- 1Laboratory of Experimental Virology, Department of Medical Microbiology, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands
- 2Infectious Diseases Department, Liège University Hospital, Liège, Belgium
Combination antiretroviral therapy (ART) suppresses human immunodeficiency virus (HIV) replication and improves immune function, but is unable to eradicate the virus. Therefore, development of an HIV cure has become one of the main priorities of the HIV research field. The main obstacle for an HIV cure is the formation of latent viral reservoirs, where the virus is able to “hide” despite decades of therapy, just to reignite active replication once therapy is stopped. Revealing HIV hiding places is thus central to HIV cure research, but the absence of markers of these reservoir cells greatly complicates the search for a cure. Identification of one or several marker(s) of latently infected cells would represent a significant step forward toward a better description of the cell types involved and improved understanding of HIV latency. Moreover, it could provide a “handle” for selective therapeutic targeting of the reservoirs. A number of cellular markers of HIV reservoir have recently been proposed, including immune checkpoint molecules, CD2, and CD30. CD32a is perhaps the most promising of HIV reservoir markers as it is reported to be associated with a very prominent enrichment in HIV DNA, although this finding has been challenged. In this review, we provide an update on the current knowledge about HIV reservoir markers. We specifically highlight studies that characterized markers of persistently infected cells in the lymphoid tissues.
Introduction
Antiretroviral therapy (ART) allows the clinical management of the vast majority of human immunodeficiency virus (HIV) infected individuals, resulting in the prevention of AIDS and in the drastic reduction of the virus transmission risk. Unfortunately, ART is not curative. Interruption of therapy almost invariably leads to a quick viral rebound to pre-ART levels (1–6). Moreover, chronic immune activation and inflammation despite ART contribute to the excessive risk of non-AIDS events in ART-treated individuals (7). The etiology of persistent immune activation in the context of successful ART is considered multifactorial and involves microbial translocation, co-infections, metabolic disorders, regulatory T cell (Treg) deficiency, and decreased thymic function (8–11).
Presence of latent HIV reservoirs forms the major obstacle to an HIV cure. There is currently no consensus definition of the viral reservoirs due to their heterogeneous and dynamic nature. Viral reservoirs are commonly defined as cell types or anatomical sites that support the long-term persistence of replication-competent virus (12–14). This definition limits the HIV reservoirs to proviruses capable of triggering viral rebound after ART interruption (15). However, there is evidence that some defective proviruses that cannot reignite infection may still elicit immune activation through the production of viral proteins or novel antigens and thus participate in residual HIV pathogenesis (16–19). The eradication of this fraction of the HIV reservoir should therefore also be considered in the context of a durable cure. This concept led to extension of the HIV reservoir definition to all infected cells (20–22).
The definition of the HIV latent reservoir was initially restricted to transcriptionally silent proviruses (12). However, HIV post-integration latency is a multifactorial phenomenon controlled by mechanisms that operate not only at the transcriptional but also posttranscriptional levels (e.g., splicing and nuclear export of HIV RNA, translation, viral particle assembly, and maturation) (23, 24). These multiple blocks to virus production (25, 26) explain why subsets of latently infected cells in individuals on long-term ART can express HIV RNA and proteins (27–29). Such latently infected but transcription-competent cells have been defined as active HIV reservoirs (30–32). HIV latency has thus to be seen as a continuum with blocks to productive infection at different stages of the viral replication cycle in different populations of latently infected cells (33, 34). It has therefore been suggested to avoid the word “latent” when describing these cellular HIV reservoirs (22), and the term “leaky latency” has been proposed (Conference on Retroviruses and Opportunistic Infections 2015). However, it might be more practical to use a wider definition of HIV latency, integrating all the transcriptional and post-transcriptional blocks to virus production (34). Interestingly, those blocks are not “frozen” but can evolve over time. For instance, DNA methylation of the HIV promoter progressively increases during ART, possibly strengthening HIV latency (35). HIV reservoir expression also leads to clearance of infected cells due to immune surveillance or viral cytotoxicity, as suggested by a recent longitudinal HIV sequencing study (36). Prolonged ART is consequently associated with enrichment for proviral sequences that exhibit multiple features of deeper latency, such as positioning of the provirus in intergenic regions, in opposite orientation to host genes, and in either proximity to or increased distance from host transcription start sites and accessible chromatin (37).
In summary, a complex interplay of viral and host factors is continuously reshaping the HIV reservoirs. Moreover, this likely varies in different tissues, which are characterized by different anatomic properties and enriched for specific CD4+ T-cell subsets. It is therefore not surprising that mechanisms underlying HIV latency might differ between the peripheral blood and the tissues, as recently proposed (38). Viral and host markers of HIV reservoirs have been the focus of two recent reviews (39, 40). Here, we provide an update on some recent developments in the quest to distinguish the HIV reservoir cells from the plethora of cells in blood and tissues of an infected individual.
Heterogeneous Nature of HIV Reservoirs
Understanding the cell types and anatomical sites that harbor latent HIV proviruses is essential for the design of therapeutic strategies to eradicate HIV from an infected individual. The developmental profile of a memory T cell has been increasingly well conceptualized, and is currently best characterized as a linear program during which highly immature, long-lasting memory T cells progressively transition toward more mature, differentiated and short-lived effector-memory cells (41, 42). HIV can persist in stem cell-like memory (TSCM), central (TCM), transitional (TTM), and effector (TEM) memory CD4+ T cells, in addition to naïve (TN) CD4+ T cells (43–47). Although the frequency of HIV-infected cells is lower in TN compared to TCM, Zerbato et al. demonstrated that TN harbor a large inducible reservoir of replication-competent HIV, suggesting that TN cells may constitute an important HIV reservoir (48). Confirming these results, Venanzi Rullo et al. recently reported a large proportion of intact proviruses in TN from two ART-treated subjects (49). Hiener et al. showed that, among memory CD4+ T cells, TEM contain the larger proportion of intact HIV genomes (50). These studies indicate that the advent of improved culture- or PCR-based assays to measure the different reservoirs does seriously challenge the dogma that resting CD4+ memory T cells constitute the main HIV reservoir.
T cells can also be categorized according to functional properties, defined by specific antimicrobial properties or distinctive cytokine secretion patterns. Th1, Th2, Th17, Th9, regulatory T cell (Treg), and follicular T helper cell (Tfh) T-cell subpopulations have been identified based on functional polarization (42). Functionally polarized CD4+ T cells can support HIV persistence by distinct mechanisms related to their discrete functional profiles. Functional commitment toward a Th1 profile is possible during all stages of memory CD4+ T-cell development including the most immature and most durable subpopulations (51). Interestingly, proliferation of HIV-infected Th1 cells can play a crucial role in HIV persistence by maintaining the number of cells that harbor intact, replication-competent HIV (52). Recent evidence also highlights the contribution of long-lived Th17 cells to HIV persistence during ART, particularly in the gut (53). Th1/17 cells have been understood as a long-lasting subpopulation that may be somewhat more mature than Th17 cells, but retain the developmental program of long-lasting precursor cells for effector lymphocytes (54). CD4+ T cells enriched for Th1/17 polarization harbor high levels of HIV DNA in ART treated individuals and contribute disproportionately to the viral reservoirs that remain stable over many years of ART (45, 55).
Besides the gut and the lymph nodes (LN) that constitute major anatomical sites for HIV persistence, multiple other tissues and organs harbor HIV provirus in ART-suppressed individuals (56). Besides T cells, other cell types such as monocytes, astrocytes and tissue resident macrophages (splenic, alveolar, and microglia) may potentially serve as stable viral reservoirs in the tissues (20, 57–63). Given the difficulties in accessing tissue biopsies from humans, animal models such as humanized mouse models or SIV-infected non-human primate (NHP) models have been very helpful for examining HIV dynamics. These models have been used to show the persistence of HIV reservoirs in various tissues of HIV- or SIV-suppressed animals, with new tools allowing for demonstrating the intactness of these reservoirs (64–66).
Altogether, these studies highlight the highly heterogeneous nature of HIV reservoirs and underline the urgent need for identification of marker(s) of latently infected cells. Such markers could provide a “handle” to further dissect the complex and dynamic nature of the reservoirs and might even enable their selective targeting for eradication (67, 68). So far, no cellular marker has been discovered that is capable of identifying all HIV reservoirs. However, several HIV reservoir markers have recently been proposed and their further characterization could represent a significant step toward a better understanding of the HIV reservoirs.
Immune Checkpoint Molecules
In chronic viral infections, high antigenic loads constantly stimulate T cells, resulting in progressive loss of function termed T-cell exhaustion (69, 70). During this period, T cells demonstrate increased expression of some inhibitory receptors, known as immune checkpoint molecules (ICs), which increase the threshold for activation, leading to suppressed immune responses. Such ICs include programmed cell death-1 (PD-1), cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), lymphocyte activation gene 3 (LAG-3), T cell immunoglobulin, and ITIM domain (TIGIT), T cell immunoglobulin and mucin 3 (TIM-3), CD160, and 2B4 (CD244). Fromentin et al. showed that expression of PD-1, LAG-3, and TIGIT in CD4+ T cells was positively associated with the frequency of cells harboring integrated HIV DNA (71). More importantly, they observed that cells expressing these markers were enriched for HIV infection in several memory CD4+ T-cell subsets during ART. The majority of inducible HIV proviruses were found in memory CD4+ T cells that expressed at least one of these markers (71). More recently, the same group demonstrated a modest enrichment for HIV capsid (CA-p24)-producing cells (5.4-fold) in ART-suppressed individuals-derived cells displaying a CD45RA−, α4β1+, TIGIT+ phenotype (72). In line with these studies, Fromentin et al. recently reported that ex vivo HIV reactivation from latency is enhanced by a PD-1 blockade in CD4+ T cells from ART-suppressed individuals (73), although another group could not demonstrate such effect (74). In addition, Hurst et al. found that the expression of ICs measured prior to ART predicts time to viral rebound following ART interruption, confirming that T-cell exhaustion markers can identify latently infected cells with a higher tendency to viral transcription (75).
LN PD-1+/Tfh cells, as well as their circulating counterpart in peripheral blood (CXCR3+CD4+), represent a major cellular location for persistent virus in untreated and treated HIV-infected individuals, as demonstrated by the Perreau group (76–78). Notably, Tfh are localized to the germinal centers within the LN B-cell follicles, which are an immunologically privileged site with restricted CTL function (79). This may explain why PD-1+/Tfh cells are enriched in replication-competent HIV as compared to any other PD-1 negative memory CD4+ T-cell population isolated from blood or LN (77). However, intracellular concentrations of a number of antiretroviral drugs have been shown to be reduced in LN compared to peripheral blood (80–82), providing another possible reason for the persistence of infectious virus in the LNs. Interestingly, McGary et al. reported that CTLA-4+PD-1− memory CD4+ T cells significantly contribute to SIV persistence in ART-treated rhesus macaques (83). CTLA-4+PD-1− memory CD4+ T cells, a subset comprised predominantly of Tregs, were shown to be enriched for SIV DNA in several tissues and to contain replication-competent and infectious virus. In contrast to PD-1+/Tfh cells, SIV-enriched CTLA-4+PD-1− Treg cells localized outside the LN B-cell follicle. Moreover, in HIV-infected individuals on ART, CTLA-4+PD-1− memory CD4+ T cells harboring HIV DNA also persisted outside the B-cell follicle and contributed to the long-term latent viral reservoir (83).
Better definition of the parameters that dictate exhaustion is promoting the development of methods that prevent functionally inferior responses or “revitalize” them (84). IC inhibitors are already currently used to treat malignancies. This treatment would work for HIV infection in two ways: through boosting HIV-specific CD8+ T-cell effector function and through reactivating HIV expression from latency (85, 86). However, the case reports and clinical trials investigating the use of IC blockers have produced conflicting results (87, 88). Toxicity also appears as an important concern (89).
CD2, CD30, and CD20
Using an in vitro primary CD4+ T-cell model for HIV post-integration latency (90), Iglesias-Ussel et al. studied the expression profile of latently infected CD4+ T cells (91). Among the markers identified in this study, CD2 was selected for further analysis. Resting memory CD4+ CD2high T cells harbored higher HIV DNA copy numbers compared with the other cell subsets, although the fold enrichment was limited (5.7-fold). Iglesias-Ussel et al. further showed that CD4+ CD2high T cells could be stimulated to express high levels of HIV RNA, although no evidence was provided that infectious viruses could be produced (91). In contrast to the other markers described in this review and for which some tissue data is available, the relevance of CD2 still has to be confirmed in lymphoid tissues.
CD30 is expressed in Hodgkin and other aggressive lymphomas and on a small fraction of lymphocytes in healthy individuals (92). CD30 expression can be triggered by infections with several viruses, including human T-cell lymphotropic virus and Epstein-Barr virus (92). It is known for more than 20 years that triggering CD30 may play an important role in both HIV replication and HIV-infected CD4+ T-cell death (93). More recently, Hogan et al. demonstrated that CD30+CD4+ T cells were significantly enriched for cell-associated HIV RNA but not for HIV DNA in several individuals regardless of ART use (94). CD30 expression and HIV transcriptional activity co-localized in gut-associated lymphoid tissue from ART-treated or ART-naïve individuals. Following ex vivo culture of peripheral blood cells in the presence of brentuximab vedotin, an anti-CD30 antibody-drug conjugate, they observed a significant reduction in total HIV DNA. These data suggested that CD30 may be a relevant HIV therapeutic target as it is not expressed on a vast majority of cells (93). Subsequently, the same group studied the impact of 3 doses of brentuximab vedotin therapy for Hodgkin lymphoma on HIV persistence and immune phenotype in an individual on long-term ART (95). An encouraging but transient reduction of CD4+ T cells expressing CD30 was observed, accompanied by reductions in CD4+ T-cell–associated HIV RNA, low-level plasma viremia, and to lesser extent HIV DNA levels (95). The reason for the lack of a sustained effect of brentuximab vedotin on CD30-expressing T cells is currently unknown.
Very recently, Serra-Peinado et al. reported a higher percentage of HIV RNA+ cells among cells expressing CD20 (96). CD20 is a known B-cell surface marker but is dimly expressed on a small subpopulation of CD4+ T cells. Although the contribution of CD20dimCD4+ T cells to the total pool of HIV RNA+ cells was modest (median of 18.6% in ART-treated and 25.0% in viremic HIV-infected individuals), ex vivo treatment of primary peripheral blood mononuclear cells (PBMCs) from ART-suppressed individuals with the anti-CD20 monoclonal antibody Rituximab appeared to reduce the pool of HIV RNA+ cells when combined with latency-reversing agents (96). Further studies are needed to elucidate whether Rituximab treatment is also capable of reducing the total or intact HIV DNA reservoir.
CD32a
CD32a, also known as Fc gamma receptor IIa (FcγIIa), is the low-affinity receptor for the immunoglobulin G Fc fragment that is highly expressed on myeloid cells and expressed on a small subset of T cells (97, 98). CD32a was recently proposed by Descours et al. as a marker of HIV reservoir cells (99). Notably, CD32 has already been identified by an earlier study among several surface markers overexpressed in latently infected vs. uninfected CD4+ T cells in a primary CD4+ T-cell model for HIV latency (91). In contrast to all the markers described above that are associated with very limited HIV DNA enrichment, an extremely high (~1,000-fold) enrichment in HIV DNA was observed by Descours et al. in CD4+ T cells with high CD32a expression as compared to CD32−CD4+ T cells (99). They also demonstrated an enrichment for replication-competent proviruses in these cells, although not to a higher degree than the enrichment for total HIV DNA. If confirmed, these findings would represent a milestone in the efforts to develop a cure for HIV infection. However, a number of subsequent reports questioned whether CD32a would be a bona fide marker of the HIV latent reservoir (100–105), which decreased the initial enthusiasm. Some studies observed limited enrichment for HIV DNA in CD32+ cells in some participants but not in others (101, 102). Other reports could not demonstrate any enrichment for total HIV DNA or for replication-competent proviruses in CD32+ cells (100, 103–105).
The key for this controversy likely resides in the technical difficulty to obtain a sufficiently pure population of bona fide CD32+CD4+ T cells. The frequency of CD32+ cells among antigen-presenting cells (APCs) is much higher than among CD4+ T cells. Therefore, even if the residual APC contamination of CD4+ T cells is low in general, APCs will be disproportionally overrepresented in the CD32+ fraction, as reported by several groups (101, 103) and also observed in our initial attempts to purify CD32+CD4+ cells (Figure 1A) (106). The excess of residual non-T cells in the sorted CD32+ fraction can easily obscure the enrichment for HIV DNA in CD32+CD4+ cells. Alternatively, certain cell sorting strategies and/or settings might result in the isolation of T-B cell doublets or conjugates instead of bona fide CD32+CD4+ T cells, as reported by two groups (105, 107). Because another CD32 isoform, CD32b, is highly expressed on the surface of B cells, and consequently on T-B cell doublets or conjugates, the latter cells may be preferentially recognized during FACS sorting for CD32+CD4+ cells as most anti-CD32 antibodies do not discriminate between the CD32a and CD32b isoforms. The extracellular domains of these two proteins are very similar, and no CD32a-specific antibody is available yet. Depending on the sorting settings and/or antibody titers, bona fide CD32+CD4+ T cells, which likely express less CD32 molecules per cell than T-B cell doublets/conjugates, might be missed, unless specific efforts are made to thoroughly deplete the CD32+ non-T cells.
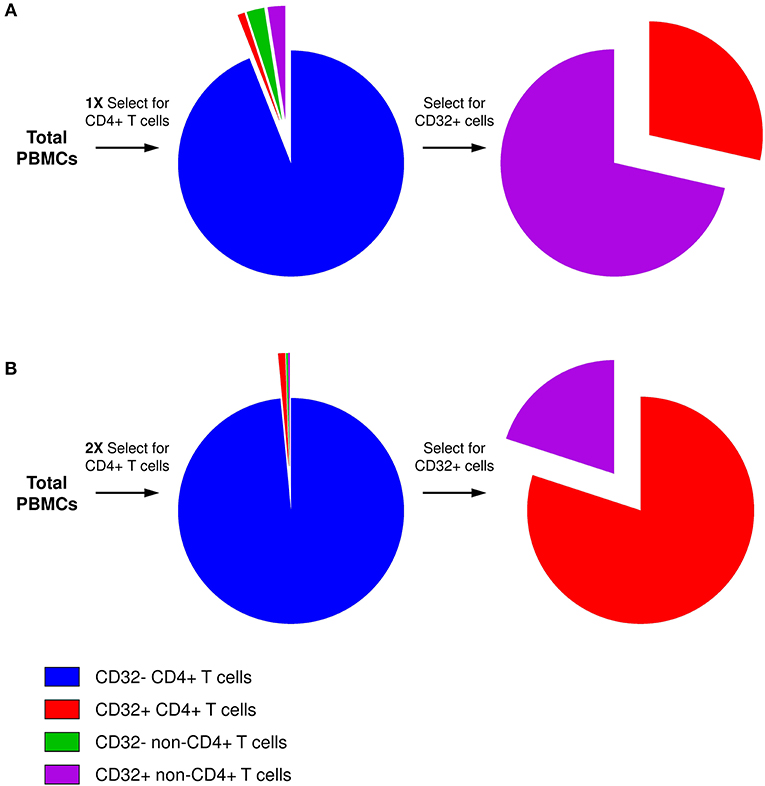
Figure 1. Presence of residual non-CD4+ T cells in the CD32+ cell fraction. (A) After one round of selection for CD4+ T cells, although the residual non-CD4+ T-cell contamination is low in general, the percentage of CD32+ cells among non-T cells is much higher than among CD4+ T cells. Therefore, after selection for CD32-expressing cells, the majority of CD32+ cells will be non-T cells. (B) Two consecutive rounds of CD4+ T-cell selection result in much purer CD4+ T-cell population, and the majority of CD4+ T cells is maintained even in the CD32+ fraction.
To overcome these problems, we performed two consecutive rounds of CD4+ T-cell negative selection by magnetic cell sorting before we started with CD32+ cell isolation. This sequential cell purification strategy could efficiently deplete the vast majority of contaminating non-T cells, including any T-B cell doublets, which allowed us to obtain a purified population of CD32+CD4+ cells (Figure 1B) (106). Remarkably, we demonstrated a progressive increase in HIV DNA enrichment in CD32+CD4+ cells with further purification of these cells, and in the purified population we measured a very prominent enrichment for HIV DNA (average, 292-fold) (106), confirming the results of Descours et al. (99). Interestingly, several groups found that the majority of peripheral blood CD32+CD4+ T cells express the activation marker HLA-DR (100–102, 106, 108, 109). Although it has been shown previously that HIV can establish latent infection in activated CD4+ T cells (110, 111) and that the peripheral blood HIV DNA load is higher in activated compared to resting cells in ART-treated individuals (112), the existence of latently infected CD4+ cells that are activated, and therefore relatively short-lived, suggests continuous replenishment of this component of the reservoir by cellular proliferation (113). At present, however, high HLA-DR expression on CD32+CD4+ cells should be interpreted with caution, as it cannot be ruled out that the HLA-DR signal may partly originate from the non-T cells within the contaminating CD32+ T-non-T cell doublets or conjugates, as recently demonstrated by Thornhill et al. (107).
Persistence of CD32+ cells has also been studied in tissues. Abdel-Mohsen et al. showed that a high percentage of HIV-expressing cells localized within the B-cell follicle co-express CD32a, whereas CD32a single-positive cells were rarely observed (100). These RNA in situ hybridization (ISH) experiments showing the co-expression of CD32a RNA and HIV RNA in lymphoid tissue support the notion that CD32a is associated with transcriptionally active tissue reservoirs. Similar results were recently obtained by Vásquez et al. in the gut tissue (114). By using ISH-based methods, they demonstrated co-expression of HIV and CD32a mRNA, with most of the CD3+CD32+ cells in the gut co-expressing HIV RNA. Moreover, Thornhill et al. recently found that the T cells within the CD32+ T-B cell doublets in the gut and tonsil tissues of ART-treated individuals were associated with a Tfh phenotype (107). As Tfh cells have been previously shown to harbor high levels of HIV RNA and infectious virus in LN (77), this may provide a clue why CD32+ cells are associated with HIV transcription in tissues.
Two groups studied co-expression of CD32a and other HIV reservoir markers in tissues. Noto et al. analyzed HIV transcription in LN memory CD4+ T-cell populations sorted for CD32a and PD-1 expression (108). The CD32+ and PD-1+ CD4+ T-cell populations overlapped to a large extent and CD32+PD-1+ cells were significantly enriched for HIV RNA compared to CD32−PD-1−, CD32+PD-1−, and CD32−PD-1+ cells. In the LNs, CD32+ and PD-1+ CD4+ T cells expressed high levels of the HIV co-receptors CCR5 and CXCR4, potentially making these cells a preferential target for HIV infection, as well as higher levels of activation markers, which can explain why these cells are characterized by a high level of HIV transcription (108). Hogan et al. sorted rectal biopsy cells for the expression of CD32a and CD30 (94). The highest HIV RNA and DNA levels from rectal tissue-derived cells were measured in cells co-expressing CD30 and CD32. These two studies demonstrate that different HIV reservoir markers can be co-expressed on CD4+ T cells, raising the possibility that there is some overlap in the reservoirs identified by these markers.
Taken together, these studies suggest an association between CD32a expression and active HIV transcription in tissues. In contrast, in peripheral blood we found no enrichment for cell-associated HIV RNA in CD32+CD4+ cells, and significantly reduced HIV RNA/DNA ratios were measured in these cells compared to CD32−CD4+ cells (106), indicating that HIV is transcriptionally silent in most of the infected peripheral blood CD32+ cells and suggesting that CD32a indeed marks latently infected cells. These seemingly discrepant results might suggest differences in the underlying mechanisms of persistence of HIV-infected CD32+ cells between blood and tissues. Residual HIV replication in tissue compartments can be quite prominent due to poor antiretroviral drug penetration (80–82, 115, 116) and CD32a expression in CD4+ T cells is induced upon in vitro HIV infection (29, 99, 100, 102). Therefore, in tissues, CD32a could mark cells that were recently infected despite ART. Alternatively, CD32a expression may be up-regulated upon re-activation of HIV transcription in long-lived latently infected cells in tissues, although the reverse association between HIV transcription and CD32a expression in peripheral blood makes this possibility less likely. To study the association between CD32a expression and HIV transcription in tissues, it will also be important to determine the relative contribution of CD32b+ T-B cell doublets vs. bona fide CD32a+CD4+ T cells to the total pool of tissue CD32+ T cells, and to which extent these two cell types are associated with active HIV transcription vs. viral latency. Interestingly, Thornhill et al. observed a positive correlation between the frequency of Tfh cells in CD32+ T-B cell doublets and total HIV DNA in the rectum (but not terminal ileum or PBMC) (107), suggesting that these doublets might be associated with a higher level of HIV infection in some sites. As B cells are capable of transmission of infectious HIV to T cells via CD21-complement interactions or DC-SIGN-mediated virion capture (117, 118), some of the observed T-B cell doublets can even be involved in residual B-to-T-cell HIV trans-infection in tissues.
Despite the considerable interest triggered by the Descours study (99), many questions with regard to the CD32a-HIV link are still open. Mechanisms of CD32a expression in HIV-infected cells remain to be investigated, as well as the role of CD32a in HIV reservoir formation and/or persistence. Notably, the contribution of CD32+ CD4+ T cells to the HIV reservoirs seems to be highly variable from one HIV-infected individual to the other, both in peripheral blood and in tissues (94, 99, 100, 106). This may restrict the efficiency of CD32a therapeutic targeting for HIV reservoir eradication in some individuals. In general, targeting of CD32a alone is not a feasible option because this would deplete the majority of an individual's myeloid immune cells. However, advanced targeting strategies (e.g., bi- and tri-specific antibodies) can be used to guarantee high specificity of CD32+CD4+ cell targeting. Targeting downstream CD32a signaling in CD4+ T cells could also be a feasible alternative.
Conclusions
The identification of a marker of latently infected cells has long been considered the “Holy Grail” of HIV cure programs (67). Several CD4+ T-cell subsets have been shown to harbor important HIV reservoirs, including resting memory CD4+ T cells. However, this picture is getting more complex every day as numerous other cell types were found to contribute to the HIV reservoirs. Activated T cells, naïve T cells, Tregs, Tfh, and their circulating counterpart, but also tissue macrophages, appear as major sites of HIV persistence.
Although much progress has been made in recent years in identifying HIV reservoir markers, no single cell surface molecule is currently able to mark all HIV-infected cells. However, some of the putative markers, including CD32a, CD30, and ICs, will likely play an important role in future studies aimed at understanding and eradicating the HIV reservoirs. Although a significant overlap was found in the expression of different HIV reservoir markers, targeting several markers at the same time in a “multicolor” approach may significantly increase the proportion of the reservoir that is eradicated.
Author Contributions
AP, GD, and BB wrote the manuscript. AP made the figure. All authors approved the final manuscript.
Funding
This work was supported by Aidsfonds Netherlands under Grant No. 2012025 to AP, the King Baudouin Foundation Grant to BB, and the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number R01AI145045 to BB.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to Jeroen den Dunnen and Rogier Sanders for helpful discussions. GD is post-doctorate clinical master specialist for the National Fund for Scientific Research (FNRS). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank the Rotary Foundation and the Fonds Léon Fredericq.
References
1. Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. (1997) 387:183–8. doi: 10.1038/387183a0
2. Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med. (1995) 1:1284–90. doi: 10.1038/nm1295-1284
3. Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. (1999) 5:512–7. doi: 10.1038/8394
4. Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. (2003) 9:727–8. doi: 10.1038/nm880
5. Chun TW, Davey RT Jr, Ostrowski M, Shawn Justement J, Engel D, Mullins JI, et al. Relationship between pre-existing viral reservoirs and the re-emergence of plasma viremia after discontinuation of highly active anti-retroviral therapy. Nat Med. (2000) 6:757–61. doi: 10.1038/77481
6. Davey RT Jr, Bhat N, Yoder C, Chun TW, Metcalf JA, Dewar R, et al. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc Natl Acad Sci USA. (1999) 96:15109–14. doi: 10.1073/pnas.96.26.15109
7. Deeks SG, Tracy R, Douek DC. Systemic effects of inflammation on health during chronic HIV infection. Immunity. (2013) 39:633–45. doi: 10.1016/j.immuni.2013.10.001
8. Paiardini M, Muller-Trutwin M. HIV-associated chronic immune activation. Immunol Rev. (2013) 254:78–101. doi: 10.1111/imr.12079
9. Klatt NR, Chomont N, Douek DC, Deeks SG. Immune activation and HIV persistence: implications for curative approaches to HIV infection. Immunol Rev. (2013) 254:326–42. doi: 10.1111/imr.12065
11. De Voeght A, Martens H, Renard C, Vaira D, Debruche M, Simonet J, et al. Exploring the link between innate immune activation and thymic function by measuring sCD14 and TRECs in HIV patients living in Belgium. PLoS ONE. (2017) 12:e0185761. doi: 10.1371/journal.pone.0185761
12. Darcis G, Van Driessche B, Van Lint C. HIV latency: should we shock or lock? Trends Immunol. (2017) 38:217–28. doi: 10.1016/j.it.2016.12.003
13. Pasternak AO, Berkhout B. HIV reservoir: finding the right needles in a needlestack. Cell Host Microbe. (2016) 20:280–2. doi: 10.1016/j.chom.2016.08.011
14. Eisele E, Siliciano RF. Redefining the viral reservoirs that prevent HIV-1 eradication. Immunity. (2012) 37:377–88. doi: 10.1016/j.immuni.2012.08.010
15. Bruner KM, Hosmane NN, Siliciano RF. Towards an HIV-1 cure: measuring the latent reservoir. Trends Microbiol. (2015) 23:192–203. doi: 10.1016/j.tim.2015.01.013
16. Imamichi H, Dewar RL, Adelsberger JW, Rehm CA, O'Doherty U, Paxinos EE, et al. Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. Proc Natl Acad Sci USA. (2016) 113:8783–8. doi: 10.1073/pnas.1609057113
17. Pollack RA, Jones RB, Pertea M, Bruner KM, Martin AR, Thomas AS, et al. Defective HIV-1 proviruses are expressed and can be recognized by cytotoxic T lymphocytes, which shape the proviral landscape. Cell Host Microbe. (2017) 21:494–506.e4. doi: 10.1016/j.chom.2017.03.008
18. Sharaf R, Lee GQ, Sun X, Etemad B, Aboukhater LM, Hu Z, et al. HIV-1 proviral landscapes distinguish posttreatment controllers from noncontrollers. J Clin Invest. (2018) 128:4074–85. doi: 10.1172/JCI120549
19. Das AT, Pasternak AO, Berkhout B. On the generation of the MSD-Ψ class of defective HIV proviruses. Retrovirology. (2019) 16:19. doi: 10.1186/s12977-019-0481-2
20. Avettand-Fenoel V, Hocqueloux L, Ghosn J, Cheret A, Frange P, Melard A, et al. Total HIV-1 DNA, a marker of viral reservoir dynamics with clinical implications. Clin Microbiol Rev. (2016) 29:859–80. doi: 10.1128/CMR.00015-16
21. Douek DC. Disrupting T-cell homeostasis: how HIV-1 infection causes disease. AIDS Rev. (2003) 5:172–7.
22. Baxter AE, O'Doherty U, Kaufmann DE. Beyond the replication-competent HIV reservoir: transcription and translation-competent reservoirs. Retrovirology. (2018) 15:18. doi: 10.1186/s12977-018-0392-7
23. Karn J, Stoltzfus CM. Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harb Perspect Med. (2012) 2:a006916. doi: 10.1101/cshperspect.a006916
24. Khoury G, Darcis G, Lee MY, Bouchat S, Van Driessche B, Purcell DFJ, et al. The molecular biology of HIV latency. Adv Exp Med Biol. (2018) 1075:187–212. doi: 10.1007/978-981-13-0484-2_8
25. Kula A, Guerra J, Knezevich A, Kleva D, Myers MP, Marcello A. Characterization of the HIV-1 RNA associated proteome identifies Matrin 3 as a nuclear cofactor of Rev function. Retrovirology. (2011) 8:60. doi: 10.1186/1742-4690-8-60
26. Sarracino A, Gharu L, Kula A, Pasternak AO, Avettand-Fenoel V, Rouzioux C, et al. Posttranscriptional regulation of HIV-1 gene expression during replication and reactivation from latency by nuclear matrix protein MATR3. MBio. (2018) 9:e02158-18. doi: 10.1128/mBio.02158-18
27. DeMaster LK, Liu XH, VanBelzen DJ, Trinite B, Zheng LJ, Agosto LM, et al. A subset of CD4/CD8 double-negative T cells expresses HIV proteins in patients on antiretroviral therapy. J Virol. (2016) 90:2165–79. doi: 10.1128/JVI.01913-15
28. Baxter AE, Niessl J, Fromentin R, Richard J, Porichis F, Charlebois R, et al. Single-cell characterization of viral translation-competent reservoirs in HIV-infected individuals. Cell Host Microbe. (2016) 20:368–80. doi: 10.1016/j.chom.2016.07.015
29. Grau-Expósito J, Serra-Peinado C, Miguel L, Navarro J, Curran A, Burgos J, et al. A novel single-cell FISH-flow assay identifies effector memory CD4(+) T cells as a major niche for HIV-1 transcription in HIV-infected patients. MBio. (2017) 8:e00876-17. doi: 10.1128/mBio.00876-17
30. Pasternak AO, de Bruin M, Jurriaans S, Bakker M, Berkhout B, Prins JM, et al. Modest nonadherence to antiretroviral therapy promotes residual HIV-1 replication in the absence of virological rebound in plasma. J Infect Dis. (2012) 206:1443–52. doi: 10.1093/infdis/jis502
31. Van Gulck E, Bracke L, Heyndrickx L, Coppens S, Atkinson D, Merlin C, et al. Immune and viral correlates of “secondary viral control” after treatment interruption in chronically HIV-1 infected patients. PLoS ONE. (2012) 7:e37792. doi: 10.1371/journal.pone.0037792
32. Pasternak AO, Lukashov VV, Berkhout B. Cell-associated HIV RNA: a dynamic biomarker of viral persistence. Retrovirology. (2013) 10:41. doi: 10.1186/1742-4690-10-41
33. Pace MJ, Agosto L, Graf EH, O'Doherty U. HIV reservoirs and latency models. Virology. (2011) 411:344–54. doi: 10.1016/j.virol.2010.12.041
34. Pasternak AO, Berkhout B. What do we measure when we measure cell-associated HIV RNA. Retrovirology. (2018) 15:13. doi: 10.1186/s12977-018-0397-2
35. Trejbalova K, Kovarova D, Blazkova J, Machala L, Jilich D, Weber J, et al. Development of 5' LTR DNA methylation of latent HIV-1 provirus in cell line models and in long-term-infected individuals. Clin Epigenetics. (2016) 8:19. doi: 10.1186/s13148-016-0185-6
36. Pinzone MR, VanBelzen DJ, Weissman S, Bertuccio MP, Cannon L, Venanzi-Rullo E, et al. Longitudinal HIV sequencing reveals reservoir expression leading to decay which is obscured by clonal expansion. Nat Commun. (2019) 10:728. doi: 10.1038/s41467-019-08431-7
37. Einkauf KB, Lee GQ, Gao C, Sharaf R, Sun X, Hua S, et al. Intact HIV-1 proviruses accumulate at distinct chromosomal positions during prolonged antiretroviral therapy. J Clin Invest. (2019) 129:988–98. doi: 10.1172/JCI124291
38. Telwatte S, Lee S, Somsouk M, Hatano H, Baker C, Kaiser P, et al. Gut and blood differ in constitutive blocks to HIV transcription, suggesting tissue-specific differences in the mechanisms that govern HIV latency. PLoS Pathog. (2018) 14:e1007357. doi: 10.1371/journal.ppat.1007357
39. Bruel T, Schwartz O. Markers of the HIV-1 reservoir: facts and controversies. Curr Opin HIV AIDS. (2018) 13:383–8. doi: 10.1097/COH.0000000000000482
40. Sharaf RR, Li JZ. The alphabet soup of HIV reservoir markers. Curr HIV/AIDS Rep. (2017) 14:72–81. doi: 10.1007/s11904-017-0355-y
41. Jameson SC, Masopust D. Understanding subset diversity in T cell memory. Immunity. (2018) 48:214–26. doi: 10.1016/j.immuni.2018.02.010
42. Lee GQ, Lichterfeld M. Diversity of HIV-1 reservoirs in CD4+ T-cell subpopulations. Curr Opin HIV AIDS. (2016) 11:383–7. doi: 10.1097/COH.0000000000000281
43. Wightman F, Solomon A, Khoury G, Green JA, Gray L, Gorry PR, et al. Both CD31(+) and CD31− naive CD4(+) T cells are persistent HIV type 1-infected reservoirs in individuals receiving antiretroviral therapy. J Infect Dis. (2010) 202:1738–48. doi: 10.1086/656721
44. Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. (2009) 15:893–900. doi: 10.1038/nm.1972
45. Khoury G, Anderson JL, Fromentin R, Hartogenesis W, Smith MZ, Bacchetti P, et al. Persistence of integrated HIV DNA in CXCR3 + CCR6 + memory CD4+ T cells in HIV-infected individuals on antiretroviral therapy. AIDS. (2016) 30:1511–20. doi: 10.1097/QAD.0000000000001029
46. Buzon MJ, Sun H, Li C, Shaw A, Seiss K, Ouyang Z, et al. HIV-1 persistence in CD4+ T cells with stem cell-like properties. Nat Med. (2014) 20:139–42. doi: 10.1038/nm.3445
47. Gattinoni L, Lugli E, Ji Y, Pos Z, Paulos CM, Quigley MF, et al. A human memory T cell subset with stem cell-like properties. Nat Med. (2011) 17:1290–7. doi: 10.1038/nm.2446
48. Zerbato JM, McMahon DK, Sobolewski MD, Mellors JW, Sluis-Cremer N. Naive CD4+ T cells harbor a large inducible reservoir of latent, replication-competent HIV-1. Clin Infect Dis. (2019). doi: 10.1093/cid/ciz108. [Epub ahead of print].
49. Venanzi Rullo E, Cannon L, Pinzone MR, Ceccarelli M, Nunnari G, O'Doherty U. Genetic evidence that Naïve T cells can contribute significantly to the HIV intact reservoir: time to re-evaluate their role. Clin Infect Dis. (2019) ciz378. doi: 10.1093/cid/ciz378. [Epub ahead of print].
50. Hiener B, Horsburgh BA, Eden JS, Barton K, Schlub TE, Lee E, et al. Identification of genetically intact HIV-1 proviruses in specific CD4(+) T cells from effectively treated participants. Cell Rep. (2017) 21:813–22. doi: 10.1016/j.celrep.2017.09.081
51. Takeshita M, Suzuki K, Kassai Y, Takiguchi M, Nakayama Y, Otomo Y, et al. Polarization diversity of human CD4+ stem cell memory T cells. Clin Immunol. (2015) 159:107–17. doi: 10.1016/j.clim.2015.04.010
52. Lee GQ, Orlova-Fink N, Einkauf K, Chowdhury FZ, Sun X, Harrington S, et al. Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. J Clin Invest. (2017) 127:2689–96. doi: 10.1172/JCI93289
53. Gosselin A, Wiche Salinas TR, Planas D, Wacleche VS, Zhang Y, Fromentin R, et al. HIV persists in CCR6+CD4+ T cells from colon and blood during antiretroviral therapy. AIDS. (2017) 31:35–48. doi: 10.1097/QAD.0000000000001309
54. Muranski P, Restifo NP. Essentials of Th17 cell commitment and plasticity. Blood. (2013) 121:2402–14. doi: 10.1182/blood-2012-09-378653
55. Sun H, Kim D, Li X, Kiselinova M, Ouyang Z, Vandekerckhove L, et al. Th1/17 polarization of CD4 T cells supports HIV-1 persistence during antiretroviral therapy. J Virol. (2015) 89:11284–93. doi: 10.1128/JVI.01595-15
56. Couturier J, Lewis DE. HIV persistence in adipose tissue reservoirs. Curr HIV/AIDS Rep. (2018) 15:60–71. doi: 10.1007/s11904-018-0378-z
57. Abreu CM, Veenhuis RT, Avalos CR, Graham S, Queen SE, Shirk EN, et al. Infectious virus persists in CD4+ T cells and macrophages in ART-suppressed SIV-infected Macaques. J Virol. (2019) 93:e00065-19. doi: 10.1128/JVI.00065-19
58. Kumar A, Darcis G, Van Lint C, Herbein G. Epigenetic control of HIV-1 post integration latency: implications for therapy. Clin Epigenetics. (2015) 7:103. doi: 10.1186/s13148-015-0137-6
59. Avalos CR, Price SL, Forsyth ER, Pin JN, Shirk EN, Bullock BT, et al. Quantitation of productively infected monocytes and macrophages of simian immunodeficiency virus-infected macaques. J Virol. (2016) 90:5643–56. doi: 10.1128/JVI.00290-16
60. Ganor Y, Real F, Sennepin A, Dutertre CA, Prevedel L, Xu L, et al. HIV-1 reservoirs in urethral macrophages of patients under suppressive antiretroviral therapy. Nat Microbiol. (2019) 4:633–44. doi: 10.1038/s41564-018-0335-z
61. Darcis G, Coombs RW, Van Lint C. Exploring the anatomical HIV reservoirs: role of the testicular tissue. AIDS. (2016) 30:2891–3. doi: 10.1097/QAD.0000000000001281
62. Jenabian MA, Costiniuk CT, Mehraj V, Ghazawi FM, Fromentin R, Brousseau J, et al. Immune tolerance properties of the testicular tissue as a viral sanctuary site in ART-treated HIV-infected adults. AIDS. (2016) 30:2777–86. doi: 10.1097/QAD.0000000000001282
63. Wong JK, Yukl SA. Tissue reservoirs of HIV. Curr Opin HIV AIDS. (2016) 11:362–70. doi: 10.1097/COH.0000000000000293
64. Wong ME, Jaworowski A, Hearps AC. The HIV reservoir in monocytes and macrophages. Front Immunol. (2019) 10:1435. doi: 10.3389/fimmu.2019.01435
65. Frank I, Acharya A, Routhu NK, Aravantinou M, Harper JL, Maldonado S, et al. A Tat/Rev induced limiting dilution assay to measure viral reservoirs in non-human primate models of HIV infection. Sci Rep. (2019) 9:12078. doi: 10.1038/s41598-019-48354-3
66. Nixon CC, Mavigner M, Silvestri G, Garcia JV. In vivo models of human immunodeficiency virus persistence and cure strategies. J Infect Dis. (2017) 215(Suppl. 3):S142–51. doi: 10.1093/infdis/jiw637
67. Pillai SK, Deeks SG. Signature of the sleeper cell: a biomarker of HIV latency revealed. Trends Immunol. (2017) 38:457–8. doi: 10.1016/j.it.2017.04.007
68. Dahabieh MS, Battivelli E, Verdin E. Understanding HIV latency: the road to an HIV cure. Annu Rev Med. (2015) 66:407–21. doi: 10.1146/annurev-med-092112-152941
70. Chew GM, Fujita T, Webb GM, Burwitz BJ, Wu HL, Reed JS, et al. TIGIT marks exhausted T cells, correlates with disease progression, and serves as a target for immune restoration in HIV and SIV infection. PLoS Pathog. (2016) 12:e1005349. doi: 10.1371/journal.ppat.1005349
71. Fromentin R, Bakeman W, Lawani MB, Khoury G, Hartogensis W, DaFonseca S, et al. CD4+ T cells expressing PD-1, TIGIT and LAG-3 contribute to HIV persistence during ART. PLoS Pathog. (2016) 12:e1005761. doi: 10.1371/journal.ppat.1005761
72. Pardons M, Baxter AE, Massanella M, Pagliuzza A, Fromentin R, Dufour C, et al. Single-cell characterization and quantification of translation-competent viral reservoirs in treated and untreated HIV infection. PLoS Pathog. (2019) 15:e1007619. doi: 10.1371/journal.ppat.1007619
73. Fromentin R, DaFonseca S, Costiniuk CT, El-Far M, Procopio FA, Hecht FM, et al. PD-1 blockade potentiates HIV latency reversal ex vivo in CD4(+) T cells from ART-suppressed individuals. Nat Commun. (2019) 10:814. doi: 10.1038/s41467-019-08798-7
74. Bui JK, Cyktor JC, Fyne E, Campellone S, Mason SW, Mellors JW. Blockade of the PD-1 axis alone is not sufficient to activate HIV-1 virion production from CD4+ T cells of individuals on suppressive ART. PLoS ONE. (2019) 14:e0211112. doi: 10.1371/journal.pone.0211112
75. Hurst J, Hoffmann M, Pace M, Williams JP, Thornhill J, Hamlyn E, et al. Immunological biomarkers predict HIV-1 viral rebound after treatment interruption. Nat Commun. (2015) 6:8495. doi: 10.1038/ncomms9495
76. Banga R, Procopio FA, Ruggiero A, Noto A, Ohmiti K, Cavassini M, et al. Blood CXCR3+ CD4 T cells are enriched in inducible replication competent HIV in aviremic antiretroviral therapy-treated individuals. Front Immunol. (2018) 9:144. doi: 10.3389/fimmu.2018.00144
77. Banga R, Procopio FA, Noto A, Pollakis G, Cavassini M, Ohmiti K, et al. PD-1(+) and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat Med. (2016) 22:754–61. doi: 10.1038/nm.4113
78. Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, et al. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med. (2013) 210:143–56. doi: 10.1084/jem.20121932
79. Fukazawa Y, Lum R, Okoye AA, Park H, Matsuda K, Bae JY, et al. B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat Med. (2015) 21:132–9. doi: 10.1038/nm.3781
80. Rothenberger M, Nganou-Makamdop K, Kityo C, Ssali F, Chipman JG, Beilman GJ, et al. Impact of Integrase Inhibition compared to non-nucleoside inhibition on HIV reservoirs in Lymphoid Tissues. J Acquir Immune Defic Syndr. (2019) 81:355–60. doi: 10.1097/QAI.0000000000002026
81. Estes JD, Kityo C, Ssali F, Swainson L, Makamdop KN, Del Prete GQ, et al. Defining total-body AIDS-virus burden with implications for curative strategies. Nat Med. (2017) 23:1271–6. doi: 10.1038/nm.4411
82. Fletcher CV, Staskus K, Wietgrefe SW, Rothenberger M, Reilly C, Chipman JG, et al. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc Natl Acad Sci USA. (2014) 111:2307–12. doi: 10.1073/pnas.1318249111
83. McGary CS, Deleage C, Harper J, Micci L, Ribeiro SP, Paganini S, et al. CTLA-4+PD-1- Memory CD4+ T cells critically contribute to viral persistence in antiretroviral therapy-suppressed, SIV-infected rhesus macaques. Immunity. (2017) 47:776–88.e5. doi: 10.1016/j.immuni.2017.09.018
84. Yi JS, Cox MA, Zajac AJ. T-cell exhaustion: characteristics, causes and conversion. Immunology. (2010) 129:474–81. doi: 10.1111/j.1365-2567.2010.03255.x
85. Boyer Z, Palmer S. Targeting immune checkpoint molecules to eliminate latent HIV. Front Immunol. (2018) 9:2339. doi: 10.3389/fimmu.2018.02339
86. Zerbato JM, Purves HV, Lewin SR, Rasmussen TA. Between a shock and a hard place: challenges and developments in HIV latency reversal. Curr Opin Virol. (2019) 38:1–9. doi: 10.1016/j.coviro.2019.03.004
87. Guihot A, Marcelin AG, Massiani MA, Samri A, Soulié C, Autran B, et al. Drastic decrease of the HIV reservoir in a patient treated with nivolumab for lung cancer. Ann Oncol. (2018) 29:517–8. doi: 10.1093/annonc/mdx696
88. Scully EP, Rutishauser RL, Simoneau CR, Delagrèverie H, Euler Z, Thanh C, et al. Inconsistent HIV reservoir dynamics and immune responses following anti-PD-1 therapy in cancer patients with HIV infection. Ann Oncol. (2018) 29:2141–2. doi: 10.1093/annonc/mdy259
89. Day D, Hansen AR. Immune-related adverse events associated with immune checkpoint inhibitors. BioDrugs. (2016) 30:571–84. doi: 10.1007/s40259-016-0204-3
90. Marini A, Harper JM, Romerio F. An in vitro system to model the establishment and reactivation of HIV-1 latency. J Immunol. (2008) 181:7713–20. doi: 10.4049/jimmunol.181.11.7713
91. Iglesias-Ussel M, Vandergeeten C, Marchionni L, Chomont N, Romerio F. High levels of CD2 expression identify HIV-1 latently infected resting memory CD4+ T cells in virally suppressed subjects. J Virol. (2013) 87:9148–58. doi: 10.1128/JVI.01297-13
92. Biswas P, Mantelli B, Delfanti F, Ferrarini M, Poli G, Lazzarin A. CD30 ligation differentially affects CXCR4-dependent HIV-1 replication and soluble CD30 secretion in non-Hodgkin cell lines and in gamma delta T lymphocytes. Eur J Immunol. (2003) 33:3136–45. doi: 10.1002/eji.200324344
93. Romagnani S, Annunziato F, Manetti R, Almerigogna F, Biagiotti R, Giudizi MG, et al. Role for CD30 in HIV expression. Immunol Lett. (1996) 51:83–8. doi: 10.1016/0165-2478(96)02559-X
94. Hogan LE, Vasquez J, Hobbs KS, Hanhauser E, Aguilar-Rodriguez B, Hussien R, et al. Increased HIV-1 transcriptional activity and infectious burden in peripheral blood and gut-associated CD4+ T cells expressing CD30. PLoS Pathog. (2018) 14:e1006856. doi: 10.1371/journal.ppat.1006856
95. Wang CC, Thanh C, Gibson EA, Ball-Burack M, Hogan LE, Descours B, et al. Transient loss of detectable HIV-1 RNA following brentuximab vedotin anti-CD30 therapy for Hodgkin lymphoma. Blood Adv. (2018) 2:3479–82. doi: 10.1182/bloodadvances.2018024364
96. Serra-Peinado C, Grau-Expósito J, Luque-Ballesteros L, Astorga-Gamaza A, Navarro J, Gallego-Rodriguez J, et al. Expression of CD20 after viral reactivation renders HIV-reservoir cells susceptible to Rituximab. Nat Commun. (2019) 10:3705. doi: 10.1038/s41467-019-11556-4
97. Hogarth PM, Pietersz GA. Fc receptor-targeted therapies for the treatment of inflammation, cancer and beyond. Nat Rev Drug Discov. (2012) 11:311–31. doi: 10.1038/nrd2909
98. Holgado MP, Sananez I, Raiden S, Geffner JR, Arruvito L. CD32 ligation promotes the activation of CD4+ T cells. Front Immunol. (2018) 9:2814. doi: 10.3389/fimmu.2018.02814
99. Descours B, Petitjean G, López-Zaragoza JL, Bruel T, Raffel R, Psomas C, et al. CD32a is a marker of a CD4 T-cell HIV reservoir harbouring replication-competent proviruses. Nature. (2017) 543:564–7. doi: 10.1038/nature21710
100. Abdel-Mohsen M, Kuri-Cervantes L, Grau-Exposito J, Spivak AM, Nell RA, Tomescu C, et al. CD32 is expressed on cells with transcriptionally active HIV but does not enrich for HIV DNA in resting T cells. Sci Transl Med. (2018) 10:eaar6759. doi: 10.1126/scitranslmed.aar6759
101. Martin GE, Pace M, Thornhill JP, Phetsouphanh C, Meyerowitz J, Gossez M, et al. CD32-expressing CD4 T cells are phenotypically diverse and can contain proviral HIV DNA. Front Immunol. (2018) 9:928. doi: 10.3389/fimmu.2018.00928
102. Badia R, Ballana E, Castellví M, García-Vidal E, Pujantell M, Clotet B, et al. CD32 expression is associated to T-cell activation and is not a marker of the HIV-1 reservoir. Nat Commun. (2018) 9:2739. doi: 10.1038/s41467-018-05157-w
103. Pérez L, Anderson J, Chipman J, Thorkelson A, Chun TW, Moir S, et al. Conflicting evidence for HIV enrichment in CD32. Nature. (2018) 561:E9–16. doi: 10.1038/s41586-018-0493-4
104. Bertagnolli LN, White JA, Simonetti FR, Beg SA, Lai J, Tomescu C, et al. The role of CD32 during HIV-1 infection. Nature. (2018) 561:E17–9. doi: 10.1038/s41586-018-0494-3
105. Osuna CE, Lim SY, Kublin JL, Apps R, Chen E, Mota TM, et al. Evidence that CD32a does not mark the HIV-1 latent reservoir. Nature. (2018) 561:E20–8. doi: 10.1038/s41586-018-0495-2
106. Darcis G, Kootstra N, Hooibrink B, van Montfort T, Groen K, Jurriaans S, et al. CD32+CD4+ T cells are enriched in HIV DNA. In: Conference on Retroviruses and Opportunistic Infections. Seattle, WA (2019).
107. Thornhill JP, Pace M, Martin GE, Hoare J, Peake S, Herrera C, et al. CD32 expressing doublets in HIV-infected gut-associated lymphoid tissue are associated with a T follicular helper cell phenotype. Mucosal Immunol. (2019) 12:1212–9. doi: 10.1038/s41385-019-0180-2
108. Noto A, Procopio FA, Banga R, Suffiotti M, Corpataux JM, Cavassini M, et al. CD32(+) and PD-1(+) lymph node CD4 T cells support persistent HIV-1 transcription in treated aviremic individuals. J Virol. (2018) 92:e00901–18. doi: 10.1128/JVI.00901-18
109. Wittner M, Dunay GA, Kummer S, Bockhorn M, Hüfner A, Schmiedel S, et al. CD32 Expression of different memory T cell subpopulations in the blood and lymph nodal tissue of HIV patients and healthy controls correlates with immune activation. J Acquir Immune Defic Syndr. (2018) 77:345–9. doi: 10.1097/QAI.0000000000001622
110. Chavez L, Calvanese V, Verdin E. HIV latency is established directly and early in both resting and activated primary CD4 T cells. PLoS Pathog. (2015) 11:e1004955. doi: 10.1371/journal.ppat.1004955
111. van der Sluis RM, van Montfort T, Pollakis G, Sanders RW, Speijer D, Berkhout B, et al. Dendritic cell-induced activation of latent HIV-1 provirus in actively proliferating primary T lymphocytes. PLoS Pathog. (2013) 9:e1003259. doi: 10.1371/journal.ppat.1003259
112. Kaiser P, Joos B, Niederost B, Weber R, Gunthard HF, Fischer M. Productive human immunodeficiency virus type 1 infection in peripheral blood predominantly takes place in CD4/CD8 double-negative T lymphocytes. J Virol. (2007) 81:9693–706. doi: 10.1128/JVI.00492-07
113. Chun TW, Nickle DC, Justement JS, Large D, Semerjian A, Curlin ME, et al. HIV-infected individuals receiving effective antiviral therapy for extended periods of time continually replenish their viral reservoir. J Clin Invest. (2005) 115:3250–5. doi: 10.1172/JCI26197
114. Vásquez JJ, Aguilar-Rodriguez BL, Rodriguez L, Hogan LE, Somsouk M, McCune JM, et al. CD32-RNA co-localizes with HIV-RNA in CD3+ cells found within gut tissues from viremic and ART-suppressed individuals. Pathog Immun. (2019) 4:147–60. doi: 10.20411/pai.v4i1.271
115. Lorenzo-Redondo R, Fryer HR, Bedford T, Kim EY, Archer J, Pond SLK, et al. Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature. (2016) 530:51–6. doi: 10.1038/nature16933
116. Thompson CG, Rosen EP, Prince HMA, White N, Sykes C, de la Cruz G, et al. Heterogeneous antiretroviral drug distribution and HIV/SHIV detection in the gut of three species. Sci Transl Med. (2019) 11:eaap8758. doi: 10.1126/scitranslmed.aap8758
117. Moir S, Malaspina A, Li Y, Chun TW, Lowe T, Adelsberger J, et al. B cells of HIV-1-infected patients bind virions through CD21-complement interactions and transmit infectious virus to activated T cells. J Exp Med. (2000) 192:637–46. doi: 10.1084/jem.192.5.637
Keywords: HIV, biomarker, viral reservoirs, HIV latent reservoir, HIV persistence
Citation: Darcis G, Berkhout B and Pasternak AO (2019) The Quest for Cellular Markers of HIV Reservoirs: Any Color You Like. Front. Immunol. 10:2251. doi: 10.3389/fimmu.2019.02251
Received: 18 July 2019; Accepted: 05 September 2019;
Published: 20 September 2019.
Edited by:
Francesca Chiodi, Karolinska Institute (KI), SwedenReviewed by:
Jacob D. Estes, Vaccine and Gene Therapy Institute, Oregon Health & Science University, United StatesJennifer Zerbato, Peter Doherty Institute for Infection and Immunity, Australia
Copyright © 2019 Darcis, Berkhout and Pasternak. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gilles Darcis, Z2RhcmNpc0BjaHVsaWVnZS5iZQ==; Alexander O. Pasternak, YS5vLnBhc3Rlcm5ha0BhbWMudXZhLm5s
 Gilles Darcis
Gilles Darcis Ben Berkhout
Ben Berkhout Alexander O. Pasternak
Alexander O. Pasternak