- 1Department of Obstetrics, Women’s Hospital of Nanjing Medical University, Nanjing Maternity and Child Health Care Hospital, Nanjing, China
- 2State Key Laboratory of Reproductive Medicine, Institute of Toxicology, Nanjing Medical University, Nanjing, China
- 3Key Laboratory of Modern Toxicology of Ministry of Education, School of Public Health, Nanjing Medical University, Nanjing, China
- 4Department of Pathology, Women’s Hospital of Nanjing Medical University, Nanjing Maternity and Child Health Care Hospital, Nanjing, China
- 5Department of Urology, Stanford University Medical Center, Stanford, CA, United States
- 6Department of Andrology, Women’s Hospital of Nanjing Medical University, Nanjing Maternity and Child Health Care Hospital, Nanjing, China
- 7Department of Reproductive Medicine, Women’s Hospital of Nanjing Medical University, Nanjing Maternity and Child Health Care Hospital, Nanjing, China
Background: Growing evidence has indicated that epigenetic factors might be associated with the pathophysiology of idiopathic nonobstructive azoospermia (iNOA). As the most common RNA modification, N6-methyladenosine (m6A) methylation has recently attracted more attention in the regulation of spermatogenesis; however, its role in the mechanisms of iNOA is still unclear.
Objective: To determine the differential expression of mRNA and m6A methylation status in the testes of iNOA patients.
Methods: Testes tissues from diagnosed iNOA and controlled obstructive azoospermia (OA) patients were collected and grouped according to the histological examinations. Total RNA was isolated and quantified by an m6A RNA Methylation Quantification Kit. The expression level of mRNAs was detected by qRT-PCR analysis. Differentially expressed m6A genes were analyzed using the human ArrayStar m6A epitranscriptomic microarray, and bioinformatics analyses were applied.
Results: A total of 36 iNOA and 8 controlled patients were included. The global expression of m6A in the iNOA group was significantly decreased. A dosage relationship was observed between the m6A decline and the degree of impaired spermatogenesis, with the successive process of normal spermatogeneis, hypospermatogenesis (HP), maturation arrest (MA), and Sertoli cell-only syndrome (SO). Four down-expressed genes (BDNF, TMEM38B, RPL3L, and C22orf42) displayed significantly lower expression of m6A methylation. Additionally, they also showed a gradually down-expressed tendency in the three groups (OA, HP, SO/MA groups). Moreover, m6A reader EIF3A was approved to have differential expression through microarrays analysis, which was consistent with the result from the qRT-PCR test.
Conclusions: The m6A expression was gradually downregulated in the testes tissue from iNOA patients in accordance with the degree of spermatogenic dysfunction. The determined differential expression of mRNA and m6A methylation status may represent potentially novel molecular targets for the mechanism study of iNOA in the epigenetic level, which could benefit the understanding of the pathophysiology of iNOA.
Introduction
Infertility affects approximately 15% of couples of reproductive age worldwide, where male factors account for up to 50% of cases (1). As the most severe condition, nonobstructive azoospermia (NOA) contributes to 10-15% of male infertility, with an incidence rate of 1% in the adult male population (2). There are multiple identified factors that could lead to NOA, including chromosomal abnormalities (Klinefelter syndrome, Y-chromosome microdeletions, etc.), cryptorchidism, and chemotherapy or radiotherapy history. However, collectively, these reasons may account for just 30% of NOA instances (2, 3). With ineffective medical therapy and unpredictable sperm extraction rates, the remaining idiopathic NOA (iNOA) cases whose pathological results include Sertoli cell-only syndrome (SO), maturation arrest (MA), and hypospermatogenesis (HP), are the most challenging and frustrating ones in the clinical practice.
With the advancement of epigenomics, it is becoming increasingly apparent that the mechanisms of iNOA might be accompanied by epigenetic variables (4). Generally, the epigenetic modification includes DNA methylation, histone modifications, and non-coding RNAs (5, 6). Nowadays, RNA modifications have shown a critical role in the epigenetic programming of spermatogenesis, with N6-methyladenosine (m6A) being the most common (7, 8). With three well-established regulation patterns, namely m6A writers, erasers, and readers, m6A has shown its extensive function in mRNA splicing and stability, microRNA processing, and so on. In spermatogenesis research, the deletion of m6A writers, Mettl3 and Mettl14, could disrupt spermiogenesis through the experiments in vitro (9), and the ablation of Mettl3 could severely inhibit meiosis in the germ cells (10).
Furthermore, the m6A eraser ALKBH5 had an essential role in correct splicing in the round spermatids phase (11). In addition, the elevated expression of Mettl3 and Mettl14 was identified in the semen of asthenozoospermia patients, indicating the correlation between m6A and sperm quality (12). However, it is still unclear how m6A affects the spermiogenesis process; to our knowledge, there have been only a few reports so far on the subjects.
In this case-control study, we collected the testes tissues from iNOA patients and controlled obstructive azoospermia (OA) patients. Additionally, the testes tissues from iNOA patients were divided into SO/MA or HP groups, indicating the extent of spermatogenesis damage according to the pathological findings. m6A and mRNA expression profiles were then tested among all the groups by applying ArrayStar m6A microarrays, and verified by qRT-PCR. Bioinformatic analysis was also employed to predict the differentially expressed genes involved in the regulation of m6A methylation for spermatogenesis in iNOA.
Materials and methods
Testes tissue collection
Approved by the Ethics Committee of Women’s Hospital of Nanjing Medical University, Nanjing Maternity and Child Health Care Hospital (2019KY-027), this study was conducted in the reproductive medical center of Nanjing Maternity and Child Health Care Hospital from November 2019 to February 2021. For the case group, patients with male infertility who had been diagnosed with azoospermia were enrolled. The semen analysis, with a complete lack of sperm in the ejaculate at least twice, combined with hormonal evaluation, was used to determine the diagnosis of iNOA. The exclusion criteria include 1) obstructive azoospermia (OA), which diagnosis was based on the physical examination, sex hormone testing, and transrectal ultrasound performed by a skilled urologist; 2) NOA with genetic causes (abnormal karyotype, pathogenic Y-chromosomal microdeletions, etc.); 3) NOA with a detectable disorder affecting hypothalamic-pituitary-gonadal axis based on hormonal evaluation and imaging diagnoses (hypogonadotropic hypogonadism, hyperprolactinemia); 4) NOA caused by secondary testicular failures, such as cryptorchidism and iatrogenic factors (chemotherapy, radiotherapy). Besides, the OA patients who chose ICSI with testicular sperm extraction (TESE) were included in the control group, and who chose to undertake anastomosis were excluded in this study. All the participants’ testes tissues were collected using the residual tissues after the TESE operation with written informed consent.
Histological analysis
All collected testes tissues were fixed in 4% Paraformaldehyde (PFA) for 24 hours and then embedded with paraffin after dehydration. Next, samples were cut into 4-micron sections and stained with hematoxylin and eosin (H&E) using a fully automatic H&E staining machine and Dako CoverStainer (Agilent Technologies, Inc., USA). The improved Mayer hematoxylin (CS700, Dako) was used in the process. Finally, the stained slides were evaluated, and the images were analyzed under a light microscope (Eclipse 80i, Nikon, Japan) at 100× to 400× magnifications.
RNA extraction and m6A quantification of the overall m6A levels
Total RNA was isolated from each testicular sample using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The concentration and purity of total RNA samples were determined using NanoDrop 2000 (Thermo Fisher Scientific, Inc.). The total RNA samples were stored at -80˚C for subsequent experiments. The m6A quantification was conducted using the m6A RNA Methylation Quantification Kit (Epigentek) following the manufacturer’s protocol. Briefly, 200 ng total RNA sample was added to each test well, followed by the addition of capture antibody and test antibody in turn, and then the reaction was stopped with stop solution. Finally, the absorbance at 450 nm of the test wells was detected by a microplate reader to calculate the percentage of m6A in total RNA.
Analysis of related mRNA using qRT-PCR
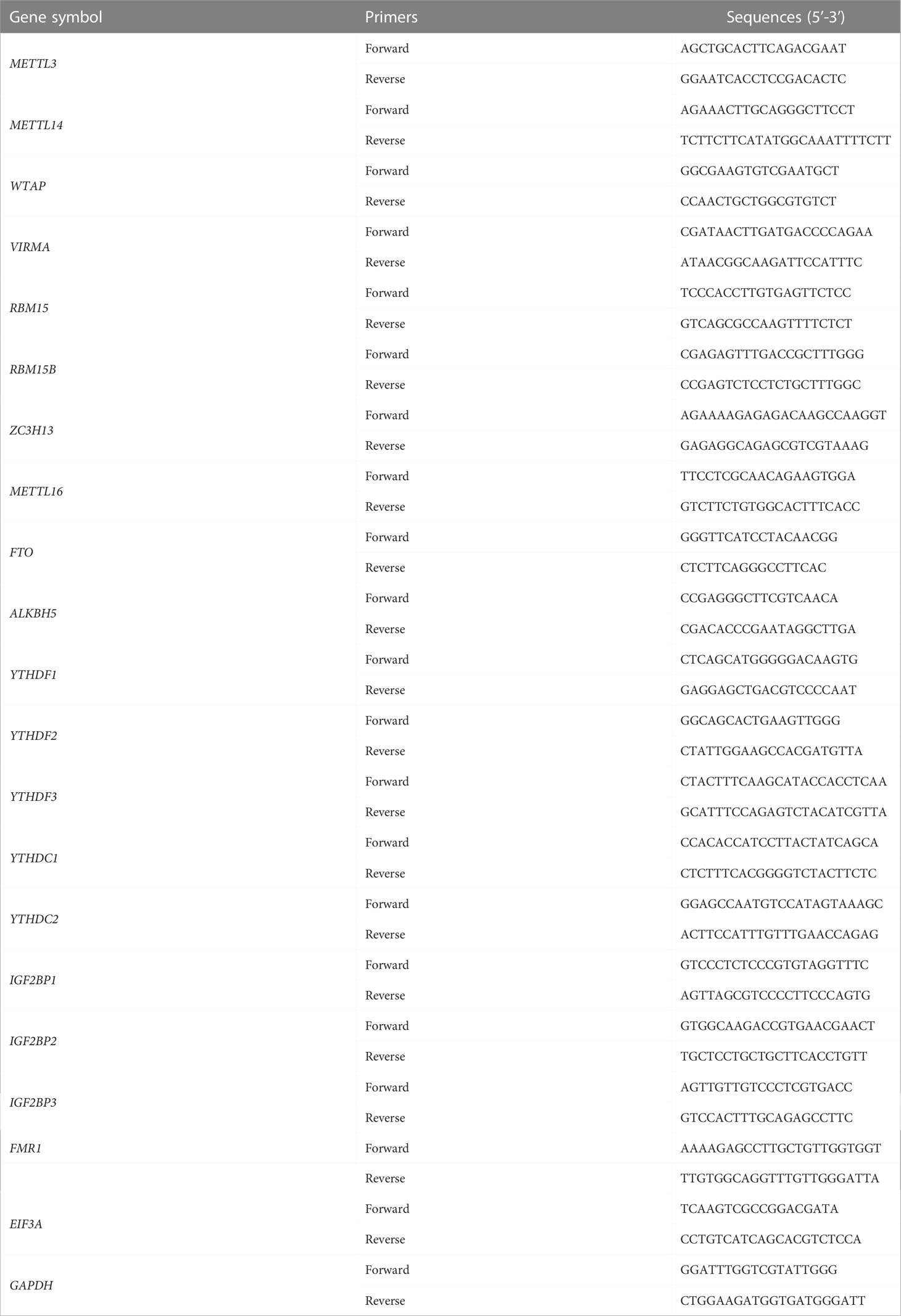
The reverse transcription step to cDNA was performed using the PrimeScript RT reagent Kit (Takara, Tokyo, Japan), and qRT-PCR analysis was conducted by SYBR® Premix Ex Taq™ on the LightCycler 480 II real-time fluorescent quantitative PCR system (Roche Diagnostics). The 2-ΔCt method was used to calculate the relative expression level of related mRNAs. GAPDH was used as an internal control for mRNA quantification. All the reactions were run in duplicate. The primers used for qRT-PCR in this study were verified by previous published papers which sequences were listed in Table 1.
ArrayStar m6A epitranscriptomic microarray analysis
The sample preparation and microarray hybridization were performed according to Arraystar’s standard protocols. Briefly, the total RNAs were immunoprecipitated with anti-m6A antibody. Next, the modified RNAs “IP” and the unmodified RNAs “Sup” were labeled with Cy5 and Cy3 using Arraystar Super RNA Labeling Kit. The synthesized cRNAs were then hybridized onto Arraystar Human mRNA&lncRNA Epitranscriptomic Microarray (8×60K, Arraystar). Then, the arrays were scanned using the Agilent Scanner G2505C system.
Agilent Feature Extraction software (version 11.0.1.1) was used to analyze microarray data. First, raw intensities of IP and Sup were normalized and log2-transformed. After Spike-in normalization, the probe signals having Present (P) or Marginal (M) QC flags in at least 3 out of 9 samples were retained as “All Targets Value” for further analyses. Then it was followed by the following procedures: 1) the “m6A methylation level” was calculated for the percentage of modification based on the IP and Sup normalized intensities; 2) the “m6A quantity” was calculated for the m6A methylation amount based on the IP normalized intensities; 3) the “expression level” was calculated based on the total of IP and Sup normalized intensities of RNA. Differentially m6A-methylated RNAs or RNAs expression among the three groups were compared by calculating the fold change (FC) with a cutoff of 2-fold (P < 0.05). Finally, the clustering analysis heatmaps and the Venn diagram were performed to show the distinguishable m6A methylation, m6A quantity, and mRNA expression among the three groups.
Bioinformatic analysis
Gene Ontology (GO) terms, which encompass molecular function, cellular structure, and cellular processes, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment analyses were performed using DAVID (https://david.ncifcrf.gov/). To assess the importance of genes whose m6A methylation level was significantly downregulated in the iNOA group, we established an evaluating system using five pieces of evidence that has hierarchical significance (i to v), based on the tendency and cooperativity in the scales of m6A methylation, m6A quantity, and mRNA expression for differentially expressed genes.
Statistical analysis
Data were expressed as mean ± SEM. Mann-Whitney U test was used for continuous variables and the Chi-square test for categorical variables (GraphPad software, San Diego, California). One-way ANOVA was used for multiple group comparisons. A P-value <0.05 was considered statistically significant (*P < 0.05).
Results
The testes tissues from 36 iNOA and 8 OA patients were collected in this study. And their demographic characteristics are listed in Table 2. The histological results included 7 SO, 17 MA, and 12 HP in iNOA patients. Among them, testes tissues from 30 iNOA patients with 6 SO, 15 MA, and 9 HP cases, and 5 randomly selected OA patients were used for the m6A quantification experiment. The tissue samples from the remaining 6 iNOA containing 1 SO, 2 MA, and 3 HP, as well as 3 OA patients were used to perform the ArrayStar m6A microarrays. No significant differences could be seen in the comparison of age and BMI in different groups, while the smoking and alcohol drinking status of OA groups were significantly lower (P < 0.05). In addition, the LH and FSH values were lower in OA and HP groups when compared with that in the SO or MA groups (P < 0.05). Also, the volumes of testes in OA groups were significantly more prominent than in other groups (P < 0.05).
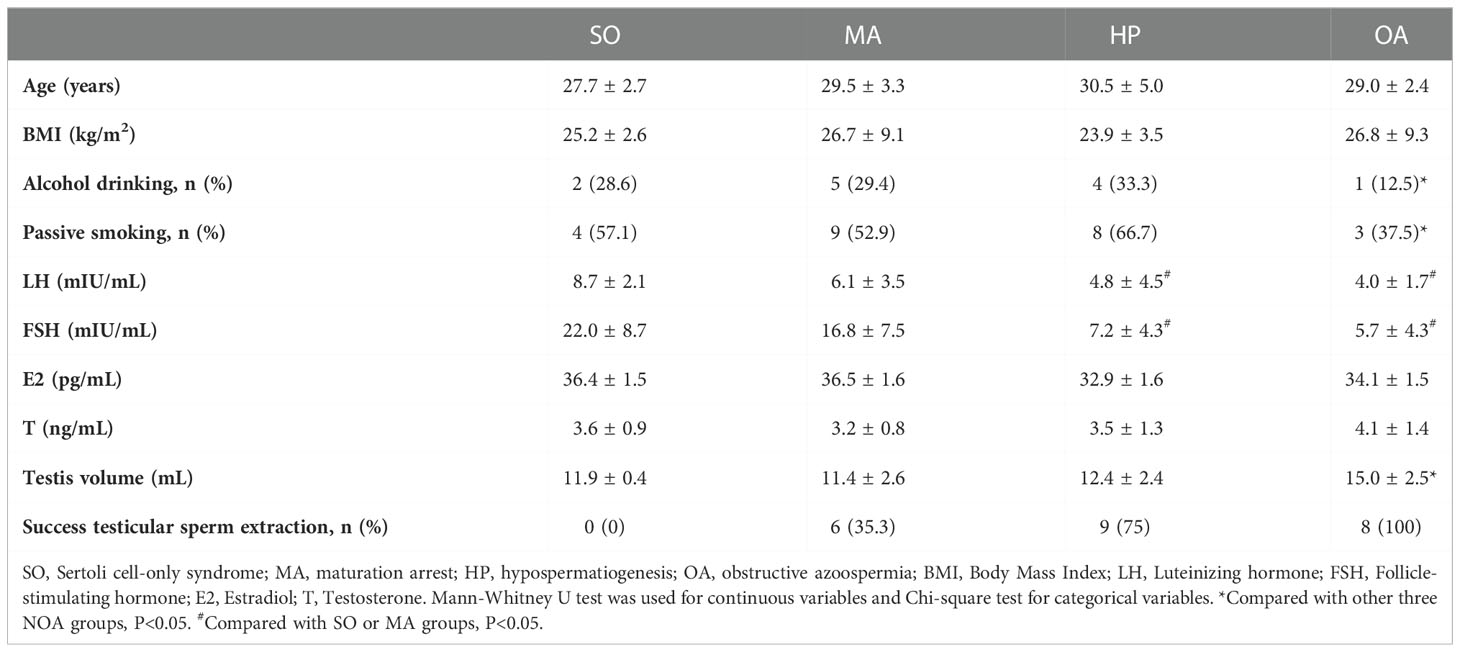
Table 2 Baseline characteristics, gonadal hormone concentrations, testes volumes and the success testicular sperm extraction rates of all the patients.
The expression of m6A was downregulated in iNOA patients’ testes
The global expression of m6A in testes from iNOA patients was significantly decreased compared to that in the OA group. After dividing the iNOA group into three subgroups according to the pathological types, it was discovered that the m6A expression gradually declined from OA to HP, MA, and SO groups, with a dosage relationship observed between the decline and the degree of spermatogenic dysfunction (Figure 1).
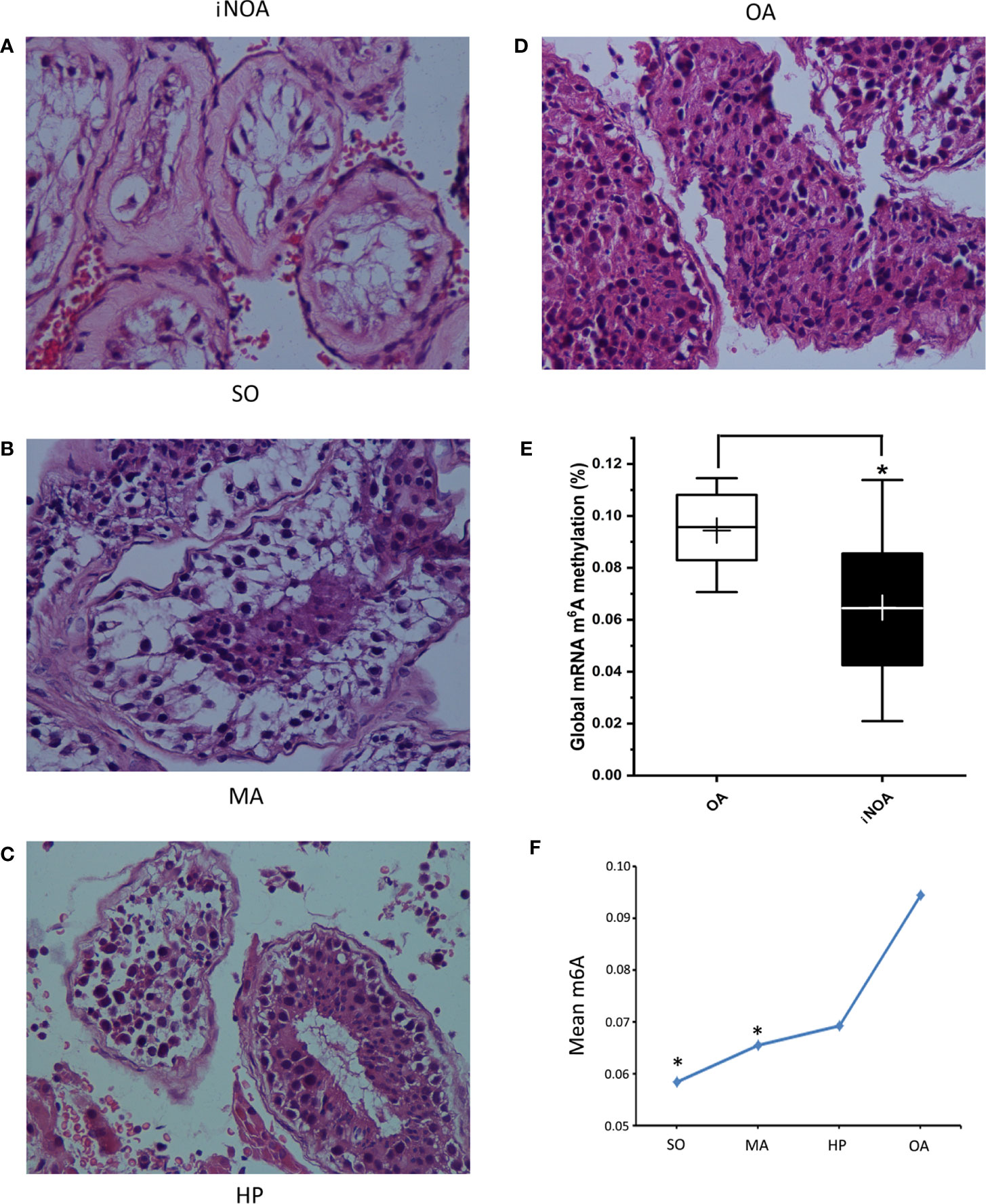
Figure 1 The iNOA group was divided into three groups according to the pathological types, namely the SO group (A), the MA group (B), and the HP group (C). (D) shows the histology of OA. All the four representative images are 400× magnifications using H&E staining. (E) shows the significantly decreased global expression of m6A in testes tissues from iNOA groups. (F) shows the a dosage relationship between the m6A expression decline and the degree of impaired spermatogenesis. SO: Sertoli cell-only syndrome, MA: maturation arrest, HP: hypospermatogenesis, OA: obstructive azoospermia. *P < 0.05 compared with OA group.
iNOA is associated with the alterations of mRNA expression and their m6A methylation status
The expression profiles of mRNA and their m6A methylation status in the SO/MA, HP, and OA groups were identified by the ArrayStar m6A microarrays. The three heatmaps showed the clustering analysis of m6A methylation, m6A quantity, and mRNA expression in the three groups (Figure 2).
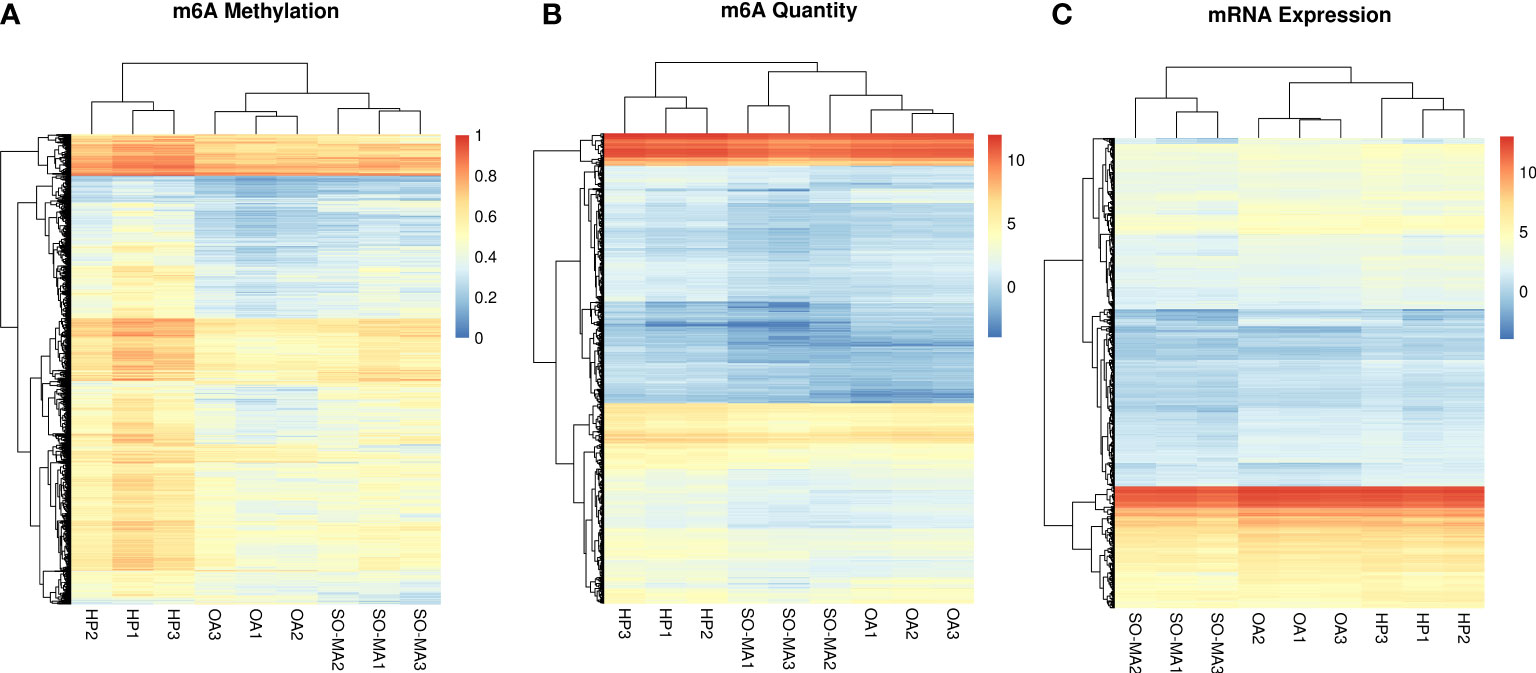
Figure 2 The clustering analysis heatmaps of m6A methylation (A), m6A quantity (B), and mRNA expression (C) in the SO/MA, HP, and OA groups.
As a result, there are 160 significantly up-expressed genes in the SO/MA group and 649 in the HP group compared to the OA group. The intersection of the two results found 31 up-expressed genes (Figure 3A). Meanwhile, 2469 significantly down-expressed genes from the SO/MA group and 230 ones from the HP group were identified compared to the OA group. Moreover, 287 significantly down-expressed genes were placed in the SO/MA group compared to the HP group. The intersection of the three results highlighted 4 down-expressed genes, which are listed in Figure 3B.

Figure 3 Bioinformatics analysis of differentially expressed genes and m6A methylation status in the three groups (the SO/MA, HP, and OA groups). (A) The Venn diagram of up-regulated mRNA gene in three groups, and the intersection area included 31 listed genes. (B) The Venn diagram of down-expressed mRNA gene in three groups, and the intersection area included four listed genes. (C) GO analysis of the 31 up-expressed and 4 down-expressed genes. (D) KEGG pathway analysis of 141 genes with significantly lower m6A methylation quantity both in the SO/MA and HP groups when compared to that in the OA group.
Furthermore, the Gene Ontology (GO) analysis was performed to investigate the potential functions of the 31 up-expressed and 4 down-expressed genes. As Figure 3C showed, the most significant GO terms related to molecular function and cellular components were protein binding, plasma membrane, and extracellular exosome. We mainly focused on the hypomethylated genes when comparing the difference in gene m6A methylation level or m6A quantity in three groups. As a result, 9 genes with significantly lower m6A methylation levels in the SO/MA group and 7 in the HP group were found compared to the OA group. In addition, there were 1937 genes with significantly lower m6A methylation quantity in the SO/MA group and 309 in the HP group compared to the OA group. The intersection of these two results included 141 genes, and their KEGG pathway analysis result was shown in Figure 3D. The most enriched KEGG pathway was the cell cycle.
To predict the genes with m6A methylation status alteration potentially involved in the mechanisms of iNOA, we further compared the genes with significantly decreased m6A methylation levels from the SO/MA and HP groups to the OA group. From Table 3, four genes (BDNF, TMEM38B, RPL3L, and C22orf42) displayed significantly lower m6A methylation status associated with decreased gene expression, which was accompanied by the gradual down-expressed tendency in the three groups, reflecting the degree of spermatogenic dysfunction.
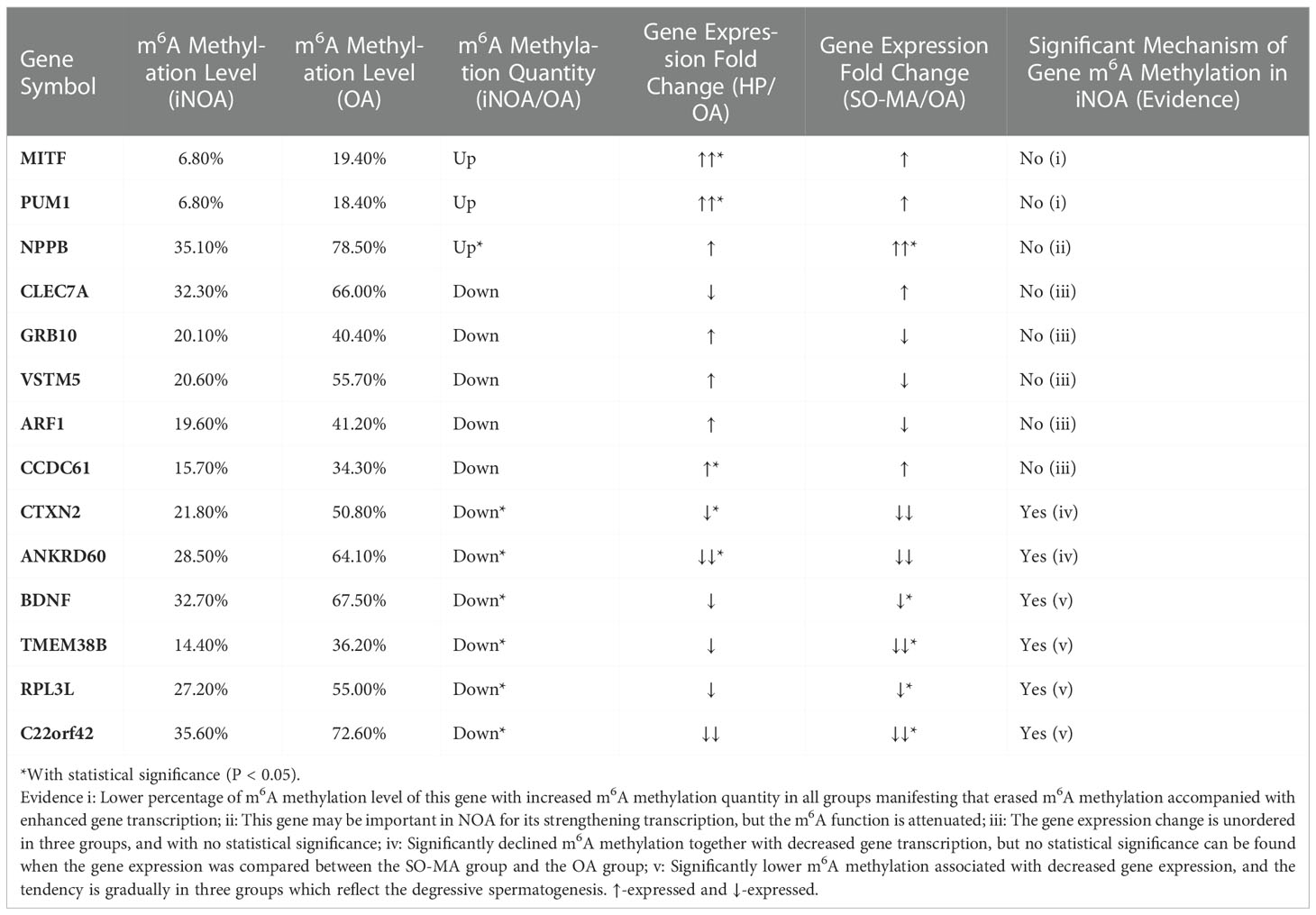
Table 3 To predict the importance of differentially expressed genes with significantly downregulated m6A Methylation levels in iNOA group using a 5-degree evidence evaluating system.
An m6A reader was upregulated in the testes of iNOA patients
Corresponding to the global decline of m6A expression in the iNOA group, the majority of m6A-related writers’ and readers’ genes were down-expressed in the iNOA groups, especially in the SO/MA group (Figure 4A). However, one m6A reader, eukaryotic translation initiation factor 3 subunit A (EIF3A), was significantly upregulated in the iNOA group, which was also verified by the qRT-PCR test. As for the significantly declined m6A writers (METTL3, WTAP, and RBM15), their ArrayStar m6A microarrays results were not testified by the qRT-PCR (Figure 4B).
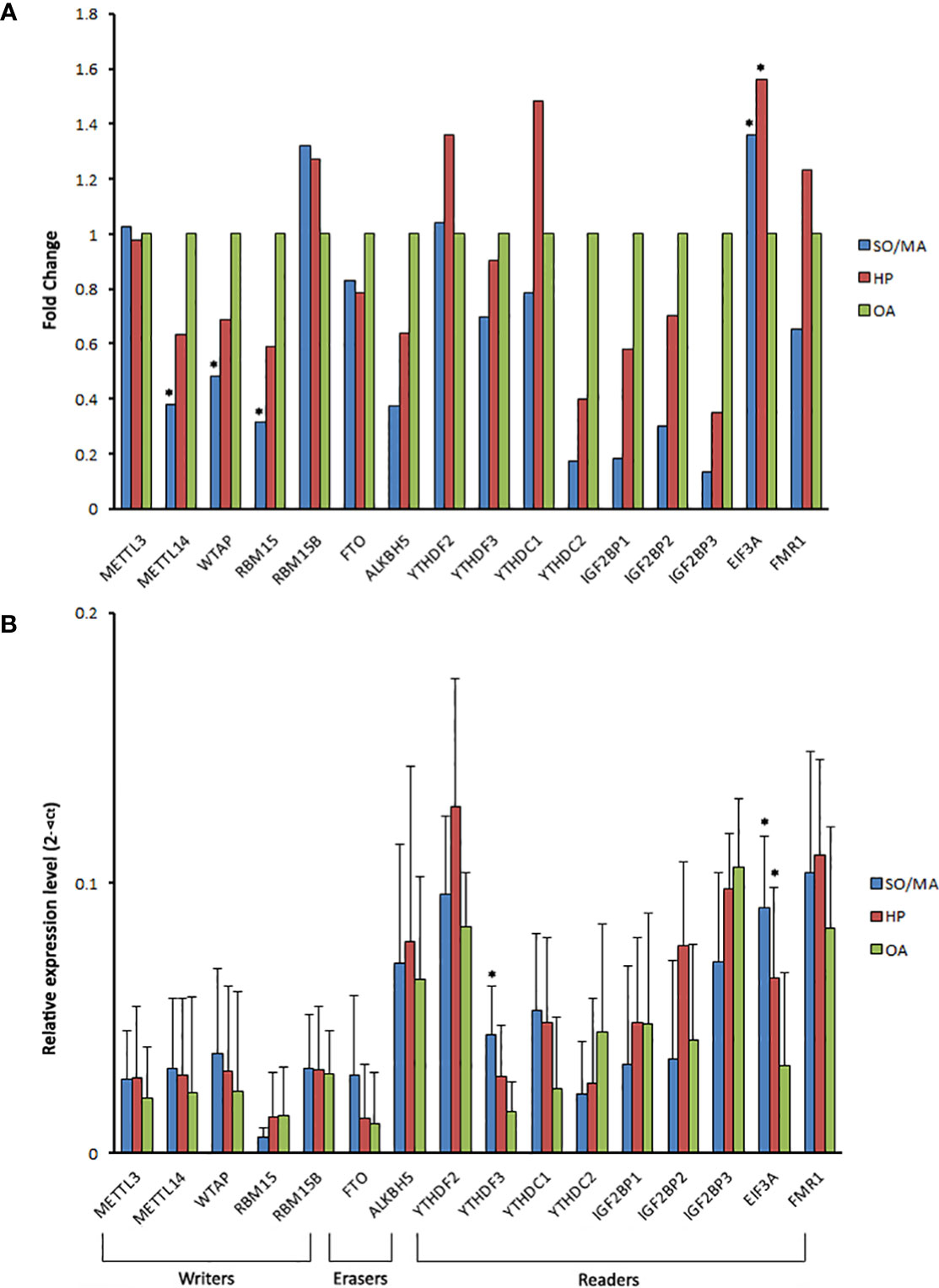
Figure 4 The gene expression of m6A related writers, erasers, and readers in the different groups. (A) The gene expression fold change of the m6A related writers, erasers, and readers in the SO/MA or HP group compared to the OA group via the detection of ArrayStar m6A microarrays. (B) The comparison of m6A related regulators’ mRNA expression between the three groups using qRT-PCR. *P < 0.05 compared with the OA group.
Discussion
m6A plays diverse functions in stem cell pluripotency regulation, posttranscriptional regulation, and mRNA splicing and stability (13, 14). Notably, there is growing evidence in support of its significance in human spermatogenesis (12, 15, 16), which is a highly organized process. A previous clinical study showed that the m6A expression in sperm RNA was upregulated in asthenozoospermia (12), in which the researchers illustrated that the abnormal expression of m6A might regulate the mRNAs of certain genes related to sperm motility. Being different from the study subject, our study explored the m6A expression in the testes tissues of iNOA patients. As is well-known, there is a distinction between the spermatids in testes and the ejected spermatozoa, which experience the capacitation in the epididymis, and have a potential contamination by the somatic cell. Overall, it could be illustrated that dynamic and varied mechanisms participate in the m6A regulation in different spermatogenesis processes for male fertility.
It is well known that, spermatogenic and even spermatogonial cells are absent in SO patients. In contrast, the MA patients include the complete MA characterized by the absence of haploid spermatids, and the incomplete MA, in which a few round or later stage spermatids could be found potentially (17). Studies have shown that m6A is dynamically present in all mRNAs from different stages of mouse spermatogenic cells, with the peak expression in pachytene spermatocytes and round spermatids (11, 18); thus, the lack of these stages of meiosis with or without spermiogenesis might lead to the decline of m6A expression. Consistently, our results also identified the dosage relationship between the m6A and different spermatogenesis conditions. The m6A expression decreased gradually within OA, HP, MA, and SO gourps, and a significant decline was presented in the MA and SO groups.
It is reported that the m6A writers, methyltransferase like 3 (METTL3) and METTL14, likely act critical roles in the spermatogonial stem cells differentiation/proliferation and the spermatids differentiation in the late stages of spermiogenesis (9, 10). METTL14 can form a component with METTL3, as well as strengthen their activities as m6A writers (9). Moreover, WTAP, a mammalian splicing factor, was reported to interact with the complex of METTL3- METTL14 and affect m6A methylation (19). In our study, METTL14 and WTAP were tested down-expressed based on the ArrayStar m6A microarrays’ results, which were not verified by the qRT-PCR. More investigations are warranted in this aspect for further studies.
Based on the findings from Kasowitz SD et al., the YTHDC1 KO mice lacked germ cells and exhibited a SO phenotype, manifesting its necessary regulations in spermatogonial differentiation (20). The YTHDC2 could affect the spermatocyte stage via interacting with the meiosis-specific protein MEIOC (21). And YTHDF2 might influence spermatogonia proliferation by affecting the stability of m6A-containing transcripts (22). However, these ‘readers’ did not show the significantly differential expressions in iNOA patients’ testes based on our analysis. It might be partially explained by the differences in the spermatogonial stem cells and spermatogenesis stages between humans and rodents (23). By applying the bioinformatic methodology, two newly-emerging fields of study from the same study group revealed that m6A methylation-related ALKBH5 and METTL3 were significantly downregulated, and YTHDF3 was upregulated considerably in the iNOA group comparing to the normal group (15, 16). However, their original data were based on Affymetrix Human Gene microarrays’ results, and further verification could provide more evidence on the functions of YTHDF3.
Our study reported that EIF3A was upregulated in the iNOA group, which was not a classic m6A reader and seldom studied in spermatogenesis. As the largest subunit of eIF3, eIF3a is a 170-kDa protein consisting of 1382 amino acids, and is a major initiation factor in mRNA translation progress (24). Moreover, eIF3a involves in cell cycle, DNA synthesis and repair regulation, and serves as a negative regulator of cell differentiation in some tissues (24–26). In carcinogenesis studies, eIF3a has been recognized as a proto-oncogene, and may be a potential anticancer drug target in the eIF family (24). Recently, researchers found that eIF3a could involve in fibrosis through regulation of the TGF-β1 signaling pathway (27); the m6A reader YTHDF3 could recruit eIF3a to facilitate FOXO3 translation, subsequently initiating autophagy (28). These findings may provide directions for subsequent studies of eIF3a in the mechanisms of iNOA. In the present study, considering that the majority of iNOA spermatogenesis states entire translational repression, we hypothesized that the compensated mechanism might be responsible for the upregulation of EIF3A in the testes of iNOA patients.
Furthermore, four genes (BDNF, TMEM38B, RPL3L, and C22orf42) were predicted to be involved in the dysregulation of m6A methylation in iNOA mechanisms in the present study. First, Brain-derived neurotrophic factor (BDNF) was detected in human spermatozoa which might influence sperm mitochondrial activity and apoptosis (29, 30). Second, Transmembrane protein 38B (TMEM38B) encodes trimeric intracellular cation channel type B, expressed in most mammalian tissues’ endoplasmic reticulum (31). Finally, Ribosomal protein L3-like gene (RPL3L) is a novel gene located near the PKD1 and TSC2 genes on chromosome 16p13.3, expressed mainly in skeletal muscle and heart tissue (32). It may be involved in the translation initiation. All these genes’ potential functions in iNOA still need further research.
There are some limitations and flaws in this study. First, the testis tissue contains other types of cells besides the spermatogenic cells, such as Leydig cells, Sertoli cells, myoid cells, macrophages, etc. Therefore, the findings might be impacted by the contamination to some extent. Second, the fixation of PFA and HE staining applied in the pathological examination might cause some limitation because the results always displayed shrinkage artifacts between seminiferous tubules and germ cells. Third, the patients’ number was relatively small for that the residual testicular tissue after sperm retrieval in clinic was always not enough for the following RNA experiments. And just because of this, the SO and MA patients were combined into one subgroup in this study. Besides, we considered that the pathology of SO and MA were homologous to some extent, and clinically their sperm retrieval rate were both very low. Nevertheless, more studies with a large population size of iNOA patients are highly desired in the future.
Conclusions
This study, to our knowledge, is a first exploration in the joint expression profiles of m6A and mRNA in the testes of iNOA patients. As shown in Figure 5, the m6A expression was gradually downregulated in the testes tissue from iNOA patients in accordance with the degree of spermatogenic dysfunction. As m6A reader, EIF3A was testified to be upregulated in iNOA patients’ testes. iNOA is associated with the alterations of mRNA expression and their m6A methylation status. Four genes displayed significantly lower m6A methylation status associated with decreased gene expression were hypothesized to be involved in the dysregulation of m6A methylation in the iNOA mechanisms. Overall, the determined differential expression of mRNA and m6A methylation status may represent potentially novel molecular targets for the mechanism study of iNOA in the epigenetic level.
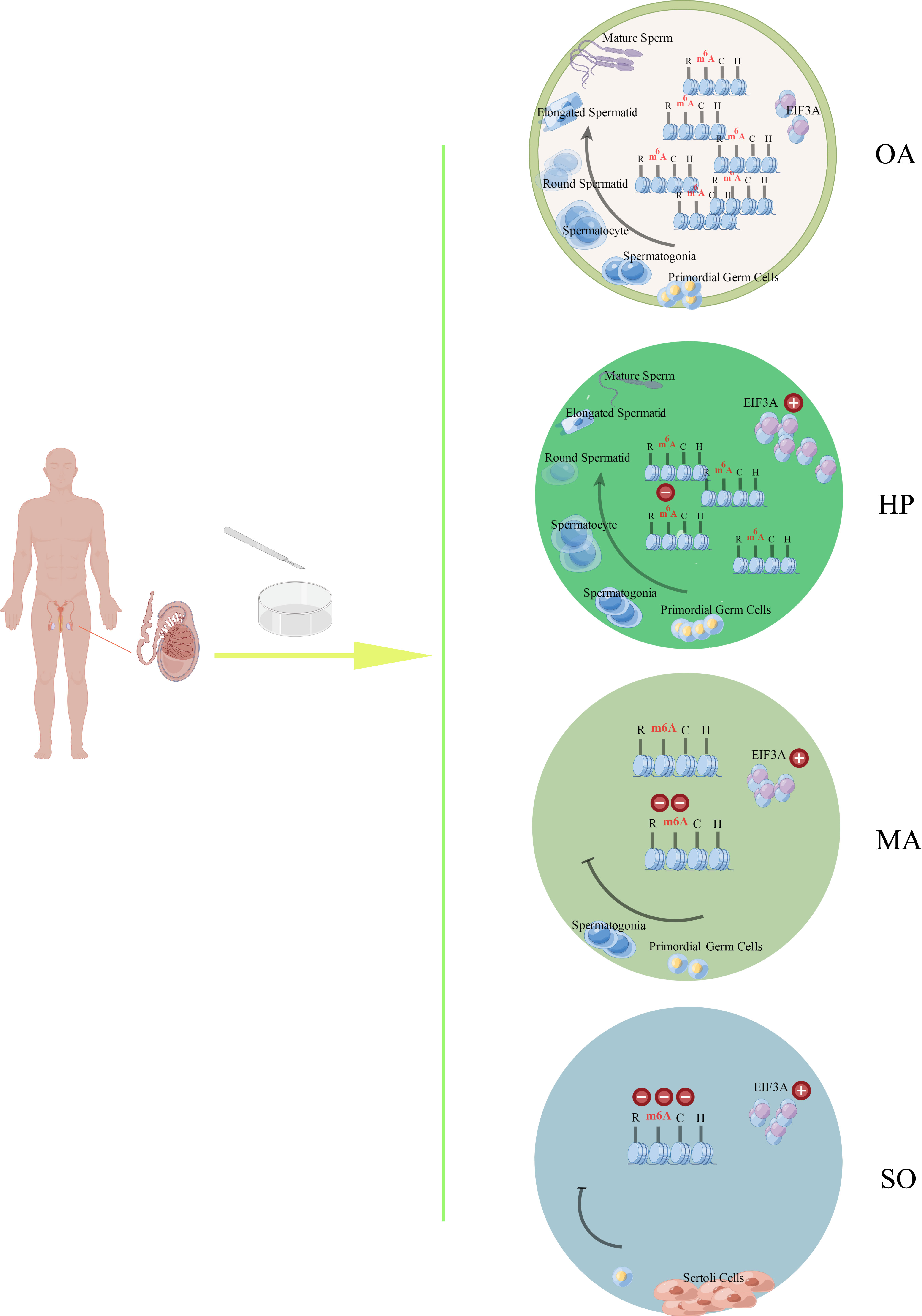
Figure 5 The schematic drawing of the main findings from this study by Figdraw. The m6A expression was gradually downregulated in the testes tissues from different groups (OA, HP, MA, and SO groups) in accordance with the degree of spermatogenic dysfunction. As m6A reader, EIF3A was testified to be upregulated in the testes from iNOA patients, which include the patients with HP, MA, or SO. OA: obstructive azoospermia (with the successive process of normal spermatogeneis), HP, hypospermatogenesis; MA, maturation arrest; SO, Sertoli cell-only syndrome.
Data availability statement
The original contributions presented in the study are included in the article/supplementary materials, further inquiries can be directed to the corresponding author/s.
Ethics statement
The studies involving human participants were reviewed and approved by The Ethics Committee of Women’s Hospital of Nanjing Medical University, Nanjing Maternity and Child Health Care Hospital. The patients/participants provided their written informed consent to participate in this study.
Author contributions
Conception and design: QT, WW, and FP Acquisition of data: QT, YL, and YZ. Analysis and interpretation of data: WW, LP, XL, and FP Drafting the manuscript: QT and FP Laboratory testing: YL, YZ, and WFW. Critical revision of the manuscript: WW, LP, XL, FP, and JL. Final approval to be published: all authors. Agreement to be accountable for all aspects: all authors. All authors contributed to the article and approved the submitted version.
Funding
The study was funded by Nanjing Health Youth Talent Training Project during the 13th Five-Year Plan (QRX17071), and supported by grant from the National Natural Science Foundation of China (81971405, 82273662), Major Project of Natural Science Research in Jiangsu Province Colleges and Universities (20KJA330001), Medical Research Project of Jiangsu Health and Health Commission (Z2019010), Priority Academic Program for the Development of Jiangsu Higher Education Institutions (Public Health and Preventive Medicine), Natural Science Foundation of Jiangsu Province (BK20221307), and Jiangsu Maternal and Child Health Association (FYX202207).
Acknowledgments
We thank all the patients who participated in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Kobori Y. Home testing for male factor infertility: A review of current options. Fertility sterility (2019) 111(5):864–70. doi: 10.1016/j.fertnstert.2019.01.032
2. Hamada AJ, Esteves SC, Agarwal A. A comprehensive review of genetics and genetic testing in azoospermia. Clinics (2013) 68 Suppl 1:39–60. doi: 10.6061/clinics/2013(sup01)06
3. Cervan-Martin M, Castilla JA, Palomino-Morales RJ, Carmona FD. Genetic landscape of nonobstructive azoospermia and new perspectives for the clinic. J Clin Med (2020) 9(2). doi: 10.3390/jcm9020300
4. Chen Q, Yan W, Duan E. Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nat Rev Genet (2016) 17(12):733–43. doi: 10.1038/nrg.2016.106
5. Carrell DT. Epigenetics of the male gamete. Fertility sterility (2012) 97(2):267–74. doi: 10.1016/j.fertnstert.2011.12.036
6. Hamatani T. Human spermatozoal RNAs. Fertility sterility (2012) 97(2):275–81. doi: 10.1016/j.fertnstert.2011.12.035
7. Chen X, Li X, Guo J, Zhang P, Zeng W. The roles of microRNAs in regulation of mammalian spermatogenesis. J Anim Sci Biotechnol (2017) 8:35. doi: 10.1186/s40104-017-0166-4
8. Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol (2017) 18(1):31–42. doi: 10.1038/nrm.2016.132
9. Lin Z, Hsu PJ, Xing X, Fang J, Lu Z, Zou Q, et al. Mettl3-/Mettl14-mediated mRNA N(6)-methyladenosine modulates murine spermatogenesis. Cell Res (2017) 27(10):1216–30. doi: 10.1038/cr.2017.117
10. Xu K, Yang Y, Feng GH, Sun BF, Chen JQ, Li YF, et al. Mettl3-mediated m(6)A regulates spermatogonial differentiation and meiosis initiation. Cell Res (2017) 27(9):1100–14. doi: 10.1038/cr.2017.100
11. Tang C, Klukovich R, Peng H, Wang Z, Yu T, Zhang Y, et al. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3’-UTR mRNAs in male germ cells. Proc Natl Acad Sci United States America (2018) 115(2):E325–33. doi: 10.1073/pnas.1717794115
12. Yang Y, Huang W, Huang JT, Shen F, Xiong J, Yuan EF, et al. Increased N6-methyladenosine in human sperm RNA as a risk factor for asthenozoospermia. Sci Rep (2016) 6:24345. doi: 10.1038/srep24345
13. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell (2017) 169(7):1187–200. doi: 10.1016/j.cell.2017.05.045
14. Meyer KD, Jaffrey SR. Rethinking m(6)A readers, writers, and erasers. Annu Rev Cell Dev Biol (2017) 33:319–42. doi: 10.1146/annurev-cellbio-100616-060758
15. Bai G, Zhai X, Liu L, Cai Z, Xiong J, Li H, et al. The molecular characteristics in different procedures of spermatogenesis. Gene (2022) 826:146405. doi: 10.1016/j.gene.2022.146405
16. Cai Z, Zhang J, Xiong J, Ma C, Yang B, Li H. New insights into the potential mechanisms of spermatogenic failure in patients with idiopathic azoospermia. Mol Hum Reprod (2020) 26(7):469–84. doi: 10.1093/molehr/gaaa033
17. Krausz C, Riera-Escamilla A, Moreno-Mendoza D, Holleman K, Cioppi F, Algaba F, et al. Genetic dissection of spermatogenic arrest through exome analysis: Clinical implications for the management of azoospermic men. Genet medicine: Off J Am Coll Med Genet (2020) 22(12):1956–66. doi: 10.1038/s41436-020-0907-1
18. Gui Y, Yuan S. Epigenetic regulations in mammalian spermatogenesis: RNA-m(6)A modification and beyond. Cell Mol Life sciences: CMLS (2021) 78(11):4893–905. doi: 10.1007/s00018-021-03823-9
19. Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res (2014) 24(2):177–89. doi: 10.1038/cr.2014.3
20. Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD, et al. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PloS Genet (2018) 14(5):e1007412. doi: 10.1371/journal.pgen.1007412
21. Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res (2017) 27(9):1115–27. doi: 10.1038/cr.2017.99
22. Qi M, Sun H, Guo Y, Zhou Y, Gu X, Jin J, et al. m(6) a reader protein YTHDF2 regulates spermatogenesis by timely clearance of phase-specific transcripts. Cell proliferation (2022) 55(1):e13164. doi: 10.1111/cpr.13164
23. Fayomi AP, Orwig KE. Spermatogonial stem cells and spermatogenesis in mice, monkeys and men. Stem Cell Res (2018) 29:207–14. doi: 10.1016/j.scr.2018.04.009
24. Yin JY, Zhang JT, Zhang W, Zhou HH, Liu ZQ. eIF3a: A new anticancer drug target in the eIF family. Cancer Lett (2018) 412:81–7. doi: 10.1016/j.canlet.2017.09.055
25. Dong Z, Liu LH, Han B, Pincheira R, Zhang JT. Role of eIF3 p170 in controlling synthesis of ribonucleotide reductase M2 and cell growth. Oncogene (2004) 23(21):3790–801. doi: 10.1038/sj.onc.1207465
26. Liu Z, Dong Z, Yang Z, Chen Q, Pan Y, Yang Y, et al. Role of eIF3a (eIF3 p170) in intestinal cell differentiation and its association with early development. Differentiation; Res Biol Diversity (2007) 75(7):652–61. doi: 10.1111/j.1432-0436.2007.00165.x
27. Wu YH, Li XW, Li WQ, Li XH, Li YJ, Hu GY, et al. Fluorofenidone attenuates bleomycin-induced pulmonary fibrosis by inhibiting eukaryotic translation initiation factor 3a (eIF3a) in rats. Eur J Pharmacol (2016) 773:42–50. doi: 10.1016/j.ejphar.2016.01.006
28. Hao W, Dian M, Zhou Y, Zhong Q, Pang W, Li Z, et al. Autophagy induction promoted by m(6)A reader YTHDF3 through translation upregulation of FOXO3 mRNA. Nat Commun (2022) 13(1):5845. doi: 10.1038/s41467-022-32963-0
29. Safari H, Khanlarkhani N, Sobhani A, Najafi A, Amidi F. Effect of brain-derived neurotrophic factor (BDNF) on sperm quality of normozoospermic men. Hum Fertil (Camb) (2018) 21(4):248–54. doi: 10.1080/14647273.2017.1346301
30. Zheng L, Li C, Sun Y, Liu Z, Zhou X. Expression of brain-derived neurotrophic factor in mature spermatozoa from fertile and infertile men. Clinica chimica acta; Int J Clin Chem (2011) 412(1-2):44–7. doi: 10.1016/j.cca.2010.08.045
31. Lv F, Xu XJ, Wang JY, Liu Y, Asan, Wang JW, et al. Two novel mutations in TMEM38B result in rare autosomal recessive osteogenesis imperfecta. J Hum Genet (2016) 61(6):539–45. doi: 10.1038/jhg.2016.11
Keywords: m6A, methylation, azoospermia, spermatogenesis, male infertility
Citation: Tang Q, Wu W, Lu Y, Zhou Y, Wu W, Li J, Pan L, Ling X and Pan F (2022) Joint analysis of m6A and mRNA expression profiles in the testes of idiopathic nonobstructive azoospermia patients. Front. Endocrinol. 13:1063929. doi: 10.3389/fendo.2022.1063929
Received: 07 October 2022; Accepted: 30 November 2022;
Published: 15 December 2022.
Edited by:
Qing Chen, Army Medical University, ChinaReviewed by:
Jun Wang, Jilin Agriculture University, ChinaMaria Schubert, Centre of Reproductive Medicine and Andrology, Germany
Copyright © 2022 Tang, Wu, Lu, Zhou, Wu, Li, Pan, Ling and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Pan, ZmVuZ3Bhbm1kQDEyNi5jb20=
†These authors have contributed equally to this work
‡ORCID: Feng Pan, orcid.org/0000-0001-5164-9799
 Qiuqin Tang1†
Qiuqin Tang1† Wei Wu
Wei Wu Yijie Zhou
Yijie Zhou Jinhui Li
Jinhui Li Xiufeng Ling
Xiufeng Ling Feng Pan
Feng Pan