Abstract
Innate immunity is the primary defense mechanism against infection in metazoans. However, aberrant upregulation of innate immune-signaling pathways can also be detrimental to the host. The p38 MAPK/PMK-1 innate immune-signaling pathway has been demonstrated to play essential roles in cellular defenses against numerous infections in metazoans, including Caenorhabditis elegans. However, the negative regulators that maintain the homeostasis of this important innate immune pathway remain largely understudied. By screening a focused RNAi library against the kinome of C. elegans, we identified RIOK-1, a human RIO kinase homolog, as a novel suppressor of the p38 MAPK/PMK-1 signal pathway. We demonstrated that the suppression of riok-1 confers resistance to Aeromonas dhakensis infection in C. elegans. Using quantitative real time-PCR and riok-1 reporter worms, we found the expression levels of riok-1 to be significantly upregulated in worms infected with A. dhakensis. Our genetic epistasis analysis suggested that riok-1 acts on the upstream of the p38 MAPK/pmk-1 genetic pathway. Moreover, the suppression of riok-1 enhanced the p38 MAPK signal, suggesting that riok-1 is a negative regulator of this innate pathway in C. elegans. Our epistatic results put riok-1 downstream of skn-1, which encodes a p38 MAPK downstream transcription factor and serves as a feedback loop to the p38 MAPK pathway during an A. dhakensis infection. In conclusion, riok-1 is proposed as a novel innate immune suppressor and as a negative feedback loop model involving p38 MAPK, SKN-1, and RIOK-1 in C. elegans.
Introduction
Coordination of the innate immune response in intestinal epithelial cells maintains host–microbial interaction and tissue homeostasis (1). In humans, overactivated or aberrantly triggered immune responses are associated with inflammatory diseases (2). The activation of the immune response is required for hosts to survive challenges from invading pathogens. By contrast, an inhibitory mechanism that functions to lessen the damage caused by pathogens limits collateral injury due to immune activation. For example, the ASK-1–MKK3/6–p38 MAPK signal pathway in mammals is a pivotal regulator of inflammatory cytokine production in response to pathogen detection at the mucosal surface, and the activation of the pathway is tightly regulated via negative regulatory circuits (3). The disruption of the p38 MAPK pathway has been closely associated with the pathogenesis of several human diseases, such as inflammatory diseases, cancers, and autoimmune diseases (3). Conceptually, immune tolerance to nonpathogenic bacteria may be beneficial for host survival. The coexistence of activation and suppression mechanisms for the immune response is like a balance between yin and yang to achieve homeostasis.
The p38 MAPK has evolutionarily conserved roles in the control of cellular responses to microbial and abiotic stress (3). The homolog of the ASK–1–MKK3/6–p38 MAPK signal pathway in mammals is NSY-1–SEK-1–PMK-1 MAPK in Caenorhabditis elegans. In the aging model, p38 MAPKK/SEK-1 is activated by NADPH oxidase-generated ROS, and the activation of the pathway promotes longevity in response to stress and is also negatively regulated by memo-1 to maintain homeostasis (4). In addition, p38 MAPK/PMK-1-mediated resistance to bacterial infection has been well established in a C. elegans infection model (5). However, the regulatory mechanisms that ensure homeostasis of this pathway in response to infection are not very well understood. Previous studies have showed that a regulatory mechanism for p38 MAPK/PMK-1 pathway exists. MOM-4 has been reported to be an activator of the p38 MAPK immune pathway in response to Pseudomonas aeruginosa infection (6). By contrast, VHP-1, an MAP kinase phosphatase, is the only reported negative regulator of the p38 MAPK/PMK-1 innate immune pathway in C. elegans (7). We believe other unknown components that regulate the complex p38 MAPK/PMK-1 innate immune pathway exist and are yet to be discovered.
Kinases comprise one of the largest and most important protein families. The genes encoding kinases constitute 2% of genes in a variety of eukaryotic genomes and play major roles in modulating most cellular processes (8–10). The collection of kinases in an organism is termed the “kinome,” a concept that has been widely used to study life science from a general viewpoint (10, 11). The kinome of C. elegans has been reported to contain 418 protein kinases, 20 atypical kinases, and 25 kinase fragments or pseudogenes, which are about 80% homologous to human kinases (8, 9, 11). Here, we used the model organism C. elegans to study the immune response to a bacterial infection.
Caenorhabditis elegans is attractive as a model because of its suitability for studying innate immunity in a host (12, 13), conveniently related to gene analysis and observation, and its short life span. Moreover, it is rational to study immune homeostasis in organisms like bacterivorous C. elegans that live in microbe-rich environments and must defend against invading pathogens and tolerate food-source bacteria. In our study model, A. dhakensis was selected as an infectious pathogen in vivo because of its well-established virulence to C. elegans (14, 15) and, therefore, the presence of respective strategies by which to combat each other.
In the screening of the C. elegans kinome, we discovered that the loss of riok-1 confers resistance to virulent Aeromonas, indicating its potential role as an immune repressor. RIOK-1, a human RIO (right open reading frame) kinase homolog that is universally present in both invertebrate and vertebrate animals and encodes an atypical protein kinase, which is known to participate in cell cycle control and ribosomal RNA processing (16–21). Previous studies have found that riok-1 and riok-2 are required for EGFR- and PI3K-mediated tumorigenesis in glioblastoma (22). However, the role of RIOK-1 in innate immunity against infection remains unclear. The present study further identified RIOK-1 as an immune suppressor that specifically regulates the p38 MAPK pathway.
In the present study, we identified riok-1 as a novel innate immune suppressor and proposed a negative feedback loop model among p38 MAPK, SKN-1, and RIOK-1 in C. elegans. Bacterial infection activates the p38 MAPK pathway, which transcribes the expression of riok-1 by skn-1, and the activation of riok-1 results in downregulation of the p38 MAPK pathway.
Materials and Methods
Bacteria and C. elegans Strain
The C. elegans wild-type Bristol N2 strain, riok-1-mutant VC2676 riok-1(gk1101), tissue-specific RNAi strain NL2099 rrf-3(pk1426), NR222 rde-1(ne219);kzIs9[lin-26p:nls:GFP + lin-26p:rde-1 + rol-6(su1006)], VP303 rde-1(ne219);kbIs7[nhx-2p:rde-1 + rol-6(su1006)], TU3311 uIs60[unc-119p:YFP + unc-119p:sid-1], and p38 MAPK pathway mutants, sek-1(km4), pmk-1(km25), were provided by the Caenorhabditis Genetics Center (CGC). The p38 MAPK gain-of-function mutant nsy-1(ums8) was a gift from Dr. Read Pukkila-Worley (23). riok-1 transcriptional reporter riok-1p:mcherry and translational reporter riok-1p:riok-1:mcherry were generated in this research. The animals were maintained on NGM plates using Escherichia coli strain OP50 as the food source. The A. dhakensis strain AAK1 used in the study was a clinical isolate from a patient with septicemia and necrotizing fasciitis (24). The C. elegans RNAi feeding clones were obtained from Ahringer’s C. elegans RNAi library and were kept in an RNAi vector pL4440 in E. coli strain HT115 (25).
Screen of RNAi Kinome Library
The kinome RNAi library was collected from Ahringer’s C. elegans RNAi library followed by the C. elegans kinome database (8, 9, 11). Three hundred and four kinase RNAi clones from RNAi library were selected according to 438 C. elegans kinases. The excluded 134 kinases were either missing in the RNAi library or unable to culture. The RNAi clones were cultured in LB broth with 1 mM IPTG (to induce siRNA production), tetracycline, and ampicillin in 96 wells at 37°C overnight and were then mixed with 20–25 N2 L1 worms in 160 µl of M9 culture medium at 25°C for another 40 h. Ten microliters of A. dhakensis AAK1, which was incubated with LB broth at 37°C at a concentration of OD600 2.0, was added to each well. The total numbers of live worms were counted after 2 days of culture at 25°C. The survival rates were calculated by measuring the numbers of live worms among the total worms as a percentage.
Life Span Assay of C. elegans With RNAi
A plate assay was conducted to measure the life span of worms with bacterial infection. Briefly, the eggs were separated from the adult worms with a sodium hypochlorite/NaOH solution, and the resulting eggs were synchronized in M9 medium. The synchronized L1 larvae worms were seeded onto NGI plates (NGM plates with 1 mM IPTG) that were cultured with E. coli HT115 containing the desired RNAi target on the empty vector pL4440. The L1 worms grew at 20°C until the L4 stage. These L4 stage animals were transferred to plates together with E. coli HT115 either carrying RNAi plasmids or the empty vector together with A. dhakensis AAK1 at a ratio of 1:1. The animals were then incubated at 20°C. The worms were monitored for death events daily. Animals that escaped from the plate or died due to internal hatching were censored. Censored animals were included in the statistical analysis only until the day of the censoring event.
Life Span Assay of C. elegans With Infection
Synchronized worms were grown on NGM plates covered with E. coli OP50 until the L4 stage. About 50 nematodes were transferred to fresh plates of OP50 or pathogens that were cultured in LB for 18 h at 37°C to achieve an OD600 2.0 for tests. The worms were monitored for death events daily. Animals that escaped from the plate or died due to internal hatching were censored. Censored animals were included in the statistical analysis only until the day of the censoring event.
Measurement of Gene Expression
The expression of riok-1, p38 MAPK downstream genes, and nhr-23 as an internal control was measured using quantitative real-time-PCR (qRT-PCR) as described previously (15). Approximately 2,000 worms were collected for RNA extraction. An RNA sample (2.0 µg) for each experimental group was converted to cDNA via reverse transcription. All qRT-PCRs were carried out using the FastStart Universal SYBR Green Master (Rox) according to the manufacturer’s specifications and analyzed on a StepOnePlus Real-Time-PCR System. The expression level of each target gene was collected as a ΔCt value, where Ct was equal to the number of PCR cycles required to amplify a given gene from a cDNA population. The fold-change values were estimated using the following equation: fold change = 2[−ΔCt (AAK1)]/2[−ΔCt (OP50)]. Changes in the expression genes were initially measured as ΔCt values which were subsequently normalized against a housekeeping gene nhr-23. Primers for qRT-PCR included riok-1 (forward primer: CGA AAG ATT GCT ATG CAC ACG; reverse primer: CTT CCT CTG TTC CCG TTT CTC), riok-1 isoform a (forward primer: CCG GCT CCG TTG CTT AAA; reverse primer: TCT CAT ATC GCG CAC CAA AC), riok-1 isoform b (forward primer: GGC CAT GTA CTT GTG ATG GA; reverse primer: TCT CAT TTA TCC ACA CCT CTT GG), K08D8.5 (forward primer: GAA TCT TTC GGA GCC CTA CTA C; reverse primer: CGT TCC CTG AGG AAC ATT GA), C32H11.12 (forward primer: CGA GCC AGG AGG TTA TCT TTA C; reverse primer: GTC CGT CCC GAT GTT GAT TT), C29F3.7 (forward primer: CGT ATC TTG GAA CAG GAC TTC A; reverse primer: CAG CCC AGG AAT CAC CAA TA), F08G5.6 (forward primer: TGT CCA TTT CCA TCG CAT ACT; reverse primer: CGG TAG ATT GCT AAT GGG TTC T), T24B8.5 (forward primer: TGT AAC GAA GCA GAT GTT AGA AGT G; reverse primer: TGG CTC TGC AGT TGT ACC A), nhr-23 (forward primer: GCC GAA GAT GAT GCC GAG AT; reverse primer: GTC GCA GTG TCA AGA ATC CC).
Construction of the riok-1 Reporter
To create riok-1p:mCherry and riok-1p:mCherry:riok-1, a 2,000-bp region 5' occupying the ATG site and the riok-1 cDNA fused with mCherry (pCG150) were constructed and then cloned into pPBCN39 using the BP/LR gateway system. unc-119 worms were microinjected, and the offspring of movers with florescence were picked and confirmed with sequencing.
Tissue-Specific RNAi
Synchronized NL2099, NR222, VP303, and TU3311 L1 larvae were cultured on plates seeding E. coli HT115 with L4440, riok-1 RNAi, and tissue-specificity control RNAi act-5, unc-73, and bli-1 at 20°C until the L4 stage. RNAi knockdowns were restricted specifically in whole body (NL2099), epidermis (NR222), intestine (VP303), and neurons (TU3311). These L4 stage animals were transferred to plates together with E. coli HT115 either carrying RNAi plasmids or the L4440 plasmid together with A. dhakensis AAK1 at a ratio of 1:1. The animals were then incubated at 20°C and the survival was observed.
p-p38 Western Blot
Analysis of the p38 MAPK activation of C. elegans was performed using a Western blot method described previously with some modifications (26). Briefly, approximately 2,000 worms were collected to extract the total protein with a RIPA buffer containing inhibitors for protease and phosphatase. Monoclonal antibody to phospho-p38 MAPK (Cell Signaling Technology) and monoclonal antibody to tubulin (Sigma-Aldrich) were used for detection.
Statistical Analysis
All experiments were performed a minimum of three times independently. Nematode survival assays were assessed using the Kaplan–Meier method, and survival differences were tested using the log-rank test. The results of the statistical analysis between two values were compared with a Student’s t-test. Statistical analysis among three or more values for one independent variable was done with a matched one-way ANOVA test. Statistical significance was set at P < 0.05.
Results
Suppressing Expression of riok-1 Confers Resistance Against A. dhakensis Infection in C. elegans
We created a focus RNAi library (Figure 1A) of the C. elegans kinome according to the kinome database (8, 9, 11). Our library contained 304 kinase RNAi clones that covered 69.41% of currently known kinases (Figure 1B; Table S1 in Supplementary Material). We knocked down the expression of kinases using specific RNAi and observed their influence on the survival of worms with A. dhakensis infection. The results were grouped with the survival rate of worms and are partially listed in Tables 1A,B. The screening results were compatible with previous findings suggesting that the repression of kinases, such as aak-1 (27), dkf-2 (28), and sek-1 (5, 29), participates in immune pathway-altered survival shortening of worms with Aeromonas infection (Table 1B). Among these kinases, the repression of riok-1, one of the atypical serine kinases known to be associated with rRNA maturation, significantly improved the resistance of C. elegans with A. dhakensis infection (Table 1A). RIOK-1 was conserved from C. elegans to mammals (17), and its activation led to tumorigenesis in glioblastoma cancer cells as reported in Ref. (22). The N2 worms treated with riok-1 RNAi enhanced the resistance to A. dhakensis strain AAK1 in the plate-killing assay (Figure 1C). The riok-1 knockdown worms were significantly resistant to A. dhakensis in comparison to wild-type worms (P < 0.001). A previous study showed that different assays may result in different outcomes in a C. elegans infection model (30). Our study showed similar survival results in the plate assay (Figure 1C) and the screening liquid assay (Table 1A; Table S1 in Supplementary Material), showing that worms with riok-1 suppression mediated by RNAi were more resistant to A. dhakensis infection.
Figure 1
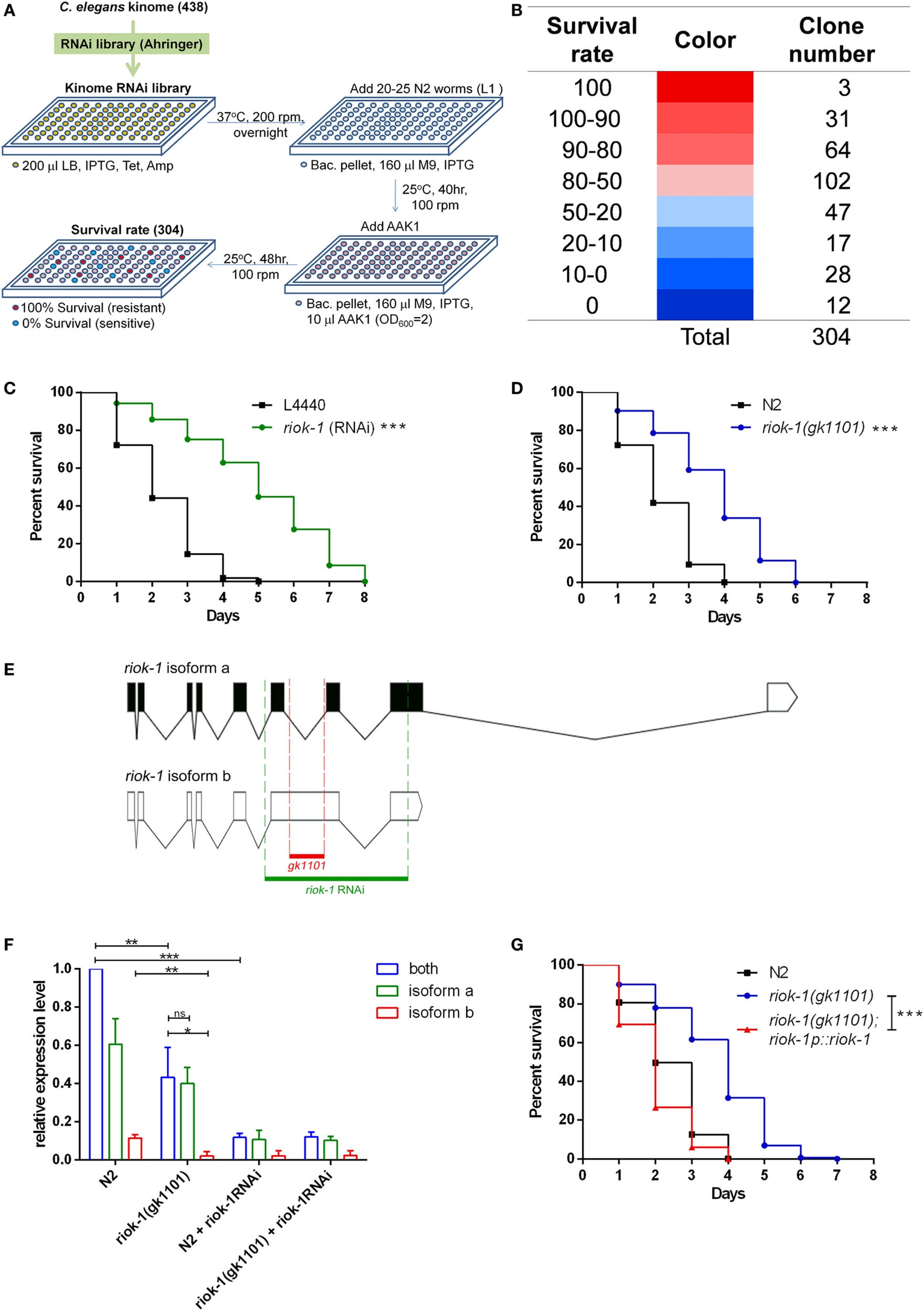
Loss of riok-1 confers resistance to Aeromonas dhakensis infection in Caenorhabditis elegans through screening an RNAi kinome library. Through screening the RNAi kinome library in C. elegans, worms with riok-1 knockdown showed resistance to A. dhakensis infection. (A) Flowchart showing the screening of the RNAi kinome library in C. elegans. Three hundred and four kinase RNAi clones from the RNAi library were selected according to 438 C. elegans kinases. The excluded 134 kinases were either missing in the RNAi library or unable to culture. (B) Hit numbers at each level classified by the survival rate of worms in the RNAi kinome library. (C) Compared to worms with empty vector control L4440, worms with RNAi-mediated knockdown of riok-1 showed resistance to A. dhakensis (***P < 0.001). (D)riok-1(gk1101) worms showed resistance to A. dhakensis compared to wild-type N2 worms (***P < 0.001). (E) The location of riok-1(gk1101) (in red) and the target region of riok-1 RNAi (in green) labeled on riok-1 isoforms. (F) Expression level of riok-1 isoforms by quantitative real time-PCR (qRT-PCR) in wild-type N2 worms and riok-1(gk1101)-mutant worms with or without treatment with riok-1 RNAi (***P < 0.001, **P < 0.01, and *P < 0.05). (G) The complement of riok-1 in riok-1 (gk1101) mutants reversed the resistant phenotype of riok-1(gk1101) worms (***P < 0.001). IPTG, isopropyl β-D-1-thiogalactopyranoside 1 mM; Tet, tetracycline 15 µg/ml; Amp, ampicillin 50 µg/ml; M9, worm M9 buffer.
Table 1
| A | Sequence name | Gene | Survival rate | SD | p-Value |
|---|---|---|---|---|---|
| RNAi control vector pL4440 | 28.39 | 6.67 | – | ||
| Gene depression confers resistance to Aeromonas. dhakensis | M01B12.5 | riok-1 | 100.00 | 0.00 | <0.001 |
| C04G2.2 | 100.00 | 0.00 | <0.001 | ||
| C01H6.9 | hasp-1 | 100.00 | 0.00 | <0.001 | |
| R06C7.8 | bub-1 | 98.81 | 2.06 | <0.001 | |
| B0511.4 | tag-344 | 98.41 | 2.75 | <0.001 | |
| ZC404.9 | gck-2 | 98.33 | 2.89 | <0.001 | |
| Y54E10B_152.b | mek-2 | 98.15 | 3.21 | <0.001 | |
| T01H8.1 | rskn-1 | 97.62 | 2.20 | <0.001 | |
| T09B4.7 | 97.60 | 2.21 | <0.001 | ||
| F42A10.4 | efk-1 | 97.29 | 2.53 | <0.001 | |
| K06H7.1 | 96.97 | 5.25 | <0.001 | ||
| F46F5.2 | 96.93 | 2.95 | <0.001 | ||
| Y106G6A.1 | 96.90 | 2.69 | <0.001 | ||
| F59A3.8 | 96.83 | 5.50 | <0.001 | ||
| Y65B4A_185.a | 96.30 | 6.42 | <0.001 | ||
| F23C8.7 | 96.00 | 6.93 | <0.01 | ||
| F58A3.2 | egl-15 | 95.49 | 4.78 | <0.001 | |
| Y48G1B_55.a | 95.42 | 5.39 | <0.001 | ||
| F52F12.3 | mom-4 | 95.24 | 8.25 | <0.01 | |
| F35G12.3 | sel-5 | 95.24 | 8.25 | <0.01 | |
| C34F11.5 | 95.15 | 5.69 | <0.001 | ||
| T13H10.1 | kin-5 | 94.48 | 3.53 | <0.001 | |
| F23C8.8 | 93.94 | 10.50 | <0.01 | ||
| K03E5.3 | cdk-2 | 92.64 | 2.55 | <0.001 | |
| K06H7.8 | 92.50 | 6.61 | <0.001 | ||
| F46F6.2 | pkn-1 | 91.83 | 7.08 | <0.01 | |
| C05C12.1 | 91.44 | 5.79 | <0.001 | ||
| F49B2.5 | src-2 | 91.07 | 6.89 | <0.01 | |
| F59A6.4 | 90.97 | 12.06 | <0.01 | ||
| K08H2.5 | 90.77 | 7.03 | <0.01 | ||
| F26E4.5 | 90.70 | 3.31 | <0.001 | ||
| W04G5.6 | kin-23 | 90.48 | 16.50 | <0.05 | |
| C44C8.6 | mak-2 | 90.45 | 8.29 | <0.01 | |
| Y38F1A.10 | max-2 | 90.14 | 7.80 | <0.01 | |
|
|
|||||
| B | Sequence name | Gene | Survival rate | SD | p-Value |
|
|
|||||
| RNAi control vector pL4440 | 28.39 | 6.67 | – | ||
| Gene depression confers resistance to A. dhakensis | R03G5.2 | sek-1 | 9.87 | 6.54 | <0.05 |
| F09A5.2 | 9.76 | 1.17 | <0.001 | ||
| F22D6.5 | prpf-4 | 9.18 | 7.95 | <0.05 | |
| C25A8.5 | 9.17 | 5.46 | <0.05 | ||
| F57B9.8 | 8.38 | 7.70 | <0.05 | ||
| C08H9.5 | old-1 | 7.34 | 6.53 | <0.05 | |
| C50H2.7 | 7.14 | 3.89 | <0.01 | ||
| F16B12.5 | 6.71 | 5.97 | <0.05 | ||
| F52B5.2 | 6.56 | 6.92 | <0.05 | ||
| D2023.6 | 6.55 | 2.22 | <0.001 | ||
| F12F3.2 | 6.06 | 10.50 | 0.058 | ||
| R11G1.4 | sax-1 | 5.68 | 5.89 | <0.01 | |
| C07G1.3 | pct-1 | 5.33 | 6.84 | <0.05 | |
| B0218.5 | 5.00 | 8.66 | <0.05 | ||
| C34G6.5 | cdc-7 | 4.89 | 4.84 | <0.01 | |
| F55C5.7 | rskd-1 | 4.44 | 7.70 | <0.05 | |
| W07G4.3 | 4.29 | 4.17 | <0.01 | ||
| C49C8.1 | 4.17 | 4.17 | <0.01 | ||
| F22B3.8 | 4.07 | 3.57 | <0.001 | ||
| R90.1 | 2.56 | 4.44 | <0.01 | ||
| T19A5.2 | gck-1 | 2.47 | 4.28 | <0.01 | |
| F26A1.4 | 2.15 | 3.72 | <0.001 | ||
| PAR2.3 | aak-1 | 1.67 | 2.89 | <0.001 | |
| F49C5.4 | 1.67 | 2.89 | <0.001 | ||
| F13B9.4 | 1.08 | 1.86 | <0.001 | ||
| C24G6.2 | 1.04 | 1.80 | <0.001 | ||
| T25B9.4 | 0.95 | 1.65 | <0.001 | ||
| B0496.3 | unc-82 | 0.93 | 1.60 | <0.001 | |
| ZC449.3 | sek-3 | 0.00 | 0.00 | <0.001 | |
| Y39H10A_224.a | chk-1 | 0.00 | 0.00 | <0.001 | |
| Y39G8B.f | 0.00 | 0.00 | <0.001 | ||
| W08D2.8 | kin-21 | 0.00 | 0.00 | <0.001 | |
| T25E12.4 | dkf-2 | 0.00 | 0.00 | <0.001 | |
| R02C2.2 | kin-34 | 0.00 | 0.00 | <0.001 | |
| F57F5.5 | pkc-1 | 0.00 | 0.00 | <0.001 | |
| F49E11.1 | mbk-2 | 0.00 | 0.00 | <0.001 | |
| F47F2.1 | 0.00 | 0.00 | <0.001 | ||
| F11D5.3 | ddr-2 | 0.00 | 0.00 | <0.001 | |
| F09G2.1 | 0.00 | 0.00 | <0.001 | ||
| C44C10.7 | 0.00 | 0.00 | <0.001 | ||
Excerpt from the kinome RNAi screen.
The table shows a section of the screening results of the kinome RNAi. For the full screening results of the kinome RNAi, please see Table S1 in Supplementary Material.
(A) The RNAi knockdown targets of kinase that caused higher survival rates in A. dhakensis strain AAK1-infected C. elegans.
(B) The RNAi knockdown targets of kinase that caused lower survival rates in A. dhakensis strain AAK1-infected C. elegans.
The resistance to A. dhakensis in riok-1(gk1101)-mutant worms was similar to that observed in worms treated with riok-1 RNAi (P < 0.001) (Figure 1D). Of note, we found the life span of riok-1 mutant to be shorter than that of riok-1 knockdown worms under A. dhakensis infection. It seems that worms with riok-1 knockdown are more resistant to A. dhakensis infection than with riok-1 mutants. The difference may be explained by the fact that the riok-1(gk1101) worm is not a riok-1 null mutant that still maintains isoform a, one of the two isoforms of riok-1 (Figure 1E). The resistance of worms with riok-1 knockdown by RNAi is stronger than that of riok-1(gk1101) worms, which is correlated with the lower-expression level of riok-1 in worms measured by qRT-PCR (Figure 1F). The expression level of riok-1 isoform b was significantly lower in riok-1(gk1101) than in wild-type N2 worms (P < 0.01). Nevertheless, overexpressed riok-1 in the riok-1(gk1101) mutant reversed the resistant phenotype of the riok-1(gk1101) mutant upon A. dhakensis infection (Figure 1G). Taken together, these results showed that suppressed riok-1 expression confers resistance against A. dhakensis infection in C. elegans.
Expression Level of riok-1 Is Increased Under A. dhakensis Infection in C. elegans
In order to monitor the expression level and pattern of riok-1 in C. elegans, we generated the riok-1 transcriptional reporter (riok-1p:mCherry) and translational reporter (riok-1p:mCherry:riok-1), respectively. In these riok-1 reporter worms, riok-1 expression was increased in a time-dependent manner after A. dhakensis was added (Figures 2A,C). The riok-1 transcriptional and translational levels were compatible and are summarized in Figures 2B,D. The expression level of riok-1 as measured by qRT-PCR is also compatible with those observed in reporter worms microscopically (Figure 2E). Taken together, A. dhakensis AAK1 infection can induce riok-1 expression in C. elegans.
Figure 2
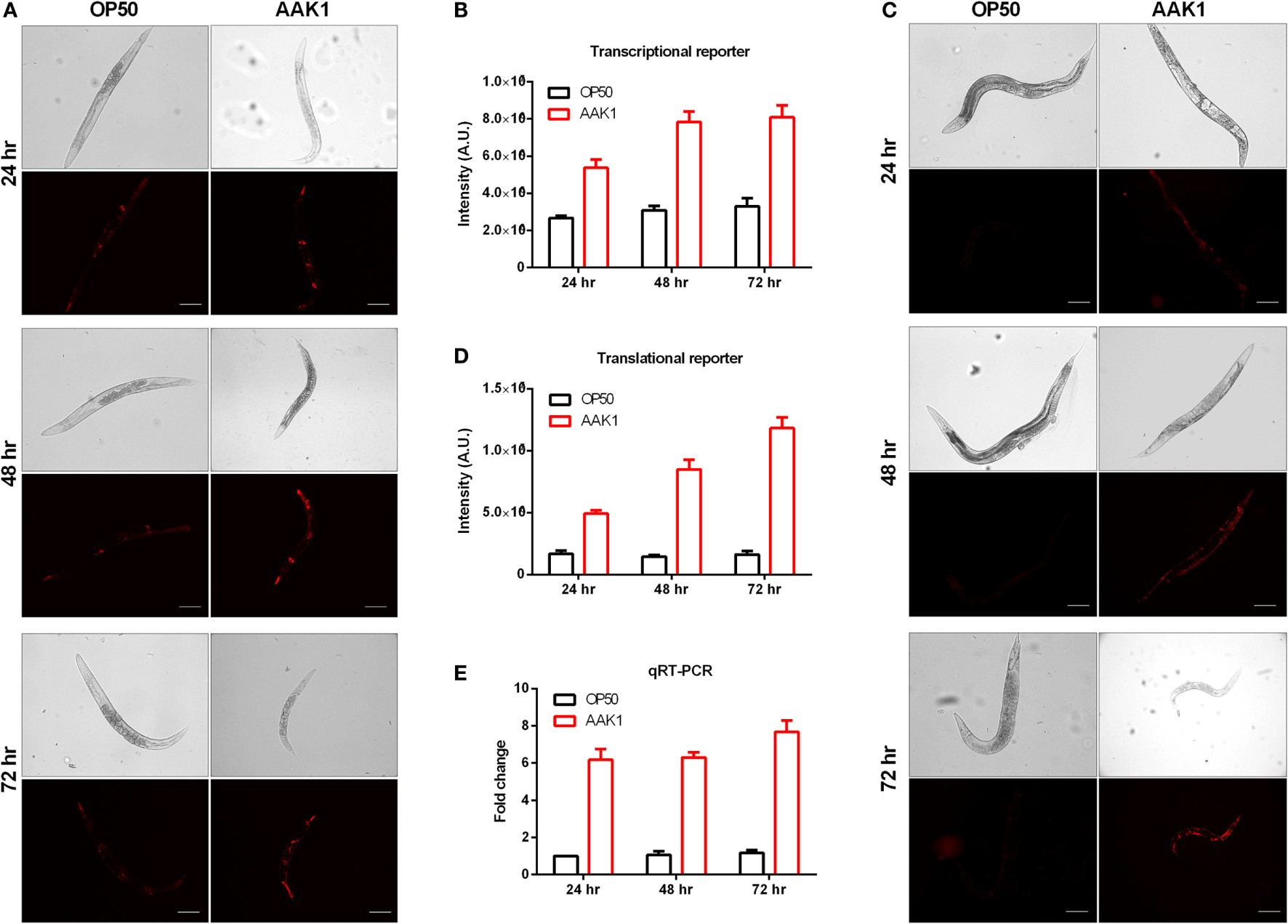
Expression level of riok-1 induced upon Aeromonas dhakensis infection. (A)riok-1 expression in the riok-1 transcriptional reporter strain (riok-1p:mCherry) (B) Quantification of riok-1 expression in riok-1 transcriptional reporter worms. (C)riok-1 expression in the translational reporter strain (riok-1p:mCherry:riok-1) (D) Quantification of riok-1 translation in the reporter strain. (E)riok-1 level as determined by quantitative real time-PCR (qRT-PCR). The scale bars in (A,C) are all 100 µm. The statistical analyses of OP50 and AAK1 at each time point in (B,D,E) are all P < 0.001.
Knockdown of riok-1 in Intestine and Neurons Conferred Resistance to A. dhakensis in C. elegans
We found that the expression of riok-1 was increased by A. dhakensis infection. The constitutive expression of riok-1 appeared in specific parts of C. elegans. Our data showed that riok-1 is expressed in many neurons, in the intestine, in the pharynx, and probably in spermatheca in the riok-1 transcriptional reporter worms (riok-1p::mCherry) (Figure 3A; Figures S2A, S2C, and S2D in Supplementary Material) as well as the translational reporters (riok-1p::mCherry::riok-1) (Figure 3B; Figure S2B in Supplementary Material), as described in previous reports (31, 32). In order to determine the site of action of riok-1 and clarify the tissue specificity of riok-1 in terms of A. dhakensis resistance, the life span assay of worms treated with riok-1 RNAi restricted to specific tissues was observed. The specificity of tissue-specific RNAi worm strains was confirmed with tissue-specific RNAi clones, which acted only in the corresponding tissues and demonstrated specific phenotypes (33) (Figure S2E in Supplementary Material). With tissue-specific RNAi worms, the knockdown of riok-1 in the whole body (Figure 3C), intestine (Figure 3D), and neurons (Figure 3E) enhanced resistance to A. dhakensis infection in C. elegans. By contrast, suppressed riok-1 expression in the epidermis did not alter the life span of worms with A. dhakensis AAK1 infection (Figure 3F).
Figure 3
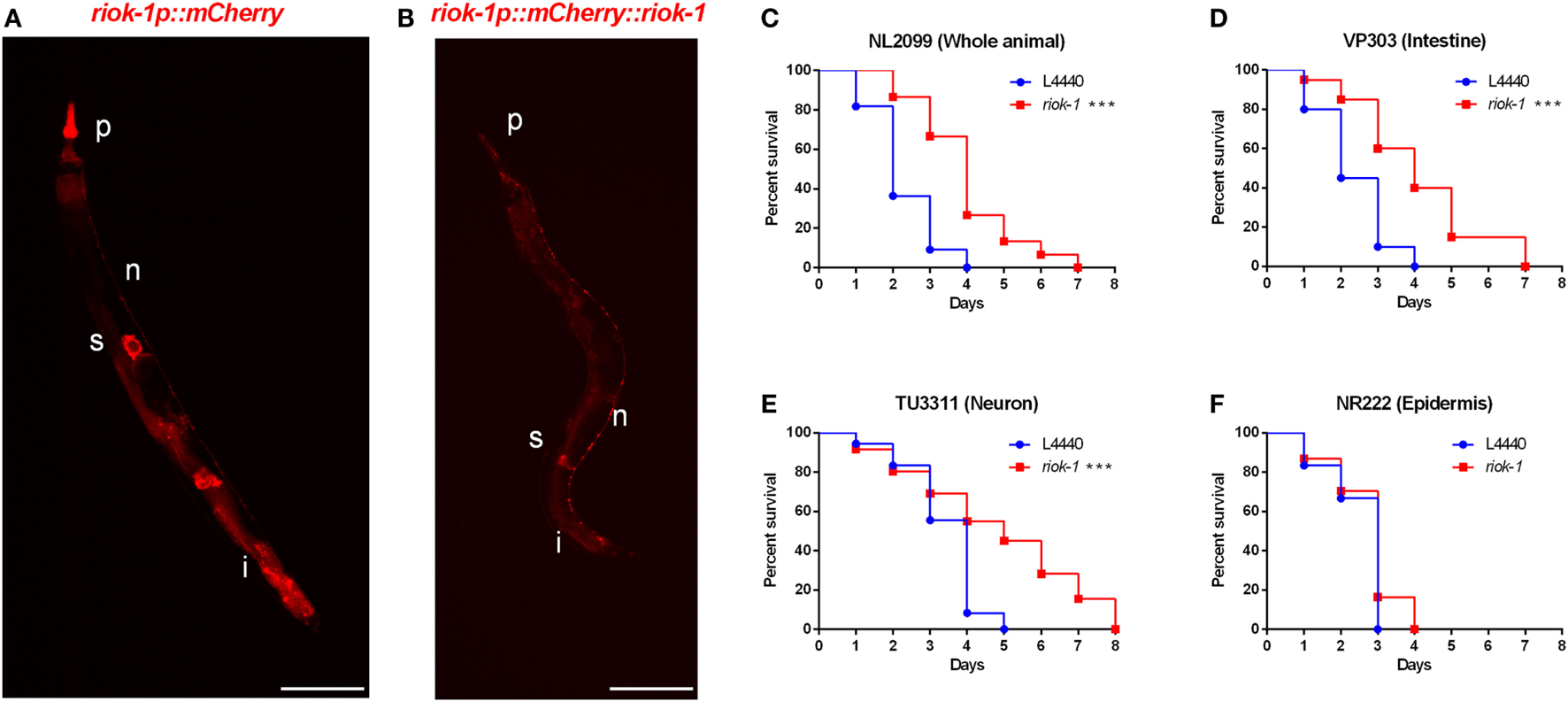
Tissue-specific knockdown of riok-1 in the intestines and neurons resulted in similar resistance to Aeromonas dhakensis as in worms with general riok-1 knockdown. (A)riok-1 is expressed in the pharynx (P), intestines (i), spermatheca (s), and pan-neuronally (n) in riok-1 transcriptional reporter worms. (B)riok-1 is expressed in the pharynx (p), intestines (i), spermatheca (s), and pan-neuronally (n) in translational reporter worms. (C–F)A. dhakensis resistance was seen in worms infected by A. dhakensis AAK1 with tissue-specific knockdown of riok-1 in the whole body (C), intestines (D), and neurons (E) in contrast to non-resistance to A. dhakensis among those with riok-1 knockdown in the epidermis (F) ***P < 0.001. The scale bars in (A,B) are both 100 µm.
Progeny Deficiency Is Not Associated With Resistance to A. dhakensis Infection Mediated by Suppressing riok-1
Previous studies have shown that germ-cell loss extends the life span of C. elegans (34, 35). Our observation was compatible with previous findings, showing that riok-1 deletion confers progeny reduction (32). To clarify whether the progeny deficiency caused by riok-1 RNAi or the riok-1 mutant confers longer life span in A. dhakensis infection, glp-4(bn2) worms were tested. The glp-4 gene is critical for germ line proliferation, and glp-4(bn2) worms are characteristic of germ line loss with progeny deficiency at 25°C. Gaining resistance from loss of fertility against pathogen infection has been widely reported (36). Nevertheless, our results showed that the survival curve of glp-4(bn2) worms infected by A. dhakensis was similar to that observed among N2 worms with or without riok-1 knockdown mediated by RNAi (Figure S3 in Supplementary Material). These findings suggest that progeny deficiency is not associated with A. dhakensis resistance mediated by riok-1 suppression in C. elegans.
RIOK-1 Is a Suppresser of the p38 MAPK Immune Pathway
Our results showed that progeny deficiency did not contribute to A. dhakensis resistance in riok-1-deficient worms (Figure S3 in Supplementary Material). We also found tissue-specific knockdown of riok-1 in the intestine and neurons, which are two major organs where innate immunity present in C. elegans increased the resistance to A. dhakensis infection (Figures 3D,E). We then hypothesized that riok-1 may regulate innate immunity in C. elegans. In order to understand the relationship between riok-1 and innate immunity in C. elegans, the major innate immune-signaling pathways were screened. The screening showed that worms with suppressed riok-1 expression had increased susceptibility upon A. dhakensis infection when pmk-1 was downregulated with RNAi, which was similar to the susceptibility of worms with pmk-1 RNAi (Figure 4A). The results indicate that the p38 MAPK pathway is epistatic to riok-1 while the other immune pathways are not (Figure S4 in Supplementary Material). Further tests using mutant worms of the p38 MAPK-signaling pathway showed similar results. The knockdown of riok-1 in p38 MAPK pmk-1 (Figure 4B) and MAPKK sek-1 (Figure 4C) mutants led to hypersensitivity to A. dhakensis. The nsy-1(ums8) (a p38 MAPK gain-of-function allele)-mutant worms treated with riok-1 RNAi did not gain additional resistance to A. dhakensis infection (Figure 4D). A. dhakensis AAK1 induced the activation of the p38 MAPK pathway based on the evidence of p38 phosphorylation in a Western blot analysis (Figure 4E). In addition, the knockdown of riok-1 induced p38 phosphorylation (Figure 4E) and the increased expression of an array of p38 MAPK downstream genes (Figures 4F–J). These results suggest that riok-1 lies upstream of the p38 MAPK pathway and that it also plays a role in negative regulation of this pathway.
Figure 4
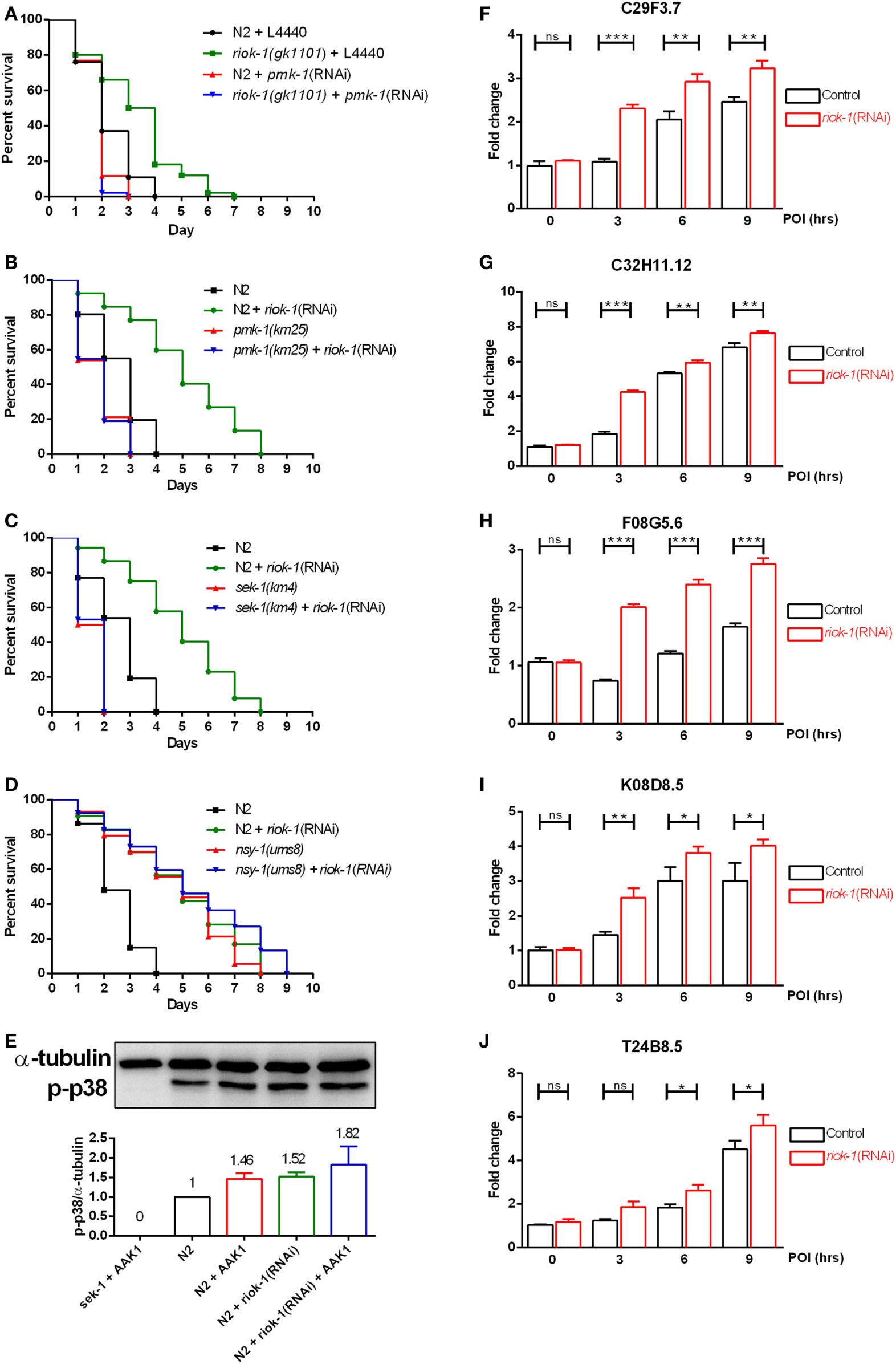
riok-1 negatively regulates the p38 MAPK pathway. (A) Life spans of wild-type N2 and riok-1(gk1101) worm knockdown with pmk-1 RNAi were both shortened. (B) MAPK (pmk-1) mutants, (C) MAPKK (sek-1) mutants, and (D) MAPKKK (nsy-1) gain-of-function mutants with riok-1 knockdown using RNAi showed similar life spans when compared with those without RNAi knockdown under A. dhakensis infection. (E)A. dhakensis infection and RNAi-mediated riok-1 knockdown induced p38 phosphorylation as shown in a Western blot analysis. (F–J) Increasing expression of p38 MAPK downstream genes C29F3.7 (F), C32H11.12 (G), F08G5.6 (H), K08D8.5 (I), and T24B8.5 (J) as determined using qRT-PCR in worms with riok-1 knockdown mediated by RNAi under A. dhakensis infection. For the unprocessed data of western blot analysis in (E), please see the Figure S6 in Supplementary Material. ***P < 0.001, **P < 0.01, and *P < 0.05.
RIOK-1 Feeds Back Negatively to the p38 MAPK Pathway Through Transcriptional Factor skn-1 After A. dhakensis Infection in C. elegans
Previous studies have shown that p38 MAPK downstream gene skn-1, a transcription factor that is able to bind to the promoter region of riok-1, promotes riok-1 expression in the intestines of C. elegans (32). SKN-1 has also been proven to be an important component in the intestinal immune system of worms (37). Our results were comparable with a previous finding indicating that riok-1 expression is affected by the p38 MAPK downstream transcription factor skn-1 (32). Worms with a suppressed riok-1 expression had more resistance after A. dhakensis infection in contrast to those treated with skn-1 RNAi. By contrast, the enhancement of resistance to A. dhakensis infection mediated by riok-1 knockdown was decreased once skn-1 was suppressed (Figure 5A). Taken together, these results suggest that skn-1, the p38 MAPK downstream gene, is also epistatic to riok-1. Consistent with the findings reported by Mendes et al. (32), the intestinal florescence in riok-1 transcriptional reporter worms was decreased when the worms were treated with skn-1 RNAi (Figure 5C), in contrast to the RNAi control with the RNAi empty vector L4440 (Figure 5B). The expression of riok-1 in the transcriptional reporter worms co-treated with skn-1 RNAi and A. dhakensis was similar to those treated with skn-1 RNAi alone (Figure 5E) and was lower than that found in the control (Figure 5D). The quantification of florescence intensity in riok-1 transcriptional reporter worms under different conditions is summarized (Figure 5F). In addition, the expression level of riok-1 in the p38 MAPK nsy-1(ums8) gain-of-function mutants was similar to those in the wild-type N2 worms and was not increased additionally by A. dhakensis in the nsy-1(ums8) worms (Figure 5G). The results also indicated that A. dhakensis could not induce riok-1 expression directly without passing through the p38 MAPK-signaling pathway. In addition, the riok-1 expression level induced by A. dhakensis AAK1 infection in the wild-type N2 worms was significantly higher than those in the pmk-1(km25) mutants, suggesting the coexistence of another pathway that is independent of p38 MAPK-regulated activation of riok-1 expression (Figure 5H). Briefly, the results show that A. dhakensis-induced riok-1 expression occurs partially through the p38 MAPK pathway. The activation of the pathway is skn-1-dependent, and riok-1 suppresses immunity by targeting the p38 MAPK pathway, participating in a feedback circuit to regulate the immune system in C. elegans.
Figure 5
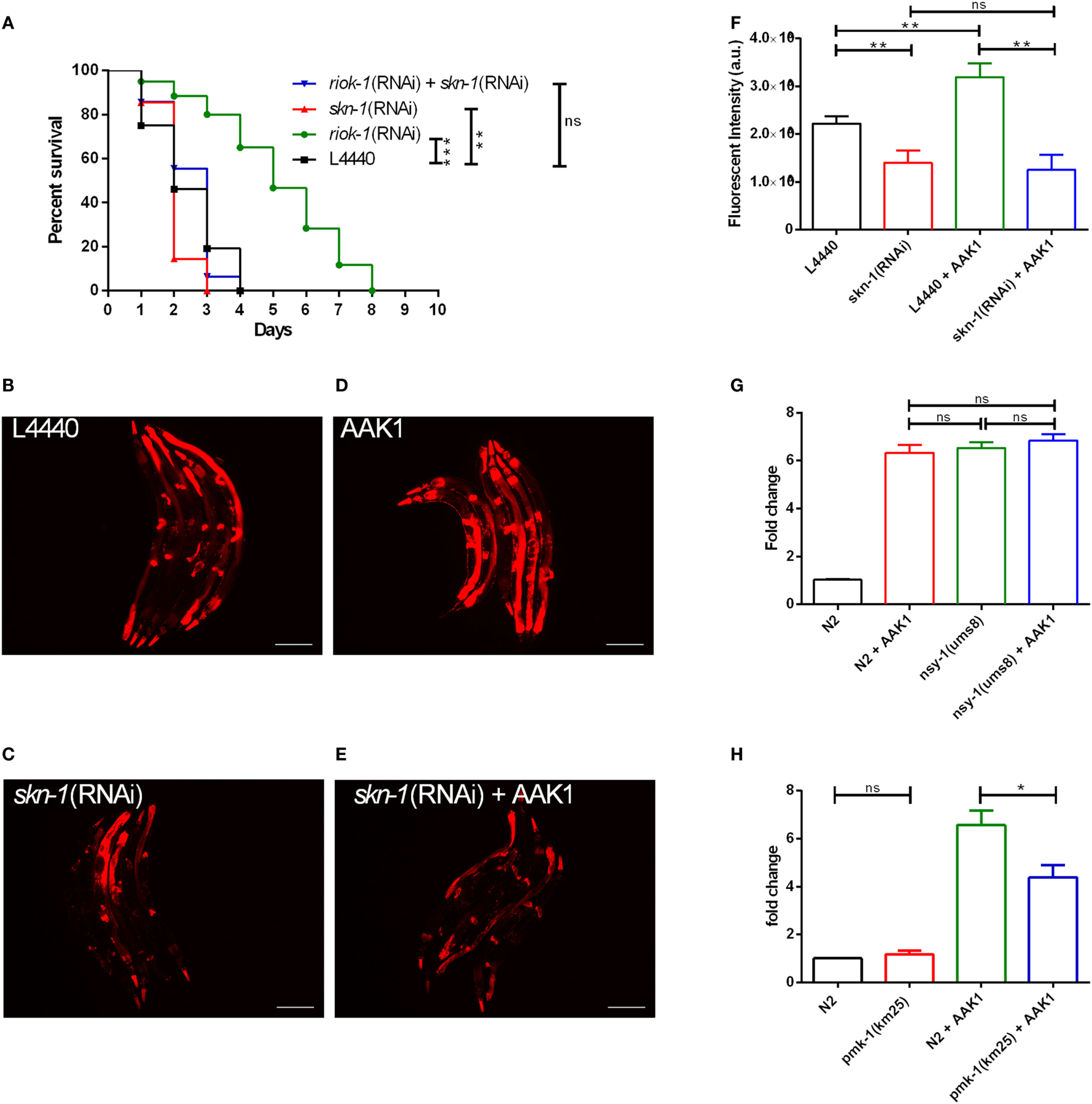
The increase in riok-1 is skn-1-dependent in Caenorhabditis elegans with Aeromonas dhakensis infection. (A) Single knockdown of skn-1 or simultaneous knockdown of skn-1 and riok-1 resulted in a decrease in the life span in C. elegans with A. dhakensis infection. (B)riok-1 expression in riok-1 transcriptional reporter worms with RNAi control vector L4440. (C)riok-1 expression in riok-1 transcriptional reporter worms with RNAi knockdown of skn-1. (D)riok-1 expression in riok-1 transcriptional reporter worms with A. dhakensis AAK1 infection. (E)riok-1 expression in riok-1 transcriptional reporter worms with RNAi knockdown of skn-1 and A. dhakensis AAK1 infection. (F) A summary of the florescence quantification of (B–E). (G) The expression level of riok-1 as determined using quantitative real time-PCR (qRT-PCR) in the p38 gain-of-function-mutant worms nsy-1(ums8) with and without A. dhakensis AAK1 infection was similar to those in wild-type N2 worms with A. dhakensis AAK1 infection. (H) The expression levels of riok-1 as determined using qRT-PCR in the p38 MAPK/pmk-1-mutant worms pmk-1(km25) with A. dhakensis AAK1 infection were lower than in wild-type N2 worms with A. dhakensis AAK1 infection. The scale bars in (B–E) are all 100 µm. ***P < 0.001, **P < 0.01, and *P < 0.05.
RIOK-1 Deficiency in C. elegans Increases the Susceptibility to Different Gram-Negative Bacterial Infections
Our study proved riok-1 to be a negative regulator of the p38 MAPK immune pathway in C. elegans. In order to understand whether the riok-1-p38 MAPK pathway is a universal mechanism against bacterial infection, resistance to different bacterial species, including Salmonella typhimurium (38, 39), enterohemorrhagic E. coli (26), and P. aeruginosa (23, 40, 41), which have been reported to activate the p38 MAPK pathway in C. elegans, was studied. Our survival analysis showed that riok-1 knockdown mediated by RNAi in C. elegans conferred resistance not only to A. dhakensis AAK1 (Figure 6A) but also to enterohemorrhagic E. coli EDL933 (Figure 6B), P. aeruginosa PA14 (Figure 6C), and S. typhimurium LT2 (Figure 6D). The results demonstrated that the riok-1–p38 MAPK pathway plays an important role in immunity against Gram-negative bacterial infection in C. elegans.
Figure 6
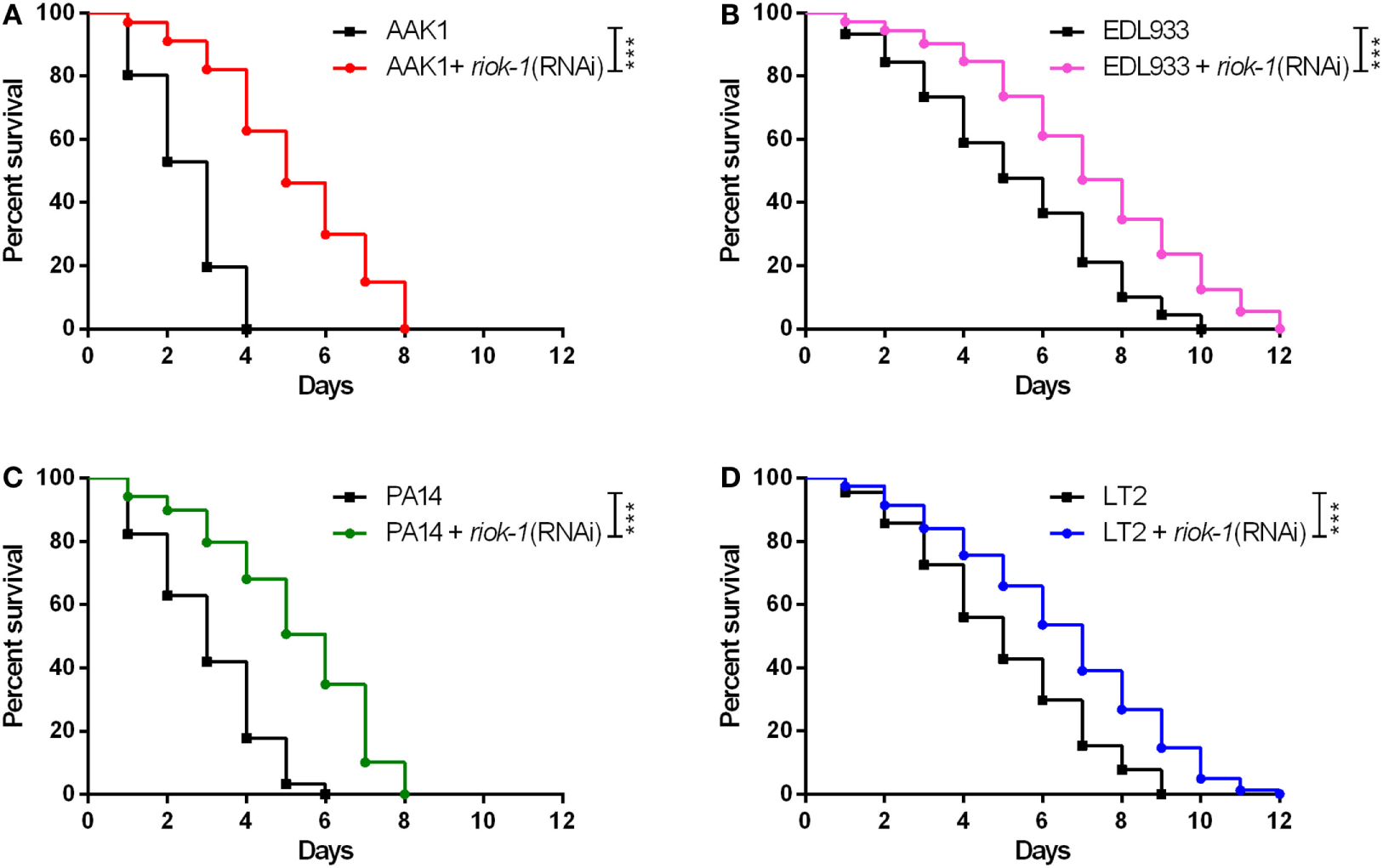
Suppressed expression of riok-1 increases the resistance to pathogens in Caenorhabditis elegans. Knockdown of riok-1 with RNAi in C. elegans confers resistance to (A)Aeromonas dhakensis AAK1, (B) enterohemorrhagic Escherichia coli EDL933, (C)Pseudomonas aeruginosa PA14, and (D)Salmonella typhimurium LT2. The statistical analyses of pathogens with and without riok-1 RNAi are all ***P < 0.001.
According to our results, we propose a negative feedback loop model among p38 MAPK, SKN-1, and RIOK-1 in C. elegans (Figure 7). A. dhakensis infection activates the p38 MAPK pathway in C. elegans. It also induces riok-1 expression by the p38 MAPK pathway through the transcription of skn-1, and riok-1 suppresses the p38 MAPK pathway in a feedback loop in order to maintain immune homeostasis.
Figure 7
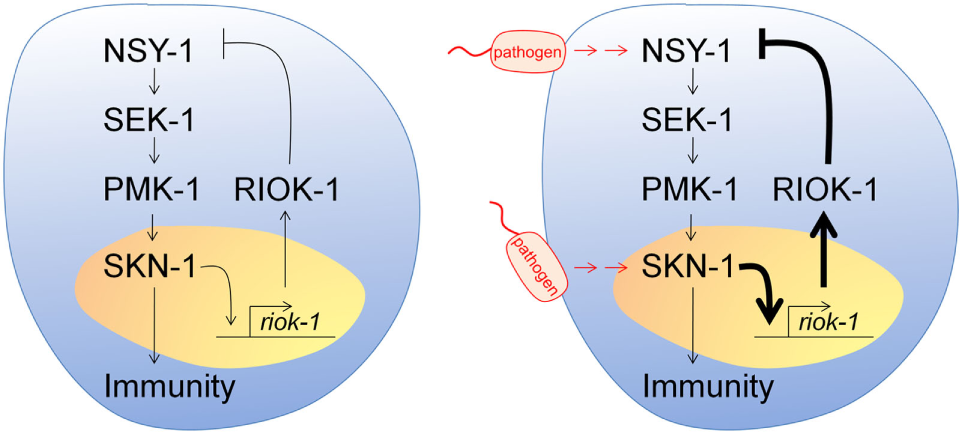
Model of a p38 MAPK, SKN-1, and RIOK-1 feedback loop upon infection in Caenorhabditis elegans. Pathogens activate the p38 MAPK pathway and increase the expression of riok-1 through the transcription of skn-1. RIOK-1 has an important role in a feedback loop that regulates the expression of the p38 MAPK pathway in order to maintain immune homeostasis.
Discussion
In this study, we screened the kinases that may participate in a defensive mechanism against Aeromonas infection in a focused RNAi kinome library in C. elegans (8, 9, 11). RIOK-1 as a negative regulator of immunity against infection conferred resistance to Aeromonas in C. elegans when it was suppressed. The resistant group may include those acting as pathogen-dependent receptors or suppressors of host immunity, such as riok-1. By contrast, candidate genes might be associated with enhancers of immunity against infection and will make worms hypersensitive to infection when their expression is suppressed (Table 1B).
From an evolutionary perspective, immune homeostasis is an important survival strategy, particularly for creatures such as bacterivorous nematodes that live in environments rich with microbes that must differentiate pathogens from potential food sources. Improper activation of immune responses in humans underlies many disorders, such as inflammatory bowel disease, cancer, and autoimmune diseases (3, 42). A mechanism of feedback control for a hyperactivated immune pathway has been discovered in flies and C. elegans (4, 7, 43). Here, we discovered that riok-1 is a novel immune suppresser of the infection-activated p38 MAPK pathway in C. elegans and plays an important role in achieving immune homeostasis.
A previous study showed that the aberrant activation of p38 MAPK-dependent immune responses is toxic to C. elegans (23). Feedback control of hyperactivation of the p38 immune pathway to avoid negative physiological consequences has been reported. For example, MEMO-1 was complexed with RHO-1/RhoA/GTPase and loss of memo-1 resulted in enhanced interaction of RHO-1 with BLI-3/NADPH oxidase, thereby stimulating ROS production that signals the p38 MAPK pathway to promote stress resistance and longevity (4). In another example, infection was associated with an increase in p38 MAPK/PMK-1 activity (5), which was negatively regulated by MAPK phosphatase VHP-1 (7). However, the knockdown of vhp-1 with RNAi did not increase resistance to A. dhakensis when compared with the control in our study (Figure S5 in Supplementary Material). The results suggested that the p38 MAPK-negative regulator vhp-1, although important in P. aeruginosa PA14 infection (7), may not participate in resistance against A. dhakensis infection in C. elegans. Herein, we identified riok-1, which regulates the p38 MAPK pathway in a pattern distinguished from memo-1 and vhp-1. However, the exact mechanism through which riok-1 functions as a suppressor of the p38 MAPK immune pathway remains unclear and requires further study.
The expression sites of riok-1 suggest where riok-1 regulates immunity against infection. Our research demonstrated that riok-1 is expressed in neurons and in the intestines. Previous studies have shown that neurons can modulate the innate immune response of intestine cells in response to bacterial infection in C. elegans (44). The neuronal products modulated the daf-2/daf-16 insulin-like pathway in the intestines of C. elegans. By contrast, no epistatic relationship between riok-1 and daf-2 was discovered in our study (Figure S4A in Supplementary Material). In addition, the knockdown of skn-1 decreased the expression of riok-1 predominately in the intestine (Figure 5C), indicating that the p38 MAPK–skn-1 immune pathway is not operative in neurons.
In summary, we discovered that riok-1 is a novel innate immune suppressor and proposed a negative feedback model among the p38 MAPK, SKN-1, and RIOK-1 loop in C. elegans in order to maintain immune homeostasis. Further study of RIOK-1 may bring new insights into defects in the p38 MAPK pathway underlying human diseases and may reveal new therapeutic opportunities for bacterial infection.
Statements
Author contributions
P-LC, Y-WC, W-CK, and C-SC conceived and designed the experiments. Y-WC performed all the experiments. Y-WC, P-LC, and C-SC analyzed the data. Y-WC, C-SC, and P-LC wrote the paper. C-SC, W-CK, and P-LC reviewed and edited the paper.
Funding
This study was partially supported by grants from the Ministry of Science and Technology of Taiwan (MOST 105-2628-B-006-017-MY3), the Ministry of Health and Welfare (MOHW106-TDU-B-211-113003), and the National Cheng Kung University Hospital (NCKUH-10605020) to P-LC and MOST 106-2321-B-006-005 to C-SC.
Acknowledgments
We are grateful for the assistance from the C. elegans core facility Taiwan and comments and helpful discussions from the Taiwan C. elegans research community. We also acknowledge the Caenorhabditis Genetics Center (CGC) and the WormBase. We also thank Miranda Loney and Savana Moore for editing the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at https://www.frontiersin.org/articles/10.3389/fimmu.2018.00774/full#supplementary-material.
References
1
Peterson LW Artis D . Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol (2014) 14(3):141–53.10.1038/nri3608
2
Xavier RJ Podolsky DK . Unravelling the pathogenesis of inflammatory bowel disease. Nature (2007) 448(7152):427–34.10.1038/nature06005
3
Kyriakis JM Avruch J . Mammalian MAPK signal transduction pathways activated by stress and inflammation: a 10-year update. Physiol Rev (2012) 92(2):689–737.10.1152/physrev.00028.2011
4
Ewald CY Hourihan JM Bland MS Obieglo C Katic I Moronetti Mazzeo LE et al NADPH oxidase-mediated redox signaling promotes oxidative stress resistance and longevity through memo-1 in C. elegans. Elife (2017) 6:e19493.10.7554/eLife.19493
5
Kim DH Feinbaum R Alloing G Emerson FE Garsin DA Inoue H et al A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science (2002) 297(5581):623–6.10.1126/science.1073759
6
Xu A Shi G Liu F Ge B . Caenorhabditis elegans mom-4 is required for the activation of the p38 MAPK signaling pathway in the response to Pseudomonas aeruginosa infection. Protein Cell (2013) 4(1):53–61.10.1007/s13238-012-2080-z
7
Kim DH Liberati NT Mizuno T Inoue H Hisamoto N Matsumoto K et al Integration of Caenorhabditis elegans MAPK pathways mediating immunity and stress resistance by MEK-1 MAPK kinase and VHP-1 MAPK phosphatase. Proc Natl Acad Sci U S A (2004) 101(30):10990–4.10.1073/pnas.0403546101
8
Manning G . Genomic overview of protein kinases. WormBook (2005):1–19.10.1895/wormbook.1.60.1
9
Manning G Plowman GD Hunter T Sudarsanam S . Evolution of protein kinase signaling from yeast to man. Trends Biochem Sci (2002) 27(10):514–20.10.1016/S0968-0004(02)02179-5
10
Lehmann S Bass JJ Szewczyk NJ . Knockdown of the C. elegans kinome identifies kinases required for normal protein homeostasis, mitochondrial network structure, and sarcomere structure in muscle. Cell Commun Signal (2013) 11(71):17.10.1186/1478-811X-11-71
11
Plowman GD Sudarsanam S Bingham J Whyte D Hunter T . The protein kinases of Caenorhabditis elegans: a model for signal transduction in multicellular organisms. Proc Natl Acad Sci U S A (1999) 96(24):13603–10.10.1073/pnas.96.24.13603
12
Kim D . Studying host–pathogen interactions and innate immunity in Caenorhabditis elegans. Dis Model Mech (2008) 1(4–5):205–8.10.1242/dmm.000265
13
Marsh EK May RC . Caenorhabditis elegans, a model organism for investigating immunity. Appl Environ Microbiol (2012) 78(7):2075–81.10.1128/AEM.07486-11
14
Chen PL Wu CJ Tsai PJ Tang HJ Chuang YC Lee NY et al Virulence diversity among bacteremic Aeromonas isolates: ex vivo, animal, and clinical evidences. PLoS One (2014) 9(11):e111213.10.1371/journal.pone.0111213
15
Chen PL Chen YW Ou CC Lee TM Wu CJ Ko WC et al A disease model of muscle necrosis caused by Aeromonas dhakensis infection in Caenorhabditis elegans. Front Microbiol (2016) 7:2058.10.3389/fmicb.2016.02058
16
LaRonde-LeBlanc N Wlodawer A . The RIO kinases: an atypical protein kinase family required for ribosome biogenesis and cell cycle progression. Biochim Biophys Acta (2005) 1754(1–2):14–24.10.1016/j.bbapap.2005.07.037
17
Laronde-Leblanc N Guszczynski T Copeland T Wlodawer A . Structure and activity of the atypical serine kinase Rio1. FEBS J (2005) 272(14):3698–713.10.1111/j.1742-4658.2005.04796.x
18
Widmann B Wandrey F Badertscher L Wyler E Pfannstiel J Zemp I et al The kinase activity of human Rio1 is required for final steps of cytoplasmic maturation of 40S subunits. Mol Biol Cell (2012) 23(1):22–35.10.1091/mbc.E11-07-0639
19
Ferreira-Cerca S Kiburu I Thomson E LaRonde N Hurt E . Dominant Rio1 kinase/ATPase catalytic mutant induces trapping of late pre-40S biogenesis factors in 80S-like ribosomes. Nucleic Acids Res (2014) 42(13):8635–47.10.1093/nar/gku542
20
Turowski TW Lebaron S Zhang E Peil L Dudnakova T Petfalski E et al Rio1 mediates ATP-dependent final maturation of 40S ribosomal subunits. Nucleic Acids Res (2014) 42(19):12189–99.10.1093/nar/gku878
21
Iacovella MG Golfieri C Massari LF Busnelli S Pagliuca C Dal Maschio M et al Rio1 promotes rDNA stability and downregulates RNA polymerase I to ensure rDNA segregation. Nat Commun (2015) 6:6643.10.1038/ncomms7643
22
Read RD Fenton TR Gomez GG Wykosky J Vandenberg SR Babic I et al A kinome-wide RNAi screen in Drosophila glia reveals that the RIO kinases mediate cell proliferation and survival through TORC2-Akt signaling in glioblastoma. PLoS Genet (2013) 9(2):e1003253.10.1371/journal.pgen.1003253
23
Cheesman HK Feinbaum RL Thekkiniath J Dowen RH Conery AL Pukkila-Worley R . Aberrant activation of p38 MAP kinase-dependent innate immune responses is toxic to Caenorhabditis elegans. G3 (Bethesda) (2016) 6:541–9.10.1534/g3.115.025650/-/DC1
24
Wu CJ Wang HC Chen CS Shu HY Kao AW Chen PL et al Genome sequence of a novel human pathogen, Aeromonas aquariorum. J Bacteriol (2012) 194(15):4114–5.10.1128/JB.00621-12
25
Kamath RS Fraser AG Dong Y Poulin G Durbin R Gotta M et al Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature (2003) 421(6920):231–7.10.1038/nature01278
26
Chou TC Chiu HC Kuo CJ Wu CM Syu WJ Chiu WT et al Enterohaemorrhagic Escherichia coli O157:H7 Shiga-like toxin 1 is required for full pathogenicity and activation of the p38 mitogen-activated protein kinase pathway in Caenorhabditis elegans. Cell Microbiol (2013) 15(1):82–97.10.1111/cmi.12030
27
Narbonne P Roy R . Inhibition of germline proliferation during C. elegans dauer development requires PTEN, LKB1 and AMPK signalling. Development (2006) 133(4):611–9.10.1242/dev.02232
28
Ren M Feng H Fu Y Land M Rubin CS . Protein kinase D is an essential regulator of C. elegans innate immunity. Immunity (2009) 30(4):521–32.10.1016/j.immuni.2009.03.007
29
Hoeven R McCallum KC Cruz MR Garsin DA . Ce-Duox1/BLI-3 generated reactive oxygen species trigger protective SKN-1 activity via p38 MAPK signaling during infection in C. elegans. PLoS Pathog (2011) 7(12):e1002453.10.1371/journal.ppat.1002453
30
Feinbaum RL Urbach JM Liberati NT Djonovic S Adonizio A Carvunis AR et al Genome-wide identification of Pseudomonas aeruginosa virulence-related genes using a Caenorhabditis elegans infection model. PLoS Pathog (2012) 8(7):e1002813.10.1371/journal.ppat.1002813
31
Weinberg F Schulze E Fatouros C Schmidt E Baumeister R Brummer T . Expression pattern and first functional characterization of riok-1 in Caenorhabditis elegans. Gene Expr Patterns (2014) 15(2):124–34.10.1016/j.gep.2014.05.005
32
Mendes TK Novakovic S Raymant G Bertram SE Esmaillie R Nadarajan S et al Investigating the role of RIO protein kinases in Caenorhabditis elegans. PLoS One (2015) 10(2):e0117444.10.1371/journal.pone.0117444
33
Zhuang JJ Hunter CP . Tissue specificity of Caenorhabditis elegans enhanced RNA interference mutants. Genetics (2011) 188(1):235–7.10.1534/genetics.111.127209
34
Alper S McElwee MK Apfeld J Lackford B Freedman JH Schwartz DA . The Caenorhabditis elegans germ line regulates distinct signaling pathways to control lifespan and innate immunity. J Biol Chem (2010) 285(3):1822–8.10.1074/jbc.M109.057323
35
Berman JR Kenyon C . Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell (2006) 124(5):1055–68.10.1016/j.cell.2006.01.039
36
TeKippe M Aballay A . C. elegans germline-deficient mutants respond to pathogen infection using shared and distinct mechanisms. PLoS One (2010) 5(7):e11777.10.1371/journal.pone.0011777
37
Inoue H Hisamoto N An JH Oliveira RP Nishida E Blackwell TK et al The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev (2005) 19(19):2278–83.10.1101/gad.1324805
38
Aballay A Drenkard E Hilbun LR Ausubel FM . Caenorhabditis elegans innate immune response triggered by Salmonella enterica requires intact LPS and is mediated by a MAPK signaling pathway. Curr Biol (2003) 13(1):47–52.10.1016/S0960-9822(02)01396-9
39
Tenor JL McCormick BA Ausubel FM Aballay A . Caenorhabditis elegans-based screen identifies Salmonella virulence factors required for conserved host-pathogen interactions. Curr Biol (2004) 14(11):1018–24.10.1016/j.cub.2004.05.050
40
Shivers RP Pagano DJ Kooistra T Richardson CE Reddy KC Whitney JK et al Phosphorylation of the conserved transcription factor ATF-7 by PMK-1 p38 MAPK regulates innate immunity in Caenorhabditis elegans. PLoS Genet (2010) 6(4):e1000892.10.1371/journal.pgen.1000892
41
Block DH Twumasi-Boateng K Kang HS Carlisle JA Hanganu A Lai TY et al The developmental intestinal regulator ELT-2 controls p38-dependent immune responses in adult C. elegans. PLoS Genet (2015) 11(5):e1005265.10.1371/journal.pgen.1005265
42
Waetzig GH Seegert D Rosenstiel P Nikolaus S Schreiber S . p38 mitogen-activated protein kinase is activated and linked to TNF-alpha signaling in inflammatory bowel disease. J Immunol (2002) 168(10):5342–51.10.4049/jimmunol.168.10.5342
43
Aggarwal K Silverman N . Positive and negative regulation of the Drosophila immune response. BMB Rep (2008) 41(4):267–77.10.5483/BMBRep.2008.41.4.267
44
Kao CY Los FC Aroian RV . Nervous about immunity: neuronal signals control innate immune system. Nat Immunol (2008) 9(12):1329–30.10.1038/ni1208-1329
Summary
Keywords
riok-1 , p38 MAPK/pmk-1, skn-1 , innate immunity, immune suppressor, Aeromonas dhakensis , Caenorhabditis elegans
Citation
Chen Y-W, Ko W-C, Chen C-S and Chen P-L (2018) RIOK-1 Is a Suppressor of the p38 MAPK Innate Immune Pathway in Caenorhabditis elegans. Front. Immunol. 9:774. doi: 10.3389/fimmu.2018.00774
Received
28 November 2017
Accepted
28 March 2018
Published
17 April 2018
Volume
9 - 2018
Edited by
Janice C. Telfer, University of Massachusetts Amherst, United States
Reviewed by
Gert Jansen, Erasmus University Rotterdam, Netherlands; Hai-Peng Liu, Xiamen University, China
Updates
Copyright
© 2018 Chen, Ko, Chen and Chen.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chang-Shi Chen, cschen@mail.ncku.edu.tw; Po-Lin Chen, cplin@mail.ncku.edu.tw
Specialty section: This article was submitted to Comparative Immunology, a section of the journal Frontiers in Immunology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.