- Animal Health Research Center (CISA-INIA), Madrid, Spain
Most ligands and receptors from the tumor necrosis factor (TNF) superfamily play very important roles in the immune system. In particular, many of these molecules are essential in the regulation of B cell biology and B cell-mediated immune responses. Hence, in mammals, it is known that many TNF family members play a key role on B cell development, maturation, homeostasis, activation, and differentiation, also influencing the ability of B cells to present antigens or act as regulators of immune responses. Evolutionarily, jawed fish (including cartilaginous and bony fish) constitute the first animal group in which an adaptive immune response based on B cells and immunoglobulins is present. However, until recently, not much was known about the expression of TNF ligands and receptors in these species. The sequences of many members of the TNF superfamily have been recently identified in different species of jawed fish, thus allowing posterior analysis on the role that these ligands and receptors have on B cell functionality. In this review, we summarize the current knowledge on the impact that the TNF family members have in different aspects of B cell functionality in fish, also providing an in depth comparison with functional aspects of TNF members in mammals, that will permit a further understanding of how B cell functionality is regulated in these distant animal groups.
Introduction
In mammals, the tumor necrosis factor (TNF) ligand superfamily (TNFSF) signal through members of the tumour necrosis factor receptor superfamily (TNFRSF) to activate signaling pathways which play biological roles in development, organogenesis, cell death, and survival. Since the discovery of the first TNFSF members, TNF (1) and lymphotoxin α (Ltα) (2), more than 40 years ago, 17 additional TNFSF members and 29 cognate receptors have been identified in humans (3). With the completion of the large-scale sequencing of the human and mouse genomes, it is assumed that almost all TNFSF and TNFRSF members have now been identified in mammals (3, 4). Thus, ligands within the TNFSF which include 4-1BBL, a proliferation-inducing ligand (APRIL), B cell-activating factor of the TNF family (BAFF), CD27L, CD30L, CD40L, EDA1, EDA2, Fas ligand (FasL), GITRL, LIGHT, Ltα, Ltβ, OX40L, RANK ligand, TL1A, TNF, TNF-like weak inducer of apoptosis and proliferation, and TNF-related apoptosis-inducing ligand (TRAIL) are key effector proteins in the orchestration of innate and adaptive immune responses [reviewed in Ref. (3) and summarized in Table 1].
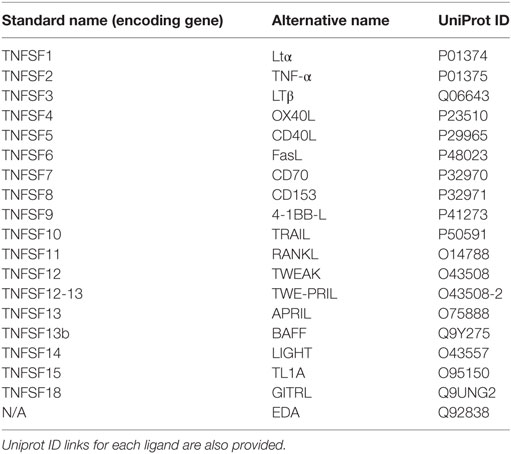
Table 1. Relation of TNF superfamily ligands (TNFSF) present in human indicating their standard name within the TNF superfamily and their alternative (most common) name.
TNFSF ligands are type II membrane-bound proteins with an intracellular N terminal and an extracellular C terminal domain. Among the 19 known ligands described in human and mouse, 11 encode for a proteolytic cleavage site that generates biologically active soluble forms (5). Within the C terminus, they contain the TNF homology domain (THD), which presents a weak degree of conservation (20–30%) between ligand members, and is typically formed by 10 β-strands (6). The THD folds into what has been called an antiparallel β-sandwich (3) exhibiting a compact jellyroll topology (4). The THD structure has the ability to assemble each TNF ligand into conical trimers which allow the ligands to bind respective receptors to initiate signaling (6).
To date, 29 TNFRs have been identified in mammals, namely, 4-1BB, BAFFR, B cell maturation antigen (BCMA), CD27, CD30, CD40, decoy receptor 3 (DcR3), death receptor 3, DR6, EDAR, Fas, Fn14, glucocorticoid-induced TNFR, herpesvirus entry mediator (HVEM), LTbR, OPG, OX40, RANK, receptor expressed in lymphoid tissues (RELT), transmembrane activator and calcium-modulating cyclophilin ligand interactor (TACI), TNFR1, TNFR2, TRAILR1, TRAILR2, TRAILR3, TRAILR4, TROY, and XEDAR [reviewed in Ref. (7)]. The main feature of these TNFRs is a cysteine-rich domain (CRD) formed of three disulfide bonds surrounding a core motif of CXXCXXC creating an elongated molecule. There is an important variation in the number of CRDs among family members, from BAFFR or BCMA containing only one CRD to CD30 containing six CRDs. These receptors are type I membrane proteins, with the exceptions of BAFFR, BCMA, TACI, and XEDAR, which are type III membrane proteins, and OPG and DcR3, which are secreted (4, 8). The determination of the X-ray crystal structures of some ligands bound to the extracellular domain of their receptors has been primordial to characterize the binding mechanisms between them (9, 10). These analyses have revealed that those receptors with several CRDs adopt an elongated structure and bind at the interface between two ligand monomers, whereas single CRD receptors are more compact and contact a single ligand monomer in a trimeric ligand (5, 11, 12). Generally, one trimeric ligand engages three monomeric receptors, a key event for the activation of intracellular signaling pathways. By conducting a systematic flow cytometry-based assay, Bossen et al. elegantly demonstrated the mechanisms that regulate TNFSF–TNFRSF interactions in mouse and human (8). They found that TNFSF ligands bound from one to five different receptors, while most receptors bound from one to three ligands. Strikingly, they observed that, although containing the classical CRD structure, DR6, RELT, TROY, and nerve growth factor receptor (NGFR) did not bind to any of the TNFSF, thus suggesting that they either bind to other ligands or function in a ligand-independent manner. In this sense, it was later shown that although NGFR has the classic CRD structure, it binds a structurally different kind of ligands, the neurotrophins (13). This information regarding the different ligands that signal through each receptor in mammals is summarized in Table 2.
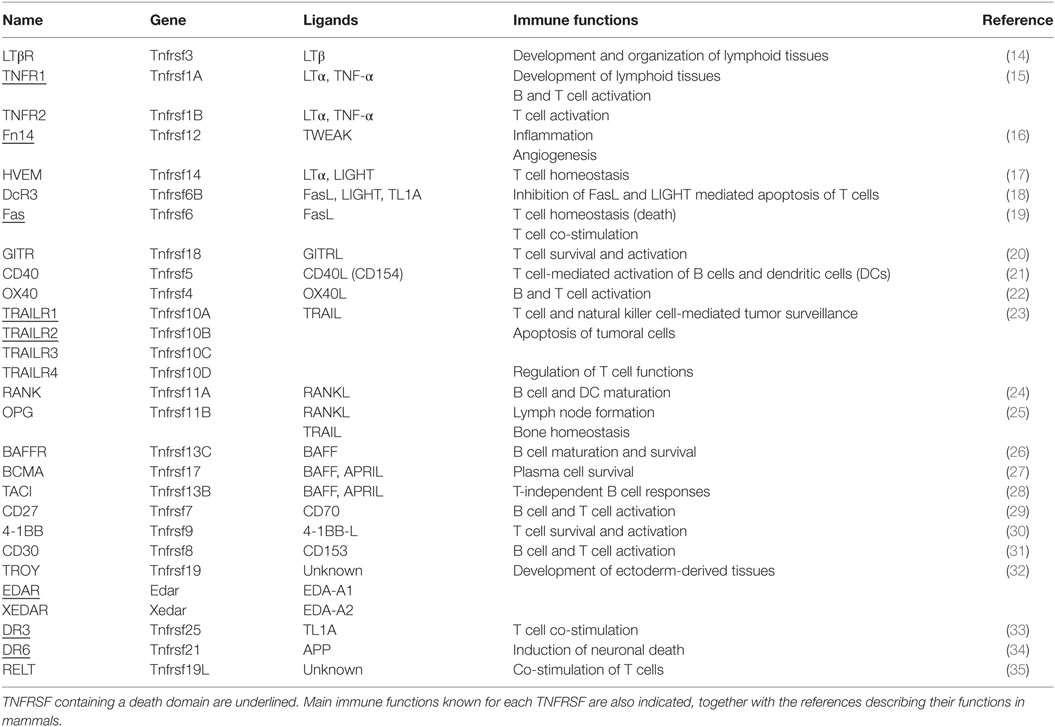
Table 2. Functional relation of the tumour necrosis factor receptor superfamily (TNFRSF) and their cognate ligands (TNFSF).
Evolution of TNFSF and TNFRSF Members
The appearance and further specialization of the adaptive immune response is a hallmark of the successful evolution of vertebrates. The adaptive immune system is based on the presence of recombination-activating gene (RAG)-recombined B cell receptors (BCR) and T cell receptors (TCR) on the surface of B cells and T cells, respectively, and the major histocompatibility complex (MHC). Molecular studies have shown that adaptive immunity arose early on vertebrate evolution, between the divergences of cyclostomes (lampreys) and cartilaginous fish, around 450 million years ago, by diversification and recombination of gene clusters on a span of time of 20 million years (36). This event is known as the “big bang” theory of the appearance of the adaptive immune response, since it occurred in the very short period of time in which jawed fish appeared and is thought to be linked to genome duplication events (37). Thus, jawed fish were the most ancient animal group where all these elements were found, while jawless fish (Agnathans) seemed to have none of them (38). However, the posterior discovery of a lymphoid cell-based adaptive immune system in Agnathans, in which immune receptors recombined in a similar way to that of the BCR (39), pushed the origin of the adaptive immune response earlier in evolution. Despite this, the “big bang” theory for the origin and development of acquire immunity still prevails.
Interestingly, Collette et al. postulated that the divergence of the TNFSF and TNFRSF members parallels the emergence of the adaptive immune response (7). Since 11 out of the 19 human TNFSF members are clustered within the MHC and paralogous regions on chromosomes 1, 6, 9, and 19, the authors suggest that this disposition might be a consequence of the ancestral arrangement of a proto-TNFSF cluster, before en bloc duplication of the proto-MHC region in a vertebrate ancestor 500–800 million years ago (40). Remarkably, the different number of TNFSF and TNFRSF members found in the different species within vertebrates correlates with the number of rounds of genome duplication during evolution (4), thus supporting the idea that genome duplication created paralogous clusters. This hypothesis is further supported by phylogenetic analysis that indicates an ancient evolutionary origin of TNFSF ligand and receptor genes that precedes the appearance of vertebrates (7, 41).
As many invertebrate and vertebrate genomes are now available, the discovery of TNFSF and TNFRSF orthologs and paralogs has greatly increased. Consequently, recent phylogenetic studies on invertebrate TNFSF ligands and receptors have been key to better understand the appearance of TNF molecules in metazoans and to further support the hypothesis of their divergent evolution [reviewed in Ref. (4)]. The most primitive TNF superfamily member that has been functionally characterized is Eiger, a TNFSF homolog found on the fruit fly (Drosophila melanogaster) (42). Eiger binds to a cognate TNFRSF member, called Wengen, which contains an intracellular death domain, thus inducing cell death through the activation of signaling pathways similar to those activated by mammalian TNFRSFs (43, 44). A TNFSF member named MjTNF has also been characterized in another invertebrate, the marine arthropod kuruma shrimp (Marsupenaeus japonicas) (45). This protein contains a predicted transmembrane region and a THD, and shares 30.7% sequence identity with Drosophila Eiger. Two molluscan TNFSF members containing transmembrane regions and THDs were identified in the disk abalone, Haliotis discus discus. One was designated AbTNF-α (46) and the other AbFas ligand (47). Within the genome of the equinoderm purple sea urchin (Strongylocentrotus purpuratus), four different TNFSF genes have been found, identified as potential gene orthologs of TNFSF14 (LIGHT), TNFSF15 (TL1A), and two separate genes resembling EDA (48, 49). In parallel, several TNF superfamily receptors were also identified on these invertebrate organisms [summarized in Ref. (4)]. As most of the invertebrate TNFSF members are constitutively expressed and most TNFRSFs are phylogenetically related to EDAR, a major role in development and organogenesis with restricted immunoregulatory properties is foreseen in these organisms.
By contrast, in teleost fish, the first animal group comprising all the elements of the adaptive arm of the immune system, a diversification of the TNFSF and TNFRSF has occurred. To date, 13 different TNFSF and 13 TNFRSF homologs have been identified, together with new members of the TNFSF that are novel to this animal group (4, 41, 50–54) supporting the hypothesis of a diversification of the TNF superfamily with the appearance of the adaptive immune response. Interestingly, recent studies in sarcopterygian fish (African lungfish) revealed a different TNFSF and TNFRSF gene pattern to that seen in teleost (55). African lungfish is an extant representative of the closest ancestral lineage to all tetrapods that presents organized lymphoid structures which cannot be found in other fish species (56). Several TNFSF were reported in this study, but additional analyses are needed to further characterize their identity and functionality. In parallel, many TNFRSF members were identified, and although most of them were present in both teleost and lungfish, only the latter presented homologue sequences for the TNF receptors HVEM (TNFRSF14), 4-1BB (TNFRSF9), and OPG (TNFRSF11B) (55). These TNF superfamily members have been shown to regulate T cell homeostasis and activation, as well as development of lymphoid structures, such as lymph nodes (LNs) (15, 17, 25, 30, 31). These results suggest that an expansion of TNF molecules could have occurred in the African lungfish, conferring a phenotypical advantage that was positively selected, thus leading to the appearance of organized lymphoid structures, which might play a key role on T cell activation. Therefore, throughout the evolution of vertebrates, the expansion of TNFSF and TNFRSF has led to the appearance of new members involved in the regulation of novel immune functions (4, 57), which were coopted under selective pressure, being this crucial for the evolution of the adaptive immune system (7, 41).
Role of TNFSF Ligands on B Cell Regulation
In mammals, many TNFSF ligands have been shown to play essential roles on the regulation of the functionality of B cells. These TNFSF ligands may influence all aspects of B cell biology from development, maturation, survival, proliferation, activation, and differentiation [reviewed in Ref. (58)] (Figure 1), thus playing a fundamental role on B cell-mediated immune responses.
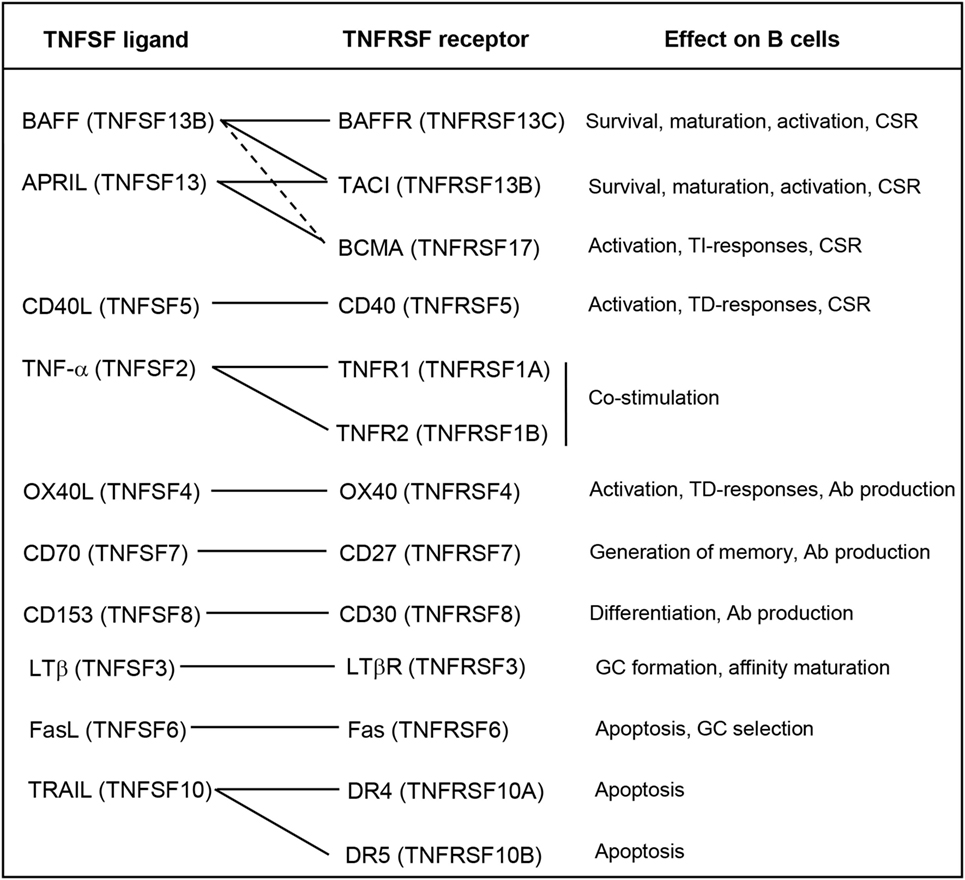
Figure 1. Roles of the TNFSF on B cell functions. Known interactions between ligands and receptors are indicated with solid lines, or dashed lines if the interactions are of weak affinity. Abbreviations: Ab, antibody; CSR, class-switch recombination; GC, germinal center; TD, thymus dependent; TI, thymus independent.
Among TNFSF members, BAFF (TNFSF13B) and APRIL (TNFSF13) are probably the cytokines that seem to play a prevailing role on the regulation of B cell activity [reviewed in Ref. (59)]. These two cytokines exist as membrane-bound and soluble forms, being both forms biologically active (60). Both BAFF and APRIL bind to and signal through BCMA (TNFRSF17) and TACI (TNFRSF13B), whereas BAFF also binds to BAFFR (TNFRSF13C). These receptors are mainly expressed in B cells, and their specific activation leads to different outcomes of the B cell response (61). BAFF-mediated survival signals through BAFFR are necessary for immature B cells to become mature circulating B cells and for peripheral B cell survival (62). These signals regulate the size of the B cell compartment, especially that of conventional B2 cells, since the absence of BAFF does not affect the maturation or survival of innate-like B cells, such as marginal zone (MZ) or B1 cells (63). In fact, there is some evidence to suggest that the maintenance of the B1 B cell compartment is controlled by APRIL signaling through TACI (64, 65), which is highly expressed on the surface MZ B cells and B1 cells (66). In this context, BAFF and APRIL signaling through TACI have been shown to induce class-switch recombination (CSR) in response to thymus-independent (TI) antigens (Ags) (28).
Other TNFSF members can induce CSR on B cells in response to thymus-dependent (TD) Ags; such as, for example, CD40L (TNFSF5) and OX40L (TNFSF4) (21, 22). CD40L is expressed mainly by activated T cells during TD responses, thus mediating the co-stimulation of BCR-activated B cells, which express the receptor CD40 (TNFRSF5), usually within the germinal center (GC). As a consequence, co-stimulated B cells in the GC enhance their proliferation and undergo somatic hypermutation (SHM) to increase their affinity and CSR to switch from producing IgM to producing immunoglobulin isotypes with higher Ag affinity such as IgA, IgE, or IgG [reviewed in Ref. (21)]. In this context, part of these CD40L-induced proliferating B cells also differentiates to antibody (Ab)-secreting plasma cells (PCs), since CD40L and IL-21 synergistically induce the expression of B lymphocyte-induced maturation protein-1 (67), a transcription factor which is the master regulator of terminal differentiation to PCs (68). Moreover, BAFF and APRIL signaling through BAFFR and TACI can contribute to enhance PC differentiation triggered by CD40L (69). On terminally differentiated PCs, signaling through BCMA is highly expressed on PCs and is needed to promote their survival (70). Both BAFF and APRIL can signal through BCMA, although it shows much higher affinity for APRIL than BAFF (71). Concerning OX40L, this cytokine is expressed mainly on activated B cells while its receptor OX40 is expressed on activated CD4+ T cells. Their interaction triggers a bidirectional co-stimulation of both B and T cells during TD responses. Furthermore, cross-linking of OX40L on B cells by OX40 has been shown to greatly enhance B cell proliferation and Ig production (22).
One of the most studied TNFSF members is TNF-α, a well-known pro-inflammatory cytokine able to promote cell death (72). However, TNF-α has also been shown to play an important role on B cell functionality (73). TNF-α binds to two different receptors, TNFR1, which is ubiquitously expressed on almost all cell types, and TNFR2, whose expression is limited to the central nervous system and the immune system, especially found on T cells (15). TNF-α expression is rapidly and strongly upregulated in vitro or in vivo in the presence of many types of Ags or inflammatory mediators (15). In addition, TNF-α is produced by T cells after TCR engagement (74) and by B cells after TI BCR cross-linking and also after CD40 ligation by T cell-derived CD40L (75). In this context, TNF-α provides co-stimulatory signals which increase the proliferation and Ab production of B cells after Ag encounter, being very important for the polyclonal expansion needed within primary responses (15).
After BCR engagement, expression of CD70 (TNFSF7) is also induced on B cells. Ligation of CD70 with its ligand CD27 delivers signals to enhance proliferation, inhibit B cell differentiation to PCs, trigger SHM, and promote the generation of memory B cells (76). However, it has also been shown that ligation of CD70 in the presence of co-stimulatory T cell signals such as CD40L can promote B cell differentiation into Ab-producing PCs (77).
Recent studies have shown that BCR cross-linking increases the sensitivity of B cells to TRAIL (TNFSF10)-mediated cell death. It has been demonstrated that this effect can be reverted by ligation of CD40 on B cells, while B1 cells, which are involved in TI responses showed very high sensitivity to TRAIL-induced death. These data suggested that TRAIL is involved in B cell differentiation and survival at the GC reaction, and in Ab affinity maturation (78). Another member playing a similar role is Fas ligand (FasL) (TNFSF6), which induces apoptosis after ligation of its receptor (Fas) on the surface of the target cell (79). BCR activation induces the expression of Fas on the surface of B cells, making them more susceptible of FasL-mediated apoptosis. During the GC reaction, CD40 ligation protects B cells from Fas-induced apoptosis, thus contributing to the selection of B cells bearing a high-affinity BCR (80). LTβ has also been demonstrated to play an important role in the formation of GCs and also on Ab affinity maturation (81). Finally, CD153 (TNFSF8) also plays a role on B cells since the binding to its receptor (CD30) on T cells modulates B cell differentiation and CSR mediated by reverse signaling induced by CD30+ activated T cells (82).
The Adaptive Immune System in Fish
The adaptive immune system, characterized by an Ag-specific combinatorial immune response (36), first appeared in jawed fish. Thus, evolutionarily, cartilaginous fish (sharks, skates, and rays) are the first animal group in which the adaptive immune system, based on immunoglobulin superfamily members, namely, BCR, TCR and MHC, and RAG 1 and 2 genes are present (38).
Due to the anatomical differences between fish and mammals (i.e., humans), significant differences are found in the distribution and functionality of primary and secondary lymphoid organs, such as the absence of LN or bone marrow (BM) in fish (56, 83). The fish spleen functions as the major secondary lymphoid organ, as it happens in mammals, and since fish lack LN, the spleen has been shown as the most important tissue for Ag trapping (84).
Regarding hematopoiesis, fish do not have a conventional BM as it is described in the mammalian immune system. In cartilaginous fish, the Leydig organ and the epigonal organ are believed to be the equivalents of mammalian BM (85). Both are reticular structures that contain large numbers of immature leukocytes, including neutrophils, eosinophils, and other granulocytes, as well as lymphocyte aggregates with scattered PCs. Either one or both of these tissues have been demonstrated to be present in all cartilaginous species examined (83). The expression of RAG-1 and B-cell-specific transcription factors strongly supports a lymphopoietic role for these tissues (86). In the case of bony fish (teleost), the anterior part of the kidney (head kidney/anterior kidney) has no renal functions and has been shown to assume hematopoietic functions (87). B cell development at the anterior kidney has been proven by the expression of RAG-1/2 (88, 89), TdT (90), and the transcription factor Ikaros (91), and the posterior cellular analysis defining the B cell subsets residing within the kidney (92). As in mammalian BM (93), anterior kidney also stores Ig-secreting long-lived PCs (94).
The thymus is similar to that found in mammals, composed by a cortex and a medulla, and is responsible for the production of T cells (95). TCR cell surface expression has been shown in teleost (96) but remains to be described in cartilaginous fish. However, the expression of all the TCR genes identified in mammals (α, β, δ, and γ) has been reported in cartilaginous fish (97).
Fish B Cells
B cells are one of the most important elements of the adaptive immune response, since they are able to produce specific high-affinity immunoglobulins against pathogens (Abs), and also generate memory B cells which will protect the organism against future infections (98). In fish, there are important differences concerning the isotypes of immunoglobulins produced by B cells in comparison to mammals.
Cartilaginous fish B cells produce three types of immunoglobulins IgM, IgW, and IgNAR (immunoglobulin new antigen receptor) (38). In these species, IgM is orthologous to mammalian IgM, while IgW has been postulated as the orthologous of mammalian IgD (99), although its function is still unknown. IgNAR is a shark-specific heavy chain (H) homodimer which does not associate with light chains (L) (100). IgNAR is produced by a different B cell subset than that expressing IgM and although the specific function for IgNAR remains unclear, IgNAR responses have been shown to be TD and show high specificity for the Ag (101).
Teleost fish species produce three types of Igs, namely, IgM, IgD, and IgT/Z (102). The latter was first described in 2005 in both rainbow trout (Oncorhynchus mykiss) (designated as IgT for teleost) (103) and in zebrafish (Danio rerio) where it was designated as IgZ (104). Since then, it has been described in most teleost species (105, 106), while it seems absent in others such as channel catfish (Ictalurus punctatus) and medaka fish (Oryzias latipes) (107). While the most abundant B cell type in the main lymphoid organs in teleost is IgD+IgM+, as it has been described for mammalian naïve mature B cells (108), IgT+ B cells constitute a different lineage in teleost, which do not co-express any other surface Ig (109). These cells are more abundant in mucosal surfaces than in the main lymphoid organs and consequently IgT-expressing B cells have been cataloged as B cells specialized in mucosal responses (109–111). Despite this, IgT responses have also been reported outside the mucosal compartments, thus the function of IgT in teleost immune responses is still largely unknown. In addition, IgD+IgM− (IgD single) cells have been identified in rainbow trout gills (112) and channel catfish blood (113) although their function is still unknown. Moreover, IgD−IgM+ (IgM single) cells exhibiting an antibody-secreting cell (ASC) phenotype have been shown to inhabit in the peritoneal cavity of vaccinated rainbow trout (114). These observations suggest that the B cell compartment in fish is composed by different cell subsets which most probably differ in functionality and/or cytokine-mediated regulation.
Interestingly, teleost B cells have been shown to possess strong phagocytic and microbicidal activities (115). It was later shown that is ability is preserved in mammalian innate B1 cells from the peritoneal cavity of mice (116) strongly suggesting that fish B cells are less evolved than mammalian B2 cells and still retain features of the innate immune system, which could be consistent with a posterior appearance of specific B1 and B2 B cell lineages throughout evolution. Further evidence supporting this hypothesis has been collected from studies showing the capacity of fish B cells to respond to pro-inflammatory stimuli (117), the expression of innate B cell markers CD9 and CD63 (118) and toll-like receptors (TLRs) (119) or the synthesis of antimicrobial peptides (117).
TNFSF Ligands in Fish: At the Dawn of Acquired Immunity?
As mentioned earlier, Collette et al. proposed that the divergence of the TNFSF and TNFRSF families parallels the emergence of the adaptive immune system, after performing a detailed analysis of the phylogenetic relations between the TNFSF and TNFRSF members of invertebrate and vertebrate species (7). As the identification of TNFSF orthologs and paralogs in fish has been rapidly increasing in the recent years, as can be inferred from the work undertaken by Glenney and Wiens (4, 41), studying the function of these cytokines and their receptors in fish is becoming a fascinating research topic to better understand the evolution and regulation of the adaptive immune system. In this review, we illustrate the information available regarding the effect of TNFSF ligands in fish, focusing specifically on those with a presumed role on fish B cells.
B Cell-Activating Factor of the TNF Family
The homologue sequences to mammalian BAFF have been reported in many teleost fish species including rainbow trout (41), zebrafish (120), mefugu (Takifugu obscurus) (121), Japanese sea perch (Lateolabrax japonicus) (122), grass carp (Ctenopharyngodon idella) (123), yellow grouper (Epinephelus awoara) (124), miiuy croaker (Miichthys miiuy) (125), tongue sole (Cynoglossus semilaevis) (126), Nile tilapia (Oreochromis niloticus) (127), rock bream (Oplegnathus fasciatus) (128), and also in cartilaginous fish such as white-spotted catshark (Chiloscyllium plagiosum) (129), spiny dogfish (Squalus acanthias) (130), and small-spotted catshark (Scyliorhinus canicula) (131) (summarized in Table 3). Interestingly, some studies have revealed that many cartilaginous and bony fish species have more than one BAFF gene (51, 54), representing two distinct groups. In mammals, BAFF is considered the master regulator of B cell development and function. Besides having been identified in many cartilaginous and bony fish species, it has been found in all vertebrate groups including birds, amphibians and reptiles (132), suggesting that BAFF has been essential throughout evolution for the development of peripheral mature B cells.
Recombinant BAFF proteins have been generated in some of these fish species, such as, for example, the cartilaginous fish white-spotted catshark (129), and the teleost zebrafish (120), fugu (121), Japanese sea perch (122), yellow grouper (124), tongue sole (126), rock bream (128), or tilapia (127), to carry out functional studies. These studies have proven an increase on the number of leukocytes mediated by BAFF, although the authors did not clarify whether this was due to a promotion of cell survival or an increase on cell proliferation, and they did not demonstrate if the surviving/proliferating fish leukocytes were in fact B cells. In some of these studies, the authors tested the effect of recombinant BAFF on mouse splenic B cells co-stimulated with anti-IgM (120, 121, 124, 127, 129). In all these experiments, fish recombinant BAFF promoted the survival of mouse B cells, which indicates that the molecular mechanism underlying BAFF-mediated B cell survival is preserved throughout species, from cartilaginous fish to mammals. However, specific studies on the impact of BAFF on B cells from lower vertebrates are needed to reveal new aspects about the evolution of B cell homeostasis and activation. Recent studies from our group showed the role of BAFF on teleost B cells, using rainbow trout as a model (53, 159). In these studies, we determined that teleost BAFF recapitulated mammalian BAFF regulatory aspects on B cells. Teleost BAFF promoted the survival of B cells but did not induced significant proliferation, and also increased the levels of ASCs which consequently increased IgM secretion and upregulated the expression of surface MHC II, in a similar trend to that seen in mammals (132). The similarity between teleost and mammalian BAFF functions strengthens the hypothesis of BAFF as a key master regulator of B cell functionality throughout evolution. In mammals, BAFF is produced by macrophages, DCs, stimulated neutrophils, and at low levels by T cells, but is never produced by resting B cells (60, 160, 161). Strikingly, one of our findings was that not only myeloid cells produced BAFF but also that specific subsets of splenic and peritoneal B cells were able to produce BAFF in rainbow trout (53, 159). Interestingly, in mammals, B cells from B-cell chronic lymphocytic leukemia (162) or non-Hodgkin’s lymphoma (163), as well as B cells from patients with autoimmune disorders, namely, rheumatoid arthritis (164), systemic lupus erythematosus (165), and primary Sjogren’s syndrome (166) also express BAFF, which rescues them from apoptosis in an autocrine loop. Thus, our studies seemed to have revealed a primitive mechanism through which B cells would produce BAFF at the steady-state to regulate their homeostasis and function in teleost. It seems that this regulatory mechanism throughout evolution was later assumed by other immune myeloid cells, probably after the appearance of lymphoid follicles, although in mammals, it can reemerge in various B cell disorders such as autoimmune diseases (ADs) or B cell malignancies. In a parallel study, we were also able to report for the first time an upregulation of BAFF transcription in peritoneal IgM+ B cells from fish immunized intra-peritoneally (i.p.) with viral hemorrhagic septicemia virus (VHSV) (114). In addition, it has been very recently shown that in vivo transgenic overexpression of BAFF induced an increased production of IgD, IgM, and IgZ in zebrafish (167). This implies that BAFF is not only involved on the homeostasis of peripheral B cell compartments but also plays an important role on the activation of different B cell subsets present in teleost.
A Proliferation-Inducing Ligand
Although APRIL is probably the most important B cell-regulating TNFSF together with BAFF, very little is known about fish APRIL. In fact, although BAFF homologs have been identified in many cartilaginous and bony fish (summarized in Table 3), only a few APRIL homologue sequences have been found in teleost, specifically in zebrafish, channel catfish, Atlantic salmon, rainbow trout (41), and grass carp (134) (Table 3). In addition, while BAFF is present in all vertebrates, APRIL is missing in cartilaginous fish, birds, and several bony fish (168). This has encouraged evolutionary immunologists to hypothesize that since BAFF and APRIL have structural and functional similarities and share receptors, the absence of APRIL on cartilaginous fish or the loss of APRIL in avian species could have been functionally compensated by BAFF. From the functional point of view, teleost APRIL has been shown to be mostly expressed in lymphoid tissues (spleen, head kidney) as well as in some mucosal tissues (skin, intestine) but at very low levels (41, 114, 134). After a bacterial or a viral challenge, the transcription of APRIL was quickly upregulated in immune tissues of grass carp (134), and recombinant APRIL promoted the survival of total splenocytes in zebrafish (169), indicating that this TNF ligand plays some role on the activation of the immune response. In addition, a study by our group showed that APRIL promoted the survival of peritoneal IgM+ B cells in rainbow trout (114). Since APRIL signaling through BCMA is key for the survival of PCs (70), it is tempting to hypothesize that those teleost species in which APRIL is found to have obtained a phenotypical advantage, the long-term survival of peripheral subsets of Ab-secreting PCs, that would have been positively selected.
BAFF- and APRIL-Like Molecule (BALM)
Searching through the rainbow trout EST databases, Glenney and Wiens reported in 2007 the identification of an additional sequence with high similarity values with BAFF but containing a D–E loop characteristic of APRIL, which was subsequently designated BALM. This new TNFSF ligand was also found in fugu and three spined stickleback (41). This study also determined that BALM gene was mainly expressed in lymphoid tissues. In a more recent study, BALM orthologs have been found in cartilaginous fish (168) (Table 3), in addition to a BAFF-like sequence showing homology to BAFF, APRIL and BALM present in lampreys (168). In this study, the authors described that BALM is absent in all tetrapods, and there is a selective deletion of this gene in zebrafish. These data point to BALM as an ancestral BAFF-like ligand, which appeared early in evolution and was then lost when BAFF and APRIL acquired divergent functions. However, the presence of an ancient BAFF-like homolog in lampreys indicates that further research is needed to classify these closely related TNFSF members, and clarify their evolution and biological functions. From the functional point of view, it has been shown that teleost BALM can promote B cell survival and proliferation in rainbow trout kidney (170). Moreover, its transcription is upregulated on peritoneal lymphocytes after i.p. administration of VHSV (114), and it has also been demonstrated that there is a significant correlation between the expression of BALM and the progress of the proliferative kidney disease in rainbow trout (170). Although very little is yet known about this ligand, these data indicate that it plays a very important role in the activation of B cells on the responses against different types of pathogens in lower vertebrates.
CD40L and OX40L
CD40L homologue sequences have been identified in several teleost species, such as rainbow trout, Atlantic salmon, zebrafish, fugu, and spotted green pufferfish (41) and more recently in one cartilaginous fish, the small-spotted catshark (131) (summarized in Table 3). In teleost, CD40L is mainly expressed in spleen, head kidney, and gills from resting animals (41, 171, 172). Furthermore, T cell mitogens such as PHA and ConA upregulated the transcription of CD40L on the spleen, head kidney, and gills of Atlantic salmon suggesting these may be special locations for T-B cell cooperation during TD immune responses (172). Interestingly, the production of CD40L by T cells was significantly increased in zebrafish immunized with TD Ags, and the upregulation of CD40L in vivo increased the levels of serum IgM on those animals (171). This last set of data by Yong-Feng Gong et al. elegantly demonstrated that the interaction between T and B cells through ligation of CD40 elicits TD immune responses in fish. Studies on the small-spotted catshark paralleled these results, showing that CD40L was mainly expressed in the spleen, as well as in the mucosal tissues, such as gills and gut (131). The transcription of CD40L was significantly upregulated after the addition of T cell polyclonal activators to cultures of PBL in vitro. Altogether, these data suggest that CD40L produced by T cells plays a very important role on TD immune responses. Since fish lack LNs and do not form lymphoid follicles, it would be of great interest to study how and where T–B cell interactions take place to better understand the evolutionary origins of TD immune responses.
To date, only one sequence with certain homology to OX40L has been reported in zebrafish (41), designated as Dr_TNF-New. This sequence was most similar to human OX40L and to Xenopus TNF-α. However, to the light of these results, it cannot be stated that true OX40L homologs exist in fish, and further studies are required its existence. In mammals, cross-linking of OX40L on B cells by OX40 expressed on activated CD4+ T cells promotes B cell proliferation (22). Since fish B cells seem to maintain many functions of innate B1 cells, and B–T cell interaction might be less abundant than that seen in mammals, it is tempting to hypothesize that the absence of OX40L in fish may be compensated by CD40L on the development of TD immune responses.
TNF-α
TNF-α is probably the most studied TNFSF ligand, due to its potential roles in development, cell proliferation, apoptosis, inflammation, and immunity [reviewed in Ref. (173)]. Hirono et al. first described a fish homolog of TNF-α in the Japanese flounder (135). Since then, TNF-α homologs have been identified in a plethora of teleost species (summarized in Table 3), such as Japanese flounder (135), sea bream (139), channel catfish (140), turbot (143), sea bass (144), mandarin fish (145), striped trumpeter (148), tongue sole (150), Atlantic salmon (146), goldfish (147), bluefin tuna (149), common carp (141, 142), and rainbow trout (136–138).
Interestingly, several teleost species present multiple isoforms of TNF-α. Two copies were initially found in rainbow trout (137) and four within the common carp (141, 142). Given that both species are tetraploid, the presence of multiple TNF-α isoforms was not surprising. However, with the discovery of at least two TNF-α genes within non-tetraploid fish species, such as bluefin tuna (149), orange-spotted grouper (174), zebrafish, and medaka (151), it has become clear that teleost have two different groups of TNF-α genes. Furthermore, the analysis of the zebrafish and medaka genomes (151) showed that members from the two different groups of TNF-α genes were found on different chromosomes, with conserved genes around them, thus indicating that the presence of these two groups is a consequence of a duplication event that occurred within bony fish.
Although both groups of TNF-α molecules can be found in various fish genomes, the role played by them in the immune response remains to be determined. On those species containing a single copy, it has been shown that TNF-α is ubiquitously expressed in all tissues analyzed from unstimulated fish such as Japanese flounder, sea bream, or mandarin fish (135, 139, 145). Results regarding the activation of TNF-α expression after treatment with lipopolysaccharide (LPS) were consistent between species; as its expression on the head kidney was significantly upregulated after treatment with LPS both in vitro and in vivo. Although TNF-α expression was also upregulated in response to viral or bacterial infections, some differences between species have been observed. For example, in turbot, virus induced higher TNF-α expression than bacteria in kidney cells (although the response was shorter in time) (143), while a very recent study showed the opposite result in tongue sole kidney, spleen, and blood cells from virus or bacteria immunized virus (150). In any case, macrophage-driven inflammation was activated in both studies. This suggests that although TNF regulation seems to be very similar among fish species, the interaction between host and pathogen might shift the spatiotemporal expression of TNF-α, to adapt the response to each specific pathogen. A nice example of this can be seen in the three striped trumpeter, which in response to the infection with the ectoparasite Chondracanthus goldsmidi significantly upregulates the expression of TNF-α not only in the head kidney but also in the gills, the attachment site of the parasite (148), thus demonstrating the adaptation of the host response to different pathogens.
Regarding the species that present two copies of TNF-α, some differences were found for such isoforms on each species. Both TNF-α1 and TNF-α2 were constitutively expressed in a number of tissues of healthy Atlantic salmon (146). However, incubation of head kidney leukocytes with bacteria upregulated the expression of TNF-α2 but not TNF-α1, suggesting different activation pathways for each TNF molecule. In goldfish, TNF-α2 showed higher expression levels than TNF-α1, and TNF-α2 was also able to activate primary macrophages (147). Something similar was observed in bluefin tuna, where TNF-α2 mRNA was significantly higher than TNF-α1 in blood leukocytes (149). In addition, the expression of TNF-α2 was increased by stimulation with B cell, T cell, or macrophage activators, while the expression of TNF-α1 was not affected. These observations led the authors to propose that while TNF-α1 is a ubiquitous cytokine involved in cell survival and apoptosis, TNF-α2 is an inducible form which may be a key regulator of the innate immune response. In mammals, TNFR1 and TNFR2 exert different functions, being the former involved in cell death and the latter in the regulation of the immune response (15). Little is known about TNFR in fish, so it would be of great interest to study whether the different TNF-α isoforms found in fish signal through different receptors.
Of special interest is the case of the rainbow trout where an additional TNF-α isoform was found (138): TNF-α1 and TNF-α2 are expressed constitutively in some tissues, such as head kidney, and gills, and they induced a pro-inflammatory response on a rainbow trout macrophage cell line (137). By contrast, LPS induced a much faster, stronger, and longer upregulation of TNF-α2 expression when compared with TNF-α1, pointing to the fact that TNF-α1 could be involved in cell survival and apoptosis, while TNF-α2 is an inducible form involved in the activation of the immune response. Moreover, the third form (TNF-α3) identified, showed low identity with the two forms previously described (138). Its basal expression was the lowest among the three molecules, but it was the most responsive of them against different stimuli, showing a strong upregulation of its expression at very early time points after stimulation of primary leukocytes with LPS or T cell mitogens. In addition, TNF-α3 induced the expression of many pro-inflammatory mediators and immune regulators (138). This suggests that TNF-α3 might be in fact a strong amplifier of the early inflammatory and immune responses.
In mammals, TNF-α has been proven as an important co-stimulator of B cells for their polyclonal expansion on primary immune responses (15). Fish B cells retain many innate immune features, such as phagocytic capacities (115) and expression of multiple TLRs (119), and their responses are likely to be TI since no GCs are observed. Thus, fish B cells responses seem to be based in polyclonal activation of Ag-reactive pools of cells (175). Since the direct effect of recombinant TNF-α has not been tested on fish B cells, we think it is paramount to study if this TNFSF ligand can improve B cell-mediated Ab responses.
LTβ
Initially, homologous sequences of mammalian LTβ were found in zebrafish and fugu (142), designated as TNF-new (TNF-N). Posterior investigations identified homologs of this gene in medaka and zebrafish (151). Phylogenetic analysis of these sequences with known vertebrate sequences showed a closer relationship of these sequences to TNFSF3 (LTβ) than to TNFSF1 and TNFSF2. This was further supported with the inclusion of Xenopus sequences, which are available for all TNFSF1, 2, and 3 (50). Interestingly, in rainbow trout, two different isoforms were isolated, and named LT-β1 and LT-β2 (50) (Table 3). The authors hypothesized that the presence of two isoforms was a result of the additional genome duplication event that took place in salmonids around 400 million years ago (176, 177). LT-β1 was expressed in the spleen, head kidney, intestine, and gills, while LT-β2 was expressed only in the gills of rainbow trout (50). This might indicate a different role for each LTβ in the homeostasis of B and T cells. In mammals, LTβ has been involved in the formation of LN, organization of lymphoid structures, and the formation of GCs (81). The absence of LN or GC reactions in fish may explain why LTβ has not yet been identified in other cartilaginous or bony fish species. Nevertheless, immunological assays are required to clarify whether this cytokine is needed in fish, as well as its potential role in central and peripheral immune tissues in rainbow trout, the only fish in which the presence of LTβ has been shown to date (50).
Fas Ligand
Fas ligand is a well-known TNFSF ligand that controls the extrinsic apoptosis pathway (178). In fish, homologue sequences of FasL have been reported in some teleost, namely, zebrafish (152), rainbow trout (41), Japanese flounder (153), Nile tilapia (154), rock bream (155), and Japanese pufferfish (Takifugu rubripes) (133) (Table 3). Homologue sequences have also been identified in cartilaginous fish (52). In all the species studied, FasL was highly expressed on spleen and head kidney, which are secondary lymphoid tissues in fish. In some species, such as rainbow trout and tilapia, it was also expressed on the gills and gut mucosal tissues (41, 154). Functional studies have determined that the T cell mitogen PHA was able to upregulate the expression of FasL in PHA-bound lymphocytes from Japanese flounder (153) while LPS was not able to do so, thus showing that activated T cells are responsible for production of FasL. In line with this, mammalian FasL has been widely reported to be expressed in Ag-activated CD8+ and CD4+ T cells and natural killer cells (80). A similar upregulation of FasL has been observed in rock bream after a viral challenge, suggesting that FasL may also play a role on the regulation of antiviral immune responses (155). On the other hand, Fas is expressed at low levels on resting B cells but is upregulated after Ag activation (79), making these cells more susceptible to FasL-mediated cell death. Thus, it has been proposed that FasL–Fas interaction controls the size of the B cell compartment in homeostasis, while during the immune response it seems to be responsible for the elimination of activated B cells after the generation of ASCs and memory B cells, and the subsequent clearance of the pathogen (179). Recombinant tilapia FasL has been shown to induce apoptosis on Fas-expressing HeLa cell lines (154), demonstrating its functionality. Unfortunately, the effect of FasL on fish B cells has not yet been addressed, so further investigations are needed to understand the impact of FasL on the B cell compartment in physiological or pathological conditions.
TNF-Related Apoptosis-Inducing Ligand
TNF-related apoptosis-inducing ligand sequences have been characterized in several teleost fish, such as grass carp (157), zebrafish (152), rainbow trout (41), pufferfish (41), and mandarin fish (158) (summarized in Table 3). In humans, there is only a single TRAIL gene while two genes have been reported in avian species (41). Eimon et al. identified four TRAIL genes in zebrafish (41, 152). Hence, the authors proposed that the ancestral gene was duplicated before the divergence of ray-finned and lobe-finned fishes (around 400 million years ago) (152). In line with this, three genes with homology to TRAIL have been identified in fugu (133). This study also showed that each of these homologs presented a very different gene organization forming distinct groups (133). In a thorough study based on the search of immune genes in leukocytes from jawless fish, a sequence showing homology to TRAIL was identified in the inshore hagfish (Eptatretus burgeri) (156), pointing to a conserved role of this cytokine throughout evolution. In all the teleost fish analyzed, TRAIL was expressed mainly in spleen and kidney but also in mucosal tissues, such as gills, skin, and/or intestine, depending on the species. In zebrafish, overexpression of TRAIL has been reported to induce cell death by activation of the extrinsic apoptosis pathway machinery (152). In parallel, mandarin fish recombinant TRAIL-induced apoptosis of HeLa cells (158) although it was previously reported that the extracellular domain of the death receptors (DR) for TRAIL in zebrafish differs from those in human DR4 and DR5 (152), suggesting that cross-reactivity with mammalian DRs could only be achieved with certain fish TRAIL proteins. In mammals, TRAIL is involved in B cell differentiation at the GC reaction, and although fish lack secondary lymphoid structures and do not form GCs, it would be of great interest to investigate what is the role of TRAIL on B cells from lower vertebrates.
Concluding Remarks
A plethora of studies in mammals have revealed that TNF ligand–receptor interactions elicit complex and divergent functions during the immune response. TNFSF ligands have the ability to induce both cell death or cell activation/co-stimulation and although some of these molecular mechanisms have been elucidated, the origin of these complex interactions and their multiple functionalities are not yet fully understood. TNFSF ligands play a key role in the immune response, and many researchers have taken advantage of this to develop new therapeutic strategies based on the modulation of TNF ligands. In fact, several anti-TNF neutralizing Abs are already available or under clinical trials for the treatment of inflammatory and ADs. Moreover, many immunologists are considering the option of using engineered TNFSF ligands as vaccine adjuvants. For instance, DNA vectors encoding for CD40L multi-trimers are being tested as adjuvants for a HIV vaccine, based on their positive effects on B cell co-stimulation during the vaccination process, thus generating high-affinity Abs against the virus. Hence, this is a fascinating research topic that is becoming of great importance within the field of immunotherapy. Most of the questions that have not been answered yet could be explained through the analysis of the evolution of the TNF superfamily of ligands and receptors. From an evolutionary point of view, it is widely accepted that acquired immunity first appeared in fish. This animal taxon possesses unique immune features, and recent evidences suggest that we can learn much about the evolution and functionality of TNF ligands in fish. For instance, they retain a unique TNF cytokine, BALM, which has become extinct in tetrapods, and although its function is still unknown, many evidences point to the fact that this molecule could be key to understand the molecular and functional divergence of BAFF and APRIL. On the other hand, fish express several isotypes of some TNF ligands (i.e., TNF-α) while mammals only express one. In fish, these ligand isotypes show different expression patterns, activation profiles, and immune functions. Thus, analyzing these isotypes could provide us with vital information about their original function, or about how and why some members of the TNF family were lost or acquired throughout evolution. Moreover, this analysis could also help to elucidate the convergence or divergence of some immune functions played by TNF ligands.
Author Contributions
CT and AG reviewed the bibliography and wrote the manuscript.
Conflict of Interest Statement
The authors declare the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work was supported by project AGL2014-54456-JIN from the Spanish Ministry of Economy and Competitiveness (MINECO) and by the European Research Council (ERC Consolidator Grant 2016 725061 TEMUBLYM).
Abbreviations
Ab, antibody; AD, autoimmune disease; Ag, antigen; APRIL, a proliferation-inducing ligand; BAFF, B cell-activating factor of the TNF family; BAFFR, BAFF receptor; BCMA, B cell maturation antigen; BCR, B cell receptor; Blimp-1, B lymphocyte-induced maturation protein-1; BM, bone marrow; CRD, cysteine-rich domain; CSR, class-switch recombination; DcR3, decoy receptor 3; DR3, death receptor 3; DC, dendritic cell; DD, death domain; EDA, ectodysplasin; EDAR, EDA receptor; FasL, Fas ligand; GC, germinal center; GITR, glucocorticoid-induced TNFR; GITRL, GITR ligand; HVEM, herpesvirus entry mediator; LPS, lipopolysaccharide; LIGHT, homology with lymphotoxin, inducible expression, competing for GpD of herpes virus, that binds to the HVEM, and is expressed on activated T lymphocytes; LN, lymph node; LT, lymphotoxin; MHC, major histocompatibility complex; MZ, marginal zone; NGFR, nerve growth factor receptor; NHL, non-Hodgkin’s lymphoma; NK, natural killer; PC, plasma cell; RA, rheumatoid arthritis; RANK, receptor activator of NF-κB; RANKL, RANK ligand; RELT, receptor expressed in lymphoid tissues; SHM, somatic hypermutation; SLE, systemic lupus erythematosus; SS, Sjögren’s syndrome; TACI, transmembrane activator and calcium-modulating cyclophilin ligand interactor; TCR, T cell receptor; TD, thymus dependent; THD, TNF homology domain; TI, thymus independent; TLR, toll-like receptor; TNF receptor superfamily, tumour necrosis factor receptor superfamily; TNF ligand superfamily, tumour necrosis factor (ligand) superfamily; TRAIL, TNF-related apoptosis-inducing ligand; TWEAK, TNF-like weak inducer of apoptosis and proliferation.
References
1. Carswell EA, Old LJ, Kassel RL, Green S, Fiore N, Williamson B. An endotoxin-induced serum factor that causes necrosis of tumors. Proc Natl Acad Sci U S A (1975) 72:3666–70. doi:10.1073/pnas.72.9.3666
2. Granger GA, Shacks SJ, Williams TW, Kolb WP. Lymphocyte in vitro cytotoxicity: specific release of lymphotoxin-like materials from tuberculin-sensitive lymphoid cells. Nature (1969) 221:1155–7. doi:10.1038/2211155a0
3. Ware CF. The TNF superfamily. Cytokine Growth Factor Rev (2003) 14:181–4. doi:10.1016/S1359-6101(03)00032-7
4. Wiens GD, Glenney GW. Origin and evolution of TNF and TNF receptor superfamilies. Dev Comp Immunol (2011) 35:1324–35. doi:10.1016/j.dci.2011.03.031
5. Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell (2001) 104:487–501. doi:10.1016/S0092-8674(01)00237-9
6. Bodmer JL, Schneider P, Tschopp J. The molecular architecture of the TNF superfamily. Trends Biochem Sci (2002) 27:19–26. doi:10.1016/S0968-0004(01)01995-8
7. Collette Y, Gilles A, Pontarotti P, Olive D. A co-evolution perspective of the TNFSF and TNFRSF families in the immune system. Trends Immunol (2003) 24:387–94. doi:10.1016/S1471-4906(03)00166-2
8. Bossen C, Ingold K, Tardivel A, Bodmer JL, Gaide O, Hertig S, et al. Interactions of tumor necrosis factor (TNF) and TNF receptor family members in the mouse and human. J Biol Chem (2006) 281:13964–71. doi:10.1074/jbc.M601553200
9. Banner DW, D’arcy A, Janes W, Gentz R, Schoenfeld HJ, Broger C, et al. Crystal structure of the soluble human 55 kd TNF receptor-human TNF beta complex: implications for TNF receptor activation. Cell (1993) 73:431–45. doi:10.1016/0092-8674(93)90132-A
10. Liu Y, Hong X, Kappler J, Jiang L, Zhang R, Xu L, et al. Ligand-receptor binding revealed by the TNF family member TALL-1. Nature (2003) 423:49–56. doi:10.1038/nature01543
11. Kim HM, Yu KS, Lee ME, Shin DR, Kim YS, Paik SG, et al. Crystal structure of the BAFF-BAFF-R complex and its implications for receptor activation. Nat Struct Biol (2003) 10:342–8. doi:10.1038/nsb925
12. Nelson CA, Warren JT, Wang MW, Teitelbaum SL, Fremont DH. RANKL employs distinct binding modes to engage RANK and the osteoprotegerin decoy receptor. Structure (2012) 20:1971–82. doi:10.1016/j.str.2012.08.030
13. Tcherpakov M, Bronfman FC, Conticello SG, Vaskovsky A, Levy Z, Niinobe M, et al. The p75 neurotrophin receptor interacts with multiple MAGE proteins. J Biol Chem (2002) 277:49101–4. doi:10.1074/jbc.C200533200
14. De Togni P, Goellner J, Ruddle NH, Streeter PR, Fick A, Mariathasan S, et al. Abnormal development of peripheral lymphoid organs in mice deficient in lymphotoxin. Science (1994) 264:703–7. doi:10.1126/science.8171322
15. Naude PJ, Den Boer JA, Luiten PG, Eisel UL. Tumor necrosis factor receptor cross-talk. FEBS J (2011) 278:888–98. doi:10.1111/j.1742-4658.2011.08017.x
16. Winkles JA. The TWEAK-Fn14 cytokine-receptor axis: discovery, biology and therapeutic targeting. Nat Rev Drug Discov (2008) 7:411–25. doi:10.1038/nrd2488
17. Steinberg MW, Cheung TC, Ware CF. The signaling networks of the herpesvirus entry mediator (TNFRSF14) in immune regulation. Immunol Rev (2011) 244:169–87. doi:10.1111/j.1600-065X.2011.01064.x
18. Hsieh SL, Lin WW. Decoy receptor 3: an endogenous immunomodulator in cancer growth and inflammatory reactions. J Biomed Sci (2017) 24:39. doi:10.1186/s12929-017-0347-7
19. O’Reilly LA, Kruse EA, Puthalakath H, Kelly PN, Kaufmann T, Huang DC, et al. MEK/ERK-mediated phosphorylation of Bim is required to ensure survival of T and B lymphocytes during mitogenic stimulation. J Immunol (2009) 183:261–9. doi:10.4049/jimmunol.0803853
20. Ronchetti S, Zollo O, Bruscoli S, Agostini M, Bianchini R, Nocentini G, et al. GITR, a member of the TNF receptor superfamily, is costimulatory to mouse T lymphocyte subpopulations. Eur J Immunol (2004) 34:613–22. doi:10.1002/eji.200324804
21. Elgueta R, Benson MJ, De Vries VC, Wasiuk A, Guo Y, Noelle RJ. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol Rev (2009) 229:152–72. doi:10.1111/j.1600-065X.2009.00782.x
22. Stuber E, Neurath M, Calderhead D, Fell HP, Strober W. Cross-linking of OX40 ligand, a member of the TNF/NGF cytokine family, induces proliferation and differentiation in murine splenic B cells. Immunity (1995) 2:507–21. doi:10.1016/1074-7613(95)90031-4
23. Finnberg N, El-Deiry WS. TRAIL death receptors as tumor suppressors and drug targets. Cell Cycle (2008) 7:1525–8. doi:10.4161/cc.7.11.5975
24. Perlot T, Penninger JM. Development and function of murine B cells lacking RANK. J Immunol (2012) 188:1201–5. doi:10.4049/jimmunol.1102063
25. Boyce BF, Xing L. The RANKL/RANK/OPG pathway. Curr Osteoporos Rep (2007) 5:98–104. doi:10.1007/s11914-007-0024-y
26. Ng LG, Sutherland AP, Newton R, Qian F, Cachero TG, Scott ML, et al. B cell-activating factor belonging to the TNF family (BAFF)-R is the principal BAFF receptor facilitating BAFF costimulation of circulating T and B cells. J Immunol (2004) 173:807–17. doi:10.4049/jimmunol.173.2.807
27. Peperzak V, Vikstrom I, Walker J, Glaser SP, Lepage M, Coquery CM, et al. Mcl-1 is essential for the survival of plasma cells. Nat Immunol (2013) 14:290–7. doi:10.1038/ni0813-877c
28. He B, Santamaria R, Xu W, Cols M, Chen K, Puga I, et al. The transmembrane activator TACI triggers immunoglobulin class switching by activating B cells through the adaptor MyD88. Nat Immunol (2010) 11:836–45. doi:10.1038/ni.1914
29. Borst J, Hendriks J, Xiao Y. CD27 and CD70 in T cell and B cell activation. Curr Opin Immunol (2005) 17:275–81. doi:10.1016/j.coi.2005.04.004
30. Shuford WW, Klussman K, Tritchler DD, Loo DT, Chalupny J, Siadak AW, et al. 4-1BB costimulatory signals preferentially induce CD8+ T cell proliferation and lead to the amplification in vivo of cytotoxic T cell responses. J Exp Med (1997) 186:47–55. doi:10.1084/jem.186.1.47
31. Kennedy MK, Willis CR, Armitage RJ. Deciphering CD30 ligand biology and its role in humoral immunity. Immunology (2006) 118:143–52. doi:10.1111/j.1365-2567.2006.02354.x
32. Drew CF, Lin CM, Jiang TX, Blunt G, Mou C, Chuong CM, et al. The Edar subfamily in feather placode formation. Dev Biol (2007) 305:232–45. doi:10.1016/j.ydbio.2007.02.011
33. Bittner S, Ehrenschwender M. Multifaceted death receptor 3 signaling-promoting survival and triggering death. FEBS Lett (2017) 591:2543–55. doi:10.1002/1873-3468.12747
34. Nikolaev A, Mclaughlin T, O’Leary DD, Tessier-Lavigne M. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature (2009) 457:981–9. doi:10.1038/nature07767
35. Sica GL, Zhu G, Tamada K, Liu D, Ni J, Chen L. RELT, a new member of the tumor necrosis factor receptor superfamily, is selectively expressed in hematopoietic tissues and activates transcription factor NF-kappaB. Blood (2001) 97:2702–7. doi:10.1182/blood.V97.9.2702
36. Marchalonis JJ, Schluter SF. A stochastic model for the rapid emergence of specific vertebrate immunity incorporating horizontal transfer of systems enabling duplication and combinational diversification. J Theor Biol (1998) 193:429–44. doi:10.1006/jtbi.1998.0711
37. Schluter SF, Bernstein RM, Bernstein H, Marchalonis JJ. ‘Big bang’ emergence of the combinatorial immune system. Dev Comp Immunol (1999) 23:107–11.
38. Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet (2010) 11:47–59. doi:10.1038/nrg2703
39. Pancer Z, Amemiya CT, Ehrhardt GR, Ceitlin J, Gartland GL, Cooper MD. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature (2004) 430:174–80. doi:10.1038/nature02740
40. Abi-Rached L, Gilles A, Shiina T, Pontarotti P, Inoko H. Evidence of en bloc duplication in vertebrate genomes. Nat Genet (2002) 31:100–5. doi:10.1038/ng855
41. Glenney GW, Wiens GD. Early diversification of the TNF superfamily in teleosts: genomic characterization and expression analysis. J Immunol (2007) 178:7955–73. doi:10.4049/jimmunol.178.12.7955
42. Moreno E, Yan M, Basler K. Evolution of TNF signaling mechanisms: JNK-dependent apoptosis triggered by Eiger, the Drosophila homolog of the TNF superfamily. Curr Biol (2002) 12:1263–8. doi:10.1016/S0960-9822(02)00954-5
43. Igaki T, Kanda H, Yamamoto-Goto Y, Kanuka H, Kuranaga E, Aigaki T, et al. Eiger, a TNF superfamily ligand that triggers the Drosophila JNK pathway. EMBO J (2002) 21:3009–18. doi:10.1093/emboj/cdf306
44. Narasimamurthy R, Geuking P, Ingold K, Willen L, Schneider P, Basler K. Structure-function analysis of Eiger, the Drosophila TNF homolog. Cell Res (2009) 19:392–4. doi:10.1038/cr.2009.16
45. Mekata T, Sudhakaran R, Okugawa S, Inada M, Kono T, Sakai M, et al. A novel gene of tumor necrosis factor ligand superfamily from kuruma shrimp, Marsupenaeus japonicus. Fish Shellfish Immunol (2010) 28:571–8. doi:10.1016/j.fsi.2009.12.020
46. De Zoysa M, Jung S, Lee J. First molluscan TNF-alpha homologue of the TNF superfamily in disk abalone: molecular characterization and expression analysis. Fish Shellfish Immunol (2009) 26:625–31. doi:10.1016/j.fsi.2008.10.004
47. De Zoysa M, Nikapitiya C, Moon DO, Whang I, Kim GY, Lee J. A novel Fas ligand in mollusk abalone: molecular characterization, immune responses and biological activity of the recombinant protein. Fish Shellfish Immunol (2009) 27:423–32. doi:10.1016/j.fsi.2009.06.019
48. Hibino T, Loza-Coll M, Messier C, Majeske AJ, Cohen AH, Terwilliger DP, et al. The immune gene repertoire encoded in the purple sea urchin genome. Dev Biol (2006) 300:349–65. doi:10.1016/j.ydbio.2006.08.065
49. Robertson AJ, Croce J, Carbonneau S, Voronina E, Miranda E, Mcclay DR, et al. The genomic underpinnings of apoptosis in Strongylocentrotus purpuratus. Dev Biol (2006) 300:321–34. doi:10.1016/j.ydbio.2006.08.053
50. Kono T, Zou J, Bird S, Savan R, Sakai M, Secombes CJ. Identification and expression analysis of lymphotoxin-beta like homologues in rainbow trout Oncorhynchus mykiss. Mol Immunol (2006) 43:1390–401. doi:10.1016/j.molimm.2005.07.037
51. Secombes CJ. What’s new in fish cytokine research? Fish Shellfish Immunol (2016) 53:1–3. doi:10.1016/j.fsi.2016.03.035
52. Secombes CJ, Wang T, Bird S. Chapter 5 – vertebrate cytokines and their evolution A2. In: Malagoli D, editor. The Evolution of the Immune System. Modena: Academic Press (2016). p. 87–150.
53. Granja AG, Tafalla C. Different IgM(+) B cell subpopulations residing within the peritoneal cavity of vaccinated rainbow trout are differently regulated by BAFF. Fish Shellfish Immunol (2017). doi:10.1016/j.fsi.2017.10.003
54. Jacobson G, Muncaster S, Mensink K, Forlenza M, Elliot N, Broomfield G, et al. Omics and cytokine discovery in fish: presenting the yellowtail kingfish (Seriola lalandi) as a case study. Dev Comp Immunol (2017) 75:63–76. doi:10.1016/j.dci.2017.04.001
55. Tacchi L, Larragoite ET, Munoz P, Amemiya CT, Salinas I. African lungfish reveal the evolutionary origins of organized mucosal lymphoid tissue in vertebrates. Curr Biol (2015) 25:2417–24. doi:10.1016/j.cub.2015.07.066
56. Zapata A, Diez B, Cejalvo T, Gutierrez-De Frias C, Cortes A. Ontogeny of the immune system of fish. Fish Shellfish Immunol (2006) 20:126–36. doi:10.1016/j.fsi.2004.09.005
57. Zhang P, Gu Z, Li WH. Different evolutionary patterns between young duplicate genes in the human genome. Genome Biol (2003) 4:R56. doi:10.1186/gb-2003-4-9-r56
58. Figgett WA, Vincent FB, Saulep-Easton D, Mackay F. Roles of ligands from the TNF superfamily in B cell development, function, and regulation. Semin Immunol (2014) 26:191–202. doi:10.1016/j.smim.2014.06.001
59. Vincent FB, Saulep-Easton D, Figgett WA, Fairfax KA, Mackay F. The BAFF/APRIL system: emerging functions beyond B cell biology and autoimmunity. Cytokine Growth Factor Rev (2013) 24:203–15. doi:10.1016/j.cytogfr.2013.04.003
60. Nardelli B, Belvedere O, Roschke V, Moore PA, Olsen HS, Migone TS, et al. Synthesis and release of B-lymphocyte stimulator from myeloid cells. Blood (2001) 97:198–204. doi:10.1182/blood.V97.1.198
61. Schneider P. The role of APRIL and BAFF in lymphocyte activation. Curr Opin Immunol (2005) 17:282–9. doi:10.1016/j.coi.2005.04.005
62. Mackay F, Figgett WA, Saulep D, Lepage M, Hibbs ML. B-cell stage and context-dependent requirements for survival signals from BAFF and the B-cell receptor. Immunol Rev (2010) 237:205–25. doi:10.1111/j.1600-065X.2010.00944.x
63. Gross JA, Dillon SR, Mudri S, Johnston J, Littau A, Roque R, et al. TACI-Ig neutralizes molecules critical for B cell development and autoimmune disease. Impaired B cell maturation in mice lacking BLyS. Immunity (2001) 15:289–302. doi:10.1016/S1074-7613(01)00183-2
64. Castigli E, Scott S, Dedeoglu F, Bryce P, Jabara H, Bhan AK, et al. Impaired IgA class switching in APRIL-deficient mice. Proc Natl Acad Sci U S A (2004) 101:3903–8. doi:10.1073/pnas.0307348101
65. Sindhava VJ, Scholz JL, Stohl W, Cancro MP. APRIL mediates peritoneal B-1 cell homeostasis. Immunol Lett (2014) 160:120–7. doi:10.1016/j.imlet.2014.01.018
66. Stadanlick JE, Kaileh M, Karnell FG, Scholz JL, Miller JP, Quinn WJ III, et al. Tonic B cell antigen receptor signals supply an NF-kappaB substrate for prosurvival BLyS signaling. Nat Immunol (2008) 9:1379–87. doi:10.1038/ni.1666
67. Ding BB, Bi E, Chen H, Yu JJ, Ye BH. IL-21 and CD40L synergistically promote plasma cell differentiation through upregulation of Blimp-1 in human B cells. J Immunol (2013) 190:1827–36. doi:10.4049/jimmunol.1201678
68. Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity (2002) 17:51–62. doi:10.1016/S1074-7613(02)00335-7
69. Castigli E, Wilson SA, Elkhal A, Ozcan E, Garibyan L, Geha RS. Transmembrane activator and calcium modulator and cyclophilin ligand interactor enhances CD40-driven plasma cell differentiation. J Allergy Clin Immunol (2007) 120:885–91. doi:10.1016/j.jaci.2007.06.012
70. O’Connor BP, Raman VS, Erickson LD, Cook WJ, Weaver LK, Ahonen C, et al. BCMA is essential for the survival of long-lived bone marrow plasma cells. J Exp Med (2004) 199:91–8. doi:10.1084/jem.20031330
71. Day ES, Cachero TG, Qian F, Sun Y, Wen D, Pelletier M, et al. Selectivity of BAFF/BLyS and APRIL for binding to the TNF family receptors BAFFR/BR3 and BCMA. Biochemistry (2005) 44:1919–31. doi:10.1021/bi048227k
72. Wajant H, Pfizenmaier K, Scheurich P. Tumor necrosis factor signaling. Cell Death Differ (2003) 10:45–65. doi:10.1038/sj.cdd.4401189
73. Rieckmann P, Tuscano JM, Kehrl JH. Tumor necrosis factor-alpha (TNF-alpha) and interleukin-6 (IL-6) in B-lymphocyte function. Methods (1997) 11:128–32. doi:10.1006/meth.1996.0396
74. Goldfeld AE, Mccaffrey PG, Strominger JL, Rao A. Identification of a novel cyclosporin-sensitive element in the human tumor necrosis factor alpha gene promoter. J Exp Med (1993) 178:1365–79. doi:10.1084/jem.178.4.1365
75. Boussiotis VA, Nadler LM, Strominger JL, Goldfeld AE. Tumor necrosis factor alpha is an autocrine growth factor for normal human B cells. Proc Natl Acad Sci U S A (1994) 91:7007–11. doi:10.1073/pnas.91.15.7007
76. Tesselaar K, Xiao Y, Arens R, Van Schijndel GM, Schuurhuis DH, Mebius RE, et al. Expression of the murine CD27 ligand CD70 in vitro and in vivo. J Immunol (2003) 170:33–40. doi:10.4049/jimmunol.170.1.33
77. Arens R, Nolte MA, Tesselaar K, Heemskerk B, Reedquist KA, Van Lier RA, et al. Signaling through CD70 regulates B cell activation and IgG production. J Immunol (2004) 173:3901–8. doi:10.4049/jimmunol.173.6.3901
78. Guerreiro-Cacais AO, Levitskaya J, Levitsky V. B cell receptor triggering sensitizes human B cells to TRAIL-induced apoptosis. J Leukoc Biol (2010) 88:937–45. doi:10.1189/jlb.0510246
79. Krammer PH. CD95’s deadly mission in the immune system. Nature (2000) 407:789–95. doi:10.1038/35037728
80. Strasser A, Jost PJ, Nagata S. The many roles of FAS receptor signaling in the immune system. Immunity (2009) 30:180–92. doi:10.1016/j.immuni.2009.01.001
81. Futterer A, Mink K, Luz A, Kosco-Vilbois MH, Pfeffer K. The lymphotoxin beta receptor controls organogenesis and affinity maturation in peripheral lymphoid tissues. Immunity (1998) 9:59–70. doi:10.1016/S1074-7613(00)80588-9
82. Cerutti A, Schaffer A, Goodwin RG, Shah S, Zan H, Ely S, et al. Engagement of CD153 (CD30 ligand) by CD30+ T cells inhibits class switch DNA recombination and antibody production in human IgD+ IgM+ B cells. J Immunol (2000) 165:786–94. doi:10.4049/jimmunol.165.2.786
83. Rumfelt LL, Mckinney EC, Taylor E, Flajnik MF. The development of primary and secondary lymphoid tissues in the nurse shark Ginglymostoma cirratum: B-cell zones precede dendritic cell immigration and T-cell zone formation during ontogeny of the spleen. Scand J Immunol (2002) 56:130–48. doi:10.1046/j.1365-3083.2002.01116.x
84. Zapata A, Amemiya CT. Phylogeny of lower vertebrates and their immunological structures. Curr Top Microbiol Immunol (2000) 248:67–107.
85. Zapata A. Ultrastructure of elasmobranch lymphoid tissue. 2. Leydig’s and epigonal organs. Dev Comp Immunol (1981) 5:43–52. doi:10.1016/S0145-305X(81)80006-7
86. Rumfelt LL, Avila D, Diaz M, Bartl S, Mckinney EC, Flajnik MF. A shark antibody heavy chain encoded by a nonsomatically rearranged VDJ is preferentially expressed in early development and is convergent with mammalian IgG. Proc Natl Acad Sci U S A (2001) 98:1775–80. doi:10.1073/pnas.98.4.1775
87. Zapata AG, Torroba M, Vicente A, Varas A, Sacedon R, Jimenez E. The relevance of cell microenvironments for the appearance of lympho-haemopoietic tissues in primitive vertebrates. Histol Histopathol (1995) 10:761–78.
88. Hansen JD, Kaattari SL. The recombination activation gene 1 (RAG1) of rainbow trout (Oncorhynchus mykiss): cloning, expression, and phylogenetic analysis. Immunogenetics (1995) 42:188–95. doi:10.1007/BF00191224
89. Hansen JD, Kaattari SL. The recombination activating gene 2 (RAG2) of the rainbow trout Oncorhynchus mykiss. Immunogenetics (1996) 44:203–11. doi:10.1007/BF02602586
90. Hansen JD. Characterization of rainbow trout terminal deoxynucleotidyl transferase structure and expression. TdT and RAG1 co-expression define the trout primary lymphoid tissues. Immunogenetics (1997) 46:367–75. doi:10.1007/s002510050290
91. Hansen JD, Strassburger P, Du Pasquier L. Conservation of a master hematopoietic switch gene during vertebrate evolution: isolation and characterization of Ikaros from teleost and amphibian species. Eur J Immunol (1997) 27:3049–58. doi:10.1002/eji.1830271143
92. Zwollo P, Cole S, Bromage E, Kaattari S. B cell heterogeneity in the teleost kidney: evidence for a maturation gradient from anterior to posterior kidney. J Immunol (2005) 174:6608–16. doi:10.4049/jimmunol.174.11.6608
93. Ellyard JI, Avery DT, Phan TG, Hare NJ, Hodgkin PD, Tangye SG. Antigen-selected, immunoglobulin-secreting cells persist in human spleen and bone marrow. Blood (2004) 103:3805–12. doi:10.1182/blood-2003-09-3109
94. Bromage ES, Kaattari IM, Zwollo P, Kaattari SL. Plasmablast and plasma cell production and distribution in trout immune tissues. J Immunol (2004) 173:7317–23. doi:10.4049/jimmunol.173.12.7317
95. Bowden TJ, Cook P, Rombout JH. Development and function of the thymus in teleosts. Fish Shellfish Immunol (2005) 19:413–27. doi:10.1016/j.fsi.2005.02.003
96. Romano N, Abelli L, Mastrolia L, Scapigliati G. Immunocytochemical detection and cytomorphology of lymphocyte subpopulations in a teleost fish Dicentrarchus labrax. Cell Tissue Res (1997) 289:163–71. doi:10.1007/s004410050862
97. Rast JP, Anderson MK, Strong SJ, Luer C, Litman RT, Litman GW. Alpha, beta, gamma, and delta T cell antigen receptor genes arose early in vertebrate phylogeny. Immunity (1997) 6:1–11. doi:10.1016/S1074-7613(00)80237-X
98. Kurosaki T, Aiba Y, Kometani K, Moriyama S, Takahashi Y. Unique properties of memory B cells of different isotypes. Immunol Rev (2010) 237:104–16. doi:10.1111/j.1600-065X.2010.00939.x
99. Ohta Y, Flajnik M. IgD, like IgM, is a primordial immunoglobulin class perpetuated in most jawed vertebrates. Proc Natl Acad Sci U S A (2006) 103:10723–8. doi:10.1073/pnas.0601407103
100. Greenberg AS, Avila D, Hughes M, Hughes A, Mckinney EC, Flajnik MF. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature (1995) 374:168–73. doi:10.1038/374168a0
101. Dooley H, Flajnik MF. Shark immunity bites back: affinity maturation and memory response in the nurse shark, Ginglymostoma cirratum. Eur J Immunol (2005) 35:936–45. doi:10.1002/eji.200425760
102. Sunyer JO. Evolutionary and functional relationships of B cells from fish and mammals: insights into their novel roles in phagocytosis and presentation of particulate antigen. Infect Disord Drug Targets (2012) 12:200–12. doi:10.2174/187152612800564419
103. Hansen JD, Landis ED, Phillips RB. Discovery of a unique Ig heavy-chain isotype (IgT) in rainbow trout: implications for a distinctive B cell developmental pathway in teleost fish. Proc Natl Acad Sci U S A (2005) 102:6919–24. doi:10.1073/pnas.0500027102
104. Danilova N, Bussmann J, Jekosch K, Steiner LA. The immunoglobulin heavy-chain locus in zebrafish: identification and expression of a previously unknown isotype, immunoglobulin Z. Nat Immunol (2005) 6:295–302. doi:10.1038/ni1166
105. Gambon-Deza F, Sanchez-Espinel C, Magadan-Mompo S. Presence of an unique IgT on the IGH locus in three-spined stickleback fish (Gasterosteus aculeatus) and the very recent generation of a repertoire of VH genes. Dev Comp Immunol (2010) 34:114–22. doi:10.1016/j.dci.2009.08.011
106. Ryo S, Wijdeven RH, Tyagi A, Hermsen T, Kono T, Karunasagar I, et al. Common carp have two subclasses of bonyfish specific antibody IgZ showing differential expression in response to infection. Dev Comp Immunol (2010) 34:1183–90. doi:10.1016/j.dci.2010.06.012
107. Mashoof S, Criscitiello MF. Fish immunoglobulins. Biology (Basel) (2016) 5:45. doi:10.3390/biology5040045
108. Rajewsky K. Clonal selection and learning in the antibody system. Nature (1996) 381:751–8. doi:10.1038/381751a0
109. Zhang YA, Salinas I, Li J, Parra D, Bjork S, Xu Z, et al. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat Immunol (2010) 11:827–35. doi:10.1038/ni.1913
110. Xu Z, Parra D, Gomez D, Salinas I, Zhang YA, Von Gersdorff Jorgensen L, et al. Teleost skin, an ancient mucosal surface that elicits gut-like immune responses. Proc Natl Acad Sci U S A (2013) 110:13097–102. doi:10.1073/pnas.1304319110
111. Xu Z, Takizawa F, Parra D, Gomez D, Von Gersdorff Jorgensen L, Lapatra SE, et al. Mucosal immunoglobulins at respiratory surfaces mark an ancient association that predates the emergence of tetrapods. Nat Commun (2016) 7:10728. doi:10.1038/ncomms10728
112. Castro R, Bromage E, Abos B, Pignatelli J, Gonzalez Granja A, Luque A, et al. CCR7 is mainly expressed in teleost gills, where it defines an IgD+IgM- B lymphocyte subset. J Immunol (2014) 192:1257–66. doi:10.4049/jimmunol.1302471
113. Edholm ES, Bengten E, Stafford JL, Sahoo M, Taylor EB, Miller NW, et al. Identification of two IgD+ B cell populations in channel catfish, Ictalurus punctatus. J Immunol (2010) 185:4082–94. doi:10.4049/jimmunol.1000631
114. Soleto I, Abos B, Castro R, Gonzalez L, Tafalla C, Granja AG. The BAFF/APRIL axis plays an important role in virus-induced peritoneal responses in rainbow trout. Fish Shellfish Immunol (2017) 64:210–7. doi:10.1016/j.fsi.2017.03.023
115. Li J, Barreda DR, Zhang YA, Boshra H, Gelman AE, Lapatra S, et al. B lymphocytes from early vertebrates have potent phagocytic and microbicidal abilities. Nat Immunol (2006) 7:1116–24. doi:10.1038/ni1389
116. Parra D, Rieger AM, Li J, Zhang YA, Randall LM, Hunter CA, et al. Pivotal advance: peritoneal cavity B-1 B cells have phagocytic and microbicidal capacities and present phagocytosed antigen to CD4+ T cells. J Leukoc Biol (2012) 91:525–36. doi:10.1189/jlb.0711372
117. Abos B, Wang T, Castro R, Granja AG, Leal E, Havixbeck J, et al. Distinct differentiation programs triggered by IL-6 and LPS in teleost IgM(+) B cells in the absence of germinal centers. Sci Rep (2016) 6:30004. doi:10.1038/srep30004
118. Castro R, Abos B, Gonzalez L, Aquilino C, Pignatelli J, Tafalla C. Molecular characterization of CD9 and CD63, two tetraspanin family members expressed in trout B lymphocytes. Dev Comp Immunol (2015) 51:116–25. doi:10.1016/j.dci.2015.03.002
119. Abos B, Castro R, Pignatelli J, Luque A, Gonzalez L, Tafalla C. Transcriptional heterogeneity of IgM+ cells in rainbow trout (Oncorhynchus mykiss) tissues. PLoS One (2013) 8:e82737. doi:10.1371/journal.pone.0082737
120. Liang Z, Kong Y, Luo C, Shen Y, Zhang S. Molecular cloning, functional characterization and phylogenetic analysis of B-cell activating factor in zebrafish (Danio rerio). Fish Shellfish Immunol (2010) 29:233–40. doi:10.1016/j.fsi.2010.03.006
121. Ai H, Shen Y, Min C, Pang S, Zhang J, Zhang S, et al. Molecular structure, expression and bioactivity characterization of TNF13B (BAFF) gene in mefugu, Takifugu obscurus. Fish Shellfish Immunol (2011) 30:1265–74. doi:10.1016/j.fsi.2011.03.020
122. Cui XW, Li JF, Xiao W, Xuan Y, Tian AY, Xu XZ, et al. Molecular cloning, expression and functional analysis of TNF13b (BAFF) in Japanese sea perch, Lateolabrax japonicus. Int Immunopharmacol (2012) 12:34–41. doi:10.1016/j.intimp.2011.10.009
123. Pandit NP, Shen Y, Wang W, Chen Y, Li J. Identification of TNF13B (BAFF) gene from grass carp (Ctenopharyngodon idella) and its immune response to bacteria and virus. Dev Comp Immunol (2013) 39:460–4. doi:10.1016/j.dci.2013.01.004
124. Xiao W, Long W, Liu GY, Sui CL, Guo XR, Tian A, et al. Molecular cloning, expression and functional analysis of B-cell activating factor (BAFF) in yellow grouper, Epinephelus awoara. Mol Immunol (2014) 59:64–70. doi:10.1016/j.molimm.2014.01.005
125. Meng F, Sun Y, Xu T. Comparative genomic of the BAFF and BAFF-like genes and immune response to bacteria of miiuy croaker (Miichthys miiuy). Fish Shellfish Immunol (2015) 43:191–9. doi:10.1016/j.fsi.2014.12.022
126. Sun Y, Sun L. CsBAFF, a teleost B cell activating factor, promotes pathogen-induced innate immunity and vaccine-induced adaptive immunity. PLoS One (2015) 10:e0136015. doi:10.1371/journal.pone.0136015
127. Liu H, Zhang J, Li J, Song J, Zhang S. Molecular structure, distribution, and immunology function of TNFSF13B (BAFF) in Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol (2016) 51:240–50. doi:10.1016/j.fsi.2016.02.026
128. Godahewa GI, Perera NC, Umasuthan N, Wan Q, Whang I, Lee J. Molecular characterization and expression analysis of B cell activating factor from rock bream (Oplegnathus fasciatus). Dev Comp Immunol (2016) 55:1–11. doi:10.1016/j.dci.2015.10.004
129. Ren W, Pang S, You F, Zhou L, Zhang S. The first BAFF gene cloned from the cartilaginous fish. Fish Shellfish Immunol (2011) 31:1088–96. doi:10.1016/j.fsi.2011.09.013
130. Li R, Dooley H, Wang T, Secombes CJ, Bird S. Characterisation and expression analysis of B-cell activating factor (BAFF) in spiny dogfish (Squalus acanthias): cartilaginous fish BAFF has a unique extra exon that may impact receptor binding. Dev Comp Immunol (2012) 36:707–17. doi:10.1016/j.dci.2011.11.010
131. Li R, Redmond AK, Wang T, Bird S, Dooley H, Secombes CJ. Characterisation of the TNF superfamily members CD40L and BAFF in the small-spotted catshark (Scyliorhinus canicula). Fish Shellfish Immunol (2015) 47:381–9. doi:10.1016/j.fsi.2015.09.033
132. Mackay F, Schneider P. Cracking the BAFF code. Nat Rev Immunol (2009) 9:491–502. doi:10.1038/nri2572
133. Biswas G, Kinoshita S, Kono T, Hikima J, Sakai M. Evolutionary evidence of tumor necrosis factor super family members in the Japanese pufferfish (Takifugu rubripes): comprehensive genomic identification and expression analysis. Mar Genomics (2015) 22:25–36. doi:10.1016/j.margen.2015.03.003
134. Pandit NP, Shen YB, Chen Y, Wang WJ, Li JL. Molecular characterization, expression, and immunological response analysis of the TWEAK and APRIL genes in grass carp, Ctenopharyngodon idella. Genet Mol Res (2014) 13:10105–20. doi:10.4238/2014.December.4.5
135. Hirono I, Nam BH, Kurobe T, Aoki T. Molecular cloning, characterization, and expression of TNF cDNA and gene from Japanese flounder Paralichthys olivaceus. J Immunol (2000) 165:4423–7. doi:10.4049/jimmunol.165.8.4423
136. Laing KJ, Wang T, Zou J, Holland J, Hong S, Bols N, et al. Cloning and expression analysis of rainbow trout Oncorhynchus mykiss tumour necrosis factor-alpha. Eur J Biochem (2001) 268:1315–22. doi:10.1046/j.1432-1327.2001.01996.x
137. Zou J, Wang T, Hirono I, Aoki T, Inagawa H, Honda T, et al. Differential expression of two tumor necrosis factor genes in rainbow trout, Oncorhynchus mykiss. Dev Comp Immunol (2002) 26:161–72. doi:10.1016/S0145-305X(01)00058-1
138. Hong S, Li R, Xu Q, Secombes CJ, Wang T. Two types of TNF-alpha exist in teleost fish: phylogeny, expression, and bioactivity analysis of type-II TNF-alpha3 in rainbow trout Oncorhynchus mykiss. J Immunol (2013) 191:5959–72. doi:10.4049/jimmunol.1301584
139. Garcia-Castillo J, Pelegrin P, Mulero V, Meseguer J. Molecular cloning and expression analysis of tumor necrosis factor alpha from a marine fish reveal its constitutive expression and ubiquitous nature. Immunogenetics (2002) 54:200–7. doi:10.1007/s00251-002-0451-y
140. Zou J, Peddie S, Scapigliati G, Zhang Y, Bols NC, Ellis AE, et al. Functional characterisation of the recombinant tumor necrosis factors in rainbow trout, Oncorhynchus mykiss. Dev Comp Immunol (2003) 27:813–22. doi:10.1016/S0145-305X(03)00077-6
141. Saeij JP, Stet RJ, De Vries BJ, Van Muiswinkel WB, Wiegertjes GF. Molecular and functional characterization of carp TNF: a link between TNF polymorphism and trypanotolerance? Dev Comp Immunol (2003) 27:29–41. doi:10.1016/S0145-305X(02)00064-2
142. Savan R, Sakai M. Presence of multiple isoforms of TNF alpha in carp (Cyprinus carpio L.): genomic and expression analysis. Fish Shellfish Immunol (2004) 17:87–94. doi:10.1016/j.fsi.2003.11.001
143. Ordas MC, Costa MM, Roca FJ, Lopez-Castejon G, Mulero V, Meseguer J, et al. Turbot TNFalpha gene: molecular characterization and biological activity of the recombinant protein. Mol Immunol (2007) 44:389–400. doi:10.1016/j.molimm.2006.02.028
144. Nascimento DS, Pereira PJ, Reis MI, Do Vale A, Zou J, Silva MT, et al. Molecular cloning and expression analysis of sea bass (Dicentrarchus labrax L.) tumor necrosis factor-alpha (TNF-alpha). Fish Shellfish Immunol (2007) 23:701–10. doi:10.1016/j.fsi.2007.02.003
145. Xiao J, Zhou ZC, Chen C, Huo WL, Yin ZX, Weng SP, et al. Tumor necrosis factor-alpha gene from mandarin fish, Siniperca chuatsi: molecular cloning, cytotoxicity analysis and expression profile. Mol Immunol (2007) 44:3615–22. doi:10.1016/j.molimm.2007.03.016
146. Morrison RN, Zou J, Secombes CJ, Scapigliati G, Adams MB, Nowak BF. Molecular cloning and expression analysis of tumour necrosis factor-alpha in amoebic gill disease (AGD)-affected Atlantic salmon (Salmo salar L.). Fish Shellfish Immunol (2007) 23:1015–31. doi:10.1016/j.fsi.2007.04.003
147. Grayfer L, Walsh JG, Belosevic M. Characterization and functional analysis of goldfish (Carassius auratus L.) tumor necrosis factor-alpha. Dev Comp Immunol (2008) 32:532–43. doi:10.1016/j.dci.2007.09.009
148. Covello JM, Bird S, Morrison RN, Battaglene SC, Secombes CJ, Nowak BF. Cloning and expression analysis of three striped trumpeter (Latris lineata) pro-inflammatory cytokines, TNF-alpha, IL-1beta and IL-8, in response to infection by the ectoparasitic, Chondracanthus goldsmidi. Fish Shellfish Immunol (2009) 26:773–86. doi:10.1016/j.fsi.2009.03.012
149. Kadowaki T, Harada H, Sawada Y, Kohchi C, Soma G, Takahashi Y, et al. Two types of tumor necrosis factor-alpha in bluefin tuna (Thunnus orientalis) genes: molecular cloning and expression profile in response to several immunological stimulants. Fish Shellfish Immunol (2009) 27:585–94. doi:10.1016/j.fsi.2008.12.006
150. Li MF, Zhang J. CsTNF1, a teleost tumor necrosis factor that promotes antibacterial and antiviral immune defense in a manner that depends on the conserved receptor binding site. Dev Comp Immunol (2016) 55:65–75. doi:10.1016/j.dci.2015.10.010
151. Kinoshita S, Biswas G, Kono T, Hikima J, Sakai M. Presence of two tumor necrosis factor (tnf)-alpha homologs on different chromosomes of zebrafish (Danio rerio) and medaka (Oryzias latipes). Mar Genomics (2014) 13:1–9. doi:10.1016/j.margen.2013.10.004
152. Eimon PM, Kratz E, Varfolomeev E, Hymowitz SG, Stern H, Zha J, et al. Delineation of the cell-extrinsic apoptosis pathway in the zebrafish. Cell Death Differ (2006) 13:1619–30. doi:10.1038/sj.cdd.4402015
153. Kurata O, Iwasaki T, Matsuyama T, Nakayasu C, Wada S, Hatai K. Lymphocytes with T-cell-like properties express the Fas ligand in the Japanese flounder Paralichthys olivaceus. Fish Shellfish Immunol (2011) 30:509–14. doi:10.1016/j.fsi.2010.11.030
154. Ma TY, Wu JY, Gao XK, Wang JY, Zhan XL, Li WS. Molecular cloning, functional identification and expressional analyses of FasL in tilapia, Oreochromis niloticus. Dev Comp Immunol (2014) 46:448–60. doi:10.1016/j.dci.2014.06.003
155. Jung MH, Nikapitiya C, Song JY, Lee JH, Lee J, Oh MJ, et al. Gene expression of pro- and anti-apoptotic proteins in rock bream (Oplegnathus fasciatus) infected with megalocytivirus (family Iridoviridae). Fish Shellfish Immunol (2014) 37:122–30. doi:10.1016/j.fsi.2014.01.012
156. Suzuki T, Shin IT, Kohara Y, Kasahara M. Transcriptome analysis of hagfish leukocytes: a framework for understanding the immune system of jawless fishes. Dev Comp Immunol (2004) 28:993–1003. doi:10.1016/j.dci.2004.04.005
157. Chang MX, Nie P, Xie HX, Wang GL, Gao Y. Characterization and expression analysis of TNF-related apoptosis inducing ligand (TRAIL) in grass carp Ctenopharyngodon idella. Vet Immunol Immunopathol (2006) 110:51–63. doi:10.1016/j.vetimm.2005.09.003
158. Gao Y, Chang MX, Sun BJ, Nie P. TRAIL in the mandarin fish Siniperca chuatsi: gene and its apoptotic effect in HeLa cells. Fish Shellfish Immunol (2008) 24:55–66. doi:10.1016/j.fsi.2007.08.001
159. Tafalla C, Gonzalez L, Castro R, Granja AG. B cell-activating factor regulates different aspects of B cell functionality and is produced by a subset of splenic B cells in teleost fish. Front Immunol (2017) 8:295. doi:10.3389/fimmu.2017.00295
160. Craxton A, Magaletti D, Ryan EJ, Clark EA. Macrophage- and dendritic cell-dependent regulation of human B-cell proliferation requires the TNF family ligand BAFF. Blood (2003) 101:4464–71. doi:10.1182/blood-2002-10-3123
161. Scapini P, Nardelli B, Nadali G, Calzetti F, Pizzolo G, Montecucco C, et al. G-CSF-stimulated neutrophils are a prominent source of functional BLyS. J Exp Med (2003) 197:297–302. doi:10.1084/jem.20021343
162. Kern C, Cornuel JF, Billard C, Tang R, Rouillard D, Stenou V, et al. Involvement of BAFF and APRIL in the resistance to apoptosis of B-CLL through an autocrine pathway. Blood (2004) 103:679–88. doi:10.1182/blood-2003-02-0540
163. He B, Chadburn A, Jou E, Schattner EJ, Knowles DM, Cerutti A. Lymphoma B cells evade apoptosis through the TNF family members BAFF/BLyS and APRIL. J Immunol (2004) 172:3268–79. doi:10.4049/jimmunol.172.5.3268
164. Nakajima K, Itoh K, Nagatani K, Okawa-Takatsuji M, Fujii T, Kuroki H, et al. Expression of BAFF and BAFF-R in the synovial tissue of patients with rheumatoid arthritis. Scand J Rheumatol (2007) 36:365–72. doi:10.1080/03009740701286615
165. Chu VT, Enghard P, Schurer S, Steinhauser G, Rudolph B, Riemekasten G, et al. Systemic activation of the immune system induces aberrant BAFF and APRIL expression in B cells in patients with systemic lupus erythematosus. Arthritis Rheum (2009) 60:2083–93. doi:10.1002/art.24628
166. Daridon C, Devauchelle V, Hutin P, Le Berre R, Martins-Carvalho C, Bendaoud B, et al. Aberrant expression of BAFF by B lymphocytes infiltrating the salivary glands of patients with primary Sjogren’s syndrome. Arthritis Rheum (2007) 56:1134–44. doi:10.1002/art.22458
167. Zhang L, Liu C, Zhou X, Xie Y, Su L, Geng Q, et al. Transgenic overexpression of BAFF regulates the expression of immune-related genes in zebrafish, Danio rerio. J Genet (2016) 95:751–60. doi:10.1007/s12041-016-0690-6
168. Das S, Sutoh Y, Hirano M, Han Q, Li J, Cooper MD, et al. Characterization of lamprey BAFF-like gene: evolutionary implications. J Immunol (2016) 197:2695–703. doi:10.4049/jimmunol.1600799
169. Min C, Liang Z, Cui X, Wang Q, Chen Y, Zhang S. Molecular cloning and partial functional characterization of a proliferation inducing ligand (APRIL) in zebrafish (Danio rerio). Dev Comp Immunol (2012) 37:202–6. doi:10.1016/j.dci.2011.12.002
170. Granja AG, Holland JW, Pignatelli J, Secombes CJ, Tafalla C. Characterization of BAFF and APRIL subfamily receptors in rainbow trout (Oncorhynchus mykiss). Potential role of the BAFF/APRIL axis in the pathogenesis of proliferative kidney disease. PLoS One (2017) 12:e0174249. doi:10.1371/journal.pone.0174249
171. Gong YF, Xiang LX, Shao JZ. CD154-CD40 interactions are essential for thymus-dependent antibody production in zebrafish: insights into the origin of costimulatory pathway in helper T cell-regulated adaptive immunity in early vertebrates. J Immunol (2009) 182:7749–62. doi:10.4049/jimmunol.0804370
172. Lagos LX, Iliev DB, Helland R, Rosemblatt M, Jorgensen JB. CD40L – a costimulatory molecule involved in the maturation of antigen presenting cells in Atlantic salmon (Salmo salar). Dev Comp Immunol (2012) 38:416–30. doi:10.1016/j.dci.2012.07.011
173. Sedger LM, McDermott MF. TNF and TNF-receptors: from mediators of cell death and inflammation to therapeutic giants – past, present and future. Cytokine Growth Factor Rev (2014) 25:453–72. doi:10.1016/j.cytogfr.2014.07.016
174. Lam FW, Wu SY, Lin SJ, Lin CC, Chen YM, Wang HC, et al. The expression of two novel orange-spotted grouper (Epinephelus coioides) TNF genes in peripheral blood leukocytes, various organs, and fish larvae. Fish Shellfish Immunol (2011) 30:618–29. doi:10.1016/j.fsi.2010.12.011
175. Arkoosh MR, Kaattari SL. Development of immunological memory in rainbow trout (Oncorhynchus mykiss). I. An immunochemical and cellular analysis of the B cell response. Dev Comp Immunol (1991) 15:279–93. doi:10.1016/0145-305X(91)90021-P
176. Bailey GS, Poulter RT, Stockwell PA. Gene duplication in tetraploid fish: model for gene silencing at unlinked duplicated loci. Proc Natl Acad Sci U S A (1978) 75:5575–9. doi:10.1073/pnas.75.11.5575
177. Grimholt U. Whole genome duplications have provided teleosts with many roads to peptide loaded MHC class I molecules. BMC Evol Biol (2018) 18:25. doi:10.1186/s12862-018-1138-9
178. Ashkenazi A, Dixit VM. Death receptors: signaling and modulation. Science (1998) 281:1305–8. doi:10.1126/science.281.5381.1305
Keywords: TNFSF, TNFRSF, fish, B cells, evolution, immunity
Citation: Tafalla C and Granja AG (2018) Novel Insights on the Regulation of B Cell Functionality by Members of the Tumor Necrosis Factor Superfamily in Jawed Fish. Front. Immunol. 9:1285. doi: 10.3389/fimmu.2018.01285
Received: 05 March 2018; Accepted: 22 May 2018;
Published: 07 June 2018
Edited by:
Brian Dixon, University of Waterloo, CanadaReviewed by:
Irene Salinas, University of New Mexico, United StatesAntonio Figueras, Consejo Superior de Investigaciones Científicas (CSIC), Spain
Steve Bird, University of Waikato, New Zealand
Copyright: © 2018 Tafalla and Granja. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aitor G. Granja, YWl0b3IuZ29uemFsZXpAaW5pYS5lcw==
 Carolina Tafalla
Carolina Tafalla Aitor G. Granja
Aitor G. Granja