- 1Anhui Province Key Laboratory of Major Autoimmune Diseases, Department of Epidemiology and Biostatistics, School of Public Health, Anhui Medical University, Hefei, China
- 2Department of Infectious Diseases, The First Affiliated Hospital of Anhui Medical University, Hefei, China
- 3Department of Rheumatology, The First Affiliated Hospital of University of Science and Technology of China, Hefei, China
Long non-coding RNAs (lncRNAs) are increasingly recognized to play important roles in multiple autoimmune diseases. This study aimed to evaluate the association of four lncRNAs (ANRIL, lnc-DC, MALAT1, ZFAS1) genes single nucleotide polymorphisms (SNPs) with susceptibility to rheumatoid arthritis (RA) patients, as well as their expression levels. Seventeen SNPs of the four lncRNAs were genotyped in a cohort of 660 RA patients and 710 controls using improved multiple ligase detection reaction (iMLDR). The lncRNAs expressions in peripheral blood mononuclear cells (PBMCs) from 120 RA patients and 120 controls were detected by qRT-PCR. No significant differences were found for the allele and genotype frequencies distribution of ANRIL SNPs (rs1412830, rs944796, rs61271866, rs2518723, rs3217992), lnc-DC SNPs (rs7217280, rs10515177), MALAT1 SNPs (rs619586, rs4102217, rs591291, rs11227209, rs35138901), ZFAS1 SNPs (rs237742, rs73116127, rs6125607, rs6125608) between RA patients and normal controls (all P > 0.05). The genotype effects of dominant and recessive models were also evaluated, but no significant association was found. In addition, our results demonstrated that the rs944796 G allele, rs2518723 T allele, rs3217992 T allele frequencies were significantly associated with anti-CCP in RA patients (all P < 0.05). The haplotype CGTA frequency for ZFAS1 was significantly higher in RA patients (P = 0.036). Compared with normal controls, the expression levels of ANRIL, lnc-DC, MALAT1, ZFAS1 in PBMCs were significantly reduced in RA patients (all P < 0.001). Moreover, ZFAS1 expression was negatively associated with CRP in RA patients (P = 0.002). In summary, ANRIL, lnc-DC, MALAT1, and ZFAS1 genes SNPs were not associated with RA susceptibility, while altered ANRIL, lnc-DC, MALAT1, ZFAS1 levels in RA patients suggested that these lncRNAs might play a role in RA.
Introduction
Rheumatoid arthritis (RA) is known as a common autoimmune, inflammation disease characterized by systemic manifestations of immune and inflammatory response including marginal bone erosion, inflammatory joint fluid, synovitis, and destruction of articular cartilage (1, 2). Several researches have indicated that the incidences of RA in different ethnic groups, geographical areas are different, and the RA prevalence is approximately 1% around the world (3, 4). It has been revealed that genetic susceptibilities, abnormal metabolic enzymes, aberrant immune response are involved in RA development (5, 6). Latterly, a number of single nucleotide polymorphisms (SNPs), the majority of which are located in the non-coding intervals, are gradually identified to be associated with the susceptibility of this disease according to genome-wide association studies (7, 8).
Long non-coding RNAs (lncRNAs), which are defined as RNAs longer than 200 nucleotides in length, have no or little protein-coding capacity (9). LncRNAs are reported to be involved in a variety of autoimmunity- and inflammation- related processes, and regulate gene expression in multiple mechanisms including alternative splicing, epigenetics, small RNA sponging (10, 11). Increasing studies have been performed to explore the potential role of lncRNAs on the pathogenesis of autoimmune diseases, such as RA and systemic lupus erythematosus (SLE) (12–14). Our recent study demonstrated that the lnc0640, lnc5150 expression levels were alternated among RA patients, and lnc0640 rs13039216 TT genotype was statistically associated with RA susceptibility (14). Another previous study suggested that aberrant lncRNA expression level in peripheral blood mononuclear cells (PBMCs) could be a potential biomarker for RA diagnosing (13).
Recently, lncRNA ANRIL (antisense non-coding RNA in the INK4 locus) had attracted attention in autoimmune diseases, as it had been implicated in regulation of immune, inflammatory response (15). ANRIL expression was found to be regulated through STAT1 signaling pathway, which participated in immune regulation by induction of the pro-inflammatory cytokine TNF-γ (16). In addition, another study indicated that ANRIL expression level in PBMCs was decreased in RA by lncRNA array (13). Dendritic cell (DC) was a specific antigen presenting cell which link the innate and adaptive immune responses, and was thought to drive the activation of self-peptide-reactive inflammatory T cells, follicular helper T cells and consequently B cells for secreting autoantibodies in RA (17). Lnc-DC was a specialized, highly expressed lncRNA in DCs, and had the ability to regulate Th17 differentiation, DCs to stimulate T cell activation, and the production of interleukin 12 (IL-12) (9). LncRNA MALAT1 (metastasis-associated lung adenocarcinoma transcript-1) had been shown to play a role in the development of autoimmune diseases. SLE patients had increased MALAT1 level in PBMCs compared with normal individuals, and knockdown of MALAT-1 significantly suppressed IL-21 level in monocytes (18). In RA, Pan et al. found that knockdown of MALAT1 could inhibit the apoptosis of fibroblast-like synoviocytes (FLS) and lead to the activation of phosphoinositide 3-kinase (PI3K)/AKT signaling pathway (19). In another study, lncRNA ZFAS1 (zinc finger antisense 1) was shown to participate in RA-FLS migration and invasion by interacting with miR-27a and suppressing miR-27a expression, and ZFAS1 expression level was statistically evaluated in FLS of RA patients (20).
These studies demonstrated that ANRIL, lnc-DC, MALAT1, ZFAS1 might be involved in the occurrence and development of RA. However, no studies regarding the relationship between these lncRNAs genetic variation and RA have been reported. Thus, in the present study, we explored the associations of these lncRNAs genes SNPs with RA risk, as well as these lncRNAs expressions in PBMCs of RA patients and normal controls.
Materials and Methods
Patients and Normal Controls
In this study, case-control studies were performed in unrelated ethnic Han Chinese population. A total of 1,370 subjects including 660 RA patients and 710 normal controls were consecutively enrolled to investigate the association between ANRIL, lnc-DC, MALAT1, ZFAS1 genes polymorphisms and RA susceptibility. Then, 120 RA patients and 120 normal controls were included to detect these lncRNAs expression levels. RA patients were selected from the First Affiliated Hospital of University of Science and Technology of China, and the First Affiliated Hospital of Anhui Medical University. The diagnosis of these patients was according to the 1987 American College of Rheumatology revised criteria (21). The normal controls, who were recruited from the healthy blood donors in the same region, did not have no a history of RA, or other inflammatory/autoimmune diseases, cancer. Disease Activity Score 28 (DAS 28) was used to evaluate RA disease activity (22). The demographic data of all subjects were collected, and the following clinical data of RA patients were retrieved from the medical records: complements 3 (C3), complements 4 (C4), erythrocyte sedimentation rate (ESR), C-reactive protein (CRP), anti-cyclic citrullinated peptide (anti-CCP), and rheumatoid factor (RF). After informed consent was obtained, peripheral blood samples and data were collected from RA patients and normal controls. This study protocol was approved by the Medical Ethics Committee of Anhui Medical University.
SNP Selection, DNA Extraction, and Genotyping
The genetic and location information were obtained from two public databases, LNCipedia.org (v4.0) and Genome Browser Gateway (UCSC). We selected the tagSNPs with a minor allele frequency (MAF) ≥ 0.05 in CHB capturing all the common SNPs located in the chromosome locus transcribed into these lncRNAs (ANRIL, lnc-DC, MALAT1, ZFAS1) and their flanking 2,000 bp region through using genotype data of Han Chinese in Beijing from Ensembl genome browser 85 and CHBS_1000 g. The selection was conducted through linkage disequilibrium (LD) analysis with r2 threshold > 0.8 by using Haploview 4.0 software (Cambridge, MA, USA). In addition, the existing studies about these lncRNA genes polymorphisms were also reviewed. Finally, we selected six tagSNPs (rs1412830, rs7044859, rs944796, rs61271866, rs2518723, rs3217992) in ANRIL, two tagSNPs (rs7217280, rs10515177) in lnc-DC, five tagSNPs (rs619586, rs4102217, rs591291, rs11227209, rs35138901) in MALAT1, four tagSNPs (rs237742, rs73116127, rs6125607, rs6125608) in ZFAS1 for genotyping in the present study.
The genomic DNA was extracted from the peripheral blood leukocytes by the Flexi Gene-DNA Kit (Qiagen, Valencia, CA). Improved multiple ligase detection reaction (iMLDR) genotyping assay, with technical support from the Center for Genetic & Genomic Analysis, Genesky Biotechnologies (Inc., Shanghai), was used for genotyping. Those individuals with 100% genotyping success rate for the above SNPs were included for final analysis.
Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
PBMCs were isolated from 5 ml anticoagulated peripheral blood, and stored at −80°C until processed. Total RNA in PBMCs was extracted with TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) and the concentration of RNA was quantified using NanoDrop 2000 spectrophotometer (Thermo Scientific, USA). Then, the PrimeScriptTM RT reagent Kit (Takara Bio Inc., Japan) was used to reverse-transcribed total RNA into cDNA.
The expression levels of ANRIL, lnc-DC, MALAT1, ZFAS1 in PBMCs were detected by qRT-PCR with SYBR Green (SYBR Premix Ex Taq II, Takara Bio Inc., Japan). This experiment was performed on QuantStudio 12K Flex Real-Time PCR System (Applied Biosystems, Foster City, CA, USA), and according to the following cycle conditions: 95°C for 1 min, followed by 42 cycles at 95°C for 10 s, 60°C for 30 s and 72°C for 1 min. The relative expression levels of lncRNAs were calculated by comparison with housekeeping gene β-actin in the same sample as internal control, and expressed using 2−ΔΔCt method normalized to endogenous control (23).
Statistical Analysis
Statistical analysis was performed with the SPSS 23.0 (SPSS Inc., IL, USA). We performed Hardy-Weinberg equilibrium test by Chi-square (χ2) among normal controls. For the associations of genotype, allele distribution frequencies of each SNP with RA were estimated by logistic regression analyses. The lncRNAs levels were shown as median value and interquartile range, and the differences in lncRNAs levels between two groups, three groups were analyzed by Mann-Whitney U-test, Kruskal-Wallis H-test, respectively. The correlations between lncRNAs levels and several experimental indexes of RA patients were analyzed by Spearman rank correlation coefficient test. Dominant model, recessive mode was used for statistical analysis, and haplotype analysis was conducted with the SHEsis software (24). P value (two-sided) <0.05 was considered as statistically significant. False discovery rate (FRD) was used for multiple testing in SNP analysis.
Results
Association of lncRNAs Genes Polymorphisms With RA Susceptibility
We included 546 females and 114 males in RA patients for genotyping with a median age of 51, while there were 574 females and 136 males with a median age of 49 in normal controls. The observed genotype frequencies of rs7044859 was not conform to Hardy-Weinberg equilibrium, thus we excluded this SNP in finally analysis. The results of allele and genotype frequencies of all SNPs were summarized in Table 1.
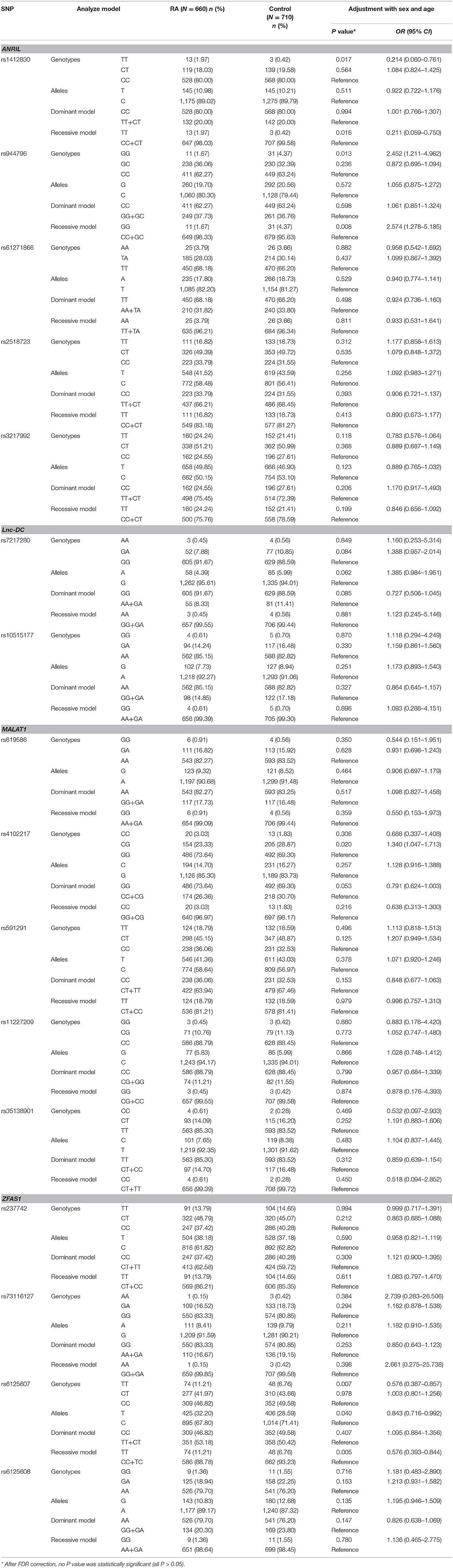
Table 1. Genotypes and alleles frequencies of lncRNAs genes polymorphisms in RA patients and normal controls.
In ANRIL gene, the rs1412830 TT genotype frequency was significantly increased in RA patients in comparison to normal controls, while the rs944796 GG genotype frequency was significantly decreased (TT vs. CC: P = 0.017; GG vs. CC: P = 0.013, respectively). In addition, an increased risk of rs1412830 variant, as well as a decreased risk of rs944796, was observed under the recessive model (TT vs. CC+CT: P = 0.016; GG vs. CC+GC: P = 0.008, respectively). However, these significant associations were disappeared after multiple testing by FDR correction (all P > 0.05). Comparing the genotype and allele frequencies of the ZFAS1 rs6125607 polymorphism among RA patients and normal controls, we found that TT genotype and T allele frequencies were significantly higher in RA patients than normal controls (TT vs. CC: P = 0.007; T vs. C: P = 0.040, respectively), and an increased risk of rs6125607 polymorphism existed in recessive model (TT vs. CC+TC: P = 0.005). After FDR correction, these differences were not statistically significant (TT vs. CC: P = 0.181; T vs. C: P = 0.496, TT vs. CC+TC: P = 0.080, respectively). Similarly, no significant associations between lnc-DC, MALAT1 genes polymorphism and RA susceptibility were found (Table 1).
To examine the potential genetic association between the genotype, allele frequencies of ANRIL, lnc-DC, MALAT1, ZFAS1 genes and anti-CCP, RF in RA patients, we performed a case-only analysis (Table S1). In ANRIL gene, the rs944796 G allele, rs2518723 T allele frequencies were significantly increased in RA patients with anti-CCP-positive when compared to patients with anti-CCP-negative (all P < 0.05), while rs3217992 T allele frequency was reduced (P = 0.039) (Table 2). No significant differences existed in allele and genotype frequencies of lnc-DC, MALAT1, ZFAS1 genes.
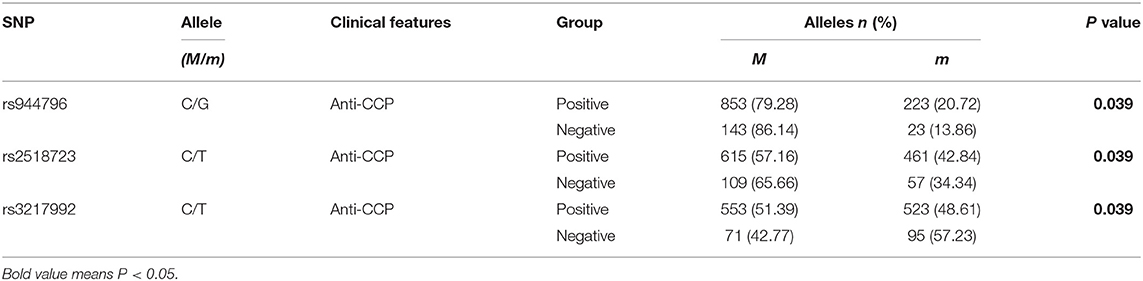
Table 2. The positive findings of associations between ANRIL gene polymorphisms and anti-CCP of RA patients.
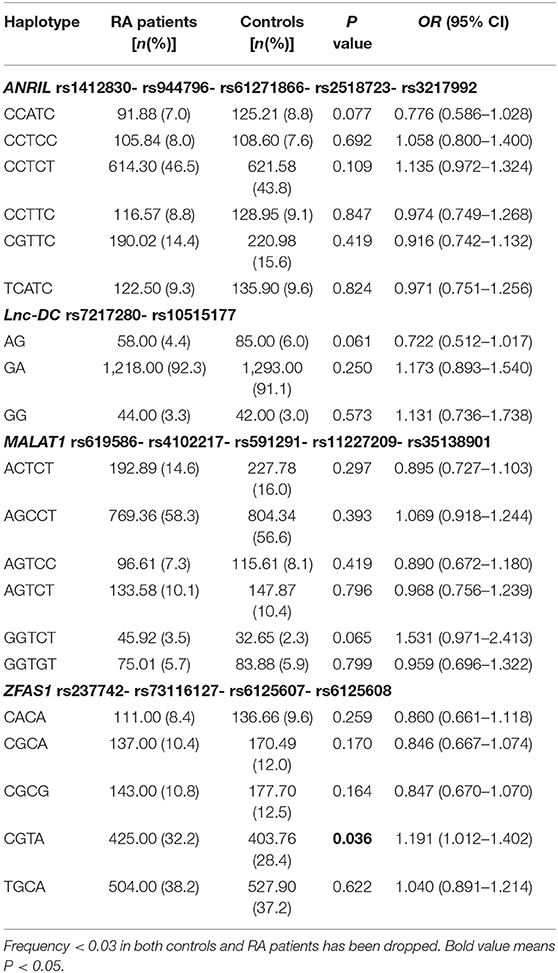
Haplotype Analysis
Six main haplotypes (CCATC, CCTCC, CCTCT, CCTTC, CGTTC, TCATC) for ANRIL, three main haplotypes (AG, GA, GG) for lnc-DC, six main haplotypes (ACTCT, AGCCT, AGTCC, AGTCT, GGTCT, GGTGT) for MALAT1 and five main haplotypes (CACA, CGCA, CGCG, CGTA, TGCA) for ZFAS1 were detected by SHEsis software (Table 3). The results demonstrated that the haplotype CGTA frequency was significantly higher in RA patients than normal controls (OR = 1.191, 95% CI: 1.012–1.402, P = 0.036).
LncRNAs Expression Levels in PBMCs From RA Patients and Normal Controls
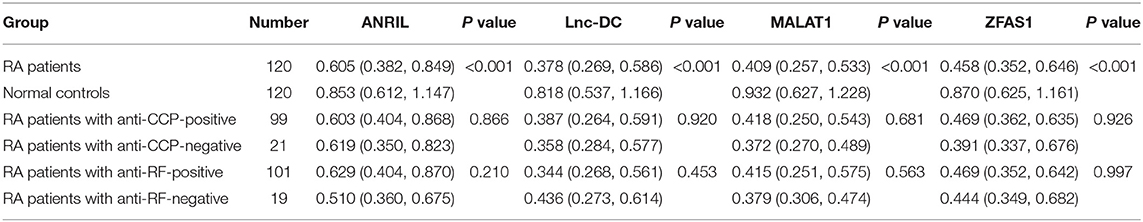
We further analyzed the association of ANRIL, lnc-DC, MALAT1, ZFAS1 levels with RA patients by qRT-PCR. As shown in Table 4, the expression levels of ANRIL, lnc-DC, MALAT1, ZFAS1 in PBMCs were significantly reduced in RA patients than normal controls (all P < 0.001). However, the differences in these lncRNAs levels between anti-CCP-positive RA patients and anti-CCP-negative RA patients, as well as RA patients with RF-positive and RF-negative RA patients, were not statistically significant.
The correlation of ANRIL, lnc-DC, MALAT1, ZFAS1 expression levels with clinical parameters, disease activity of RA patients were also analyzed, and the results shown that the expression level of ZFAS1 was negatively associated with CRP in RA patients (P = 0.002). However, there were no significant correlations of these lncRNAs levels with DAS28 of RA patients (Table 5). The potential influence of main medical therapies including glucocorticoids, disease-modifying antirheumatic drugs (DMARDs), biologics on lncRNAs expression levels in RA patients were assessed in this study. Similarly, no significant association was found (Table 6).
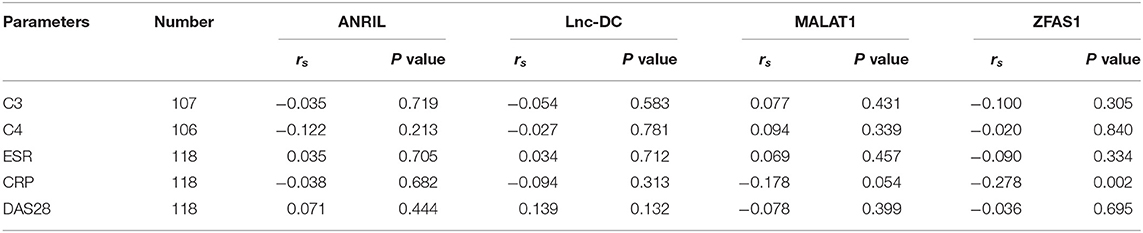
Table 5. Association of lncRNAs expression levels with clinical parameters, disease activity of RA patients.
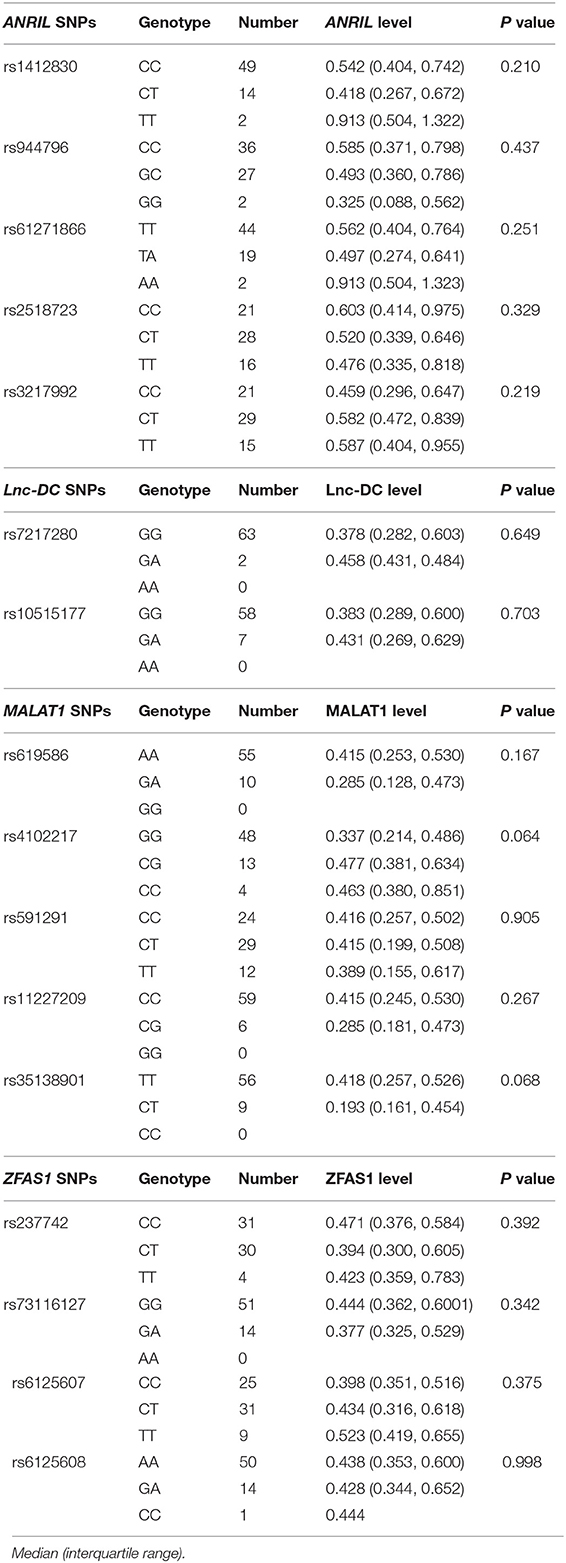
Associations Between lncRNAs Genes Polymorphisms With Their Levels in RA Patients
To examine the associations between the respective genotype frequencies of these lncRNAs genes with their expression levels in RA patients, we included 65 patients for analysis. However, there were no significant differences regarding these lncRNAs expression levels between their disparate genotypes of RA patients (Table 7).
Discussion
To date, the exact pathogenic mechanism of RA remains largely unknown, although several pro-inflammatory cytokines including tumor necrosis factor-α (TNF-α), IL-6, IL-1b have been reported to related to the occurrence of RA (25–27). Previous studies have shown that lncRNAs had distinct and specific roles in the activation and differentiation modulating of immune cell, and lncRNAs played an important role in autoimmune diseases (28). A study detected lncRNA transcription in CD14+ monocytes isolated from peripheral blood cells of RA patients before and after anti-IL-6R (tocilizumab) or anti-TNF-α (adalimumab) therapy by a microarray-based experiment. They observed that 7,419 lncRNAs expression levels were altered by either IL-6 or TNF-α, 85 of which exhibited were significant changed (29). These results suggested that lncRNAs were very important in the molecular pathophysiology of RA. In the present study, our results demonstrated that lower expression levels of ANRIL, lnc-DC, MALAT1, ZFAS1 existed in RA patients, and ANRIL, lnc-DC, MALAT1, ZFAS1 genes were not related to RA susceptibility.
ANRIL gene was located in the chromosome 9p21 region, and it was the well-defined genetic risk locus related to several diseases such as coronary artery disease (CAD), diabetes, and breast cancer (30–32). Our results implied that ANRIL rs1412830, rs944796 variant might associated with RA susceptibility, while the significant associations were disappeared after multiple testing. However, we found that ANRIL rs944796 G, rs2518723 T, rs3217992 T allele frequencies were significantly associated with anti-CCP in RA patients, this suggested to us that ANRIL gene variation might be involved in the RA development. In addition, disease-associated SNPs resided in this region had been reported to change the expression of ANRIL, demonstrating that altered ANRIL expression might be involved in predisposition to these disorders (33). Two SNPs (rs10757278 and rs1333045) in ANRIL, which had been highlighted as potential causal variants for the association with CAD, were reported to be associated with abnormal ANRIL expression level in Peripheral blood (34, 35). Moreover, our results demonstrated that compared with normal controls, ANRIL expression level was significantly decreased in PBMC from RA patients. We further explored the influence of the five SNPs on ANRIL level in PBMC from RA patients, unfortunately, there were no significant differences regarding ANRIL level between disparate genotypes of these SNPs.
In a previous study, the authors discovered a new lncRNA (named lnc-DC) located on chromosome 17 region, which near signal transducer and activator of transcription 3 (STAT3) gene (36). There were increasing researches to explore the contribution of lnc-DC in autoimmune diseases. Shaker et al. found that serum level of lnc-DC in multiple sclerosis (MS) patients were significantly increased, and serum lnc-DC level maybe used to as a potential novel biomarkers for MS diagnosis (37). One of our recent studies shown that the lnc-DC expression level was significantly decreased in PBMCs from SLE patients than controls, while lnc-DC rs10515177 variant was not associated with SLE susceptibility (38). Similarly, the lnc-DC expression level was significantly lower in PBMC from RA patients than normal controls in the present study. We also explore the potential association of two SNP (rs7217280, rs10515177) in lnc-DC with RA susceptibility, however, no significant relationship was found. Our study provided the first evidence that lnc-DC might be involved in the development of RA, and the specific roles of lnc-DC genetic variation in pathophysiology of RA need to be further explored.
MALAT1 was expressed on chromosome 11q13, and widely expressed in multiple normal tissues such as reproductive, endocrine and immune systems with an important role in autoimmune diseases including RA, SLE, MS (18, 19, 37, 39). Quercetin is a dietary antioxidant, which has been shown to be effective in the treatment of arthritis in pre-clinical studies, and Pan et al. tried to analyze the mechanisms responsible for the quercetin-induced FLS apoptosis in RA patients (19). Their data indicated that quercetin induced FLS apoptosis in RA patients via upregulating MALAT1, and MALAT1 promoted apoptosis by inhibiting the activation of the phosphoinositide 3-kinase (PI3K)/AKT pathway. In this study, decreased MALAT1 expression level in PBMC from RA patients was firstly reported, while MALAT1 genetic variation was not correlated with RA risk. Our findings provided new evidence that MALAT1 might be involved in RA development.
ZFAS1, located at chromosomal band 20q13.13, was reported as an important player to regulate the development of human cancers including glioma, lung, ovary, gastric, and breast cancer (40–42). In addition, ZFAS1 was found to promote chondrocytes proliferation, migration, and inhibit apoptosis and matrix synthesis in osteoarthritis (OA), and ZFAS1 expression level was downregulated in OA chondrocytes in comparison to mild chondrocytes (43). Another study by Xiao et al. also found more than five times ZFAS1 level in the healthy appearing area of cartilage compared with the pathology area in human knee osteoarthritis (44). Similarly, our results demonstrated that the expression of ZFAS1 in PBMCs was significantly reduced in RA patients than normal controls, and associated with CRP in RA. In the present study, we also analyzed the potential relationship between rs237742, rs73116127, rs6125607, rs6125608 variants in ZFAS1 and genetic susceptibility to RA, and no difference achieved statistical significance. However, haplotype analysis implied that the haplotype CGTA frequency for ZFAS1 was significantly higher in RA patients than normal controls. These founding would help improve our understanding of the roles of ZFAS1 genetic variants in the pathogenesis of RA.
In conclusion, our study provided the first evidence that ANRIL, lnc-DC, MALAT1, ZFAS1 genes polymorphisms might not be associated with RA susceptibility in the Chinese population. However, several SNPs in ANRIL were related to anti-CCP in RA. In addition, alternations of ANRIL, lnc-DC, MALAT1, ZFAS1 levels and significant correlations of ZFAS1 level with CRP in RA patient demonstrated that these lncRNAs might be regarded as an auxiliary biomarker for RA diagnosis, as well as used to distinguish RA serotypes.
However, some limitations existed in our study should be acknowledged. Firstly, this study does not eliminate the potential influence of ethnic background, environmental factor. Secondly, we are not able to assess the associations between these lncRNAs levels and disease severity, clinical variables, and therapeutic schedule of RA patients over a long period in this case-control study. Finally, our sample size may not be sufficient, and lead to the low power of this study. Hence, replication studies with larger sample size, different ethnic populations are awaited to further explore the exact role of these lncRNAs in RA.
Ethics Statement
This study was approved by the Ethical Committee of Anhui Medical University (Hefei, Anhui, China). All the study subjects provided informed consent to participate in this study. All studies on humans described in the present manuscript were carried out with the approval of the responsible ethics committee and in accordance with national law and the Helsinki Declaration of 1975 (in its current, revised form).
Author Contributions
T-PZ, H-FP, and D-QY designed the study. T-PZ and B-QZ conducted the experiment. S-ST and Y-GF performed the statistical analyses. T-PZ drafted the manuscript. X-ML participated in the collection of samples. H-FP and D-QY contributed to manuscript revision. All the authors approved the final submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81872687, 81573222).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.02529/full#supplementary-material
References
1. Cooles FA, Isaacs JD. Pathophysiology of rheumatoid arthritis. Curr Opin Rheumatol. (2011) 23:233–40. doi: 10.1097/BOR.0b013e32834518a3
2. Hötker U, Henniger M, Rehart S. Perioperative management in patients with rheumatoid arthritis. Aktuelle Rheumatologie. (2016) 41:448–54. doi: 10.1055/s-0042-112358
3. Pan HF, Leng RX, Wu GC, Ye DQ. Advance in epidemiologic studies on major autoimmune diseases. Chin J Dis Control Prev. (2018) 22:1093–5. doi: 10.16462/j.cnki.zhjbkz.2018.11.001
4. Rossini M, Rossi E, Bernardi D, Viapiana O, Gatti D, Idolazzi L, et al. Prevalence and incidence of rheumatoid arthritis in Italy. Rheumatol Int. (2014) 34:659–64. doi: 10.1007/s00296-014-2974-6
5. Taneja V. Cytokines pre-determined by genetic factors are involved in pathogenesis of Rheumatoid arthritis. Cytokine. (2015) 75:216–21. doi: 10.1016/j.cyto.2014.11.028
6. Zhang TP, Lv TT, Xu SZ, Pan HF, Ye DQ. Association of interleukin-10 gene single nucleotide polymorphisms with rheumatoid arthritis in a Chinese population. Postgrad Med J. (2018) 94:284–8. doi: 10.1136/postgradmedj-2017-135441
7. Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. (2010) 42:508–14. doi: 10.1038/ng.582
8. Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. (2014) 506:376–81. doi: 10.1038/nature12873
9. Zhao CN, Mao YM, Liu LN, Li XM, Wang DG, Pan HF. Emerging role of lncRNAs in systemic lupus erythematosus. Biomed Pharmacother. (2018) 106:584–92. doi: 10.1016/j.biopha.2018.06.175
10. Özgür E, Mert U, Isin M, Okutan M, Dalay N, Gezer U. Differential expression of long non-coding RNAs during genotoxic stress-induced apoptosis in HeLa and MCF-7 cells. Clin Exp Med. (2013) 13:119–26. doi: 10.1007/s10238-012-0181-x
11. Li Z, Li X, Jiang C, Qian W, Tse G, Chan MTV, et al. Long non-coding RNAs in rheumatoid arthritis. Cell Prolif. (2018) 51:e12404. doi: 10.1111/cpr.12404
12. Xue Z, Cui C, Liao Z, Xia S, Zhang P, Qin J, et al. Identification of LncRNA Linc00513 containing lupus-associated genetic variants as a novel regulator of interferon signaling pathway. Front Immunol. (2018) 9:2967. doi: 10.3389/fimmu.2018.02967
13. Song J, Kim D, Han J, Kim Y, Lee M, Jin EJ. PBMC and exosome-derived Hotair is a critical regulator and potent marker for rheumatoid arthritis. Clin Exp Med. (2015) 15:121–6. doi: 10.1007/s10238-013-0271-4
14. Zhang TP, Zhang Q, Wu J, Zhao YL, Wang JB, Leng RX, et al. The expression levels of long non-coding RNAs lnc0640 and lnc5150 and its gene single-nucleotide polymorphisms in rheumatoid arthritis patients. J Cell Biochem. (2018) 119:10095–106. doi: 10.1002/jcb.27346
15. Rezazadeh M, Gharesouran J, Moradi M, Noroozi R, Omrani MD, Taheri M, et al. Association study of ANRIL genetic variants and multiple sclerosis. J Mol Neurosci. (2018) 65:54–9. doi: 10.1007/s12031-018-1069-3
16. Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, Heintzman N, et al. 9p21 DNA variants associated with coronary artery disease impair interferon-γ signalling response. Nature. (2011) 470:264–8. doi: 10.1038/nature09753
17. Wehr P, Purvis H, Law SC, Thomas R. Dendritic cells, T cells and their interaction in rheumatoid arthritis. Clin Exp Immunol. (2019)196:12–27. doi: 10.1111/cei.13256
18. Yang H, Liang N, Wang M, Fei Y, Sun J, Li Z, et al. Long non-coding RNA MALAT-1 is a novel inflammatory regulator in human systemic lupus erythematosus. Oncotarget. (2017) 8:77400–6. doi: 10.18632/oncotarget.20490
19. Pan F, Zhu L, Lv H, Pei C. Quercetin promotes the apoptosis of fibroblast-like synoviocytes in rheumatoid arthritis by upregulating lncRNA MALAT1. Int J Mol Med. (2016) 38:1507–14. doi: 10.3892/ijmm.2016.2755
20. Ye Y, Gao X, Yang N. LncRNA ZFAS1 promotes cell migration and invasion of fibroblast-like synoviocytes by suppression of miR-27a in rheumatoid arthritis. Human Cell. (2018) 31:14–21. doi: 10.1007/s13577-017-0179-5
21. Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. (1988) 31:315–24. doi: 10.1002/art.1780310302
22. Turunen S, Koivula MK, Melkko J, Alasaarela E, Lehenkari P, Risteli J. Different amounts of protein-bound citrulline and homocitrulline in foot joint tissues of a patient with anti-citrullinated protein antibody positive erosive rheumatoid arthritis. J Transl Med. (2013) 11:224. doi: 10.1186/1479-5876-11-224
23. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 [-Delta Delta C(T)] method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
24. Li Z, Zhang Z, He Z, Tang W, Li T, Zeng Z, et al. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn). Cell Res. (2009) 19:519–23. doi: 10.1038/cr.2009.33
25. Yamanaka H. TNF as a target of inflammation in rheumatoid arthritis. Endocr Metab Immune Disord Drug Targets. (2015) 15:129–34. doi: 10.2174/1871530315666150316121808
26. Solus JF, Chung CP, Oeser A, Li C, Rho YH, Bradley KM, et al. Genetics of serum concentration of IL-6 and TNFα in systemic lupus erythematosus and rheumatoid arthritis: a candidate gene analysis. Clin Rheumatol. (2015) 34:1375–82. doi: 10.1007/s10067-015-2881-6
27. Jahid M, Rehan-Ul-Haq, Chawla D, Avasthi R, Ahmed RS. Association of polymorphic variants in IL1B gene with secretion of IL-1β protein and inflammatory markers in north Indian rheumatoid arthritis patients. Gene. (2018) 641:63–7. doi: 10.1016/j.gene.2017.10.051
28. Sigdel KR, Cheng A, Wang Y, Duan L, Zhang Y. The emerging functions of long non-coding RNA in immune cells: autoimmune diseases. J Immunol Res. (2015) 2015:848790. doi: 10.1155/2015/848790
29. Müller N, Döring F, Klapper M, Neumann K, Schulte DM, Türk K, et al. Interleukin-6 and tumour necrosis factor-alpha differentially regulate lincRNA transcripts in cells of the innate immune system in vivo in human subjects with rheumatoid arthritis. Cytokine. (2014) 68:65–8. doi: 10.1016/j.cyto.2014.03.004
30. Xie Y, Zhao D, Dong P, Wang H, Li D, Lai L. Effects of ANRIL polymorphisms on the likelihood of coronary artery disease: a meta-analysis. J Cell Biochem. (2018) 120:6113–9. doi: 10.1002/jcb.27898
31. Kong Y, Sharma RB, Nwosu BU, Alonso LC. Islet biology, the CDKN2A/B locus and type 2 diabetes risk. Diabetologia. (2016) 59:1579–93. doi: 10.1007/s00125-016-3967-7
32. Khorshidi HR, Taheri M, Noroozi R, Sarrafzadeh S, Sayad A, Ghafouri-Fard S. ANRIL genetic variants in Iranian breast cancer patients. Cell J. (2017) 19:72–8. doi: 10.22074/cellj.2017.4496
33. Cunnington MS, Santibanez Koref M, Mayosi BM, Burn J, Keavney B. Chromosome 9p21 SNPs associated with multiple disease phenotypes correlate with ANRIL expression. PLoS Genet. (2010) 6:e1000899. doi: 10.1371/journal.pgen.1000899
34. Jarinova O, Stewart AFR, Roberts R, Wells G, Lau P, Naing T, et al. Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arterioscler Thromb Vasc Biol. (2009) 29:1671–7. doi: 10.1161/ATVBAHA.109.189522
35. Liu Y, Sanoff HK, Cho H, Burd CE, Torrice C, Mohlke KL, et al. INK4/ARF transcript expression is associated with chromosome 9p21 variants linked to atherosclerosis. PLoS ONE. (2009) 4:e5027. doi: 10.1371/journal.pone.0005027
36. Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, et al. The STAT3-binding long non-coding RNA lnc-DC controls human dendritic cell differentiation. Science. (2014) 344:310–3. doi: 10.1126/science.1251456
37. Shaker OG, Mahmoud RH, Abdelaleem OO, Ibrahem EG, Mohamed AA, Zaki OM, et al. LncRNAs, MALAT1 and lnc-DC as potential biomarkers for multiple sclerosis diagnosis. Biosci Rep. (2018) 4:BSR20181335. doi: 10.1042/BSR20181335
38. Li J, Wu GC, Zhang TP, Yang XK, Chen SS, Li LJ, et al. Association of long non-coding RNAs expression levels and their gene polymorphisms with systemic lupus erythematosus. Sci Rep. (2017) 7:15119. doi: 10.1038/s41598-017-15156-4
39. Chen R, Liu Y, Zhuang H, Yang B, Hei K, Xiao M, et al. Quantitative proteomics reveals that long non-coding RNA MALAT1 interacts with DBC1 to regulate p53 acetylation. Nucleic Acids Res. (2017) 45:9947–59. doi: 10.1093/nar/gkx600
40. Liu F, Gao H, Li S, Ni X, Zhu Z. Long non-coding RNA ZFAS1 correlates with clinical progression and prognosis in cancer patients. Oncotarget. (2017) 8:61561–9. doi: 10.18632/oncotarget.18633
41. Dong D, Mu Z, Zhao C, Sun M. ZFAS1: a novel tumor-related long non-coding RNA. Cancer Cell Int. (2018) 18:125. doi: 10.1186/s12935-018-0623-y
42. Askarian-Amiri ME, Crawford J, French JD, Smart CE, Smith MA, Clark MB, et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. (2011) 17:878–91. doi: 10.1261/rna.2528811
43. Ye D, Jian W, Feng J, Liao X. Role of long non-coding RNA ZFAS1 in proliferation, apoptosis and migration of chondrocytes in osteoarthritis. Biomed Pharmacother. (2018) 104:825–31. doi: 10.1016/j.biopha.2018.04.124
Keywords: ANRIL, lnc-DC, MALAT1, ZFAS1, single nucleotide polymorphisms, rheumatoid arthritis
Citation: Zhang T-P, Zhu B-Q, Tao S-S, Fan Y-G, Li X-M, Pan H-F and Ye D-Q (2019) Long Non-coding RNAs Genes Polymorphisms and Their Expression Levels in Patients With Rheumatoid Arthritis. Front. Immunol. 10:2529. doi: 10.3389/fimmu.2019.02529
Received: 11 January 2019; Accepted: 11 October 2019;
Published: 31 October 2019.
Edited by:
David Stephen Pisetsky, Duke University, United StatesReviewed by:
Betty P. Tsao, Medical University of South Carolina, United StatesMohammad Taheri, Shahid Beheshti University of Medical Sciences, Iran
Ruiping Liu, Changzhou Second People's Hospital, China
Copyright © 2019 Zhang, Zhu, Tao, Fan, Li, Pan and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hai-Feng Pan, cGFuaGFpZmVuZzE5ODJAc2luYS5jb20=; cGFuaGFpZmVuZ0BhaG11LmVkdS5jbg==; Dong-Qing Ye, eWRxYWhtdUAxMjYuY29t; eWRxQGFobXUuZWR1LmNu
†These authors have contributed equally to this work as co-first authors
 Tian-Ping Zhang1,2†
Tian-Ping Zhang1,2† Hai-Feng Pan
Hai-Feng Pan Dong-Qing Ye
Dong-Qing Ye