- 1Immunology Laboratory, CSIR-Institute of Microbial Technology, Chandigarh, India
- 2Gastroenterology Division, Washington University in St. Louis, St. Louis, MO, United States
- 3Immunology Division, Texas Biomedical Research Institute, San Antonio, TX, United States
- 4Department of Microbiology, School of Medicine, The University of Alabama at Birmingham, Birmingham, AL, United States
- 5Center for Biomedical Engineering, Indian Institute of Technology-Ropar, Rupnagar, India
The gut microbiota significantly regulates the development and function of the innate and adaptive immune system. The attribute of immunological memory has long been linked only with adaptive immunity. Recent evidence indicates that memory is also present in the innate immune cells such as monocytes/macrophages and natural killer cells. These cells exhibit pattern recognition receptors (PRRs) that recognize microbe- or pathogen-associated molecular patterns (MAMPs or PAMPs) expressed by the microbes. Interaction between PRRs and MAMPs is quite crucial since it triggers the sequence of signaling events and epigenetic rewiring that not only play a cardinal role in modulating the activation and function of the innate cells but also impart a sense of memory response. We discuss here how gut microbiota can influence the generation of innate memory and functional reprogramming of bone marrow progenitors that helps in protection against infections. This article will broaden our current perspective of association between the gut microbiome and innate memory. In the future, this knowledge may pave avenues for development and designing of novel immunotherapies and vaccination strategies.
Introduction
The host immune system has the two major arms of protection. The first is innate immunity, which is characterized by non-specific and rapid response against the infectious agent. The second is adaptive, whose hallmark is specificity and memory (1). In the absence of adaptive immune response, innate immunity takes charge of mounting a successful defense response in many organisms (e.g., invertebrates, plants) including mammals (2). Innate immune memory is an emerging concept initially coined by Netea and colleagues that defines the rapid protective response of innate cells to heterologous infections (3–5). This is accompanied by the epigenetic reprogramming that modulates their gene expression and metabolic state and thereby affects the physiology and function of innate immune cells (6, 7). Notably, innate immune memory does not lead to permanent changes in the genome of cells such as mutations and rearrangement of genes, a characteristic of adaptive immune cells (T cells, B cells) (6). Recent findings showed the presence of previously encountered memories even in the non-immune cells of the host (8, 9).
The key players mediating the communication of host and microbes are the sensors, known as pattern recognition receptors (PRRs), expressed by innate immune cells such as dendritic cells (DCs), monocytes/macrophages, and natural killer (NK) cells (10–13). These PRRs recognize microbe- or pathogen-associated molecular patterns (MAMPs or PAMPs) (14–16) PRRs mainly include the families of toll-like receptors (TLRs), nucleotide-binding oligomerization domain (NOD)–like receptors (NLRs), C-type lectin receptors (CLRs), and RIG-I–like receptors (RLRs) (10, 11). The microbiota recognition via these PRRs may induce the memory response upon primary exposure (17, 18). This immunological memory in innate cells has been associated with non-specific vaccination effects. For instance, NK cells and monocytes derived from BCG-immunized individuals displayed a heightened immune response upon re-stimulation and heterologous infections (19, 20). Further, there is evidence of innate memory induction by polio and measles vaccines in humans (21). However, much remains to be studied in the context of gut microbiota–induced innate memory.
Gut microbiota has been established to be a crucial regulator of immune cell development and function (10, 22). The gut microbes and mammals have coevolved and cohabitated for millions of years and exhibit a high degree of mutualism (23). While the microbes get a habitat and nourishment from the host, these microbes return the favor by regulating various host physiological functions, including dietary digestion, and imparting protective immunity against pathogens (24, 25). Further, gut commensal–mediated competition for habitat site, nutrients, or secretion of antimicrobial peptides aids in the maintenance of homeostasis (26, 27). Additionally, signals derived from gut microbes are suggested to tune the immune cells for pro- and anti-inflammatory responses that may affect the susceptibility to diseases (22, 28). Likewise, germ-free mice have been shown to possess immune defects and impaired defense systems (29).
In a healthy state, the immune system reacts against the pathogenic microbes via activation of the inflammatory response, while being tolerant of beneficial microbiota (24, 30). For instance, bacterial phyla such as Bifidobacteria and Lactobacillus are considered beneficial and thus classified as “symbionts.” On the other hand, few species of Escherichia coli are viewed as opportunistic pathogens (pathobionts) (31, 32). Thus, the intestinal immune system requires a careful surveillance system to constantly monitor the flora communities in the lumen for maintaining the host defense. It is well-documented that T cell homeostasis and differentiation and their function are extensively modulated by the gut bacteria (33). For example, Bacteroides fragilis and segmented filamentous bacteria (SFB) have been reported to induce Tregs and Th17 cell differentiation, respectively, in the intestine, thus affecting the host response to infections (34, 35). It is still unclear how the gut microbial population, and its components, could reprogram the innate immune cells to exhibit memory responses.
Given the importance of gut microbiota, characterization and understanding of the involved microbial factors that determine the innate immune memory response is crucial for constructing novel therapeutic interventions (3, 7). This review provides current knowledge of gut microbial signatures and their interaction with the innate cells in imparting them the “memory” characteristics. It would be beneficial to develop immunotherapies and vaccination strategies that can generate memory features in innate cells to efficiently combat pathogens. Here, we discuss and hypothesize the possible impact of gut microbiota in inducing the beneficial innate memory response in the host (Figure 1).
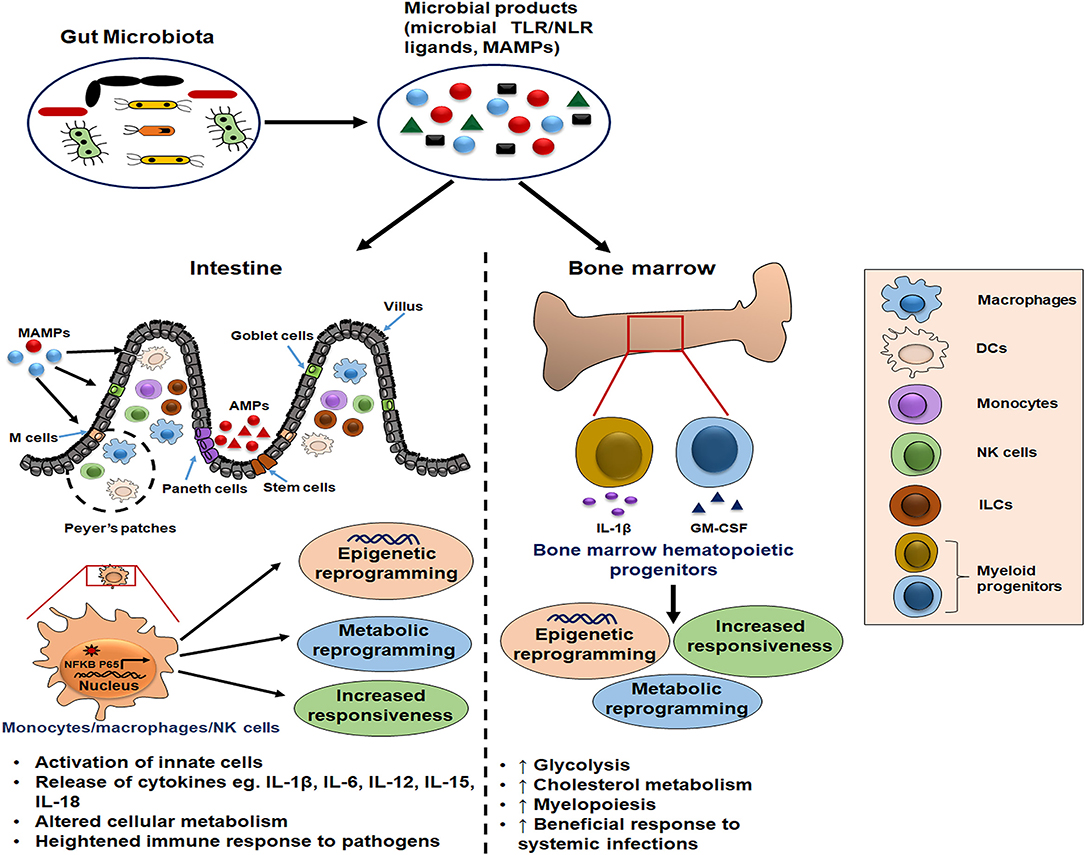
Figure 1. Schematic illustration of gut microbiota as potential inducer of innate memory. The gut microbial products serve as a source of microbe-associated molecular patterns (MAMPs) that bind pattern recognition receptors (PRRs) on innate cells such as monocytes/macrophages and natural killer (NK) cells. Further, this cell activation is accompanied by the epigenetic and metabolic reprogramming which is responsible for their increased cytokine release and heightened immune response upon the subsequent pathogenic exposure. Moreover, these microbial ligands reach the bone marrow through blood circulation and condition the hematopoietic progenitors to induce long-term memory traits and enhance myelopoiesis for mounting the beneficial inflammatory response during systemic infections.
Prospective Link Between Gut Microbiota and Innate Immune Memory
The presence of microbiota-derived ligands/products/metabolites affects the differentiation and function of myeloid and lymphoid lineage innate cells via PRRs (36–38). Innate immune memory has been seen to be an attribute of myeloid cells (monocytes/macrophages), innate lymphoid cells (ILCs) including NK cells, and bone marrow progenitors (39). It is mediated by the transcriptional changes in genes or a specific locus and epigenetic rewiring of these cells upon the primary exposure (39). Consequently, the secondary response to the subsequent infections is enhanced, rapid, and nonspecific (Figure 2). This phenomenon also exists in the bone marrow progenitors, indicating the systemic effects of gut microbiota (40), and the induced memory may persist from weeks up to months (20, 41).
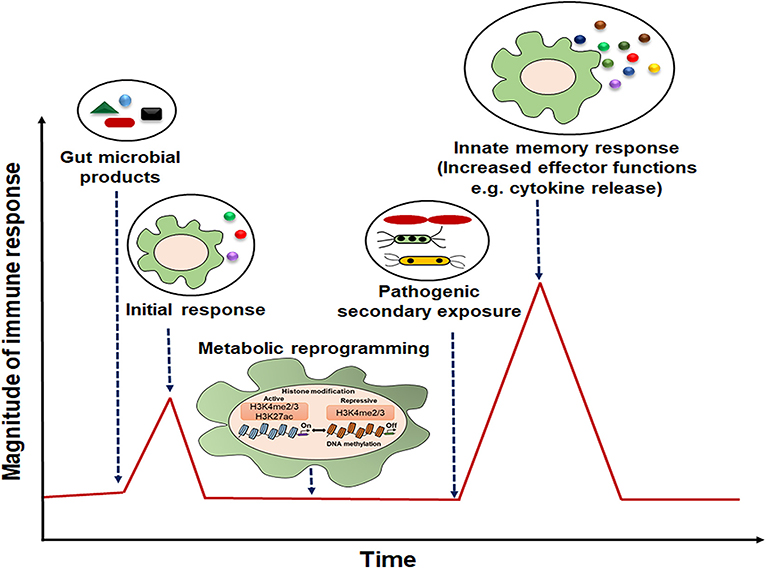
Figure 2. Representative model of innate immune memory response. After initial exposure to gut microbial components, innate cells with “memory” traits respond rapidly with high magnitude of immune response to the secondary stimulation.
Training of PRRs expressing innate cells with gut microbial/non-microbial ligands is required as a protective mechanism independent of adaptive immunity during secondary infection/pathogenic exposures (42). For instance, administration of unmethylated CpG oligodeoxynucleotides prior to infection confers protection in a sepsis and meningitis model (43). Further, polysaccharide β-glucan has been reported to impart defense against Staphylococcus infection (44, 45). Other microbial components such as peptidoglycan that are expressed on numerous bacteria generate innate memory in Toxoplasma infection (46). In addition, cytokines such as IL-18, IL-12, IL-6, IL-23, IL-1β, and IL-15 have been demonstrated to generate memory response in innate cells (47, 48). Several studies established the existence of NK cell memory that leads to their increased activation upon second stimulation (43, 48). Moreover, DCs from protectively immunized mice demonstrated memory response against a fungal pathogen. These DCs showed increased IFN signaling pathway activation and specific histone (H3K4me3 and H3K27me3) modifications (49).
Importantly, commensals in the gut are involved in the production of immunomodulatory metabolites that comprise short-chain fatty acids (SCFAs) such as butyrate, acetate, and propionate (50–52). Further, commensals such as Bacteroides, Lactobacillus, and Bifidobacteria synthesize secondary bile acids that are derived from the metabolism of primary bile acids (53–55). Binding of these bioactive molecules to the receptors on the innate cells regulate their metabolism and functions (51, 56).
SCFAs serve as inhibitors of histone deacetylases (HDACs) in innate cells such as DCs and macrophages (57–59). Moreover, it was shown that SCFAs boost the population of myeloid precursors, resulting in protection against infection (60, 61). Additionally, secondary bile acids are known to modulate gut microbial composition (62). Further, they influence the function of innate cells by inhibiting NF-κB activation (63, 64). These findings suggest the possible epigenetic regulation of these cells via metabolites in the process of innate memory formation.
Emerging evidence suggests that diet-induced microbial changes in the gut could lead to the long-lasting rewiring of the innate cells. Interestingly, western diet (WD) has been linked to enhanced innate immune response. It is shown to induce epigenetic and transcriptional reprogramming of myeloid progenitors via the NLRP3 inflammasome and IL-1R signaling (65). In this case, WD-induced dysbalanced cholesterol biosynthesis led to the accumulation of mevalonate, which is implicated in the generation of innate memory. Indeed, a few studies indicate that maternal diet during pregnancy can have a pronounced effect in shaping the offspring microbiome (66, 67). Further, the gut may be a source of bacteria present in breast milk (68, 69). Of note, microbial exposure during pregnancy enhances the ILC3 and F4/80 (+) CD11c (+) mononuclear cell population in the gut of neonates along with the reprogramming of their intestinal transcriptional profiles. This effect is attributed to the transfer of maternal antibodies that retains the microbial signatures (70). In addition, infections during pregnancy can induce maternal immune activation (MIA), which can lead to the generation of immune cells with “memory” phenotype via epigenomic changes (71, 72). This can result in the condition of hyperimmune activation and neuropsychiatric diseases later in life.
Apart from the diet, stress is another emerging factor that can elicit innate memory phenotype (73, 74). Host cells secrete an array of small molecules upon stress or any cellular damage that can activate PRRs (75). These molecules, termed danger-associated molecular patterns (DAMPs), resemble PAMPs and thus are potential inducers of innate immune memory in cells (75). Social stress also releases IL-6, IL-1β, and TNF-α cytokines (73). Exposure to stressors has been demonstrated to trigger gut microbe–mediated release of MAMPs in blood circulation (76). Further, stressors implicate modification of H3K9 histones and the activation of transcription factor ATF7 (77).
Gut microbiota has been reported to play a key role in the induction of innate immune memory and protect against infections in both vertebrates and invertebrates (78, 79). These robust, non-specific memory effects mediated by MAMP–microbiota interaction may contribute to the development of effective vaccines and therapies that rely on boosting the host innate defense. Understanding the phenomenon and the involved mechanisms can be utilized to train these innate cells and enhance their function against pathogenic infections in the host. Further, this would aid the design and development of novel therapies to treat diseases (80).
Mechanisms of Innate Memory Induction by Microbial Components
Recent studies have highlighted the array of mechanisms through which microbes imprint memory phenotype in innate cells. Transcriptional changes are the hallmark of memory imprints in innate cells, which involve chromatin modifications; specifically, the activation and expression of inflammatory genes take place many times higher than the basal level. This happens via the enhanced accessibility of DNA to enhancers/promoters, increased histone methylation, and acetylation along with enzyme RNA polymerase activity (81). These events are accompanied by transcription factor (NF-κB, STAT molecules, AP-1) translocation and activation (82–84). After the initial exposure to microbial ligands, DNA methylation/histone modifications continue to trigger rapid response upon re-exposure. Further, various immunological pathways such as STAT1, JNK, and MAPK are activated in the process of innate memory generation. For instance, MAPK activates the ATF7 transcription factor and decreases the repression of histones by recruiting the histone H3K9 dimethyltransferase (85). For instance, BCG changes the transcriptional signatures of hematopoietic stem cells (HSCs) to promote myelopoiesis and generate more potent macrophages that can protect against subsequent Mycobacterium tuberculosis infection (9). Of interest, a diet enriched in low-density-lipoprotein cholesterol such as WD elicits the expansion of HSCs along with the reprogramming of granulocyte monocyte precursor cells (GMPs) through the activation of NLRP3 inflammasome and possibly includes gut dysbiosis (65).
Importantly, cells with innate memory were reported to exhibit increased size, granularity, and activated phenotype (86). They are usually deprived of acetylation and lack active transcription. However, their inflammatory cytokine gene promoters are marked with histone methylation (H3K4), conferring them the attribute of rapid response upon re-exposure (87). These epigenetically rewired innate cells may be sustained in the host niche as seen in the case of NK cells and monocytes (40, 88). Interestingly, microbiota absence in mice impaired the histone modification in NK cells, rendering them unable to trigger a protective inflammatory response against viral infection (78).
Interestingly, various cellular metabolic pathways are involved in triggering and maintaining these epigenetic modifications (89, 90). Induction of memory features by β-glucan (microbial polysaccharide) is accompanied by a metabolic shift to aerobic glycolysis; this is referred to as the “Warburg effect” (91). Several studies reported that genes of the mTOR-HIF1α pathway were induced in β-glucan triggered monocytes that have undergone epigenetic changes, i.e., H3K4me3 and H3K27ac (7, 91). Moreover, TCA cycle metabolites such as mevalonate, succinate, and fumarate induce the activation of genes required to generate innate memory (92, 93). The pathway of cholesterol synthesis and mevalonate through the activation of IGF1 receptor and mTOR and further enrichment of histone H3K4me3 generated innate memory (93). There are also studies indicating the metabolic shift that leads to enhanced aerobic glycolysis, cholesterol synthesis, and NAD+/NADH ratio in cells (7, 91). Further, another metabolite, acetyl-CoA, has been shown to induce histone acetylation of genes related to glycolytic enzymes, such as phosphofructokinase, hexokinase 2, and lactate dehydrogenase (LDH), thus increasing glycolysis and inducing memory phenotype (94). In fact, β-glucan has also been shown to access bone marrow and act on myeloid-biased long-term HSCs (40). It is accompanied by changes in lipid metabolism, IL-1β signaling, and activation of the GM-CSF/CD131 axis. This β-glucan–mediated training is sufficient to protect against secondary challenges and recover from chemotherapy-induced myelosuppression.
The occurrence of innate immune memory relies on factors such as dose/amount and duration of initial inflammatory stimulus or PAMP. For example, a single low dose of lipopolysaccharide (LPS) was able to induce more release of proinflammatory molecules upon re-exposure and thus induce innate memory (86). In another study, a similar phenomenon was observed via epigenomic changes such as histone H3K4me1 modification. On the contrary, four-time administration of LPS led to the tolerant phenotype in cells (95). Moreover, Ifrim et al. reported that moderate to high doses of PRR ligands such as flagellin (10 μg/ml), LPS (100 μg/ml), and poly I:C (100 μg/ml) led to tolerance (86). On the contrary, a low to moderate dose of β-glucan (1 μg/ml) and muramyl dipeptide (MDP) (10 μg/ml) was able to elicit memory generation.
PRR Mediated Regulation of Innate Memory by Gut Microbiota
Gut microbiota is a source of ligands that serve as MAMPs and activate innate cells expressing PRRs. In the homeostatic condition, the aberrant PRR activation is limited by various mechanisms such as the mucus layer (96); secretion of antimicrobial peptides, e.g., defensins (97); regenerating islet-derived protein 3 gamma (RegIIIγ) release by Paneth cells (98); secretory IgA (99); and inhibitory TLR signaling (100–102). Further, the PRR expression is context-dependent and varies in cell types to control the deleterious inflammatory response (16, 103). Interestingly, it is seen that the administration of Lactobacillus plantarum protected well against viral infections (104, 105). Further, there is emerging evidence that gut microbiota is known to affect innate memory phenotype at the distant mucosal sites or peripheral tissues. Yao et al. reported the immunological memory phenotype and protective functions in alveolar macrophages after respiratory virus infection (106).
In the context of memory, initial stimulation of innate cells by MAMPs serves as a factor for “priming” and functionally reprogramming of these cells to mediate heightened non-specific response to subsequent pathogenic exposure (39). Gut microbial components, mainly peptidoglycan, flagellin, β-glucan, and lipoproteins, may induce memory phenotype in the innate cells, which could underline their potential as an effective adjuvant for vaccination studies (107, 108).
Peptidoglycan: It is an important component of the bacterial cell membrane envelope, not present in the eukaryotic host. It is found in both gram-positive and gram-negative commensals (109). Further, the synthesis of peptidoglycan is ubiquitous in gut bacteria that is recognized by NOD receptors (110). For instance, NOD-1 binds only γ-d-glutamyl-meso-diaminopimelic acid (DAP)–containing muropeptides, whereas NOD-2 recognizes the MDP component (34). Further, a report demonstrated that NOD-2 induced H3K4me3 epigenetic modification in monocytes, a feature linked to innate memory (111). Moreover, NOD receptor activation triggers the inflammasomes to secrete cytokines such as IL-1β and IL-18, which is implicated in memory generation (112).
Flagellin: It is the essential component of many commensals and pathogens that activates TLR-5 signaling on innate cells (113). CD103+ DCs in the intestine recognize flagellin and secrete IL-23, which in turn triggers the ILCs to secrete IL-22 and thus facilitates innate defense (114).
β-glucan: This cross-linked glucan particle is commonly found in fungi and some bacterial cell walls (115). It is reported to induce long-term memory response in macrophages and bone marrow progenitors (40). This is accompanied by accumulation of metabolite mevalonate and Warburg effect in innate cells (91, 93). β-glucan is known to bind dectin-1 receptor (44, 116).
LPS: It is a glycolipid majorly found in the outer membrane of gram-negative bacteria and induces activation of TLR-4 and downstream NF-κB signaling (117). Earlier studies have shown that pretreatment with LPS prevents subsequent infection (118, 119). Further, LPS derived from gut microbes such as Bacteriodetes species is a potent activator of innate response (120), although the dose of LPS is a crucial factor to determine the induction of either memory or tolerant phenotype in cells (86).
Amongst the PRRs, TLRs are the most extensively studied transmembrane or intracellular glycoproteins that recognize a variety of microbial ligands or MAMPs as discussed above (10, 11). TLRs trigger signaling pathways, which leads to the secretion of cytokines and gene transcription in monocytes/macrophages and bone marrow progenitors (10). Triggering of TLR-2 by microbial lipoteichoic acids, lipoproteins, lipopeptides, and glycolipids activates NF-κB and is known to impart protection in intestinal acute inflammation (121). Additionally, initial priming of macrophages with TLR-2 and NOD-2 has been shown to confer protection against acute infection (105). This rapid response to continuously exposed mucosal tissue appears to be an essential part of host defense. In addition, a study demonstrated that stimulation of TLR-3 on macrophages with dsRNA could lessen the symptoms of DSS-induced colitis (122). Another study demonstrated the protective response in colitis upon TLR-3 activation by dsRNA of lactic acid–producing commensals (123).
In a healthy intestine, sensing of microbial LPS by TLR-4 is required to defend against invading pathogens (14). At homeostasis, there is a low level of TLR-4 expression, which gets elevated in inflammatory conditions and diseases. This results in the activation of innate immunity to restrict pathogenic exposure (124). Moreover, TLR-5 binds the flagellated bacteria and imparts protection to Enterobacter and Salmonella infections in the host (125, 126). Furthermore, another intracellular receptor, TLR-9, binds to the unmethylated CpG dinucleotides, which are abundant in commensals (127). TLR-9 activation stimulates the secretion of many proinflammatory cytokines including IL-12, which is considered a crucial cytokine to induce innate memory phenotype. Notably, it is the pathogenic microbe, not commensals, that breaches the gut lining and activate the basolateral TLR-9 receptor (128, 129). Notably, host genetic variations in the microbiota composition and pathogenic exposure could impact the commensal-mediated immune memory induction.
Another important class of PRRs involved in the education of innate cells is intracellular NLRs (130). Interestingly, commensal recognition by NLRs maintains homeostasis in the gut, while the pathogenic species of Salmonella and Helicobacter pylori trigger the inflammatory response (131, 132). NOD-1 and NOD-2 expressed in monocytes/macrophages are known to recognize microbial peptidoglycans DAP and MDP, respectively (34). NLR activation initiates the signaling pathways such as p38, MAPK, and NF-κB and elicits the release of cytokines (such as IL-1β and IL-18) that are known to induce innate memory phenotype (47, 48, 133). A study revealed that gut microbiota–derived peptidoglycan enhanced the pathogen-clearing capacity of bone marrow–derived neutrophils. The MDP fragments from the gut translocated to the bone marrow and triggered the neutrophil activation via NOD-1 signaling (134). This raises the possible hypothesis that gut microbial components reach peripheral sites and affect their epigenetic programming to imprint the memory phenotype (40).
Noticeably, these PRRs' stimulation may also lead to the tolerogenic phenotype, but this relies on the nature and duration of exposure to the initial microbial stimuli (135). Thus, these PRRs mediated modulation of innate cells should be monitored and utilized to generate the protective memory phenotype and ultimately the rapid, heightened, and efficient response against invading pathogens.
Gut Microbiota Influences Functional Rewiring of Bone Marrow Progenitors
The impact of microbiota in the immediate sites of colonization such as the intestine, skin epithelium, and respiratory mucosa is quite plausible (27). However, its role in the primary site of hematopoiesis (bone marrow) may have significant immunological relevance. Long-term innate immune memory can be apparent in either the persistence of reprogrammed monocytes/macrophages in different tissues or modulation of bone marrow progenitors. This can be easily hypothesized, from the fact that LPS treatment in germ-free mice led to an elevation in the level of inflammatory cytokines and neutrophil recruitment and thus imparted systemic immunity (118). Concordantly, microbiota-derived peptidoglycan trigger NOD-1 receptor in peripheral blood neutrophils and boosted their anti-bacterial activity (134).
Monocytes, macrophages, and DCs that play a crucial role in shaping the immune response fall under myeloid-derived cells. Myeloid-derived cells originate in the bone marrow and then populate all lymphoid and non-lymphoid tissues. The myeloid cellular system has a non-redundant capacity to act in concert during the elimination process of pathogens and re-establishing tissue integrity (136). Recent reports demonstrated that gut bacteria, especially gram-negative bacteria, regulate granulopoietic events (137–140). Goris et al. showed that germ-free and polymyxin-treated mice have a lower number of bone marrow progenitor cells (141). Further, these germ-free mice complemented with fecal matter from wild type showed the reversion of the myelopoiegenic capability of precursors in generating colony-forming unit–granulocyte/macrophage (CFU–GM) colonies (142).
Interestingly, naïve mice with bone marrow transferred from the SFB and Clostridium spp. colonized mice demonstrated protection from Entamoeba histolytica infection. This is due to the expansion of marrow GMPs and increased expression of the epigenetic mediator JMJD3 in GMPs. SFB also altered the bone marrow DCs such that they have an enhanced capacity to secrete IL-23 (143). Further, adoptive transfer of DCs from SFB-supplemented mice to SFB-deficient mice was sufficient to protect against E. histolytica infection (143). Additionally, IL-1β and GM-CSF cytokines released from peripheral sites have been shown to confer the innate memory trait to the bone marrow cells (40, 65, 144).
The F4/80hi macrophages have an embryonic origin, while F4/80lo leukocytes have a hematopoietic origin (145, 146). To test the contribution of gut microbiota in the promotion of myelopoiesis, germ-free and specific pathogen–free (SPF) mice were administered a thymidine analog, 5-ethynyl-2′-deoxyuridine (EdU). In comparison to SPF mice, germ-free mice showed reduced uptake of EdU in both F4/80hi and F4/80lo phagocytes (61). This observation highlighted that commensals play a prominent role in the preservation of both the HSC-derived myeloid and splenic yolk sac–derived cells along with the inflammatory monocytes (145, 147). Further, NOD-1 is known to be responsible for mediating myeloid cell longevity. A study has shown decreased levels of NOD-1 ligand (DAP) in mice with antibiotic-altered microbiota, and upon NOD-1 stimulation, they found an abundance of IL-17–secreting lymphocytes in the intestine, which relay the microbial detection for systemic control of the phagocyte life span (148).
Cross Talk of Gut Microbiota and ILCs
ILCs are known as the subset of innate leukocytes of lymphoid morphology that are mainly located in the mucosa. They lack antigen-specific rearranged receptors and have been grouped into NK cells and three classes as ILC1, ILC2, and ILC3 based on the type of cytokines they produce (149). Non-specific memory NK cells generated upon cytokine stimulation have been shown to exhibit a strong immune response to infections.
Amongst ILCs, group 3 ILCs have a key role in the innate immune response to invading pathogens in the gut. They are the prominent source of IL-22 and other antimicrobial proteins (AMPs) in the lamina propria of the intestine. IL-22 binds to receptors present on the intestinal epithelial cells (IECs) and induce the secretion of AMPs (RegIIIγ and RegIIIβ), which eventually limit the colonization of pathogens such as Citrobacter rodentium (150–152). Further, the functions of ILC3 are influenced by the gut microbial metabolites (153). Gut microbiota also aids in the conditioning and development of ILCs (154, 155). Further, germ-free and antibiotic-treated mice have a relatively diminished subset of NCR+ RORγt+ ILC population (156–158). A recent study revealed that TLR-2 agonists can directly bind to human RORγt+ ILC, inducing the secretion of IL-2, which further triggers the release of IL-22, a cytokine known to be implicated in antimicrobial defense (159). It would be interesting to see whether the interaction between gut microbiota and these ILCs imparts them the memory traits.
IECs as a Mediator of Microbiota and Immune Cell Interaction
IECs stand as a single-cell barrier between the intestinal microbiota and the submucosal immune cells. When the IEC barrier senses microbial pathogens, it reinforces its integrity and thus protects against pathogen invasion (160). IECs expressing TLRs are stimulated by commensal-derived ligands, triggering the release of proinflammatory cytokines such as IL-6 and TNF-α (14). Further, gut-derived metabolite butyrate binds the IECs and triggers the innate sensors such as NLRP3 inflammasome to secrete IL-18 (112, 161). Commensal bacteria–derived peptidoglycan has been shown to trigger NOD-1 receptor in IECs. This event is accompanied by the production of defensin molecule and chemokine CCL20 secretion, which compel the generation of isolated lymphoid follicles (ILFs), which are the site of B cell recruitment and immune response generation (162). Moreover, there are intestinal mononuclear phagocytes (iMPs), residing in the intestinal sub-epithelium (163). Although the ontology of iMPs is still in debate, they comprise macrophages and DCs that regulate intestinal homeostasis (164). It is possible that these cells with copious expression of several PRRs upon stimulation by gut microbial components get memory signatures and persist in secondary organs.
Intestinal epithelial stem cells (IESCs) are crucial cells in the gut that have the capacity to differentiate into IECs. Gut microbiota composition has a significant impact on the IESC activity and renewal of intestinal epithelium, which is located at proximity to the lumen (165). Interestingly, SCFAs such as butyrate serve as a source of energy for IECs in metabolic processes and act to inhibit HDAC activity in IESCs (166, 167). Although it not very conclusive as of now to state the particular species of bacteria that is specifically responsible for such regulation, it is clear that some microbial metabolites stimulate the Wnt/β-catenin pathway and maintain the IESCs (168). Other signaling pathways, namely the JAK and STAT pathways, are very important in the bacteria-modulated epithelium homeostasis via stem cell regulation (165, 169, 170). These cells may serve as the potential niche for the generation of long-term innate memory.
Future Direction and Concluding Remarks
The development of non-specific innate immune memory appears to be a crucial evolutionary phenomenon to benefit and protect the host against a variety of pathogens. Gut microbiota performs the intricate function of immune system maturation in neonates (171). Thus, the induction of memory in innate cells during this process appears to be a part of the host–microbiome co-adaptation to mediate a prompt response to the infectious stimulus.
It is evident that there would be circumstances in which the innate memory can lead to the deleterious systemic inflammatory response. If that persists for a long time, and activates innate cells in conditions such as sepsis, it can lead to tissue damage or immune paralysis. Further, the differential generation of innate memory in various organs, its duration in various immune cells, and the signaling pathways induced by gut microbial components need to be investigated in the future.
In conclusion, the process of innate memory generation should be considered as an effective approach to boost host defense and well-managed to minimize side effects while being favorable to the host. Beneficial commensals or derived products that induce the effective innate memory with regulated inflammatory response can be utilized as potential novel therapeutics to treat infections and diseases.
Author Contributions
SNe and JA contributed to the conception of the idea, design, and conceptualization of the manuscript. SN, DD, SP, SNa, and JA contributed to the review of literature, drafting, and revision of the manuscript.
Funding
This study was supported by the Council of Scientific and Industrial Research (CSIR) and Department of Biotechnology (DBT), India.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Muller V, de Boer RJ, Bonhoeffer S, Szathmary E. An evolutionary perspective on the systems of adaptive immunity. Biol Rev Camb Philos Soc. (2018) 93:505–28. doi: 10.1111/brv.12355
2. Medzhitov R, Janeway C Jr. Innate immune recognition: mechanisms and pathways. Immunol Rev. (2000) 173:89–97. doi: 10.1034/j.1600-065X.2000.917309.x
3. Netea MG, Quintin J, van der Meer JW. Trained immunity: a memory for innate host defense. Cell Host Microbe. (2011) 9:355–61. doi: 10.1016/j.chom.2011.04.006
4. Netea MG. Training innate immunity: the changing concept of immunological memory in innate host defence. Eur J Clin Invest. (2013) 43:881–4. doi: 10.1111/eci.12132
5. Kurtz J, Franz K. Innate defence: evidence for memory in invertebrate immunity. Nature. (2003) 425:37–8. doi: 10.1038/425037a
6. De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. (2007) 130:1083–94. doi: 10.1016/j.cell.2007.08.019
7. Saeed S, Quintin J, Kerstens HH, Rao NA, Aghajanirefah A, Matarese F, et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science. (2014) 345:1251086. doi: 10.1126/science.1251086
8. Hamada A, Torre C, Drancourt M, Ghigo E. Trained immunity carried by non-immune cells. Front Microbiol. (2018) 9:3225. doi: 10.3389/fmicb.2018.03225
9. Kaufmann E, Sanz J, Dunn JL, Khan N, Mendonca LE, Pacis A, et al. BCG educates hematopoietic stem cells to generate protective innate immunity against tuberculosis. Cell. (2018) 172:176–90.e19. doi: 10.1016/j.cell.2017.12.031
10. Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. (2011) 30:16–34. doi: 10.3109/08830185.2010.529976
11. Pahari S, Kaur G, Negi S, Aqdas M, Das DK, Bashir H, et al. Reinforcing the functionality of mononuclear phagocyte system to control tuberculosis. Front Immunol. (2018) 9:193. doi: 10.3389/fimmu.2018.00193
12. Pahari S, Negi S, Aqdas M, Arnett E, Schlesinger LS, Agrewala JN. Induction of autophagy through CLEC4E in combination with TLR4: an innovative strategy to restrict the survival of Mycobacterium tuberculosis. Autophagy. (2019) 8:1–23. doi: 10.1080/15548627.2019.1658436
13. Negi S, Pahari S, Das DK, Khan N, Agrewala JN. Curdlan limits mycobacterium tuberculosis survival through STAT-1 regulated nitric oxide production. Front Microbiol. (2019) 10:1173. doi: 10.3389/fmicb.2019.01173
14. Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. (2004) 118:229–41. doi: 10.1016/j.cell.2004.07.002
15. Gewirtz AT, Simon PO Jr, Schmitt CK, Taylor LJ, Hagedorn CH, O'Brien AD, et al. Salmonella typhimurium translocates flagellin across intestinal epithelia, inducing a proinflammatory response. J Clin Invest. (2001) 107:99–109. doi: 10.1172/JCI10501
16. Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol. (2001) 167:1882–5. doi: 10.4049/jimmunol.167.4.1882
17. Mills KH. TLR-dependent T cell activation in autoimmunity. Nat Rev Immunol. (2011) 11:807–22. doi: 10.1038/nri3095
18. Kleinnijenhuis J, Quintin J, Preijers F, Joosten LA, Ifrim DC, Saeed S, et al. Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc Natl Acad Sci USA. (2012) 109:17537–42. doi: 10.1073/pnas.1202870109
19. Kleinnijenhuis J, Quintin J, Preijers F, Joosten LA, Jacobs C, Xavier R, et al. BCG-induced trained immunity in NK cells: Role for non-specific protection to infection. Clin Immunol. (2014) 155:213–9. doi: 10.1016/j.clim.2014.10.005
20. Kleinnijenhuis J, Quintin J, Preijers F, Benn CS, Joosten LA, Jacobs C, et al. Long-lasting effects of BCG vaccination on both heterologous Th1/Th17 responses and innate trained immunity. J Innate Immun. (2014) 6:152–8. doi: 10.1159/000355628
21. Benn CS, Netea MG, Selin LK, Aaby P. A small jab - a big effect: nonspecific immunomodulation by vaccines. Trends Immunol. (2013) 34:431–9. doi: 10.1016/j.it.2013.04.004
22. Negi S, Pahari S, Bashir H, Agrewala JN. Gut microbiota regulates mincle mediated activation of lung dendritic cells to protect against Mycobacterium tuberculosis. Front Immunol. (2019) 10:1142. doi: 10.3389/fimmu.2019.01142
23. Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. (2008) 320:1647–51. doi: 10.1126/science.1155725
24. Slack E, Hapfelmeier S, Stecher B, Velykoredko Y, Stoel M, Lawson MA, et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science. (2009) 325:617–20. doi: 10.1126/science.1172747
25. Bron PA, van Baarlen P, Kleerebezem M. Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nat Rev Microbiol. (2011) 10:66–78. doi: 10.1038/nrmicro2690
26. Moens E, Veldhoen M. Epithelial barrier biology: good fences make good neighbours. Immunology. (2012) 135:1–8. doi: 10.1111/j.1365-2567.2011.03506.x
27. Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. (2013) 13:321–35. doi: 10.1038/nri3430
28. Khan N, Vidyarthi A, Nadeem S, Negi S, Nair G, Agrewala JN. Alteration in the gut microbiota provokes susceptibility to tuberculosis. Front Immunol. (2016) 7:529. doi: 10.3389/fimmu.2016.00529
29. Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. (2009) 9:313–23. doi: 10.1038/nri2515
30. Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. (2012) 489:231–41. doi: 10.1038/nature11551
31. Klijn A, Mercenier A, Arigoni F. Lessons from the genomes of bifidobacteria. FEMS Microbiol Rev. (2005) 29:491–509. doi: 10.1016/j.fmrre.2005.04.010
32. Carvalho FA, Barnich N, Sivignon A, Darcha C, Chan CH, Stanners CP, et al. Crohn's disease adherent-invasive Escherichia coli colonize and induce strong gut inflammation in transgenic mice expressing human CEACAM. J Exp Med. (2009) 206:2179–89. doi: 10.1084/jem.20090741
33. Kuhn KA, Stappenbeck TS. Peripheral education of the immune system by the colonic microbiota. Semin Immunol. (2013) 25:364–9. doi: 10.1016/j.smim.2013.10.002
34. Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. (2010) 107:12204–9. doi: 10.1073/pnas.0909122107
35. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. (2009) 139:485–98. doi: 10.1016/j.cell.2009.09.033
36. Gorjifard S, Goldszmid RS. Microbiota-myeloid cell crosstalk beyond the gut. J Leukoc Biol. (2016) 100:865–79. doi: 10.1189/jlb.3RI0516-222R
37. Weaver LK, Minichino D, Biswas C, Chu N, Lee JJ, Bittinger K, et al. Microbiota-dependent signals are required to sustain TLR-mediated immune responses. JCI Insight. (2019) 4:124370. doi: 10.1172/jci.insight.124370
38. McCoy KD, Burkhard R, Geuking MB. The microbiome and immune memory formation. Immunol Cell Biol. (2019) 97:625–35. doi: 10.1111/imcb.12273
39. Gourbal B, Pinaud S, Beckers GJM, Van Der Meer JWM, Conrath U, Netea MG. Innate immune memory: an evolutionary perspective. Immunol Rev. (2018) 283:21–40. doi: 10.1111/imr.12647
40. Mitroulis I, Ruppova K, Wang B, Chen LS, Grzybek M, Grinenko T, et al. Modulation of myelopoiesis progenitors is an integral component of trained immunity. Cell. (2018) 172:147–61.e12. doi: 10.1016/j.cell.2017.11.034
41. Quintin J, Saeed S, Martens JHA, Giamarellos-Bourboulis EJ, Ifrim DC, Logie C, et al. Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe. (2012) 12:223–32. doi: 10.1016/j.chom.2012.06.006
42. Pahari S, Kaur G, Aqdas M, Negi S, Chatterjee D, Bashir H, et al. Bolstering immunity through pattern recognition receptors: a unique approach to control tuberculosis. Front Immunol. (2017) 8:906. doi: 10.3389/fimmu.2017.00906
43. Ribes S, Meister T, Ott M, Redlich S, Janova H, Hanisch UK, et al. Intraperitoneal prophylaxis with CpG oligodeoxynucleotides protects neutropenic mice against intracerebral Escherichia coli K1 infection. J Neuroinflammation. (2014) 11:14. doi: 10.1186/1742-2094-11-14
44. Marakalala MJ, Williams DL, Hoving JC, Engstad R, Netea MG, Brown GD. Dectin-1 plays a redundant role in the immunomodulatory activities of β-glucan-rich ligands in vivo. Microbes Infect. (2013) 15:511–5. doi: 10.1016/j.micinf.2013.03.002
45. Di Luzio NR, Williams DL. Protective effect of glucan against systemic Staphylococcus aureus septicemia in normal and leukemic mice. Infect Immun. (1978) 20:804–10.
46. Krahenbuhl JL, Sharma SD, Ferraresi RW, Remington JS. Effects of muramyl dipeptide treatment on resistance to infection with Toxoplasma gondii in mice. Infect Immun. (1981) 31:716–22.
47. Romee R, Rosario M, Berrien-Elliott MM, Wagner JA, Jewell BA, Schappe T, et al. Cytokine-induced memory-like natural killer cells exhibit enhanced responses against myeloid leukemia. Sci Transl Med. (2016) 8:357ra123. doi: 10.1126/scitranslmed.aaf2341
48. Romee R, Schneider SE, Leong JW, Chase JM, Keppel CR, Sullivan RP, et al. Cytokine activation induces human memory-like NK cells. Blood. (2012) 120:4751–60. doi: 10.1182/blood-2012-04-419283
49. Hole CR, Wager CML, Castro-Lopez N, Campuzano A, Cai H, Wozniak KL, et al. Induction of memory-like dendritic cell responses in vivo. Nat Commun. (2019) 10:2955. doi: 10.1038/s41467-019-10486-5
50. Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. (2016) 16:341–52. doi: 10.1038/nri.2016.42
51. Levy M, Thaiss CA, Elinav E. Metabolites: messengers between the microbiota and the immune system. Genes Dev. (2016) 30:1589–97. doi: 10.1101/gad.284091.116
52. Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. (2012) 489:242–9. doi: 10.1038/nature11552
53. Kitahara M, Takamine F, Imamura T, Benno Y. Clostridium hiranonis sp. nov., a human intestinal bacterium with bile acid 7alpha-dehydroxylating activity. Int J Syst Evol Microbiol. (2001) 51(Pt 1):39–44. doi: 10.1099/00207713-51-1-39
54. Jia W, Xie G, Jia W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol. (2018) 15:111–28. doi: 10.1038/nrgastro.2017.119
55. Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. (2006) 47:241–59. doi: 10.1194/jlr.R500013-JLR200
56. Wang G, Huang S, Wang Y, Cai S, Yu H, Liu H, et al. Bridging intestinal immunity and gut microbiota by metabolites. Cell Mol Life Sci. (2019) 76:3917–37. doi: 10.1007/s00018-019-03190-6
57. Singh N, Thangaraju M, Prasad PD, Martin PM, Lambert NA, Boettger T, et al. Blockade of dendritic cell development by bacterial fermentation products butyrate and propionate through a transporter (Slc5a8)-dependent inhibition of histone deacetylases. J Biol Chem. (2010) 285:27601–8. doi: 10.1074/jbc.M110.102947
58. Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci USA. (2014) 111:2247–52. doi: 10.1073/pnas.1322269111
59. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. (2014) 20:159–66. doi: 10.1038/nm.3444
60. Balmer ML, Schurch CM, Saito Y, Geuking MB, Li H, Cuenca M, et al. Microbiota-derived compounds drive steady-state granulopoiesis via MyD88/TICAM signaling. J Immunol. (2014) 193:5273–83. doi: 10.4049/jimmunol.1400762
61. Khosravi A, Yanez A, Price JG, Chow A, Merad M, Goodridge HS, et al. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe. (2014) 15:374–81. doi: 10.1016/j.chom.2014.02.006
62. Wang H, Chen J, Hollister K, Sowers LC, Forman BM. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol Cell. (1999) 3:543–53. doi: 10.1016/S1097-2765(00)80348-2
63. Wang YD, Chen WD, Yu D, Forman BM, Huang W. The G-protein-coupled bile acid receptor, Gpbar1 (TGR5), negatively regulates hepatic inflammatory response through antagonizing nuclear factor κ light-chain enhancer of activated B cells (NF-κB) in mice. Hepatology. (2011) 54:1421–32. doi: 10.1002/hep.24525
64. Vavassori P, Mencarelli A, Renga B, Distrutti E, Fiorucci S. The bile acid receptor FXR is a modulator of intestinal innate immunity. J Immunol. (2009) 183:6251–61. doi: 10.4049/jimmunol.0803978
65. Christ A, Gunther P, Lauterbach MAR, Duewell P, Biswas D, Pelka K, et al. Western Diet Triggers NLRP3-Dependent Innate Immune Reprogramming. Cell. (2018) 172:162–75.e14. doi: 10.1016/j.cell.2017.12.013
66. Chu DM, Antony KM, Ma J, Prince AL, Showalter L, et al. The early infant gut microbiome varies in association with a maternal high-fat diet. Genome Med. (2016) 8:77. doi: 10.1186/s13073-016-0330-z
67. Myles IA, Fontecilla NM, Janelsins BM, Vithayathil PJ, Segre JA, Datta SK. Parental dietary fat intake alters offspring microbiome and immunity. J Immunol. (2013) 191:3200–9. doi: 10.4049/jimmunol.1301057
68. Rodriguez JM. The origin of human milk bacteria: is there a bacterial entero-mammary pathway during late pregnancy and lactation? Adv Nutr. (2014) 5:779–84. doi: 10.3945/an.114.007229
69. Latuga MS, Stuebe A, Seed PC. A review of the source and function of microbiota in breast milk. Semin Reprod Med. (2014) 32:68–73. doi: 10.1055/s-0033-1361824
70. Gomez de Aguero M, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, et al. The maternal microbiota drives early postnatal innate immune development. Science. (2016) 351:1296–302. doi: 10.1126/science.aad2571
71. Tang B, Jia H, Kast RJ. Thomas EA. Epigenetic changes at gene promoters in response to immune activation in utero. Brain Behav Immun. (2013) 30:168–75. doi: 10.1016/j.bbi.2013.01.086
72. Choi GB, Yim YS, Wong H, Kim S, Kim H, Kim SV, et al. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science. (2016) 351:933–9. doi: 10.1126/science.aad0314
73. Ramirez K, Shea DT, McKim DB, Reader BF, Sheridan JF. Imipramine attenuates neuroinflammatory signaling and reverses stress-induced social avoidance. Brain Behav Immun. (2015) 46:212–20. doi: 10.1016/j.bbi.2015.01.016
74. Salam AP, Borsini A, Zunszain PA. Trained innate immunity: a salient factor in the pathogenesis of neuroimmune psychiatric disorders. Mol Psychiatry. (2018) 23:170–6. doi: 10.1038/mp.2017.186
75. Seong SY, Matzinger P. Hydrophobicity: an ancient damage-associated molecular pattern that initiates innate immune responses. Nat Rev Immunol. (2004) 4:469–78. doi: 10.1038/nri1372
76. Fleshner M. Stress-evoked sterile inflammation, danger associated molecular patterns (DAMPs), microbial associated molecular patterns (MAMPs) and the inflammasome. Brain Behav Immun. (2013) 27:1–7. doi: 10.1016/j.bbi.2012.08.012
77. Maekawa T, Kim S, Nakai D, Makino C, Takagi T, Ogura H, et al. Social isolation stress induces ATF-7 phosphorylation and impairs silencing of the 5-HT 5B receptor gene. EMBO J. (2010) 29:196–208. doi: 10.1038/emboj.2009.318
78. Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. (2012) 37:171–86. doi: 10.1016/j.immuni.2012.05.020
79. Rodrigues J, Brayner FA, Alves LC, Dixit R, Barillas-Mury C. Hemocyte differentiation mediates innate immune memory in Anopheles gambiae mosquitoes. Science. (2010) 329:1353–5. doi: 10.1126/science.1190689
80. Mulder WJM, Ochando J, Joosten LAB, Fayad ZA, Netea MG. Therapeutic targeting of trained immunity. Nat Rev Drug Discov. (2019) 18:553–66. doi: 10.1038/s41573-019-0025-4
81. Smale ST, Tarakhovsky A, Natoli G. Chromatin contributions to the regulation of innate immunity. Annu Rev Immunol. (2014) 32:489–511. doi: 10.1146/annurev-immunol-031210-101303
82. Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. (2010) 32:317–28. doi: 10.1016/j.immuni.2010.02.008
83. Smale ST, Natoli G. Transcriptional control of inflammatory responses. Cold Spring Harb Perspect Biol. (2014) 6:a016261. doi: 10.1101/cshperspect.a016261
84. Barozzi I, Simonatto M, Bonifacio S, Yang L, Rohs R, Ghisletti S, et al. Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Mol Cell. (2014) 54:844–57. doi: 10.1016/j.molcel.2014.04.006
85. Yoshida K, Maekawa T, Zhu Y, Renard-Guillet C, Chatton B, Inoue K, et al. The transcription factor ATF7 mediates lipopolysaccharide-induced epigenetic changes in macrophages involved in innate immunological memory. Nat Immunol. (2015) 16:1034–43. doi: 10.1038/ni.3257
86. Ifrim DC, Quintin J, Joosten LA, Jacobs C, Jansen T, Jacobs L, et al. Trained immunity or tolerance: opposing functional programs induced in human monocytes after engagement of various pattern recognition receptors. Clin Vaccine Immunol. (2014) 21:534–45. doi: 10.1128/CVI.00688-13
87. Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, et al. Latent enhancers activated by stimulation in differentiated cells. Cell. (2013) 152:157–71. doi: 10.1016/j.cell.2012.12.018
88. Yanez A, Hassanzadeh-Kiabi N, Ng MY, Megias J, Subramanian A, Liu GY, et al. Detection of a TLR2 agonist by hematopoietic stem and progenitor cells impacts the function of the macrophages they produce. Eur J Immunol. (2013) 43:2114–25. doi: 10.1002/eji.201343403
89. Arts RJ, Joosten LA, Netea MG. Immunometabolic circuits in trained immunity. Semin Immunol. (2016) 28:425–30. doi: 10.1016/j.smim.2016.09.002
90. Dominguez-Andres J, Joosten LA, Netea MG. Induction of innate immune memory: the role of cellular metabolism. Curr Opin Immunol. (2019) 56:10–6. doi: 10.1016/j.coi.2018.09.001
91. Cheng SC, Quintin J, Cramer RA, Shepardson KM, Saeed S, Kumar V, et al. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. (2014) 345:1250684. doi: 10.1126/science.1250684
92. Arts RJ, Novakovic B, Ter Horst R, Carvalho A, Bekkering S, Lachmandas E, et al. Glutaminolysis and fumarate accumulation integrate immunometabolic and epigenetic programs in trained immunity. Cell Metab. (2016) 24:807–19. doi: 10.1016/j.cmet.2016.10.008
93. Bekkering S, Arts RJW, Novakovic B, Kourtzelis I, van der Heijden C, Li Y, et al. Metabolic Induction of Trained Immunity through the Mevalonate Pathway. Cell. (2018) 172:135–46.e9. doi: 10.1016/j.cell.2017.11.025
94. Benit P, Letouze E, Rak M, Aubry L, Burnichon N, Favier J, et al. Unsuspected task for an old team: succinate, fumarate and other Krebs cycle acids in metabolic remodeling. Biochim Biophys Acta. (2014) 1837:1330–7. doi: 10.1016/j.bbabio.2014.03.013
95. Wendeln AC, Degenhardt K, Kaurani L, Gertig M, Ulas T, Jain G, et al. Innate immune memory in the brain shapes neurological disease hallmarks. Nature. (2018) 556:332–8. doi: 10.1038/s41586-018-0023-4
96. McGuckin MA, Linden SK, Sutton P, Florin TH. Mucin dynamics and enteric pathogens. Nat Rev Microbiol. (2011) 9:265–78. doi: 10.1038/nrmicro2538
97. Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. (2003) 3:710–20. doi: 10.1038/nri1180
98. Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. (2006) 313:1126–30. doi: 10.1126/science.1127119
99. Mestecky J, Russell MW. Specific antibody activity, glycan heterogeneity and polyreactivity contribute to the protective activity of S-IgA at mucosal surfaces. Immunol Lett. (2009) 124:57–62. doi: 10.1016/j.imlet.2009.03.013
100. Castellaneta A, Sumpter TL, Chen L, Tokita D, Thomson AW. NOD2 ligation subverts IFN-α production by liver plasmacytoid dendritic cells and inhibits their T cell allostimulatory activity via B7-H1 up-regulation. J Immunol. (2009) 183:6922–32. doi: 10.4049/jimmunol.0900582
101. Mueller T, Terada T, Rosenberg IM, Shibolet O, Podolsky DK. Th2 cytokines down-regulate TLR expression and function in human intestinal epithelial cells. J Immunol. (2006) 176:5805–14. doi: 10.4049/jimmunol.176.10.5805
102. Carvalho FA, Aitken JD, Gewirtz AT, Vijay-Kumar M. TLR5 activation induces secretory interleukin-1 receptor antagonist (sIL-1Ra) and reduces inflammasome-associated tissue damage. Mucosal Immunol. (2011) 4:102–11. doi: 10.1038/mi.2010.57
103. Cario E, Podolsky DK. Differential alteration in intestinal epithelial cell expression of toll-like receptor 3 (TLR3) and TLR4 in inflammatory bowel disease. Infect Immun. (2000) 68:7010–7. doi: 10.1128/IAI.68.12.7010-7017.2000
104. Wells JM. Immunomodulatory mechanisms of lactobacilli. Microb Cell Fact. (2011) 10(Suppl. 1):S17. doi: 10.1186/1475-2859-10-S1-S17
105. Santecchia I, Vernel-Pauillac F, Rasid O, Quintin J, Gomes-Solecki M, Boneca IG, et al. Innate immune memory through TLR2 and NOD2 contributes to the control of Leptospira interrogans infection. PLoS Pathog. (2019) 15:e1007811. doi: 10.1371/journal.ppat.1007811
106. Yao Y, Jeyanathan M, Haddadi S, Barra NG, Vaseghi-Shanjani M, Damjanovic D, et al. Induction of autonomous memory alveolar macrophages requires T cell help and is critical to trained immunity. Cell. (2018) 175:1634–50.e17. doi: 10.1016/j.cell.2018.09.042
107. Jackson EM, Herbst-Kralovetz MM. Intranasal vaccination with murabutide enhances humoral and mucosal immune responses to a virus-like particle vaccine. PLoS ONE. (2012) 7:e41529. doi: 10.1371/journal.pone.0041529
108. Honko AN, Sriranganathan N, Lees CJ, Mizel SB. Flagellin is an effective adjuvant for immunization against lethal respiratory challenge with Yersinia pestis. Infect Immun. (2006) 74:1113–20. doi: 10.1128/IAI.74.2.1113-1120.2006
109. Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. (2010) 2:a000414. doi: 10.1101/cshperspect.a000414
110. Irazoki O, Hernandez SB, Cava F. Peptidoglycan muropeptides: release, perception, and functions as signaling molecules. Front Microbiol. (2019) 10:500. doi: 10.3389/fmicb.2019.00500
111. Miao EA, Alpuche-Aranda CM, Dors M, Clark AE, Bader MW, Miller SI, et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol. (2006) 7:569–75. doi: 10.1038/ni1344
112. Levy M, Thaiss CA, Zeevi D, Dohnalova L, Zilberman-Schapira G, Mahdi JA, et al. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell. (2015) 163:1428–43. doi: 10.1016/j.cell.2015.10.048
113. Hajam IA, Dar PA, Shahnawaz I, Jaume JC, Lee JH. Bacterial flagellin-a potent immunomodulatory agent. Exp Mol Med. (2017) 49:e373. doi: 10.1038/emm.2017.172
114. Kinnebrew MA, Buffie CG, Diehl GE, Zenewicz LA, Leiner I, Hohl TM, et al. Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity. (2012) 36:276–87. doi: 10.1016/j.immuni.2011.12.011
115. Garcia-Valtanen P, Guzman-Genuino RM, Williams DL, Hayball JD, Diener KR. Evaluation of trained immunity by β-1, 3 (d)-glucan on murine monocytes in vitro and duration of response in vivo. Immunol Cell Biol. (2017) 95:601–10. doi: 10.1038/icb.2017.13
116. Schorey JS, Lawrence C. The pattern recognition receptor Dectin-1: from fungi to mycobacteria. Curr Drug Targets. (2008) 9:123–9. doi: 10.2174/138945008783502430
117. Chow JC, Young DW, Golenbock DT, Christ WJ, Gusovsky F. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J Biol Chem. (1999) 274:10689–92. doi: 10.1074/jbc.274.16.10689
118. Williams AE, Edwards L, Humphreys IR, Snelgrove R, Rae A, Rappuoli R, et al. Innate imprinting by the modified heat-labile toxin of Escherichia coli (LTK63) provides generic protection against lung infectious disease. J Immunol. (2004) 173:7435–43. doi: 10.4049/jimmunol.173.12.7435
119. Breyne K, Steenbrugge J, Demeyere K, Vanden Berghe T, Meyer E. Preconditioning with lipopolysaccharide or lipoteichoic acid protects against Staphylococcus aureus mammary infection in mice. Front Immunol. (2017) 8:833. doi: 10.3389/fimmu.2017.00833
120. Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. (2005) 308:1635–8. doi: 10.1126/science.1110591
121. Frosali S, Pagliari D, Gambassi G, Landolfi R, Pandolfi F, Cianci R. How the intricate interaction among toll-like receptors, microbiota, and intestinal immunity can influence gastrointestinal pathology. J Immunol Res. (2015) 2015:489821. doi: 10.1155/2015/489821
122. Vijay-Kumar M, Wu H, Aitken J, Kolachala VL, Neish AS, Sitaraman SV, et al. Activation of toll-like receptor 3 protects against DSS-induced acute colitis. Inflamm Bowel Dis. (2007) 13:856–64. doi: 10.1002/ibd.20142
123. Kawashima T, Kosaka A, Yan H, Guo Z, Uchiyama R, Fukui R, et al. Double-stranded RNA of intestinal commensal but not pathogenic bacteria triggers production of protective interferon-β. Immunity. (2013) 38:1187–97. doi: 10.1016/j.immuni.2013.02.024
124. Furuta T, Kikuchi T, Akira S, Watanabe N, Yoshikawa Y. Roles of the small intestine for induction of toll-like receptor 4-mediated innate resistance in naturally acquired murine toxoplasmosis. Int Immunol. (2006) 18:1655–62. doi: 10.1093/intimm/dxl099
125. Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. (2001) 410:1099–103. doi: 10.1038/35074106
126. Uematsu S, Jang MH, Chevrier N, Guo Z, Kumagai Y, Yamamoto M, et al. Detection of pathogenic intestinal bacteria by Toll-like receptor 5 on intestinal CD11c+ lamina propria cells. Nat Immunol. (2006) 7:868–74. doi: 10.1038/ni1362
127. Vilaysane A, Muruve DA. The innate immune response to DNA. Semin Immunol. (2009) 21:208–14. doi: 10.1016/j.smim.2009.05.006
128. Gribar SC, Sodhi CP, Richardson WM, Anand RJ, Gittes GK, Branca MF, et al. Reciprocal expression and signaling of TLR4 and TLR9 in the pathogenesis and treatment of necrotizing enterocolitis. J Immunol. (2009) 182:636–46. doi: 10.4049/jimmunol.182.1.636
129. Lee J, Mo JH, Katakura K, Alkalay I, Rucker AN, Liu YT, et al. Maintenance of colonic homeostasis by distinctive apical TLR9 signalling in intestinal epithelial cells. Nat Cell Biol. (2006) 8:1327–36. doi: 10.1038/ncb1500
130. Palm NW, Medzhitov R. Pattern recognition receptors and control of adaptive immunity. Immunol Rev. (2009) 227:221–33. doi: 10.1111/j.1600-065X.2008.00731.x
131. Keestra AM, Winter MG, Auburger JJ, Frassle SP, Xavier MN, Winter SE, et al. Manipulation of small Rho GTPases is a pathogen-induced process detected by NOD1. Nature. (2013) 496:233–7. doi: 10.1038/nature12025
132. Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, Moran AP, et al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol. (2004) 5:1166–74. doi: 10.1038/ni1131
133. Sahoo M, Ceballos-Olvera I, del Barrio L, Re F. Role of the inflammasome, IL-1β, and IL-18 in bacterial infections. ScientificWorldJournal. (2011) 11:2037–50. doi: 10.1100/2011/212680
134. Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. (2010) 16:228–31. doi: 10.1038/nm.2087
135. Finlay CM, Stefanska AM, Walsh KP, Kelly PJ, Boon L, Lavelle EC, et al. Helminth products protect against autoimmunity via innate type 2 cytokines IL-5 and IL-33, which promote eosinophilia. J Immunol. (2016) 196:703–14. doi: 10.4049/jimmunol.1501820
136. Soehnlein O, Lindbom L. Phagocyte partnership during the onset and resolution of inflammation. Nat Rev Immunol. (2010) 10:427–39. doi: 10.1038/nri2779
137. MacVittie TJ, Walker RI. Canine granulopoiesis: alterations induced by suppression of gram-negative flora. Exp Hematol. (1978) 6:639–47.
138. Chang CF, Pollard M. Effects of microbial flora on levels of colonay stimulating factor in serums of irradiated CFW mice. Proc Soc Exp Biol Med. (1973) 144:177–80. doi: 10.3181/00379727-144-37551
139. Joshi JH, Entringer MA, Robinson WA. Bacterial stimulation of serum colony-stimulating activity and neutrophil production in germ-free mice. Proc Soc Exp Biol Med. (1979) 162:44–7. doi: 10.3181/00379727-162-40615
140. Staber FG, Tarcsay L, Dukor P. Modulations of myelopoiesis in vivo by chemically pure preparations of cell wall components from gram-negative bacteria: effects at different stages. Infect Immun. (1978) 20:40–9.
141. Goris H, de Boer F, van der Waaij D. Myelopoiesis in experimentally contaminated specific-pathogen-free and germfree mice during oral administration of polymyxin. Infect Immun. (1985) 50:437–41
142. Nicaise P, Gleizes A, Sandre C, Forestier F, Kergot R, Quero AM, et al. Influence of intestinal microflora on murine bone marrow and spleen macrophage precursors. Scand J Immunol. (1998) 48:585–91. doi: 10.1046/j.1365-3083.1998.00487.x
143. Burgess SL, Buonomo E, Carey M, Cowardin C, Naylor C, Noor Z, et al. Bone marrow dendritic cells from mice with an altered microbiota provide interleukin 17A-dependent protection against Entamoeba histolytica colitis. MBio. (2014) 5:e01817. doi: 10.1128/mBio.01817-14
144. Walachowski S, Tabouret G, Fabre M, Foucras G. Molecular analysis of a short-term model of β-glucans-trained immunity highlights the accessory contribution of GM-CSF in priming mouse macrophages response. Front Immunol. (2017) 8:1089. doi: 10.3389/fimmu.2017.01089
145. Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. (2012) 336:86–90. doi: 10.1126/science.1219179
146. Hoeffel G, Ginhoux F. Ontogeny of tissue-resident macrophages. Front Immunol. (2015) 6:486. doi: 10.3389/fimmu.2015.00486
147. Sieweke MH, Allen JE. Beyond stem cells: self-renewal of differentiated macrophages. Science. (2013) 342:1242974. doi: 10.1126/science.1242974
148. Hergott CB, Roche AM, Tamashiro E, Clarke TB, Bailey AG, Laughlin A, et al. Peptidoglycan from the gut microbiota governs the lifespan of circulating phagocytes at homeostasis. Blood. (2016) 127:2460–71. doi: 10.1182/blood-2015-10-675173
149. Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells–a proposal for uniform nomenclature. Nat Rev Immunol. (2013) 13:145–9. doi: 10.1038/nri3365
150. Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. (2008) 14:282–9. doi: 10.1038/nm1720
151. Pickert G, Neufert C, Leppkes M, Zheng Y, Wittkopf N, Warntjen M, et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med. (2009) 206:1465–72. doi: 10.1084/jem.20082683
152. Sonnenberg GF, Monticelli LA, Elloso MM, Fouser LA, Artis D. CD4(+) lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity. (2011) 34:122–34. doi: 10.1016/j.immuni.2010.12.009
153. Lee JS, Cella M, McDonald KG, Garlanda C, Kennedy GD, Nukaya M, et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol. (2011) 13:144–51. doi: 10.1038/ni.2187
154. Minton K. ILC3s take control in small intestine. Nat Rev Immunol. (2019) 19:353. doi: 10.1038/s41577-019-0166-z
155. Miani M, Le Naour J, Waeckel-Enee E, Verma SC, Straube M, Emond P, et al. Gut microbiota-stimulated innate lymphoid cells support β-Defensin 14 expression in pancreatic endocrine cells, preventing autoimmune diabetes. Cell Metab. (2018) 28:557–572.e6. doi: 10.1016/j.cmet.2018.06.012
156. Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. (2008) 29:958–70. doi: 10.1016/j.immuni.2008.11.001
157. Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, Johner C, et al. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol. (2009) 10:83–91. doi: 10.1038/ni.1684
158. Vonarbourg C, Mortha A, Bui VL, Hernandez PP, Kiss EA, Hoyler T, et al. Regulated expression of nuclear receptor RORgammat confers distinct functional fates to NK cell receptor-expressing RORgammat(+) innate lymphocytes. Immunity. (2010) 33:736–51. doi: 10.1016/j.immuni.2010.10.017
159. Crellin NK, Trifari S, Kaplan CD, Satoh-Takayama N, Di Santo JP, Spits H. Regulation of cytokine secretion in human CD127(+) LTi-like innate lymphoid cells by Toll-like receptor 2. Immunity. (2010) 33:752–64. doi: 10.1016/j.immuni.2010.10.012
160. Goto Y, Kiyono H. Epithelial barrier: an interface for the cross-communication between gut flora and immune system. Immunol Rev. (2012) 245:147–63. doi: 10.1111/j.1600-065X.2011.01078.x
161. Levy M, Blacher E, Elinav E. Microbiome, metabolites and host immunity. Curr Opin Microbiol. (2017) 35:8–15. doi: 10.1016/j.mib.2016.10.003
162. Bouskra D, Brezillon C, Berard M, Werts C, Varona R, Boneca IG, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. (2008) 456:507–10. doi: 10.1038/nature07450
163. Bain CC, Mowat AM. Intestinal macrophages - specialised adaptation to a unique environment. Eur J Immunol. (2011) 41:2494–8. doi: 10.1002/eji.201141714
164. Varol C, Zigmond E, Jung S. Securing the immune tightrope: mononuclear phagocytes in the intestinal lamina propria. Nat Rev Immunol. (2010) 10:415–26. doi: 10.1038/nri2778
165. Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. (2009) 137:1343–55. doi: 10.1016/j.cell.2009.05.014
166. Donohoe DR, Garge N, Zhang X, Sun W, O'Connell TM, Bunger MK, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. (2011) 13:517–26. doi: 10.1016/j.cmet.2011.02.018
167. Kaiko GE, Ryu SH, Koues OI, Collins PL, Solnica-Krezel L, Pearce EJ, et al. The colonic crypt protects stem cells from microbiota-derived metabolites. Cell. (2016) 167:1137. doi: 10.1016/j.cell.2016.10.034
168. Peck BCE, Shanahan MT, Singh AP, Sethupathy P. Gut microbial influences on the mammalian intestinal stem cell niche. Stem Cells Int. (2017) 2017:5604727. doi: 10.1155/2017/5604727
169. Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. (2009) 23:2333–44. doi: 10.1101/gad.1827009
170. Cronin SJ, Nehme NT, Limmer S, Liegeois S, Pospisilik JA, Schramek D, et al. Genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science. (2009) 325:340–3. doi: 10.1126/science.1173164
Keywords: gut microbiota, macrophages, monocytes, innate immunity, innate memory
Citation: Negi S, Das DK, Pahari S, Nadeem S and Agrewala JN (2019) Potential Role of Gut Microbiota in Induction and Regulation of Innate Immune Memory. Front. Immunol. 10:2441. doi: 10.3389/fimmu.2019.02441
Received: 25 June 2019; Accepted: 01 October 2019;
Published: 25 October 2019.
Edited by:
Lia Danelishvili, Oregon State University, United StatesReviewed by:
Antonio Molinaro, University of Naples Federico II, ItalyRaphael Chevre, Ludwig Maximilian University of Munich, Germany
Copyright © 2019 Negi, Das, Pahari, Nadeem and Agrewala. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Javed N. Agrewala, amFncmV3YWxhQGlpdHJwci5hYy5pbg==
 Shikha Negi
Shikha Negi Deepjyoti Kumar Das
Deepjyoti Kumar Das Susanta Pahari
Susanta Pahari Sajid Nadeem
Sajid Nadeem Javed N. Agrewala
Javed N. Agrewala