- 1Department of Biochemistry and Molecular Biology, Snyder Institute for Chronic Diseases, University of Calgary, Calgary, AB, Canada
- 2Department of Medicine, Snyder Institute for Chronic Diseases, University of Calgary, Calgary, AB, Canada
- 3Department of Oncology, University of Calgary, Calgary, AB, Canada
- 4Department of Pathology and Laboratory Medicine, University of Calgary, Calgary, AB, Canada
- 5Calgary Laboratory Services, Foothills Medical Centre, Calgary, AB, Canada
The MS4A gene family in humans includes CD20 and at least 15 other genes. CD20 exists as homo-oligomers in the plasma membrane, however different MS4A proteins expressed in the same cell may hetero-oligomerize. Given the importance of CD20 in B-cell function and as a therapeutic target, we sought to explore the potential for CD20 hetero-oligomerization with other MS4A proteins. We investigated expression in primary human B-cells of the four MS4A genes previously shown to be expressed in human B-cell lines (MS4A4A, MS4A6A, MS4A7, MS4A8B), as well as two genes comprising the closely related TMEM176 gene family, with a view to identifying candidates for future investigation at the protein level. TMEM176A and TMEM176B transcripts were either not detected, or were detected at relatively low levels in a minority of donor B-cell samples. MS4A4A and MS4A8B transcripts were not detected in any normal B-cell sample. MS4A6A and MS4A7 transcripts were detected at low levels in most samples, however the corresponding proteins were not at the plasma membrane when expressed as GFP conjugates in BJAB cells. We also examined expression of these genes in chronic lymphocytic leukemia (CLL), and found that it was similar to normal B-cells with two exceptions. First, whereas MS4A4A expression was undetected in normal B-cells, it was expressed in 1/14 CLL samples. Second, compared to expression levels in normal B-cells, MS4A6A transcripts were elevated in 4/14 CLL samples. In summary, none of the MS4A/TMEM176 genes tested was expressed at high levels in normal or in most CLL B-cells. MS4A6A and MS4A7 were expressed at low levels in most B-cell samples, however the corresponding proteins may not be positioned at the plasma membrane. Altogether, these data suggest that CD20 normally does not form hetero-oligomers with other MS4A proteins and that there are unlikely to be other MS4A proteins in CLL that might provide useful alternate therapeutic targets.
Introduction
MS4A is a family of tetraspanning membrane proteins, most of which remain uncharacterized (1, 2). Phylogenetic analysis indicated that the genes encoding these proteins first appeared and expanded in early vertebrates, with rapid diversification such that homologs of some of the human MS4A genes are not found in the mouse and vice versa (3). The founding member of this family, MS4A1 (CD20), is expressed at high levels only in B-lymphocytes, is associated with the B-cell antigen receptor (BCR) (4), functions in BCR-activated calcium influx (5), and is required for optimal humoral immunity (6). Expression of CD20 first occurs during the pre-B stage of development and is lost upon terminal differentiation to plasma cells. CD20 is also expressed at high levels in B-cell lymphomas, and is an effective target for therapeutic depletion of pathogenic B-cells in various disease settings (7–9). Rituximab and other CD20-directed antibody therapeutics bind to epitopes on one or both of the two small extracellular loops between the transmembrane spanning regions (10–13). The short length of these loops brings bound antibodies close to the plasma membrane, a property that is likely to contribute to the efficacy of CD20 as a target (9). Antibody-mediated stabilization of CD20 in lipid rafts (14, 15) appears to allow enhanced complement activation (16), and the slow rate of internalization of antibody-bound CD20 allows prolonged display of Fc regions to Fc receptor-bearing effector cells (17–19). Some or all of these factors are likely to be shared by other MS4A proteins, which could thus be a source of potentially novel immunotherapeutic targets.
CD20 exists as homo-oligomers in the plasma membrane (4, 20), however it has been suggested that different MS4A proteins expressed in the same cell may hetero-oligomerize with one another. Indeed, hetero-oligomerization of murine MS4a4B and MS4a6B has been reported (21), and unusual MS4A genes with fused repeating MS4A sequences occur in some species (3), suggesting that hetero-oligomerization may be a general feature of MS4A proteins. Given the importance of human CD20 to BCR signaling, humoral immunity, and immunotherapeutic depletion of B-cells, and the potential for other MS4A proteins to hetero-oligomerize with CD20, we sought to identify non-CD20 MS4A genes expressed in primary human B-cells. Most of the known MS4A genes have expression that is restricted to other tissues, particularly myeloid-lineage cells, testes, lung, and intestinal tissues; however, MS4A4A, MS4A6A, MS4A7, and MS4A8B were reported to be expressed in human B-cell lines (2). We investigated the quantitative expression of these 4 MS4A genes, as well as two genes comprising the closely related TMEM176 gene family (3), in human blood and tonsil B-cells with a view to identifying candidates for future investigation at the protein level. A secondary goal was to identify MS4A proteins that might provide alternate therapeutic targets for chronic lymphocytic leukemia (CLL), a B-cell malignancy in which CD20 is expressed at much lower levels than in B-cell lymphomas (22–24). We therefore also investigated expression of MS4A and TMEM176 genes in a group of untreated CLL patients.
Materials and Methods
Cell Isolation
All samples in this study were obtained by approval of the Conjoint Health Research Ethics Board at the University of Calgary. Blood samples from healthy volunteer donors were obtained from laboratory personnel in the Immunology Research Group, University of Calgary. Tonsils were obtained from patients undergoing tonsillectomy at the Alberta Children’s Hospital. Whole blood from patients with CLL was obtained anonymously as leftover samples from the Flow Cytometry Laboratory of Calgary Laboratory Services at Foothills Medical Center; only samples with dim CD20 expression and ≥96% CD5+ B-cells were included in the study. All samples were processed within 24 h of collection. B-cells were purified from whole blood using the RosetteSep™ Human B-cell Enrichment Cocktail (StemCell Technologies, Vancouver, BC, Canada) and from tonsils using a modified RosetteSep™ method (25). CD5+ B-cells were prepared from normal blood B-cells using the EasySep Positive Selection kit (StemCell Technologies) with PE-conjugated anti-CD5 (BioLegend, Mississauga, ON, Canada). CD27 positive selection was achieved using CD27-PE with the EasySep PE magnetic selection kit. A minimum of 105 highly purified cells was collected for each population. T cells were purified from whole blood using the Pan T Cell Isolation Kit II (Miltenyi Biotec, Auburn, CA, USA).
RNA Isolation and Real-Time PCR
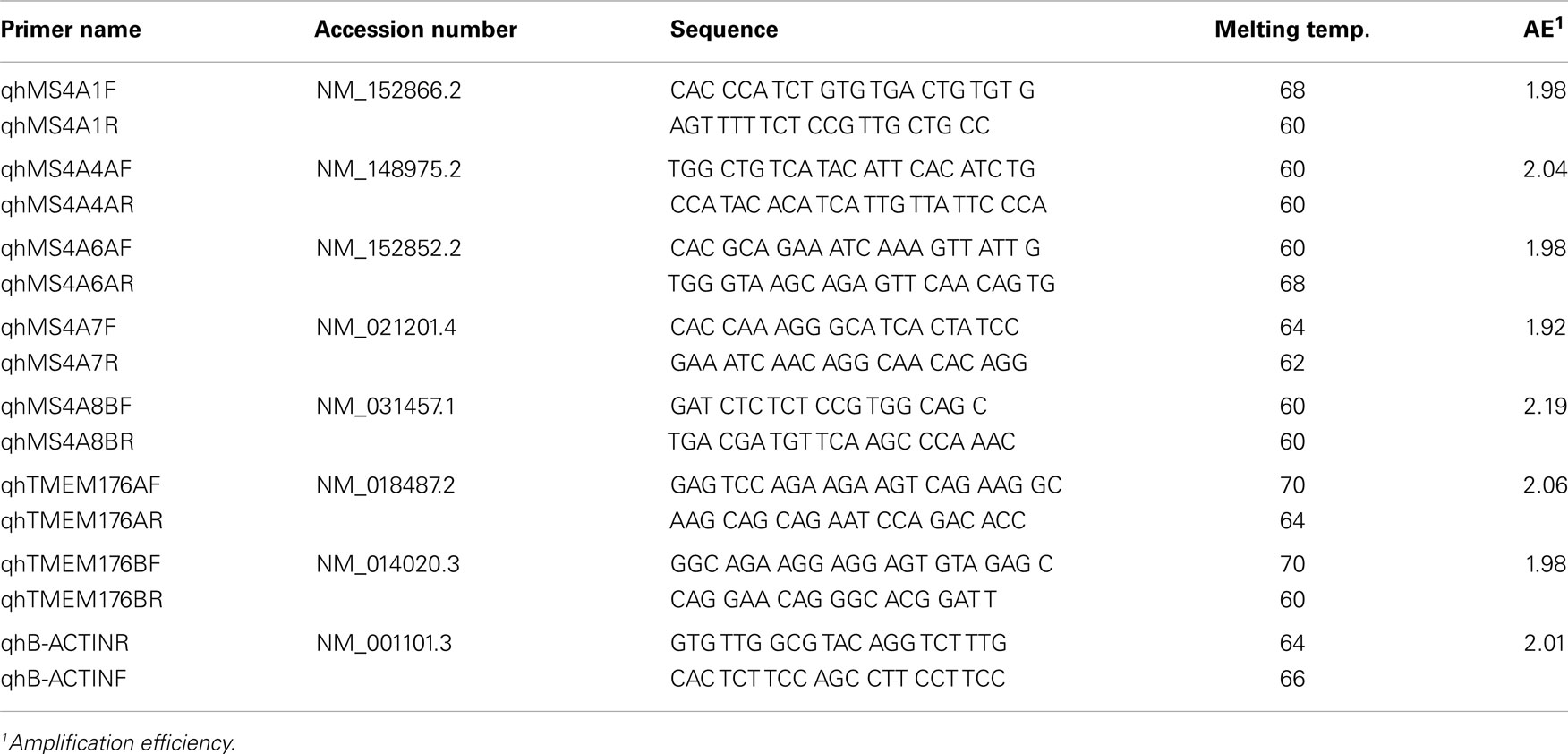
Total RNA from blood and tonsil tissue was extracted using Trizol (Invitrogen, Burlington, ON, Canada). RNA from a variety of human tissues was purchased from Clontech, Mountain View, CA. 1 μg of total RNA was converted to cDNA using SuperScript III Reverse Transcriptase (Invitrogen, Burlington, ON, Canada) and oligo dT primers in a volume of 20 μl. The cDNA sample was diluted to 100 and 1 μl of this was used as template for quantitative real-time PCR (qPCR) using SybrGreen (Applied Biosystems, Streetsville, ON, Canada), the primer sets shown in Table 1, and the ABI 7000 machine (Applied Biosystems) in the 96-well format. Reactions were performed in duplicate under the following conditions: 95 °C for 10 min followed by 40 cycles of 60 °C for 1 min and 95 °C for 15 s. Each run was concluded with a melting curve analysis to verify the specificity of the PCR product. Amplification of specific products was further validated during PCR optimization by gel purification and sequencing. Amplification efficiencies, calculated using 10-fold dilutions (10−1 to 10−5) of cDNA prepared from tonsil B-cell RNA, were close to 2.0 in every case (Table 1). Data were normalized to an endogenous control, ACTB (β-actin). mRNA levels in each sample were thus expressed as the difference between threshold values (ΔCT) for the gene transcript of interest and ACTB, using the equation Ratio = (Etarget)ΔCP target(control − sample)/(Eref)ΔCP ref(control − sample) (26).
CDNA Constructs and Transfections
The human MS4A4A, MS4A6A, MS4A7, and MS4A8B coding sequences were cloned and inserted into the multicloning site (MCS) of the pEGFP-C1 vector (Clontech, Mountain View, CA, USA). BJAB cells were electroporated at 340 V and 950 μF (Gene Pulser II, Bio-Rad, Hercules, CA, USA) with 20 μg of cDNA. GFP-positive cell lines were generated in 1 mg/ml Geneticin (Life Technologies). BJAB cells stably expressing GFP-CD20 were previously established (15).
Immunofluorescence Microscopy
Cells were incubated with antibodies either before or after fixation in 4% paraformaldehyde (PFA, Electron Microscopy Sciences, Hatfield, PA, USA) at room temperature for 15 min. Antibodies against EEA1, golgin 95, golgin 97, and giantin were provided by Dr. Marvin Fritzler (University of Calgary, AB, Canada). CD20 mAb 2H7 was provided by Dr. J. Ledbetter (Bristol-Myers Squibb, Seattle, WA, USA). Cy3-conjugated F(ab′)2 goat anti-mouse IgG was purchased from Jackson ImmunoResearch Laboratories Inc. (West Grove, PA, USA). Nuclei were stained with 4′-6-Diamidino-2-phenylindole (DAPI, Sigma). Fluorescence imaging was done with a Leica DM RXA microscope (Rockleigh, NJ, USA) attached to a 14-bit cooled CCD camera (Princeton Instruments, Monmouth Junction, NJ, USA). Digital deconvolution was performed using the nearest neighbor algorithm, Microtome for Windows (Vaytek, Fairfield, IA, USA).
Results
The MS4A4A, MS4A6A, MS4A7, and MS4A8B genes were selected for analysis in this study on the basis of evidence that they are expressed in B-cell lines (2). MS4A genes with expression known to be limited to myeloid-lineage cells or non-hematopoietic tissues were excluded. MS4A1 (CD20), which is expressed at high levels in mature B-cells, was included for comparison. In addition, we examined expression of the MS4A-related genes, TMEM176A and TMEM176B, which have not previously been assessed in human B-cells. The levels of expression were assessed using quantitative RT-PCR (qPCR), with validation of the primer sets done by confirming the expected sizes and sequences of the amplified products. Amplification of each gene was compared to that of ACTB (β-actin) and shown on the y-axes in Figures 1 and 2 as Relative mRNA.
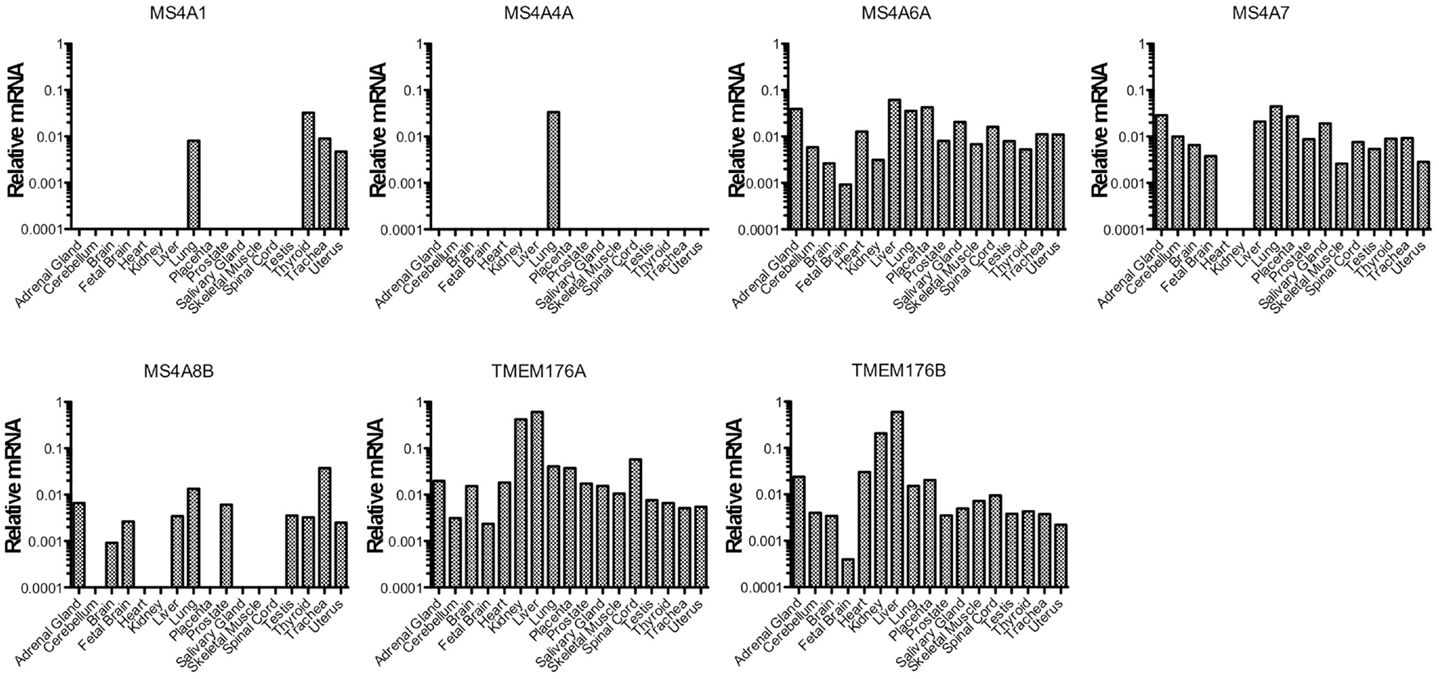
Figure 1. MS4A/TMEM176 mRNA levels in non-hematopoietic human tissues. cDNA was prepared from tissue RNA samples in the human total RNA master panel II (Clontech). Quantitative PCR was performed to amplify MS4A1, MS4A4A, MS4A6A, MS4A7, MS4A8B, TMEM176A, TMEM176B, and ACTB cDNA using primer sets with equivalent amplification efficiencies (Table 1). Vertical axes show mRNA levels relative to ACTB.
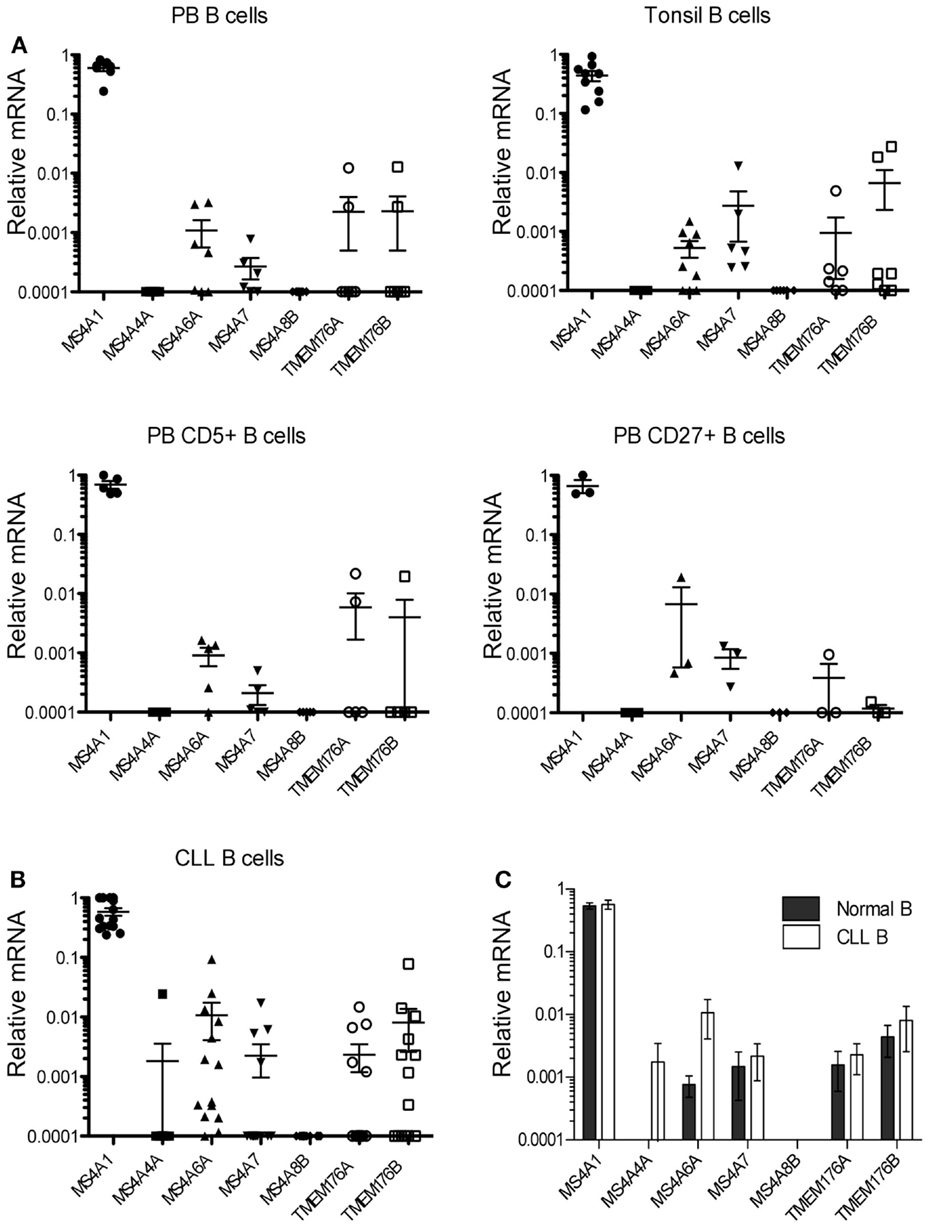
Figure 2. MS4A/TMEM176 mRNA levels in normal and CLL B-cells. cDNA was prepared from freshly isolated normal B-cell samples and CLL B-cell samples. Quantitative PCR was performed to amplify MS4A1, MS4A4A, MS4A6A, MS4A7, MS4A8B, TMEM176A, TMEM176B, and ACTB cDNA as in Figure 1. Vertical axes show mRNA levels relative to ACTB. Each data point represents one donor or patient sample. (A) peripheral blood (PB) B-cells isolated from seven healthy donors, B-cells isolated from tonsillar tissue from seven patients, CD5+ B-cells sorted from five donor blood samples, CD27+ B-cells sorted from three donor blood samples, and (B) 14 CLL B-cell samples. Means and SEM are indicated by the horizontal bars. (C) Quantitative PCR data from the 14 blood and tonsil B-cell samples were compared with data from the 14 CLL B-cell samples, with means and SEM shown. Data were analyzed by Mann–Whitney U and there was no statistical significance between the groups (p > 0.05).
We first examined the expression of the MS4A and TMEM176 genes in non-hematopoietic tissues (Figure 1). Detection of MS4A1 (CD20) transcripts in the lung, trachea, thyroid, and uterus is probably indicative of blood contamination or infiltration of these tissues. Among non-hematopoietic tissues, MS4A4A detection was limited to the lung. Considering that MS4A1 (CD20) transcripts were also amplified from the lung, MS4A4A may be expressed on a hematopoietic cell type in that tissue. MS4A8B was expressed most highly in the lung and trachea, consistent with a previous report (27), and was also expressed in a few other tissues but with a more restricted profile than the remaining genes, MS4A6A, MS4A7, TMEM176A, and TMEM176B, which were detected broadly in non-hematopoietic tissues (Figure 1).
We then examined expression of the MS4A4A and TMEM176 genes in B-cells isolated from multiple donor samples of peripheral blood and tonsil tissues. As expected, MS4A1 (CD20) transcripts were always detected at high levels (Figure 2A). Surprisingly, however, we detected only relatively low levels of transcripts for any of the six new MS4A/TMEM176 genes in either tonsil or peripheral blood B-cells. Indeed, MS4A4A and MS4A8B transcripts were not detected in any sample (Figure 2A). MS4A6A and MS4A7 transcripts were detected in most B-cell samples but at two to four orders of magnitude lower than MS4A1 (CD20). TMEM176A and TMEM176B transcripts were found in a minority of blood and tonsil B-cell samples. The expression of MS4A and TMEM176 genes in CD5+ blood B-cells and CD27+ (memory) blood B-cells was examined with similar findings: no detection of MS4A4A or MS4A8B, and low or no detection of transcripts from the remaining genes (Figure 2A).
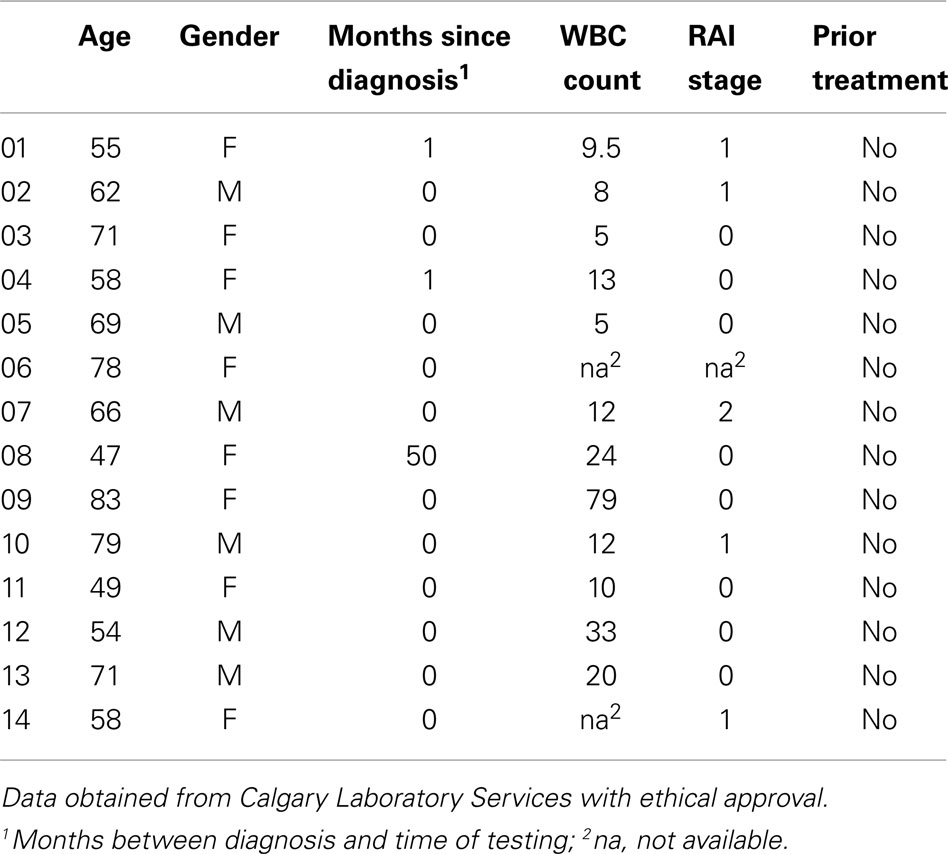
Data were obtained for 14 blood samples obtained from CLL patients (Table 2) and tested similarly for the expression of the MS4A4A and TMEM176 genes (Figure 2B). MS4A1 (CD20) transcripts were detected at high levels in every sample, as expected (24). MS4A4A expression, which was undetected in normal B-cells, was detected in a single sample. Notably, MS4A6A transcripts were detected at higher levels in 4/14 of CLL samples than in all but one of the 14 normal blood or tonsil B-cell samples, the single exception being one of the blood CD27+ samples. However, the differences in MS4A6A expression between CLL and normal B-cells were not statistically significant (Figure 2C). MS4A8B was not detected in any CLL sample. Expression of MS4A7 and the TMEM176 genes was either low or undetected in CLL samples, as it was in normal B-cells.
Statistical analysis confirmed that the levels of all MS4A and TMEM176 transcripts in CLL were not different than levels found in normal B-cells (Figure 2C).
The data obtained from normal primary B-cells indicated some expression of MS4A6A and MS4A7 in the majority of the samples tested (Figure 2A). In order to assess the potential for the corresponding MS4A proteins to hetero-oligomerize with CD20, we expressed them as GFP fusion proteins in the human BJAB cell line and examined their subcellular localization by fluorescence microscopy. GFP-tagged MS4A4A, MS4A8B, and CD20 were expressed for comparison. Cells were fixed and nuclei stained with DAPI. GFP-MS4A4A and GFP-MS4A8B, like GFP-CD20, appeared to be expressed at the plasma membrane (Figure 3A) and this was confirmed by visualizing the plasma membrane using anti-CD20 (Figure 3C). In contrast, GFP-MS4A6A and GFP-MS4A7 were localized intracellularly in the perinuclear space (Figure 3A). To characterize the intracellular compartment, we stained early endosomes, cis-Golgi, cis/medial Golgi, and trans-Golgi compartments using antibodies detecting EEA1, golgin 97, golgin 95, and giantin, respectively. Both GFP-MS4A6A and GFP-MS4A7 co-localized with giantin, indicating that they were concentrated in the trans-Golgi complex (Figure 3B); no colocalization with EEA1, golgin 97, or golgin 95 was observed (data not shown).
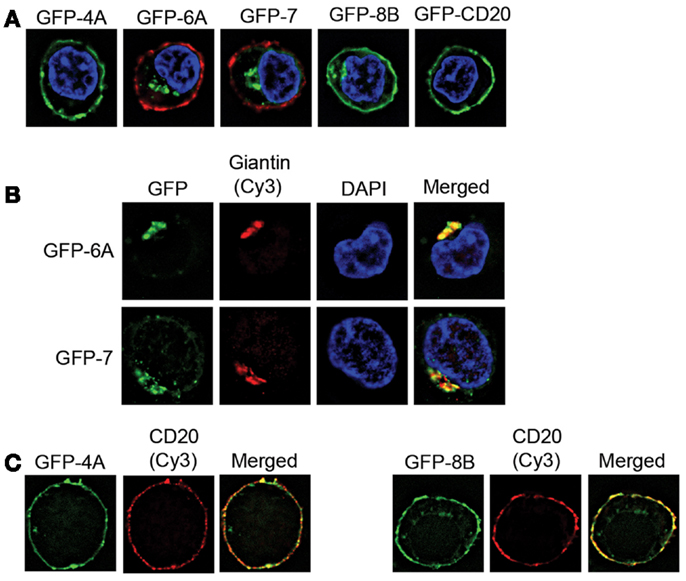
Figure 3. Intracellular localization of GFP-tagged MS4A6A and MS4A7. BJAB cell lines were derived that express GFP-tagged MS4A4A, MS4A6A, MS4A7, MS4A8B, and CD20. (A) Immunofluorescence images are of individual cells representative of>100 cells observed in at least two independent experiments for each cell line. Cells were fixed and nuclei stained with DAPI. In the case of GFP-MS4A6A and GFP-MS4A7 the cells were counterstained with mouse anti-human CD20 (2H7) and Cy3-conjugated F(ab′)2 anti-mouse IgG to delineate the plasma membrane. (B) Cells expressing GFP-MS4A6A or GFP-MS4A7 were fixed, permeabilized, and stained with rabbit anti-giantin and Cy3-conjugated anti-rabbit IgG to mark the trans-Golgi. Control experiments with non-specific primary antibody confirmed the specificity of the fluorescent signals obtained. Single-labeled control samples were imaged separately to confirm negligible bleed-through of fluorophores to the other channel. Nuclei were stained with DAPI. Results are representative of>100 cells observed in at least two independent experiments. (C) Cells expressing GFP-MS4A4A or GFP-MS4A8B were stained for CD20 as in(A).
Discussion
This report describes the results of our investigation into the expression of MS4A4A, MS4A6A, MS4A7, MS4A8B, and both of the MS4A-related TMEM176 genes in normal B-cells and in CLL. Unlike MS4A1 (CD20), which is consistently and highly expressed in both normal and CLL B-cells at the mRNA level, the other MS4A/TMEM176 genes tested were either not expressed or expressed at relatively low and/or variable levels. MS4A4A and MS4A8B genes were not expressed in normal blood or tonsillar B-cells; this was a surprising result given a previous report of their expression in B-cell lines (2), however, antibodies generated against the corresponding proteins confirmed that MS4A4A and MS4A8B are not expressed in mature human B-cells (not shown).
MS4A6A and MS4A7 transcripts were detected at low levels, generally 200–400 times lower than MS4A1 (CD20). A recent global analysis of RNA and protein levels showed that they are significantly correlated in ∼40% of comparisons, higher than previous estimates but clearly not absolute (28). Although the overall correlation is relatively poor, unstable mRNAs tend not to generate stable proteins except in restricted categories of proteins such as those regulating RNA processing (28). Thus, it is unlikely that very low levels of mRNAs would generate high levels of membrane proteins. We could not examine endogenous MS4A6 and MS4A7 as reliable antibodies were not available, however we found that neither of these proteins localized to the plasma membrane when expressed as GFP fusion proteins in a human B-cell line. This is unlikely to be an artifact of GFP fusion as a similarly constructed GFP-CD20 protein was properly localized to the plasma membrane, as were GFP-MS4A4A and GFP-MS4A8B. Known intracellular localization of MS4A3 (HTm4) provides a precedent for intracellular localization of some other MS4A proteins (29). It is possible that MS4A6A and/or MS4A7 are mobilized to the plasma membrane under certain conditions, however, our results suggest that neither MS4A6A or MS4A7 is found in sufficient abundance or in the right subcellular compartment to normally form hetero-oligomers with CD20 or to be considered as an alternate therapeutic target for B-cell depletion.
TMEM176A and TMEM176B transcripts were undetected or detected at only trace levels in most samples of normal blood or tonsillar B-cells. From this it is clear that neither corresponding protein is normally expressed in all mature human B-cells, although expression could be upregulated under certain conditions. Similar to MS4A6A and MS4A7, tissue expression was broad for both TMEM176 genes suggesting generalized rather than cell-type specific functions, and negating their potential utility as therapeutic targets.
The expression profiles of the tested MS4A and TMEM176 genes in CLL were generally similar to that in normal B-cells, however there were a few CLL samples in which expression levels of one of the MS4A genes was unusually high. MS4A4A, which was never detected in blood or tonsil B-cells, was expressed at a high level in one CLL sample. This sample was from a patient who presented with RAI stage II CLL at the time of diagnosis when the sample was collected (Table 2), however studies on much larger, defined patient populations would need to be performed to determine the significance, if any, of this observation. MS4A6A was more highly expressed in about 1/3 of CLL samples than in any of the normal B-cell samples, but this difference did not reach statistical significance (Figure 2C). Our study was not designed to test the potential correlation of differential MS4A/TMEM176 gene expression with clinical indicators of prognosis and no conclusions about the significance of these observations can be made at this time. However, testing the protein expression of MS4A4A and MS4A6A in a large cohort of defined CLL samples would be an important direction when antibodies suitable for flow cytometry become available.
In conclusion, we searched for and did not find evidence for MS4A4A, MS4A6A, MS4A7, MS4A8B, or TMEM176 transcripts likely to generate proteins in sufficient quantity to form hetero-oligomers with the abundant CD20 molecules expressed in all normal mature B-cells. MS4A4A and MS4A8B were not detected. MS4A6A and MS4A7 transcripts were detected in most normal B samples but at very low levels and the corresponding proteins are probably not normally expressed at the plasma membrane. TMEM176A and TMEM176B transcripts were only detected in a minority of B-cell samples. Available data for other MS4A genes shows expression restricted to myeloid-lineage cells, testis, lung, and intestinal tissues. Therefore, in the absence of evidence of other MS4A proteins co-expressed with CD20 at the plasma membrane, it is most likely that CD20 normally exists in homo-oligomeric form and that alternate therapeutic targets for B-cells are unlikely to be found in the MS4A family. Novel targets are more likely to be found among members of the unrelated but structurally similar tetraspanin superfamily (30). Although broad tissue expression of tetraspanin proteins such as CD9 and CD81 limits their potential in this regard, high expression of CD37 in B-cells makes it a promising target for CLL and other B-cell cancers (31–34).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge Laurie Kennedy, Laurie Robertson, and Yiping Liu for expert assistance with flow cytometric cell sorting, and Dr. Marvin Fritzler for generously providing antibodies. Research was supported by a Limited Term grant from the Alberta Cancer Board and by operating grant no. 15095 from the Canadian Institutes of Health Research (www.cihr-irsc.gc.ca). Jonathan Zuccolo was supported by a graduate studentship from the Alberta Cancer Research Institute. Jeremy A. Bau was supported by a summer studentship from the Alberta Heritage Foundation for Medical Research.
References
1. Ishibashi K, Suzuki M, Sasaki S, Imai M. Identification of a new multigene four-transmembrane family (MS4A) related to CD20, HTm4 and beta subunit of the high-affinity IgE receptor. Gene (2001) 264(1):87–93. doi:10.1016/S0378-1119(00)00598-9
2. Liang Y, Tedder TF. Identification of a CD20-, FcepsilonRIbeta-, and HTm4-related gene family: sixteen new MS4A family members expressed in human and mouse. Genomics (2001) 72(2):119–27. doi:10.1006/geno.2000.6472
3. Zuccolo J, Bau J, Childs SJ, Goss GG, Sensen CW, Deans JP. Phylogenetic analysis of the MS4A and TMEM176 gene families. PLoS ONE (2010) 5(2):e9369. doi:10.1371/journal.pone.0009369
4. Polyak MJ, Li H, Shariat N, Deans JP. CD20 homo-oligomers physically associate with the B cell antigen receptor. Dissociation upon receptor engagement and recruitment of phosphoproteins and calmodulin-binding proteins. J Biol Chem (2008) 283(27):18545–52. doi:10.1074/jbc.M800784200
5. Li H, Ayer LM, Lytton J, Deans JP. Store-operated cation entry mediated by CD20 in membrane rafts. J Biol Chem (2003) 278(43):42427–34. doi:10.1074/jbc.M308802200
6. Kuijpers TW, Bende RJ, Baars PA, Grummels A, Derks IA, Dolman KM, et al. CD20 deficiency in humans results in impaired T cell-independent antibody responses. J Clin Invest (2010) 120(1):214–22. doi:10.1172/JCI40231
7. Becker YT, Samaniego-Picota M, Sollinger HW. The emerging role of rituximab in organ transplantation. Transpl Int (2006) 19(8):621–8. doi:10.1111/j.1432-2277.2006.00345.x
8. Pescovitz MD. Rituximab, an anti-cd20 monoclonal antibody: history and mechanism of action. Am J Transplant (2006) 6(5 Pt 1):859–66. doi:10.1111/j.1600-6143.2006.01288.x
9. Lim SH, Beers SA, French RR, Johnson PW, Glennie MJ, Cragg MS. Anti-CD20 monoclonal antibodies: historical and future perspectives. Haematologica (2010) 95(1):135–43. doi:10.3324/haematol.2008.001628
10. Polyak MJ, Deans JP. Alanine-170 and proline-172 are critical determinants for extracellular CD20 epitopes; heterogeneity in the fine specificity of CD20 monoclonal antibodies is defined by additional requirements imposed by both amino acid sequence and quaternary structure. Blood (2002) 99(9):3256–62. doi:10.1182/blood.V99.9.3256
11. Teeling JL, Mackus WJ, Wiegman LJ, van den Brakel JH, Beers SA, French RR, et al. The biological activity of human CD20 monoclonal antibodies is linked to unique epitopes on CD20. J Immunol (2006) 177(1):362–71.
12. Niederfellner G, Lammens A, Mundigl O, Georges GJ, Schaefer W, Schwaiger M, et al. Epitope characterization and crystal structure of GA101 provide insights into the molecular basis for type I/II distinction of CD20 antibodies. Blood (2011) 118(2):358–67. doi:10.1182/blood-2010-09-305847
13. Cragg MS. CD20 antibodies: doing the time warp. Blood (2011) 118(2):219–20. doi:10.1182/blood-2011-04-346700
14. Deans JP, Robbins SM, Polyak MJ, Savage JA. Rapid redistribution of CD20 to a low density detergent-insoluble membrane compartment. J Biol Chem (1998) 273(1):344–8. doi:10.1074/jbc.273.1.344
15. Li H, Ayer LM, Polyak MJ, Mutch CM, Petrie RJ, Gauthier L, et al. The CD20 calcium channel is localized to microvilli and constitutively associated with membrane rafts: antibody binding increases the affinity of the association through an epitope-dependent cross-linking-independent mechanism. J Biol Chem (2004) 279(19):19893–901. doi:10.1074/jbc.M400525200
16. Cragg MS, Morgan SM, Chan HT, Morgan BP, Filatov AV, Johnson PW, et al. Complement-mediated lysis by anti-CD20 mAb correlates with segregation into lipid ‘rafts’. Blood (2003) 101:1045–52. doi:10.1182/blood-2002-06-1761
17. Press OW, Farr AG, Borroz KI, Anderson SK, Martin PJ. Endocytosis and degradation of monoclonal antibodies targeting human B-cell malignancies. Cancer Res (1989) 49(17):4906–12.
18. Press OW, Howell-Clark J, Anderson S, Bernstein I. Retention of B-cell-specific monoclonal antibodies by human lymphoma cells. Blood (1994) 83(5):1390–7.
19. Beers SA, French RR, Chan HT, Lim SH, Jarrett TC, Vidal RM, et al. Antigenic modulation limits the efficacy of anti-CD20 antibodies: implications for antibody selection. Blood (2010) 115(25):5191–201. doi:10.1182/blood-2010-01-263533
20. Bubien JK, Zhou LJ, Bell PD, Frizzell RA, Tedder TF. Transfection of the CD20 cell surface molecule into ectopic cell types generates a Ca2+ conductance found constitutively in B lymphocytes. J Cell Biol (1993) 121(5):1121–32. doi:10.1083/jcb.121.5.1121
21. Howie D, Nolan KF, Daley S, Butterfield E, Adams E, Garcia-Rueda H, et al. MS4A4B is a GITR-associated membrane adapter, expressed by regulatory T cells, which modulates T cell activation. J Immunol (2009) 183(7):4197–204. doi:10.4049/jimmunol.0901070
22. Almasri NM, Duque RE, Iturraspe J, Everett E, Braylan RC. Reduced expression of CD20 antigen as a characteristic marker for chronic lymphocytic leukemia. Am J Hematol (1992) 40(4):259–63. doi:10.1002/ajh.2830400404
23. Golay J, Lazzari M, Facchinetti V, Bernasconi S, Borleri G, Barbui T, et al. CD20 levels determine the in vitro susceptibility to rituximab and complement of B-cell chronic lymphocytic leukemia: further regulation by CD55 and CD59. Blood (2001) 98(12):3383–9. doi:10.1182/blood.V98.12.3383
24. Sarro SM, Unruh TL, Zuccolo J, Sanyal R, Luider JM, Auer-Grzesiak IA, et al. Quantification of CD20 mRNA and protein levels in chronic lymphocytic leukemia suggests a post-transcriptional defect. Leuk Res (2010) 34(12):1670–3. doi:10.1016/j.leukres.2010.06.031
25. Zuccolo J, Unruh TL, Deans JP. Efficient isolation of highly purified tonsil B lymphocytes using RosetteSep with allogeneic human red blood cells. BMC Immunol (2009) 10:30. doi:10.1186/1471-2172-10-30
26. Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res (2001) 29(9):e45. doi:10.1093/nar/29.9.e45
27. Bangur CS, Johnson JC, Switzer A, Wang YH, Hill B, Fanger GR, et al. Identification and characterization of L985P, a CD20 related family member over-expressed in small cell lung carcinoma. Int J Oncol (2004) 25(6):1583–90.
28. Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature (2011) 473(7347):337–42. doi:10.1038/nature10098
29. Donato JL, Ko J, Kutok JL, Cheng T, Shirakawa T, Mao XQ, et al. Human HTm4 is a hematopoietic cell cycle regulator. J Clin Invest (2002) 109(1):51–8. doi:10.1172/JCI200214025
30. Sala-Valdes M, Ailane N, Greco C, Rubinstein E, Boucheix C. Targeting tetraspanins in cancer. Expert Opin Ther Targets (2012) 16(10):985–97. doi:10.1517/14728222.2012.712688
31. Robak T, Robak P, Smolewski P. TRU-016, a humanized anti-CD37 IgG fusion protein for the potential treatment of B-cell malignancies. Curr Opin Investig Drugs (2009) 10(12):1383–90.
32. Zhao X, Lapalombella R, Joshi T, Cheney C, Gowda A, Hayden-Ledbetter MS, et al. Targeting CD37-positive lymphoid malignancies with a novel engineered small modular immunopharmaceutical. Blood (2007) 110(7):2569–77. doi:10.1182/blood-2006-12-062927
33. van Spriel AB. Tetraspanins in the humoral immune response. Biochem Soc Trans (2011) 39(2):512–7. doi:10.1042/BST0390512
Keywords: MS4A, TMEM176, CD20, B lymphocytes, CLL
Citation: Zuccolo J, Deng L, Unruh TL, Sanyal R, Bau JA, Storek J, Demetrick DJ, Luider JM, Auer-Grzesiak IA, Mansoor A and Deans JP (2013) Expression of MS4A and TMEM176 genes in human B lymphocytes. Front. Immunol. 4:195. doi: 10.3389/fimmu.2013.00195
Received: 15 May 2013; Accepted: 02 July 2013;
Published online: 15 July 2013.
Edited by:
Michael R. Gold, The University of British Columbia, CanadaReviewed by:
Sherri L. Christian, Memorial University of Newfoundland, CanadaAnnemiek Van Spriel, Nijmegen Centre for Molecular Life Sciences, Netherlands
Copyright: © 2013 Zuccolo, Deng, Unruh, Sanyal, Bau, Storek, Demetrick, Luider, Auer-Grzesiak, Mansoor and Deans. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Julie P. Deans, Department of Biochemistry and Molecular Biology, University of Calgary, 3330 Hospital Drive NW, Calgary, AB T2N 4N1, Canada e-mail:amRlYW5zQHVjYWxnYXJ5LmNh

 Lili Deng1
Lili Deng1