- 1Department of Biological Sciences, National University of Singapore, Singapore, Singapore
- 2School of Biological Sciences, University of Northern Colorado, Greeley, CO, United States
Snake venom research has focused on front-fanged venomous snakes because of the high incidence of human morbidity and mortality from envenomations and larger venom yields of these species, while venoms from rear-fanged snakes have been largely neglected. Rear-fanged snakes (RFS) are a phylogenetically diverse collection of species that feed on a variety of prey and show varying prey capture strategies, from constriction to envenomation. In general, RFS venoms share many toxin families with front-fanged snakes, and venoms generally are either a neurotoxic three-finger toxin (3FTx)-dominated venom or an enzymatic metalloproteinase-dominated venom. These venoms have also been discovered to contain several unique venom protein families. New venom protein superfamilies in RFS venoms include matrix metalloproteinases, distinct from but closely related to snake venom metalloproteinases, veficolins, and acid lipases. Specialized three-finger toxins that target select prey taxa have evolved in some RFS venoms, and this prey capture strategy has appeared in multiple RFS species, from Old World Boiga to New World Spilotes and Oxybelis. Though this same protein superfamily is commonly found in the venoms of elapid (front-fanged) snakes, no elapid 3FTxs appear to show prey-specific toxicity (with the exception of perhaps Micrurus). Neofunctionalization of Spilotes sulphureus 3FTx genes has even resulted in the evolution within a single venom of 3FTxs selectively neurotoxic to different prey taxa (mammals or lizards), allowing this non-constricting RFS to take larger mammalian prey. The large number of 3FTx protein sequences available, together with a growing database of RFS venom 3FTxs, make possible predictions concerning structure-function relationships among these toxins and the basis of selective toxicity of specific RFS venom 3FTxs. Rear-fanged snake venoms are therefore of considerable research interest due to the evolutionary novelties they contain, providing insights into the evolution of snake venom proteins and potential predator-prey coevolution in a broader phylogenetic context. Because of the limited complexity of these venoms, they represent a more tractable source to inform about the biological roles of specific venom proteins that are found in the venoms of this rich diversity of snakes.
Introduction
Venomous snakes and their venoms have instilled both fear and fascination in humans, and they have especially inspired the interest of scientists over the years as unparalleled examples of trophic adaptation. The evolution of venom, venom glands, and specialized maxillary teeth greatly contributed to the radiation of colubroid snakes during the Cenozoic era, generating the diversity of snakes present today (Kardong, 1979; Savitsky, 1980; Vidal, 2002; Jackson, 2003; Vonk et al., 2008; Fry et al., 2012a). There are over 3,700 recognized extant snake species (Uetz et al., 2018); however, the minority of these, <20%, are known to have venoms that result in medically significant bites to humans (Uetz et al., 2018; www.toxinology.com1). Snakebites from venomous snakes that are of medical concern are predominately front-fanged snakes from the families Viperidae and Elapidae, but a large number of snakes from various other families, previously classified as the single family Colubridae, have been discovered to be rear-fanged and also venom-producing (reviewed in Mackessy, 2002; Saviola et al., 2014;Junqueira-de-Azevedo et al., 2016).
Front-fanged snakes (FFS) occur in three families, but rear-fanged snake (RFS) species are phylogenetically diverse, presently assigned to at least three (and sometimes five) distinct families: Colubridae, Homalopsidae, and Lamprophiidae (Vonk et al., 2008; Pyron et al., 2013). Front-fanged snakes have tubular fangs positioned anteriorly in the upper jaw and a venom apparatus that includes an encapsulated reservoir with compressor glandulae (Viperidae) or adductor externus superficialis (Elapidae) muscles inserted directly onto the venom gland capsule (Kochva, 1962). Rear-fanged snakes exhibit a venom apparatus that is morphological variable (Taub, 1967), but typically it is without a storage reservoir or attaching muscles (Figure 1A; see also Kardong, 2002). As we have noted previously (Saviola et al., 2014), the venom-producing gland of RFS is referred to as the Duvernoy's venom gland, a name that recognizes the distinctive nature of this venom gland (secretory epithelial structure, mode of secretion/storage, mechanics of venom delivery, etc.) while acknowledging the clear embryonic, evolutionary and biochemical homologies with FFS venom glands. Snakes generally have numerous oral glands (Kardong, 2002), but the Duvernoy's venom gland, which is a serous secretory gland, is histologically distinct from the closely adjacent supralabial gland, which is mucosecretory (Figure 1B). A medial venom gland duct conducts secreted venom to one-several, often enlarged, posterior maxillary teeth, and these may be bladed, grooved and/or ungrooved (Figures 1C–G; see also Young and Kardong, 1996; Jackson, 2003). Because of the lack of storage reservoir and musculature, venom yields from RFS are considerably less in comparison to FFS, and it is difficult to obtain large quantities of venom for protein characterization. However, venom yields have been improved with the use of ketamine and pilocarpine to sedate snakes and induce gland secretions (Rosenberg, 1992; Hill and Mackessy, 1997).
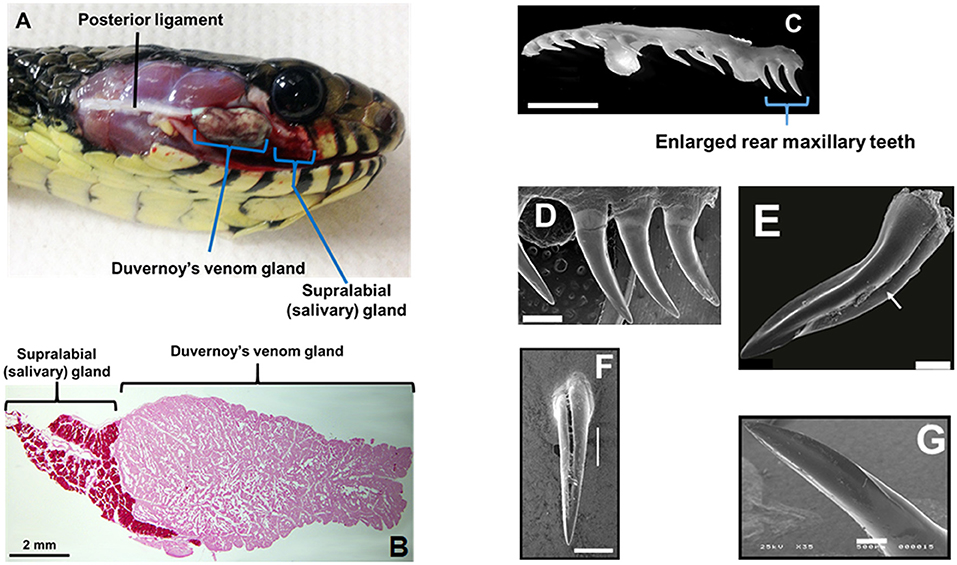
Figure 1. Duvernoy's venom gland and enlarged maxillary teeth of rear-fanged snakes. (A) Spilotes sulfureus Duvernoy's venom gland in situ. Note that no muscles are directly associated with the gland—venom secretion is realized via compression of the gland between the skin and the contracting jaw adductor muscles. (B) Duvernoy's venom gland and supralabial gland of S. sulfureus. (C) Left maxilla (ventro-lateral view) of S. sulfureus showing three serially enlarged rear teeth. Bar = 5 mm. (D) SEM micrograph of enlarged rear teeth of S. sulfureus. Bar = 1 mm. (E) SEM micrograph of the enlarged rear maxillary fang of Erythrolamprus aesculapii. Note deep anterior groove (arrow) and both anterior and posterior blades. Bar = 0.2 mm. (F) Rear maxillary tooth of Boiga irregularis (SEM); note deep anterior groove. Bar = 0.5 mm. (G) Enlarged rear maxillary tooth of Hydrodynastes gigas showing anterior-facing blade (SEM). Enlarged rear maxillary tooth of Hydrodynastes gigas showing anterior-facing blade (SEM). Bar = 0.5 mm. (A–D) From Modahl et al. (2018b); (E) from Sánchez et al. (2019); (F,G) from Mackessy (2010b).
For some RFS, what appears superficially to be a less well-developed venom system is still capable of producing lethal envenomations in humans. The previously underestimated venom of the RFS Dispholidus typus resulted in the death of eminent herpetologist Karl P. Schmidt in the late 1950s (Pope, 1958; Pla et al., 2017b). Although the large majority of RFS are unable to deliver lethal toxin quantities or even venom yields great enough to result in systemic envenomations in humans (Weinstein et al., 2013), at least three species (D. typus, Thelotornis capensis, and Rhabdophis tigrinus) have caused human fatalities, and bites by two additional species (Philodryas olfersii and Tachymenis peruviana) have resulted in serious human envenomations (Kuch and Mebs, 2002; Mackessy, 2002; Prado-Franceschi and Hyslop, 2002; Weinstein et al., 2011). For two of these genera, Dispholidus and Rhabdophis, antivenom is currently manufactured and is the recommended management of snakebites from these species (Weinstein et al., 2013). Increasing awareness of severe, at times fatal, envenomations from RFS has led to a slowly growing interest in their venoms. Additionally, advances in the sensitivity of research technologies has resulted in the ability to profile venoms with relatively little starting material.
Current –omic technologies have made venom characterization attainable for organisms with small amounts of venom and venom gland tissue. As a result of these technologies, databases are rapidly growing from transcriptomic and proteomic studies. Genomic and transcriptomic fields have become more inclusive with the affordability of next-generation sequencing (NGS), making these technologies available for non-model organisms. There are now a number of snake genome references (Castoe et al., 2013; Vonk et al., 2013; Yin et al., 2016; Perry et al., 2018), and the number of NGS generated venom gland transcriptomes from snakes is also on the rise (Durban et al., 2011; Rokyta et al., 2011, 2015; Aird et al., 2013; Margres et al., 2013), including for RFS species (McGivern et al., 2014; Zhang et al., 2015; Campos et al., 2016; Pla et al., 2017a,b; Modahl et al., 2018a,b). The sequencing depth of NGS allows for lowly expressed transcripts in snake venom glands that were previously difficult to obtain with expressed sequence tags (ESTs) to now be observed (Campos et al., 2016). In addition to NGS advancements, the increasing sensitivity of mass spectrometry instruments has made it possible to characterize venom proteomes with small amounts of venom (~100–200 ng). The integration of these –omic technologies together has led to comprehensive venom profiles of RFS venoms. These profiles are now readily generated, are affordable, and demonstrate more accurate evolutionary overviews of venom compositional diversification.
These more complete venom gland transcriptomes and venom proteomes have revealed common patterns of toxin expression and secretion for RFS (Junqueira-de-Azevedo et al., 2016), as well as identified new venom proteins that had previously not been recognized as venom components in FFS species (OmPraba et al., 2010; Ching et al., 2012; Fry et al., 2012b; Campos et al., 2016). Further, these venoms have been shown to possess toxins with unique activities, such as prey-specific toxicity (Mackessy et al., 2006; Pawlak et al., 2006, 2009; Heyborne and Mackessy, 2013; Modahl et al., 2018b). Venom proteins from RFS show distinct gene organization and evolutionary trajectories (Pawlak and Kini, 2008; Dashevsky et al., 2018), making these generally neglected venoms ideal models to study venom as trophic adaptations. By examining a large and divergent clade of snakes, we can begin to address and answer questions as to how venom proteins acquire their functionalities and what biological roles they provide.
Rear-Fanged Snake Venoms: Known Proteins
Even as phylogenetically diverse as venomous snakes are, there are some venom proteins commonly observed in all snake venoms. These include three-finger toxins (3FTxs), snake venom metalloproteinases (SVMPs), C-type lectins (CTLs), and cysteine-rich secretory proteins (CRiSPs) (Mackessy, 2010b; Junqueira-de-Azevedo et al., 2016). Phospholipase A2s (PLA2) are ubiquitous in FFS venoms (Mackessy, 2010b), but have been found to be abundant in only a few RFS species (Hill and Mackessy, 2000; Huang and Mackessy, 2004). Overall venom composition observed for snakes is one of two types, either a venom is dominated by smaller, usually neurotoxic 3FTxs, such as those in elapid (cobras, mambas, kraits, etc.) venoms, or a venom that consists primarily of larger enzymatic proteins, such as SVMPs, as in the case of viper and pit viper venoms (Mackessy, 2010b). Interestingly, RFS venoms can have either an elapid-like neurotoxic or viper-like enzymatic composition (McGivern et al., 2014). A summary of the presence and absence of venom proteins in RFS venoms can be found in a colubrid –omics review (Junqueira-de-Azevedo et al., 2016).
Another commonality that is observed for all venomous snakes is the large number of proteoforms present in each venom. This large number of isoforms is a result of multiple gene duplication events and single nucleotide polymorphisms (SNPs), which through neofunctionalization has generated toxins with different activities (Casewell et al., 2013; Modahl et al., 2018b). This has allowed for RFS, which tend to have less complex venom (Peichoto et al., 2012), to expand toxin functionalities.
Three-Finger Toxins
One venom protein superfamily that is prevalent in snake venoms, with an incredibly vast range of activities, are the three-finger toxins (3FTxs). Three-finger toxins can be neurotoxic, acting as antagonists of nicotinic acetylcholine receptors (nAChRs) (Chang and Lee, 1963; Nirthanan and Gwee, 2004; Bourne et al., 2005), muscarinic acetylcholine receptors (mAChRs) (Karlsson et al., 2000; Chung et al., 2002), adrenergic receptors (Rajagopalan et al., 2007), GABA receptors (Rosso et al., 2015), or even bind to and alter the activation of ion channels (de Weille et al., 1991; Rivera-Torres et al., 2016; Yang et al., 2016). These toxins can also be anticoagulants, inhibiting platelet aggregation (Kini et al., 1988; McDowell et al., 1992). Regardless of activity, all 3FTxs maintain a conserved structural scaffold of three β-stranded loops, crosslinked by four disulfide bridges. This forms the “three-finger” arrangement, appearing like three fingers of a hand. Three-finger toxins are non-enzymatic, small proteins consisting of 60–85 amino acid residues, and occur either as monomers, which is most common, or as dimers (Kini and Doley, 2010).
Elapid venoms are the most well-known sources of 3FTxs (Fry et al., 2003b), but 3FTxs have been documented in the venoms of many RFS species (Fry et al., 2003a, 2008; Pawlak et al., 2006, 2009; Heyborne and Mackessy, 2013; Junqueira-de-Azevedo et al., 2016) and can make up large portions of these venoms, as much as 84–92% of the total venom composition (Pla et al., 2017a; Modahl et al., 2018b). Rear-fanged snake genera that have abundant proteins in the molecular mass range of 3FTxs include Boiga, Spilotes, Trimorphodon, Oxybelis, Leioheterodon, Psammophis, Rhamphiophis, and Thelotornis (Figure 2A). Of these snakes mentioned, 3FTxs have been sequenced and functionally characterized from the venoms of Boiga dendrophila (Lumsden et al., 2005; Pawlak et al., 2006), Boiga irregularis (Pawlak et al., 2009), Oxybelis fulgidus (Heyborne and Mackessy, 2013), and Spilotes sulphureus (Modahl et al., 2018b). Transcripts for 3FTx have been found in the venom gland, or in the venom, of Boiga cynodon, Boiga nigriceps, Trimorphodon biscutatus, Ahaetulla prasina, Leioheterodon madagascariensis, Thrasops jacksonii, and Psammophis mossambicus (Fry et al., 2008; Modahl and Mackessy, 2016; Dashevsky et al., 2018; Modahl et al., 2018a), among others.
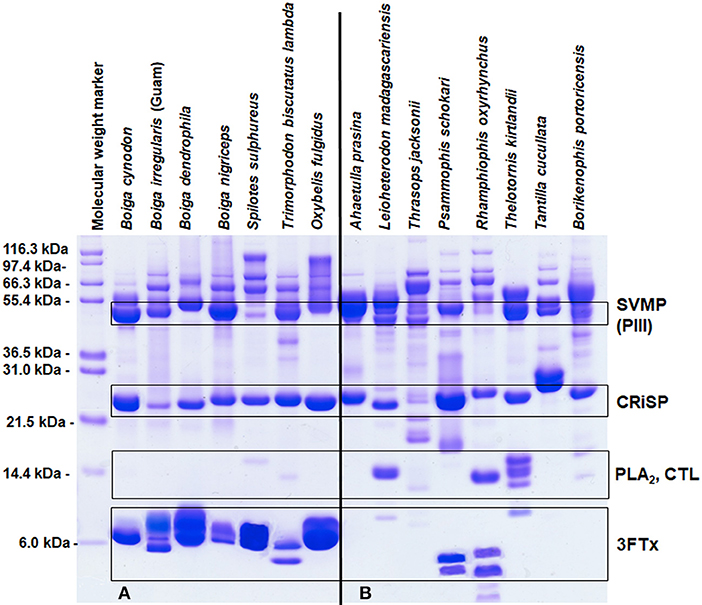
Figure 2. Reduced SDS-PAGE protein profiles for various rear-fanged snake venoms. Rear-fanged snakes tend to have either three-finger toxin-dominated venoms (A) or venoms rich in metalloproteinases (B). Molecular masses are indicated on the left, and putative venom protein identities are listed on the right. 3FTx, three-finger toxin; CRiSP, cysteine-rich secretory protein; CTL, C-type lectin; PLA2, phospholipase A2; and SVMP (PIII), snake venom metalloproteinase type PIII. Modified from Modahl et al. (2018b).
Three-finger toxins were one of the first venom proteins characterized pharmacologically in RFS venoms and were reported to exhibit postsynaptic neurotoxicity, similar to elapid α-neurotoxins (Broaders et al., 1999; Fry et al., 2003a; Lumsden et al., 2005). Several 3FTxs from RFS venoms have been the only well-characterized venom proteins with taxon-specific toxicities, showing preferential binding to lizard or bird nAChRs, in direct relation to snake diet (Pawlak et al., 2006, 2009). Several elapid venoms have shown differential toxicity toward bird or rodent neuromuscular preparations (e.g., Hart et al., 2012, 2013), but these studies were on crude venoms and did not include toxicity assays on native prey species. Sequences and structures of 3FTxs isolated from RFS have an elongated N-terminal segment, and tend to be larger in size than observed in FFS venoms (Lumsden et al., 2007; Pawlak et al., 2009; Heyborne and Mackessy, 2013; Modahl et al., 2018b). It is currently unknown how this longer N-terminal region is involved in receptor binding.
Several of these 3FTxs are also N-terminally blocked by a pyroglutamic acid residue (Broaders et al., 1999; Fry et al., 2003a; Pawlak et al., 2006; Heyborne and Mackessy, 2013). This pyroglutamic acid blockage especially makes these proteins hard to sequence by Edman degradation, and obtaining venom protein transcript sequences has been the most successful approach to study 3FTx amino acid residue diversity in RFS (Fry et al., 2008; McGivern et al., 2014; Modahl and Mackessy, 2016; Pla et al., 2017a; Dashevsky et al., 2018; Modahl et al., 2018b). Rear-fanged snake 3FTxs demonstrate low amino acid sequence identity with 3FTxs from elapids (usually <50%), and all RFS 3FTxs are members of the “non-conventional” toxin classification, characterized by an additional fifth disulfide bond in the first loop (Nirthanan et al., 2003). This type of 3FTx also occurs in elapid venoms, where it was first characterized in cobra (Naja) venoms (Carlsson, 1975; Utkin et al., 2001), but they are the only 3FTx type present in RFS venoms. This suggests that the non-conventional cysteine pattern may be the more basal arrangement for 3FTxs, and provides insight into the evolution of 3FTxs (cf. Fry et al., 2003b). The incredible diversity of RFS 3FTxs with low identity to elapid 3FTxs, combined with the binding selectivity exhibited by some of these toxins, provides a database of proteins to explore how venom proteins target prey and adapt toxicity, and will be discussed more in another section below.
Metalloproteinases
Snake venom metalloproteinases occur primarily in viperid venoms (Mackessy, 2010b), and similar to 3FTxs, are also a large multigene family, exhibiting a diversity of activities. These enzymes have effects including hemorrhage, coagulopathy, fibrinolysis, apoptosis, and the activation of factor X and prothrombin (Takeya et al., 1993). Many SVMPs function by degrading endothelial cell membrane components or target proteins involved in coagulation, such as fibrinogen or platelet receptors (Takeda et al., 2012). As part of the metzincin superfamily of proteinases, they are characterized by the presence of the Zn2+-binding motif HEXXHXXGXXH at the catalytic site. Snake venom metalloproteinases are closely related to mammalian ADAM (a disintegrin and metalloproteinase) and ADAMTS (ADAM with thrombospondin type-1 motif), but differ in domain organization (Fox and Serrano, 2005; Takeda et al., 2012). The SVMP P-I class has only a catalytic metalloproteinase domain present, P-IIs contain a metalloproteinase domain followed by a disintegrin domain, and P-IIIs have a metalloproteinase, disintegrin-like, and cysteine-rich domain (Hite et al., 1994; Fox and Serrano, 2005).
In some RFS venoms, SVMPs are the most abundant toxins, making up 62–70% of the total venom composition (Modahl et al., 2018a). SVMP-dominated venoms are found in RFS species such as A. prasina (Modahl et al., 2018a), B. portoricensis (Weldon and Mackessy, 2012; Modahl et al., 2018a), D. typus (Kamiguti et al., 2000; Pla et al., 2017b), Hydrodynastes gigas (Hill and Mackessy, 2000), Hypsiglena torquata (McGivern et al., 2014), Phalotris mertensi (Campos et al., 2016), Pseudoboa neuwiedii (Torres-Bonilla et al., 2018), Thamnodynastes strigatus (Ching et al., 2012), Thamnophis sirtalis (Perry et al., 2018), and several Philodryas species (Ching et al., 2006; Peichoto et al., 2012; Urra et al., 2015) (Figure 2B). Potent SVMPs are observed in the venoms of snakes from the genus Philodryas (Assakura et al., 1994; Rocha et al., 2006; Sánchez et al., 2014), and venoms from these species in particular have been commonly reported to induced hemorrhage, myonecrosis and edema (Peichoto et al., 2005; Nery et al., 2014; Sánchez et al., 2014; Oliveira et al., 2017). The proteolytic activity of Philodryas venoms is inhibited by metal chelators (Assakura et al., 1992; Acosta et al., 2003; Peichoto et al., 2005, 2012; Rocha and Furtado, 2007), suggesting that SVMPs are resulting in these clinical snakebite symptomologies; in some species, serine proteinases may also be involved (Assakura et al., 1994; Peichoto et al., 2005; Ching et al., 2006). In comparison to venom from the pit viper species Bothrops jararaca, proteinase activity was 25 times greater for P. baroni venom (Sánchez et al., 2014), and also greater for P. olfersii and P. patagoniensis venoms (Carreiro da Costa et al., 2008). This demonstrates that the SVMP enzymatic activity from RFS venoms can be even more impressive than some FFS species. Philodryas sp. have been noted to share similar SVMP epitopes with snakes of the genus Bothrops (Assakura et al., 1992; Tancioni et al., 2004; Carreiro da Costa et al., 2008) and antivenom produced from Bothrops venoms have been observed to neutralize the systemic envenomation effects of Philodryas (Rocha et al., 2006).
Several SVMPs from RFS venoms have been purified and characterized, including patagonfibrase from P. patagoniensis (Peichoto et al., 2007), alsophinase from Borikenophis (previously the genus Alsophis) portoricensis (Weldon and Mackessy, 2012), and several from P. olfersii (Assakura et al., 1994). These characterized SVMPs are most noted for fibrin(ogen)olytic activity, preferentially degrading the α-chain of fibrinogen over the β-chain, however selectivity was variable, but none degraded the γ-chain (Assakura et al., 1994; Peichoto et al., 2007; Weldon and Mackessy, 2012). Patagonfibrase also impaired platelet aggregation induced by collagen and ADP (Peichoto et al., 2007). Both patagonfibrase and alsophinase were shown to cause hemorrhage and edema in mice, once again suggestive of the significant role SVMPs play in producing the envenomation symptomologies from these snakes (Peichoto et al., 2007, 2011; Weldon and Mackessy, 2012).
Only SVMPs of the P-III class has been reported in RFS (Saviola et al., 2014; Junqueira-de-Azevedo et al., 2016), with the exception of one truncated SVMP of a new class (Campos et al., 2016). This new type of SVMP was identified in the RFS P. mertensi, in which a P-III SVMP was discovered to be truncated in the middle of the disintegrin-like domain by a nonsense mutation in the gene; the resulting truncated SVMP was also observed in the venom (Campos et al., 2016). Rear-fanged SVMPs appear to have such nucleotide mutations frequently; within the venom gland transcriptome of B. portoricensis, a nucleotide substitution resulted in the elimination of the conserved stop codon and produced SVMP transcripts with a 9-residue extension at the C-terminus (Modahl et al., 2018a). P-IIIs were the first class of SVMPs recruited as a venom toxin (Casewell et al., 2011), and these RFS toxin genes and transcripts provide insight into the evolution of SVMPs and where alterations in ancestral gene sequences occurred, allowing the domains of this superfamily of proteins to diversify.
Cysteine-Rich Secretory Proteins
Cysteine-rich secretory proteins are common in many reptile venoms (Mackessy, 2002; Sunagar et al., 2012), but very little is currently known about their exact biological targets as venom components. These non-enzymatic proteins all share a conserved 16 cysteine residue pattern, forming eight disulfide bonds (Mackessy and Heyborne, 2010). CRiSPs lack proteolytic, hemorrhage and coagulant activity (Lodovicho et al., 2017). The few snake venom CRiSPs that have been characterized have been found to inhibit various ion channels (Nobile et al., 1996; Brown et al., 1999; Yamazaki et al., 2002; Wang et al., 2006) or induce inflammation, activating the complement system (Lodovicho et al., 2017).
CRiSPs occur in a wide range of RFS venoms (Hill and Mackessy, 2000; Peichoto et al., 2012), and are likely at least present at the transcript level for all species (Junqueira-de-Azevedo et al., 2016). Few CRiSPs have been purified and characterized from RFS venoms, but the CRiSP patagonin was purified from the venom of Philodryas patagoniensis and demonstrated necrotic activity toward murine gastrocnemius muscle when injected intramuscularly at doses of 43 μg and greater (Peichoto et al., 2009). It was suggested that patagonin was potentially binding to ion channels. Patagonin had no effect on the aggregation of human platelets and no proteolytic activity toward azocasein or fibrinogen (Peichoto et al., 2009). Helicopsin, a CRiSP from the RFS Helicops angulatus, was found have robust neurotoxic activity, causing respiratory paralysis in mice (Estrella et al., 2011). However, tigrin, a CRiSP isolated from the venom of RFS R. tigrinus tigrinus, was found to have no effect on high potassium or caffeine-induced contraction of helical strips of endothelium-free rat-tail arterial smooth muscle (Yamazaki et al., 2002), suggesting a lack of any neurotoxicity. The biological roles of CRiSPs in venoms still remains a bit unclear, but given the wide occurrence of these proteins in venoms, their conserved sequence and cysteine scaffold, and moderate-high levels of abundance in RFS venoms (often one of the most abundant venom components; Pla et al., 2017a; Modahl et al., 2018a), they likely serve important biological roles.
Non-ubiquitous and/or Minor Venom Components
Other proteins that have been found in RFS venom proteomes include serine proteinases, phospholipase A2s (type IA), acetylcholinesterases, and C-type lectins. Serine proteinases dominate many viper venoms (Mackessy, 2010b) and are responsible for both promoting and inhibiting blood coagulation, via activation of coagulation factors to induction of platelet aggregation or direct action on fibrinogen (Braud et al., 2000). In RFS venoms, serine proteinases have been identified in the venoms of P. olfersii (Assakura et al., 1994; Ching et al., 2006), P. patagoniensis (Peichoto et al., 2005), and P. mertensi (Campos et al., 2016), but overall these enzymes appear to be uncommon in this group of snakes. Phospholipase A2s (type IA) are also uncommon in RFS venoms, but for some species, such as T. biscutatus lambda, PLA2 enzymatic activity is detectable from crude venom and a PLA2 has even been purified from this venom (Hill and Mackessy, 2000; Huang and Mackessy, 2004). Front-fanged snake venoms usually contain many PLA2 isoforms and this large venom protein superfamily has a wide range of pharmacological effects, including neurotoxicity, myotoxic, cardiotoxic, anticoagulant, hemolytic, and hypotensive activities (Kini, 2003). PLA2 enzymes from RFS appear to be more similar to PLA2 sequences from elapid venoms (Huang and Mackessy, 2004; Fry et al., 2008), and perhaps serve a predigestive or prey capture role in RFS venoms, but they are not an abundant venom component. Acetylcholinesterase is another enzyme that has been detected at low levels in some RFS venoms, such as Boiga species (Broaders and Ryan, 1997; Hill and Mackessy, 2000) and Leptophis ahaetulla marginatus (Sánchez et al., 2018), but it is also not broadly distributed in RFS venoms (Mackessy, 2002; Junqueira-de-Azevedo et al., 2016).
C-type lectins (CTL) do appear to be a ubiquitous component of RFS venoms, and they are consistently present as transcripts in venom glands of RFS species (Junqueira-de-Azevedo et al., 2016). CTLs are non-enzymatic proteins capable of binding reversibly and non-covalently to carbohydrates, inducing hemagglutination by binding surface glycoconjugates on erythrocytes (Lu et al., 2005; Morita, 2005). Lectins have been isolated from many natural sources, and are more common components of viper than elapid venoms (Mackessy, 2010b). For FFS species, CTL abundance appears to be variable in snake venoms (Durban et al., 2011) and has only been detected at a transcript level in the venom gland of some species (e.g., Vonk et al., 2013). A CTL from the RFS Cerberus rynchops shared 79% sequence identity to a CTL from the elapid Bungarus multicinctus (OmPraba et al., 2010), and CTL transcripts from the venom gland of P. olfersii were also found to be more similar to those of elapids than viperids (Ching et al., 2006). All lectins have a carbohydrate recognition domain responsible for the glycan interaction activity, and the EPD triplet and conserved calcium-binding region indicated mannose specificity for the CTL discovered in venom from the RFS C. rynchops. Interestingly, CTLs in RFS have various glycan binding motifs, including EPD, QPD, EPN, RPS, QVE, and EPK (Fry et al., 2008; Junqueira-de-Azevedo et al., 2016). The variability in binding motifs in various FFS species suggests that these genes have greatly diversified in colubroids and might provide different functionalities in RFS venoms.
Rear-Fanged Snake Venoms: New Protein Families
New venom protein families have been identified from RFS venoms. These previously unknown venom proteins can make up 26% of the total venom gland transcripts in these species (Cerberus rhynchops: OmPraba et al., 2010) or almost 50% of the expressed toxins in the venom gland (T. strigatus: Ching et al., 2012), demonstrating that these newly recognized venom proteins are not just minor venom components and likely are biologically relevant as venom toxins. However, there have been transcripts for many toxins identified in the venom gland of RFS species that have been found to be absent from the venom (Fry et al., 2008; Junqueira-de-Azevedo et al., 2016; Modahl et al., 2018a). Additionally, toxin homologs are expressed at low-moderate levels in other tissues and are not restricted to the venom gland (Hargreaves et al., 2014; Junqueira-de-Azevedo et al., 2015; Reyes-Velasco et al., 2015), and mistaking these non-toxin genes as venom proteins can lead to confounding evolutionary analyses. Expression of genes belonging to venom toxins is higher in venom gland tissues in comparison to other organ tissues, and examining these expression profiles has been a successful approach to identifying true venom proteins and what could potentially be new toxins (Hargreaves et al., 2014; Campos et al., 2016; Perry et al., 2018). It is still critical that when characterizing new toxins, any transcriptome study is accompanied by a venom proteome to confirm the presence of the toxin in the venom. Ribonuclease, lipocalin, phospholipase A2 (type IIE) and vitelline membrane outer layer proteins have been suggested to be new toxins from RFS species (Fry et al., 2012b), but have yet to be confirmed and functionally characterized in these venoms. Therefore, only new venom protein families with detected presence in RFS venoms will be discussed below. These new toxins include veficolins, matrix metalloproteinases and acid lipases.
Veficolins
A new venom protein family, named veficolins (venom ficolins), was discovered in the venom of C. rynchops (OmPraba et al., 2010), one of only two RFS species of the family Homalopsidae that has been studied. OmPraba et al. used a combined transcriptomic and proteomic approach to characterize C. rynchops venom, confirming both veficolin gene expression in the venom gland and its presence in the venom. Transcripts for veficolins have been identified in the venom glands of other RFS species (Fry et al., 2012b; Junqueira-de-Azevedo et al., 2016; Pla et al., 2017a; Modahl et al., 2018b), but C. rynchops venom has been the only species reported with these toxins in the venom proteome (OmPraba et al., 2010). Veficolins share amino acid sequence similarities to mammalian ficolins with collagen-like and fibrinogen-like domains. It is possible that these venom proteins can induce platelet aggregation and/or initiate complement activation when delivered into prey (OmPraba et al., 2010), aiding in prey capture. However, the two veficolins from C. rynchops venom have yet to be experimentally characterized.
Matrix Metalloproteinases
Snake venom matrix metalloproteinases (svMMPs) were first identified as a new venom protein present in the venom of RFS R. tigrinus tigrinus (Komori et al., 2006). Transcripts for svMMPs appear to be abundant in the venom glands of Dipsadinae RFS, including T. strigatus (Ching et al., 2012) and Erythrolamprus miliaris (Junqueira-de-Azevedo et al., 2016), and they are closely related to but functionally distinct from SVMPs. The expression of svMMPs in the venom gland of T. strigatus was considerably higher than that of SVMPs: 46.2–8.2%, respectively (Ching et al., 2012). Snake venom matrix metalloproteinases were also detected in the 2-D electrophoresis venom profile of P. mertensi (Campos et al., 2016). RFS svMMP genes cluster with a single MMP-9 ancestral gene, regardless of the presence or absence of ancillary domains (Junqueira-de-Azevedo et al., 2016), suggestive of a separate recruitment event relative to SVMPs. Matrix metalloproteinases are also zinc-dependent enzymes that degrade extracellular matrix proteins, such as collagens, elastin, proteoglycans, and laminins (Ra and Parks, 2007). These enzymes have been well-recognized for their roles in many physiological and pathological processes, including organ growth, wound healing, bone remodeling, immunity modulation, tumor invasion, and metastasis (Vu and Werb, 2000; Parks et al., 2004; Page-McCaw et al., 2007). They likely serve tissue-degrading roles in RFS venoms.
Acid Lipases
Recently, another new venom protein, snake venom acid lipase (svLIPA) was identified in the venom of the RFS P. mertensi (Campos et al., 2016). The venom gland transcriptome of this species revealed a highly expressed transcript coding for a lysosomal acid lipase protein; P. mertensi venom also exhibited lipase activity (Campos et al., 2016), and the recombinantly expressed svLIPA transcript produced a functional protein. Campos et al. supported their identification of svLIPA as a new venom protein based on its presence in P. mertensi venom, presence in the venom of the FFS Micrurus corallinus (Correa-Netto et al., 2011), high transcript abundance in the venom gland of P. mertensi in comparison to other body tissues, and previous reports of its transcription in the oral glands of other snakes (Hargreaves et al., 2014). Acid lipases hydrolyze cholesteryl esters and triglycerides to free cholesterol and fatty acids, aiding in cell metabolism and immunity (Gomaraschi et al., 2019). As with svMMPs, lipases could serve predigestive roles in RFS venoms.
Rear-Fanged Snake Venoms: New Activities
One of the greatest hurdles to overcome when functionally characterizing a venom protein is acquiring enough material to perform assays. Experimental characterization of minor venom components or toxins from venomous animals with low venom yields is usually done by first producing enough of the required protein from a recombinant expression system, such as Escherichia coli, Pichia pastoris, or mammalian cells (Gomes et al., 2016). However, toxins have proven difficult to express in these systems due to being rich in disulfide bonds, leading to protein aggregates and misfolding, and they can be toxic to expressing cells as well (Saez et al., 2014). It is also possible to chemically synthesize toxins, but this is only successful for smaller toxins. With FFS species, venom yields are much larger, so it is possible to avoid these challenges by purifying venom proteins directly from the venom. This has led to a characterization bias of only venom proteins that are abundant and/or that originate from snake species with large venom yields. Because of this, only a few proteins have been characterized from RFS venoms. For the toxins that have been explored, interesting new activities and targets have emerged, especially in relation to snake diet, prey capture strategies and the biological roles RFS venoms provide. Venoms from these species are an additional and highly productive resource for examining the functional diversity of prevalent protein superfamilies.
Three-Finger Toxins
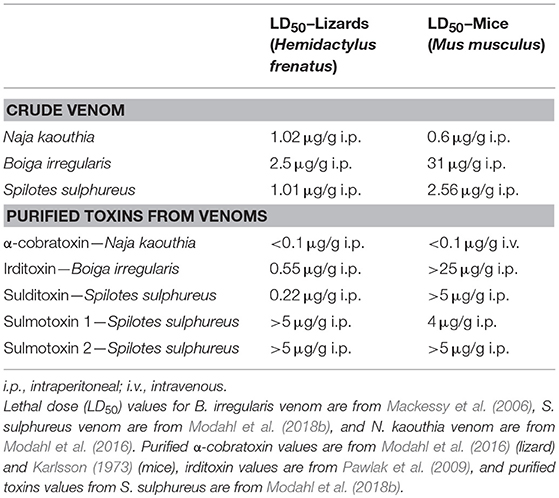
Crude venoms from several RFS species show drastic toxicity differences depending on the model organism used for lethal dose (LD50) experiments (Mackessy, 2002). Philodryas patagoniensis venom was tested on pigeons (Columba livia domestica), guinea pigs (Cavia porcellus), rabbits (Oryctolagus cuniculus), and frogs (Leptodactylus sp.), and pigeons were the most sensitive to this venom (Martins, 1907). Boiga irregularis venom LD50 values were determined for domestic chickens (Gallus domesticus), geckos (Hemidactylus sp.), skinks (Carlia sp.), and mice (Mus musculus), and it was found that crude venom was much more toxic to birds and lizards than to mammals (Table 1) (Mackessy et al., 2006). Venom from S. sulphureus was also more toxic toward geckos (Hemidactylus sp.) than mice (M. musculus), even at 22-fold higher mass-adjusted doses in mice (Table 1) (Modahl et al., 2018b). This impressive toxicity in geckos of S. sulphureus venom is equivalent to that observed for venom from the highly toxic FFS Naja kaouthia (Table 1). The enhanced toxicity of crude venom toward different taxa has led to explorations into these venoms to discover components responsible. In most cases, 3FTxs have been identified to be the taxon-specific targeting toxins in these venoms, first characterized in the venoms of RFS species in the genus Boiga, commonly referred to as cat snakes.
From B. dendrophila venom, two 3FTxs have been characterized with postsynaptic neurotoxicity, boigatoxin-A (Lumsden et al., 2005) and denmotoxin, B. dendrophila monomeric toxin (Pawlak et al., 2006). Purified denmotoxin was found to bind to postsynaptic nAChRs in chick muscle preparations 100-fold more readily in comparison to mice nAChRs. This is also consistent with the lack of B. dendrophila crude venom toxic effects after injections into mice of doses up to 20 μg/g (Pawlak et al., 2006). Boigatoxin-A shows 78% identity to denmotoxin, but was only tested on chick neuromuscular junctions, so it is unknown if this toxin would display taxa targeting specificity. For the venom of B. irregularis, taxon-specific toxicity was found to be a result of the 3FTx complex irditoxin, B. irregularis dimeric toxin (Pawlak et al., 2009). Irditoxin inhibited postsynaptic nAChRs in chick biventer cervicis muscle preparations, but was three orders of magnitude less effective at the mammalian neuromuscular junction. This corresponds with the in vivo toxicity of irditoxin, which was nontoxic in mammals at doses up to 25 μg/g, but has an LD50 value of 0.55 μg/g in Hemidactylus geckos and 0.22 μg/g in chickens (G. domesticus) (Table 1; Pawlak et al., 2009). Irditoxin was the first identified covalently linked 3FTx heterodimeric complex; previous 3FTx complexes were observed to be non-covalent homodimers, examples including κ-bungarotoxins (Dewan et al., 1994). Both of these rear-fanged cat snake species, B. dendrophila and B. irregularis, are arboreal snakes with diets primarily of birds and lizards, especially as juveniles (Greene, 1989). Therefore, the 3FTx taxon-specific toxicity appears to be correlated to prey items these snakes commonly eat.
Taxon-specific 3FTxs have also been isolated in two New World species, O. fulgidus (Heyborne and Mackessy, 2013) and S. sulphureus (Modahl et al., 2018b). Fulgimotoxin, O. fulgidus monomeric toxin, has an LD50 of 0.28 μg/g in Anolis lizards, but was found to be nontoxic to mice at mass-adjusted doses more than 15 times the observed lizard LD50 (Heyborne and Mackessy, 2013). Oxybelis fulgidus is an arboreal snake that has a diet of birds and lizards (Robert, 1982), supporting the link between taxon-specific 3FTxs and snake diet. Sulditoxin, S. sulphureus dimeric toxin, was identified as the lizard-specific 3FTx complex in S. sulphureus venom, and has a LD50 value of 0.22 μg/g in Hemidactylus geckos, but is non-toxic to mammals up to doses of 5 μg/g (Table 1) (Modahl et al., 2018b). Sulditoxin was the second heterodimeric 3FTx complex discovered in RFS venoms, and interestingly shares the same cysteine residue pattern as irditoxin, likely using the same additional cysteines in the first and second loops (residues 17 and 42, respectively) to form an intermolecular disulfide linkage of the two subunits. A second taxon-specific toxin exists in S. sulphureus venom, but this toxin demonstrated specific toxicity toward mammals instead of lizards. This mammal-specific toxin was found to be a monomer and was named sulmotoxin 1, S. sulphureus monomeric toxin (Table 1; Modahl et al., 2018b). The third most abundant 3FTx in S. sulphureus venom, sulmotoxin 2, did not show taxon specificity. Spilotes sulphureus are arboreal snakes that are generalist predators, feeding on birds, lizards, amphibians, and small mammals (Andrade et al., 2017). The venom of S. sulphureus has two taxa specific 3FTxs present, and this correlates with its diet, which includes a diversity of prey.
Gene trees of RFS 3FTx sequences suggest that taxon-specific targeting convergently evolved in these separate snake species, at least for the dimeric complexes (irditoxin and sulditoxin), because even though these sequences share the same cysteine pattern, they cluster separately (Modahl et al., 2018b); and the two irditoxin subunit sequences are not common to all species within the Boiga genus (Dashevsky et al., 2018). It is also possible that these genes could have been lost in some Boiga species, which has been suggested for adaptive PLA2s in the rattlesnake clade (Dowell et al., 2016). Until genome sequences are obtained, the evolutionary histories of these genes are difficult to assess, especially given the challenges with assembling de novo venom gland transcriptomes. Currently, most toxin sequences are obtained from venom gland NGS, and assemblers that are commonly used struggle to assemble toxin genes because of the multitude of similar isoforms and high expression levels of these toxins (Macrander et al., 2015; Holding et al., 2018; Modahl et al., 2019).
Selection analyses have revealed conflicting trends in RSF 3FTx evolution, where positive selection was reported previously (Sunagar et al., 2013), and it has since been observed that selection is variable for different 3FTx clades (Dashevsky et al., 2018). Some RFS 3FTxs are evolving under negative selection, especially sequences of subunits that form the dimeric complexes, where residues that are responsible for subunit interactions must be maintained (Dashevsky et al., 2018). With the current shortage of 3FTx sequences in databases from RFS species and absence of genomes, it is difficult to fully recognize the evolutionary complexities for this group toxins. However, by continuing to study these sequences from phylogenetically diverse species that are converging on the same trophic adaptation, it is possible to begin to explore how a venom protein with a conserved structure, like 3FTxs, can evolve specific activities and alter its mechanism of action. These toxins are therefore ideal to examine protein structure-function relationships.
When lizard-specific 3FTx sequences from RFS species were aligned, conserved motifs were observed in the central loop, where residues critical to nAChR receptor binding are found for FFS 3FTxs erabutoxin-a (Pillet et al., 1993) and α-cobratoxin (Antil et al., 1999). These conserved sequences were identified as CYTLY and WAVK for residues 34–38 and 46–49, respectively (Heyborne and Mackessy, 2013). These two motifs were hypothesized to be involved in toxin specificity to lizard nAChRs. Sulmotoxin 1, which is mammal specific, exhibits CYNLY, a threonine to asparagine substitution, and WTVK, an alanine to threonine substitution, motifs in this region (Modahl et al., 2018b), supporting this hypothesis, because sulmotoxin 1 lacks lizard toxicity. The size of the central loops between sulmotoxin, denmotoxin, and fulgimotoxin is also variable (Figure 3A), with longer central loops observed in lizard-specific 3FTxs, in addition to a proline in this region, followed by a negatively charged amino acid (either an aspartic acid or glutamic acid, or both; Figure 3B). Site-directed mutagenesis studies have yet to be performed for these taxa-specific 3FTxs and would help to elucidate the residues responsible for taxon-specific receptor binding. Key residues involved in receptor interactions could also be different than what is known for elapid 3FTxs, and identification of novel receptor-ligand interactions could also provide insight into how covalent 3FTx complexes (irditoxin or sulditoxin) interact with nAChRs.
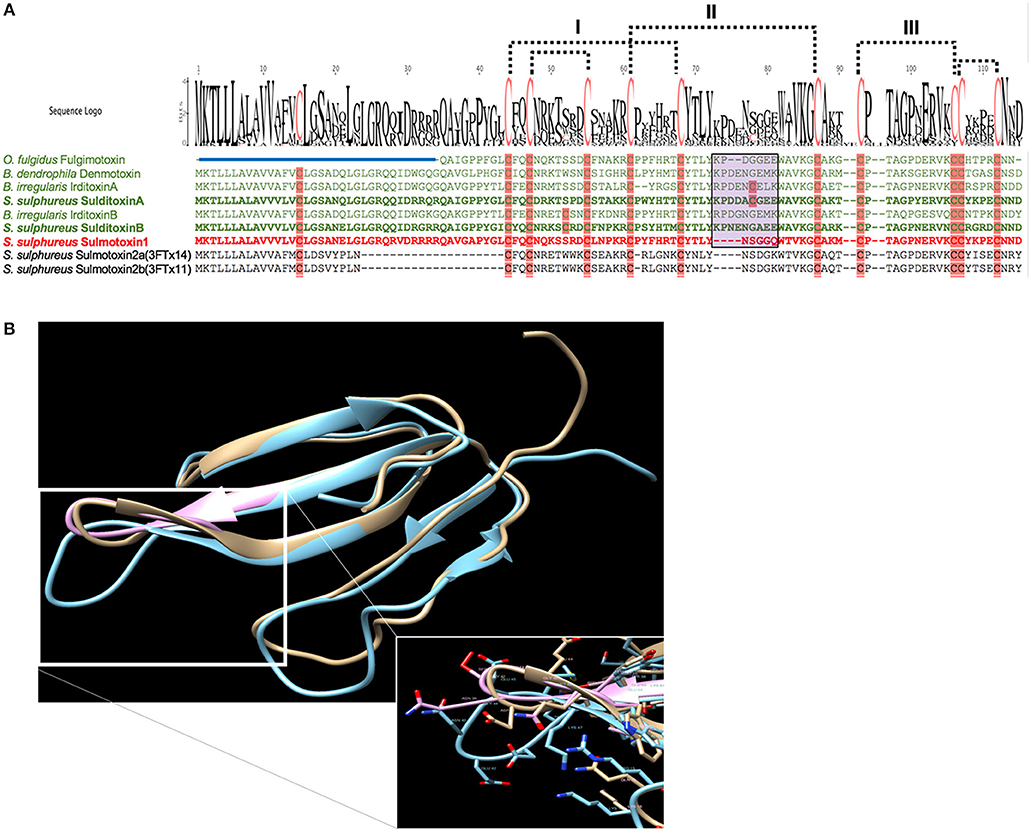
Figure 3. Taxon-specific three-finger toxin (3FTx) sequences (A) and structures (B). (A) Characterized lizard specific 3FTxs are shown in green, and sulmotoxin 1, a mammal-specific 3FTx, is in red. Sulmotoxin 2 is labeled twice because of the presence of two isoforms. Cysteines are highlighted in pink. The five disulfides characteristic of nonconventional 3FTxs are shown with dotted lines. The numbers above the alignments note the arrangement of the three 3FTx structural loops. Amino acid variation observed within the end of the central structural loop is highlighted in purple for characterized toxins. The signal peptides are indicated by the blue bar above all sequences except fulgimotoxin. (B) Structure of monomeric 3FTxs sulmotoxin 1 is displayed in pink, fulgimotoxin (C0HJD3.1) in orange, and denmotoxin (Q06ZW0.1) in blue. Differences in amino acids and the resulting structural alterations within the second/central 3FTx loop of lizard-specific monomeric 3FTxs are highlighted in insert. From Modahl et al. (2018b).
Metalloproteinases
All metalloproteinases that have been characterized from RFS venoms are of the P-III class and commonly exhibit fibrinogenolytic activity, with rapid degradation of the α-chain of fibrinogen and lower activity toward β- and γ-chains (Assakura et al., 1992; Peichoto et al., 2007; Weldon and Mackessy, 2012). Several SVMPs purified from the venom of P. olfersii degraded the α-chain of fibrinogen, with some also hydrolyzing the β-chain, but with differences in selectivity and rate (Assakura et al., 1994). In the venom of P. patagoniensis, the isolated SVMP named patagonfibrase was found to be the venom component responsible for the hydrolysis of the α-chain of fibrinogen, but not the β-chain (Peichoto et al., 2007), and for B. portoricensis, the SVMP alsophinase was found to hydrolyzed the α-subunit of fibrinogen almost instantly, and slightly degraded the β-chain after a 60 min incubation (Weldon and Mackessy, 2012). Although no individual SVMPs have been purified from the venom of A. prasina, there are SVMPs in this venom that act at a more rapid rate than those from the venom of B. portoricensis, hydrolyzing both α- and β-chains of fibrinogen (Figure 4A) even faster than what is observed with crude Prairie Rattlesnake (Crotalus viridis viridis) venom (Figure 4B; Modahl et al., 2018a). SVMPs of the P-III class are also found in venoms of elapid snakes but do not typically degrade the β-chain of fibrinogen and only target the α-chain (Evans, 1981; Guo et al., 2007; Sun and Bao, 2010). These studies demonstrate the underappreciated potency and functional diversity of SVMPs in RFS venoms.
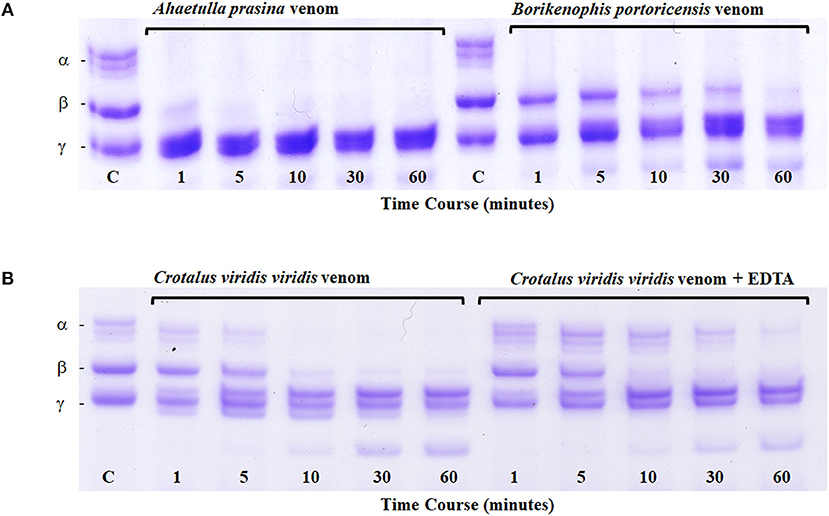
Figure 4. Fibrinogen digest assay. Ahaetulla prasina and Borikenophis portoricensis crude venoms (20 μg) were incubated with human fibrinogen over a 60-min time course (A). The control, labeled C, was removed before the addition of any venom and shows all intact fibrinogen subunits. Fibrinogen subunits are labeled α, β, and γ. Crotalus viridis viridis crude venom incubations with and without EDTA demonstrate the effects of active and inhibited metalloproteinase activity, respectively (B). From Modahl et al. (2018a) used with permission of Elsevier.
Isoforms of SVMP P-IIIs have been observed to have distinct coagulation mechanisms in humans, birds, and small rodents, with some isoforms more specialized for rats or chickens (Bernardoni et al., 2014). There have been several studies that suggest SVMPs from RFS venoms might also have taxon selectivity. Higher molecular mass proteins isolated from the venom of P. olfersii were found to produce an irreversible blockade in chick biventer cervicis preparations, but had no effect on mouse phrenic nerve-diaphragm preparations (Prado-Franceschi et al., 1996). A separate study of P. olfersii venom revealed that all large molecular mass protein spots on a 2-D gel were SVMPs (Ching et al., 2006). Venom from P. patagoniensis, induced a time-dependent neuromuscular blockade of chick biventer cervicis preparations, but had no effect of mouse phrenic nerve-diaphragm preparations (Carreiro da Costa et al., 2008). Although not currently tested, SVMPs in this venom might also be responsible for the taxon-specific neuromuscular effects, given that this is a closely related species and that the venom lacks 3FTxs (Harvey and Mackessy, unpubl. obs.). An SVMP-dominated pit viper venom (from Bothrops insularis) was shown to have differential effects on mouse phrenic nerve diaphragm and chick biventer cervicis muscle preparations (Cogo et al., 1993). In FFS venoms, variations in SVMP P-III isoform expression have been observed in rattlesnake populations that are locally adapted to prey (Margres et al., 2017), and a similar relationship between SVMP variants and prey targeting has been hypothesized for Bothrops neuwiedi venom SVMPs (Bernardoni et al., 2014). More work in this area is needed to determine if SVMPs from RFS venoms are indeed taxon-specific venom components and linked to snake diet. Dietary studies show that for Philodryas species, amphibians and lizards are the primary prey taken (Marques and Hartmann, 2005; López and Giraudo, 2008) and isolated SVMPs have yet to be evaluated on these taxa. In addition, it would be important to determine what molecular interactions mediate SVMP taxon-specific toxicity.
SVMPs from RFS could therefore provide interesting models for future work focused on structure-function relationships. How do some SVMPs degrade both α- and β-chains while others do not? How can SVMPs be selective to prey type? Venoms from RFS species are ideal for addressing these questions because these venoms are overall less complex. For instance, vipers have several different classes of SVMPs, as well as serine proteinases that can also degrade fibrinogen, making it difficult to study specific toxin activity without multiple purification steps. Because of the abundance and extensive occurrence of P-III SVMPs in RFS venoms, these proteins can provide insight into the evolution of this toxin family and the biological roles that SVMPs provide in venoms.
Rear-Fanged Snake Venoms: Biological Roles
Recognized biological roles of venom proteins include: (1) a trophic role—use in prey capture by affecting prey locomotor ability by immobilization and preventing escape, by inducing quiescence in prey, and/or by causing rapid death; (2) participating in digestion, such as by accelerating digestion of bulky prey items; (3) helping to lubricate prey during ingestion; and (4) functioning as a form of defense (Kardong, 2002). It is likely that prey capture is the primary role, as there have been multiple publications demonstrating a strong link between venom and snake diet (Mackessy, 1985, 1988; da Silva and Aird, 2001; Mackessy et al., 2003; Li et al., 2005a; Pawlak et al., 2006, 2009; Barlow et al., 2009; Modahl et al., 2018b). Venom has been observed to be variable in composition (Chippaux et al., 1991), and this variation has been associated with dietary differences from geographic locality (Daltry et al., 1996; Creer et al., 2003; Smiley-Walters et al., 2017; Sousa et al., 2017) and ontogenetic dietary shifts (Mackessy, 1985, 1988; Andrade and Abe, 1999; Mackessy et al., 2006; Cipriani et al., 2017).
Venoms from both FFS and RFS species have been found to show differential toxicity toward prey items (da Silva and Aird, 2001; Mackessy et al., 2006; Gibbs and Mackessy, 2009), and population-level correlations between venomous snakes' venoms and their sympatric prey have been proposed (Margres et al., 2017; Smiley-Walters et al., 2017). The toxicity of venoms from four species of the FFS genus Echis to a natural scorpion prey was found to be associated with the degree of utilization of arthropods in the diet for each Echis species (Barlow et al., 2009), and a similar trend was observed for Sistrurus rattlesnakes that vary in the extent of amphibian, lizard and mammal prey taken for each species (Gibbs and Mackessy, 2009). Further, snakes with a greater dietary breadth appear to have greater toxin diversity within their venoms (Pahari et al., 2007; Calvete et al., 2012; but see Zancolli et al., 2019). The opposite also appears to be true if there is a reduction in dietary diversity or when venom no longer plays a pivotal role in prey capture. The sea snake Aipysurus eydouxii that feeds exclusively on fish eggs has atrophied venom glands, a loss of venom neurotoxicity and a reduction in the diversity of toxins in the venom of this species (Li et al., 2005b); a similar trend is seen in the terrestrial elapid Brachyurophis (Goodyear and Pianka, 2008). One of the advantages of exploring RFS venoms is that various species feed on a variety of specific prey types and show varying prey capture strategies, from constriction to envenomation, making these venoms ideal for predator-prey evolutionary studies.
Prey Capture
Predatory strategy, venom vs. constriction, can drive venom evolution and snake behavior, favoring the evolution of venom that allows for a greater diversity and larger prey to be taken with minimum prey retaliation, and transitioning away from reliance on constriction (Savitsky, 1980). Boiga irregularis has the lizard-specific 3FTx complex irditoxin (Table 1) in abundance within its venom, and lizard prey is bitten and held by these snakes (Mackessy et al., 2006). However, mammalian prey is constricted by B. irregularis, and this is likely because this venom lacks mammal-specific toxins, as the crude venom murine LD50 value is >30 μg/g (Table 1) (Mackessy et al., 2006). Spilotes sulphureus venom has two taxon-specific 3FTxs, one that is lizard-specific and one that is mammal-specific (Table 1), and both are abundant in the venom; constriction is not observed for S. sulphureus (Boos, 2001; Modahl et al., 2018b). Larger mammalian prey are metabolically more favorable to consume than smaller lizard prey, and sulmotoxin, the mammal-specific 3FTx, appears to be a more recent adaptation facilitating mammalian prey capture. In the case of the FFS snake Naja kaouthia, a species that also does not use constriction, this highly toxic snake has one main 3FTx, α-cobratoxin, that is lethal toward both mammal and lizard prey (Table 1; Modahl et al., 2016). These examples demonstrate two different venom evolutionary strategies to target a greater diversity of prey, one venom with a single 3FTx that has evolved lethal toxicity to many prey species, and a second venom in which toxin gene duplication and neofunctionalization has resulted in separate 3FTxs that are selectively toxic toward different prey types. Again, toxin evolution in rear-fanged snake venoms can provide key insights into venom evolution and predatory strategies, as these snakes vary greatly in predatory behavior and types of prey consumed.
When atypical prey items are taken, venom proteins with unique activities may be discovered. This has been the case for several FFS species, such as snakes that consume other snakes. The King Cobra, Ophiophagus hannah, is a species that consumes other snakes and was found to have a toxin from a new venom protein superfamily, ohanin, in its venom (Pung et al., 2005). The Long-glanded Coral Snake, Calliophis bivirgatus, also a snake-eating snake, was found to have a 3FTx with unusual activity toward sodium ion channels (Yang et al., 2016). Snakes of the genus Bungarus, which also commonly feed on other snakes, exhibit unique 3FTx homodimer complexes (κ-bungarotoxins; Dewan et al., 1994). Rear-fang snakes have even more dietary specialists, including species that feed on scorpions, spiders or centipedes, and so it is expected that novel toxins remain to be discovered in venoms of this diverse group of colubroid snakes.
Therefore, it is likely that venoms from RFS species potentially have many new and currently undiscovered potent toxins. Experimental studies have found that only about half of the total venom expended by the RFS B. irregularis is delivered into the viscera of prey, and the other half remains embedded in the skin (Hayes et al., 1993). But even with these lower yields delivered into prey tissue, effects are still observed as prey that is removed without being consumed by the snake may become sluggish and eventually die after many minutes or several hours (Hayes et al., 1993). In combination with a less rapid venom delivery system, these snakes also generally have less complex venom (Mackessy, 2010b; Peichoto et al., 2012), resulting in venom toxins that are optimized for specific types of prey. Venoms from most species of RFS are uncharacterized, and because of historic biases, most LD50 work utilizes domestic murine models due to availability of mice, genetic uniformity and presumed closer applicability to humans. However, by excluding other models, such as lizards, other vertebrates and invertebrates, one can overlook the biological potency of specialized venom proteins that may have been selected for toxic effects on non-mammalian prey (cf. Richards et al., 2012; Smiley-Walters et al., 2018).
Digestion
Metalloproteinases in rattlesnake venoms have been suggested to facilitate efficient digestion of prey at suboptimal temperatures or when large prey are consumed (Mackessy, 1988, 2010a); however, several studies have indicated that envenomation does not increase digestive efficacy (McCue, 2007; Chu et al., 2009). In the case of many rattlesnake species, the extent of SVMP activity in a venom is negatively correlated with overall venom toxicity (Mackessy, 2010a). This has led to rattlesnake venoms being characterized as type I (SVMP-dominated venoms that are less toxic) or type II (venom that is more toxic but with low metalloproteinase activity; Mackessy, 2010a). A similar dichotomy is found in many RFS venoms, where a venom is either dominated by toxic 3FTxs or enzymatic SVMPs (McGivern et al., 2014). For the RFS venoms that are heavily dominated by SVMPs, these venom components likely also aid in prey predigestion, as similar fibrinogenolytic activity is observed for SVMPs from both rattlesnakes and RFS species (Figure 4; Modahl et al., 2018a). Studying RFS venoms can reveal parallels to trends seen in FFS venoms, emphasizing the important of specific venom proteins in facilitating prey handling in diverse species of venomous snakes, regardless of the venom delivery system.
Future Research
Advancements and integrations of research technologies now allow much more detailed approaches to characterize unknown venoms and individual toxins. Transcriptomes assembled from venom glands provide custom databases to be paired with proteomics and make it possible to identify proteins in a venom even when they are currently missing from public databases (Modahl et al., 2019). For the recently characterized sulditoxin, without the species-specific NGS transcriptome, interpretation of the MS/MS spectra of the trypsin digested toxin by searching against public domain databases did not result in identification of this isolated toxin. By including the NGS venom gland transcriptome, the identification of the exact transcript and thus the full amino acid sequence was readily achieved (Modahl et al., 2018b). Previously, a similar situation produced negative results; from the analysis of RFS Rhamphiophis oxyrhynchus venom, a neurotoxin was isolated and partially characterized, but it showed no similarities to any toxins in public databases at that time, and the exact venom protein classification could not be determined (Lumsden et al., 2007). Unique or unusual toxin sequences still may not be present in public databases, and hence species-specific venom gland transcriptome databases are critical for identification (Campos et al., 2016; Modahl et al., 2018b, 2019).
Newer technologies have allowed for modifications to various characterization approaches. For venom gland transcriptome assemblies, venom protein identities are usually based on known key word searches for toxins. More recently, several machine learning programs have been developed to identify unknown toxins from large transcriptomic datasets (Gacesa et al., 2016; Macrander et al., 2018). Additionally, sequencing other snake tissues besides venom glands has provided insight into how gene expression can help in the identification of true toxins, what mechanisms result in toxin high expression in the venom gland in comparison to other tissues, and what gene homologs are present in other tissues (Hargreaves et al., 2014; Junqueira-de-Azevedo et al., 2015; Reyes-Velasco et al., 2015). Genomes of RFS are also useful to identify toxin gene duplication events (Perry et al., 2018). They help to provide support to approaches to identify where toxin genes originate and selection pressures they experience.
Because rear-fanged venomous snakes encompass such a large diversity of colubroid snakes, venom evolution can be studied on a broader evolutionary scale, addressing such questions as the effect of phylogeny on venom evolution or dietary specialization. There is an extreme range of toxicity of RFS venoms to humans, with some species being life-threating and others being harmless. This gradient of toxicity can be used for explorations into venom in terms of the biological roles of individual venom proteins, such as how toxicity and prey specificity can develop.
Venoms from rear-fanged snakes show many parallels to those of FFS, but one area that has yet to be explored is the level of venom variation within a single RFS species. The venom of B. irregularis has been found to exhibit ontogenetic variation related to diet, and venoms from different populations (Indonesia vs. Guam) show demonstrably different toxin compositions (Mackessy et al., 2006; Pla et al., 2017a). Conversely, individuals from the same populations of B. portoricensis and A. prasina showed very little venom variation (Modahl et al., 2018a), but population-level variation in venom composition is currently unknown for RFS. Variation in rear-fanged snake venoms deserves more attention, as it can help to uncover the mechanisms behind commonly observed venom variation, which has been an area of controversy. Studies are also still lacking at the level of posttranslational modifications of venom proteins, for most venomous snakes, and how this contributes to overall venom diversity.
Another neglected area of research is the interactive or synergistic potential of toxins. Because studying purified toxins usually requires a reductionist approach, few studies have attempted to evaluate interactions between toxins. Dimeric toxins, such as sulditoxin and irditoxin, consist of two dissimilar 3FTxs, but the importance of dimeric associations to specific pharmacological activity is unknown. Interactive complexes are likely between venom toxins, including RFS toxins, and the generally lower complexity of these venoms should make such investigations more tractable. Venoms from RFS therefore have the potential to contribute importantly to our understanding of many phenomena still outstanding in toxinology, and there are multitude opportunities for investigations of these venoms, from basic compositional analyses to detailed structure-function studies to NGS-based investigations of regulation and evolution of venom expression systems.
Author Contributions
CM wrote the original draft. SM edited, contributed to, and modified the manuscript. CM and SM created the figures, formulated the original concepts, and edited the final versions of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
1. ^Toxinology Department, Women's and Children's Hospital, Adelaide, Australia.
References
Acosta, O., Leiva, L. C., Peichoto, M. E., Marunak, S., Teibler, P., and Rey, L. (2003). Hemorrhagic activity of the Duvernoy's gland secretion of the xenodontine colubrid Philodryas patagoniensis from the north-east region of Argentina. Toxicon 41, 1007–1012. doi: 10.1016/S0041-0101(03)00074-6
Aird, S. D., Watanabe, Y., Villar-Briones, A., Roy, M. C., Terada, K., and Mikheyev, A. S. (2013). Quantitative high-throughput profiling of snake venom gland transcriptomes and proteomes (Ovophis okinavensis and Protobothrops flavoviridis). BMC Genomics 14:790. doi: 10.1186/1471-2164-14-790
Andrade, D. V., and Abe, A. S. (1999). Relationship of venom ontogeny and diet in Bothrops. Herpetologica 55, 200–204.
Andrade, H., Almeida, R. P. S., and Dias, E. J. R. (2017). Spilotes Sulphureus Wagler, 1824 (Squamata: Colubridae): Review of Distribution and First Record in the State of Sergipe, Northeastern Brazil. Available online at: https://checklist.pensoft.net/article/19657/ (accessed May 16, 2019).
Antil, S., Servent, D., and Ménez, A. (1999). Variability among the sites by which curaremimetic toxins bind to Torpedo acetylcholine receptor, as revealed by identification of the functional residues of α-cobratoxin. J. Biol. Chem. 274, 34851–34858. doi: 10.1074/jbc.274.49.34851
Assakura, M. T., Reichl, A. P., and Mandelbaum, F. R. (1994). Isolation and characterization of five fibrin(ogen)olytic enzymes from the venom of Philodryas olfersii (green snake). Toxicon 32, 819–831. doi: 10.1016/0041-0101(94)90007-8
Assakura, M. T., Salomão, M. G., Puorto, G., and Mandelbaum, F. R. (1992). Hemorrhagic, fibrinogenolytic and edema-forming activities of the venom of the colubrid snake Philodryas olfersii (green snake). Toxicon 30, 427–438. doi: 10.1016/0041-0101(92)90539-H
Barlow, A., Pook, C. E., Harrison, R. A., and Wüster, W. (2009). Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc. Biol. Sci. 276, 2443–2449. doi: 10.1098/rspb.2009.0048
Bernardoni, J. L., Sousa, L. F., Wermelinger, L. S., Lopes, A. S., Prezoto, B. C., Serrano, S. M., et al. (2014). Functional variability of snake venom metalloproteinases: adaptive advantages in targeting different prey and implications for human envenomation. PLoS ONE 9:e109651. doi: 10.1371/journal.pone.0109651
Boos, H. E. A. (2001). The Snakes of Trinidad and Tobago. College Station, TX: Texas AandM University Press.
Bourne, Y., Talley, T. T., Hansen, S. B., Taylor, P., and Marchot, P. (2005). Crystal structure of a Cbtx-AChBP complex reveals essential interactions between snake alpha-neurotoxins and nicotinic receptors. EMBO J. 24, 1512–1522. doi: 10.1038/sj.emboj.7600620
Braud, S., Bon, C., and Wisner, A. (2000). Snake venom proteins acting on hemostasis. Biochimie 82, 851–859. doi: 10.1016/S0300-9084(00)01178-0
Broaders, M., Faro, C., and Ryan, M. F. (1999). Partial purification of acetylcholine receptor binding components from the Duvernoy's secretions of blanding's tree snake (Boiga blandingi) and the mangrove snake (Boiga dendrophila). J. Nat. Toxins 8, 155–166.
Broaders, M., and Ryan, M. F. (1997). Enzymatic properties of the Duvernoy's secretion of Blanding's tree snake (Boiga blandingi) and of the mangrove snake (Boiga dendrophila). Toxicon 35, 1143–1148. doi: 10.1016/S0041-0101(96)00207-3
Brown, R. L., Haley, T. L., West, K. A., and Crabb, J. W. (1999). Pseudechetoxin: a peptide blocker of cyclic nucleotide-gated ion channels. Proc. Natl. Acad. Sci. U.S.A. 96, 754–759. doi: 10.1073/pnas.96.2.754
Calvete, J. J., Ghezellou, P., Paiva, O., Matainaho, T., Ghassempour, A., Goudarzi, H., et al. (2012). Snake venomics of two poorly known Hydrophiinae: comparative proteomics of the venoms of terrestrial Toxicocalamus longissimus and marine Hydrophis cyanocinctus. J. Proteomics 75, 4091–4101. doi: 10.1016/j.jprot.2012.05.026
Campos, P. F., Andrade-Silva, D., Zelanis, A., Paes Leme, A. F., Rocha, M. M. T., Menezes, M. C., et al. (2016). Trends in the evolution of snake toxins underscored by an integrative omics approach to profile the venom of the colubrid Phalotris mertensi. Genome Biol. Evol. 8, 2266–2287. doi: 10.1093/gbe/evw149
Carlsson, F. H. (1975). Snake venom toxins. The primary structure of protein S4C11. A neurotoxin homologue from the venom of forest cobra (Naja melanoleuca). Biochim. Biophys. Acta 400, 310–321. doi: 10.1016/0005-2795(75)90186-5
Carreiro da Costa, R. S., Prudencio, L., Ferrari, E. F., Souza, G. H., de Mello, S. M., Prianti Junior, A. C., et al. (2008). Neuromuscular action of venom from the South American colubrid snake Philodryas patagoniensis. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 148, 31–38. doi: 10.1016/j.cbpc.2008.03.006
Casewell, N. R., Wagstaff, S. C., Harrison, R. A., Renjifo, C., and Wüster, W. (2011). Domain loss facilitates accelerated evolution and neofunctionalization of duplicate snake venom metalloproteinase toxin genes. Mol. Biol. Evol. 28, 2637–2649. doi: 10.1093/molbev/msr091
Casewell, N. R., Wüster, W., Vonk, F. J., Harrison, R. A., and Fry, B. G. (2013). Complex cocktails: the evolutionary novelty of venoms. Trends Ecol. Evol. 28, 219–229. doi: 10.1016/j.tree.2012.10.020
Castoe, T. A., de Koning, A. P. J., Hall, K. T., Card, D. C., Schield, D. R., Fujita, M. K., et al. (2013). The Burmese python genome reveals the molecular basis for extreme adaptation in snakes. Proc. Natl. Acad. Sci. U.S.A. 110, 20645–20650. doi: 10.1073/pnas.1314475110
Chang, C. C., and Lee, C. Y. (1963). Isolation of neurotoxins from the venom of Bungarus multicinctus and their modes of neuromuscular blocking action. Arch. Int. Pharmacodyn. Ther. 144, 241–257.
Ching, A. T., Paes Leme, A. F., Zelanis, A., Rocha, M. M. T., Furtado, M. F. D., Silva, D. A., et al. (2012). Venomics profiling of Thamnodynastes strigatus unveils matrix metalloproteinases and other novel proteins recruited to the toxin arsenal of rear-fanged snakes. J. Proteome Res. 11, 1152–1162. doi: 10.1021/pr200876c
Ching, A. T. C., Rocha, M. M. T., Paes Leme, A. F., Pimenta, D. C., Furtado, M. F. D., Serrano, S. M. T., et al. (2006). Some aspects of the venom proteome of the Colubridae snake Philodryas olfersii revealed from a Duvernoy's (venom) gland transcriptome. FEBS Lett. 580, 4417–4422. doi: 10.1016/j.febslet.2006.07.010
Chippaux, J. P., Williams, V., and White, J. (1991). Snake venom variability: methods of study, results and interpretation. Toxicon 29, 1279–1303. doi: 10.1016/0041-0101(91)90116-9
Chu, C.-W., Tsai, T.-S., Tsai, I.-H., Lin, Y.-S., and Tu, M.-C. (2009). Prey envenomation does not improve digestive performance in Taiwanese pit vipers (Trimeresurus gracilis and T. stejnegeri stejnegeri). Compar. Biochem. Physiol. A 152, 579–585. doi: 10.1016/j.cbpa.2009.01.006
Chung, C., Wu, B. N., Yang, C. C., and Chang, L. S. (2002). Muscarinic toxin-like proteins from Taiwan banded krait (Bungarus multicinctus) venom: purification, characterization and gene organization. Biol. Chem. 383, 1397–1406. doi: 10.1515/BC.2002.158
Cipriani, V., Debono, J., Goldenberg, J., Jackson, T. N. W., Arbuckle, K., Dobson, J., et al. (2017). Correlation between ontogenetic dietary shifts and venom variation in Australian brown snakes (Pseudonaja). Compar. Biochem. Physiol. C Toxicol. Pharmacol. 197, 53–60. doi: 10.1016/j.cbpc.2017.04.007
Cogo, J. C., Prado-Franceschi, J., Cruz-Höfling, M. A., Corrado, A. P., and Rodrigues-Simioni, L. (1993). Effect of Bothrops insularis venom on the mouse and chick nerve-muscle preparation. Toxicon 31, 1237–1247. doi: 10.1016/0041-0101(93)90397-2
Correa-Netto, C., Junqueira-de-Azevedo Ide, I., Silva, D. A., Ho, P. L., Leitão-de-Araujo, M., Alves, M. L., et al. (2011). Snake venomics and venom gland transcriptomic analysis of Brazilian coral snakes, Micrurus altirostris and M. corallinus. J. Proteomics 74, 1795–1809. doi: 10.1016/j.jprot.2011.04.003
Creer, S., Malhotra, A., Thorpe, R. S., Stocklin, R. S., Favreau, P. S., and Hao Chou, W. S. (2003). Genetic and ecological correlates of intraspecific variation in pitviper venom composition detected using matrix-assisted laser desorption time-of-flight mass spectrometry (MALDI-TOF-MS) and isoelectric focusing. J. Mol. Evol. 56, 317–329. doi: 10.1007/s00239-002-2403-4
da Silva, N. J., and Aird, S. D. (2001). Prey specificity, comparative lethality and compositional differences of coral snake venoms. Compar. Biochem. Physiol. C Toxicol. Pharmacol. 128, 425–456. doi: 10.1016/S1532-0456(00)00215-5
Daltry, J. C., Wüster, W., and Thorpe, R. S. (1996). Diet and snake venom evolution. Nature 379, 537–540.
Dashevsky, D., Debono, J., Rokyta, D., Nouwens, A., Josh, P., and Fry, B. G. (2018). Three-finger toxin diversification in the venoms of cat-eye snakes (Colubridae: Boiga). J. Mol. Evol. 86, 531–545. doi: 10.1007/s00239-018-9864-6
de Weille, J. R., Schweitz, H., Maes, P., Tartar, A., and Lazdunski, M. (1991). Calciseptine, a peptide isolated from black mamba venom, is a specific blocker of the L-type calcium channel. Proc. Natl. Acad. Sci. U.S.A. 88, 2437–2440. doi: 10.1073/pnas.88.6.2437
Dewan, J. C., Grant, G. A., and Sacchettini, J. C. (1994). Crystal structure of kappa-bungarotoxin at 2.3-A resolution. Biochemistry 33, 13147–13154. doi: 10.1021/bi00248a026
Dowell, N. L., Giorgianni, M. W., Kassner, V. A., Selegue, J. E., Sánchez, E. E., and Carroll, S. B. (2016). The deep origin and recent loss of venom toxin genes in rattlesnakes. Curr. Biol. 26, 2434–2445. doi: 10.1016/j.cub.2016.07.038
Durban, J., Juárez, P., Angulo, Y., Lomonte, B., Flores-Diaz, M., Alape-Girón, A., et al. (2011). Profiling the venom gland transcriptomes of Costa Rican snakes by 454 pyrosequencing. BMC Genomics 12:259. doi: 10.1186/1471-2164-12-259
Estrella, A., Sánchez, E. E., Galán, J. A., Tao, W. A., Guerrero, B., Navarrete, L. F., et al. (2011). Characterization of toxins from the broad-banded water snake Helicops angulatus (Linnaeus, 1758): isolation of a cysteine-rich secretory protein, helicopsin. Arch. Toxicol. 85, 305–313. doi: 10.1007/s00204-010-0597-6
Evans, H. J. (1981). Cleavage of the Aα-chain of fibrinogen and the α-polymer of fibrin by the venom of spitting cobra (Naja nigricollis). Biochim. Biophys. Acta 660, 219–226. doi: 10.1016/0005-2744(81)90163-7
Fox, J. W., and Serrano, S. M. (2005). Structural considerations of the snake venom metalloproteinases, key members of the M12 reprolysin family of metalloproteinases. Toxicon 45, 969–985. doi: 10.1016/j.toxicon.2005.02.012
Fry, B. G., Casewell, N. R., Wüster, W., Vidal, N., Young, B., and Jackson, T. N. (2012a). The structural and functional diversification of the Toxicofera reptile venom system. Toxicon 60, 434–448. doi: 10.1016/j.toxicon.2012.02.013
Fry, B. G., Lumsden, N. G., Wüster, W., Wickramaratna, J. C., Hodgson, W. C., and Kini, R. M. (2003a). Isolation of a neurotoxin (alpha colubritoxin) from a nonvenomous colubrid: Evidence for early origin of venom in snakes. J. Mol. Evol. 57, 446–452. doi: 10.1007/s00239-003-2497-3
Fry, B. G., Scheib, H., Junqueira de Azevedo, I., de, L., Silva, D. A., and Casewell, N.R. (2012b). Novel transcripts in the maxillary venom glands of advanced snakes. Toxicon 59, 696–708. doi: 10.1016/j.toxicon.2012.03.005
Fry, B. G., Scheib, H., van der Weerd, L., Young, B., McNaughtan, J., Ramjan, S. F. R., et al. (2008). Evolution of an arsenal: structural and functional diversification of the venom system in the advanced snakes (Caenophidia). Mol. Cell. Proteomics 7:215. doi: 10.1074/mcp.M700094-MCP200
Fry, B. G., Wüster, W., Kini, R. M., Brusic, V., Khan, A., Venkataraman, D., et al. (2003b). Molecular evolution and phylogeny of elapid snake venom three-finger toxins. J. Mol. Evol. 57, 110–129. doi: 10.1007/s00239-003-2461-2
Gacesa, R., Barlow, D., and Long, P. (2016). Machine learning can differentiate venom toxins from other proteins having non-toxic physiological functions. PeerJ Comput. Sci. 2:e90. doi: 10.7717/peerj-cs.90
Gibbs, H. L., and Mackessy, S. P. (2009). Functional basis of a molecular adaptation: prey-specific toxic effects of venom from Sistrurus rattlesnakes. Toxicon 53, 672–679. doi: 10.1016/j.toxicon.2009.01.034
Gomaraschi, M., Bonacina, F., and Norata, G. D. (2019). Lysosomal acid lipase: from cellular lipid handler to immunometabolic target. Trends Pharmacol. Sci. 40, 104–115. doi: 10.1016/j.tips.2018.12.006
Gomes, A. R., Byregowda, S. M., Veeregowda, B. M., and Balamurugan, V. (2016). An overview of heterologous expression host systems for the production of recombinant proteins. Adv. Anim. Vet. Sci. 4, 346–356. doi: 10.14737/journal.aavs/2016/4.7.346.356
Goodyear, S. E., and Pianka, E. R. (2008). Sympatric ecology of five species of fossorial snakes (Elapidae) in Western Australia. J. Herpetol. 42, 279–285. doi: 10.1670/07-1391.1
Greene, H. W. (1989). Ecological, evolutionary, and conservation implications of feeding biology in Old World cat snakes, genus Boiga (Colubridae). Proc. Calif. Acad. Sci. 46, 193–207.
Guo, X.-X., Zeng, L., Lee, W.-H., Zhang, Y., and Jin, Y. (2007). Isolation and cloning of a metalloproteinase from king cobra snake venom. Toxicon 49, 954–965. doi: 10.1016/j.toxicon.2007.01.003
Hargreaves, A. D., Swain, M. T., Logan, D. W., and Mulley, J. F. (2014). Testing the Toxicofera: comparative transcriptomics casts doubt on the single, early evolution of the reptile venom system. Toxicon 92, 140–156. doi: 10.1016/j.toxicon.2014.10.004
Hart, A. J., Isbister, G. K., and Hodgson, W. C. (2012). In vitro neurotoxic effects of Pseudechis spp. venoms: a comparison of avian and murine skeletal muscle preparations. Toxicon 63:112–115. doi: 10.1016/j.toxicon.2012.12.002
Hart, A. J., Isbister, G. K., O'Donnell, P., Williamson, N. A., and Hodgson, W. C. (2013). Species differences in the neuromuscular activity of post-synaptic neurotoxins from two Australian black snakes (Pseudechis porphyriacus and Pseudechis colletti). Toxicol. Lett. 219, 262–268. doi: 10.1016/j.toxlet.2013.03.026
Hayes, W. K., Lavin-Murcio, P., and Kardong, K. V. (1993). Delivery of Duvernoy's secretion into prey by the brown tree snake, Boiga irregularis (Serpentes:Colubridae). Toxicon 31, 881–887. doi: 10.1016/0041-0101(93)90223-6
Heyborne, W. H., and Mackessy, S. P. (2013). Identification and characterization of a taxon-specific three-finger toxin from the venom of the Green Vinesnake (Oxybelis fulgidus; family Colubridae). Biochimie 95, 1923–1932. doi: 10.1016/j.biochi.2013.06.025
Hill, R. E., and Mackessy, S. P. (1997). Venom yields from several species of colubrid snakes and differential effects of ketamine. Toxicon 35, 671–678. doi: 10.1016/S0041-0101(96)00174-2
Hill, R. E., and Mackessy, S. P. (2000). Characterization of venom (Duvernoy's secretion) from twelve species of colubrid snakes and partial sequence of four venom proteins. Toxicon 38, 1663–1687. doi: 10.1016/S0041-0101(00)00091-X
Hite, L. A., Jia, L. G., Bjarnason, J. B., and Fox, J. W. (1994). cDNA sequences for four snake venom metalloproteinases: structure, classification, and their relationship to mammalian reproductive proteins. Arch. Biochem. Biophys. 308, 182–191. doi: 10.1006/abbi.1994.1026
Holding, M. L., Margres, M. J., Mason, A. J., Parkinson, C. L., and Rokyta, D. R. (2018). Evaluating the performance of de novo assembly methods for venom-gland transcriptomics. Toxins (Basel) 10:249. doi: 10.3390/toxins10060249
Huang, P., and Mackessy, S. P. (2004). Biochemical characterization of phospholipase A2 (trimorphin) from the venom of the Sonoran Lyre Snake Trimorphodon biscutatus lambda (family Colubridae). Toxicon 44, 27–36. doi: 10.1016/j.toxicon.2004.03.027
Jackson, K. (2003). The evolution of venom-delivery systems in snakes. Zool. J. Linn. Soc. 137, 337–354. doi: 10.1046/j.1096-3642.2003.00052.x
Junqueira-de-Azevedo, I. L., Bastos, C. M. V., Ho, P. L., Luna, M. S., Yamanouye, N., and Casewell, N. R. (2015). Venom-related transcripts from Bothrops jararaca tissues provide novel molecular insights into the production and evolution of snake venom. Mol. Biol. Evol. 32, 754–766. doi: 10.1093/molbev/msu337
Junqueira-de-Azevedo, I. L., Campos, P. F., Ching, A. T., and Mackessy, S. P. (2016). Colubrid venom composition: an -omics perspective. Toxins 8:E230. doi: 10.3390/toxins8080230
Kamiguti, A. S., Theakston, R. D., Sherman, N., and Fox, J. W. (2000). Mass spectrophotometric evidence for P-III/P-IV metalloproteinases in the venom of the Boomslang (Dispholidus typus). Toxicon 38, 1613–1620. doi: 10.1016/S0041-0101(00)00089-1
Kardong, K. V. (2002). Colubrid snakes and Duvernoy's “venom” glands. J. Toxicol. 21, 1–19. doi: 10.1081/TXR-120004739
Karlsson, E. (1973). Chemistry of some potent animal toxins. Experientia 29, 1319–1327. doi: 10.1007/BF01922797
Karlsson, E., Jolkkonen, M., Mulugeta, E., Onali, P., and Adem, A. (2000). Snake toxins with high selectivity for subtypesof muscarinic acetylcholine receptors. Biochimie 82, 793–806. doi: 10.1016/S0300-9084(00)01176-7
Kini, R. M. (2003). Excitement ahead: structure, function and mechanism of snake venom phospholipase A2 enzymes. Toxicon 42, 827–840. doi: 10.1016/j.toxicon.2003.11.002
Kini, R. M., and Doley, R. (2010). Structure, function and evolution of three-finger toxins: mini proteins with multiple targets. Toxicon 56, 855–867. doi: 10.1016/j.toxicon.2010.07.010
Kini, R. M., Haar, N. C., and Evans, H. J. (1988). Non-enzymatic inhibitors of coagulation and platelet aggregation from Naja nigricollis venom are cardiotoxins. Biochem. Biophys. Res. Commun. 150, 1012–1016 doi: 10.1016/0006-291X(88)90729-2
Kochva, E. (1962). On the lateral jaw musculature of the solenoglypha with remarks on some other snakes. J. Morphol. 110, 227–284. doi: 10.1002/jmor.1051100209
Komori, K., Konishi, M., Maruta, Y., Toriba, M., Sakai, A., Matsuda, A., et al. (2006). Characterization of a novel metalloproteinase in duvernoy's gland of Rhabdophis tigrinus tigrinus. J. Toxicol. Sci. 31, 157–168. doi: 10.2131/jts.31.157
Kuch, U., and Mebs, D. (2002). Envenomations by colubrid snakes in Africa, Europe, and the Middle East. J. Toxicol. 21, 159–179. doi: 10.1081/TXR-120004745
Li, M., Fry, B. G., and Kini, R. M. (2005a). Eggs-only diet: Its implications for the toxin profile changes and ecology of the marbled sea snake (Aipysurus eydouxii). J. Mol. Evol. 60, 81–89. doi: 10.1007/s00239-004-0138-0
Li, M., Fry, B. G., and Kini, R. M. (2005b). Putting the brakes on snake venom evolution: the unique molecular evolutionary patterns of Aipysuras eydouxii (Marbled sea snake) phospholipase A2 toxins. Mol. Biol. Evol. 22, 934–941. doi: 10.1093/molbev/msi077
Lodovicho, M. E., Costa, T. R., Bernardes, C. P., Menaldo, D. L., Zoccal, K. F., Carone, S. E., et al. (2017). Investigating possible biological targets of Bj-CRP, the first cysteine-rich secretory protein (CRISP) isolated from Bothrops jararaca snake venom. Toxicol. Lett. 265, 156–169. doi: 10.1016/j.toxlet.2016.12.003
López, M. S., and Giraudo, A. R. (2008). Ecology of the snake Philodryas patagoniensis (Serpentes, Colubridae) from Northeast Argentina. J. Herpetol. 42, 474–480. doi: 10.1670/07-087.1
Lu, Q., Navdaev, A., Clemetson, J. M., and Clemetson, K. J. (2005). Snake venom C-type lectins interacting with platelet receptors. Structure-function relationships and effects on haemostasis. Toxicon 45, 1089–1098. doi: 10.1016/j.toxicon.2005.02.022
Lumsden, N. G., Banerjee, Y., Kini, R. M., Kuruppu, S., and Hodgson, W. C. (2007). Isolation and characterization of rufoxin, a novel protein exhibiting neurotoxicity from venom of the psammophiine, Rhamphiophis oxyrhynchus (Rufous beaked snake). Neuropharmacology 52, 1065–1070. doi: 10.1016/j.neuropharm.2006.11.002
Lumsden, N. G., Fry, B. G., Ventura, S., Kini, R. M., and Hodgson, W. C. (2005). Pharmacological characterisation of a neurotoxin from the venom of Boiga dendrophila (mangrove catsnake). Toxicon 45, 329–334. doi: 10.1016/j.toxicon.2004.11.003
Mackessy, S. P., Williams, K., and Ashton, K. (2003). Characterization of the venom of the midget faded rattlesnake (Crotalus viridis concolor): a case of venom paedomorphosis? Copeia 2003, 769–782. doi: 10.1643/HA03-037.1
Mackessy, S. P. (1985). Fractionation of red diamond rattlesnake (Crotalus ruber ruber) venom: protease, phosphodiesterase, L-amino acid oxidase activities and effects of metal ions on protease activity. Toxicon 23:337–340. doi: 10.1016/0041-0101(85)90157-6
Mackessy, S. P. (1988). Venom ontogeny in the pacific rattlesnakes Crotalus viridis helleri and C. v. oreganus. Copeia 1, 92–101. doi: 10.2307/1445927
Mackessy, S. P. (2002). Biochemistry and pharmacology of colubrid snake venoms. J. Toxicol. 21, 43–83. doi: 10.1081/TXR-120004741
Mackessy, S. P. (2010a). Evolutionary trends in venom composition in the western rattlesnakes (Crotalus viridis sensu lato): toxicity vs. tenderizers. Toxicon 55, 1463–1474. doi: 10.1016/j.toxicon.2010.02.028
Mackessy, S. P. (2010b). “The field of reptile toxinology: snakes, lizards and their venoms,” in Handbook of Venoms and Toxins of Reptiles, ed S. P. Mackessy (Boca Raton, FL: CRC Press;Taylor and Francis Group, 2–23.
Mackessy, S. P., and Heyborne, W. H. (2010). “Cysteine-rich secretory proteins in reptile venoms,” in Handbook of Venoms and Toxins of Reptiles, ed S. P. Mackessy (Boca Raton, FL.: CRC Press; Taylor and Francis Group, 325–334.
Mackessy, S. P., Sixberry, N. M., Heyborne, W. H., and Fritts, T. (2006). Venom of the brown treesnake, Boiga irregularis: ontogenetic shifts and taxa-specific toxicity. Toxicon 47, 537–548. doi: 10.1016/j.toxicon.2006.01.007
Macrander, J., Broe, M., and Daly, M. (2015). Multi-copy venom genes hidden in de novo transcriptome assemblies, a cautionary tale with the snakelocks sea anemone Anemonia sulcata (Pennant, 1977). Toxicon 108, 184–188. doi: 10.1016/j.toxicon.2015.09.038
Macrander, J., Panda, J., Janies, D., Daly, M., and Reitzel, A. M. (2018). Venomix: a simple bioinformatic pipeline for identifying and characterizing toxin gene candidates from transcriptomic data. PeerJ 6:e5361. doi: 10.7717/peerj.5361
Margres, M., Aronow, K., Loyacano, J., and Rokyta, D. (2013). The venom-gland transcriptome of the eastern coral snake (Micrurus fulvius) reveals high venom complexity in the intragenomic evolution of venoms. BMC Genomics 14:531. doi: 10.1186/1471-2164-14-531
Margres, M. J., Wray, K. P., Hassinger, A. T. B., Ward, M. J., McGivern, J. J., Moriarty Lemmon, E., et al. (2017). Quantity, not quality: rapid adaptation in a polygenic trait proceeded exclusively through expression differentiation. Mol. Biol. Evol. 34, 3099–3110. doi: 10.1093/molbev/msx231
Marques, O., and Hartmann, P. (2005). Diet and habitat use of two sympatric species of Philodryas (Colubridae), in south Brazil. Amphibia-Reptilia 26, 25–31. doi: 10.1163/1568538053693251
Martins, N. (1907). Das opisthoglyphas brazileiras e seu veneno. Col. Trab. Inst. Butantan 1, 427–496.
McCue, M. D. (2007). Prey envenomation does not improve digestive performance in Western Diamondback Rattlesnakes (Crotalus atrox). J. Exp. Zool. 307A, 568–577. doi: 10.1002/jez.411
McDowell, R. S., Dennis, M. S., Louie, A., Shuster, M., Mulkerrin, M. G., and Lazarus, R. A. (1992). Mambin, a potent glycoprotein IIb-IIIa antagonist and platelet aggregation inhibitor structurally related to the short neurotoxins. Biochemistry 31, 4766–4772. doi: 10.1021/bi00135a004
McGivern, J. J., Wray, K. P., Margres, M. J., Couch, M. E., Mackessy, S. P., and Rokyta, D. R. (2014). RNA-seq and high-definition mass spectrometry reveal the complex and divergent venoms of two rear-fanged colubrid snakes. BMC Genomics 15:1061. doi: 10.1186/1471-2164-15-1061
Modahl, C. M., Durban, J., and Mackessy, S. P. (2019). “Exploring toxin evolution: venom protein transcript sequencing and transcriptome-guided high-throughput proteomics,” in Methods in Molecular Biology: Snake and Spider Toxins, ed A. Priel (New York, NY: Springer Nature). doi: 10.1007/978-1-4939-9845-6
Modahl, C. M., Frietze, S., and Mackessy, S. P. (2018a). Transcriptome-facilitated proteomic characterization of rear-fanged snake venoms reveal abundant metalloproteinases with enhanced activity. J. Proteomics 187, 223–234. doi: 10.1016/j.jprot.2018.08.004
Modahl, C. M., and Mackessy, S. P. (2016). Full-length venom protein cDNA sequences from venom-derived mRNA: exploring compositional variation and adaptive multigene evolution. PLoS Negl. Trop. Dis. 10:e0004587. doi: 10.1371/journal.pntd.0004587
Modahl, C. M., Mrinalini, Frietze, S., and Mackessy, S. P. (2018b). Adaptive evolution of distinct prey-specific toxin genes in rear-fanged snake venom. Proc. Biol. Sci. 285:20181003. doi: 10.1098/rspb.2018.1003
Modahl, C. M., Mukherjee, A. K., and Mackessy, S. P. (2016). An analysis of venom ontogeny and prey-specific toxicity in the Monocled Cobra (Naja kaouthia). Toxicon 119, 8–20. doi: 10.1016/j.toxicon.2016.04.049
Morita, T. (2005). Structures and functions of snake venom CLPs (C-type lectin-like proteins) with anticoagulant-, procoagulant-, and platelet-modulating activities. Toxicon 45, 1099–1114. doi: 10.1016/j.toxicon.2005.02.021
Nery, M. D., de Aquino, H. D., Ribeiro, R. T. M., Nery, E. A., de Menezes, D. B., Ximenes, R. M., et al. (2014). Edematogenic and myotoxic activities induced by snake venom of Philodryas nattereri from the Northeast of Brazil. Fundam. Toxicol. Sci. 1, 7–13. doi: 10.2131/fts.1.7
Nirthanan, S., Gopalakrishnakone, P., Gwee, M. C. E., Khoo, H. E., and Kini, R. M. (2003). Non-conventional toxins from elapid venoms. Toxicon 41, 397–407. doi: 10.1016/S0041-0101(02)00388-4
Nirthanan, S., and Gwee, M. C. E. (2004). Three-finger α-neurotoxins and the nicotinic acetylcholine receptor, forty years on. J. Pharmacol. Sci. 94, 1–17. doi: 10.1254/jphs.94.1
Nobile, M., Noceti, F., Prestipino, G., and Possani, L. D. (1996). Helothermine, a lizard venom toxin, inhibits calcium current in cerebellar granules. Exp. Brain Res. 110, 15–20. doi: 10.1007/BF00241369
Oliveira, J. S., Sant'Anna, L. B., Oliveira Junior, M. C., Souza, P. R. M., Andrade-Souza, A. S., Ribeiro, W., et al. (2017). Local and hematological alterations induced by Philodryas olfersii snake venom in mice. Toxicon 132, 9–17. doi: 10.1016/j.toxicon.2017.03.013
OmPraba, G., Chapeaurouge, A., Doley, R., Devi, K. R., Padmanaban, P., Venkatraman, C., et al. (2010). Identification of a novel family of snake venom proteins veficolins from Cerberus rynchops using a venom gland transcriptomics and proteomics approach. J. Proteome Res. 9, 1882–1893. doi: 10.1021/pr901044x
Page-McCaw, A., Ewald, A. J., and Werb, Z. (2007). Matrix metalloproteinases and the regulation of tissue remodeling. Nat. Rev. Mol. Cell Biol. 8:221. doi: 10.1038/nrm2125
Pahari, S., Bickford, D., Fry, B. G., and Kini, R. M. (2007). Expression pattern of three-finger toxin and phospholipase A2 genes in the venom glands of two sea snakes, Lapemis curtus and Acalyptophis peronii: comparison of evolution of these toxins in land snakes, sea kraits and sea snakes. BMC Evol. Biol. 7:175. doi: 10.1186/1471-2148-7-175
Parks, W. C., Wilson, C. L., and López-Boado, Y. S. (2004). Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat. Rev. Immunol. 4, 617–629. doi: 10.1038/nri1418
Pawlak, J., and Kini, R. M. (2008). Unique gene organization of colubrid three-finger toxins: complete cDNA and gene sequences of denmotoxin, a bird-specific toxin from colubrid snake Boiga dendrophila (Mangrove Catsnake). Biochimie 90, 868–877. doi: 10.1016/j.biochi.2008.02.016
Pawlak, J., Mackessy, S. P., Fry, B. G., Bhatia, M., Mourier, G., Fruchart-Gaillard, C., et al. (2006). Denmotoxin, a three-finger toxin from the colubrid snake Boiga dendrophila (mangrove catsnake) with bird-specific activity. J. Biol. Chem. 281, 29030–29041. doi: 10.1074/jbc.M605850200
Pawlak, J., Mackessy, S. P., Sixberry, N. M., Stura, E. A., Le Du, M. H., Ménez, R., et al. (2009). Irditoxin, a novel covalently linked heterodimeric three-finger toxin with high taxon-specific neurotoxicity. FASEB J. 23, 534–545. doi: 10.1096/fj.08-113555
Peichoto, M. E., Leiva, L. C., Guaimas Moya, L. E., Rey, L., and Acosta, O. (2005). Duvernoy's gland secretion of Philodryas patagoniensis from the northeast of Argentina: its effects on blood coagulation. Toxicon 45, 527–534. doi: 10.1016/j.toxicon.2004.12.016
Peichoto, M. E., Mackessy, S. P., Teibler, P., Tavares, F. L., Burckhardt, P. L., Breno, M. C., et al. (2009). Purification and characterization of a cysteine-rich secretory protein from Philodryas patagoniensis snake venom. Compar. Biochem. Physiol. C Toxicol. Pharmacol. 150, 79–84. doi: 10.1016/j.cbpc.2009.03.002
Peichoto, M. E., Tavares, F. L., Santoro, M. L., and Mackessy, S. P. (2012). Venom proteomes of South and North American opisthoglyphous (Colubridae and Dipsadidae) snake species: a preliminary approach to understanding their biological roles. Compar. Biochem. Physiol. D Genomics Proteomics 7, 361–369. doi: 10.1016/j.cbd.2012.08.001
Peichoto, M. E., Teibler, P., Mackessy, S. P., Leiva, L., Acosta, O., Gonçalves, L. R., et al. (2007). Purification and characterization of patagonfibrase, a metalloproteinase showing alpha-fibrinogenolytic and hemorrhagic activities, from Philodryas patagoniensis snake venom. Biochim. Biophys. Acta 1770, 810–819. doi: 10.1016/j.bbagen.2006.12.014
Peichoto, M. E., Zychar, B. C., Tavares, F. L., de Camargo Gonçalves, L. R., Acosta, O., and Santoro, M. L. (2011). Inflammatory effects of patagonfibrase, a metalloproteinase from Philodryas patagoniensis (Patagonia Green Racer; Dipsadidae) venom. Exp. Biol. Med. 236, 1166–1172. doi: 10.1258/ebm.2011.011125
Perry, B. W., Card, D. C., McGlothlin, J. W., Pasquesi, G. I. M., Adams, R. H., Schield, D. R., et al. (2018). Molecular adaptations for sensing and securing prey and insight into amniote genome diversity from the garter snake genome. Genome Biol. Evol. 10, 2110–2129. doi: 10.1093/gbe/evy157
Pillet, L., Trémeau, O., Ducancel, F., Drevet, P., Zinn-Justin, S., Pinkasfeld, S., et al. (1993). Genetic engineering of snake toxins. Role of invariant residues in the structural and functional properties of a curaremimetic toxin, as probed by site-directed mutagenesis. J. Biol. Chem. 268, 909–916.
Pla, D., Petras, D., Saviola, A. J., Modahl, C. M., Sanz, L., Pérez, A., et al. (2017a). Transcriptomics-guided bottom-up and top-down venomics of neonate and adult specimens of the arboreal rear-fanged Brown Treesnake, Boiga irregularis, from Guam. J. Proteomics 174, 71–84. doi: 10.1016/j.jprot.2017.12.020
Pla, D., Sanz, L., Whiteley, G., Wagstaff, S. C., Harrison, R. A., Casewell, N. R., et al. (2017b). What killed Karl Patterson Schmidt? Combined venom gland transcriptomic, venomic and antivenomic analysis of the South African green tree snake (the boomslang), Dispholidus typus. Biochim. Biophys. Acta 1861, 814–823. doi: 10.1016/j.bbagen.2017.01.020
Pope, C. H. (1958). Fatal bite of captive african rear-fanged snake (Dispholidus). Copeia 1958, 280–282. doi: 10.2307/1439959
Prado-Franceschi, J., and Hyslop, S. (2002). South American colubrid envenomations. J. Toxicol. 21, 117–158. doi: 10.1081/TXR-120004744
Prado-Franceschi, J., Hyslop, S., Cogo, J. C., Andrade, A. L., Assakura, M., Cruz-Höfling, M. A., et al. (1996). The effects of Duvernoy's gland secretion from the xenodontine colubrid Philodryas olfersii on striated muscle and the neuromuscular junction: partial characterization of a neuromuscular fraction. Toxicon 34, 459–466. doi: 10.1016/0041-0101(95)00146-8
Pung, Y. F., Wong, P. T., Kumar, P. P., Hodgson, W. C., and Kini, R. M. (2005). Ohanin, a novel protein from king cobra venom, induces hypolocomotion and hyperalgesia in mice. J. Biol. Chem. 280, 13137–13147. doi: 10.1074/jbc.M414137200
Pyron, R. A., Burbink, F. T., and Wiens, J. J. (2013). A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol. Biol. 13:93. doi: 10.1186/1471-2148-13-93
Ra, H.-J., and Parks, W. C. (2007). Control of matrix metalloproteinase catalytic activity. Matrix Biol. 26, 587–596. doi: 10.1016/j.matbio.2007.07.001
Rajagopalan, N., Pung, Y. F., Zhu, Y. Z., Wong, P. T., Kumar, P. P., and Kini, R. M. (2007). Beta-cardiotoxin: a new three-finger toxin from Ophiophagus hannah (king cobra) venom with beta-blocker activity. FASEB J. 21, 3685–3695. doi: 10.1096/fj.07-8658com
Reyes-Velasco, J., Card, D. C., Andrew, A. L., Shaney, K. J., Adams, R. H., Schield, D. R., et al. (2015). Expression of venom gene homologs in diverse python tissues suggests a new model for the evolution of snake venom. Mol. Biol. Evol. 32, 173–183. doi: 10.1093/molbev/msu294
Richards, D.P., Barlow, A., and Wuster, W. (2012). Venom lethality and diet: differential responses of natural prey and model organisms to the venom of the saw-scaled vipers (Echis). Toxicon. 59, 110–116. doi: 10.1016/j.toxicon.2011.10.015
Rivera-Torres, I. O., Jin, T. B., Cadene, M., Chait, B. T., and Poget, S. F. (2016). Discovery and characterisation of a novel toxin from Dendroaspis angusticeps, named Tx7335, that activates the potassium channel KcsA. Sci. Rep. 6, 23904–23904. doi: 10.1038/srep23904
Robert, W. H. (1982). Trophic Relationships and Foraging Strategies of some New World Tree Snakes (Leptophis, Oxybelis, Uromacer). Amph. Reptil. 3, 71–80. doi: 10.1163/156853882X00185
Rocha, M. M. T., and Furtado, M. F. D. (2007). Analysis of biological activities from Philodryas olfersii (Lichtenstein) and P. patagoniensis (Girard) venoms (Serpents, Colubridae). Rev. Bras. Zool. 24, 410–418. doi: 10.1590/S0101-81752007000200019
Rocha, M. M. T., Paixao-Cavalcante, D., Tambourgi, D. V., and Furtado, M. F. D. (2006). Duvernoy's gland secretion of Philodryas olfersii and Philodryas patagoniensis (Colubridae): neutralization of local and systemic effects by commercial bothropic antivenom (Bothrops genus). Toxicon 47, 95–103. doi: 10.1016/j.toxicon.2005.10.005
Rokyta, D. R., Wray, K. P., Lemmon, A. R., Lemmon, E. M., and Caudle, B. S. (2011). A high-throughput venom-gland transcriptome for the Eastern Diamondback Rattlesnake (Crotalus adamanteus) and evidence for pervasive positive selection across toxin classes. Toxicon 57, 657–671. doi: 10.1016/j.toxicon.2011.01.008
Rokyta, D. R., Wray, K. P., McGivern, J. J., and Margres, M. J. (2015). The transcriptomic and proteomic basis for the evolution of a novel venom phenotype within the Timber Rattlesnake (Crotalus horridus). Toxicon 98, 34–48. doi: 10.1016/j.toxicon.2015.02.015
Rosenberg, H. I. (1992). An improved method for collecting secretion from Duvernoy's gland of colubrid snakes. Copeia 1992, 244–246. doi: 10.2307/1446561
Rosso, J. P., Schwarz, J. R., Diaz-Bustamante, M., Céard, B., Gutiérrez, J. M., Kneussel, M., et al. (2015). MmTX1 and MmTX2 from coral snake venom potently modulate GABA(A) receptor activity. Biophys. J. 108:434a. doi: 10.1016/j.bpj.2014.11.2369
Saez, N. J., Nozach, H., Blemont, M., and Vincentelli, R. (2014). High throughput quantitative expression screening and purification applied to recombinant disulfide-rich venom proteins produced in E. coli. J. Vis. Exp. 89:e51464. doi: 10.3791/51464
Sánchez, M. N., Teibler, G. P., López, C. A., Mackessy, S. P., and Peichoto, M. E. (2018). Assessment of the potential toxicological hazard of the Green Parrot Snake (Leptophis ahaetulla marginatus): characterization of its venom and venom-delivery system. Toxicon 148, 202–212. doi: 10.1016/j.toxicon.2018.04.027
Sánchez, M. N., Teibler, G. P., Sciani, J. M., Casafús, M. G., Maruñak, S. L., Mackessy, S. P., et al. (2019). Unveiling toxicological aspects of venom from the Aesculapian False Coral Snake Erythrolamprus aesculapii. Toxicon 164, 71–81. doi: 10.1016/j.toxicon.2019.04.007
Sánchez, M. N., Timoniuk, A., Marunak, S., Teibler, P., Acosta, O., and Peichoto, M. E. (2014). Biochemical and biological analysis of Philodryas baroni (Baron's Green Racer; Dipsadidae) venom: relevance to the findings of human risk assessment. Hum. Exp. Toxicol. 33, 22–31. doi: 10.1177/0960327113493302
Saviola, A. J., Peichoto, M. E., and Mackessy, S. P. (2014). Rear-fanged snake venoms: an untapped source of novel compounds and potential drug leads. Toxin Rev. 33, 1–17. doi: 10.3109/15569543.2014.942040
Savitsky, A. H. (1980). The role of venom delivery strategies in snake evolution. Evolution 34, 1194–1204. doi: 10.1111/j.1558-5646.1980.tb04065.x
Smiley-Walters, S. A., Farrell, T. M., and Gibbs, H. L. (2017). Evaluating local adaptation of a complex phenotype: reciprocal tests of pigmy rattlesnake venoms on treefrog prey. Oecologia 184, 739–748. doi: 10.1007/s00442-017-3882-8
Smiley-Walters, S. A., Farrell, T. M., and Gibbs, H. L. (2018). The importance of species: pygmy rattlesnake venom toxicity differs between native prey and related non-native species. Toxicon 144, 42–47. doi: 10.1016/j.toxicon.2018.01.022
Sousa, L. F., Portes-Junior, J. A., Nicolau, C. A., Bernardoni, J. L., Nishiyama, M. Y. Jr., Amazonas, D. R., et al. (2017). Functional proteomic analyses of Bothrops atrox venom reveals phenotypes associated with habitat variation in the Amazon. J. Proteomics 159, 32–46. doi: 10.1016/j.jprot.2017.03.003
Sun, Q. Y., and Bao, J. (2010). Purification, cloning and characterization of a metalloproteinase from Naja atra venom. Toxicon 56, 1459–1469. doi: 10.1016/j.toxicon.2010.08.013
Sunagar, K., Jackson, T., Undheim, E., Ali, S., Antunes, A., and Fry, B. G. (2013). Three-fingered RAVERs: Rapid accumulation of variations in exposed residues of snake venom toxins. Toxins 5, 2172–2208. doi: 10.3390/toxins5112172
Sunagar, K., Vasconcelos, V., Johnson, W. E., O'Brien, S. J., and Antunes, A. (2012). Evolution of CRISPs associated with toxicoferan-reptilian venom and mammalian reproduction. Mol. Biol. Evol. 29, 1807–1822. doi: 10.1093/molbev/mss058
Takeda, S., Takeya, H., and Iwanaga, S. (2012). Snake venom metalloproteinases: structure, function and relevance to the mammalian ADAM/ADAMTS family proteins. Biochim. Biophys. Acta 1824, 164–176. doi: 10.1016/j.bbapap.2011.04.009
Takeya, H., Miyata, T., Nishino, N., Omori-Satoh, T., and Iwanaga, S. (1993). “[24] Snake venom hemorrhagic and nonhemorrhagic metalloendopeptidases,” in Methods in Enzymology: Proteolytic Enzymes in Coagulation, Fibrinolysis, and Complement Activation Part B: Complement Activation, Fibrinolysis, and Nonmammalian Blood Coagulation Factors and Inhibitors, eds L. Lorand and K. G. Mann (Cambridge, MA: Academic Press, 365–378.
Tancioni, I., Butera, D., Spencer, P. A., Takehara, H., Fernandes, I., and Moura-da-Silva, A. M. (2004). Phylogenetic conservation of a snake venom metalloproteinase epitope recognized by a monoclonal antibody that neutralizes hemorrhagic activity. Toxicon 42, 809–816. doi: 10.1016/j.toxicon.2003.10.011
Taub, A. M. (1967). Comparative histological studies on Duvernoy's gland of colubrid snakes. Bull. Am. Mus. Nat. Hist. 138, 6–13.
Torres-Bonilla, K. A., Andrade-Silva, D., Serrano, S. M. T., and Hyslop, S. (2018). Biochemical characterization of venom from Pseudoboa neuwiedii (Neuwied's false boa; Xenodontinae; Pseudoboini). Compar. Biochem. Physiol. C Toxicol. Pharmacol. 213, 27–38. doi: 10.1016/j.cbpc.2018.06.003
Uetz, P., Freed, P., and Hosek, J. (2018). The Reptile Database. Available online at: http://www.reptile-database.org/
Urra, F. A., Pulgar, R., Gutiérrez, R., Hodar, C., Cambiazo, V., and Labra, A. (2015). Identification and molecular characterization of five putative toxins from the venom gland of the snake Philodryas chamissonis (Serpentes: Dipsadidae). Toxicon 108, 19–31. doi: 10.1016/j.toxicon.2015.09.032
Utkin, Y. N., Kukhtina, V. V., Maslennikov, I. V., Eletsky, A. V., Starkov, V. G., Weise, C., et al. (2001). First tryptophan-containing weak neurotoxin from cobra venom. Toxicon 39, 921–927. doi: 10.1016/S0041-0101(00)00223-3
Vidal, N. (2002). Colubroid systematics: evidence for an early appearance of the venom apparatus followed by extensive evolutionary tinkering. J. Toxicol. 21, 21–41. doi: 10.1081/TXR-120004740
Vonk, F. J., Admiraal, J. F., Jackson, K., Reshef, R., de Bakker, M. A., Vanderschoot, K., et al. (2008). Evolutionary origin and development of snake fangs. Nature 454, 630–633. doi: 10.1038/nature07178
Vonk, F. J., Casewell, N. R., Henkel, C. V., Heimberg, A. M., Jansen, H. J., McCleary, R. J., et al. (2013). The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl. Acad. Sci. U.S.A. 110, 20651–20656. doi: 10.1073/pnas.1314702110
Vu, T. H., and Werb, Z. (2000). Matrix metalloproteinases: effectors of development and normal physiology. Genes Dev. 14, 2123–2133. doi: 10.1101/gad.815400
Wang, F., Li, H., Liu, M., Song, H., Han, H., Wang, Q., et al. (2006). Structural and functional analysis of natrin, a venom protein that targets various ion channels. Biochem. Biophys. Res. Commun. 351, 443–448. doi: 10.1016/j.bbrc.2006.10.067
Weinstein, S. A., Warrell, D. A., White, J., and Keyler, D. E. (2011). “Venomous” Bites From Non-Venomous Snakes: A Critical Analysis of Risk and Management of “Colubrid” Snake Bites. London: Elsevier.
Weinstein, S. A., White, J., Keyler, D. E., and Warrell, D. A. (2013). Non-front-fanged colubroid snakes: a current evidence-based analysis of medical significance. Toxicon 69, 103–113. doi: 10.1016/j.toxicon.2013.02.003
Weldon, C. L., and Mackessy, S. P. (2012). Alsophinase, a new P-III metalloproteinase with alpha-fibrinogenolytic and hemorrhagic activity from the venom of the rear-fanged Puerto Rican Racer Alsophis portoricensis (Serpentes: Dipsadidae). Biochimie 94, 1189–1198. doi: 10.1016/j.biochi.2012.02.006
Yamazaki, Y., Koike, H., Sugiyama, Y., Motoyoshi, K., Wada, T., Hishinuma, S., et al. (2002). Cloning and characterization of novel snake venom proteins that block smooth muscle contraction. Eur. J. Biochem. 269, 2708–2715. doi: 10.1046/j.1432-1033.2002.02940.x
Yang, D. C., Deuis, J. R., Dashevsky, D., Dobson, J., Jackson, T. N. W., Brust, A., et al. (2016). The Snake with the scorpion's sting: novel three-finger toxin sodium channel activators from the venom of the long-glanded blue coral snake (Calliophis bivirgatus). Toxins 8:303. doi: 10.3390/toxins8100303
Yin, W., Wang, Z. J., Li, Q. Y., Lian, J. M., Zhou, Y., Lu, B. Z., et al. (2016). Evolutionary trajectories of snake genes and genomes revealed by comparative analyses of five-pacer viper. Nat. Commun. 7:13107. doi: 10.1038/ncomms13107
Young, B. A., and Kardong, K. V. (1996). Dentitional surface features in snakes (Reptilia: Serpentes). Amph. Reptil. 17, 261–276. doi: 10.1163/156853896X00432
Zancolli, G., Calvete, J. J., Cardwell, M. D., Greene, H. W., Hayes, W. K., Hegarty, M. J., et al. (2019). When one phenotype is not enough: divergent evolutionary trajectories govern venom variation in a widespread rattlesnake species. Proc. R. Soc. B. Biol. Sci. 286:20182735. doi: 10.1098/rspb.2018.2735
Keywords: evolution, metalloproteinase, neofunctionalization, proteomics, three-finger toxin, toxin, transcriptomics
Citation: Modahl CM and Mackessy SP (2019) Venoms of Rear-Fanged Snakes: New Proteins and Novel Activities. Front. Ecol. Evol. 7:279. doi: 10.3389/fevo.2019.00279
Received: 29 March 2019; Accepted: 09 July 2019;
Published: 23 July 2019.
Edited by:
Kartik Sunagar, Indian Institute of Science (IISc), IndiaReviewed by:
Wolfgang Wuster, Bangor University, United KingdomBryan Fry, University of Queensland, Australia
Copyright © 2019 Modahl and Mackessy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephen P. Mackessy, stephen.mackessy@unco.edu
 Cassandra M. Modahl
Cassandra M. Modahl Stephen P. Mackessy
Stephen P. Mackessy