- 1Center for Oncology and Cell Biology, The Feinstein Institute for Medical Research, Manhasset, NY, USA
- 2Department of Pathology, Immunology, and Laboratory Medicine, University of Florida, Gainesville, FL, USA
- 3Department of Medicine, The Hofstra Northwell School of Medicine, Manhasset, NY, USA
- 4Department of Molecular Medicine, The Hofstra Northwell School of Medicine, Manhasset, NY, USA
B6.Sle1.Sle2.Sle3 (B6.TC) lupus-prone mice carrying the NZB allele of Cdkn2c, encoding for the cyclin-dependent kinase inhibitor P18INK4, accumulate B-1a cells due to a higher rate of proliferative self-renewal. However, it is unclear whether this affects primarily early-appearing B-1a cells of fetal origin or later-appearing B-1a cells that emerge from bone marrow. B-1a cells are the major source of natural autoantibodies, and it has been shown that their protective nature is associated with a germline-like sequence, which is characterized by few N-nucleotide insertions and a repertoire skewed toward rearrangements predominated during fetal life, VH11 and VH12. To determine the nature of B-1a cells expanded in B6.TC mice, we amplified immunoglobulin genes by PCR from single cells in mice. Sequencing showed a significantly higher proportion of B-1a cell antibodies that display fewer N-additions in B6.TC mice than in B6 control mice. Following this lower number of N-insertions within the CDR-H3 region, the B6.TC B-1a cells display shorter CDR-H3 length than B6 B-1a cells. The absence of N-additions is a surrogate for fetal origin, as TdT expression starts after birth in mice. Therefore, our results suggest that the B-1a cell population is not only expanded in autoimmune B6.TC mice but also qualitatively different with the majority of cells from fetal origin. Accordingly, our sequencing results also demonstrated the overuse of VH11 and VH12 in autoimmune B6.TC mice as compared to B6 controls. These results suggest that the development of lupus autoantibodies in these mice is coupled with skewing of the B-1a cell repertoire and possible retention of protective natural antibodies.
Introduction
Murine B-1a cells are a unique B-lymphocyte lineage characterized by phenotypic, functional, and ontologic characteristics (1, 2). B-1a cells are defined by surface marker expression of IgMhiIgDloCD45RloCD5+CD43+CD19hi and are found in the peritoneal cavity, spleen, and bone marrow (3, 4). Functionally, B-1a cells exhibit unique signaling characteristics (4–6), are potent antigen-presenting cells (7), and spontaneously produce 80–90% of natural serum IgM in mice (8). Natural IgM is non-immune, low-affinity immunoglobulin (Ig) that is both polyreactive and autoreactive. It functions in infection, atherosclerosis, B cell homeostasis, inflammation, and autoimmunity [reviewed in Ref. (9)]. Minimal N-region addition contributes to the germline-like nature of natural IgM. Furthermore, natural IgM manifests biased variable heavy chain (VH) gene usage in favor of VH11 and VH12, which are specific for phosphatidylcholine (PtC), a major component of cell membrane phospholipids (3, 10–13). This unique germline structure of natural IgM is established during the early fetal and neonatal development of B-1a cells (8).
The polyreactive nature of natural IgM provides initial defense against both bacterial and viral pathogens, which affords the organism protection during the period preceding generation of high-affinity antigen-specific antibodies produced by germinal center B-2 cells (3, 4, 14–17). The autoreactive quality of natural IgM has been shown to aid in the elimination of excess autoantigens through the removal of apoptotic cells and noxious molecular debris, thereby maintaining homeostasis and preventing inflammation (9, 18). These autoreactive natural antibodies are often directed against cell membrane components, such as PtC and phosphorylcholine (PC), which is the polar head group of PtC and is a major microbial cell wall determinant (19). Interestingly, such components are closely related to those also present on pathogens, which suggest that the natural autoreactive repertoire also react with common pathogens (20).
While this cross-reactivity of natural autoreactive antibodies with pathogens is beneficial, it highlights the importance of regulating B-1a cell expansion. Accumulation of B-1a cells has been shown in the NZM2410 (NZB × NZW F1 hybrid) mouse model of systemic lupus erythematosus (SLE) (21, 22). However, the role of B-1a cells in lupus is still unclear (23). Some studies have demonstrated a role for B-1a cells via production of IL-10 (22), increase in antigen presentation (22), or with overexpression of osteopontin, resulting in expansion of B-1a cells and increased anti-dsDNA antibody production (24). In other models, however, B-1a cells do not contribute to disease (25, 26).
The expansion of B-1a cells in the NZM2410 model was traced to the Sle2c1 lupus susceptibility locus, which contains Ckdn2c (27, 28). The Ckdn2c gene encodes for p18INK4c, which is a cyclin-dependent kinase inhibitor that controls progression through G1 of the cell cycle (28). These studies demonstrated that the expansion of B-1a cells is intrinsic. B6. Sle2c1 B-1a cells showed increased proliferation at rest, as well as increased resistance to cell death. In addition, B-1a cell reconstitution from fetal liver, adult bone marrow, and adult spleen was higher in B6. Sle2c1 than in control C57/BL6 (B6) mice following lethal irradiation (21). Furthermore, comparison of p18−/− mice with B6. Sle2c1 mice demonstrated that both produced autoantibodies; however, the amount produced by p18−/− mice was greater. This demonstrates that the control of the B-1a cell population depends on the amount of p18. B6. Sle2c1 mouse B cells have fourfold less Ckdn2c than normal mice, whereas p18−/− mice completely lack Ckdn2c (28). Together, these results demonstrate an important role for p18 in B-1a cell numbers, which in turn affects the production of autoantibodies and development of autoimmunity. However, the origin of B-1a cell expansion in B6.TC, B6.Slec1, and p18−/− mice could be due to an increase in proliferation of early-appearing fetal-derived B-1a cells or heightened production of later-appearing bone marrow-derived B-1a cells. As the repertoires of early- and later-appearing B-1a cells differ, these two possibilities can be distinguished. Herein, we investigated whether significant changes to the natural IgM repertoire occur in triple congenic B6.Sle1.Sle2.Sle3 (B6.TC) lupus-prone mice. These mice carry the Sle2c1 locus that drives B-1a cell expansion and present clinical autoimmune pathology that has been described for the NZM2410 pathology (29). B6.TC mice carry the NZM2410 susceptibility loci on a B6 genetic background (>95%) that includes both heavy and light immunoglobulin chains, which allow to directly compare the lupus-prone B6.TC mice to the control B6 mice. Specifically, we found that the expansion of B-1a cells in B6.TC mice is associated with repertoire skewing toward VH11 and VH12 usage.
Materials and Methods
Mice
B6.NZM-Sle1NZM2410/AegSle2NZM2410/AegSle3NZM2410/Aeg/LmoJ (B6.TC) congenic mice have been previously described (29). B6.TC lupus-prone and C57BL6/J (B6) control mice were cared for and handled in accordance with National Institutes of Health and institutional guidelines.
Single-Cell Sequencing and Analysis
Peritoneal washout cells were obtained from 8-week-old wild-type B6 mice and 8-week-old B6.TC mice (two each). The cells were stained with fluorescence-labeled antibodies to CD45R/B220 (clone RA3-6B2), CD5 (clone 53-7.3), and CD23 (clone B3B4) (BD Biosciences). B-1a cells (CD45Rlo/CD5+/CD23−) were then purified using an Influx cell sorter (BD Biosciences). Post-sort reanalysis of B cell populations showed them to be ≥98% pure. Peritoneal B-1a cells were sorted into a 96-well plate containing 20 μl of lysis buffer per well (dH2O, RNase Out, 5× SuperScript III Buffer, DTT, IgePAL, and Carrier RNA). Reverse transcription was performed (42°C – 10 min, 25°C – 10 min, 50°C – 60 min, 94°C – 5 min, hold at 4°C) after addition of random hexamers, dNTP mix, and SuperScript III reverse transcriptase. Semi-nested PCR was performed using the cDNA diluted 1:1. Using Qiagen’s HotStart Taq Plus, 2.5 μl of cDNA was used for the first PCR reaction with previously described primers (30). The product from this first PCR reaction was then diluted 1:100 and 2 μl of the diluted product was added to the second PCR reaction. The products were purified and then sequenced (Genewiz) using the forward primer. Sequences were then analyzed using an online sequence analysis tool for VDJ sequences (IMGT, the international ImMunoGeneTics information system).
Statistics
Comparisons were conducted between the pooled B6.TC and B6 sequences and the two strains using Graphpad Prism 6.0 with two-tailed tests, as indicated in the figure legends.
Results
Lupus-Prone Triple Congenic Mice Display an Increase in Duplicate Sequences
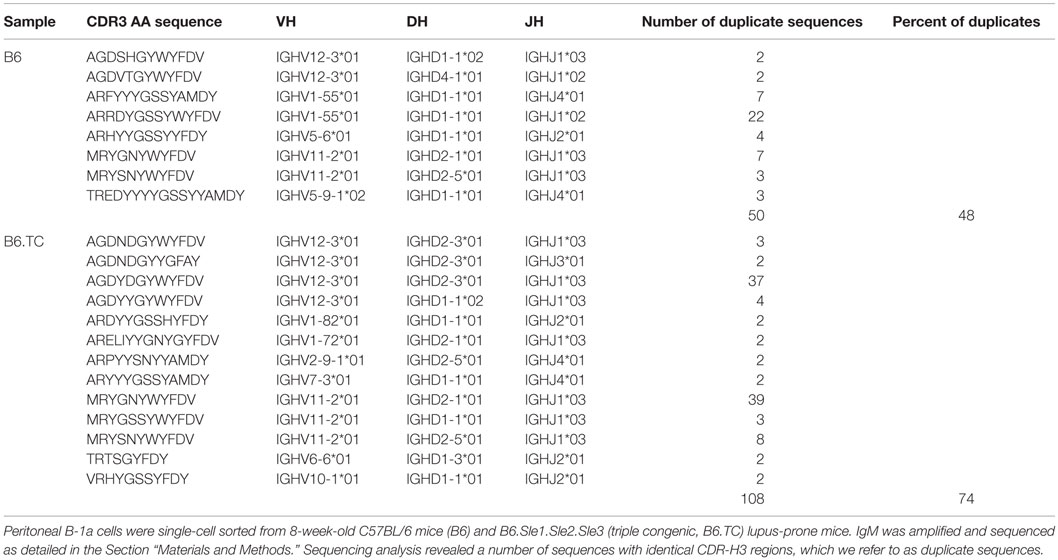
The B-1a cell repertoire of B6.TC mice was compared to control B6 mice. Repertoire analysis was performed by PCR amplification of the VHDJH region from individual peritoneal B-1a cells, which were previously shown to accumulate in B6.TC mice (21, 28). Interestingly, the B-1a cells from B6.TC mice had a significantly larger number of IgM sequences with identical VH, DH, and JH segments as well as identical CDR3 regions than B6 mice (Table 1, B6.TC: 108 out of 146 total sequences; B6: 50 out of 105 total sequences; p = 0.0335, Mann–Whitney test). As stated in previously published work (30), it cannot be determined whether these sequences containing identical VH, DH, JH, and CDR3 regions result from a single clonal expansion or from analysis of independent cells with identical rearrangements. Therefore, we will refer to such sequences as duplicate sequences instead of clones. Furthermore, VH usage within the duplicate sequences differed significantly between B6 and B6.TC mice. As shown in Table 1, the duplicate sequences with the highest frequency in B6 mice utilized VH1–55 (58%), whereas the most frequent duplicate sequences in B6.TC mice utilized VH11 and VH12 (43 and 46%, respectively). While both VH11 and VH12 utilization (20 and 8%, respectively) was observed in B6 mice, this percentage of duplicate sequences was significantly less than that seen in B6.TC mice (Figure 1). These results suggest that the accumulation of peritoneal B-1a cells seen in B6.TC mice might be influenced by autoantigen since there is an expansion of B-1a cells utilizing VH11 and VH12, which are specific for PtC, a major component of cell membrane phospholipids.
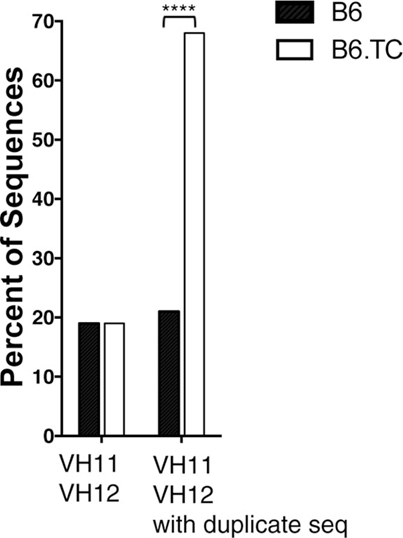
Figure 1. Percent of VH11 and VH12 representation. Peritoneal B-1a cells were single-cell sorted from 8-week-old B6 control mice and B6.TC lupus-prone mice. IgM was amplified and sequenced as detailed in the Section “Materials and Methods.” The percent of sequences that utilized VH11 and VH12 are shown for all unique sequences (left, B6 n = 62; B6.TC n = 47), and all sequences obtained, including the duplicates (right, B6 n = 105; B6.TC n = 146). Statistical analysis was performed using chi-square analysis (2 × 2 using the VH of interest and all others as the two categories), VH11/VH12 with duplicate sequences ****p < 0.0001.
VH–DH–JH Usage Shows Differences between B6 and B6.TC Mouse Repertoires
For analysis of VH, DH, and JH usage, we evaluated the repertoire in two ways. First, we analyzed only sequences with unique CDR-H3 regions by removing all duplicate sequences. In the second method, we analyzed all sequences, which included the duplicate sequences.
When analyzing only sequences with unique CDR-H3 regions, we found overall similarity in DH and JH usage with only one major significant difference in VH usage. Among VH gene segments, VH1 was expressed significantly less frequently by B6.TC B-1a cells (26%) as compared to B6 B-1a cells (48%) (p = 0.0034, chi-square test) (Figure 2A).
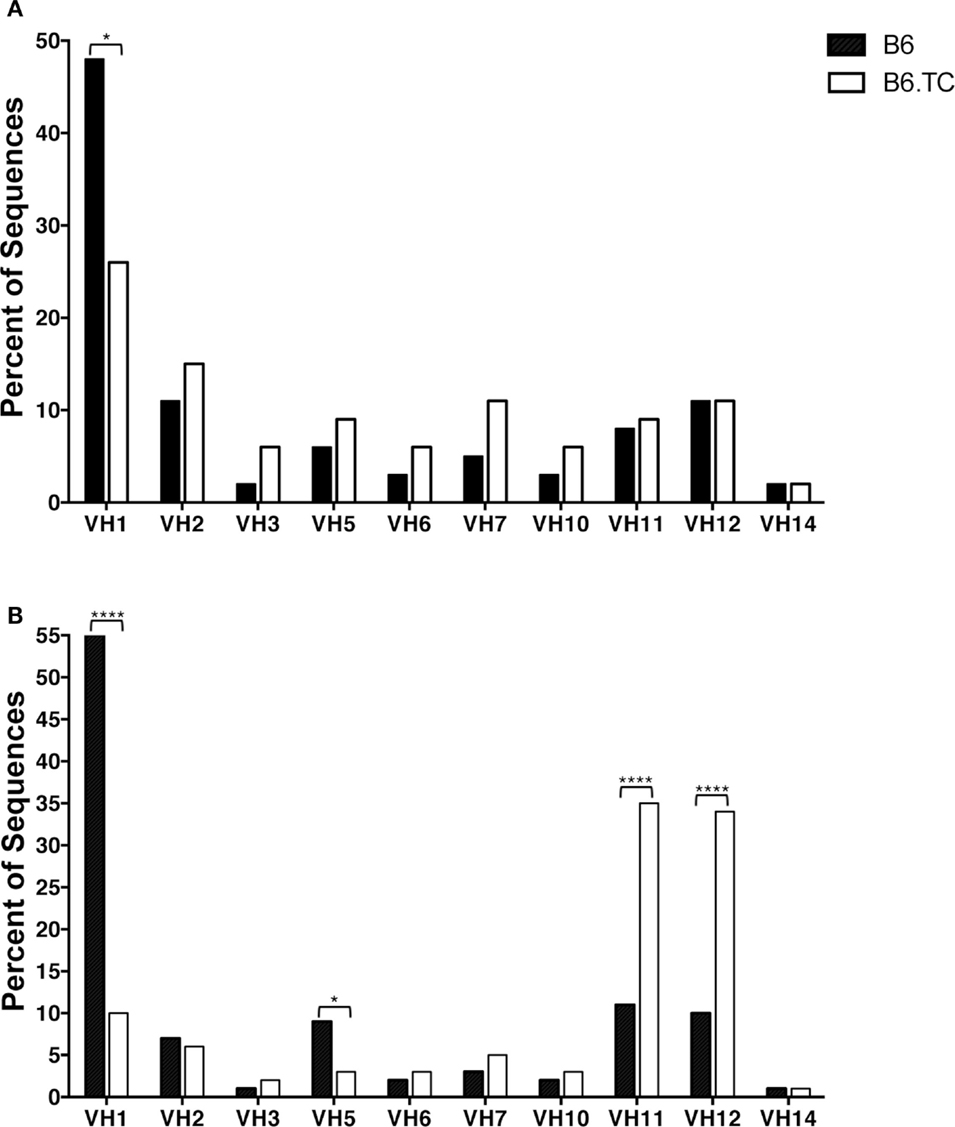
Figure 2. VH analysis of IgM from C57BL/6 and triple congenic peritoneal B-1a cells. Immunoglobulins were amplified by PCR from single-cell sorted peritoneal B-1a cells obtained from B6 and B6.TC mice and evaluated for the variable (V) segment heavy chain usage. The percent of cells (sequences) expressing the V segment usage is displayed. Chi-squared test was used to determine significance. (A) Analysis of sequences with only unique CDR-H3 regions (B6, n = 62; B6.TC n = 47). (B) Analysis of all sequences obtained, including the duplicate sequences (B6, n = 105; B6.TC n = 146).
We found numerous differences in VH–DH–JH usage when analyzing all sequences, including the duplicate sequences (Figure 2B). VH1 and VH5 were expressed significantly less frequently by B6.TC B-1a cells (10 and 3%, respectively), as compared to B6 B-1a cells (55 and 9%, respectively) (p < 0.0001 and p = 0.0397, chi-square test). Conversely, VH11 and VH12 were utilized significantly more frequently by B6.TC B-1a cells (35 and 34%, respectively), as compared to B6 B-1a cells (11 and 10%, respectively) (p < 0.0001 and p < 0.0001, chi-square test). Among DH gene segments, no difference was observed for the unique sequences between strains (Figure 3A). When all sequences were compared, DFL16.1 was expressed less frequently, and DSP was expressed more frequently by B6.TC B-1a cells (17 and 74%, respectively) as compared to B6 B-1a cells (54 and 32%, respectively) (p < 0.0001 and p < 0.0001, chi-square test) (Figure 3B). As for JH segments, no difference was observed between strains among JH gene segments in unique sequences (Figure 4A). However, when all sequences were compared, JH1 was expressed more frequently (74%) (p < 0.0001), and JH2 and JH4 were expressed less frequently (11 and 8%, respectively) (p = 0.0293 and p < 0.0001, respectively) by B6.TC B-1a cells as compared to B6 B-1a cells (45, 21, and 28%, respectively) (Figure 4B). Thus, distinct VH, DH, and JH gene segment usage separated B6.TC from B6 peritoneal B-1a cells across all sequences.
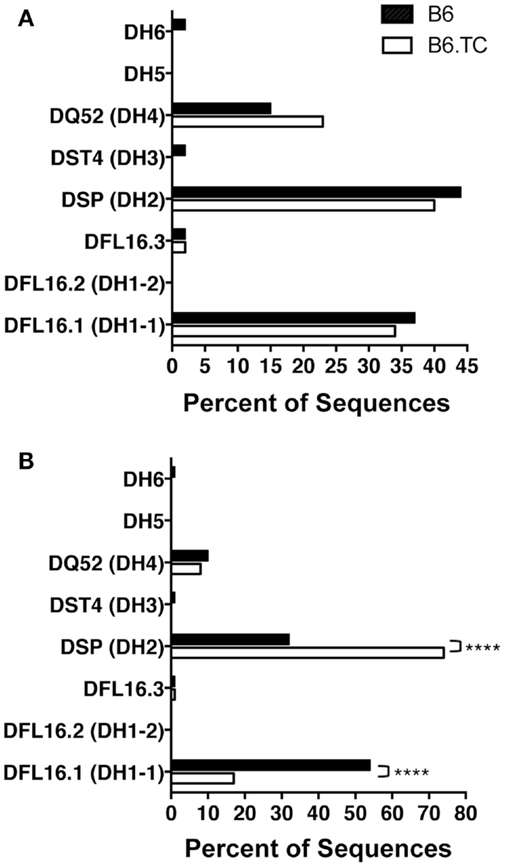
Figure 3. DH analysis of IgM from C57BL/6 and triple congenic mouse peritoneal B-1a cells. Immunoglobulins were amplified by PCR from single-cell sorted peritoneal B-1a cells obtained from B6 and B6.TC mice and evaluated for the diversity (D) segment heavy chain usage. The percent of cells (sequences) expressing the D segment usage is displayed. Chi-squared test was used to determine significance. (A) Analysis of sequences with only unique CDR-H3 regions (B6, n = 62; B6.TC n = 47). (B) Analysis of all sequences obtained, including the duplicate sequences (B6, n = 105; B6.TC n = 146).
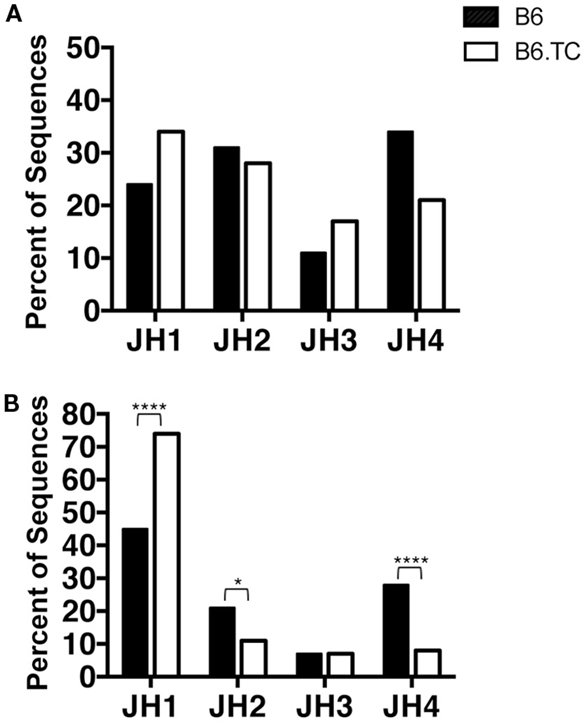
Figure 4. JH analysis of IgM from C57BL/6 and triple congenic mouse peritoneal B-1a cells. Immunoglobulins were amplified by PCR from single-cell sorted peritoneal B-1a cells obtained from B6 and B6.TC mice and evaluated for the junction (J) segment heavy chain usage. The percent of cells (sequences) expressing the J segment usage is displayed. Chi-squared test was used to determine significance. (A) Analysis of sequences with only unique CDR-H3 regions (B6, n = 62; B6.TC n = 47). (B) Analysis of all sequences obtained, including the duplicate sequences (B6, n = 105; B6.TC n = 146).
The B-1a Cell Repertoire Is Less Diverse in B6.TC Mice as Compared to B6 Mice
N-region addition provides diversity to the CDR-H3 region of antibodies via random insertion of nucleotides at the V–D and D–J junctions by the enzyme TdT. It is well-documented that peritoneal B-1a cells have limited N-addition due to the lack of TdT expression during fetal development (31). We analyzed N-addition at the D–J and V–D junctions and determined CDR3 length. No significant differences were found when analyzing sequences with only unique CDR-H3 regions (Table 2). In contrast, analysis of all sequences, including the duplicates, demonstrated significant differences between B-1a cells from B6.TC and B6 mice. We found that the number of N-additions at the D–J or V–D junctions of B6.TC B-1a cells was significantly less than B6 B-1a cells (p < 0.0001 and p = 0.0120, respectively) (Table 2). B6.TC B-1a cells were also found to contain significantly fewer N-additions when analyzing the sum of the two junctions as compared to B6 B-1a cells (p < 0.0001). We further examined CDR-H3 length and found, consistent with the differences in N-addition between B6.TC and B6 B-1a cells, the average CDR-H3 lengths differed significantly when analyzing all sequences (p = 0.0044), but did not differ when analyzing only sequences with unique CDR-H3 regions (Table 2). These results demonstrate that the B6.TC B-1a cell population expresses immunoglobulin that is less diverse due to fewer N-region additions as compared to the B6 peritoneal B-1a cell population.
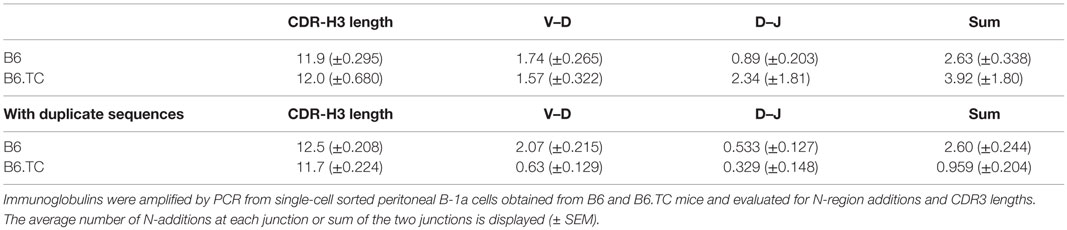
Table 2. N-region addition and CDR3 length analysis of IgM from C57BL/6 and triple congenic mouse peritoneal B-1a cells.
We then focused the analysis of N-addition on sequences utilizing VH11 and VH12, which are overrepresented in the B6.TC B-1a cells when considering all sequences, including the duplicate sequences. Interestingly, the number of B6.TC B-1a cell sequences lacking N-additions at both junctions in cells utilizing VH11 and VH12 (98 and 100%, respectively) was significantly different than B6 B-1a cell sequences (92 and 10%, respectively) (p = 0.0381 and p < 0.0001, respectively, by chi-square test), particularly with respect to VH12, although the number of B6 VH11/VH12 sequences was small (n = 12/n = 10). These results are summarized in Figure 5.
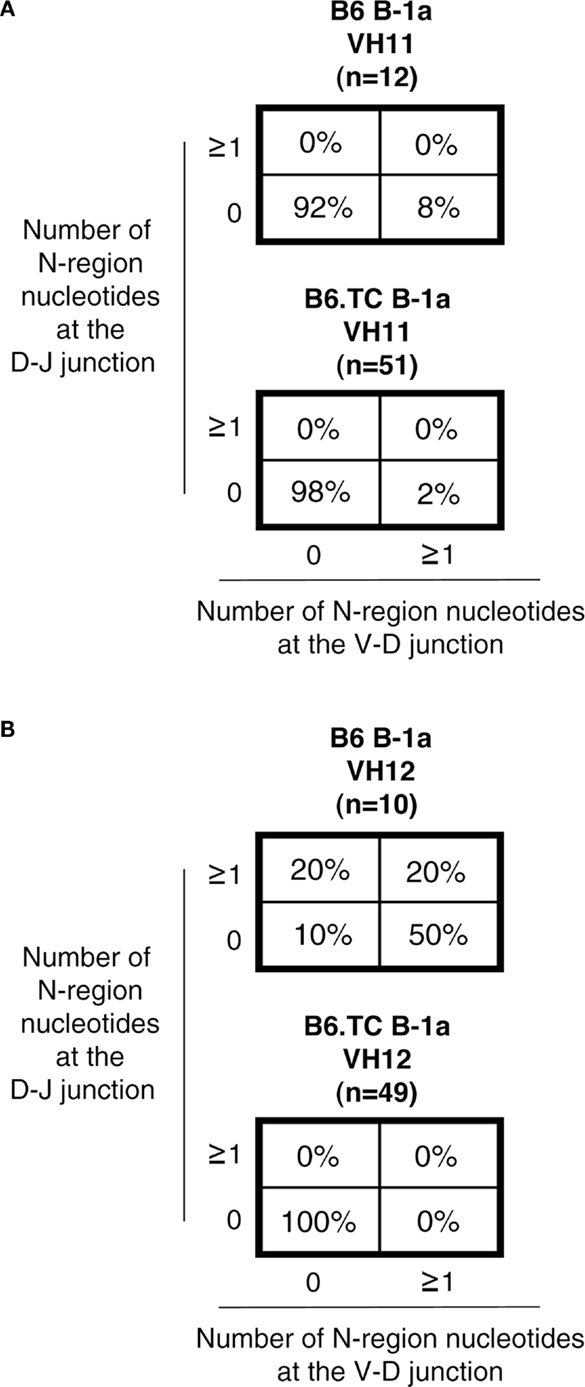
Figure 5. N-region addition analysis of IgM from C57BL/6 and triple congenic mouse peritoneal B-1a cells utilizing VH11 and VH12. Immunoglobulins were amplified by PCR from single-cell sorted peritoneal B-1a cells obtained from B6 and B6.TC mice and evaluated for N-region additions. The percent of sequences with 0 N-additions at both junctions, one or more N-additions at both junctions, 0 N-additions at V–D and one or more at D–J junctions, or 0 N-additions at D–J and one or more at V–D junctions is shown. (A) N-addition analysis of B-1a cells utilizing VH11 (B6 n = 12; B6.TC n = 51). (B) N-addition analysis of B-1a cells utilizing VH12 (B6 n = 10; B6.TC n = 49).
CDR-H3s Are More Charged in B6.TC than B6 B-1a Cells
Autoreactive antibodies, and in particular anti-dsDNA antibodies, are often enriched for charged amino acids in their CDR-H3 loop region (32–34). This region normally contains neutral, hydrophilic amino acids, which is due partly to usage of certain DH and JH sequences and use of the D reading frame I (32, 35). Amino acid changes within the loop region are also affected by N-region insertions (32). Upon evaluation of B6.TC B-1a cell CDR-H3 charge, we found that the average charge was increased in B6.TC B-1a cells (−0.213) over B6 B-1a cells (−0.142); however, this difference did not reach significance in the analysis of unique sequences (Figure 6A). When analyzing all sequences (including the duplicates), again the average charge of the CDR-H3 loop region was increased in B6.TC B-1a cells (−0.298) over B6 B-1a cells (−0.201) (p = 0.0015) (Figure 6B). Furthermore, comparing the average charge of the CDR-H3 loop region of B-1a cells utilizing VH11 and VH12 also demonstrated a greater charge in B6.TC B-1a cells (−0.362) than B6 B-1a cells (−0.290) (p = 0.0197) (Figure 6C). These results correlate with the differences observed in N-region addition between B6.TC and B6 B-1a cells utilizing VH11 and VH12.
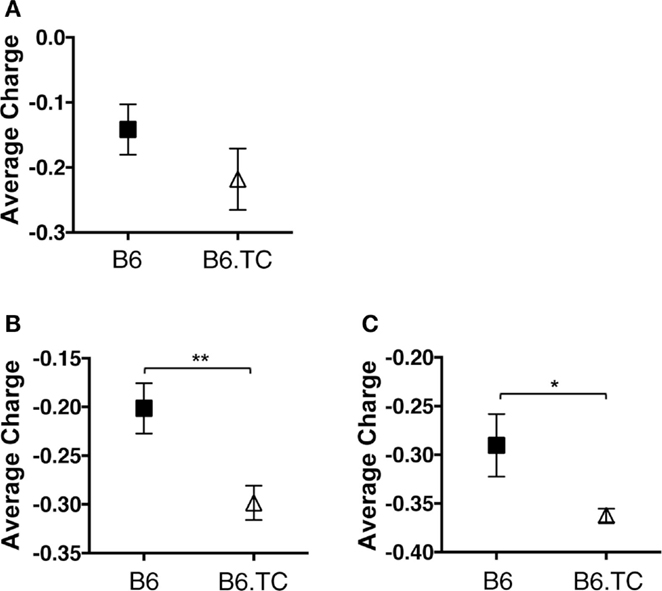
Figure 6. Average charge of CDR-H3 loop region of IgM from C57BL/6 and triple congenic mouse peritoneal B-1a cells. Immunoglobulins were amplified by PCR from single-cell sorted peritoneal B-1a cells obtained from B6 and B6.TC mice. IgM was amplified and sequenced as detailed in the Section “Materials and Methods.” The average charge of the CDR-H3 loop region of IgM is shown. (A) Analysis of sequences with only unique CDR-H3 regions (B6, n = 62; B6.TC n = 47). (B) Analysis of all sequences obtained, including the duplicate sequences (B6, n = 105; B6.TC n = 146). (C) Analysis of all VH11 and VH12 sequences, including the duplicate sequences (B6, n = 22; B6.TC n = 100).
Discussion
Primary repertoire analysis of B-1a cells from 8-week-old B6.Sle1.Sle2.Sle3 (B6.TC) lupus-prone mice demonstrated a large number of sequences that express identical CDR-H3 regions as compared to B-1a cells from healthy 8-week-old C57BL/6 (B6). This analysis demonstrates a significant increase in identical VH, DH, JH usage in B6.TC mice. Although it is not possible to determine whether the duplicate sequences observed herein result from a single clonal expansion or from analysis of multiple cells with identical rearrangements, it has been well-documented over the years that B-1 cells have a limited repertoire (11, 14, 36–38), can undergo clonal expansion (39–42), and are self-replenishing (8). Therefore, these duplicate sequences are most likely due to expansion of single B-1a cells. Further analysis, including the duplicate sequences, reveals that the B6.TC B-1a cell repertoire displays early fetal/neonatal-like characteristics, which consists of an increase in use of JH1 [Figure 4B; Ref. (43)], few N-additions at both the V–D and D–J junctions, and a shorter average CDR-H3 length (Table 2). In addition, the B6.TC repertoire overused VH11 and VH12 as compared to B6 (Figures 1 and 2). Interestingly, VH11 and VH12 rearrangements are utilized almost exclusively by B-1a cells and target the cell membrane component PtC (19). Studies have shown VH11 in particular is a VH gene utilized during fetal development but not during adult development (44, 45). More recently, Yang et al. have shown overuse of VH11 in the normal healthy peritoneal B-1a cell pool (38). Our results demonstrate the most common CDR3 in peritoneal B-1a cells from our normal healthy 2-month old B6 mice is ARRDYGSSYWYFDV (VH1-55, DH1-1, JH1). Examining Yang et al’s most common CDR3 in peritoneal B-1a cells from their normal healthy 2-month old B6 mice, it is ARFYYYGSSYAMDY, (VH1-55, DH1-1, JH4), which does not share the exact same CDR3 as ours but does share the same VH and DH region. Our second most common CDR3 sequences (two are tied for second place) are identical to Yang et al’s first and second most common CDR3 sequences ARFYYYGSSYAMDY and MRYGNYWYFDV (VH11-2, D2-8, JH1), respectively. The rank order of the sequences we identified is very similar to that of Yang et al. with only minor differences. Together, these results indicate that the B-1a cell repertoire in B6.TC mice reflects fetal rearrangements to a much greater extent than the B6 B-1a cell repertoire.
The mechanism for this selection toward fetal rearrangements in B6.TC mice is unknown; however, it can be speculated that the Sle2c1 lupus susceptibility locus, which contains Ckdn2c and results in less p18 expression, could lead to a difference in expansion of B-1a cells with different BCR signaling requirements. It is possible the VH11 and VH12 specificities require a different level of BCR signaling, which the reduced level of p18 might provide, thereby allowing for increased proliferation of VH11 and VH12 expressing B-1a cells. In this view, Ckdn2c does not encode B-1a cell expansion on its own but does so in collaboration with BCR signals of requisite intensity. Our data suggest that such signals might be provided by self-antigen binding of PtC-specific immunoglobulins in preference to other elements of the B-1a cell repertoire.
Various studies have shown that B-1a cell-derived natural IgM provides protection against not only infection but also autoimmunity (9, 18, 46). The role of IgM in protection against autoimmunity was recently demonstrated in a mouse model lacking secretory IgM. This study demonstrated that natural IgM is required to control the accumulation of autoantibodies via its ability to regulate B cell development and selection (46). In mice lacking secretory IgM, the number of peritoneal B-1a cells was significantly decreased and of the B-1a cells present, there was little VH11 expression, which correlated with a lack of PtC-binding B-1a cells (46). These findings would seem to be at odds with previous studies demonstrating that mice with the Sle2c1 lupus susceptibility locus have an expansion of peritoneal B-1a cells and yet they develop autoimmunity (28). Furthermore, results presented herein reveal that the B-1a cell repertoire in B6.TC mice is significantly more skewed toward VH11 and VH12 than control B6 mice (Figures 1 and 2). Together, these studies raise the question as to why the B6.TC mice are not protected against autoimmunity if they have an expansion of B-1a cells producing protective natural IgM. As demonstrated by Nguyen et al., selection of the B cell repertoire is affected by the presence of IgM (46). Herein, we demonstrate the expanded B-1a cells in mice carrying the Sle2c1 lupus susceptibility locus are skewed toward a different specificity than control B6 mice. Together, these studies suggest that altering the pool of natural IgM disrupts the balance of antibodies, which enable selection of a healthy non-autoreactive repertoire. In other words, alteration of the natural IgM repertoire in the TC.B6 mice could then lead to selection of an autoreactive repertoire instead of a non-autoreactive repertoire, despite natural IgM being present. Furthermore, the role of B-1a cells in autoimmunity may not be limited to the antibodies they produce. B-1a cells have been shown to be potent antigen-presenting cells, which could also contribute to autoimmunity (7, 47, 48). In addition, B-1a cells have immunoregulatory functions through the secretion of IL-10 (49), ability to produce adenosine (50, 51), and ability to class switch in sites of inflammation (52).
The greater expansion/overuse of VH11 and VH12 might not help in the regulation of autoimmunity; however, it might afford increased protection from sepsis. It has been shown that the mice lacking secretory IgM are more susceptible to sepsis induced by cecal ligation and puncture (53). Interestingly, these mice could be rescued by injection of anti-PtC antibody, but not anti-PC antibody (53). Future studies could provide insight into whether the B6.TC lupus-prone mice might be more protected against bacterial sepsis. Such resistance to bacteria has been shown for mice with the Sle3 lupus susceptibility locus (54); however, a role for B cells in such resistance has yet to be investigated.
The results presented herein demonstrate that the available B-1a cell repertoire present in 8-week-old B6.TC lupus-prone mice is more characteristic of an early fetal/neonatal B cell repertoire than that of B-1a cells from healthy aged-matched B6 mice. Thus, B6.TC-enhanced B-1a cell expansion is established early on and affects developing B-1a cells in a BCR-specific manner. Nonetheless, further analysis is required to determine the mechanism of B-1a cell expansion, and in particular, whether certain BCR specificities have a growth advantage in B6.TC mice. Overall, our results together with previous studies suggest the development of natural IgM that is protective against both bacterial infections and autoimmunity might require a balance of repertoire specificities. Previously published studies suggest this balance is greatly influenced by the IgM repertoire present (46). Further repertoire analyses of healthy and autoimmune models will help uncover factors that might affect this balance of protection against both infection and autoimmunity.
Ethics Statement
Experiments using animals were conducted under approved Institutional Animal Care and Use Committee (IACUC) at the University of Florida.
Author Contributions
Conceptualization and methodology, LM, NH, and TR; investigation, NH, LZ, and LM; writing – original draft, NH; writing – review and editing, LM, TR, and NH; funding acquisition, resources, and supervision, LM and TR.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work was supported by Public Health Service grants AI029690 (TR) and AI058150 (LM) awarded by the National Institutes of Health.
References
1. Herzenberg LA, Tung JW. B cell lineages: documented at last! Nat Immunol (2006) 7:225–6. doi: 10.1038/ni0306-225
2. Rothstein TL, Griffin DO, Holodick NE, Quach TD, Kaku H. Human B-1 cells take the stage. Ann N Y Acad Sci (2013) 1285:97–114. doi:10.1111/nyas.12137
3. Hardy RR, Hayakawa K. B cell development pathways. Annu Rev Immunol (2001) 19:595–621. doi:10.1146/annurev.immunol.19.1.595
4. Rothstein TL. Cutting edge commentary: two B-1 or not to be one. J Immunol (2002) 168:4257–61. doi:10.4049/jimmunol.168.9.4257
5. Wong SC, Chew WK, Tan JE, Melendez AJ, Francis F, Lam KP. Peritoneal CD5+ B-1 cells have signaling properties similar to tolerant B cells. J Biol Chem (2002) 277:30707–15. doi:10.1074/jbc.M202460200
6. Holodick NE, Tumang JR, Rothstein TL. Continual signaling is responsible for constitutive ERK phosphorylation in B-1a cells. Mol Immunol (2009) 46:3029–36. doi:10.1016/j.molimm.2009.06.011
7. Zhong X, Gao W, Degauque N, Bai C, Lu Y, Kenny J, et al. Reciprocal generation of Th1/Th17 and T(reg) cells by B1 and B2 B cells. Eur J Immunol (2007) 37:2400–4. doi:10.1002/eji.200737296
8. Baumgarth N. The double life of a B-1 cell: self-reactivity selects for protective effector functions. Nat Rev Immunol (2011) 11:34–46. doi:10.1038/nri2901
9. Ehrenstein MR, Notley CA. The importance of natural IgM: scavenger, protector and regulator. Nat Rev Immunol (2010) 10:778–86. doi:10.1038/nri2849
10. Feeney AJ. Lack of N regions in fetal and neonatal mouse immunoglobulin V-D-J junctional sequences. J Exp Med (1990) 172:1377–90. doi:10.1084/jem.172.5.1377
11. Pennell CA, Arnold LW, Haughton G, Clarke SH. Restricted Ig variable region gene expression among Ly-1+ B cell lymphomas. J Immunol (1988) 141:2788–96.
12. Hardy RR, Carmack CE, Shinton SA, Riblet RJ, Hayakawa K. A single VH gene is utilized predominantly in anti-BrMRBC hybridomas derived from purified Ly-1 B cells. Definition of the VH11 family. J Immunol (1989) 142:3643–51.
13. Wang H, Clarke SH. Positive selection focuses the VH12 B-cell repertoire towards a single B1 specificity with survival function. Immunol Rev (2004) 197:51–9. doi:10.1111/j.0105-2896.2004.0098.x
14. Su SD, Ward MM, Apicella MA, Ward RE. The primary B cell response to the O/core region of bacterial lipopolysaccharide is restricted to the Ly-1 lineage. J Immunol (1991) 146:327–31.
15. Ochsenbein AF, Fehr T, Lutz C, Suter M, Brombacher F, Hengartner H, et al. Control of early viral and bacterial distribution and disease by natural antibodies. Science (1999) 286:2156–9. doi:10.1126/science.286.5447.2156
16. Baumgarth N, Herman OC, Jager GC, Brown LE, Herzenberg LA, Chen J. B-1 and B-2 cell-derived immunoglobulin M antibodies are nonredundant components of the protective response to influenza virus infection. J Exp Med (2000) 192:271–80. doi:10.1084/jem.192.2.271
17. Haas KM, Poe JC, Steeber DA, Tedder TF. B-1a and B-1b cells exhibit distinct developmental requirements and have unique functional roles in innate and adaptive immunity to S. pneumoniae. Immunity (2005) 23:7–18. doi:10.1016/j.immuni.2005.04.011
18. Gronwall C, Vas J, Silverman GJ. Protective roles of natural IgM antibodies. Front Immunol (2012) 3:66. doi:10.3389/fimmu.2012.00066
19. Mercolino TJ, Arnold LW, Hawkins LA, Haughton G. Normal mouse peritoneum contains a large population of Ly-1+ (CD5) B cells that recognize phosphatidyl choline. Relationship to cells that secrete hemolytic antibody specific for autologous erythrocytes. J Exp Med (1988) 168:687–98. doi:10.1084/jem.168.2.687
20. Baumgarth N, Tung JW, Herzenberg LA. Inherent specificities in natural antibodies: a key to immune defense against pathogen invasion. Springer Semin Immunopathol (2005) 26:347–62. doi:10.1007/s00281-004-0182-2
21. Xu Z, Butfiloski EJ, Sobel ES, Morel L. Mechanisms of peritoneal B-1a cells accumulation induced by murine lupus susceptibility locus Sle2. J Immunol (2004) 173:6050–8. doi:10.4049/jimmunol.173.10.6050
22. Mohan C, Morel L, Yang P, Wakeland EK. Accumulation of splenic B1a cells with potent antigen-presenting capability in NZM2410 lupus-prone mice. Arthritis Rheum (1998) 41:1652–62. doi:10.1002/1529-0131(199809)41:9<1652::AID-ART17>3.0.CO;2-W
23. Xu Z, Morel L. Contribution of B-1a cells to systemic lupus erythematosus in the NZM2410 mouse model. Ann N Y Acad Sci (2015) 1362:215–23. doi:10.1111/nyas.12607
24. Iizuka J, Katagiri Y, Tada N, Murakami M, Ikeda T, Sato M, et al. Introduction of an osteopontin gene confers the increase in B1 cell population and the production of anti-DNA autoantibodies. Lab Invest (1998) 78:1523–33.
25. Reap EA, Sobel ES, Cohen PL, Eisenberg RA. Conventional B cells, not B-1 cells, are responsible for producing autoantibodies in lpr mice. J Exp Med (1993) 177:69–78. doi:10.1084/jem.177.1.69
26. Atencio S, Amano H, Izui S, Kotzin BL. Separation of the New Zealand Black genetic contribution to lupus from New Zealand Black determined expansions of marginal zone B and B1a cells. J Immunol (2004) 172:4159–66. doi:10.4049/jimmunol.172.7.4159
27. Xu Z, Potula HH, Vallurupalli A, Perry D, Baker H, Croker BP, et al. Cyclin-dependent kinase inhibitor Cdkn2c regulates B cell homeostasis and function in the NZM2410-derived murine lupus susceptibility locus Sle2c1. J Immunol (2011) 186:6673–82. doi:10.4049/jimmunol.1002544
28. Potula HH, Xu Z, Zeumer L, Sang A, Croker BP, Morel L. Cyclin-dependent kinase inhibitor Cdkn2c deficiency promotes B1a cell expansion and autoimmunity in a mouse model of lupus. J Immunol (2012) 189:2931–40. doi:10.4049/jimmunol.1200556
29. Morel L, Croker BP, Blenman KR, Mohan C, Huang G, Gilkeson G, et al. Genetic reconstitution of systemic lupus erythematosus immunopathology with polycongenic murine strains. Proc Natl Acad Sci U S A (2000) 97:6670–5. doi:10.1073/pnas.97.12.6670
30. Kantor AB, Merrill CE, Herzenberg LA, Hillson JL. An unbiased analysis of V(H)-D-J(H) sequences from B-1a, B-1b, and conventional B cells. J Immunol (1997) 158:1175–86.
31. Gregoire KE, Goldschneider I, Barton RW, Bollum FJ. Ontogeny of terminal deoxynucleotidyl transferase-positive cells in lymphohemopoietic tissues of rat and mouse. J Immunol (1979) 123:1347–52.
32. Zemlin M, Ippolito GC, Zemlin C, Link J, Monestier M, Schroeder HW Jr. Adult lupus-prone MRL/MpJ2+ mice express a primary antibody repertoire that differs in CDR-H3 length distribution and hydrophobicity from that expressed in the C3H parental strain. Mol Immunol (2005) 42:789–98. doi:10.1016/j.molimm.2004.07.049
33. Radic MZ, Mackle J, Erikson J, Mol C, Anderson WF, Weigert M. Residues that mediate DNA binding of autoimmune antibodies. J Immunol (1993) 150:4966–77.
34. Kirkham PM, Schroeder HW Jr. Antibody structure and the evolution of immunoglobulin V gene segments. Semin Immunol (1994) 6:347–60. doi:10.1006/smim.1994.1045
35. Gu H, Kitamura D, Rajewsky K. B cell development regulated by gene rearrangement: arrest of maturation by membrane-bound D mu protein and selection of DH element reading frames. Cell (1991) 65:47–54. doi:10.1016/0092-8674(91)90406-O
36. Forster I, Rajewsky K. Expansion and functional activity of Ly-1+ B cells upon transfer of peritoneal cells into allotype-congenic, newborn mice. Eur J Immunol (1987) 17:521–8. doi:10.1002/eji.1830170414
37. Hayakawa K, Carmack CE, Hyman R, Hardy RR. Natural autoantibodies to thymocytes: origin, VH genes, fine specificities, and the role of Thy-1 glycoprotein. J Exp Med (1990) 172:869–78. doi:10.1084/jem.172.3.869
38. Yang Y, Wang C, Yang Q, Kantor AB, Chu H, Ghosn EE, et al. Distinct mechanisms define murine B cell lineage immunoglobulin heavy chain (IgH) repertoires. Elife (2015) 4:e09083. doi:10.7554/eLife.09083
39. Forster I, Gu H, Rajewsky K. Germline antibody V regions as determinants of clonal persistence and malignant growth in the B cell compartment. EMBO J (1988) 7:3693–703.
40. Tarlinton D, Stall AM, Herzenberg LA. Repetitive usage of immunoglobulin VH and D gene segments in CD5+ Ly-1 B clones of (NZB x NZW)F1 mice. EMBO J (1988) 7:3705–10.
41. Tornberg UC, Holmberg D. B-1a, B-1b and B-2 B cells display unique VHDJH repertoires formed at different stages of ontogeny and under different selection pressures. EMBO J (1995) 14:1680–9.
42. Stall AM, Farinas MC, Tarlinton DM, Lalor PA, Herzenberg LA, Strober S, et al. Ly-1 B-cell clones similar to human chronic lymphocytic leukemias routinely develop in older normal mice and young autoimmune (New Zealand Black-related) animals. Proc Natl Acad Sci U S A (1988) 85:7312–6. doi:10.1073/pnas.85.19.7312
43. Gu H, Forster I, Rajewsky K. Sequence homologies, N sequence insertion and JH gene utilization in VHDJH joining: implications for the joining mechanism and the ontogenetic timing of Ly1 B cell and B-CLL progenitor generation. EMBO J (1990) 9:2133–40.
44. Wasserman R, Li YS, Shinton SA, Carmack CE, Manser T, Wiest DL, et al. A novel mechanism for B cell repertoire maturation based on response by B cell precursors to pre-B receptor assembly. J Exp Med (1998) 187:259–64. doi:10.1084/jem.187.2.259
45. Hardy RR. Development of VH11+ B cells: a model for selection of B cells producing natural autoantibodies. Curr Dir Autoimmun (2003) 6:196–211. doi:10.1159/000066862
46. Nguyen TT, Elsner RA, Baumgarth N. Natural IgM prevents autoimmunity by enforcing B cell central tolerance induction. J Immunol (2015) 194:1489–502. doi:10.4049/jimmunol.1401880
47. Zhong X, Tumang JR, Gao W, Bai C, Rothstein TL. PD-L2 expression extends beyond dendritic cells/macrophages to B1 cells enriched for V(H)11/V(H)12 and phosphatidylcholine binding. Eur J Immunol (2007) 37:2405–10. doi:10.1002/eji.200737461
48. Zhong X, Lau S, Bai C, Degauque N, Holodick NE, Steven SJ, et al. A novel subpopulation of B-1 cells is enriched with autoreactivity in normal and lupus-prone mice. Arthritis Rheum (2009) 60:3734–43. doi:10.1002/art.25015
49. O’Garra A, Chang R, Go N, Hastings R, Haughton G, Howard M. Ly-1 B (B-1) cells are the main source of B cell-derived interleukin 10. Eur J Immunol (1992) 22:711–7. doi:10.1002/eji.1830220314
50. Kaku H, Cheng KF, Al-Abed Y, Rothstein TL. A novel mechanism of B cell-mediated immune suppression through CD73 expression and adenosine production. J Immunol (2014) 193:5904–13. doi:10.4049/jimmunol.1400336
51. Wang H, Shin DM, Abbasi S, Jain S, Kovalchuk AL, Beaty N, et al. Expression of plasma cell alloantigen 1 defines layered development of B-1a B-cell subsets with distinct innate-like functions. Proc Natl Acad Sci U S A (2012) 109:20077–82. doi:10.1073/pnas.1212428109
52. Enghard P, Humrich JY, Chu VT, Grussie E, Hiepe F, Burmester GR, et al. Class switching and consecutive loss of dsDNA-reactive B1a B cells from the peritoneal cavity during murine lupus development. Eur J Immunol (2010) 40:1809–18. doi:10.1002/eji.200940050
53. Boes M, Prodeus AP, Schmidt T, Carroll MC, Chen J. A critical role of natural immunoglobulin M in immediate defense against systemic bacterial infection. J Exp Med (1998) 188:2381–6. doi:10.1084/jem.188.12.2381
Keywords: B cells, B-1 cells, autoimmunity, lupus erythematosus, systemic, repertoire analysis, mouse model, natural antibodies
Citation: Holodick NE, Zeumer L, Rothstein TL and Morel L (2016) Expansion of B-1a Cells with Germline Heavy Chain Sequence in Lupus Mice. Front. Immunol. 7:108. doi: 10.3389/fimmu.2016.00108
Received: 29 January 2016; Accepted: 10 March 2016;
Published: 24 March 2016
Edited by:
George C. Tsokos, Harvard Medical School, USAReviewed by:
Subbarao Bondada, University of Kentucky, USAYoshiteru Sasaki, Kyoto University, Japan
Copyright: © 2016 Holodick, Zeumer, Rothstein and Morel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nichol E. Holodick, bmhvbG9kaWNrQGljbG91bGQuY29t;
Laurence Morel, bW9yZWxAdWZsLmVkdQ==
 Nichol E. Holodick
Nichol E. Holodick Leilani Zeumer2
Leilani Zeumer2 Thomas L. Rothstein
Thomas L. Rothstein Laurence Morel
Laurence Morel