- 1Department of Experimental Medicine, University of Perugia, Perugia, Italy
- 2Center for Translational Medicine, International Clinical Research Centre, St. Anne's University Hospital Brno, Brno, Czechia
Microbial Causes of Respiratory Tract Infections
The respiratory system is the first point of contact with airborne microbial compounds. Consequently, lung mucosal immunity has been extensively studied to understand the mechanisms of host resistance to respiratory infections. The lungs exhibit highly active innate and adaptive mucosal immune mechanisms: they are infiltrated with a wide spectrum of immune cells in steady state and possess the capacity to recruit vast numbers of infiltrating cells upon infection or encounter with inflammatory stimuli. Despite the existence of such protective mechanisms, respiratory tract infections (RTIs) with epidemic and pandemic potential are one of the most common causes of morbidity and mortality worldwide. In recent years, studies using new lung culture systems, such as air liquid interface (ALI), spheroids, tissue explants and advances in DNA sequencing technology have helped identify that the upper and lower respiratory tracts represent distinct biomes in terms of their commensal microorganism colonization, immune barriers and host defense mechanisms (1–3). Most lower respiratory tract infections (LRTIs) cause bronchitis, bronchiolitis and pneumonia as a result of Streptococcus pneumonia or Haemophilus Influenzae infection. In children, respiratory viruses are responsible for an enormous amount of serious LRTIs (4, 5). In addition, most upper respiratory tract infections are of viral etiology (6). Fungal infections of the lower respiratory tract are also typically caused by pathogenic dimorphic fungi (7). In addition, opportunistic fungi as Aspergillus fumigatus commonly cause pneumonia. There is an extraordinary need to better understand human respiratory tract infections, as LRTI represent one of the ten most common causes of death in the world (8).
Technical limitations are inherent with pneumonia animal models and in vitro lung infections modeled using immortalized cell lines. In particular, for in vivo models, lung anatomy, namely the distribution of the bronchial glands, differs between rodents and humans, and complex processes such as mucus production, or organization of the epithelial barrier are not accurately reproduced experimentally. For in vitro lung infections, it is not possible to reproduce in vivo-like architecture, the microenvironment, the pulmonary cell complexity in composition. Moreover, bronchial epithelial cells lack cilia and tight junctions. Although lung epithelial barrier cell signaling is today more deeply understood, it has still not been fully evaluated in reproducible lung infection models.
Recent advances in the stem-cell field, including the generation of protocols allowing tissue differentiation from induced pluripotent stem cells (iPSCs), have provided new opportunities to study host–pathogen interactions in a human experimental system that maintains controlled tissue complexity. For this reason, recently developed techniques now allow for innovative and more meaningful investigations of 3D human lung tissue. Here we outline the complexity of the epithelial barrier to opportunistic microbes and the new 2D and 3D lung models of infection, and explain how these models may be used to improve our knowledge on epithelial cell signaling events upon infection.
Complexity of the Lung Epithelial Barrier
Epithelial cells represent the first point of contact for opportunistic microbes or pathogens in the respiratory tract (9). The lung mucosa senses infection through pattern recognition receptors expressed by the airway epithelia (10–12), alveolar cells (13, 14), and mesenchymal stem cells (15, 16). Several cell types then orchestrate mucosal barrier immunity: club cells, ciliated cells, basal cells, goblet cells and neuroendocrine cells as tuft cells decorate the proximal airways, while type-1 and type-2 alveolar cells populate the distal epithelium. The lungs can also be divided into a conducting zone and a respiratory zone, which are populated by different progenitor cell types. The conducting zone is abundant in basal cells (17, 18), airway secretory club cells and lineage-negative epithelial cells (18). The respiratory zone is mainly populated by alveolar type II cells (AEC II) that can proliferate and act as progenitor cells, replacing AECII and AECI cells (19).
The complex barrier functions executed by the lung epithelial layer, including mucociliary clearance and antimicrobial production, cooperate to clear inhaled pathogens. Unsurprisingly, gaining a clear understanding of the lung epithelial barrier has been restrained by this described complexity of the lung organization and its underlying cell types. Early research strategies based on immortalized airway epithelial cells or lung primary cells thus may not replicate the conditions where inhaled microorganisms become pathogens that trigger infections.
Developing 2D and 3D Tools to Mimic Lung Structure
In vivo lung epithelial barrier experiments are challenging: dissecting the roles of individual cell types is complex due to the heterogeneity of the lung and the lack of specific cell markers. Much research has thus utilized the 2D ALI system and immortalized, lung cancer-derived cell culture approaches to study airway epithelial barrier–pathogen interactions in vitro (20, 21). The ALI system has been successfully used to differentiate progenitor cells, such as primary bronchial epithelial cells, into the corresponding airway tract upon exposure to the appropriate culture conditions (22). For example, basal cells cultured in an ALI system, can differentiate into a pseudo-stratified epithelium containing ciliated, goblet and basal cells (23). So far, this method has helped elucidate the transcriptomic profile of basal cells (24) and the impact of virus-infected basal cells on epithelium development (25). The limits of these “conventional” approaches, however, are namely the lack of tissue architecture; indeed, they are not able to faithfully recapitulate the phenotypic and morphological characteristics of the native epithelium (Table 1).
To overcome such limitations of the ALI 2D system, an ALI 3D culture system has been developed, in which human bronchial epithelial cells are cultured on a permeable membrane submerged in media supplemented with stromal cells or growth factors (26).
A precursor to lung organoid culture was the development of a “Lung-on-a-Chip”: a microphysciological device that replicates on a chip the functional unit of the breathing lung. This system is based on communication between alveolar and endothelial cells through a microporous elastomeric membrane. The alveolar cells located in the upper chamber, are exposed to air and fed by endothelial cells grown in the bottom layer. Although this Lung-on-a-Chip system has been used for drug discovery and toxicology studies (27), it is not possible to recreate the lung architecture, which has a central role in various physiological functions (Table 1) (28).
Over the past 15 years, new in vitro strategies have facilitated the production of miniature 3D structures, known as “mini organs” or organoids. Organoids are multi-cellular, stem-cell-derived systems in which cells spontaneously self-organize into properly differentiated, functional cell types that resemble in vivo counterparts and recapitulate the key features of the entire organ (Table 1). Organoids can help dissecting the role of individual cell types because are deprived of immune cells and endothelial cells. The overall approach is based on using hydrogels containing a gelatinous mixture, such as laminin and collagen, to mimic the extracellular matrix and self-organizing iPSCs. The human adult lung stem cells that are essential for epithelial renewal and tissue repair have proven capable of generating such 3D human lung organoids when cultured in the appropriate differentiating conditions. Adult stem cells, once isolated from the peripheral tissue, are usually cultured in enriched medium in the presence of a hydrogel scaffold that provides structural support and mediates instructive signaling for cell polarization, retention and mobilization. This organoid medium needs to be changed every 4 days, and the organoids must be passaged every 2 weeks. After 3 weeks of culture, starting from adult stem cells, the first small organoids reproduce the faithful microanatomy of the lung and recapitulate some specific lung functions (Figure 1) (29). The protocols using human tissue stem cells and iPSCs have been developed further to study organ-related pathologies and ontogeny (30–32). Over the past decade, although lung organoids have been used for much translational research, such as lung engraftment (33), the main application has been in vitro disease modeling.
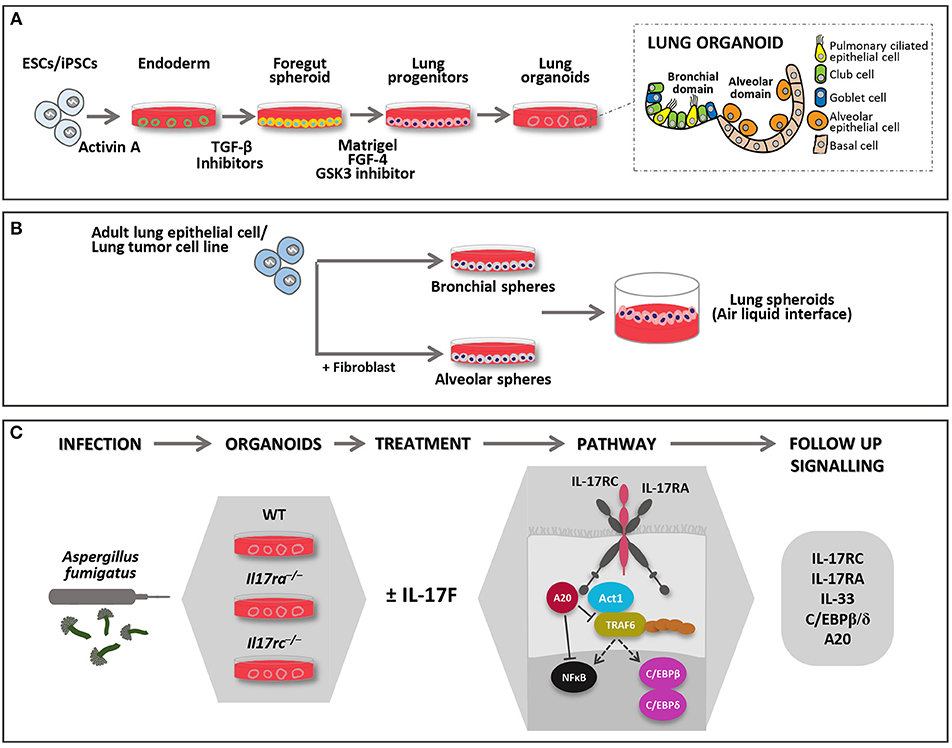
Figure 1. Models of 3D lung organoid infections. Lung organoids can be developed from iPSCs or adult stem cells (upper panel). (A) iPSCs derived lung organoids: cells are differentiated into endoderm by Activin A and further to anterior foregut followed by lung progenitor spheroids through the activation and inhibition of several signaling pathways. The progenitor spheroids are further embedded in matrigel to develop 3D lung organoids when supplemented with appropriate growth factors, which resemble lung tissue in morphology and function. They form the bronchial and alveolar like domains of the lungs and has both functional (epithelial cells) and supportive (basal mesenchymal cells) pulmonary tissue. (B) Adult lung progenitor cells can form spheroids, which can be further cultured on ALI to mimic lung environment. Spheroids do not form the exact morphology and lack some of the functional cell types. They can either form alveolar or bronchial branch based on various protocols, some of them require co-culture with support cells. (C) In the lower panel, the description of an experimental model of RTIs where lung organoids derived from different genotypes may be injected with Aspergillus fumigatus. The model may be used to study IL-17R signaling pathways in 3D system where the complex role of IL-17F may be studied. FGF-4 (Fibroblast growth factor 4); GSK3 (Glycogen synthase kinase 3); TGF-β (Transforming growth factor beta 1).
Using Organoids to Model RTIs
Access to the organoid lumen for experimental perturbation is challenging; thus, many researchers add bacteria to the supernatants of organoid-derived 2D cultures to monitor host–pathogen interactions. Cutting-edge technologies, however, now permit microinjection of microbes into the organoid lumen (34), allowing host–microbiota interplay within the 3D structure. More recently, a high-throughtput organoid microinjector system has been developed that can deliver microbial communities into the organoid lumen (35).
The first tissue 3D organoid models used to study host–pathogen interactions were intestinal organoids (36). To date, lung organoids have been used in microbial infection studies to understand the molecular mechanisms of epithelial renewal upon viral infection (37) and to study the cytokine profile released in response to pattern recognition receptor activation by Pseudomonas aeruginosa (38). Here, wild-type and transgenic lung organoids were treated with bacterial flagellar hook proteins eliciting IL-1β and IL-6 release (38). Another study using organoids infected with Cryptosporidium oocytes provided deep understanding of the microbial life cycle and showed that the parasite is able to infect secretory and non-secretory cells, triggering the Type I interferon release from epithelial cells (39). Of note, even though organoids have reproduced the 3D lung architecture during infection, a role for immune cells has not yet been evaluated.
While the above-mentioned studies have been based on the use of 3D organoids differentiated from murine adult stem cells, several efforts are ongoing to generate organoids from immature lung epithelial and iPSCs. iPSCs are obtained by transfecting and reprogramming adult somatic cells with pluripotency transcription factors (40).
As well as studying host–pathogen interactions, lung organoids have proven valuable in understanding cystic fibrosis pathology (41, 42). Here, major breakthroughs have been achieved through using iPSCs derived from patients carrying genetic mutations to generate organoids. These organoids modeling cystic fibrosis have permitted drug testing directly on patient cells with affected organ properties (43). Further studies that aim to elucidate the molecular nature of the protective immune barriers in the lower and upper respiratory tracts will rapidly advance the rational design of novel therapeutics targeting such important diseases. Future studies using patient-specific organoids may permit bio-banking and the development of personalized medicines and targeted therapies for opportunistic pulmonary infections.
Using Lung Organoids to Delineate IL-17R Signaling in Epithelial Lung Cells
Recent studies have identified the importance and complexity of interleukin-17 receptor (IL-17R) signaling by lung epithelial cells and highlighted the need for deeper investigations into the regulatory network activated by IL-17 cytokines in acute or chronic inflammation. Thus far, studies have shown that high IL-17R expression on lung epithelial cells has a prominent role in the innate immune defense against pulmonary fungal pathogens, including Blastomyces dermatitidis (44) and Aspergillus fumigatus (45). These epithelial cells may orchestrate innate antifungal immunity by first up-regulating the number of lymphocytes that secrete interleukin-17A (IL-17A) and granulocyte-macrophage colony-stimulating factor (GM-CSF) (44). They then respond to secreted IL-17 via the IL-17R, which regulates the secretion of antimicrobial peptides and chemokines that recruit neutrophils. IL-17R expressed on lung club cells orchestrates neutrophil recruitment and Klebsiella pneumonia resistance (46).
IL-17A and IL-17F homodimers both bind the IL-17R subunits IL-17RA and IL-17RC (47–49).
Also, the human IL-17A/F heterodimer may bind the complex IL-17R (50) since it may mimic the IL-17A as well as IL-17F behaving as a two-face cytokine. During Aspergillus fungal infection, the fungus increases IL-17F expression, which subsequently induces IL-33 and IL-17RC expression on lung epithelial cells, especially in the context of IL-17RA deficiency (45).
A polarized lung epithelium is required for IL-17R expression and innate immune functions, such as mucus production (51). Because organoid cultures recapitulate tissue polarity, they thus provide an exciting possibility of using lung organoids to comprehensively investigate IL-17R signaling in the lung. Improving our understanding of IL-17R signaling by lung epithelial cells is likely to offer new opportunities to develop and test therapeutics for inflammatory diseases and identify new molecular targets to improve resistance to infections. Such work is important given that human studies have demonstrated the importance of IL-17-driven immunity in LRTI infections, with mutations in IL-17RA or IL-17RC conferring increased susceptibility to RTIs (52, 53). In addition, the immune-free organoid microenvironment favors IL-17RC signaling studies, as epithelial cells express high levels of IL-17RC compared to immune cells; furthermore, the best characterized IL-17A-targeted cells are non-immune cells, including epithelial cells and mesenchymal cells of the lung (51).
Another important area of research in which lung organoids are anticipated to be of value is in deciphering IL-17F function in asthma. It is produced by multiple cell types including bronchial epithelial cells. IL-17F mediates asthma via IL-17R binding on bronchial epithelial cells, eosinophils, fibroblasts and airway smooth muscle cells (54). A certain extent of pulmonary IL-17F, however, is also released by immune cells, which are not differentiated in 3D-organoids. This limitation may be compensated by using exogenous IL-17F or by co-culturing organoids with IL-17F-producing cells. It would be interesting to study the independent role of epithelial IL-17F in modulating airway remodeling, asthma and steroid resistance in 3D cultures. In addition, transgenic lung organoids for IL-17R subunits expression may be useful to better disentangle IL-17F receptor signaling (Figure 1). The need to elucidate the function of the receptor subunits comes from the evidence that IL-17RA or IL-17RC mutations have been also described in human fungal infections (52, 53). Clearly, there is a need to recapitulate the 3D structure of lung organoids with the appropriate cell mixture in order to properly investigate pulmonary intercellular networks and immune receptor signaling pathways as IL-17R.
Author Contributions
GP, AL, MA, IT, and SSJ critically read, analyzed, and discussed the literature and conceived the outline of the manuscript. JF and TZ wrote the manuscript. All the authors edited the manuscript and provided valuable discussions and criticisms.
Funding
This research was supported by European Social Fund and European Regional Development Fund—Project MAGNET (No. CZ.02.1.01/0.0/0.0/15_003/0000492), by Ministry of Health of the Czechia, grant no. NV18-06-00529 to JF and by the Italian Grant Programma per Giovani Ricercatori—Rita Levi Montalcini 2013 PGR13XNIDJ to TZ. All rights reserved.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Insight Editing London for his assistance in the preparation of the text and Dr. Cristina Massi-Benedetti for digital art and editorial assistance.
References
1. Dickson RP. The microbiome and critical illness. Lancet Respir Med. (2016) 4:59–72. doi: 10.1016/S2213-2600(15)00427-0
2. Zscheppang K, Berg J, Hedtrich S, Verheyen L, Wagner DE, Suttorp N, et al. Human pulmonary 3D models for translational research. Biotechnol J. (2018) 13:1700341. doi: 10.1002/biot.201700341
3. Moffatt MF, Cookson WO. The lung microbiome in health and disease. Clin Med. (2017) 17:525–9. doi: 10.7861/clinmedicine.17-6-525
4. Pavia AT. Viral infections of the lower respiratory tract: old viruses, new viruses, and the role of diagnosis. Clin Infect Dis. (2011) 52(Suppl. 4):S284–9. doi: 10.1093/cid/cir043
5. de Benedictis FM, Bush A. Recurrent lower respiratory tract infections in children. BMJ. (2018) 362:k2698. doi: 10.1136/bmj.k2698
6. Hijano DR, Maron G, Hayden RT. Respiratory viral infections in patients with cancer or undergoing hematopoietic cell transplant. Front Microbiol. (2018) 9:3097. doi: 10.3389/fmicb.2018.03097
7. Nemecek JC, Wüthrich M, Klein BS. Global control of dimorphism and virulence in fungi. Science. (2006) 312:583–8. doi: 10.1126/science.1124105
8. Reddy KS. Global burden of disease study 2015 provides GPS for global health 2030. Lancet. (2016) 388:1448–9. doi: 10.1016/S0140-6736(16)31743-3
9. Whitsett JA, Alenghat T. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat Immunol. (2015) 16:27–35. doi: 10.1038/ni.3045
10. Hauber HP, Tulic MK, Tsicopoulos A, Wallaert B, Olivenstein R, Daigneault P, et al. Toll-like receptors 4 and 2 expression in the bronchial mucosa of patients with cystic fibrosis. Can Respir J. (2005) 12:13–8. doi: 10.1155/2005/648984
11. Muir A, Soong G, Sokol S, Reddy B, Gomez MI, Van Heeckeren A, et al. Toll-like receptors in normal and cystic fibrosis airway epithelial cells. Am J Respir Cell Mol Biol. (2004) 30:777–83. doi: 10.1165/rcmb.2003-0329OC
12. Sha Q, Truong-Tran AQ, Plitt JR, Beck LA, Schleimer RP. Activation of airway epithelial cells by toll-like receptor agonists. Am J Respir Cell Mol Biol. (2004) 31:358–64. doi: 10.1165/rcmb.2003-0388OC
13. Armstrong L, Medford AR, Uppington KM, Robertson J, Witherden IR, Tetley TD, et al. Expression of functional toll-like receptor-2 and−4 on alveolar epithelial cells. Am J Respir Cell Mol Biol. (2004) 31:241–5. doi: 10.1165/rcmb.2004-0078OC
14. Ritter M, Mennerich D, Weith A, Seither P. Characterization of Toll-like receptors in primary lung epithelial cells: strong impact of the TLR3 ligand poly(I:C) on the regulation of Toll-like receptors, adaptor proteins and inflammatory response. J Inflamm. (2005) 2:16. doi: 10.1186/1476-9255-2-16
15. Cheung MB, Sampayo-Escobar V, Green R, Moore ML, Mohapatra S, Mohapatra SS. Respiratory syncytial virus-infected mesenchymal stem cells regulate immunity via interferon beta and indoleamine-2,3-Dioxygenase. PloS One. (2016) 11:e0163709. doi: 10.1371/journal.pone.0163709
16. Zhou P, Liu Z, Li X, Zhang B, Wang X, Lan J, et al. Migration ability and Toll-like receptor expression of human mesenchymal stem cells improves significantly after three-dimensional culture. Biochem Biophys Res Commun. (2017) 491:323–8. doi: 10.1016/j.bbrc.2017.07.102
17. Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci USA. (2009) 106:12771–5. doi: 10.1073/pnas.0906850106
18. Lee JH, Bhang DH, Beede A, Huang TL, Stripp BR, Bloch KD, et al. Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis. Cell. (2014) 156:440–55. doi: 10.1016/j.cell.2013.12.039
19. Rawlins EL, Ostrowski LE, Randell SH, Hogan BL. Lung development and repair: contribution of the ciliated lineage. Proc Natl Acad Sci USA. (2007) 104:410–7. doi: 10.1073/pnas.0610770104
20. Kesimer M, Kirkham S, Pickles RJ, Henderson AG, Alexis NE, Demaria G, et al. Tracheobronchial air-liquid interface cell culture: a model for innate mucosal defense of the upper airways? Am J Physiol Lung Cell Mol Physiol. (2009) 296:L92–100. doi: 10.1152/ajplung.90388.2008
21. Lee MK, Yoo JW, Lin H, Kim YS, Kim DD, Choi YM, et al. Air-liquid interface culture of serially passaged human nasal epithelial cell monolayer for in vitro drug transport studies. Drug Deliv. (2005) 12:305–11. doi: 10.1080/10717540500177009
22. Prytherch Z, Job C, Marshall H, Oreffo V, Foster M, BéruBé K. Tissue-specific stem cell differentiation in an in vitro airway model. Macromol Biosci. (2011) 11:1467–77. doi: 10.1002/mabi.201100181
23. Walters MS, Gomi K, Ashbridge B, Moore MA, Arbelaez V, Heldrich J, et al. Generation of a human airway epithelium derived basal cell line with multipotent differentiation capacity. Respir Res. (2013) 14:135. doi: 10.1186/1465-9921-14-135
24. Hackett NR, Shaykhiev R, Walters MS, Wang R, Zwick RK, Ferris B, et al. The human airway epithelial basal cell transcriptome. PLoS ONE. (2011) 6:e18378. doi: 10.1371/journal.pone.0018378
25. Persson BD, Jaffe AB, Fearns R, Danahay H. Respiratory syncytial virus can infect basal cells and alter human airway epithelial differentiation. PLoS ONE. (2014) 9:e102368. doi: 10.1371/journal.pone.0102368
26. Whitcutt MJ, Adler KB, Wu R. A biphasic chamber system for maintaining polarity of differentiation of cultured respiratory tract epithelial cells. Vitro Cell Dev Biol. (1988) 24:420–8. doi: 10.1007/BF02628493
27. Huh D, Leslie DC, Matthews BD, Fraser JP, Jurek S, Hamilton GA, et al. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci Transl Med. (2012) 4:159ra147. doi: 10.1126/scitranslmed.3004249
28. Huh DD. A human breathing lung-on-a-chip. Ann Am Thorac Soc. (2015) 12(Suppl. 1):S42-44. doi: 10.1513/AnnalsATS.201410-442MG
29. Huang SX, Islam MN, O'Neill J, Hu Z, Yang YG, Chen YW, et al. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat Biotechnol. (2014) 32:84–91. doi: 10.1038/nbt.2754
30. Pollard BS, Pollard HB. Induced pluripotent stem cells for treating cystic fibrosis: State of the science. Pediatr Pulmonol. (2018) 53:S12–29. doi: 10.1002/ppul.24118
31. Chen YW, Huang SX, de Carvalho ALRT, Ho SH, Islam MN, Volpi S, et al. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat Cell Biol. (2017) 19:542–9. doi: 10.1038/ncb3510
32. Dye BR, Hill DR, Ferguson MA, Tsai YH, Nagy MS, Dyal R, et al. in vitro generation of human pluripotent stem cell derived lung organoids. Elife. (2015) 4:e05098. doi: 10.7554/eLife.05098
33. Dye BR, Dedhia PH, Miller AJ, Nagy MS, White ES, Shea LD, et al. A bioengineered niche promotes in vivo engraftment and maturation of pluripotent stem cell derived human lung organoids. Elife. (2016) 5:e19732. doi: 10.7554/eLife.19732
34. Bertaux-Skeirik N, Feng R, Schumacher MA, Li J, Mahe MM, Engevik AC, et al. CD44 plays a functional role in Helicobacter pylori-induced epithelial cell proliferation. PLoS Pathog. (2015) 11:e1004663. doi: 10.1371/journal.ppat.1004663
35. Williamson IA, Arnold JW, Samsa LA, Gaynor L, DiSalvo M, Cocchiaro JL, et al. A high-throughput organoid microinjection platform to study gastrointestinal microbiota and luminal physiology. Cell Mol Gastroenterol Hepatol. (2018) 6:301–19. doi: 10.1016/j.jcmgh.2018.05.004
36. Bartfeld S. Modeling infectious diseases and host-microbe interactions in gastrointestinal organoids. Dev Biol. (2016) 420:262–70. doi: 10.1016/j.ydbio.2016.09.014
37. Quantius J, Schmoldt C, Vazquez-Armendariz AI, Becker C, El Agha E, Wilhelm J, et al. Influenza virus infects epithelial stem/progenitor cells of the distal lung: impact on Fgfr2b-driven epithelial repair. PLoS Pathog. (2016) 12:e1005544. doi: 10.1371/journal.ppat.1005544
38. Shen Y, Chen L, Wang M, Lin D, Liang Z, Song P, et al. Flagellar hooks and hook protein FlgE participate in host microbe interactions at immunological level. Sci Rep. (2017) 7:1433. doi: 10.1038/s41598-017-01619-1
39. Heo I, Dutta D, Schaefer DA, Iakobachvili N, Artegiani B, Sachs N, et al. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat Microbiol. (2018) 3:814–23. doi: 10.1038/s41564-018-0177-8
40. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. (2006) 126:663–76. doi: 10.1016/j.cell.2006.07.024
41. Ikpa PT, Bijvelds MJ, de Jonge HR. Cystic fibrosis: toward personalized therapies. Int J Biochem Cell Biol. (2014) 52:192–200. doi: 10.1016/j.biocel.2014.02.008
42. Mou H, Zhao R, Sherwood R, Ahfeldt T, Lapey A, Wain J, et al. Generation of multipotent lung and airway progenitors from mouse ESCs and patient-specific cystic fibrosis iPSCs. Cell Stem Cell. (2012) 10:385–97. doi: 10.1016/j.stem.2012.01.018
43. Wong AP, Bear CE, Chin S, Pasceri P, Thompson TO, Huan LJ, et al. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat Biotechnol. (2012) 30:876–82. doi: 10.1038/nbt.2328
44. Hernández-Santos N, Wiesner DL, Fites JS, McDermott AJ, Warner T, Wüthrich M, et al. Lung epithelial cells coordinate innate lymphocytes and immunity against pulmonary fungal infection. Cell Host Microbe. (2018) 23:511–22 e515. doi: 10.1016/j.chom.2018.02.011
45. De Luca A, Pariano M, Cellini B, Costantini C, Villella VR, Jose SS, et al. The IL-17F/IL-17RC axis promotes respiratory allergy in the proximal airways. Cell Rep. (2017) 20:1667–80. doi: 10.1016/j.celrep.2017.07.063
46. Chen K, Eddens T, Trevejo-Nunez G, Way EE, Elsegeiny W, Ricks DM, et al. IL-17 Receptor signaling in the lung epithelium is required for mucosal chemokine gradients and pulmonary host defense against K. pneumoniae. Cell Host Microbe. (2016) 20:596–605. doi: 10.1016/j.chom.2016.10.003
47. Aggarwal S, Gurney AL. IL-17: prototype member of an emerging cytokine family. J Leukoc Biol. (2002) 71:1–8. doi: 10.1189/jlb.71.1.1
48. Ely LK, Fischer S, Garcia KC. Structural basis of receptor sharing by interleukin 17 cytokines. Nat Immunol. (2009) 10:1245–51. doi: 10.1038/ni.1813
49. Toy D, Kugler D, Wolfson M, Vanden Bos T, Gurgel J, Derry J, et al. Cutting edge: interleukin 17 signals through a heteromeric receptor complex. J Immunol. (2006) 177:36–9. doi: 10.4049/jimmunol.177.1.36
50. Goepfert A, Lehmann S, Wirth E, Rondeau JM. The human IL-17A/F heterodimer: a two-faced cytokine with unique receptor recognition properties. Sci Rep. (2017) 7:8906. doi: 10.1038/s41598-017-08360-9
51. Tsai HC, Velichko S, Hung LY, Wu R. IL-17A and Th17 cells in lung inflammation: an update on the role of Th17 cell differentiation and IL-17R signaling in host defense against infection. Clin Dev Immunol. (2013) 2013:267971. doi: 10.1155/2013/267971
52. Ling Y, Cypowyj S, Aytekin C, Galicchio M, Camcioglu Y, Nepesov S, et al. Inherited IL-17RC deficiency in patients with chronic mucocutaneous candidiasis. J Exp Med. (2015) 212:619–31. doi: 10.1084/jem.20141065
53. Puel A, Cypowyj S, Bustamante J, Wright JF, Liu L, Lim HK, et al. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science. (2011) 332:65–8. doi: 10.1126/science.1200439
Keywords: IL-17 immunity, epithelial barrier, lung infection, Aspergillus fumigatus, lung organoids
Citation: Paolicelli G, Luca AD, Jose SS, Antonini M, Teloni I, Fric J and Zelante T (2019) Using Lung Organoids to Investigate Epithelial Barrier Complexity and IL-17 Signaling During Respiratory Infection. Front. Immunol. 10:323. doi: 10.3389/fimmu.2019.00323
Received: 31 August 2018; Accepted: 07 February 2019;
Published: 28 February 2019.
Edited by:
François Trottein, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Nicolas Riteau, UMR7355 Immunologie et Neurogénétique Expérimentales et Moléculaires (INEM), FranceCopyright © 2019 Paolicelli, Luca, Jose, Antonini, Teloni, Fric and Zelante. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Teresa Zelante, dGVyZXNhLnplbGFudGVAdW5pcGcuaXQ=
†These authors have contributed equally to this work
 Giuseppe Paolicelli
Giuseppe Paolicelli Antonella De Luca
Antonella De Luca Shyam S. Jose
Shyam S. Jose Martina Antonini1
Martina Antonini1 Jan Fric
Jan Fric Teresa Zelante
Teresa Zelante