- Department of Medical Specialties, Division of Hematology, Geneva University Hospitals, University of Geneva, Geneva, Switzerland
Recent reports give insights into the role of the T-box transcription factors, T-bet and Eomesodermin (Eomes), in NK cell biology. In this mini-review, we recapitulate the initial reports that delineate T-bet and Eomes as master regulators of NK cell development, maturation, and function. We discuss how T-bet and Eomes expression is regulated during NK cell development and peripheral maturation. Furthermore, we summarize the current literature on the role of T-bet and Eomes in the transcriptional regulation of NK cell function and review possible effects of T-box transcription factor anomalies during aging, infection, cancer, and after hematopoietic stem cell transplantation. We discuss how the current data argue in favor of a model of T-bet and Eomes synergy in transcriptional regulation of NK cell function and identify T-box transcription factors as potential targets for therapeutic interventions.
Introduction
The phylogenetically conserved family of T-box transcription factors, which share T-box DNA-binding domains, is critically involved in developmental processes in vertebrates. The T-box protein in T cells (T-bet) is a tyrosine- and serine-phosphorylated protein encoded by the Tbx21 gene that is expressed only in cells of hematopoietic origin. T-bet was originally identified in T lymphocytes as the key transcription factor involved in interferon-gamma (IFN-γ) production that commits CD4 T cells to the Th1 lineage (1). Eomesodermin (Eomes), another T-box transcription factor sharing homology with T-bet, was originally described as a key player in vertebrate embryogenesis (2). More recently, Eomes and T-bet have been reported to coordinate the differentiation of CD8 T cells into effector cells (3–5) as well as their transition to the memory cell pool (6, 7). T-bet and Eomes are therefore considered as master regulators of T cell function. The bulk of mature murine (6, 8, 9) and human (10–12) NK cells express high levels of T-bet and Eomes, but until recently, their impact on NK cell function was not known. In the present work, we summarize the current knowledge about the role of T-bet and Eomes in NK cell development, peripheral maturation, and function.
T-bet and Eomes in NK Cell Development
The first evidence for a role of T-bet in NK cell biology came from the observation that T-bet deficient (T-bet−/−) mice have slightly higher NK cell numbers in the bone marrow but reduced numbers of NK cells in spleen, liver, and peripheral blood (13). Because many NK cells in T-bet−/− mice expressed an immature CD27posCD11bpos phenotype, it was suggested that T-bet played a role in NK cell maturation without being essential for the early stages of NK cell development. Because Eomes−/− mice die in an early embryonic stage, the role of Eomes in NK cell development has initially been assessed only in compound mutant Eomes+/− Tbx21−/− mice (6). Interestingly, Eomes+/− Tbx21−/− mice displayed a severely exacerbated defect in the NK cell compartment compared to mice only lacking T-bet, suggesting a distinct, but complimentary function for Eomes in NK cell development. Importantly, the loss of one allele of Eomes results in a severe downregulation of CD122 (6), the beta-chain of IL-2R and IL-15R, which is essential for IL-15 signaling and NK cell development. Chromatin Immunoprecipitation (ChIP) assays showed that Eomes regulated CD122 transcription (6) for which T-bet appeared to be unnecessary (13). More recently, the role of Eomes has been studied in mice harboring floxed alleles of Eomes and expressing hematopoietic-restricted Cre recombinase under control of Vav regulatory elements (Eomesflox/floxVav–Cre+ mice), which restricts the Eomes-inactivation to cells of the hematopoietic lineage (8). Deletion of Eomes resulted in a severe reduction of NK cells in spleen and blood whereas only a modest reduction in NK cell numbers was observed in liver, lymph node, and bone marrow. Deletion of Eomes and T-bet in Tbx21−/−Eomesflox/floxVav–Cre+ mice resulted in complete absence of NK cells in all organs (8). Hence, T-bet and Eomes are essential for normal NK cell development, but in the absence of either T-bet or Eomes, an incomplete development may still occur suggesting that the two T-box transcription factors share several functions.
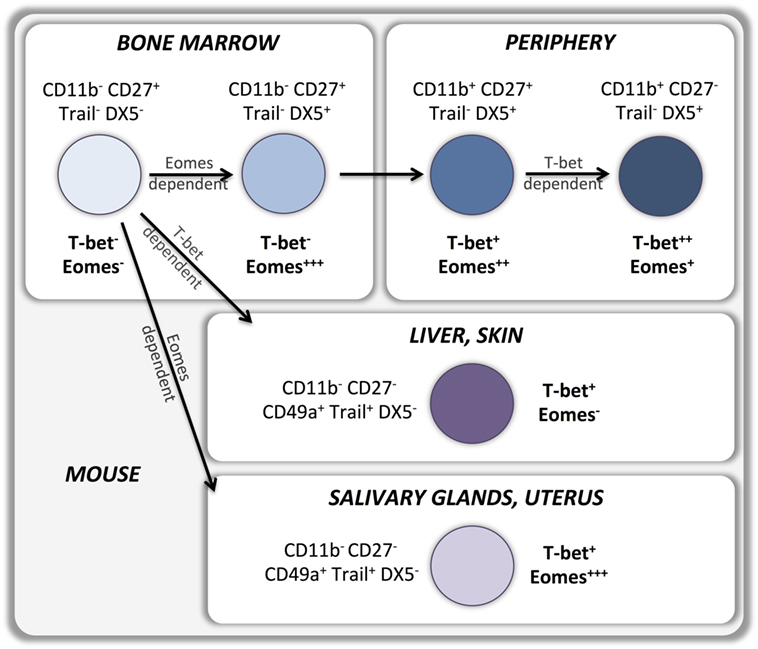
The analysis of the contribution of Eomes and T-bet to NK cell development also led to the identification of an ontologically distinct subset of innate lymphocyte (ILC) cells residing in the liver. Lymphocytes expressing NK cell markers in murine liver contain up to 40% of Eomes-negative cells that express high levels of T-bet (8, 9, 14). Hepatic EomesnegT-bethigh NK cells display an immature phenotype characterized by the expression of Trail and lack of expression of DX5 (TrailposDX5neg) (8). Initial experiments suggested that EomesnegT-bethighTrailposDX5neg cells represented an intermediate developmental stage that could differentiate into mature EomesposTrailnegDX5pos cells (8). However, experiments performed with Eomes-negative cells isolated from Eomes-GFP reporter mice demonstrated that EomesnegT-bethighTrailposDX5neg cells are in fact an ontologically and functionally different subset of ILCs differentiating in the liver (14).
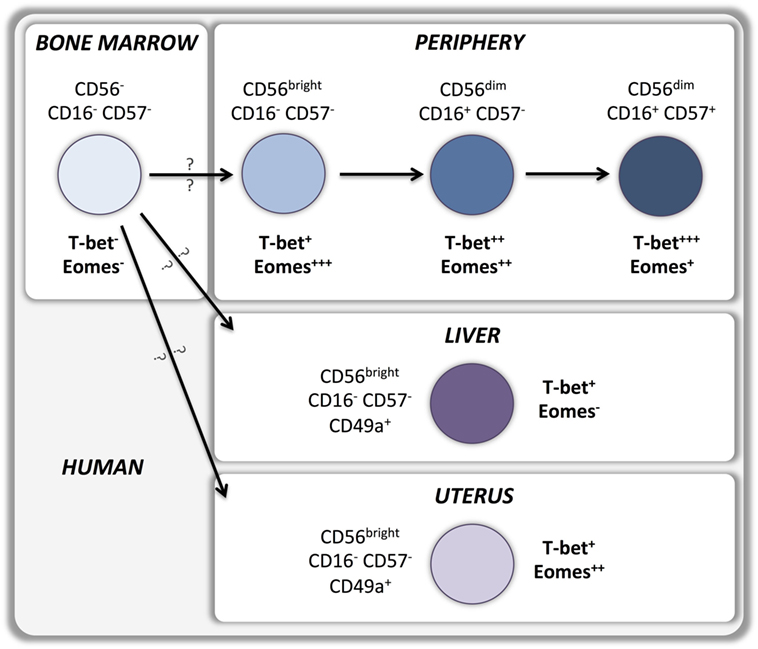
According to the current model, type 1 ILCs differentiate into two developmentally distinct lineages, type 1 helper innate lymphoid cells (hILC1s) and conventional NK cells (cNKs), which can be discriminated by the T-box transcription factors expressed (Figure 1). hILC1 differentiate in the liver when T-bet is upregulated and Eomes transcription is suppressed (14). Conversely, Eomes expression directs ILC1 development toward bone marrow-derived conventional NK cells that express relatively low levels of T-bet (15). Whether similar developmental pathways exist in human NK cells is still unknown (Figure 2). A recent study identified a T-betposEomesneg CD49apos NK cell subset in human liver, absent from hepatic venous or peripheral blood, with a CD56brightCD16negCD57negperforinneg phenotype that may represent the human equivalent of murine T-betpos intrahepatic hILC1 (16).
In recent years, other ontologically distinct tissue residing NK cell subsets have been identified in salivary glands, skin, and uterus. Similarly to liver residing cells, NK cells isolated from these tissues display an immature CD49aposDX5neg phenotype associated with the expression of markers of tissue residency. However T-box transcription factors expression varies with the tissues the NK cells reside in, which may point at distinct developmental pathways. Skin residing NK cells do not express Eomes and strictly depend on T-bet for their development (17), suggesting a developmental relationship with liver residing NK cells. Conversely, salivary gland (18) and uterine (17, 19–21) NK cells express high levels of Eomes and are T-bet-independent for their development and appear therefore to be a more distinct NK cell lineage (Figure 1).
Little is known on the mechanisms that induce cells to upregulate or repress T-box transcription factors in different organs. By contrast, several cell intrinsic mechanisms regulating Eomes and T-bet expression have been elucidated to date. First, T-bet and Eomes regulate each other’s expression during NK cell development, and levels of Eomes expression correlate inversely with levels of T-bet in developing NK cells suggesting that active repressive mechanisms regulate the balance of T-bet/Eomes expression (14). This hypothesis has been confirmed by showing that T-bet−/− NK cells express high levels of Eomes whereas transgenic NK cells overexpressing T-bet display low levels of Eomes (14). In addition, expression of T-bet and Eomes has been shown to be strictly related with expression of other transcription factors crucial for NK cell development. T-bet expression is induced by the transcription factors ETS1 (V-Ets Avian Erythroblastosis Virus E26 Oncogene Homolog 1) (22), TOX1 (thymocyte selection-associated HMG box protein TOX-1) (23), and TOX2 (24). Conversely, Eomes expression depends on the bZIP transcription factor Nfil3 (Nuclear factor, interleukin 3 regulated, also known as E4BP4), which binds to the regulatory regions of the Eomes gene to promote its transcription (25). Nfil3 deficiency interferes with the development of the EomesposTRAILnegDX5pos bone marrow-derived NK cells while hepatic TbetposEomesnegTRAILposDX5neg cells are unaffected (17, 26, 27). Interestingly, tissue-resident Eomespos NK cells localized in salivary glands and in the uterus can develop in the absence of Nfil3 (17, 18, 20, 27), suggesting the existence of alternative molecular mechanisms for the induction of Eomes transcription.
The only direct available information about the impact of T-box transcription factors deficiency on human NK cell development comes from a study describing a patient with a rare autosomal recessive microcephaly syndrome related to a translocation between chromosomes 3p and 10q leading to the silencing of the Eomes transcript (28). The fact that the infant displayed a normal distribution of T, B, and NK cells suggests that human NK cell development is possible in the absence of Eomes, although no information is available about the patient’s NK cell functionality.
T-bet and Eomes in NK Cell Peripheral Maturation
Upregulation of T-bet and Eomes expression during development is maintained by most peripheral NK cells in mice (6) as well as in humans (10). Indeed, sustained expression of T-bet and Eomes in the periphery is necessary to maintain the NK cell maturation status while deletion of both T-box transcription factors results in reversion into an immature phenotype (8). T-bet expression is upregulated, and Eomes expression is downregulated during maturation of CD11bposCD27pos murine NK cells to the CD11bposCD27neg stage (14) (Figure 1). Importantly, T-bet appears to be essential for completion of this final maturation step as it controls the repression of CD27 and c-kit expression as well as the upregulation of S1P5 and KLRG1 (8, 13, 29–31). Therefore, bone marrow-derived EomesposTRAILnegDX5pos NK cells can develop in the absence of T-bet, but are unable to undergo the terminal stages of maturation. Part of this terminal maturation process seems to be mediated by T-bet induction of the PR domain zinc finger protein 1 (Blimp-1) (30) and the zinc finger E-box-binding homeobox 2 (ZEB2) (32) transcription factors. Conversely, Forkhead box protein O1 (FOXO1) inhibits NK terminal maturation through repression of T-bet (33). Similar patterns of T-bet and Eomes expression exist in human NK cells that also upregulate T-bet and downregulate Eomes during peripheral maturation (Figure 2). Cytokine-producing CD56bright NK cells express higher levels of Eomes and lower levels of T-bet than cytotoxic CD56dim NK cells (10–12, 34, 35). Moreover, terminally differentiated CD57posCD56dim NK cells express the highest levels of T-bet and the lowest levels of Eomes (11, 12). Accordingly, upregulation of Killer-cell immunoglobulin-like receptors (KIRs) during maturation is associated with a decrease of Eomes and an increase of T-bet levels (11), which appears to be independent of the fact whether KIRs are licensing or not (Pradier et al., unpublished observations submitted for the present Frontiers Immunology Research Topic).
T-bet and Eomes in NK Cell Function
Chromatin Immunoprecipitation assays combined with the analysis of T-bet- and Eomes-deficient mice have partially uncovered the role of T-box transcription factors in NK cell biology. Similarly to what previously reported in CD4+ Th1 cells, ChIP experiments identified IFN-γ as a target gene of T-bet in NK cells (13). By contrast, no evidence of Eomes binding to the IFN-γ promoter has been reported. Conditions that induce IFN-γ production, such as stimulation with IL-12 plus IL-15 or with IL-12 plus IL-18, also induce upregulation of T-bet (8, 13) and Eomes (12). Murine NK cells are still able to produce IFN-γ in vivo in the absence of T-bet, Eomes, or both (8, 13), but the maintenance of IFN-γ production is impaired in the absence of T-bet (13). In addition, T-bet and Eomes expression correlates positively with IFN-γ production in vitro in mice (36) as well as in humans (12, 37, 38). Furthermore, NK cells are less cytotoxic in the absence of T-bet (13, 39), which is possibly caused by a decreased production of perforin and granzyme B (8, 13). Murine studies suggest that T-bet but not Eomes is directly involved in the production of cytotoxic molecules (8). Accordingly, we found a positive correlation between T-bet levels and perforin production in human NK cells and no relationship between Eomes levels and expression of cytotoxic molecules (12). Collectively, these findings support a model in which T-bet and Eomes cooperatively regulate IFN-γ production in NK cells while T-bet seems to be the crucial regulator of their cytotoxic activity.
T-bet and Eomes in NK Cell Biology in Health and Disease
Given their impact in NK cell development, peripheral maturation and function, alterations in T-bet and Eomes expression could account for NK cell abnormalities in pathological conditions in which NK cells exert an essential role, such as infections and cancer. Reduction in T-bet and Eomes levels in NK cells occurs during aging and is associated with an impaired NK cell cytotoxicity (40). Interestingly, T-bet and Eomes downregulation in aged mice is not related to a cell intrinsic defect but is induced by the aged environment pointing to a cell extrinsic induction of a senescent phenotype.
The role of T-bet and Eomes expression in NK cells has been investigated in several disease models. NK cells activated during murine cytomegalovirus or vaccinia virus infection do not undergo terminal maturation in the absence of T-bet (13, 41, 42), which concords with the typical role of T-bet in NK cell differentiation (13). However, despite the fact that this led to a significant reduction of NK cell virus-specific cytotoxicity early after infection (13, 42), the viral load remained unchanged suggesting that the NK cell activity in T-bet−/− mice is still sufficient to control viral replication. Similar sufficient in vivo NK cell responses have been reported after infection of T-bet−/− mice with Listeria monocytogenes (43) or with Toxoplasma Gondii (44).
Murine models of cancer have illustrated the impact of T-box transcription factors in NK cell antitumor responses. Peng and coworkers used a transgenic prostate adenocarcinoma mouse model to demonstrate that although T-bet deficiency only had a very limited impact on primary tumor development, it significantly affected the ability to control tumor spread (45). This observation, subsequently confirmed in a murine model of metastatic cancer (46), led to the conclusion that in vivo NK activity in metastasized cancer strongly depends on T-bet expression. Although NK cells are still capable to infiltrate metastatic tumors in the absence of T-bet, their survival and capacity to terminally differentiate into fully competent, cytotoxic CD27negKLRG1pos NK cells appears to be diminished (31, 46). It has been show that in vivo IL-15 administration overcomes the defect of T-bet−/− NK cells by inducing differentiation of EomeshighKLRG1pos NK cells that are able to efficiently control metastatic pulmonary colorectal cancer, suggesting that IL-15 induced Eomes upregulation may compensate for the lack of T-bet inducing expansion of phenotypically and functionally mature NK cells.
Important insights into the relationship between T-bet and Eomes expression in NK cells and cancer come from the work of Gill and coworkers who identified the downregulation of T-bet and Eomes as the molecular signature of NK cell exhaustion in a murine NK adoptive transfer model of lymphoma (36). Importantly, downregulation of T-box transcription factors appeared to be not only the consequence of the NK cells’ exposure to tumor cells but also of their homeostatic proliferation induced by the treatment-induced lymphopenic environment. Lymphopenia occurs frequently after cancer chemotherapy as well as after conditioning regimens for hematopoietic stem cell transplantation (HSCT). Indeed, we found the same exhausted phenotype in human NK cells isolated from patients undergoing HSCT (12). Similar to what had been observed in mice, T-bet and Eomes downregulation after HSCT was associated with impaired NK function and lower levels of T-bet in NK cells were associated with reduced patient overall survival (12). Surprisingly, improved survival associated with higher levels of T-bet in NK cells was not the consequence of improved cancer control but the result of a reduced non-relapse mortality, which suggests that sustained T-bet and Eomes expression in NK cell could participate to prevent the development of transplant related complications after HSCT. This hypothesis is supported by a recent study showing that adoptively transferred IL-12/15/18-preactivated NK cells, which do not undergo exhaustion and maintain high levels of Eomes and T-bet expression, suppressed acute Graft-versus-Host-Disease in a murine model of HSCT (47). These results suggest that T-bet and Eomes expression may also modulate NK cell function in immunopathological settings, similarly to what recently shown in multiple sclerosis patients (35).
Concluding Remarks
Recent findings clarifying the role of the two T-box transcription factors T-bet and Eomes in NK cells have considerably increased our knowledge of NK cell biology. Notably, they led to the characterization of previously unknown NK cells developmental pathways. Furthermore, they led to the identification of a molecular signature of NK cell exhaustion, which may represent a future target for immunomodulatory therapies.
Author Contributions
FS wrote the manuscript and designed the figures. AP and ER critically discussed the work and edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
FS is supported by grants from the Geneva University Hospitals’ Clinical Research Center and the Dubois-Ferrière-Dinu-Lipatti Foundation. ER is supported by grants from the Swiss National Science Foundation and the Swiss Cancer Research.
References
1. Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell (2000) 100(6):655–69. doi:10.1016/S0092-8674(00)80702-3
2. Ryan K, Garrett N, Mitchell A, Gurdon J. Eomesodermin, a key early gene in Xenopus mesoderm differentiation. Cell (1996) 87(6):989–1000. doi:10.1016/S0092-8674(00)81794-8
3. Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science (2003) 302(5647):1041–3. doi:10.1126/science.1090148
4. Intlekofer AM, Banerjee A, Takemoto N, Gordon SM, DeJong CS, Shin H, et al. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science (2008) 321(5887):408–11. doi:10.1126/science.1159806
5. Cruz-Guilloty F, Pipkin ME, Djuretic IM, Levanon D, Lotem J, Lichtenheld MG, et al. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J Exp Med (2009) 206(1):51–9. doi:10.1084/jem.20081242
6. Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol (2005) 6(12):1236–44. doi:10.1038/ni1268
7. Banerjee A, Gordon SM, Intlekofer AM, Paley MA, Mooney EC, Lindsten T, et al. Cutting edge: the transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J Immunol (2010) 185(9):4988–92. doi:10.4049/jimmunol.1002042
8. Gordon SM, Chaix J, Rupp LJ, Wu J, Madera S, Sun JC, et al. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity (2012) 36(1):55–67. doi:10.1016/j.immuni.2011.11.016
9. Paley MA, Gordon SM, Bikoff EK, Robertson EJ, Wherry EJ, Reiner SL. Technical advance: fluorescent reporter reveals insights into eomesodermin biology in cytotoxic lymphocytes. J Leukoc Biol (2013) 93(2):307–15. doi:10.1189/jlb.0812400
10. Knox JJ, Cosma GL, Betts MR, McLane LM. Characterization of T-bet and eomes in peripheral human immune cells. Front Immunol (2014) 5:217. doi:10.3389/fimmu.2014.00217
11. Luetke-Eversloh M, Cicek BB, Siracusa F, Thom JT, Hamann A, Frischbutter S, et al. NK cells gain higher IFN-gamma competence during terminal differentiation. Eur J Immunol (2014) 44(7):2074–84. doi:10.1002/eji.201344072
12. Simonetta F, Pradier A, Bosshard C, Masouridi-Levrat S, Chalandon Y, Roosnek ENK. Cell functional impairment after allogeneic hematopoietic stem cell transplantation is associated with reduced levels of T-bet and eomesodermin. J Immunol (2015) 195(10):4712–20. doi:10.4049/jimmunol.1501522
13. Townsend MJ, Weinmann AS, Matsuda JL, Salomon R, Farnham PJ, Biron CA, et al. T-bet regulates the terminal maturation and homeostasis of NK and Vα14i NKT cells. Immunity (2004) 20(4):477–94. doi:10.1016/S1074-7613(04)00076-7
14. Daussy C, Faure F, Mayol K, Viel S, Gasteiger G, Charrier E, et al. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J Exp Med (2014) 211(3):563–77. doi:10.1084/jem.20131560
15. Pikovskaya O, Chaix J, Rothman NJ, Collins A, Chen Y-H, Scipioni AM, et al. Cutting edge: eomesodermin is sufficient to direct type 1 innate lymphocyte development into the conventional NK lineage. J Immunol (2016):1502396. doi:10.4049/jimmunol.1502396
16. Marquardt N, Beziat V, Nystrom S, Hengst J, Ivarsson MA, Kekalainen E, et al. Cutting edge: identification and characterization of human intrahepatic CD49a+ NK cells. J Immunol (2015) 194(6):2467–71. doi:10.4049/jimmunol.1402756
17. Sojka DK, Plougastel-Douglas B, Yang L, Pak-Wittel MA, Artyomov MN, Ivanova Y, et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. Elife (2014) 3:e01659. doi:10.7554/eLife.01659
18. Cortez VS, Fuchs A, Cella M, Gilfillan S, Colonna M. Cutting edge: salivary gland NK cells develop independently of Nfil3 in steady-state. J Immunol (2014) 192(10):4487–91. doi:10.4049/jimmunol.1303469
19. Tayade C, Fang Y, Black GP, Paffaro VA Jr, Erlebacher A, Croy BA. Differential transcription of Eomes and T-bet during maturation of mouse uterine natural killer cells. J Leukoc Biol (2005) 78(6):1347–55. doi:10.1189/jlb.0305142
20. Boulenouar S, Doisne J-M, Sferruzzi-Perri A, Gaynor LM, Kieckbusch J, Balmas E, et al. The residual innate lymphoid cells in nFil3-deficient mice support suboptimal maternal adaptations to pregnancy. Front Immunol (2016) 7:43. doi:10.3389/fimmu.2016.00043
21. Montaldo E, Vacca P, Chiossone L, Croxatto D, Loiacono F, Martini S, et al. Unique Eomes+ innate lymphoid cell subsets are present in uterus and decidua during early pregnancy. Front Immunol (2016) 6:646. doi:10.3389/fimmu.2015.00646
22. Ramirez K, Chandler KJ, Spaulding C, Zandi S, Sigvardsson M, Graves BJ, et al. Gene deregulation and chronic activation in natural killer cells deficient in the transcription factor ETS1. Immunity (2012) 36(6):921–32. doi:10.1016/j.immuni.2012.04.006
23. Yun S, Lee SH, Yoon SR, Kim MS, Piao ZH, Myung PK, et al. TOX regulates the differentiation of human natural killer cells from hematopoietic stem cells in vitro. Immunol Lett (2011) 136(1):29–36. doi:10.1016/j.imlet.2010.11.008
24. Vong QP, Leung WH, Houston J, Li Y, Rooney B, Holladay M, et al. TOX2 regulates human natural killer cell development by controlling T-BET expression. Blood (2014) 124(26):3905–13. doi:10.1182/blood-2014-06-582965
25. Male V, Nisoli I, Kostrzewski T, Allan DS, Carlyle JR, Lord GM, et al. The transcription factor E4bp4/Nfil3 controls commitment to the NK lineage and directly regulates Eomes and Id2 expression. J Exp Med (2014) 211(4):635–42. doi:10.1084/jem.20132398
26. Crotta S, Gkioka A, Male V, Duarte JH, Davidson S, Nisoli I, et al. The transcription factor E4BP4 is not required for extramedullary pathways of NK cell development. J Immunol (2014) 192(6):2677–88. doi:10.4049/jimmunol.1302765
27. Seillet C, Huntington ND, Gangatirkar P, Axelsson E, Minnich M, Brady HJ, et al. Differential requirement for Nfil3 during NK cell development. J Immunol (2014) 192(6):2667–76. doi:10.4049/jimmunol.1302605
28. Baala L, Briault S, Etchevers HC, Laumonnier F, Natiq A, Amiel J, et al. Homozygous silencing of T-box transcription factor EOMES leads to microcephaly with polymicrogyria and corpus callosum agenesis. Nat Genet (2007) 39(4):454–6. doi:10.1038/ng1993
29. Jenne CN, Enders A, Rivera R, Watson SR, Bankovich AJ, Pereira JP, et al. T-bet-dependent S1P5 expression in NK cells promotes egress from lymph nodes and bone marrow. J Exp Med (2009) 206(11):2469–81. doi:10.1084/jem.20090525
30. Kallies A, Carotta S, Huntington ND, Bernard NJ, Tarlinton DM, Smyth MJ, et al. A role for Blimp1 in the transcriptional network controlling natural killer cell maturation. Blood (2011) 117(6):1869–79. doi:10.1182/blood-2010-08-303123
31. Malaise M, Rovira J, Renner P, Eggenhofer E, Sabet-Baktach M, Lantow M, et al. KLRG1+ NK cells protect T-bet-deficient mice from pulmonary metastatic colorectal carcinoma. J Immunol (2014) 192(4):1954–61. doi:10.4049/jimmunol.1300876
32. van Helden MJ, Goossens S, Daussy C, Mathieu AL, Faure F, Marcais A, et al. Terminal NK cell maturation is controlled by concerted actions of T-bet and Zeb2 and is essential for melanoma rejection. J Exp Med (2015) 212(12):2015–25. doi:10.1084/jem.20150809
33. Deng Y, Kerdiles Y, Chu J, Yuan S, Wang Y, Chen X, et al. Transcription factor Foxo1 is a negative regulator of natural killer cell maturation and function. Immunity (2015) 42(3):457–70. doi:10.1016/j.immuni.2015.02.006
34. Killig M, Friedrichs B, Meisig J, Gentilini C, Bluthgen N, Loddenkemper C, et al. Tracking in vivo dynamics of NK cells transferred in patients undergoing stem cell transplantation. Eur J Immunol (2014) 44(9):2822–34. doi:10.1002/eji.201444586
35. McKay FC, Gatt PN, Fewings N, Parnell GP, Schibeci SD, Basuki MA, et al. The low EOMES/TBX21 molecular phenotype in multiple sclerosis reflects CD56+ cell dysregulation and is affected by immunomodulatory therapies. Clin Immunol (2016) 163:96–107. doi:10.1016/j.clim.2015.12.015
36. Gill S, Vasey AE, De Souza A, Baker J, Smith AT, Kohrt HE, et al. Rapid development of exhaustion and down-regulation of eomesodermin limit the antitumor activity of adoptively transferred murine natural killer cells. Blood (2012) 119(24):5758–68. doi:10.1182/blood-2012-03-415364
37. Yu J, Wei M, Becknell B, Trotta R, Liu S, Boyd Z, et al. Pro- and antiinflammatory cytokine signaling: reciprocal antagonism regulates interferon-gamma production by human natural killer cells. Immunity (2006) 24(5):575–90. doi:10.1016/j.immuni.2006.03.016
38. Luetke-Eversloh M, Hammer Q, Durek P, Nordstrom K, Gasparoni G, Pink M, et al. Human cytomegalovirus drives epigenetic imprinting of the IFNG locus in NKG2Chi natural killer cells. PLoS Pathog (2014) 10(10):e1004441. doi:10.1371/journal.ppat.1004441
39. Svensson A, Nordstrom I, Sun JB, Eriksson K. Protective Immunity to genital herpes simpex virus type 2 infection is mediated by T-bet. J Immunol (2005) 174(10):6266–73. doi:10.4049/jimmunol.174.10.6266
40. Shehata HM, Hoebe K, Chougnet CA. The aged nonhematopoietic environment impairs natural killer cell maturation and function. Aging Cell (2015) 14(2):191–9. doi:10.1111/acel.12303
41. Robbins SH, Tessmer MS, Van Kaer L, Brossay L. Direct effects of T-bet and MHC class I expression, but not STAT1, on peripheral NK cell maturation. Eur J Immunol (2005) 35(3):757–65. doi:10.1002/eji.200425797
42. Matsui M, Moriya O, Yoshimoto T, Akatsuka T. T-bet is required for protection against vaccinia virus infection. J Virol (2005) 79(20):12798–806. doi:10.1128/JVI.79.20.12798-12806.2005
43. Way SS, Wilson CB. Cutting edge: immunity and IFN-production during Listeria monocytogenes infection in the absence of T-bet. J Immunol (2004) 173(10):5918–22. doi:10.4049/jimmunol.173.10.5918
44. Harms Pritchard G, Hall AO, Christian DA, Wagage S, Fang Q, Muallem G, et al. Diverse roles for T-bet in the effector responses required for resistance to infection. J Immunol (2015) 194(3):1131–40. doi:10.4049/jimmunol.1401617
45. Peng SL, Townsend MJ, Hecht JL, White IA, Glimcher LH. T-bet regulates metastasis rate in a murine model of primary prostate cancer. Cancer Res (2004) 64(2):452–5. doi:10.1158/0008-5472.CAN-03-3401
46. Werneck MBF, Lugo-Villarino G, Hwang ES, Cantor H, Glimcher LH. T-Bet plays a key role in NK-mediated control of melanoma metastatic disease. J Immunol (2008) 180(12):8004–10. doi:10.4049/jimmunol.180.12.8004
Keywords: natural killer cells, T-box transcription factors, T-bet, Eomes
Citation: Simonetta F, Pradier A and Roosnek E (2016) T-bet and Eomesodermin in NK Cell Development, Maturation, and Function. Front. Immunol. 7:241. doi: 10.3389/fimmu.2016.00241
Received: 29 April 2016; Accepted: 07 June 2016;
Published: 20 June 2016
Edited by:
Emanuela Marcenaro, University of Genova, ItalyReviewed by:
Guido Ferlazzo, University of Messina, ItalyElisa Montaldo, San Raffaele Telethon Institute for Gene Therapy (SR-TIGET), Italy
Copyright: © 2016 Simonetta, Pradier and Roosnek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Federico Simonetta, ZmVkZXJpY28uc2ltb25ldHRhQGhjdWdlLmNo
 Federico Simonetta
Federico Simonetta Amandine Pradier
Amandine Pradier Eddy Roosnek
Eddy Roosnek