- Xenolaüs Genetics LLC, Los Angeles, CA, USA
T-cell ontogeny optimizes the α/β T-cell receptor (TCR) repertoire for recognition of major histocompatibility complex (MHC) class-I/II genetic polymorphism, and co-evolution of TCR germline V-gene segments and the MHC must entail somatic diversity generated in the third complimentary determining regions (CDR3α/β); however, it is still not clear how. Herein, a conspicuous structural link between the V-Jα used by several different TCR [all in complex with the same MHC molecule (HLA-A2)], and a conserved MHC motif (a.a., R65-X-X-K-A-X-S-Q72) is described. We model this R65-joint in detail, and show that the same TCR’s CDR3α loop maintains its CDR2α loop at a distance of ~4 Å from polymorphic amino acid (a.a.) positions of the α-2 helix in all but one of the analyzed crystal structures. Indeed, the pitch of docked TCRs varies as their twist/tilt/sway maintains the R65-joint and peptide contacts. Thus, the R65-joint appears to have poised the HLA-A lineage toward alloreactivity.
Introduction
The same DNA-recombinase system (RAG-1/-2) used in B cells for the generation of variants of the canonical immunoglobulin (Ig) cell-surface receptor is used in T cells to generate a vast diverse repertoire of T-cell receptor (TCR) variants; these variants of the TCR are clonally distributed on T cells, as are sIg on B cells (1). By contrast, within any given individual, the number of possible major histocompatibility complex (MHC) (HLA in human) components of the TCR ligand is limited by two (at most) different alleles of any given HLA heavy-chain gene (1–3). The most enigmatic phenomenon involving TCR and the MHC concerns a very high relative frequency of T cells with exquisite sensitivity to minor changes in the peptide component of pHLA, which nevertheless proliferate against allogeneic pHLA. Because allo-HLA is not present in the thymus, and as such the TCR repertoire cannot be selected against different individuals’ HLA molecules, there exists a high precursor frequency of T cells that cross-react against allo-HLA bearing targets (1–10). Thus, there is a potent biological capacity in the apparent absence of any stimulus, except during gestation. Here, we describe how somatically distinct CDR3α (with one exception) achieves a germline-encoded mean interface of 3.94 ± 0.23 Å between CDR2α and a discreet polymorphic region of HLA-A. Together with bioinformatics evidence, this R65-joint indicates that adaptive immunity is constrained by an apparent need for precise alloreactivity (11).
Results and Discussion
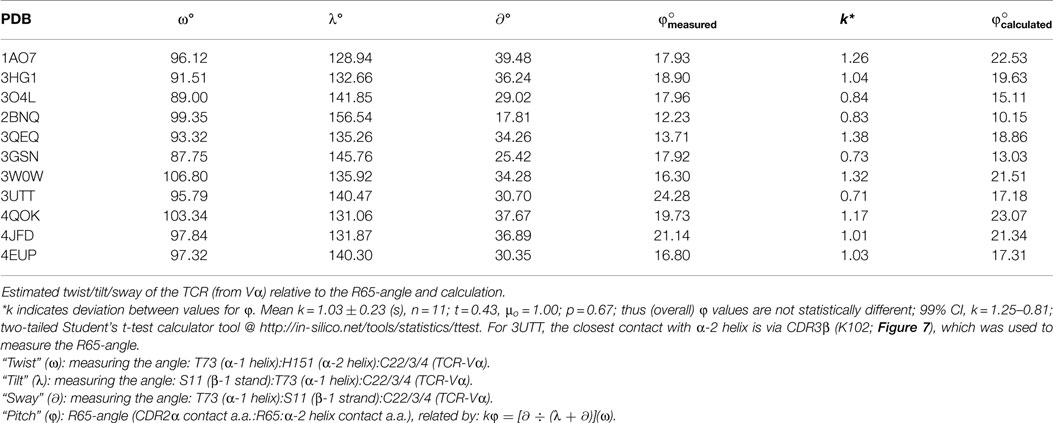
Shown in Figure 1A is our analysis of the CDR1 and CDR2 contacts made by several distinct TCR across different TCR:pHLA structures available in the Protein data bank (PDB). All of these structures involve HLA-A*0201 and each has a distinct peptide component. As can be seen from the closest contacts made by the TCR, one can classify these TCR as alpha-dominant, alpha/beta, and in one case, beta-dominant, on the basis of these interactions. Indirectly, this corroborates the role of the CDR3 regions in selective binding of any given TCR for the peptide component (12–16). Theoretically, TCR bearing CDR3 regions that did not disrupt these CDR1/2 interactions with the α-helices of the HLA groove during fetal life would have been repetitively engaged with thymic antigen presenting cells, and such clones would be deleted (1, 4, 9).
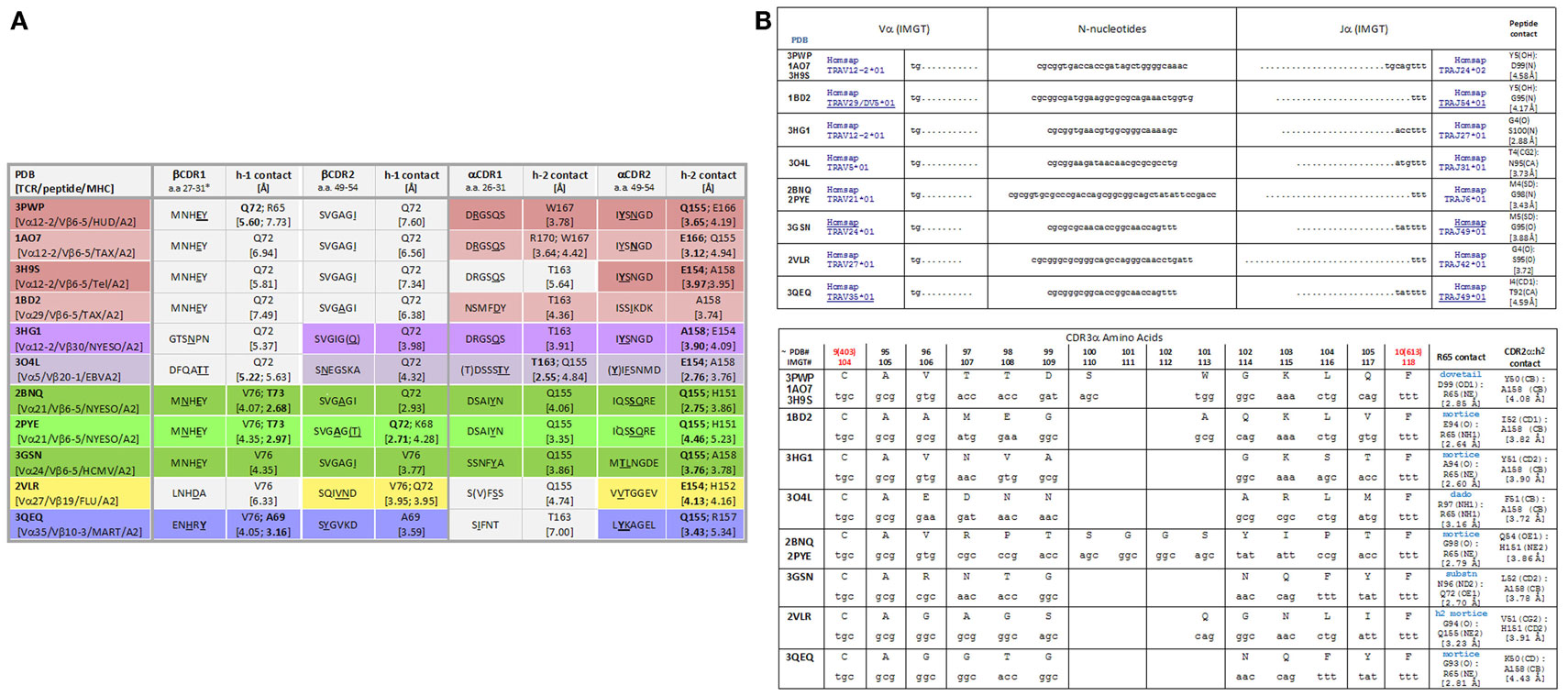
Figure 1. (A) CDR1/CDR2 contacts with MHC amino acids among TCR:pHLA-A2 crystallographic structures. The colors indicate the alpha-dominant (rose shades), alpha/beta (lavender, green, and yellow), and beta-dominant (blue) modes of binding. [(B), top] nucleotide sequences for all CDR3α of TCR in the indicated PDB files. TCRA were reverse translated then subjected to joint analysis. [(B), bottom] CDR3α joints (consensus IMGT numbering is shown with PDB numbering as indicated); contacts were measured with VMD software (www.ks.uiuc.edu). Characterization of R65 contacts was by analysis of “surf representations” (i.e., space-filled modeling) as shown in figures below. For multiple structures involving the same TCR, contacts in one structure are shown, i.e., for 1AO7 and 2BNQ.
Bioinformatics Analysis
Protein data bank files available for TCR:pHLA-A2 solved crystallographic structures (as listed in Figure 1A) were used to obtain the most likely nucleotide codons of the TCRA chain by reverse translation using the algorithms available at the SMS.1 Identification of Vα and Jα usage (IMGT/V-Quest) and junctional analysis (IMGT/JunctionAnalysis) among these TCR were performed by the IMGT algorithms2 and the results are shown in Figure 1B. Notice, all the CDR3α joints use extensive N-nucleotide additions (a hallmark of TCRVA somatic DNA rearrangements) to create a diverse set of amino acid sequences used within the solved structures. With 54 Vα and 61 Jα, TCRVA is unique among antigen receptors, and continuous rearrangement at TCRA ensures pHLA selects TCR (1). Here, we have undertaken a comprehensive analysis of each of the TCR:pHLA-A2 structures to examine the contacts made between each CDR3α loop and pHLA-A2 after we noticed that alpha-dominant, alpha/beta, and beta-dominant TCR binding all involved CDR3α contact with the MHC. Shown in Figures 2A–F is this conspicuous contact that all CDR3α make with the α-1 helix of HLA-A2. Note that all CDR3α make closest contact at the same motif centered on amino acid (a.a.) R65; 2VLR is the exception (Figures 2E,F).
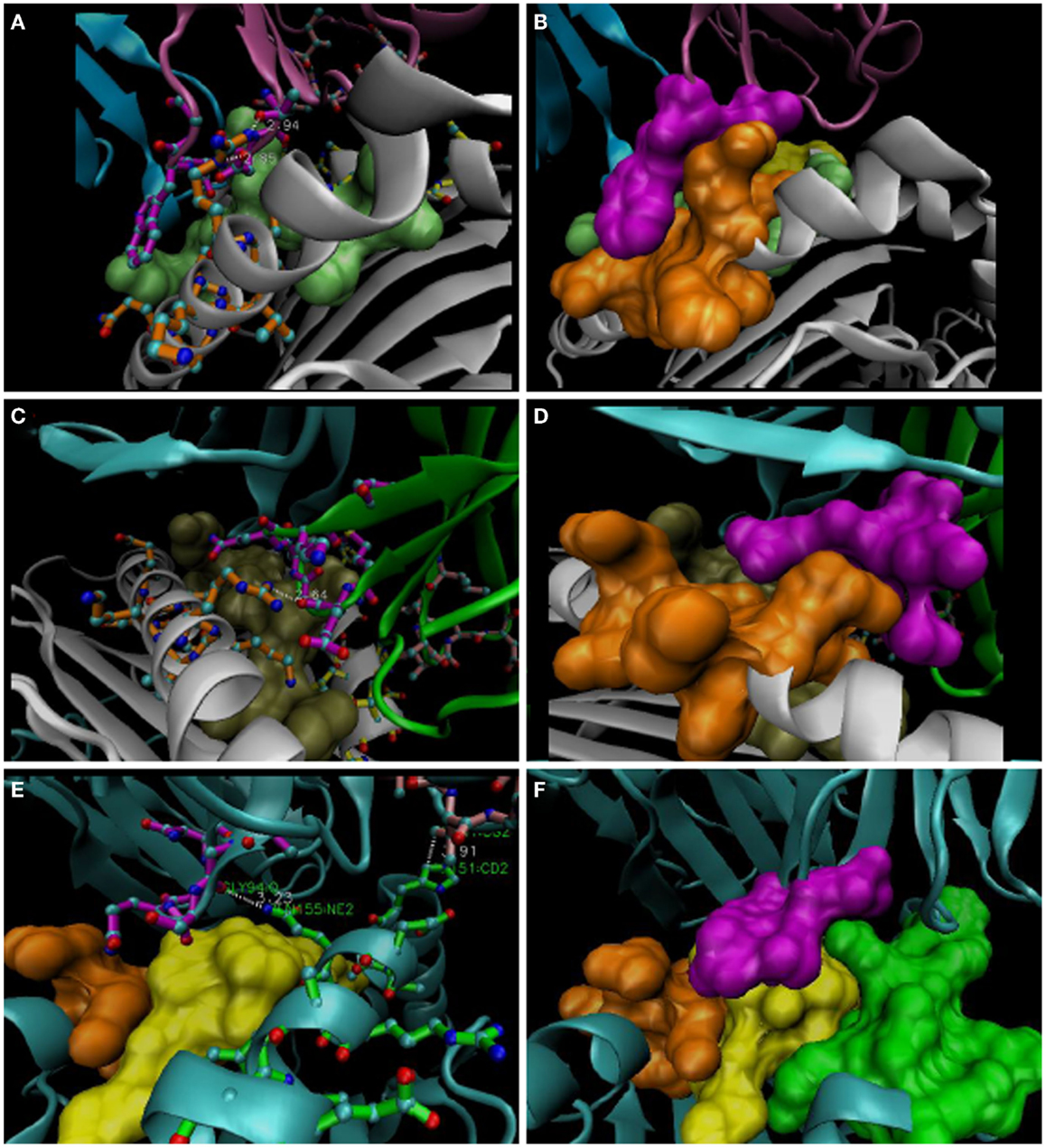
Figure 2. Representative R65-Joints of these TCR:pMHC. (A,B) 1AO7; (C,D) 1BD2; (E,F) 2VLR. VMD software used to isolate structures and make bond measurements; “licorice” representations are shown at left and “surf” representations are shown on the right. Docking CDR3α a.a. are in magenta (Vα: in magenta, 1AO7; green, 1BD2; cyan, 2VLR); the R65 motif is in orange, and the H151-A158 region is in green (bottom panels). Peptides are lime, tan, and yellow for three structures, respectively. Note the W101 dovetail of 1AO7 with salt-bridges to R65 mediated by the CDR3α loop (A,B). TCR represented by the 1BD2 file (see Figure 1B) utilizes a concave mortise, wherein R65 also forms salt bridges. 2VLR’s CDR3α contacts Q155 in a different strategy (see text).
R65-Joint
As shown in Figure 2 (compiled in Figure 1B), individual CDR3α rearrangements lead to structurally distinct types of contact with the R65 motif, principally, projection-type (dovetail), concave-type (mortise), or flat-type (dado), all best appreciated with space-filled models. For example, the dovetail joint of the A6 TCR (in 1AO7, 3PWP, and 3H9S complexes) fits W101 into the complimentary slot made by the side-chains of the R65 motif, i.e., within the α-helical secondary structure of the α-1 helix (Figures 2A,B). W101 is located on the lateral side of the CDR3α loop (i.e., the arm of the parabolic loop that faces away from the groove), and close contacts with α-1 are mediated by the arm of CDR3α that faces into the groove, i.e., ~3 Å contacts involving salt bridges (R65NE:D99OD1; R65NH2:T98OG1). Interestingly, the closest contact with the peptide also involves D99, i.e., Y5OH:D99N (peptide contacts listed in Figure 1B).
Mortise
Looking further into the R65-motif connections demonstrates the use of a mortise, i.e., a CDR3α lock for the R65 key. As illustrated in Figures 2C,D, this is the most common type of contact and involves salt bridge formation between an acidic group(s) in CDR3α and one or more N of R65. One such joint involves N-H-N contact (dado-type of 304L); also, one of the contacts is shifted to Q72 by the 3GSN TCR (Figure 1B), and the 2VLR TCR is in less contact with α-1 helix (i.e., ~5 Å to R65); however, contacts the α-2 helix via a strikingly congruent mortise involving Q155 (Figures 2E,F). Indeed, 2VLR’s CDR3α seems like an alternative solution among these structures.
CDR2α/α-2 Helix Interface
The R65-joint is consistent with a range of TCR twist/tilt/sway (rotations about the plane of the pHLA top face) such that ~4 Å juxtaposition of CDR2α over HLA a.a. 151–158 is achieved (Figure 3A). Alignment of distant HLA-A alleles with A*0201 (Figure 3B) reveals that H151 of A2 is R151 in A-74, A-31, A-33, A-29, A-30, A-32, A-23, and A-80. Also, polymorphic is A158 of A2, which is V158 in A-36 and A-1. Other a.a. 151–158 α-2 polymorphisms are not oriented toward the TCR due to the α-helix. While they might influence allogeneic peptide identity, and thus indirectly the R65-joint (see below), A158V and H151R clearly define the interface. Since closer contacts would be expected for those CDR2α contacting A158 when the two –CH3 groups replace two –H on the pos. 158 a.a. Cβ, i.e., V158 (as found in HLA alleles, A-1 and A-36), and too, H151R could decrease contact distances (a longer side chain), it follows that all of these TCR maintain the R65-joint and the marginal contact with the α-2 helix by some shared mechanism. Moreover, it leads to an apparent steric consideration with respect to which allotypes are recognized by a given TCR (see Figure 4).
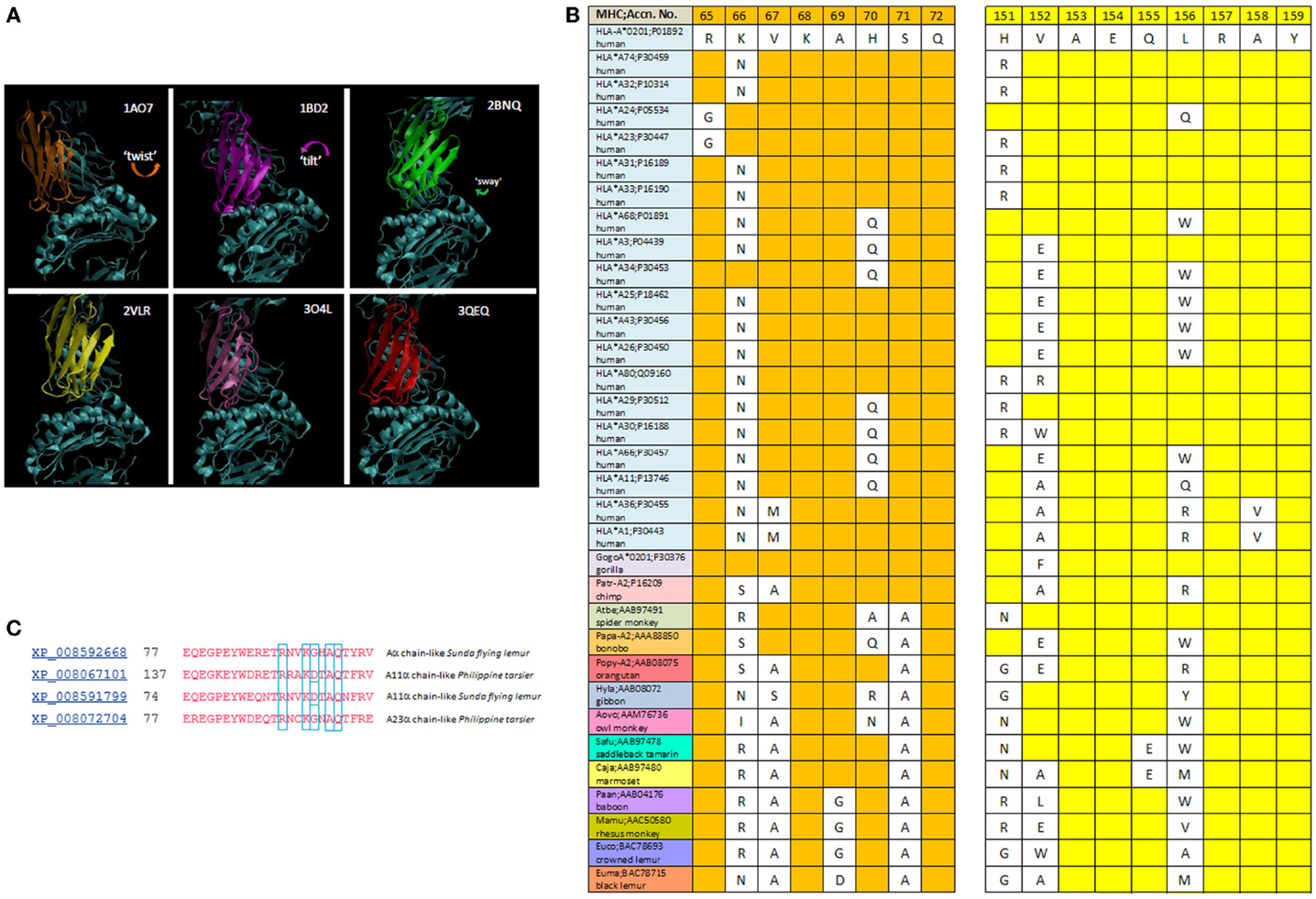
Figure 3. (A) The twist/tilt/sway of TCR-Vα relative to pHLA-A2; PDB files are denoted for each structure. TCR display a diversity of rotation in-plane to the groove {twist} with or without rotation perpendicular to the groove {tilting}; and this includes parallel (side-to-side) variation {sway}. Vα of each different TCR are colored; Vβ and pHLA-A2 are in cyan. (B) Alignment of the R65 motif and CDR2α-contact region among HLA-A alleles and non-human primate MHC A-like proteins. Sequences are from NCBI (blink analysis) with the HLA-A*0201 sequence as query (www.ncbi.nlm.nih.gov). (C) NCBI (blast) of HLA-A2 against prosimians taxid; alignment of different alleles (see text).
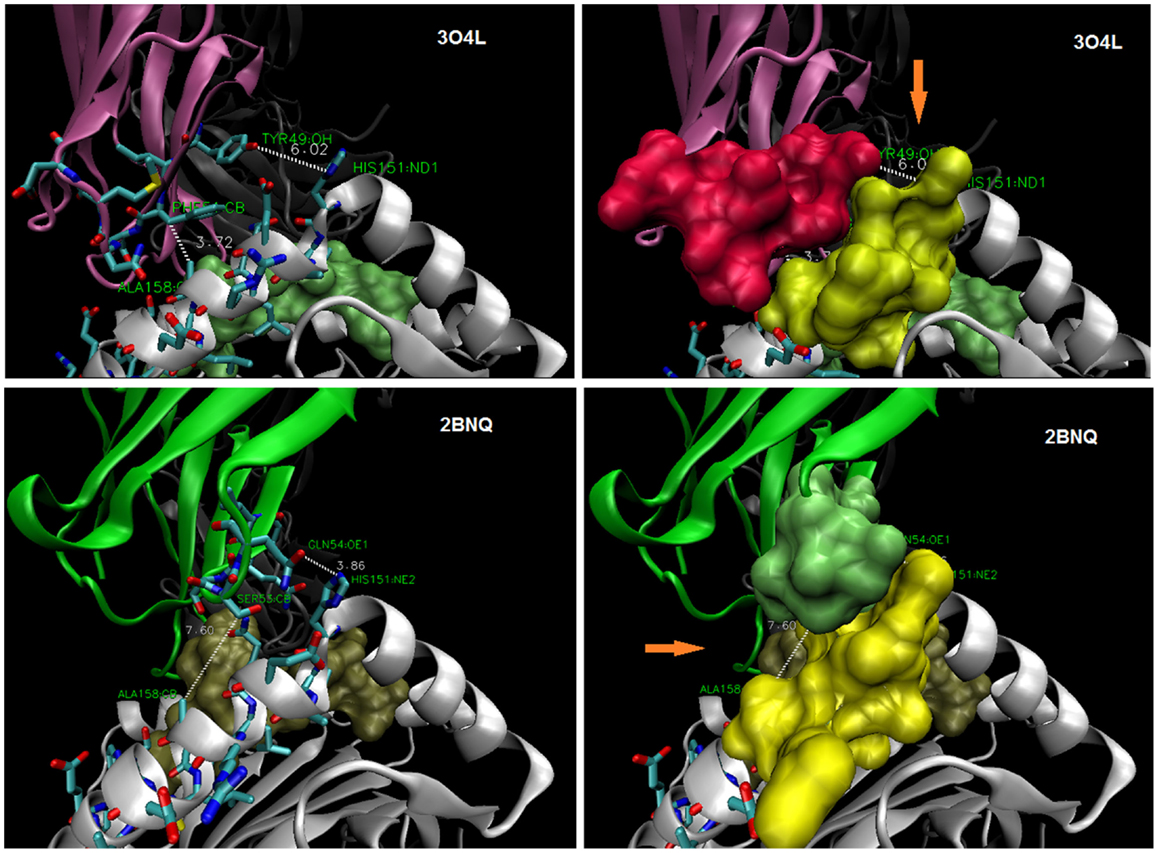
Figure 4. TCR display interfaces with ~4 Å contact at either A158 (top) or at H151 (bottom). Thus, a steric site opposite (orange arrow) is available for occupancy by a larger side-chain (~6–7 Å). HLA-A30 (R151) bearing a permissive peptide would be expected to dock with 3O4L-like TCR, but not 2BNQ-like TCR; HLA-A1 (V158) with 2BNQ-like, but not 3O4L-like TCR. CDR2α and α2-helix (a.a. 151–158) side chains as stick (left), and space-filled (right); peptides are shown as space-filled.
Conservation
R65-X-X-K-A/G-X-S/A-Q72 is conserved in nearly all primate MHC A-like molecules (black lemurs are exceptions, with an A69D disruption; Figure 3B). Interestingly, baboon, rhesus, and crowned lemur have an A69G substitution, but this would substantively conserve motif structure. HLA-A24, -23 (as shown) do not have R65, but interestingly, variants of both do, e.g., A*2424, and A*2429. PDB 3W0W (TCR:HIV-1, Nef peptide:A-2402) has a mortise involving CDR3α Q94-G-G-K97 contact with E62 of the α-1 helix. This shifts the across-the-groove joint, but the CDR2α/α-2 interface range is maintained (see Figure 6) in 3W0W, the TCR is more twisted than in any of the HLA-A2 complexes (see below). More interesting (Figure 3C) is the apparent disruption of the R65 motif in alleles of Tarsius syrichta and the colugo, Galeopterus variegatus, as this puts the motif in a common ancestor (11), some 79.6 Mya (Cretaceous), i.e., well before Paleocene-Eocene, when lemuriforms and tarsiiforms are thought to have diverged (17, 18).
Role of the Peptide in the R65-Joint
As shown in Figure 5, the peptide contacts CDR3α in a fashion compatible with the angle between the R65-joint and the polymorphic contacts with MHC, viz., the CDR2α/α-2 helix interface. Within the structures examined here is displayed a consistent peptide interaction with what could be described as the arm of the CDR3α loop that faces away from R65. The closest contact of this nature among the examined complexes is in 3HG1, which is interesting because this peptide assumes an extended (less bulged) structure, and the angle between the CDR2α contact residue (alpha carbon), the R65 alpha carbon, and the α2-helix contact residue (alpha carbon) (viz., the CDR2α:R65:α-2 angle) is the largest amongst the structures at 18.90°(Figure 5). Interestingly, there is no direct correlation between this angle and the closeness of peptide contact (i.e., when we compare all the structures). However, the CDR2α:R65:α-2 angle does correlate with the overall orientation of the TCR on pHLA-A. For example, 2BNQ with a “flat” angle at 12.23° is tilted similarly to 3O4L, but is more twisted than 3O4L (Figure 3A); thus, the lack of “twist” for 3O4L correlates with its increased R65-angle, 17.96°, as would be the expected geometry. However, 3QEQ and 3W0W have about the same “tilt,” but 3W0W is quite more twisted; here, more “twist” correlated with an increased R65-angle. Therefore, twisting (ω) of the TCR in the plane of the groove seems dependent on the side-to-side sway (∂) parallel to the groove in its exact relationship to tilting (λ), i.e., toward the α-1 helix, at least with respect to increasing or decreasing the R65-angle, or pitch (φ). A plausible formula for the mechanism, based upon our estimates of these parameters, is the following (see Figure 6 and Table 1, for compiled data).
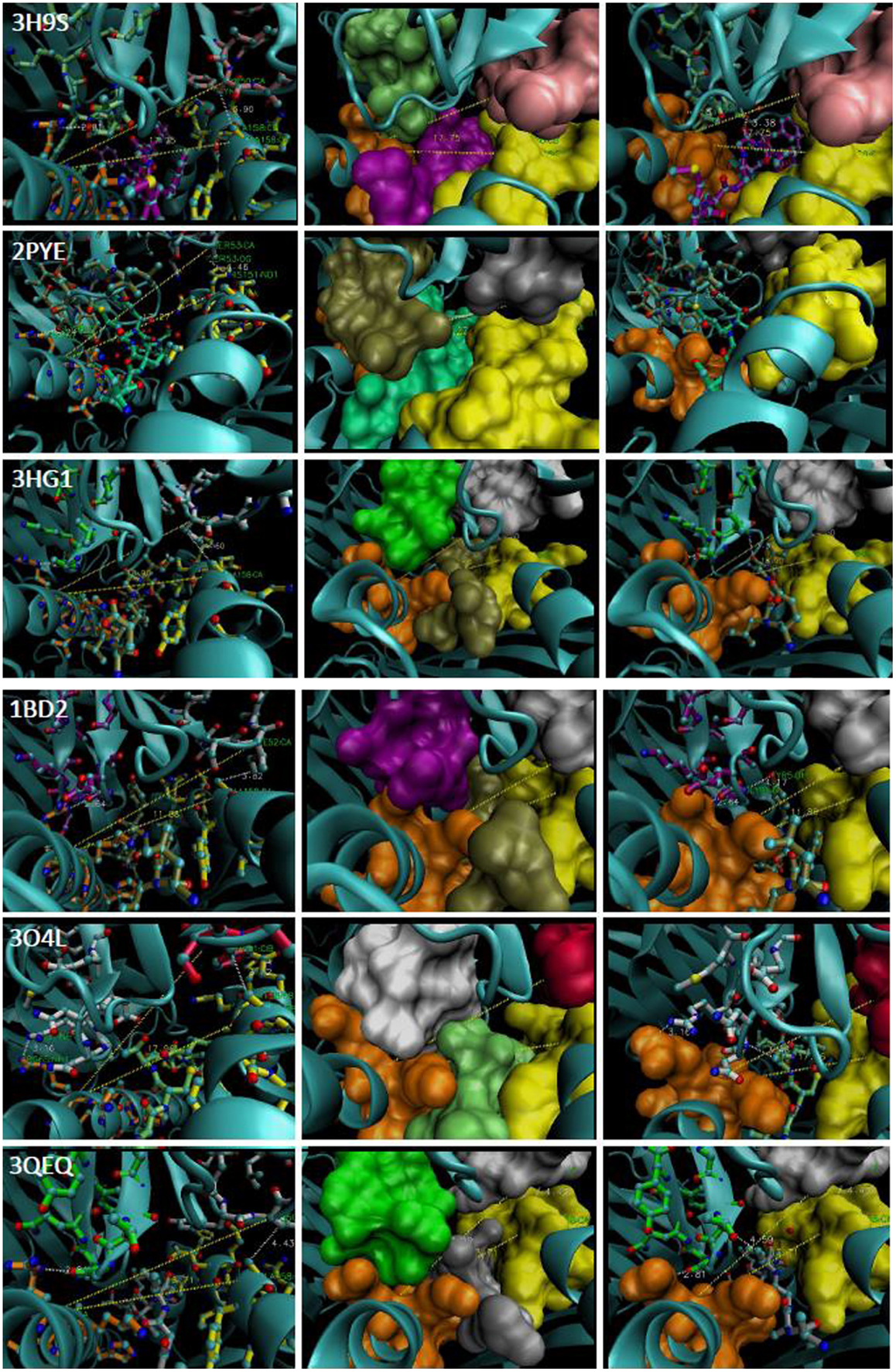
Figure 5. The CDR2α:R65:α-2 angle of representative TCR:pHLA-A2. PDB file is denoted in far left panels and middle and right panels reflect different views of each complex. The R65 motif is in orange and the H151-A158 region is in yellow. Contacting CDR3α a.a., lime (3H9S), tan (2PYE), green (3HG1), magenta (1BD2), white (3O4L), light green (3QEQ). CDR2α: pink (3H9S), silver (2PYE), white (3HG1), white (1BD2), rose (3O4L), and white (3QEQ); cyan ribbon alpha carbon backbones. Peptides: magenta (3H9S), foam (2PYE), tan (3HG1), tan (1BD2), lime (3O4L), and silver (3QEQ). The R65-angle was measured with VMD (shown as yellow trace). CDR3α:peptide contacts are shown as white trace.
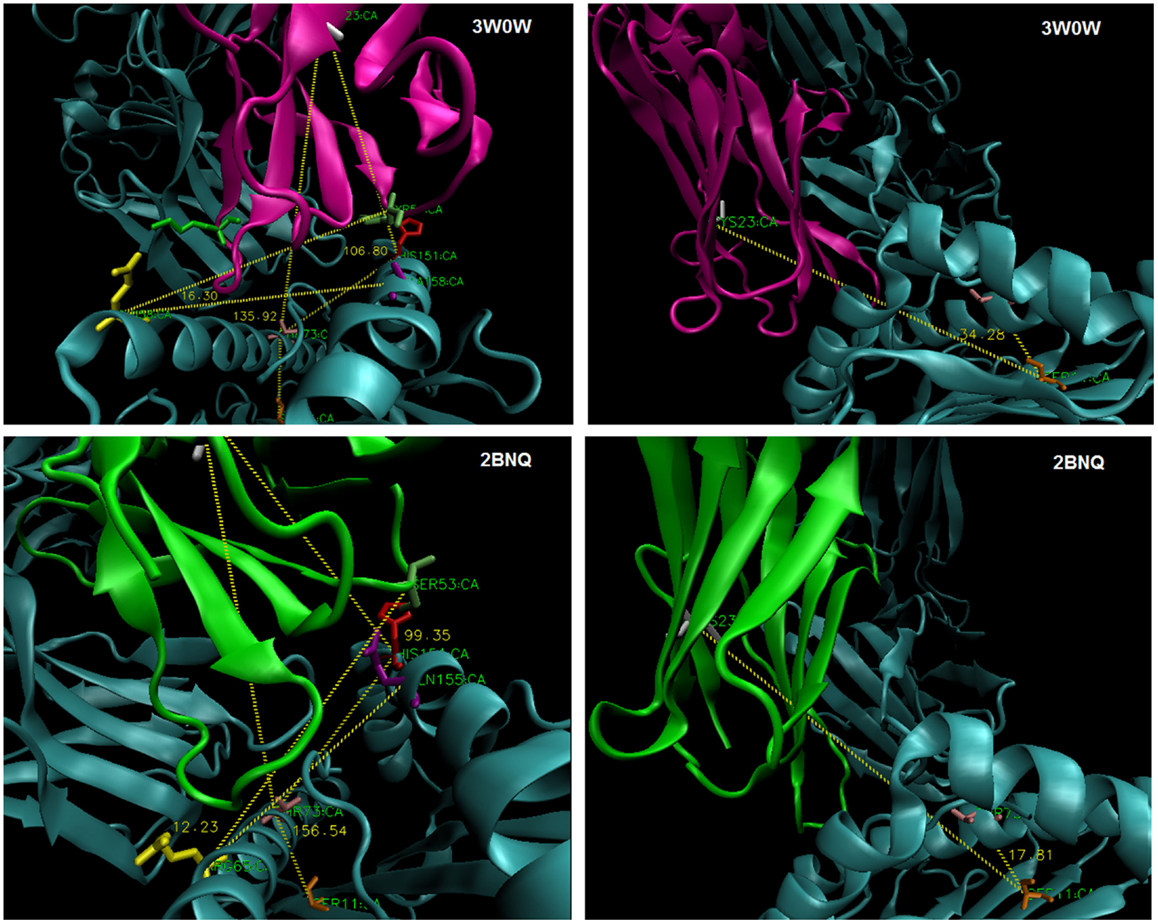
Figure 6. Estimating TCR twist/tilt/sway: (left) measuring an angle across the groove to C22/3/4 of TCR-Vα (“twist”) and perpendicular to the groove to C22/3/4 (“tilt”); (right) measuring an angle parallel to the groove (“sway”).
Angles and contacts for PDB files not previously shown: 1AO7: Y50:R65:A158@17.93°, 2BNQ:S53:R65:Q155@12.23°, 4QOK: Y51:R65:A158@19.73°, 3GSN: I52:Q72:A158@17.92°, 4JFD: Y51:R65:A158@21.14°, 3UTT:K102β:R65:H151@24.28°, and 4EUP: Y52:R65:A158@16.80°.
One testable (19–21) idea is that peptide contacts stabilize dynamics and the CDR2α/α-2 helix interface. Perhaps, a “transition state,” involving key TCR interactions with the MHC, exists initially, followed by peptide interactions with the TCR being “scanned” in a two-step mechanism (22, 23). Alternatively, the TCR may “scan-clamp,” where peptide interactions come first (24–26), or peptide and MHC contacts might occur at the same time (14). Importantly, the R65-joint mechanism is not incompatible with any of these ideas; indeed, different rearrangements might utilize different dynamics to get to the same structural geometry.
The corollary that the R65-angle of these obviously selected TCR reflects deleted (not-selected) thymocytes yielding closer or more distant CDR2α/α-2 helix contacts is intriguing. In other words, a mature T-cell alloreactive capacity is selected-for via CDR3α that can do the R65-joint. Clearly, exceptions are 2VLR (as discussed), and notably 3UTT, wherein CDR3β assumes the ~4 Å contact with the α-2 helix, at H151. In this structure, the closest CDR3α contact is ~5 Å from Q155 (Figure 7). Thus, in the case of 3UTT, the interface of the TCR with a.a. 151–158 polymorphic positions appears to have been directly selected-for, i.e., the other TCR utilizes the indirect across-the-groove Vα geometry described herein.
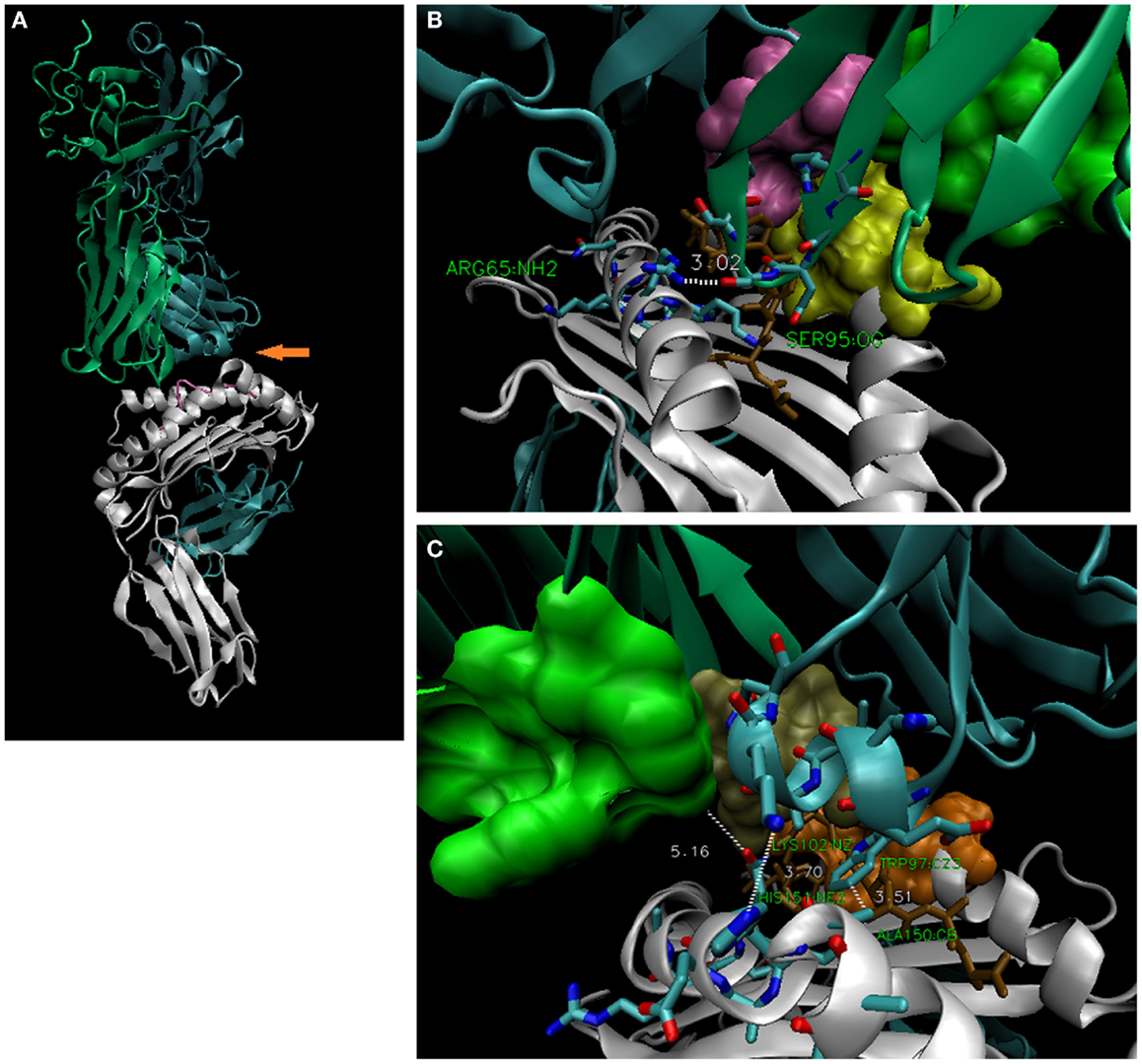
Figure 7. The 3UTT TCR contacts R65 via CDR3α, but contacts the α-2 helix via CDR3β. (A) Overall structure of 3UTT; orange arrow indicates unusual (here) across-the-groove contact between CDR3β and H151 [3.70 Å; bottom right, (C)]. Note CDR3α makes “usual” R65-joint [3.02 Å; upper right, (B)]. Like 2VLR, this seems an alternate strategy to the same distance of TCR contacts with the polymorphic a.a. 151–158 subregion. In this case, the interface was directly selected via the rearranged CDR3β; in all others, it is indirect via the described geometry; actual frequency for these TCR strategies is not known.
Conclusion
The idea that CDR2 and/or CDR1 have “co-evolved” with the MHC with the product being conserved/predictable contacts between them (4) has been disputed (27–29). For instance, co-receptors have been suggested as the true selective agents (28), and TCR have been selected in MHC knock-out mice independently of MHC (29). Nevertheless, CDR1/2 and MHC are clearly germ-line encoded, and any observations of conservative interactions across phylogeny are indeed evidence for “co-evolution” per se; what particular mechanism of thymic selection dictates it is still debatable. However, it must be considered that the somatic mechanism of CDR3 has had to entail with MHC polymorphism for some 400 My (30); and indeed, that the TCR repertoire is inherently alloreactive (1, 11).
The analysis presented here suggests a novel structural mechanism for MHC control of TCR diversity, and may help explain the enigmatic biology of T-cell alloreactivity. Thus, somatic CDR3α appears selected for TCR contact with allo-HLA-A by virtue of the R65-joint geometry explained herein, manifest in the TCR repertoire as the germline CDR2α/α-2 helix interface. Seemingly unusual 3UTT, wherein the interface is apparently directly selected-for via CDR3β, still utilized the R65-joint (S95O:R65NH2, 3.02 Å), and crucially maintained ~4 Å contact at the same α-2 helix position (βK102N2:H151NE2, 3.70 Å). Indeed, in both the 2VLR and 3UTT structures, TCR strategies for maintaining contact with the α-2 helix polymorphic positions seem like exceptions to the rule. Although, to be clear, the actual relative frequency of these different strategies within the TCR repertoire is not known. Nevertheless, the consistent use of the R65-joint geometry, even among these available structures, certainly hints at a rather straightforward hypothesis. Thus, TCR with CDR3α’s yielding TCR:pHLA-A2 complexes with the CDR2α/α-2 helix interface below or above ~4 Å (exception being 3UTT-like TCR) are proposed to be theoretically not selected. That surviving thymocytes turn out to be the best TCR bearers for protective immunity is assumed (this seems essential); what is clear, is that part of the immune system does respond directly against allo-HLA class I molecules for a biologically apparent reason. Indeed, R65-joint bioinformatics (as indicated) are consistent with the emergence of HLA-C and KIR genes (10). Maternal uNK cells induce fetal trophoblast-mediated re-modeling of the maternal circulation; yet, HLA-C:KIR is restricted to the higher primates (31). The structural R65 motif in a shared prosimian ancestor (11, 17, 18), that KIR and TCR bind to overlapping sites on pHLA-A molecules (10); pseudogenes and orphan receptors in extant human KIR genes (10, 31); and the balance of inflammatory/non-inflammatory cytokines (32), all tempt speculation that the R65-joint had/has a role in pregnancy. Finally, while several elegant mechanisms have been described for maintaining maternal tolerance against the fetal paternal allotype (31, 32); the R65-joint might facilitate fetal CD8 T cells to “reject” infiltrating maternal cells via the unshared HLA-A allele, perhaps in the second or third trimesters (33). Obviously, as gestation became more prolonged in primates, alleles containing the motif could have been favored.
Conflict of Interest Statement
The author declares that he has no conflicts of interest regarding this research, manuscript, or its publication; and is solely responsible for the research and the writing of the manuscript.
Acknowledgments
This manuscript would not have been possible without the dedicated work of those publishing PDB coordinates, sequences, and data analysis tools; on the community of science, thank you.
Footnotes
References
2. Davis MM, Bjorkman PA. T-cell antigen receptor genes and T-cell recognition. Nature (1988) 334:395–402. doi:10.1038/334395a0
3. Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature (1996) 384:134–41. doi:10.1038/384134a0
4. Scott-Browne JP, White J, Kappler JW, Gapin L, Marrack P. Germline-encoded amino acids in the αβ T cell receptor control thymic selection. Nature (2009) 458:1043–6. doi:10.1038/nature07812
5. Stewart-Jones G, Wadle A, Hombach A, Shenderov E, Held G, Fischer E, et al. Rational development of high-affinity T-cell receptor-like antibodies. Proc Natl Acad Sci U S A (2009) 106:5784–8. doi:10.1073/pnas.0901425106
6. Adams JJ, Narayanan S, Liu B, Birnbaum ME, Kruse AC, Bowerman NA, et al. T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity (2011) 35:681–93. doi:10.1016/j.immuni.2011.09.013
7. Borbulevych OY, Piepenbrink KH, Baker BM. Conformational melding permits a conserved binding geometry in TCR recognition of foreign and self-molecular mimics. J Immunol (2011) 186:2950–8. doi:10.4049/jimmunol.1003150
8. Ishizuka J, Stewart-Jones GB, van der Merwe A, Bell JI, McMichael AJ, Jones EY. The structural dynamics and energetics of an immunodominant T cell receptor are programmed by its Vbeta domain. Immunity (2008) 28:171–82. doi:10.1016/j.immuni.2007.12.018
9. Labrecque N, Baldwin T, Lesage S. Molecular and genetic parameters defining T-cell clonal selection. Immunol Cell Biol (2011) 89:16–26. doi:10.1038/icb.2010.119
10. Parham P, Norman PJ, Abi-Rached L, Guethlien LA. Human-specific evolution of killer cell immunoglobulin-like receptor recognition of major histocompatibility complex class I molecules. Philos Trans R Soc Lond B Biol Sci (2012) 367:800–11. doi:10.1098/rstb.2011.0266
12. Zoete V, Irving M, Ferber M, Cuendet MA, Michielin O. Structure-based, rational design of T cell receptors. Front Immunol (2013) 4:268. doi:10.3389/fimmu.2013.00268
13. Malecek K, Grigoryan A, Zhong S, Gu WJ, Johnson LA, Rosenberg SA, et al. Specific increase in potency via structure-based design of a TCR. J Immunol (2014) 193:2587–99. doi:10.4049/jimmunol.1302344
14. Cole DK, Miles KM, Madura F, Holland CJ, Schauenburg AJ, Godkin AJ, et al. T-cell receptor (TCR)-peptide specificity overrides affinity-enhancing TCR-major histocompatibility complex interactions. J Biol Chem (2014) 289:628–38. doi:10.1074/jbc.M113.522110
15. Rangarajan S, Mariuzza RA. T cell receptor bias for MHC: co-evolution or co-receptors? Cell Mol Life Sci (2014) 71:3059–68. doi:10.1007/s00018-014-1600-9
16. Birnbaum ME, Mendoza JL, Sethi DK, Dong S, Glanville J, Dobbins J, et al. Deconstructing the peptide-MHC specificity of T cell recognition. Cell (2014) 157:1073–87. doi:10.1016/j.cell.2014.03.047
17. Gingerich PD. Temporal scaling of molecular evolution in primates and other mammals. Mol Biol Evol (1986) 3:205–21.
18. Beard KC. The oldest North American primate and mammalian biogeography during the Paleocene-Eocene thermal maximum. Proc Natl Acad Sci U S A (2008) 105:3815–8. doi:10.1073/pnas.0710180105
19. Wang J, Reinherz EL. A new angle on TCR activation. Immunity (2011) 35:658–60. doi:10.1016/j.immuni.2011.11.008
20. Depoil D, Dustin ML. Force and affinity in ligand discrimination by the TCR. Trends Immunol (2014) 35:597–603. doi:10.1016/j.it.2014.10.007
21. Robert P, Aleksic M, Dushek O, Cerundolo V, Bongrand P, van der Merwe PA. Kinetics and mechanics of two-dimensional interactions between T cell receptors and different activating ligands. Biophys J (2012) 102:248–57. doi:10.1016/j.bpj.2011.11.4018
22. Wu LC, Tuot DS, Lyons DS, Garcia KC, Davis MM. Two-step binding mechanism for T-cell receptor recognition of peptide MHC. Nature (2002) 418:552–6. doi:10.1038/nature00920
23. Housset D, Malissen B. What do TCR-pMHC crystal structures teach us about MHC restriction and alloreactivity? Trends Immunol (2003) 24:429–37. doi:10.1016/S1471-4906(03)00180-7
24. Tynan FE, Burrows SR, Buckle AM, Clements CS, Borg NA, Miles JJ, et al. T cell receptor recognition of a “super-bulged” major histocompatibility complex class I-bound peptide. Nat Immunol (2005) 6:1114–22. doi:10.1038/ni1257
25. Sethi DK, Schubert DA, Anders AK, Heroux A, Bonsor DA, Thomas CP, et al. A highly tilted binding mode by a self-reactive T cell receptor results in altered engagement of peptide and MHC. J Exp Med (2011) 208:91–102. doi:10.1084/jem.20100725
26. Cole DK, Sami M, Scott DR, Rizkallah PJ, Borbulevych OY, Todorov PT, et al. Increased peptide contacts govern high affinity binding of a modified TCR whilst maintaining a native pMHC docking mode. Front Immunol (2013) 4:168. doi:10.3389/fimmu.2013.00168
27. Holland SJ, Bartok I, Attaf M, Genolet R, Luescher IF, Kotsiou E, et al. The T-cell receptor is not hardwired to engage MHC ligands. Proc Natl Acad Sci U S A (2012) 109:E3111–8. doi:10.1073/pnas.1210882109
28. Van Laethem F, Tikhonova AN, Pobezinsky LA, Tai X, Kimura MY, Le Saout C, et al. Lck availability during thymic selection determines the recognition specificity of the T cell repertoire. Cell (2013) 154:1326–41. doi:10.1016/j.cell.2013.08.009
29. Van Laethem F, Sarafova SD, Park JH, Tai X, Pobezinsky L, Guinter TI, et al. Deletion of CD4 and CD8 coreceptors permits generation of alphabetaT cells that recognize antigens independently of the MHC. Immunity (2007) 27:735–50. doi:10.1016/j.immuni.2007.10.007
30. Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature (1998) 394:744–51. doi:10.1038/29457
31. Parham P, Norman PJ, Abi-Rached L, Hilton HG, Guethlein LA. Immunogenetics of human placentation. Placenta (2012) 33:S71–80. doi:10.1016/j.placenta.2011.11.020
32. Leno-Durán E, Muñoz-Fernández R, Garcia Olivares E, Tirado-González I. Liaison between natural killer cells and dendritic cells in human gestation. Cell Mol Immunol (2014) 11:449–55. doi:10.1038/cmi.2014.36
Keywords: immunogenetics, TCR, MHC, HLA, alloreactivity, placentation, primate, evolution
Citation: Murray JS (2015) An old twist in HLA-A: CDR3α hook up at an R65-joint. Front. Immunol. 6:268. doi: 10.3389/fimmu.2015.00268
Received: 19 January 2015; Accepted: 14 May 2015;
Published: 27 May 2015
Edited by:
Nilabh Shastri, University of California Berkeley, USAReviewed by:
Edward John Collins, The University of North Carolina at Chapel Hill, USAJia-Huai Wang, Dana-Farber Cancer Institute, USA
Copyright: © 2015 Murray. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joseph S. Murray, Xenolaüs Genetics LLC, 10880 Wilshire Blvd., Suite 1101, Los Angeles, CA 90024, USA,anNtdXJyYXlAeGVub2xhdXNnZW5ldGljcy5jb20=
 Joseph S. Murray
Joseph S. Murray