- 1Unit of Clinical and Experimental Immunology, Humanitas Clinical and Research Center, Milan, Italy
- 2Department of Medical Biotechnologies and Translational Medicine (BioMeTra), University of Milan, Milan, Italy
In the context of allogeneic transplant platforms, human leukocyte antigen (HLA)-haploidentical hematopoietic stem cell transplantation (haplo-HSCT) represents one of the latest and most promising curative strategies for patients affected by high-risk hematologic malignancies. Indeed, this platform ensures a suitable stem cell source immediately available for virtually any patents in need. Moreover, the establishment in recipients of a state of immunologic tolerance toward grafted hematopoietic stem cells (HSCs) remarkably improves the clinical outcome of this transplant procedure in terms of overall and disease free survival. However, the HLA-mismatch between donors and recipients has not been yet fully exploited in order to optimize the Graft vs. Leukemia effect. Furthermore, the efficacy of haplo-HSCT is currently hampered by several life-threatening side effects including the onset of Graft vs. Host Disease (GvHD) and the occurrence of opportunistic viral infections. In this context, the quality and the kinetic of the immune cell reconstitution (IR) certainly play a major role and several experimental efforts have been greatly endorsed to better understand and accelerate the post-transplant recovery of a fully competent immune system in haplo-HSCT. In particular, the IR of innate immune system is receiving a growing interest, as it recovers much earlier than T and B cells and it is able to rapidly exert protective effects against both tumor relapses, GvHD and the onset of life-threatening opportunistic infections. Herein, we review our current knowledge in regard to the kinetic and clinical impact of Natural Killer (NK), γδ and Innate lymphoid cells (ILCs) IRs in both allogeneic and haplo-HSCT. The present paper also provides an overview of those new therapeutic strategies currently being implemented to boost the alloreactivity of the above-mentioned innate immune effectors in order to ameliorate the prognosis of patients affected by hematologic malignancies and undergone transplant procedures.
Introduction
Allogeneic (allo-) hematopoietic stem cell transplantation (HSCT) represents the best curative approach for patients affected by high-risk hematologic malignancies and several genetic disorders (1). In the absence of human leukocyte antigen (HLA)-identical siblings, HLA-haploidentical (haplo) related donors are a source of hematopoietic stem cells (HSCs) immediately available for almost any patients in need (2).
However, the first developed protocols of haplo-HSCT were mainly associated with graft rejection, high degree of treatment-related mortality (TRM) and severe graft-vs.-host-disease (GvHD) due to the partial HLA-mismatch between donors and recipients. This poor clinical outcome was also worsened by the increased risk of developing opportunistic infections, a phenomenon associated with a delayed immune-reconstitution (IR) following the transplant. On the other hand, HLA-mismatch remarkably boosted the so-called Graft-vs.-Leukemia (GvL) effect that eradicates those malignant cells surviving conditioning regimes (3, 4). Hence, the mechanisms inducing both GvHD and GvL rely on immunologic alloreactivity that, indeed, represents the bad and good side of the same coin in both allogeneic and haplo-HSCT. The possibility to improve GvL while limiting life-threatening side effects have firmly driven the development of new clinical protocols of haplo-HSCT delivering better clinical outcomes. In this context, a better understanding of both kinetics and mechanisms of IR is key to improve the prognosis of patients undergone haplo-HSCT and limit its side effects (5–13).
Several lines of evidences clearly showed that a full recovery of adaptive immune responses in transplanted patients take long time. Indeed, adaptive B- and T-cell effector-functions are either lacking or not completely competent for several months after haplo-HSCT, thus leaving the patients in a deadly condition of immune-deficiency. On the other hand, innate immune cells reconstitute early after haplo-HSCT, thus ensuring a certain degree of immune-protections in the first days/weeks after the transplant (3, 14). In particular, neutrophils and monocytes recirculate at levels similar to those of healthy individuals 1 month after the infusion of HSCs, while innate lymphocyte IR starts from the 2nd week after the transplant (15–17). Nonetheless, quite a few cell compartments of innate immunity are greatly impaired in their functions early after haplo-HSCT (18, 19). This scenario enforced the implementation of graft manipulations in allo-and haplo-HSCT setting (i.e., αβ T and/or B cell depletion) able to preserve Natural Killer (NK), gamma-delta (γδ) T and innate lymphoid (ILCs), thus avoiding a prolonged immune suppression and speeding their IR early after the transplant (Table 1) (12, 26–28). In particular, NK and γδ T cells have been shown to recover faster in those recipients receiving αβ T cell depleted grafts rather than the conventional CD34pos conventional counterparts in the context of the haplo-HSCT setting (25, 28, 29).
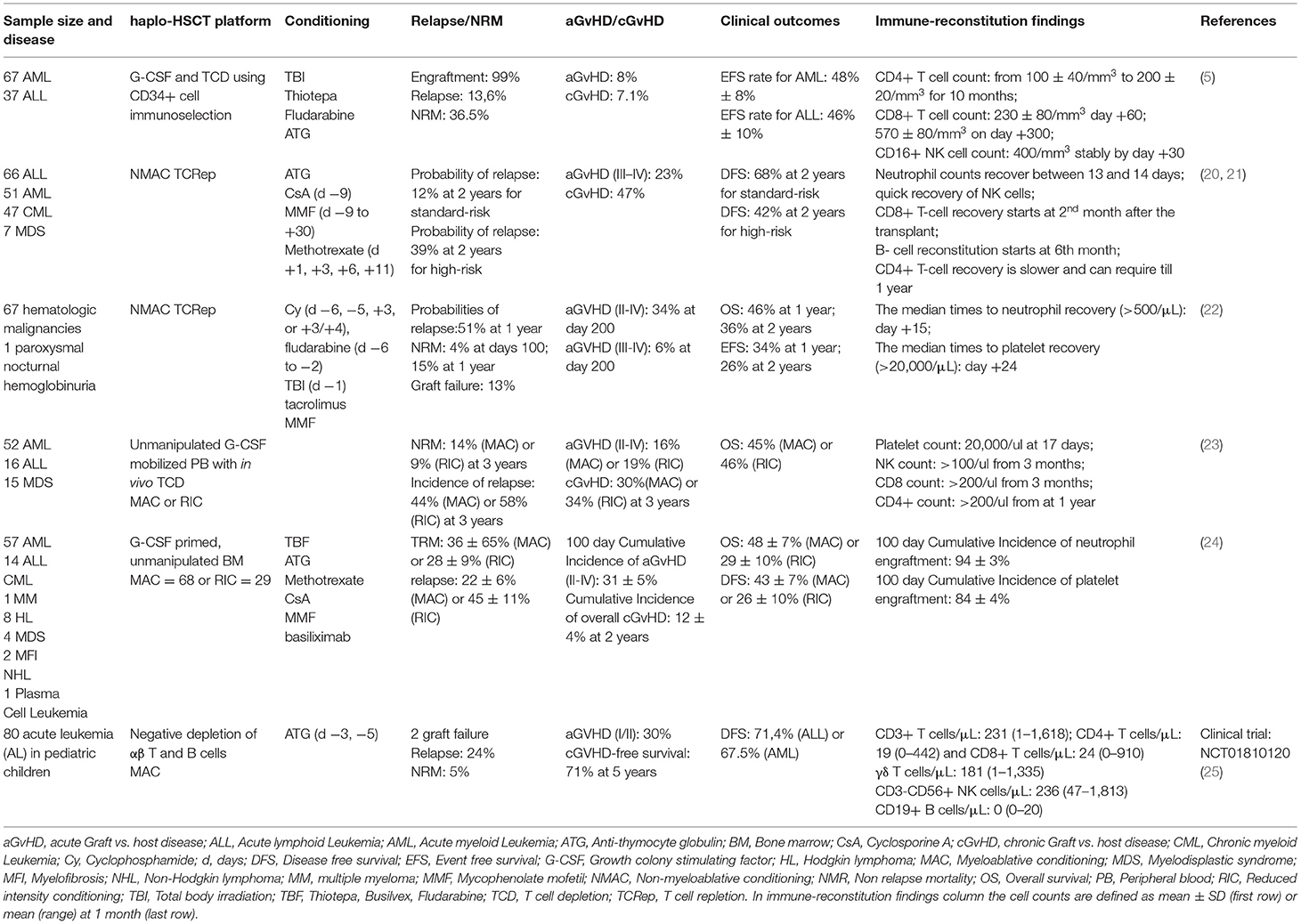
Table 1. Main results of different haplo-HSCT protocols with relative clinical outcomes and immunological recovery.
Graft vs. Host Diseases and Opportunistic Infections
One of the main complications affecting the positive clinical outcomes of allo-HSCT is still represented by the donor-derived alloreactive T cell responses against host tissues, a phenomenon inducing the onset of GvHD mainly affecting skin, gastrointestinal tract and liver (30, 31). Moreover, the different expression of tissue antigens between donors and recipients together with the clinical setting of induced immune-deficiency in recipients represent additional factors that remarkably worsen the impact of GvHD (32). In order to limit T cell alloreactivity, several haplo-HSCT platforms have been developed over the recent years (summarized in Table 1), including T-cell depleted (TCD) and T-cell replete (TCRep) approaches (5, 22, 25). Although the infusion of TCD grafts coupled with a mega-dose of CD34pos peripheral blood HSCs (on average 10 × 106 cells/kg body weight) ensures high engraftment rates associated with potent GvL effect and reduced GvHD, the small number of residual T lymphocytes administered in recipients are still able to induce high degrees of TRM and to delay IR with a subsequent increased rates of opportunistic infection onsets (5, 33). Hence, alternative and more efficient TCRep approaches able to better target alloreactive T cells have been developed in haplo-HSCT setting. These new protocols employ the infusion of high doses of post-transplant cyclophosphamide (PT-Cy), an immune-suppressant drug that is able to deplete in vivo all alloreactive and proliferating T cells (34). This new PT-Cy TCRep strategy showed since from the beginning very good clinical outcomes in term of engraftment, decreased GvHD and a faster kinetic of IR. Indeed, while donor T cell infused at the time of the transplant mediates a strong GvL in the first days soon after the administration of HSCs, the removal of those alloreactive and proliferating donor-derived T cells clones by PT-Cy limited the onset of GvHD afterward. These TCRep protocols have been then further optimized by infusing colony-stimulation factor (G-CSF)-primed grafts, by depleting in vivo selective T cell populations and by using a combination of other immune-suppressive agents (24, 35, 36).
Both the induced clinical condition of immune-deficiency early after allo- and haplo- HSCT and the delayed/aberrant IR facilitate the occurrence of opportunistic infections that greatly affect the quality and duration of life. Human cytomegalovirus (HCMV) is one of the most aggressive opportunistic microbes in allogeneic transplant including haplo-HSCT. Indeed, while HCMV infection is often asymptomatic or associated with mild flu-like symptoms in immune-competent hosts, its reactivation or de novo infection occurs in more than 50% of patients undergone haplo-HSCT within the first 3 months after the procedure and it remains a major cause of morbidity and mortality especially in TCD procedures (22, 37–45). Although the efficacy of the novel antiviral therapies decreased the incidence of HCMV infections/reactivations (46), this still represents one of main complications of allo-HSCT (47). In this regard, a careful selection of donors is recommended particularly within the haplo-HSCT setting, since their mismatch with the HCMV-serostatus of recipients greatly impacts the incidence and the virulence of HCMV reactivation (47). In particular, HCMV-seropositive recipients receiving a graft from HCMV-seronegative donors have the highest risks to develop HCMV reactivations. On the other hand, administering grafts from HCMV-seropositive donors increases the degree of OS in HCMV-seropositive patients receiving myeloablative conditioning (40). Hence, also the type of conditioning regimens plays a role in HCMV reactivations after allo-HSCT. The protective effect of HCMV-seropositive donors toward HCMV-seropositive recipient is also associated with the transfer of anti-HCMV specific T cell immunity (48). The frequency of primary infections in HCMV-seronegative recipients receiving a transplant from a HCMV-seronegative donor is very low since the reactivating viral strains generally origin from recipients, while their control is mediated by donor-derived alloreactive immune cells (45, 49, 50). However, a few other studies denied any significant impact of donor serostatus on HCMV reactivation in recipients undergone allo-HSCT (51, 52), thus leaving this important matter open for further discussion and clinical investigations. HCMV infections/reactivations also greatly affects the pattern of IR of both adaptive (53, 54) and innate immune cells (55, 56). Hence, it is conceivable that the kinetic of ILCs, NK and γδ T cell IR after haplo-HSCT as well as their effector-functions are somewhat influenced by HCMV infections/reactivations (55–58).
Innate Lymphoid Cells
ILCs are a heterogeneous population of non-B and non-T lymphocytes that originate from common lymphoid progenitors. Since they lack adaptive antigen receptors, ILCs are able to rapidly produce and secrete pro-inflammatory and regulatory cytokines in response to local injuries, inflammation, infections or commensal microbiota perturbations (59–61). Similar to T cells, ILCs have been grouped into cytotoxic and helper lymphocytes and classified into three distinct sub-populations on the basis of their cytokines production and of the transcription factors involved in their development. These cell subsets are named ILC1, ILC2, and ILC3 and functionally mirror the CD4pos T helper (Th)1, Th2, and Th17 cells, respectively. More recently, also NK cells have been grouped within ILC family and resemble the functions of CD8pos cytotoxic T cells (59, 62–65).
ILC1 are mainly involved in interferon-γ (IFN-γ) production and represent potent effectors against bacterial and viral infections (61, 66–68). Despite sharing these functions with NK cells, ILC1 are currently considered a distinct subpopulation in terms of phenotype, function and development. Indeed, ILC1 are generally poorly cytotoxic and, unlike NK cells, are found at high frequency in tonsil and gut epithelium (i.e., intraepithelial ILC1) (69). Instead, ILC1 are rare in peripheral blood (PB) where they can be easily distinguished from NK cells due to their lack of CD56 and CD94 surface expression (63, 70, 71). ILC2 are also mostly tissue-resident lymphocytes and their effector-functions are triggered by interleukin (IL)-25 and IL-33 produced by epithelial cells or other immune cells in response to parasite infections or to allergen exposure. Following activation, ILC2 produce and secrete type 2 cytokines including IL-4, IL-5, IL-9, and IL-13 (62, 72–75). Moreover, ILC2 contribute to the resolution of inflammation by producing amphiregulin (AREG), a member of the epidermal growth factor that helps repairing damaged tissues (76). ILC3 are mainly resident in the gut lamina propria but have been also found in skin, lung and liver (77). Two different ILC3 subsets have been identified based on the expression of the Natural Cytotoxic Receptor (NCR) NKp44 in humans and NKp46 in mice. Both NCRpos/ and NCRneg/ILC3 subsets are able to produce IL-17, a cytokine crucial for fungal infection resistance. NCRpos/ILC3 can also secrete IL-22, an important cytokine that regulates the homeostasis of gut epithelium, prevents the dissemination of commensal bacteria and limits inflammatory response (78). Another subset of lymphocytes grouped within ILC family is represented by the so-called lymphoid tissue-inducer (LTi) cells that are mainly involved in lymphoid organogenesis in fetal life. However, LTi-like cells are present also in adult life where they facilitate the generation of secondary lymphoid organs (79). LTi/LTi-like cells also produce IL-22 and initiate protective immune responses against extracellular bacteria. However, these latter lymphocytes have been grouped separately from ILC3 since they have a unique transcriptional profile and are generated from distinct progenitors (80). Moreover, LTi/LTi-like cells are endowed with specialized functions related to adaptive immunity as they are involved in T and B cell development (79).
Despite their differences in term of phenotype and functions, several lines of evidence indicates that the helper-ILCs (i.e., ILC1, ILC2, and ILC3) have high degrees of cell plasticity, as each one of these three subsets can give rise to other members of the same family if cultured with the proper cytokine stimulation (81). Moreover, recent findings indicate that, although ILC1, ILC2, and ILC3 are mainly tissue-resident, they might traffic through the different organs by recirculating in the bloodstream. Indeed, gut-resident ILC2 can migrate into the lung and other peripheral tissues in response to helminthes or upon IL-25 stimulation to either fight the parasite infections or to contribute to tissue repair (82). This experimental evidence suggests that helper-ILCs, other than exerting anti-microbial responses and tissue remodeling in those organs where they reside under homeostatic conditions, can also mediate a protective role against tissue damage in different anatomic compartments following exposure to inflammatory stimuli. In the context of allo-HSCT, this phenomenon is highly relevant in the mucositis induced by chemo/radiotherapies, in the development of GvHD and in response to infections. However, little is known about the role(s) played by ILCs in the pathogenesis of hematologic malignancies as well as in the clinical outcomes of transplantation. Indeed, very few studies have addressed the role of immune- reconstituting ILC in the context of allo-HSCT (83), while their functions in haplo-HSCT remain still completely unexplored. Thus, in the next paragraphs we will summarize the evidence on ILCs in allo-HSCT setting.
Immune-Reconstitution of Innate Lymphoid Cells
It has been recently disclosed that ILCs have a great clinical impact in patients affected by Acute Myeloid Leukemia (AML) either at disease onset or after chemo/radiotherapy and allo-HSCT (84, 85). In particular, there is a great reduction of circulating ILCs in AML, a phenomenon associated with a relative increase of ILC1 and a decrease of NCRpos/ILC3. The overall frequencies of PB NCRpos/ILC3 but not the ones of ILC1 are restored to normal levels in AML responders to chemotherapy. These quantitative changes of circulating ILCs in AML patients mirror their impaired abilities in producing IFN-γ and type 2 cytokines (85). Taken together, these data suggest that either leukemia burden or disease relapse markedly affect ILC development, a phenomenon also confirmed in vitro by co-culturing ILC precursors with AML blasts (86).
It has been also reported that conditioning regimens prior allo-HSCT deplete circulating ILCs that then undergo in recipients through a slow process of IR taking at last 6 months for a complete recover. In this setting, reconstituting ILCs show an increased expression of markers associated with tissue homing, such as the skin-homing receptors CLA and CCR10, the gut-homing molecules α4β7 and CCR6, the activation/tissue-residence marker CD69 and the cell proliferation nuclear protein Ki-67 (84). After 3 months from the transplant, the levels of circulating ILC2 are still strongly decreased compared to those of healthy subjects, while NCRpos/ILC3 outnumber the other ILC subsets (84). These data suggest that ILC3 play a major role in ILC IR after allo-HSCT. In line with this working hypothesis, a study showed that the high amounts of IL-22 produced by ILC3 can enhance both thymic regeneration and a more rapid T cell IR in a IL2−/− mouse model receiving a TCD allo-HSCT (87).
It has been also reported that both conditioning regimens and different source of HSCs affects ILC IR after the transplant. This is of great importance in those children affected by severe combined immune deficiency (SCID) and carrying mutations of genes either encoding the common γ-chain subunit of IL-2 receptor or the tyrosine kinase JAK3. These patients lack all ILC subsets and experience an effective T cell IR following allo-HSCT only in the presence of myeloablative conditioning regimens (88). Instead, the administrations of cyclosporine or corticosteroids do not affect ILC IR (84). Another study showed in an in vitro setting that ILC3 IR is hampered by both pre- and post-transplant treatments with the mobilizing agent G-CSF (89). Moreover, it has been also reported that the generation of ILCs (especially NCRpos/ILC3) is much higher when culturing in vitro HSCs from bone marrow (BM) and umbilical cord blood rather than their counterparts from PB following mobilization with G-CSF (89).
Innate Lymphoid Cells and Graft vs. Host Disease
Several lines of evidence demonstrated that ILCs play a key role in limiting the onset of GvHD after allo-HSCT. In particular, it has been shown in murine models that ILC3 have a great impact in protecting recipient gut epithelial cells from alloreactive responses exerted by donor immune cells. This phenomenon is mediated by the ILC3 high production of IL-22 (90). Indeed, IL-22 deficient mice undergone allo-HSCT suffer from severe intestinal GvHD and intestinal barrier disruption, while the administration of IL-22 in transplanted wild type animals limits the onset of intestinal GvHD and enhances both intestinal stem cell recovery and epithelial cell regeneration (91). In humans, increased frequencies of circulating NCRpos/ILC3 early after allo-HSCT correlate with a lower incidence of intestinal GvHD. Notably, the ability to secrete high amounts of IL-22 by NCRpos/ILC3 exerts a key role in the regeneration of the mucosal gut barrier after immune depletion following allo-HSCT, thus protecting from GvHD onset (92, 93). Moreover, higher expressions on recipients' circulating ILCs of both CD69 and α4β7 markers before the transplant reduce the risk of developing GvHD and can serve as good prognostic factors (84). Even increased frequencies of CD69pos/ILC1 are associated with lower incidence of severe cutaneous GvHD since these cells express high levels of the skin homing markers CLA and CCR10. It has been also reported in murine models that type 2 cytokines play a protective role in GvHD development (92). Another reported mechanism protecting from GvHD is the ability of ILC2 to produce AREG that, in turn, boosts epithelial cell regeneration after the tissue damage induced by the conditioning regimens (76).
Innate Lymphoid Cells and Opportunistic Viral Infections
Although the role of ILCs in controlling infections in immune-competent individuals seem marginal, studies in immune-deficient mice showed that these innate lymphocytes can fight different pathogens (83, 94). However, very little if nothing is known in regard to their functional role in allo- and in haplo-HSCT setting. Since both T and B cell IR start to be effective and functional relevant only after a few months after haplo-HSCT, innate immune system certainly plays a key role in controlling opportunistic infections early after the transplant (19, 48, 95, 96). In this regard, while NK cells represent an immediate available source of IFN-γ in the bloodstream, ILC1 can provide large amounts of the same pro-inflammatory cytokine in tissues as reported in murine models of CMV, influenza, and Sendai infections (97, 98). Unlike ILC1, ILC2 are mainly involved in tissue damage repair during the resolution of the inflammatory process rather than in controlling the opportunistic infections (76, 99). Indeed, the proliferation and effector-functions of ILC2 are inhibited by both type I and II IFN that are largely produced during the course of viral infections (75, 100). Thus, high levels of IFN-γ produced by tissue-residence ILC1 not only control viral replication but also limit the dysregulation of ILC2 homeostasis.
Natural Killer Cells
NK cells are innate lymphocytes playing a major role in the immune-surveillance mainly against cancer and viral infections without a prior antigen sensitization and through the signal delivered by large families of inhibitory and activating NK cell receptors (aNKRs and iNKRs) (101).
iNKRs recognize, as their natural ligands, “self” HLA-I molecules expressed on the surface of all nucleated cells, ensuring both the recognition of autologous targets and a certain threshold of immunologic tolerance especially at tissue levels. On contrary, tumor-transformed, viral infected, and heterologous cells lack or have reduced or express heterologous HLA-I molecules, respectively. NK cells can recognize these abnormalities on “non-self” and threatening targets due to the impaired or missing binding with iNKRs, whose downstream signaling is normally dominant over the activating stimuli driven by aNKRs in NK cells (“missing-self hypothesis”). The absence of this dominant inhibition shifts the balance toward NK cell activation via the engagement of aNKRs that binds their putative ligands on heterologous cell targets. These mechanisms trigger NK cell release of cytotoxic granules (i.e., perforin and granzymes) and secretion of anti-viral/pro-inflammatory cytokines for the clearance of both tumor and viral-infected cells (102–105).
The repertoire of NKRs is highly variable among different individuals and in different anatomic compartment and it is influenced by genetic factors, environmental exposure to non-self targets and tissue microenvironments (106, 107). Moreover, the phenotypic profiles of NK cells also depends by the so called “education/licensing” process that dictates the avidities of the interactions between iNKRs and their putative HLA ligands (108). The main classes of NKRs specific for HLA-I molecules include Killer Ig-like Receptors (KIRs) that recognize different HLA-A, -B, and -C allotypes (109) as well as the C-type lectin receptors CD94/NKG2A and CD94/NKG2C that bind the non-classical HLA-E molecules (110, 111). KIRs (known as CD158 molecules) represent a highly polymorphic family of NKRs that serve as regulators of development, tolerance and activation of NK cells (112). Interestingly, KIR superfamily includes both activating and inhibitory forms sharing homology in the extracellular domain, while differing for their cytoplasmatic tails. Activating KIRs (aKIRs) are characterized by a short intracellular domains that interact with adaptor signaling molecules carrying an Immunoreceptor Tyrosine-Based Activating Motif (ITAM) such as DAP-12 (113). On contrary, long cytoplasmatic tails containing Tyrosine-Based Inhibitory Motif (ITIM) distinguish inhibitory receptors (iKIRs) (109, 113, 114).
Similarly, the inhibitory C-type lectin receptor CD94/NKG2A is characterized by long intracellular tail containing ITIM motifs, while the trans-membrane domain of CD94/NKG2C interacts with the ITAM-containing adaptor molecule DAP-12 driving NK cell activation (115, 116). Among the other aNKRS driving the activation of NK cells there are the NCRs NKp30, NKp44, NKp46 together with the co-receptor NKp80 and 2B4 (117, 118).
CD16 (FCγRIII) is an immunoglobulin (Ig) receptor that, upon binding with the Fc portion of IgG antibodies, induces series of potent activating signals through the adaptor molecules CD3ζ and FcεRγ containing the activation ITAM motif. This down-stream pathway mediates the so-called antibody-dependent cell mediated cytotoxicity (ADCC) (119). The sequential expressions of CD16 together with KIRs, NCRs and C-type lectin receptors characterize the developmental stages, the effector-functions and the education of NK cells (120). The main steps of NK cell ontogenesis take place in BM niche starting from CD34pos HSCs but, differently from helper ILCs, these innate lymphocytes are mainly enriched in PB (121). Indeed, under homeostatic conditions, NK cells account up to 10% of total circulating lymphocytes and represent an heterogeneous population that can be subdivided into two main subsets according to the surface expression of CD56 and CD16 (122). CD56bright/CD16neg−low (CD56br) NK cells represent 5–15% of total circulating NK cells and are considered regulatory lymphocytes, as they produce high amounts of chemokines/cytokines and are involved in the cross-talk with other immune cells such as dendritic cells (DCs) and monocytes/macrophages (123–125). On the other hand, CD56dim/CD16pos (CD56dim) NK cells are the largest NK cell subset in PB (up to 95%) and mainly exert cytototoxic functions via the secretion of lytic granules (104, 126–128). CD56br and CD56dim are also considered two sequential stages of NK cell maturation with the latter subset being the terminally-differentiation one characterized by shortest telomere length (120, 121, 129, 130). CD56br NK cells usually show high levels of CD94/NKG2A, while almost lack KIRs (131). On contrary, CD56dim NK cells acquire KIR expression and loose CD94/NKG2A, thus being fully licensed end-stage effector cells (115, 132). Despite intense efforts in better disclosing human NK cell ontogenesis, the mechanisms tuning the appearance of NKRs and the different NK cell developmental stages remain to be elucidated (120).
NK Cell Immune-Reconstitution
Given the ability of NK cells to promptly mount effective alloreactive responses against tumor cells and pathogens, their kinetic and quality of IR certainly play important roles in determining the clinical outcome of allo- and haplo-HSCT. Indeed, delayed recoveries of these donor-derived alloreactive innate lymphocytes result in poor clinical outcomes of transplants (133, 134). As a matter of fact, NK cells are the first lymphocytes to appear soon after allo-and haplo-HSCTs and are essential for a better engraftment, to avoid tumor relapse and to limit the onsets of both GvHD and opportunistic viral infection. Moreover, the possibility to follow human NK cell IR in this unique in vivo setting is key in disclosing the several unknown mechanisms and patterns of their ontogenesis and differentiation (19, 135, 136). Regardless of the graft sources, NK cell chimerism in recipient is completely donor dependent after one month from haplo-HSCT. However, although the frequencies and absolute counts of circulating NK cells reach normal levels after few weeks post-transplant, their maturation and achievement of efficient effector-functions takes much longer (6, 15, 19, 130, 135, 137). Similar results have been observed also in recipients receiving HLA-matched HSCT, where reconstituting NK cells remain immature for more than 6 months after the infusion of HSCs. These phenomena are associated with functional defects that do not ensure an optimal protection against HCMV infections/reactivations, GvHD onset and tumor relapse in the first year after HSCT (138).
Reconstituting NK cells derive from CD34pos progenitors rather than from already mature NK cells infused with the graft. Indeed, the PT-Cy eliminates proliferating alloreactive NK cells in haplo-HSCT as they have an even higher proliferation rate compared to T cells in the first days after the graft infusion and before the Cy administration. The 2nd wave of proliferating donor-derived NK cells occurs after 15 days from haplo-HSCT and these new innate lymphocytes display an immature phenotype, thus confirming that they are de novo generated from donor HSCs (96). Indeed, CD56br NK cell subset appears much earlier that terminally differentiated CD56dim, while the NK cell surface distribution of both CD56 and CD16 return similar to that of healthy donors only several months later (6, 19, 96, 120, 139–141). Unexpectedly though, we recently reported that the subset of reconstituting donor-derived NK cells expanded at the highest frequency in the first weeks after haplo-HSCT is characterized by an unconventional CD56dim/CD16neg−low phenotype (unCD56dim). This neglected NK cell population is present at very low frequency under homeostatic conditions, but plays a key role in the IR and in the clinical outcome of haplo-HSCT. In particular, although armed to be cytotoxic and carrying large amounts of perforin and granzymes, unCD56dim NK cells are highly defective in their killing activities due to the transient high expression of CD94/NKG2A receptor. Hence, this C-lectin type receptor functions as an inhibitory checkpoint that renders donor-derived unCD56dim NK cells anergic against residual tumor cells, recipients T cells and Antigen Presenting Cells (APC). This NK cell status early after haplo-HSCT makes recipients more at risk to undergo tumor relapse and to develop acute (a) GvHD. Similar transient high surface levels of CD94/NKG2A have been observed also on terminally-differentiated and cytotoxic CD56dim NK cells that start to reconstitute from the 2nd month after the transplants and subsequent to the appearance of unCD56dim NK cells (19, 135, 139, 142). This gained knowledge paved the ground for a novel therapeutic approach targeting CD94/NKG2A in order to unleash NK cell cytotoxicity in haplo-HSCT.
NK Cells and Graft vs. Host Diseases
The HLA-mismatch between donor and recipient cells allow donor-derived and alloreactive NK cells to both limit the onset of GvHD and to prevent graft rejection in allo- and haplo-HSCT (143, 144). Indeed, several studies directly correlated an efficient NK cell IR in allogeneic transplant with the reduced incidence of relapse as well as with decreased rates of opportunistic infections in the presence of lower TRM and increased OS (134, 145, 146). In contrast, low frequencies of NK cells in the first weeks after allo-HSCT are associated with increased non-relapse mortality, shorter OS and higher degrees of opportunistic infections (133, 145). This clinical evidence underline the importance of NK cell IR in shaping the clinical outcomes of allogeneic transplants and its possible exploitation for developing novel therapeutic strategies (2, 135, 147). However, the exact NK cell-mediated mechanism preventing GvHD onset are not yet fully elucidated. One working hypothesis is that alloreactive NK cells could limit GvHD by killing donor T cells via the NKG2D-mediated recognition of stress-induced NKG2D-ligands on activated T lymphocytes (148, 149). Another study claimed that high frequency of NK cells in the first weeks after HSCT might prevent T cell proliferation through IL-10 production (150). Conversely, it has been also reported that NK-cell production of pro-inflammatory IFN-γ could promote tissue damage and consequent GvHD (151). Notably, also the quality of NK cells IR greatly affects the occurrence of GvHD after allo-HSCT. Indeed, higher surface levels of CD94/NKG2A on NK cells have been reported to limit aGvHD in vivo by inhibiting T cell proliferation and activation (152). Furthermore, increased frequencies of CD94/NKG2Cpos NK cells are associated with a lower incidence of GvHD in allo-HSCT (153).
Even the NK cell maturation stage is important, as a recent report showed that those haplo-HSCT recipients developing GvHD display a more differentiated and activated NK cell phenotype (154). This evidence has been also further corroborated by other studies reporting that a reduction of circulating CD56br NK cells in the first 2 months after allo-HSCT is associated with higher incidence of aGvHD. This latter clinical correlation was so evident in the recruited cohorts of patients receiving allo-HSCT to be proposed as an early prognostic factor to predict GvHD (141, 143). Moreover, a higher ratio of T/NK during IR after phase correlates with a higher risk to develop both acute and chronic GvHD in haplo-HSCT (8).
Remarkably, the potential clinical benefits of reconstituting NK cells in haplo-HSCT might be influenced by pre- and post-conditioning treatments. In this regard, many studies performing adoptive transfer of NK cells after haplo-HSCT showed a reduced risk in aGvHD induction (151). Moreover, GvHD prophylaxis with Mycophenolate Mofetil has been demonstrated to inhibit NK cell proliferation and effector-functions (155, 156), thus affecting the NK cell mediated control of GvHD and opportunistic infections.
NK Cells and Viral Infections
The occurrence of an optimal quantitative and qualitative NK cell IR in haplo-HSCT is key for hampering the onset of life-threatening opportunistic infections. Indeed, lower frequencies of circulating donor-derived NK cells are associated with higher susceptibilities to develop viral infections, mainly HCMV (157). In turn, HCMV infections/reactivations are also able to influence NK cell homeostasis and differentiation by inducing the expansion of the CD56neg/CD16pos (CD56neg) NK cell subset (158, 159). While poorly represented in healthy individuals, CD56neg NK cells are present at high frequencies in active and chronic HIV-1 and HCV infections (58, 160, 161) and display impaired effector-functions due to their abnormal repertoire of NKRs (162, 163). Indeed, CD56neg NK cells are defective in the clearance of viral infections and express markers of cell exhaustion of their surface (164, 165). However, the ontogenesis and the impact of CD56neg NK cells in determining the clinical outcomes of allo- and haplo-HSCT are still being debated. Recent studies revealed that HCMV infections/reactivations are beneficial rather than detrimental on NK cell recovery upon haplo-HSCT. In particular, it has been reported that this virus can accelerate NK cell maturation and shape their NKR repertoire in haplo-HSCT by inducing the expansion of terminally-differentiated and alloreactive CD56dim NK cells which, in turn, exert potent GvL effects (166). Indeed, upon HCMV infections/reactivations, CD56dim NK cells acquire a mature NKG2Cpos/CD57pos/NKG2Aneg/KIRpos phenotype, thus becoming fully licensed to efficiently exert anti-viral and anti-tumor properties (i.e., production of IFN-γ and Tumor Necrosis Factor (TNF)-α) (167–169). On the contrary, NK cells from haplo-HSCT patients that do not experience HCMV infections/reactivations retain an immature phenotype characterized by high expressions of CD94/NKG2A (170).
These HCMV-induced NKG2Cpos/CD57pos/NKG2Aneg/KIRpos/CD56dim NK cell subset can persist even after 1 year from haplo-HSCT and show higher effector-functions when re-encountering the same antigen or following a proper activation with specific pro-inflammatory cytokines. These data suggest that HCMV infections/reactivations drive the expansion of NK cells with adaptive properties (167, 170–172). Similar features have been reported in murine models in vivo, where the murine CMV (MCMV) infection is responsible for the expansion of the so-called “memory-like” NK (ml-NK) cells that specifically recognize the viral glycoprotein m157 through the activating receptor Ly49H (173, 174). However, neither a univocal phenotype nor the receptor(s) able to specifically bind HCMV antigens have been clearly defined in human ml-NK cell and this is a matter currently being highly investigated in several models in vitro and ex vivo. In this regard, NKG2C has been proposed as the best putative candidate binding HCMV antigens, since those NK cells expressing this aNKR are the ones preferentially expanded following this viral infection (175, 176). In this regard, it has also been reported that the HCMV-encoded UL40 protein stabilizes HLA-E surface expression on target cells, thus favoring the recognition of viral-infected via the NKG2C/HLA-E interactions (159, 177). Moreover, another study claimed that proliferation/expansion of NKG2Cpos NK cells requires additional signaling pathways including the one mediated by IL-12 produced by autologous monocyte (178). Despite all the above-mentioned experimental evidence, the primary role of NKG2C in the homeostasis and functional relevance of ml-NK cells is still unclear. Indeed, other subsets of NKG2Cneg/KIRpos NK cells are also expanded in response to HCMV infection and they are able as well to recognize viral-infected cells (106), thus suggesting the existence of additional aNKRs (i.e., KIRs) involved in the expansion of human ml-NK cells (179). Furthermore, NKG2C-deficient individuals can mount equivalent adaptive NK cell response against HCMV (180). In agreement, in patients receiving cord blood grafts from NKG2C−/− donors, HCMV infection is still able to promote NK cell maturation in the absence of this activating C-lectin type molecule. This latter experimental evidence further supports the current working hypothesis that other NKRs such aKIRs play a central role in the generation of ml-NK cells (181).
More recently, other studies demonstrated that the generation of ml-NK cells is associated with epigenetic reprogramming through a specific reconfiguration of adaptor molecules including tyrosine kinase SYK, the intracellular adaptor EAT-2, and the transmembrane adaptor protein FcεRγ. The gene expression of these three factors is regulated by the transcription factor promyelocytic leukemia zinc finger (PLZF), which is downregulated in the majority of ml-NK cells upon HCMV infections. As a matter of fact, the reduced expression of at least one of the above-mentioned signaling proteins is observed in the 50% of the HCMV-seropositive donors. Moreover, the reduced levels of PLZF also decreases the expression of IL-12 and IL-18 receptors, thus lowering NK cell responsiveness to these pro-inflammatory cytokines. The lack of FcεRγ, SYK, and EAT-2 in mature CD56dim NK cells is also correlated with the expansion of NKG2Cpos NK cells upon HCMV infection (182–185). In this regard, CD56neg NK cells expanded in those patients receiving umbilical cord blood transplant and experiencing HCMV infection/reactivation are characterized by the downregulation of FcεRγ (186). In addition to the downregulation of PLZF, FcεRγ, SYK, and EAT-2, ml-NK cells share with cytotoxic CD8pos T cells similar genome-wide DNA methylation patterns (182), thus suggesting the existence of epigenetic determination programs associated with HCMV infections. Notably and similar to memory Th1 lymphocytes, the increased production of IFN-γ by ml-NK cells correlates with a stable demethylation of conserved non-coding sequence 1 of the IFNG locus (187).
Although there is a phenotypic heterogeneity of ml-NK cells following HCMV exposure, their rapid maturation in response to the viral challenges could favor not only the control of infection, but also NK cell alloreactivity against residual tumor cells (188). Hence, HCMV infection can represent a “natural” tool to generate ml-NK cells to then use for adoptive cellular immunotherapies (132, 189, 190). In this regard, newborn mice challenged with MCMV showed that ml-NK cells undergo expansion, release cytokines and provide a protective anti-tumor immune response in adoptive cell transfers (173). In humans, the expansion and the functional relevance of NKG2Cpos ml-NK cells in HSCT recipients experiencing de novo viral infection or undergone HCMV reactivations also depends from donor serostatus. Indeed, the in vivo expanded NKG2Cpos ml-NK showed higher cytokine productions in those recipients receiving grafts from HCMV-seropositive donors compared to their counterparts originated from grafts of HCMV-seronegative donors. However, NKG2Cpos ml-NK cells also expand in the absence of detectable HCMV viremia when both donor and recipient are HCMV-seropositive. These data suggest that also human NKG2Cpos ml-NK cells are transplantable and require exposure to either active or latent (subclinical) HCMV antigens in the recipients for the expansion of alloreactive NK cells from seropositive donors (191). Moreover, NKG2Cpos ml-NK cells are able to produce high levels of IFN-γ following in vitro co-culture with K562 erytroleukemia cell line, thus supporting their high potential in GvL effect (192). Consistent with these findings, the adoptive transfer of donor-derived or cytokine-induced (i.e., activation with IL-12, IL-15, IL-18) ml-NK cells induces in the recipients affected by refractory AML the expansion of NK cells producing high levels of IFN-γ when encountering tumor cell targets (172).
Taken together these results suggest that ml-NK cells can be potentially exploited in order to both better control HCMV infection/reactivation and to enhance GvL (193).
γδ T Cells
γδ T cells are a group of unconventional T cells that bridge the gap between innate and adaptive immunity. Similar to αβ T cells, γδ T cells develop in the thymus and express a somatically rearranged T cell receptor (TCR) consisting of a TCR-γ and a TCR-δ chains (65, 194–196). In humans, γδ T cells normally account for the 1–10% of circulating T lymphocytes, while in mucosal tissues and skin they constitute the major subset of resident T cells (194, 196). Different γδ T cell subsets can be identified based on the Vδ expression (Vδ1, Vδ2, Vδ3, and Vδ5) (195, 197). Under homeostatic conditions, 95% of circulating γδ T cells express Vδ2 TCR paired with Vγ9 chain, whereas in mucosa and skin γδ T cells mostly express Vδ1 or Vδ3 TCRs paired with various Vγ chains (195, 198–200).
γδ T cells are rapid responders to pathogens and tumor-transformed cells, since they do not require further peripheral maturation or extensive clonal expansion to initiate their effector-functions (194). Therefore, γδ T cells allow a prompt immune-surveillance in a MHC-independent manner through the recognition of a diverse array of antigens including peptides, sulfatides and phospholipids (194, 196, 199, 201, 202). Moreover, the γδ TCR can bind CD1d expressed by APC loaded with glycolipids and microbial lipids (203). In addition to their TCR, γδ T cells express an array of pattern-recognition receptors, such as toll-like receptors (TLRs) (201, 204), activating and inhibitory NKRs (201, 205, 206), the NCRs NKp30 and NKp44 (206, 207), the aNKR DNAM-1, the Fc receptor CD16 as well as the C-type lectin-like receptors NKG2D and CD94/NKG2A (195, 206, 208, 209). The presence of such receptor repertoire suggests a tight regulation of the TCR-mediated activity through an interplay between activating and inhibitory signaling downstream pathways (206).
Upon their activation, γδ T cells secrete high levels of Th1 cytokines (i.e., IFN-γ and TNF-α) modulating the responses of other neighboring immune effectors which, in turn, induce monocyte-derived DC maturation/activation and enhance antigen-specific αβ T cell responses (194, 195). Moreover, γδ T cells are able to directly lyse target cells by the release of granzymes and perforin and the engagement of FAS and TRAIL death receptors (195, 197, 210). As consequence of their high heterogeneity, γδ T cells are implied in diverse biological functions. First, these cells exert anti-tumor activities against various types of solid tumors and hematological malignancies (211). Since they represent the most abundant population among epithelial-resident lymphocytes in mucosal tissues and skin, γδ T cells are also the first line of defense against pathogens in these anatomic compartments (211, 212). Finally, several γδ T cell subtypes are involved in the induction of transplant immune-tolerance both in solid organ transplantation and in allo-HSCT (211, 213).
γδ T Cell Immune-Reconstitution
The growing interests on the role of γδ T cells IR in HSCT arose from their potential ability to perform GvL effects and fight opportunistic infections in the absence of GvHD (205, 211, 214). Indeed, pediatric and adult patients undergone haplo-HSCT and showing a long-term disease-free survival (DFS) were coupled with high frequencies of circulating γδ T cells (215, 216). γδ T cells are also the predominant T cell population reconstituting early after haplo-HSCT, with the Vδ2 cells showing a faster recovery compared with B and T lymphocytes in the PB of recipients receiving αβ and CD19 depleted grafts. In particular, it has been shown that the recovery of the complimentary determinant region 3 (CDR3) of the TCR δ chain is almost completed after 2 months from haplo-HSCT (25, 28, 29, 48, 95, 217). In the context of allo-HSCT, the majority of both donor-derived Vδ1 and Vδ2 cell subset recovering in the first weeks have a CD27pos/CD45RAneg Central Memory (CM) phenotype and contribute to ensure an early protection against viruses, bacteria and residual tumor cells that survived the conditioning regimes (65). The current working hypothesis of a peripheral expansion of graft-derived mature γδ T cells is further supported by experimental evidence indicating that the same γδ T cell clones found in the donor are present in the recipient after the transplant (218). Later on, within a range of 14–60 days post-transplantation, the frequency of CM γδ T cells progressively decreases and it is counterbalanced by increase frequencies of naïve CD27pos/CD45RApos γδ T cells originated from donor infused HSCs (28, 65). This latter de novo generation of reconstituting γδ T cells is confirmed by the fact that, while the repertoire of the γ and δ chains is qualitatively comparable between donors and recipients, their clonotype is different (57).
γδ T cell IR after allo- and haplo-HSCT can be influenced by different variables including the conditioning regimen, the administration of immuno-suppressive agents, the GvHD prophylaxis and the onset of opportunistic infections (211). In this regard, it has been reported that stem cell mobilization with G-CSF in allo-HSCT induces higher frequencies of Vδ1 T cells endowed with potent alloreactivity against AML blasts (214). Moreover, also donor/recipient characteristics (i.e., gender, age, disease type, and graft source) affect γδ T cell IR too. Indeed, patients receiving a transplant from either matched related (MRD) or haplo-related donors have significant differences in the recovery of γδ T cells compared to matched unrelated donor (MUD) (215).
γδ T Cells and Graft vs. Host Diseases
It has been reported in allogenic HSCT that patients developing aGvHD show an increased frequency of reconstituting γδ T cells (219). However, this evidence has been denied by more recent findings indicating that absolute counts of γδ T cells do not influence the incidence and the severity of GvHD (65, 215). Instead, higher frequencies of donor-derived γδ T cells in the grafts seem to protect against the development of severe aGvHD (220). Similarly, patients receiving a TCD haplo-HSCT and showing increased frequencies of γδ T cells undergo longer DFS and OS compared to those with normal/decreased immune-reconstituting γδ T cells. These data corroborate the current consensus stating that γδ T cells can facilitate GvL effect without inducing GvHD (196, 216, 221, 222).
γδ T Cells in Viral Infections
The occurrence of high frequencies of reconstituting γδ T cells early after haplo-HSCT also protect from bacterial infections and show a decreased incidence of both viral and fungal infections (215). Indeed, pediatric patients, receiving αβ TCD grafts in haplo-HSCT setting have both reduced numbers of γδ T cells at day 30 post-transplant and higher incidence of HCMV infections/reactivations (65). At the same time, opportunistic infections can also shape the homeostasis and maturation of these cells (28, 55, 57, 195, 223). Indeed, patients undergoing allo-HSCT and experiencing HCMV reactivations display a preferential proliferation of specific Vδ1 and Vδ3 T cell clones, thus suggesting that γδ T cells are capable of adaptive responses through an oligoclonal selection of specific TCR repertoires (57). In particular, HCMV reactivation in haplo-HSCT patients has been associated with a specific expansion of terminally differentiated cytotoxic Vδ1 T expressing the effector memory CD45RApos/CD27neg (TEMRA) phenotype (28). This HCMV-induced expansion of TEMRA γδ T cells also enhance their anti-tumor functions both against hematological (28, 223) and solid (224) tumor cell targets in vitro. Taken together, these results suggest that the adoptive transfer of HCMV-specific Vδ1-donor γδ T cells can be used as a possible alternative to the common infusion of HCMV-specific αβ T cells (225). Indeed, this novel approach could promote viral immunity, protect from HCMV-related complications while contribute to prevent from leukemic relapses (214).
Novel Therapeutic Strategies to Improve IR Upon HSCT
The early protection and the limited side effects following HSCT render innate immune system a particularly attractive tool for adoptive cell therapy strategies. In this context, several approaches have been recently developed to improve NK and γδ T cell IR and to enhance their reactivity against cancer. These new therapeutic strategies include the targeting of checkpoint inhibitors, the stimulation with activating cytokines and genetic engineering of immune cells (Table 2) (Figure 1).
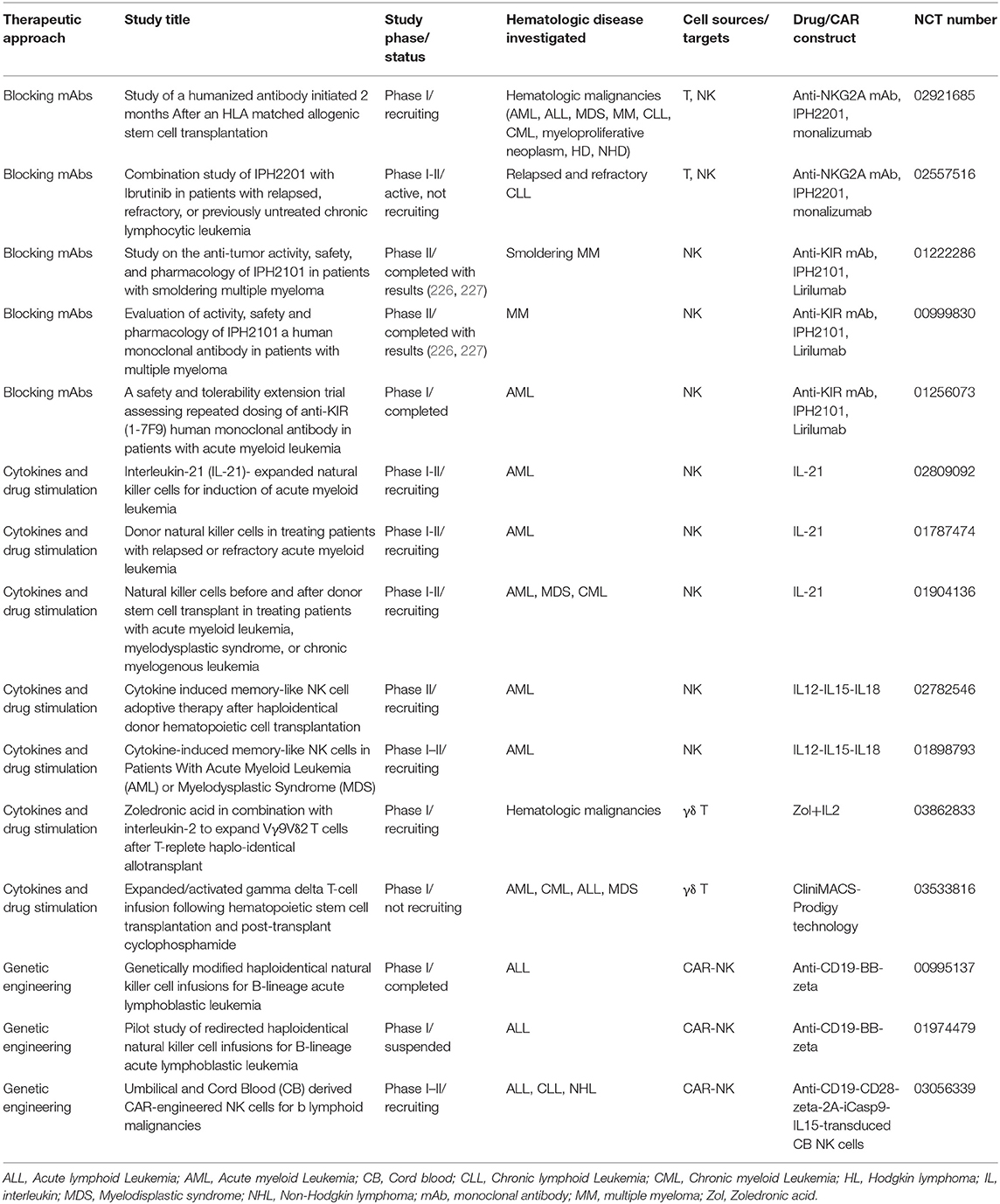
Table 2. Clinical trials targeting NK/γδ T cells in HSCT to cure patients with hematologic malignancies.
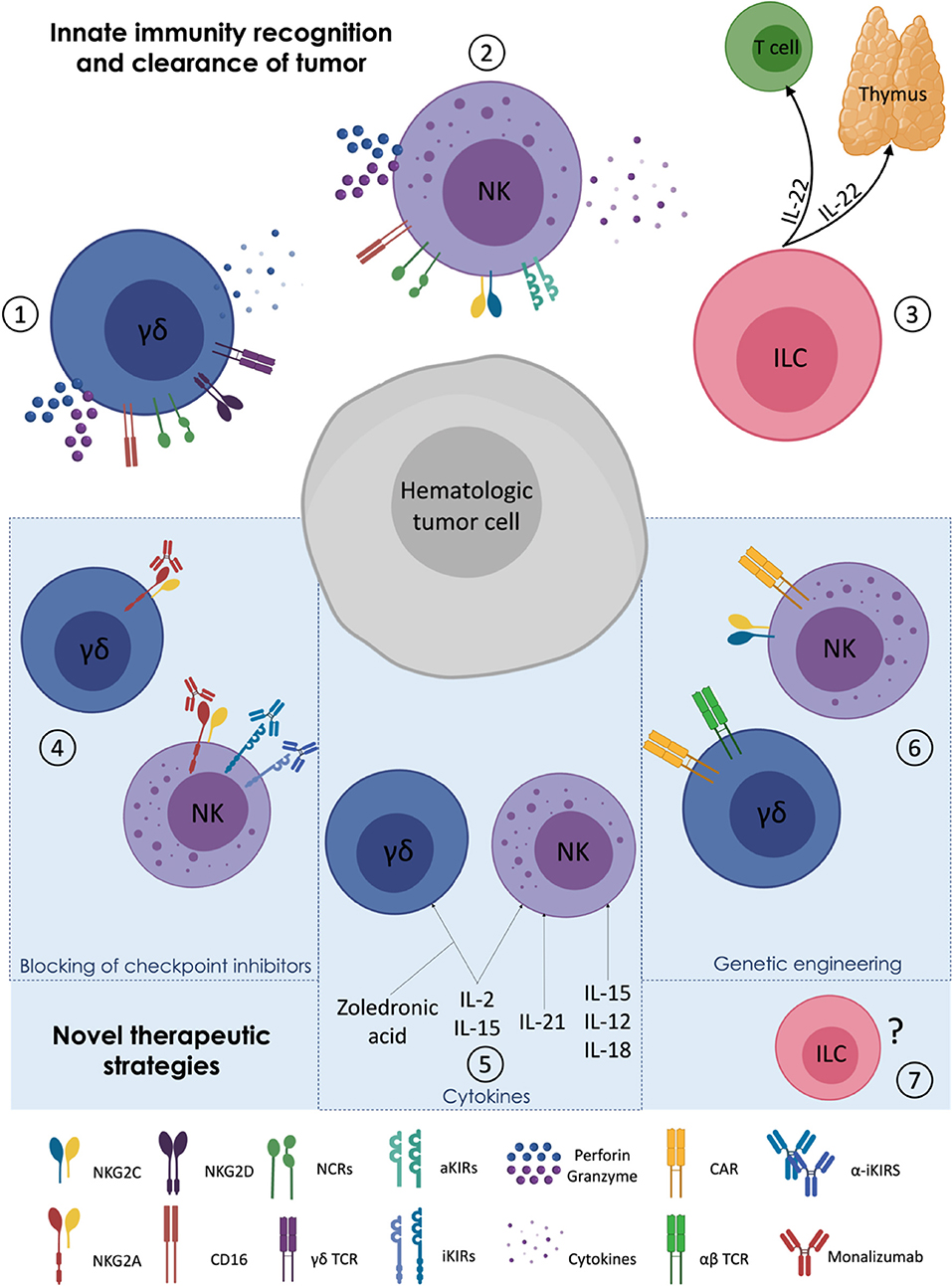
Figure 1. Targeting γδ T, Natural Killer, and Innate Lymphoid cells in haplo-HSCT. (1–3) MHC-independent activation of innate immune cells: γδ T lymphocytes (1) and NK cells (2) can kill hematologic tumors by direct cytotoxicity and cytokine secretion. Innate Lymphoid cells (ILCs and ILC3 in particular) (3) play an indirect role in the clearance of tumors cells by improving both thymic regeneration and T cell maturation via their secretion of IL-22. (4–6) Novel therapeutic strategies implemented to enhance NK and γδ T cell alloreactivity against cancer: administration of monoclonal antibodies (mAbs) against NK cell inhibitory checkpoints (4); use of cytokines and zoledronic acid to activate γδ T cells (5); CAR editing and genetic engineering of γδ T and CAR on NK cells (6). (7) Ad hoc manipulation/editing/engineering of ILCs in transplant setting have not yet been explored.
Checkpoint Inhibitor
NK cells and γδ T lymphocytes share several receptors including NCRs and iNKRs as CD94/NKG2A (195, 206, 209). The use of monoclonal antibodies (mAbs) against inhibitory immune checkpoints represents a promising therapeutic approach for both hematologic and solid tumors (228, 229). Of particular relevance, the blockade of NKG2A binding to HLA-E has been demonstrated to unleash the effector-functions of both T and NK cells in different kind of tumors (230–233). These encouraging results have driven the development of humanized IgG4 anti-NKG2A mAb (IPH2201, monalizumab), currently under investigation in many clinical trials for the treatment of solid tumors (clinicaltrials.gov) (Table 2). Conversely, only one phase I clinical study is now investigating the potential role of IPH2201 in hematologic malignancies after HLA-identical transplantation (NCT02921685). In this regard, our recent data demonstrate that there is a clear clinic indication to extend the IPH2201 administration early after haplo-HSCT, thus targeting those hypo-functional NK cells expressing high levels of NKG2A with the aim of enhancing their alloreactivity (19). Moreover, given the fast recovery of γδ T lymphocytes following haplo-HSCT, the post-transplant infusion of IPH2201 could also positively impact their anti-tumor responses in synergy with NK cells before the acquisition of a full functional competence of the adaptive immune response (i.e., T and B cells).
Among other receptors regulating NK cell missing-self responses, KIRs cover an important place. Indeed, their clinical impact have been firstly shown in AML patients undergoing haplo-HSCT where the mismatch between KIRs and their ligands in the recipient has been exploited to promote alloreactive NK cell-mediated GvL effect (135). In this context, therapeutic anti-KIR mAb (IPH2101, 1-7F9, lirilumab) has been generated and its administration showed positive outcomes in AML and multiple myeloma (MM) patients (Table 2) (226, 227).
Cytokines
As anticipated, NK cell anti-tumor responses are finely governed by an array of NKRs tuning their balance between inhibition and activation. This gained knowledge allowed to implement several protocols of in vitro NK cell manipulation that use cytokines to regulate the aNKR repertoire, thus boosting their killing ability against tumor targets (Table 2).
IL-2 and IL-15 represent the first molecules used to induce the proliferation and increase the cytotoxic potential of both T and NK cells for adoptive cell transfer therapies in different tumor settings (234). Later on, IL-21, another cytokine involved in NK cell maturation (235) also gained clinical relevance for the treatment of hematologic malignancies. Indeed, a recent phase I clinical trial using K562-based feeder cells expressing membrane-bound chimeras of IL-21 (mbIL21) was conducted in patients affected by AML/myelodysplastic syndrome and demonstrated that the infusion of ex vivo–expanded NK cells from BM haplo-donor could control tumor relapse without major toxicity (236). Other clinical trials exploiting the same technology are currently ongoing for AML in haplo-HSCT setting (Table 2) (NCT02809092, NCT01787474, NCT01904136). In order to optimize NK cell expansion and effector-functions, other experimental approaches also tested the combination of different cytokines. In particular, the stimulation with IL-15, IL-12, and IL-18 together drive the expansion of a particular subset of NK cells displaying adaptive traits similar to those of ml-NK cells re-challenged by HCMV (172). The adoptive transfer of these donor-derived and cytokine-induced ml-NK cells in patients affected by refractory AML is associated with higher levels of IFN-γ encountering and eliminating tumor cell targets (172). Two clinical trials are currently administering cytokine-induced ml-NK cells in AML patients undergone haplo-HSCT (Table 2) (NCT02782546, NCT01898793).
The combination of IL-2 and IL-15 either alone or in synergy with other stimulant agents have been extensively used also to expand γδ T cells (237). In this regard, one of the more promising protocols is represented by in vivo post-transplant administration of Zoledronic Acid (ZA) that improves the cytotoxicity of γδ T cells against leukemic cells (Table 2). This latter strategy relies on the use of ZA and IL-15 to expand terminally-differentiated and anti-tumor CD45ROneg/CD27neg effector memory (TEMRA) Vδ2 cells. In this setting, the use of IL-15 is meant also to simultaneously boost the cytotoxicity and the proliferation of NK cells, thus targeting the two main anti-cancer effectors at the same time (28, 238, 239). In haplo-HSCT platforms, two very recent phase I studies propose to expand/activate γδ T cell prior (NCT03533816) or after (NCT03862833) cell infusion to provide innate GvL responses and to limit the onset of GvHD (Table 2).
Genetic Engineering
Genetic manipulation of immune cells allows the generation of highly specific anti-tumor effectors effectively targeting several tumor antigens. The introduction of chimeric antigen receptor (CAR)-T cells in HSCT opened new insight for the treatment of hematologic malignancies. Despite the very good clinical outcomes given by autologous CAR-T cell therapies against several tumors (240–243), the occurrence of life-threatening side effects such as tumor relapses (240, 244) and higher frequencies of GvHD and cytokine release syndrome onsets (245) have arisen major limitations in the use of allogeneic CAR-T cells. In this regard, engineering CAR-NK and CAR-γδ T cells may provide alternative procedures to improve their anti-tumor potentials, while overcoming allogeneic CAR-T cell therapy obstacles (Table 2) (139, 246–249). Notably, CAR-NK cells and CAR-γδ T cells retain the expression of their NKR repertoire and γδ TCR, respectively (214, 250). Hence, they can recognize tumor targets by their native receptors independently from CAR-restriction, thus reducing antigen-driven escape of tumor cells and further increasing their killing activities. CAR-NK cells are also characterized by relatively short life-span. If this latter feature certainly limits NK cell cytotoxicity over the time after transplantation, it can then prevent long-term side effects (such as cytopenia) that are observed upon CAR-T cell infusion (251).
A multitude of preclinical studies have tested the efficacy of CAR-NK cells against a variety of target antigens such as CD19 (252, 253) and CD20 (254, 255) for hematological malignancies as well as solid tumors. Another methodology used to promote the persistence of CAR-NK cell is to incorporate genes for IL-2 (256, 257) or IL-15 (258) within the CAR construct to constantly provide cytokine support to the CAR-transduced cells. In particular, this approach showed improved tumor control and prolonged survival in a mouse model of Raji lymphoma (258). These encouraging pre-clinical data opened new insights for the transfer of such protocols into human clinical trials such as the one that is optimizing the dose of IL-15-transduced CAR-NK cells for the treatment of B cell lymphoma (Table 2) (NCT03056339). Finally, genetic engineered CAR-NK cells mimicking ml-NK cells have been obtained redirecting NKG2C-mediated NK cell responses against cells expressing HLA-E. This protocol allows to overcome the dominant NKG2A-mediated inhibition, while boosting CAR-redirected NK cell activation via NKG2C (259).
Besides NK cells, also γδ T cells have been engineered against tumor targets using CAR technology (260). However, although CAR-γδ T cells were firstly introduced in 2004 (249), relatively few studies report their benefic potential in the treatment of hematologic and solid tumors.
Among these trials, PB-derived Vg9Vd2 T cells transduced with retroviral vectors encoding either disialoganglioside GD2- or CD19-specific CARs showed a higher capacity to secrete antigen-specific IFN-γ and to exert potent cytotoxicity against GD2pos neuroblastoma cells and CD19pos leukemic blasts in vitro (249). Furthermore, γδ T cells can be also transduced with exogenous αβ TCR directed against tumor associated antigens (214, 261). However, no clinical trials using CAR-γδ T cells have been initiated yet.
Concluding Remarks
Great efforts have been put in place to ameliorate the clinical outcome of allo-HSCT, to find an ideal donor for every patient in need and to limit the life-threatening complication of this transplant procedure. The development of haplo-HSCT platforms certainly represents a great step forward on these matters, although quite a few side effects, including the occurrence of GvHD and opportunistic infections, still affect the quality and the duration of life of these patients. In this regard, the quantity and quality of IR play a central role and require a deep understanding of all the mechanisms tuning the kinetic and the effector-functions of those immune cells that can better control the onset of tumor relapse, GvHD, and opportunistic infections. In this context, innate immune responses are key as they act immediately after the transplant. Several experimental and clinical studies clearly highlighted the importance to boost both adaptive and innate IR, ameliorate anti-tumor alloreactivity and develop alternative immunotherapy weapons against cancer.
The advances of current technologies have optimized the ex vivo expansion/activation of immune effectors and have selectively targeted checkpoint inhibitors also in the field of haplo-HSCT, where NK cells and γδ T lymphocytes early provide protection against cancers. Although helper ILCs could theoretically play a key role against tumors, the investigations of their clinical and functional impacts following HSCT are still in their infancy and must be deeper exploited. Our challenges and clinical perspectives over the next decade rely on our ability to give answers to the several important biological questions we still have on these matters.
Author Contributions
EZ, MC, CD, and DM wrote the manuscript and approved the final version.
Funding
This work was supported by Fondazione Cariplo (2015/0603 to DM), Associazione Italiana per la Ricerca sul Cancro (IG 2018-21567 to DM), and Intramural Research Funding of Istituto Clinico Humanitas (to DM). EZ was a recipient of the Nella Orlandini fellowship from the Associazione Italiana per la Ricerca sul Cancro (2018-20870). CD was a recipient of the post-doctoral fellowships from the Fondazione Umberto Veronesi (2017-1464, 2018-1974, 2019-2563).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Esiashvili N, Pulsipher MA, Hematopoietic stem cell transplantation. In: Merchant TE, Kortmann RD, editors. Pediatric Radiation Oncology. Cham: Springer International Publishing (2018). p. 301–11.
2. Locatelli F, Pende D, Maccario R, Mingari MC, Moretta A, Moretta L. Haploidentical hemopoietic stem cell transplantation for the treatment of high-risk leukemias: how NK cells make the difference. Clin Immunol. (2009) 133:171–8. doi: 10.1016/j.clim.2009.04.009
3. Bosch M, Khan FM, Storek J. Immune reconstitution after hematopoietic cell transplantation. Curr Opin Hematol. (2012) 19:324–35. doi: 10.1097/MOH.0b013e328353bc7d
4. Mackall C, Fry T, Gress R, Peggs K, Storek J, Toubert AB, et al. Background to hematopoietic cell transplantation, including post transplant immune recovery. Bone Marrow Transplant. (2009) 44:457–62. doi: 10.1038/bmt.2009.255
5. Aversa F, Terenzi A, Tabilio A, Falzetti F, Carotti A, Ballanti S, et al. Full haplotype-mismatched hematopoietic stem-cell transplantation: a phase II study in patients with acute leukemia at high risk of relapse. J Clin Oncol. (2005) 23:3447–54. doi: 10.1200/JCO.2005.09.117
6. Nguyen S, Dhedin N, Vernant JP, Kuentz M, Al Jijakli A, Rouas-Freiss N, et al. NK-cell reconstitution after haploidentical hematopoietic stem-cell transplantations: immaturity of NK cells and inhibitory effect of NKG2A override GvL effect. Blood. (2005) 105:4135–42. doi: 10.1182/blood-2004-10-4113
7. Chen X, Hale GA, Barfield R, Benaim E, Leung WH, Knowles J, et al. Rapid immune reconstitution after a reduced-intensity conditioning regimen and a CD3-depleted haploidentical stem cell graft for paediatric refractory haematological malignancies. Br J Haematol. (2006) 135:524–32. doi: 10.1111/j.1365-2141.2006.06330.x
8. Chang YJ, Zhao XY, Huang XJ. Effects of the NK cell recovery on outcomes of unmanipulated haploidentical blood and marrow transplantation for patients with hematologic malignancies. Biol Blood Marrow Transplant. (2008) 14:323–34. doi: 10.1016/j.bbmt.2007.12.497
9. Chang YJ, Zhao XY, Huo MR, Xu LP, Liu DH, Liu KY, et al. Influence of lymphocyte recovery on outcome of haploidentical transplantation for hematologic malignancies. Medicine. (2009) 88:322–30. doi: 10.1097/MD.0b013e3181c167e2
10. Federmann B, Hagele M, Pfeiffer M, Wirths S, Schumm M, Faul C, et al. Immune reconstitution after haploidentical hematopoietic cell transplantation: impact of reduced intensity conditioning and CD3/CD19 depleted grafts. Leukemia. (2011) 25:121–9. doi: 10.1038/leu.2010.235
11. Ciurea SO, Mulanovich V, Jiang Y, Bassett R, Rondon G, McMannis J, et al. Lymphocyte recovery predicts outcomes in cord blood and T cell-depleted haploidentical stem cell transplantation. Biol Blood Marrow Transplant. (2011) 17:1169–75. doi: 10.1016/j.bbmt.2010.11.020
12. Oevermann L, Lang P, Feuchtinger T, Schumm M, Teltschik HM, Schlegel P, et al. Immune reconstitution and strategies for rebuilding the immune system after haploidentical stem cell transplantation. Ann N Y Acad Sci. (2012) 1266:161–70. doi: 10.1111/j.1749-6632.2012.06606.x
13. Park BG, Park CJ, Jang S, Chi HS, Kim DY, Lee JH, et al. Reconstitution of lymphocyte subpopulations after hematopoietic stem cell transplantation: comparison of hematologic malignancies and donor types in event-free patients. Leuk Res. (2015) 39:1334–41. doi: 10.1016/j.leukres.2015.09.010
14. Williams KM, Gress RE. Immune reconstitution and implications for immunotherapy following haematopoietic stem cell transplantation. Best Pract Res Clin Haematol. (2008) 21:579–96. doi: 10.1016/j.beha.2008.06.003
15. Chang YJ, Zhao XY, Huang XJ. Immune reconstitution after haploidentical hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. (2014) 20:440–9. doi: 10.1016/j.bbmt.2013.11.028
16. Chang YJ, Zhao XY, Huo MR, Xu LP, Liu DH, Liu KY, et al. Immune reconstitution following unmanipulated HLA-mismatched/haploidentical transplantation compared with HLA-identical sibling transplantation. J Clin Immunol. (2012) 32:268–80. doi: 10.1007/s10875-011-9630-7
17. Storek J, Dawson MA, Storer B, Stevens-Ayers T, Maloney DG, Marr KA, et al. Immune reconstitution after allogeneic marrow transplantation compared with blood stem cell transplantation. Blood. (2001) 97:3380–9. doi: 10.1182/blood.V97.11.3380
18. Zimmerli W, Zarth A, Gratwohl A, Speck B. Neutrophil function and pyogenic infections in bone marrow transplant recipients. Blood. (1991) 77:393–9. doi: 10.1182/blood.V77.2.393.bloodjournal772393
19. Roberto A, Di Vito C, Zaghi E, Mazza MC, Capucetti A, Calvi M, et al. The early expansion of anergic NKG2A(pos)/CD56(dim)/CD16(neg) natural killer represents a therapeutic target in haploidentical hematopoietic stem cell transplantation. Haematologica. (2018) 103:1390–402. doi: 10.3324/haematol.2017.186619
20. Huang XJ, Liu DH, Liu KY, Xu LP, Chen H, Han W, et al. Haploidentical hematopoietic stem cell transplantation without in vitro T-cell depletion for the treatment of hematological malignancies. Bone Marrow Transplant. (2006) 38:291–7. doi: 10.1038/sj.bmt.1705445
21. McCurdy SR, Luznik L. Immune reconstitution after T-cell replete HLA-haploidentical transplantation. Semin Hematol. (2019) 56:221–6. doi: 10.1053/j.seminhematol.2019.03.005
22. Luznik L, O'Donnell PV, Symons HJ, Chen AR, Leffell MS, Zahurak M, et al. HLA-haploidentical bone marrow transplantation for hematologic malignancies using nonmyeloablative conditioning and high-dose, posttransplantation cyclophosphamide. Biol Blood Marrow Transplant. (2008) 14:641–50. doi: 10.1016/j.bbmt.2008.03.005
23. Lee KH, Lee JH, Lee JH, Kim DY, Seol M, Lee YS, et al. Reduced-intensity conditioning therapy with busulfan, fludarabine, and antithymocyte globulin for HLA-haploidentical hematopoietic cell transplantation in acute leukemia and myelodysplastic syndrome. Blood. (2011) 118:2609–17. doi: 10.1182/blood-2011-02-339838
24. Arcese W, Picardi A, Santarone S, De Angelis G, Cerretti R, Cudillo L, et al. Haploidentical, G-CSF-primed, unmanipulated bone marrow transplantation for patients with high-risk hematological malignancies: an update. Bone Marrow Transplant. (2015) 50(Suppl 2):S24–30. doi: 10.1038/bmt.2015.91
25. Locatelli F, Merli P, Pagliara D, Li Pira G, Falco M, Pende D, et al. Outcome of children with acute leukemia given HLA-haploidentical HSCT after αβ T-cell and B-cell depletion. Blood. (2017) 130:677–85. doi: 10.1182/blood-2017-04-779769
26. Bollard CM, Rooney CM, Heslop HE. T-cell therapy in the treatment of post-transplant lymphoproliferative disease. Nat Rev Clin Oncol. (2012) 9:510–9. doi: 10.1038/nrclinonc.2012.111
27. Maschan M, Shelikhova L, Ilushina M, Kurnikova E, Boyakova E, Balashov D, et al. TCR-alpha/beta and CD19 depletion and treosulfan-based conditioning regimen in unrelated and haploidentical transplantation in children with acute myeloid leukemia. Bone Marrow Transplant. (2016) 51:668–74. doi: 10.1038/bmt.2015.343
28. Airoldi I, Bertaina A, Prigione I, Zorzoli A, Pagliara D, Cocco C, et al. γδ T-cell reconstitution after HLA-haploidentical hematopoietic transplantation depleted of TCR- αβ+/CD19+ lymphocytes. Blood. (2015) 125:2349–58. doi: 10.1182/blood-2014-09-599423
29. Lang P, Feuchtinger T, Teltschik HM, Schwinger W, Schlegel P, Pfeiffer M, et al. Improved immune recovery after transplantation of TCRαβ/CD19-depleted allografts from haploidentical donors in pediatric patients. Bone Marrow Transplant. (2015) 50(Suppl 2):S6–10. doi: 10.1038/bmt.2015.87
30. Vogelsang GB, Lee L, Bensen-Kennedy DM. Pathogenesis and treatment of graft-versus-host disease after bone marrow transplant. Annu Rev Med. (2003) 54:29–52. doi: 10.1146/annurev.med.54.101601.152339
31. Zeiser R, Blazar BR. Acute graft-versus-host disease - biologic process, prevention, and therapy. N Engl J Med. (2017) 377:2167–79. doi: 10.1056/NEJMra1609337
33. Aversa F. T cell depleted haploidentical transplantation: positive selection. Pediatr Rep. (2011) 3(Suppl 2):e14. doi: 10.4081/pr.2011.s2.e14
34. Robinson TM, O'Donnell PV, Fuchs EJ, Luznik L. Haploidentical bone marrow and stem cell transplantation: experience with post-transplantation cyclophosphamide. Semin Hematol. (2016) 53:90–7. doi: 10.1053/j.seminhematol.2016.01.005
35. Mancusi A, Ruggeri L, Velardi A. Haploidentical hematopoietic transplantation for the cure of leukemia: from its biology to clinical translation. Blood. (2016) 128:2616–23. doi: 10.1182/blood-2016-07-730564
36. Lee CJ, Savani BN, Mohty M, Labopin M, Ruggeri A, Schmid C, et al. Haploidentical hematopoietic cell transplantation for adult acute myeloid leukemia: a position statement from the Acute Leukemia Working Party of the European Society for Blood and Marrow Transplantation. Haematologica. (2017) 102:1810–22. doi: 10.3324/haematol.2017.176107
37. Dupont L, Reeves MB. Cytomegalovirus latency and reactivation: recent insights into an age old problem. Rev Med Virol. (2016) 26:75–89. doi: 10.1002/rmv.1862
38. Sinclair J, Sissons P. Latency and reactivation of human cytomegalovirus. J Gen Virol. (2006) 87:1763–79. doi: 10.1099/vir.0.81891-0
39. Landolfo S, Gariglio M, Gribaudo G, Lembo D. The human cytomegalovirus. Pharmacol Ther. (2003) 98:269–97. doi: 10.1016/S0163-7258(03)00034-2
40. Ljungman P, Brand R, Hoek J, de la Camara R, Cordonnier C, Einsele H, et al. Infectious diseases working party of the european group for, and T marrow, donor cytomegalovirus status influences the outcome of allogeneic stem cell transplant: a study by the European group for blood and marrow transplantation. Clin Infect Dis. (2014) 59:473–81. doi: 10.1093/cid/ciu364
41. Venton G, Crocchiolo R, Furst S, Granata A, Oudin C, Faucher C, et al. Risk factors of Ganciclovir-related neutropenia after allogeneic stem cell transplantation: a retrospective monocentre study on 547 patients. Clin Microbiol Infect. (2014) 20:160–6. doi: 10.1111/1469-0691.12222
42. Crocchiolo R, Bramanti S, Vai A, Sarina B, Mineri R, Casari E, et al. Infections after T-replete haploidentical transplantation and high-dose cyclophosphamide as graft-versus-host disease prophylaxis. Transpl Infect Dis. (2015) 17:242–9. doi: 10.1111/tid.12365
43. Ciurea SO, Mulanovich V, Saliba RM, Bayraktar UD, Jiang Y, Bassett R, et al. Improved early outcomes using a T cell replete graft compared with T cell depleted haploidentical hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. (2012) 18:1835–44. doi: 10.1016/j.bbmt.2012.07.003
44. Raiola AM, Dominietto A, Ghiso A, Di Grazia C, Lamparelli T, Gualandi F, et al. Unmanipulated haploidentical bone marrow transplantation and posttransplantation cyclophosphamide for hematologic malignancies after myeloablative conditioning. Biol Blood Marrow Transplant. (2013) 19:117–22. doi: 10.1016/j.bbmt.2012.08.014
45. Blyth E, Withers B, Clancy L, Gottlieb D. CMV-specific immune reconstitution following allogeneic stem cell transplantation. Virulence. (2016) 7:967–80. doi: 10.1080/21505594.2016.1221022
46. Marty FM, Ljungman P, Chemaly RF, Maertens J, Dadwal SS, Duarte RF, et al. Letermovir prophylaxis for cytomegalovirus in hematopoietic-cell transplantation. N Engl J Med. (2017) 377:2433–44. doi: 10.1056/NEJMoa1706640
47. Matthes-Martin S, Lion T, Aberle SW, Fritsch G, Lawitschka A, Bittner B, et al. Pre-emptive treatment of CMV DNAemia in paediatric stem cell transplantation: the impact of recipient and donor CMV serostatus on the incidence of CMV disease and CMV-related mortality. Bone Marrow Transplant. (2003) 31:803–8. doi: 10.1038/sj.bmt.1703927
48. Roberto A, Castagna L, Zanon V, Bramanti S, Crocchiolo R, McLaren JE, et al. Role of naive-derived T memory stem cells in T-cell reconstitution following allogeneic transplantation. Blood. (2015) 125:2855–64. doi: 10.1182/blood-2014-11-608406
49. Nichols WG, Corey L, Gooley T, Davis C, Boeckh M. High risk of death due to bacterial and fungal infection among cytomegalovirus. (CMV)-seronegative recipients of stem cell transplants from seropositive donors: evidence for indirect effects of primary CMV infection. J Infect Dis. (2002) 185:273–82. doi: 10.1086/338624
50. Gandhi MK, Wills MR, Okecha G, Day EK, Hicks R, Marcus RE, et al. Late diversification in the clonal composition of human cytomegalovirus-specific CD8+ T cells following allogeneic hemopoietic stem cell transplantation. Blood. (2003) 102:3427–38. doi: 10.1182/blood-2002-12-3689
51. Crough T, Khanna R. Immunobiology of human cytomegalovirus: from bench to bedside. Clin Microbiol Rev. (2009) 22:76–98. doi: 10.1128/CMR.00034-08
52. Crocchiolo R, Castagna L, Furst S, Devillier R, Sarina B, Bramanti S, et al. The patient's CMV serological status affects clinical outcome after T-cell replete haplo-HSCT and post-transplant cyclophosphamide. Bone Marrow Transplant. (2016) 51:1134–6. doi: 10.1038/bmt.2016.69
53. Itzykson R, Robin M, Moins-Teisserenc H, Delord M, Busson M, Xhaard A, et al. Cytomegalovirus shapes long-term immune reconstitution after allogeneic stem cell transplantation. Haematologica. (2015) 100:114–23. doi: 10.3324/haematol.2014.113415
54. Lugthart G, van Ostaijen-Ten Dam MM, Jol-van der Zijde CM, van Holten TC, Kester MG, Heemskerk MH, et al. Early cytomegalovirus reactivation leaves a specific and dynamic imprint on the reconstituting T cell compartment long-term after hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. (2014) 20:655–61. doi: 10.1016/j.bbmt.2014.01.018
55. Knight A, Madrigal AJ, Grace S, Sivakumaran J, Kottaridis P, Mackinnon S, et al. The role of Vdelta2-negative γδ T cells during cytomegalovirus reactivation in recipients of allogeneic stem cell transplantation. Blood. (2010) 116:2164–72. doi: 10.1182/blood-2010-01-255166
56. Storek J. Immunological reconstitution after hematopoietic cell transplantation - its relation to the contents of the graft. Exp Opin Biol Ther. (2008) 8:583–97. doi: 10.1517/14712598.8.5.583
57. Ravens S, Schultze-Florey C, Raha S, Sandrock I, Drenker M, Oberdorfer L, et al. Human γδ T cells are quickly reconstituted after stem-cell transplantation and show adaptive clonal expansion in response to viral infection. Nat Immunol. (2017) 18:393–401. doi: 10.1038/ni.3686
58. Lugli E, Marcenaro E, Mavilio D. NK cell subset redistribution during the course of viral infections. Front Immunol. (2014) 5:390. doi: 10.3389/fimmu.2014.00390
59. Diefenbach A, Colonna M, Koyasu S. Development, differentiation, and diversity of innate lymphoid cells. Immunity. (2014) 41:354–65. doi: 10.1016/j.immuni.2014.09.005
60. Eberl G, Colonna M, Di Santo JP, McKenzie AN. Innate lymphoid cells. Innate lymphoid cells: a new paradigm in immunology. Science. (2015) 348:aaa6566. doi: 10.1126/science.aaa6566
61. Klose CS, Artis D. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat Immunol. (2016) 17:765–74. doi: 10.1038/ni.3489
62. Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells: 10 years on. Cell. (2018) 174:1054–66. doi: 10.1016/j.cell.2018.07.017
63. Trabanelli S, Gomez-Cadena A, Salome B, Michaud K, Mavilio D, Landis BN, et al. Human innate lymphoid cells. (ILCs): toward a uniform immune-phenotyping. Cytomet Clin Cytom. (2018) 94:392–9. doi: 10.1002/cyto.b.21614
64. Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells–a proposal for uniform nomenclature. Nat Rev Immunol. (2013) 13:145–9. doi: 10.1038/nri3365
65. Park M, Im HJ, Lee YJ, Park N, Jang S, Kwon SW, et al. Reconstitution of T and NK cells after haploidentical hematopoietic cell transplantation using αβ T cell-depleted grafts and the clinical implication of γδ T cells. Clin Transplant. (2018) 32:e13147. doi: 10.1111/ctr.13147
66. Abt MC, Lewis BB, Caballero S, Xiong H, Carter RA, Susac B, et al. Innate immune defenses mediated by two ILC subsets are critical for protection against acute clostridium difficile infection. Cell Host Microbe. (2015) 18:27–37. doi: 10.1016/j.chom.2015.06.011
67. Klose CSN, Flach M, Mohle L, Rogell L, Hoyler T, Ebert K, et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. (2014) 157:340–56. doi: 10.1016/j.cell.2014.03.030
68. Shah SV, Manickam C, Ram DR, Reeves RK. Innate lymphoid cells in HIV/SIV infections. Front Immunol. (2017) 8:1818. doi: 10.3389/fimmu.2017.01818
69. Fuchs A, Vermi W, Lee JS, Lonardi S, Gilfillan S, Newberry RD, et al. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-gamma-producing cells. Immunity. (2013) 38:769–81. doi: 10.1016/j.immuni.2013.02.010
70. Roan F, Stoklasek TA, Whalen E, Molitor JA, Bluestone JA, Buckner JH, et al. CD4+ Group 1 innate lymphoid cells. (ILC) form a functionally distinct ILC subset that is increased in systemic sclerosis. J Immunol. (2016) 196:2051–62. doi: 10.4049/jimmunol.1501491
71. Lim AI, Di Santo JP. ILC-poiesis: ensuring tissue ILC differentiation at the right place and time. Eur J Immunol. (2019) 49:11–18. doi: 10.1002/eji.201747294
72. Koyasu S, Moro K, Tanabe M, Takeuchi T. Natural helper cells: a new player in the innate immune response against helminth infection. Adv Immunol. (2010) 108:21–44. doi: 10.1016/B978-0-12-380995-7.00002-1
73. Rauber S, Luber M, Weber S, Maul L, Soare A, Wohlfahrt T, et al. Resolution of inflammation by interleukin-9-producing type 2 innate lymphoid cells. Nat Med. (2017) 23:938–44. doi: 10.1038/nm.4373
74. Gasteiger G, Fan X, Dikiy S, Lee SY, Rudensky AY. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science. (2015) 350:981–5. doi: 10.1126/science.aac9593
75. Moro K, Kabata H, Tanabe M, Koga S, Takeno N, Mochizuki M, et al. Interferon and IL-27 antagonize the function of group 2 innate lymphoid cells and type 2 innate immune responses. Nat Immunol. (2016) 17:76–86. doi: 10.1038/ni.3309
76. Monticelli LA, Osborne LC, Noti M, Tran SV, Zaiss DM, Artis D. IL-33 promotes an innate immune pathway of intestinal tissue protection dependent on amphiregulin-EGFR interactions. Proc Natl Acad Sci USA. (2015) 112:10762–7. doi: 10.1073/pnas.1509070112
77. Montaldo E, Juelke K, Romagnani C. Group 3 innate lymphoid cells. (ILC3s): origin, differentiation, and plasticity in humans and mice. Eur J Immunol. (2015) 45:2171–82. doi: 10.1002/eji.201545598
78. Glatzer T, Killig M, Meisig J, Ommert I, Luetke-Eversloh M, Babic M, et al. RORgammat(+) innate lymphoid cells acquire a proinflammatory program upon engagement of the activating receptor NKp44. Immunity. (2013) 38:1223–35. doi: 10.1016/j.immuni.2013.05.013
79. Zhong C, Zheng M, Zhu J. Lymphoid tissue inducer-A divergent member of the ILC family. Cytokine Growth Factor Rev. (2018) 42:5–12. doi: 10.1016/j.cytogfr.2018.02.004
80. Ishizuka IE, Chea S, Gudjonson H, Constantinides MG, Dinner AR, Bendelac A, et al. Single-cell analysis defines the divergence between the innate lymphoid cell lineage and lymphoid tissue-inducer cell lineage. Nat Immunol. (2016) 17:269–76. doi: 10.1038/ni.3344
81. Colonna M. Innate lymphoid cells: diversity, plasticity, and unique functions in immunity. Immunity. (2018) 48:1104–17. doi: 10.1016/j.immuni.2018.05.013
82. Huang Y, Mao K, Chen X, Sun MA, Kawabe T, Li W, et al. S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Science. (2018) 359:114–9. doi: 10.1126/science.aam5809
83. Vacca P, Montaldo E, Croxatto D, Moretta F, Bertaina A, Vitale C, et al. NK cells and other innate lymphoid cells in hematopoietic stem cell transplantation. Front Immunol. (2016) 7:188. doi: 10.3389/fimmu.2016.00188
84. Munneke JM, Bjorklund AT, Mjosberg JM, Garming-Legert K, Bernink JH, Blom B, et al. Activated innate lymphoid cells are associated with a reduced susceptibility to graft-versus-host disease. Blood. (2014) 124:812–21. doi: 10.1182/blood-2013-11-536888
85. Trabanelli S, Curti A, Lecciso M, Salome B, Riether C, Ochsenbein A, et al. CD127+ innate lymphoid cells are dysregulated in treatment naive acute myeloid leukemia patients at diagnosis. Haematologica. (2015) 100:e257–60. doi: 10.3324/haematol.2014.119602
86. Vitale C, Ambrosini P, Montaldo E, Ballerini F, Moretta L, Mingari MC. IL-1β-releasing human acute myeloid leukemia blasts modulate natural killer cell differentiation from CD34+ precursors. Haematologica. (2015) 100:e42–5. doi: 10.3324/haematol.2014.110494
87. Dudakov JA, Hanash AM, Jenq RR, Young LF, Ghosh A, Singer NV, et al. Interleukin-22 drives endogenous thymic regeneration in mice. Science. (2012) 336:91–5. doi: 10.1126/science.1218004
88. Vely F, Barlogis V, Vallentin B, Neven B, Piperoglou C, Ebbo M, et al. Evidence of innate lymphoid cell redundancy in humans. Nat Immunol. (2016) 17:1291–9. doi: 10.1038/ni.3553
89. Moretta F, Petronelli F, Lucarelli B, Pitisci A, Bertaina A, Locatelli F, et al. The generation of human innate lymphoid cells is influenced by the source of hematopoietic stem cells and by the use of G-CSF. Eur J Immunol. (2016) 46:1271–8. doi: 10.1002/eji.201546079
90. Hanash AM, Dudakov JA, Hua G, O'Connor MH, Young LF, Singer NV, et al. Interleukin-22 protects intestinal stem cells from immune-mediated tissue damage and regulates sensitivity to graft versus host disease. Immunity. (2012) 37:339–50. doi: 10.1016/j.immuni.2012.05.028
91. Lindemans CA, Calafiore M, Mertelsmann AM, O'Connor MH, Dudakov JA, Jenq RR, et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. (2015) 528:560–4. doi: 10.1038/nature16460
92. Blazar BR, Murphy WJ, Abedi M. Advances in graft-versus-host disease biology and therapy. Nat Rev Immunol. (2012) 12:443–58. doi: 10.1038/nri3212
93. Sabat R, Ouyang W, Wolk K. Therapeutic opportunities of the IL-22-IL-22R1 system. Nat Rev Drug Discov. (2014) 13:21–38. doi: 10.1038/nrd4176
94. Rankin LC, Girard-Madoux MJ, Seillet C, Mielke LA, Kerdiles Y, Fenis A, et al. Complementarity and redundancy of IL-22-producing innate lymphoid cells. Nat Immunol. (2016) 17:179–86. doi: 10.1038/ni.3332
95. Roberto A, Castagna L, Gandolfi S, Zanon V, Bramanti S, Sarina B, et al. B-cell reconstitution recapitulates B-cell lymphopoiesis following haploidentical BM transplantation and post-transplant CY. Bone Marrow Transplant. (2015) 50:317–9. doi: 10.1038/bmt.2014.266
96. Russo A, Oliveira G, Berglund S, Greco R, Gambacorta V, Cieri N, et al. NK cell recovery after haploidentical HSCT with posttransplant cyclophosphamide: dynamics and clinical implications. Blood. (2018) 131:247–62. doi: 10.1182/blood-2017-05-780668
97. Weizman OE, Adams NM, Schuster IS, Krishna C, Pritykin Y, Lau C, et al. ILC1 confer early host protection at initial sites of viral infection. Cell. (2017) 171:795–808.e12. doi: 10.1016/j.cell.2017.09.052
98. Vashist N, Trittel S, Ebensen T, Chambers BJ, Guzman CA, Riese P. Influenza-activated ILC1s contribute to antiviral immunity partially influenced by differential GITR expression. Front Immunol. (2018) 9:505. doi: 10.3389/fimmu.2018.00505
99. Gorski SA, Hahn YS, Braciale TJ. Group 2 innate lymphoid cell production of IL-5 is regulated by NKT cells during influenza virus infection. PLoS Pathog. (2013) 9:e1003615. doi: 10.1371/journal.ppat.1003615
100. Duerr CU, McCarthy CD, Mindt BC, Rubio M, Meli AP, Pothlichet J, et al. Type I interferon restricts type 2 immunopathology through the regulation of group 2 innate lymphoid cells. Nat Immunol. (2016) 17:65–75. doi: 10.1038/ni.3308
101. Lanier LL. NK cell recognition. Annu Rev Immunol. (2005) 23:225–74. doi: 10.1146/annurev.immunol.23.021704.115526
102. Shifrin N, Raulet DH, Ardolino M. NK cell self tolerance, responsiveness and missing self recognition. Semin Immunol. (2014) 26:138–44. doi: 10.1016/j.smim.2014.02.007
103. Hudspeth K, Donadon M, Cimino M, Pontarini E, Tentorio P, Preti M, et al. Human liver-resident CD56(bright)/CD16(neg) NK cells are retained within hepatic sinusoids via the engagement of CCR5 and CXCR6 pathways. J Autoimm. (2016) 66:40–50. doi: 10.1016/j.jaut.2015.08.011
104. Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, et al. Innate or adaptive immunity? The example of natural killer cells. Science. (2011) 331:44–9. doi: 10.1126/science.1198687
105. Gupta N, Arthos J, Khazanie P, Steenbeke TD, Censoplano NM, Chung EA, et al. Targeted lysis of HIV-infected cells by natural killer cells armed and triggered by a recombinant immunoglobulin fusion protein: implications for immunotherapy. Virology. (2005) 332:491–7. doi: 10.1016/j.virol.2004.12.018
106. Beziat V, Liu LL, Malmberg JA, Ivarsson MA, Sohlberg E, Bjorklund AT, et al. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood. (2013) 121:2678–88. doi: 10.1182/blood-2012-10-459545
107. Farag SS, Fehniger TA, Ruggeri L, Velardi A, Caligiuri MA. Natural killer cell receptors: new biology and insights into the graft-versus-leukemia effect. Blood. (2002) 100:1935–47. doi: 10.1182/blood-2002-02-0350
108. Brodin P, Hoglund P. Beyond licensing and disarming: a quantitative view on NK-cell education. Eur J Immunol. (2008) 38:2934–7. doi: 10.1002/eji.200838760
109. Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. (2005) 5:201–14. doi: 10.1038/nri1570
110. Braud VM, Allan DS, O'Callaghan CA, Soderstrom K, D'Andrea A, Ogg GS, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. (1998) 391:795–9. doi: 10.1038/35869
111. Wada H, Matsumoto N, Maenaka K, Suzuki K, Yamamoto K. The inhibitory NK cell receptor CD94/NKG2A and the activating receptor CD94/NKG2C bind the top of HLA-E through mostly shared but partly distinct sets of HLA-E residues. Eur J Immunol. (2004) 34:81–90. doi: 10.1002/eji.200324432
112. Campbell KS, Purdy AK. Structure/function of human killer cell immunoglobulin-like receptors: lessons from polymorphisms, evolution, crystal structures and mutations. Immunology. (2011) 132:315–25. doi: 10.1111/j.1365-2567.2010.03398.x
113. Moretta A, Sivori S, Vitale M, Pende D, Morelli L, Augugliaro R, et al. Existence of both inhibitory. (p58) and activatory. (p50) receptors for HLA-C molecules in human natural killer cells. J Exp Med. (1995) 182:875–84. doi: 10.1084/jem.182.3.875
114. Manser AR, Weinhold S, Uhrberg M. Human KIR repertoires: shaped by genetic diversity and evolution. Immunol Rev. (2015) 267:178–96. doi: 10.1111/imr.12316
115. Zaghi E, Calvi M, Marcenaro E, Mavilio D, Di Vito C. Targeting NKG2A to elucidate natural killer cell ontogenesis and to develop novel immune-therapeutic strategies in cancer therapy. J Leukoc Biol. (2019) 105:1243–51. doi: 10.1002/JLB.MR0718-300R
116. Hsu KC, Liu XR, Selvakumar A, Mickelson E, O'Reilly RJ, Dupont B. Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J Immunol. (2002) 169:5118–29. doi: 10.4049/jimmunol.169.9.5118
117. Pegram HJ, Andrews DM, Smyth MJ, Darcy PK, Kershaw MH. Activating and inhibitory receptors of natural killer cells. Immunol Cell Biol. (2011) 89:216–24. doi: 10.1038/icb.2010.78
118. Kruse PH, Matta J, Ugolini S, Vivier E. Natural cytotoxicity receptors and their ligands. Immunol Cell Biol. (2014) 92:221–9. doi: 10.1038/icb.2013.98
119. Bryceson YT, March ME, Ljunggren HG, Long EO. Activation, coactivation, and costimulation of resting human natural killer cells. Immunol Rev. (2006) 214:73–91. doi: 10.1111/j.1600-065X.2006.00457.x
120. Di Vito C, Mikulak J, Mavilio D. On the way to become a Natural Killer cell. Front Immunol. (2019) 10:1812. doi: 10.3389/fimmu.2019.01812
121. Freud AG, Caligiuri MA. Human natural killer cell development. Immunol Rev. (2006) 214:56–72. doi: 10.1111/j.1600-065X.2006.00451.x
122. Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol. (2001) 22:633–40. doi: 10.1016/S1471-4906(01)02060-9
123. Mikulak J, Di Vito C, Zaghi E, Mavilio D. Host immune responses in HIV-1 infection: the emerging pathogenic role of siglecs and their clinical correlates. Front Immunol. (2017) 8:314. doi: 10.3389/fimmu.2017.00314
124. Mattiola I, Pesant M, Tentorio PF, Molgora M, Marcenaro E, Lugli E, et al. Priming of human resting NK cells by autologous M1 macrophages via the engagement of IL-1β, IFN-β, and IL-15 pathways. J Immunol. (2015) 195:2818–28. doi: 10.4049/jimmunol.1500325
125. Nash WT, Teoh J, Wei H, Gamache A, Brown MG. Know thyself: NK-cell inhibitory receptors prompt self-tolerance, education, and viral control. Front Immunol. (2014) 5:175. doi: 10.3389/fimmu.2014.00175
126. Castagna L, Mavilio D. Re-discovering NK cell allo-reactivity in the therapy of solid tumors. J Immunother Cancer. (2016) 4:54. doi: 10.1186/s40425-016-0159-4
127. Caligiuri MA. Human natural killer cells. Blood. (2008) 112:461–9. doi: 10.1182/blood-2007-09-077438
128. Moretta A. The dialogue between human natural killer cells and dendritic cells. Curr Opin Immunol. (2005) 17:306–11. doi: 10.1016/j.coi.2005.03.004
129. Fali T, Papagno L, Bayard C, Mouloud Y, Boddaert J, Sauce D, et al. New insights into lymphocyte differentiation and aging from telomere length and telomerase activity measurements. J Immunol. (2019) 202:1962–9. doi: 10.4049/jimmunol.1801475
130. Romagnani C, Juelke K, Falco M, Morandi B, D'Agostino A, Costa R, et al. CD56brightCD16- killer Ig-like receptor- NK cells display longer telomeres and acquire features of CD56dim NK cells upon activation. J Immunol. (2007) 178:4947–55. doi: 10.4049/jimmunol.178.8.4947
131. Yokoyama WM, Kim S. Licensing of natural killer cells by self-major histocompatibility complex class. Immunol Rev. (2006) 214:143–54. doi: 10.1111/j.1600-065X.2006.00458.x
132. Cichicki F, Schlums H, Theorell J, Tesi B, Miller JS, Ljunggren HG, et al. Diversification and functional specialization of human NK cell subsets. Curr Top Microbiol Immunol. (2016) 395:63–94. doi: 10.1007/82_2015_487
133. Hattori N, Saito B, Sasaki Y, Shimada S, Murai S, Abe M, et al. Status of natural killer cell recovery in day 21 bone marrow after allogeneic hematopoietic stem cell transplantation predicts clinical outcome. Biol Blood Marrow Transplant. (2018) 24:1841–7. doi: 10.1016/j.bbmt.2018.05.007
134. Minculescu L, Marquart HV, Friis LS, Petersen SL, Schiodt I, Ryder LP, et al. Early natural killer cell reconstitution predicts overall survival in T cell-replete allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. (2016) 22:2187–93. doi: 10.1016/j.bbmt.2016.09.006
135. Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. (2002) 295:2097–100. doi: 10.1126/science.1068440
136. Moretta L, Locatelli F, Pende D, Marcenaro E, Mingari MC, Moretta A, Killer Ig-like receptor-mediated control of natural killer cell alloreactivity in haploidentical hematopoietic stem cell transplantation. Blood. (2011) 117:764–71. doi: 10.1182/blood-2010-08-264085
137. Ullah MA, Hill GR, Tey SK. Functional reconstitution of natural killer cells in allogeneic hematopoietic stem cell transplantation. Front Immunol. (2016) 7:144. doi: 10.3389/fimmu.2016.00144
138. Pical-Izard C, Crocchiolo R, Granjeaud S, Kochbati E, Just-Landi S, Chabannon C, et al. Reconstitution of natural killer cells in HLA-matched HSCT after reduced-intensity conditioning: impact on clinical outcome. Biol Blood Marrow Transplant. (2015) 21:429–39. doi: 10.1016/j.bbmt.2014.11.681
139. Vago L, Forno B, Sormani MP, Crocchiolo R, Zino E, Di Terlizzi S, et al. Temporal, quantitative, and functional characteristics of single-KIR-positive alloreactive natural killer cell recovery account for impaired graft-versus-leukemia activity after haploidentical hematopoietic stem cell transplantation. Blood. (2008) 112:3488–99. doi: 10.1182/blood-2007-07-103325
140. Nguyen S, Kuentz M, Vernant JP, Dhedin N, Bories D, Debre P, et al. Involvement of mature donor T cells in the NK cell reconstitution after haploidentical hematopoietic stem-cell transplantation. Leukemia. (2008) 22:344–52. doi: 10.1038/sj.leu.2405041
141. Huenecke S, Cappel C, Esser R, Pfirrmann V, Salzmann-Manrique E, Betz S, et al. Development of three different NK cell subpopulations during immune reconstitution after pediatric allogeneic hematopoietic stem cell transplantation: prognostic markers in GvHD and viral infections. Front Immunol. (2017) 8:109. doi: 10.3389/fimmu.2017.00109
142. Stabile H, Nisti P, Pagliara D, Locatelli F, Santoni A, Gismondi A. Response to comment on Multifunctional human CD56low CD16low NK cells are the prominent subset in bone marrow of both pediatric healthy donors and leukemic patients. Haematologica. (2015) 100:e332–3. doi: 10.3324/haematol.2014.116053
143. Ullrich E, Salzmann-Manrique E, Bakhtiar S, Bremm M, Gerstner S, Herrmann E, et al. Relation between Acute GVHD and NK cell subset reconstitution following allogeneic stem cell transplantation. Front Immunol. (2016) 7:595. doi: 10.3389/fimmu.2016.00595
144. Savani BN, Mielke S, Adams S, Uribe M, Rezvani K, Yong AS, et al. Rapid natural killer cell recovery determines outcome after T-cell-depleted HLA-identical stem cell transplantation in patients with myeloid leukemias but not with acute lymphoblastic leukemia. Leukemia. (2007) 21:2145–52. doi: 10.1038/sj.leu.2404892
145. Kim SY, Lee H, Han MS, Shim H, Eom HS, Park B, et al. Post-transplantation natural killer cell count: a predictor of acute graft-versus-host disease and survival outcomes after allogeneic hematopoietic stem cell transplantation. Clin Lymphoma Myeloma Leuk. (2016) 16:527–35.e2. doi: 10.1016/j.clml.2016.06.013
146. de Koning C, Plantinga M, Besseling P, Boelens JJ, Nierkens S. Immune reconstitution after allogeneic hematopoietic cell transplantation in children. Biol Blood Marrow Transplant. (2016) 22:195–206. doi: 10.1016/j.bbmt.2015.08.028
147. Moretta A, Locatelli F, Moretta L. Human NK cells: from HLA class I-specific killer Ig-like receptors to the therapy of acute leukemias. Immunol Rev. (2008) 224:58–69. doi: 10.1111/j.1600-065X.2008.00651.x
148. Olson JA, Leveson-Gower DB, Gill S, Baker J, Beilhack A, Negrin RS. NK cells mediate reduction of GVHD by inhibiting activated, alloreactive T cells while retaining GVT effects. Blood. (2010) 115:4293–301. doi: 10.1182/blood-2009-05-222190
149. Lang PA, Lang KS, Xu HC, Grusdat M, Parish IA, Recher M, et al. Natural killer cell activation enhances immune pathology and promotes chronic infection by limiting CD8+ T-cell immunity. Proc Natl Acad Sci USA. (2012) 109:1210–5. doi: 10.1073/pnas.1118834109
150. ChanYLT Zuo J, Inman C, Croft W, Begum J, Croudace J, et al. NK cells produce high levels of IL-10 early after allogeneic stem cell transplantation and suppress development of acute GVHD. Eur J Immunol. (2018) 48:316–29. doi: 10.1002/eji.201747134
151. Simonetta F, Alvarez M, Negrin RS. Natural killer cells in graft-versus-host-disease after allogeneic hematopoietic cell transplantation. Front Immunol. (2017) 8:465. doi: 10.3389/fimmu.2017.00465
152. Hu LJ, Zhao XY, Yu XX, Lv M, Han TT, Han W, et al. Quantity and quality reconstitution of NKG2A(+) natural killer cells are associated with graft-versus-host disease after allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transplant. (2019) 25:1–11. doi: 10.1016/j.bbmt.2018.08.008
153. Kordelas L, Steckel NK, Horn PA, Beelen DW, Rebmann V. The activating NKG2C receptor is significantly reduced in NK cells after allogeneic stem cell transplantation in patients with severe graft-versus-host disease. Int J Mol Sci. (2016) 17:1797. doi: 10.3390/ijms17111797
154. Willem C, Makanga DR, Guillaume T, Maniangou B, Legrand N, Gagne K, et al. Impact of KIR/HLA incompatibilities on NK cell reconstitution and clinical outcome after T cell-replete haploidentical hematopoietic stem cell transplantation with posttransplant cyclophosphamide. J Immunol. (2019) 202:2141–52. doi: 10.4049/jimmunol.1801489
155. Ohata K, Espinoza JL, Lu X, Kondo Y, Nakao S. Mycophenolic acid inhibits natural killer cell proliferation and cytotoxic function: a possible disadvantage of including mycophenolate mofetil in the graft-versus-host disease prophylaxis regimen. Biol Blood Marrow Transplant. (2011) 17:205–13. doi: 10.1016/j.bbmt.2010.08.014
156. Bremm M, Huenecke S, Zimmermann O, Pfirrmann V, Quaiser A, Bonig H, et al. In-vitro influence of mycophenolate mofetil. (MMF) and Ciclosporin A. (CsA) on cytokine induced killer. (CIK) cell immunotherapy. J Transl Med. (2016) 14:264. doi: 10.1186/s12967-016-1024-4
157. Orange JS. Human natural killer cell deficiencies and susceptibility to infection. Microbes Infect. (2002) 4:1545–58. doi: 10.1016/S1286-4579(02)00038-2
158. De Angelis C, Mancusi A, Ruggeri L, Capanni M, Urbani E, Velardi A, et al. Expansion of CD56-negative, CD16-positive, KIR-expressing natural killer cells after T cell-depleted haploidentical hematopoietic stem cell transplantation. Acta Haematol. (2011) 126:13–20. doi: 10.1159/000323661
159. Della Chiesa M, Sivori S, Carlomagno S, Moretta L, Moretta A. Activating KIRs and NKG2C in Viral Infections: toward NK Cell Memory? Front Immunol. (2015) 6:573. doi: 10.3389/fimmu.2015.00573
160. Bjorkstrom NK, Ljunggren HG, Sandberg JK. CD56 negative NK cells: origin, function, and role in chronic viral disease. Trends Immunol. (2010) 31:401–6. doi: 10.1016/j.it.2010.08.003
161. Mavilio D, Benjamin J, Daucher M, Lombardo G, Kottilil S, Planta MA, et al. Natural killer cells in HIV-1 infection: dichotomous effects of viremia on inhibitory and activating receptors and their functional correlates. Proc Natl Acad Sci USA. (2003) 100:15011–6. doi: 10.1073/pnas.2336091100
162. Mavilio D, Lombardo G, Benjamin J, Kim D, Follman D, Marcenaro E, et al. Characterization of CD56-/CD16+ natural killer. (NK) cells: a highly dysfunctional NK subset expanded in HIV-infected viremic individuals. Proc Natl Acad Sci USA. (2005) 102:2886–91. doi: 10.1073/pnas.0409872102
163. Brunetta E, Hudspeth KL, Mavilio D. Pathologic natural killer cell subset redistribution in HIV-1 infection: new insights in pathophysiology and clinical outcomes. J Leukoc Biol. (2010) 88:1119–30. doi: 10.1189/jlb.0410225
164. Milush JM, Lopez-Verges S, York VA, Deeks SG, Martin JN, Hecht FM, et al. CD56negCD16(+) NK cells are activated mature NK cells with impaired effector function during HIV-1 infection. Retrovirology. (2013) 10:158. doi: 10.1186/1742-4690-10-158
165. Fogli M, Mavilio D, Brunetta E, Varchetta S, Ata K, Roby G, et al. Lysis of endogenously infected CD4+ T cell blasts by rIL-2 activated autologous natural killer cells from HIV-infected viremic individuals. PLoS Pathog. (2008) 4:e1000101. doi: 10.1371/journal.ppat.1000101
166. Zhao XY, Luo XY, Yu XX, Zhao XS, Han TT, Chang YJ, et al. Recipient-donor KIR ligand matching prevents CMV reactivation post-haploidentical T cell-replete transplantation. Br J Haematol. (2017) 177:766–81. doi: 10.1111/bjh.14622
167. Foley B, Cooley S, Verneris MR, Pitt M, Curtsinger J, Luo X, et al. Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood. (2012) 119:2665–74. doi: 10.1182/blood-2011-10-386995
168. Lopez-Verges S, Milush JM, Schwartz BS, Pando MJ, Jarjoura J, York VA, et al. Expansion of a unique CD57(+)NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc Natl Acad Sci USA. (2011) 108:14725–32. doi: 10.1073/pnas.1110900108
169. Muccio L, Bertaina A, Falco M, Pende D, Meazza R, Lopez-Botet M, et al. Analysis of memory-like natural killer cells in human cytomegalovirus-infected children undergoing αβ+T and B cell-depleted hematopoietic stem cell transplantation for hematological malignancies. Haematologica. (2016) 101:371–81. doi: 10.3324/haematol.2015.134155
170. Della Chiesa M, Muccio L, Moretta A. CMV induces rapid NK cell maturation in HSCT recipients. Immunol Lett. (2013) 155:11–3. doi: 10.1016/j.imlet.2013.09.020
171. O'Sullivan TE, Sun JC, Lanier LL. Natural killer cell memory. Immunity. (2015) 43:634–45. doi: 10.1016/j.immuni.2015.09.013
172. Romee R, Rosario M, Berrien-Elliott MM, Wagner JA, Jewell BA, Schappe T, et al. Cytokine-induced memory-like natural killer cells exhibit enhanced responses against myeloid leukemia. Sci Transl Med. (2016) 8:357ra123. doi: 10.1126/scitranslmed.aaf2341
173. Sun JC, Lopez-Verges S, Kim CC, DeRisi JL, Lanier LL. NK cells and immune “memory”. J Immunol. (2011) 186:1891–7. doi: 10.4049/jimmunol.1003035
174. Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. (2002) 296:1323–6. doi: 10.1126/science.1070884
175. Guma M, Angulo A, Vilches C, Gomez-Lozano N, Malats N, Lopez-Botet M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood. (2004) 104:3664–71. doi: 10.1182/blood-2004-05-2058
176. Guma M, Budt M, Saez A, Brckalo T, Hengel H, Angulo A, et al. Expansion of CD94/NKG2C+ NK cells in response to human cytomegalovirus-infected fibroblasts. Blood. (2006) 107:3624–31. doi: 10.1182/blood-2005-09-3682
177. Hammer Q, Ruckert T, Borst EM, Dunst J, Haubner A, Durek P, et al. Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat Immunol. (2018) 19:453–63. doi: 10.1038/s41590-018-0082-6
178. Rolle A, Pollmann J, Ewen EM, Le VT, Halenius A, Hengel H, et al. IL-12-producing monocytes and HLA-E control HCMV-driven NKG2C+ NK cell expansion. J Clin Invest. (2014) 124:5305–16. doi: 10.1172/JCI77440
179. Charoudeh HN, Terszowski G, Czaja K, Gonzalez A, Schmitter K, Stern M. Modulation of the natural killer cell KIR repertoire by cytomegalovirus infection. Eur J Immunol. (2013) 43:480–7. doi: 10.1002/eji.201242389
180. Liu LL, Landskron J, Ask EH, Enqvist M, Sohlberg E, Traherne JA, et al. Critical Role of CD2 Co-stimulation in adaptive natural killer cell responses revealed in NKG2C-deficient humans. Cell Rep. (2016) 15:1088–99. doi: 10.1016/j.celrep.2016.04.005
181. Della Chiesa M, Falco M, Bertaina A, Muccio L, Alicata C, Frassoni F, et al. Human cytomegalovirus infection promotes rapid maturation of NK cells expressing activating killer Ig-like receptor in patients transplanted with NKG2C−/− umbilical cord blood. J Immunol. (2014) 192:1471–9. doi: 10.4049/jimmunol.1302053
182. Schlums H, Cichocki F, Tesi B, Theorell J, Beziat V, Holmes TD, et al. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity. (2015) 42:443–56. doi: 10.1016/j.immuni.2015.02.008
183. Zhang T, Scott JM, Hwang I, Kim S. Cutting edge: antibody-dependent memory-like NK cells distinguished by FcRgamma deficiency. J Immunol. (2013) 190:1402–6. doi: 10.4049/jimmunol.1203034
184. Lee J, Zhang T, Hwang I, Kim A, Nitschke L, Kim M, et al. Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity. (2015) 42:431–42. doi: 10.1016/j.immuni.2015.02.013
185. Gleimer M, von Boehmer H, Kreslavsky T. PLZF controls the expression of a limited number of genes essential for NKT cell function. Front Immunol. (2012) 3:374. doi: 10.3389/fimmu.2012.00374
186. Muccio L, Falco M, Bertaina A, Locatelli F, Frassoni F, Sivori S, et al. Late Development of FcepsilonRgamma(neg) adaptive natural killer cells upon human cytomegalovirus reactivation in umbilical cord blood transplantation recipients. Front Immunol. (2018) 9:1050. doi: 10.3389/fimmu.2018.01050
187. Luetke-Eversloh M, Hammer Q, Durek P, Nordstrom K, Gasparoni G, Pink M, et al. Human cytomegalovirus drives epigenetic imprinting of the IFNG locus in NKG2Chi natural killer cells. PLoS Pathog. (2014) 10:e1004441. doi: 10.1371/journal.ppat.1004441
188. Elmaagacli AH, Steckel NK, Koldehoff M, Hegerfeldt Y, Trenschel R, Ditschkowski M, et al. Early human cytomegalovirus replication after transplantation is associated with a decreased relapse risk: evidence for a putative virus-versus-leukemia effect in acute myeloid leukemia patients. Blood. (2011) 118:1402–12. doi: 10.1182/blood-2010-08-304121
189. Green ML, Leisenring WM, Xie H, Walter RB, Mielcarek M, Sandmaier BM, et al. CMV reactivation after allogeneic HCT and relapse risk: evidence for early protection in acute myeloid leukemia. Blood. (2013) 122:1316–24. doi: 10.1182/blood-2013-02-487074
190. Ito S, Pophali P, Co W, Koklanaris EK, Superata J, Fahle GA, et al. CMV reactivation is associated with a lower incidence of relapse after allo-SCT for CML. Bone Marrow Transplant. (2013) 48:1313–6. doi: 10.1038/bmt.2013.49
191. Foley B, Cooley S, Verneris MR, Curtsinger J, Luo X, Waller EK, et al. Human cytomegalovirus. (CMV)-induced memory-like NKG2C(+) NK cells are transplantable and expand in vivo in response to recipient CMV antigen. J Immunol. (2012) 189:5082–8. doi: 10.4049/jimmunol.1201964
192. Jin F, Lin H, Gao S, Wang H, Yan H, Guo J, et al. Characterization of IFNgamma-producing natural killer cells induced by cytomegalovirus reactivation after haploidentical hematopoietic stem cell transplantation. Oncotarget. (2017) 8:51–63. doi: 10.18632/oncotarget.13916
193. Capuano C, Pighi C, Battella S, Santoni A, Palmieri G, Galandrini R. Memory NK cell features exploitable in anticancer immunotherapy. J Immunol Res. (2019) 2019:8795673. doi: 10.1155/2019/8795673
194. Vantourout P, Hayday A. Six-of-the-best: unique contributions of γδ T cells to immunology. Nat Rev Immunol. (2013) 13:88–100. doi: 10.1038/nri3384
195. Pistoia V, Tumino N, Vacca P, Veneziani I, Moretta A, Locatelli F, et al. Human γδ T-cells: from surface receptors to the therapy of high-risk leukemias. Front Immunol. (2018) 9:984. doi: 10.3389/fimmu.2018.00984
196. Minculescu L, Sengelov H. The role of gamma delta T cells in haematopoietic stem cell transplantation. Scand J Immunol. (2015) 81:459–68. doi: 10.1111/sji.12289
197. Zhao Y, Niu C, Cui J. Gamma-delta (γδ) T cells: friend or foe in cancer development? J Transl Med. (2018) 16:3. doi: 10.1186/s12967-018-1491-x
198. Carding SR, Egan PJ. Gammadelta T cells: functional plasticity and heterogeneity. Nat Rev Immunol. (2002) 2:336–45. doi: 10.1038/nri797
199. Bonneville M, O'Brien RL, Born WK. Gammadelta T cell effector functions: a blend of innate programming and acquired plasticity. Nat Rev Immunol. (2010) 10:467–78. doi: 10.1038/nri2781
200. Braza MS, Klein B, Fiol G, Rossi JF. γδ T-cell killing of primary follicular lymphoma cells is dramatically potentiated by GA101, a type II glycoengineered anti-CD20 monoclonal antibody. Haematologica. (2011) 96:400–7. doi: 10.3324/haematol.2010.029520
201. Scheper W, Sebestyen Z, Kuball J. Cancer Immunotherapy Using γδT cells: dealing with diversity. Front Immunol. (2014) 5:601. doi: 10.3389/fimmu.2014.00601
202. Adams EJ, Gu S, Luoma AM. Human gamma delta T cells: evolution and ligand recognition. Cell Immunol. (2015) 296:31–40. doi: 10.1016/j.cellimm.2015.04.008
203. Kabelitz D, Glatzel A, Wesch D. Antigen recognition by human γδ T lymphocytes. Int Arch Allergy Immunol. (2000) 122:1–7. doi: 10.1159/000024353
204. Wesch D, Peters C, Oberg HH, Pietschmann K, Kabelitz D. Modulation of γδ T cell responses by TLR ligands. Cell Mol Life Sci. (2011) 68:2357–70. doi: 10.1007/s00018-011-0699-1
205. Scheper W, Grunder C, Straetemans T, Sebestyen Z, Kuball J. Hunting for clinical translation with innate-like immune cells and their receptors. Leukemia. (2014) 28:1181–90. doi: 10.1038/leu.2013.378
206. Correia DV, Lopes A, Silva-Santos B. Tumor cell recognition by γδ T lymphocytes: T-cell receptor vs. NK-cell receptors. Oncoimmunology. (2013) 2:e22892. doi: 10.4161/onci.22892
207. Correia DV, Fogli M, Hudspeth K, da Silva MG, Mavilio D, Silva-Santos B. Differentiation of human peripheral blood Vdelta1+ T cells expressing the natural cytotoxicity receptor NKp30 for recognition of lymphoid leukemia cells. Blood. (2011) 118:992–1001. doi: 10.1182/blood-2011-02-339135
208. Silva-Santos B, Serre K, Norell H. γδ T cells in cancer. Nat Rev Immunol. (2015) 15:683–91. doi: 10.1038/nri3904
209. Bhagat G, Naiyer AJ, Shah JG, Harper J, Jabri B, Wang TC, et al. Small intestinal CD8+TCRγδ+NKG2A+ intraepithelial lymphocytes have attributes of regulatory cells in patients with celiac disease. J Clin Invest. (2008) 118:281–93. doi: 10.1172/JCI30989
210. Todaro M, D'Asaro M, Caccamo N, Iovino F, Francipane MG, Meraviglia S, et al. Efficient killing of human colon cancer stem cells by γδ T lymphocytes. J Immunol. (2009) 182:7287–96. doi: 10.4049/jimmunol.0804288
211. Hu Y, Cui Q, Luo C, Luo Y, Shi J, Huang H. A promising sword of tomorrow: human γδ T cell strategies reconcile allo-HSCT complications. Blood Rev. (2016) 30:179–88. doi: 10.1016/j.blre.2015.11.002
212. Zheng J, Liu Y, Lau YL, Tu W. γδ-T cells: an unpolished sword in human anti-infection immunity. Cell Mol Immunol. (2013) 10:50–7. doi: 10.1038/cmi.2012.43
213. Daniele N, Scerpa MC, Caniglia M, Bernardo ME, Rossi C, Ciammetti C, et al. Transplantation in the onco-hematology field: focus on the manipulation of αβ and γδ T cells. Pathol Res Pract. (2012) 208:67–73. doi: 10.1016/j.prp.2011.10.006
214. Handgretinger R, Schilbach K. The potential role of γδ T cells after allogeneic HCT for leukemia. Blood. (2018) 131:1063–72. doi: 10.1182/blood-2017-08-752162
215. Perko R, Kang G, Sunkara A, Leung W, Thomas PG, Dallas MH. Gamma delta T cell reconstitution is associated with fewer infections and improved event-free survival after hematopoietic stem cell transplantation for pediatric leukemia. Biol Blood Marrow Transplant. (2015) 21:130–6. doi: 10.1016/j.bbmt.2014.09.027
216. Godder KT, Henslee-Downey PJ, Mehta J, Park BS, Chiang KY, Abhyankar S, et al. Long term disease-free survival in acute leukemia patients recovering with increased γδ T cells after partially mismatched related donor bone marrow transplantation. Bone Marrow Transplant. (2007) 39:751–7. doi: 10.1038/sj.bmt.1705650
217. Bertaina A, Merli P, Rutella S, Pagliara D, Bernardo ME, Masetti R, et al. HLA-haploidentical stem cell transplantation after removal of αβ+ T and B cells in children with nonmalignant disorders. Blood. (2014) 124:822–6. doi: 10.1182/blood-2014-03-563817
218. Hirokawa M, Horiuchi T, Kawabata Y, Kitabayashi A, Miura AB. Reconstitution of γδ T cell repertoire diversity after human allogeneic hematopoietic cell transplantation and the role of peripheral expansion of mature T cell population in the graft. Bone Marrow Transplant. (2000) 26:177–85. doi: 10.1038/sj.bmt.1702478
219. Viale M, Ferrini S, Bacigalupo A. TCR gamma/delta positive lymphocytes after allogeneic bone marrow transplantation. Bone Marrow Transplant. (1992) 10:249–53.
220. Pabst C, Schirutschke H, Ehninger G, Bornhauser M, Platzbecker U. The graft content of donor T cells expressing gamma delta TCR+ and CD4+foxp3+ predicts the risk of acute graft versus host disease after transplantation of allogeneic peripheral blood stem cells from unrelated donors. Clin Cancer Res. (2007) 13:2916–22. doi: 10.1158/1078-0432.CCR-06-2602
221. Lamb LS, Henslee-Downey PJ, Parrish RS, Godder K, Thompson J, Lee C, et al. Increased frequency of TCR gamma delta + T cells in disease-free survivors following T cell-depleted, partially mismatched, related donor bone marrow transplantation for leukemia. J Hematother. (1996) 5:503–9. doi: 10.1089/scd.1.1996.5.503
222. Lamb LS, Gee AP, Hazlett LJ, Musk P, Parrish RS, O'Hanlon TP, et al. Influence of T cell depletion method on circulating γδ T cell reconstitution and potential role in the graft-versus-leukemia effect. Cytotherapy. (1999) 1:7–19. doi: 10.1080/0032472031000141295
223. Scheper W, van Dorp S, Kersting S, Pietersma F, Lindemans C, Hol S, et al. γδT cells elicited by CMV reactivation after allo-SCT cross-recognize CMV and leukemia. Leukemia. (2013) 27:1328–38. doi: 10.1038/leu.2012.374
224. Halary F, Pitard V, Dlubek D, Krzysiek R, de la Salle H, Merville P, et al. Shared reactivity of V{delta}2(neg) {gamma}{delta} T cells against cytomegalovirus-infected cells and tumor intestinal epithelial cells. J Exp Med. (2005) 201:1567–78. doi: 10.1084/jem.20041851
225. Feuchtinger T, Opherk K, Bethge WA, Topp MS, Schuster FR, Weissinger EM, et al. Adoptive transfer of pp65-specific T cells for the treatment of chemorefractory cytomegalovirus disease or reactivation after haploidentical and matched unrelated stem cell transplantation. Blood. (2010) 116:4360–7. doi: 10.1182/blood-2010-01-262089
226. Benson DM, Hofmeister CC, Padmanabhan S, Suvannasankha A, Jagannath S, Abonour R, et al. A phase 1 trial of the anti-KIR antibody IPH2101 in patients with relapsed/refractory multiple myeloma. Blood. (2012) 120:4324–33. doi: 10.1182/blood-2012-06-438028
227. Vey N, Bourhis JH, Boissel N, Bordessoule D, Prebet T, Charbonnier A, et al. A phase 1 trial of the anti-inhibitory KIR mAb IPH2101 for AML in complete remission. Blood. (2012) 120:4317–23. doi: 10.1182/blood-2012-06-437558
228. Chiossone L, Vienne M, Kerdiles YM, Vivier E. Natural killer cell immunotherapies against cancer: checkpoint inhibitors and more. Semin Immunol. (2017) 31:55–63. doi: 10.1016/j.smim.2017.08.003
229. Muntasell A, Ochoa MC, Cordeiro L, Berraondo P, Lopez-Diaz de Cerio A, Cabo M, et al. Targeting NK-cell checkpoints for cancer immunotherapy. Curr Opin Immunol. (2017) 45:73–81. doi: 10.1016/j.coi.2017.01.003
230. Ruggeri L, Urbani E, Andre P, Mancusi A, Tosti A, Topini F, et al. Effects of anti-NKG2A antibody administration on leukemia and normal hematopoietic cells. Haematologica. (2016) 101:626–33. doi: 10.3324/haematol.2015.135301
231. Moretta A, Vitale M, Sivori S, Bottino C, Morelli L, Augugliaro R, et al. Human natural killer cell receptors for HLA-class I molecules. Evidence that the Kp43. (CD94) molecule functions as receptor for HLA-B alleles. J Exp Med. (1994) 180:545–55. doi: 10.1084/jem.180.2.545
232. McWilliams EM, Mele JM, Cheney C, Timmerman EA, Fiazuddin F, Strattan EJ, et al. Therapeutic CD94/NKG2A blockade improves natural killer cell dysfunction in chronic lymphocytic leukemia. Oncoimmunology. (2016) 5:e1226720. doi: 10.1080/2162402X.2016.1226720
233. Godal R, Bachanova V, Gleason M, McCullar V, Yun GH, Cooley S, et al. Natural killer cell killing of acute myelogenous leukemia and acute lymphoblastic leukemia blasts by killer cell immunoglobulin-like receptor-negative natural killer cells after NKG2A and LIR-1 blockade. Biol Blood Marrow Transplant. (2010) 16:612–21. doi: 10.1016/j.bbmt.2010.01.019
234. Fujisaki H, Kakuda H, Shimasaki N, Imai C, Ma J, Lockey T, et al. Expansion of highly cytotoxic human natural killer cells for cancer cell therapy. Cancer Res. (2009) 69:4010–7. doi: 10.1158/0008-5472.CAN-08-3712
235. Parrish-Novak J, Dillon SR, Nelson A, Hammond A, Sprecher C, Gross JA, et al. Interleukin 21 and its receptor are involved in NK cell expansion and regulation of lymphocyte function. Nature. (2000) 408:57–63. doi: 10.1038/35040504
236. Ciurea SO, Schafer JR, Bassett R, Denman CJ, Cao K, Willis D, et al. Phase 1 clinical trial using mbIL21 ex vivo-expanded donor-derived NK cells after haploidentical transplantation. Blood. (2017) 130:1857–68. doi: 10.1182/blood-2017-05-785659
237. Wang RN, Wen Q, He WT, Yang JH, Zhou CY, Xiong WJ, et al. Optimized protocols for γδ T cell expansion and lentiviral transduction. Mol Med Rep. (2019) 19:1471–80. doi: 10.3892/mmr.2019.9831
238. Bertaina A, Zorzoli A, Petretto A, Barbarito G, Inglese E, Merli P, et al. Zoledronic acid boosts γδ T-cell activity in children receiving αβ+ T and CD19(+) cell-depleted grafts from an HLA-haplo-identical donor. Oncoimmunology. (2017) 6:e1216291. doi: 10.1080/2162402X.2016.1216291
239. de Witte MA, Sarhan D, Davis Z, Felices M, Vallera DA, Hinderlie P, et al. Early reconstitution of NK and γδ T cells and its implication for the design of post-transplant immunotherapy. Biol Blood Marrow Transplant. (2018) 24:1152–62. doi: 10.1016/j.bbmt.2018.02.023
240. Walsh Z, Yang Y, Kohler ME. Immunobiology of chimeric antigen receptor T cells and novel designs. Immunol Rev. (2019) 290:100–13. doi: 10.1111/imr.12794
241. Louis CU, Savoldo B, Dotti G, Pule M, Yvon E, Myers GD, et al. Antitumor activity and long-term fate of chimeric antigen receptor-positive T cells in patients with neuroblastoma. Blood. (2011) 118:6050–6. doi: 10.1182/blood-2011-05-354449
242. Ali SA, Shi V, Maric I, Wang M, Stroncek DF, Rose JJ, et al. T cells expressing an anti-B-cell maturation antigen chimeric antigen receptor cause remissions of multiple myeloma. Blood. (2016) 128:1688–700. doi: 10.1182/blood-2016-04-711903
243. Fry TJ, Shah NN, Orentas RJ, Stetler-Stevenson M, Yuan CM, Ramakrishna S, et al. CD22-targeted CAR T cells induce remission in B-ALL that is naive or resistant to CD19-targeted CAR immunotherapy. Nat Med. (2018) 24:20–8. doi: 10.1038/nm.4441
244. Hay KA, Gauthier J, Hirayama AV, Voutsinas JM, Wu Q, Li D, et al. Factors associated with durable EFS in adult B-cell ALL patients achieving MRD-negative CR after CD19 CAR T-cell therapy. Blood. (2019) 133:1652–63. doi: 10.1182/blood-2018-11-883710
245. Klingemann H. Are natural killer cells superior CAR drivers? Oncoimmunology. (2014) 3:e28147. doi: 10.4161/onci.28147
246. Hu Y, Tian ZG, Zhang C. Chimeric antigen receptor. (CAR)-transduced natural killer cells in tumor immunotherapy. Acta Pharmacol Sin. (2018) 39:167–76. doi: 10.1038/aps.2017.125
247. Mehta RS, Rezvani K. Chimeric antigen receptor expressing natural killer cells for the immunotherapy of cancer. Front Immunol. (2018) 9:283. doi: 10.3389/fimmu.2018.00283
248. Di Vito C, Mikulak J, Zaghi E, Pesce S, Marcenaro E, Mavilio D. NK cells to cure cancer. Semin Immunol. (2019) 41:101272. doi: 10.1016/j.smim.2019.03.004
249. Rischer M, Pscherer S, Duwe S, Vormoor J, Jurgens H, Rossig C. Human γδ T cells as mediators of chimaeric-receptor redirected anti-tumour immunity. Br J Haematol. (2004) 126:583–92. doi: 10.1111/j.1365-2141.2004.05077.x
250. Ingegnere T, Mariotti FR, Pelosi A, Quintarelli C, De Angelis B, Tumino N, et al. Human CAR NK cells: a new non-viral method allowing high efficient transfection and strong tumor cell killing. Front Immunol. (2019) 10:957. doi: 10.3389/fimmu.2019.00957
251. Rezvani K. Adoptive cell therapy using engineered natural killer cells. Bone Marrow Transplant. (2019) 54:785–8. doi: 10.1038/s41409-019-0601-6
252. Imai C, Iwamoto S, Campana D. Genetic modification of primary natural killer cells overcomes inhibitory signals and induces specific killing of leukemic cells. Blood. (2005) 106:376–83. doi: 10.1182/blood-2004-12-4797
253. Shimasaki N, Fujisaki H, Cho D, Masselli M, Lockey T, Eldridge P, et al. A clinically adaptable method to enhance the cytotoxicity of natural killer cells against B-cell malignancies. Cytotherapy. (2012) 14:830–40. doi: 10.3109/14653249.2012.671519
254. Muller T, Uherek C, Maki G, Chow KU, Schimpf A, Klingemann HG, et al. Expression of a CD20-specific chimeric antigen receptor enhances cytotoxic activity of NK cells and overcomes NK-resistance of lymphoma and leukemia cells. Cancer Immunol Immunother. (2008) 57:411–23. doi: 10.1007/s00262-007-0383-3
255. Chu Y, Hochberg J, Yahr A, Ayello J, van de Ven C, Barth M, et al. Targeting CD20+ aggressive B-cell non-hodgkin lymphoma by anti-CD20 CAR mRNA-modified expanded natural killer cells in vitro and in NSG mice. Cancer Immunol Res. (2015) 3:333–44. doi: 10.1158/2326-6066.CIR-14-0114
256. Tam YK, Maki G, Miyagawa B, Hennemann B, Tonn T, Klingemann HG. Characterization of genetically altered, interleukin 2-independent natural killer cell lines suitable for adoptive cellular immunotherapy. Hum Gene Ther. (1999) 10:1359–73. doi: 10.1089/10430349950018030
257. Jochems C, Hodge JW, Fantini M, Fujii R, Morillon YM II, Greiner JW, et al. An NK cell line. (haNK) expressing high levels of granzyme and engineered to express the high affinity CD16 allele. Oncotarget. (2016) 7:86359–73. doi: 10.18632/oncotarget.13411
258. Liu E, Tong Y, Dotti G, Shaim H, Savoldo B, Mukherjee M, et al. Cord blood NK cells engineered to express IL-15 and a CD19-targeted CAR show long-term persistence and potent antitumor activity. Leukemia. (2018) 32:520–31. doi: 10.1038/leu.2017.226
259. Oei VYS, Siernicka M, Graczyk-Jarzynka A, Hoel HJ, Yang W, Palacios D, et al. Intrinsic functional potential of NK-cell subsets constrains retargeting driven by chimeric antigen receptors. Cancer Immunol Res. (2018) 6:467–80. doi: 10.1158/2326-6066.CIR-17-0207
260. Mirzaei HR, Mirzaei H, Lee SY, Hadjati J, Till BG. Prospects for chimeric antigen receptor. (CAR) γδ T cells: a potential game changer for adoptive T cell cancer immunotherapy. Cancer Lett. (2016) 380:413–23. doi: 10.1016/j.canlet.2016.07.001
Keywords: innate lymphocytes, haploidentical hematopoietic stem cell transplantation, immune-reconstitution, natural killer cells, innate lymphoid cells, γδ T cells, alloreactivity
Citation: Zaghi E, Calvi M, Di Vito C and Mavilio D (2019) Innate Immune Responses in the Outcome of Haploidentical Hematopoietic Stem Cell Transplantation to Cure Hematologic Malignancies. Front. Immunol. 10:2794. doi: 10.3389/fimmu.2019.02794
Received: 31 July 2019; Accepted: 14 November 2019;
Published: 28 November 2019.
Edited by:
Antonella Mancusi, University of Perugia, ItalyReviewed by:
Sergio Rutella, Nottingham Trent University, United KingdomFederico Simonetta, Stanford University, United States
Copyright © 2019 Zaghi, Calvi, Di Vito and Mavilio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Domenico Mavilio, ZG9tZW5pY28ubWF2aWxpb0BodW1hbml0YXMuaXQ=
 Elisa Zaghi
Elisa Zaghi Michela Calvi
Michela Calvi Clara Di Vito
Clara Di Vito Domenico Mavilio
Domenico Mavilio