- 1Laboratory of Immunology, Department of Biological Sciences, Indian Institute of Science Education and Research (IISER), Bhopal, India
- 2Department of Biotechnology and Bioinformatics, Yogi Vemana University, Kadapa, India
- 3Department of Microbiology, Yogi Vemana University, Kadapa, India
- 4WPI Immunology Frontier Research Center, Osaka University, Osaka, Japan
Innate sensors play a critical role in the early innate immune responses to invading pathogens through sensing of diverse biochemical signatures also known as pathogen associated molecular patterns (PAMPs). These biochemical signatures primarily consist of a major family of biomolecules such as proteins, lipids, nitrogen bases, and sugar and its complexes, which are distinct from host molecules and exclusively expressed in pathogens and essential to their survival. The family of sensors known as pattern recognition receptors (PRRs) are germ-line encoded, evolutionarily conserved molecules, and consist of Toll-like receptors (TLRs), RIG-I-like receptors (RLRs), NOD-like receptors (NLRs), C-type lectin-like receptors (CLRs), and DNA sensors. Sensing of PAMP by PRR initiates the cascade of signaling leading to the activation of transcription factors, such as NF-κB and interferon regulatory factors (IRFs), resulting in a variety of cellular responses, including the production of interferons (IFNs) and pro-inflammatory cytokines. In this review, we discuss sensing of different types of glycosylated PAMPs such as β-glucan (a polymeric sugar) or lipopolysaccharides, nucleic acid, and so on (sugar complex PAMPs) by different families of sensors, its role in pathogenesis, and its application in development of potential vaccine and vaccine adjuvants.
Introduction
The complexity of carbohydrates exhibits enormous diversity in structure and function in the living system. Carbohydrates play crucial role in fundamental biological processes such as energy requirements and modification of basic macromolecules like proteins and lipids to make them functional macromolecules for cellular functions such as cellular communication, cell-type specificity, cell signaling, and so on. The diversification of carbohydrate structure is because of branching and complexing of monomers or polymers or through formation of glycoconjugates such as glycoproteins and glycolipids. These glycoconjugates are fundamental components of both the host and pathogens and play an important role in host–pathogen interactions and provide the molecular basis for discrimination and elicitation of appropriate immune responses in the host (1–3). Pathogen associated molecular patterns (PAMPs) are molecular signature of pathogens recognized by the host germ-line encoded pattern recognition receptors (PRRs), which are classified as toll-like receptors (TLRs), retinoic acid-inducible gene (RIG)-I-like receptors (RLRs), C-type lectin receptors (CLRs), and nucleotide oligomerization domain (NOD)-like receptors (NLRs), and DNA sensors (4–8). Most of the PAMPs such lipopolysaccharides (LPS) from gram-negative bacteria, bacterial/viral nucleic acid including single-stranded RNA (ssRNA), double-stranded RNA (dsRNA), CpG rich DNA, β-glucans from fungus, and trehalose dimycolate (TDM) from Mycobacterium contain sugar as integral part of structure. PAMPs, which either constituted of sugar or contain sugar as a component of the complex structure can be given a new terminology as sugar complexed PAMPs (SCPs). The SCPs group of PAMPs is a largest group of PAMPs and almost all the PAMPs are grouped into it except pure lipid and protein such as flagellin. SCPs constitute essential structural and functional moieties of the pathogen that are essential for infection and establishment of disease in the host. On other hand these SCPs also play an important role in vaccine biology. The sensing of SCPs in various compartments of cells by different PRRs activates an array of biochemical reactions, leading to the activation of transcription factors such as nuclear factor kappa B (NF-κB) and interferon (IFN) regulatory factors (IRFs) for induction of inflammatory cytokine and type-I IFNs respectively. Additionally, these innate immune responses also play a pivotal role in initiation of pathogen-specific adaptive immunity via T and B lymphocytes. In this review, we will discuss different kinds of SCPs, their recognition by PRRs, and role in disease, and how these PAMPs can be explored for therapeutic application as vaccines and/or vaccine adjuvants.
Sensing of SCPs by PRRs
Large numbers of SCPs from different classes of pathogens (bacteria, virus, and fungi) are reported and sensing of these PAMPs by different families of PRRs is described below in detail.
Sensing of SCPs by TLRs
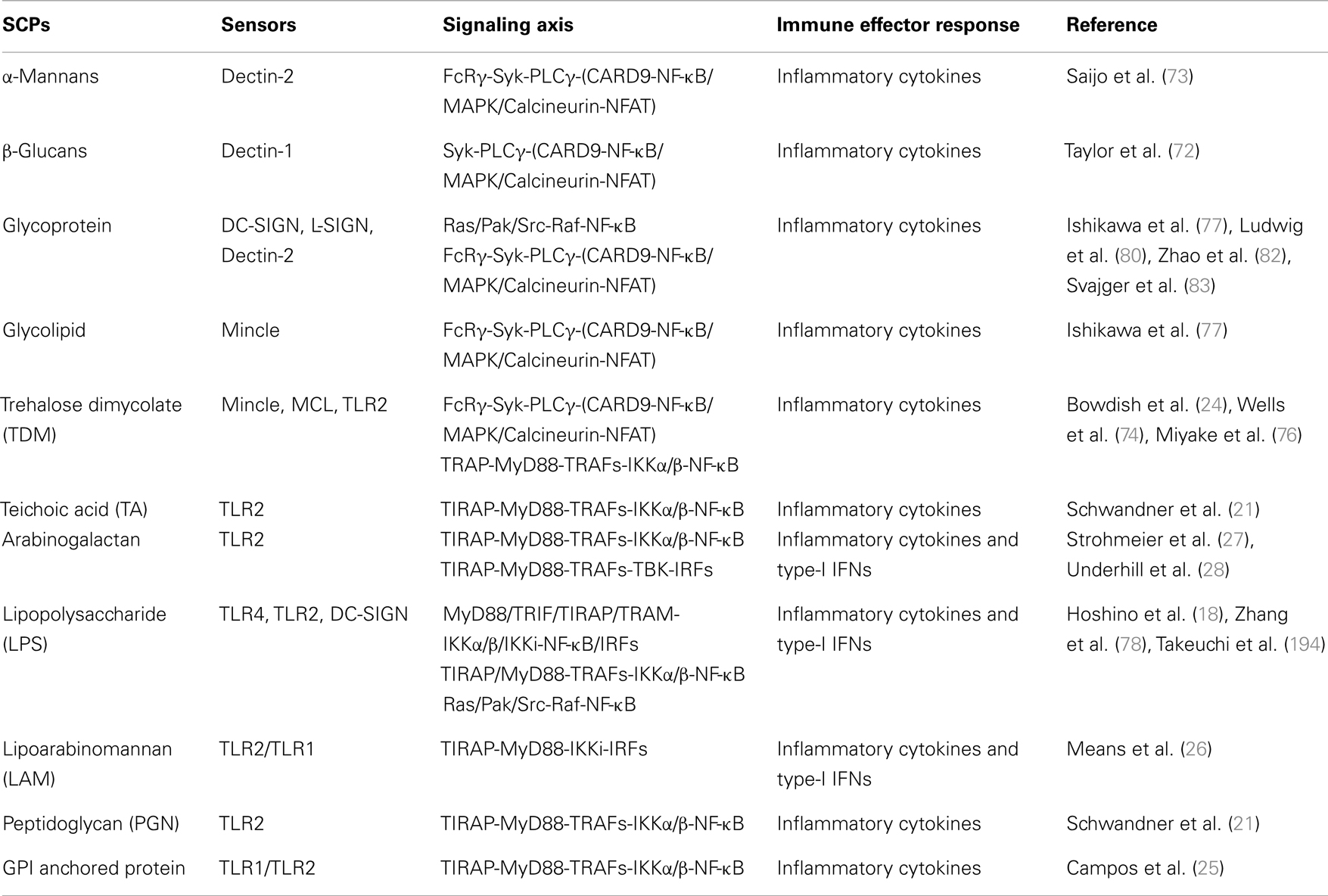
Toll-like receptors sense various kinds of SCPs such as LPS, ssRNA, and hypomethylated dsDNA (CpG) in different cellular compartments of the cells. The sensing of SCPs by TLRs is described in several review and listed in Tables 1 and 2 (7, 9–11). The recently discovered TLR13 is involved in sensing of bacterial 23S rRNA (12). TLR1, TLR2, TLR4, and TLR6 are plasma membrane localized TLRs, mainly sense hydrophobic SCPs (Table 1) such as LPS, while TLR3, TLR7, TLR8, and TLR9 are endosome localized TLRs, sense hydrophilic SCPs (Table 2) such as 5′-ppp-ssRNA (9, 13). TLRs are comprised of N-terminal ligand binding extra cellular domain (ECD) with 19–25 leucine rich repeat (LRRs) motifs, single transmembrane domain, and C-terminal intracellular toll/interleukin receptor (TIR) domain (14, 15). LRRs are consists of “xLxxLxLxx” where “L” stands for leucine and “x” stands for any amino acid, and it is important for recognition of PAMPs. Binding of PAMPs to LRRs leads to receptor dimerization that induces conformational changes to TIR domain for recruitment of an adaptor and activation of the signaling cascade (13, 15). Sensing of PAMPs by TLR2 induces oligomerization with TLR1 or TLR6, while TLR3, TLR7, TLR8, and TLR9 form homodimers; however, the sensing mechanism for TLR13 is not well known. It is possible that TLR13 forms a homodimer for the activation of a downstream signaling pathway. The glycolipid LPS, which is structurally constituted of lipid A, central oligosaccharide, and antigenic O-polysaccharide (16) is sensed by TLR4 (17, 18) (Table 1) along with co-receptors known as myeloid differential factor 2 (MD-2) and cluster CD14 (17, 19, 20). TLR2 senses a large class of bacterial SCPs such as lipoteichoic acid (LTA) (21–23), teichoic acid (TA) (21), TDM (24), peptidoglycan (PGN) (21), glycophosphatidylinositol (GPI) anchored proteins (25), lipoarabinomannan (LAM) (26), and arabinogalactan (AG) (27, 28). Nucleic acids, the pentose sugar containing PAMPs are sensed by endosomal TLRs. The bacterial and viral DNA, which are hypomethylated compared to the host DNA, sensed by TLR9 (29–31). The viral dsRNA and ssRNA are sensed by TLR3 and TLR7, respectively (32–34). Recently, human TLR8 was shown to sense viral ssRNA-derived from human immunodeficiency virus (HIV) (35, 36).
Sensing of SCPs by RLRs
Retinoic acid-inducible gene-I-like receptors, retinoic acid-inducible gene I (RIG-I), melanoma-differentiation-associated gene 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2) (37), sense RNA SCPs (Table 2). Studies on RIG-I and MDA5 knockout mice demonstrate that RIG-I and MDA5 sense viral RNA from different classes of RNA viruses (38, 39) and induce antiviral immune response; however, LGP2 functions as a regulatory molecule for RIG-I and MDA5 mediated sensing of viral RNA (40, 41). RIG-I and MDA5 are comprised of two N-terminal caspase activation and recruitment domains (CARD), middle ATPase containing DExD helicase domain (DExD helicase), and a C-terminal domain (CTD). The N-terminal CARD of RIG-I and MDA5 is necessary for recruitment of adaptor and downstream signaling. RIG-I mainly senses small size (<1 kb) RNA containing 5′ppp-ssRNA moieties (42), which could be ssRNA (43, 44) or dsRNA (45), while MDA5 mainly recognizes larger size RNA (>1 kb) from various kinds of viruses and discussed in reviews (46). MDA5 recognizes internal duplex structure of RNA, while RIG-I recognizes the terminus of RNA (47).
Sensing of SCPs by NLRs
NOD-like receptors are cytosolic PRR consisting of 22 and 24 receptors, in humans and mice, respectively (48) and categorized into four subfamilies, NLR-A, NLR-B, NLR-C, and NLR-P, where A, B, C, and P represent acidic trans-activating domain (AD), baculoviral inhibitor of apoptosis repeats (BIR), CARD, and pyrin domain (PYD) respectively. Although these NLRs sense various kinds of PAMPs (7), only SCP-sensing NLRs are in the scope of this review. Nucleotide oligomerization domain (NOD) 1 and 2, belongs to NLR-C family, sense PGN (Table 2) from bacteria (49–51), are comprised of C-terminal LRRs, central oligomerization domain, and N-terminal CARD. NOD1 senses d-gamma-Glu-meso-DAP dipeptide (iE-DAP) (49, 52); however, NOD2 senses MurNAc-l-Ala-d-isoGln (MDP) of bacteria (49, 50). NOD1 and NOD2 receptors sense a large class of bacteria and discussed in detail in several reviews (7, 9). The PYD is containing NLRP1, NLRP3, and HIN domain containing cytosolic DNA sensor absent in melanoma 2 (AIM2) sense various SCPs from bacteria (MDP, RNA, and DNA), virus (RNA and DNA), and fungi (β-glucan and mannans) as an oligomeric complex known as inflammasome (53, 54). Bacterial, viral, and fungal PAMPs can activate different NLR inflammasome complexes and AIM2 inflammasome complex (54–59). NLRP3 inflammasome is the most studied inflammasome senses β 1, 3 d-glucan (Table 2) and regulates innate and adaptive immunity, particularly B-lymphocytes mediated antibodies (Abs) responses (58). NLRP1 inflammasome senses MDP, but the exact mechanism is not well understood (55). AIM2 (also known as PYHIN4) is a member of PYHIN (pyrin and HIN-200 family) and plays an important role in sensing of microbial DNA (Table 2) is comprised of N-terminal PYD and C-terminal HIN domain. The HIN domain sense dsDNA and PYD interacts with PYD of apoptosis-associated speck-like protein containing a CARD (ASC). The complex of DNA, AIM2, and ASC activates inactive caspase-1 and forms an inflammasome complex for production of interleukin (IL)-1 family cytokines (56, 57, 59).
Sensing of SCPs by CLR
C-type lectin-like receptors sense SCPs from various kinds of pathogens, such as parasites (60), fungi (8, 61), bacteria, and viruses (62, 63) consist of more than 1000 members (60, 64). The number of extracellular carbohydrate recognition domain (CRD) and their cellular localization classifies CLRs to type-I transmembrane, type-II transmembrane, and soluble CLRs (65); where type-I CLRs contain multiple CRD and type-II CLRs contains single CRD (66). Furthermore, based on domain organization and sequence homology, CLRs are grouped into 17 clusters (67). Among these clusters DCs associate C-type lectin (Dectin)-1 and 2 clusters are the most widely explored (8, 68, 69). Dectin-1 senses fungal β-glucan (Table 1) (8, 70–72), which is mediated through hydrophobic entrapment of β-glucan at a groove between Trp221 and His223 residue of Dectin-1 (71). Structurally Dectin-1 is comprised of a CRD that connected to transmembrane domain through a stalk region, which contains a small intracellular tail, where immunoreceptor tyrosine-based activation motif (ITAM)-like (YxxL) and tri-acidic (DED) motifs exist. “Y” represents tyrosine, “x” can be any amino acid, and “D” and “E” are glutamic acid and aspartic acid, respectively (8). Dectin-2 senses α-mannans (Table 1); mice lacking Dectin-2, challenged with α-mannans derived from Candida albicans do not induce cytokines suggesting that Dectin-2 plays a pivotal role in sensing α-mannans (73). Structurally, Dectin-2 is comprised of one CTLD domain, and a short transmembrane domain that is associated with ITAM containing Fc receptor γ-chain (FcRγ) receptor. Mincle and macrophage C-type lectin (MCL; also known as Clec4d) receptor (Table 1) sense TDM (74–76). Similar to Dectin-2, Mincle also contain one CTLD, a short transmembrane domain, and an attached ITAM containing FcRγ receptor; however, while MCL is a gene duplication product of Mincle and structurally belongs to the same CLR subgroup (76). Recently, it has been reported that opportunistic skin fungal pathogen Malassezia is sensed by Mincle and Dectin-2, where Mincle senses glucosyl-glycolipid and mannosyl-glycolipid and Dectin-2 senses O-linked mannobiose-rich glycoprotein; however, Mincle and Dectin-2 deficient mice were unresponsive to these ligands suggesting that Mincle and Dectin-2 sense distinct ligands from fungus Malassezia and induce host immune response (77). DC-SIGN (also known as CD209) a type-II CLR senses various kinds of SCPs (Table 1) such as LPS (78), G-glycoprotein from respiratory syncytial virus (RSV), E1 and E2 glycoproteins from hepatitis C virus (HCV), and glycoprotein-140 (gp-140) from HIV (79, 80). Furthermore administration of LPS in mice promotes differentiation of monocytes to DC-SIGN receptor rich DCs (81), suggesting that DC-SIGN plays a crucial role in recognition of microbial pathogens. Liver/lymph-node specific ICAM-3 grabbing non-integrin (L-SIGN; also known as CD209L) senses E2 glycoprotein from HCV (82). Sensing of glycoproteins by DC-SIGN and L-SIGN mediates viral attachment and leads to internalization of the virus into non-lysosomal compartments in host cells (80, 82, 83). MBL, the soluble CLR, senses microbial pathogens through their sugar moieties in a non-specific manner and promotes complement fixation, for which MBL forms a multiprotein complex known as collectins (84).
PRR Signaling
After sensing of SCPs, PRRs recruit different adaptors for effector responses, which are mainly elicited through secretion of innate cytokines such as type I IFNs and inflammatory cytokines.
TLR Signaling
Sensing of SCPs by TLR induces conformational changes and oligomerization of receptors, which leads to the recruitment of various adaptor(s) molecules to the TIR domain, which include myeloid differentiation primary response gene 88 (MyD88), TIR domain containing adaptor-inducing IFN-β (TRIF), TIR-containing adaptor protein (TIRAP), and TRIF-related adaptor molecule (TRAM). The signaling pathways activated by different TLRs are MyD88-dependent and MyD88-independent (Figure 1) and described in several reviews in detail (7, 10, 11, 85). TLR signaling is regulated at three different levels, which include recruitment of adaptors, stability of signaling intermediates, and transcription regulation (86). A recent study shows that vitamin-D also plays a crucial role in regulation of TLR signaling (87).
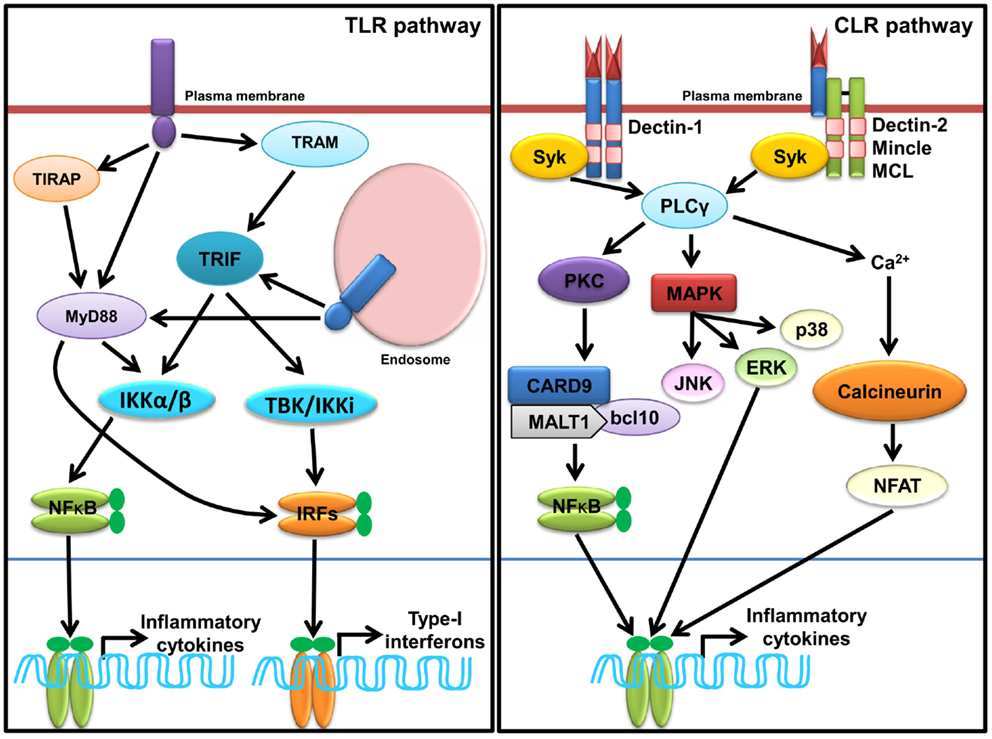
Figure 1. Sensing of SCPs through membrane associated PRRs, TLRs, and CLRs. In macrophages and dendritic cells plasma membrane localized TLRs (TLR1, TLR2, TLR4, and TLR6) and endosomal localized TLRs (TLR3, TLR7, TLR8, and TLR9) and TLR13 sense various SCPs as homo or heterodimer (shown in Table 1). TLR recruits MyD88, TRIFs, TIRAP, and TRAM adaptors for induction of inflammatory cytokines and type-I IFNs through activation of NF-κB and IRFs (IRF3 and IRF7) via IKKα/β and TBK1/IKK1, respectively. Dectin-1, Dectin-2, Mincle, and MCL are representative CLRs of Dectin-1 and Dectin-2 clusters. Sensing of various SCPs by Dectin-1 and Dectin-2 (shown in Table 1) recruits, Syk adaptor which further recruits PLCγ and activate cascade of signaling for induction of inflammatory cytokines via NF-κB and MAP kinases.
RLR Signaling
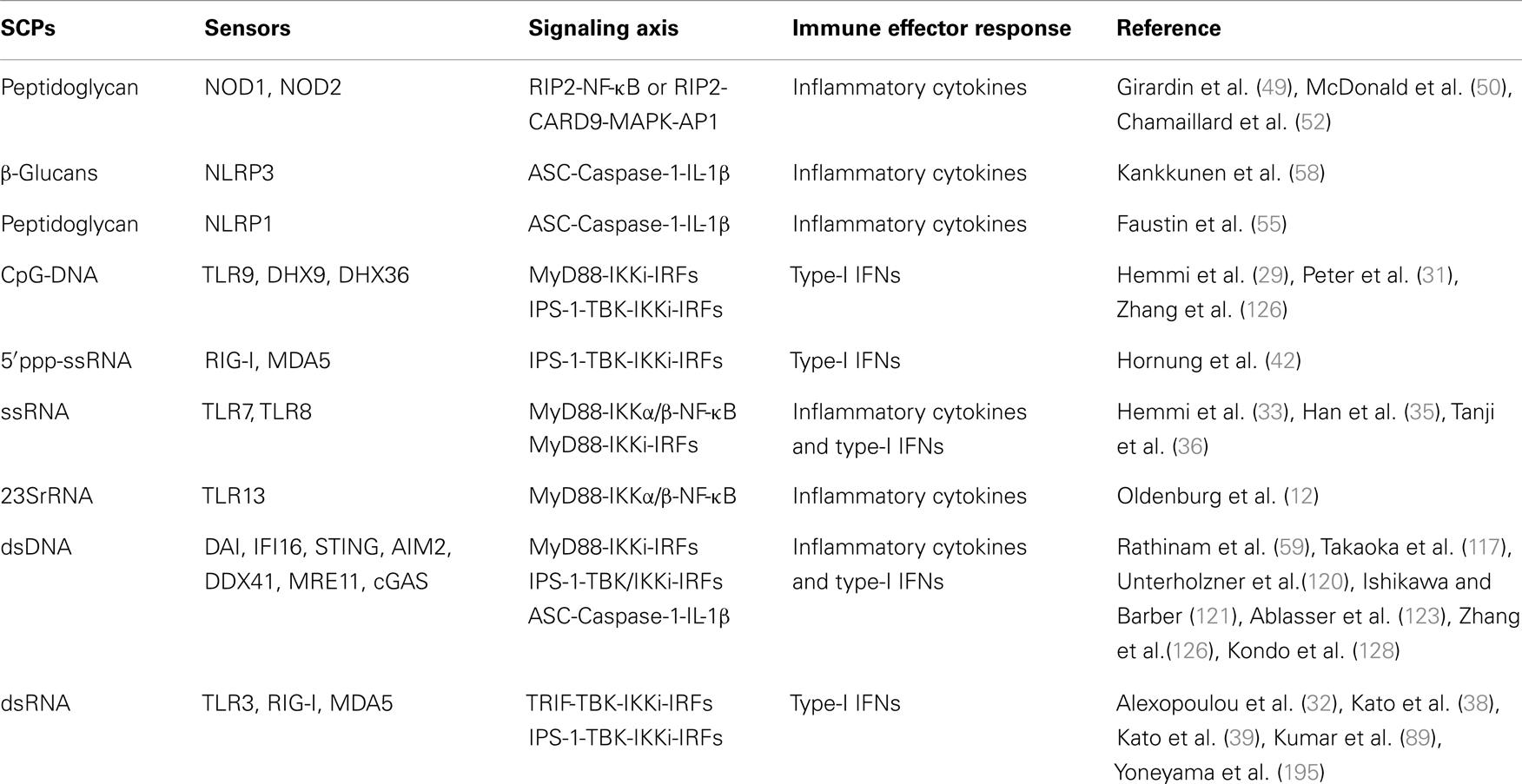
Ligation of RNA to RIG-I and MDA5 promotes receptor oligomerization and recruitment of an adaptor IPS-1/MAVS/VISA/Cardiff (47, 88–90). Encounter of viral RNA with these sensors leads to the activation of sensors though accessibility of CARD for recruitment of CARD containing IPS-1 through K63 linked ubiquitination. The downstream signaling and its regulation are discussed in several reviews (37, 91–94). Recently, in vitro studies show that recognition of RNA by MDA5 requires CTD domains and forms head to tail oligomer strand around the RNA, leaving two CARD exposed outside, which oligomerize and recruit IPS-1, forming a string-like structure around the oligomerized MDA5 assembly (47). IPS-1 activates IKK-related kinase TBK1 and IKKi/IKKɛ, which activates transcription factor IRFs (mainly IRF1, IRF3, and IRF7), and subsequent transcription of type-I IFNs (Figure 2). It has been shown that IPS-1 is localized in mitochondria and peroxisomes and signals through IRF3 and IRF1, respectively (95). Furthermore, IPS-1 also induces production of inflammatory cytokines and requires tumor necrosis factor receptor type-1 associated death domain protein (TRADD), Fas associated protein with death domain (FADD), caspase-8, and caspase-10 (96, 97). RLR signaling is tightly regulated at multiple levels that include regulation of RNA sensing by LGP2 (40, 41), post-translational modification (PTM) of IPS-1, RIG-I, and MDA5 (98, 99), disruption of RLRs (RIG-I/MDA5)-IPS-1 signaling axis (100), destabilization of IPS-1, modulation of mitochondrial dynamics, and IPS-1 activation and regulation of downstream signaling molecules (99).
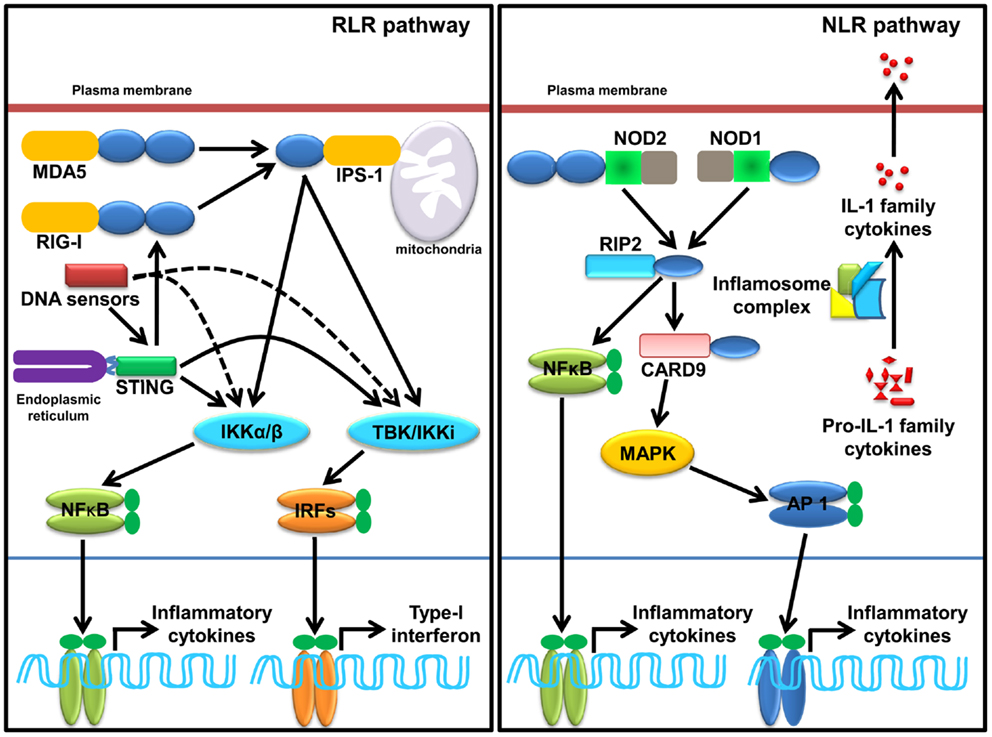
Figure 2. Sensing of SCPs through cytosolic PRRs, RLRs, NLRs, and cytosolic DNA sensors. RLRs family member, RIG-I and MDA5 sense viral dsRNA (shown in Table 2), recruits an adaptor IPS-1 for induction of type-I IFNs and inflammatory cytokines. STING, a molecule plays a pivotal role in sensing of RNA. Several DNA sensors induce inflammatory cytokine and type-I IFNs in STING dependent and independent manner. NLRs family member, NOD1, NOD2, and inflammasome (mainly NLRP1 and NLRP3 and AIM2) induces inflammatory cytokine. NOD1 and NOD2 sense various SCPs (shown in Table 2) recruit an adaptor RIP2 and CARD9 for induction of inflammatory cytokines. Activation of inflammasome complex after stimulation with appropriate SCPs (shown in Table 2) process an inactive, proIL-1 family cytokine to active IL-1 family cytokine.
NLR Signaling
Ligand binding leads to self-oligomerization of NOD1 and NOD2 and recruits an adaptor Receptor-interacting serine/threonine-protein kinase 2 (RIP2). RIP2 is a serine threonine kinase and activates transforming growth factor activated kinase-1 (TAK-1) that subsequently activates NF-κB and mitogen-activated protein kinases (MAPKs) such as p38 and c-Jun N-terminal kinases (JNKs) (Figure 2). MAPKs is activated through an adaptor CARD9 (7, 101–104). NLR such as NLRP1 and NLRP3 induce IL-1 family cytokines, where ASC functions as a signaling adaptor and induces caspase-1-dependent maturation of IL-1 family cytokines and plays a crucial role in inflammatory responses and pyroptosis (103, 105, 106). Among all known inflammasome, NLRP3 is the most investigated inflammasome and it can be activated by multiple factors such as components of pathogenic microbes, tissue damage, autophagy, type-I IFNs, T cells, metabolic dysregulation, miRNA, and so on (107–110). Furthermore, viruses also enhance the negative regulatory mechanism of hosts to promote survival within the host (111). Activation of NLRP1 induces pyroptosis in hematopoietic cells and inhibition of NLRP1 may play a protective role in individuals having blood disorders such as anemia and leukopenia and suffering from bacterial septic shock (112). Distinct from NLRP1 and NLRP3, inflammasome AIM2 activates caspase-1 and NF-κB in an ASC dependent manner (56).
CLR Signaling
Sensing of SCPs by CLRs activates multiple signaling cascades through their own ITAMs or interacting with ITAM-motif containing adaptor proteins such as FcRγ. Signaling cascades lead to activation of NF-κB through a spleen tyrosine kinase (Syk) and CARD9 dependent pathway(s). Binding of ligands to Dectin-1 promotes phosphorylation of YxxL motifs through Syk that promotes recruitment of adaptor CARD9, which exists as a complex B cell lymphoma-10 (Bcl-10) and mucosa-associated lymphoid tissue lymphoma gene-1 (MALT-1), and subsequently activates IKK kinase complex (Figure 2). The activated IKK complex activates canonical NF-κB subunits p65 and c-Rel to induce production of multiple cytokines such as IL-6, IL-10, IL-23, IL-1β, and TNF-α through a subset of T cells such as Th1, Th2, Th17, and THF (113–115). In DCs, assembling of CARD9/Bcl-10/MALT-1 complex is dependent on paclitaxel-lipopolysaccharide complex (PLC)-γ2 receptor (8, 116). Alternatively, non-canonical activation of NF-κB takes place through Ras-RAF1 mediated phosphorylation of p65 at serine 279. Binding of activated p65 with histone acetyl transferase CBP/p300 promotes cytokines synthesis; however, its binding with b-Rel inhibits the NF-κB activity (66, 113, 115). For downstream signaling, Dectin-2 and Mincle interact with signaling adaptor FcRγ through positively charged amino acids that promote phosphorylation of ITAMs of FcRγ and subsequent recruitment of Syk. Furthermore, Dectin-2 activates NF-κB (p65–p50) and Mincle activates NF-κB in a CARD9 dependent manner (Figure 2) (115).
SCPs and Cytosolic DNA Sensors
DNA-dependent activator of IFN-regulatory factors (DAI) is the first identified cytosolic DNA sensor that recognizes dsDNA and induces production of type-I IFNs (117); however, DAI-knockout cells display normal responses to poly(dA:dT), a synthetic analog of B-DNA, suggesting the existence of multiple sensors for cytosolic dsDNA (118). Nucleic acid binding protein known as leucine rich repeat Fli-I interacting protein (LRRFIPI) is a cytosolic sensor of AT rich B-DNA and GC rich Z-DNA and induces production of IFN-β, and the sensing is dependent on β-Catenin (119). Absent in melanoma 2 (AIM2), a member of the hematopoietic IFN-inducible nuclear protein HIN-200 family, has been recognized as a cytosolic DNA sensor (57) and plays a crucial role against cytosolic bacteria and DNA viruses (59). IFN-γ-inducible protein-16 (IFI16; also known as p204) induces type-I IFNs in stimulator of IFN genes (STING) dependent manner (120). STING is an endoplasmic reticulum (ER) localized dsDNA sensor and induces activation of transcription factor NF-κB and IRF3 for downstream signaling (Figure 2) (121). Sensing of dsDNA by STING is positively regulated by IFN-inducible TRIM-56, which leads to production of IFN-β (122). Recently, a nucleotidyltransferase family member known as cyclic guanosine monophosphate-adenosine monophosphate (cyclic-GMP-AMP, or cGAMP) synthase was identified as a cytosolic DNA sensor that senses DNA in a STING dependent manner. During this process cGAMP synthase (cGAS) binds to microbial DNA and synthesizes secondary messenger cGAMP that interacts with STING and activates IRF3, which subsequently leads to transcription of type-I IFNs. The crystal structure of cGAS alone and bound with DNA, ATP, and GTP demonstrates that cGAS is structurally similar to dsRNA sensor 2′–5′ oligoadenylate synthase (OAS1), but it contains a unique zinc-thumb structure through which it recognizes microbial B-DNA (123–125). DEAH (Asp-Glu-Ala-His) box containing polypeptide 9 and 36 (also known as DHX9 and DHX36) senses hypomethylated CpG DNA in Myd88 dependent manner; however, DDX41, a newly identified PRR, senses dsDNA in STING dependent manner and induces IFN-β (126). DDX41 is negatively regulated through E3 ubiquitin ligase TRIM21, where TRIM21 ubiquitinates DDX41 at K9, K48, and K115 positions, which leads to its degradation (127). Meiotic recombination 11 homolog A (MRE11) in association with DNA repair protein 50 (RAD50) recognizes various kinds of dsDNA in cytosol and induces production of type-I IFNs in a STING-IRF3 dependent manner (128). Although stimulation of granulocyte-monocyte-colony stimulating factor (GM-CSF) induces bone marrow-derived DCs (GM-DCs) with herpes simplex virus (HSV-1) and Listeria monocytogenes enhances type-I IFNs, siRNA mediated silencing of MRE11 does not abrogate the response, suggesting that further studies are required to understand the role of MRE11 in innate immunity (128). It can be concluded that sensing of DNA and RNA SCPs by cytosolic PRRs is more complicated than thought. Although several sensors have been discovered, the general consensus is that there may be some more cytosolic nucleic acid sensors that sense the respective DNA.
SCPs Mediated Immune Dysregulation/Disease
Sensing of PAMPs by PRRs initiates innate immune responses through secretion of inflammatory cytokines, and subsequent B and T lymphocyte mediated humoral and cellular immune responses are essential for eradication of invading pathogens. However, excessive or hyper activation of innate and adaptive immune responses could be detrimental to the host (129, 130). SCPs are known to induce lethal immune response in the host, such as septic shock, which is characterized by a storm of inflammatory cytokines in the host after infection with gram bacteria. The LPS (also known as endotoxin) induces activation of myeloid and non-myeloid lineage of cells (131) and leads to cytokine mediated immune shock that is generally known as septic shock; the mechanism of LPS mediated immune stimulation is largely studied in laboratory animals, which is a reflective process for humans. Administration of LPS in mice induces inflammatory lesions through excessive secretion of TNF-α, IL-1β from monocytes, and macrophages that ultimately culminates in septic shock mediated death (132). LPS induces lethal effects due to TLR4-CD14-MD12 receptor complex mediated recruitment of multiple signaling adaptors (MyD88, TRAF6, TRAM, and TRIF). Septic shock is associated with progression of kidney disease with incident microalbuminuria and induces degeneration of neurons in individuals with type-I diabetes, which ultimately leads to a condition known as diabetic neuropathy (133). LPS is also shown to aggravate polyclonal B cell activation, which is a similar kind of condition that occurs in systemic lupus erythematosus (SLE). The SLE kind of effect from LPS is exerted through increased level of IgG and IgM serotype and concurrent decrease of IgA specific isotopes (134). Receptors for advanced glycation (RAGE) are involved in negative regulation of LPS induced septic shock that is mediated through reduction in LPS induced inflammatory cytokines (135). The other glycolipid constituent LTA from bacteria induces inflammatory responses that are similar to LPS and plays a critical role in bacteria pathogenesis (130).
TLR9 ligand, CpG-ODNs induces liver injury in d-Galactosamine sensitized mice that is mediated through excessive induction of TNF-α leading to mitochondria-mediated apoptosis of hepatic cells and ultimately culminates in death of the animal (129). The effect of CpG-ODNs is due to the organization of constitutive subunits, that is differential for different kind of CpG-ODNs, and some time it can be fatal to the host. Likewise, DC-SIGN receptor sensed gp120 from HIV virus inhibits secretion of inflammatory cytokine IL-6 and phosphorylation and signal transducer and activator of transcription-3 (STAT-3) that ultimately leads down regulation of host antiviral innate immune response and that further assist for viral internalization in host cell into non-endosomal compartments (136). Similarly, glycoprotein E1 and E2 from HCV modulate host cell entry factors and escape virus from neutralizing Abs. Hepatitis E virus (HEV) originated whole virion particles down regulate RIG-I mediated type-I IFNs production and employ IPS-1 in restricting antiviral inflammatory response (137). The HN series of influenza virus H1N1 can escape immune response through antigenic drift that is enabled by hemagglutinin (HA) head (138). It can be suggested that pathogens utilizes different strategies for down regulating host defense mechanism through targeting major innate immune pathways (129, 136–138). Herewith, these are the few examples to comprehend the role of SCPs in immune dysregulation and development of disease; however, proteins PAMPs are prime components involved in immune escape mechanism, which is beyond the scope of this review.
Role of SCPs in Vaccine Biology
The term vaccine was coined by Edward Jenner for cowpox, used for protection from small pox in humans. Formulation of vaccine can be synthetic or biological, and exerts its effect through activation of both arms of the immune system, namely, the innate and adaptive immune system (Figure 3) (139). Therapeutics agents or antigen alone do not mount appropriate immune responses for development of immunity against infection; therefore, an additional immune boosting agent known as adjuvant is required, which provides enough non-specific immune response to elicit appropriate immune response against antigenic determinants of the pathogen and protects the host from infection. Microbial PAMPs are potential candidates as vaccine adjuvants, which mainly activate innate immune cells such as macrophages and DCs. The DCs are pivotal for immunity as they link innate and adaptive immunity (140). How appropriately the immune system will be activated by a particular PAMP depends on its structural diversity and complexity. SCPs such as β-glucan (sensed by Dectin-1 and NLRP3) are made from the same basic unit of sugar but the bonds between monomeric subunits vary, which diversifies the structure. The β-glucan polymer with β 1,6 glycosidic bonds in polymer chain and β 1,3 glycosidic bonds at branching point is shown to be associated with immune stimulatory property (141). Formulation of β-glucan adjuvant with candidosis vaccine consists of α, β mannan-tetanus toxoid complex and enhances protection in mice against pathogenic C. albicans (142). Furthermore, β-glucan also functions as a vaccine candidate for immune clearance of fungal pathogens. Its formulation with immune adjuvant known as MF59 protects mice from vaginal candidosis infection through anti-β 1,3 d-glucan Abs (143). In addition to adjuvant and vaccine for infectious disease, β-glucan is also used for cancer therapeutics for example yeast derived β-glucan adjuvant potentially enhance antitumor effect of MUCIN-1 Abs in mice. Adjuvant and vaccine properties of β-glucan are exerted through induction of innate cytokines such as IL-6, IL-12, TGF-β, and IL-1β, and Th1 and Th2 polarization through NLRP3, MyD88, and Syk (141–143). Aforementioned studies suggest that β-glucan can be used as a potential therapeutic candidate for both infectious and non-infectious disease. Mannans (α-mannan) from fungus are Dectin-2 ligands, impart their immune response through CARD9 dependent signaling, and are involved in activation of Th1 cells (73). C. albicans derived mannans with α 1,6 glycosidic backbone and α 1,2 or α 1,3 glycosidic branching as well β 1,2 linked oligomer in conjugation with bovine serum albumin protects mice from fungal infection. This study suggests that similar to β-glucan, α-mannans also possess vaccine adjuvant property and hence they can be a potential vaccine candidate for protection from fungal infection (144).
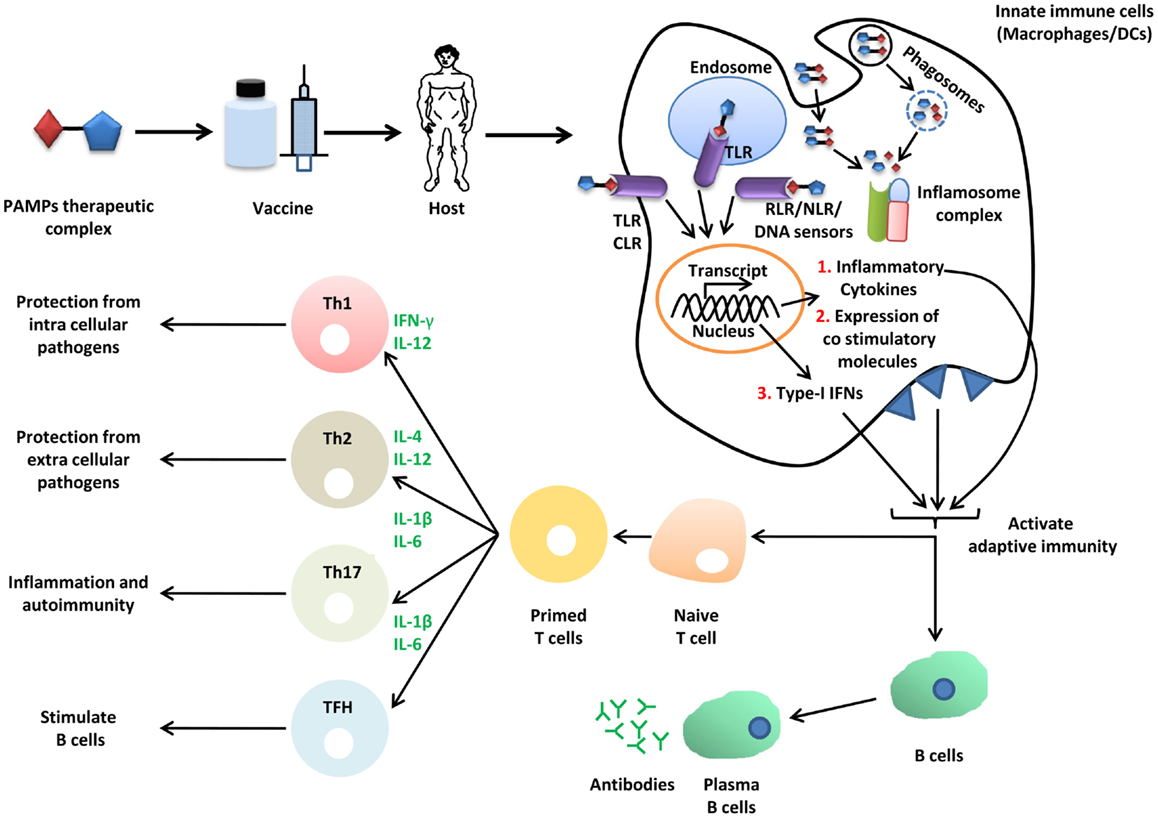
Figure 3. Role of SCPs in vaccine biology. Immunization of host (mice or human) with vaccine or vaccine adjuvant formulation PTC (PAMPs therapeutic complex) activates innate immune pathways (TLRs, CLRs, RLRs, NLRs, and DNA sensors), induce secretion of inflammatory cytokines and type-I IFNs. These cytokines further activate adaptive immune components through B and T lymphocytes. Adaptive immune memory cells protects host from infection.
For decades, LPS was known for its adjuvant property; however, the endotoxic nature renders the use of LPS in immunotherapy (145). The modified derivative of LPS known as monophosphoryl lipid A (MPLA) is less toxic and retains its immunostimulatory activity, and is used as an immunotherapeutic vaccine adjuvant (146). Co-administration of MPLA along with basic fibroblast growth factor (bFGF) enhances IgG and IFN-γ level in the serum, and show antitumor activity in mice. Similarly, formulation of MPLA with IFN-γ induces secretion of IL-12 from DCs and shows antitumor activity in mice (147). The antitumor effects of bFGF growth factor and IFN-γ are exerted through cytokine induced by DCs for activation of cytotoxic T lymphocytes (CTL). In addition, these effects are very cell-specific; for example, antitumor effects of IFN-γ are mediated through CD4+ T cells and selectively eliminate human leukocyte antigen (HLA) positive tumor cells (148). Furthermore, a combination of MPLA and alum is used in humans as vaccine adjuvant and it has been shown to induce CD8+ T cell mediated immune responses. Alum induces long-term CD8+ T cells; however, due to programed death (PD)-1 molecule, differentiation of CD8+ T memory cells to effector cytotoxic T cells is inhibited. It has been shown that in the combination of MPLA and alum, MPLA inhibits PD1 activity through IL-6 that ultimately leads to differentiation of alum primed CD8+ T cells into effector CTLs, suggesting its application for protection from microbial pathogens (149). A polymeric form of LPS known as SP-LPS has immuno-modulatory functions without toxicity. The conjugate of cancer drug paclitaxel and SP-LPS known as PLC shows promising antitumor effects through induction of apoptosis. Additionally, the apoptotic bodies generated during apoptosis further stimulate antigen presenting cells (APCs) and promote more efficient tumor clearance (150). Another synthetic derivative of LPS known as glucopyranosyl lipid (GLA) is shown to function as immune adjuvant. GLA, along with Mycobacterium tuberculosis (MTB) antigen ID83 [combination of factor of virulence (Rv2608, Rv3620) and latency Rv1813)], induces protection against MTB infection in mice. Furthermore, a GLA and MTB Ag ID3 formulation induces Th1 cytokines, suggesting that this combination can be used in humans for protection against MTB infection (151).
Mincle and TLR2 agonist TDM (also known as MTB cord factor) is shown to function as immune adjuvant. Co-administration of TDM with type III pneumococcal polysaccharide (SSS-III) in mice augments Abs response to SSS-III antigen and further protects from pneumonia infection (152). A recent genetic study demonstrated that adjuvant action of TDM activates adaptive immune responses through MCL; however, Mincle is not essential (76). The use of TDM is limited due to its toxic nature; a synthetic analog of TDM known as trehalose-6,6-dibehenate (TDB) does not show cytotoxic effects in mice. The combination of dimethyl dioctadecyl ammonium (DDA) and TDB generates a complex DDA-TDB adjuvant; immunization of mice with DDA-TDB along with MTB fusion protein (Ag85B-ESAT) shows MTB specific Th1 adaptive immune response through elevation of IFN-γ secreting CD4+T cells and production of isotype Abs IgG2b against the antigen (153–155), suggesting that DDA-TDB can serve as potential vaccine adjuvant against MTB. The size of DDA-TDB vesicle governs antigenic release time and overall it governs cell mediated adaptive immunity; however, the production of Abs is independent of vesicle size (156). Furthermore, TDB requires MyD88 for induction of IL-1β secretion and also activates adaptive immune response through Th17 cells (156). An in vitro study using mice bone marrow-derived DCs shows that TDB activates NLRP3 inflammasome in a caspase-1-dependent manner for production of IL-1β (157).
Lipoarabinomannan (sensed by TLR2 and Mincle) is shown to elicit protective immune response against Mycobacterium pathogenesis. Stimulation of cattle peripheral blood mononuclear cells (PBMCs) with LAM isolated from M. avium along with Freund’s incomplete adjuvant (FIA) induces secretion of IFN-γ. The immune stimulatory function of LAM is exerted through a MyD88 directed CARD9/NF-κB pathway and NLRP3 inflammasome activation, suggesting that M. avium LAM could be used as a potential vaccine candidate against bovine tuberculosis (158). PGN of bacteria is not a potent immune adjuvant; however, peptidoglycan monomer (PGM) induces appropriate immune responses without any pyrogenic and toxic effects (159). Mice sensitized with PGM adjuvant and ovalbumin antigen (OVN) formulation enhance secretion of IFN-γ and IL-4 cytokines and specific anti-OVN Abs. Similarly, PGM derived from Staphylococcus known as A170PG protect mice from Staphylococcus infection (160). The adjuvant effects of PGM are initiated through TLR2 and culminate in activation of Th1 and Th2 mediated adaptive immune responses. Most of the studies related to PGM adjuvant are performed using OVN antigen (159–161). Therefore further studies are required with a wide range of antigens from pathogens to establish the adjuvant properties of PGM.
Peptidoglycan teichoic acid (PG-TA) and LTA from Bacillus subtilis are non-pyrogenic at low concentration and show adjuvant activity by enhancing the number of granulocyte-monocyte-colony forming cells in bone marrow in mice. The immune stimulatory property of these SCPs is possibly mediated through GM-CSF (162), but the application of PG-TA and LTA as an immune adjuvant is limited due to poor immunostimulatory functions. The synthetic adjuvant OK-PSA prepared from the cell wall of non-virulent Streptococcus pyogenes strain OK-432 is structurally similar to LTA and shows potent antitumor effect in mice. Antitumor effects of OK-PSA are mediated through induction of IFN-γ, TNF-α, IL-2, IL-12, and IL-18 cytokines from Th1 cells (163), suggesting its utility as a potent immune adjuvant. Similarly, GPI anchored proteins (sensed by TLR2/TLR1) are also shown to possess adjuvant activity (164). Immunization of mice with enzymatically cleaved GPI anchored proteins along with membrane molecules from Schistosoma mansoni potentially protects mice from worm infection. The adjuvant effect of GPI anchored proteins induces Th1/Th2 responses via IFN-γ and TNF-α production (164), suggesting the potential application of GPI anchored proteins as an adjuvant for protection from parasite infection.
The unmethylated CpG rich DNA from bacteria and virus is a well-established deoxy pentose sugar containing SCPs. The cellular responses to CpG are mediated through its sensing by the TLR9 receptor (29). The synthetic analogs of CpG DNA known as CpG oligodeoxynucleotides (CpG-ODNs) are functionally similar to bacterial CpG DNA and show potent immune stimulatory property through induction of innate cytokines (IL-6 and type-I IFNs), improve antigen presentation by DCs, and enhance humoral and cellular immune responses (165–168). The CpG-ODNs are structurally different from bacterial DNA; backbone of bacterial DNA contains phosphodiester while CpG-ODN has a phosphorothioated (PS) backbone, which protects them from DNAse in cells. CpG-ODN contains poly G tail at 3′ and 5′ ends, due to which CpG-ODNs form high molecular weight aggregates, which is responsible for enhanced cellular uptake and potent immune responses (169, 170). Based on sequence, CpG-ODNs are classified as type A, B, and C (171, 172); however, another report shows five types of CpG-ODNs (173). Type A CpG-ODNs contain phosphodiester backbone, a central CpG dinucleotides palindrome, and a PS 3′ poly G string, activates natural killer (NK) cells, and induces high IFN-α production from pDCs but weakly induces NF-κB dependent inflammatory cytokines. Type B CpG-ODNs contain full PS backbone with one or more CpG dinucleotides and are weak inducers of cytokine IFN-α. Type C CpG-ODNs (with complete PS backbone and CpG palindrome motif) show combined property of both A and B class CpG-ODNs and efficiently stimulate B and NK cells and also induce IFN-α from pDCs. CpG-ODNs from all classes have been used as immune adjuvants. Immunization of mice with hepatitis B virus surface antigen (HBsAg) along with type B CpG-ODN (ODN 1826) enhanced HBsAg-specific IgG2a Abs (174). Similarly, CpG-ODN 1758 enhances production of tumor surface associated Id-38C13 antigen-specific IgG2a Abs in mice (175). CpG-ODN 7909 enhances hepatitis B virus (HBV) induced PBMCs proliferation (176). A study shows that CpG-ODN stimulated secretion of type-I IFNs from DCs is a dose and time dependent mechanism (177). In addition to these applications, CpG-ODNs have a wide range of application in therapeutics and discussed in several reviews (167, 178, 179).
HN series of influenza virus such as H1N1 and H5N1 lyse PBMCs in IFN-α dependent manner (180). Priming of the host immune system prior to infection can potentially protect the host from pathogenesis from influenza. Immunization of mice with β-propiolactone inactivated (this inactivation does not affect ssRNA genome of virus) H5N1 and H1N1 virus known as whole inactivated virus (WIV) protects mice from viral infection. The immune protection is mediated through TLR7 directed activation Th1 and Th2 response, as mice lacking TLR7 are susceptible to H5N1 infection (181). Furthermore, in vitro stimulation of human respiratory bronchial epithelial cells with GNR (gold nano rods) complexed 5′ppp-ssRNA activates RIG-I and MDA5 mediated antiviral immune memory in these cells, which further protects from infection with pandemic influenza H1N1 and common influenza A/Solomon Island/03/06 (182), suggesting the potential application of viral 5′ppp-ssRNA and WIV as vaccine candidates for protection from influenza pandemics. Poly(I:C), the synthetic analog of viral dsRNA (sensed by TLR3, RIG-I, and MDA5), shows potential antiviral immune response (183). Co-administration of poly-l-Lysine and carboxymethylcellulose along with poly(I:C) and with Plasmodium falciparum circumsporozoite protein (CSP) antigen increase the frequency of activation of CD8+ Th1 cells and anti-CSP-Abs production (184). Furthermore, immunization of mice with CpG-ODNs or poly(I:C) and alum adjuvant with HBV vaccine rAdSS1 [adenoviral vector encoding HBV S (1–223aa) and pre S (1–47aa) fusion gene] shows immune protection through CD4+ T cells and CD8+ T cells via IFN-γ and IL-12 production and inducing cytotoxic effects, suggesting their potential application in vaccine biology for protection from HBV (185, 186). Since it is known that poly(I:C) and CpG-ODNs are potent immune adjuvant, their use in combination has been shown as potential immune adjuvant functions. Mice immunized with poly(I:C) or CpG-ODN 2006 with low dose of salmonid alphavirus (SAV) antigen preparations show potent induction of type-I IFNs and further protection from SAV (187). Recently in mice and ferrets, it was shown that oral administration of an enteric capsule containing adenovirus vector expressing HA from H5N1 virus and a dsRNA adjuvant enhances antiviral immunity through anti-HA Abs; however, there is no Abs against adjuvant. Furthermore, this combination ensures the stability and slow release of viral HA antigen and dsRNA into the gastrointestinal tract, suggesting the potential application of enteric capsules as a vaccine candidate against avian influenza; however, further research work is required to understand the mechanism for immune activation (188).
Carbohydrate based targeting of CLRs (189) in APCs is an interesting strategy for therapeutic application of synthetic glucans and glycopolymer. Virus utilizes DC-SIGN for immune escape can be targeting through synthetic glyconanoparticles such as ManLAM (190–192). DC-SIGN mediated internalized glucans further activate CD4+ and CD8+ cells of adaptive immune system. Furthermore the strategy can be utilized for non-pathogenic disease like cancer (191) These synthetic glycopolymers can mimic viral glycoprotein and can be utilized efficiently as antiviral therapeutic agents (193). In summary, there are potential roads of adjuvant based vaccine development using formulations of both intact pathogen and pathogen derived SCPs or their synthetic derivatives.
Future Perspectives
The complexity of the transcription networks that operate during SCP sensing and subsequent immune response is a major spotlight of present research. Although there is enormous progress in the field of innate immune sensing of pathogenic SCPs, there is still a lot to understand about the evolving dynamicity of protein–sugar complex (glycoproteins). Glycoproteins are the most dynamic PAMPs that pathogens utilize to escape from immune recognition and therapeutic targets. Synthetic drugs or Abs targeting protein epitopes can be masked by pathogens through protein PTM; for example, the HN series of influenza virus has evolved immune escape mechanism through the glycosylation in HA head, and during recurrence these viruses show drug resistance. Other pathogens also utilize such strategies against therapeutic targets; for example, evolution of drug resistant MTB. Therefore, it is all about the glycobiome diversity and dynamicity that pathogens explore as a survival mechanism. Though sugars are the simplest biomolecules, the bonding flexibility with the same sugar, other sugar, lipids, and proteins generates very complex structures. Understanding the sugar moieties dynamicity in pathogens is very crucial for identification of therapeutic targets. It is also necessary to understand what carbohydrate metabolic pathways are specific to pathogen survival and are not present in the host. It is also interesting to implement bio-informatics prediction method to identify putative PTM sites in structural and non-structural proteins of viruses. Pathogens target particular cells types in the host, so understanding the epigenetic changes that are modulated by pathogens in infected cells but are not occurring in uninfected cells can provide new insights in designing potential therapeutic targets. Mutation incorporated in viral genomes, genetic polymorphism of the host, and dynamicity of carbohydrate metabolism in both the host and pathogens generate a condition that decides latent or virulent stage of pathogens; for example, a third of the world population is infected with MTB, but virulence cannot be seen in all individuals. Therefore, it is very important to consider multiple factors while designing a therapeutic agent, which mainly includes comparative glycobiome analysis of the host and the pathogen and analysis of genetic makeup of the host.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work is supported by Himanshu Kumar research grants number SR/S2/RJN-55/2009 and BT/PR6009/GBD/27/382/2012 from Department of Science and technology (DST) and Rapid Grant for Young Investigators (RGYI), Department of Biotechnology (DBT), Government of India. Himanshu Kumar and D. Vijaya Raghava Prasad is supported by Ramanujan fellowship funded by DST, Government of India.
Abbreviations
Abs, antibodies; AG, arabinogalactan; AIM2, absent in melanoma2; APCs, antigen presenting cells; ASC, apoptosis-associated speck-like protein containing a CARD; Bcl-10, B cell lymphoma-10; bFGF, basic fibroblast growth factor; BIR, baculoviral inhibitor of apoptosis repeats; CARD, caspase activation and recruitment domain; cGAMP, cyclic guanosine monophosphate-adenosine monophosphate; cGAS, cyclic-GMP-AMP synthase; CLRs, c-type lectin receptors; CRD, carbohydrate recognition domain; CSP, circumsporozoite protein; CTD, c-terminal domain; CTL, cytotoxic T lymphocytes; DAI, DNA-dependent activator of IFN-regulatory factors; DC-SIGN, DCs specific intracellular adhesion molecule (ICAM)-3 grabbing non-integrin; DDA, dimethyl dioctadecyl ammonium; Dectin, DCs associate C-type lectin; ECD, extra cellular domain; FADD, fas associated protein with death domain; FcRγ, Fc receptor γ-chain; FIA, Freund’s incomplete adjuvant; GLA, glucopyranosyl lipid; GM-CSF, granulocyte macrophage colony stimulating factor; GNR, gold nano rods; GPI, glycophosphatidylinositol; HA, hemagglutinin; HBsAg, hepatitis B virus (HBV) surface antigen; HCV, hepatitis C virus; HEV, hepatitis E virus; HIV, human immunodeficiency virus; HLA, human leukocyte antigen; HSV, herpes simplex virus; iE-DAP, d-gamma-Glu-meso-DAP dipeptide; IFI16, IFN-γ-inducible protein-16; IRAKs, interleukin-1 receptor-associated kinases; IRFs, interferon (IFN) regulatory factors; ISGs, IFN stimulatory genes; ISRE, IFN stimulatory response elements; ITAMs; immunoreceptor tyrosine-based activation motifs; JNKs, c-Jun N-terminal kinases; LAM, lipoarabinomannan; LGP2, laboratory of genetics and physiology 2; LPS, lipopolysaccharide; LRRFIPI, leucine rich repeat (LRR) Fli-I interacting protein; L-SIGN, liver/lymph-node specific ICAM-3grabbing non-integrin; LTA, lipoteichoic acid; MALT-1, mucosa-associated lymphoid tissue lymphoma gene-1; MAPKs, mitogen-activated protein kinases; MCL, macrophage C-type lectin; MDA5, melanoma-differentiation-associated gene 5; MD-2, myeloid differential factor 2; MDP, MurNAc-l-Ala-d-isoGln; MRE 11, meiotic recombination 11 homolog A; MTB, Mycobacterium tuberculosis; MyD88, myeloid differentiation primary response gene 88; NF-κB, nuclear factor kappa B; NLRPs, NLR family pyrin domain containing proteins; NLRs, nucleotide oligomerization domain (NOD) like receptors; ODNs, oligodeoxynucleotides; OAS1, oligoadenylate synthase 1; OVN, ovalbumin antigen; PAMPs, pathogen associated molecular patterns; PBMCs, peripheral blood mononuclear cells; pDCs, plasmacytoid dendritic cells (DCs); PD, programed death; PGN, peptidoglycan; PG-TA, peptidoglycan teichoic acid; PLC, paclitaxol lipopolysaccharide complex; PRRs, pattern recognition receptors; PS, phosphorothioated; PYD, pyrin domain; RAD50, DNA repair protein 50; RAGE, receptors for advanced glycation; RD, repressor domain; RIP2, receptor-interacting serine/threonine-protein kinase 2; RSV, respiratory syncytial virus; SAV, salmonid alpha virus; SCPs, sugar complex PAMPs; SLE, systemic lupus erythematosus; STING, stimulator of IFN genes; Syk, spleen tyrosine kinase; SIII, type III pneumococcal polysaccharide; TA, teichoic acid; TAK-1, transforming growth factor activated kinase-1; TBK1, TANK-binding kinase-1; TDB, trehalose-6, 6-dibehenate; TDM, trehalose dimycolate; TIRAP, TIR-containing adaptor protein; TIR, toll/interleukin receptor; TLRs, toll-like receptors; TNF, tumor necrosis factor; TRADD, tumor necrosis factor receptor type-1associated death domain protein; TRAF6, TNF receptor-associated factor 6; TRAM, TRIF-related adaptor molecule; TRIF, TIR domain containing adaptor-inducing IFN-β; TRIM, tripartite motif-containing protein; WIV, whole inactivated virus.
References
1. Lloyd DH, Viac J, Werling D, Reme CA, Gatto H. Role of sugars in surface microbe-host interactions and immune reaction modulation. Vet Dermatol (2007) 18:197–204. doi:10.1111/j.1365-3164.2007.00594.x
2. Kolarich D, Lepenies B, Seeberger PH. Glycomics, glycoproteomics and the immune system. Curr Opin Chem Biol (2012) 16:214–20. doi:10.1016/j.cbpa.2011.12.006
3. Rabinovich GA, Van Kooyk Y, Cobb BA. Glycobiology of immune responses. Ann N Y Acad Sci (2012) 1253:1–15. doi:10.1111/j.1749-6632.2012.06492.x
4. Franchi L, Warner N, Viani K, Nunez G. Function of Nod-like receptors in microbial recognition and host defense. Immunol Rev (2009) 227:106–28. doi:10.1111/j.1600-065X.2008.00734.x
5. Barber GN. Cytoplasmic DNA innate immune pathways. Immunol Rev (2011) 243:99–108. doi:10.1111/j.1600-065X.2011.01051.x
6. Kato H, Takahasi K, Fujita T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol Rev (2011) 243:91–8. doi:10.1111/j.1600-065X.2011.01052.x
7. Kumar S, Ingle H, Prasad DV, Kumar H. Recognition of bacterial infection by innate immune sensors. Crit Rev Microbiol (2013) 39:229–46. doi:10.3109/1040841X.2012.706249
8. Plato A, Willment JA, Brown GD. C-type lectin-like receptors of the dectin-1 cluster: ligands and signaling pathways. Int Rev Immunol (2013) 32:134–56. doi:10.3109/08830185.2013.777065
9. Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol (2010) 11:373–84. doi:10.1038/ni.1863
10. Lim KH, Staudt LM. Toll-like receptor signaling. Cold Spring Harb Perspect Biol (2013) 5:a011247. doi:10.1101/cshperspect.a011247
11. O’Neill LA, Golenbock D, Bowie AG. The history of Toll-like receptors – redefining innate immunity. Nat Rev Immunol (2013) 13:453–60. doi:10.1038/nri3446
12. Oldenburg M, Kruger A, Ferstl R, Kaufmann A, Nees G, Sigmund A, et al. TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science (2012) 337:1111–5. doi:10.1126/science.1220363
13. Jin MS, Lee JO. Structures of the toll-like receptor family and its ligand complexes. Immunity (2008) 29:182–91. doi:10.1016/j.immuni.2008.07.007
14. Gay NJ, Keith FJ. Drosophila Toll and Il-1 receptor. Nature (1991) 351:355–6. doi:10.1038/351355b0
15. Botos I, Segal DM, Davies DR. The structural biology of Toll-like receptors. Structure (2011) 19:447–59. doi:10.1016/j.str.2011.02.004
16. Heinrichs DE, Yethon JA, Whitfield C. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol Microbiol (1998) 30:221–32. doi:10.1046/j.1365-2958.1998.01063.x
17. Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science (1998) 282:2085–8. doi:10.1126/science.282.5396.2085
18. Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, et al. Cutting edge: toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol (1999) 162:3749–52.
19. Shimazu R, Akashi S, Ogata H, Nagai Y, Fukudome K, Miyake K, et al. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J Exp Med (1999) 189:1777–82. doi:10.1084/jem.189.11.1777
20. Beutler B. Tlr4: central component of the sole mammalian LPS sensor. Curr Opin Immunol (2000) 12:20–6. doi:10.1016/S0952-7915(99)00046-1
21. Schwandner R, Dziarski R, Wesche H, Rothe M, Kirschning CJ. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by toll-like receptor 2. J Biol Chem (1999) 274:17406–9. doi:10.1074/jbc.274.25.17406
22. Triantafilou M, Manukyan M, Mackie A, Morath S, Hartung T, Heine H, et al. Lipoteichoic acid and toll-like receptor 2 internalization and targeting to the Golgi are lipid raft-dependent. J Biol Chem (2004) 279:40882–9. doi:10.1074/jbc.M400466200
23. Knapp S, Von Aulock S, Leendertse M, Haslinger I, Draing C, Golenbock DT, et al. Lipoteichoic acid-induced lung inflammation depends on TLR2 and the concerted action of TLR4 and the platelet-activating factor receptor. J Immunol (2008) 180:3478–84.
24. Bowdish DM, Sakamoto K, Kim MJ, Kroos M, Mukhopadhyay S, Leifer CA, et al. MARCO, TLR2, and CD14 are required for macrophage cytokine responses to mycobacterial trehalose dimycolate and Mycobacterium tuberculosis. PLoS Pathog (2009) 5:e1000474. doi:10.1371/journal.ppat.1000474
25. Campos MA, Almeida IC, Takeuchi O, Akira S, Valente EP, Procopio DO, et al. Activation of Toll-like receptor-2 by glycosylphosphatidylinositol anchors from a protozoan parasite. J Immunol (2001) 167:416–23.
26. Means TK, Wang S, Lien E, Yoshimura A, Golenbock DT, Fenton MJ. Human toll-like receptors mediate cellular activation by Mycobacterium tuberculosis. J Immunol (1999) 163:3920–7.
27. Strohmeier GR, Fenton MJ. Roles of lipoarabinomannan in the pathogenesis of tuberculosis. Microbes Infect (1999) 1:709–17. doi:10.1016/S1286-4579(99)80072-0
28. Underhill DM, Ozinsky A, Smith KD, Aderem A. Toll-like receptor-2 mediates mycobacteria-induced proinflammatory signaling in macrophages. Proc Natl Acad Sci U S A (1999) 96:14459–63. doi:10.1073/pnas.96.25.14459
29. Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, et al. A toll-like receptor recognizes bacterial DNA. Nature (2000) 408:740–5. doi:10.1038/35047123
30. Lund J, Sato A, Akira S, Medzhitov R, Iwasaki A. Toll-like receptor 9-mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. J Exp Med (2003) 198:513–20. doi:10.1084/jem.20030162
31. Peter ME, Kubarenko AV, Weber AN, Dalpke AH. Identification of an N-terminal recognition site in TLR9 that contributes to CpG-DNA-mediated receptor activation. J Immunol (2009) 182:7690–7. doi:10.4049/jimmunol.0900819
32. Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature (2001) 413:732–8. doi:10.1038/35099560
33. Hemmi H, Kaisho T, Takeuchi O, Sato S, Sanjo H, Hoshino K, et al. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol (2002) 3:196–200. doi:10.1038/ni758
34. Diebold SS, Kaisho T, Hemmi H, Akira S, Reis E, Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science (2004) 303:1529–31. doi:10.1126/science.1093616
35. Han XB, Li X, Yue SC, Anandaiah A, Hashem F, Reinach PS, et al. Epigenetic regulation of tumor necrosis factor alpha (TNF alpha) release in human macrophages by HIV-1 single-stranded RNA (ssRNA) is dependent on TLR8 signaling. J Biol Chem (2012) 287:13778–86. doi:10.1074/jbc.M112.342683
36. Tanji H, Ohto U, Shibata T, Miyake K, Shimizu T. Structural reorganization of the Toll-like receptor 8 dimer induced by agonistic ligands. Science (2013) 339:1426–9. doi:10.1126/science.1229159
37. Yoneyama M, Fujita T. Function of RIG-I-like receptors in antiviral innate immunity. J Biol Chem (2007) 282:15315–8. doi:10.1074/jbc.R700007200
38. Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity (2005) 23:19–28. doi:10.1016/j.immuni.2005.04.010
39. Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature (2006) 441:101–5. doi:10.1038/nature04734
40. Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, et al. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci U S A (2010) 107:1512–7. doi:10.1073/pnas.0912986107
41. Bruns AM, Pollpeter D, Hadizadeh N, Myong S, Marko JF, Horvath CM. ATP hydrolysis enhances RNA recognition and antiviral signal transduction by the innate immune sensor, laboratory of genetics and physiology 2 (LGP2). J Biol Chem (2013) 288:938–46. doi:10.1074/jbc.M112.424416
42. Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, et al. 5’-Triphosphate RNA is the ligand for RIG-I. Science (2006) 314:994–7. doi:10.1126/science.1132505
43. Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5’-phosphates. Science (2006) 314:997–1001. doi:10.1126/science.1132998
44. Wang Y, Ludwig J, Schuberth C, Goldeck M, Schlee M, Li H, et al. Structural and functional insights into 5’-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat Struct Mol Biol (2010) 17:781–7. doi:10.1038/nsmb.1863
45. Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol (2004) 5:730–7. doi:10.1038/ni1087
46. Loo YM, Gale M Jr. Immune signaling by RIG-I-like receptors. Immunity (2011) 34:680–92. doi:10.1016/j.immuni.2011.05.003
47. Wu B, Peisley A, Richards C, Yao H, Zeng X, Lin C, et al. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell (2013) 152:276–89. doi:10.1016/j.cell.2012.11.048
48. Ting JP, Lovering RC, Alnemri ES, Bertin J, Boss JM, Davis BK, et al. The NLR gene family: a standard nomenclature. Immunity (2008) 28:285–7. doi:10.1016/j.immuni.2008.02.005
49. Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jehanno M, Viala J, et al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science (2003) 300:1584–7. doi:10.1126/science.1084677
50. McDonald C, Inohara N, Nunez G. Peptidoglycan signaling in innate immunity and inflammatory disease. J Biol Chem (2005) 280:20177–80. doi:10.1074/jbc.R500001200
51. Kufer TA, Kremmer E, Adam AC, Philpott DJ, Sansonetti PJ. The pattern-recognition molecule Nod1 is localized at the plasma membrane at sites of bacterial interaction. Cell Microbiol (2008) 10:477–86.
52. Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol (2003) 4:702–7. doi:10.1038/ni945
53. Davis BK, Wen H, Ting JP. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu Rev Immunol (2011) 29:707–35. doi:10.1146/annurev-immunol-031210-101405
54. Bauernfeind F, Hornung V. Of inflammasomes and pathogens – sensing of microbes by the inflammasome. EMBO Mol Med (2013) 5:814–26. doi:10.1002/emmm.201201771
55. Faustin B, Lartigue L, Bruey JM, Luciano F, Sergienko E, Bailly-Maitre B, et al. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol Cell (2007) 25:713–24. doi:10.1016/j.molcel.2007.01.032
56. Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature (2009) 458:514–8. doi:10.1038/nature07725
57. Jones JW, Kayagaki N, Broz P, Henry T, Newton K, O’Rourke K, et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc Natl Acad Sci U S A (2010) 107:9771–6. doi:10.1073/pnas.1003738107
58. Kankkunen P, Teirila L, Rintahaka J, Alenius H, Wolff H, Matikainen S. (1,3)-Beta-glucans activate both dectin-1 and NLRP3 inflammasome in human macrophages. J Immunol (2010) 184:6335–42. doi:10.4049/jimmunol.0903019
59. Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol (2010) 11:395–402. doi:10.1038/ni.1864
60. Vazquez-Mendoza A, Carrero JC, Rodriguez-Sosa M. Parasitic infections: a role for C-type lectins receptors. Biomed Res Int (2013) 2013:456352. doi:10.1155/2013/456352
61. Vautier S, MacCallum DM, Brown GD. C-type lectin receptors and cytokines in fungal immunity. Cytokine (2012) 58:89–99. doi:10.1016/j.cyto.2011.08.031
62. Pluddemann A, Mukhopadhyay S, Gordon S. The interaction of macrophage receptors with bacterial ligands. Expert Rev Mol Med (2006) 8:1–25. doi:10.1017/S1462399406000159
63. Lambert AA, Gilbert C, Richard M, Beaulieu AD, Tremblay MJ. The C-type lectin surface receptor DCIR acts as a new attachment factor for HIV-1 in dendritic cells and contributes to trans- and cis-infection pathways. Blood (2008) 112:1299–307. doi:10.1182/blood-2008-01-136473
64. Osorio F, Reis e Sousa C. Myeloid C-type lectin receptors in pathogen recognition and host defense. Immunity (2011) 34:651–64. doi:10.1016/j.immuni.2011.05.001
65. Redelinghuys P, Brown GD. Inhibitory C-type lectin receptors in myeloid cells. Immunol Lett (2011) 136:1–12. doi:10.1016/j.imlet.2010.10.005
66. Geijtenbeek TB, Gringhuis SI. Signalling through C-type lectin receptors: shaping immune responses. Nat Rev Immunol (2009) 9:465–79. doi:10.1038/nri2569
67. Zelensky AN, Gready JE. The C-type lectin-like domain superfamily. FEBS J (2005) 272:6179–217. doi:10.1111/j.1742-4658.2005.05031.x
68. Huysamen C, Brown GD. The fungal pattern recognition receptor, Dectin-1, and the associated cluster of C-type lectin-like receptors. FEMS Microbiol Lett (2009) 290:121–8. doi:10.1111/j.1574-6968.2008.01418.x
69. Kerscher B, Willment JA, Brown GD. The Dectin-2 family of C-type lectin-like receptors: an update. Int Immunol (2013) 25:271–7. doi:10.1093/intimm/dxt006
70. Brown GD, Gordon S. Immune recognition – a new receptor for beta-glucans. Nature (2001) 413:36–7. doi:10.1038/35092620
71. Dennehy KM, Brown GD. The role of the beta-glucan receptor Dectin-1 in control of fungal infection. J Leukoc Biol (2007) 82:253–8. doi:10.1189/jlb.1206753
72. Taylor PR, Tsoni SV, Willment JA, Dennehy KM, Rosas M, Findon H, et al. Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat Immunol (2007) 8:31–8. doi:10.1038/ni1408
73. Saijo S, Ikeda S, Yamabe K, Kakuta S, Ishigame H, Akitsu A, et al. Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity (2010) 32:681–91. doi:10.1016/j.immuni.2010.05.001
74. Wells CA, Salvage-Jones JA, Li X, Hitchens K, Butcher S, Murray RZ, et al. The macrophage-inducible C-type lectin, Mincle, is an essential component of the innate immune response to Candida albicans. J Immunol (2008) 180:7404–13.
75. Lobato-Pascual A, Saether PC, Dahle MK, Gaustad P, Dissen E, Fossum S, et al. Rat macrophage C-type lectin is an activating receptor expressed by phagocytic cells. PLoS ONE (2013) 8:e57406. doi:10.1371/journal.pone.0057406
76. Miyake Y, Toyonaga K, Mori D, Kakuta S, Hoshino Y, Oyamada A, et al. C-type lectin MCL is an FcRgamma-coupled receptor that mediates the adjuvanticity of mycobacterial cord factor. Immunity (2013) 38:1050–62. doi:10.1016/j.immuni.2013.03.010
77. Ishikawa T, Itoh F, Yoshida S, Saijo S, Matsuzawa T, Gonoi T, et al. Identification of distinct ligands for the C-type lectin receptors Mincle and Dectin-2 in the pathogenic fungus Malassezia. Cell Host Microbe (2013) 13:477–88. doi:10.1016/j.chom.2013.03.008
78. Zhang P, Snyder S, Feng P, Azadi P, Zhang SS, Bulgheresi S, et al. Role of N-acetylglucosamine within core lipopolysaccharide of several species of gram-negative bacteria in targeting the DC-SIGN (CD209). J Immunol (2006) 177:4002–11.
79. Turville S, Wilkinson J, Cameron P, Dable J, Cunningham AL. The role of dendritic cell C-type lectin receptors in HIV pathogenesis. J Leukoc Biol (2003) 74:710–8. doi:10.1189/jlb.0503208
80. Ludwig IS, Lekkerkerker AN, Depla E, Bosman F, Musters RJ, Depraetere S, et al. Hepatitis C virus targets DC-SIGN and L-SIGN to escape lysosomal degradation. J Virol (2004) 78:8322–32. doi:10.1128/JVI.78.15.8322-8332.2004
81. Cheong C, Matos I, Choi JH, Dandamudi DB, Shrestha E, Longhi MP, et al. Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209(+) dendritic cells for immune T cell areas. Cell (2010) 143:416–29. doi:10.1016/j.cell.2010.09.039
82. Zhao LJ, Wang W, Ren H, Qi ZT. Interaction of L-SIGN with hepatitis C virus envelope protein E2 up-regulates Raf-MEK-ERK pathway. Cell Biochem Biophys (2013) 66:589–97. doi:10.1007/s12013-012-9505-4
83. Svajger U, Anderluh M, Jeras M, Obermajer N. C-type lectin DC-SIGN: an adhesion, signalling and antigen-uptake molecule that guides dendritic cells in immunity. Cell Signal (2010) 22:1397–405. doi:10.1016/j.cellsig.2010.03.018
84. Brummer E, Stevens DA. Collectins and fungal pathogens: roles of surfactant proteins and mannose binding lectin in host resistance. Med Mycol (2010) 48:16–28. doi:10.3109/13693780903117473
85. Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol (2004) 4:499–511. doi:10.1038/nri1391
86. Kondo T, Kawai T, Akira S. Dissecting negative regulation of Toll-like receptor signaling. Trends Immunol (2012) 33:449–58. doi:10.1016/j.it.2012.05.002
87. Chen Y, Liu W, Sun T, Huang Y, Wang Y, Deb DK, et al. 1,25-Dihydroxyvitamin D promotes negative feedback regulation of TLR signaling via targeting microRNA-155-SOCS1 in macrophages. J Immunol (2013) 190:3687–95. doi:10.4049/jimmunol.1203273
88. Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol (2005) 6:981–8. doi:10.1038/ni1243
89. Kumar H, Kawai T, Kato H, Sato S, Takahashi K, Coban C, et al. Essential role of IPS-1 in innate immune responses against RNA viruses. J Exp Med (2006) 203:1795–803. doi:10.1084/jem.20060792
90. Kaji R, Kiyoshima-Shibata J, Nagaoka M, Nanno M, Shida K. Bacterial teichoic acids reverse predominant IL-12 production induced by certain Lactobacillus strains into predominant IL-10 production via TLR2-dependent ERK activation in macrophages. J Immunol (2010) 184:3505–13. doi:10.4049/jimmunol.0901569
91. Kawai T, Akira S. Toll-like receptor and RIG-I-like receptor signaling. Ann N Y Acad Sci (2008) 1143:1–20. doi:10.1196/annals.1443.020
92. Kawai T, Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int Immunol (2009) 21:317–37. doi:10.1093/intimm/dxp017
93. Bruns AM, Horvath CM. Activation of RIG-I-like receptor signal transduction. Crit Rev Biochem Mol Biol (2012) 47:194–206. doi:10.3109/10409238.2011.630974
94. Suthar MS, Diamond MS, Gale M Jr. West Nile virus infection and immunity. Nat Rev Microbiol (2013) 11:115–28. doi:10.1038/nrmicro2950
95. Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell (2010) 141:668–81. doi:10.1016/j.cell.2010.04.018
96. Takahashi K, Kawai T, Kumar H, Sato S, Yonehara S, Akira S. Roles of caspase-8 and caspase-10 in innate immune responses to double-stranded RNA. J Immunol (2006) 176:4520–4.
97. Michallet MC, Meylan E, Ermolaeva MA, Vazquez J, Rebsamen M, Curran J, et al. TRADD protein is an essential component of the RIG-like helicase antiviral pathway. Immunity (2008) 28:651–61. doi:10.1016/j.immuni.2008.03.013
98. Arimoto K, Takahashi H, Hishiki T, Konishi H, Fujita T, Shimotohno K. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc Natl Acad Sci U S A (2007) 104:7500–5. doi:10.1073/pnas.0611551104
99. Eisenacher K, Krug A. Regulation of RLR-mediated innate immune signaling – it is all about keeping the balance. Eur J Cell Biol (2012) 91:36–47. doi:10.1016/j.ejcb.2011.01.011
100. Xu L, Xiao N, Liu F, Ren H, Gu J. Inhibition of RIG-I and MDA5-dependent antiviral response by gC1qR at mitochondria. Proc Natl Acad Sci U S A (2009) 106:1530–5. doi:10.1073/pnas.0811029106
101. Park JH, Kim YG, McDonald C, Kanneganti TD, Hasegawa M, Body-Malapel M, et al. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J Immunol (2007) 178:2380–6.
102. Hasegawa M, Fujimoto Y, Lucas PC, Nakano H, Fukase K, Nunez G, et al. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-kappaB activation. EMBO J (2008) 27:373–83. doi:10.1038/sj.emboj.7601962
103. Shaw PJ, Lamkanfi M, Kanneganti TD. NOD-like receptor (NLR) signaling beyond the inflammasome. Eur J Immunol (2010) 40:624–7. doi:10.1002/eji.200940211
104. Barber GN. Innate immune DNA sensing pathways: STING, AIMII and the regulation of interferon production and inflammatory responses. Curr Opin Immunol (2011) 23:10–20. doi:10.1016/j.coi.2010.12.015
105. Juliana C, Fernandes-Alnemri T, Kang S, Farias A, Qin F, Alnemri ES. Non-transcriptional priming and deubiquitination regulate NLRP3 inflammasome activation. J Biol Chem (2012) 287:36617–22. doi:10.1074/jbc.M112.407130
106. Satoh T, Kambe N, Matsue H. NLRP3 activation induces ASC-dependent programmed necrotic cell death, which leads to neutrophilic inflammation. Cell Death Dis (2013) 4:e644. doi:10.1038/cddis.2013.169
107. Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature (2008) 456:264–8. doi:10.1038/nature07383
108. Guarda G, Braun M, Staehli F, Tardivel A, Mattmann C, Forster I, et al. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity (2011) 34:213–23. doi:10.1016/j.immuni.2011.02.006
109. Beynon V, Quintana FJ, Weiner HL. Activated human CD4+CD45RO+ memory T-cells indirectly inhibit NLRP3 inflammasome activation through downregulation of P2X7R signalling. PLoS ONE (2012) 7:e39576. doi:10.1371/journal.pone.0039576
110. Mao K, Chen S, Chen M, Ma Y, Wang Y, Huang B, et al. Nitric oxide suppresses NLRP3 inflammasome activation and protects against LPS-induced septic shock. Cell Res (2013) 23:201–12. doi:10.1038/cr.2013.6
111. Chen S, Sun B. Negative regulation of NLRP3 inflammasome signaling. Protein Cell (2013) 4:251–8. doi:10.1007/s13238-013-2128-8
112. Masters SL, Gerlic M, Metcalf D, Preston S, Pellegrini M, O’Donnell JA, et al. NLRP1 inflammasome activation induces pyroptosis of hematopoietic progenitor cells. Immunity (2012) 37:1009–23. doi:10.1016/j.immuni.2012.08.027
113. Gringhuis SI, Den Dunnen J, Litjens M, Van Der Vlist M, Wevers B, Bruijns SC, et al. Dectin-1 directs T helper cell differentiation by controlling noncanonical NF-kappaB activation through Raf-1 and Syk. Nat Immunol (2009) 10:203–13. doi:10.1038/ni.1692
114. Gringhuis SI, Wevers BA, Kaptein TM, Van Capel TM, Theelen B, Boekhout T, et al. Selective C-Rel activation via Malt1 controls anti-fungal T(H)-17 immunity by dectin-1 and dectin-2. PLoS Pathog (2011) 7:e1001259. doi:10.1371/journal.ppat.1001259
115. Sancho D, Reis e Sousa C. Signaling by myeloid C-type lectin receptors in immunity and homeostasis. Annu Rev Immunol (2012) 30:491–529. doi:10.1146/annurev-immunol-031210-101352
116. Xu S, Huo J, Lee KG, Kurosaki T, Lam KP. Phospholipase Cgamma2 is critical for Dectin-1-mediated Ca2+ flux and cytokine production in dendritic cells. J Biol Chem (2009) 284:7038–46. doi:10.1074/jbc.M806650200
117. Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature (2007) 448:501–5. doi:10.1038/nature06013
118. Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature (2008) 451:725–9. doi:10.1038/nature06537
119. Yang P, An H, Liu X, Wen M, Zheng Y, Rui Y, et al. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat Immunol (2010) 11:487–94. doi:10.1038/ni.1876
120. Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol (2010) 11:997–1004. doi:10.1038/ni.1932
121. Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature (2008) 455:674–8. doi:10.1038/nature07317
122. Tsuchida T, Zou JA, Saitoh T, Kumar H, Abe T, Matsuura Y, et al. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity (2010) 33:765–76. doi:10.1016/j.immuni.2010.10.013
123. Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, et al. cGAS produces a 2’-5’-linked cyclic dinucleotide second messenger that activates STING. Nature (2013) 498:380–4. doi:10.1038/nature12306
124. Civril F, Deimling T, De Oliveira Mann CC, Ablasser A, Moldt M, Witte G, et al. Structural mechanism of cytosolic DNA sensing by cGAS. Nature (2013) 498:332–37. doi:10.1038/nature12305
125. Sun LJ, Wu JX, Du FH, Chen X, Chen ZJJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the Type I interferon pathway. Science (2013) 339:786–91. doi:10.1126/science.1232458
126. Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol (2011) 12:959–65. doi:10.1038/ni.2091
127. Zhang Z, Bao M, Lu N, Weng L, Yuan B, Liu YJ. The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat Immunol (2013) 14:172–8. doi:10.1038/ni.2492
128. Kondo T, Kobayashi J, Saitoh T, Maruyama K, Ishii KJ, Barber GN, et al. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proc Natl Acad Sci U S A (2013) 110:2969–74. doi:10.1073/pnas.1222694110
129. Yi AK, Yoon H, Park JE, Kim BS, Kim HJ, Martinez-Hernandez A. CpG DNA-mediated induction of acute liver injury in D-galactosamine-sensitized mice: the mitochondrial apoptotic pathway-dependent death of hepatocytes. J Biol Chem (2006) 281:15001–12. doi:10.1074/jbc.M601337200
130. Zadeh M, Khan MW, Goh YJ, Selle K, Owen JL, Klaenhammer T, et al. Induction of intestinal pro-inflammatory immune responses by lipoteichoic acid. J Inflamm (Lond) (2012) 9:7. doi:10.1186/1476-9255-9-7
131. Ulevitch RJ, Tobias PS. Receptor-dependent mechanisms of cell stimulation by bacterial-endotoxin. Annu Rev Immunol (1995) 13:437–57. doi:10.1146/annurev.iy.13.040195.002253
132. Shapira L, Soskolne WA, Houri Y, Barak V, Halabi A, Stabholz A. Protection against endotoxic shock and lipopolysaccharide-induced local inflammation by tetracycline: correlation with inhibition of cytokine secretion. Infect Immun (1996) 64:825–8.
133. Nymark M, Pussinen PJ, Tuomainen AM, Forsblom C, Groop PH, Lehto M, et al. Serum lipopolysaccharide activity is associated with the progression of kidney disease in Finnish patients with Type 1 diabetes. Diabetes Care (2009) 32:1689–93. doi:10.2337/dc09-0467
134. Cavallo T, Granholm NA. Bacterial lipopolysaccharide induces long-lasting IgA deficiency concurrently with features of polyclonal B cell activation in normal and in lupus-prone mice. Clin Exp Immunol (1991) 84:134–8.
135. Yamamoto Y, Harashima A, Saito H, Tsuneyama K, Munesue S, Motoyoshi S, et al. Septic shock is associated with receptor for advanced glycation end products ligation of LPS. J Immunol (2011) 186:3248–57. doi:10.4049/jimmunol.1002253
136. Sarkar R, Mitra D, Chakrabarti S. HIV-1 gp120 protein downregulates Nef induced IL-6 release in immature dendritic cells through interplay of DC-SIGN. PLoS ONE (2013) 8:e59073. doi:10.1371/journal.pone.0059073
137. Devhare PB, Chatterjee SN, Arankalle VA, Lole KS. Analysis of antiviral response in human epithelial cells infected with hepatitis E virus. PLoS ONE (2013) 8:e63793. doi:10.1371/journal.pone.0063793
138. Medina RA, Stertz S, Manicassamy B, Zimmermann P, Sun X, Albrecht RA, et al. Glycosylations in the globular head of the hemagglutinin protein modulate the virulence and antigenic properties of the H1N1 influenza viruses. Sci Transl Med (2013) 5:187ra170. doi:10.1126/scitranslmed.3005996
139. Demento SL, Siefert AL, Bandyopadhyay A, Sharp FA, Fahmy TM. Pathogen-associated molecular patterns on biomaterials: a paradigm for engineering new vaccines. Trends Biotechnol (2011) 29:294–306. doi:10.1016/j.tibtech.2011.02.004
140. Kaisho T. Pathogen sensors and chemokine receptors in dendritic cell subsets. Vaccine (2012) 30:7652–7. doi:10.1016/j.vaccine.2012.10.043
141. Driscoll M, Hansen R, Ding C, Cramer DE, Yan J. Therapeutic potential of various beta-glucan sources in conjunction with anti-tumor monoclonal antibody in cancer therapy. Cancer Biol Ther (2009) 8:214–21.
142. Lipinski T, Fitieh A, St Pierre J, Ostergaard HL, Bundle DR, Touret N. Enhanced immunogenicity of a tricomponent mannan tetanus toxoid conjugate vaccine targeted to dendritic cells via Dectin-1 by incorporating beta-glucan. J Immunol (2013) 190:4116–28. doi:10.4049/jimmunol.1202937
143. Pietrella D, Rachini A, Torosantucci A, Chiani P, Brown AJ, Bistoni F, et al. A beta-glucan-conjugate vaccine and anti-beta-glucan antibodies are effective against murine vaginal candidiasis as assessed by a novel in vivo imaging technique. Vaccine (2010) 28:1717–25. doi:10.1016/j.vaccine.2009.12.021
144. Liu M, Machova E, Nescakova Z, Medovarska I, Clemons KV, Martinez M, et al. Vaccination with mannan protects mice against systemic aspergillosis. Med Mycol (2012) 50:818–28. doi:10.3109/13693786.2012.683539
145. Johnson AG, Gaines S, Landy M. Studies on the O antigen of Salmonella typhosa. V. Enhancement of antibody response to protein antigens by the purified lipopolysaccharide. J Exp Med (1956) 103:225–46. doi:10.1084/jem.103.2.225
146. Johnson AG, Tomai M, Solem L, Beck L, Ribi E. Characterization of a nontoxic monophosphoryl lipid A. Rev Infect Dis (1987) 9(Suppl 5):S512–6. doi:10.1093/clinids/9.Supplement_5.S512
147. Zhong Z, Wei X, Qi B, Xiao W, Yang L, Wei Y, et al. A novel liposomal vaccine improves humoral immunity and prevents tumor pulmonary metastasis in mice. Int J Pharm (2010) 399:156–62. doi:10.1016/j.ijpharm.2010.07.053
148. ten Brinke A, Van Schijndel G, Visser R, De Gruijl TD, Zwaginga JJ, Van Ham SM. Monophosphoryl lipid A plus IFNgamma maturation of dendritic cells induces antigen-specific CD8+ cytotoxic T cells with high cytolytic potential. Cancer Immunol Immunother (2010) 59:1185–95. doi:10.1007/s00262-010-0843-z
149. MacLeod MK, McKee AS, David A, Wang J, Mason R, Kappler JW, et al. Vaccine adjuvants aluminum and monophosphoryl lipid A provide distinct signals to generate protective cytotoxic memory CD8 T cells. Proc Natl Acad Sci U S A (2011) 108:7914–9. doi:10.1073/pnas.1104588108
150. Roy A, Chandra S, Mamilapally S, Upadhyay P, Bhaskar S. Anticancer and immunostimulatory activity by conjugate of paclitaxel and non-toxic derivative of LPS for combined chemo-immunotherapy. Pharm Res (2012) 29:2294–309. doi:10.1007/s11095-012-0756-y
151. Coler RN, Bertholet S, Moutaftsi M, Guderian JA, Windish HP, Baldwin SL, et al. Development and characterization of synthetic glucopyranosyl lipid adjuvant system as a vaccine adjuvant. PLoS ONE (2011) 6:e16333. doi:10.1371/journal.pone.0016333
152. Baker PJ, Fauntleroy MB, Stashak PW, Hiernaux JR, Cantrell JL, Rudbach JA. Adjuvant effects of trehalose dimycolate on the antibody response to type III pneumococcal polysaccharide. Infect Immun (1989) 57:912–7.
153. Holten-Andersen L, Doherty TM, Korsholm KS, Andersen P. Combination of the cationic surfactant dimethyl dioctadecyl ammonium bromide and synthetic mycobacterial cord factor as an efficient adjuvant for tuberculosis subunit vaccines. Infect Immun (2004) 72:1608–17. doi:10.1128/IAI.72.3.1608-1617.2004
154. Davidsen J, Rosenkrands I, Christensen D, Vangala A, Kirby D, Perrie Y, et al. Characterization of cationic liposomes based on dimethyldioctadecylammonium and synthetic cord factor from M. tuberculosis (trehalose 6,6’-dibehenate)-a novel adjuvant inducing both strong CMI and antibody responses. Biochim Biophys Acta (2005) 1718:22–31. doi:10.1016/j.bbamem.2005.10.011
155. Henriksen-Lacey M, Devitt A, Perrie Y. The vesicle size of DDA:TDB liposomal adjuvants plays a role in the cell-mediated immune response but has no significant effect on antibody production. J Control Release (2011) 154:131–7. doi:10.1016/j.jconrel.2011.05.019
156. Desel C, Werninghaus K, Ritter M, Jozefowski K, Wenzel J, Russkamp N, et al. The Mincle-activating adjuvant TDB induces MyD88-dependent Th1 and Th17 responses through IL-1R signaling. PLoS ONE (2013) 8:e53531. doi:10.1371/journal.pone.0053531
157. Schweneker K, Gorka O, Schweneker M, Poeck H, Tschopp J, Peschel C, et al. The mycobacterial cord factor adjuvant analogue trehalose-6,6’-dibehenate (TDB) activates the Nlrp3 inflammasome. Immunobiology (2013) 218:664–73. doi:10.1016/j.imbio.2012.07.029
158. Colavecchia SB, Jolly A, Fernandez B, Fontanals AM, Fernandez E, Mundo SL. Effect of lipoarabinomannan from Mycobacterium avium subsp avium in Freund’s incomplete adjuvant on the immune response of cattle. Braz J Med Biol Res (2012) 45:139–46. doi:10.1590/S0100-879X2012007500012
159. Halassy B, Krstanovic M, Frkanec R, Tomasic J. Adjuvant activity of peptidoglycan monomer and its metabolic products. Vaccine (2003) 21:971–6. doi:10.1016/S0264-410X(02)00547-9
160. Capparelli R, Nocerino N, Medaglia C, Blaiotta G, Bonelli P, Iannelli D. The Staphylococcus aureus peptidoglycan protects mice against the pathogen and eradicates experimentally induced infection. PLoS ONE (2011) 6:e28377. doi:10.1371/journal.pone.0028377
161. Habjanec L, Frkanec R, Halassy B, Tomasic J. Effect of liposomal formulations and immunostimulating peptidoglycan monomer (PGM) on the immune reaction to ovalbumin in mice. J Liposome Res (2006) 16:1–16. doi:10.1080/08982100500528537
162. Himanen JP, Pyhala L, Olander RM, Merimskaya O, Kuzina T, Lysyuk O, et al. Biological-activities of lipoteichoic acid and peptidoglycan teichoic-acid of Bacillus-subtilis-168 (Marburg). J Gen Microbiol (1993) 139:2659–65. doi:10.1099/00221287-139-11-2659
163. Okamoto M, Ohe G, Oshikawa T, Furuichi S, Nishikawa H, Tano T, et al. Enhancement of anti-cancer immunity by a lipoteichoic-acid-related molecule isolated from a penicillin-killed group A Streptococcus. Cancer Immunol Immunother (2001) 50:408–16. doi:10.1007/s002620100207
164. Martins VP, Pinheiro CS, Figueiredo BC, Assis NR, Morais SB, Caliari MV, et al. Vaccination with enzymatically cleaved GPI-anchored proteins from Schistosoma mansoni induces protection against challenge infection. Clin Dev Immunol (2012) 2012:962538. doi:10.1155/2012/962538
165. Krieg AM, Yi AK, Matson S, Waldschmidt TJ, Bishop GA, Teasdale R, et al. Cpg motifs in bacterial-DNA trigger direct B-cell activation. Nature (1995) 374:546–9. doi:10.1038/374546a0
166. Klinman DM, Yi AK, Beaucage SL, Conover J, Krieg AM. CpG motifs present in bacterial DNA rapidly induce lymphocytes to secrete interleukin 6, interleukin 12, and interferon gamma. Proc Natl Acad Sci U S A (1996) 93:2879–83. doi:10.1073/pnas.93.7.2879
167. Bode C, Zhao G, Steinhagen F, Kinjo T, Klinman DM. CpG DNA as a vaccine adjuvant. Expert Rev Vaccines (2011) 10:499–511. doi:10.1586/erv.10.174
168. Desmet CJ, Ishii KJ. Nucleic acid sensing at the interface between innate and adaptive immunity in vaccination. Nat Rev Immunol (2012) 12:479–91. doi:10.1038/nri3247
169. Dalpke AH, Zimmermann S, Albrecht I, Heeg K. Phosphodiester CpG oligonucleotides as adjuvants: polyguanosine runs enhance cellular uptake and improve immunostimulative activity of phosphodiester CpG oligonucleotides in vitro and in vivo. Immunology (2002) 106:102–12. doi:10.1046/j.1365-2567.2002.01410.x
170. Wu CC, Lee J, Raz E, Corr M, Carson DA. Necessity of oligonucleotide aggregation for toll-like receptor 9 activation. J Biol Chem (2004) 279:33071–8. doi:10.1074/jbc.M311662200
171. Krug A, Rothenfusser S, Hornung V, Jahrsdorfer B, Blackwell S, Ballas ZK, et al. Identification of CpG oligonucleotide sequences with high induction of IFN-alpha/beta in plasmacytoid dendritic cells. Eur J Immunol (2001) 31:2154–63. doi:10.1002/1521-4141(200107)31:7<2154::AID-IMMU2154>3.0.CO;2-U
172. Marshall JD, Fearon KL, Higgins D, Hessel EM, Kanzler H, Abbate C, et al. Superior activity of the type C class of ISS in vitro and in vivo across multiple species. DNA Cell Biol (2005) 24:63–72. doi:10.1089/dna.2005.24.63
173. Vollmer J, Krieg AM. Immunotherapeutic applications of CpG oligodeoxynucleotide TLR9 agonists. Adv Drug Deliv Rev (2009) 61:195–204. doi:10.1016/j.addr.2008.12.008
174. Davis HL, Weeranta R, Waldschmidt TJ, Tygrett L, Schorr J, Krieg AM. CpG DNA is a potent enhancer of specific immunity in mice immunized with recombinant hepatitis B surface antigen. J Immunol (1998) 160:870–6.
175. Weiner GJ, Liu HM, Wooldridge JE, Dahle CE, Krieg AM. Immunostimulatory oligodeoxynucleotides containing the CpG motif are effective as immune adjuvants in tumor antigen immunization. Proc Natl Acad Sci U S A (1997) 94:10833–7. doi:10.1073/pnas.94.20.10833
176. Angel JB, Cooper CL, Clinch J, Young CD, Chenier A, Parato KG, et al. CpG increases vaccine antigen-specific cell-mediated immunity when administered with hepatitis B vaccine in HIV infection. J Immune Based Ther Vaccines (2008) 6:4. doi:10.1186/1476-8518-6-4
177. Jeske S, Pries R, Wollenberg B. CpG-induced IFN-alpha production of plasmacytoid dendritic cells: time and dosage dependence and the effect of structural modifications to the CpG backbone. Nucleic Acid Ther (2013) 23:118–24. doi:10.1089/nat.2012.0384
178. Klinman DM. Adjuvant activity of CpG oligodeoxynucleotides. Int Rev Immunol (2006) 25:135–54. doi:10.1080/08830180600743057
179. Coffman RL, Sher A, Seder RA. Vaccine adjuvants: putting innate immunity to work. Immunity (2010) 33:492–503. doi:10.1016/j.immuni.2010.10.002
180. Ato M, Takahashi Y, Fujii H, Hashimoto S, Kaji T, Itamura S, et al. Influenza A whole virion vaccine induces a rapid reduction of peripheral blood leukocytes via interferon-alpha-dependent apoptosis. Vaccine (2013) 31:2184–90. doi:10.1016/j.vaccine.2013.02.016
181. Geeraedts F, Goutagny N, Hornung V, Severa M, De Haan A, Pool J, et al. Superior immunogenicity of inactivated whole virus H5N1 influenza vaccine is primarily controlled by Toll-like receptor signalling. PLoS Pathog (2008) 4:e1000138. doi:10.1371/journal.ppat.1000138
182. Chakravarthy KV, Bonoiu AC, Davis WG, Ranjan P, Ding H, Hu R, et al. Gold nanorod delivery of an ssRNA immune activator inhibits pandemic H1N1 influenza viral replication. Proc Natl Acad Sci U S A (2010) 107:10172–7. doi:10.1073/pnas.0914561107
183. Wong JP, Saravolac EG, Sabuda D, Levy HB, Kende M. Prophylactic and therapeutic efficacies of poly(IC.LC) against respiratory influenza A virus infection in mice. Antimicrob Agents Chemother (1995) 39:2574–6. doi:10.1128/AAC.39.11.2574
184. Tewari K, Flynn BJ, Boscardin SB, Kastenmueller K, Salazar AM, Anderson CA, et al. Poly(I:C) is an effective adjuvant for antibody and multi-functional CD4+ T cell responses to Plasmodium falciparum circumsporozoite protein (CSP) and alphaDEC-CSP in non human primates. Vaccine (2010) 28:7256–66. doi:10.1016/j.vaccine.2010.08.098
185. Chen H, Chuai X, Deng Y, Wen B, Wang W, Xiong S, et al. Optimisation of prime-boost immunization in mice using novel protein-based and recombinant vaccinia (Tiantan)-based HBV vaccine. PLoS ONE (2012) 7:e43730. doi:10.1371/journal.pone.0043730
186. Chuai X, Chen H, Wang W, Deng Y, Wen B, Ruan L, et al. Poly(I:C)/alum mixed adjuvant priming enhances HBV subunit vaccine-induced immunity in mice when combined with recombinant adenoviral-based HBV vaccine boosting. PLoS ONE (2013) 8:e54126. doi:10.1371/journal.pone.0054126
187. Thim HL, Iliev DB, Christie KE, Villoing S, McLoughlin MF, Strandskog G, et al. Immunoprotective activity of a Salmonid Alphavirus vaccine: comparison of the immune responses induced by inactivated whole virus antigen formulations based on CpG class B oligonucleotides and poly I:C alone or combined with an oil adjuvant. Vaccine (2012) 30:4828–34. doi:10.1016/j.vaccine.2012.05.010
188. Scallan CD, Tingley DW, Lindbloom JD, Toomey JS, Tucker SN. An adenovirus-based vaccine with a double-stranded RNA adjuvant protects mice and ferrets against H5N1 avian influenza in oral delivery models. Clin Vaccine Immunol (2013) 20:85–94. doi:10.1128/CVI.00552-12
189. Lepenies B, Lee J, Sonkaria S. Targeting C-type lectin receptors with multivalent carbohydrate ligands. Adv Drug Deliv Rev (2013). doi:10.1016/j.addr.2013.05.007
190. Cruz LJ, Tacken PJ, Pots JM, Torensma R, Buschow SI, Figdor CG. Comparison of antibodies and carbohydrates to target vaccines to human dendritic cells via DC-SIGN. Biomaterials (2012) 33:4229–39. doi:10.1016/j.biomaterials.2012.02.036
191. Garcia-Vallejo JJ, Unger WWJ, Kalay H, Van Kooyk Y. Glycan-based DC-SIGN targeting to enhance antigen cross-presentation in anticancer vaccines. Oncoimmunology (2013) 2(2):e23040. doi:10.4161/onci.23040
192. van Kooyk Y, Unger WW, Fehres CM, Kalay H, Garcia-Vallejo JJ. Glycan-based DC-SIGN targeting vaccines to enhance antigen cross-presentation. Mol Immunol (2013) 55:143–5. doi:10.1016/j.molimm.2012.10.031
193. Ribeiro-Viana R, Sanchez-Navarro M, Luczkowiak J, Koeppe JR, Delgado R, Rojo J, et al. Virus-like glycodendrinanoparticles displaying quasi-equivalent nested polyvalency upon glycoprotein platforms potently block viral infection. Nat Commun (2012) 3:1303. doi:10.1038/ncomms2302
Keywords: innate immunity, innate sensors, sugar associated PAMPs, disease pathogenesis, vaccinology
Citation: Mahla RS, Reddy MC, Prasad DVR and Kumar H (2013) Sweeten PAMPs: role of sugar complexed PAMPs in innate immunity and vaccine biology. Front. Immunol. 4:248. doi: 10.3389/fimmu.2013.00248
Received: 09 July 2013; Accepted: 09 August 2013;
Published online: 02 September 2013.
Edited by:
Paul A. Ramsland, Burnet Institute, AustraliaReviewed by:
Ashley Mansell, Monash Institute of Medical Research, AustraliaBernd Lepenies, Max Planck Society, Germany
Copyright: © 2013 Mahla, Reddy, Prasad and Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Himanshu Kumar, Laboratory of Immunology, Department of Biological Sciences, Indian Institute of Science Education and Research (IISER), Indore By-pass Road, Bhauri, Bhopal 462030, Madhya Pradesh, India e-mail:aGt1bWFyQGlpc2VyYi5hYy5pbg==