- 1Laboratory on Thymus Research, Oswaldo Cruz Institute, Oswaldo Cruz Foundation (Fiocruz), Rio de Janeiro, Brazil
- 2Graduate Program in Cell and Developmental Biology, Institute of Biomedical Sciences, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil
Multiple signaling pathways control every aspect of cell behavior, organ formation, and tissue homeostasis throughout the lifespan of any individual. This review takes an ontogenetic view focused on the large superfamily of TGF-β/bone morphogenetic protein ligands to address thymus morphogenesis and function in T cell differentiation. Recent findings on a role of GDF11 for reversing aging-related phenotypes are also discussed.
Introduction
The adaptive immune system evolved as a complex set of defense mechanisms amplified by the specificity properties of antigen receptor-bearing B and T lymphocytes (1). Following blood trafficking into the thymus, bone marrow-derived lymphoid progenitors become committed to T cell lineage development. Within this organ, cell specialization occurs gradually in a manner that T cell development results in the generation of conventional CD4 and CD8 αβ T cells along with natural killer T cell (NKT; an innate-like T cell subpopulation), regulatory T cell (Treg), and γδ T cell subsets (2). Classically, commitment to T cell lineage was found to rely on the Delta-class Notch ligand Delta-like 4 (DLL4) and the interleukin-7 (IL-7) along with kit and flt3 ligands at stages usually prior to TCRβ chain assembling (3–6). Branching into distinct paths can be observed throughout the mainstream developmental pathway, from the double-negative (DN; CD4−CD8−) T cell precursors to the highly expanded double-positive (DP; CD4+CD8+) cells, and the resulting mature single-positive (SP; CD4+CD8− or CD4−CD8+) stages. Thus, at specific niches, the thymus provides to developing T cells signals that trigger a series of ordered events leading to cell proliferation, TCR gene rearrangements, and selective checkpoints along with massive cell death (7). Altogether, these events culminate in a proper repertoire of distinct and specialized mature thymocyte subpopulations able to emigrate to the periphery. In this review paper, we highlight the role of members of the large transforming growth factor-β (TGF-β) superfamily (Box 1) during thymic ontogeny, thymic epithelial cell (TEC) differentiation and function, as well as T cell maturation. Lastly, we discuss recent information on a possible regenerative potential of TGF-β ligands to rescue aging-related thymus atrophy.
Box 1. Multiple roads for signaling by TGF-β superfamily members.
The TGF-β superfamily comprises TGF-β1–3, bone morphogenetic proteins (BMPs), growth and differentiation factors (GDFs), Nodal, activins/inhibins, Müllerian inhibiting substance (MIS)/anti-Müllerian hormone (AMH), and Lefty. These ligands were initially grouped accordingly to the functional roles observed following their original identification (8–11). As it became clear that most ligands play multiple functions depending on cell type, developmental stage, or tissue conditions, they are now classified by sequence similarity and the downstream pathway they activate (12). Each family member has an overall basic structure, in which inactive forms are produced with an N-terminal secretion peptide and a large propeptide domain known as latency-associated peptide (LAP). Cleavage of the propeptide domain by proprotein convertases releases a mature domain at the C-terminus, which eventually dimerizes (13). The propeptide domain has major regulatory roles. It influences protein stability and functions as chaperone during secretion, also mediating diffusion through interactions with the extracellular matrix and inhibiting the active peptide form even after cleavage (14–16).
Signaling by TGF-β superfamily members occurs through a similar mechanism, but operates with distinct components. Ligands bind single-pass transmembrane receptor serine/threonine kinases, which relay the signal for intracellular effectors capable of translocating into the nucleus to modulate gene transcription (Figure 1). More specifically, these receptors are classified into two structurally similar types. Ligand binding occurs only through type II receptors, which then recruit and phosphorylate type I receptors [e.g., Ref. (17, 18)]. Type II receptors, such as ActRII (Acvr2a) or ActRIIB (Acvr2b), may take part in many distinct pathways or may be specific for a given group of ligands, such as AMHR2 (Amhr2) for MIS/AMH, BMPRII (Bmpr2) for most BMPs and Gdf9, and TβRII (Tgfbr2) for TGF-βs (19, 20). Type I receptors are also known as activin receptor-like kinases (ALKs) due to their sequence similarity to activin receptors (21). These receptors are usually specific to a more restricted set of ligands. For instance, Nodal, Gdf1, Gdf11, activins, and inhibins bind ActRII to recruit Alk4 (Acvr1b) and Alk7 (Acvr1c) or they bind ActRIIB to recruit either Alk4, Alk7, or Alk5 (Tgfbr1) (19). Together, type II and type I receptors form a heterotetrameric complex, in which the type I receptor further phosphorylates intracellular effectors of the Smad family (22). Depending on the ligand/receptor complex they are responding to, receptor-activated Smads (R-Smads) can be subdivided into two groups: a BMP-related set gathers Smad1, Smad5, and Smad9 (formerly Smad8), whereas Smad2 and Smad3 are responsive to TGF-β-related signals (Figure 1). An N-terminal MH1 domain negatively regulates the MH2 domain, being indispensable for Smad translocation into the nucleus and DNA binding (23–25). However, these functional properties do not hold true for all R-Smads. In particular, Smad2 seems to interact to DNA only indirectly (24).
A common mediator Smad (co-Smad), or Smad4, integrates signals from both branches by associating with the R-Smads (Figure 1). They form transcriptional complexes able to translocate into the nucleus (26–28). Nuclear transportation of Smads depends on accessory proteins, particularly importins, exportins, and nucleoporins (29, 30). The presence of DNA molecules harboring Smad-binding elements favors heterodimerization between R-Smads and co-Smad (28). They ultimately associate with cell-type-specific transcription factors and co-activators to regulate a plethora of target genes (31).
Regulation of Smad activity occurs through multiple mechanisms (32). Two inhibitory Smads (I-Smads) impair signaling by competing with R-Smads for receptors or by co-Smad interaction (33). For instance, Smad6 forms stable interactions with type I receptors, blocking phosphorylation of Smad2 and Smad1, but not Smad3 (34, 35). Similarly, Smad7, the other I-Smad member, also binds type I receptors and suppresses further phosphorylation by targeting them for proteasome-dependent degradation (35, 36). The available literature on the molecular interactions of TGF-β superfamily members is vast, but not in the scope of this review. Further information can be found elsewhere (33, 37–40).
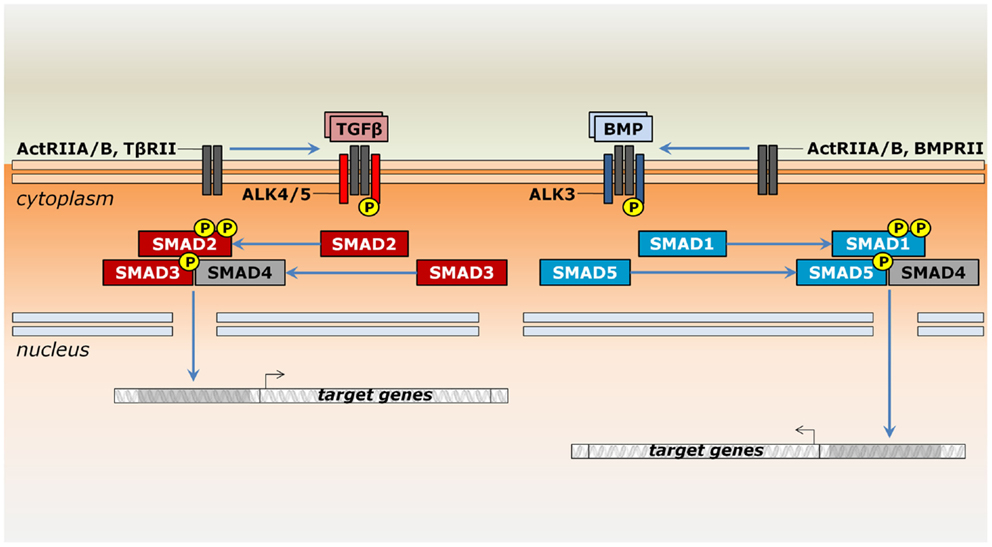
Figure 1. Signaling by ligands of the TGF-β superfamily in the thymus. Members of the TGF-β superfamily may signal by either the TGF-β (reddish) or the BMP branch (bluish). Upon binding to type II serine/threonine receptors occurs the recruitment of type I receptors, which further phosphorylate Smad proteins. Whereas ActRIIA and ActRIIB may be shared between both pathways, TβRII and BMPRII are specific to TGF-β and BMP signaling, respectively. In general, Smad2 and Smad3 relay signals from the Alk4, Alk5, and Alk7 receptors, while Smad1, Smad5, and Smad8/9 are phosphorylated by Alk2, Alk3, and Alk6 receptors. However, Alk2, Alk6, and Alk7 are not expressed during thymocyte maturation. Modulation of gene expression occurs after Smad complex translocates into the nucleus and depends on the interaction with additional protein complexes (not shown).
TGF-β Signaling and Thymus Formation
Organogenesis relies on well-organized interactions between distinct germ layers and differentiating cell types controlled by intricate molecular hierarchies. Thymus development occurs from common parathyroid bilateral rudiments in the epithelial endodermal lining of the third pharyngeal pouch around embryonic days (E) 9.0–9.5 in mice and early week 5 in humans (Figure 2A) (41–44). As growth continues through E10.5 in mice and early week 6 in humans, the contact between the third pharyngeal pouch and the third pharyngeal cleft ectoderm determines paired organ primordia with stratified epithelium and a central lumen lined by precursors of medullary thymic epithelial cells (mTECs). These cells are characterized by the expression of both claudin-3/4 and cytokeratin-5 (K5) (46, 47). Further development of thymic medulla also depends on the successful establishment of the cortical region, as observed in mice with arrested T cell development (48). Within each primordium, a dorso-rostralmost domain expressing Gcm2 gives rise to a parathyroid gland from E9.5 in mice or as early as the onset of week 6 in humans, whereas a ventro-caudalmost domain identified by Foxn1 expression produces a thymic lobe from E11.25 in mice or mid-week 6 in humans (Figure 2B) (44, 49–52). Epithelial cell proliferation fills the pharyngeal pouch lumen by forming cord-like structures with smaller lumina, similar to branching morphogenetic events in other organs (Figure 2C) (47). In this context, activation of Foxn1 blocks the respiratory development (53) and, along with subsequent colonization by lymphocyte precursors, seems to be responsible to produce a concentric medulla less densely cellular than the surrounding cortex (47). Fetal liver-derived lymphocyte progenitors colonize the embryonic thymus from E11.5 in mice and week 8 in humans (54, 55), whereas short-term apoptotic events around E12.0 disconnect the developing anlagen from the embryonic pharynx (41). The rudiments migrate downwards at different paces, gradually resolving the Gcm2- and Foxn1-restricted domains into two morphologically distinct structures enclosed by neural crest-derived mesenchyme (Figures 2C–F) (51, 56). Parathyroid primordia usually lag behind and move toward the tracheal region dorsally to the thyroid gland, whereas thymic rudiments move ventrally and more caudally into the thoracic cavity (Figures 2D,E). The thymic primordia ultimately fuse at the midline to produce a bi-lobed organ above the developing heart (Figure 2F). Unlike mice, humans exhibit superior parathyroid glands derived from the fourth pharyngeal pouch (Figure 2) (43), whereas organogenesis of the human thymus is essentially similar to mice both morphologically and molecularly (44). Each of these morphogenetic events during thymus organogenesis is controlled by a multitude of signals, including members of the TGF-β superfamily.
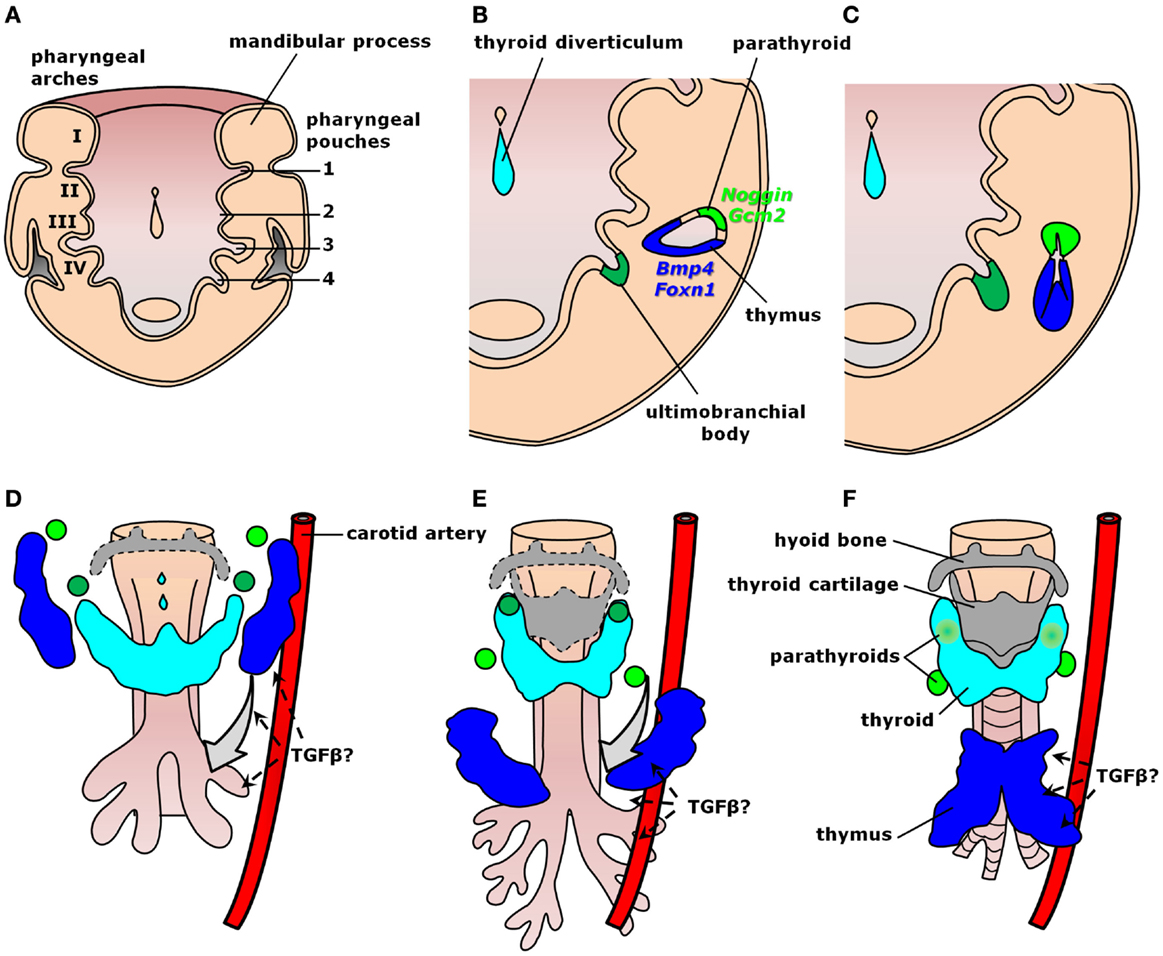
Figure 2. Signaling by TGF-β superfamily members during thymus organogenesis. Schematic representation of thymus formation at different stages of development. (A–C) Thymus specification viewed dorsally at the ventral half of the pharyngeal region. (A) The common parathyroid–thymus primordium arises from the third pharyngeal pouch endoderm. (B) Within each anlage, mTEC precursors line a central lumen surrounded by a dorso-rostralmost domain expressing the BMP-antagonist Noggin and the parathyroid specific gene Gcm2 (light green), whereas the ventro-caudalmost domain expresses Bmp4 and the thymus-specific gene Foxn1 (blue). (C) Each primordium grows in size while proliferating cells fill the rudiment lumen, later colonized by lymphocyte precursors to produce an inner medulla. (D–F) Thymus migration toward the heart. The inferior parathyroid (light green) and the thymus (blue) primordia are gradually resolved as they migrate downwards. (D) TGF-β cues from the endothelium of pharyngeal blood vessels (e.g., carotid arteries) seem to orient thymic and parathyroid migration toward their final location. (E) The third pharyngeal pouch-derived thymic and the inferior parathyroid rudiments pass by the primordia of the superior parathyroid (dark green), which migrate only a short distance downward the tracheal region. (F) Fusion of the thymic primordia occurs at the midline just above the developing heart (not shown) [modified from Ref. (45)].
Thymus Specification and Thymic Epithelial Cell Differentiation
Early production of Bmp4 by the endoderm, the surrounding neural crest-derived mesenchyme, and the overlying ectoderm of the third pharyngeal arch and cleft raised the possibility that bone morphogenetic protein (BMP) signals may trigger thymus and parathyroid formation (57). However, conditional inactivation of Bmp4 in both pharyngeal endoderm and mesenchyme using a Foxg1–Cre line had no effect in organ induction, but resulted in abnormal morphogenesis (see below) (58). This could be the result of a short-time window of 24 h necessary to establish the prospective thymic and parathyroid domains as observed in chicken embryos (59). Indeed, Patel et al. have observed using a Bmp4lacZ-reporter line that the onset of Bmp4 production occurred at E9.5 in the ventral pharynx close to the third pouch entrance, but not in the pouch endoderm or mesenchyme proper (57). Expression in these tissues was later achieved and expanded to the overlying ectoderm (57). The realization that endoderm patterning occurs before primitive gut and pharyngeal pouch formation still hampers the identification of signals responsible for thymus specification in vivo and other members of the TGF-β superfamily may also be at play (60). Particularly, activin A is required to induce definitive endoderm prior to the differentiation of third pharyngeal pouch endoderm in vitro (61). Since gene targeting of some superfamily ligands or their receptors results in embryonic lethality (62–64), new conditional mutants should be produced taking into consideration that gene deletion may have to occur earlier and at different embryonic compartments than previously thought.
The possibility that thymus induction depends on synergistic effects of TGF-β superfamily ligands with non-superfamily signals is a likely case (59). Endoderm-derived undifferentiated epithelial cells comprise a homogeneous population phenotypically defined as cytokeratin (K)5+K8+EpCAM+MTS24+ in the thymic primordium of mouse embryos at E12.0 (65). When a single progenitor cell labeled with enhanced yellow fluorescent protein (eYFP) was microinjected into an unlabeled syngeneic thymus rudiment with the same age, and transplanted under the kidney capsule, both cortical and medullary portions showed scattered eYFP+ TECs also positive for region-specific markers after 4 weeks, revealing that common bipotent progenitors are able to produce both epithelial lineages during embryogenesis (65). Recently, thymic epithelial progenitor cells (TEPCs) bearing stem-cell features were also identified in the thymus of adult mice as a MHCIIlowα6 integrinhighSca-1high subset (66). They mature in a highly complex stepwise process not fully understood, ultimately producing cortical TECs (cTECs) or mTECs (67).
Cortical TECs are sparsely distributed and may be identified as CD45−EpCAM+Ly51(CD249)+Ulex europaeus lectin 1 (UEA-1)−K5−K8+ cells with high levels of both MHC II and the proteasome subunit β5t (68–71). Considering the TGF-β-related pathways, cells from neonatal mice express both the Acvr2a (ActRII) and Acvr2b (ActRIIB) genes for the common receptors, in addition to Acvr1 (Alk2), Bmpr1a (Alk3), and Bmpr2 (BMPRII) for the BMP-specific receptors, and the TGF-β-specific type I receptors, Alk4 (Acvr1b) and Alk5 (Tgfbr1), and type II receptor TβRII (Tgfbr2) (71, 72). This set of receptor genes allows cTEC to respond to both signaling branches of the TGF-β superfamily, even though the BMP receptor, Bmpr1b (Alk6), and the TGF-β receptor, Acvr1c (Alk7), are not present. Yet, expression of subunit genes Inha and Inhbb for inhibins and activins, Bmp2 and Bmp4, and Tgfb1 and Tgfb3 makes possible the existence of an autocrine circuitry for thymic homeostasis, and indicate that these factors might influence early thymopoiesis (71, 72).
In the thymic medulla, mTECs are characterized by a CD45−EpCAM+Ly51−K5+K8− phenotype with variable levels of UEA-1, MHCII, CD80, and Aire (67). These distinct expression profiles seem to take part in the differentiation program in which MHCIIhighCD80high mature mTECs expressing Aire are responsible for the production of numerous peripheral self-antigens in the thymus, a critical event for central tolerance (67, 73–77). Hence, SP cells that strongly interact with self peptides through MHC molecules (pMHC) arrest migration, exhibit sustained TCR activation, persistent high levels of cytosolic Ca2+, and early caspase activation, leading to macrophage-dependent phagocytosis (78, 79). Surprisingly, thymocyte apoptosis triggers the production of all three TGF-β ligands by dendritic cells (DC), macrophages, and TECs in the medullary region of neonate or adult thymuses, a phenotype that was partially impaired in Bim mutants (80). In addition, apoptosis-driven production of TGF-β signals resulted in an increased generation of thymic regulatory T (tTreg) cells (see below) (80). Interestingly, mTECs are the cell type in the thymus that express most ligand genes of the TGF-β superfamily and their cognate receptors – Inha and Inhbb, Bmp2, Bmp3, Bmp4, Bmp5, Bmp6, and Gdf6/Bmp13, Gdf3, Gdf6/Bmp13, Gdf8/myostatin, Gdf10, Gdf11, and Gdf15, Lefty1 and Lefty2, Tgfb1, Tgfb2, and Tgfb3 along with Acvr2a (ActRII), and Acvr2b (ActRIIB), Acvr1 (Alk2), Bmpr1a (Alk3), and Bmpr2 (BMPRII) for BMP/growth and differentiation factor (GDF) signaling, and Acvr1b (Alk4) and Tgfbr1 (Alk5) for the TGF-β/Activin/Nodal pathway, in addition to the type III receptor gene Tdgf1 (Cripto) (71, 72, 81, 82).
The possibility that members of the TGF-β superfamily produced by mTECs may influence T cell differentiation or impact thymus physiology cannot be ruled out and remains to be thoroughly investigated. For instance, despite the previously identified BMP ligands in mTECs – Bmp3/osteogenin, Bmp5, Bmp6, and Bmp13 – there is no available functional information regarding their activities in the thymus to our knowledge. It is known, on the other hand, that Bmp6 exerts an antiproliferative effect in peripheral CD19+ B cells and induces apoptosis in CD27+ memory B cells (83). By contrast, Tgfbr2 deficiency in differentiating T cells increased apoptosis of TCRβhighCD4+ and TCRβhighCD8+ mature SP cells after anti-CD3 treatment or of TCRβhighOT-II T cells after antigen-dependent stimulation, thus revealing that TGF-β signals might be involved in thymocyte-negative selection (84). Interestingly, loss of Tgfbr2 in TECs using a Foxn1–Cre mouse line resulted in an expansion of the mTEC compartment – especially MHCIIhigh cells – without affecting cTEC cellularity and the morphology of the corticomedullary junction (85). Indeed, other lymphocyte-derived signals than TGF-β ligands are known to influence mTEC maturation, a phenomenon that is largely known as “thymic cross-talk” (86).
Signaling by TGF-β superfamily members appears to play a secondary role in regulating a master regulator of thymus development and function. Inactivation of the transcription factor Foxn1 results in an athymic phenotype despite the formation of an epithelial anlagen during embryogenesis (49, 87). Expression of Foxn1 in thymic primordia is anticipated by the production of Bmp4 and Wnt4 in the epithelium and the adjacent mesenchyme of the third pharyngeal pouch from E10.5 in mice and from mid-week 6 in human embryos (23, 57, 88). Accordingly, in vitro treatment of fetal thymic organ culture (FTOC) with BMP4 or overexpression of Wnt4 in a TEC cell line upregulated the expression of Foxn1 (88, 89). However, conditional inactivation of Bmp4 in the pharyngeal endoderm and mesenchyme did not affect Foxn1 expression (58), similarly to transgenic embryos expressing the BMP-antagonist Noggin in TECs (90). In turn, information on blockage of Wnt4 and its effect over the expression of Foxn1 is limited. In particular, Talaber et al. have shown that a single administration of dexamethasone caused the reduction of both Wnt4 and Foxn1 levels (91). Interestingly, conditional deletion of β-catenin in mTECs using a BK5–CreERT line resulted in Foxn1 downregulation (92). Altogether, the available evidence suggests that induction or maintenance of such an essential transcription factor in the thymic epithelia relies on an intricate molecular hierarchy with a key participation for BMP and WNT signals, which may provide some kind of redundancy for TEC differentiation and function.
With a great potential for translational medicine, differentiation of TEPCs from mouse or human embryonic stem cells (ESCs) can be achieved under culture conditions by the addition of selected growth factors, including TGF-β superfamily ligands. For instance, Lai and Jin have initially reported that incubation with Fgf7, Bmp4, Egf, and Fgf10 produced K5+K8+EpCAM+ cells from mouse ESCs (93). These cells were able to further differentiate into medullary K5+K8− and cortical K5−K8+ TECs when transplanted with CD4−CD8−CD45+ thymocytes under the kidney capsule and sustain normal T cell maturation (93). In humans, an Activin A-dependent inductive stepwise process first differentiate ESCs into definitive endoderm (94), and later into SOX2+FOXA2+CDX2− anterior foregut endodermal cells by the concurrent inhibition of BMP and Activin/TGF-β signaling using Noggin and the type I receptor-specific inhibitor SB-431542, respectively (95). Further development into TEPC may be achieved by relatively similar approaches, generally modulating retinoic acid, canonical Wnt, and BMP level (61, 96).
Thymus Colonization by Lymphoid Precursors
Colonization of the thymic primordia occurs through intermittent cell flow based on chemokine-dependent mechanisms (55, 97–100). It begins discretely prior to organ vascularization with T cell-restricted progenitors that are unable to definitely populate the thymus (55, 97). Cell influx is transiently interrupted during thymus migration to the thoracic cavity (42). Then, a second wave of cell colonization brings multipotent T cell- and NK-cell progenitors before birth (55). The most significant chemokines currently identified for attracting early T lineage progenitors (ETPs) to the developing avascularized thymus are CCL25 and CCL21 (98). Curiously, whereas CCL25 is produced by both Foxn1-positive TECs and the adjacent parathyroid primordium, CCL21 is expressed only by Gcm2-positive cells (99, 101). These ligands signal, respectively, through the CCR9 and CCR7 receptors present in CD45+ ETPs (102–105). However, it is still poor defined whether members of the TGF-β superfamily directly or indirectly influence or are modulated by these chemokines during thymus colonization. In particular, Gordon et al. observed delayed ETP homing into Bmp4-deficient thymic primordia at E11.5, but no significant differences in CCL25 expression in relation to wild-type thymus (58). The relationship with CCL21, other chemokines and their cognate receptors in the embryonic thymus, if present, remains to be determined. Of note, many pathological conditions and morphogenetic events show participation of TGF-βs, BMPs/GDFs, and activins/inhibins in the modulation of chemokine production and vice versa (106–113).
Interaction of immigrating lymphocyte progenitors with the thymic stroma is critical for adult thymus organization, but not for TEC differentiation during embryonic development. Using CD3ϵ transgenic mouse embryos, known to exhibit arrested T cell maturation at the triple negative (TN) CD3−CD4−CD8−CD44+CD25− ETP stage (114, 115), Jenkinson et al. have shown that K5+K8+ bipotent TEPCs normally differentiate into functional K5+K8− medullary and K5−K8+ cortical TECs, although adult thymus in these transgenic animals exhibit persistent flat organization with morphologically abnormal cortex (115, 116). In particular, transfer of normal bone marrow cells into RAG2−/−; tgϵ26 chimeric mice, in which bone marrow cells from mice mutant for the recombination activating gene 2 (RAG2) were previously transplanted into newborn tgϵ26 mice, rescued thymic organization and cellularity in the adult (48).
Thymus Migration
The subsequent migration of the thymus into the thoracic cavity also relies on signaling by members of the TGF-β superfamily and depends on neural crest cells. Despite a minor contribution in thymus cellularity, forced production of the BMP-antagonist Noggin in the caudal hindbrain prior to neural crest migration using B2-NC:Noggin transgenic mice culminated in thymic hypoplasia or aplasia later in development (117). Indeed, Bmp2 induces Cdc42-dependent actin cytoskeleton reorganization and filopodia formation in neural crest cells, consequently affecting their subsequent migration (118). Moreover, conditional loss of Bmp4 in mice expressing Foxg1–Cre impaired the separation between correctly patterned parathyroid and thymus, which also exhibited a partially compromised capsule (58). Yet, based on observations performed for thyroid migration (119), Gordon and Manley have proposed that the downward migration of the thymus may be driven by signals from the pharyngeal blood vessels, more specifically the carotid arteries (Figures 2D–F) (42). Remarkably, mouse embryos with cardiac neural crest cells deficient for the type I receptor Alk5 (Tgfbr1) show defective cardiac outflow development, with atypical branching of carotid arteries and failed migration of still connected parathyroid and capsule-encased thymus (120). This raises the possibility that the directional cue for thymus migration might be Alk5 ligand (e.g., TGF-β1–3 or Gdf11), possibly secreted or released through the endothelium (Figures 2D–F). By contrast, conditional inactivation of Tgfbr2 in TECs by a Foxn1–Cre mouse line does not affect thymus final positioning (121). Although producing distinct phenotypes, each signaling branch by members of the TGF-β superfamily is involved in the downward migration of thymic primordia and reveals a critical, but still poorly understood role for the neural crest-derived capsule during thymus organogenesis. Neural crest cells may also differentiate into endothelial cells, pericytes, and smooth muscle cells, and were found to persist in adult mice up to the onset of thymus involution (122).
Thymus Organization and Maturation of T Cells Under TGF-β Superfamily Signals
The adult thymus exhibits two gross anatomical regions easily identified by their histological staining patterns. The peripheral cortex harbors more immature and mostly small thymocytes, and is darker-stained due to a higher cell density. A corticomedullary junction supplied by numerous septal blood vessels makes the transition between the cortex and the central medulla. This latter region is paler due to cell size and a lower T cell density (123–125). A capsule of connective tissue encases the organ. It consists of an outer layer rich in type I collagen and an inner layer of reticular fibers containing type III collagen, and projects type I collagen-containing septa into the parenchyma, partially subdividing the thymus into smaller lobules (126).
Signals from members of the TGF-β superfamily have a major influence on T cell differentiation and thymus homeostasis. As secreted molecules, they may be locally produced by thymic stromal cells and act over developing T cells as paracrine factors or be produced by the thymocytes themselves and work autocrinely. Alternatively, factors from the developing T cells may similarly operate over stromal cells to support thymus homeostasis. However, thymocytes do not express most members of the TGF-β superfamily and the ones present vary in expression as cells differentiate (Figure 3). Similar changes are also found for receptor genes (127). Such differences in gene expression occur during T cell maturation, but also when comparing the same stage from fetal and adult thymuses (127–131). Nevertheless, provision of soluble growth factors seems to rely mostly to stromal cells, particularly TECs (71, 127). It is still possible that members of the TGF-β superfamily also act over large distances, being produced by other organs and reaching the thymus through the circulatory system (132). The importance of endocrine stimuli for intrathymic T cell maturation has been largely investigated (133), but whether a given TGF-β ligand exerts long-range effects over thymopoiesis remains to be properly addressed.
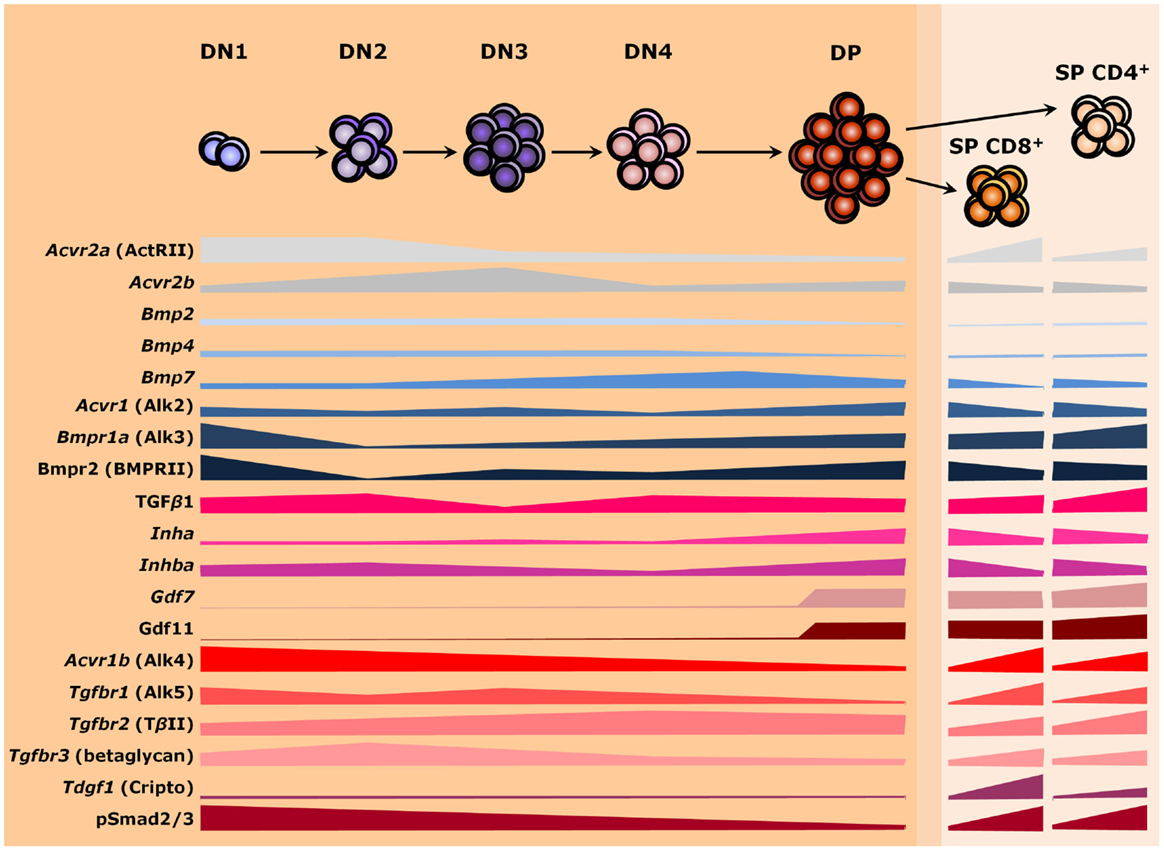
Figure 3. TGF-β superfamily during thymopoiesis. Levels of selected ligands, receptors, and Smad intracellular effectors during the differentiation of αβ T lymphocytes. Common receptors between the TGF-β and the BMP branches are colored in shades of gray, whereas components of the BMP and the TGF-β pathways are colored in shades of blue and red, respectively. The darker orange region of the scheme represents the thymic cortex, whereas light orange represents the thymic medulla. A thin corticomedullary region is represented in between the cortex and medulla. Omitted components are either not present during thymocyte maturation or no information is available at present. DN, double-negative; DP, double-positive; SP, single-positive.
Changes in phosphorylation levels of Smad2/3 (pSmad2/3) and Smad1/5/8 (pSmad1/5/8), respectively, used as read-outs for the activities of TGF-β/Activin/Nodal and BMP/GDF signaling, follow differences in the expression of respective cognate receptors as thymocytes mature (134, 135). Thymocytes differentiate in a stepwise process that involves the somatic rearrangement of T cell receptor (TCR) genes while migrating in close contact with stromal cells and the extracellular matrix (ECM) throughout thymic compartments (136, 137). In this process, a major group of αβ TCR-bearing T cells are produced, which ultimately function by recognizing peptide antigens presented by class I or class II major histocompatibility complexes (MHC I or MHC II, respectively) on the surface of host cells (138). Alternatively, a distinct lineage of T cells bearing γδ TCR chains develop, which recognize a quite unique group of molecules (139). Noteworthy, intrathymic lineage restriction and cell fate are determined not only by the type of TCR and its avidity for self-antigens but also by the acquisition of co-receptors that relay signals to intracellular effectors during T cell activation. Hence, generation of distinct cell types are tightly controlled as thymocyte progresses through thymic niches (7). Herein, we will point out some key aspects of the expression and influence of TGF-β superfamily signaling molecules on the distinct paths of thymocyte development: from CD4−CD8− DN T cell precursors (further subdivided in DN1 to DN4 stages based on the surface expression of CD44 and CD25) to the highly expanded immature CD4+CD8+ DP cells, and upon the mature CD4+CD8− or CD4−CD8+ SP cells.
DN1 to DN2 Cells
Entry of bone marrow-derived Lin−cKithighCD44+CD25− cells, or ETPs, into the thymus occurs through the corticomedullary junction. In this intermediate region, these immature cells with T cell–B cell–myeloid potential come into contact with K5+K8+ bipotent TEPCs and mature T cells (67, 124, 140). They subsequently move into the thymus cortex toward the subcapsular zone as DN cells, as defined by the lack of CD4 and CD8 co-receptors (7, 141). In the cortex, developing thymocytes then upregulate CD25 – the α chain of the IL-2 receptor – to become CD44+CD25+ DN2 cells, which undergo Dβ to Jβ recombination of the β-chain locus (142, 143). This DN1 to DN2 transition is accompanied by a strong downregulation of Bmpr1a (Alk3) and Bmpr2 (BMPRII) expression (127, 129). Most cells at this stage present high levels of pSmad2 along with Alk4 (Acvr1b) and ActRII (Acvr2a) on their cell surface, although a few cells also exhibit Alk5 (Tgfbr1) and TβRII (Tgfbr2) receptors (134). DN cells also express the type III co-receptor betaglycan/TβRIII (Tgfbr3), with highest levels at DN3 cells (144). Betaglycan seems to increase the binding strength of some ligands with their cognate receptors, therefore potentializing their effects (145–147). Thymocytes express no Bmpr1b (Alk6), Acvr1 (Alk2), and Acvr1c (Alk7) during thymopoiesis (127, 129). Yet, high levels of inhibin βA subunit (Inhba) and TGF-β1 (Tgfb1) contrast with reduced levels of the inhibin α subunit (Inha), Bmp2, Bmp4, and Bmp7 at the DN2 stage (81, 82, 127, 130). When Inha mutants were used for E14.0 FTOC, a partial arrest at the DN2 stage impaired further T cell maturation (148). Likewise, antibody-dependent blocking of betaglycan in E14.0 FTOC resulted in a reduction of both DN2 and DP cells (144). By contrast, addition of TGF-β1 or TGF-β2 in E14.0 FTOC strongly inhibited T cell development by mainly impairing the differentiation of DN1 cells into DN2 (149). A slightly less strong impact after BMP4 treatment of E15.0–E15.5 FTOC or suspension cultures of fetal thymocytes resulted in cell cycle arrest at the DN1 stage without induction of apoptosis (89, 150). The use of BMP4-treated chimeric human–mouse FTOC produced similar findings (81), revealing a conserved role for Bmp4 during evolution. Besides, partial redundancy between BMP ligands also seems to occur in the thymus, since treatment of FTOC with BMP2, but not with BMP7, similarly affected the production of DP cells (150).
DN2 to DN3 Cells
Following T cell differentiation into CD44−/lowCD25+ DN3 cells, Vβ to DJβ recombination gives rise to the β chain of the pre-TCR (143). At this stage, the levels of Inha, Bmp2, and Bmp4 remain relatively low, Bmp7 becomes upregulated up to the CD3−CD8+ intermediate single-positive (ISP) stage, and expression of Inhba and Tgfb1 declines (82, 127, 130). Levels of Alk4 (Acvr1c), Alk5 (Tgfbr1), and ActRII (Acvr2a) gradually reduce as thymocytes mature, in contrast to TβRII, which is slowly upregulated – at this stage, Alk4 and Alk5 are co-expressed (134). Expression of Bmpr1a (Alk3) and Bmpr2 (BMPRII) presents a small recovery at the DN3 and DN4 stages (127, 129). Nevertheless, conditional inactivation of Bmp7 in the hematopoietic lineage using a vav-iCre line had no significant impact on T cell differentiation and total cell numbers, likely because endoderm-derived cTECs and mTECs may supply enough Bmp7 or other redundant factor for the mutant thymocytes (71, 82, 150). In particular, subcapsular cTECs, cortical DCs, and mTECs express Bmp2 and Bmp4 (71, 81, 82). Activation of the Bmp4 pathway in stromal cells indirectly impacts the DN to DP transition, as revealed by reconstitution experiments with thymocyte-depleted stroma treated with BMP4 or untreated stroma with BMP4-treated DN cells (89). Of note, although highly expressed up to the transition from DN2 to DN3, being downregulated up to the DP stage, and sustained at low levels at SP subsets (127), the gene referred as Bmp1 is a procollagen C-proteinase involved in ventral body wall closure during embryogenesis. To our knowledge, there is no available functional information regarding its role during thymopoiesis, except that it was also found in cTECs and mTECs (71, 151).
DN3 to DN4 Cells
Should rearrangements result in unproductive β chains, DN3 cells undergo apoptosis and are phagocytized by cortical macrophages or DCs in a process termed β-selection (143, 152). Otherwise, successful recombination leads to a reduction in CD25 expression and the expansion of CD44−CD25− DN4 thymocytes (153, 154). Both activin A and inhibin A similarly stimulate the DN3 to DN4 transition, as revealed in FTOC from wild-type fetuses at E14.0. However, treatment with activin A led to higher numbers of mature CD24lowCD8+TCRβhigh T cells at the expense of CD4+ cells, in contrast to inhibin A treatment, which stimulated the transition from DN4 to DP cells (148).
DP Cells
Rearrangement of the TCRα chain occurs at the DP stage and cells move from the cortical zone toward the thymic medulla (143, 155). During this migration, cTECs present self peptides through MHC molecules (pMHC) to the TCR of intermingling DP thymocytes in a process known as positive selection, in which interactions of low-avidity drive clones to survive and continue maturation (156). At the DP stage, Alk4 (Acvr1b), Alk5 (Tgfbr1), and ActRII (Acvr2a) reach their lowest levels, but the number of cells concomitantly presenting Alk5 and pSmad2 increases in relation to Alk4-positive cells (134). By contrast, Bmpr1a (Alk3) and Bmpr2 (BMPRII) are highly expressed (127, 129). Two members of the GDF subgroup, Gdf7 and Gdf11, seem to be induced in DP cells and sustained at SP stages, with CD4+ T cells presenting relatively higher levels than CD8+ T cells (127). Gdf7 signals through BMP-specific receptors as Alk3 and BMPRII, whereas Gdf11 binds TGF-β-related receptors, as Alk4 and Alk5 (157–159). Their roles on T cell function are largely obscure, if any. Mouse mutants for Gdf7 exhibit variable hydrocephalus and fail to produce a class of commissural neurons (160). Male mutants are sterile due to impaired differentiation and branching morphogenesis of the seminal vesicle, with no other affected reproductive structure (161). In turn, mutants for Gdf11 show homeotic transformations due to a delayed trunk to tail transition (162, 163). They die after birth because of renal defects, which may vary from hypoplasia to complete bilateral agenesis (164). Curiously, oral infection with Gram-negative bacteria, Aggregatibacter actinomycetemcomitans, in rats led to a chronic upregulation of Gdf11 expression among other cytokines in both peripheral CD45RA+CD4+ T cells and B cells (165). At present, however, little is known on the effects of GDFs over thymopoiesis.
SP Cells
Still in the cortex, differentiating thymocytes start to lose the expression of either CD4 or CD8 and migrate toward the medulla. The choice for either CD4 or CD8 SP lineage seems to occur at a transitional step defined as CD4+CD8low and depends on TCR interaction with the MHC class II or class I, respectively (166, 167). Additionally, it also relies on the triggering of a transcriptional machinery that operates distinctly for final differentiation (165, 166). Noteworthy, the SP cells sustain Bmpr1a (Alk3) and Bmpr2 (BMPRII) expression, and upregulate Alk5 (Tgfbr1) and TβRII (Tgfbr2), which lead to increased levels of pSmad2 (84, 127, 129, 134). At this stage, fine-tuning of TGF-β signaling may occur by type III co-receptors – CD4+CD8− cells upregulate Tgfbr3 (betaglycan), whereas CD4−CD8+ cells exhibit higher levels of Cripto (Tdgf1) (127, 144). Genetic loss of Tgfbr3 in FTOC resulted in decreased numbers of both DP and SP cells, probably related to the high rates of apoptosis in DN, DP, and CD4+ SP subsets (144). An apoptotic phenotype was also observed in the liver of Tgfbr3 mutants (168). However, a functional role for Cripto during thymopoiesis is currently unresolved, despite its importance for TGF-β binding and inhibition (169). Mutants for this gene present a strong deleterious phenotype during gastrulation and die shortly afterward (170, 171). Modulation of TGF-β family members, their receptors, and co-receptors at the DP stage is therefore associated with the terminal differentiation of thymocytes.
Impact of TGF-β Signals on the Differentiation of Thymic Regulatory T Cells
Regulatory T (Treg) cells have the ability to suppress autoreactive T cells, and they can originate from the thymus or be induced in the periphery (172). Thymus-derived Treg (tTreg) arise in the thymus from SP CD4+ T cells that escape negative selection during maturation by presenting TCR signals of variable affinities (80, 172–174). More specifically, TCRs with high avidity for self-antigens trigger a new upregulation of CD25 (IL-2 receptor α chain) and therefore exhibit an increased responsiveness to IL-2, ultimately inducing the expression of the transcription factor forkhead box P3 (Foxp3) through a STAT5-dependent mechanism (175–177). Foxp3 is the critical transcription factor for Treg cell lineage, as its loss abolishes tTreg cells and lead to systemic autoimmunity and death (178, 179). Conversely, forced expression of Foxp3 in CD25−CD45RBhighCD4+ SP cells transferred into severe combined immunodeficiency (SCID) hosts suppressed exacerbated inflammation (180). Unlike previously thought (181), however, expression of Foxp3 in developing tTreg cells induced apoptosis instead of cell survival. Cell death is prevented by limiting concentrations of γc-mediated survival signals enough to sustain only fewer than one million Foxp3+ cells (182).
Signals from members of TGF-β superfamily also play important roles over the differentiation and survival of tTreg cells. In particular, conditional loss of Tgfbr1 (Alk5) in thymocytes seems to be involved in tTreg specification, since a Lck–Cre mouse line completely blocked differentiation of tTreg cells in neonatal mice, whereas later inactivation of Tgfbr1 by a Foxp3–Cre line produced no differences in tTreg numbers as compared to wild-type mice (80, 183). In addition, the intrathymic injection of an anti-TGF-β antibody suppressed Foxp3 expression in a TCR transgenic CD4+CD25− SP cells (80). Of note, impaired Alk5 signaling induced by the Lck–Cre line caused no significant impact on CD4+ and CD8+ SP cell numbers (183). A later increase in Treg cells induced in the periphery (pTreg) in these mutant mice relied on IL-2 signaling, since ablation of this cytokine produced no detectable cells in organs, such as the spleen and liver (183). Similarly, thymocyte deficiency of Tgfbr2 from a CD4–Cre mouse line resulted in reduced numbers of tTreg cells due to Bim-dependent apoptosis likely independent of γc-signaling, without affecting TCR-βhighCD4+Foxp3− mature T cells in neonatal mice (84). Unlike Tfgbr1-mutant thymocytes, conditional deletion of Tgfbr2 also resulted in low numbers of pTreg cells (84). Induction of pTreg cells relies on the Smad3-dependent upregulation of Foxp3 triggered by activation of both TCR and TGF-β signaling and facilitated by retinoic acid, which increased pSmad3 accessibility to regulatory sequences of the Foxp3 promoter and concurrently counteracted the suppressing effects of a c-Jun N-terminal Kinase (JNK) inhibitor (184, 185). Genetic analyses of the regulatory CNS1 region of Foxp3, which contains binding sites for NFAT, Smad3, and RAR/RXR, revealed that tTreg cell development occurs independently of its activation, whereas its chromosomal deletion largely impaired the production of pTreg cells in secondary lymphoid organs (184–186). In accordance to the different requirements revealed for tTreg in comparison to pTreg populations, TGF-β1 is essential for the peripheral differentiation and maintenance of pTreg cells, but seems to be dispensable for tTreg maturation (187).
Taking into consideration the upregulation of all three TGF-β ligands by stromal cells upon thymocyte apoptosis in the thymus, along with recent findings regarding mutants for distinct TGF-β-specific receptors (80, 84, 183), it is possible that TGF-β ligands may play a redundant yet underestimated role in the immune system. Noteworthy, mutants for TGF-β2 and TGF-β3 also exhibit perinatal mortality, a characteristic that complicates the examination of their role in adults (188–190). Although at first sight, the phenotypes observed in these mutants were generally non-overlapping, some particular structures showed similar defects between single mutants (e.g., cleft palate in either TGF-β2 and TGF-β3 mutants) or exclusive abnormalities in compound mutants, such as abnormal brain vascular morphogenesis and impaired midline fusion along with earlier embryonic lethality in Tgfb1RGE;Tgfb3 and Tgfb2;Tgfb3 compound mutants, respectively (191, 192). However, development of tTreg was never evaluated in these compound mutants. An alternative explanation may consider the participation of a previously unappreciated ligand of the TGF-β superfamily in the differentiation of tTreg cells. Whether this is indeed the case, this candidate ligand should probably signal through Alk5 and TβRII receptors to phosphorylate Smad2 and Smad3 intracellular effectors. Thereby, likely ligands to be thoroughly evaluated due to their expression pattern and receptor affinity are Gdf11 and Gdf8/myostatin – curiously two members that showed redundancy in patterning the axial skeleton as revealed by Gdf11;Mstn double mutants. Unfortunately, examination of fetal thymus morphology and T cell differentiation using FTOC was not performed in these mutants (193).
Noteworthy, TGF-β signals also regulate the thymic development of IL-17-producing cells. A subset of γδ T cells acquire the capacity to produce IL-17 inside the thymus via a TGF-β1-dependent machinery, and both Tgfβ1−/− and Smad3−/− mice were shown to be completely devoid of IL-17-producing γδ T cells (194). Additionally, NKT17 cells comprise a thymic-derived IL-17-producing, CD1d-restricted, and glycolipid antigen-reactive T cell subset (195, 196). These cells express high levels of TβRII and depend on TGF-β signals for differentiation and survival within the thymus and in the periphery (197, 198).
A TGF-β Member for Thymus Rejuvenation?
Aging is an inherent process of living beings, normally associated with gradual loss of function and structure over time – accumulation of reactive species, DNA damage, abnormally folded proteins, and telomere shortening are just some of the molecular changes that may be followed by increased apoptosis, cell transformation, or other cellular event that will ultimately lead to death (199). Although this negative scenario was initially thought to be irreversible, numerous evidences point out that at least in part it is possible to slow down or eventually reverses some specific aging phenotypes. Taking the thymus as example, aging is easily recognizable by a sharp decrease in cellularity of both lymphoid and stromal compartments, whereas the number of thymic adipocytes inversely increases (200, 201). Ultimately, these thymic changes lead to a reduction of naïve T cells in the periphery along with an increase of memory T cells, which reflects in the organism ability to respond to both infection and tumorigenesis (202).
Multiple factors may trigger thymic involution, including the production of sex steroid hormones from puberty, increased calorie intake, or diminished levels of some growth factors and cytokines, such as fibroblast growth factor 7 (FGF7)/keratinocyte growth factor (KGF), insulin-like growth factor (IGF-1), growth hormone (GH), interleukin-7 (IL-7), and IL-22 (203). Modulation of each of them is able to rescue the aged thymic phenotype and restore the immune function at some level (204–210). However, some of these strategies may be inefficient, invasive, non-specific, or produce undesirable side effects to be used in humans (211). A quest for thymic rejuvenation therapies therefore faces daunting challenges in the clinic. Of particular interest, forced expression of Foxn1 was shown to effectively reprogram fibroblasts into TECs or regenerate fully involuted thymuses at many different experimental setups, both in vitro or in vivo (212–214). In this context, signals that control Foxn1 expression might be used to restore the integrity of the thymic epithelial niche and subsequently flourish thymopoisesis in the elderly. In this scenario, administration of soluble factors, such as ligands of the TGF-β superfamily, may be used as regenerative drugs.
Recent findings have revealed that levels of some circulating factors vary with age and that heterochronic parabiosis, i.e., a surgical procedure that connects the circulatory systems of animals with different ages, was able to reverse age-related phenotypes as cardiac hypertrophy (132). These authors further identified the TGF-β member Gdf11 as responsible for restoring cardiac function in old mice, a finding that was further expanded to other systems. In particular, daily treatment of old mice with recombinant GDF11 improved skeletal muscle mass and strength, as well as the integrity of brain vasculature and cognitive function (215, 216). In culture, Gdf11 promoted osteoblastogenesis while inhibiting adipogenesis in bone marrow-derived cells (217). Administration of GDF11 in endothelial progenitor cells triggered cell sprouting and migration, also revealing a role in the formation of blood vessels (218).
Whether Gdf11 or other circulating factor can be used as a rejuvenating cytokine for the thymus remains to be thoroughly assessed. Indeed, Gdf11 is expressed in the thymus of young mice (132), whereas the levels of its non-exclusive receptors, Alk4 and Alk5, vary in thymocytes and TECs, as previously discussed. Of note, however, therapy with Gdf11 produced some side effects in mice (219), and a recent study by Egerman et al. has recently questioned the aforementioned observations (220). Whereas these controversial data on Gdf11 await further investigation, it is noteworthy that heterochronic parabiosis did not reverse thymic involution, but caused atrophy with mild effects on T cell subpopulations of young mice and a reduction in the number of CD4+CD25+Foxp3+ regulatory T cells in old partners to the level of the young pair (221). Although a putative rejuvenating factor for the thymus still awaits to be determined, this controversial matter helps to bring the debate on the role of TGF-β superfamily members for the thymus function.
Concluding Remarks
Although the differentiation of T cells is mainly driven by the rearrangement of TCR genes, many members of the TGF-β superfamily exert critical roles in their stepwise progression during thymic migration. Historically, special attention had been given to the activity of TGF-β ligands in the induction of Treg cells and tolerance to self-antigens, as well as to BMP signaling on thymus organogenesis. However, other members are also produced by developing thymocytes, thymic stromal cells, or may circulate throughout the body by the blood stream and reach the thymus. These ligands signal through the same limited sets of type I and type II receptors to produce dissimilar outcomes either by affecting distinct stages or cell types (e.g., thymocytes versus TECs). How such TGF-β superfamily ligands affect T cell maturation, thymus proper physiology, or its involution remain poorly understood and should be the focus of future research. In addition, a scenario in which a TGF-β superfamily member or its inhibitor acts to rejuvenate the aged thymus may be a likely case for future research.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work benefited from data assembled by the ImmGen consortium and was supported by grants from the Brazilian Research Council/Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Rio de Janeiro State Research Council/Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) (to ADJ and VC-A). LV-F received a Masters’ fellowship from CNPq.
References
1. Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet (2010) 11:47–59. doi:10.1038/nrg2703
2. Cowan JE, Jenkinson WE, Anderson G. Thymus medulla fosters generation of natural Treg cells, invariant γδ T cells, and invariant NKT cells: what we learn from intrathymic migration. Eur J Immunol (2015) 45:652–60. doi:10.1002/eji.201445108
3. Agosti V, Corbacioglu S, Ehlers I, Waskow C, Sommer G, Berrozpe G, et al. Critical role for Kit-mediated Src kinase but not PI 3-kinase signaling in pro T and pro B cell development. J Exp Med (2004) 199:867–78. doi:10.1084/jem.20031983
4. Fry TJ, Sinha M, Milliron M, Chu YW, Kapoor V, Gress RE, et al. Flt3 ligand enhances thymic-dependent and thymic-independent immune reconstitution. Blood (2004) 104:2794–800. doi:10.1182/blood-2003-11-3789
5. Besseyrias V, Fiorini E, Strobl LJ, Zimber-strobl U, Dumortier A, Koch U, et al. Hierarchy of Notch–Delta interactions promoting T cell lineage commitment and maturation. J Exp Med (2007) 204:331–43. doi:10.1084/jem.20061442
6. Magri M, Yatim A, Benne C, Balbo M, Henry A, Serraf A, et al. Notch ligands potentiate IL-7-driven proliferation and survival of human thymocyte precursors. Eur J Immunol (2009) 39:1231–40. doi:10.1002/eji.200838765
7. Petrie HT, Zúñiga-Pflücker JC. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu Rev Immunol (2007) 25:649–79. doi:10.1146/annurev.immunol.23.021704.115715
8. Urist MR. Bone: formation by autoinduction. Science (1965) 150:893–9. doi:10.1126/science.150.3698.893
9. De Larco JE, Todaro GJ. Growth factors from murine sarcoma virus-transformed cells. Proc Natl Acad Sci U S A (1978) 75:4001–5. doi:10.1073/pnas.75.8.4001
10. Roberts AB, Lamb LC, Newton DL, Sporn MB, De Larco JE, Todaro GJ. Transforming growth factors: isolation of polypeptides from virally and chemically transformed cells by acid/ethanol extraction. Proc Natl Acad Sci U S A (1980) 77:3494–8. doi:10.1073/pnas.77.6.3494
11. Frolik CA, Dart LL, Meyers CA, Smith DM, Sporn MB. Purification and initial characterization of a type beta transforming growth factor from human placenta. Proc Natl Acad Sci U S A (1983) 80:3676–80. doi:10.1073/pnas.80.12.3676
12. Wrana JL. Signaling by the TGFβ superfamily. Cold Spring Harb Perspect Biol (2013) 5:a011197. doi:10.1101/cshperspect.a011197
13. Constam DB. Regulation of TGFβ and related signals by precursor processing. Semin Cell Dev Biol (2014) 32:85–97. doi:10.1016/j.semcdb.2014.01.008
14. Degnin C, Jean F, Thomas G, Christian JL. Cleavages within the prodomain direct intracellular trafficking and degradation of mature bone morphogenetic protein-4. Mol Biol Cell (2004) 15:5012–20. doi:10.1091/mbc.E04-08-0673
15. Nelsen SM, Christian JL. Site-specific cleavage of BMP4 by furin, PC6, and PC7. J Biol Chem (2009) 284:27157–66. doi:10.1074/jbc.M109.028506
16. Li Z, Kawasumi M, Zhao B, Moisyadi S, Yang J. Transgenic over-expression of growth differentiation factor 11 propeptide in skeleton results in transformation of the seventh cervical vertebra into a thoracic vertebra. Mol Reprod Dev (2010) 77(11):990–7. doi:10.1002/mrd.21252
17. Wrana JL, Attisano L, Wieser R, Ventura F, Massagué J. Mechanism of activation of the TGF-beta receptor. Nature (1994) 370:341–7. doi:10.1038/370341a0
18. Kretzschmar M, Liu F, Hata A, Doody J, Massagué J. The TGF-β family mediator Smad1 is phosphorylated directly and activated functionally by the BMP receptor kinase. Genes Dev (1997) 11:984–95. doi:10.1101/gad.11.8.984
19. Massagué J, Gomis RR. The logic of TGFβ signaling. FEBS Lett (2006) 580:2811–20. doi:10.1016/j.febslet.2006.04.033
20. Mueller TD, Nickel J. Promiscuity and specificity in BMP receptor activation. FEBS Lett (2012) 586:1846–59. doi:10.1016/j.febslet.2012.02.043
21. Ten Dijke P, Ichijo H, Franzén P, Schulz P, Saras J, Toyoshima H, et al. Activin receptor-like kinases: a novel subclass of cell-surface receptors with predicted serine/threonine kinase activity. Oncogene (1993) 8:2879–87.
22. Wu G, Chen YG, Ozdamar B, Gyuricza CA, Chong PA, Wrana JL, et al. Structural basis of Smad2 recognition by the Smad anchor for receptor activation. Science (2000) 287:92–7. doi:10.1126/science.287.5450.92
23. Shi Y, Wang YF, Jayaraman L, Yang H, Massagué J, Pavletich NP. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-beta signaling. Cell (1998) 94:585–94. doi:10.1016/S0092-8674(00)81600-1
24. Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, et al. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell (1998) 1:611–7. doi:10.1016/S1097-2765(00)80061-1
25. Makkar P, Metpally RPR, Sangadala S, Reddy BVB. Modeling and analysis of MH1 domain of Smads and their interaction with promoter DNA sequence motif. J Mol Graph Model (2009) 27:803–12. doi:10.1016/j.jmgm.2008.12.003
26. Xiao Z, Latek R, Lodish HF. An extended bipartite nuclear localization signal in Smad4 is required for its nuclear import and transcriptional activity. Oncogene (2003) 22:1057–69. doi:10.1038/sj.onc.1206212
27. Chacko BM, Qin BY, Tiwari A, Shi G, Lam S, Hayward LJ, et al. Structural basis of heteromeric smad protein assembly in TGF-beta signaling. Mol Cell (2004) 15:813–23. doi:10.1016/j.molcel.2004.07.016
28. Baburajendran N, Jauch R, Tan CYZ, Narasimhan K, Kolatkar PR. Structural basis for the cooperative DNA recognition by Smad4 MH1 dimers. Nucleic Acids Res (2011) 39:8213–22. doi:10.1093/nar/gkr500
29. Chen X, Xu L. Specific nucleoporin requirement for Smad nuclear translocation. Mol Cell Biol (2010) 30:4022–34. doi:10.1128/MCB.00124-10
30. Cautain B, Hill R, de Pedro N, Link W. Components and regulation of nuclear transport processes. FEBS J (2015) 282:445–62. doi:10.1111/febs.13163
31. Morikawa M, Koinuma D, Miyazono K, Heldin C-H. Genome-wide mechanisms of Smad binding. Oncogene (2012) 32:1–7. doi:10.1038/onc.2012.191
32. Ross S, Hill CS. How the Smads regulate transcription. Int J Biochem Cell Biol (2008) 40:383–408. doi:10.1016/j.biocel.2007.09.006
33. Shi Y, Massagué J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell (2003) 113:685–700. doi:10.1016/S0092-8674(03)00432-X
34. Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, et al. Smad6 inhibits signalling by the TGF-beta superfamily. Nature (1997) 389:622–6. doi:10.1038/39355
35. Hanyu A, Ishidou Y, Ebisawa T, Shimanuki T, Imamura T, Miyazono K. The N domain of Smad7 is essential for specific inhibition of transforming growth factor-beta signaling. J Cell Biol (2001) 155:1017–27. doi:10.1083/jcb.200106023
36. Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, et al. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGFβ receptor for degradation. Mol Cell (2000) 6:1365–75. doi:10.1016/S1097-2765(00)00134-9
37. Feng X-H, Derynck R. Specificity and versatility in Tgf-beta signaling through Smads. Annu Rev Cell Dev Biol (2005) 21:659–93. doi:10.1146/annurev.cellbio.21.022404.142018
38. Wu MY, Hill CS. Tgf-beta superfamily signaling in embryonic development and homeostasis. Dev Cell (2009) 16:329–43. doi:10.1016/j.devcel.2009.02.012
39. Massagué J, Xi Q. TGF-β control of stem cell differentiation genes. FEBS Lett (2012) 586:1953–8. doi:10.1016/j.febslet.2012.03.023
40. Xie F, Zhang Z, van Dam H, Zhang L, Zhou F. Regulation of TGF-β superfamily signaling by SMAD mono-ubiquitination. Cells (2014) 3:981–93. doi:10.3390/cells3040981
41. Gordon J, Wilson VA, Blair NF, Sheridan J, Farley A, Wilson L, et al. Functional evidence for a single endodermal origin for the thymic epithelium. Nat Immunol (2004) 5:546–53. doi:10.1038/ni1064
42. Gordon J, Manley NR. Mechanisms of thymus organogenesis and morphogenesis. Development (2011) 138:3865–78. doi:10.1242/dev.059998
43. Sadler TW. Langman’s Medical Embryology. 12th ed. Philadelphia, PA: Lippincott Williams & Wilkins (2012).
44. Farley AM, Morris LX, Vroegindeweij E, Depreter MLG, Vaidya H, Stenhouse FH, et al. Dynamics of thymus organogenesis and colonization in early human development. Development (2013) 140:2015–26. doi:10.1242/dev.087320
45. Manley NR, Capecchi MR. Hox group 3 paralogs regulate the development and migration of the thymus, thyroid, and parathyroid glands. Dev Biol (1998) 195:1–15. doi:10.1006/dbio.1997.8827
46. Hamazaki Y, Fujita H, Kobayashi T, Choi Y, Scott HS, Matsumoto M, et al. Medullary thymic epithelial cells expressing Aire represent a unique lineage derived from cells expressing claudin. Nat Immunol (2007) 8:304–11. doi:10.1038/ni1438
47. Muñoz JJ, Cejalvo T, Tobajas E, Fanlo L, Cortés A, Zapata AG. 3D immunofluorescence analysis of early thymic morphogenesis and medulla development. Histol Histopathol (2015) 30:589–99. doi:10.14670/HH-30.589
48. Van Ewijk W, Holländer G, Terhorst C, Wang B. Stepwise development of thymic microenvironments in vivo is regulated by thymocyte subsets. Development (2000) 127:1583–91.
49. Nehls M, Kyewski B, Messerle M, Waldschütz R, Schüddekopf K, Smith AJ, et al. Two genetically separable steps in the differentiation of thymic epithelium. Science (1996) 272:886–9. doi:10.1126/science.272.5263.886
50. Günther T, Chen ZF, Kim J, Priemel M, Rueger JM, Amling M, et al. Genetic ablation of parathyroid glands reveals another source of parathyroid hormone. Nature (2000) 406:199–203. doi:10.1038/35018111
51. Gordon J, Bennett AR, Blackburn CC, Manley NR. Gcm2 and Foxn1 mark early parathyroid- and thymus-specific domains in the developing third pharyngeal pouch. Mech Dev (2001) 103:141–3. doi:10.1016/S0925-4773(01)00333-1
52. Liu Z, Farley A, Chen L, Kirby BJ, Kovacs CS, Blackburn CC, et al. Thymus-associated parathyroid hormone has two cellular origins with distinct endocrine and immunological functions. PLoS Genet (2010) 6:e1001251. doi:10.1371/journal.pgen.1001251
53. Dooley J, Erickson M, Roelink H, Farr AG. Nude thymic rudiment lacking functional Foxn1 resembles respiratory epithelium. Dev Dyn (2005) 233:1605–12. doi:10.1002/dvdy.20495
54. Haynes BF, Heinly CS. Early human T cell development: analysis of the human thymus at the time of initial entry of hematopoietic stem cells into the fetal thymic microenvironment. J Exp Med (1995) 181:1445–58. doi:10.1084/jem.181.4.1445
55. Ramond C, Berthault C, Burlen-Defranoux O, de Sousa AP, Guy-Grand D, Vieira P, et al. Two waves of distinct hematopoietic progenitor cells colonize the fetal thymus. Nat Immunol (2014) 15:27–35. doi:10.1038/ni.2782
56. Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development (2000) 127:1607–16.
57. Patel SR, Gordon J, Mahbub F, Blackburn CC, Manley NR. Bmp4 and Noggin expression during early thymus and parathyroid organogenesis. Gene Expr Patterns (2006) 6:794–9. doi:10.1016/j.modgep.2006.01.011
58. Gordon J, Patel SR, Mishina Y, Manley NR. Evidence for an early role for BMP4 signaling in thymus and parathyroid morphogenesis. Dev Biol (2010) 339:141–54. doi:10.1016/j.ydbio.2009.12.026
59. Neves H, Dupin E, Parreira L, Le Douarin NM. Modulation of Bmp4 signalling in the epithelial-mesenchymal interactions that take place in early thymus and parathyroid development in avian embryos. Dev Biol (2012) 361:208–19. doi:10.1016/j.ydbio.2011.10.022
60. Moore-Scott BA, Opoka R, Lin S-CJ, Kordich JJ, Wells JM. Identification of molecular markers that are expressed in discrete anterior-posterior domains of the endoderm from the gastrula stage to mid-gestation. Dev Dyn (2007) 236:1997–2003. doi:10.1002/dvdy.21204
61. Sun X, Xu J, Lu H, Liu W, Miao Z, Sui X, et al. Directed differentiation of human embryonic stem cells into thymic epithelial progenitor-like cells reconstitutes the thymic microenvironment in vivo. Cell Stem Cell (2013) 13:230–6. doi:10.1016/j.stem.2013.06.014
62. Conlon FL, Lyons KM, Takaesu N, Barth KS, Kispert A, Herrmann B, et al. A primary requirement for nodal in the formation and maintenance of the primitive streak in the mouse. Development (1994) 120:1919–28.
63. Zhang H, Bradley A. Mice deficient for BMP2 are nonviable and have defects in amnion/chorion and cardiac development. Development (1996) 122:2977–86.
64. Lawson KA, Dunn NR, Roelen BAJ, Zeinstra LM, Davis AM, Wright CVE, et al. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev (1999) 13:424–36. doi:10.1101/gad.13.4.424
65. Rossi SW, Jenkinson WE, Anderson G, Jenkinson EJ. Clonal analysis reveals a common progenitor for thymic cortical and medullary epithelium. Nature (2006) 441:988–91. doi:10.1038/nature04813
66. Wong K, Lister NL, Barsanti M, Lim JMC, Hammett MV, Khong DM, et al. Multilineage potential and self-renewal define an epithelial progenitor cell population in the adult thymus. Cell Rep (2014) 8:1198–209. doi:10.1016/j.celrep.2014.07.029
67. Danzl NM, Jeong S, Choi Y, Alexandropoulos K. Identification of novel thymic epithelial cell subsets whose differentiation is regulated by RANKL and Traf6. PLoS One (2014) 9:e86129. doi:10.1371/journal.pone.0086129
68. Farr AG, Anderson SK. Epithelial heterogeneity in the murine thymus: fucose-specific lectins bind medullary epithelial cells. J Immunol (1985) 134:2971–7.
69. Klug DB, Carter C, Crouch E, Roop D, Conti CJ, Richie ER. Interdependence of cortical thymic epithelial cell differentiation and T-lineage commitment. Proc Natl Acad Sci U S A (1998) 95:11822–7. doi:10.1073/pnas.95.20.11822
70. Murata S, Sasaki K, Kishimoto T, Niwa S, Hayashi H, Takahama Y, et al. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science (2007) 316:1349–53. doi:10.1126/science.1141915
71. St-Pierre C, Brochu S, Vanegas JR, Dumont-Lagacé M, Lemieux S, Perreault C. Transcriptome sequencing of neonatal thymic epithelial cells. Sci Rep (2013) 3:1860. doi:10.1038/srep01860
72. Schluns KS, Grutkoski PS, Cook JE, Engelmann GL, Le PT. Human thymic epithelial cells produce TGF-beta 3 and express TGF-beta receptors. Int Immunol (1995) 7:1681–90. doi:10.1093/intimm/7.10.1681
73. Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science (2002) 298:1395–401. doi:10.1126/science.1075958
74. Anderson MS, Venanzi ES, Chen Z, Berzins SP, Benoist C, Mathis D. The cellular mechanism of Aire control of T cell tolerance. Immunity (2005) 23:227–39. doi:10.1016/j.immuni.2005.07.005
75. Derbinski J, Gäbler J, Brors B, Tierling S, Jonnakuty S, Hergenhahn M, et al. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J Exp Med (2005) 202:33–45. doi:10.1084/jem.20050471
76. Ohigashi I, Zuklys S, Sakata M, Mayer CE, Zhanybekova S, Murata S, et al. Aire-expressing thymic medullary epithelial cells originate from β5t-expressing progenitor cells. Proc Natl Acad Sci U S A (2013) 110:9885–90. doi:10.1073/pnas.1301799110
77. Derbinski J, Schulte A, Kyewski B, Klein L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol (2001) 2:1032–9. doi:10.1038/ni723
78. Melichar HJ, Ross JO, Herzmark P, Hogquist KA, Robey EA. Distinct temporal patterns of T cell receptor signaling during positive versus negative selection in situ. Sci Signal (2013) 6:ra92. doi:10.1126/scisignal.2004400
79. Dzhagalov IL, Chen KG, Herzmark P, Robey EA. Elimination of self-reactive T cells in the thymus: a timeline for negative selection. PLoS Biol (2013) 11:e1001566. doi:10.1371/journal.pbio.1001566
80. Konkel JE, Jin W, Abbatiello B, Grainger JR, Chen W. Thymocyte apoptosis drives the intrathymic generation of regulatory T cells. Proc Natl Acad Sci U S A (2014) 111:E465–73. doi:10.1073/pnas.1320319111
81. Cejalvo T, Sacedón R, Hernández-López C, Diez B, Gutierrez-Frías C, Valencia J, et al. Bone morphogenetic protein-2/4 signalling pathway components are expressed in the human thymus and inhibit early T-cell development. Immunology (2007) 121:94–104. doi:10.1111/j.1365-2567.2007.02541.x
82. Passa O, Tsalavos S, Belyaev NN, Petryk A, Potocnik AJ, Graf D. Compartmentalization of bone morphogenetic proteins and their antagonists in lymphoid progenitors and supporting microenvironments and functional implications. Immunology (2011) 134:349–59. doi:10.1111/j.1365-2567.2011.03495.x
83. Kersten C, Sivertsen EA, Hystad ME, Forfang L, Smeland EB, Myklebust JH. BMP-6 inhibits growth of mature human B cells; induction of Smad phosphorylation and upregulation of Id1. BMC Immunol (2005) 6:9. doi:10.1186/1471-2172-6-9
84. Ouyang W, Beckett O, Ma Q, Li MO. Transforming growth factor-beta signaling curbs thymic negative selection promoting regulatory T cell development. Immunity (2010) 32:642–53. doi:10.1016/j.immuni.2010.04.012
85. Hauri-Hohl M, Zuklys S, Holländer GA, Ziegler SF. A regulatory role for TGF-β signaling in the establishment and function of the thymic medulla. Nat Immunol (2014) 15:554–61. doi:10.1038/ni.2869
86. Marrella V, Poliani PL, Notarangelo LD, Villa A. Rag defects and thymic stroma: lessons from animal models. Front Immunol (2014) 5:529. doi:10.3389/fimmu.2014.00259
87. Nehls M, Pfeifer D, Schorpp M, Hedrich H, Boehm T. New member of the winged-helix protein family disrupted in mouse and rat nude mutations. Nature (1994) 372:103–7. doi:10.1038/372103a0
88. Balciunaite G, Keller MP, Balciunaite E, Piali L, Zuklys S, Mathieu YD, et al. Wnt glycoproteins regulate the expression of FoxN1, the gene defective in nude mice. Nat Immunol (2002) 3:1102–8. doi:10.1038/ni850
89. Tsai PT, Lee RA, Wu H. BMP4 acts upstream of FGF in modulating thymic stroma and regulating thymopoiesis. Blood (2003) 102:3947–53. doi:10.1182/blood-2003-05-1657
90. Bleul CC, Boehm T. BMP signaling is required for normal thymus development. J Immunol (2005) 175:5213–21. doi:10.4049/jimmunol.175.8.5213
91. Talaber G, Kvell K, Varecza Z, Boldizsar F, Parnell SM, Jenkinson EJ, et al. Wnt-4 protects thymic epithelial cells against dexamethasone-induced senescence. Rejuvenation Res (2011) 14:241–8. doi:10.1089/rej.2010.1110
92. Liang C-C, You L-R, Yen JJ, Liao N-S, Yang-Yen H-F, Chen C-M. Thymic epithelial β-catenin is required for adult thymic homeostasis and function. Immunol Cell Biol (2013) 91:511–23. doi:10.1038/icb.2013.34
93. Lai L, Jin J. Generation of thymic epithelial cell progenitors by mouse embryonic stem cells. Stem Cells (2009) 27:3012–20. doi:10.1002/stem.238
94. D’Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol (2005) 23:1534–41. doi:10.1038/nbt1163
95. Green MD, Chen A, Nostro M-C, d’Souza SL, Schaniel C, Lemischka IR, et al. Generation of anterior foregut endoderm from human embryonic and induced pluripotent stem cells. Nat Biotechnol (2011) 29:267–72. doi:10.1038/nbt.1788
96. Parent AV, Russ HA, Khan IS, Laflam TN, Metzger TC, Anderson MS, et al. Generation of functional thymic epithelium from human embryonic stem cells that supports host T cell development. Cell Stem Cell (2013) 13:219–29. doi:10.1016/j.stem.2013.04.004
97. Douagi I, Andre I, Ferraz JC, Cumano A. Characterization of T cell precursor activity in the murine fetal thymus: evidence for an input of T cell precursors between days 12 and 14 of gestation. Eur J Immunol (2000) 30:2201–10. doi:10.1002/1521-4141(2000)30:8<2201:AID-IMMU2201>3.0.CO;2-2
98. Liu C, Ueno T, Kuse S, Saito F, Nitta T, Piali L, et al. The role of CCL21 in recruitment of T-precursor cells to fetal thymi. Blood (2005) 105:31–9. doi:10.1182/blood-2004-04-1369
99. Liu C, Saito F, Liu Z, Lei Y, Uehara S, Love P, et al. Coordination between CCR7- and CCR9-mediated chemokine signals in prevascular fetal thymus colonization. Blood (2006) 108:2531–9. doi:10.1182/blood-2006-05-024190
100. Calderón L, Boehm T. Three chemokine receptors cooperatively regulate homing of hematopoietic progenitors to the embryonic mouse thymus. Proc Natl Acad Sci U S A (2011) 108:7517–22. doi:10.1073/pnas.1016428108
101. Bleul CC, Boehm T. Chemokines define distinct microenvironments in the developing thymus. Eur J Immunol (2000) 30:3371–9. doi:10.1002/1521-4141(2000012)30:12<3371:AID-IMMU3371>3.0.CO;2-L
102. Zaballos A, Gutiérrez J, Varona R, Ardavín C, Márquez G. Cutting edge: identification of the orphan chemokine receptor GPR-9-6 as CCR9, the receptor for the chemokine TECK. J Immunol (1999) 162:5671–5.
103. Youn BS, Kim CH, Smith FO, Broxmeyer HE. TECK, an efficacious chemoattractant for human thymocytes, uses GPR-9-6/CCR9 as a specific receptor. Blood (1999) 94:2533–6.
104. Yoshida R, Nagira M, Kitaura M, Imagawa N, Imai T, Yoshie O. Secondary lymphoid-tissue chemokine is a functional ligand for the CC chemokine receptor CCR7. J Biol Chem (1998) 273:7118–22. doi:10.1074/jbc.273.12.7118
105. Jenkinson WE, Rossi SW, Parnell SM, Agace WW, Takahama Y, Jenkinson EJ, et al. Chemokine receptor expression defines heterogeneity in the earliest thymic migrants. Eur J Immunol (2007) 37:2090–6. doi:10.1002/eji.200737212
106. Hillyer P, Mordelet E, Flynn G, Male D. Chemokines, chemokine receptors and adhesion molecules on different human endothelia: discriminating the tissue-specific functions that affect leucocyte migration. Clin Exp Immunol (2003) 134:431–41. doi:10.1111/j.1365-2249.2003.02323.x
107. Lambrecht S, Smith V, De Wilde K, Coudenys J, Decuman S, Deforce D, et al. Growth differentiation factor 15, a marker of lung involvement in systemic sclerosis, is involved in fibrosis development but is not indispensable for fibrosis development. Arthritis Rheumatol (2014) 66:418–27. doi:10.1002/art.38241
108. Kempf T, Zarbock A, Widera C, Butz S, Stadtmann A, Rossaint J, et al. GDF-15 is an inhibitor of leukocyte integrin activation required for survival after myocardial infarction in mice. Nat Med (2011) 17:581–8. doi:10.1038/nm.2354
109. Carrillo-García C, Prochnow S, Simeonova IK, Strelau J, Hölzl-Wenig G, Mandl C, et al. Growth/differentiation factor 15 promotes EGFR signalling, and regulates proliferation and migration in the hippocampus of neonatal and young adult mice. Development (2014) 141:773–83. doi:10.1242/dev.096131
110. Zhao X, Huang Y, Huang Y, Lei P, Peng J, Wu S, et al. Transforming growth factor-beta1 upregulates the expression of CXC chemokine receptor 4 (CXCR4) in human breast cancer MCF-7 cells. Acta Pharmacol Sin (2010) 31:347–54. doi:10.1038/aps.2009.204
111. Yu S, Crawford D, Tsuchihashi T, Behrens TW, Srivastava D. The chemokine receptor CXCR7 functions to regulate cardiac valve remodeling. Dev Dyn (2011) 240:384–93. doi:10.1002/dvdy.22549
112. Park BY, Hong CS, Sohail FA, Saint-Jeannet JP. Developmental expression and regulation of the chemokine CXCL14 in Xenopus. Int J Dev Biol (2009) 53:535–40. doi:10.1387/ijdb.092855bp
113. Sierra-Filardi E, Nieto C, Domínguez-Soto A, Barroso R, Sánchez-Mateos P, Puig-Kroger A, et al. CCL2 shapes macrophage polarization by GM-CSF and M-CSF: identification of CCL2/CCR2-dependent gene expression profile. J Immunol (2014) 192:3858–67. doi:10.4049/jimmunol.1302821
114. Wang B, Biron C, She J, Higgins K, Sunshine MJ, Lacy E, et al. A block in both early T lymphocyte and natural killer cell development in transgenic mice with high-copy numbers of the human CD3E gene. Proc Natl Acad Sci U S A (1994) 91:9402–6. doi:10.1073/pnas.91.20.9402
115. Holländer GA, Wang B, Nichogiannopoulou A, Platenburg PP, van Ewijk W, Burakoff SJ, et al. Developmental control point in induction of thymic cortex regulated by a subpopulation of prothymocytes. Nature (1995) 373:350–3. doi:10.1038/373350a0
116. Jenkinson WE, Rossi SW, Jenkinson EJ, Anderson G. Development of functional thymic epithelial cells occurs independently of lymphostromal interactions. Mech Dev (2005) 122:1294–9. doi:10.1016/j.mod.2005.08.003
117. Ohnemus S, Kanzler B, Jerome-Majewska LA, Papaioannou VE, Boehm T, Mallo M. Aortic arch and pharyngeal phenotype in the absence of BMP-dependent neural crest in the mouse. Mech Dev (2002) 119:127–35. doi:10.1016/S0925-4773(02)00345-3
118. Liu Y, Jin Y, Li J, Seto E, Kuo E, Yu W, et al. Inactivation of Cdc42 in neural crest cells causes craniofacial and cardiovascular morphogenesis defects. Dev Biol (2013) 383:239–52. doi:10.1016/j.ydbio.2013.09.013
119. Alt B, Elsalini OA, Schrumpf P, Haufs N, Lawson ND, Schwabe GC, et al. Arteries define the position of the thyroid gland during its developmental relocalisation. Development (2006) 133:3797–804. doi:10.1242/dev.02550
120. Wang J, Nagy A, Larsson J, Dudas M, Sucov HM, Kaartinen V. Defective ALK5 signaling in the neural crest leads to increased postmigratory neural crest cell apoptosis and severe outflow tract defects. BMC Dev Biol (2006) 6:51. doi:10.1186/1471-213X-6-51
121. Hauri-Hohl MM, Zuklys S, Keller MP, Jeker LT, Barthlott T, Moon AM, et al. TGF-beta signaling in thymic epithelial cells regulates thymic involution and postirradiation reconstitution. Blood (2008) 112:626–34. doi:10.1182/blood-2007-10-115618
122. Foster K, Sheridan J, Veiga-Fernandes H, Roderick K, Pachnis V, Adams R, et al. Contribution of neural crest-derived cells in the embryonic and adult thymus. J Immunol (2008) 180:3183–9. doi:10.4049/jimmunol.180.5.3183
123. Crivellato E, Vacca A, Ribatti D. Setting the stage: an anatomist’s view of the immune system. Trends Immunol (2004) 25:210–7. doi:10.1016/j.it.2004.02.008
124. Pearse G. Normal structure, function and histology of the thymus. Toxicol Pathol (2006) 34:504–14. doi:10.1080/01926230600865549
125. Gameiro J, Nagib P, Verinaud L. The thymus microenvironment in regulating thymocyte differentiation. Cell Adh Migr (2010) 4:382–90. doi:10.4161/cam.4.3.11789
126. Berrih S, Savino W, Cohen S. Extracellular matrix of the human thymus: immunofluorescence studies on frozen sections and cultured epithelial cells. J Histochem Cytochem (1985) 33:655–64. doi:10.1177/33.7.3891843
127. Heng TSP, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol (2008) 9:1091–4. doi:10.1038/ni1008-1091
128. Hager-Theodorides AL, Outram SV, Shah DK, Sacedon R, Shrimpton RE, Vicente A, et al. Bone morphogenetic protein 2/4 signaling regulates early thymocyte differentiation. J Immunol (2002) 169:5496–504. doi:10.4049/jimmunol.169.10.5496
129. Hager-Theodorides AL, Ross SE, Sahni H, Mishina Y, Furmanski AL, Crompton T. Direct BMP2/4 signaling through BMP receptor IA regulates fetal thymocyte progenitor homeostasis and differentiation to CD4+CD8+ double-positive cell. Cell Cycle (2014) 13:324–33. doi:10.4161/cc.27118
130. Licona P, Chimal-Monroy J, Soldevila G. Inhibins are the major activin ligands expressed during early thymocyte development. Dev Dyn (2006) 235:1124–32. doi:10.1002/dvdy.20707
131. Mingueneau M, Kreslavsky T, Gray D, Heng T, Cruse R, Ericson J, et al. The transcriptional landscape of αβ T cell differentiation. Nat Immunol (2013) 14:619–32. doi:10.1038/ni.2590
132. Loffredo FS, Steinhauser ML, Jay SM, Gannon J, Pancoast JR, Yalamanchi P, et al. Growth differentiation factor 11 is a circulating factor that reverses age-related cardiac hypertrophy. Cell (2013) 153:828–39. doi:10.1016/j.cell.2013.04.015
133. Savino W. Intrathymic T cell migration is a multivectorial process under a complex neuroendocrine control. Neuroimmunomodulation (2010) 17:142–5. doi:10.1159/000258708
134. Rosendahl A, Speletas M, Leandersson K, Ivars F, Sideras P. Transforming growth factor-beta- and Activin-Smad signaling pathways are activated at distinct maturation stages of the thymopoeisis. Int Immunol (2003) 15:1401–14. doi:10.1093/intimm/dxg139
135. Yoshioka Y, Ono M, Osaki M, Konishi I, Sakaguchi S. Differential effects of inhibition of bone morphogenic protein (BMP) signalling on T-cell activation and differentiation. Eur J Immunol (2012) 42:749–59. doi:10.1002/eji.201141702
136. Sanos SL, Nowak J, Fallet M, Bajenoff M. Stromal cell networks regulate thymocyte migration and dendritic cell behavior in the thymus. J Immunol (2011) 186:2835–41. doi:10.4049/jimmunol.1003563
137. Savino W, Mendes-Da-Cruz DA, Smaniotto S, Silva-Monteiro E, Villa-Verde DMS. Molecular mechanisms governing thymocyte migration: combined role of chemokines and extracellular matrix. J Leukoc Biol (2004) 75:951–61. doi:10.1189/jlb.1003455
138. Von Boehmer H. Selection of the T-cell repertoire: receptor-controlled checkpoints in T-cell development. Adv Immunol (2004) 84:201–38. doi:10.1016/S0065-2776(04)84006-9
139. Ferreira LMR. Gammadelta T cells: innately adaptive immune cells? Int Rev Immunol (2013) 32:223–48. doi:10.3109/08830185.2013.783831
140. Luc S, Luis TC, Boukarabila H, Macaulay IC, Buza-Vidas N, Bouriez-Jones T, et al. The earliest thymic T cell progenitors sustain B cell and myeloid lineage potential. Nat Immunol (2012) 13:412–9. doi:10.1038/ni.2255
141. Ciofani M, Zúñiga-Pflücker JC. The thymus as an inductive site for T lymphopoiesis. Annu Rev Cell Dev Biol (2007) 23:463–93. doi:10.1146/annurev.cellbio.23.090506.123547
142. Schmitt TM, Ciofani M, Petrie HT, Zúñiga-Pflücker JC. Maintenance of T cell specification and differentiation requires recurrent notch receptor-ligand interactions. J Exp Med (2004) 200:469–79. doi:10.1084/jem.20040394
143. Dudley EC, Petrie HT, Shah LM, Owen MJ, Hayday AC. T cell receptor beta chain gene rearrangement and selection during thymocyte development in adult mice. Immunity (1994) 1:83–93. doi:10.1016/1074-7613(94)90102-3
144. Aleman-Muench GR, Mendoza V, Stenvers K, Garcia-Zepeda EA, Lopez-Casillas F, Raman C, et al. Betaglycan (TβRIII) is expressed in the thymus and regulates T cell development by protecting thymocytes from apoptosis. PLoS One (2012) 7:e44217. doi:10.1371/journal.pone.0044217
145. López-Casillas F, Wrana JL, Massagué J. Betaglycan presents ligand to the TGF beta signaling receptor. Cell (1993) 73:1435–44. doi:10.1016/0092-8674(93)90368-Z
146. López-Casillas F, Payne HM, Andres JL, Massagué J. Betaglycan can act as a dual modulator of TGF-β access to signaling receptors: mapping of ligand binding and GAG attachment sites. J Cell Biol (1994) 124:557–68. doi:10.1083/jcb.124.4.557
147. Eickelberg O, Centrella M, Reiss M, Kashgarian M, Wells RG. Betaglycan inhibits TGF-β signaling by preventing type I-type II receptor complex formation: glycosaminoglycan modifications alter betaglycan function. J Biol Chem (2002) 277:823–9. doi:10.1074/jbc.M105110200
148. Licona-Limón P, Alemán-Muench G, Chimal-Monroy J, Macías-Silva M, García-Zepeda EA, Matzuk MM, et al. Activins and inhibins: novel regulators of thymocyte development. Biochem Biophys Res Commun (2009) 381:229–35. doi:10.1016/j.bbrc.2009.02.029
149. Plum J, De Smedt M, Leclercq G, Vandekerckhove B. Influence of TGF-beta on murine thymocyte development in fetal thymus organ culture. J Immunol (1995) 154:5789–98.
150. Graf D, Nethisinghe S, Palmer DB, Fisher AG, Merkenschlager M. The developmentally regulated expression of Twisted gastrulation reveals a role for bone morphogenetic proteins in the control of T cell development. J Exp Med (2002) 196:163–71. doi:10.1084/jem.20020276
151. Suzuki N, Labosky PA, Furuta Y, Hargett L, Dunn R, Fogo AB, et al. Failure of ventral body wall closure in mouse embryos lacking a procollagen C-proteinase encoded by Bmp1, a mammalian gene related to Drosophila tolloid. Development (1996) 122:3587–95.
152. Surh CD, Sprent J. T-cell apoptosis detected in situ during positive and negative selection in the thymus. Nature (1994) 372:100–3. doi:10.1038/372100a0
153. Yamasaki S, Ishikawa E, Sakuma M, Ogata K, Sakata-Sogawa K, Hiroshima M, et al. Mechanistic basis of pre-T cell receptor-mediated autonomous signaling critical for thymocyte development. Nat Immunol (2006) 7:67–75. doi:10.1038/ni1290
154. Crompton T, Moore M, MacDonald HR, Malissen B. Double-negative thymocyte subsets in CD3ζ chain-deficient mice: absence of HSA+CD44-CD25- cells. Eur J Immunol (1994) 24:1903–7. doi:10.1002/eji.1830240828
155. Petrie HT, Livak F, Schatz DG, Strasser A, Crispe IN, Shortman K. Multiple rearrangements in T cell receptor alpha chain genes maximize the production of useful thymocytes. J Exp Med (1993) 178:615–22. doi:10.1084/jem.178.2.615
156. Morris GP, Allen PM. How the TCR balances sensitivity and specificity for the recognition of self and pathogens. Nat Immunol (2012) 13:121–8. doi:10.1038/ni.2190
157. Oh SP, Yeo C-Y, Lee Y, Schrewe H, Whitman M, Li E. Activin type IIA and IIB receptors mediate Gdf11 signaling in axial vertebral patterning. Genes Dev (2002) 16:2749–54. doi:10.1101/gad.1021802
158. Mazerbourg S, Sangkuhl K, Luo CW, Sudo S, Klein C, Hsueh AJW. Identification of receptors and signaling pathways for orphan bone morphogenetic protein/growth differentiation factor ligands based on genomic analyses. J Biol Chem (2005) 280:32122–32. doi:10.1074/jbc.M504629200
159. Andersson O, Reissmann E, Ibáñez CF. Growth differentiation factor 11 signals through the transforming growth factor-beta receptor ALK5 to regionalize the anterior-posterior axis. EMBO Rep (2006) 7:831–7. doi:10.1038/sj.embor.7400752
160. Lee KJ, Mendelsohn M, Jessell TM. Neuronal patterning by BMPs: a requirement for GDF7 in the generation of a discrete class of commissural interneurons in the mouse spinal cord. Genes Dev (1998) 12:3394–407. doi:10.1101/gad.12.21.3394
161. Settle S, Marker P, Gurley K, Sinha A, Thacker A, Wang Y, et al. The BMP family member Gdf7 is required for seminal vesicle growth, branching morphogenesis, and cytodifferentiation. Dev Biol (2001) 234:138–50. doi:10.1006/dbio.2001.0244
162. McPherron AC, Lawler AM, Lee SJ. Regulation of anterior/posterior patterning of the axial skeleton by growth/differentiation factor 11. Nat Genet (1999) 22:260–4. doi:10.1038/10320
163. Jurberg AD, Aires R, Varela-Lasheras I, Nóvoa A, Mallo M. Switching axial progenitors from producing trunk to tail tissues in vertebrate embryos. Dev Cell (2013) 25:451–62. doi:10.1016/j.devcel.2013.05.009
164. Esquela AF, Lee S-J. Regulation of metanephric kidney development by growth/differentiation factor 11. Dev Biol (2003) 257:356–70. doi:10.1016/S0012-1606(03)00100-3
165. Li Y, Messina C, Bendaoud M, Fine DH, Schreiner H, Tsiagbe VK. Adaptive immune response in osteoclastic bone resorption induced by orally administered Aggregatibacter actinomycetemcomitans in a rat model of periodontal disease. Mol Oral Microbiol (2010) 25:275–92. doi:10.1111/j.2041-1014.2010.00576.x
166. Singer A, Adoro S, Park J-H. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat Rev Immunol (2008) 8:788–801. doi:10.1038/nri2416
167. Adoro S, McCaughtry T, Erman B, Alag A, Van Laethem F, Park J-H, et al. Coreceptor gene imprinting governs thymocyte lineage fate. EMBO J (2012) 31:366–77. doi:10.1038/emboj.2011.388
168. Stenvers KL, Tursky ML, Harder KW, Kountouri N, Amatayakul-Chantler S, Grail D, et al. Heart and liver defects and reduced transforming growth factor beta2 sensitivity in transforming growth factor beta type III receptor-deficient embryos. Mol Cell Biol (2003) 23:4371–85. doi:10.1128/MCB.23.12.4371-4385.2003
169. Gray PC, Shani G, Aung K, Kelber J, Vale W. Cripto binds transforming growth factor beta (TGF-beta) and inhibits TGF-beta signaling. Mol Cell Biol (2006) 26:9268–78. doi:10.1128/MCB.01168-06
170. Ding J, Yang L, Yan YT, Chen A, Desai N, Wynshaw-Boris A, et al. Cripto is required for correct orientation of the anterior-posterior axis in the mouse embryo. Nature (1998) 395:702–7. doi:10.1038/27215
171. Xu C, Liguori G, Persico MG, Adamson ED. Abrogation of the Cripto gene in mouse leads to failure of postgastrulation morphogenesis and lack of differentiation of cardiomyocytes. Development (1999) 126:483–94. doi:10.1006/dbio.1998.8862
172. Ohkura N, Kitagawa Y, Sakaguchi S. Development and maintenance of regulatory T cells. Immunity (2013) 38:414–23. doi:10.1016/j.immuni.2013.03.002
173. Lee HM, Bautista JL, Scott-Browne J, Mohan JF, Hsieh CSA. Broad range of self-reactivity drives thymic regulatory T cell selection to limit responses to self. Immunity (2012) 37:475–86. doi:10.1016/j.immuni.2012.07.009
174. Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat Rev Immunol (2014) 14:377–91. doi:10.1038/nri3667
175. Zorn E, Nelson EA, Mohseni M, Porcheray F, Kim H, Litsa D, et al. IL-2 regulates FOXP3 expression in human CD4+CD25+ regulatory T cells through a STAT-dependent mechanism and induces the expansion of these cells in vivo. Blood (2006) 108:1571–9. doi:10.1182/blood-2006-02-004747
176. Zeiser R, Negrin RS. Interleukin-2 receptor downstream events in regulatory T cells: implications for the choice of immunosuppressive drug therapy. Cell Cycle (2008) 7:458–62. doi:10.4161/cc.7.4.5454
177. Burchill MA, Yang J, Vang KB, Moon JJ, Chu HH, Lio C-WJ, et al. Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity (2008) 28:112–21. doi:10.1016/j.immuni.2007.11.022
178. Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet (2001) 27:68–73. doi:10.1038/83784
179. Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol (2003) 4:330–6. doi:10.1038/ni904
180. Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science (2003) 299:1057–61. doi:10.1126/science.1079490
181. Gavin MA, Rasmussen JP, Fontenot JD, Vasta V, Manganiello VC, Beavo JA, et al. Foxp3-dependent programme of regulatory T-cell differentiation. Nature (2007) 445:771–5. doi:10.1038/nature05543
182. Tai X, Erman B, Alag A, Mu J, Kimura M, Katz G, et al. Foxp3 transcription factor is proapoptotic and lethal to developing regulatory T cells unless counterbalanced by cytokine survival signals. Immunity (2013) 38:1116–28. doi:10.1016/j.immuni.2013.02.022
183. Liu Y, Zhang P, Li J, Kulkarni AB, Perruche S, Chen W. A critical function for TGF-beta signaling in the development of natural CD4+CD25+Foxp3+ regulatory T cells. Nat Immunol (2008) 9:632–40. doi:10.1038/ni.1607
184. Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat Immunol (2008) 9:194–202. doi:10.1038/ni1549
185. Xu L, Kitani A, Stuelten C, McGrady G, Fuss I, Strober W. Positive and negative transcriptional regulation of the Foxp3 gene is mediated by access and binding of the Smad3 protein to enhancer I. Immunity (2010) 33:313–25. doi:10.1016/j.immuni.2010.09.001
186. Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, Zheng Y, et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature (2012) 482:395–9. doi:10.1038/nature10772
187. Marie JC, Letterio JJ, Gavin M, Rudensky AY. TGF-beta1 maintains suppressor function and Foxp3 expression in CD4+CD25+ regulatory T cells. J Exp Med (2005) 201:1061–7. doi:10.1084/jem.20042276
188. Kaartinen V, Voncken JW, Shuler C, Warburton D, Bu D, Heisterkamp N, et al. Abnormal lung development and cleft palate in mice lacking TGF-beta 3 indicates defects of epithelial-mesenchymal interaction. Nat Genet (1995) 11:415–21. doi:10.1038/ng1295-415
189. Sanford LP, Ormsby I, Gittenberger-de Groot AC, Sariola H, Friedman R, Boivin GP, et al. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development (1997) 124:2659–70.
190. Proetzel G, Pawlowski SA, Wiles MV, Yin M, Boivin GP, Howles PN, et al. Transforming growth factor-beta 3 is required for secondary palate fusion. Nat Genet (1995) 11:409–14. doi:10.1038/ng1295-409
191. Dünker N, Krieglstein K. Tgfβ2-/-Tgfβ3-/- double knockout mice display severe midline fusion defects and early embryonic lethality. Anat Embryol (Berl) (2002) 206:73–83. doi:10.1007/s00429-002-0273-6
192. Mu Z, Yang Z, Yu D, Zhao Z, Munger JS. TGFβ1 and TGFβ3 are partially redundant effectors in brain vascular morphogenesis. Mech Dev (2008) 125:508–16. doi:10.1016/j.mod.2008.01.003
193. McPherron AC, Huynh TV, Lee S-J. Redundancy of myostatin and growth/differentiation factor 11 function. BMC Dev Biol (2009) 9:24. doi:10.1186/1471-213X-9-24
194. Do J, Fink PJ, Li L, Spolski R, Robinson J, Leonard WJ, et al. Cutting edge: spontaneous development of IL-17-producing gamma delta T cells in the thymus occurs via a TGF-beta 1-dependent mechanism. J Immunol (2010) 184:1675–9. doi:10.4049/jimmunol.0903539
195. Michel M-L, Mendes-da-Cruz D, Keller AC, Lochner M, Schneider E, Dy M, et al. Critical role of ROR-γt in a new thymic pathway leading to IL-17-producing invariant NKT cell differentiation. Proc Natl Acad Sci U S A (2008) 105:19845–50. doi:10.1073/pnas.0806472105
196. Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol (2013) 14:1146–54. doi:10.1038/ni.2731
197. Monteiro M, Almeida CF, Agua-Doce A, Graca L. Induced IL-17-producing invariant NKT cells require activation in presence of TGF-β and IL-1β. J Immunol (2013) 190:805–11. doi:10.4049/jimmunol.1201010
198. Havenar-Daughton C, Li S, Benlagha K, Marie JC. Development and function of murine ROR t+ iNKT cells are under TGF-signaling control. Blood (2012) 119:3486–94. doi:10.1182/blood-2012-01-401604
199. Rando TA, Chang HY. Aging, rejuvenation, and epigenetic reprogramming: resetting the aging clock. Cell (2012) 148:46–57. doi:10.1016/j.cell.2012.01.003
200. Douek DC, McFarland RD, Keiser PH, Gage EA, Massey JM, Haynes BF, et al. Changes in thymic function with age and during the treatment of HIV infection. Nature (1998) 396:690–5. doi:10.1038/25374
201. Coelho VDM, Bunbury A, Rangel LB, Giri B, Weeraratna A, Morin PJ, et al. Fat-storing multilocular cells expressing CCR5 increase in the thymus with advancing age: potential role for CCR5 ligands on the differentiation and migration of preadipocytes. Int J Med Sci (2010) 7:1–14. doi:10.7150/ijms.7.1
202. Müller L, Pawelec G. Aging and immunity – impact of behavioral intervention. Brain Behav Immun (2014) 39:8–22. doi:10.1016/j.bbi.2013.11.015
203. Ventevogel MS, Sempowski GD. Thymic rejuvenation and aging. Curr Opin Immunol (2013) 25:516–22. doi:10.1016/j.coi.2013.06.002
204. Sutherland JS, Goldberg GL, Hammett MV, Uldrich AP, Berzins SP, Heng TS, et al. Activation of thymic regeneration in mice and humans following androgen blockade. J Immunol (2005) 175:2741–53. doi:10.4049/jimmunol.175.4.2741
205. Hirakata A, Okumi M, Griesemer AD, Shimizu A, Nobori S, Tena A, et al. Reversal of age-related thymic involution by an LHRH agonist in miniature swine. Transpl Immunol (2010) 24:76–81. doi:10.1016/j.trim.2010.08.001
206. Berent-Maoz B, Montecino-Rodriguez E, Signer RAJ, Dorshkind K. Fibroblast growth factor-7 partially reverses murine thymocyte progenitor aging by repression of Ink4a. Blood (2012) 119:5715–21. doi:10.1182/blood-2011-12-400002
207. Min D, Panoskaltsis-Mortari A, Kuro-o M, Holländer GA, Blazar BR, Weinberg KI. Sustained thymopoiesis and improvement in functional immunity induced by exogenous KGF administration in murine models of aging. Blood (2007) 109:2529–37. doi:10.1182/blood-2006-08-043794
208. Morrhaye G, Kermani H, Legros J-J, Baron F, Beguin Y, Moutschen M, et al. Impact of growth hormone (GH) deficiency and GH replacement upon thymus function in adult patients. PLoS One (2009) 4:e5668. doi:10.1371/journal.pone.0005668
209. Taub DD, Murphy WJ, Longo DL. Rejuvenation of the aging thymus: growth hormone-mediated and ghrelin-mediated signaling pathways. Curr Opin Pharmacol (2010) 10:408–24. doi:10.1016/j.coph.2010.04.015
210. Phillips JA, Brondstetter TI, English CA, Lee HE, Virts EL, Thoman ML. IL-7 gene therapy in aging restores early thymopoiesis without reversing involution. J Immunol (2004) 173:4867–74. doi:10.4049/jimmunol.173.8.4867
211. Velardi E, Dudakov JA, Van den Brink MRM. Clinical strategies to enhance thymic recovery after allogeneic hematopoietic stem cell transplantation. Immunol Lett (2013) 155:31–5. doi:10.1016/j.imlet.2013.09.016
212. Zook EC, Krishack PA, Zhang S, Zeleznik-Le NJ, Firulli AB, Witte PL, et al. Overexpression of Foxn1 attenuates age-associated thymic involution and prevents the expansion of peripheral CD4 memory T cells. Blood (2011) 118:5723–31. doi:10.1182/blood-2011-03-342097
213. Bredenkamp N, Nowell CS, Blackburn CC. Regeneration of the aged thymus by a single transcription factor. Development (2014) 141:1627–37. doi:10.1242/dev.103614
214. Bredenkamp N, Ulyanchenko S, O’Neill KE, Manley NR, Vaidya HJ, Blackburn CC. An organized and functional thymus generated from FOXN1-reprogrammed fibroblasts. Nat Cell Biol (2014) 16(9):902–8. doi:10.1038/ncb3023
215. Sinha M, Jang YC, Oh J, Khong D, Wu EY, Manohar R, et al. Restoring systemic GDF11 levels reverses age-related dysfunction in mouse skeletal muscle. Science (2014) 344:649–52. doi:10.1126/science.1251152
216. Katsimpardi L, Litterman NK, Schein PA, Miller CM, Loffredo FS, Wojtkiewicz GR, et al. Vascular and neurogenic rejuvenation of the aging mouse brain by young systemic factors. Science (2014) 344:630–4. doi:10.1126/science.1251141
217. Zhang Y, Shao J, Wang Z, Yang T, Liu S, Liu Y, et al. Growth differentiation factor 11 is a protective factor for osteoblastogenesis by targeting PPARgamma. Gene (2014) 557(2):6–11. doi:10.1016/j.gene.2014.12.039
218. Finkenzeller G, Stark GB, Strassburg S. Growth differentiation factor 11 supports migration and sprouting of endothelial progenitor cells. J Surg Res (2015) 198(1):1–7. doi:10.1016/j.jss.2015.05.001
219. Suragani RNVS, Cadena SM, Cawley SM, Sako D, Mitchell D, Li R, et al. Transforming growth factor-β superfamily ligand trap ACE-536 corrects anemia by promoting late-stage erythropoiesis. Nat Med (2014) 20:408–14. doi:10.1038/nm.3512
220. Egerman MA, Cadena SM, Gilbert JA, Meyer A, Nelson HN, Swalley SE, et al. GDF11 increases with age and inhibits skeletal muscle regeneration. Cell Metab (2015) 22(1):1–11. doi:10.1016/j.cmet.2015.05.010
Keywords: TGF-β, BMP, thymus, thymopoiesis, T cell development
Citation: Jurberg AD, Vasconcelos-Fontes L and Cotta-de-Almeida V (2015) A tale from TGF-β superfamily for thymus ontogeny and function. Front. Immunol. 6:442. doi: 10.3389/fimmu.2015.00442
Received: 20 June 2015; Accepted: 14 August 2015;
Published: 10 September 2015
Edited by:
Geraldo Aleixo Passos, University of São Paulo, BrazilReviewed by:
Natalio Garbi, University of Bonn, GermanyAmanda Freire Assis, University of São Paulo, Brazil
Copyright: © 2015 Jurberg, Vasconcelos-Fontes and Cotta-de-Almeida. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arnon Dias Jurberg and Vinícius Cotta-de-Almeida, Laboratory on Thymus Research, Oswaldo Cruz Foundation, Oswaldo Cruz Institute, Avenida Brasil 4365, Pavilhão Leônidas Deane, 5º andar – Sala 510, Manguinhos, Rio de Janeiro 21040-360., Rio de Janeiro, Brazil,YWp1cmJlcmdAaW9jLmZpb2NydXouYnIsYWp1cmJlcmdAZ21haWwuY29t;dmNhQGlvYy5maW9jcnV6LmJy
 Arnon Dias Jurberg
Arnon Dias Jurberg Larissa Vasconcelos-Fontes
Larissa Vasconcelos-Fontes Vinícius Cotta-de-Almeida
Vinícius Cotta-de-Almeida