- Faculty of Dentistry, Department of Oral Biology, University of Oslo, Oslo, Norway
Commensal microbes are currently in the limelight in biomedical research because they play an important role in health and disease. Humans harbor an enormous diversity of commensals in various parts of the body, including the gastrointestinal and respiratory tracts. Advancement in metagenomic and other omic approaches, and development of suitable animal models have provided an unprecedented appreciation into the diversity of commensals, and the intricacies of their intimate communication with the host immune system. Most studies have focused on the host–commensal interaction in the gut, while less is known on this relationship in other sites of the body, such as the respiratory tract. In this article, we review emerging data from human and animal studies on the host responses to respiratory commensals, immune cross-reactivity between commensals and pathogens, and use of commensals as a vaccine delivery system. A better understanding of the delicate interplay between commensals and host may aid in efforts to develop effective vaccines and therapeutics.
Introduction
Microbial commensalism is traditionally defined as a microbe–host relationship in which microbes benefit from the host, but the latter remains largely unaffected. The commensal microbiota has co-evolved with humans for eons and effectively colonizes various body sites, including the intestinal and respiratory tracts. With the advent of new experimental tools, it is becoming clearer that this commensal–host relationship is much more complex and sophisticated than previously thought. Metagenomic and other omic approaches and the use of germ-free and gnotobiotic animals have provided cutting-edge evidence that gut commensals exert a profound impact on the overall health of the body that is crucial for maintaining homeostasis between the host and commensals. These commensals are not only endowed with unique roles in food digestion, immune system development, production of vitamins and hormones, but also in the production of neurotransmitters/neuromodulators, and development of the central nervous system (1–3). Dysbiosis, the disruption of host–commensal homeostasis, can result in a large array of human diseases, such as obesity, diabetes, allergy, and inflammatory bowel disease (1, 2). Of note, some of the commensals with high pathogenic potential are termed as commensal pathogens or pathobionts (4). The contribution of gut commensals in health and disease is widely studied. Limited information is, however, available on the role of commensals colonizing the respiratory tract, which harbors a complex microbial community subjected to continuous exposure from the external environment. Recent studies using humans and mice have begun to shed light on the functional roles played by respiratory commensals in immunity and immunomodulation (5–12). Data are also accumulating on the potential of respiratory commensals in designing novel prophylactic strategies due to their ability to act as a vaccine delivery system and to show immune cross-reactivity with pathogens (7, 13–17).
In this article, we review current literature on the interaction between host and commensal microbes in the respiratory tract. Specifically, we focus on host immunity to respiratory tract commensals and on the use of commensal microbes for prophylactic purposes. A better understanding of the cross-talk between commensals and host holds promise for developing effective and safe prophylactic and therapeutic strategies.
The Respiratory Tract Microbiota
The respiratory tract consists of the upper respiratory tract (URT) and the lower respiratory tract (LRT). The major passages and structures of the URT extend from the nasal and oral cavities to the throat, whereas the LRT includes the trachea and the lung. Although gene sequencing using 16S/18S ribosomal RNA has enabled culture-independent characterization of the URT microbiota, relatively fewer studies have been conducted to study the LRT microbiota in healthy individuals (18). Colonization of the URT by commensals depends upon host and environmental factors, such as mode of child delivery (vaginal vs cesarean), dietary habits (breast milk vs formula), use of antibiotics, and to a lesser extent on host genetics (19, 20). Healthy full-term neonates in their first hours of life carry a large number of microbes in their respiratory tract that are considered to be of maternal origin. During and following the first week of life, predominant microbial genera colonizing the URT include Staphylococcus, Corynebacterium, and Dolosigranulum. This is followed by a later increase in Moraxella and Streptococcus (20, 21). Early colonization by Corynebacterium, Dolosigranulum, and Moraxella are particularly correlated with respiratory health (20). In healthy adult individuals, the nasal cavity and nasopharynx harbor a community of microorganisms represented by Staphylococcus, Propionibacterium, Corynebacterium, Streptococcus, Dolosigranulum, and Moraxella, while the oropharynx is mainly colonized by Streptococcus, Rothia, Veillonella, and Prevotella (21, 22). Commonly found fungal genera in the URT consist of Aspergillus, Penicillium, and Candida, whereas viral genera include Anellovirus and Herpesvirus (23, 24).
The LRT has long been thought to be sterile. The historical reasons for such an assumption were recently reviewed, and several arguments put forward strongly suggest that sterility of the lungs is a highly unlikely scenario (25). The premise upon which this assumption was installed encompasses microbiological assays that are culture-dependent, and that have been biased toward the identification of respiratory pathogens. With the help of next-generation sequencing, Hilty et al. for the first time reported the occurrence of a diverse microbial community in the lungs of healthy individuals (26). This has been followed by various studies using metagenomics that show the existence of a lung microbiota (18). As the composition of the commensal microbiota in the URT and LRT is similar, it has been maintained that the presence of a LRT microbiota merely stems from microaspiration and mucosal dispersion of microbes from the URT. Sample collection from the lung is challenging because pulmonary samples are liable to contamination by the URT microbiota while performing bronchoscopy. Recovery of low quantities of microbial biomass in the samples also poses challenges for microbiological analysis. In healthy adults, cough, mucociliary clearance, and host defenses contribute to keep a low number of microorganisms in the lungs. In respiratory diseases, this situation is changed to one in which conditions for microbial growth are favored, resulting in expansion of the microbiota (1, 25). These findings confirm the existence of the microbiota in the lung, but whether the microbiota is microaspirated or dispersed from the URT remains elusive. However, exclusive presence of certain species in the LRT, such as Tropheryma whipplei, supports the notion that microbes found in the LRT samples are, at least in part, not derived from the URT (27).
The Respiratory Mucosal Immune System
The mucosal respiratory immune system provides a podium for immune reactions due to its proximity with the external environment. The mucosal layer is lined by ciliated pseudostratified columnar epithelium in the nasal cavity, larynx, trachea, and bronchi, whereas simple squamous epithelium is found in pulmonary alveoli. The epithelial layer is interspersed with mucus-producing goblet cells (G), which secrete mucus that prevents the entry of pathogens by entrapping them (28). The presence of microfold (M) cells is well-described in the gut-associated lymphoid tissue and mucosa-associated lymphoid tissue of the gastrointestinal tract where they transport antigens across the epithelial cell layer from the intestinal lumen to the lamina propria (29). In the respiratory epithelium, this is not firmly established. Previous studies have reported that pathogens like Mycobacterium tuberculosis and reovirus can gain access to the body from the murine lung via M cells (30, 31). Furthermore, Id2−/− mice that are deficient in lymphoid tissues reveal a similar frequency of M cells in the nasal epithelia and generate significantly higher antigen-specific antibody responses compared with Id2+/− control mice (32). These results indicate that respiratory M cells represent an alternative gateway for antigen transport and sampling in mice. On the other hand, although the occurrence of isolated lymphoid follicles localized in contact with bronchial epithelium in children indicates the presence of cells in the epithelium that have an M cell-like function, further studies are required to arrive at a definitive conclusion about M cells in humans (33). The epithelial layer confers physical resistance to the invading pathogens and produces antibacterial peptides, such as β-defensins and cathelicidins (34). The lymphoid tissues associated with the mucosal layer include nasopharynx-associated lymphoid tissue and bronchi-associated lymphoid tissue and contain a variety of immune cells, such as T cells, B cells, and dendritic cells (DCs) for induction, regulation, and effector function of mucosal immune responses (35, 36). In the respiratory tract, DCs are present beneath the epithelial layer and capture microbial antigens by projecting their dendrites through intercellular spaces (36, 37). DCs upregulate costimulatory molecules (CD40, CD80, CD83, and CD86), produce multiple cytokines (IL-12, IL-10, IL-23, IL-6, and IL-23), and migrate via afferent lymph vessels to the lung-draining mediastinal lymph nodes to present the captured antigens to naive T cells. T cell subsets, including CD4+ and CD8+ T cells are primed by DCs and transform into effector T cells that exit the lymph nodes via efferent lymph vessels and migrate to effector sites. CD4+ T cells perform their effector function by secreting cytokines such as IFN-γ, IL-10, and IL-17, whereas CD8+ T cells do so via granzyme/perforin and Fas–FasL pathway (37). After encountering their cognate antigens, B cells in the lymph nodes differentiate into plasma cells that produce antibodies like secretory IgA (38, 39).
Host Immunity to Respiratory Commensals
Germ-free and gnotobiotic animals have proven to be critical tools to study the dynamic relationship of the immune system with the microbiota. Although most of the host responses induced by the microbiota have been attributed to microbial residents of the gut, depletion of the microbiota in germ-free animals is not restricted to the gut, but extends to all organs. Thus, attempts have been made to deplete the microbiota at specific sites to better understand the contribution of the specific microbiota in immunity and pathology. Unfortunately, efficient animal models to specifically study the respiratory tract microbiota are currently not available. The majority of the data on host–respiratory commensal interactions stems from in vitro studies involving bacterial commensals and immune cells isolated from humans and mice. Recent studies provide evidence that bacterial commensals have an impact on modulation of inflammatory responses and suppression/killing of pathogens in the respiratory tract (6, 8–12). Larsen et al. analyzed the phenotypic and functional changes in human DCs in response to respiratory bacterial commensals (Prevotella spp. and Veillonella spp.) and pathogens (Actinomyces spp.) (6). Upon stimulation with commensals, monocyte-derived DCs exhibited higher expression of CD40, CD80, and CD86 and enhanced production of pro- and anti-inflammatory cytokines (IL-12p70, IL-23, and IL-10) compared with the unstimulated DCs. The expression level of the costimulatory molecules on DCs was similar between the commensals and pathogens, whereas commensals produced 3–5 times lower levels of cytokines compared with the pathogenic bacteria (6). Addition of Prevotella spp. into cultures of DCs pulsed with the pathogenic species Haemophilus influenzae partially reduced the production of IL-12p70, but not of IL-23 and IL-10, by the DCs (6). This indicates that Prevotella spp. can suppress Th1 immunity specific to H. influenzae.
A recent study evaluated the impact of commensals, including Streptococcus mitis, Streptococcus salivarius, Streptococcus gordonii, and Streptococcus sanguinis on pro-inflammatory responses in oral keratinocytes by the pathogens, Aggregatibacter actinomycetemcomitans, and Fusobacterium nucleatum (12). The production of IL-8, a chemokine that induces chemotaxis in granulocytes, was significantly higher in human epithelial cell lines infected with the pathogenic bacteria than by the cells exposed to commensals. Upon co-infection with both commensal and pathogenic bacteria, the commensals reduced the pathogen-induced IL-8 secretion (12). Similar modulation of the IL-8 response by S. salivarius strain K12 was demonstrated in relation to the pathogen S. pyogenes (9). More importantly, intragastric administration of live S. salivarius strain JIM8772 led to a significant reduction in intestinal inflammatory reactions in a mouse model of TNBS-induced colitis, compared with control buffer (11). This suggests an immunomodulatory and protective role for this commensal in inflammatory conditions. The mechanisms by which commensals exert their effect is yet to be elucidated. Overall, these data indicate that commensal bacteria modulate host responses induced by pathogens, which is in line with previous studies showing a positive impact of using S. salivarius strains as probiotics against streptococcal pharyngitis and halitosis (40). Anti-inflammatory properties of commensal bacteria have the potential to be used to treat inflammatory and autoimmune diseases.
Little is known about the interaction of the host with commensal viruses and fungi that inhabit the respiratory system. Latent infection with herpesviruses can lead to opportunistic infections in immunocompromised individuals (41). Recent findings, however, highlight a new role for these viruses in increasing host resistance to bacterial infections. Infection with herpesviruses in mice results in chronic production of large quantities of IFN-γ and activation of macrophages that confer protection from subsequent infection with Listeria monocytogenes and Yersinia pestis (42). On the other hand, the opportunisitic fungus Aspergillus fumigatus causes severe infections in patients with a compromised immune system (43). Aspergillus spp. first encounter alveolar macrophages in the lung and are cleared by them via an NADPH oxidase-dependent pathway (44). A strong Th1 response is central to protective immunity against A. fumigatus infection, but the role of IL-17 in immunity to aspergillosis is poorly understood (45). In vivo neutralization of IL-17 in non-immunocompromised mice led to increased fungal loads in the lungs when subjected to A. fumigatus infection, suggesting a protective role for IL-17 (46). To understand the contribution of IL-17 in humans, Chai et al. examined the IL-17 response to fungal infection using human peripheral blood mononuclear cells (PBMCs) and clinical samples from patients with invasive aspergillosis (47). Incubation of live A. fumigatus with human PBMCs resulted in enhanced Th1, but not Th17, responses compared with the PBMC culture without fungal stimulation. This finding was in line with the low levels of IL-17 in the bronchoalveolar lavage fluid and serum of patients with invasive aspergillosis (47). Thus, in contrast to mice, human immunity to A. fumigatus does not appear to be dependent on IL-17 responses. There is a need to know more about the immune responses to these commensals to determine if they are to be used for prophylactic purposes, which is important for developing vaccines against opportunistic infections in patients with compromised immune system.
Cross-Reactivity Between Commensals and Pathogens
A high degree of antigenic similarity between phylogenetically related commensal and pathogenic bacteria supports the hypothesis that natural immunity against pathogens in most adults is the outcome of repeated colonization by commensals that share epitopes during childhood and youth. This hypothesis is anchored on various studies that have demonstrated serological and cell-mediated cross-recognition between respiratory commensals and pathogens, particularly commensal streptococci and Streptococcus pneumoniae (7, 15–17, 48–52) (Figure 1). Recently, Skov Sorensen et al. performed a comprehensive analysis of genetic and antigenic similarities between a range of commensal streptococci and S. pneumoniae and found that 74% of 66 commensal S. mitis strains produced capsule via the Wzy/Wzx pathway, which is in contrast with the previous assumption that capsule production by S. pneumoniae distinguishes them from commensal streptococci that lack capsule (15). Furthermore, double immunodiffusion experiments showed that rabbit antisera raised with different strains of S. mitis and S. oralis exhibited cross-reactivity with capsular polysaccharide antigens of S. pneumoniae (15). These findings raise significant questions regarding misidentification of S. pneumoniae due to their capsular similarity with commensal streptococci, which can have consequences for effective vaccination in infants and elderly. Moreover, it is worth exploring the immune responses against S. mitis and its relationship with S. pneumoniae in humans. Studies have shown that salivary IgA antibodies in infants react to S. mitis antigens, but it is unclear whether the antibodies are elicited by S. mitis or by cross-reactive streptococcal pathogens (53–55). Ongoing studies in our laboratory focus at the antibody-mediated cross-reactivity between S. mitis and pneumococcal antigens. In addition, commensal Neisseria lactamica spp. and pathogenic Neisseria meningitidis have been reported to share major outer membrane cross-reactive antigens (16, 17). Future studies need to assess whether shared immune reactivity between respiratory commensals and pathogens, e.g., S. mitis and S. pneumoniae, could be translated into cross-protection against pathogenic S. pneumoniae. Immune cross-reactivity is not confided to antibodies. T cells specific to commensals can cross-react with pathogens as well. Recently, we examined the phenotype and function of human memory CD4+ T helper (Th) cells reactive with antigens from S. mitis by isolation of memory CD4+ Th cells from healthy human donors and construction of T cell libraries (7). Our findings demonstrated that Th17 cells that were isolated on the basis of their reactivity with S. mitis antigens also responded to pneumococcal antigens (7). Since Th17 cells play an important role in clearance of S. pneumoniae, S. mitis-specific Th17 cross-reactivity with S. pneumoniae might be used as a way forward to devise a prophylactic strategy against pneumococcal infections.
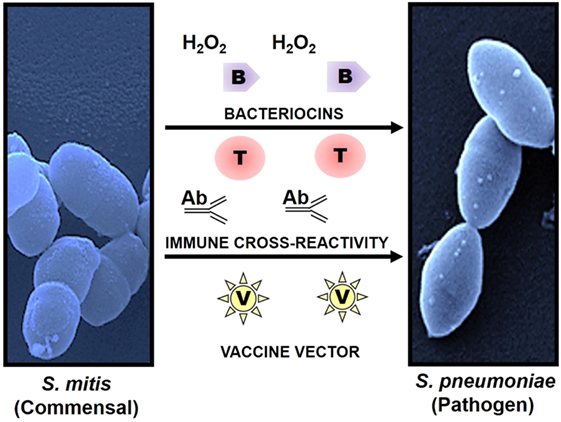
Figure 1. Use of commensals as a prophylactic strategy against respiratory pathogens. Commensals showing high homology with pathogenic species have the potential to induce cross-reactive immune responses and to act as a vaccine delivery system. Commensal Streptococcus mitis induces cross-reactive antibody (Ab) and T cell (T) responses against pathogen Streptococcus pneumoniae. Production of bacteriocins (B) and hydrogen peroxide (H2O2) by S. mitis elicits direct protection against pathogens like S. pneumoniae. In addition, S. mitis has been used as a potent vector (V) for delivery of heterologous bacterial and viral antigens, which confers robust and antigen-specific immunity against respiratory infections. Scanning electron microscopy images of S. mitis NCTC12261 (left), and S. pneumoniae TIGR4 (right).
It may not be out of place to mention here that commensals, such as S. mitis, can exert direct protection against respiratory pathogens, including S. pneumoniae, by producing hydrogen peroxide (H2O2) and bacteriocins that have an inhibitory effect on the pathogens (12, 56) (Figure 1).
Commensals as a Vaccine Delivery System
Some mucosal vaccines, such as polio vaccine, elicit antigen-specific immune responses against a variety of pathogens (57, 58). Since commensals and pathogens colonize the same mucosal surfaces in the respiratory tract, commensals have gained interest as a vaccine delivery system for combating respiratory infections. Previous studies have tested the efficacy of commensal bacteria such as S. gordonii and Lactobacillus lactis as vectors of antigen delivery due to their commensal nature and ease in genetic manipulation. While S. gordonii offers a platform to express various heterologous antigens (e.g., diphtheria toxin antigens), it fails to mount a robust protective response against the recombinant antigens (59, 60). Similarly, a number of mouse studies using L. lactis have been conducted to examine the expression of heterologous antigens and to analyze antigen-specific immune responses (61). Intranasal immunization of mice with recombinant L. lactis expressing human papillomavirus type 16 (HPV-16) E7 protein elicited antigen-specific T cell responses characterized by IFN-γ compared to the non-immunized mice or mice with wild-type Lactobacillus (61). Overall, these commensals have shown a potential as vaccine delivery vehicles, but their ability to confer robust protective immunity is limited. This might be due to poor colonizing capacity of Lactobacillus and low abundance of S. gordonii (62, 63). Two recent studies have evaluated the potential of S. mitis as a vaccine vector for M. tuberculosis and human immunodeficiency virus (HIV) antigens (13, 14). Oral administration of recombinant S. mitis expressing HIV envelope protein in germ-free mice resulted in effective and persistent colonization and was associated with strong antigen-specific IgA and IgG, but non-responsive T cell, responses (14). Similar colonization and antibody responses (IgA and IgG) were observed when gnotobiotic piglets received S. mitis recombined with M. tuberculosis protein (Ag85b) through the oral route (13). These findings suggest that S. mitis has the potential to function as a vaccine vector, which may be attributed to its persistent colonization, abundance in the oral cavity, stable expression of the genes encoding vaccine candidates, and robust antibody responses (Figure 1). However, it has to be noted that these studies have been conducted in germ-free or gnotobiotic animals that may not mimic what happens under natural conditions and in humans, thus warranting the use of animal models with intact microbiota to arrive at a more realistic conclusion.
Conclusion and Future Perspectives
With advancement in sequencing techniques, it is now possible to critically examine and analyze various microbiological and immunological aspects of respiratory tract commensals. From the microbial side, studies that move from the identification of genus or species to include the collection of genes (metagenomics) or transcripts (metatranscriptomics) in the microbiota are likely to contribute to a better understanding of their role in health and disease, and to reveal possible targets for intervention. To date, metagenomic and metatranscriptomic results are still limited to few studies (1, 64). In respect to host–microbial interactions, recent studies have shown that respiratory commensals modulate host immunity by suppressing proinflammatory responses elicited during infections and autoimmune diseases. Controlling excessive immune responses is beneficial to the host as uncontrolled responses can be pathological. On the flip side, immune suppression can make the host prone to infections. Therefore, a delicate balance between host response and microbial activity is critical for maintaining homeostasis in the body. Furthermore, immune cross-reactivity between commensals and pathogens has the potential to generate protective immunity against pathogens. Future studies are required to address the following questions: (1) Can immune cross-reactivity between commensals and pathogens be translated into cross-protection against pathogens? (2) What are the cross-reactive antigens that can be used as potential vaccine candidates? (3) What are the specific agonists produced by commensals to activate different immune cells? (4) How can we develop animal models that specifically lack respiratory tract microbiota? (5) Does an alteration in composition and/or function of the respiratory microbiota lead to the development of respiratory diseases? (6) Can the therapeutic manipulation of the respiratory microbiota be used as a tool to curb diseases? Answers to these questions can lead to better designing of vaccines and drugs against respiratory pathogens.
Author Contributions
SS is the primary author of this manuscript. FP and SS produced the included figure. All authors assisted in the conception of this review, acquisition of relevant literature, and editing the manuscript. All authors gave approval of the final version to be published.
Conflict of Interest Statement
The authors declare that this work was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Rabia Khan for her proofreading of this manuscript.
Funding
This work was supported by a grant from the Norwegian Research Council (Grant number—241011).
References
1. Man WH, de Steenhuijsen Piters WA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol (2017) 15:259–70. doi:10.1038/nrmicro.2017.14
2. Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol (2016) 16:341–52. doi:10.1038/nri.2016.42
4. Henriques-Normark B, Normark S. Commensal pathogens, with a focus on Streptococcus pneumoniae, and interactions with the human host. Exp Cell Res (2010) 316:1408–14. doi:10.1016/j.yexcr.2010.03.003
5. Rangel-Moreno J, Carragher DM, de la Luz Garcia-Hernandez M, Hwang JY, Kusser K, Hartson L, et al. The development of inducible bronchus-associated lymphoid tissue depends on IL-17. Nat Immunol (2011) 12:639–46. doi:10.1038/ni.2053
6. Larsen JM, Steen-Jensen DB, Laursen JM, Sondergaard JN, Musavian HS, Butt TM, et al. Divergent pro-inflammatory profile of human dendritic cells in response to commensal and pathogenic bacteria associated with the airway microbiota. PLoS One (2012) 7:e31976. doi:10.1371/journal.pone.0031976
7. Engen SA, Valen Rukke H, Becattini S, Jarrossay D, Blix IJ, Petersen FC, et al. The oral commensal Streptococcus mitis shows a mixed memory Th cell signature that is similar to and cross-reactive with Streptococcus pneumoniae. PLoS One (2014) 9:e104306. doi:10.1371/journal.pone.0104306
8. Sliepen I, Van Damme J, Van Essche M, Loozen G, Quirynen M, Teughels W. Microbial interactions influence inflammatory host cell responses. J Dent Res (2009) 88:1026–30. doi:10.1177/0022034509347296
9. Cosseau C, Devine DA, Dullaghan E, Gardy JL, Chikatamarla A, Gellatly S, et al. The commensal Streptococcus salivarius K12 downregulates the innate immune responses of human epithelial cells and promotes host-microbe homeostasis. Infect Immun (2008) 76:4163–75. doi:10.1128/IAI.00188-08
10. Kaci G, Lakhdari O, Dore J, Ehrlich SD, Renault P, Blottiere HM, et al. Inhibition of the NF-kappaB pathway in human intestinal epithelial cells by commensal Streptococcus salivarius. Appl Environ Microbiol (2011) 77:4681–4. doi:10.1128/AEM.03021-10
11. Kaci G, Goudercourt D, Dennin V, Pot B, Dore J, Ehrlich SD, et al. Anti-inflammatory properties of Streptococcus salivarius, a commensal bacterium of the oral cavity and digestive tract. Appl Environ Microbiol (2014) 80:928–34. doi:10.1128/AEM.03133-13
12. Santagati M, Scillato M, Patane F, Aiello C, Stefani S. Bacteriocin-producing oral streptococci and inhibition of respiratory pathogens. FEMS Immunol Med Microbiol (2012) 65:23–31. doi:10.1111/j.1574-695X.2012.00928.x
13. Daifalla N, Cayabyab MJ, Xie E, Kim HB, Tzipori S, Stashenko P, et al. Commensal Streptococcus mitis is a unique vector for oral mucosal vaccination. Microbes Infect (2015) 17:237–42. doi:10.1016/j.micinf.2014.11.002
14. Xie E, Kotha A, Biaco T, Sedani N, Zou J, Stashenko P, et al. Oral delivery of a novel recombinant Streptococcus mitis vector elicits robust vaccine antigen-specific oral mucosal and systemic antibody responses and T cell tolerance. PLoS One (2015) 10:e0143422. doi:10.1371/journal.pone.0143422
15. Skov Sørensen UB, Yao K, Yang Y, Tettelin H, Kilian M. Capsular polysaccharide expression in commensal Streptococcus species: genetic and antigenic similarities to Streptococcus pneumoniae. MBio (2016) 7(6):e01844-16. doi:10.1128/mBio.01844-16
16. Troncoso G, Sanchez S, Moreda M, Criado MT, Ferreiros CM. Antigenic cross-reactivity between outer membrane proteins of Neisseria meningitidis and commensal Neisseria species. FEMS Immunol Med Microbiol (2000) 27:103–9. doi:10.1111/j.1574-695X.2000.tb01419.x
17. Troncoso G, Sanchez S, Criado MT, Ferreiros CM. Analysis of Neisseria lactamica antigens putatively implicated in acquisition of natural immunity to Neisseria meningitidis. FEMS Immunol Med Microbiol (2002) 34:9–15. doi:10.1111/j.1574-695X.2002.tb00597.x
18. Segal LN, Blaser MJ. A brave new world: the lung microbiota in an era of change. Ann Am Thorac Soc (2014) 11(Suppl 1):S21–7. doi:10.1513/AnnalsATS.201306-189MG
19. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A (2010) 107:11971–5. doi:10.1073/pnas.1002601107
20. Bosch AA, Levin E, van Houten MA, Hasrat R, Kalkman G, Biesbroek G, et al. Development of upper respiratory tract microbiota in infancy is affected by mode of delivery. EBioMedicine (2016) 9:336–45. doi:10.1016/j.ebiom.2016.05.031
21. Vissers M, de Groot R, Ferwerda G. Severe viral respiratory infections: are bugs bugging? Mucosal Immunol (2014) 7:227–38. doi:10.1038/mi.2013.93
22. Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol (2012) 13:R42. doi:10.1186/gb-2012-13-6-r42
23. Wang Y, Zhu N, Li Y, Lu R, Wang H, Liu G, et al. Metagenomic analysis of viral genetic diversity in respiratory samples from children with severe acute respiratory infection in China. Clin Microbiol Infect (2016) 22:458.e1–9. doi:10.1016/j.cmi.2016.01.006
24. Eidi S, Kamali SA, Hajari Z, Fata A, Farid Hosseini R, Naseri A, et al. Nasal and indoors fungal contamination in healthy subjects. Health Scope (2016) 5(1):e30033. doi:10.17795/jhealthscope-30033
25. Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Falkowski NR, Huffnagle GB, et al. Bacterial topography of the healthy human lower respiratory tract. MBio (2017) 8(1):e02287–16. doi:10.1128/mBio.02287-16
26. Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered microbial communities in asthmatic airways. PLoS One (2010) 5:e8578. doi:10.1371/journal.pone.0008578
27. Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med (2013) 187:1067–75. doi:10.1164/rccm.201210-1913OC
28. Vareille M, Kieninger E, Edwards MR, Regamey N. The airway epithelium: soldier in the fight against respiratory viruses. Clin Microbiol Rev (2011) 24:631. doi:10.1128/CMR.00012-11
29. Corr SC, Gahan CCGM, Hill C. M-cells: origin, morphology and role in mucosal immunity and microbial pathogenesis. FEMS Immunol Med Microbiol (2008) 52:2–12. doi:10.1111/j.1574-695X.2007.00359.x
30. Morin MJ, Warner A, Fields BN. A pathway for entry of reoviruses into the host through M-cells of the respiratory-tract. J Exp Med (1994) 180:1523–7. doi:10.1084/jem.180.4.1523
31. Teitelbaum R, Schubert W, Gunther L, Kress Y, Macaluso F, Pollard JW, et al. The M cell as a portal of entry to the lung for the bacterial pathogen Mycobacterium tuberculosis. Immunity (1999) 10:641–50. doi:10.1016/S1074-7613(00)80063-1
32. Kim DY, Sato A, Fukuyama S, Sagara H, Nagatake T, Kong IG, et al. The airway antigen sampling system: respiratory M cells as an alternative gateway for inhaled antigens. J Immunol (2011) 186:4253–62. doi:10.4049/jimmunol.0903794
33. Heier I, Malmstrom K, Sajantila A, Lohi J, Makela M, Jahnsen FL. Characterisation of bronchus-associated lymphoid tissue and antigen-presenting cells in central airway mucosa of children. Thorax (2011) 66:151–6. doi:10.1136/thx.2010.149591
34. Tecle T, Tripathi S, Hartshorn KL. Review: defensins and cathelicidins in lung immunity. Innate Immun (2010) 16:151–9. doi:10.1177/1753425910365734
35. Kiyono H, Fukuyama S. NALT- versus Peyer’s-patch-mediated mucosal immunity. Nat Rev Immunol (2004) 4:699–710. doi:10.1038/nri1439
36. Randall TD. Bronchus-associated lymphoid tissue (BALT) structure and function. Adv Immunol (2010) 107:187–241. doi:10.1016/B978-0-12-381300-8.00007-1
37. Condon TV, Sawyer RT, Fenton MJ, Riches DWH. Lung dendritic cells at the innate-adaptive immune interface. J Leukoc Biol (2011) 90:883–95. doi:10.1189/jlb.0311134
38. Rayamajhi M, Delgado C, Condon TV, Riches DW, Lenz LL. Lung B cells promote early pathogen dissemination and hasten death from inhalation anthrax. Mucosal Immunol (2012) 5:444–54. doi:10.1038/mi.2012.21
39. Kato A, Hulse KE, Tan BK, Schleimer RP. B-lymphocyte lineage cells and the respiratory system. J Allergy Clin Immunol (2013) 131:933–57. doi:10.1016/j.jaci.2013.02.023
40. Tagg JR, Dierksen KP. Bacterial replacement therapy: adapting ‘germ warfare’ to infection prevention. Trends Biotechnol (2003) 21:217–23. doi:10.1016/S0167-7799(03)00085-4
41. Meyding-Lamade U, Strank C. Herpesvirus infections of the central nervous system in immunocompromised patients. Ther Adv Neurol Disord (2012) 5:279–96. doi:10.1177/1756285612456234
42. Barton ES, White DW, Cathelyn JS, Brett-McClellan KA, Engle M, Diamond MS, et al. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature (2007) 447:326–9. doi:10.1038/nature05762
43. Hohl TM, Feldmesser M. Aspergillus fumigatus: principles of pathogenesis and host defense. Eukaryot Cell (2007) 6:1953–63. doi:10.1128/EC.00274-07
44. Grimm MJ, Vethanayagam RR, Almyroudis NG, Dennis CG, Khan AN, D’Auria AC, et al. Monocyte- and macrophage-targeted NADPH oxidase mediates antifungal host defense and regulation of acute inflammation in mice. J Immunol (2013) 190:4175–84. doi:10.4049/jimmunol.1202800
46. Werner JL, Metz AE, Horn D, Schoeb TR, Hewitt MM, Schwiebert LM, et al. Requisite role for the Dectin-1 beta-glucan receptor in pulmonary defense against Aspergillus fumigatus. J Immunol (2009) 182:4938–46. doi:10.4049/jimmunol.0804250
47. Chai LYA, van de Veerdonk F, Marijnissen RJ, Cheng SC, Khoo AL, Hectors M, et al. Anti-Aspergillus human host defence relies on type 1 T helper (Th1), rather than type 17 T helper (Th17), cellular immunity. Immunology (2010) 130:46–54. doi:10.1111/j.1365-2567.2009.03211.x
48. Lund E. Antigenic relationship between Pneumococci and non-hemolytic Streptococci. Acta Pathol Microbiol Scand (1950) 27:110–8. doi:10.1111/j.1699-0463.1950.tb05200.x
49. Lee CJ, Koizumi K, Henrichsen J, Perch B, Lin CS, Egan W. Capsular polysaccharides of nongroupable Streptococci that cross-react with pneumococcal group-19. J Immunol (1984) 133:2706–11.
50. Morch E. Capsular antigens common to a Streptococcus strain and some pneumococcus types. Acta Pathol Microbiol Scand (1945) 22:159–64. doi:10.1111/j.1699-0463.1945.tb05037.x
51. Yurchak AM, Austrian R. Serologic and genetic relationships between pneumococci and other respiratory streptococci. Trans Assoc Am Physicians (1966) 79:368–75.
52. Kawai T, Paster BJ, Komatsuzawa H, Ernst CW, Goncalves RB, Sasaki H, et al. Cross-reactive adaptive immune response to oral commensal bacteria results in an induction of receptor activator of nuclear factor-kappaB ligand (RANKL)-dependent periodontal bone resorption in a mouse model. Oral Microbiol Immunol (2007) 22:208–15. doi:10.1111/j.1399-302X.2007.00348.x
53. Nogueira RD, Sesso MLT, Borges MCL, Mattos-Graner RO, Smith DJ, Ferriani VPL. Salivary IgA antibody responses to Streptococcus mitis and Streptococcus mutans in preterm and fullterm newborn children. Arch Oral Biol (2012) 57:647–53. doi:10.1016/j.archoralbio.2011.11.011
54. Borges MC, Sesso ML, Roberti LR, de Menezes Oliveira MA, Nogueira RD, Geraldo-Martins VR, et al. Salivary antibody response to streptococci in preterm and fullterm children: a prospective study. Arch Oral Biol (2015) 60:116–25. doi:10.1016/j.archoralbio.2014.08.003
55. Wirth KA, Bowden GH, Kirchherr JL, Richmond DA, Sheridan MJ, Cole MF. Humoral immunity to commensal oral bacteria in human infants: evidence that Streptococcus mitis biovar 1 colonization induces strain-specific salivary immunoglobulin A antibodies. ISME J (2008) 2:728–38. doi:10.1038/ismej.2008.26
56. Prevaes SMPJ, de Winter-de Groot KM, Janssens HM, Piters WAAD, Tramper-Stranders GA, Wyllie AL, et al. Development of the nasopharyngeal microbiota in infants with cystic fibrosis. Am J Respir Crit Care Med (2016) 193:504–15. doi:10.1164/rccm.201509-1759OC
57. Sabin AB. Oral poliovirus vaccine – history of its development and use and current challenge to eliminate poliomyelitis from the world. J Infect Dis (1985) 151:420–36. doi:10.1093/infdis/151.3.420
58. Nisini R, Biselli R, Matricardi PM, Fattorossi A, D’Amelio R. Clinical and immunological response to typhoid vaccination with parenteral or oral vaccines in two groups of 30 recruits. Vaccine (1993) 11:582–6. doi:10.1016/0264-410X(93)90237-R
59. Lee SF, March RJ, Halperin SA, Faulkner G, Gao L. Surface expression of a protective recombinant pertussis toxin S1 subunit fragment in Streptococcus gordonii. Infect Immun (1999) 67:1511–6.
60. Lee CW, Lee SF, Halperin SA. Expression and immunogenicity of a recombinant diphtheria toxin fragment A in Streptococcus gordonii. Appl Environ Microbiol (2004) 70:4569–74. doi:10.1128/AEM.70.8.4569-4574.2004
61. Bermúdez-Humarán LG, Kharrat P, Chatel JM, Langella P. Lactococci and lactobacilli as mucosal delivery vectors for therapeutic proteins and DNA vaccines. Microb Cell Fact (2011) 10(Suppl 1):S4. doi:10.1186/1475-2859-10-S1-S4
62. Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Distribution of selected bacterial species on intraoral surfaces. J Clin Periodontol (2003) 30:644–54. doi:10.1034/j.1600-051X.2003.00376.x
63. Eren AM, Borisy GG, Huse SM, Mark Welch JL. Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci U S A (2014) 111:E2875–84. doi:10.1073/pnas.1409644111
Keywords: host, commensal, immunity, lungs, vaccine
Citation: Shekhar S, Schenck K and Petersen FC (2018) Exploring Host–Commensal Interactions in the Respiratory Tract. Front. Immunol. 8:1971. doi: 10.3389/fimmu.2017.01971
Received: 14 September 2017; Accepted: 20 December 2017;
Published: 17 January 2018
Edited by:
Mats Bemark, University of Gothenburg, SwedenReviewed by:
Irving Coy Allen, Virginia Tech, United StatesRita Carsetti, Bambino Gesù Ospedale Pediatrico (IRCCS), Italy
Copyright: © 2018 Shekhar, Schenck and Petersen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fernanda Cristina Petersen, Zi5jLnBldGVyc2VuQG9kb250LnVpby5ubw==
 Sudhanshu Shekhar
Sudhanshu Shekhar Karl Schenck
Karl Schenck Fernanda Cristina Petersen
Fernanda Cristina Petersen