- Department of Medicine, Section of Rheumatology, Gwen Knapp Center for Lupus and Immunology Research, The University of Chicago, Chicago, IL, USA
The essential events of B-cell development are the stochastic and sequential rearrangement of immunoglobulin heavy (Igμ) and then light chain (Igκ followed by Igλ) loci. The counterpoint to recombination is proliferation, which both maintains populations of pro-B cells undergoing Igμ recombination and expands the pool of pre-B cells expressing the Igμ protein available for subsequent Igκ recombination. Proliferation and recombination must be segregated into distinct and mutually exclusive developmental stages. Failure to do so risks aberrant gene translocation and leukemic transformation. Recent studies have demonstrated that proliferation and recombination are each affected by different and antagonistic receptors. The IL-7 receptor drives proliferation while the pre-B-cell antigen receptor, which contains Igμ and surrogate light chain, enhances Igκ accessibility and recombination. Remarkably, the principal downstream proliferative effectors of the IL-7R, STAT5 and cyclin D3, directly repress Igκ accessibility through very divergent yet complementary mechanisms. Conversely, the pre-B-cell receptor represses cyclin D3 leading to cell cycle exit and enhanced Igκ accessibility. These studies reveal how cell fate decisions can be directed and reinforced at each developmental transition by single receptors. Furthermore, they identify novel mechanisms of Igκ repression that have implications for gene regulation in general.
Introduction
Development of a diverse repertoire of peripheral B cells is dependent on the appropriate and ordered progression of B-lymphopoiesis. This process occurs through discrete developmental stages driven by the sequential rearrangement and expression of genes encoding the immunoglobulin heavy (Igμ) and then light chains (Igκ or Igλ). Successful expression of a functional Igμ capable of pairing with surrogate light chain (SLC) components and Igα/Igβ to form the pre-B-cell receptor (pre-BCR) at the cell surface is associated with a proliferative burst that expands the pool of pre-B cells expressing Igμ prior to cell cycle exit and the rearrangement of Igκ. Proliferation and recombination must remain mutually exclusive to maintain genomic integrity and prevent excessive cell death or oncogenesis through aberrant translocations. Recent work has begun to uncover the molecular mechanisms dictating these developmental stages. Of particular interest, is the integration and opposition of the IL-7R and pre-BCR signaling pathways along with the effect of downstream epigenetic modifications on Igκ loci rearrangement and early B-cell proliferation.
B-Cell Development
Interactions with bone marrow (BM) stromal cells induce the differentiation of common lymphoid progenitor cells (CLPs), capable of generating B and T cells, into multipotential precursor–progenitor (pre–pro) B cells (1, 2). At this stage, initial Igμ rearrangements occur at diversity (DH) and joining (JH) gene segments (3). Pre–pro-B cells are not committed to the B-cell lineage as some developing T cells bear Igμ DHJH rearrangements. Within IL-7 rich niches of the BM, pre–pro-B cells commit to the B-cell lineage through differentiation into progenitor (pro)-B cells expressing CD19 (4–6). IL-7 provides critical proliferative and survival signals needed to maintain the pool of pro-B cells. The hallmark event of pro-B cells is the completion of Igμ rearrangement with the addition of a variable (VH) region to the DHJH segment. This process of recombination is mediated by the semi-random induction of double-stranded DNA breaks by the recombinase activating gene (Rag)-1 and Rag-2 proteins at recombination signal sequences (RSS) followed by non-homologous end joining (NHEJ) (7). Rag-mediated recombination of the antigen receptor loci is an essential and defining feature of B- and T-lymphopoiesis. Genetic mutation of the Rag genes results in severe combined immunodeficiency (SCID) in humans and mice (8–10).
Progression to the pre-B-cell stage of development is marked by the expression of a functional Igμ, due to in-frame rearrangement at one locus, which can pair with SLC components, VpreB and λ5, to form the pre-BCR at the cell surface (11). Early events following the expression of the pre-BCR serve to expand in number B-cell populations that have successfully rearranged Igμ (12). Not all Igμ chains effectively pair with SLC and therefore the pre-BCR checkpoint shapes the repertoire of Igμ chains selected into the small pre-B-cell pool (13). In mice deficient in SLC, cells that escape by rearranging immunoglobulin light chain are preferentially autoreactive (14). Furthermore, conferring defined self-reactivity rescues SLC deficiency (15). However, it is not clear if this means that the pre-BCR censors autoreactivity or if autoreactivity, and ligation by self-antigen, is required to complement SLC deficiency.
Following poly-clonal expansion, late (small) pre-B cells migrate away from proliferation-inducing IL-7 rich niches of the BM, exit cell cycle, and begin to rearrange Igκ genes (6). Final pairing of translated Igμ and Igκ form the antigen-specific BCR on immature B cells which are then subjected to the mechanisms of tolerance that diminish autoreactivity in the naïve repertoire. Although the necessity of the IL-7R and pre-BCR for B-lymphopoiesis has long been appreciated, recent work has begun to detail the molecular mechanisms and downstream interplay of these pathways that drive B-cell development.
IL-7R and Pro-B Cells Fate
Signaling through the IL-7R, which is a heterodimer of the IL-7Rα chain and the common γ chain, is essential for proliferation and survival of pro- and pre-B cells. In vitro culture assays demonstrated that pro-B cells and not pre–pro-B cells proliferate in response to IL-7 (4). Accordingly, IL-7Rα-deficient mice demonstrate a significant impairment in B-lymphopoiesis beginning at the pro-B-cell stage (16–18). However, IL-7-deficient mice display a less severe defect in pro-B-cell development suggesting the IL-7Rα chain may participate in an additional signaling complex that compensates for loss of IL-7-induced signaling (17). Nonetheless, although pairing of IL-7Rα with alternative complexes may provide some compensation to IL-7-induced signaling, it is clear that the downstream components of the IL-7R pathway determine the pro-B-cell fate.
Through pairing with Janus kinase (JAK) 3 and JAK1, the IL-7R, upon activation, recruits and activates signal transducer and activator of transcription (STAT) 5a and b (19). STAT5 is critical for the biological effects of the IL-7R. B-cell development in mice deficient in both STAT5a and b is blocked at the pro-B stage, similar to IL-7Rα-deficient mice (20). Accordingly, constitutive activation (CA) of STAT5 in mice mostly restores B-lymphopoiesis in the absence of IL-7R signaling, while in humans, CA-STAT5 gene mutations have been identified in patients with acute lymphoblastic leukemia (21–23). Activated STAT5 primarily drives proliferation by inducing expression of the gene encoding cyclin D3, Ccnd3 (23, 24). Pairing of cyclin D family members with cyclin-dependent kinases 4 and 6 (CDK4/6) during G1 activates retinoblastoma protein (Rb) family members and E2f transcription factors to induce upregulation of cell cycle genes and suppress cell cycle inhibitors p27Kip1 and p21Cip1 (25). Although both cyclin D2 and D3 are expressed during B-cell development, only cyclin D3 can be found in complexes with CDK4/6 in pro-B cells (26). Moreover, a defect in early B-cell development is found only in Ccnd3−/− mice, while Ccnd2−/− mice display a later defect in peripheral B-cell proliferation (24, 27, 28). In addition to proliferative signals, STAT5 maintains survival of developing B cells through induction of several pro-survival genes including Mcl1, Bcl2, and Pim1 (22, 29, 30). Therefore, IL-7R-mediated activation of STAT5 represents a critical event in the expansion and stability of early B cells populations.
Pro-B cells are both proliferating and rearranging Igμ genes (4). Recent studies have provided some insights into how these incompatible processes are segregated to distinct populations within the pro-B-cell pool (31, 32). For example, it has been demonstrated that the core machineries of recombination and proliferation are antagonistic. The Rag proteins are expressed in G0/G1 and are degraded in dividing cells at the transition from G1 to S phase (33). Cyclin A/CDK2 complexes induce cell cycle entry and inhibit the accumulation of Rag-2, while several CDK inhibitors, including p21Cip1, p27Kip1, and p57Kip2 induce Rag-2 expression (34). This is because the cyclin A/CDK2 complex phosphorylates threonine 490 of Rag-2 targeting it for degradation by Skp2 (35). Mutation of threonine 490 results in persistence of Ig recombination in proliferating cells and increases the prevalence of chromosomal translocations and lymphoid malignancies (36). Impaired NHEJ accompanied with defective DNA-damage-induced apoptosis also increases the occurrence of leukemogenesis. Mice with combined deficiencies of the pro-apoptotic protein p53 with either XRCC4 or Ku80, both members of the NHEJ machinery, develop IgH–Myc translocations that promote pro-B leukemia (37, 38). Therefore, separation of proliferation and recombination is crucial to the avoidance of excessive B cells’ death or development of B-cell leukemia.
It is also now clear that the pro-B-cell compartment is not homogeneous but contains subpopulations of cells that express relatively high or low levels of the IL-7R. Furthermore, in these populations, IL-7R expression levels correlate with intracellular-activated STAT5 (39). These findings suggest a dynamic model where pro-B cells shift from proliferation to recombination through the oscillation of IL-7R expression (Figure 1). In contrast to oscillating between IL-7R high and low states, it is also possible that pro-B cells sequentially progress through IL-7R high and low stages. The mechanism driving IL-7R downregulation in pro-B cells, however, is still unknown. One possibility is through asymmetric cell division, where the accumulation of IL-7R toward IL-7-producing stromal cells results in distal daughter cells inheriting less IL-7R on their surface, therein, providing a transient decrease in STAT5 activation and the initiation of VH–DHJH rearrangement.
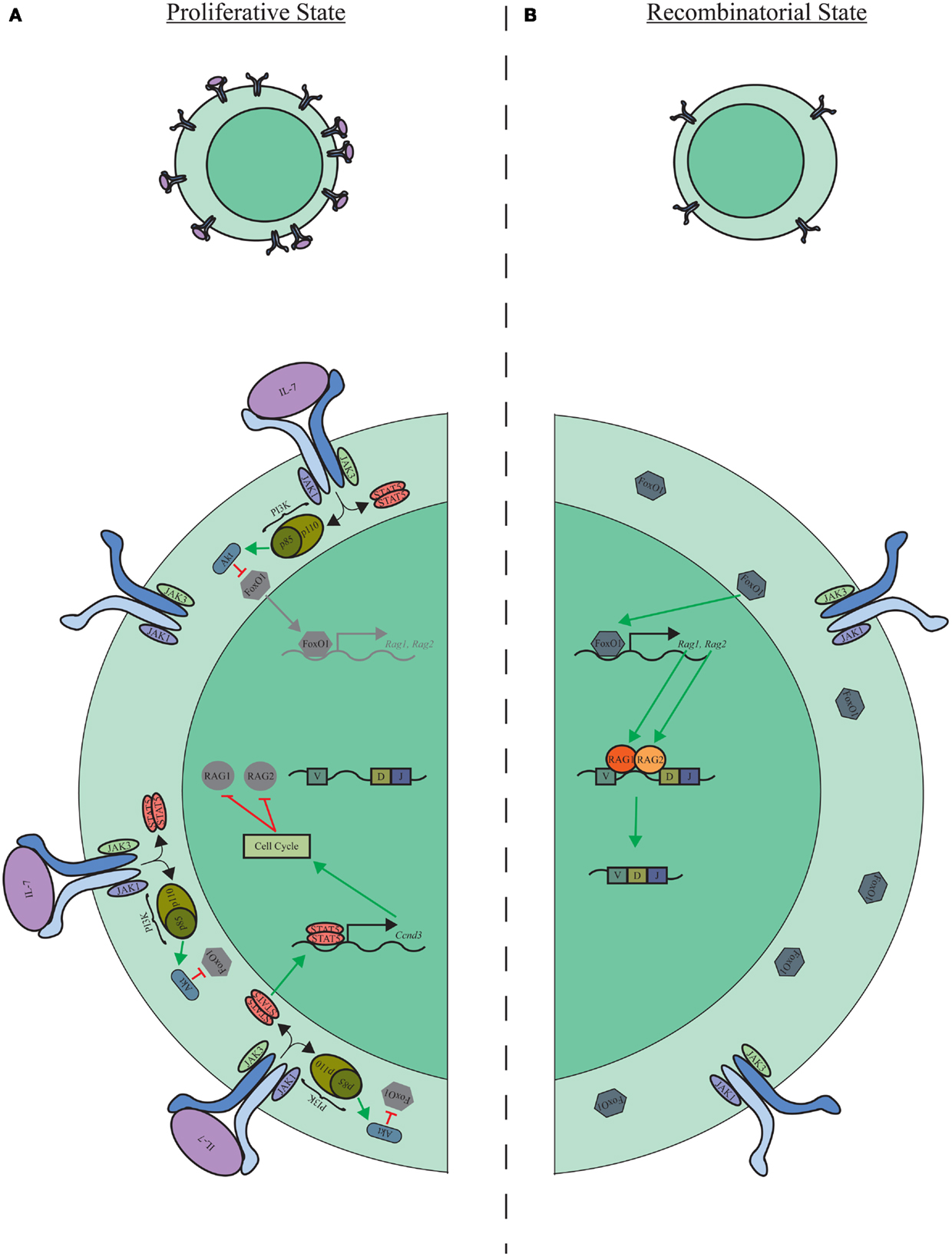
Figure 1. Proliferative and recombinatorial states of pro-B cells. (A) Elevated levels of IL-7R expression and signaling activate STAT5 and PI3K/Akt signaling modules, which enforce the proliferative program of pro-B cells while suppressing Igμ recombination. (B) Down modulation of the IL-7R is associated with a loss of proliferative signaling through STAT5 and PI3K/Akt and release of FoxO1, Rag-1, and Rag-2 suppression allowing progression of Igμ recombination.
Pre-BCR, Proliferation, and Igκ Rearrangement of Pre-B Cells
Large Pre-B Cells
Cells transition to the pre-B-cell stage when Igμ pairs with SLC components, VpreB and λ5, along with the signaling module Igα/Igβ to form the pre-BCR at the cell surface. Initial expression of the pre-BCR is associated with a proliferative burst of early pre-B cells, also known as large pre-B cells, to expand the population of cells expressing a functional Igμ. Proper expression of the pre-BCR is critical to development as deficiencies of Igα, Igβ, or surface Igμ completely arrest B-lymphopoiesis while rearrangement and expression of Igκ inefficiently rescues SLC deficiency (40–43). Activation of the pre-BCR requires the non-immunoglobulin domain of λ5, which mediates aggregation of the receptor (44–46). Although receptor aggregation is required, it is not clear if receptor aggregation is an intrinsic property of λ5 or if the SLC enables recognition of one or more selecting ligands within the BM (44, 47). Putative selecting ligands identified within the BM including heparin sulfate and galectin-1 have been suggested as natural ligands (48–50).
Concurrent to pre-BCR expression, large pre-B cells maintain IL-7R expression. It is within large pre-B cells that an additional downstream target of IL-7R signaling important for B-cell development, the phosphoinositide 3-kinase (PI3K) pathway, plays a role (51, 52). The absence of PI3K has a definitive effect on peripheral B-cell proliferation, and selective deletion of the regulatory subunit p85α or the combined catalytic subunits p110α and p110δ result in impairment of B-lymphopoiesis (53–55). However, the effects of PI3K on early B-cell proliferation appear to be within the initial proliferative events of pre-B cells, not pro-B cells. Deficiencies in p85α or PTEN, a negative regulator of PI3K does not affect the number of pro-B cells in cycle, and the defect in development in p110α- and p110δ-deficient mice begins at the pre-B-cell stage (26, 52). Compared to cycling pro-B cells, large pre-B cells are indeed larger in size and display a heightened rate of proliferation (4). PI3K may be required in large pre-B cells to support increased protein synthesis and rapid cell division through increased glucose uptake and glycolytic activity by activated Akt, downstream of PI3K (56–58). Coincidently, Akt is capable of enhancing survival by inhibiting pro-apoptotic pathways through direct repression of BAD and also indirectly by suppressing FoxO transcription factors, which induce Bim (59–62).
The pre-BCR is expressed on large pre-B cells and therefore has been thought to enhance proliferation in response to IL-7R signaling. Among, the signaling pathways common to the BCR and the IL-7R in the periphery, PI3K was an attractive candidate for any synergy that might occur between the two receptors. However, the pre-BCR does not efficiently couple to PI3K. Transfection of Rag-2−/− pro-B cells in the presence of IL-7 with a prearranged, functional Igμ resulting in pre-BCR expression does not increase phospho-Akt activation and phospho-Akt levels are similar in pro and large pre-B cells (52). Furthermore, deletions of the genes encoding several pre-BCR downstream signaling components, including BLNK (SLP-65), Btk, and phospholipase Cγ2 (PLCγ2), result in a developmental block at the cycling pre-B-cell stage (63–65). Finally, re-expression of BLNK in deficient cells induces cell cycle arrest and Igκ rearrangement (66). These observations indicate that the pre-BCR signals cell cycle exit rather than proliferation.
Therefore, the mechanisms driving the pre-B-cell proliferative burst remain unclear. It is possible that in pre-B cells, the pre-BCR has two signaling states, one pro-proliferative and one anti-proliferative (52, 67). However, the downstream effectors of such a pre-BCR-dependent proliferative pathway have yet to be identified. Alternatively, signaling mechanisms occurring independently of the pre-BCR could enhance IL-7R-mediated proliferation.
In addition to driving proliferation, signals through the IL-7R, and the downstream activation of STAT5, potently repress Igκ recombination (68). Activated STAT5 binds as a tetramer to a critical E-box-containing enhancer region of Igκ, the intronic enhancer (Eκi), and tetrameric binding enables recruitment of the polycomb repressive complex (PRC2), which represses accessibility of the Igκ region (69). Additionally, PI3K–Akt activation by the IL-7R represses recombination through indirect downregulation of Rag proteins (52). FoxO transcription factors induce Rag-1 and Rag-2 expression, however, repression of FoxO by the PI3K–Akt module inhibits Rag protein expression and inhibits recombination (70, 71). Therefore, beyond the intrinsic regulation of Rag proteins by the cell cycle machinery as described above, in large pre-B cells, IL-7R signaling through STAT5, and the PI3K–Akt module, further enforce proliferation while suppressing pre-BCR-induced recombination.
Small Pre-B Cells
The transition from highly proliferative large pre-B cells to small resting pre-B cells undergoing Igκ recombination is a pivotal point in normal B-lymphopoiesis. This transition is controlled by the signaling cascades downstream of the IL-7R and pre-BCR (Figure 2). As described below, the pre-BCR orchestrates Igκ recombination, but cannot do so while the IL-7R is transmitting signals (23, 52, 68). First cells must escape IL-7 signaling, presumably through migration toward IL-7 low niches of the BM (6). Interestingly, upregulation of the interferon regulatory factor (IRF)-4 by the pre-BCR induces the expression of the chemokine receptor CXCR4 (68). The potential presence of the CXCR4 ligand, CXCL12, outside of IL-7 niches, may provide a mechanism by which early events of the pre-BCR enables movement into relatively IL-7-deficient niches and transition from proliferation-inducing signals (IL-7R) to those driving recombination (pre-BCR).
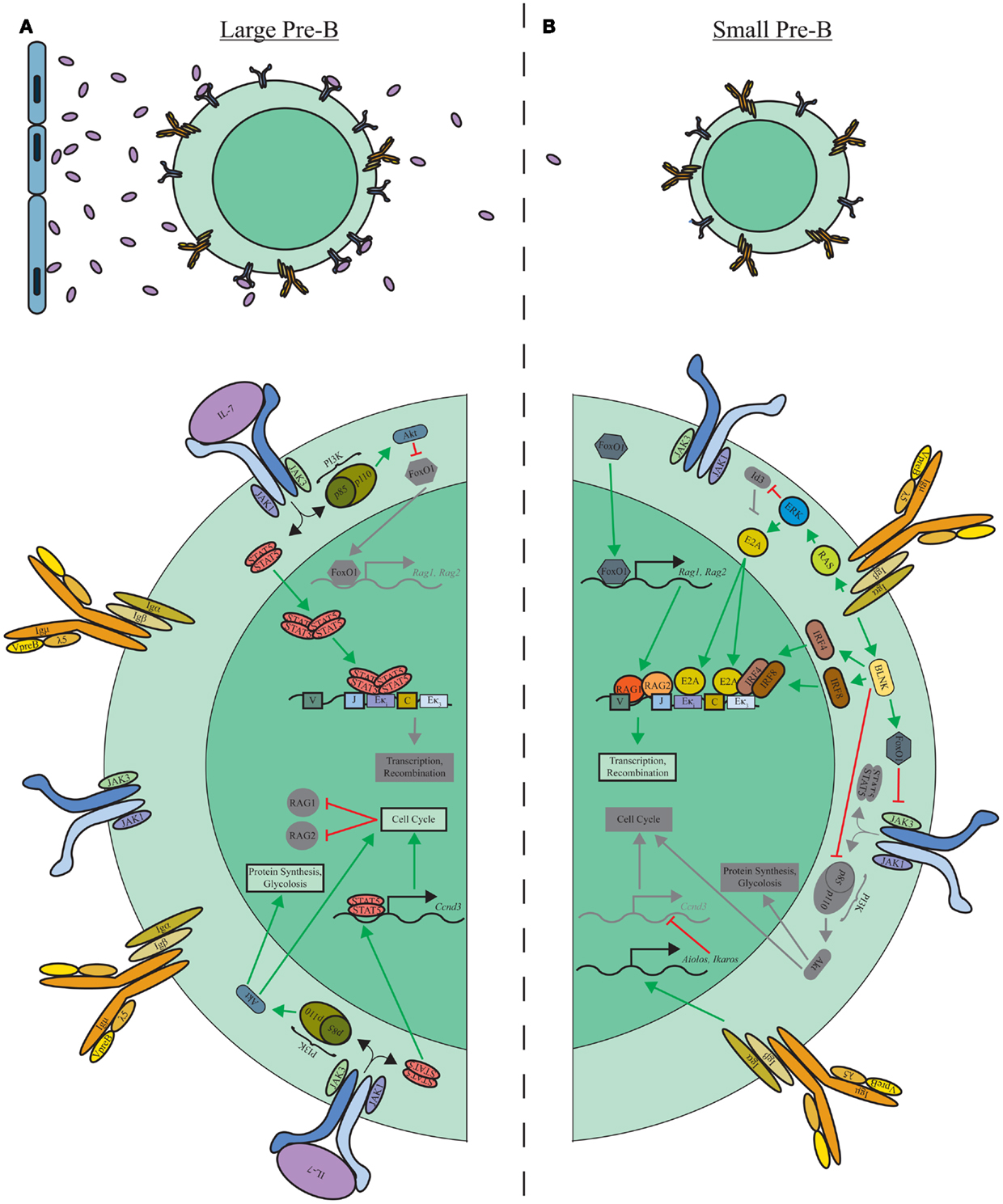
Figure 2. IL-7R and pre-BCR mediated transition of large pre-B to small pre-B cells. (A) Localization of large pre-B cells near IL-7-producing stromal cells maintains IL-7R-induced proliferation through STAT5 and PI3K/Akt signaling modules. Additionally, tetrameric STAT5 reinforces inhibition of Igκ recombination through direct binding to Eκi. (B) Migration away from IL-7-rich niches limits IL-7R signaling allowing pre-BCR-induced Ras/ERK and BLNK signaling modules to promote E2A and IRF4/IRF8 induction. Binding of these transcription factors to Igκ enhancer elements enables recombination in small pre-B cells. Additionally, the BLNK module, along with Aiolos and Ikaros, downstream of the pre-BCR inhibit proliferation by repressing IL-7R expression, PI3K/Akt activation, and Ccnd3 transcription.
The opening of the Igκ locus by the pre-BCR is predominately accomplished through activation of the Ras/Erk pathway (23, 72). Activated Erk induces E2A and inhibits the E2A repressor Id3 leading to an accumulation of free E2A within the nucleus (23, 73) that then binds the Eκi and the Igκ 3′ enhancer (Eκ3) (23). Escape from IL-7 signaling relieves tetrameric STAT5 occupancy of Eκi, allowing E2A to bind, which promotes accessibility of the Igκ loci for transcription and recombination (69). Genetic targeting of the E-boxes within Eκi has demonstrated the importance of E2A recruitment in Igκ recombination (74).
In addition to de-repressing Igκ, loss of IL-7R signaling enhances specific pre-BCR-dependent and -independent mechanisms important for Igκ recombination. Loss of IL-7R-induced PI3K–Akt activation results in increased FoxO expression. FoxO1 directly binds the Rag-1 and -2 genes and induces their expression (70). FoxO also binds and induces expression of the Syk and BLNK genes (52). The Syk/BLNK module induces the transcription factors IRF4 and 8, which bind the 3′ Igκ enhancer (Eκ3) and enhance Igκ accessibility (68, 75, 76). Furthermore, downstream of BLNK, activation of p38 MAP kinase further enhances FoxO activation thereby setting up a feed-forward loop that reinforces commitment to Igκ recombination (52).
Pre-B-cell receptor signals additionally repress the proliferative program. FoxO1 represses surface expression of IL-7R in pre-B cells, while BLNK inhibits PI3K/Akt activation (52, 71). Pre-BCR signals also induce the expression of the transcription factors Aiolos and Ikaros (77, 78). These factors impede cell cycle by repression of Myc and cyclin D3 gene expression (23, 78). Accordingly, conditional deletion of Ikaros at the pro-B-cell stage of development results in a severe block in B-lymphopoiesis with an accumulation of cycling large pre-B cells (79). Ikaros might have a direct role in Igκ recombination although the mechanisms remain to be defined (79). Collectively, downstream of the IL-7R and pre-BCR, these networks of feed-forward and feed-back mechanisms mediate the transition from proliferation to recombination and ensure sharp demarcation between each developmental state (80).
Epigenetic Regulation of Igκ Accessibility and Recombination
IL-7R and Pre-BCR Imposed Regulation of Igκ Accessibility
Chromatin structure and accessibility are fundamental to B-cell development. Recent evidence indicates that, at least in part, accessibility of Ig genes is determined by post-translational epigenetic modifications of regional histone cores. Accessibility to recombination correlates with transcription (81) and indeed the primary effectors of epigenetic remodeling are transcription factors. It has become apparent that both STAT5 and E2A regulate Igκ accessibility by determining the epigenetic landscape of the locus in pre-B cells (Figure 3). Initially, tetrameric STAT5, downstream of the IL-7R, recruits the histone methyltransferase Ezh2, which decorates the Igκ locus with repressive histone 3 lysine 27 trimethylation (H3K27me3) marks (69). Following release from STAT5-mediated repression of Igκ, E2A can access Eκi, and marks the flanking Jκ and Cκ segments with activating H3K4 trimethylation (H3K4me3) and H4 acetylation (H4Ac) to promote an open chromatin structure (69, 82).
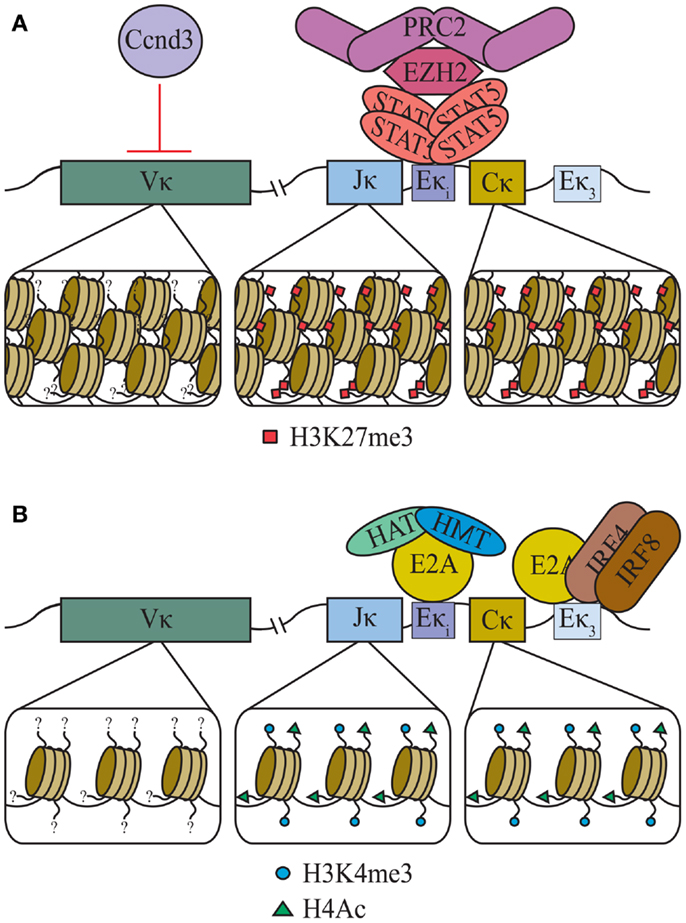
Figure 3. Epigenetic regulation of the Igκ loci. (A) In large pre-B cells, downstream of the IL-7R, tetrameric STAT5 directly binds at Eκi as a tetrameric complex. This both inhibits E2A binding and recruits the methyltransferase EZH2 and polycomb repressive complex 2 (PRC2) which decorates Jκ and Cκ with H3K27me3. Additionally, through an unknown mechanism, Cyclin D3 (Ccnd3) restricts Vκ segments’ accessibility. (B) Loss of IL-7R signaling in small pre-B cells leads to a loss of tetrameric STAT5 at Eκi which allows E2A binding and the recruitment of histone methyltransferases (HMT) and histone acetyltransferases (HAT). The resulting H3K4me3 and H4Ac marks open Jκ and Cκ to transcription and recombination.
Interestingly, the above mechanisms of epigenetic regulation apply only to Jκ and Cκ and do not extend to the extensive Vκ regions (69). In fact, the Vκ regions are relatively devoid of any measured post-translational histone modifications identified for Cκ and Jκ [unpublished data and (83)]. Surprisingly, Vκ transcription is repressed by cyclin D3, through mechanisms that do not involve direct DNA binding (26). Instead, it appears that nuclear matrix-associated cyclin D3, and not that fraction associated with CDK4/6, represses Vκ. The mechanisms by which cyclin D3 regulates Vκ transcription are not known, but might include controlling access to RNA polymerase II or nuclear positioning (84, 85). Regardless of mechanism, repression of Vκ accessibility by cyclin D3 provides a direct link between cell cycle transit and repression of Igκ recombination.
Rag-Mediated Recombination Depends upon Epigenetic Modifications
Recombination events at Igκ are also dependent on an open chromatin structure for accessibility of Rag proteins to RSS sites. RAG-mediated cleavage at RSS sites is restricted by a closed nucleosome structure (86–88). Histone modifications associated with open chromatin structures, including H3K4me3, histone 3 lysine 36 trimethylation (H3K36me3), H3Ac, and H4Ac correlate with recombination (89–91). Additionally, the recruitment of Rag-2 is dependent on the Rag-2 PHD domain binding to H3K4me3 (92, 93). The epigenetic regulation of JκCκ, and the recruitment of RAG-2 to the marks of open chromatin, is consistent with current concepts that the JκCκ region serves as the site of recombination (94). Furthermore, the JκCκ region is anchored to the nuclear matrix and anchoring is necessary for efficient Igκ recombination (95). This suggests that the recombination platform is relatively fixed and Vκ segments are recruited to it.
Although histone modifications at Jκ and Cκ have been associated with recombination and Rag-2 recruitment in vivo, there is no direct evidence that these modifications alone are capable of inducing RSS accessibility. In fact, in vitro experiments have demonstrated that hyperacetylation of histones is unable to overcome nucleosome-induced restriction of RSS sites and allow Rag-mediated recombination (87, 96). However, these extracellular in vitro experiments may lack additional lineage or stage-specific factors needed to translate epigenetic modifications into open chromatin. One such factor might be the SWI/SNF complex which can read specific epigenetic marks and open immunoglobulin gene loci for recombination (83, 97).
Concluding Remarks
Recent observations have revealed that the IL-7R and the pre-BCR regulate complex networks of signaling and transcription cascades that direct and reinforce either pre-B-cell proliferation or Igκ recombination. Central to understanding these networks is the clear demonstration that the IL-7R induces proliferation and represses Igκ recombination and these biological activities are diametrically opposed by the pre-BCR. However, several questions still remain. For instance, if IL-7R signaling is constant in pro- and pre-B cells, and the pre-BCR does not provide a proliferative signal, what then is driving the large pre-B-cell proliferative burst? Additionally, although much effort has begun to describe how fate-determining transcription factors and epigenetic modifiers prime the required epigenetic landscape, little is known about the “readers” of these marks that impose and specify B-cell developmental events. The precise relationships between Igκ transcription and recombination are unclear. Moreover, in the absence of epigenetic modifications, how is Vκ accessibility regulated? Further research into the molecular mechanisms that target and regulate the recombinatorial machinery to specific sites of the Ig loci will be critical for understanding normal and pathogenic B-lymphopoiesis.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work is supported by National Institute of Health (NIH)/National Institute of General Medical Sciences (NIGMS) grant numbers: 5R01GM088847, 5R01GM101090, and 5F32GM103143.
References
1. Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell (1997) 91:661–72. doi: 10.1016/S0092-8674(00)80453-5
2. Izon D, Rudd K, Demuth W, Pear WS, Clendenin C, Lindsley RC, et al. A common pathway for dendritic cell and early B cell development. J Immunol (2001) 167:1387–92.
3. Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol (2006) 24:541–70. doi:10.1146/annurev.immunol.23.021704.115830
4. Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med (1991) 173:1213–25. doi:10.1084/jem.173.5.1213
5. Schlissel MS. Regulating antigen-receptor gene assembly. Nat Rev Immunol (2003) 3:890–9. doi:10.1038/nri1225
6. Tokoyoda K, Egawa T, Sugiyama T, Choi BI, Nagasawa T. Cellular niches controlling B lymphocyte behavior within bone marrow during development. Immunity (2004) 20:707–18. doi:10.1016/j.immuni.2004.05.001
7. Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem (2002) 71:101–32. doi:10.1146/annurev.biochem.71.090501.150203
8. Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell (1992) 68:869–77. doi:10.1016/0092-8674(92)90030-G
9. Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell (1992) 68:855–67. doi:10.1016/0092-8674(92)90029-C
10. de Miranda NF, Bjorkman A, Pan-Hammarstrom Q. DNA repair: the link between primary immunodeficiency and cancer. Ann N Y Acad Sci (2011) 1246:50–63. doi:10.1111/j.1749-6632.2011.06322.x
11. Nishimoto N, Kubagawa H, Ohno T, Gartland GL, Stankovic AK, Cooper MD. Normal pre-B cells express a receptor complex of mu heavy chains and surrogate light-chain proteins. Proc Natl Acad Sci U S A (1991) 88:6284–8. doi:10.1073/pnas.88.14.6284
12. Melchers F. The pre-B-cell receptor: selector of fitting immunoglobulin heavy chains for the B-cell repertoire. Nat Rev Immunol (2005) 5:578–84. doi:10.1038/nri1649
13. Melchers F, Ten Boekel E, Seidl T, Kong XC, Yamagami T, Onishi K, et al. Repertoire selection by pre-B-cell receptors and B-cell receptors, and genetic control of B-cell development from immature to mature B cells. Immunol Rev (2000) 175:33–46. doi:10.1111/j.1600-065X.2000.imr017510.x
14. Keenan RA, De Riva A, Corleis B, Hepburn L, Licence S, Winkler TH, et al. Censoring of autoreactive B cell development by the pre-B cell receptor. Science (2008) 321:696–9. doi:10.1126/science.1157533
15. Eschbach C, Bach MP, Fidler I, Pelanda R, Kohler F, Rajewsky K, et al. Efficient generation of B lymphocytes by recognition of self-antigens. Eur J Immunol (2011) 41:2397–403. doi:10.1002/eji.201041344
16. Peschon JJ, Morrissey PJ, Grabstein KH, Ramsdell FJ, Maraskovsky E, Gliniak BC, et al. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J Exp Med (1994) 180:1955–60. doi:10.1084/jem.180.5.1955
17. von Freeden-Jeffry U, Vieira P, Lucian LA, Mcneil T, Burdach SE, Murray R. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J Exp Med (1995) 181:1519–26. doi:10.1084/jem.181.4.1519
18. Corcoran AE, Riddell A, Krooshoop D, Venkitaraman AR. Impaired immunoglobulin gene rearrangement in mice lacking the IL-7 receptor. Nature (1998) 391:904–7. doi:10.1038/36122
19. O’Shea JJ, Plenge R. JAK and STAT signaling molecules in immunoregulation and immune-mediated disease. Immunity (2012) 36:542–50. doi:10.1016/j.immuni.2012.03.014
20. Yao Z, Cui Y, Watford WT, Bream JH, Yamaoka K, Hissong BD, et al. Stat5a/b are essential for normal lymphoid development and differentiation. Proc Natl Acad Sci U S A (2006) 103:1000–5. doi:10.1073/pnas.0507350103
21. Weber-Nordt RM, Egen C, Wehinger J, Ludwig W, Gouilleux-Gruart V, Mertelsmann R, et al. Constitutive activation of STAT proteins in primary lymphoid and myeloid leukemia cells and in Epstein-Barr virus (EBV)-related lymphoma cell lines. Blood (1996) 88:809–16.
22. Goetz CA, Harmon IR, O’Neil JJ, Burchill MA, Farrar MA. STAT5 activation underlies IL7 receptor-dependent B cell development. J Immunol (2004) 172:4770–8.
23. Mandal M, Powers SE, Ochiai K, Georgopoulos K, Kee BL, Singh H, et al. Ras orchestrates exit from the cell cycle and light-chain recombination during early B cell development. Nat Immunol (2009) 10:1110–7. doi:10.1038/ni.1785
24. Cooper AB, Sawai CM, Sicinska E, Powers SE, Sicinski P, Clark MR, et al. A unique function for cyclin D3 in early B cell development. Nat Immunol (2006) 7:489–97. doi:10.1038/ni1324
25. Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev (1999) 13:1501–12. doi:10.1101/gad.13.12.1501
26. Powers SE, Mandal M, Matsuda S, Miletic AV, Cato MH, Tanaka A, et al. Subnuclear cyclin D3 compartments and the coordinated regulation of proliferation and immunoglobulin variable gene repression. J Exp Med (2012) 209:2199–213. doi:10.1084/jem.20120800
27. Lam EW, Glassford J, Banerji L, Thomas NS, Sicinski P, Klaus GG. Cyclin D3 compensates for loss of cyclin D2 in mouse B-lymphocytes activated via the antigen receptor and CD40. J Biol Chem (2000) 275:3479–84. doi:10.1074/jbc.275.5.3479
28. Solvason N, Wu WW, Parry D, Mahony D, Lam EW, Glassford J, et al. Cyclin D2 is essential for BCR-mediated proliferation and CD5 B cell development. Int Immunol (2000) 12:631–8. doi:10.1093/intimm/12.5.631
29. Malin S, McManus S, Cobaleda C, Novatchkova M, Delogu A, Bouillet P, et al. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat Immunol (2010) 11:171–9. doi:10.1038/ni.1827
30. Bednarski JJ, Nickless A, Bhattacharya D, Amin RH, Schlissel MS, Sleckman BP. RAG-induced DNA double-strand breaks signal through Pim2 to promote pre-B cell survival and limit proliferation. J Exp Med (2012) 209:11–7. doi:10.1084/jem.20112078
31. Bassing CH, Alt FW. The cellular response to general and programmed DNA double strand breaks. DNA Repair (2004) 3:781–96. doi:10.1016/j.dnarep.2004.06.001
32. Zhang Y, Gostissa M, Hildebrand DG, Becker MS, Boboila C, Chiarle R, et al. The role of mechanistic factors in promoting chromosomal translocations found in lymphoid and other cancers. Adv Immunol (2010) 106:93–133. doi:10.1016/S0065-2776(10)06004-9
33. Lin WC, Desiderio S. Cell cycle regulation of V(D)J recombination-activating protein RAG-2. Proc Natl Acad Sci U S A (1994) 91:2733–7. doi:10.1073/pnas.91.7.2733
34. Lee J, Desiderio S. Cyclin A/CDK2 regulates V(D)J recombination by coordinating RAG-2 accumulation and DNA repair. Immunity (1999) 11:771–81. doi:10.1016/S1074-7613(00)80151-X
35. Jiang H, Chang FC, Ross AE, Lee J, Nakayama K, Desiderio S. Ubiquitylation of RAG-2 by Skp2-SCF links destruction of the V(D)J recombinase to the cell cycle. Mol Cell (2005) 18:699–709. doi:10.1016/j.molcel.2005.05.011
36. Zhang L, Reynolds TL, Shan X, Desiderio S. Coupling of V(D)J recombination to the cell cycle suppresses genomic instability and lymphoid tumorigenesis. Immunity (2011) 34:163–74. doi:10.1016/j.immuni.2011.02.003
37. Difilippantonio MJ, Zhu J, Chen HT, Meffre E, Nussenzweig MC, Max EE, et al. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature (2000) 404:510–4. doi:10.1038/35006670
38. Gao Y, Ferguson DO, Xie W, Manis JP, Sekiguchi J, Frank KM, et al. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature (2000) 404:897–900. doi:10.1038/35009138
39. Johnson K, Chaumeil J, Micsinai M, Wang JM, Ramsey LB, Baracho GV, et al. IL-7 functionally segregates the pro-B cell stage by regulating transcription of recombination mediators across cell cycle. J Immunol (2012) 188:6084–92. doi:10.4049/jimmunol.1200368
40. Kitamura D, Roes J, Kuhn R, Rajewsky K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature (1991) 350:423–6. doi:10.1038/350423a0
41. Gong S, Nussenzweig MC. Regulation of an early developmental checkpoint in the B cell pathway by Ig beta. Science (1996) 272:411–4. doi:10.1126/science.272.5260.411
42. Pelanda R, Braun U, Hobeika E, Nussenzweig MC, Reth M. B cell progenitors are arrested in maturation but have intact VDJ recombination in the absence of Ig-alpha and Ig-beta. J Immunol (2002) 169:865–72.
43. Shimizu T, Mundt C, Licence S, Melchers F, Martensson IL. VpreB1/VpreB2/lambda 5 triple-deficient mice show impaired B cell development but functional allelic exclusion of the IgH locus. J Immunol (2002) 168:6286–93.
44. Ohnishi K, Melchers F. The nonimmunoglobulin portion of lambda5 mediates cell-autonomous pre-B cell receptor signaling. Nat Immunol (2003) 4:849–56. doi:10.1038/ni959
45. Bankovich AJ, Raunser S, Juo ZS, Walz T, Davis MM, Garcia KC. Structural insight into pre-B cell receptor function. Science (2007) 316:291–4. doi:10.1126/science.1139412
46. Vettermann C, Herrmann K, Albert C, Roth E, Bosl MR, Jack HM. A unique role for the lambda5 nonimmunoglobulin tail in early B lymphocyte development. J Immunol (2008) 181:3232–42.
47. Kohler F, Hug E, Eschbach C, Meixlsperger S, Hobeika E, Kofer J, et al. Autoreactive B cell receptors mimic autonomous pre-B cell receptor signaling and induce proliferation of early B cells. Immunity (2008) 29:912–21. doi:10.1016/j.immuni.2008.10.013
48. Gauthier L, Rossi B, Roux F, Termine E, Schiff C. Galectin-1 is a stromal cell ligand of the pre-B cell receptor (BCR) implicated in synapse formation between pre-B and stromal cells and in pre-BCR triggering. Proc Natl Acad Sci U S A (2002) 99:13014–9. doi:10.1073/pnas.202323999
49. Bradl H, Wittmann J, Milius D, Vettermann C, Jack HM. Interaction of murine precursor B cell receptor with stroma cells is controlled by the unique tail of lambda 5 and stroma cell-associated heparan sulfate. J Immunol (2003) 171:2338–48.
50. Mourcin F, Breton C, Tellier J, Narang P, Chasson L, Jorquera A, et al. Galectin-1-expressing stromal cells constitute a specific niche for pre-BII cell development in mouse bone marrow. Blood (2011) 117:6552–61. doi:10.1182/blood-2010-12-323113
51. Corcoran AE, Smart FM, Cowling RJ, Crompton T, Owen MJ, Venkitaraman AR. The interleukin-7 receptor alpha chain transmits distinct signals for proliferation and differentiation during B lymphopoiesis. EMBO J (1996) 15:1924–32.
52. Ochiai K, Maienschein-Cline M, Mandal M, Triggs JR, Bertolino E, Sciammas R, et al. A self-reinforcing regulatory network triggered by limiting IL-7 activates pre-BCR signaling and differentiation. Nat Immunol (2012) 13:300–7. doi:10.1038/ni.2210
53. Fruman DA, Snapper SB, Yballe CM, Davidson L, Yu JY, Alt FW, et al. Impaired B cell development and proliferation in absence of phosphoinositide 3-kinase p85alpha. Science (1999) 283:393–7. doi:10.1126/science.283.5400.393
54. Suzuki H, Terauchi Y, Fujiwara M, Aizawa S, Yazaki Y, Kadowaki T, et al. Xid-like immunodeficiency in mice with disruption of the p85alpha subunit of phosphoinositide 3-kinase. Science (1999) 283:390–2. doi:10.1126/science.283.5400.390
55. Okkenhaug K, Bilancio A, Farjot G, Priddle H, Sancho S, Peskett E, et al. Impaired B and T cell antigen receptor signaling in p110delta PI 3-kinase mutant mice. Science (2002) 297:1031–4. doi:10.1126/science.1073560
56. Gottlob K, Majewski N, Kennedy S, Kandel E, Robey RB, Hay N. Inhibition of early apoptotic events by Akt/PKB is dependent on the first committed step of glycolysis and mitochondrial hexokinase. Genes Dev (2001) 15:1406–18. doi:10.1101/gad.889901
57. Rathmell JC, Fox CJ, Plas DR, Hammerman PS, Cinalli RM, Thompson CB. Akt-directed glucose metabolism can prevent Bax conformation change and promote growth factor-independent survival. Mol Cell Biol (2003) 23:7315–28. doi:10.1128/MCB.23.20.7315-7328.2003
58. Doughty CA, Bleiman BF, Wagner DJ, Dufort FJ, Mataraza JM, Roberts MF, et al. Antigen receptor-mediated changes in glucose metabolism in B lymphocytes: role of phosphatidylinositol 3-kinase signaling in the glycolytic control of growth. Blood (2006) 107:4458–65. doi:10.1182/blood-2005-12-4788
59. Datta K, Bellacosa A, Chan TO, Tsichlis PN. Akt is a direct target of the phosphatidylinositol 3-kinase. Activation by growth factors, v-src and v-Ha-ras, in Sf9 and mammalian cells. J Biol Chem (1996) 271:30835–9. doi:10.1074/jbc.271.48.30835
60. Essafi A, Fernandez De Mattos S, Hassen YA, Soeiro I, Mufti GJ, Thomas NS, et al. Direct transcriptional regulation of Bim by FoxO3a mediates STI571-induced apoptosis in Bcr-Abl-expressing cells. Oncogene (2005) 24:2317–29. doi:10.1038/sj.onc.1208421
61. Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene (2005) 24:7410–25. doi:10.1038/sj.onc.1209086
62. Huntington ND, Labi V, Cumano A, Vieira P, Strasser A, Villunger A, et al. Loss of the pro-apoptotic BH3-only Bcl-2 family member Bim sustains B lymphopoiesis in the absence of IL-7. Int Immunol (2009) 21:715–25. doi:10.1093/intimm/dxp043
63. Jumaa H, Wollscheid B, Mitterer M, Wienands J, Reth M, Nielsen PJ. Abnormal development and function of B lymphocytes in mice deficient for the signaling adaptor protein SLP-65. Immunity (1999) 11:547–54. doi:10.1016/S1074-7613(00)80130-2
64. Bai L, Chen Y, He Y, Dai X, Lin X, Wen R, et al. Phospholipase Cgamma2 contributes to light-chain gene activation and receptor editing. Mol Cell Biol (2007) 27:5957–67. doi:10.1128/MCB.02273-06
65. Xu S, Lee KG, Huo J, Kurosaki T, Lam KP. Combined deficiencies in Bruton tyrosine kinase and phospholipase Cgamma2 arrest B-cell development at a pre-BCR+ stage. Blood (2007) 109:3377–84. doi:10.1182/blood-2006-07-036418
66. Flemming A, Brummer T, Reth M, Jumaa H. The adaptor protein SLP-65 acts as a tumor suppressor that limits pre-B cell expansion. Nat Immunol (2003) 4:38–43. doi:10.1038/ni862
67. Sandoval GJ, Graham DB, Bhattacharya D, Sleckman BP, Xavier RJ, Swat W. Cutting edge: cell-autonomous control of IL-7 response revealed in a novel stage of precursor B cells. J Immunol (2013) 190:2485–9. doi:10.4049/jimmunol.1203208
68. Johnson K, Hashimshony T, Sawai CM, Pongubala JM, Skok JA, Aifantis I, et al. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity (2008) 28:335–45. doi:10.1016/j.immuni.2007.12.019
69. Mandal M, Powers SE, Maienschein-Cline M, Bartom ET, Hamel KM, Kee BL, et al. Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nat Immunol (2011) 12:1212–20. doi:10.1038/ni.2136
70. Amin RH, Schlissel MS. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat Immunol (2008) 9:613–22. doi:10.1038/ni.1612
71. Herzog S, Hug E, Meixlsperger S, Paik JH, Depinho RA, Reth M, et al. SLP-65 regulates immunoglobulin light chain gene recombination through the PI(3)K-PKB-Foxo pathway. Nat Immunol (2008) 9:623–31. doi:10.1038/ni.1616
72. Shaw AC, Swat W, Davidson L, Alt FW. Induction of Ig light chain gene rearrangement in heavy chain-deficient B cells by activated Ras. Proc Natl Acad Sci U S A (1999) 96:2239–43. doi:10.1073/pnas.96.5.2239
73. Kee BL, Quong MW, Murre C. E2A proteins: essential regulators at multiple stages of B-cell development. Immunol Rev (2000) 175:138–49. doi:10.1111/j.1600-065X.2000.imr017514.x
74. Inlay MA, Tian H, Lin T, Xu Y. Important roles for E protein binding sites within the immunoglobulin kappa chain intronic enhancer in activating Vkappa Jkappa rearrangement. J Exp Med (2004) 200:1205–11. doi:10.1084/jem.20041135
75. Lu R, Medina KL, Lancki DW, Singh H. IRF-4,8 orchestrate the pre-B-to-B transition in lymphocyte development. Genes Dev (2003) 17:1703–8. doi:10.1101/gad.1104803
76. Ma S, Pathak S, Trinh L, Lu R. Interferon regulatory factors 4 and 8 induce the expression of Ikaros and Aiolos to down-regulate pre-B-cell receptor and promote cell-cycle withdrawal in pre-B-cell development. Blood (2008) 111:1396–403. doi:10.1182/blood-2007-08-110106
77. Thompson EC, Cobb BS, Sabbattini P, Meixlsperger S, Parelho V, Liberg D, et al. Ikaros DNA-binding proteins as integral components of B cell developmental-stage-specific regulatory circuits. Immunity (2007) 26:335–44. doi:10.1016/j.immuni.2007.02.010
78. Ma S, Pathak S, Mandal M, Trinh L, Clark MR, Lu R. Ikaros and Aiolos inhibit pre-B-cell proliferation by directly suppressing c-Myc expression. Mol Cell Biol (2010) 30:4149–58. doi:10.1128/MCB.00224-10
79. Heizmann B, Kastner P, Chan S. Ikaros is absolutely required for pre-B cell differentiation by attenuating IL-7 signals. J Exp Med (2013) 210:2823–32. doi:10.1084/jem.20131735
80. Clark MR, Mandal M, Ochiai K, Singh H. Orchestrating B cell lymphopoiesis through interplay of IL-7 receptor and pre-B cell receptor signalling. Nat Rev Immunol (2013) 14:69–80. doi:10.1038/nri3570
81. Amin RH, Cado D, Nolla H, Huang D, Shinton SA, Zhou Y, et al. Biallelic, ubiquitous transcription from the distal germline Ig{kappa} locus promoter during B cell development. Proc Natl Acad Sci U S A (2009) 106:522–7. doi:10.1073/pnas.0808895106
82. Beck K, Peak MM, Ota T, Nemazee D, Murre C. Distinct roles for E12 and E47 in B cell specification and the sequential rearrangement of immunoglobulin light chain loci. J Exp Med (2009) 206:2271–84. doi:10.1084/jem.20090756
83. Du H, Ishii H, Pazin MJ, Sen R. Activation of 12/23-RSS-dependent RAG cleavage by hSWI/SNF complex in the absence of transcription. Mol Cell (2008) 31:641–9. doi:10.1016/j.molcel.2008.08.012
84. Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, et al. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science (2002) 296:158–62. doi:10.1126/science.1068768
85. Fitzsimmons SP, Bernstein RM, Max EE, Skok JA, Shapiro MA. Dynamic changes in accessibility, nuclear positioning, recombination, and transcription at the Ig kappa locus. J Immunol (2007) 179:5264–73.
86. Kwon J, Imbalzano AN, Matthews A, Oettinger MA. Accessibility of nucleosomal DNA to V(D)J cleavage is modulated by RSS positioning and HMG1. Mol Cell (1998) 2:829–39. doi:10.1016/S1097-2765(00)80297-X
87. Golding A, Chandler S, Ballestar E, Wolffe AP, Schlissel MS. Nucleosome structure completely inhibits in vitro cleavage by the V(D)J recombinase. EMBO J (1999) 18:3712–23. doi:10.1093/emboj/18.13.3712
88. Baumann M, Mamais A, Mcblane F, Xiao H, Boyes J. Regulation of V(D)J recombination by nucleosome positioning at recombination signal sequences. EMBO J (2003) 22:5197–207. doi:10.1093/emboj/cdg487
89. McMurry MT, Krangel MS. A role for histone acetylation in the developmental regulation of VDJ recombination. Science (2000) 287:495–8. doi:10.1126/science.287.5452.495
90. Perkins EJ, Kee BL, Ramsden DA. Histone 3 lysine 4 methylation during the pre-B to immature B-cell transition. Nucleic Acids Res (2004) 32:1942–7. doi:10.1093/nar/gkh523
91. Abarrategui I, Krangel MS. Noncoding transcription controls downstream promoters to regulate T-cell receptor alpha recombination. EMBO J (2007) 26:4380–90. doi:10.1038/sj.emboj.7601866
92. Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity (2007) 27:561–71. doi:10.1016/j.immuni.2007.09.005
93. Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature (2007) 450:1106–10. doi:10.1038/nature06431
94. Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol (2011) 11:251–63. doi:10.1038/nri2941
95. Xu Y, Davidson L, Alt FW, Baltimore D. Deletion of the Ig kappa light chain intronic enhancer/matrix attachment region impairs but does not abolish V kappa J kappa rearrangement. Immunity (1996) 4:377–85. doi:10.1016/S1074-7613(00)80251-4
96. McBlane F, Boyes J. Stimulation of V(D)J recombination by histone acetylation. Curr Biol (2000) 10:483–6. doi:10.1016/S0960-9822(00)00449-8
Keywords: B cells, lymphopoiesis, recombination, proliferation, epigenetics
Citation: Hamel KM, Mandal M, Karki S and Clark MR (2014) Balancing proliferation with Igκ recombination during B-lymphopoiesis. Front. Immunol. 5:139. doi: 10.3389/fimmu.2014.00139
Received: 17 January 2014; Paper pending published: 28 February 2014;
Accepted: 19 March 2014; Published online: 02 April 2014.
Edited by:
Ananda L. Roy, Tufts University School of Medicine, USAReviewed by:
Roberta Pelanda, National Jewish Health and University of Colorado, USABonnie B. Blomberg, University of Miami Miller School of Medicine, USA
Copyright: © 2014 Hamel, Mandal, Karki and Clark. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marcus R. Clark, Department of Medicine, Section of Rheumatology, Gwen Knapp Center for Lupus and Immunology Research, The University of Chicago, 924 East 57th Street, Chicago, IL 60637, USA e-mail:bWNsYXJrQG1lZGljaW5lLmJzZC51Y2hpY2Fnby5lZHU=
 Keith M. Hamel
Keith M. Hamel Malay Mandal
Malay Mandal Marcus R. Clark
Marcus R. Clark