- 1Stanford Immunology, Stanford University School of Medicine, Stanford, CA, USA
- 2Department of Medicine, Stanford University School of Medicine, Stanford, CA, USA
Natural killer (NK) cells are a unique lymphocyte lineage with remarkable agility in the rapid destruction of virus-infected cells. They are also the most poorly understood class of lymphocyte. A spectrum of activating and inhibitory receptors at the NK cell surface leads to an unusual and difficult-to-study mechanism of cellular recognition, as well as a very high capacity for diversity at the single-cell level. Here, we review the evidence for the role of NK cells in the earliest stage of human viral infection, and in its prevention. We argue that single-cell diversity is a logical evolutionary adaptation for their position in the immune response and contributes to their ability to kill virus-infected cells. Finally, we look to the future, where emerging single-cell technologies will enable a new generation of rigorous and clinically relevant studies on NK cells accounting for all of their unique and diverse characteristics.
Natural killer (NK) cells were discovered in 1975 (1, 2) on the basis of their ability to selectively lyse leukemic cell lines. Originally called “null cells” because they were believed not to express cell surface receptors, they were eventually recognized as a third lineage of lymphocyte distinct from T and B cells (3). Recently, the role of NK cells as the founding member of a growing group known as innate lymphoid cells has also been appreciated (4).
Natural killer cells are a unique lineage with remarkable agility. They rapidly detect and destroy virus-infected, malignant, and stressed cells (5, 6). They are also the most poorly understood class of lymphocyte, due in part to their unusual and difficult-to-study mechanism of cellular recognition. The NK cell surface contains a spectrum of activating and inhibitory receptors (7). It is the integration and balance of signals from these receptors that determine a cell’s activation status (8).
This array of receptors also generates the opportunity for vast diversity in the NK repertoire. Examination of this diversity has been limited: studying a large number of parameters on a single cell is technologically challenging. Yet, it may be a critical functional feature of the NK cell repertoire.
In many of the ways in which immune cells are conventionally categorized, NK cells are intermediates. They borrow, share, and combine functional features of other cell types to form their own recognition paradigm. This helps to explain their enigmatic nature and argues that they occupy a unique evolutionary niche. Here, we review this recognition paradigm, with a focus on NK responses to viruses, and argue that single-cell diversity enhances their ability to fulfill this “middleman” role. We also discuss current and future studies, where single-cell technology will allow a much more detailed and nuanced dissection of the roles and promise of NK cells in the antiviral response.
NK Cells have Lifetimes of Intermediate Length
In comparison to their classic innate and adaptive counterparts, NK cells take an intermediate position in estimates of cellular lifetime (Figure 1). In deuterium incorporation studies, human T cells have the longest estimated leukocyte half-lives, at 1–8 years for naive T cells and 1–12 months for memory T cells (9, 10). Estimated half-lives for CD27− naive B cells, CD27+ memory B cells, and plasma cells are somewhat more limited, at 22, 11, and 40 days, respectively (11, 12). For NK cells, half-lives are shorter, estimated at 7 days (13). Yet, they still outlast their innate counterparts, with monocytes’ half-lives estimated at 71 h (14) and neutrophils’ reported between 5 and 90 h (15). It is important to note that only actively dividing cells incorporate deuterium, and thus these studies may not account for long-lived, non-dividing cells. They are also based entirely in peripheral blood; cell kinetics in tissues may follow completely different patterns. Yet, with these caveats in mind, these data suggest that NK cells fall midway on the spectrum of cellular lifetime. These estimates may also serve as a reasonable proxy for the amount of time they require to respond to viral infection.
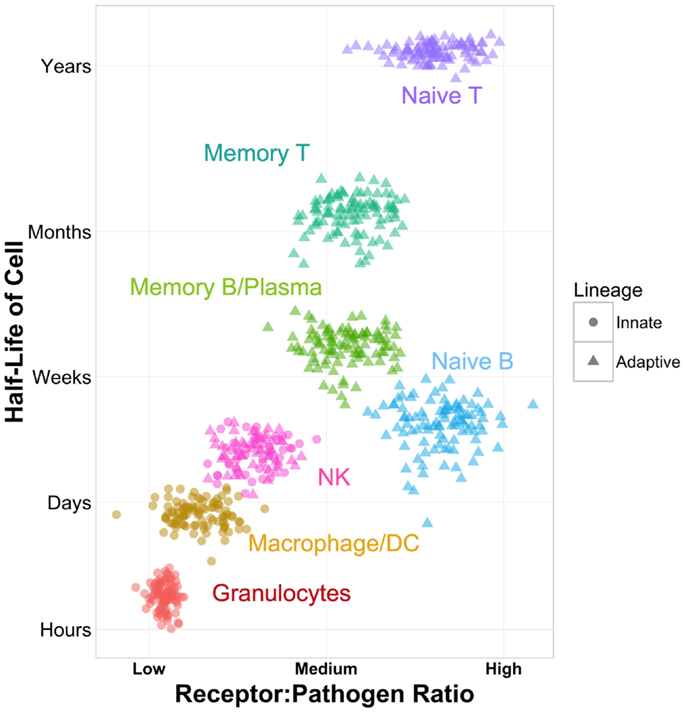
Figure 1. Schematic of NK cells as immune intermediates in cell lineage, ratio of receptors to potential pathogens, and cellular half-life. Half-life values are approximated based on published data (9–15).
NK Cells are Intermediate in Their Mechanism of Recognition
Cells of the innate system typically recognize non-self entities through specific receptor–ligand interactions. The toll-like receptor (TLR) system and other pattern recognition receptors, including RIG-I-like receptors, NOD-like receptors, and C-type lectin receptors, are designed to recognize specific features of pathogens or other danger signals (16, 17). Recognition is fast and digital, having only on/off states. It triggers phagocytosis, the release of inflammatory cytokines, and regulation of downstream adaptive responses. With 10 TLRs identified in humans to date and other pattern recognition receptor classes in the same order of magnitude or fewer, the innate paradigm is characterized by a low ratio of receptors per potential pathogen (Figure 1).
While the innate system has evolved to recognize broad classes of pathogens through a limited set of receptors, the adaptive system takes the converse approach of employing a broad set of receptors that each recognize a very limited set of pathogens. In T and B cells, V(D)J recombination forms specialized, antigen-specific receptors. When triggered in combination with costimulation, these receptors initiate proliferation, cytotoxicity, production of antibodies, and release of cytokines. The total potential diversity of the human T cell receptor (TCR) is estimated at 1015–1020 sequences (18, 19). The adaptive system is therefore characterized by a high ratio of receptors to potential pathogens (Figure 1).
Natural killer cells take cues from both extremes. They express a spectrum of receptors, which can be either activating or inhibitory, through which they integrate signals to determine their activation status. These include killer immunoglobulin-like receptors (KIR), C-type lectin-like receptors, signaling lymphocyte activation molecule (SLAM) family receptors, and natural cytotoxicity receptors, among others (20). Unlike other cell types, NK activation is analog: increased activating or decreased inhibitory signals can tip the balance toward activation, but typically no specific receptor–ligand interaction is required. In fact, CD16 is the only receptor that has been shown to be capable of activating NK cells independently (21). In addition, NK cells are the only lymphocytes that can be activated purely through soluble signals. Proliferation, cytotoxicity, and a migratory phenotype can be induced by cytokines alone without specific antigen signal or presentation (22, 23).
Thus, the lifespan of NK cells, their mechanism of recognition, and in addition, recent evidence for memory in NK cell responses (24–26) suggest that they play a truly intermediate role between innate and adaptive responses (Figure 1).
Single-Cell Diversity: A Logical Adaptation for Middlemen
The diversity in a T cell or B cell repertoire is captured in a single receptor. NK cells, by contrast, are diverse at a single-cell level. Each cell expresses a combination of receptors drawn from a vast potential pool. Our early estimates, based on 28 markers, place the total number of combinations in human peripheral blood on the order of 104 (27). With deeper sampling, measurement in a greater diversity of tissues, and improvements in technology allowing increased dimensionality, this estimate will certainly increase. A modest increase to measuring 40 NK cell receptors on a single cell would yield 1012 theoretical combinations. In comparison, T cells have been estimated to have 2.5 × 107 unique TCR sequences in humans (28). As mentioned above, their total potential diversity is estimated at 1015–1020 sequences (18, 19).
Thus, both theoretically and empirically, NK cells are highly diverse, but several orders of magnitude less so than T cells. Yet, T cell (and B cell) diversity is rigid, modifiable primarily by population-level expansion and contraction. The antigen-specific TCR or BCR, while optimizable via somatic hypermutation or affinity maturation, is pre-formed following exit from the bone marrow or thymus. For adaptive cells tasked with developing a slow but reliable memory response, it is a logical tradeoff to expend energy on selecting a vast but inflexible repertoire.
Natural killer diversity compensates for its lack of breadth with its flexibility, a logical evolutionary adaptation for cells that must act quickly. NK diversity can be modulated with vastly more agility and at many different levels. NK cells can up- or downregulate receptors to adjust repertoire diversity at the timescale of cellular processes rather than cellular division (29), one cell at a time rather than by population. Many NK cell receptors, especially the KIRs, are defined by vast population-level genetic diversity (30). This can be compounded at multiple levels: selective transcription and translation, post-translational modifications, protein trafficking, cytokine signaling, cell–cell interactions, and epigenetic modifications (31–33). NK cell diversity is thus much more responsive to both genetic and environmental determinants. This responsiveness may prove to be a critical feature for maintaining flexibility in the earliest lymphocyte response.
NK Cells in Viral Susceptibility
Because of their uniquely centered position in the immune system, their associations with improved control of viral infections (34), and recent evidence of their potential for a memory response, much interest has been focused on whether NK cells can be harnessed as part of a strategy for the initial prevention of viral infection (Table 1). Prevention is an especially significant goal for chronic incurable diseases, such as HIV. Mechanisms by which NK cells could successfully prevent viral infections include localization of NK cells to barriers of viral entry, efficient cytotoxicity, swift production of IFN-γ for recruitment and activation of downstream adaptive responses, and production of β-chemokines that can block viral entry (35).
Directly studying the immune correlates of human viral susceptibility is challenging. It requires either tracking uninfected people over long periods of time, a difficult logistical hurdle, or infecting people in a controlled environment, an ethical quandary. Several approaches have therefore been used to provide indirect evidence for the importance of NK cells in human viral susceptibility.
First, much has been learned from studies of immunodeficient subjects with deficiencies in NK cell frequency or function. In addition to several mutations known to exert their effects on NK cells in relative isolation, at least 46 primary immunodeficiencies are also associated with NK cell defects. These subjects’ overarching and unifying feature is the unusual susceptibility to herpesviruses (54), especially HSV-1, EBV, VSV, and HPV. These findings suggest that NK cells play a non-redundant role in preventing the establishment of these infections.
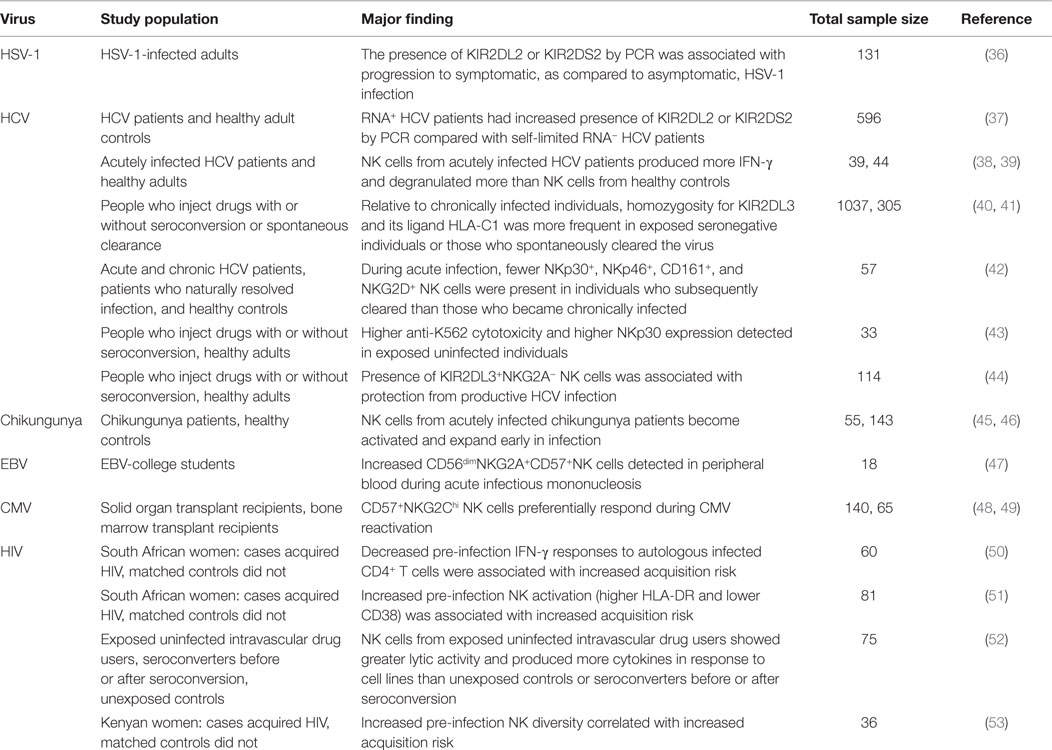
Second, many studies identify NK-relevant genetic correlates associated with disease-afflicted individuals versus healthy controls. These retrospective studies are not direct measures of acquisition probability, but do provide evidence of possible associations. The presence of KIR2DL2 and KIR2DS2 has been associated with progression to infection in both HSV-1 (36) and HCV (37).
A third approach has been to examine the characteristics and strength of NK responses during acute infection and sometimes draw correlations with disease outcome. In these studies, blood is typically drawn at the time of clinical presentation. This approach provides an imprecise approximation of the earliest events in infection but has the distinct advantage of allowing functional immunological studies. These studies have identified several early-stage markers of NK cell activation in viral infection. In HCV, KIR2DL3 in combination with HLA-C1 at both the genetic and cellular level has emerged as an important correlate of protection from infection (40, 41, 44). In addition, NKG2C+ NK cells have been shown to specifically respond to CMV reactivation in transplant recipients (48, 49).
A fourth type of study measures the immune activity of individuals who remain seronegative despite behavior that grants high probability of viral exposure, providing clues as to what is assumed to be an effective protective immune response. The primary issue with these studies is that this form of resistance is not fully understood, and the highly exposed immune state of these individuals (typically sex workers or people who inject drugs) may not be representative of other routes of acquisition.
Finally, a handful of studies, so far only in the HIV field, have enrolled and tracked large cohorts at risk of infection, banking blood samples before and after detection of infection, and performed functional studies on pre-infection samples. While impressive in scope, their conclusions have unfortunately not been entirely clear. While the ability of NK cells to secrete IFN-γ in response to HIV-infected cells was associated with decreased risk of infection (50), generalized NK cell activation (as measured primarily by expression of CD38 and/or HLA-DR) was associated with increased risk of HIV acquisition (51). Thus, depending on its nature, NK cell activation has been both positively and negatively associated with increased risk of HIV acquisition.
Our recent study took a different approach. In addition to measuring activation status and individual markers, we calculated a diversity score for the NK cell repertoire of each donor before the onset of HIV infection. We showed that no single marker was predictive of HIV acquisition risk, but that higher NK cell diversity was associated with increased risk of HIV acquisition in a cohort of Kenyan women (53). This raises the intriguing possibility that NK repertoire diversification is detrimental to the NK cell response. As it represents an apparent state of ramification and inflexibility, high NK diversity may signal a higher risk of exposure and/or less resistant state of immunity.
All of these studies involve clinical cohorts that are limited in size. The brute-force approach of obtaining additional validation cohorts will be a necessary step in bolstering these results. However, targeted single-cell approaches that incorporate leading edge technologies and account for the increasingly appreciated diversity of the NK repertoire are in rapid development. These approaches hold great promise in bringing a new level of understanding to the role of NK cells in viral susceptibility.
New NK Cell-Focused Approaches in Single-Cell Technology
Innovative microchip-based approaches have begun to uncover broad diversity in single-cell function. In a single-cell tracking system, the distribution of average kill times on a per-NK-cell basis was skewed, with a few “serial killers” performing the majority of the killing (55, 56). Furthermore, increased cytotoxicity has been shown to result from simultaneous interaction of NK cells with multiple targets (57) as well as IL-2 activation (23). NKG2A+ NK cells have also been shown to be superior to NKG2A− NK cells in terms of dynamic migration, conjugation, spreading, and killing. “Serial killers” were also more common in this population (58).
Natural killer cell-focused mass cytometry, which has been pioneered by our group and others (27, 53, 59, 60), also holds great promise in further defining the role of NK diversity in viral acquisition at a single-cell level. This technology will be especially powerful when used in conjunction with humanized mice (61), presenting the next frontier in detailed tracking of all stages of the human NK cell response to viral infection.
Furthermore, single-cell dissection of functional diversity, especially in the context of a viral response, will greatly improve understanding of the processes by which NK cells may contribute to the prevention of infection. The generation of more single-cell data will also help to alleviate the statistical issues associated with small sample sizes in clinical studies.
Many questions remain about the functional consequences of NK cell diversity. Does a differentiated NK cell respond better and faster when re-encountering its initial stimulus? What is the spectrum of antigens that can drive NK cell diversification? Does a diverse NK cell repertoire correspond to a less diverse T and B cell repertoire? In addition, most studies have focused only on peripheral blood, but tissue-specific functions of NK cells are increasingly being appreciated (62).
Together, single-cell studies offer great promise in defining the scope of the human NK repertoire and its significance in the context of viral infection. This understanding will be essential in order to optimally harness the NK cells in the next generation of NK cell-based vaccines and therapeutics.
Author Contributions
CB and DSA wrote the manuscript, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the members of the Blish laboratory for critical feedback.
Funding
Funding for this work was provided by National Science Foundation training grant DGE-114740 (DSA), Ruth L. Kirschstein National Research Service Award 1F31AI118469-01 (DA), Beckman Young Investigator Award (CB), NIH Directors’ New Innovator Award DP2AI11219301 (CB), and the Tashia and John Morgridge Endowed Faculty Scholar Award in Pediatric Translational Medicine, Child Health Research Institute at Stanford (CB).
References
1. Herberman RB, Nunn ME, Lavrin DH. Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic acid allogeneic tumors. I. Distribution of reactivity and specificity. Int J Cancer (1975) 16:216–29. doi: 10.1002/ijc.2910160204
2. Kiessling R, Klein E, Wigzell H. “Natural” killer cells in the mouse. I. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Specificity and distribution according to genotype. Eur J Immunol (1975) 5:112–7. doi:10.1002/eji.1830050208
3. Lanier LL, Le AM, Phillips JH, Warner NL, Babcock GF. Subpopulations of human natural killer cells defined by expression of the Leu-7 (HNK-1) and Leu-11 (NK-15) antigens. J Immunol (1983) 131:1789–96.
4. Artis D, Spits H. The biology of innate lymphoid cells. Nature (2015) 517:293–301. doi:10.1038/nature14189
5. Pahl J, Cerwenka A. Tricking the balance: NK cells in anti-cancer immunity. Immunobiology (2015):1–10. doi:10.1016/j.imbio.2015.07.012
6. Waggoner SN, Reighard SD, Gyurova IE, Cranert SA, Mahl SE, Karmele EP, et al. Roles of natural killer cells in antiviral immunity. Curr Opin Virol (2015) 16:15–23. doi:10.1016/j.coviro.2015.10.008
7. Lanier LL. NK cell recognition. Annu Rev Immunol (2005) 23:225–74. doi:10.1146/annurev.immunol.23.021704.115526
8. Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol (2008) 9:495–502. doi:10.1038/ni1581
9. Farber DL, Yudanin NA, Restifo NP. Human memory T cells: generation, compartmentalization and homeostasis. Nat Rev Immunol (2014) 14:24–35. doi:10.1038/nri3567
10. Vrisekoop N, den Braber I, de Boer AB, Ruiter AFC, Ackermans MT, van der Crabben SN, et al. Sparse production but preferential incorporation of recently produced naive T cells in the human peripheral pool. Proc Natl Acad Sci U S A (2008) 105:6115–20. doi:10.1073/pnas.0709713105
11. Bernasconi NL, Traggiai E, Lanzavecchia A. Maintenance of serological memory by polyclonal activation of human memory B cells. Science (2002) 298:2199–202. doi:10.1126/science.1076071
12. Macallan DC, Wallace DL, Zhang Y, Ghattas H, Asquith B, de Lara C, et al. B-cell kinetics in humans: rapid turnover of peripheral blood memory cells. Blood (2005) 105:3633–40. doi:10.1182/blood-2004-09-3740
13. Zhang Y, Wallace DL, de Lara CM, Ghattas H, Asquith B, Worth A, et al. In vivo kinetics of human natural killer cells: the effects of ageing and acute and chronic viral infection. Immunology (2007) 121:258–65. doi:10.1111/j.1365-2567.2007.02573.x
14. Whitelaw DM. Observations on human monocyte kinetics after pulse labeling. Cell Prolif (1972) 5(4):311–7. doi:10.1111/j.1365-2184.1972.tb00369.x/pdf
15. Tak T, Tesselaar K, Pillay J, Borghans JAM, Koenderman L. What’s your age again? Determination of human neutrophil half-lives revisited. J Leukoc Biol (2013) 94:595–601. doi:10.1189/jlb.1112571
16. Goubau D, Deddouche S, Sousa CRE. Cytosolic sensing of viruses. Immunity (2013) 38:855–69. doi:10.1016/j.immuni.2013.05.007
17. Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell (2010) 140:805–20. doi:10.1016/j.cell.2010.01.022
18. Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature (1988) 334:395–402. doi:10.1038/334395a0
19. Zarnitsyna VI, Evavold BD, Schoettle LN, Blattman JN, Antia R. Estimating the diversity, completeness, and cross-reactivity of the T cell repertoire. Front Immunol (2013) 4:485. doi:10.3389/fimmu.2013.00485
20. Watzl C, Long EO. Signal transduction during activation and inhibition of natural killer cells. Curr Protoc Immunol (2010) Chapter 11:Unit11.9B. doi:10.1002/0471142735.im1109bs90
21. Bryceson YT, March ME, Ljunggren H-G, Long EO. Synergy among receptors on resting NK cells for the activation of natural cytotoxicity and cytokine secretion. Blood (2006) 107:159–66. doi:10.1182/blood-2005-04-1351
22. Long EO, Sik Kim H, Liu D, Peterson ME, Rajagopalan S. Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu Rev Immunol (2013) 31:227–58. doi:10.1146/annurev-immunol-020711-075005
23. Olofsson PE, Forslund E, Vanherberghen B, Chechet K, Mickelin O, Ahlin AR, et al. Distinct migration and contact dynamics of resting and IL-2-activated human natural killer cells. Front Immunol (2014) 5:80. doi:10.3389/fimmu.2014.00080
24. Paust S, Gill HS, Wang B-Z, Flynn MP, Moseman EA, Senman B, et al. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat Immunol (2010) 11:1127–35. doi:10.1038/ni.1953
25. Reeves RK, Li H, Jost S, Blass E, Li H, Schafer JL, et al. Antigen-specific NK cell memory in rhesus macaques. Nat Immunol (2015) 16:927–32. doi:10.1038/ni.3227
26. Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature (2009) 457:557–61. doi:10.1038/nature07665
27. Horowitz A, Strauss-Albee DM, Leipold M, Kubo J, Nemat-Gorgani N, Dogan OC, et al. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med (2013) 5:208ra145. doi:10.1126/scitranslmed.3006702
28. Arstila TP, Casrouge A, Baron V, Even J, Kanellopoulos J, Kourilsky P. A direct estimate of the human alphabeta T cell receptor diversity. Science (1999) 286:958–61. doi:10.1126/science.286.5441.958
29. Hromadnikova I, Pirkova P, Sedlackova L. Influence of in vitro IL-2 or IL-15 alone or in combination with Hsp-70-derived 14-mer peptide (TKD) on the expression of NK cell activatory and inhibitory receptors. Mediators Inflamm (2013) 2013:1–12. doi:10.1155/2013/405295
30. Parham P. The genetic and evolutionary balances in human NK cell receptor diversity. Semin Immunol (2008) 20:311–6. doi:10.1016/j.smim.2008.10.002
31. Lee J, Zhang T, Hwang I, Kim A, Nitschke L, Kim M, et al. Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity (2015) 42:431–42. doi:10.1016/j.immuni.2015.02.013
32. Luetke-Eversloh M, Hammer Q, Durek P, Nordström K, Gasparoni G, Pink M, et al. Human cytomegalovirus drives epigenetic imprinting of the IFNG locus in NKG2Chi natural killer cells. PLoS Pathog (2014) 10:e1004441. doi:10.1371/journal.ppat.1004441
33. Schlums H, Cichocki F, Tesi B, Theorell J, Béziat V, Holmes TD, et al. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity (2015) 42:443–56. doi:10.1016/j.immuni.2015.02.008
34. Jost S, Altfeld M. Control of human viral infections by natural killer cells. Annu Rev Immunol (2013) 31:163–94. doi:10.1146/annurev-immunol-032712-100001
35. Fehniger TA, Herbein G, Yu H, Para MI, Bernstein ZP, O’Brien WA, et al. Natural killer cells from HIV-1+ patients produce C-C chemokines and inhibit HIV-1 infection. J Immunol (1998) 161:6433–8.
36. Estefanía E, Gómez Lozano N, Portero F, De Pablo R, Solís R, Sepúlveda S, et al. Influence of KIR gene diversity on the course of HSV-1 infection: resistance to the disease is associated with the absence of KIR2DL2 and KIR2DS2. Tissue Antigens (2007) 70:34–41. doi:10.1111/j.1399-0039.2007.00844.x
37. Paladino N, Flores AC, Marcos CY, Fainboim H, Theiler G, Arruvito L, et al. Increased frequencies of activating natural killer receptors are associated with liver injury in individuals who do not eliminate hepatitis C virus. Tissue Antigens (2007) 69:109–11. doi:10.1111/j.1399-0039.2006.762_7.x
38. Amadei B, Urbani S, Cazaly A, Fisicaro P, Zerbini A, Ahmed P, et al. Activation of natural killer cells during acute infection with hepatitis C virus. Gastroenterology (2010) 138:1536–45. doi:10.1053/j.gastro.2010.01.006
39. Pelletier S, Drouin C, Bédard N, Khakoo SI, Bruneau J, Shoukry NH. Increased degranulation of natural killer cells during acute HCV correlates with the magnitude of virus-specific T cell responses. J Hepatol (2010) 53:805–16. doi:10.1016/j.jhep.2010.05.013
40. Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science (2004) 305:872–4. doi:10.1126/science.1097670
41. Knapp S, Warshow U, Hegazy D, Brackenbury L, Guha IN, Fowell A, et al. Consistent beneficial effects of killer cell immunoglobulin-like receptor 2DL3 and group 1 human leukocyte antigen-C following exposure to hepatitis C virus. Hepatology (2010) 51:1168–75. doi:10.1002/hep.23477
42. Alter G, Jost S, Rihn S, Reyor LL, Nolan BE, Ghebremichael M, et al. Reduced frequencies of NKp30+NKp46+, CD161+, and NKG2D+ NK cells in acute HCV infection may predict viral clearance. J Hepatol (2011) 55:278–88. doi:10.1016/j.jhep.2010.11.030
43. Golden-Mason L, Cox AL, Randall JA, Cheng L, Rosen HR. Increased natural killer cell cytotoxicity and NKp30 expression protects against hepatitis C virus infection in high-risk individuals and inhibits replication in vitro. Hepatology (2010) 52:1581–9. doi:10.1002/hep.23896
44. Thoens C, Berger C, Trippler M, Siemann H, Lutterbeck M, Broering R, et al. KIR2DL3+NKG2A- natural killer cells are associated with protection from productive hepatitis C virus infection in people who inject drugs. J Hepatol (2014) 61:475–81. doi:10.1016/j.jhep.2014.04.020
45. Petitdemange C, Becquart P, Wauquier N, Béziat V, Debré P, Leroy EM, et al. Unconventional repertoire profile is imprinted during acute chikungunya infection for natural killer cells polarization toward cytotoxicity. PLoS Pathog (2011) 7:e1002268. doi:10.1371/journal.ppat.1002268.t001
46. Thanapati S, Das R, Tripathy AS. Phenotypic and functional analyses of NK and NKT-like populations during the early stages of chikungunya infection. Front Microbiol (2015) 6:895. doi:10.3389/fmicb.2015.00895
47. Hendricks DW, Balfour HH, Dunmire SK, Schmeling DO, Hogquist KA, Lanier LL. Cutting edge: NKG2C(hi)CD57+ NK cells respond specifically to acute infection with cytomegalovirus and not Epstein-Barr virus. J Immunol (2014) 192:4492–6. doi:10.4049/jimmunol.1303211
48. Foley B, Cooley S, Verneris MR, Curtsinger J, Luo X, Waller EK, et al. Human cytomegalovirus (CMV)-induced memory-like NKG2C+ NK cells are transplantable and expand in vivo in response to recipient CMV antigen. J Immunol (2012) 189:5082–8. doi:10.4049/jimmunol.1201964
49. Lopez-Vergès S, Milush JM, Schwartz BS, Pando MJ, Jarjoura J, York VA, et al. Expansion of a unique CD57+NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc Natl Acad Sci U S A (2011) 108:14725–32. doi:10.1073/pnas.1110900108
50. Naranbhai V, Altfeld M, Abdool Karim Q, Ndung’u T, Abdool Karim SS, Carr WH, et al. Natural killer cell function in women at high risk for HIV acquisition: insights from a microbicide trial. AIDS (2012) 26:1745–53. doi:10.1097/QAD.0b013e328357724f
51. Naranbhai V, Abdool Karim SS, Altfeld M, Samsunder N, Durgiah R, Sibeko S, et al. Innate immune activation enhances HIV acquisition in women, diminishing the effectiveness of tenofovir microbicide gel. J Infect Dis (2012) 206:993–1001. doi:10.1093/infdis/jis465
52. Scott-Algara D, Truong LX, Versmisse P, David A, Luong TT, Nguyen NV, et al. Cutting edge: increased NK cell activity in HIV-1-exposed but uninfected Vietnamese intravascular drug users. J Immunol (2003) 171:5663–7. doi:10.4049/jimmunol.171.11.5663
53. Strauss-Albee DM, Fukuyama J, Liang EC, Yao Y, Jarrell JA, Drake AL, et al. Human NK cell repertoire diversity reflects immune experience and correlates with viral susceptibility. Sci Transl Med (2015) 7:297ra115. doi:10.1126/scitranslmed.aac5722
54. Orange JS. Natural killer cell deficiency. J Allergy Clin Immunol (2013) 132:515–25. doi:10.1016/j.jaci.2013.07.020
55. Choi PJ, Mitchison TJ. Imaging burst kinetics and spatial coordination during serial killing by single natural killer cells. Proc Natl Acad Sci U S A (2013) 110(16):6488–93. doi:10.1073/pnas.1221312110
56. Vanherberghen B, Olofsson PE, Forslund E, Sternberg-Simon M, Khorshidi MA, Pacouret S, et al. Classification of human natural killer cells based on migration behavior and cytotoxic response. Blood (2013) 121:1326–34. doi:10.1182/blood-2012-06-439851
57. Christakou AE, Ohlin M, Vanherberghen B, Khorshidi MA, Kadri N, Frisk T, et al. Live cell imaging in a micro-array of acoustic traps facilitates quantification of natural killer cell heterogeneity. Integr Biol (Camb) (2013) 5:712–9. doi:10.1039/c3ib20253d
58. Forslund E, Sohlberg E, Enqvist M, Olofsson PE, Malmberg KJ, Onfelt B. Microchip-based single-cell imaging reveals that CD56dimCD57-KIR-NKG2A+ NK cells have more dynamic migration associated with increased target cell conjugation and probability of killing compared to CD56dimCD57-KIR-NKG2A-NK cells. J Immunol (2015) 195:3374–81. doi:10.4049/jimmunol.1500171
59. Horowitz A, Guethlein LA, Nemat-Gorgani N, Norman PJ, Cooley S, Miller JS, et al. Regulation of adaptive NK cells and CD8 T cells by HLA-C correlates with allogeneic hematopoietic cell transplantation and with Cytomegalovirus reactivation. J Immunol (2015) 195(9):4524–36. doi:10.4049/jimmunol.1401990
60. Strauss-Albee DM, Horowitz A, Parham P, Blish CA. Coordinated regulation of NK receptor expression in the maturing human immune system. J Immunol (2014) 193:4871–9. doi:10.4049/jimmunol.1401821
61. Paust S, Bettini M. Daring to learn from humanized mice. Blood (2015) 125:3829–31. doi:10.1182/blood-2015-04-639435
Keywords: natural killer cells, lymphocyte diversity, mass cytometry, viral susceptibility, single-cell technology
Citation: Strauss-Albee DM and Blish CA (2016) Human NK Cell Diversity in Viral Infection: Ramifications of Ramification. Front. Immunol. 7:66. doi: 10.3389/fimmu.2016.00066
Received: 07 December 2015; Accepted: 12 February 2016;
Published: 03 March 2016
Edited by:
Emanuela Marcenaro, University of Genova, ItalyReviewed by:
Todd A. Fehniger, Washington University School of Medicine, USARaffaele Badolato, University of Brescia, Italy
Copyright: © 2016 Strauss-Albee and Blish. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Catherine A. Blish, Y2JsaXNoQHN0YW5mb3JkLmVkdQ==
 Dara M. Strauss-Albee
Dara M. Strauss-Albee Catherine A. Blish
Catherine A. Blish