- 1Laboratório de Agentes Infecciosos e Vetores, Programa de Pós-graduação em Patologia Investigativa, Centro das Ciências Biológicas e da Saúde, Universidade Federal do Oeste da Bahia, Bahia, Brazil
- 2Programa de Pós-graduação em Biologia e Biotecnologia de Microrganismos, Universidade Estadual de Santa Cruz, Bahia, Brazil
The Flavivirus genus is composed by viral serocomplexes with relevant global epidemiological impact. Many areas of the world present both, vector fauna and geographical conditions compatible with co-circulation, importing, emergence, and epidemics of flaviviruses of different serocomplexes. In this study, we aimed to identify both, immunological determinants and patterns of immune response possibly involved in flavivirus serocomplex cross-protection. We searched B and T cells epitopes which were thoroughly shown to be involved in flavivirus immunological control. Such epitopes were analyzed regarding their conservation, population coverage, and location along flavivirus polyprotein. We found that epitopes capable of eliciting flavivirus cross-protective immunity to a wide range of human populations are concentrated in proteins E, NS3, and NS5. Such identification of both, immunological determinants and patterns of immune response involved in flavivirus cross-protective immunity should be considered in future vaccine development. Moreover, cross-reactive epitopes presented in this work may be involved in dynamics of diseases caused by flaviviruses worldwide.
Introduction
The Flavivirus genus of the Flaviviridae family is composed by over than 70 viral species with relevant global epidemiological impact. Flaviviruses viral particles have icosahedral capsid, are enveloped and present a single stranded genomic RNA of positive sense (1). The entry of flaviviruses is mainly based on endocytosis mediated by clathrin-coated pits and transport by an endocytic compartment. The low-pH environment within endosomes triggers conformational changes in the envelope glycoprotein (E), leading to membrane fusion of the viral envelope with the endosomal membrane and subsequent release of the nucleocapsid into the cytosol. The genomic RNA is translated into a viral polyprotein, which is cleaved by viral and cellular proteases, to originate flavivirus proteins (1). Three of these proteins are structural components of viral particle, called structural proteins. The E protein is the major antigen of the viral envelope. In addition to E protein, flavivirus particles contains also another envelope protein (M) and the capsid protein (C) (1). The polyprotein also originates non-structural proteins, which are not present on viral particle, but have important roles in viral replication and pathogenesis. There are seven non-structural proteins: NS1, NS2A, NS2B, NS3, NS4A, NS4B e NS5. After protein synthesis, the assembly of viral particles takes place in the endoplasmic reticulum. Immature viral particles are then sent to Golgi complex to be maturated by furin activity. Mature viral particles are then transported through vesicles to the plasma membrane and released by exocytosis (1).
The Flavivirus genus is involved in important chapters of both, World and Brazilian epidemiology histories. Yellow fever virus (YFV), Dengue virus (DENV) and Zika virus (ZIKV) were sequentially imported to Brazil and South America. Cases of yellow fever, caused by YFV, were first reported in Brazil in 1685. Thousands of yellow fever cases were noticed until the beginning of the twentieth century, when the Aedes aegypti mosquitoe was identified as the main vector of the disease. Vector eradication together with vaccination contributed to elimination of yellow fever from Brazilian urban areas (2). In 1981, the first epidemic caused by DENV was confirmed in Brazil (3). Since then, many epidemics have reached the country, usually related to the introduction of a new serotype or the change of the dominant serotype in a given region (3, 4). Such flavivirus is responsible for approximately 390 million of infections annually by one of its four serotypes (DENV1-4) (5). Thus, DENV is considered as the flavivirus of highest epidemiologic relevance of the world. But recently, ZIKV which was a pathogen previously associated with mild infections in Africa and Asia, was associated to microcephaly and Guillain-Barré syndrome in the Americas (6). The first cases of ZIKV infection were reported in early 2015, in the Brazilian Northeast. The virus was very quickly spreaded and 141 suspected cases of microcephaly were reported in Northeastern states, in addition to many cases of spontaneous abortions and stillbirths (6–9). In 2016 yellow fever cases were reported again in Brazil, with a total of 1,127 cases and 331 confirmed deaths until April 2018 (10). There is a number of additional flaviviruses with worldwide epidemiological importance or which are highly pathogenic. Examples are West Nile virus (WNV), Japanese encephalitis virus (JEV), Saint Louis encephalitis virus (SLEV) and Rocio virus (ROCV) (11). Such pathogens are vector-borne viruses, which are capable of traveling over long distances, carried by both, the human and/or the arthropod hosts. The risk of importing, emergence and epidemics caused by additional flaviviruses is clear in both, Brazil and other tropical regions of the world.
Humoral immune response to flaviviruses is complex and is involved in both, viral clearance and pathogenesis. An exacerbation of the disease severity mediated by antibodies, known as antibody-dependent enhancement (ADE), is observed in some cases of DENV infection. Immunoglobulins produced in a first infection by a specific serotype cross-react with viral particles present in a second infection caused by a different serotype. Viral particles are targeted to Fc-γ receptors bearing cells by non-neutralizing antibodies, which facilitates penetration and replication of DENV (1, 11). Increased viral replication leads to increased viral loads, exacerbated inflammation, increased release of inflammatory cytokines and vasoactive amines, a phenomenon known as cytokine storm (11, 12). ADE has also been described between DENV and ZIKV in vitro (8, 13–15). This phenomenon does not explain by itself the occurrence of severe dengue, but contribute to it in a relevant way. Investigations on ADE between different species of flavivirus have been reported, both in vitro (8, 13–16) and in vivo (17, 18). Most of the residues exposed at the external surface of flaviviral envelope are not conserved and are specific to each virus. Antibodies directed against envelope proteins potently neutralize the autologous and closely related viruses only (16, 19–21). This criterion of cross-neutralization by polyclonal antibodies has led to the classification of flaviviruses into serocomplexes (22). At present, the literature does not show evidence that antibodies are capable of conferring anti-flavivirus cross-protective immunity by themselves.
On the other hand, cellular immune responses to flaviviruses were shown to be involved in cross-protective immunity (23–28). Non-structural proteins, especially NS3 and NS5, are the main targets of T cells involved in flavivirus cross-protection (27, 29). The use of such proteins as subunit vaccine antigens was well documented in the literature. However, such vaccines were not tested in a serocomplex cross-protection context. Recent reports showed the importance of non-structural proteins in autologous (27, 30–32) or heterologous protection (23), regarding immunization with live attenuated viruses or natural infections. In contrast to structural proteins, it seems there are conserved residues in non-structural proteins which are capable of eliciting flavivirus cross-protective T cell immunity.
The Americas, Africa, Middle East, South East Asia and Europe have at least one Flavivirus serocomplex in circulation (23). Many areas of the world present both, vector fauna and geographical conditions compatible with co-circulation, importing, emergence and epidemics of flaviviruses. The encounter with multiple flaviviruses in a lifetime is increasingly likely. However, flavivirus cross-protective immunity is not well understood. In this study we aimed to find both, immunological determinants, and patterns of immune response possibly involved in flavivirus cross-protective immunity. We searched B and T cells epitopes which were thoroughly shown to be involved in flavivirus immunological control. Such epitopes were analyzed regarding their conservation along with different flavivirus serocomplexes. The epitopes were also studied regarding their population coverage, considering HLA allele frequencies of different human populations. Finally, epitopes with highest conservancy and population coverage were studied regarding their position along flavivirus polyprotein. Results presented in this study indicate that epitopes capable of eliciting flavivirus cross-protective immunity to a wide range of human populations are concentrated in proteins E, NS3, and NS5. Such identification of both, immunological determinants and patterns of immune response involved in flavivirus cross-protective immunity should be considered in future vaccine development. In addition, cross-reactive epitopes presented in this work may be involved in dynamics of diseases caused by flaviviruses worldwide.
Methods
Study Design
In this study we aimed to find both, immunological determinants and patterns of immune response possibly involved in flavivirus cross-protective immunity. As shown in Figure 1, we carried out a literature search of flavivirus epitopes involved in protective immunity. Such epitopes were analyzed with regard to their conservancy, population coverage and location in flavivirus polyprotein.
Flavivirus Polyprotein Database
A database was built with polyprotein sequences from flaviviruses of different serocomplexes isolated in different continents of the world. Viruses taken in consideration were those of highest epidemiological relevance in Brazil and worldwide: Dengue virus (DENV), Zika virus (ZIKV), Yellow fever virus (YFV), West Nile virus (WNV), Japanese encephalitis virus (JEV), Saint Louis encephalitis virus (SLEV), Rocio virus (ROCV), Cacipocore virus (CPCV), Ilhéus virus (ILHV), and Iguape virus (IGUV). Sequences in FASTA format were retrieved from the National Center for Biotechnology Information (NCBI) protein database from September-2017 to September-2018 (http://www.ncbi.nlm.nih.gov/protein/). We aimed to recover sequences from all listed flavivirus species, isolated in all continents, with a limit of three sequences per country of each viral species. We noticed that for some species there were few deposits. In these cases, all available sequences were selected. Additional criteria for selecting sequences were: (i) complete annotation of structural and non-structural proteins within deposited polyprotein sequence and (ii) absence of undefined amino acid into sequence. The database consisted of a total of 325 polyprotein amino acid sequences (Supplemental Material 1).
Search of T Cell Epitopes Involved in Flavivirus Protective Immunity
We carried out a literature search for reports of experimental characterization of protective epitopes. CD4+ and CD8+ T cell epitopes which were related to protective immune response to flavivirus were searched in reports available at Pubmed from September-2017 to September-2018 (https://www.ncbi.nlm.nih.gov/pubmed/). Criteria for selecting research articles were: (i) evaluation of epitope binding to HLA; (ii) determination of cytokine production pattern for both, CD4+ and CD8+ T cells; (iii) use of at least two different methods for determining cytokine production pattern and (iv), clear association with control of flavivirus infection. This last criterion involves validation of epitopes associated with survival under challenge experiments using humanized animals (transgenic mice expressing HLA), control of viral load under challenge experiments using humanized animals, infection of humans without disease and identification of important epitopes in immune responses elicited by well-known protective vaccines. Words used in the search were: CD4+ T cell, CD8+ T cell, epitopes, protection, immunity, Dengue virus, Zika virus, Yellow Fever virus, West Nile virus, Japanese encephalitis virus, Saint Louis encephalitis virus, Rocio virus, Cacipocore virus, Ilheus virus, and Iguape virus.
Search of B Cell Epitopes Targeted by Neutralizing Antibodies
B cell epitopes were searched at Immune Epitope Database and Analysis Resource-IEDB (http://www.iedb.org) (34). The search was carried out until September 30, 2018, using the following conditions: any epitope at the epitope field; flavivirus at the antigen field, humans in the host field; positive assays only, B cells assay, in vitro and/or in vivo neutralization and 3D structure at the assay field; any MHC restriction at the MHC restriction field and infectious disease at the disease field.
Conservancy Analysis of Selected Epitopes
The IEDB conservancy analysis tool (http://tools.iedb.org/conservancy) was used to determine the conservancy of the selected epitopes among the flavivirus sequences in the database previously constructed, as previously described (34, 35). Only epitopes at least 90% conserved among all sequences were selected.
Population Coverage Analyses
Epitopes selected after conservancy analysis were submitted to population coverage analysis, by using the IEDB population coverage calculation tool (http://tools.immuneepitope.org/tools/population/iedb_input), as previously described (35, 36).
Localization of Selected Epitopes Along Flavivirus Polyprotein
Epitopes with high conservancy were submitted to location analysis along flavivirus polyprotein. Preliminary localization was retrieved from NCBI annotations. Then, 3D protein models of E, NS3 and NS5, deposited at the Protein Data Bank- PDB (https://www.rcsb.org/), were used for fine localization. Pymol (http://www.pymol.org/) was used to highlight selected epitopes along the proteins 3D models, as previously described (33, 35).
Results
Selection and Localization of Epitopes Which Are Targets for Neutralizing Antibodies
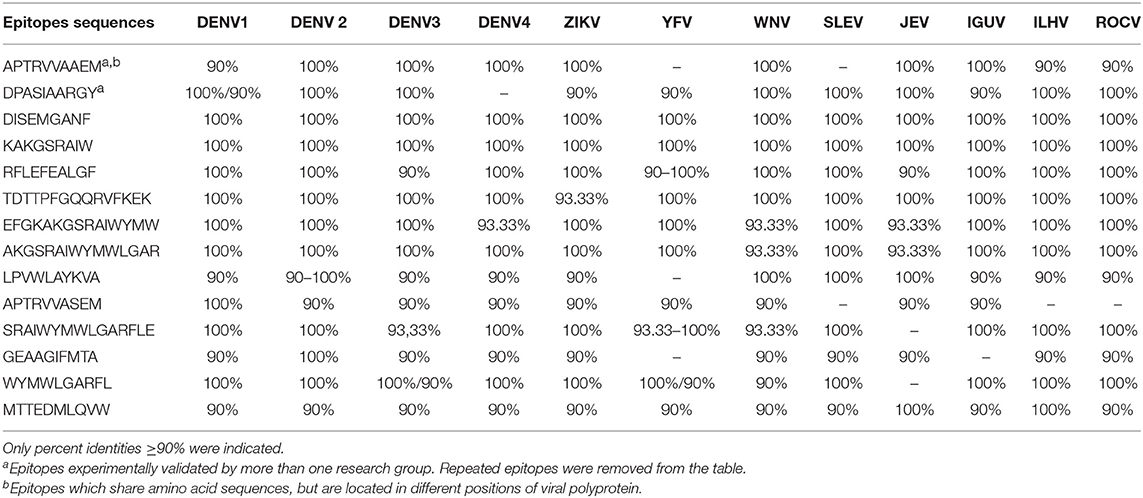
From over than 2,000 epitopes retrieved from search carried out as described in materials and methods, 19 were selected after conservancy analyses. One of the epitopes is contained inside a larger epitope as a consensus, in the same location. Thus, 18 epitopes are shown in Table 1. Such epitopes are discontinuous. From these, seven epitopes were shown to be 100% conserved and were selected for structural analyses. All selected epitopes are located in flavivirus envelope glycoprotein, mainly in domains II and III, as shown in Figure 2. Our results indicate that the seven neutralizing epitopes selected are highly conserved among flaviviruses and may be involved in cross-protection and restriction of virus circulation in some regions of the world.
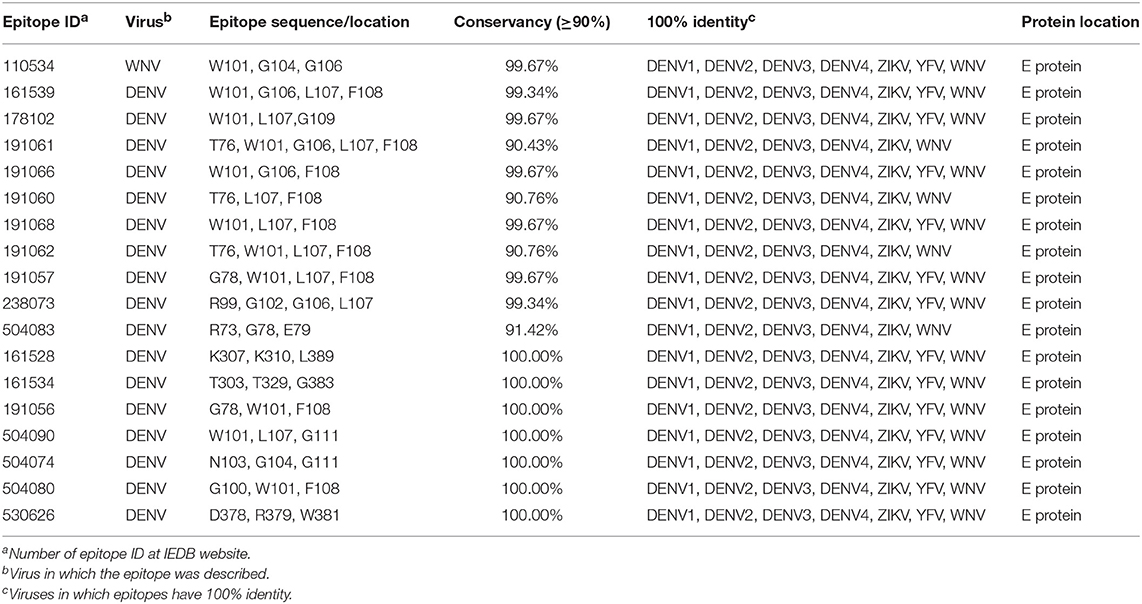
Table 1. Discontinuous B lymphocyte epitope selected from IEDB website and, posteriorly, chosen through conservancy analyzes using cut off ≥90%.
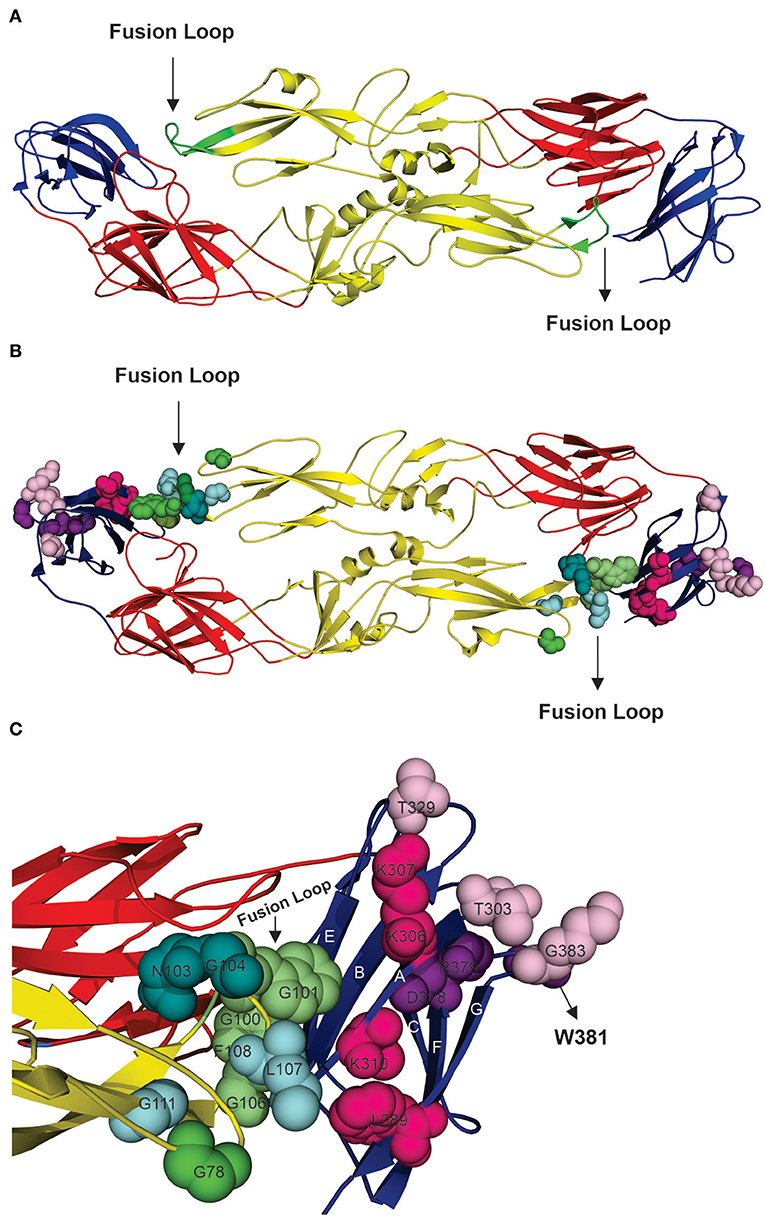
Figure 2. Structural analysis of highly conserved neutralizing epitopes in flaviviruses. (A) Model of DENV envelope glycoprotein arranged as a dimer [(33), PDB number 1UZG]. Domain I is shown in red, domain II is shown in yellow, and domain III is shown in blue, and the fusion loop is shown in green. (B) Overview of locations of the neutralizing epitopes in E protein. (C) Approximate view of locations of epitopes which are targets for neutralizing antibodies. Epitopes shown to be located in domain II are: G78, W101, F108 (shown in green), W101, L107, G111 (shown in cyan), N103, G104, G111 (shown in deepteal cyan) and G100, W101, F108 (shown in lime green). Note that these epitopes are overlapping and that are they concentrated at the fusion loop and share W101 and G111. Domain III presents the epitopes: T303, T329, G383 (shown in light pink), K307, K310, L389 (shown in hot pink) and D378, R379, W381 (shown in violet purple).
Selection of T Cell Epitopes Which May Be Involved in Flavivirus Serocomplex Cross-Protective Immunity
Nineteen research articles were selected reporting 529 epitopes. Such antigen determinants were experimentally characterized and associated with protective immunity. From the 529 epitopes 15 were selected after conservancy analysis. All selected epitopes are class I HLA ligands. Their amino acid sequences, HLA ligands, locations and percent conservations are shown in Table 2. Three epitopes showed conservation higher than 99% among all flavivirus polyproteins analyzed. In addition, virus-specific identity was also analyzed and is shown in Table 3. Epitope identity was shown to be constant in most of cases for each flavivirus. Together, these results indicate that there are fifteen epitopes associated with protective immunity which are highly conserved among flavivirus serocomplexes.
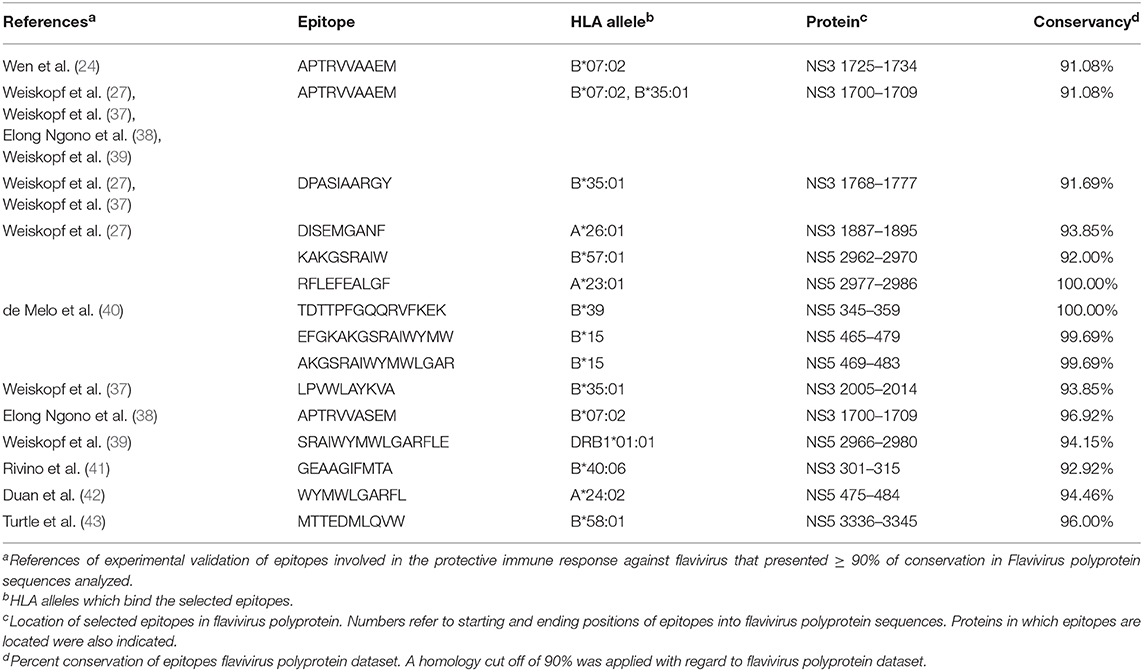
Table 2. T cell epitopes selected from literature review as contributing to flavivirus control and showing at least 90% conservation in the polyprotein dataset.
Population Coverage Analyses of Conserved T Cell Epitopes
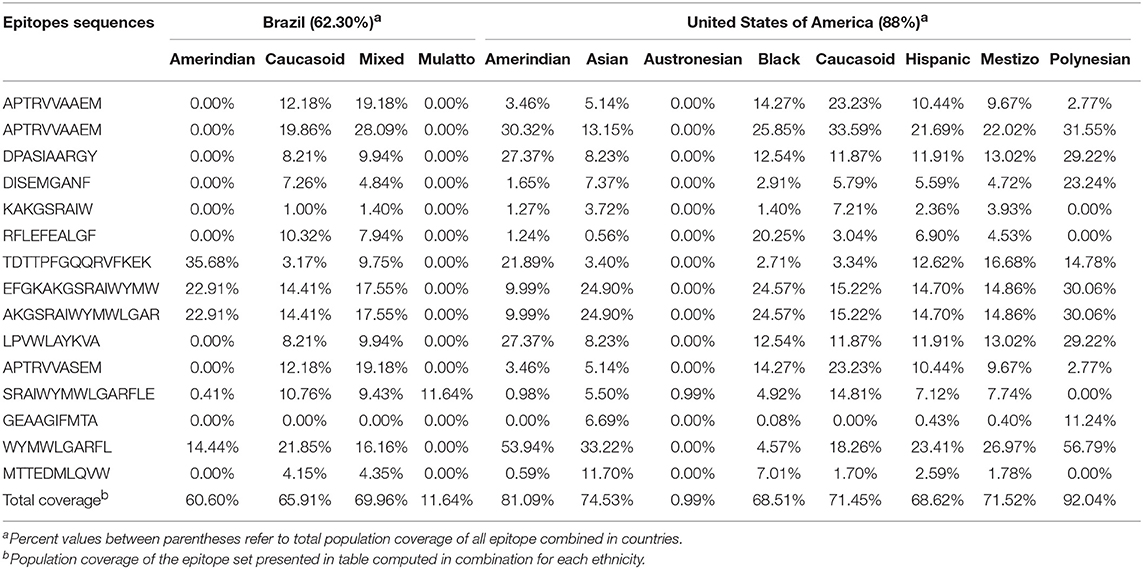
As shown in conservancy analyses, we selected fifteen epitopes with at least 90% identity among all flaviviruses. Such epitopes were shown to present relevant population coverages with regard to the most prevalent HLA alleles and main ethnicities in Brazil and United States, as shown in Table 4. In addition, population coverage for selected epitopes were shown to be also relevant for most of world regions, as shown in Table 5. Our results indicate that those highly conserved epitopes bind to HLA molecules of different human populations worldwide, with high population coverage.
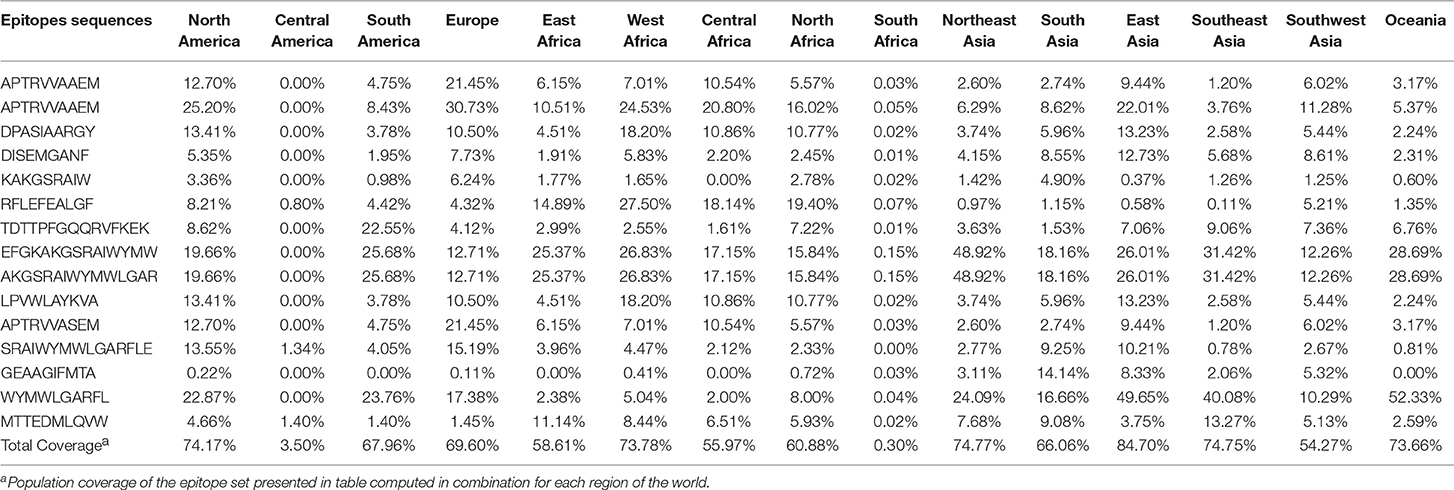
Table 5. Population coverage of selected epitopes for populations of different regions of the world.
Localization of Selected T Cell Epitopes Along Flavivirus Polyprotein
All of selected epitopes were concentrated in flavivirus non-structural proteins 3 and 5 (NS3 and NS5, respectively). Thus, we used NS5 and NS3 3D models to study the locations of the epitopes. From fifteen epitopes, six were located in Helicase domain of the NS3 protein (NS3H), and eight were located in the RNA-dependent RNA polymerases domain of the NS5 protein (NS5p) (Figure 3). These results indicate that NS3H and NS5p concentrate most of relevant T cell epitopes in the context of flavivirus cross-protective immunity.
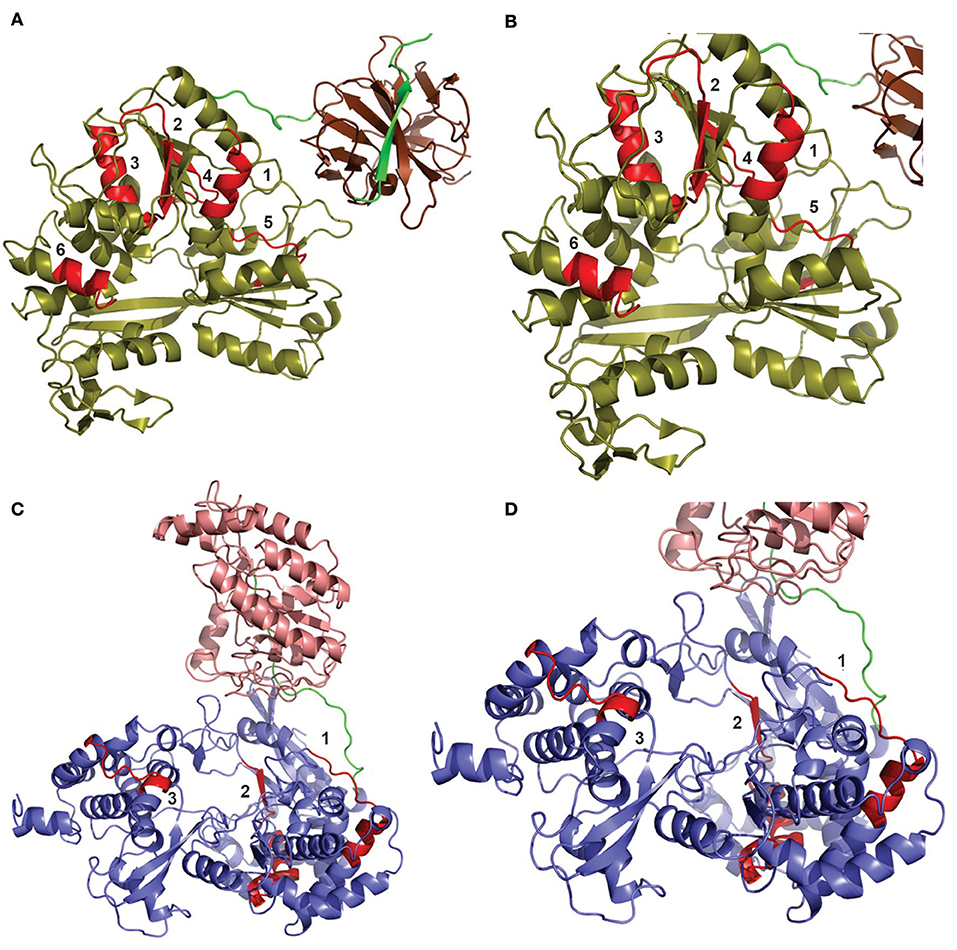
Figure 3. Distribution of T cell epitopes ≥90% conserved among flavivirus polyproteins. The locations of epitopes are shown in two different models: DENV NS3 protein (A,B) and ZIKV NS5 protein (C,D). (A) The helicase domain of the DENV NS3 protein is shown in olive, while its protease domain is shown in chocolate brown. Six epitopes (shown in red) were shown to be located in NS3, all of them located in the protease domain. (B) Zoomed view of NS3 protein to detail epitopes. The numbers refer to the following epitopes, respectively: (1) APTRVVAAEM or APTRVVASEM, (1) APTRVVAAEM, (2) DPASIAARGY, (3) GEAAGIFMTA, (4) DISEMGANF, and (5) LPVWLAYKVA. (C) The methyltransferase domain of the NS5 protein is shown in light salmon, while its RNA polymerase domain is shown in slate blue. Eight epitopes (shown in red) were shown to be located in the NS5 protein, all of them in the RNA polymerase domain. (D) Approximate view of NS5 protein to detail epitopes. Some epitopes present overlapping. Thus, we show three different regions: Region 1 is composed by one epitope TDTTPFGQQRVFKEK. Region 2 contain six epitopes: KAKGSRAIW, RFLEFEALGF, EFGKAKGSRAIWYMW, AKGSRAIWYMWLGAR, SRAIWYMWLGARFLE, and WYMWLGARFL. Region 3 is composed of only one epitope: MTTEDMLQVW.
Discussion
In this study, we aimed to identify both, immunological determinants and patterns of immune response possibly involved in flavivirus serocomplex cross-protection. Our premise was based on epidemiology of flaviviruses: most of them have both, vectoring and geographic conditions to co-circulate all together in the Americas, Africa, Asia and Oceania (44–49). However, it does not happen. In addition, recent studies showed evidences of cross-protective immunity induced by vaccines or sequential infections (23, 24, 26, 50–54). We searched B and T cells epitopes, which were thoroughly shown to be involved in flavivirus infection control. We found a relevant number of epitopes which are capable of eliciting flavivirus protective immunity and are also highly conserved among serocomplexes. In addition, such epitopes cover a wide range of human populations and are concentrated in proteins E, NS3, and NS5. For such epitopes, mechanisms involved in virus control targeting the E protein are all related to antibody-mediated virus neutralization. On the other hand, mechanisms involved in virus control by targeting NS3 and NS5 proteins are mainly related to cytotoxicity mediated by CD8+ T limphocytes.
The glycoprotein E is involved in binding of the viral particle to host cell receptors. In addition, such protein is also involved in membrane fusion. The E glycoprotein is arranged in three domains (DI, DII, and DIII). Domains I (DI) is central, surrounded by DII and DIII (55). The hydrophobic fusion loop is located in DII. It is highly conserved in flaviviruses serocomplexes due to its important role in mediating membrane fusion (55). The fusion loop has the same conformation in both protein forms trimer and dimer. In trimer, three amino acids are conserved in all flaviviruses (Trp 101, Leu 107, and Phe 108) and are exposed on molecular surface. Such amino acids have an important role in holding the fusion loop structure (33). Replacement of the Trp101 by an alanine prevents membrane fusion (56). In addition, natural infection by DENV was hypothesized to generate anti-DENV antibodies with cross-neutralizing activity against the fusion loop (57). All of discontinuous epitopes found in our study are located in E protein and most of them present these three conserved amino acids. We hypothesize that B cell epitopes presented in our study remain highly conserved across flavivirus serocomplexes due to their location at fusion loop.
The role of DIII in the flaviviral cycle is still unclear, although frequently related to a receptor binding function (55, 58, 59). It was previously shown that antibodies targeting different regions in DIII are able to strongly neutralize WNV. The regions are the N-terminal linker region (residues 302 to 309) and three strand-connecting loops, namely, BC (residues 330 to 333), DE (residues 365 to 368), and FG (residues 389 to 391) (60). In our study we found three discontinuous epitopes which are located in those regions: T303 T329 G383, K307 K310 L389, and D378 R379 W381. These epitopes revealed 100% conservancy among all 325 sequences analyzed. All of the selected B cell epitopes were show to be important in DENV neutralization (57, 61–63). Of course, a physic validation regarding cross-neutralizing activity remains to be carried out. However, positions highlighted in Figure 2 are located in the same structures of flaviviruses with known envelope glycoprotein structure. Our results indicate that such epitopes are important targets to be considered in flavivirus serocomplex cross-immunity due to their key roles in viral life cycle.
With regard to conservation of T cell-specific immunological determinants among flavivirus serocomplexes, the YVF lost the highest amount of 100% conserved T cell epitopes (see Table 3). This is in accordance with the highest genetic distance of YFV serocomplex with regard to other flavivirus serocomplexes (23). An important explanation for the epidemiology of flaviviruses may arise from this observation. For example, vaccination against YFV in Brazil is not currently related to protection against DENV and ZIKV epidemics. However, DENV and JEV serocomplexes share most of epitopes identified in this work. It seems immunity to DENV precludes viruses of JEV serocomplex of circulating in South America. Recent studies showed evidences of cross-protective immunity between DENV and JEV serocomplexes (23, 54). The same does not occur in Central America, Africa and Asia. Interestingly, Central America and South Africa present a low population coverage of the selected T cell epitopes. Nevertheless, population coverage of class-I HLA specific epitopes presented in this work does not seem to explain the whole scenario. For example, the population of United States is 88% covered by the set of epitopes, but there is co-circulation of DENV and JEV serocomplexes in that country. In addition, most of regions in Africa and Asia, which have also a high population coverage of our set of epitopes, present co-circulation of DENV and JEV serocomplexes. The explanation for this is probably related to class-II HLA restricted epitopes, from which only two were selected according to our criteria. Such epitopes did not present a high conservation among all serocomplexes and probably confer cross-protection in specific combinations of viruses and serocomplexes, as previously described (23). It was recently shown that cross-reactive epitopes can promote recall from a pool of flavivirus serocomplex cross-reactive memory CD4+ T cells (23). However, such epitopes are not enough conserved in order to induce cross-protection among all serocomplexes.
Cross-protective immunity into a given serocomplex seems to be more easily achieved (27, 50–53, 64, 65). Such cross-protection depends on recall of serocomplex cross-reactive memory CD4+ T cells. There is not a high number of highly conserved class-II HLA restricted epitopes among all serocomplexes. Nevertheless, we identified two peptides capable of binding class-II HLA, involved in anti-YFV protective immune response which are highly conserved among all flavivirus serocomplexes. Thus, it seems achieving multivalent protection among serocomplexes depends on overcoming immunodominance. Highly conserved class-II HLA restricted epitopes do not seem to be immunodominant in order to promote recall of serocomplex cross-reactive memory CD4+ T cells in a multivalent way.
With regard to class-I HLA restricted epitopes, we found fifteen highly conserved peptides which were thoroughly shown to be involved in flavivirus infection control. The peptides APTRVVAAEM, DPASIAARGY, DISEMGANF, KAKGSRAIW, and RFLEFEALGF are located in the NS3 or NS5 proteins. They bind to a subset of multifunctional CD8+ T limphocytes capable of producing IFN-γ and TNF-α against DENV infection (24, 27). In addition, the peptide APTRVVAAEM is involved in cross-protection against DENV and ZIKV. The epitopes TDTTPFGQQRVFKEK, EFGKAKGSRAIWYMW, and AKGSRAIWYMWLGAR are concentrated in the NS5 protein and contain multiple HLA-I and -II binding motifs which are involved in elicitation of protective immune memory against YFV (40). The epitope MTTEDMLQVW is involved in protection against JEV (43). The remaining selected epitopes (see Table 2) were also related to protection against DENV. All of the selected epitopes are involved in induction of anti-flavivirus CD8+ T cells. Some of the listed studies report generation os memory cells, which could also be recalled in order to favor cross-protective immunity.
Conserved epitopes could be enriched in artificial or natural immunizations. For example, sequential infection or administration of different anti-flavivirus vaccines would recall cross-reactive memory T CD4+ and T CD8+ lymphocytes. It is important to stress that approved vaccines represent three serocomplexes: DENV, JEV and YFV. However, most of those epitopes which are important for recalling cross-reactive T cells, are concentrated in non-structural proteins. The only anti-DENV vaccine currently approved for use in humans is a chimera between DENV and YFV. Its structural antigens are representative of DENV proteins, while non-structural antigens are representative of YFV proteins. In addition, the JEV vaccine is based on inactivated viruses. Enhancing the recall of cross-protective memory T cells would ideally happen with the use of live attenuated viruses or other vaccine approach capable of providing epitopes of non-structural proteins.
Data Availability Statement
All datasets generated for this study are included in the manuscript/Supplementary Files.
Author Contributions
JA conceived the study and wrote the paper. LS, LG, and WL analyzed data and wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to FAPESB for providing undergraduation fellowship to LS. We are also grateful to CNPq for providing postdoc fellowship to LG.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.02260/full#supplementary-material
References
1. Apte-Sengupta S, Sirohi D, Kuhn RJ. Coupling of replication and assembly in flaviviruses. Curr Opin Virol. (2014) 9:134–42. doi: 10.1016/j.coviro.2014.09.020
2. Febre amarela: Guia para Profissionais da saúde. 1st ed. Brasil: Ministério da Saúde, Secretaria de Atenção à Saúde (2017).
3. Diretrizes Nacionais para a Prevenção e Controle de Epidemias de Dengue. 1st ed. Brasil: Ministério da Saúde, Secretaria de Vigilância em Saúde, Departamento de Vigilância Epidemiológica (2009).
5. Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature. (2013) 496:504–7. doi: 10.1038/nature12060
6. Areas with Zika|Zika virus|CDC. Areas with Zika|Zika virus|CDC. Available online at: http://www.cdc.gov/zika/geo/ (accessed March 15, 2016).
7. de Brito CAA, Cordeiro MT. One year after the Zika virus outbreak in Brazil: from hypotheses to evidence. Rev Soc Bras Med Trop. (2016) 49:537–43. doi: 10.1590/0037-8682-0328-2016
8. Priyamvada L, Quicke KM, Hudson WH, Onlamoon N, Sewatanon J, Edupuganti S, et al. Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc Natl Acad Sci USA. (2016) 113:7852–7. doi: 10.1073/pnas.1607931113
9. Wikan N, Smith DR. Zika virus: history of a newly emerging arbovirus. Lancet Infect Dis. (2016) 16:e119–26. doi: 10.1016/S1473-3099(16)30010-X
10. De Mello M, Rezende D, Adelino R, Alves A, Campos F, Iani DM, et al. Persistence of Yellow fever virus outside the Amazon Basin, causing epidemics in Southeast Brazil, from 2016 to 2018. (2018) 12:e0006538. doi: 10.1371/journal.pntd.0006538
11. Whitehead SS, Blaney JE, Durbin AP, Murphy BR. Prospects for a dengue virus vaccine. Nat Rev Microbiol. (2007) 5:518–28. doi: 10.1038/nrmicro1690
12. Rothman AL. Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nat Rev Immunol. (2011) 11:532–43. doi: 10.1038/nri3014
13. Castanha PM, Nascimento EJM, Cynthia B, Cordeiro MT, de Carvalho OV, de Mendonça LR, et al. Dengue virus (DENV)-specific antibodies enhance Brazilian Zika virus (ZIKV) infection. J Infect Dis. (2016) 215:781–5. doi: 10.1093/infdis/jiw638
14. Dejnirattisai W, Supasa P, Wongwiwat W, Rouvinski A, Barba-Spaeth G, Duangchinda T, et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat Immunol. (2016) 17:1102–8. doi: 10.1038/ni.3515
15. Paul LM, Carlin ER, Jenkins MM, Tan AL, Barcellona CM, Nicholson CO, et al. Dengue virus antibodies enhance Zika virus infection. Clin Transl Immunol. (2016) 5:e117. doi: 10.1038/cti.2016.72
16. Mansfield KL, Horton DL, Johnson N, Li L, Barrett ADT, Smith DJ, et al. Flavivirus-induced antibody cross-reactivity. J Gen Virol. (2011) 92:2821–9. doi: 10.1099/vir.0.031641-0
17. Terzian ACB, Schanoski AS, Mota de O MT, da Silva RA, Estofolete CF, Colombo TE, et al. Viral load and cytokine response profile does not support antibody-dependent enhancement in dengue-primed zika virus–infected patients. Clin Infect Dis. (2017) 65:1260–5. doi: 10.1093/cid/cix558
18. Raboni SM, Bonfim C, Almeida BM, Zanluca C, Koishi AC, Rodrigues PRVP, et al. Flavivirus cross-reactivity in serological tests and Guillain-Barré syndrome in a hematopoietic stem cell transplant patient: a case report. Transpl Infect Dis. (2017) 19:e12700. doi: 10.1111/tid.12700
19. Fibriansah G, Lok SM. The development of therapeutic antibodies against dengue virus. Antiviral Res. (2016) 128:7–19. doi: 10.1016/j.antiviral.2016.01.002
20. Dai L, Wang Q, Qi J, Shi Y, Yan J, Gao GF. Molecular basis of antibody-mediated neutralization and protection against flavivirus. IUBMB Life. (2016) 68:783–91. doi: 10.1002/iub.1556
21. Rey FA, Stiasny K, Vaney M, Dellarole M, Heinz FX. The bright and the dark side of human antibody responses to flaviviruses: lessons for vaccine design. EMBO Rep. (2017) 19:e45302. doi: 10.15252/embr.201745302
22. Calisher CH, Karabatsos N, Dalrymple JM, Shope RE, Porterfield JS, Westaway EG, et al. Antigenic relationships between flaviviruses as determined by cross-neutralization tests with polyclonal antisera. J Gen Virol. (1989) 70(Pt 1):37–43. doi: 10.1099/0022-1317-70-1-37
23. Saron WAA, Rathore APS, Ting L, Ooi EE, Low J, Abraham SN, et al. Flavivirus serocomplex cross-reactive immunity is protective by activating heterologous memory CD4 T cells. Sci Adv. (2018) 4:eaar4297. doi: 10.1126/sciadv.aar4297
24. Wen J, Tang WW, Sheets N, Ellison J, Sette A, Kim K, et al. Identification of Zika virus epitopes reveals immunodominant and protective roles for dengue virus cross-reactive CD8(+) T cells. Nat Microbiol. (2017) 2:17036. doi: 10.1038/nmicrobiol.2017.36
25. Reynolds CJ, Suleyman OM, Ortega-Prieto AM, Skelton JK, Bonnesoeur P, Blohm A, et al. T cell immunity to Zika virus targets immunodominant epitopes that show cross-reactivity with other Flaviviruses. Sci Rep. (2018) 8:672. doi: 10.1038/s41598-017-18781-1
26. Regla-Nava JA, Elong Ngono A, Viramontes KM, Huynh AT, Wang YT, Nguyen AVT, et al. Cross-reactive Dengue virus-specific CD8+ T cells protect against Zika virus during pregnancy. Nat Commun. (2018) 9:3042. doi: 10.1038/s41467-018-05458-0
27. Weiskopf D, Angelo MA, Bangs DJ, Sidney J, Paul S, Peters B, et al. The human CD8+ T cell responses induced by a live attenuated tetravalent dengue vaccine are directed against highly conserved epitopes. J Virol. (2015) 89:120–8. doi: 10.1128/JVI.02129-14
28. Weiskopf D, Bangs DJ, Sidney J, Kolla RV, De Silva AD, de Silva AM, et al. Dengue virus infection elicits highly polarized CX3CR1+ cytotoxic CD4+ T cells associated with protective immunity. Proc Natl Acad Sci USA. (2015) 112:E4256–63. doi: 10.1073/pnas.1505956112
29. Sironi M, Forni D, Clerici M, Cagliani R. Nonstructural proteins are preferential positive selection targets in zika virus and related flaviviruses. PLoS Negl Trop Dis. (2016) 10:e0004978. doi: 10.1371/journal.pntd.0004978
30. Amorim JH, dos Santos Alves RP, Bizerra R, Araújo Pereira S, Ramos Pereira L, Nascimento Fabris DL, et al. Antibodies are not required to a protective immune response against dengue virus elicited in a mouse encephalitis model. Virology. (2016) 487:41–9. doi: 10.1016/j.virol.2015.10.006
31. Rivino L, Kumaran EAP, Jovanovic V, Nadua K, Teo EW, Pang SW, et al. Differential targeting of viral components by CD4+ versus CD8+ T lymphocytes in dengue virus infection. J Virol. (2013) 87:2693–706. doi: 10.1128/JVI.02675-12
32. Rivino L, Kumaran EA, Thein T-L, Too CT, Hao Gan VC, Hanson BJ, et al. Virus-specific T lymphocytes home to the skin during natural dengue infection. Sci Transl Med. (2015) 7:278ra35. doi: 10.1126/scitranslmed.aaa0526
33. Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. (2004) 427:313–9. doi: 10.1038/nature02165
34. Bui H-H, Sidney J, Li W, Fusseder N, Sette A. Development of an epitope conservancy analysis tool to facilitate the design of epitope-based diagnostics and vaccines. BMC Bioinformatics. (2007) 8:361. doi: 10.1186/1471-2105-8-361
35. dos Santos Franco L, Oliveira Vidal P, Amorim JH. In silico design of a Zika virus non-structural protein 5 aiming vaccine protection against zika and dengue in different human populations. J Biomed Sci. (2017) 24:88. doi: 10.1186/s12929-017-0395-z
36. Bui H-H, Sidney J, Dinh K, Southwood S, Newman MJ, Sette A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinformatics. (2006) 7:153. doi: 10.1186/1471-2105-7-153
37. Weiskopf D, Angelo MA, de Azeredo EL, Sidney J, Greenbaum JA, Fernando AN, et al. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc Natl Acad Sci USA. (2013) 110:E2046–53. doi: 10.1073/pnas.1305227110
38. Elong Ngono A, Chen HW, Tang WW, Joo Y, King K, Weiskopf D, et al. Protective role of cross-reactive CD8 T cells against dengue virus infection. EbioMed. (2016) 13:284–93. doi: 10.1016/j.ebiom.2016.10.006
39. Weiskopf D, Yauch LE, Angelo MA, John DV, Jason A, Sidney J, et al. Insights into HLA restricted T cell responses in a novel mouse model of dengue virus infection point towards new implications for vaccine design. J Immunol. (2012) 187:4268–79. doi: 10.4049/jimmunol.1101970
40. de Melo AB, Nascimento EJM, Braga-Neto U, Dhalia R, Silva AM, Oelke M, et al. T-cell memory responses elicited by yellow fever vaccine are targeted to overlapping epitopes containing multiple HLA-I and -II binding motifs. PLoS Negl Trop Dis. (2013) 7:e1938. doi: 10.1371/journal.pntd.0001938
41. Rivino L, Tan AT, Chia A, Kumaran EAP, Grotenbreg GM, MacAry PA, et al. Defining CD8+ T cell determinants during human viral infection in populations of Asian ethnicity. J Immunol. (2013) 191:4010–9. doi: 10.4049/jimmunol.1301507
42. Duan ZL, Liu HF, Huang X, Wang SN, Yang JL, Chen XY, et al. Identification of conserved and HLA-A*2402-restricted epitopes in Dengue virus serotype 2. Virus Res. (2015) 196:5–12. doi: 10.1016/j.virusres.2014.10.022
43. Turtle L, Bali T, Buxton G, Chib S, Chan S, Soni M, et al. Human T cell responses to Japanese encephalitis virus in health and disease. J Exp Med. (2016) 213:1331–52. doi: 10.1084/jem.20151517
44. Gubler, DJ, and St. John, AL Dengue Viruses*. In: Reference Module in Biomedical Sciences. Elsevier (2014). doi: 10.1016/b978-0-12-801238-3.02556-3
45. Gubler DJ. Dengue, Urbanization and globalization: the Unholy Trinity of the 21st Century. Trop Med Health. (2011) 39:S3–11. doi: 10.2149/tmh.2011-S05
46. Chancey C, Grinev A, Volkova E, Rios M. The global ecology and epidemiology of west nile virus. Biomed Res Int. (2015) 2015:1–20. doi: 10.1155/2015/376230
47. Campbell G, Hills S, Fischer M, Jacobson J, Hoke C, Hombach J, et al. Estimated global incidence of Japanese encephalitis: Bull World Health Organ. (2011) 89:766–74. doi: 10.2471/BLT.10.085233
48. Konishi E, Kitai Y, Tabei Y, Nishimura K, Harada S. Natural Japanese encephalitis virus infection among humans in west and east Japan shows the need to continue a vaccination program. Vaccine. (2010) 28:2664–70. doi: 10.1016/j.vaccine.2010.01.008
49. Jentes ES, Poumerol G, Gershman MD, Hill DR, Lemarchand J, Lewis RF, et al. The revised global yellow fever risk map and recommendations for vaccination, 2010: consensus of the Informal WHO Working Group on Geographic Risk for Yellow Fever. Lancet Infect Dis. (2011) 11:622–32. doi: 10.1016/S1473-3099(11)70147-5
50. Amarilla AA, Fumagalli MJ, Figueiredo ML, Lima-Junior DS, Santos-Junior NN, Alfonso HL, et al. Ilheus and Saint Louis encephalitis viruses elicit cross-protection against a lethal Rocio virus challenge in mice. PLoS ONE. (2018) 13:e0199071. doi: 10.1371/journal.pone.0199071
51. Hadinegoro SR, Arredondo-García JL, Capeding MR, Deseda C, Chotpitayasunondh T, Dietze R, et al. Efficacy and long-term safety of a dengue vaccine in regions of endemic disease. N Engl J Med. (2015) 373:1195–206. doi: 10.1056/NEJMoa1506223
52. Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S, et al. Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet. (2012) 380:1559–67. doi: 10.1016/S0140-6736(12)61428-7
53. Villar L, Dayan GH, Arredondo-García JL, Rivera DM, Cunha R, Deseda C, et al. Efficacy of a tetravalent dengue vaccine in children in Latin America. N Engl J Med. (2014) 372:141103114505002. doi: 10.1056/NEJMoa1411037
54. Li J, Gao N, Fan D, Chen H, Sheng Z, Fu S, et al. Cross-protection induced by Japanese encephalitis vaccines against different genotypes of Dengue viruses in mice. Sci Rep. (2016) 6:19953. doi: 10.1038/srep19953
55. Cockburn JJB, Navarro Sanchez ME, Fretes N, Urvoas A, Staropoli I, Kikuti CM, et al. Mechanism of dengue virus broad cross-neutralization by a monoclonal antibody. Structure. (2012) 20:303–14. doi: 10.1016/j.str.2012.01.001
56. Melo MN, Sousa FJR, Carneiro FA, Castanho MARB, Valente AP, Almeida FCL, et al. Interaction of the dengue virus fusion peptide with membranes assessed by NMR: the essential role of the envelope protein Trp101 for membrane fusion. J Mol Biol. (2009) 392:736–46. doi: 10.1016/j.jmb.2009.07.035
57. Smith SA, de Alwis AR, Kose N, Harris E, Ibarra KD, Kahle KM, et al. The potent and broadly neutralizing human dengue virus-specific monoclonal antibody 1C19 reveals a unique cross-reactive epitope on the bc loop of domain II of the envelope protein. MBio. (2013) 4:1–12. doi: 10.1128/mBio.00873-13
58. Sukupolvi-Petty S, Austin SK, Purtha WE, Oliphant T, Nybakken GE, Schlesinger JJ, et al. Type- and subcomplex-specific neutralizing antibodies against domain III of dengue virus type 2 envelope protein recognize adjacent epitopes. J Virol. (2007) 81:12816–26. doi: 10.1128/JVI.00432-07
59. Crill WD, Roehrig JT. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J Virol. (2001) 75:7769–73. doi: 10.1128/JVI.75.16.7769-7773.2001
60. Nybakken GE, Oliphant T, Johnson S, Burke S, Diamond MS, Fremont DH. Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature. (2005) 437:764–9. doi: 10.1038/nature03956
61. Lin HE, Tsai WY, Liu IJ, Li PC, Liao MY, Tsai JJ, et al. Analysis of epitopes on dengue virus envelope protein recognized by monoclonal antibodies and polyclonal human sera by a high throughput assay. PLoS Negl Trop Dis. (2012) 6:e1447. doi: 10.1371/journal.pntd.0001447
62. Tsai W-Y, Lai C-Y, Wu Y-C, Lin H-E, Edwards C, Jumnainsong A, et al. High-avidity and potently neutralizing cross-reactive human monoclonal antibodies derived from secondary dengue virus infection. J Virol. (2013) 87:12562–75. doi: 10.1128/JVI.00871-13
63. Shi X, Deng Y, Wang H, Ji G, Tan W, Jiang T, et al. A bispecific antibody effectively neutralizes all four serotypes of dengue virus by simultaneous blocking virus attachment and fusion. MAbs. (2016) 8:574–84. doi: 10.1080/19420862.2016.1148850
64. Li S-H, Li X-F, Zhao H, Jiang T, Deng Y-Q, Yu X-D, et al. Cross protection against lethal West Nile virus challenge in mice immunized with recombinant E protein domain III of Japanese encephalitis virus. Immunol Lett. (2011) 138:156–60. doi: 10.1016/j.imlet.2011.04.003
Keywords: epitopes, flavivirus, serocomplex, cross-protection, immune response
Citation: dos Santos Franco L, Gushi LT, Luiz WB and Amorim JH (2019) Seeking Flavivirus Cross-Protective Immunity. Front. Immunol. 10:2260. doi: 10.3389/fimmu.2019.02260
Received: 29 March 2019; Accepted: 06 September 2019;
Published: 20 September 2019.
Edited by:
Daniela Weiskopf, La Jolla Institute for Immunology (LJI), United StatesReviewed by:
Sandeep Kumar Dhanda, La Jolla Institute for Immunology (LJI), United StatesDaniel M. Altmann, Imperial College London, United Kingdom
Copyright © 2019 dos Santos Franco, Gushi, Luiz and Amorim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jaime Henrique Amorim, amFpbWUuYW1vcmltQHVmb2IuZWR1LmJy
†These authors have contributed equally to this work
 Lorrany dos Santos Franco
Lorrany dos Santos Franco Letícia Tsieme Gushi
Letícia Tsieme Gushi Wilson Barros Luiz2
Wilson Barros Luiz2 Jaime Henrique Amorim
Jaime Henrique Amorim