- 1Department of Veterinary Medical Science, University of Parma, Parma, Italy
- 2Microbiome Research Hub, University of Parma, Parma, Italy
- 3APC Microbiome Institute and School of Microbiology, Bioscience Institute, National University of Ireland, Cork, Ireland
- 4Laboratory of Probiogenomics, Department of Chemistry, Life Sciences, and Environmental Sustainability, University of Parma, Parma, Italy
Since bifidobacteria are among the pioneering colonizers of the human infant gut, their interaction with their host is believed to start soon following birth. Several members of the Bifidobacterium genus are purported to exert various health-promoting effects at local and systemic levels, e.g., limiting pathogen colonization/invasion, influencing gut homeostasis, and influencing the immune system through changes in innate and/or adaptive immune responses. This has promoted extensive research efforts to shed light on the precise mechanisms by which bifidobacteria are able to stimulate and interact with the host immune system. These studies uncovered a variety of secreted or surface-associated molecules that act as essential mediators for the establishment of a bifidobacteria-host immune system dialogue, and that allow interactions with mucosa-associated immune cells. Additionally, the by-products generated from bifidobacterial carbohydrate metabolism act as vectors that directly and indirectly trigger the host immune response, the latter by stimulating growth of other commensal microorganisms such as propionate- or butyrate-producing bacteria. This review is aimed to provide a comprehensive overview on the wide variety of strategies employed by bifidobacteria to engage with the host immune system.
Introduction
The gastrointestinal tract (GIT) of humans, and by extension of mammals, harbors an extremely dense, and complex community of microorganisms. Until recently, this microbial ensemble, collectively forming the so-called gut microbiota, was estimated to exceed the total number of the host cells by about 10-fold (1, 2). However, this number has been recently revised to a 1:1 ratio (3, 4). Despite this downward revision, the gut microbiota remains the most complex of the human-associated microbial communities and rendering the intestine one of the most intricate bacterial habitats in the biosphere. Millions of years of co-evolution between the mammalian host and its intestinal microbial ecosystem has led to the establishment of various trophic interactions, including a mutualistic relationship in which the host ensures nourishment and a suitable environment for the microbial community, while the microbiota, in turn, provides the host with multiple physiological and metabolic functions (5, 6). Beyond the provision of nutrition for enterocytes and degradation of non-digestible food compounds, the gut microbiota is also involved in a continuous dialogue with the immune system promoting the maintenance of the intestinal barrier, influencing bowel homeostasis and functionality, stimulating optimal immune responses, and providing protection against pathogen colonization (6, 7). Functional and physiological alterations of the intestinal barrier are generally associated with damage of the barrier itself, leading to a broad range of both intestinal and systemic diseases, including inflammatory bowel diseases (IBD), colorectal cancer, or celiac disease (8, 9). Among the multitude of gut bacteria that have been shown to positively influence the immune system, many studies have focused their interest on bifidobacteria as commensal microorganisms that are able to stimulate and modulate specific pathways, influencing both innate and adaptive host immune responses (10, 11). However, current knowledge on the molecular mechanisms that are responsible for the cross talk between bifidobacteria and the host immune system remain superficial and incomplete.
In this review, we will discuss current knowledge on immune-modulatory activities exerted by bifidobacteria in the intestinal environment. Particularly, we will detail how specific bifidobacterial extracellular structures (are believed to) modulate the host immune response. Furthermore, we will discuss certain interactions between bifidobacteria and other intestinal microbes affect the host immune system.
Taxonomic and Ecological Diversity of the Bifidobacterium Genus
Bifidobacteria are Gram-positive, anaerobic, non-motile, non-sporulating, saccharolytic, and Y-shaped microorganisms with a high G + C DNA content. They were described for the first time by Tissier in 1899, following their isolation from a fecal sample of a breast-fed infant. Bifidobacteria are classified as members of the Bifidobacterium genus, which forms a deep-branching lineage within the Actinobacteria phylum (12, 13). Currently, the Bifidobacterium genus encompasses 82 recognized taxa, representing 73 species and nine subspecies (14–17). A comparative genomic analysis based on 72 sequenced bifidobacterial type strains identified the presence of 261 Bifidobacterium-specific clusters of orthologous genes (COGs) shared by these genomes, collectively called the bifidobacterial core genome (18). The subsequent concatenation of the core gene protein sequences allowed the construction of a very robust phylogenetic tree highlighting the division of the Bifidobacterium genus into 10 phylogenetic groups, encompassing Bifidobacterium adolescentis, Bifidobacterium boum, Bifidobacterium pullorum, Bifidobacterium asteroides, Bifidobacterium longum, Bifidobacterium psychraerophilum, Bifidobacterium bifidum, Bifidobacterium pseudolongum, Bifidobacterium bombi, and Bifidobacterium tissieri group (18).
Bifidobacteria have been isolated from many ecological niches such as fermented milk (18), sewage (19), human blood (20), the oral cavity (21), and the gastrointestinal tract of mammals, birds, and insects (14, 16, 17, 22). Despite this apparent broad ecological distribution, the capability of bifidobacteria to adapt to different ecological niches is species-dependent. In this context, it has been demonstrated that certain bifidobacteria, such as B. longum, B. adolescentis, B. pseudolongum, and B. bifidum, display a cosmopolitan lifestyle, while other bifidobacterial taxa seem to be adapted to a gut ecosystem of a specific animal, for example Bifidobacterium angulatum in cows, Bifidobacterium cuniculi in rabbits and Bifidobacterium gallinarum in chickens (23, 24). The differential ecological success to adapt to various ecological niches very much relies on the genetic heritage of each bifidobacterial species. In this context, comparison of the genome sequences from the type strains of 47 Bifidobacterium (sub)species allowed the reconstruction of the bifidobacterial pan- and core-genome (25). The subsequent functional annotation of the core-genome by means of the EggNog database, indicated that a large part of the bifidobacterial core genes encode carbohydrate metabolism functions, thus suggesting that carbohydrate degradation plays an important role in bifidobacterial colonization of their ecological niches. Conversely, the pan-genome analysis revealed the identification of Truly Unique Genes (TUGs) corresponding to genes present in just one of the examined bifidobacterial genomes. Although the majority of TUGs cannot be attributed to a specific function, a portion of the TUGs were assigned to carbohydrate metabolism and transport functions, thus supporting the notion that the bifidobacterial gene repertoire for carbohydrate degradation plays an essential role in the differential ecological adaptation abilities among members of the Bifidobacterium genus (25). Similarly, in silico prediction of the bifidobacterial pan-secretome, i.e., the ensemble of genes encoding secreted proteins responsible for metabolism of nutrients, revealed the existence of species-specific bifidobacterial secretomes, which were predicted for the metabolism of particular glycans (26). These findings emphasize the correlation between the genetic repertoire of a Bifidobacterium species and their cosmopolitan or specialized lifestyle (26).
Development of the Human Gut Microbiota and Associated Bifidobacterial Community
Despite the broad ecological distribution of bifidobacteria, certain human-associated bifidobacteria have gained particular importance as they are purported to exert health-promoting activities, especially in the context of the infant gut microbiota (27). For a long time, it was thought that the establishment of the human intestinal microbiota occurred immediately following birth. However, recent studies have disputed the dogma of a sterile in utero environment, providing evidence of a microbial presence in the placenta, amniotic fluid and umbilical cord in healthy full-term pregnancies, though these findings are very controversial (28–31). It is well-accepted that the very first microbial colonizers of the infant gut are represented by facultative anaerobes, including several members of the Enterobacteriaceae family that deplete the oxygen in the intestine (32). Following the removal of oxygen, the infant gut is extensively colonized by strictly anaerobic microorganisms, among which bifidobacteria are considered to be major players (33–35). However, the total load and diversity of bifidobacteria in the infant gut are strongly dependent on environmental factors such as mode of delivery (natural or C-section delivery), type of feeding (breast or formula milk), duration of gestation (full-term or premature birth), and antibiotic administration (29, 36–39). In this context, it has been demonstrated that naturally delivered and breast-fed infants generally possess a higher relative abundance and diversity in terms of bifidobacterial populations when compared to the gut microbiota of those infants born by Cesarean section and fed with formula milk (29, 40). Weaning and the concomitant transition to a solid and more varied diet cause an increase in the overall composition of the infant gut microbiota, reaching its peak in adult life. The exact age that marks the passage from an infant-like to an adult-like gut microbiota is not universally fixed, but it is generally thought to happen around 3 years (41, 42). Although at this age the gut microbiota seems to have reached the stability typical of an adult intestinal community, some microbial taxa are still far from reaching a steady state. Indeed, some differences in the microbial intestinal community are found between pre-adolescence and adulthood (43). In adult life, levels of bifidobacteria are reduced (compared to that during infancy), but remain stable over time until old age (43). The latter is characterized by a general decline in species diversity coupled with a decrease in bifidobacterial abundance (44). Similar to the changes in the overall microbial biodiversity and complexity that are observed at various ages, also the bifidobacterial community composition is subject to modifications. In this context, analyses of the bifidobacterial population by either culture or metagenomic approaches revealed stage-of-life-specific bifidobacterial strains. Indeed, Bifidobacterium breve, B. bifidum, and B. longum subsp. infantis are typically found in the gut of breastfed infants, while adult life is generally associated with higher abundance of B. adolescentis and B. catenulatum species (45, 46). Conversely, B. longum subsp. longum seems to have a ubiquitous distribution across the human lifespan (47).
Immunomodulatory Effects Elicited by Bifidobacterial Extracellular Molecules Involved in Host-Microbe Interaction
Microorganisms have evolved specific strategies to interact with the host. Despite the health-promoting effects exerted by bifidobacteria, the molecular mechanisms exploited by these microorganisms to colonize the gut, adhere to the host intestinal epithelium and elicit an immune response are still largely unknown. In this context, certain extracellular structures, excreted enzymes and/or bioactive metabolites seem to play a pivotal role in host-microbe interaction, thereby modulating the immune system. In the following sections, key examples of these extracellular structures will be discussed in detail (Figure 1A; Table 1).
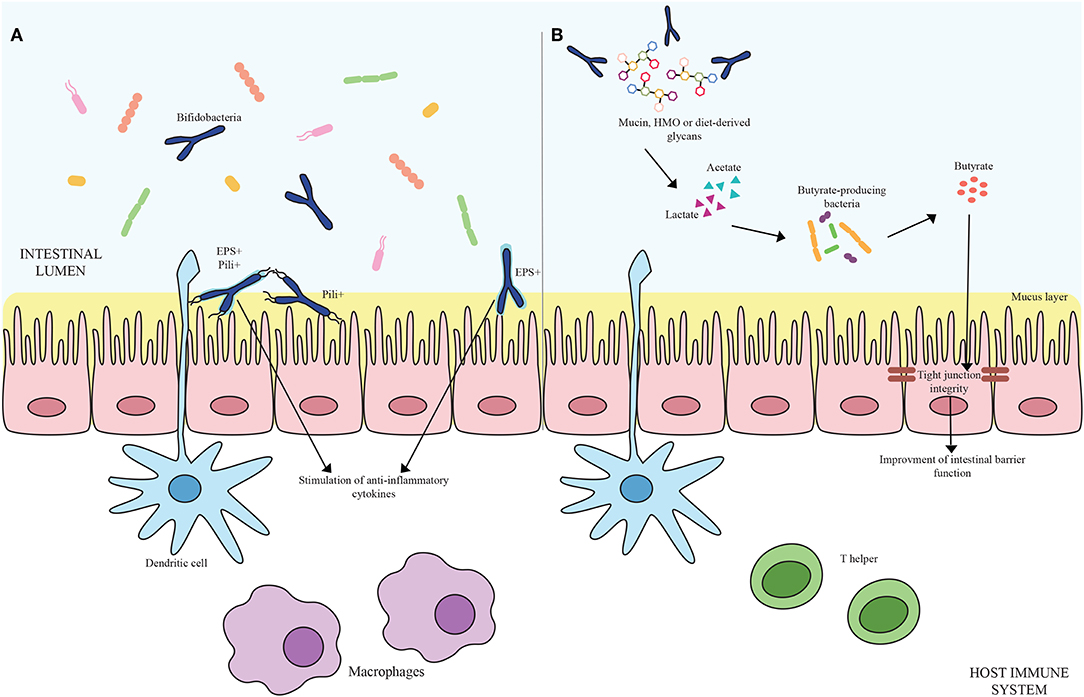
Figure 1. (A) Shows bifidobacterial extracellular structures-mediated interaction with the host immune system. Pili are depicted as black appendages (Pili+) while exopolysaccharides structures (EPS+) are displayed as light blue layers around the cells. Blue bifidobacterial shapes correspond to bifidobacterial strains. (B) Exhibits the cross-feeding effects between species of the Bifidobacterium genus and butyrate-producing bacteria. Acetate and lactate produced by bifidobacterial species by degrading mucin, HMO or diet-derived glycans become carbon sources for butyrate-producing microorganisms, stimulating a butyrogenic effect. At the bottom of the image, host immune system cells are represented.
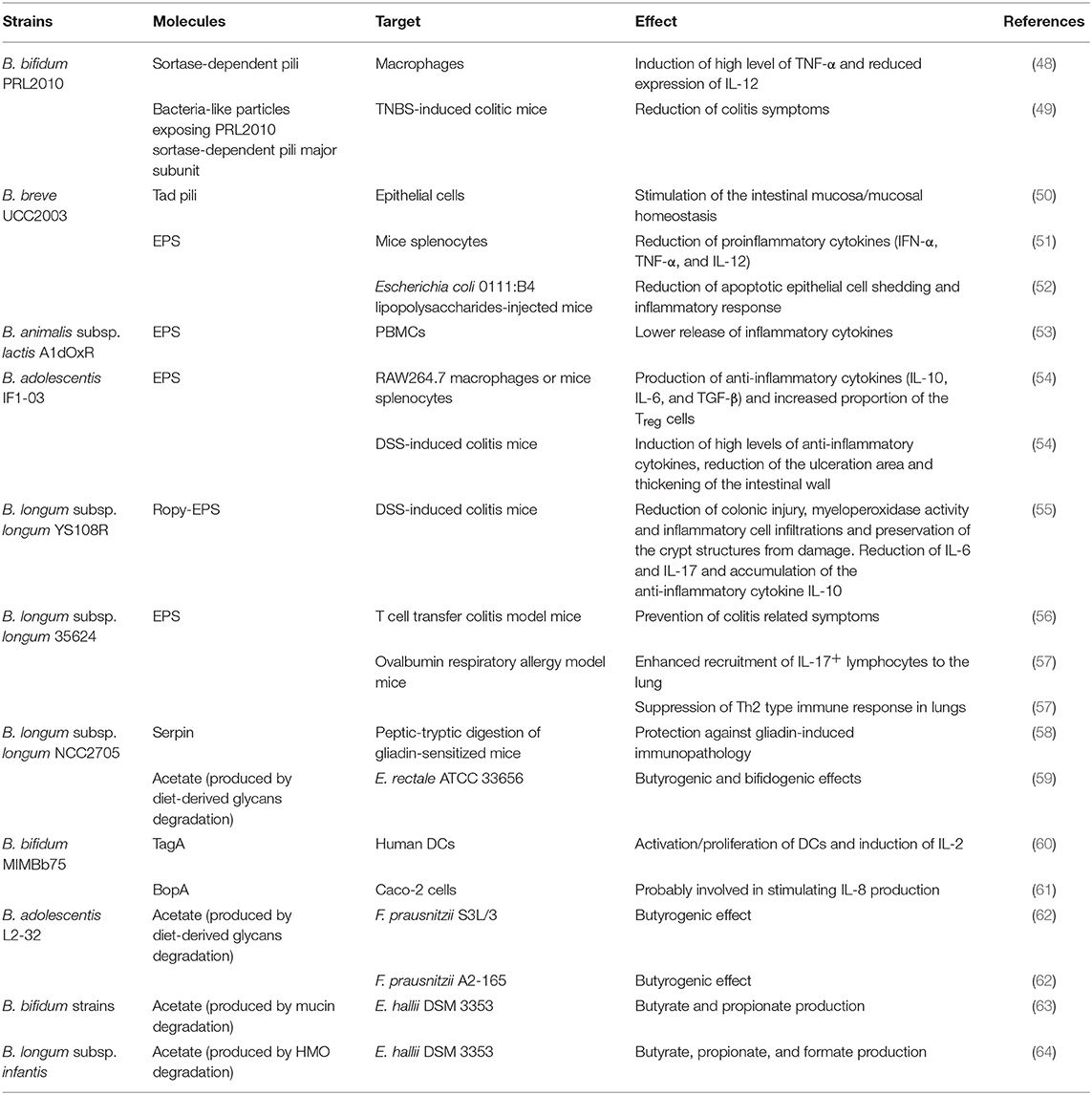
Table 1. Bifidobacterial extracellular molecules and/or metabolites exerting an immunological effect.
Pili
Pili (or fimbriae) are described as long proteinaceous appendages produced by bacteria, protruding from the extracellular cell surface, and that may be involved in microbe-host interactions by promoting adhesion to the intestinal epithelium or facilitating aggregation with other bacterial cells (65–67). In bifidobacteria, two different types of pili have been described, i.e., the sortase-dependent pili and the type IVb pili, which are also known as tight adherence pili (Tad pili) (68, 69).
To support the notion that these fibers play a role in host-microbe interaction, it was demonstrated that bifidobacterial sortase-dependent pili biosynthesis is only activated under particular conditions, such as during in vivo colonization, in vitro contact between bifidobacterial cells and human cell lines, and/or exposure to certain extracellular matrix proteins (48, 68–70). Genes encoding the biosynthetic enzymes for a particular sortase-dependent pilus are generally clustered together in a genetic locus, encompassing a gene encoding a major pilin protein, as well as one or two genes involved in the synthesis of (one or two) ancillary pilins and a third gene implicated in the expression of a pilus-specific sortase (66, 71). The sortase is a transpeptidase able to cross-link the pilus building blocks and covalently connect the resulting polymer to the cell wall surface (71). Comparison of the sequenced genomes from different bifidobacterial species revealed the presence of various sortase-dependent pilus-encoding loci in terms of number and genetic sequence variability (68). Indeed, the number of these pilus-encoding loci was observed to vary from a total absence typical of some Bifidobacterium actinocoloniforme, B. longum subsp. longum, and B. longum subsp. infantis strains to up to seven pilus-encoding loci found in the genome of B. dentium Bd1 (66, 68). Moreover, the GC content deviation and different codon usage observed in bifidobacterial sortase-dependent loci together with their frequent localization near transposons indicate that these genetic elements have been acquired through horizontal gene transfer events (68, 72).
Mediating adhesion and interaction with the host is not the only role exerted by bifidobacterial sortase-dependent pili. Indeed, they are also implicated in microbe-microbe interactions and in stimulating/modulating the host immune system. In this context, a case study focused on B. bifidum PRL2010 demonstrated that sortase-dependent pili have a pioneering role in favoring the aggregation between bacterial cells of a heterogeneous population, enhancing the colonization of host intestinal mucosa (70). In the same way, a related study highlighted that sortase-dependent pili produced by B. bifidum PRL2010 can activate various signals in macrophages inducing local high levels of TNF-α cytokines, but a reduced expression of other proinflammatory cytokines, such as IL-12, associated with systemic response (48), thus favoring initial cross-talk between this bifidobacterial strain and host immune cells without causing a detrimental inflammatory cascade response. In this context, as bifidobacteria are among the first colonizers of the human gut (29) and considering that the immune system of newborns is largely immature, the proinflammatory stimuli provoked by pili of B. bifidum PRL2010 (and possibly other infant-associated bifidobacteria) may be an essential starting point to prime the immune system (48). To further emphasize the immune-modulatory features exerted by B. bifidum PRL2010 pili, an in vivo study based on the administration of bacteria-like particles (BLP-FimPPRL2010) that expose the B. bifidum PRL2010 sortase-dependent pili major subunit 3 days before the treatment of mice model with 2,4,6-trinitrobenzensulfonic acid (TNBS) to induce colitis, clearly showed a reduction of biological/clinical markers associated with colitis symptoms when compared to the control. In addition, administration of BLP-FimPPRL2010 causes TNF-α overexpression coupled with a significantly reduction of IL-10 response in TNBS-induced colitic mice. Thus, alerting the immune system and enhancing the host reaction to inflammation, which is a typical condition of colitis (49).
Another pilus type produced by bifidobacterial species are the so-called Tight ADherence (Tad) pili. These structures were first described in the pathogenic Gram-negative Actinobacillus actinomycetemcomitans, where these pili were shown to mediate adhesion to the host cell surface, supporting colonization and pathogenesis through the formation of biofilms (73, 74). The gene cluster responsible for the biogenesis of Tad pili encodes an ATPase, two transmembrane proteins and a septum site-determining protein that together constitute the pilus production, secretion, and positioning apparatus in addition to a peptidase able to post-translationally cleave the hydrophobic leader peptide of prepilins and pseudopilins, the precursors of Tad pili structural proteins (74). The Tad pilus-encoding gene cluster is highly conserved in bifidobacterial genomes (75), although this pilus type has only been characterized in detail in B. breve UCC2003 (50, 68, 69). Transcriptomic and gene mutagenesis experiments revealed that B. breve UCC2003 tad locus are expressed only in vivo conditions emphasizing its fundamental role in colonization and persistent adhesion to the host intestinal cells (69, 74). Moreover, in a recent study, the capability of the B. breve UCC2003 Tad pili to promote in vivo colonic epithelial proliferation was observed 5 days post-administration of the strain to murine model (50). Further analyses, exploiting advanced functional genomic approaches demonstrated, both under in vivo and in vitro conditions, that the epithelial proliferation response is specifically provoked by the TadE protein. Therefore, bifidobacterial Tad pili may contribute to the maturation of epithelial cells of newborns, stimulating growth of their thin intestinal mucosa and contributing to host mucosal homeostasis (50).
Extracellular Polysaccharides
The envelope of a wide range of bacteria is surrounded by one or more glycan layers known as capsular polysaccharides (CPS) or exopolysaccharides (EPS). Based on their monosaccharide composition and biosynthesis mechanism mediated by glycosidic linkages, bacterial CPS/EPS (NB. since it is hard to distinguish these, we will refer to these polysaccharides as EPS for ease) can either be classified as homopolysaccharides, when composed by the repetition of a single type of monosaccharide, or as heteropolysaccharides when formed by two or more types of monosaccharide subunits (76). In silico analyses performed on bifidobacterial sequenced genomes showed the lack of a “consensus” functional-structural organization in the eps-encoding clusters, highlighting consistent inter/intra-species variability in terms of both length and number of genes (77, 78). The scientific community has in recent years increased its interest in EPS producers as these extracellular polymers have been reported to exert a crucial role in human health by promoting adhesion to the intestinal mucosa, as well as by modulating the intestinal microbiota composition, and conferring selective advantage to bacteria through protection to adverse conditions such as presence of bile salts or pH insults (51, 79). In addition, some of these microbial biopolymers are receiving renewed interest due to their involvement in promoting human health (51, 78, 79). In this regard, an in vitro experiment aimed at evaluating the level of pro- and anti-inflammatory cytokines stimulation when EPS purified from 10 bifidobacterial strains were co-cultivated with human peripheral blood mononuclear cells (PBMC) revealed that the differentiation of T cells toward T-helper(Th)1 (IL-12/IL-10), Th2 (IL-10/TNF-α), and Th17 (IL-1β/IL-12) effector cells is highly influenced by the physical-chemical features of the particular EPS used in the experiment. This finding suggests that a positive correlation exists between the composition, structure and size of a given EPS polymer and the corresponding elicited immune response (80). In support of this discovery, relevant results were obtained by a further in vitro study where PBMCs were co-incubated with the EPS purified from three isogenic B. animalis subsp. lactis strains, i.e., A1, A1dOx, and A1dOxR strains. The three EPS polymers differ in their molecular mass with the highest molecular weight held by the A1dOxR EPS. Analysis of cytokine profiles revealed that the A1dOxR extracellular polymer induces the lower release of both pro- and anti-inflammatory cytokine by PBMCs when compared to other bifidobacterial strains (53, 81). Similar results were obtained when the immunoregulatory effects elicited by two B. adolescentis strains (IF1-03 and IF1-11) were evaluated. In detail, the two B. adolescentis strains differ in the exopolysaccharide-mediated enterocyte adhesion/aggregation phenotypes, with B. adolescentis IF1-03 possessing the high-molecular-weight EPS. Specifically, in vitro co-culturing experiments of the two B. adolescentis strains with RAW264.7 macrophages or mice splenocytes showed that while strain IF1-03 stimulated production of anti-inflammatory cytokines such as IL-10, IL-6, and TGF-β, it also increased the proportion of T-regulatory (Treg) cells. Notably, B. adolescentis IF1-11 induced higher levels of pro-inflammatory cytokines influencing the CD4+ T cells differentiation into Th17 cells. This observation was further confirmed by in vivo administration of the two above mentioned strains individually to Dextran Sodium Sulfate (DSS)-induced colitis mice. In this case, B. adolescentis IF1-03 not only induced high levels of anti-inflammatory cytokines, but it was also shown to contribute to the reduction of the area of ulceration and thickening of the intestinal wall (54). In the same manner, a recent study demonstrated differential alleviative effects in DSS-induced colitis mice when they were individually treated with three different B. longum subsp. longum strains, i.e., HAN4-25, which is unable to produce EPS, C11A10B producing non-ropy EPS and the producing ropy-EPS strain YS108R, obtained as a spontaneous single nucleic acid mutant from C11A10B. Particularly, B. longum subsp. longum YS108R reduced the colitis-induced colonic injury, decreasing the myeloperoxidase activity, preserving the crypt structures from damage and moderating the inflammatory cell infiltrations. Moreover, the administration of YS108R correlated with a reduced IL-6 and IL-17A concentration in parallel with an increased accumulation of the anti-inflammatory cytokine IL-10 in the serum coupled with lower IL-6 colonic expression level (55). All together these finding corroborates the notion that EPS-mediated immune response is largely influenced by the physicochemical nature of these polymers (53, 54, 81).
Moreover, similar results were observed when splenocytes isolated from naïve mice were co-cultivated with the EPS-producing B. breve UCC2003 strain (EPS+) or the two EPS-deficient B. breve UCC2003 strains (EPS−) obtained by means of an insertion or deletion in the eps locus of this bifidobacterial strain. In this regard, splenocytes stimulated with EPS+ strain evoked lower expression of proinflammatory cytokines IFN-α, TNF-α, and IL-12 compared to the EPS−. This finding was further confirmed through analysis of intracellular cytokine expression in splenocytes isolated from naïve mice orally fed with EPS+ and EPS− strains, evidencing significantly altered cytokine profiles between the two experimental conditions. Specifically, a lower proinflammatory cytokine induction was observed in both adaptive and innate immune response cell populations in case of EPS+ administration. Altogether these findings suggest that the B. breve UCC2003 EPS layer plays a crucial role in the persistence of the strain in the host intestine, reducing the risk of immune clearance against this microbial strain (51). In addition, B. breve UCC2003 EPS was shown to exert beneficial effects by reducing apoptotic epithelial cell shedding, which is a condition that occurs in case of IBD patients, where it causes high levels of apoptotic extrusion of small intestinal epithelial cells (IECs) from villi (52). It was previously described that the genome of B. breve UCC2003 possesses two adjacent, oppositely orientated gene sets, i.e., esp1 and eps2. The expression of these two transcriptional units, involved in the production of two distinct EPS (EPS1 and EPS2), depends on the orientation of a single promoter. Therefore, only one of the two types of extracellular polymer can be produced at a given time (82). Based on these observations, B. breve UCC2003 EPS-positive, B. breve UCC2003 EPSdel (a deletion mutant unable to express neither EPS1 nor EPS2) and B. breve UCC2003Inv (able to express only EPS2) were administered to mice model, which were then treated with an intraperitoneal injection of lipopolysaccharides (LPS) from Escherichia coli 0111:B4 inducing cell shedding (52). Interestingly, when colonized with B. breve UCC2003del or B. breve UCC2003Inv, no significant protection against cell shedding was observed when compared to the control PBS (Phosphate Buffered Saline)-gavage mice, thus underscoring the strain variant specificity in exerting protective effects. Moreover, evaluation of transcriptional changes in murine small intestine samples from control (PBS) and colonized groups (B. breve UCC2003 EPS-positive and B. breve UCC2003del) revealed down-regulation of genes related to the apoptotic cascade in mice treated with B. breve UCC2003 EPS-positive bacteria. In this context, the EPS of B. breve UCC2003 appears to downregulate inflammatory and apoptotic responses, thus reducing cell shedding events (52). Another bifidobacterial strain exerting beneficial effects is represented by B. longum subsp. longum 35624. The analysis of its genome revealed the presence of a gene cluster (eps624) encoding the EPS biosynthetic machinery. Despite the fact that the genome comparative analysis showed that B. longum subsp. longum 35624 EPS-specifying cluster is in the same location in other B. longum subsp. longum genomes, a different genetic composition of the eps624 was observed when compared to other analyzed B. longum subsp. longum strains (83). Based on these findings, further studies were carried out to explore the role played by the EPS in influencing the host immune response. A first study used B. longum subsp. longum 35624 strain and its derivatives: the exopolysaccharide-negative mutant (EPSneg) with an insertion mutation in the eps cluster and the EPScomp where the EPS production was restored by genetic complementation of the EPSneg strain. Co-culture of human PBMCs or monocyte-derived dendritic cells (MDDCs) with one of these strains revealed that increased proinflammatory cytokine secretion is specifically related to the lack of EPS, with a marked increase of IL-17. In the same way, administration of these strains individually to mice of a T cell transfer colitis model underlined the ability of the two EPS-producing strains to prevent disease related symptoms, while EPSneg does not show any protection against the development of colitis, with increased recruitment of IL-17+ lymphocytes to the gut (56). Moreover, the intranasal administration of EPS-producing B. longum 35624 and the EPSneg strains to ovalbumin respiratory allergy model mice, resulted in enhanced recruitment of IL-17+ lymphocytes to the murine lung. In this context, a further study showed the capability of B. longum 35624 EPS to suppress the Th2 type immune response within the lungs of ovalbumin sensitized mice when they were intranasally treated with purified B. longum 35624 EPS (57). All together these observations emphasize that the EPS of B. longum 35624 plays an important role in reducing the proinflammatory response.
Serpins
Serpins (Serine protease inhibitors) represent a superfamily of proteins acting as eukaryotic-type serine protease inhibitors. These prokaryotic enzymes, synthetized by particular members of the intestinal community, are involved in the regulation of a wide range of protease-mediated processes (84, 85). Analyses of the bifidobacterial genome sequences revealed that the presence of serpin-like gene is not ubiquitous but restricted to B. breve, B. longum subsp. longum, B. longum subsp. infantis, B. longum subsp. suis, B. cuniculi, Bifidobacterium scardovii, and B. dentium species (86). Moreover, an in vitro study underlined that the activation of bifidobacterial serpin-encoding genes is driven by specific serine proteases. Transcriptional profiling experiments carried out on RNA extracted from B. breve 210B cells treated with different proteases highlighted that the highest up-regulation of the serpin-encoding locus is reached after exposure to papain (86). Conversely, a different bifidobacterial strain, B. longum NCC2705, synthetizes a serpin able to act as an efficient inhibitor of human neutrophil elastase (HNE) and, to a lesser extent, pancreatic elastase (87). Considering that the release of serine proteases may occur following intestinal inflammation caused by bacterial infection or intestinal tissue damage typical of inflammatory bowel diseases or ulcerative colitis, the production of serpins may elicit anti-inflammatory activity counteracting the negative effects of (high levels of) human serine proteases. Moreover, the self-produced serpins may assist bifidobacteria to protect themselves against host-derived proteases and survive in a competitive environment (86, 88). To further emphasize the anti-inflammatory role of these prokaryotic enzymes, a recent in vivo study demonstrated the efficacy of a specific B. longum NCC2705 serpin in the prevention of gluten-related immunopathology. Administration of the wild-type B. longum NCC2705 (srp+) to sensitized NOD/DQ8 mice with peptic-tryptic digestion of gliadin, which is one of the main gluten protein fractions, favored protection against the development of gliadin-induced immunopathology when compared with the administration of the knockout B. longum NCC2705 (srp−) that is unable to synthetize serpins. Because the aberrant effects of gluten-related immunopathology are strongly relieved in mice treated with the wild-type strain, it can be speculated that serpins are involved in immune regulation, maintenance of barrier function and inhibition of elastases released during inflammation (58).
Other Extracellular Molecules Involved in the Bifidobacterial-Host Immune System Interaction
In addition to pili, EPS and serpins, other bifidobacterial-associated extracellular proteins have been implicated in impacting on the host immune system. For instance, TagA is a protein expressed on the outer surface of B. bifidum MIMBb75 formed by two active domains: a lytic murein transglycosylase (LT) and a cysteine- and histidine-dependent amidohydrolase/peptidase (CHAP). This protein acts as a peptidoglycan lytic enzyme that when co-cultured with human DCs was shown to cause activation/proliferation of the DCs itself and the induction of IL-2 (60). B. bifidum MIMBb75 also produces the surface-associated protein named BopA, which has been reported to not only enhance adhesion of B. bifidum MIMBb75 to Caco-2 cell lines, but also to stimulate production of IL-8 by Caco-2 cells (61). However, subsequently the role of BopA in mediating adherence of B. bifidum to epithelial cells has been re-evaluated. In this setting, the gene encoding BopA was cloned and expressed in E. coli without the BopA-associated lipobox and hydrophobic signal peptide, resulting in a reduced capability of BopA to adhere to Caco-2 and HT-29 cells. Previous results may therefore have been compromised by an unspecific effect caused by higher hydrophobicity of the BopA (lipo)proteins (89).
Bifidobacterial Metabolism Influences Intestinal Immune Homeostasis and Inflammatory Response Through a Microbe-Microbe Cross-Feeding Activities
The ability to degrade complex carbohydrates that cannot be metabolized by the host is one of the key genetic features of bifidobacteria (90, 91). The nondigestible carbohydrates degraded by these commensal microorganisms include diet-derived sugars (e.g., glucans, xylans, pectins, fructans, cellulose, resistant starch) as well as host-derived glycans, encompassing Human Milk Oligosacchardes (HMO) and mucins, which are glycoproteins covering the gut epithelium and secreted by the goblet cells of the GIT, in the form of O-linked and/or N-linked glycans (90). The genetic arsenal required for the utilization of the aforementioned compounds is generally organized in gene clusters including glycosyl hydrolases (GH)-encoding genes coupled with gene sequences responsible for the assembly of sugar-specific ATP-binding cassette (ABC) transporters, permeases, proton symporters, and phosphoenolpyruvate-phosphotransferase systems (72, 90). Specifically, the degradation of complex carbohydrates requires dedicated extracellular and/or intracellular GHs to generate mono- and/or oligo-saccharides, which are then internalized by the transport systems. After reaching the cytoplasm, those carbohydrates that still need to be processed may be subject to further modification such as deacetylation, deamination, epimerization, and phosphorylation to produce phosphorylated monosaccharides before entering the final central metabolic route, known as the “bifid shunt,” where simple and complex sugars eventually converge for energy production (92). At the end of this specific pathway, 2.5 ATP molecules per Mol of glucose are theoretically obtained together with 1 Mol of lactate and 1.5 Mol of acetate. Particular interest has focused on the production of Short Chain Fatty Acids (SCFAs) by the intestinal microbiota as they contribute to the host health through increasing calcium and magnesium absorption, regulating bowel functions, providing nutritional support to colonocytes, decreasing the luminal pH and thus preventing overgrowth of pH-sensitive pathogenic bacteria and finally influencing the host immune system (93, 94). Structurally defined as saturated aliphatic organic acids, major SCFAs at the site of intestine are represented by acetate, propionate and butyrate. Among the SCFAs produced in the human colon, butyrate displays a key role in the gut health stimulating the development of the intestinal cells. Indeed, being the preferred energy source for intestinal epithelial cells, butyrate consumption improves host IEC integrity promoting tight junctions, cell proliferation, and mucin production by Goblet cells (93, 95). Moreover, butyrate is involved in guiding the immune system toward an anti-inflammatory response (93, 96, 97). For instance, butyrate exhibits anti-inflammatory properties inducing the production of TGF-β, IL-18, and IL-10 cytokines by both antigen presenting cells and IECs coupled with the stimulation of the differentiation of naïve T cells to Treg cells (98, 99) (Figure 1B).
Although bifidobacteria are unable of directly synthetizing butyrate, as acetate-producing microorganisms, they may indirectly influence the activity and composition of the butyrogenic members of the gut microbiota and the immune response through a mutual beneficial cross-feeding interaction. Indeed, bifidobacteria can operate as primary players degrading complex carbohydrates to produce acetate which is then consumed by secondary degraders that generate butyrate, thus stimulating a butyrogenic effect. Most of butyrate producers in the human colon belong to the Firmicutes phylum, specifically to the clostridial clusters IV and XIVa encompassing Faecalibacterium prausnitzii, Eubacterium rectale, Eubacterium hallii, Roseburia spp. (Roseburia hominis, Roseburia intestinalis, and Roseburia inulinivorans) and Anaerostipes spp. (Anaerostipes caccae, Anaerostipes hadrus, and Anaerostipes butyraticus) (92, 100). In this context, several studies have reported on the butyrogenic effects resulting from the degradation of diet-associated carbohydrates by bifidobacteria (Table 1). Particularly, the co-culture of B. longum subsp. longum NCC2705 and E. rectale ATCC 33656 in a growth medium with arabinoxylan oligosaccharides (AXOS) as unique carbon source affected the growth yield of both strains when co-cultivated compared to the respective mono-cultures, leading to a mutual beneficial effect. Notably, B. longum NCC2705 as an arabinose substituent degrader of AXOS, possesses the AXOS-degrading enzymes whose action allow the release of xylose backbone (XOS) and acetate. On the other side, E. rectale ATCC 33656 consumes the whole (A)XOS substrate releasing xylose and arabinose. In this way, E. rectale ATCC 33656 takes advantage of acetate for butyrate production, resulting in a butyrogenic effect, while B. longum NCC 2705 consumes (A)XOS monosaccharides, leading to an increase in acetate production and ensuring its cell concentration thus stimulating a bifidogenic effect (59). Similar findings were obtained by the co-cultivation of F. prausnitzii S3L/3 and B. adolescentis L2-32 in the presence of fructooligosaccharides (FOS 95), or the co-cultivation of F. prausnitzii A2-165 and B. adolescentis L2-32 on starch, resulting in both cases in an enhanced production of butyrate when compared to the mono-cultures (62).
As mentioned above, bifidobacteria can produce acetate not only through the consumption of diet-derived carbohydrates, but also by degrading host-derived glycans. In this regard, it has been demonstrated that a cross-feeding interaction is established when B. bifidum strains are singularly co-cultured with E. hallii DSM 3353 in presence of mucin as the only carbon source (63). Gut colonization by E. hallii generally occurs in the first months after birth reaching adult abundance levels between 5 and 10 years of age (64). This microorganism is able to consume acetate and lactate to produce butyrate and propionate, thus promoting a beneficial effect on gut health during childhood. However, E. hallii does not possess genes involved in the degradation of complex host- or diet-derived glycans, relying on other microbial players for the supply of simpler carbohydrates (101). Conversely, B. bifidum is one of the prominent bifidobacterial species able to utilize mucin as carbon source thanks to the presence in their genome of genes encoding for α-fucosidases, α-sialidases, endo-α-N-acetylgalactosaminidases, and N-acetyl-β-hexosaminidases (102). The ability of B. bifidum to degrade mucin into mucin-associated mono- and oligo-saccharides coupled with the production of acetate and lactate stimulate E. hallii colonization and consumption of simple glycans released by B. bifidum as energy source and the acetate and lactate for butyrate production (63). Moreover, three-strain fermentations involving B. bifidum and E. hallii with B. breve or B. longum subsp. infantis on mucin as the sole carbon source showed the formation of propionate in both three-strain co-cultures by E. hallii, emphasizing the health-promoting effect of cross-feeding interactions as propionate acts as a precursor for gluconeogenesis in the liver and affects intestinal homeostasis with anti-inflammatory and anti-carcinogenic actions (63, 103). A similar trophic interaction was observed when the same strain, E. hallii DSM 3353, was co-cultured with B. longum subsp. infantis strains in presence of fucosyllactose (FL; either 2′- or 3-FL), which is a component of the HMO. E. hallii does not grow on HMOs as the unique carbon source. In contrast, B. longum subsp. infantis utilizes the lactose moiety of FL to produce lactate and acetate and metabolize L-fucose to 1,2-propanediol (1,2-PD). These by-products are then used by E. hallii for its growth and the formation of butyrate coupled with propionate and formate (64).
Overall, these observations strengthen the notion that bifidobacterial abilities to degrade non-digestible diet- and/or host-derived glycans coupled with subsequent production of acetate (or 1,2-PD) have an indirect role in the modulation of the host immune system, stimulating the growth and production of butyric acid by butyrate-producing intestinal microorganisms.
Conclusions
In recent decades, a growing number of studies have demonstrated that changes in the intestinal microbial composition and gut homeostasis is directly linked to intestinal disorders resulting, in the most severe cases, in IBD and colorectal cancer. The modulation of the gut microbiota through prebiotics and/or probiotics is one of the ways to counteract these dysbiosis, improving human health. In this context, bifidobacteria are generally considered as potent probiotics for their health-promoting features. Indeed, they trigger immunomodulatory responses aimed at maintaining the host intestinal homeostasis through different mechanisms such as production of extracellular structures that can interact with other intestinal microorganisms and/or the host cells coupled with the release of by-products of their metabolism that may be utilized by other commensal bacteria, such as the butyrate-producing microorganisms, to establish a cross-feeding interaction. However, despite their key role in stimulating human health and development in technology, the precise mechanism by which bifidobacteria solicit an immune response is far from being fully understood. Indeed, most studies intended to correlate bifidobacteria and their immune modulatory effects have been carried out by exploiting cell lines and/or animal models, thus preventing a comprehensive understanding of the impact that bifidobacteria may exert on the human immune system. Indeed, because of the complexity of the gut microbiota, in case of in vivo studies it is possible that the presence of other bacterial strains may influence or even reverse the bifidobacterial immunomodulatory effects. Therefore, the challenge for future studies is to deepen the knowledge about this field, overcoming the abovementioned limits.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was funded by the EU Joint Programming Initiative—A Healthy Diet for a Healthy Life (JPI HDHL, http://www.healthydietforhealthylife.eu/) to DS (in conjunction with Science Foundation Ireland [SFI], Grant number 15/JP-HDHL/3280) and to MV (in conjunction with MIUR, Italy). This study was supported by Fondazione Cariparma, under TeachInParma Project. DS is a member of The APC Microbiome Institute funded by Science Foundation Ireland (SFI), through the Irish Government's National Development Plan (Grant number SFI/12/RC/2273-P1 and SFI/12/RC/2273-P2).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. (2007) 5:e177. doi: 10.1371/journal.pbio.0050177
2. Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. (2005) 307:1915–20. doi: 10.1126/science.1104816
3. Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. (2016) 14:e1002533. doi: 10.1371/journal.pbio.1002533
4. Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell. (2016) 164:337–40. doi: 10.1016/j.cell.2016.01.013
5. Takiishi T, Fenero CIM, Camara NOS. Intestinal barrier and gut microbiota: shaping our immune responses throughout life. Tissue Barriers. (2017) 5:e1373208 doi: 10.1080/21688370.2017.1373208
6. Pickard JM, Zeng MY, Caruso R, Nunez G. Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev. (2017) 279:70–89. doi: 10.1111/imr.12567
7. Moens E, Veldhoen M. Epithelial barrier biology: good fences make good neighbours. Immunology. (2012) 135:1–8. doi: 10.1111/j.1365-2567.2011.03506.x
8. Vancamelbeke M, Vermeire S. The intestinal barrier: a fundamental role in health and disease. Expert Rev Gastroenterol Hepatol. (2017) 11:821–34. doi: 10.1080/17474124.2017.1343143
9. Nalle SC, Turner JR. Intestinal barrier loss as a critical pathogenic link between inflammatory bowel disease and graft-versus-host disease. Mucosal Immunol. (2015) 8:720–30. doi: 10.1038/mi.2015.40
10. Ruiz L, Delgado S, Ruas-Madiedo P, Sanchez B, Margolles A. Bifidobacteria and their molecular communication with the immune system. Front Microbiol. (2017) 8:2345 doi: 10.3389/fmicb.2017.02345
11. Hidalgo-Cantabrana C, Delgado S, Ruiz L, Ruas-Madiedo P, Sanchez B, Margolles A. Bifidobacteria and their health-promoting effects. Microbiol Spectr. (2017) 5:BAD-0010-2016. doi: 10.1128/microbiolspec.BAD-0010-2016
12. Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, et al. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev. (2007) 71:495–548. doi: 10.1128/MMBR.00005-07
13. Tissier H. Recherches sur la flore intestinale des nourrissons: état normal et pathologique (Thesis/dissertation). Faculté de Médecine, Paris, France (1900).
14. Alberoni D, Gaggia F, Baffoni L, Modesto MM, Biavati B, Di Gioia D. Bifidobacterium xylocopae sp. nov. and Bifidobacterium aemilianum sp. nov., from the carpenter bee (Xylocopa violacea) digestive tract. Syst Appl Microbiol. (2019) 42:205–16. doi: 10.1016/j.syapm.2018.11.005
15. Modesto M, Watanabe K, Arita M, Satti M, Oki K, Sciavilla P, et al. Bifidobacterium jacchi sp. nov., isolated from the faeces of a baby common marmoset (Callithrix jacchus). Int J Syst Evol Microbiol. (2019) 69:2477–85. doi: 10.1099/ijsem.0.003518
16. Duranti S, Lugli GA, Napoli S, Anzalone R, Milani C, Mancabelli L, et al. Characterization of the phylogenetic diversity of five novel species belonging to the genus Bifidobacterium: Bifidobacterium castoris sp. nov., Bifidobacterium callimiconis sp. nov., Bifidobacterium goeldii sp. nov., Bifidobacterium samirii sp. nov. and Bifidobacterium dolichotidis sp. nov. Int J Syst Evol Microbiol. (2019) 69:1288–98. doi: 10.1099/ijsem.0.003306
17. Lugli GA, Mangifesta M, Duranti S, Anzalone R, Milani C, Mancabelli L, et al. Phylogenetic classification of six novel species belonging to the genus Bifidobacterium comprising Bifidobacterium anseris sp. nov., Bifidobacterium criceti sp. nov., Bifidobacterium imperatoris sp. nov., Bifidobacterium italicum sp. nov., Bifidobacterium margollesii sp. nov. and Bifidobacterium parmae sp. nov. Syst Appl Microbiol. (2018) 41:173–83. doi: 10.1016/j.syapm.2018.01.002
18. Lugli GA, Milani C, Duranti S, Alessandri G, Turroni F, Mancabelli L, et al. Isolation of novel gut bifidobacteria using a combination of metagenomic and cultivation approaches. Genome Biol. (2019) 20:96. doi: 10.1186/s13059-019-1711-6
19. Biavati B, Scardovi V, Moore WEC. Electrophoretic patterns of proteins in the genus bifidobacterium and proposal of four new species. Int J Syst Evol Microbiol. (1982) 32:358–73. doi: 10.1099/00207713-32-3-358
20. Hoyles L, Inganas E, Falsen E, Drancourt M, Weiss N, McCartney AL, et al. Bifidobacterium scardovii sp. nov., from human sources. Int J Syst Evol Microbiol. (2002) 52(Pt 3):995–9. doi: 10.1099/00207713-52-3-995
21. Okamoto M, Benno Y, Leung KP, Maeda N. Bifidobacterium tsurumiense sp. nov., from hamster dental plaque. Int J Syst Evol Microbiol. (2008) 58(Pt 1):144–8. doi: 10.1099/ijs.0.65296-0
22. Trovatelli LD, Crociani F, Pedinotti M, Scardovi V. Bifidobacterium pullorum sp. nov.: a new species isolated from chicken feces and a related group of bifidobacteria isolated from rabbit feces. Arch Microbiol. (1974) 98:187–98. doi: 10.1007/BF00425281
23. Turroni F, van Sinderen D, Ventura M. Genomics and ecological overview of the genus Bifidobacterium. Int J Food Microbiol. (2011) 149:37–44. doi: 10.1016/j.ijfoodmicro.2010.12.010
24. Milani C, Mangifesta M, Mancabelli L, Lugli GA, James K, Duranti S, et al. Unveiling bifidobacterial biogeography across the mammalian branch of the tree of life. ISME J. (2017) 11:2834–47. doi: 10.1038/ismej.2017.138
25. Milani C, Lugli GA, Duranti S, Turroni F, Mancabelli L, Ferrario C, et al. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci Rep. (2015) 5:15782. doi: 10.1038/srep15782
26. Lugli GA, Mancino W, Milani C, Duranti S, Turroni F, van Sinderen D, et al. Reconstruction of the Bifidobacterial Pan-Secretome Reveals the Network of Extracellular Interactions between Bifidobacteria and the Infant Gut. Appl Environ Microbiol. (2018) 84:e00796–18. doi: 10.1128/AEM.00796-18
27. Duranti S, Lugli GA, Milani C, James K, Mancabelli L, Turroni F, et al. Bifidobacterium bifidum and the infant gut microbiota: an intriguing case of microbe-host co-evolution. Environ Microbiol. (2019). doi: 10.1111/1462-2920.14705. [Epub ahead of print].
28. Kuperman AA, Zimmerman A, Hamadia S, Ziv O, Gurevich V, Fichtman B, et al. Deep microbial analysis of multiple placentas shows no evidence for a placental microbiome. BJOG. (2019). doi: 10.1111/1471-0528.15896. [Epub ahead of print].
29. Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, et al. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev. (2017) 81:e00036–17. doi: 10.1128/MMBR.00036-17
30. Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep. (2016) 6:23129. doi: 10.1038/srep23129
31. Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med. (2014) 6:237ra65. doi: 10.1126/scitranslmed.3008599
32. Matamoros S, Gras-Leguen C, Le Vacon F, Potel G, de La Cochetiere MF. Development of intestinal microbiota in infants and its impact on health. Trends Microbiol. (2013) 21:167–73. doi: 10.1016/j.tim.2012.12.001
33. Turroni F, Milani C, Duranti S, Ferrario C, Lugli GA, Mancabelli L, et al. Bifidobacteria and the infant gut: an example of co-evolution and natural selection. Cell Mol Life Sci. (2018) 75:103–18. doi: 10.1007/s00018-017-2672-0
34. Duranti S, Lugli GA, Mancabelli L, Armanini F, Turroni F, James K, et al. Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome. (2017) 5:66. doi: 10.1186/s40168-017-0282-6
35. Milani C, Mancabelli L, Lugli GA, Duranti S, Turroni F, Ferrario C, et al. Exploring vertical transmission of bifidobacteria from mother to child. Appl Environ Microbiol. (2015) 81:7078–87. doi: 10.1128/AEM.02037-15
36. Imoto N, Morita H, Amanuma F, Maruyama H, Watanabe S, Hashiguchi N. Maternal antimicrobial use at delivery has a stronger impact than mode of delivery on bifidobacterial colonization in infants: a pilot study. J Perinatol. (2018) 38:1174–81. doi: 10.1038/s41372-018-0172-1
37. Forsgren M, Isolauri E, Salminen S, Rautava S. Late preterm birth has direct and indirect effects on infant gut microbiota development during the first six months of life. Acta Paediatr. (2017) 106:1103–9. doi: 10.1111/apa.13837
38. Sirilun S, Takahashi H, Boonyaritichaikij S, Chaiyasut C, Lertruangpanya P, Koga Y, et al. Impact of maternal bifidobacteria and the mode of delivery on Bifidobacterium microbiota in infants. Benef Microbes. (2015) 6:767–74. doi: 10.3920/BM2014.0124
39. Del Chierico F, Vernocchi P, Petrucca A, Paci P, Fuentes S, Pratico G, et al. Phylogenetic and metabolic tracking of gut microbiota during perinatal development. PLoS ONE. (2015) 10:e0137347. doi: 10.1371/journal.pone.0137347
40. O'Sullivan A, Farver M, Smilowitz JT. The influence of early infant-feeding practices on the intestinal microbiome and body composition in infants. Nutr Metab Insights. (2015) 8(Suppl. 1):1–9. doi: 10.4137/NMI.S41125
41. Mueller NT, Bakacs E, Combellick J, Grigoryan Z, Dominguez-Bello MG. The infant microbiome development: mom matters. Trends Mol Med. (2015) 21:109–17. doi: 10.1016/j.molmed.2014.12.002
42. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. (2012) 486:222–7. doi: 10.1038/nature11053
43. Arboleya S, Watkins C, Stanton C, Ross RP. Gut bifidobacteria populations in human health and aging. Front Microbiol. (2016) 7:1204. doi: 10.3389/fmicb.2016.01204
44. Salazar N, Valdes-Varela L, Gonzalez S, Gueimonde M, de Los Reyes-Gavilan CG. Nutrition and the gut microbiome in the elderly. Gut Microbes. (2017) 8:82–97. doi: 10.1080/19490976.2016.1256525
45. Matsuki T, Watanabe K, Fujimoto J, Kado Y, Takada T, Matsumoto K, et al. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl Environ Microbiol. (2004) 70:167–73. doi: 10.1128/AEM.70.1.167-173.2004
46. Turroni F, Foroni E, Pizzetti P, Giubellini V, Ribbera A, Merusi P, et al. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl Environ Microbiol. (2009) 75:1534–45. doi: 10.1128/AEM.02216-08
47. Odamaki T, Bottacini F, Kato K, Mitsuyama E, Yoshida K, Horigome A, et al. Genomic diversity and distribution of Bifidobacterium longum subsp. longum across the human lifespan. Sci Rep. (2018) 8:85. doi: 10.1038/s41598-017-18391-x
48. Turroni F, Serafini F, Foroni E, Duranti S, O'Connell Motherway M, Taverniti V, et al. Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions. Proc Natl Acad Sci USA. (2013) 110:11151–6. doi: 10.1073/pnas.1303897110
49. Duranti S, Gaiani F, Mancabelli L, Milani C, Grandi A, Bolchi A, et al. Elucidating the gut microbiome of ulcerative colitis: bifidobacteria as novel microbial biomarkers. FEMS Microbiol Ecol. (2016) 92:fiw191. doi: 10.1093/femsec/fiw191
50. O'Connell Motherway M, Houston A, O'Callaghan G, Reunanen J, O'Brien F, O'Driscoll T, et al. A Bifidobacterial pilus-associated protein promotes colonic epithelial proliferation. Mol Microbiol. (2019) 111:287–301. doi: 10.1111/mmi.14155
51. Fanning S, Hall LJ, Cronin M, Zomer A, MacSharry J, Goulding D, et al. Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc Natl Acad Sci USA. (2012) 109:2108–13. doi: 10.1073/pnas.1115621109
52. Hughes KR, Harnisch LC, Alcon-Giner C, Mitra S, Wright CJ, Ketskemety J, et al. Bifidobacterium breve reduces apoptotic epithelial cell shedding in an exopolysaccharide and MyD88-dependent manner. Open Biol. (2017) 7:160155. doi: 10.1098/rsob.160155
53. Hidalgo-Cantabrana C, Nikolic M, Lopez P, Suarez A, Miljkovic M, Kojic M, et al. Exopolysaccharide-producing Bifidobacterium animalis subsp. lactis strains and their polymers elicit different responses on immune cells from blood and gut associated lymphoid tissue. Anaerobe. (2014) 26:24–30. doi: 10.1016/j.anaerobe.2014.01.003
54. Yu R, Zuo F, Ma H, Chen S. Exopolysaccharide-producing Bifidobacterium adolescentis strains with similar adhesion property induce differential regulation of inflammatory immune response in Treg/Th17 Axis of DSS-colitis mice. Nutrients. (2019) 11:E782. doi: 10.3390/nu11040782
55. Yan S, Yang B, Zhao J, Zhao J, Stanton C, Ross RP, et al. A ropy exopolysaccharide producing strain Bifidobacterium longum subsp. longum YS108R alleviates DSS-induced colitis by maintenance of the mucosal barrier and gut microbiota modulation. Food Funct. (2019) 10:1595–608. doi: 10.1039/C9FO00014C
56. Schiavi E, Gleinser M, Molloy E, Groeger D, Frei R, Ferstl R, et al. The surface-associated exopolysaccharide of Bifidobacterium longum 35624 plays an essential role in dampening host proinflammatory responses and repressing local TH17 responses. Appl Environ Microbiol. (2016) 82:7185–96. doi: 10.1128/AEM.02238-16
57. Schiavi E, Plattner S, Rodriguez-Perez N, Barcik W, Frei R, Ferstl R, et al. Exopolysaccharide from Bifidobacterium longum subsp. longum 35624 modulates murine allergic airway responses. Benef Microbes. (2018) 9:761–73. doi: 10.3920/BM2017.0180
58. McCarville JL, Dong J, Caminero A, Bermudez-Brito M, Jury J, Murray JA, et al. A commensal Bifidobacterium longum strain prevents gluten-related immunopathology in mice through expression of a serine protease inhibitor. Appl Environ Microbiol. (2017) 83:e01323–17. doi: 10.1128/AEM.01323-17
59. Riviere A, Gagnon M, Weckx S, Roy D, De Vuyst L. Mutual cross-feeding interactions between Bifidobacterium longum subsp. longum NCC2705 and Eubacterium rectale ATCC 33656 explain the bifidogenic and butyrogenic effects of arabinoxylan oligosaccharides. Appl Environ Microbiol. (2015) 81:7767–81. doi: 10.1128/AEM.02089-15
60. Guglielmetti S, Zanoni I, Balzaretti S, Miriani M, Taverniti V, De Noni I, et al. Murein lytic enzyme TgaA of Bifidobacterium bifidum MIMBb75 modulates dendritic cell maturation through its cysteine- and histidine-dependent amidohydrolase/peptidase (CHAP) amidase domain. Appl Environ Microbiol. (2014) 80:5170–7. doi: 10.1128/AEM.00761-14
61. Guglielmetti S, Tamagnini I, Mora D, Minuzzo M, Scarafoni A, Arioli S, et al. Implication of an outer surface lipoprotein in adhesion of Bifidobacterium bifidum to Caco-2 cells. Appl Environ Microbiol. (2008) 74:4695–702. doi: 10.1128/AEM.00124-08
62. Rios-Covian D, Gueimonde M, Duncan SH, Flint HJ, de los Reyes-Gavilan CG. Enhanced butyrate formation by cross-feeding between Faecalibacterium prausnitzii and Bifidobacterium adolescentis. FEMS Microbiol Lett. (2015) 362:fnv176. doi: 10.1093/femsle/fnv176
63. Bunesova V, Lacroix C, Schwab C. Mucin cross-feeding of infant bifidobacteria and Eubacterium hallii. Microb Ecol. (2018) 75:228–38. doi: 10.1007/s00248-017-1037-4
64. Schwab C, Ruscheweyh HJ, Bunesova V, Pham VT, Beerenwinkel N, Lacroix C. Trophic interactions of infant bifidobacteria and Eubacterium hallii during L-fucose and fucosyllactose degradation. Front Microbiol. (2017) 8:95. doi: 10.3389/fmicb.2017.00095
65. Scott JR, Zahner D. Pili with strong attachments: gram-positive bacteria do it differently. Mol Microbiol. (2006) 62:320–30. doi: 10.1111/j.1365-2958.2006.05279.x
66. Foroni E, Serafini F, Amidani D, Turroni F, He F, Bottacini F, et al. Genetic analysis and morphological identification of pilus-like structures in members of the genus Bifidobacterium. Microb Cell Fact. (2011) 10(Suppl. 1.):S16. doi: 10.1186/1475-2859-10-S1-S16
67. Kline KA, Dodson KW, Caparon MG, Hultgren SJ. A tale of two pili: assembly and function of pili in bacteria. Trends Microbiol. (2010) 18:224–32. doi: 10.1016/j.tim.2010.03.002
68. Milani C, Mangifesta M, Mancabelli L, Lugli GA, Mancino W, Viappiani A, et al. The sortase-dependent fimbriome of the genus Bifidobacterium: extracellular structures with potential to modulate microbe-host dialogue. Appl Environ Microbiol. (2017) 83:e01295–17. doi: 10.1128/AEM.01295-17
69. O'Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, et al. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci USA. (2011) 108:11217–22. doi: 10.1073/pnas.1105380108
70. Turroni F, Serafini F, Mangifesta M, Arioli S, Mora D, van Sinderen D, et al. Expression of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in response to environmental gut conditions. FEMS Microbiol Lett. (2014) 357:23–33. doi: 10.1111/1574-6968.12509
71. Spirig T, Weiner EM, Clubb RT. Sortase enzymes in Gram-positive bacteria. Mol Microbiol. (2011) 82:1044–59. doi: 10.1111/j.1365-2958.2011.07887.x
72. Bottacini F, van Sinderen D, Ventura M. Omics of bifidobacteria: research and insights into their health-promoting activities. Biochem J. (2017) 474:4137–52. doi: 10.1042/BCJ20160756
73. Kachlany SC, Planet PJ, DeSalle R, Fine DH, Figurski DH. Genes for tight adherence of Actinobacillus actinomycetemcomitans: from plaque to plague to pond scum. Trends Microbiol. (2001) 9:429–37. doi: 10.1016/S0966-842X(01)02161-8
74. Tomich M, Planet PJ, Figurski DH. The tad locus: postcards from the widespread colonization island. Nat Rev Microbiol. (2007) 5:363–75. doi: 10.1038/nrmicro1636
75. Ventura M, Turroni F, Motherway MO, MacSharry J, van Sinderen D. Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol. (2012) 20:467–76. doi: 10.1016/j.tim.2012.07.002
76. Ruas M, Salazar N, de los reyes-Gavilan CG. Biosynthesis and chemical composition of exopolysaccharides produced by lactic acid bacteria. In: Ullrich M, editor. Bacterial Polysaccharides: Current Innovations and Future Trends. Norfolk: Caister Academic Press (2009).
77. Hidalgo-Cantabrana C, Sanchez B, Milani C, Ventura M, Margolles A, Ruas-Madiedo P. Genomic overview and biological functions of exopolysaccharide biosynthesis in Bifidobacterium spp. Appl Environ Microbiol. (2014) 80:9–18. doi: 10.1128/AEM.02977-13
78. Ferrario C, Milani C, Mancabelli L, Lugli GA, Duranti S, Mangifesta M, et al. Modulation of the eps-ome transcription of bifidobacteria through simulation of human intestinal environment. FEMS Microbiol Ecol. (2016) 92:fiw056. doi: 10.1093/femsec/fiw056
79. Ruas-Madiedo P, Gueimonde M, Arigoni F, de los Reyes-Gavilan CG, Margolles A. Bile affects the synthesis of exopolysaccharides by Bifidobacterium animalis. Appl Environ Microbiol. (2009) 75:1204–7. doi: 10.1128/AEM.00908-08
80. Lopez P, Monteserin DC, Gueimonde M, de los Reyes-Gavilan CG, Margolles A, Suarez A, et al. Exopolysaccharide-producing Bifidobacterium strains elicit different in vitro responses upon interaction with human cells. Food Res Int. (2012) 46:99–107. doi: 10.1016/j.foodres.2011.11.020
81. Hidalgo-Cantabrana C, Sanchez B, Moine D, Berger B, de Los Reyes-Gavilan CG, Gueimonde M, et al. Insights into the ropy phenotype of the exopolysaccharide-producing strain Bifidobacterium animalis subsp. lactis A1dOxR. Appl Environ Microbiol. (2013) 79:3870–4. doi: 10.1128/AEM.00633-13
82. Fanning S, Hall LJ, van Sinderen D. Bifidobacterium breve UCC2003 surface exopolysaccharide production is a beneficial trait mediating commensal-host interaction through immune modulation and pathogen protection. Gut Microbes. (2012) 3:420–5. doi: 10.4161/gmic.20630
83. Altmann F, Kosma P, O'Callaghan A, Leahy S, Bottacini F, Molloy E, et al. Genome analysis and characterisation of the exopolysaccharide produced by Bifidobacterium longum subsp. longum 35624. PLoS ONE. (2016) 11:e0162983. doi: 10.1371/journal.pone.0162983
84. Gettins PG. Serpin structure, mechanism, and function. Chem Rev. (2002) 102:4751–804. doi: 10.1021/cr010170+
85. Potempa J, Korzus E, Travis J. The serpin superfamily of proteinase inhibitors: structure, function, and regulation. J Biol Chem. (1994) 269:15957–60.
86. Turroni F, Foroni E, O'Connell Motherway M, Bottacini F, Giubellini V, Zomer A, et al. Characterization of the serpin-encoding gene of Bifidobacterium breve 210B. Appl Environ Microbiol. (2010) 76:3206–19. doi: 10.1128/AEM.02938-09
87. Ivanov D, Emonet C, Foata F, Affolter M, Delley M, Fisseha M, et al. A serpin from the gut bacterium Bifidobacterium longum inhibits eukaryotic elastase-like serine proteases. J Biol Chem. (2006) 281:17246–52. doi: 10.1074/jbc.M601678200
88. Alvarez-Martin P, O'Connell Motherway M, Turroni F, Foroni E, Ventura M, van Sinderen D. A two-component regulatory system controls autoregulated serpin expression in Bifidobacterium breve UCC2003. Appl Environ Microbiol. (2012) 78:7032–41. doi: 10.1128/AEM.01776-12
89. Kainulainen V, Reunanen J, Hiippala K, Guglielmetti S, Vesterlund S, Palva A, et al. BopA does not have a major role in the adhesion of Bifidobacterium bifidum to intestinal epithelial cells, extracellular matrix proteins, and mucus. Appl Environ Microbiol. (2013) 79:6989–97. doi: 10.1128/AEM.01993-13
90. Pokusaeva K, Fitzgerald GF, van Sinderen D. Carbohydrate metabolism in Bifidobacteria. Genes Nutr. (2011) 6:285–306. doi: 10.1007/s12263-010-0206-6
91. O'Callaghan A, van Sinderen D. Bifidobacteria and their role as members of the human gut microbiota. Front Microbiol. (2016) 7:925. doi: 10.3389/fmicb.2016.00925
92. De Vuyst L, Moens F, Selak M, Riviere A, Leroy F. Summer meeting 2013: growth and physiology of bifidobacteria. J Appl Microbiol. (2014) 116:477–91. doi: 10.1111/jam.12415
93. den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. (2013) 54:2325–40. doi: 10.1194/jlr.R036012
94. Ratajczak W, Ryl A, Mizerski A, Walczakiewicz K, Sipak O, Laszczynska M. Immunomodulatory potential of gut microbiome-derived short-chain fatty acids (SCFAs). Acta Biochim Pol. (2019) 66:1–12. doi: 10.18388/abp.2018_2648
95. Rios-Covian D, Ruas-Madiedo P, Margolles A, Gueimonde M, de Los Reyes-Gavilan CG, Salazar N. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol. (2016) 7:185. doi: 10.3389/fmicb.2016.00185
96. Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. (2016) 7:189–200. doi: 10.1080/19490976.2015.1134082
97. Oliphant K, Allen-Vercoe E. Macronutrient metabolism by the human gut microbiome: major fermentation by-products and their impact on host health. Microbiome. (2019) 7:91. doi: 10.1186/s40168-019-0704-8
98. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. (2013) 504:446–50. doi: 10.1038/nature12721
99. Zhang M, Zhou Q, Dorfman RG, Huang X, Fan T, Zhang H, et al. Butyrate inhibits interleukin-17 and generates Tregs to ameliorate colorectal colitis in rats. BMC Gastroenterol. (2016) 16:84. doi: 10.1186/s12876-016-0500-x
100. Riviere A, Selak M, Lantin D, Leroy F, De Vuyst L. Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front Microbiol. (2016) 7:979. doi: 10.3389/fmicb.2016.00979
101. Scott KP, Martin JC, Duncan SH, Flint HJ. Prebiotic stimulation of human colonic butyrate-producing bacteria and bifidobacteria, in vitro. FEMS Microbiol Ecol. (2014) 87:30–40. doi: 10.1111/1574-6941.12186
102. Turroni F, Bottacini F, Foroni E, Mulder I, Kim JH, Zomer A, et al. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc Natl Acad Sci USA. (2010) 107:19514–9. doi: 10.1073/pnas.1011100107
Keywords: bifidobacteria, immune system, immunomodulation, host interaction, probiotics
Citation: Alessandri G, Ossiprandi MC, MacSharry J, van Sinderen D and Ventura M (2019) Bifidobacterial Dialogue With Its Human Host and Consequent Modulation of the Immune System. Front. Immunol. 10:2348. doi: 10.3389/fimmu.2019.02348
Received: 09 August 2019; Accepted: 17 September 2019;
Published: 01 October 2019.
Edited by:
Nuno Empadinhas, University of Coimbra, PortugalReviewed by:
Ian Antheni Myles, National Institutes of Health (NIH), United StatesRan Wei, Rutgers, The State University of New Jersey, United States
Copyright © 2019 Alessandri, Ossiprandi, MacSharry, van Sinderen and Ventura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Douwe van Sinderen, ZC52YW5zaW5kZXJlbkB1Y2MuaWU=; Marco Ventura, bWFyY28udmVudHVyYUB1bmlwci5pdA==
 Giulia Alessandri
Giulia Alessandri Maria Cristina Ossiprandi
Maria Cristina Ossiprandi John MacSharry
John MacSharry Douwe van Sinderen
Douwe van Sinderen Marco Ventura
Marco Ventura