- 1 Centre d’Immunologie de Marseille-Luminy, UM2 Aix-Marseille Université, Marseille, France
- 2 INSERM, U1104, Marseille, France
- 3 CNRS, UMR7280, Marseille, France
- 4 European Molecular Biology Laboratory-European Bioinformatics Institute, Cambridge, UK
While innate immunity has been studied in insects since the time of Pasteur and Metchnikoff (Brey, 1998), research into nematode immune defenses was initiated only comparatively recently (Kurz and Ewbank, 2003). In the mid 1990s, through a biochemical approach, Yusuke Kato was able to isolate an antibacterial activity from the body fluid of the parasitic nematode Ascaris suum. This activity was ascribed to A. suum antibacterial factor (ASABF), a peptide that is particularly potent against Gram-positive bacteria (Kato, 1995). The subsequent molecular characterization of ASABF allowed the identification of six orthologous “ABF” peptides in the model nematode C. elegans, one of which, ABF-2 displays in vitro microbicidal activity against a range of bacteria and fungi (Kato and Komatsu, 1996; Kato et al., 2002; Zhang and Kato, 2003). The six ABF peptides were immediately recognized as sharing features with defensins, placing them in the class of cysteine-stabilized α-helix and β-sheet (CSαβ) peptides, the most wide-spread and conserved class of antimicrobial peptides (AMPs; Zhu, 2008). The constitutive expression of abf-1 and abf-2, the best characterized of the six corresponding genes in C. elegans, overlaps in the pharynx (Kato et al., 2002); they are likely to contribute to the breakdown of the microbes that form the nematode’s normal diet. Although they undoubtedly act upon both bacteria and fungi, here, we consider only their potential role in anti-fungal defenses. Both abf-1 and abf-2 are up-regulated by infection with the fungus Cryptococcus neoformans (Means et al., 2009), while the expression of abf-1 but not of abf-2 is induced by infection with the natural fungal pathogens Drechmeria coniospora and Harposporium sp. (Engelmann et al., 2011). Conversely, abf-2 is up-regulated by Candida albicans. It is not clear what the underlying regulatory pathways governing abf gene expression are (Pukkila-Worley et al., 2011), but one can speculate that their differential regulation reflects both the various modes of pathogen infection (Labed and Pujol, 2011), and also their potentially distinct spectra of antimicrobial activities.
This conserved family of defensin-like peptides is something of an exception, since overall, C. elegans possesses a highly derived innate immune system. It has no equivalent of NF-κB, central to immunity in many animals, and also lacks orthologs of most of the receptors known from other species to be important for triggering host defenses (Pujol et al., 2001; Gravato-Nobre and Hodgkin, 2005). Indeed, as explained more in detail below, several classes of AMPs implicated in anti-fungal defense appear to be restricted to certain nematode species, and are controlled by signal transduction cascades with a very limited phylogenetic range (Dierking et al., 2011; Labed et al., 2012). Different fungal pathogens infect either via the intestine following ingestion, or via the epidermis. They influence AMP gene expression via distinct signaling cascades, but a detailed discussion of these regulatory mechanisms is beyond the scope of this short article.
Unlike A. suum, which can grow to a length of 40 cm, an adult C. elegans measures barely 1 mm, making extraction of body fluid technically almost impossible. Many of the other putative AMP genes in C. elegans were initially identified on the basis of their differential regulation following D. coniospora infection. The first such genes were members of the nlp (for neuropeptide-like protein) and cnc (caenacin) families (Couillault et al., 2004). At the time, the former had been tentatively annotated as neuropeptides, based on their limited sequence similarity with known neuropeptides. It was, however, observed that these genes were not generally expressed in the nervous system, but rather in the epidermis (Nathoo et al., 2001). This ties in with the fact that D. coniospora spores attach to the nematode’s cuticle and then germinate, penetrating into the body of the worm via the epidermis. Much, but not all, of the response to infection is a cell-autonomous mechanism acting in the epidermis; reviewed in Labed and Pujol (2011). The infection-induced nlp genes are structurally related to the cnc genes. The two groups further share the property of being found in clusters in the genome. Interestingly, comparison of the syntenic regions in two other Caenorhabditis species, C. briggsae and C. remanei revealed that these genes are undergoing relatively rapid evolution, with clear evidence for gene duplication and gene loss, and appear to be under positive selective pressure. Indeed, over-expression of either the nlp or cnc AMPs leads to somewhat increased resistance to D. coniospora infection (Pujol et al., 2008; Zugasti and Ewbank, 2009), suggesting that they play a direct role in host defense against invasive fungi. This is further reinforced by the finding that the nlp and cnc AMP genes feature prominently among the few genes commonly up-regulated by D. coniospora and the fungi Harposporium sp. (Engelmann et al., 2011). The expression of cnc-4 and cnc-7 is also induced by virulent C. albicans (Pukkila-Worley et al., 2011), which like Harposporium sp. infects C. elegans via the intestine.
Since the initial phylogenetic studies, more nematode genome sequences have become available. Comparative analyses reinforce the notion of rapid gene evolution. For example, the single copy C. remanei gene CRE-nlp-27 corresponds to a cluster of 5 paralogs in C. elegans (Pujol et al., 2008), but 10 predicted paralogs in C. japonica (Figures 1A,B). On the other hand, there apparently has been no expansion of the cnc genes in C. japonica, whereas multiple paralogs are found in the other Caenorhabditis species (Figure 1C). We have not found orthologous genes in several other nematode species for which genome sequences are available, including Brugia malayi (Ghedin et al., 2007), Meloidogyne incognita (Abad et al., 2008), and A. suum (Jex et al., 2011). It is possible that these parasitic nematodes have a natural environment that shields them from fungal pathogens throughout their life cycle, so that they have been able to dispense with these AMP genes (Abad et al., 2008). On the other hand, there do appear to be two similar AMP-encoding genes in Drosophila, CG7738, now called CG34227, and CG17738 (Couillault et al., 2004). It is notable that the former was found to be up-regulated by fungal infection (De Gregorio et al., 2002), and the corresponding peptide was identified via the biochemical characterization of hemolymph from septically injured flies (Verleyen et al., 2006). The extremely repetitive nature of these peptides, characterized by the presence of multiple GGY and/or GGW triplets (Couillault et al., 2004), does makes identification of orthologs in more distant species problematic, and it is currently unclear how evolutionary ancient are the NLP/CNC AMPs as defense molecules. Nevertheless, the existence of CNC-like peptides in insects supports the idea that the parasitic A. suum lost this part of its immune defenses, while retaining defensin-like peptides that may be important in the bacteria-rich intestinal tract of its vertebrate hosts, and also that the primary target of CNCs may be fungi.
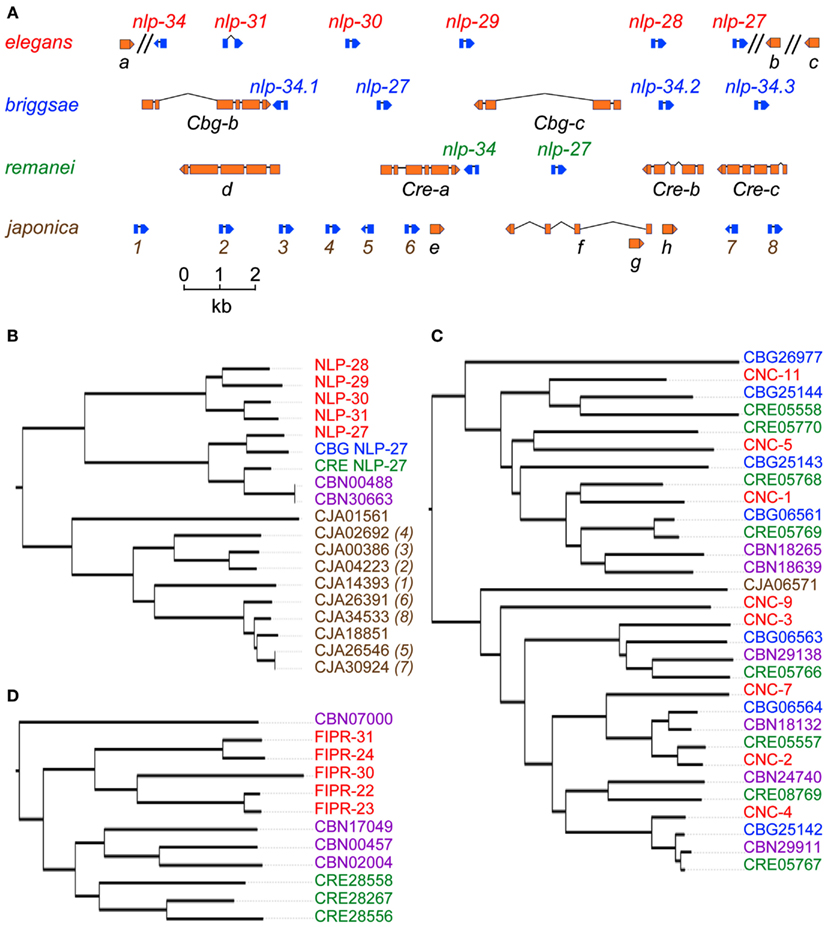
Figure 1. (A) The nlp-29 cluster in C. elegans together with syntenic regions from C. brigssae (Cbg), C. remanei (Cre), and C. japonica. AMP genes of the nlp class are shown in blue; there has been a marked expansion in C. japonica. The genes labeled “a” and “c” in C. elegans, and their orthologs (Cbg-a, Cbg-c, Cre-a, and Cre-c) are predicted to encode serpentine receptors; “b” in C. elegans is K09D9.9. Its ortholog in C. japonica is ca. 60 kb 3′ of the locus. Immediately 3′ of the gene labeled “e” in C. japonica there is the remnant of a degenerate paralog of K09D9.9. The figure is adapted from data in Wormbase WS230; it does not reproduce the predicted fusion of Cre-b and Cre-c, as this does not appear to be probable. Only the 3′ extremities of a, b, and c are shown. (B–D) Phylogenetic trees for selected members of the NLP (B), CNC (C), and FIPR (D) family peptides, including homologs in C. brenneri (CBN). A distance matrix analysis was performed using the alignment program clustalw2 to generate a guide tree via pairwise and subsequent multiple sequence alignment. This guide tree was then used to produce a true phylogenetic tree that was loaded into the Interactive Tree Of Life v2 software suite (Letunic and Bork, 2011). Partial rooted trees were extracted that corresponding to interesting features within the complete FIP/FIPR/NLP/CNC tree. For the non-elegans peptides, with the exception of CBG NLP-27 and CRE NLP-27, the corresponding gene identifier is given. The numbers in brackets for the C. japonica gene names match the corresponding genes in (A).
The ABFs, NLPs, and CNCs are not the only predicted AMPs in C. elegans. Microarray-based transcriptionally profiling of infected worms also led to the identification of seven fip (fungus-induced peptide) genes that not only were strongly up-regulated by D. coniospora, but that also fulfilled at least three of the four following criteria: (i) predicted to encode a protein of less than 100 amino acids, (ii) with a predicted signal peptide, (iii) judged by inspection to have a simple primary structure (iv) having a homolog in the immediate genomic proximity, or similar in sequence to more than one structurally related protein encoded by clustered genes. Although there is currently no direct evidence to indicate that these genes do encode genuine AMPs, the combination of the above criteria make it highly likely. A larger class of 29 genes, called fipr (fip-related) was also created for genes that shared these characteristics, but for which there was no indication of any transcriptional up-regulation after D. coniospora infection.
This distinction subsequently turned out to be somewhat premature, since an RNAseq-based analysis of the transcriptional response to infection showed that fipr-22, fipr-23, and fipr-26 are up-regulated by D. coniospora, while the expression of five others (fipr-1, 2, 16, 17, and 20) is induced by the fungus Harposporium sp. (Engelmann et al., 2011); fipr-22 and fipr-23 are also induced by C. albicans (Pukkila-Worley et al., 2011). Because of their large number, and their relatively simple structure, establishing solid phylogenetic relationships both within the nematode clade and beyond is a difficult, and on-going task. It is already clear, however, that these genes too are undergoing relatively rapid evolution, presumably a consequence of their role in host defense. For example, in C. elegans there are five paralogous fipr genes (including the infection-regulated fipr-22 and fipr-23) clustered together, for which orthologs have not been identified in C. japonica or C. briggsae (Figure 1D). A very recent study has revealed marked differences in the seasonal abundance of proliferating C. elegans and C. briggsae populations in Northern France, with C. elegans being much more prevalent in the cooler autumn than in summer, when C. briggsae predominates (Felix and Duveau, 2012). There is therefore every reason to expect that the two nematode species will not be exposed to exactly the same range of natural pathogens, since microbial communities change with the seasons, perhaps explaining the divergent evolution of their AMP gene repertoire. There is increasing evidence that the commonly used strain of C. elegans, N2, underwent a number of changes to adapt to culture under laboratory conditions (see for example, Duveau and Felix, 2012). In the future, it will be interesting to see whether wild isolates of C. elegans present variations in their AMP genes.
Most nlp genes not regulated by fungal infection encode peptides that are either known to have an endocrine signaling function, or are hypothesized to; certain have been matched to specific G-protein coupled receptors (Husson et al., 2007; Janssen et al., 2010). One cannot exclude the possibility that some of the putative AMPs from the CNC and NLP families also exert a regulatory function during the innate immune response to fungal infection. The metabolism and activity of several classes of neuropeptide, both in C. elegans and other species are influenced by neprilysins (Husson et al., 2007). Typically, these zinc metallopeptidases are found on the outer surface of animal cells. They cleave small signaling peptides and thereby block their action. Interestingly, 13 of the 27 neprilysins genes in C. elegans are down-regulated upon infection with D. coniospora or by Harposporium sp. It remains to be determined whether they act on the infection-induced NLPs, or on other classes of peptides, such as insulin-like peptides, which are also transcriptionally regulated upon fungal infection (Engelmann et al., 2011). If they do, this would add a further level of complexity to the regulation of the host response to pathogens.
In conclusion, the last decade has seen considerable advances in our understanding of the role and evolution of AMPs in C. elegans. Future studies should yield more insights into their evolutionary origins and conservation, as well as their precise mode of action and the details of their complex regulation.
Acknowledgments
Work in the Ewbank lab is supported by institutional funding from the CNRS and INSERM and program grants from the ANR. WormBase is supported by a grant from the NIH.
References
Abad, P., Gouzy, J., Aury, J.-M., Castagnone-Sereno, P., Danchin, E. G. J., Deleury, E., Perfus-Barbeoch, L., Anthouard, V., Artiguenave, F., Blok, V. C., Caillaud, M.-C., Coutinho, P. M., Dasilva, C., De Luca, F., Deau, F., Esquibet, M., Flutre, T., Goldstone, J. V., Hamamouch, N., Hewezi, T., Jaillon, O., Jubin, C., Leonetti, P., Magliano, M., Maier, T. R., Markov, G. V., McVeigh, P., Pesole, G., Poulain, J., Robinson-Rechavi, M., Sallet, E., Segurens, B., Steinbach, D., Tytgat, T., Ugarte, E., Van Ghelder, C., Veronico, P., Baum, T. J., Blaxter, M., Bleve-Zacheo, T., Davis, E. L., Ewbank, J. J., Favery, B., Grenier, E., Henrissat, B., Jones, J. T., Laudet, V., Maule, A. G., Quesneville, H., Rosso, M.-N., Schiex, T., Smant, G., Weissenbach, J., and Wincker, P. (2008). Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 26, 909–915.
Brey, P. T. (1998). “The contributions of the Pasteur school of insect immunity,” in Molecular Mechanisms of Immune Responses in Insects, eds P. T. Brey and D. Hultmark (London: Chapman & Hall), 1–39.
Couillault, C., Pujol, N., Reboul, J., Sabatier, L., Guichou, J. F., Kohara, Y., and Ewbank, J. J. (2004). TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat. Immunol. 5, 488–494.
De Gregorio, E., Spellman, P. T., Tzou, P., Rubin, G. M., and Lemaitre, B. (2002). The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 21, 2568–2579.
Dierking, K., Polanowska, J., Omi, S., Engelmann, I., Gut, M., Lembo, F., Ewbank, J. J., and Pujol, N. (2011). Unusual regulation of a STAT protein by an SLC6 family transporter in C. elegans epidermal innate immunity. Cell Host Microbe 9, 425–435.
Duveau, F., and Felix, M. A. (2012). Role of pleiotropy in the evolution of a cryptic developmental variation in Caenorhabditis elegans. PLoS Biol. 10, e1001230. doi: 10.1371/journal.pbio.1001230
Engelmann, I., Griffon, A., Tichit, L., Montanana-Sanchis, F., Wang, G., Reinke, V., Waterston, R. H., Hillier, L. W., and Ewbank, J. J. (2011). A comprehensive analysis of gene expression changes provoked by bacterial and fungal infection in C. elegans. PLoS ONE 6, e19055. doi: 10.1371/journal.pone.0019055
Felix, M. A., and Duveau, F. (2012). Population dynamics and habitat sharing of natural populations of Caenorhabditis elegans and C. briggsae. BMC Biol. 10, 59. doi: 10.1186/1741-7007-10-59
Ghedin, E., Wang, S., Spiro, D., Caler, E., Zhao, Q., Crabtree, J., Allen, J., Delcher, A., Guiliano, D., Miranda-Saavedra, D., Angiuoli, S., Creasy, T., Amedeo, P., Haas, B., El-Sayed, N., Wortman, J., Feldblyum, T., Tallon, L., Schatz, M., Shumway, M., Koo, H., Salzberg, S., Schobel, S., Pertea, M., Pop, M., White, O., Barton, G., Carlow, C., Crawford, M., Daub, J., Dimmic, M., Estes, C., Foster, J., Ganatra, M., Gregory, W., Johnson, N., Jin, J., Komuniecki, R., Korf, I., Kumar, S., Laney, S., Li, B., Li, W., Lindblom, T., Lustigman, S., Ma, D., Maina, C., Martin, D., McCarter, J., McReynolds, L., Mitreva, M., Nutman, T., Parkinson, J., Peregrín-Alvarez, J., Poole, C., Ren, Q., Saunders, L., Sluder, A., Smith, K., Stanke, M., Unnasch, T., Ware, J., Wei, A., Weil, G., Williams, D., Zhang, Y., Williams, S., Fraser-Liggett, C., Slatko, B., Blaxter, M., and Scott, A. (2007). Draft genome of the filarial nematode parasite Brugia malayi. Science 317, 1756–1760.
Gravato-Nobre, M. J., and Hodgkin, J. (2005). Caenorhabditis elegans as a model for innate immunity to pathogens. Cell. Microbiol. 7, 741–751.
Husson, S. J., Mertens, I., Janssen, T., Lindemans, M., and Schoofs, L. (2007). Neuropeptidergic signaling in the nematode Caenorhabditis elegans. Prog. Neurobiol. 82, 33–55.
Janssen, T., Lindemans, M., Meelkop, E., Temmerman, L., and Schoofs, L. (2010). Coevolution of neuropeptidergic signaling systems: from worm to man. Ann. N. Y. Acad. Sci. 1200, 1–14.
Jex, A. R., Liu, S., Li, B., Young, N. D., Hall, R. S., Li, Y., Yang, L., Zeng, N., Xu, X., Xiong, Z., Chen, F., Wu, X., Zhang, G., Fang, X., Kang, Y., Anderson, G. A., Harris, T. W., Campbell, B. E., Vlaminck, J., Wang, T., Cantacessi, C., Schwarz, E. M., Ranganathan, S., Geldhof, P., Nejsum, P., Sternberg, P. W., Yang, H., Wang, J., and Gasser, R. B. (2011). Ascaris suum draft genome. Nature 479, 529–533.
Kato, Y. (1995). Humoral defense of the nematode Ascaris suum: antibacterial, bacteriolytic and agglutinating activities in the body fluid. Zool. Sci. 12, 225–230.
Kato, Y., Aizawa, T., Hoshino, H., Kawano, K., Nitta, K., and Zhang, H. (2002). abf-1 and abf-2, ASABF-type antimicrobial peptide genes in Caenorhabditis elegans. Biochem. J. 361, 221–230.
Kato, Y., and Komatsu, S. (1996). ASABF, a novel cysteine-rich antibacterial peptide isolated from the nematode Ascaris suum. Purification, primary structure, and molecular cloning of cDNA. J. Biol. Chem. 271, 30493–30498.
Kurz, C. L., and Ewbank, J. J. (2003). Caenorhabditis elegans: an emerging genetic model for the study of innate immunity. Nat. Rev. Genet. 4, 380–390.
Labed, S., and Pujol, N. (2011). Caenorhabditis elegans antifungal defense mechanisms. J. Invasive Fungal Infect. 5, 110–117.
Labed, S. A., Omi, S., Gut, M., Ewbank, J. J., and Pujol, N. (2012). The pseudokinase NIPI-4 is a novel regulator of antimicrobial peptide gene expression. PLoS ONE 7, e33887. doi: 10.1371/journal.pone.0033887
Letunic, I., and Bork, P. (2011). Interactive Tree of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39, W475–W478.
Means, T. K., Mylonakis, E., Tampakakis, E., Colvin, R. A., Seung, E., Puckett, L., Tai, M. F., Stewart, C. R., Pukkila-Worley, R., Hickman, S. E., Moore, K. J., Calderwood, S. B., Hacohen, N., Luster, A. D., and El Khoury, J. (2009). Evolutionarily conserved recognition and innate immunity to fungal pathogens by the scavenger receptors SCARF1 and CD36. J. Exp. Med. 206, 637–653.
Nathoo, A. N., Moeller, R. A., Westlund, B. A., and Hart, A. C. (2001). Identification of neuropeptide-like protein gene families in Caenorhabditis elegans and other species. Proc. Natl. Acad. Sci. U.S.A. 98, 14000–14005.
Pujol, N., Link, E. M., Liu, L. X., Kurz, C. L., Alloing, G., Tan, M. W., Ray, K. P., Solari, R., Johnson, C. D., and Ewbank, J. J. (2001). A reverse genetic analysis of components of the Toll signalling pathway in Caenorhabditis elegans. Curr. Biol. 11, 809–821.
Pujol, N., Zugasti, O., Wong, D., Couillault, C., Kurz, C. L., Schulenburg, H., and Ewbank, J. J. (2008). Anti-fungal innate immunity in C. elegans is enhanced by evolutionary diversification of antimicrobial peptides. PLoS Pathog. 4, e1000105. doi: 10.1371/journal.ppat.1000105
Pukkila-Worley, R., Ausubel, F. M., and Mylonakis, E. (2011). Candida albicans infection of Caenorhabditis elegans induces antifungal immune defenses. PLoS Pathog. 7, e1002074. doi: 10.1371/journal.ppat.1002074
Verleyen, P., Baggerman, G., D’hertog, W., Vierstraete, E., Husson, S. J., and Schoofs, L. (2006). Identification of new immune induced molecules in the haemolymph of Drosophila melanogaster by 2D-nanoLC MS/MS. J. Insect Physiol. 52, 379–388.
Zhang, H., and Kato, Y. (2003). Common structural properties specifically found in the CSalphabeta-type antimicrobial peptides in nematodes and mollusks: evidence for the same evolutionary origin? Dev. Comp. Immunol. 27, 499–503.
Zhu, S. (2008). Discovery of six families of fungal defensin-like peptides provides insights into origin and evolution of the CSalphabeta defensins. Mol. Immunol. 45, 828–838.
Citation: Pujol N, Davis PA and Ewbank JJ (2012) The origin and function of anti-fungal peptides in C. elegans: open questions. Front. Immun. 3:237. doi: 10.3389/fimmu.2012.00237
Received: 06 July 2012; Accepted: 16 July 2012;
Published online: 01 August 2012.
Edited by:
Mark W. Robinson, Queen’s University, UKReviewed by:
Mark W. Robinson, Queen’s University, UKCopyright: © 2012 Pujol, Davis and Ewbank. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence:ZXdiYW5rQGNpbWwudW5pdi1tcnMuZnI=
 Nathalie Pujol1,2,3
Nathalie Pujol1,2,3