms of Developmental Timing in C. Elegans and Drosophila. Dev Cell (2001) 1(4):453–65. doi: 10.1016/S1534-5807(01)00060-0
3. Truman JW, Riddiford LM. Endocrine Insights Into The Evolution of Metamorphosis in Insects. Annu Rev Entomol (2002) 47(1):467–500. doi: 10.1146/annurev.ento.47.091201.145230
4. Koelle MR, Talbot WS, Segraves WA, Bender MT, Cherbas P, Hogness DS. The Drosophila EcR Gene Encodes an Ecdysone Receptor, a New Member of The Steroid Receptor Superfamily. Cell (1991) 67(1):59–77. doi: 10.1016/0092-8674(91)90572-G
5. Festucci-Buselli RA, Contim LAS, Barbosa LCA, Stuart J, Otoni WC. Review/Synth Se Biosynthesis and Potential Functions of The Ecdysteroid 20-Hydroxyecdysone — A Review. Can J Bot (2008) 86(9):978–87. doi: 10.1139/B08-049
6. Thummel CS. Ecdysone-Regulated Puff Genes 2000. Insect Biochem Mol Biol (2002) 32(2):113–20. doi: 10.1016/S0965-1748(01)00112-6
7. Song Y, Villeneuve DL, Toyota K, Iguchi T, Tollefsen KE. Ecdysone Receptor Agonism Leading to Lethal Molting Disruption in Arthropods: Review and Adverse Outcome Pathway Development. Environ Sci Technol (2017) 51(8):4142–57. doi: 10.1021/acs.est.7b00480
8. Ueda H, Hirose S. Identification and Purification of a Bombyx Mori Homologue of FTZ-F1. Nucleic Acids Res (1990) 18(24):7229–34. doi: 10.1093/nar/18.24.7229
9. Jarvela AMC, Pick L. The Function and Evolution of Nuclear Receptors in Insect Embryonic Development. Curr Top Dev Biol (2017) 125:39–70. doi: 10.1016/bs.ctdb.2017.01.003
10. Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, et al. The Nuclear Receptor Superfamily: The Second Decade. Cell (1995) 83(6):835. doi: 10.1016/0092-8674(95)90199-x
11. Bernardo TJ, Dubrovsky EB. The Drosophila Juvenile Hormone Receptor Candidates Methoprene-Tolerant (MET) and Germ Cell-Expressed (GCE) Utilize A Conserved LIXXL Motif to Bind The FTZ-F1 Nuclear Receptor. J Biol Chem (2012) 287(10):7821–33. doi: 10.1074/jbc.m111.327254
12. Pick L, Anderson WR, Shultz J, Woodard CT. The Ftz-F1 Family: Orphan Nuclear Receptors Regulated by Novel Protein–Protein Interactions. Adv Dev Biol (2006) 16(06):255–96. doi: 10.1016/S1574-3349(06)16008-1
13. Woodard CT, Baehrecke EH, Thummel CS. A Molecular Mechanism for The Stage Specificity of The Drosophila Prepupal Genetic Response to Ecdysone. Cell (1994) 79(4):607–15. doi: 10.1016/0092-8674(94)90546-0
14. Stock D, Misir V, Johnson S. Optimising FNA Processing – A Collection Fluid Allowing Giemsa, PAP and H & E Staining, and Facilitating Thinprep, Cytospin and Direct Smears and Ancillary Tests. Cytopathology (2010) 11(6):523–4. doi: 10.1046/j.1365-2303.2000.00286.x
15. Hiruma K, Riddiford LM. Regulation of Transcription Factors MHR4 and Betaftz-F1 by 20-Hydroxyecdysone During a Larval Molt in The Tobacco Hornworm. Manduca Sexta Dev Biol (2001) 232(1):265–74. doi: 10.1006/dbio.2001.0165
16. Chen J, Li TC, Pang R, Yue XZ, Hu J, Zhang WQ. Genome-Wide Screening and Functional Analysis Reveal That The Specific Microrna Nlu-Mir-173 Regulates Molting by Targeting Ftz-F1 in Nilaparvata Lugens. Front Physiol (2018) 9:1854. doi: 10.3389/fphys.2018.01854
17. Sun GC, Hirose S, Ueda H. Intermittent Expression of Bmftz-F1, A Member of The Nuclear Hormone Receptor Superfamily During Development of The Silkworm Bombyx Mori. Dev Biol (1994) 162(2):426–37. doi: 10.1006/dbio.1994.1099
18. Li C, Kapitskaya MZ, Zhu J, Miura K, Segraves W, Raikhel AS. Conserved Molecular Mechanism for The Stage Specificity of The Mosquito Vitellogenic Response to Ecdysone. Dev Biol (2000) 224(1):96–110. doi: 10.1006/dbio.2000.9792
19. Weller J, Sun GC, Zhou B, Lan Q, Hiruma K, Riddiford LM. Isolation and Developmental Expression of Two Nuclear Receptors, MHR4 and Bftz-F1, in The Tobacco Hornworm, Manduca Sexta. Insect Biochem Mol Biol (2001) 31(8):827–37. doi: 10.1016/S0965-1748(00)00188-0
20. Cruz J, Nieva C, Mané-Padrós D, Martín D, Bellés X. Nuclear Receptor Bgftz-F1 Regulates Molting and The Timing of Ecdysteroid Production During Nymphal Development in The Hemimetabolous Insect Blattella Germanica. Dev Dyn (2010) 237(11):3179–91. doi: 10.1002/dvdy.21728
21. Lin T, Zhang C, Lin L, Feng Q, Zheng S. Cloning and Expression of The Nuclear Transcription Factor SlBftz-F1 cDNA From Spodoptera Litura. Insect Sci (2011) 18(006):635–44. doi: 10.1111/j.1744-7917.2011.01412.x
22. Dubrovsky EB, Dubrovskaya VA, Bernardo T, Otte V, Difilippo R, Bryan H. The Drosophila Ftz-F1 Nuclear Receptor Mediates Juvenile Hormone Activation of E75A Gene Expression Through an Intracellular Pathway. J Biol Chem (2011) 286(38):33689. doi: 10.1074/jbc.M111.273458
23. Liu XP, Fu KY, Lü FG, Meng QW, Guo WC, Li GQ. Involvement of FTZ-F1 in The Regulation of Pupation in Leptinotarsa Decemlineata (Say). Insect Biochem Mol Biol (2014) 55:51–60. doi: 10.1016/j.ibmb.2014.10.008
24. Tan A, Palli SR. Identification and Characterization of Nuclear Receptors From The Red Flour Beetle, Tribolium Castaneum. Insect Biochem Mol Biol (2008) 38(4):430–9. doi: 10.1016/j.ibmb.2007.09.012
25. Val P, Lefrancois-Martinez AM, Veyssière G, Martinez A. SF-1 A Key Player in The Development and Differentiation of Steroidogenic Tissues. Nucl Recept (2003) 1(1):8. doi: 10.1186/1478-1336-1-8
26. Parvy JP, Blais C, Bernard F, Warren JT, Petryk A, Gilbert LI, et al. A Role for Betaftz-F1 in Regulating Ecdysteroid Titers During Post-Embryonic Development in Drosophila Melanogaster. Dev Biol (2005) 282(1):84–94. doi: 10.1016/j.ydbio.2005.02.028
27. Asahina M, Ishihara T, Jindra M, Kohara Y, Katsura I, Hirose S. The Conserved Nuclear Receptor Ftz-F1 Is Required for Embryogenesis, Moulting and Reproduction in Caenorhabditis Elegans. Genes Cells (2010) 5(9):711–23. doi: 10.1046/j.1365-2443.2000.00361.x
28. Gissendanner CR, Sluder AE. Nhr-25, The Caenorhabditis Elegans Ortholog of Ftz-F1, Is Required for Epidermal and Somatic Gonad Development. Dev Biol (2000) 221(1):259–72. doi: 10.1006/dbio.2000.9679
29. Parker KL, Rice DA, Lala DS, Ikeda Y, Luo X, Wong M, et al. Steroidogenic Factor 1: An Essential Mediator of Endocrine Development. Recent Prog Horm Res (2002) 57(1):19–36. doi: 10.1210/rp.57.1.19
30. Zhu J, Chen L, Raikhel AS. Posttranscriptional Control of The Competence Factor Bftz-F1 by Juvenile Hormone in The Mosquito Aedes Aegypti. Proc Natl Acad Sci USA (2003) 100(23):13338–43. doi: 10.1073/pnas.2234416100
31. Knapp EM, Li W, Singh V, Sun JJ. Nuclear Receptor Ftz-F1 Promotes Follicle Maturation and Ovulation Partly Via Bhlh/PAS Transcription Factor Sim. Elife (2020) 9:e54568. doi: 10.7554/elife.54568.sa1
32. Xu J, Tan A, Palli SR. The Function of Nuclear Receptors in Regulation of Female Reproduction and Embryogenesis in The Red Flour Beetle, Tribolium Castaneum. J Insect Physiol (2010) 56(10):1471–80. doi: 10.1016/j.jinsphys.2010.04.004
33. Wang Y, Kocher SD, Linksvayer TA, Grozinger CM, Page RE, Amdam GV. Regulation of Behaviorally Associated Gene Networks in Worker Honey Bee Ovaries. J Exp Biol (2012) 215(1):124–34. doi: 10.1242/jeb.060889
34. Zhu J, Li C, Sun G, Raikhel AS. The Competence Factor βftz-F1 Potentiates Ecdysone Receptor Activity Via Recruiting a P160/SRC Coactivator. Mol Cell Biol (2006) 26(24):9402–12. doi: 10.1128/MCB.01318-06
35. Qing L, Jing X, Lin H, Yuan W, Hongdan Y, Zelin D, et al. FOXL2 Down-Regulates Vitellogenin Expression at Mature Stage in Eriocheir Sinensis. Biosci Rep (2015) 35(6):e00278–8. doi: 10.1042/BSR20150151
36. Chung JS, Zmora N. Functional Studies of Crustacean Hyperglycemic Hormones (Chhs) of The Blue Crab, Callinectes Sapidus–The Expression and Release of CHH in Eyestalk and Pericardial Organ in Response to Environmental Stress. FEBS J (2010) 275(4):693–704. doi: 10.1111/j.1742-4658.2007.06231.x
37. Pamuru RR, Rosen O, Manor R, Chung JS, Zmora N, Glazer L, et al. Stimulation of Molt by RNAi of The Molt-Inhibiting Hormone in The Crayfish Cherax Quadricarinatus. Gen Comp Endocrinol (2012) 178(2):227–36. doi: 10.1016/j.ygcen.2012.05.007
38. Gao Y, Wei J, Yuan J, Zhang X, Li F, Xiang J. Transcriptome Analysis on The Exoskeleton Formation in Early Developmetal Stages and Reconstruction Scenario in Growth-Moulting in Litopenaeus Vannamei. Sci Rep (2017) 7(1):1098. doi: 10.1038/s41598-017-01220-6
39. Fu H, Jiang SF, Xiong YW. Current Status and Prospects of Farming The Giant River Prawn (Macrobrachium Rosenbergii) and The Oriental River Prawn (Macrobrachium Nipponense) in China. Aquac Res (2012) 43(7):993–8. doi: 10.1111/j.1365-2109.2011.03085.x
40. Qiao H, Jiang F, Xiong Y, Jiang S, Fu H, Li F, et al. Characterization, Expression Patterns of Molt-Inhibiting Hormone Gene of Macrobrachium Nipponense and Its Roles in Molting and Growth. PloS One (2018) 13(6):e0198861. doi: 10.1371/journal.pone.0198861
41. Yuan H, Qiao H, Fu Y, Fu H, Zhang W, Jin S, et al. RNAi Shows That Spook, The Precursor Gene of 20-Hydroxyecdysone (20e), Regulates The Molting of Macrobrachium Nipponense. J Steroid Biochem Mol Biol (2021) 213:105976. doi: 10.1016/j.jsbmb.2021.105976
42. Yang J, Zhang Y. I-TASSER Server: New Development for Protein Structure and Function Predictions. Nucleic Acids Res (2015) 43(W1):W174–81. doi: 10.1093/nar/gkv342
43. Zhang C, Freddolino PL, Zhang Y. COFACTOR: Improved Protein Function Prediction by Combining Structure, Sequence and Protein–Protein Interaction Information. Nucleic Acids Res (2017) 45(W1):W291–9. doi: 10.1093/nar/gkx366
44. Kastner P, Mark M, Chambon P. Nonsteroid Nuclear Receptors: What Are Genetic Studies Telling Us About Their Role in Real Life? Cell (1995) 83(6):859–69. doi: 10.1016/0092-8674(95)90202-3
45. Thummel CS. From Embryogenesis to Metamorphosis: The Regulation and Function of Drosophila Nuclear Receptor Superfamily Members. Cell (1995) 83(6):871–7. doi: 10.1016/0092-8674(95)90203-1
46. McEwan IJ, Escriva H, Bertrand S, Laudet V. The Evolution of The Nuclear Receptor Superfamily. Essays Biochem (2004) 40:11–26. doi: 10.1042/bse0400011
47. Solomon IH, Hager JM, Safi R, McDonnell DP, Redinbo MR, Ortlund EA. Crystal Structure of The Human LRH-1 DBD–DNA Complex Reveals Ftz-F1 Domain Positioning is Required for Receptor Activity. J Mol Biol (2005) 354(5):1091–102. doi: 10.1016/j.jmb.2005.10.009
48. Broadus J, McCabe JR, Endrizzi B, Thummel CS, Woodard CT. The Drosophila Bftz-F1 Orphan Nuclear Receptor Provides Competence for Stage-Specific Responses to The Steroid Hormone Ecdysone. Mol Cell (1999) 3(2):143–9. doi: 10.1016/S1097-2765(00)80305-6
49. Berghout HE. Drosophila Ovarian Stem Cell Establishment Is Regulated by Nuclear Hormone Receptor Ftz-F1. (2017). Available at: http://hdl.handle.net/10342/6383.
50. Borras-Castells F, Nieva C, Maestro JL, Maestro O, Belles X, Martín D. Juvenile Hormone Biosynthesis in Adult Blattella Germanica Requires Nuclear Receptors Seven-Up and FTZ-F1. Sci Rep (2017) 7(1):40234. doi: 10.1038/srep40234
51. Mello TRP, Aleixo AC, Pinheiro DG, Nunes FMF, Cristino AS, Bitondi MMG, et al. Hormonal Control and Target Genes of Ftz-F1 Expression in The Honeybee Apis Mellifera: A Positive Loop Linking Juvenile Hormone, Ftz-F1, and Vitellogenin. Insect Mol Biol (2019) 28(1):145–59. doi: 10.1111/imb.12541
52. Riddiford LM, Hiruma K, Zhou X, Nelson CA. Insights Into The Molecular Basis of The Hormonal Control of Molting and Metamorphosis From Manduca Sexta and Drosophila Melanogaster. Insect Biochem Mol Biol (2003) 33(12):1327–38. doi: 10.1016/j.ibmb.2003.06.001
53. Chen L, Reece C, O’Keefe SL, Hawryluk GWL, Engstrom MM, Hodgetts RB. Induction of The Early–Late Ddc Gene During Drosophila Metamorphosis by The Ecdysone Receptor. Mech Dev (2002) 114(1-2):95–107. doi: 10.1016/S0925-4773(02)00064-3
54. Ou Q, King-Jones K. What Goes Up Must Come Down: Transcription Factors Have Their Say in Making Ecdysone Pulses. Curr Top Dev Biol (2013) 103:35–71. doi: 10.1016/B978-0-12-385979-2.00002-2
55. Yamada M, Murata T, Hirose S, Lavorgna G, Suzuki E, Ueda H. Temporally Restricted Expression of Transcription Factor Betaftz-F1: Significance for Embryogenesis, Molting and Metamorphosis in Drosophila Melanogaster. Development (2000) 127(23):5083–92. doi: 10.1046/j.1365-2303.2000.00286.x
56. Zhang X, Kang X, Wu H, Silver K, Zhang J, Ma E, et al. Transcriptome-Wide Survey, Gene Expression Profiling and Exogenous Chemical-Induced Transcriptional Responses of Cytochrome P450 Superfamily Genes in Migratory Locust (Locusta Migratoria). Insect Biochem Mol Biol (2018) 100:66–77. doi: 10.1016/j.ibmb.2018.06.006
57. Beachum AN, Whitehead KM, McDonald SI, Phipps DN, Berghout HE, Ables ET. Orphan Nuclear Receptor Ftz-F1 (NR5A3) Promotes Egg Chamber Survival in The Drosophila Ovary. G3 (2021) 11(2):jkab003. doi: 10.1093/g3journal/jkab003
58. Lavorgna G, Karim FD, Thummel CS, Wu C. Potential Role for A FTZ-F1 Steroid Receptor Superfamily Member in The Control of Drosophila Metamorphosis. Proc Natl Acad Sci USA (1993) 90(7):3004–8. doi: 10.1073/pnas.90.7.3004
59. Cheng D, Xia Q, Duan J, Ling W, Chong H, Li Z, et al. Nuclear Receptors in Bombyx Mori: Insights Into Genomic Structure and Developmental Expression. Insect Biochem Mol Biol (2008) 38(12):1130–7. doi: 10.1016/j.ibmb.2008.09.013
60. Kusaba M. RNAi in Crop Plants. Curr Opin Biotechnol (2004) 15(2):139–43. doi: 10.1016/j.copbio.2004.02.004
61. Sun S, Wu Y, Fu H, Yang M, Ge X, Zhu J, et al. Evaluating Expression of Autophagy-Related Genes in Oriental River Prawn Macrobrachium Nipponense As Potential Biomarkers for Hypoxia Exposure. Ecotoxicol Environ Saf (2019) 171:484–92. doi: 10.1016/j.ecoenv.2018.12.079
62. Jin SB, Hu YN, Fu HT, Jiang SF, Xiong YW, Qiao H, et al. Potential Functions of Gem-Associated Protein 2-Like Isoform X1 in The Oriental River Prawn Macrobrachium Nipponense: Cloning, Qpcr, In Situ Hybridization, and Rnai Analysis. Int J Mol Sci (2019) 20(16):3995. doi: 10.3390/ijms20163995
63. Niwa R, Matsuda T, Yoshiyama T, Namiki T, Mita K, Fujimoto Y, et al. CYP306A1, A Cytochrome P450 Enzyme, Is Essential for Ecdysteroid Biosynthesis in The Prothoracic Glands of Bombyx and Drosophila. J Biol Chem (2004) 279(34):35942–9. doi: 10.1074/jbc.M404514200
64. Bai HK, Qiao H, Li FJ, Fu HT, Sun SM, Zhang WY, et al. Molecular Characterization and Developmental Expression of Vitellogenin in The Oriental River Prawn Macrobrachium Nipponense and The Effects of RNAi and Eyestalk Ablation on Ovarian Maturation. Gene (2015) 562(1):22–31. doi: 10.1016/j.gene.2014.12.008
65. Tsukimura B. Crustacean Vitellogenesis: Its Role in Oocyte Development. Am Zool (2001) 41(3):465–76. doi: 10.1093/icb/41.3.465
66. Akagi K, Sarhan M, Sultan AR, Nishida H, Koie A, Nakayama T, et al. A Biological Timer in The Fat Body Comprising Blimp-1, Bftz-F1 and Shade Regulates Pupation Timing in Drosophila Melanogaster. Development (2016) 143(13):2410–6. doi: 10.1242/dev.133595
67. Talamillo A, Herboso L, Pirone L, Pérez C, Gonzalez M, Sánchez J, et al. Scavenger Receptors Mediate The Role of Sumo and Ftz-F1 in Drosophila Steroidogenesis. PloS Genet (2013) 9:e1003473. doi: 10.1371/journal.pgen.1003473
68. Sultan ARS, Oish Y, Ueda H. Function of The Nuclear Receptor FTZ-F1 During The Pupal Stage in Drosophila Melanogaster. Dev Growth Differ (2014) 56(3):245–53. doi: 10.1111/dgd.12125
69. Cho KH, Daubnerová I, Park Y, Zitnan D, Adams ME. Secretory Competence in A Gateway Endocrine Cell Conferred by The Nuclear Receptor Bftz-F1 Enables Stage-Specific Ecdysone Responses Throughout Development in Drosophila. Dev Biol (2014) 385(2):253–62. doi: 10.1016/j.ydbio.2013.11.003
70. Bairoch A, Bucher P, Hofmann K. The PROSITE Database, Its Status in 1997. Nucleic Acids Res (1997) 25(1):217–21. doi: 10.1093/nar/25.1.217
71. Combet C, Blanchet C, Geourjon C, Deléage G. NPS@: Network Protein Sequence Analysis. Trends Biochem Sci (2000) 25(3):147–50. doi: 10.1016/S0968-0004(99)01540-6
72. Hu YN, Fu HT, Qiao H, Sun SM, Zhang WY, Jin SB, et al. Validation and Evaluation of Reference Genes for Quantitative Real-Time PCR in Macrobrachium Nipponense. Int J Mol Sci (2018) 19(8):2258. doi: 10.3390/ijms19082258
73. Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and The 2–ΔΔct Method. Methods (2001) 25(4):402–8. doi: 10.1006/meth.2001.1262
74. Qiao H, Fu H, Xiong Y, Jiang S, Sun S, Jin S, et al. Molecular Insights Into Reproduction Regulation of Female Oriental River Prawns Macrobrachium Nipponense Through Comparative Transcriptomic Analysis. Sci Rep (2017) 7(1):1–11. doi: 10.1038/s41598-017-10439-2
75. Li F, Qiao H, Fu H, Sun S, Zhang W, Jin S, et al. Identification and Characterization of Opsin Gene and Its Role in Ovarian Maturation in The Oriental River Prawn Macrobrachium Nipponense. Comp Biochem Physiol B Biochem Mol Biol (2018) 218:1–12. doi: 10.1016/j.cbpb.2017.12.016
76. Ma H, Abbas MN, Zhang K, Hu X, Xu M, Liang H, et al. 20-Hydroxyecdysone Regulates The Transcription of The Lysozyme Via Broad-Complex Z2 Gene in Silkworm, Bombyx Mori. Dev Comp Immunol (2019) 94:66–72. doi: 10.1016/j.dci.2019.01.014
77. Tian L, Guo E, Wang S, Liu S, Jiang RJ, Cao Y, et al. Developmental Regulation of Glycolysis by 20-Hydroxyecdysone and Juvenile Hormone in Fat Body Tissues of The Silkworm. Bombyx Mori. J Mol Cell Biol (2010) 2(5):255–63. doi: 10.1093/jmcb/mjq020
78. Jin X, Wu X, Zhou L, He T, Yin Q, Liu S. 20-Hydroxyecdysone-Responsive Micrornas of Insects. RNA Biol (2020) 17(10):1454–71. doi: 10.1080/15476286.2020.1775395
79. Sun S, Xuan F, Fu H, Zhu J, Ge X. Molecular Cloning and Functional Characterization of A Hexokinase From The Oriental River Prawn Macrobrachium Nipponense in Response to Hypoxia. Int J Mol Sci (2017) 18(6):1256. doi: 10.3390/ijms18061256
Keywords: 20E, Macrobrachium nipponense, MnFtz-f1, RNAi, molt, ovulation
Citation: Yuan H, Zhang W, Fu Y, Jiang S, Xiong Y, Zhai S, Gong Y, Qiao H, Fu H and Wu Y (2021) MnFtz-f1 Is Required for Molting and Ovulation of the Oriental River Prawn Macrobrachium nipponense. Front. Endocrinol. 12:798577. doi: 10.3389/fendo.2021.798577
Received: 20 October 2021; Accepted: 01 December 2021;
Published: 20 December 2021.
Edited by:
Jie Ma, University of Idaho, United StatesReviewed by:
Xiaojian Gao, Yangzhou University, ChinaZhimin Gu, Zhejiang Institute of Freshwater Fisheries, China
Chun Zhang, Hunan Normal University, China
Copyright © 2021 Yuan, Zhang, Fu, Jiang, Xiong, Zhai, Gong, Qiao, Fu and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Qiao, cWlhb2hAZmZyYy5jbg==; Hongtuo Fu, ZnVodEBmZnJjLmNu
 Huwei Yuan
Huwei Yuan Wenyi Zhang2
Wenyi Zhang2 Sufei Jiang
Sufei Jiang Hui Qiao
Hui Qiao Hongtuo Fu
Hongtuo Fu