- 1Department of Human Genetics, McGill University, Montreal, QC, Canada
- 2Complex Traits Group, McGill University, Montreal, QC, Canada
- 3Department of Medicine, McGill University, Montreal, QC, Canada
Typhoid fever and salmonellosis, which are caused by Salmonella typhi and typhimurium, respectively, are responsible for significant morbidity and mortality in both developed and developing countries. We model typhoid fever using mice infected with Salmonella typhimurium, which results in a systemic disease, whereby the outcome of infection is variable in different inbred strains of mice. This model recapitulates several clinical aspects of the human disease and allows the study of the host response to Salmonella typhimurium infection in vivo. Previous work in our laboratory has identified three loci (Ity, Ity2, and Ity3) in the wild-derived MOLF/Ei mice influencing survival after infection with Salmonella typhimurium. Fine mapping of the Ity3 locus indicated that two sub-loci contribute collectively to the susceptibility of B6.MOLF-Ity/Ity3 congenic mice to Salmonella infection. In the current paper, we provided further evidence supporting a role for Ncf2 (neutrophil cytosolic factor 2 a subunit of NADPH oxidase) as the gene underlying the Ity3.1 sub-locus. Gene expression profiling indicated that the Ity3.1 sub-locus defined a global gene expression signature with networks articulated around Ncf2. Furthermore, based on differential expression and complementation analysis using Selp (selectin-P) knock-out mice, Selp was identified as a strong candidate gene for the Ity3.2 sub-locus.
Introduction
Salmonella enterica, an intracellular Gram-negative bacterium, is the causative agent for a wide spectrum of clinical diseases with manifestations ranging from asymptomatic carriers, self-limiting gastroenteritis to fatal systemic infection (1, 2). There are over 2500 Salmonella serovars, of which, some are host adapted such as serovar Typhi and Paratyphi in humans, while others, such as Typhimurium and Enteritidis, have a broad host range and are capable of infecting multiple organisms. In humans, Salmonella typhi causes a systemic disease, typhoid fever, which has a global health burden of 26.9 million cases and 200,000 deaths annually (3). In humans, Salmonella typhimurium is the causative agent of salmonellosis, a self-limiting gastroenteritis that results from the consumption of contaminated food or water. The emergence of multi-drug resistant strains of Salmonella in recent years highlights the need for a more comprehensive understanding of the pathogenesis of Salmonella infection and for the identification of novel drug targets for vaccines and therapeutics (4).
Salmonella is a natural pathogen of mice and infection with Salmonella typhimurium results in a typhoid-like systemic disease. This murine experimental model has been used to identify several genes and pathways involved in disease pathogenesis (5–9). As there is limited genetic variation within the classical inbred strains, the use of wild-derived strains of mice, such as MOLF/Ei contributes added genetic diversity and has allowed for the identification of novel genes that play an important role in innate immunity (10–13). Classical and wild-derived strains of mice exhibit a range of susceptibilities to Salmonella infection; for example, the C57BL/6J classical inbred strain are extremely susceptible to infection with Salmonella typhimurium due to a mutation in Slc11a1 (solute carrier family 11 member 1), while the 129 sub-strains are highly resistant (14). The wild-derived mouse strain, MOLF/Ei is also susceptible to infection despite carrying functional copies of genes known to be important in Salmonella infection, such as Slc11a1 and Tlr4 (toll-like receptor 4) (6, 10).
In order to identify the genetic determinants involved in the susceptibility of MOLF/Ei mice to Salmonella infection, we have previously used linkage analysis in an F2 panel of (C57BL/6 × MOLF/Ei) mice to identify two loci linked to host defense against Salmonella typhimurium, Ity2 (Immunity to Typhimurium locus 2) and Ity3 (10, 12). The MOLF/Ei allele at the Ity2 locus improves resistance to infection, whereas MOLF/Ei allele at the Ity3 locus confers susceptibility (15). Validation and fine mapping of Ity3 locus were done using congenic B6.MOLF-Ity/Ity3 mice (12) and a panel of 12 sub-congenic mice (16). Using this approach, the Ity3 locus was refined to a 24 Mb interval and was shown to carry two sub-loci, Ity3.1 and Ity3.2 that together contribute to increased susceptibility to infection (16). The Ity3.1 sub-locus controls NADPH oxidase activity and is characterized by decreased reactive oxygen species (ROS) production, reduced inflammatory cytokine response, and increased bacterial burden. The Ity3.2 sub-locus is characterized by a hyper-responsive inflammatory cytokine phenotype after exposure to Salmonella (16). Sequencing, expression, and functional data support the candidacy of Ncf2 (neutrophil cytosolic factor 2 a subunit of NADPH oxidase) as the gene underlying the Ity3.1 sub-locus (13).
In the current study, we used global expression profiling to better understand the genetic networks that are being influenced by the Ity3 sub-loci and to identify potential candidate genes for the Ity3.2 sub-locus. We illustrate the impact of the Ity3.1 sub-locus on cell death and cytoskeletal reorganization, hematopoiesis as well as propose the candidacy of Selp (selectin P) as one of the candidate genes underlying Ity3.2 based on expression analysis, coding sequence polymorphism, and functional and allelic complementation studies.
Materials and Methods
Ethics Statement
All animals were maintained at the Animal Care Facility of McGill University according to the guidelines of the Canadian Council on Animal Care (CCAC). The animal protocol for this study was approved by the McGill University Animal Care Committee.
Animals
Classical inbred strain C57BL/6J and wild-derived MOLF/Ei mice were used to generate congenic, B6.MOLF-Ity and B6.MOLF-Ity/Ity3 and sub-congenic mice as described previously (12, 16). The susceptible Ity3 and resistant Ity mice, as well as the intermediate B6.MOLF-Ity/Ity3.RecN and B6.MOLF-Ity/Ity3.RecG mice were used for the microarray expression analysis, while the B6.MOLF-Ity and B6.MOLF-Ity/Ity3, crossed with B6.129S7-Selptm1Bay/J ordered from the Jackson Lab (Bar Harbor, ME, USA), were used for the complementation assay.
In vivo Salmonella Infection
Mice aged 7–12 weeks were infected with Salmonella typhimurium strain Keller as described previously (12, 16). Briefly, mice were inoculated with 0.2 ml of physiological saline containing 103 colony-forming units of bacteria through the caudal vein. The infectious dose was verified by serial dilutions on trypticase soy agar. Mice were either monitored for survival or euthanized at day 3 or day 5 post-infection for organ collection. The animals were monitored two to three times daily and mice showing body condition scoring <2.0 were used for clinical endpoint (17). Survival analysis was conducted using a Kaplan–Meier survival test.
Microarray Expression Analysis
RNA was extracted from the spleens of mice, which were collected before infection and at day 3 post-infection. The RNA extraction was carried out using TRIzol reagent (Invitrogen Canada, Inc., Burlington, ON, Canada). Three age-matched male mice were used per group. The concentration of RNA was determined using a NanoDrop spectrophotometer (Thermo-Fisher Scientific, Waltham, MA, USA). All hybridization and scanning of mice microarrays were carried out at the McGill University and Genome Quebec Innovation Centre, using the Illumina BeadArray technology (Illumnia Inc., San Diego, CA, USA). The expression data were analyzed using FlexArray and normalized using a Lumi algorithm (Illumnia). Following the normalization, two approaches were used to generate a list of genes differentially expressed across the various strains. First, a Cyber t-test (Baladi and Long) (18) was used to generate a list of genes differentially expressed during infection, by comparing the fold change in expression of each gene between infected and uninfected samples for each strain. These gene lists were further refined using the Benjamini Hochberg false discovery rate algorithm. Genes with an FDR p-value <0.1 and a fold change of >2 were used to generate a final list of genes, which represented genes differentially regulated in each strain upon infection.
A second approach was used to generate a list of genes that were differentially expressed at each time point, as compared to the control Ity strain. This was done by comparing the expression of genes in each strain to the control Ity strain at both day 0 and day 3 post-infection. The gene lists were further refined using the Benjamini Hochberg false discovery rate algorithm. Genes with an FDR p-value of <0.05 was used as a cut-off to characterize genes as significantly differentially regulated as compared to Ity. The gene lists generated using the two approaches were studied using a suite of online tools including DAVID (19), GeneGo (MetaCore, Thomson Reuters) and Gene Mania (20, 21).
Sequencing of Selp
Sequencing was performed on PCR-amplified cDNA from Ity and Ity3 congenics obtained by reverse transcription of Trizol spleen-extracted RNA to determine genetic variation between C57BL/6J and MOLF/Ei alleles of the Selp candidate gene. Sanger sequencing was completed at the McGill University and Génome Québec Innovation Centre.
Allelic Complementation Assay
In order to study the effect of a MOLF/Ei Selp allele on susceptibility to infection, we carried out a complementation cross. Selp knock-out mice, B6.129S2-Selptm1Hyn/J (Selp−/−) were ordered from the Jackson Laboratories (Bar Harbor, ME, USA). These mice were on C57BL/6J background with a mutant Slc11a1 allele. In order to correct for this, we crossed the Selp−/− mice to Ity as well as to the Ity3 mice. Mice were genotyped for the Selp−/− allele, and mice carrying the MOLF/Ei Slc11a1 allele along with the Selp−/− allele were inter-crossed to generate homozygous Selp−/− mice with a MOLF/Ei allele at the Slc11a1 gene. Furthermore, these mice were crossed with Ity or Ity3 mice to generate mice, which are homozygous wild-type at Slc11a1, but carry a Selp knock-out allele complemented by either a C57BL/6J or MOLF/Ei allele (Figure S1 in Supplementary Material).
Bacterial Load Enumeration
For bacterial burden quantification in the spleen and the liver, mice were euthanized using CO2 and at the required day post-infection; both organs were removed aseptically, weighed and homogenized using a Polytron (Kinematica, Bohemia, NY, USA). The resulting homogenate was diluted in 0.9% saline and plated on tryptic soy agar to determine organ bacteria burden.
Statistical Analysis
Statistical analysis was performed using Graph Pad Prism 6 (GraphPad Software, San Diego, CA, USA). One-way ANOVA with Dunnet’s multiple correction test was used to analyze the bacterial burden in the spleen. A corrected p-value <0.05 was used to establish significant differences.
Results
Ity3 Influences the Expression of Specific Genes and Pathways during Infection
The Ity3 locus is a complex QTL containing at least two sub-loci. We have previously studied the phenotypic impact of these two sub-loci (16), and in order to further characterize the genetic networks that are affected by the MOLF/Ei allele at the Ity3 locus, we have used a microarray expression approach. Using this approach, we should be able to characterize and identify candidate genes within each sub-loci and identify pathways, which affect the host immune response to Salmonella infection. We studied the splenic global expression profile of infected and uninfected congenic resistant B6.MOLF-Ity (Ity), susceptible B6.MOLF-Ity/Ity3 (Ity3), and Ity3.RecG and Ity3.RecN strains. These two sub-congenic strains were selected because they carry either the MOLF/Ei alleles at Ity3.1 (Ity3.RecG) or at Ity3.2 sub-locus (Ity3.RecN), which result in an intermediate survival phenotype after infection with Salmonella typhimurium (16). Ity, Ity3, Ity3.RecN, and Ity3.RecG mice show significant differences in spleen and liver bacterial burden by day 5 post-infection (16). To evaluate primary defects in gene expression rather than differences due to high bacterial load, we selected day 3 as the time point to be studied because there was no significant difference in the spleen bacterial burden among the four strains of mice (16). Therefore, any changes in gene expression will serve as an indicator for genes and pathways that are important in regulating the susceptibility to Salmonella infection and not a consequence of a difference in bacterial load within strains. Our aim was to identify transcriptional signatures common to both Ity3 and Ity3.RecG to define pathways controlled by the Ity3.1 locus and those similarly regulated by Ity3 and Ity3. RecN to identify transcriptional networks controlled by the Ity3.2 locus. We used two approaches to study the gene expression profiles (Figure 1).
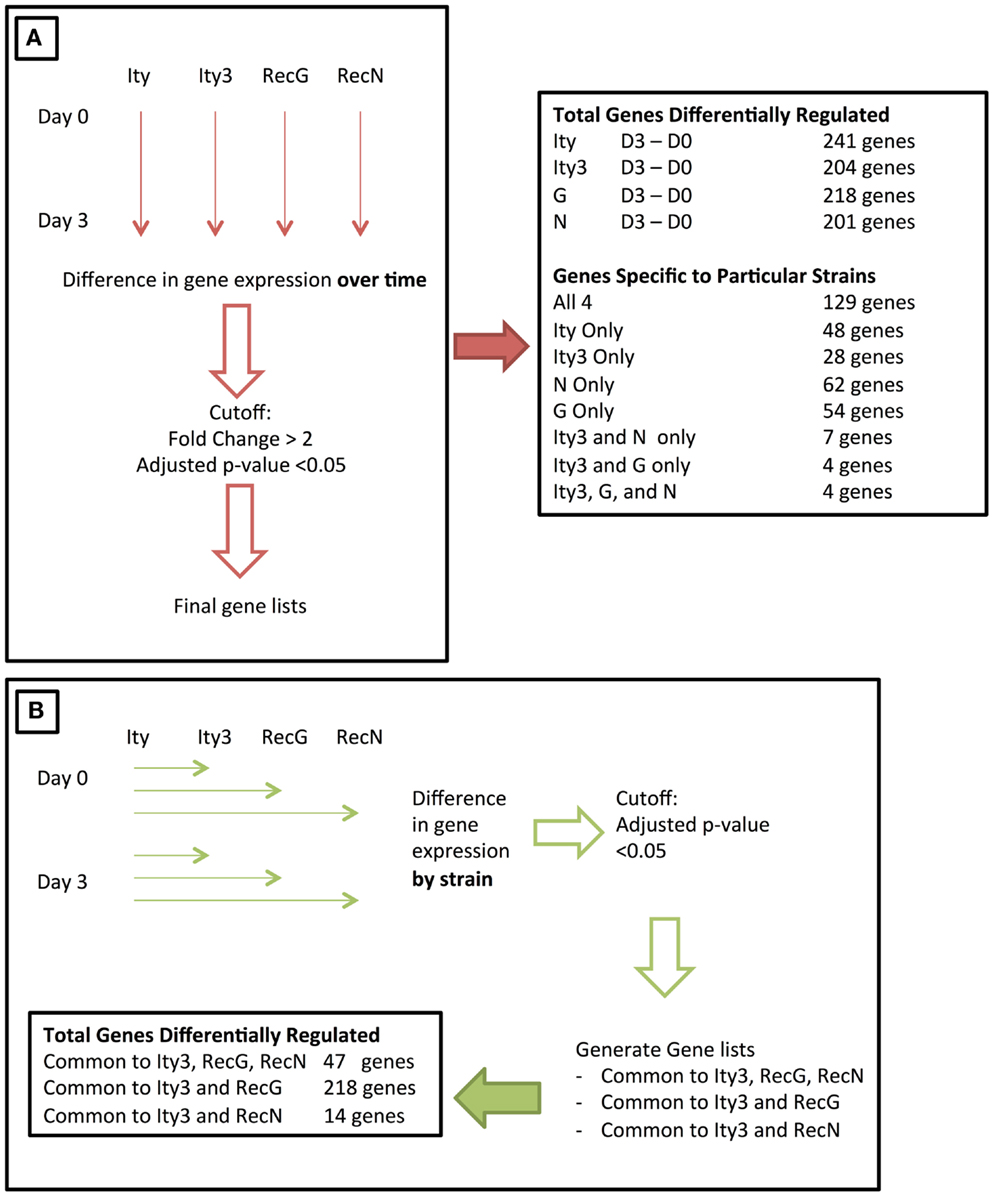
Figure 1. Figure illustrating the two approaches used to study the changes in gene expression during infection of congenic and sub-congenic mice. (A) A representation of an initial approach used for studying the change in gene expression over time. The gene expression at day 3 post-infection was compared to day 0, and the final number of genes differentially regulated is stated on the right. (B) The second approach used, where gene expression in each strain was compared to the control Ity strain, at both day 0 and day 3. The final number of genes within each category is shown at the bottom of the panel.
As an initial approach, we studied the changes in splenic gene expression in each strain over time (Day 3–Day 0). Overall, 241 (Ity), 204 (Ity3), 218 (Ity3.RecG), and 201 (Ity3.RecN) genes were differentially expressed during infection as defined by a cut-off of a fold change >2 and a p-value <0.1 (Tables S1A–D in Supplementary Material; Figure 1). We identified 129 genes that were commonly and significantly regulated during infection in all four strains including a number of pro-inflammatory genes (Table S1E in Supplementary Material). The majority of these genes were chemokines, cytokines, and other immune related genes with a significant number of genes regulated by Type I and Type II IFN including several members of the Gbp family, Stat1, Usp18 as well as others that are known to be involved in Salmonella infection (7, 15). These genes (Table S1E in Supplementary Material) define a transcriptional signature that is common during Salmonella infection and has been previously detected in different strains of mice during infection (15, 22). Additional strain-specific genes regulated during infection were detected only in Ity (40 genes), Ity3 (20 genes), Ity3.RecN (62 genes), and Ity3.RecG (54 genes) (Tables S1F–I in Supplementary Material). A large number of differentially expressed genes specific to Ity mice (Table S1F in Supplementary Material) were up-regulated in granulocytes and/or macrophages including S100a8 and S100a9 that are of particular interest as they are involved with expression of inflammatory mediators, phagocytosis, oxidative burst as well as migration of neutrophils and monocytes to the site of infection (23). The gene Clec7a (dectin 1) was differentially regulated only in Ity. Recent work has linked dectin 1/Syk kinase signaling with autophagy-dependent maturation of phagosomes (24).
In Ity3 mice, most genes that were differentially regulated were either expressed in macrophages or in megaerythrocyte precursors suggesting active extramedullary erythropoiesis in this strain (Table S1G in Supplementary Material). Interestingly, a large proportion (~40%) of genes specific to the strain Ity3.RecG are known to be down-regulated in B and T cells, as analyzed by BioGPS (25) (Table S1G in Supplementary Material). These data show the impact of Ity3 on the cellular composition of the spleen and/or changes in gene expression in specific cellular populations during infection. Very few genes were similarly regulated between Ity3 and the sub-congenic strains Ity3.RecG (4 genes), Ity3.RecN (7 genes), and Ity3.RecN and Ity3.RecG (4 genes) (Tables S1J–L in Supplementary Material).
We then used GeneGO (Thomson Reuters, NY, USA) to classify the genes differentially regulated in each strain (Tables S1A–D in Supplementary Material) into gene ontology (GO) molecular pathways, GO processes, pathways, and process networks, in order to identify the pathways differentially regulated in each strain during infection (Figure 2). The strain Ity3.RecG appeared to have fewer genes involved in various pathways and processes related to chemokine and cytokine activity and immune response as demonstrated by the lower –log2 (p-values). These results are consistent with previous observations of reduced inflammatory responses following in vivo Salmonella infection in Ity3.RecG mice (16).
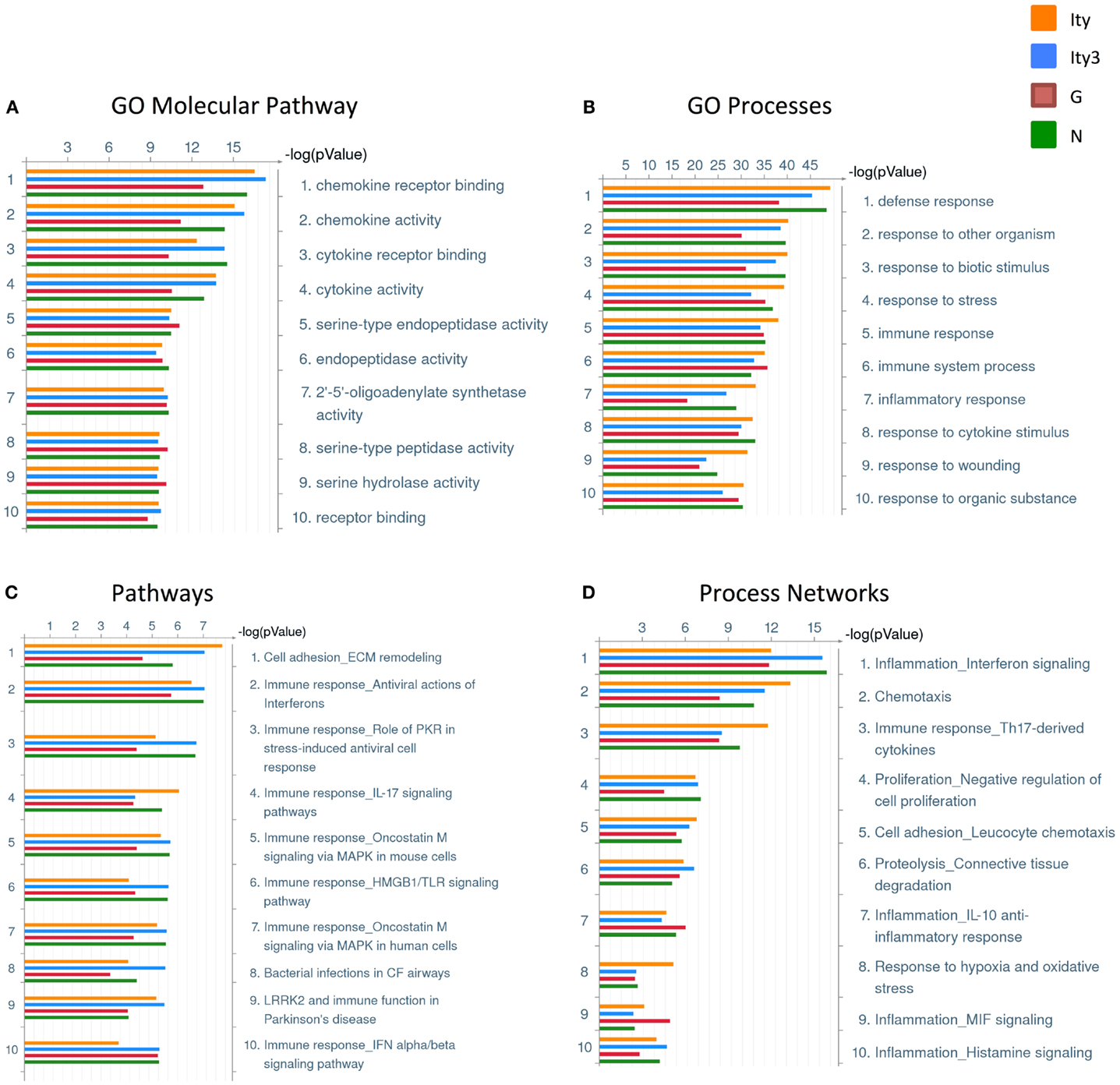
Figure 2. Pathway and process analyses of genes differentially regulated in the spleen of Ity, Ity3, Ity3.RecG, and Ity3.RecN mice during infection. Gene ontology classifications generated by clustering the genes that are differentially regulated in each strain upon infection. (A) Gene ontology (GO) molecular pathways and (B) Gene ontology (GO) processes (C) pathways (D) Process networks that are enriched in the four mouse strains. The sub-congenic strain Ity3.RecG shows a lower −log(p-value) for chemokine and cytokine receptor activity within the GO molecular pathways as well as lower −log(p-values) for response to bacteria in the GO processes. This is consistent with previous data suggesting that the Ity3.RecG mice have a diminished inflammatory response following infection compared to parental strains (16).
Ity3.1 Locus Influences the Expression of Genes Involved in Cell-Cycle Regulation and Hematopoiesis
We have previously reported that Ncf2 is a strong candidate for the Ity3.1 locus (13). To further characterize the downstream impact of Ity3.1 and Ncf2 on gene expression, we analyzed the data by evaluating variation in gene expression between Ity3, Ity3.RecG, or Ity3.RecN, and the control strain Ity at day 0 and day 3 post-infection (Figure 1B). Interestingly, 12 genes located within the introgressed Ity3 region had lower expression levels in Ity3 and Ity3.RecG, compared to both Ity and Ity3.RecN. Figure 3A shows the box plot expression pattern of one of these genes and the list is provided in Table S2A in Supplementary Material. None of these genes were differentially regulated during infection in any of the sub-congenic strains. For some of these genes, the low levels of expression detected in mouse strains Ity3 and Ity3.RecG appear to be a consequence of poor hybridization of MOLF/Ei cDNA to microarray probes as a result of high genomic variability between the MOLF/Ei and C57BL/6J strains (Figure S2 in Supplementary Material). The wild-derived inbred MOLF/Ei had been separated from the classical inbred trains by over 1 million year of evolution, and as a result they have accumulated significant sequence variability, to the order of 1 SNP every 100 bp (26, 27).
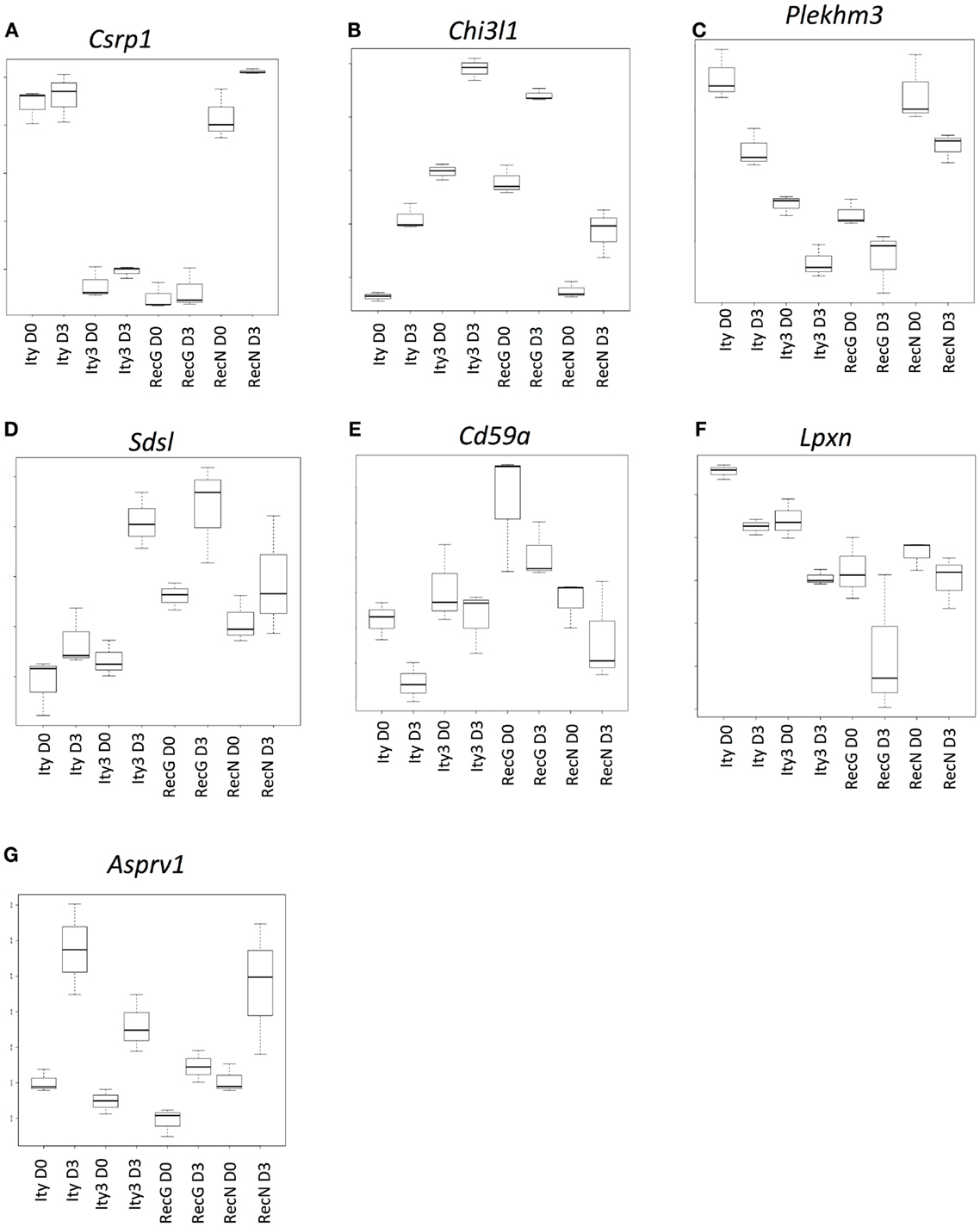
Figure 3. Genes that are under the control of the Ity3.1 locus. Sample box plots of the gene lists provided in Table S2 in Supplementary Material are shown. (A) Represents genes, which do not show any changes in expression during infection and show a similar expression pattern in Ity3 and Ity3.RecG. (B,C) Represents a sample box plot of gene expression, which show a similar regulation pattern in Ity3 and Ity3.RecG. (D–G) Also illustrate genes within Tables S2D,G in Supplementary Material, which show similar expression patterns in Ity3 and Ity3.RecG, but different from Ity and Ity3.RecN. These genes are likely under the control of the Ity3.1 locus. Csrp1 (cysteine and glycine-rich protein 1), Chi3l1 (chitinase-like 1), (Plekhm3) Pleckstrin homology domain containing, family M, member 3), Sdsl (serine dehydratase-like), Cd59a (CD59a antigen), Lpxn (leupaxin), Asprv1 (aspartic peptidase, retroviral-like 1).
A second set of genes showed similar patterns of up-regulation (Figure 3B; Table S2B in Supplementary Material) or down-regulation (Figure 3C; Table S2C in Supplementary Material) during infection in all strains, although the constitutive and induced expressions were similar in Ity3 and Ity3.RecG but significantly different from Ity and Ity3.RecN. This expression pattern highlights the impact of the MOLF/Ei allele at the Ity3 locus, as all the genes, which an expression pattern similar to Figures 3B,C carry a MOLF/Ei allele at the Ity3.1 locus (16).
Another set of genes showed higher expression during infection only in strain Ity3 and Ity3.RecG (Figures 3D,E; Tables S2D,E in Supplementary Material) as compared to Ity. In contrast to this grouping, the cluster of genes in Figure 3F exhibit lower expression levels in Ity3 and Ity3.RecG. Collectively, these groups of genes in Figures 3D–G (Tables S2D–G in Supplementary Material) exhibit a pattern of expression that is similar between Ity3 and Ity3.RecG, and the expression differences could not be attributed solely to the MOLF/Ei allele at chromosome 1, therefore, we can conclude that the expression differences are likely under the influence of the Ity3.1 locus.
Functional annotation of genes, which have an expression pattern similar between Ity3 and Ity3.RecG (Tables S2G–D in Supplementary Material) showed that a large percentage of the genes in the list plays a role in cell cycle, DNA binding, cytoskeletal reorganization, and hemopoietic and lymphoid organ development (Table S3 in Supplementary Material) (19, 28). Another major category of genes such as Ank1 and Uros are involved in heme metabolic process. These data are consistent with the observation that ROS control cell-cycle progression by influencing the presence and activity of cyclins and cyclin dependent kinases (29) and with a role for ROS in maturation and lifespan of erythroid cells (30, 31).
Ity3.1 Affects a Group of Genes That are Co-Expressed with Ncf2
In order to define the gene expression profile of the susceptible strains, we identified genes that had a similar pattern of expression in the susceptible strain, Ity3 as well as the two sub-congenic strains Ity3.RecN and Ity3.RecG. Figures 4A,B highlights two genes, Tor3a and Fam20b as examples of the expression pattern of the list of genes provided in Table S2H in Supplementary Material, which have a similar expression pattern in Ity3, Ity3.RecN, and Ity3.RecG. Only 7 of the 47 genes were within the Ity3 interval, and almost all of them were within the genomic region common to Ity3.RecN and Ity3.RecG. This gene list was classified within functional categories (Figure 4C) such as inflammatory response and regulation of angiogenesis. A large proportion of the genes were either co-expressed, co-localized, or have shared domains or predicted interactions with Ncf2 (Figure 4D). We have previously shown that the MOLF/Ei allele at the Ity3.1 locus contributed the strongest effect on susceptibility to Salmonella infection and was responsible for high bacterial burden and low ROS and cytokine production (16). The fact that a number of genes differentially regulated in Ity3, Ity3.RecN, and Ity3. RecG strains, were co-expressed with Ncf2, supports the important contribution of the Ity3.1 locus on the pathogenesis of infection in MOLF/Ei and its interaction with the other sub-locus Ity3.2 to enhance the impact of Ncf2 on ROS production.
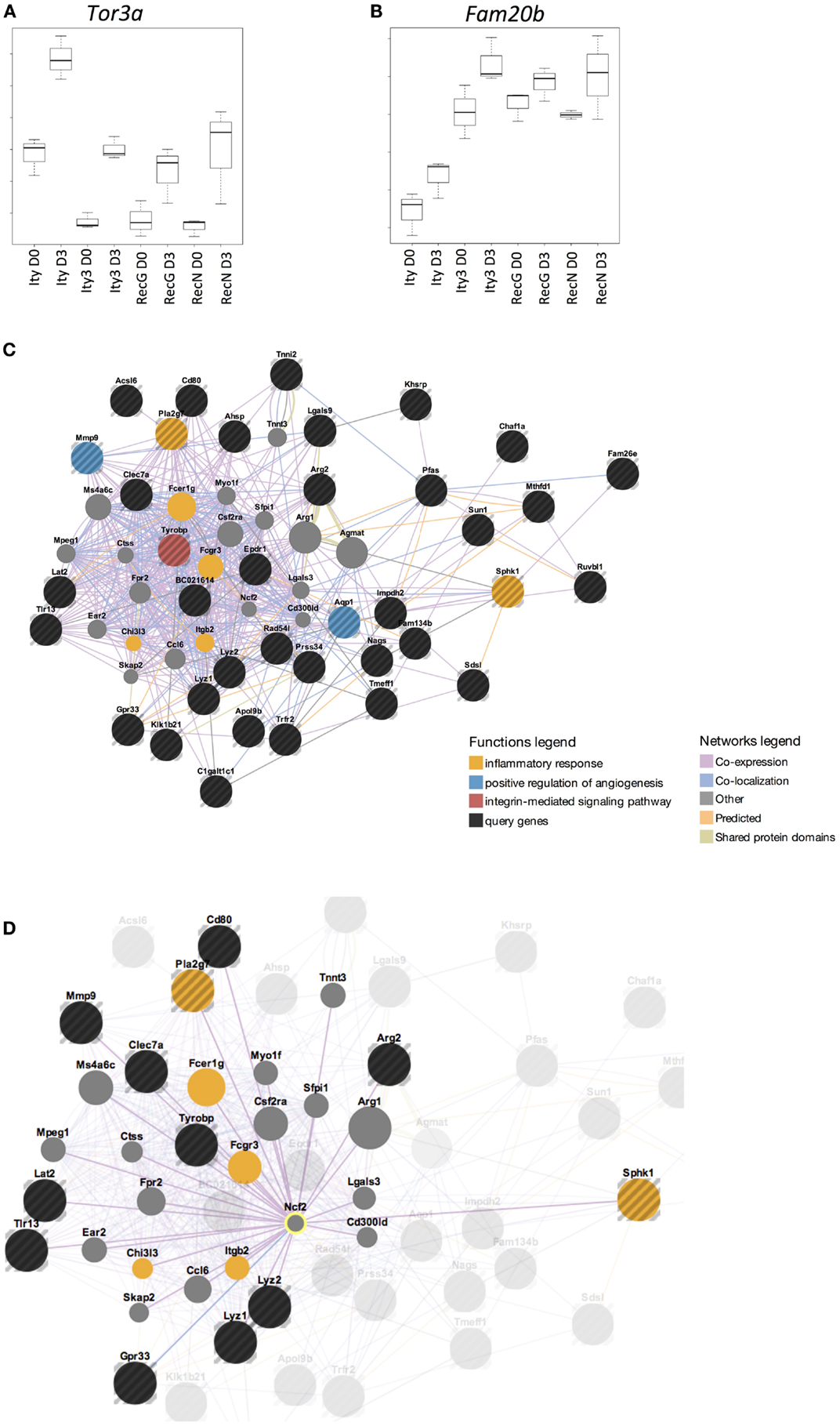
Figure 4. Genes that are differentially expressed in all susceptible and intermediate strains are under the influence of Ity3.1 sub-locus. Box plots of the expression pattern of two genes (A) Tor3a (torsin family 3, member A) and (B) Fam20b (family with sequence similarity 20, member B) are shown as examples to illustrate the expression pattern seen in the gene list provided in Table S2I in Supplementary Material. This list of genes show a similar expression in Ity3, Ity3.RecG, and Ity3.RecN and highlights the complex nature of the Ity3 locus as there are multiple genes in which expression is influenced by the combination of the two sub-loci. The genes that show a similar expression pattern in Ity3, Ity3.RecG, and Ity3.RecN were studied using GeneMania and the results are shown (C,D), with the query genes being highlighted in black. Genes, which are known to be co-expressed, co-localized, have shared domains or predicted interactions with the list of genes in Table S2I in Supplementary Material are shown. The functional categories, which are enriched within this gene list, are inflammatory response, angiogenesis, and integrin mediated signaling pathways shown in yellow, blue, and red, respectively. Genes that were not differentially expressed but important in these pathways are shown in gray. (D) Co-expression of query genes, as well as other genes within these pathways, with Ncf2 is shown.
Selp is a Candidate Gene for Ity3.2
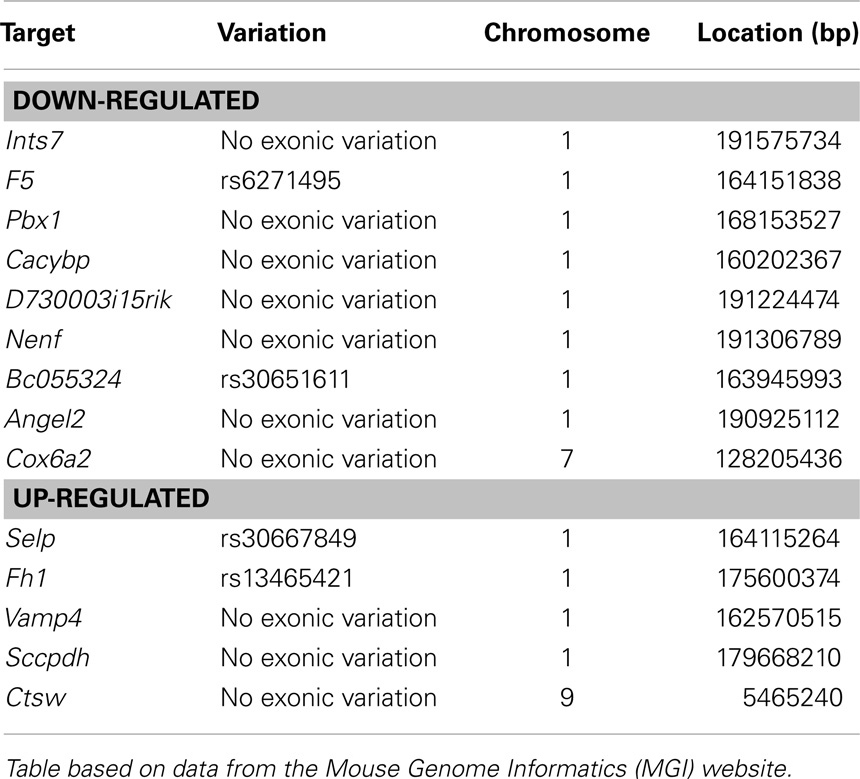
We next studied genes showing a similar regulation pattern in Ity3 and Ity3.RecN to understand the pathways differentially regulated in Ity3.2 and identify potential candidate genes for the Ity3.2 locus. There were only 14 genes that showed a similar expression pattern in Ity3 and RecN (Figure 5). Of these 14 genes, 12 are located on chromosome 1 and 6 genes (F5, Pbx1, Cacybp, Bc055342, Selp, and Vamp4) lie within the genomic region harboring Ity3.2 (Table 1). Sequence variations have been reported between the MOLF/Ei and C57BL/6J in coagulation factor F5, the cDNA BC055324 and selectin P (Selp). The coagulation factor V is synthesized by the liver and is involved in the acceleration of prothrombin to thrombin conversion (32). Coagulation Factor V deficiency leads to a bleeding disorder associated with mild to severe hemorrhagic symptoms (33). The cDNA BC055324 is poorly characterized and its function is not known. The Selp gene encodes for an adhesion molecule that mediates the recruitment of immune cells to the site of inflammation and is critical for the host immune response to infection making this gene an attractive candidate gene for Ity3.2.

Figure 5. Genes under the influence of Ity3.2. List of genes showing a similar expression pattern in Ity3 and Ity3.RecN, and different from Ity and Ity3.RecG. Ints7 (integrator complex subunit 7), F5 (coagulation factor V), Pbx1 (pre B cell leukemia homeobox 1), Cacybp (calcyclin binding protein), Nenf (neuron derived neurotrophic factor), Angel2 [angel homolog 2 (Drosophila)], Cox6a2 (cytochrome c oxidase subunit VIa polypeptide 2), Selp (selectin, platelet), Vamp4 (vesicle-associated membrane protein 4), Sccpdh [saccharopine dehydrogenase (putative)], Ctsw (cathepsin W), Fh1 (fumarate hydratase 1).
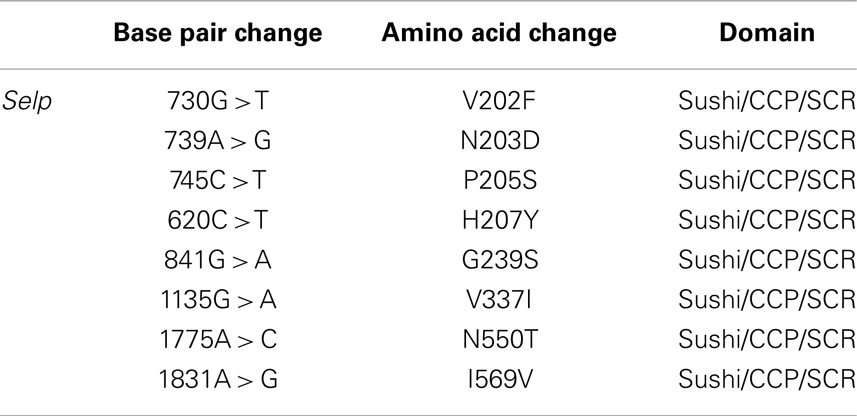
We further evaluated the candidacy of Selp as the gene underlying Ity3.2 using sequence analysis and complementation assay in vivo. Selp encodes for a protein of 768 amino acids with a C-type lectin domain, an EGF domain and 8 complement control protein (CCP) modules [or as short consensus repeats (SCRs) functional domains]. We re-sequenced the coding region of Selp in C57BL/6J and MOLF/Ei mice and identified eight SNPs (Table 2), all of which are within the less homologous CCP domains, involved in protein recognition processes (34). The amino acid proline at position 205 is well conserved across 12 mammalian species and P205S is only observed in the DBA sub-strains, which share MOLF/Ei ancestry for this region of the genome (mouse phylogeny viewer) (35). In order to validate that the sequence variation in the MOLF/Ei Selp gene has an impact on susceptibility to infection with Salmonella typhimurium and to evaluate if Selp is indeed the gene underlying Ity3.2, we used an allelic complementation assay (see breeding scheme in Figure S1 in Supplementary Material). Ity (SelpB6/B6) and Ity3 (SelpMOLF/MOLF) mice were crossed to Selp−/− knock-out mice and susceptibility to infection was assessed by survival analysis in F1 progeny with SelpMOLF/− and SelpB6/− genotypes. SelpMOLF/− mice were significantly more susceptible to infection than SelpB6/− mice and Ity controls (Figure 6A). We observed a lack of complementation in SelpMOLF/− mice with a mean survival time equivalent to Selp−/− animals (MST of 8.1 ± 0.18 and 8.3 ± 0.38, respectively), adding further support for the candidacy of Selp as the gene underlying the Ity3.2 locus (Figure 6A). Although the Selp−/− mice showed a similar susceptibility compared to the SelpMOLF/− mice in terms of survival, their tissue bacterial burden was significantly lower when compared to SelpMOLF/−, Ity3, and RecG mice (Figure 6B) suggesting that the Ity3.2 locus does not contribute significantly to the bacterial burden and that the high bacterial burden observed in SelpMOLF/− mice is rather the effect of the Ity3.1 locus.
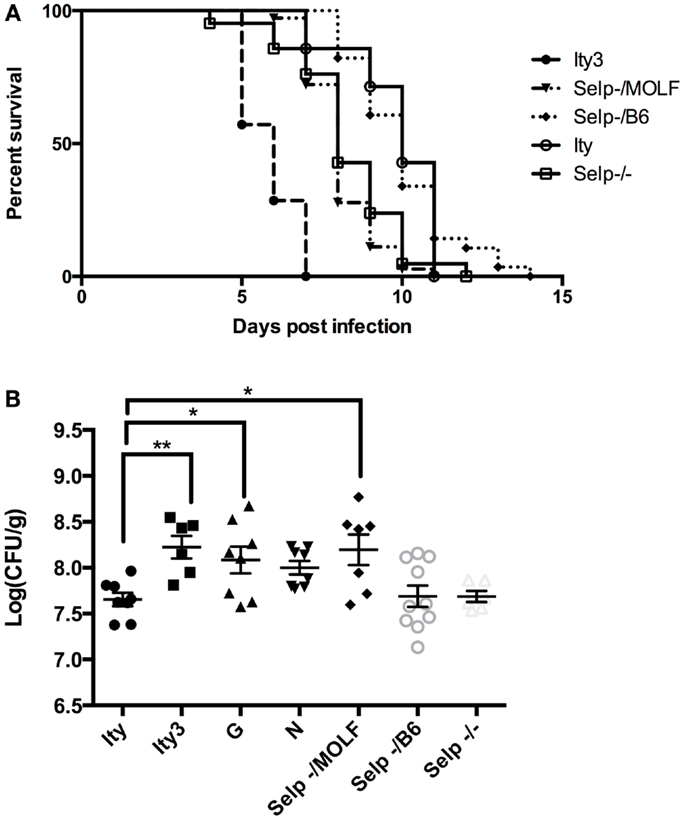
Figure 6. Allelic complementation between Ity3 congenic mouse and Selp deficient mice. In order to assess the impact of the MOLF/Ei Selp locus, Selp−/− mice were crossed with Ity and Ity3 mice to generate mice carrying the knock-out allele complemented by the MOLF/Ei allele at Selp or C57BL/6J allele at the Ity3 locus. (A) Survival curves of the congenic Ity (n = 7), Ity3 (n = 7), knock-out Selp−/− (n = 21), and Selp−/MOLF (n = 36), Selp−/B6 (n = 56) mice after infection with Salmonella typhimurium. The Selp−/− and Selp−/MOLF show a similar curve after infection with Salmonella typhimurium, both of which are more susceptible than the control Ity congenic mice and Selp−/B6, but more resistant than the mice carrying the entire Ity3 locus (Ity3). (B) Bacterial burden in the spleen of congenic Ity (n = 8) and Ity3 (n = 6), sub-congenic Ity3.RecG (n = 8), and Ity3.RecN (n = 8) and Selp−/− (n = 6) and compound heterozygous (n = 7 for Selp−/MOLF and n = 10 for Selp−/B6 mice after infection).
Discussion
The current study was specifically designed to understand the pathways that are influenced by the Ity3 locus using sub-congenic strains that exhibit different degrees of susceptibility to Salmonella infection. The global gene expression profile in the spleen was studied early during infection prior to a significant bacterial difference in the target tissue. This approach allowed us to identify networks, which are of importance in the early phases of innate immunity yet not influenced by the extent of bacterial burden in the spleen. We reported a group of genes, the majority of which are regulated by type I and type II IFN. These genes, such as the Gbp, Oas, Ifitm family members, are differentially regulated in all strains of mice during infection, and define a core transcriptional signature common to several strains of mice infected with Salmonella (15).
Additionally, we characterized a number of genes not located within the Ity3 region that were differentially expressed in the susceptible Ity3, Ity3. RecG, and Ity3. RecN strains as compared to the resistant Ity strain. We showed that these genes are also co-expressed with Ncf2, further supporting the hypothesis that there is an influence of the Ity3.1 sub-locus on other segments of the genome. These results highlight the importance of the region of distal chromosome 1 carrying Ity3, a region enriched in QTLs. Over 80 QTLs are listed at the mouse genome database (36) and a number of cis and trans eQTLs have been characterized (37). Among them, several QTLs are involved with complex inflammatory reaction, such as graft vs. host disease (38) lupus (39), modifier of LPS-response (40), and susceptibility to tuberculosis (41).
We have reported previously that Ity3 influences ROS production during infection (13) and this effect was mapped recently to a small sub-region named Ity3.1, which harbors the gene Ncf2, a subunit of the NADPH complex. (16). ROS produced by NADPH has been shown to affect a number of pathways, which are important in innate immunity including bacterial killing within the phagolysosome where ROS interact with other ions such as chloride to form toxic agents (HOCL) or can convert into hydroxyl radicals that are toxic for bacteria (42). ROS production has also been shown to influence immune cell recruitment, activation, and survival (43), and lead to translational activation of NF-κB (44). NADPH oxidase derived ROS is also a key regulator of autophagy and autophagy regulation during pathogen invasion (45). In addition, NADPH oxidase activation contributes to the recognition and removal of apoptotic neutrophils (efferocytosis) by macrophages (46–48). Therefore, an imbalance in NADPH produced ROS can lead to increased inflammation, which can be deleterious to the host.
Recent studies have shown that TNF, as well as other cytokines through NF-κB signaling induces transient increase in ROS level in endothelial cells, which results in cell surface expression of Icam and Selp (49–52). This effect has further been studied in vivo, where mice lacking the p47phox (Ncf1) subunit of the NADPH complex have reduced expression of VCAM-1, ICAM-1, SELP, and SELE in the vascular cell walls (53, 54). In the current study, we identified Selp as a candidate gene for the locus Ity3.2 and used an allelic complementation assay to provide genetic evidences that Selp is indeed a strong candidate for the Ity3.2 locus. In MOLF/Ei mice susceptibility to infection as explained by the Ity3 locus could be attributed to the individual effect of Ity3.1 and Ity3.2 sub-loci but also to the cooperation between these two sub-loci as explained by the potential impact of low activity of NADPH oxidase (16) on Selp function by reducing its expression (current paper).
We also illustrated that mice carrying a MOLF/Ei allele at the Ity3.1 locus have higher expression of a number of genes playing a role in cell cycle, DNA binding, and cytoskeletal pathways. There is a growing body of evidence discussing the link between ROS, cell-cycle progress and arrest. As discussed by Martindale (55), ROS can have multiple effects on the cell cycle, depending on the amount and type of ROS. They suggest that low doses of ROS may cause proliferation while high doses of ROS can lead to apoptosis and cell death. In our model, it is possible that reduced levels of ROS in susceptible animals could lead to cell growth arrest, hence providing a more favorable niche for bacteria to replicate.
Another pathway influenced by ROS production, is up-regulated by Ity3.1 and regroups genes involved in heme biosynthesis. Increased expression of genes within the heme biosynthesis pathway could result in increased free heme within the cells, which can act as a potent cytotoxic pro-oxidant (56). Free heme has also recently been shown to trigger necroptosis in macrophages (57). Therefore, it is possible that the increased expression of the heme biosynthesis pathway observed in susceptible mice is a mechanism that compensates for the low ROS levels.
In conclusion, our study highlights the complex nature of multi-loci interaction in the wild-derived MOLF/Ei response to Salmonella infection. We highlighted the role of low ROS and cytokine production in reduced survival of mice carrying the Ity3.1 locus, and the importance of the Ity3.2 locus, which synergistically led to increased susceptibly of the Ity3 mice. We have also shown that several pathways identified in strains Ity3, Ity3.RecN, and Ity3.RecG, are influenced by Ncf2. Furthermore, the Ity3.1 sub-locus has additional effects, which have not previously been characterized, in expression of genes involved in cell-cycle arrest and hematopoiesis. Lastly, we propose a hypothesis that the combined effects of low ROS production by the MOLF/Ei Ity3.1 locus together with the impact of Selp MOLF/Ei allele at Ity3.2 influences the host survival after infection with Salmonella typhimurium.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful for the technical assistance of Melissa Herman, Nadia Prud’homme, and Line Larivière. Rabia Tahir Khan was a recipient of a studentship award from the Fonds de la recherche en santé du Québec. Kyoko E. Yuki is a recipient of a Faculty of Medicine Internal Studentship. This work was supported by a Canadian Institutes of Health Research Grant (MOP-15461) to Danielle Malo.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Journal/10.3389/fimmu.2014.00375/abstract
References
1. Mastroeni P, Grant A. Dynamics of spread of Salmonella enterica in the systemic compartment. Microbes Infect (2013) 15:849–57. doi: 10.1016/j.micinf.2013.10.003
2. Mastroeni P, Grant A, Restif O, Maskell D. A dynamic view of the spread and intracellular distribution of Salmonella enterica. Nat Rev Microbiol (2009) 7:73–80. doi:10.1038/nrmicro2034
3. Buckle GC, Walker CL, Black RE. Typhoid fever and paratyphoid fever: systematic review to estimate global morbidity and mortality for 2010. J Glob Health (2012) 2:10401. doi:10.7189/jogh.02.010401
4. de Jong HK, Parry CM, van der Poll T, Wiersinga WJ. Host-pathogen interaction in invasive salmonellosis. PLoS Pathog (2012) 8:e1002933. doi:10.1371/journal.ppat.1002933
5. Mastroeni P, Sheppard M. Salmonella infections in the mouse model: host resistance factors and in vivo dynamics of bacterial spread and distribution in the tissues. Microbes Infect (2004) 6:398–405. doi:10.1016/j.micinf.2003.12.009
6. Sebastiani G, Olien L, Gauthier S, Skamene E, Morgan K, Gros P, et al. Mapping of genetic modulators of natural resistance to infection with Salmonella typhimurium in wild-derived mice. Genomics (1998) 47:180–6. doi:10.1006/geno.1997.5116
7. Richer E, Yuki KE, Dauphinee SM, Larivière L, Paquet M, Malo D. Impact of Usp18 and IFN signaling in Salmonella-induced typhlitis. Genes Immun (2011) 12:531–43. doi:10.1038/gene.2011.38
8. Roy MF, Riendeau N, Bédard C, Hélie P, Min-Oo G, Turcotte K, et al. Pyruvate kinase deficiency confers susceptibility to Salmonella typhimurium infection in mice. J Exp Med (2007) 204:2949–61. doi:10.1084/jem.20062606
9. Vidal S, Gros P, Skamene E. Natural resistance to infection with intracellular parasites: molecular genetics identifies Nramp1 as the Bcg/Ity/Lsh locus. J Leukoc Biol (1995) 58:382–90.
10. Sebastiani G, Blais V, Sancho V, Vogel SN, Stevenson MM, Gros P, et al. Host immune response to Salmonella enterica serovar typhimurium infection in mice derived from wild strains. Infect Immun (2002) 70:1997–2009. doi:10.1128/IAI.70.4.1997-2009.2002
11. Sebastiani G, Leveque G, Larivière L, Laroche L, Skamene E, Gros P, et al. Cloning and characterization of the murine toll-like receptor 5 (Tlr5) gene: sequence and mRNA expression studies in Salmonella-susceptible MOLF/Ei mice. Genomics (2000) 64:230–40. doi:10.1006/geno.2000.6115
12. Sancho-Shimizu V, Khan R, Mostowy S, Larivière L, Wilkinson R, Riendeau N, et al. Molecular genetic analysis of two loci (Ity2 and Ity3) involved in the host response to infection with Salmonella typhimurium using congenic mice and expression profiling. Genetics (2007) 177:1125–39. doi:10.1534/genetics.107.075523
13. Sancho-Shimizu V, Malo D. Sequencing, expression, and functional analyses support the candidacy of Ncf2 in susceptibility to Salmonella typhimurium infection in wild-derived mice. J Immunol (2006) 176:6954–61. doi:10.4049/jimmunol.176.11.6954
14. Roy MF, Malo D. Genetic regulation of host responses to Salmonella infection in mice. Genes Immun (2002) 3:381–93. doi:10.1038/sj.gene.6363924
15. Khan R, Sancho-Shimizu V, Prendergast C, Roy MF, Loredo-Osti JC, Malo D. Refinement of the genetics of the host response to Salmonella infection in MOLF/Ei: regulation of type 1 IFN and TRP3 pathways by Ity2. Genes Immun (2012) 13:175–83. doi:10.1038/gene.2011.69
16. Khan RT, Yuki KE, Malo D. Fine-mapping and phenotypic analysis of the Ity3 Salmonella susceptibility locus identify a complex genetic structure. PLoS One (2014) 9:e88009. doi:10.1371/journal.pone.0088009
17. Ullman-Cullere MH, Foltz CJ. Body condition scoring: a rapid and accurate method for assessing health status in mice. Lab Anim Sci (1999) 49:319–23.
18. Long AD, Mangalam HJ, Chan BY, Tolleri L, Hatfield GW, Baldi P. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J Biol Chem (2001) 276:19937–44. doi:10.1074/jbc.M010192200
19. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc (2009) 4:44–57. doi:10.1038/nprot.2008.211
20. Zuberi K, Franz M, Rodriguez H, Montojo J, Lopes CT, Bader GD, et al. GeneMANIA prediction server 2013 update. Nucleic Acids Res (2013) 41:W115–22. doi:10.1093/nar/gkt533
21. Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res (2010) 38:W214–20. doi:10.1093/nar/gkq537
22. Caron J, Larivière L, Nacache M, Tam M, Stevenson MM, McKerly C, et al. Influence of Slc11a1 on the outcome of Salmonella enterica serovar enteritidis infection in mice is associated with Th polarization. Infect Immun (2006) 74:2787–802. doi:10.1128/IAI.74.5.2787-2802.2006
23. Kerkhoff C, Voss A, Scholzen TE, Averill MM, Zänker KS, Bornfeldt KE. Novel insights into the role of S100A8/A9 in skin biology. Exp Dermatol (2012) 21:822–6. doi:10.1111/j.1600-0625.2012.01571.x
24. Kyrmizi I, Gresnigt MS, Akoumianaki T, Samonis G, Sidiropoulos P, Boumpas D, et al. Corticosteroids block autophagy protein recruitment in Aspergillus fumigatus phagosomes via targeting dectin-1/Syk kinase signaling. J Immunol (2013) 191:1287–99. doi:10.4049/jimmunol.1300132
25. Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol (2009) 10:R130. doi:10.1186/gb-2009-10-11-r130
26. Abe K, Noguchi H, Tagawa K, Yuzuriha M, Toyoda A, Kojima T, et al. Contribution of Asian mouse subspecies Mus musculus molossinus to genomic constitution of strain C57BL/6J, as defined by BAC-end sequence-SNP analysis. Genome Res (2004) 14:2439–47. doi:10.1101/gr.2899304
27. Ideraabdullah FY, de laCasa-Esperón E, Bell TA, Detwiler DA, Magnuson T, Sapienza C, et al. Genetic and haplotype diversity among wild-derived mouse inbred strains. Genome Res (2004) 14:1880–7. doi:10.1101/gr.2519704
28. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res (2009) 37:1–13. doi:10.1093/nar/gkn923
29. Verbon EH, Post JA, Boonstra J. The influence of reactive oxygen species on cell cycle progression in mammalian cells. Gene (2012) 511:1–6. doi:10.1016/j.gene.2012.08.038
30. Marinkovic D, Zhang X, Yalcin S, Luciano JP, Brugnara C, Huber T, et al. Foxo3 is required for the regulation of oxidative stress in erythropoiesis. J Clin Invest (2007) 117:2133–44. doi:10.1172/JCI31807
31. Xu Y, Swartz KL, Siu KT, Bhattacharyya M, Minella AC. Fbw7-dependent cyclin E regulation ensures terminal maturation of bone marrow erythroid cells by restraining oxidative metabolism. Oncogene (2013) 33(24):3161–71. doi:10.1038/onc.2013.289
32. Esmon CT, Owen WG, Duiguid DL, Jackson CM. The action of thrombin on blood clotting factor V: conversion of factor V to a prothrombin-binding protein. Biochim Biophys Acta (1973) 310:289–94. doi:10.1016/0005-2795(73)90034-2
33. Asselta R, Peyvandi F. Factor V deficiency. Semin Thromb Hemost (2009) 35:382–9. doi:10.1055/s-0029-1225760
34. Libert F, Vassart G. Structure-function relationships of the complement components. Immunol Today (1989) 10:407. doi:10.1016/0167-5699(89)90036-4
35. Wang JR, de Villena FP, McMillan L. Comparative analysis and visualization of multiple collinear genomes. BMC Bioinformatics (2012) 13(Suppl 3):S13. doi:10.1186/1471-2105-13-S3-S13
36. Blake JA, Bult CJ, Eppig JT, Kadin JA, Richardson JE, Mouse Genome Database Group. The mouse genome database: integration of and access to knowledge about the laboratory mouse. Nucleic Acids Res (2014) 42:D810–7. doi:10.1093/nar/gkt1225
37. Mozhui K, Ciobanu DC, Schikorski T, Wang X, Lu L, Williams RW. Dissection of a QTL hotspot on mouse distal chromosome 1 that modulates neurobehavioral phenotypes and gene expression. PLoS Genet (2008) 4:e1000260. doi:10.1371/journal.pgen.1000260
38. Allen RD, Dobkins JA, Harper JM, Slayback DL. Genetics of graft-versus-host disease, I. A locus on chromosome 1 influences development of acute graft-versus-host disease in a major histocompatibility complex mismatched murine model. Immunology (1999) 96:254–61. doi:10.1046/j.1365-2567.1999.00626.x
39. Kono DH, Park MS, Theofilopoulos AN. Genetic complementation in female (BXSB x NZW)F2 mice. J Immunol (2003) 171:6442–7. doi:10.4049/jimmunol.171.12.6442
40. Matesic LE, De Maio A, Reeves RH. Mapping lipopolysaccharide response loci in mice using recombinant inbred and congenic strains. Genomics (1999) 62:34–41. doi:10.1006/geno.1999.5986
41. Kramnik I, Dietrich WF, Demant P, Bloom BR. Genetic control of resistance to experimental infection with virulent Mycobacterium tuberculosis. Proc Natl Acad Sci U S A (2000) 97:8560–5. doi:10.1073/pnas.150227197
42. Yang HC, Cheng ML, Ho HY, Chiu DT. The microbicidal and cytoregulatory roles of NADPH oxidases. Microbes Infect (2011) 13:109–20. doi:10.1016/j.micinf.2010.10.008
43. Segal BH, Grimm MJ, Khan AN, Han W, Blackwell TS. Regulation of innate immunity by NADPH oxidase. Free Radic Biol Med (2012) 53:72–80. doi:10.1016/j.freeradbiomed.2012.04.022
44. Schmidt KN, Amstad P, Cerutti P, Baeuerle PA. The roles of hydrogen peroxide and superoxide as messengers in the activation of transcription factor NF-kappa B. Chem Biol (1995) 2:13–22. doi:10.1016/1074-5521(95)90076-4
45. Huang J, Canadien V, Lam GY, Steinberg BE, Dinauer MC, Magalhaes MA, et al. Activation of antibacterial autophagy by NADPH oxidases. Proc Natl Acad Sci U S A (2009) 106:6226–31. doi:10.1073/pnas.0811045106
46. Brown JR, Goldblatt D, Buddle J, Morton L, Thrasher AJ. Diminished production of anti-inflammatory mediators during neutrophil apoptosis and macrophage phagocytosis in chronic granulomatous disease (CGD). J Leukoc Biol (2003) 73:591–9. doi:10.1189/jlb.1202599
47. Frasch SC, Berry KZ, Fernandez-Boyanapalli R, Jin HS, Leslie C, Henson PM, et al. NADPH oxidase-dependent generation of lysophosphatidylserine enhances clearance of activated and dying neutrophils via G2A. J Biol Chem (2008) 283:33736–49. doi:10.1074/jbc.M807047200
48. Sanmun D, Witasp E, Jitkaew S, Tyurina YY, Kagan VE, Ahlin A, et al. Involvement of a functional NADPH oxidase in neutrophils and macrophages during programmed cell clearance: implications for chronic granulomatous disease. Am J Physiol Cell Physiol (2009) 297:C621–31. doi:10.1152/ajpcell.00651.2008
49. Min JK, Kim YM, Kim SW, Kwon MC, Kong YY, Hwang IK, et al. TNF-related activation-induced cytokine enhances leukocyte adhesiveness: induction of ICAM-1 and VCAM-1 via TNF receptor-associated factor and protein kinase C-dependent NF-kappaB activation in endothelial cells. J Immunol (2005) 175:531–40. doi:10.4049/jimmunol.175.1.531
50. Lo SK, Janakidevi K, Lai L, Malik AB. Hydrogen peroxide-induced increase in endothelial adhesiveness is dependent on ICAM-1 activation. Am J Physiol (1993) 264:L406–12.
51. Chen XL, Zhang Q, Zhao R, Ding X, Tummala PE, Medford RM. Rac1 and superoxide are required for the expression of cell adhesion molecules induced by tumor necrosis factor-alpha in endothelial cells. J Pharmacol Exp Ther (2003) 305:573–80. doi:10.1124/jpet.102.047894
52. Kim SR, Bae YH, Bae SK, Choi KS, Yoon KH, Koo TH, et al. Visfatin enhances ICAM-1 and VCAM-1 expression through ROS-dependent NF-kappaB activation in endothelial cells. Biochim Biophys Acta (2008) 1783:886–95. doi:10.1016/j.bbamcr.2008.01.004
53. Chen H, Song YS, Chan PH. Inhibition of NADPH oxidase is neuroprotective after ischemia-reperfusion. J Cereb Blood Flow Metab (2009) 29:1262–72. doi:10.1038/jcbfm.2009.47
54. Vendrov AE, Hakim ZS, Madamanchi NR, Rojas M, Madamanchi C, Runge MS. Atherosclerosis is attenuated by limiting superoxide generation in both macrophages and vessel wall cells. Arterioscler Thromb Vasc Biol (2007) 27:2714–21. doi:10.1161/ATVBAHA.107.152629
55. Martindale JL, Holbrook NJ. Cellular response to oxidative stress: signaling for suicide and survival. J Cell Physiol (2002) 192:1–15. doi:10.1002/jcp.10119
56. Balla G, Jacob HS, Balla J, Rosenberg M, Nath K, Apple F, et al. Ferritin: a cytoprotective antioxidant strategem of endothelium. J Biol Chem (1992) 267:18148–53.
Keywords: Salmonella, Ity3, innate immunity, murine model for typhoid, selectin P, Ncf2
Citation: Khan RT, Chevenon M, Yuki KE and Malo D (2014) Genetic dissection of the Ity3 locus identifies a role for Ncf2 co-expression modules and suggests Selp as a candidate gene underlying the Ity3.2 locus. Front. Immunol. 5:375. doi: 10.3389/fimmu.2014.00375
Received: 23 May 2014; Accepted: 22 July 2014;
Published online: 12 August 2014.
Edited by:
Constantino López-Macías, University of Oxford, MexicoReviewed by:
Laurel L. Lenz, National Jewish Health, USAHridayesh Prakash, University of Hyderabad, India
Copyright: © 2014 Khan, Chevenon, Yuki and Malo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Danielle Malo, McGill University, 3649 Sir William Osler Promenade, Montreal, QC H3G 0B1, Canada e-mail:ZGFuaWVsbGUubWFsb0BtY2dpbGwuY2E=
 Rabia Tahir Khan
Rabia Tahir Khan Marie Chevenon2,3
Marie Chevenon2,3 Kyoko E. Yuki
Kyoko E. Yuki Danielle Malo
Danielle Malo