- 1Laboratory of Immunogenetics, Department of Basic Health Sciences, Maringá State University (UEM), Maringá, Brazil
- 2Department of Medicine, Faculty of Medical Sciences, Campinas State University (UNICAMP), São Paulo, Brazil
Vitamin D, together with its nuclear receptor (VDR), plays an important role in modulating the immune response, decreasing the inflammatory process. Some polymorphisms of the VDR gene, such as BsmI (G>A rs1544410), ApaI (G>T rs7975232), and TaqI (T>C rs731236) could affect its stability and mRNA transcription activity, while FokI T>C (rs2228570) gives a truncated protein with three fewer amino acids and more efficiency in binding vitamin D. This study evaluated these four polymorphisms in the immunopathogenesis of leprosy in 404 patients and 432 control individuals without chronic or infectious disease in southern Brazil. When analyzing differences in the allele and genotype frequency of polymorphisms between patients (leprosy per se, multibacillary, and paucibacillary clinical forms) and controls, we found no statistically significant association. Regarding haplotype analysis, the bAt haplotype was associated with protection from leprosy per se (P = 0.004, OR = 0.34, CI = 0.16–0.71) and from the multibacillary clinical form (P = 0.005, OR = 0.30, CI = 0.13–0.70). In individuals aged 40 or more years, this haplotype has also showed protection against leprosy per se and multibacillary (OR = 0.26, CI = 0.09–0.76; OR = 0.26, CI = 0.07–0.78, respectively), while the BAt haplotype was a risk factor for leprosy per se in the same age group (OR = 1.34, CI = 1.04–1.73). In conclusion, despite having found no associations between the VDR gene polymorphisms with the development of leprosy, the haplotypes formed by the BsmI, ApaI, and TaqI polymorphisms were associated with leprosy per se and the multibacillary clinical form.
Introduction
Leprosy is a chronic granulomatous disease caused by Mycobacterium leprae, which mainly affects macrophages of the skin, Schwann cells of the peripheral nerve, and eventually other organs and systems (1). Leprosy is among the three neglected diseases with the highest prevalence worldwide. According to the World Health Organization (WHO), in 2017 its incidence reached 210,671 new cases of the disease worldwide. In Brazil, the number of new cases registered in that year was more than 26,000. This sets Brazil at the second place in the ranking of countries with the largest number of leprosy cases in the world (2).
There is considerable clinical variability among leprosy patients once M. leprae infection evokes distinct T cell responses in humans. The classification of leprosy according to immunity includes the following clinical forms: tuberculoid (TT), lepromatous (LL), borderline tuberculoid (BT), borderline borderline (BB), and borderline lepromatous (BL) (3, 4). However, based on the number of skin lesions, the patients can be classified as paucibacillary (PB) and multibacillary (MB) leprosy cases, according to WHO (5, 6). The type of immune response may determine the clinical form, as well as the resistance or susceptibility to the disease. The TT form (also classified as paucibacillary) is characterized by a small number of hypopigmented, well-bordered, anesthetic skin lesions with a low bacillary load, early peripheral nerve impairment, and a T-helper 1 (Th1)-mediated immune response. On the other hand, in form LL (therefore referred to as multibacillary), there is a prevalence of the Th2-mediated immune response, which leads to numerous infiltrated skin lesions displaying high bacillary loads, impaired peripheral nerves, and possible involvement of internal organs (7, 8). However, the predominance of one type of immune response does not mean that cytokines from the other response profiles are not being produced (9).
Many factors can modulate the type of immune response developed by the leprosy patient, such as vaccination with BCG, nutritional status, degree of exposure to M. leprae, and infections by other microorganisms. Besides, leprosy susceptibility can also be influenced by genetic factors (10). As vitamin D metabolizing enzymes and vitamin D receptors are present in many cell types, including various immune cells such as antigen-presenting cells, T cells, B cells, and monocytes, these molecules are important targets in the study of polymorphisms that can modulate the immune response against pathogens (11). Among the immunomodulatory roles of vitamin D is the inhibition of MHC class II, CD40, CD80, and CD86, which leads to the blocking of the Th1 response and activation of regulatory T cells (12). Nuclear vitamin D receptor (VDR) is an intracellular polypeptide that binds to the active vitamin D metabolite, 1,25-Dihydroxyvitamin D3 (1,25-(OH)2-D3), and then interacts with the chromatin, producing a variety of genomic effects (13, 14), such as pleiotropic regulation of human physiology, protection of the cardiac system, cancer prevention, and modulation of the immune system (15, 16). Vitamin D is a direct and indirect regulator of the immune system. The VDR are expressed in T and B cells, dendritic cells, and cells of the monocyte/macrophage lineage (17, 18). Vitamin D acts in suppressing the development of several autoimmune diseases and tissue damage (19–22). The effects of vitamin D on murines and humans include: the development of dendritic cells and T regulatory cells; inhibition of T cell proliferation; inhibition of IFN-γ and IL-17 production; and the induction of IL-4 expression (23, 24). Furthermore, the VDR activation leads to the inhibition of both maturation and proliferation of activated B cells and limits antibody production (25).
The VDR locus is at chromosome 12q13.1 and spans over 75 kb of genomic DNA. The human gene has three transcript variants which encode the Vitamin D3 receptor isoforms VDRA and VDRB1. The first exons of the VDR gene make up the leader sequence and the exons 2–9 encode the structural portion of the gene product (26–29). There are some polymorphisms located near the 3'UTR region of the VDR gene that may affect mRNA stability and translation. These polymorphisms are: BsmI G>A (rs1544410, G allele designated “b” and A allele designated “B”); ApaI G>T (rs7975232, G allele designated “a” and T allele designated “A”), and TaqI T>C (rs731236, T allele designated “T” and C allele designated “t”) (30). Another polymorphism known as FokI T>C (rs2228570, T allele designated “f” and C allele designated “F”), located within the start codon in exon 2 of VDR, gives a truncated protein with three fewer amino acids. The F allele gives rise to the variant protein, which is more efficient in mediating vitamin D action (31, 32). Therefore, considering the role that the active metabolite of VDR exert in the mechanisms of immunity, this study evaluated the association of SNPs (single nucleotide polymorphisms) of the VDR gene (FokI, BsmI, Apal, and TaqI) with the immunopathogenesis and clinical forms of leprosy.
Materials and Methods
Study Population
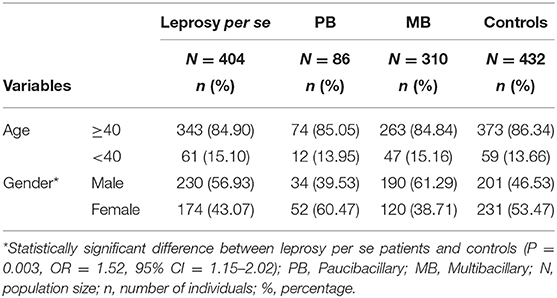
A total of 404 patients (230 men and 174 women) with leprosy from the northwestern region of Paraná, southern Brazil (22° 29′ 30″-26° 42′ 59″ S and 48° 02′ 24″-54° 37′ 38″ W), aged 10–93 years (55.00 ± 13.95), and diagnosed by clinical examination, bacilloscopy, and biopsy were evaluated. Written informed consent was obtained from the participants in this study, including the parents of participants under the age of 16.
In accordance with previous studies, investigations should not be restricted to a sub-analysis of overall leprosy, but should instead contrast multibacillary (MB) and paucibacillary (PB) individuals (6). Thus, patients were reclassified to MB (n = 310) and PB (n = 86). The control group consisted of 432 non-consanguineous individuals from the same region as the patients, and they declared that they did not present any chronic or infectious diseases. Of these, 231 were female and 201 were male, and the age of the controls ranged from 16 to 105 years (50.69 ± 18.07). The characteristics of patients and controls are described in Table 1. All participants were classified as a mixed population from southern Brazil, according to the distribution already described in populations of Paraná (33): predominantly of European origin (80.6%), with a smaller contribution of African (12.5%) and Amerindian (7.0%).
Genotyping
The genotyping of the samples with respect to the VDR gene polymorphisms: FokI T>C (rs2228570), BsmI G>A (rs1544410), ApaI G>T (rs7975232), and TaqI T>C (rs731236) was performed by PCR-RFLP (polymerase chain reaction) (34, 35) with modifications. The technique was validated by direct sequencing of 15 samples for each variant for the SNPs, using BigDye™ Terminator v3.1 Cycle Sequencing Kit (ThermoFisher). Restriction enzymes were used according to the manufacturer's recommendations: FokI # R0109S (BioLabs®Inc), TaqI # ER0671 (ThermoFisher), ApaI # ER1411 (ThermoFisher), and Mva1269I # ER0961 (BsmI) (ThermoFisher). After restriction enzyme digestion, the amplification products were subjected to 2% agarose gel electrophoresis.
Statistical Analysis
The sample size was calculated using the QUANTO software (www.biostats.usc.edu/software), aiming to reach a power of 80%. The SNPStats software (https://www.snpstats.net/start.htm) and OpenEpi program Version 3.01 (https://www.openepi.com/Menu/OE_Menu.htm) were used to determine the allelic, genotypic, and haplotypic frequencies of the VDR gene polymorphisms, and to verify the statistical differences between the groups. The association tests were performed for codominant, dominant, recessive, over dominant, and log-additive genetic inheritance models (36). Haplotype frequency estimates were carried out using expectation–maximization algorithms. Odds ratios with 95% confidence intervals were deemed necessary only for significant P-values. All tests were carried out using a significance level of 5%. Genotype frequency distributions were evaluated to ensure Hardy-Weinberg equilibrium for all genes in the populations.
Results
In this case-control study, allele and genotype frequency distributions of FokI T>C (rs2228570), BsmI G>A (rs1544410), ApaI G>T (rs7975232), and TaqI T>C (rs731236) SNPs were analyzed in a total of 404 patients with leprosy per se (of these, 310 were classified as multibacillary, 86 as paucibacillary, and 8 individuals were not classified) and 432 controls. The distribution of the genotype frequencies for all analyzed genes was consistent with the Hardy-Weinberg equilibrium (P > 0.05). To avoid bias, the gender was used as an adjustment covariate between leprosy per se and the control because of the non-pairing between the groups. Differences in the allele and genotype frequency distributions were not observed between patients (leprosy per se, MB, and PB clinical forms) and controls in linear analyses in the recessive, dominant or codominant inheritance models. There was also no statistically significant difference when comparing the PB and MB clinical forms. The FokI, BsmI, ApaI, and TaqI genotype and allele frequency distributions are summarized in Table 2.
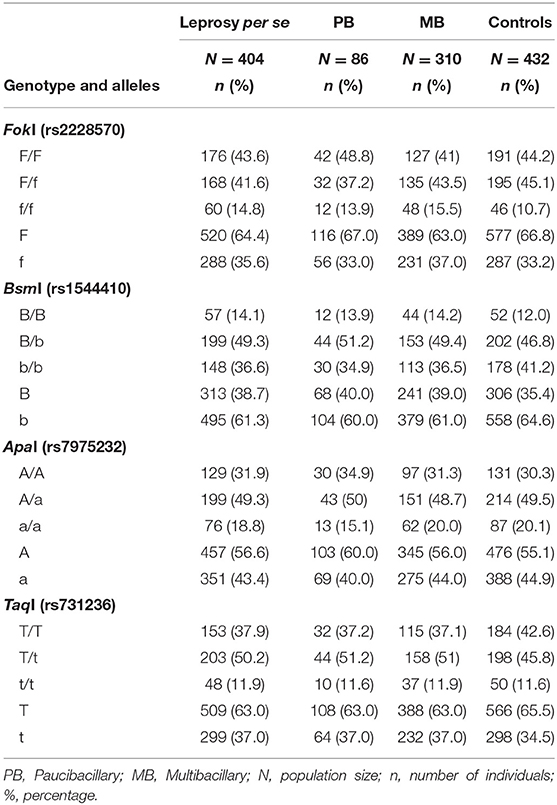
Table 2. Genotype and allele frequency distributions for FokI, BsmI, ApaI, and TaqI polymorphisms in leprosy per se, PB, and MB clinical forms patients and controls.
The BsmI, ApaI, and TaqI polymorphisms were in linkage disequilibrium (D = 0.93, 0.83, and 0.92, respectively). When we evaluated the influence of the haplotypes formed by the BsmI, ApaI, and TaqI polymorphisms of the VDR gene, the haplotype bAt was associated with protection against leprosy per se and the MB clinical form (P = 0.004, OR = 0.34, CI = 0.16–0.71; P = 0.005, OR = 0.30, CI = 0.13–0.70, respectively), as shown in Table 3. When the haplotype was associated with age, the bAt haplotype showed protection against leprosy per se and MB in individuals aged 40 or more years (OR = 0.26, CI = 0.09–0.76; OR = 0.24, CI = 0.07–0.78, respectively). Whereas, the BAt haplotype was a risk factor for leprosy per se in the same age group (OR = 1.34, CI = 1.04–1.73). The haplotype and age cross-classification interaction are shown in Table 4.
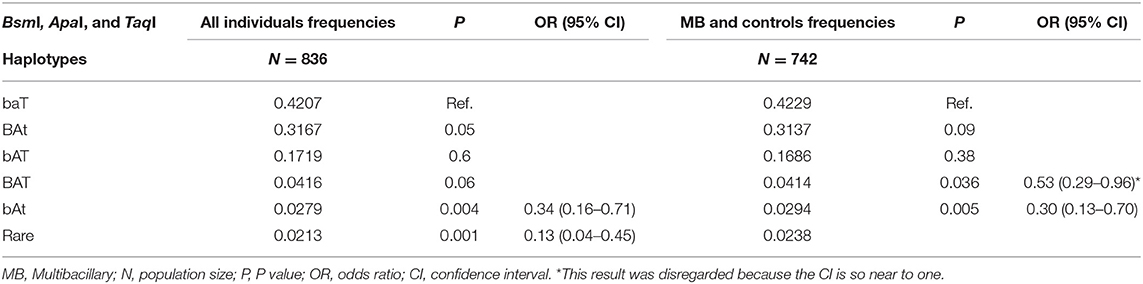
Table 3. Haplotypes formed by the BsmI, ApaI, and TaqI polymorphisms of the VDR gene evaluated in all individuals (leprosy per se patients and controls) and MB clinical form (MB patients and controls).

Table 4. Haplotypes formed by the BsmI, ApaI, and TaqI polymorphisms of the VDR gene evaluated in all individuals (leprosy per se patients and controls) and MB clinical form (MB patients and controls) and age cross-classification interaction.
Discussion
This case-control study investigated the genotypic and allelic frequencies of certain VDR gene polymorphisms in leprosy patients and controls without the disease, in order to evaluate whether these polymorphisms could act as factors of susceptibility or resistance to the disease or to a specific clinical form. When analyzing differences in the allele and genotype frequency distributions between patients (leprosy per se, MB, and PB clinical forms) and controls, we found no statistically significant association.
Consistent with our results, other studies have also found no association between the ApaI polymorphism and leprosy (37). The BsmI polymorphism has also showed no statistically significant association with leprosy in a couple of studies (37, 38). A recent meta-analysis of this polymorphism and tuberculosis showed that the b-allele was a risk factor for disease development, but this was only observed in the Asian population (39). Regarding BsmI, ApaI, and TaqI polymorphisms, it is not clear whether they have an individual effect on the expression or function of the VDR. It is possible that the associations found for these polymorphisms in the various diseases studied, if they actually exist, occurred due to a linkage disequilibrium with a polymorphism that has a functional effect on these diseases (37). A recent systematic review of VDR and leprosy suggests that such a large diversity of results would be a consequence of ethnic heterogeneity, sample size used, design of each study, and also influences from other regions of the gene that have not been studied yet, as well as the likelihood of bacillus virulence being distinct in the different geographic regions where the studies took place (40).
The results concerning haplotypes are in agreement with the hypothesis that the BsmI, ApaI, and TaqI polymorphisms are not directly related to leprosy. However, it is a significant result because it shows that these SNPs may be in linkage disequilibrium with another functional polymorphism. We have analyzed five haplotype alleles in our study, of which haplotypes 1 (baT; 42%), 2 (BAt; 32%), and 3 (bAT; 17%) were the most frequent and were similar to the ones identified by Uitterlinden et al. (41), haplotype 1 (baT; 48%), 2 (BAt; 39%), and 3 (bAT; 11%). Despite our finding of an association of the haplotype bAt with protection against leprosy per se and the MB clinical form, there is no evidence in the literature about the influence of this haplotype in any disease. Moreover, this association could be due to the low frequency of this haplotype in our individuals.
In contrast, the haplotype 2 (BAt) was associated with risk to leprosy per se when the patients were divided by age. Although studies do not agree on which VDR haplotypes are related to susceptibility or protection for various diseases, the fact that BAt shows susceptibility to leprosy can be explained due to a strong linkage disequilibrium between the BsmI, ApaI, and TaqI haplotypes and the poly(A) variable number of tandem repeats (VNTRs) in the 3′UTR of the VDR gene. The poly(A) VNTR polymorphism can be characterized as bi-allelic, and subjects can be classified as having alleles with short or long poly(A) stretches. There is a strong linkage between the haplotype 1 (baT) and the long poly(A) stretch (n = 18–24, long or L alleles), while the haplotype 2 (BAt) is in linkage to the short poly(A) stretch (n = 13–17, short or S alleles). There seems to be a trend for the BAt haplotype to display overall somewhat higher levels of mRNA expression than the baT haplotype. This could be due to a slightly higher mRNA stability and half-life, which would result in higher numbers of VDRs being present in the target cell and better response to vitamin D. Although we can assume that mRNA stability differences might be related to allelic differences, the results are not consistent among the studies (30, 41–43).
Low levels of expression, VDR stability and vitamin D3-VDR interaction were related to leprosy and its complications (44). It has been shown that some individuals with normal levels of vitamin D3, but with low levels of VDR protein, had a high bacilloscopic index and type 2 reaction (45). This fact reinforces the results obtained in the present study, which relate both the polymorphism that changes the interaction vitamin D3-VDR (FokI) and the haplotype formed by the BsmI, ApaI, and TaqI polymorphisms. Although, the FokI polymorphism has a higher consistency of results related to VDR and leprosy, the BsmI, ApaI, and TaqI polymorphisms, especially the haplotype formed by them, should be further investigated in the immunopathogenesis of leprosy.
Data Availability
This manuscript contains previously unpublished data. The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
This work was approved by the Permanent Committee of Ethics in Research with Human Beings of the State University of Maringá (COPEP—UEM) n° 2.424.046/2017.
Author Contributions
AP performed the experiments and drafted the manuscript. HA, BT, and LM analyzed the data and drafted the manuscript. LV participated in the critical revision of the manuscript. QL and JZ analyzed the data, drafted the manuscript, and participated in the critical revision. AS provided materials and participated in the experimental design. JV designed the study and finalized the manuscript.
Funding
This work was supported by the CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), Fundação Araucária do Paraná, and the Laboratory of Immunogenetics at Universidade Estadual de Maringá (Proc. no. 1589/2017-CSDUEM).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We sincerely thank all volunteers and technical staff, and CISAMUSEP in Maringá and CISMEPAR in Londrina, PR, Brazil. The article was checked with respect to the English language by the proof-reading-service.com.
References
1. Eichelmann K, González SE, Salas-Alanis JC, Ocampo-Candiani J. Leprosy. An update: definition, pathogenesis, classification, diagnosis, and treatment. Actas Dermosifiliogr. (2013) 104:554–63. doi: 10.1016/j.adengl.2012.03.028
2. World Health Organization. Global leprosy update, 2017: reducing the disease burden due to leprosy. Week Epidemiol Rec. (2018) 93:445–56.
3. Ridley DS, Jopling WH. Classification of leprosy according to immunity. A five-group system. Int J Lepr Other Mycobact Dis. (1966) 34:255–73.
5. Souza CS. Hanseníase: formas clínicas e diagnóstico. Rev Med Ribeirão Preto. (1997) 30:325–34. doi: 10.11606/issn.2176-7262.v30i3p325-334
6. Gaschignard J, Grant AV, Thuc NV, Orlova M, Cobat A, Huong NT, et al. Pauci- and multibacillary leprosy: two distinct, genetically neglected diseases. Plos Negl Trop Dis. (2016) 10:e0004345. doi: 10.1371/journal.pntd.0004345
7. Modlin RL. Th1-Th2 paradigm: insights from leprosy. J Invest Dermatol. (1994) 102:828–32. doi: 10.1111/1523-1747.ep12381958
8. Sadhu S, Mitra DK. Emerging concepts of adaptive immunity in leprosy. Front Immunol. (2018) 9:604. doi: 10.3389/fimmu.2018.00604
9. Scollard DM, Adams LB, Gillis TP, Krahenbuhl JL, Truman RW, Williams DL. The continuing challenges of leprosy. Clin Microbiol Rev. (2006) 19:338–81. doi: 10.1128/CMR.19.2.338-381.2006
10. Mazini PS, Alves HV, Reis PG, Lopes AP, Sell AM, Santos-Rosa M, et al. Gene association with leprosy: a review of published data. Front Immunol. (2016) 6:658. doi: 10.3389/fimmu.2015.00658
11. Prietl B, Treiber G, Pieber TR, Amrein K. Vitamin D and immune function. Nutrients. (2013) 5:2502–21. doi: 10.3390/nu5072502
12. Mora JR, Iwata M, von Andrian UH. Vitamin effects on the immune system. Nat Rev Immunol. (2008) 8:685–98. doi: 10.1038/nri2378
13. Pike JW. Intracellular receptors mediate the biologic action of 1,25-dihydroxyvitamin D3. Nutr Rev. (1985) 43:161–8. doi: 10.1111/j.1753-4887.1985.tb02406.x
14. Haussler MR. Vitamin D receptors: nature and function. Annu Rev Nutr. (1986) 6:527–62. doi: 10.1146/annurev.nu.06.070186.002523
15. DeLuca HF. Overview of general physiologic features and functions of vitamin D. Am J Clin Nutr. (2004) 80:1689S−96S. doi: 10.1093/ajcn/80.6.1689S
16. Karatay S, Yildirim K, Karakuzu A, Kiziltunc A, Engin RI, Eren YB, et al. Vitamin D status in patients with Behcet's disease. Clinics. (2011) 66:721–3. doi: 10.1590/s1807-59322011000500002
17. Veldman CM, Cantorna MT, DeLuca HF. Expression of 1,25-dihydroxyvitamin d3 receptor in the immune system. Arch Biochem Biophys. (2000) 374:334–8. doi: 10.1006/abbi.1999.1605
18. Mocanu V, Oboroceanu T, Zugun-Eloae F. Current status in vitamin D and regulatory T cells immunological implications. Rev Med Chir Soc Med Nat. (2013) 117:965–73.
19. Lemire JM, Archer DC, Beck L, Spiegelberg HL. Immunosuppressive actions of 1,25-dihydroxyvitamin D3: preferential inhibition of Th1 functions. J Nutr. (1995) 125:1704S−8S. doi: 10.1016/0960-0760(95)00106-A
20. Penna G, Amuchastegui S, Cossetti C, Aquilano F, Mariani R, Sanvito F, et al. Treatment of experimental autoimmune prostatitis in nonobese diabetic mice by the vitamin D receptor agonist elocalcitol. J Immunol. (2006) 177:8504–11. doi: 10.4049/jimmunol.177.12.8504
21. Joshi S, Pantalena LC, Liu XK, Gaffen SL, Liu H, Rohowsky-Kochan C, et al. 1,25-Dihydroxyvitamin D3 ameliorates Th17 autoimmunity via transcriptional modulation of interleukin-17A. Mol Cell Biol. (2011) 31:3653–69. doi: 10.1128/MCB.05020-11
22. Dankers W, Colin EM, van Hamburg JP, Lubberts E. Vitamin D in autoimmunity: molecular mechanisms and therapeutic potential. Front Immunol. (2017) 7:697. doi: 10.3389/fimmu.2016.00697
23. Cantorna MT, Snyder L, Lin YD, Yang L. Vitamin D and 1,25(OH)2D regulation of T cells. Nutrients. (2015) 7:3011–21. doi: 10.3390/nu7043011
24. Saul L, Mair I, Ivens A, Brown P, Samuel K, Campbell JDM, et al. 1,25-Dihydroxyvitamin D3 restrains CD4+ T cell priming ability of CD11c+ dendritic cells by upregulating expression of CD31. Front Immunol. (2019) 10:600. doi: 10.3389/fimmu.2019.00600
25. Røsjø E, Lossius A, Abdelmagid N, Lindstrøm JC, Kampman MT, Jørgensen L, et al. Effect of high-dose vitamin D3 supplementation on antibody responses against Epstein-Barr virus in relapsing-remitting multiple sclerosis. Mult Scler. (2017) 23:395–402. doi: 10.1177/1352458516654310
26. Baker AR, McDonnell DP, Hughes M, Crisp TM, Mangelsdorf DJ, Haussler MR, et al. Cloning and expression of full-length cDNA encoding human vitamin D receptor. Proc Natl Acad Sci USA. (1988) 85:3294–8. doi: 10.1073/pnas.85.10.3294
27. Miyamoto K, Kesterson RA, Yamamoto H, Taketani Y, Nishiwaki E, Tatsumi S, et al. Structural organization of the human vitamin D receptor chromosomal gene and its promoter. Mol Endocrinol. (1997) 11:1165–79. doi: 10.1210/mend.11.8.9951
28. Crofts LA, Hancock MS, Morrison NA, Eisman JA. Multiple promoters direct the tissue-specific expression of novel N-terminal variant human vitamin D receptor gene transcripts. Proc Natl Acad Sci USA. (1998) 95:10529–34. doi: 10.1073/pnas.95.18.10529
29. Yang CY, Leung PS, Adamopoulos IE, Gershwin ME. The implication of vitamin D and autoimmunity: a comprehensive review. Clin Rev Allergy Immunol. (2013) 45:217–26. doi: 10.1007/s12016-013-8361-3
30. Morrison NA, Qi JC, Tokita A, Kelly PJ, Crofts L, Nguyen TV, et al. Prediction of bone density from vitamin D receptor alleles. Nature. (1994) 367:284–7. doi: 10.1038/367284a0
31. Alimirah F, Peng X, Murillo G, Mehta RG. Functional significance of vitamin D receptor FokI polymorphism in human breast cancer cells. PLoS ONE. (2011) 6:e16024. doi: 10.1371/journal.pone.0016024
32. Arai H, Miyamoto K, Taketani Y, Yamamoto H, Iemori Y, Morita K, et al. A vitamin D receptor gene polymorphism in the translation initiation codon: effect on protein activity and relation to bone mineral density in Japanese women. J Bone Miner Res. (1997) 12:915–21. doi: 10.1359/jbmr.1997.12.6.915
33. Probst CM, Bompeixe EP, Pereira NF, de O Dalalio MM, Visentainer JE, Tsuneto LT, et al. HLA polymorphism and evaluation of European, African, and Amerindian contribution to the white and mulatto populations from Parana, Brazil. Hum Biol. (2000) 72:597–617. Available online at: https://www.jstor.org/stable/41465861
34. Nemenqani DM, Karam RA, Amer MG, Abd El Rahman TM. Vitamin D receptor gene polymorphisms and steroid receptor status among Saudi women with breast cancer. Gene. (2015) 558:215–9. doi: 10.1016/j.gene.2014.12.065
35. Papadopoulou A, Kouis P, Middleton N, Kolokotroni O, Karpathios T, Nicolaidou P, et al. Association of vitaminD receptor gene polymorphisms and vitamin D levels with asthma and atopy in Cypriot adolescents: a case–control study. Multidiscip Respir Med. (2015) 10:26. doi: 10.1186/s40248-015-0025-0
36. Sole X, Guino E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. (2006) 22:1928–9. doi: 10.1093/bioinformatics/btl268
37. Fitness J, Floyd S, Warndorff DK, Sichali L, Mwaungulu L, Crampin AC, et al. Large-scale candidate gene study of leprosy susceptibility in the Karonga district of northern Malawi. Am J Trop Med Hyg. (2004) 71:330–40. doi: 10.4269/ajtmh.2004.71.330
38. Sapkota BR, Macdonald M, Berrington WR, Misch EA, Ranjit C, Siddiqui MR, et al. Association of TNF, MBL, and VDR polymorphisms with leprosy phenotypes. Hum Immunol. (2010) 71:992–8. doi: 10.1016/j.humimm.2010.07.001
39. Areeshi MY, Mandal RK, Dar SA, Alshahrani AM, Ahmad A, Jawed A, et al. A reappraised meta-analysis of the genetic association between vitamin D receptor BsmI (rs1544410) polymorphism and pulmonary tuberculosis risk. Biosci Rep. (2017) 37:BSR20170247. doi: 10.1042/BSR20170247
40. Oliveira ALG, Chaves AT, Menezes CAS, Guimarães NS, Bueno LL, Fujiwara RT, et al. Vitamin D receptor expression and hepcidin levels in the protection or severity of leprosy: a systematic review. Microbes Infect. (2017) 19:311–22. doi: 10.1016/j.micinf.2017.03.001
41. Uitterlinden AG, Fang Y, Meurs JBJV, Pols HAP. Chapter 68 - Genetic vitamin D receptor polymorphisms and risk of disease. In: Feldman D, Wesley Pike J, Glorieux F, editors. Vitamin D. 2nd ed. London, UK: Academic Press (2005). p. 1121–57. doi: 10.1016/B978-012252687-9/50071-1
42. Uitterlinden AG, Fang Y, Van Meurs JB, Pols HA, Van Leeuwen JP. Genetics and biology of vitamin D receptor polymorphisms. Gene. (2004) 338:143–56. doi: 10.1016/j.gene.2004.05.014
43. Durrin LK, Haile RW, Ingles SA, Coetzee GA. Vitamin D receptor 3′-untranslated region polymorphisms: lack of effect on mRNA stability. Biochim Biophys Acta. (1999) 1453:311–20. doi: 10.1016/S0925-4439(99)00007-1
44. Singh I, Lavania M, Pathak VK, Ahuja M, Turankar RP, Singh V, et al. VDR polymorphism, gene expression and vitamin D levels in leprosy patients from North Indian population. PLoS Negl Trop Dis. (2018) 12:e0006823. doi: 10.1371/journal.pntd.0006823
Keywords: multibacillary, paucibacillary, genetic polymorphism, case-control studies, gene frequencies, biological markers
Citation: Pepineli AC, Alves HV, Tiyo BT, Macedo LC, Visentainer L, de Lima Neto QA, Zacarias JMV, Sell AM and Visentainer JEL (2019) Vitamin D Receptor Gene Polymorphisms Are Associated With Leprosy in Southern Brazil. Front. Immunol. 10:2157. doi: 10.3389/fimmu.2019.02157
Received: 31 May 2019; Accepted: 28 August 2019;
Published: 04 October 2019.
Edited by:
Abdulbari Bener, Istanbul University Cerrahpasa Faculty of Medicine, TurkeyReviewed by:
Simon John Clark, University of Manchester, United KingdomEswari Dodagatta-Marri, University of California, San Francisco, United States
Copyright © 2019 Pepineli, Alves, Tiyo, Macedo, Visentainer, de Lima Neto, Zacarias, Sell and Visentainer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jeane Eliete Laguila Visentainer, amVsdmlzZW50YWluZXJAZ21haWwuY29t
 Afonso Carrasco Pepineli1
Afonso Carrasco Pepineli1 Hugo Vicentin Alves
Hugo Vicentin Alves Bruna Tiaki Tiyo
Bruna Tiaki Tiyo Quirino Alves de Lima Neto
Quirino Alves de Lima Neto Joana Maira Valentini Zacarias
Joana Maira Valentini Zacarias Ana Maria Sell
Ana Maria Sell Jeane Eliete Laguila Visentainer
Jeane Eliete Laguila Visentainer