- 1Department of Medicine and Surgery, Pediatric Clinic, Pietro Barilla Children's Hospital, University of Parma, Parma, Italy
- 2Unit of Gastroenterology and Digestive Endoscopy, Department of Medicine and Surgery, University of Parma, Parma, Italy
Celiac disease (CD) is an immune-mediated disorder initiated by the ingestion of gluten in genetically predisposed individuals. Recent data shows that changes in the gut microbiome composition and function are linked with chronic inflammatory diseases; this might also be the case for CD. The main aim of this manuscript is to discuss our present knowledge of the relationships between gut microbiota alterations and CD and to understand if there is any role for probiotics in CD therapy. PubMed was used to search for all of the studies published from November 2009 to November 2019 using key words such as “Celiac Disease” and “Microbiota” (306 articles), “Celiac Disease” and “Gastrointestinal Microbiome” (139), and “Probiotics” and “Celiac Disease” (97 articles). The search was limited to articles published in English that provided evidence-based data. Literature analysis showed that the gut microbiota has a well-established role in gluten metabolism, in modulating the immune response and in regulating the permeability of the intestinal barrier. Promising studies suggest a possible role of probiotics in treating and/or preventing CD. Nevertheless, human trials on the subject are still scarce and lack homogeneity. A possible role was documented for probiotics in improving CD-related symptoms, modulating the peripheral immune response and altering the fecal microbiota, although the results were not consistent in all of the studies. No evidence was found that probiotic administration might prevent CD onset. Knowledge of the role of intestinal bacteria in the development of CD opens new possibilities for its treatment through probiotic administration, even though further studies are needed to better clarify whether probiotics can help treat or prevent the disease and to define which probiotics to use, at what dose and for how long.
Introduction
Celiac disease (CD) is an immune-mediated disorder initiated by the ingestion of gluten in genetically predisposed individuals (1, 2). In patients with CD, immune responses to gliadin fractions stimulate an inflammatory response. This reaction is mediated by the innate and adaptive immunity (3). The interaction between gliadin peptides and the G protein-coupled receptor CXCR3 on enterocytes triggers the release of zonulin, a potent intestinal barrier function modulator. Consequently, gliadin peptides translocate into the lamina propria and activate the immune response (4). In the lamina propria, intestinal tissue transglutaminase (tTG) reacts with gliadin peptides to deamidate them to negatively charged glutamic acid residues that are highly immunogenic. After tTG-induced deamidation, gliadin peptides activate the humoral immune response with antibodies against gliadin and tTG and to the production of pro-inflammatory cytokines, such as interferon gamma (IFNγ), interleukin-17 (IL-17) and tumor necrosis factor-alpha (TNFα) (5).
Many researches have observed that different functions of macrophages, dendritic cells and neutrophils, which are an essential part of the innate immune system, are influenced by the microbiota (6, 7). Gliadin peptides also stimulate an innate immune response in the intestinal epithelium that is characterized by increased expression of IL-15 by enterocytes, resulting in the activation of intra-epithelial lymphocytes expressing the activating receptor NK-G2D, a natural killer cell marker (8, 9).
CD does not develop unless a person has alleles that encode HLA-DQ2 or HLA-DQ8 proteins, products of two of the HLA genes. However, HLA-DQ2 and HLA-DQ8 haplotypes are not specific for CD because many people, most of whom do not have CD, carry these alleles; thus, the DQ2 and DQ8 haplotypes are necessary but not sufficient for the development of CD (10, 11). Furthermore, with the introduction of GWAS (genome-wide association studies), an additional 39 non-HLA regions of susceptibility have been associated with CD development (12, 13). Several studies have identified more genes associated with CD that are involved in immune function or related to defects in intestinal permeability or with bacterial colonization and sensing (14, 15).
The role of an environmental influence in CD pathogenesis is supported by the facts that (a) HLA and non-HLA genes explain only 55% of disease susceptibility, (b) there is a lack of 100% concordance of CD in monozygotic twins and (c) the incidence of this condition is increasing (16–18). Based on more recent epidemiological data, loss of gluten tolerance may occur at the time of its introduction into the diet or at any time in life, with several different intestinal and extra-intestinal symptoms (19–22). These findings suggest that other environmental factors must play a role in CD development. Indeed, environmental factors that influence the composition of the human gut microbiota, such as birth gestational age, type of delivery, intestinal infections and antibiotic exposure, have been associated with the risk of developing CD (23–26).
Gut bacteria are key regulators of digestion along the gastrointestinal tract and have a relevant impact in the synthesis of many nutrients and metabolites (27–29). Furthermore, the gut microbiota has a crucial immune function, inhibiting bacterial growth and maintaining intestinal epithelial integrity (30). Moreover, growing evidence has shown a critical role for commensal bacteria and their products in influencing the development, homeostasis, and function of innate and adaptive immune cells (31, 32). Recent data support the hypothesis that changes in the gut microbiome composition and function are linked with chronic inflammatory diseases; this might also be the case for CD (33). Although a gluten-free diet (GFD) influences the gut microbiota composition and diversity and thus represents a confounding factor, several studies support the hypothesis that the microbiota plays a role in the pathogenesis, clinical manifestation and risk of developing CD (34). Moreover, it has been reported that soluble CD14 (sCD14, i.e., an indicator of innate immune cell activation in response to mucosal translocation of Gram-negative bacteria) is increased in untreated patients with CD, probably because of translocation of commensal intestinal bacteria (35, 36). Finally, patients with persistent symptoms on a long-term GFD have an altered microbiota composition (37).
The evidence of intestinal dysbiosis in CD, together with the role of the gut microbiota in regulating the immune response, opens up the possibility of finding new therapeutic approaches by modulating the intestinal microbiota with the use of probiotics. The main aim of this manuscript is to discuss our present knowledge of the relationships between gut microbiota alterations and CD and to understand if there is any role for probiotics in CD therapy. PubMed was used to search for all of the studies published from November 2009 to November 2019 using key words such as “Celiac Disease” and “Microbiota” (306 articles), “Celiac Disease” and “Gastrointestinal Microbiome” (139 articles), and “Probiotics” and “Celiac Disease” (97 articles). The search was limited to articles published in English that provided evidence-based data.
Gut Microbiota and Environmental Factors
Each healthy individual has a unique gut microbiota. Core native microbiota are shaped in early life (i.e., in the first 36 months of age) by gut maturation, which is strongly influenced by environmental factors such birth gestational age, type of delivery, method of milk feeding, weaning period, lifestyle, and dietary and cultural habits (38). When a child is 2–3 years old, a relative stability in gut microbiota composition has been documented (33, 39). Gut microbiota are represented by several species of microorganisms, including bacteria, yeast, and viruses. The two major bacterial phyla are Firmicutes and Bacteroidetes, which are 90% of the whole gut microbiota (40). The Firmicutes phylum is composed of ≥ 200 different genera, and Clostridium genera are 95% of the Firmicutes phyla. Bacteroidetes consists of predominant genera such as Bacteroides and Prevotella. Actinobacteria, Proteobacteria, Fusobacteria, and Verrucomicrobia are the next most numerous phyla, which are described in a “healthy gut microbiota composition” (33, 40).
It has been observed that the HLA-DQ genotype can influence early gut microbiota composition (41). Several studies have demonstrated that the genotype of infants at family risk of developing CD, carrying the HLA-DQ2 haplotypes, influences the early gut microbiota composition. Olivares et al. (42) reported that infants with a high genetic risk have significantly higher proportions of Firmicutes and Proteobacteria and lower proportions of Actinobacteria and Bifidobacteria compared with low-risk infants. In a study including 164 healthy new-borns with ≥one first-degree relative with CD, De Palma et al. (43) showed that milk-feeding type in conjunction with HLA-DQ genotype has an impact in in establishing infants' gut microbiota.
Diet represents another key regulator of microbiome development and homeostasis. Although recent data have shown that breast-feeding has no protective effect on the development of CD, studies have reported that genotype-related differences in microbiota composition are reduced by breast-feeding (44). Moreover, human milk oligosaccharides enhance overall barrier integrity by making enterocytes less vulnerable to bacterial-induced innate immunity (45).
Some observational studies have also shown that elective cesarean delivery is linked with an increased risk of CD onset during pediatric age, suggesting the impact of dysbiosis during early life. The gut microbiota of children born by elective cesarean section, compared to vaginally delivered infants' microbiota, has reduced microbial diversity and fewer Bifidobacterium species (46, 47). According to some studies, antibiotic exposure during the first year of life has been linked with intestinal dysbiosis, reduced fecal microbial diversity, and early onset of CD (48, 49). However, there are other studies that did not confirm this result (50, 51), and a recent meta-analysis found no evidence of an association between prenatal or postnatal antibiotic exposure and CD (52).
Although it is currently recognized how these environmental factors influence the composition of the intestinal microbiota, there are no longitudinal studies that have defined whether and how the gut microbiota plays a role in the development of CD. A large, ongoing, multi-center, prospective longitudinal study called CDGEMM (Celiac Disease, Genomic, Environmental, Microbiome, and Metabolomic Study) has the goal of identifying and validating specific microbiome and metabolomic profiles able to predict loss of tolerance in children genetically at risk of autoimmunity (53).
Gut Microbiota and Implications in Celiac Disease (CD) Pathogenesis
The gut microbiota present in the human colon participates in gluten metabolism. Lactobacilli and Bifidobacterium spp. may play a role in the breakdown of gluten and its peptides to modify their immunogenic potential (54, 55). Caminero et al. (56) demonstrated that opportunistic pathogens and core gut commensals produce distinct breakdown patterns of gluten with increased or decreased immunogenicity that could influence autoimmune risk. In particular, it has been demonstrated that Lactobacilli can detoxify gliadin peptides after partial digestion by human proteases; additionally, immunogenic peptides produced by Pseudomonas aeruginosa proteases are also further degraded and rendered less immunogenic in the presence of Lactobacillus. These findings on the gluten-processing activities of specific microbial strains could suggest the use of probiotics as complementary therapy for CD.
Changes in the gut microbiota composition could also play a role in altering the intestinal barrier and increasing epithelial permeability (57). Disassembly of zonulin, that is involved in tight junctions, has a pathogenic role in increasing the intestinal permeability present in patients with CD. Some studies have reported that dysbiosis is associated with increasing zonulin release, disrupting tight junctions, and enhancing the entry of incompletely digested gliadin peptides into the lamina propria (4, 58). In addition, it is acknowledged that the gut microbiota has an important role in the regulation of host metabolism and immunity (59). Moreover, a recent study has shown how gut microbiota and their metabolites enhance the risk of developing autoimmunity through epigenetic processes (60). To find a microbial agent for disease immunomodulation, Bifidobacteria and Lactobacilli are the most studied strains. Bifidobacteria strains have been described to play a role in reducing the epithelial permeability triggered by gluten (61), in downregulating the Th1 pathway typical of CD (62), and in decreasing jejunal architecture damage (63). Furthermore, it has been reported that Escherichia coli could have a protective effect on gut barrier function (64) and that Lactobacilli strains have immunomodulatory properties (65).
In recent years, several cross-sectional studies have evaluated fecal, salivary, and duodenal microbiota associated with CD. Patients with CD show a decrease in beneficial species (Lactobacillus and Bifidobacterium) and an increase in those potentially pathogenic (Bacteroides and E. coli) in comparison with healthy subjects (66). In particular, some studies have reported that Bifidobacterium spp., Bifidobacterium longum, Clostridium histolyticum, C. lituseburense, and Faecalibacterium prausnitzii group proportions are less abundant in untreated patients with CD than in healthy controls (67–69). Ou Gangwei et al. (70) have shown that rod-shaped bacteria represented a significant fraction of the proximal small intestine microbiota in children with CD during the so-called Swedish epidemics. These bacteria present in the epithelial lining of the small intestine were observed in children with CD but not in controls. Another study analyzed the mucosa-associated microbiota of 20 children with CD, before and after a GFD regimen, and of 10 controls, with evidence of a peculiar microbial profile and a significantly higher biodiversity in the duodenal mucosa of patients with CD (71). Furthermore, Di Cagno et al. (72) demonstrated that a GFD lasting at least 2 years did not completely restore the microbiota of children with CD.
Wacklin et al. (34, 73) have shown microbiota alterations, particularly in subjects with persistent symptoms despite adherence to a long-term GFD or associated with gastrointestinal symptoms but not with dermatitis herpetiformis. These studies highlight that the alterations in gut microbiota are more pronounced in the active phase of CD, suggesting that perturbations in the interaction between the host and the microbiota could influence CD manifestation and evolution. However, the abovementioned CDGEMM study could help to understand the role that the gut microbiome show in the early steps involved in the pathogenesis of CD (53).
Probiotic Supplementation in Celiac Disease (CD)
Probiotics have shown the ability to hydrolyse immunogenic gluten peptides (56, 74–77), thus reducing their immunogenicity. Moreover, intestinal bacteria have also been implicated in modulating the immune response, directing the correct differentiation of anti-inflammatory Treg cells. In patients with CD who underwent an oral wheat challenge, circulating T cells were collected, showing an increase in both effector T cells and Treg FOXP3+ cells (86); however, these FOXP3+ T cells were found to have a significantly reduced suppressive function. Serena and colleagues suggested that this impairment may be related to the presence of an alternatively spliced isoform of FOXP3 and hypothesized that the intestinal microenvironment may have an impact in modulating this alternative splicing (60). Furthermore, studies suggest that the composition of the intestinal microbiota can affect the permeability of the intestinal mucosa (87, 88). In addition, probiotics would encourage the intestinal microbiome to improve the production of short chain fatty acids such as butyrate that can have profound effects in modulating proinflammatory activities in the colonic gut and in inducing health effects on the colonic epithelia (74).
Considering this established role of microbiota in gluten metabolism and in modulating the gut immune response, the manipulation of the microbiome via probiotic administration opens new possibilities for the treatment of CD and its related symptoms. Reports on animal models have shown promising effects of probiotics on CD. Bifidobacterium longum CECT 7347 has been found to decrease the production of inflammatory cytokines and CD 4+ T cells in rats (63) as well as ameliorate gliadin-induced enteropathy (55). Saccharomyces boulardii KK1 oral administration improved enteropathy and decreased epithelial cell CD71 expression and local cytokine production in gluten-sensitized mice (89). Administration of Lactobacillus casei was found to be effective in rescuing the normal mucosal architecture in a mouse model of gliadin-induced villous damage (65). Bifidobacterium breve prevented intestinal inflammation through the induction of intestinal IL-10-producing Th1 cells (90) and ameliorated DSS-induced colitis symptoms in mouse models as well as modulated T cell polarization toward Th2 and Tregs both in vitro and in vivo (91). In a recent study, Orlando and colleagues found that Lactobacillus rhamnosus GG administration to rats could protect the intestinal mucosa from gliadin peptide-induced damage (92).
However, despite a number of promising in vitro and in vivo animal studies on probiotic use, data from human trials are still scarce and lack homogeneity. We reviewed studies on human patients with CD published from January 2009 to December 2019 regarding the efficacy and safety of probiotic supplementation (Table 1). Although no safety issue was observed in patients with CD treated with probiotics, a few studies have shown that probiotic administration might be beneficial in improving CD-related symptoms. Smecuol et al. (78) randomized patients with CD to receive either the Bifidobacterium Infantis Natren life start strain or placebo and found a significant improvement in gastrointestinal symptoms in the group receiving probiotic, although they were not following a strict GFT. Changes in intestinal microbiota were also documented in a 2016 study from Quagliarello et al. (81) who analyzed microbial DNA extracted from the feces of 40 pediatric patients with CD before and after probiotic treatment, evidencing an increase in Actinobacteria as well as a re-establishment of the physiological Firmicutes/Bacteroidetes ratio. Similar results were obtained by Primec et al. (84) in a 2019 double-blind placebo-controlled study of 40 children with CD who were randomized to receive either probiotics or placebo for 3 months. In contrast, Harnett et al. (80) found no differences in the fecal microbiota counts or in the severity of the symptoms between the group receiving probiotic and the placebo group. Lastly, it has been documented that probiotic treatment can modulate immunological parameters such as TNF-α and peripheral T lymphocytes. Klemenak et al. (79) in a double-blinded, randomized, placebo-controlled trial, showed the positive effect of B. breve strain administration in decreasing the production of the pro-inflammatory cytokine TNF-α in children with CD on a gluten-free diet. The administration of Bifidobacterium longum CECT 7347 was also found to significantly decrease peripheral CD3+ T lymphocytes as well as cause a slight decrease in TNF- α (79). Daily oral administration of L. plantarum HEAL9 and L. paracasei 8700:2 for 6 months was related to changes in the immune response in 78 children with celiac disease autoimmunity in a 2019 study from Hakansson et al. (85) interestingly, the difference in most lymphocyte subsets found in the placebo group was similar to that found in patients with active celiac disease, indicating a progression of disease development that was not observed in the probiotic group. Two studies were conducted to investigate the ability of probiotics to prevent CD onset. The first, from Savilahti and colleagues, analyzed data taken from a trial on primary allergy prevention, including 1,223 babies treated with probiotics until the age of 6 months vs. placebo, and found no difference in the risk of developing CD during the 13-year follow-up (82). Later, Uusitalo et al. (83) conducted a multi-center study following over 6,000 genetically susceptible children for a median period of 8.7 years and found that probiotic administration did not change the risk of developing CD.
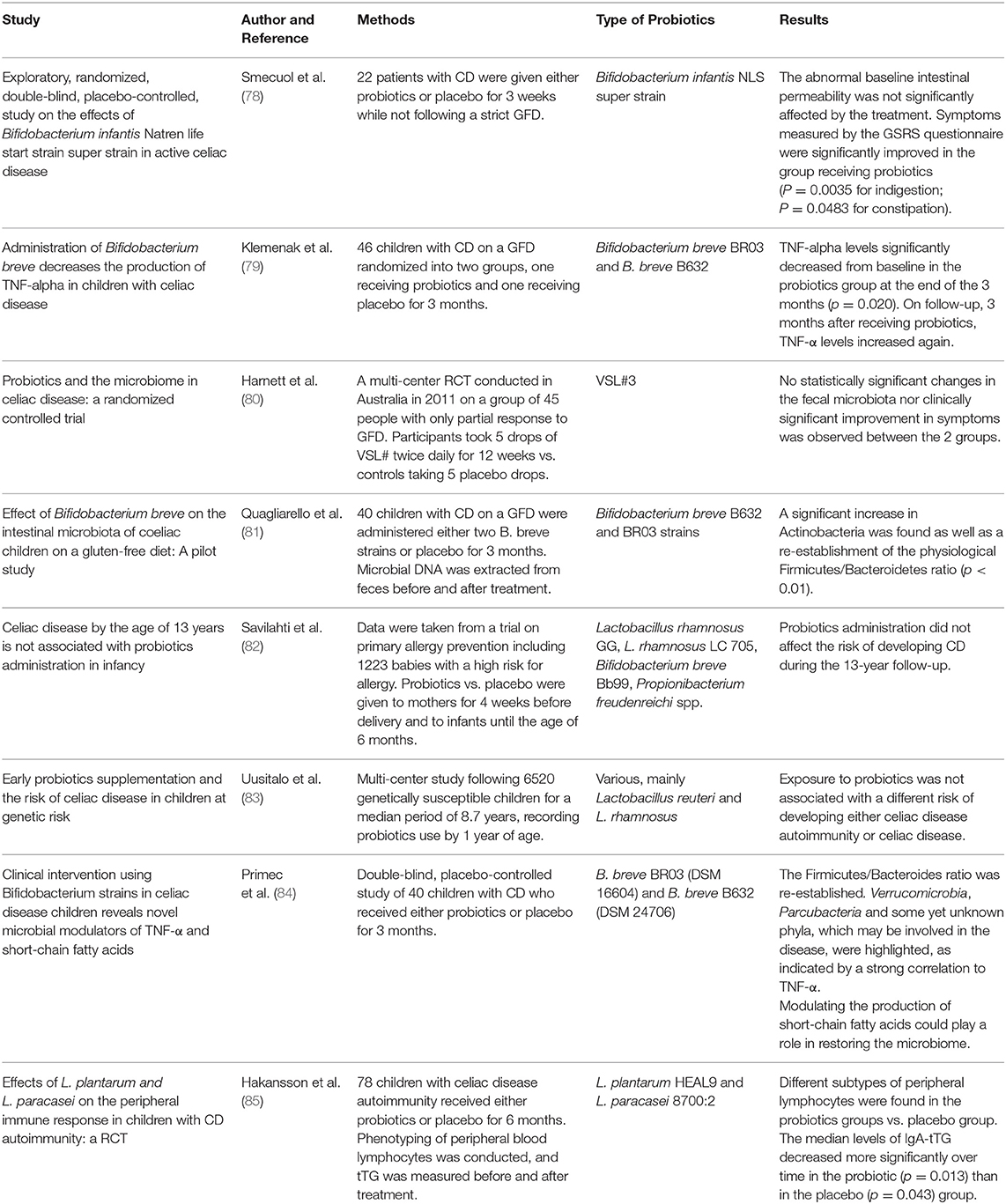
Table 1. Studies on human patients with celiac disease (CD) regarding the efficacy of probiotic supplementation.
Conclusions
Although the association between alterations in the gut microbiota and the development of CD has been demonstrated, a definite microbial signature and the exact role of dysbiosis in CD pathogenesis are not recognized. Further human studies will be needed to reach a definitive conclusion on the role of probiotics in CD using standardized probiotic formulations, dosages and periods of treatments, as well as homogeneous patient groups. Currently published data suggest the efficacy and safety of probiotic supplementation in improving CD-related symptoms (78), as well as documenting the ability of some probiotics to alter the fecal microbiota and decrease pro-inflammatory parameters such as TNF-α levels or peripheral CD3+ T lymphocyte counts (79, 81). Which probiotics are more effective, at what dose and how long they should be administered are yet to be definitively clarified. However, the encouraging data on in vitro and in vivo studies, as well as the knowledge of the mechanisms through which intestinal bacteria modulate the development of the disease, prompted us to continue the studies to achieve a better understanding of the possible role of probiotics in treating and/or preventing CD.
Author Contributions
FPec and FPer wrote the first draft of the manuscript. PG, FF, and SI performed the literature analysis and revised the first draft. GA and SE critically revised the text and made substantial scientific contributions. All authors approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
2. Lebwohl B, Sanders DS, Green PHR. Coeliac disease. Lancet. (2018) 391:70–81. doi: 10.1016/S0140-6736(17)31796-8
3. Sollid LM. Coeliac disease: dissecting a complex inflammatory disorder. Nat Rev Immunol. (2002) 2:647–55. doi: 10.1038/nri885
4. Lammers KM, Lu R, Brownley J, Lu B, Gerard C, Thomas K, et al. Gliadin induces an increase in intestinal permeability and zonulin release by binding to the chemokine receptor CXCR3. Gastroenterology. (2008) 135:194–204.e3. doi: 10.1053/j.gastro.2008.03.023
5. Gianfrani C, Auricchio S, Troncone R. Adaptive and innate immune responses in celiac disease. Immunol Lett. (2005) 99:141–5. doi: 10.1016/j.imlet.2005.02.017
6. Ohkubo T, Tsuda M, Tamura M, Yamamura M. Impaired superoxide production in peripheral blood neutrophils of germ-free rats. Scand J Immunol. (1990) 32:727–9. doi: 10.1111/j.1365-3083.1990.tb03216.x
7. Mitsuyama M, Ohara R, Amako K, Nomoto K, Yokokura T, Nomoto K. Ontogeny of macrophage function to release superoxide anion in conventional and germfree mice. Infect Immun. (1986) 52:236–9. doi: 10.1128/IAI.52.1.236-239.1986
8. Mention JJ, Ben Ahmed M, Begue B, Barbe U, Verkarre V, Asnafi V, et al. Interleukin 15: a key to disrupted intraepithelial lymphocyte homeostasis and lymphomagenesis in celiac disease. Gastroenterology. (2003) 125:730–45. doi: 10.1016/S0016-5085(03)01047-3
9. Meresse B, Chen Z, Ciszewski C, Tretiakova M, Bhagat G, Krausz TN, et al. Coordinated induction by IL15 of a TCR-independent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity. (2004) 21:357–66. doi: 10.1016/j.immuni.2004.06.020
10. Wijmenga C, Gutierrez-Achury J. Celiac disease genetics: past, present and future challenges. J Pediatr Gastroenterol Nutr. (2014) 59(Suppl. 1):S4–7. doi: 10.1097/01.mpg.0000450392.23156.10
11. Bevan S, Popat S, Braegger CP, Busch A, O'Donoghue D, Falth-Magnusson K, et al. Contribution of the MHC region to the familial risk of coeliac disease. J Med Genet. (1999) 36:687–90.
12. van Heel DA, Franke L, Hunt KA, Gwilliam R, Zhernakova A, Inouye M, et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet. (2007) 39:827–9. doi: 10.1038/ng2058
13. Trynka G, Wijmenga C, van Heel DA. A genetic perspective on coeliac disease. Trends Mol Med. (2010) 16:537–50. doi: 10.1016/j.molmed.2010.09.003
14. Leonard MM, Serena G, Sturgeon C, Fasano A. Genetics and celiac disease: the importance of screening. Expert Rev Gastroenterol Hepatol. (2015) 9:209–15. doi: 10.1586/17474124.2014.945915
15. Kumar V, Wijmenga C, Withoff S. From genome-wide association studies to disease mechanisms: celiac disease as a model for autoimmune diseases. Semin Immunopathol. (2012) 34:567–80. doi: 10.1007/s00281-012-0312-1
16. Ludvigsson JF, Rubio-Tapia A, van Dyke CT, Melton LJ III, Zinsmeister AR, Lahr BD, et al. Increasing incidence of celiac disease in a North American population. Am J Gastroenterol. (2013) 108:818–24. doi: 10.1038/ajg.2013.60
17. White LE, Merrick VM, Bannerman E, Russell RK, Basude D, Henderson P, et al. The rising incidence of celiac disease in Scotland. Pediatrics. (2013) 132:e924–31. doi: 10.1542/peds.2013-0932
18. Cristofori F, Indrio F, Miniello VL, De Angelis M, Francavilla R. Probiotics in celiac disease. Nutrients. (2018) 10:1824. doi: 10.3390/nu10121824
19. Catassi C, Kryszak D, Bhatti B, Sturgeon C, Helzlsouer K, Clipp SL, et al. Natural history of celiac disease autoimmunity in a USA cohort followed since 1974. Ann Med. (2010) 42:530–8. doi: 10.3109/07853890.2010.514285
20. Trovato CM, Montuori M, Anania C, Barbato M, Vestri AR, Guida S, et al. Are ESPGHAN “biopsy-sparing” guidelines for celiac disease also suitable for asymptomatic patients? Am J Gastroenterol. (2015) 110:1485–9. doi: 10.1038/ajg.2015.285
21. Lionetti E, Castellaneta S, Francavilla R, Pulvirenti A, Tonutti E, Amarri S, et al. Introduction of Gluten, HLA Status, and the risk of celiac disease in children. N Engl J Med. (2014) 371:1295–303. doi: 10.1056/NEJMoa1400697
22. Vriezinga SL, Auricchio R, Bravi E, Castillejo G, Chmielewska A, Crespo Escobar P, et al. Randomized feeding intervention in infants at high risk for celiac disease. N Engl J Med. (2014) 371:1304–15. doi: 10.1056/NEJMoa1404172
23. Sanz Y, De Pama G, Laparra M. Unraveling the ties between celiac disease and intestinal microbiota. Int Rev Immunol. (2011) 30:207–18. doi: 10.3109/08830185.2011.599084
24. Sandberg-Bennich S, Dahlquist G, Källén B. Coeliac disease is associated with intrauterine growth and neonatal infections. Acta Paediatr. (2002) 91:30–3. doi: 10.1111/j.1651-2227.2002.tb01635.x
25. Ivarsson A, Hernell O, Stenlund H, Persson LA. Breast-feeding protects against celiac disease. Am J Clin Nutr. (2002) 75:914–21. doi: 10.1093/ajcn/75.5.914
26. Marild K, Stephansson O, Montgomery S, Murray JA, Ludvigsson JF. Pregnancy outcome and risk of celiac disease in offspring: a nationwide case-control study. Gastroenterology. (2012) 142:39–45.e3. doi: 10.1053/j.gastro.2011.09.047
27. Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J. (2017) 474:1823–36. doi: 10.1042/BCJ20160510
28. Valdes AM, Walter J, Segal E, Spector TD. Role of the gut microbiota in nutrition and health. BMJ. (2018) 361:k2179. doi: 10.1136/bmj.k2179
29. Gibiino G, Ianiro G, Cammarota G, Gasbarrini A. The gut microbiota: its anatomy and physiology over a lifetime. Minerva Gastroenterol Dietol. (2017) 63:329–36. doi: 10.23736/S1121-421X.17.02405-9t
30. Khosravi A, Mazmanian SK. Disruption of the gut microbiome as a risk factor for microbial infections. Curr Opin Microbiol. (2013) 16:221–7. doi: 10.1016/j.mib.2013.03.009
31. Woo V, Alenghat T. Host-microbiota interactions: epigenomic regulation. Curr Opin Immunol. (2017) 44:52–60. doi: 10.1016/j.coi.2016.12.001
32. Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. (2013) 14:676–84. doi: 10.1038/ni.2640
33. Rinninella E, Raoul P, Cintoni M, Franceschi F G, Miggiano AD, Gasbarrini A, et al. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms. (2019) 7:14. doi: 10.3390/microorganisms7010014
34. Wacklin P, Kaukinen K, Tuovinen E, Collin P, Lindfors K, Partanen J, et al. The duodenal microbiota composition of adult celiac disease patients is associated with the clinical manifestation of the disease. Inflamm Bowel Dis. (2013) 19:934–41. doi: 10.1097/MIB.0b013e31828029a9
35. Ancuta P, Kamat A, Kunstman KJ, Kim EY, Autissier P, Wurcel A, et al. Microbial translocation is associated with increased monocyte activation and dementia in AIDS patients. PLoS ONE. (2008) 3:e2516. doi: 10.1371/journal.pone.0002516
36. Hoffmanova I, Sanchez D, Habova V, Andel M, Tuckova L, Tlaskalova-Hogenova H. Serological markers of enterocyte damage and apoptosis in patients with celiac disease, autoimmune diabetes mellitus and diabetes mellitus type 2. Physiol Res. (2015) 64:537–46. doi: 10.33549/physiolres.932916
37. O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. (2006) 7:688–93. doi: 10.1038/sj.embor.7400731
38. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. (2012) 486:222–7. doi: 10.1038/nature11053
39. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. (2011) 473:174–80. doi: 10.1038/nature09944
40. Laterza L, Rizzatti G, Gaetani E, Chiusolo P, Gasbarrini A. The gut microbiota and immune system relationship in human graft-versus-host disease. Mediterr J Hematol Infect Dis. (2016) 8:e2016025. doi: 10.4084/mjhid.2016.025
41. De Palma G, Capilla A, Nadal I, Nova E, Pozo T, Varea V, et al. Interplay between human leukocyte antigen genes and the microbial colonization process of the newborn intestine. Curr Issues Mol Biol. (2010) 12:1–10.
42. Olivares M, Neef A, Castillejo G, Palma GD, Varea V, Capilla A, et al. The HLA-DQ2 genotype selects for early intestinal microbiota composition in infants at high risk of developing coeliac disease. Gut. (2015) 64:406–17. doi: 10.1136/gutjnl-2014-306931
43. De Palma G, Capilla A, Nova E, Castillejo G, Varea V, Pozo T, et al. Influence of milk-feeding type and genetic risk of developing coeliac disease on intestinal microbiota of infants: the PROFICEL study. PLoS ONE. (2012) 7:e30791. doi: 10.1371/journal.pone.0030791
44. Asakuma S, Hatakeyama E, Urashima T, Yoshida E, Katayama T, Yamamoto K, et al. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J Biol Chem. (2011) 286:34583–92. doi: 10.1074/jbc.M111.248138
45. Wang C, Zhang M, Guo H, Yan J, Liu F, Chen J, et al. Human milk oligosaccharides protect against necrotizing enterocolitis by inhibiting intestinal damage via increasing the proliferation of crypt cells. Mol Nutr Food Res. (2019) 63:e1900262. doi: 10.1002/mnfr.201900262
46. Rutayisire E, Huang K, Liu Y, Tao F. The mode of delivery affects the diversity and colonization pattern of the gut microbiota during the first year of infants' life: a systematic review. BMC Gastroenterol. (2016) 16:86. doi: 10.1186/s12876-016-0498-0
47. Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, et al. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut. (2014) 63:559–66. doi: 10.1136/gutjnl-2012-303249
48. Dydensborg Sander S, Nybo Andersen AM, Murray JA, Karlstad O, Husby S, Stordal K. Association between antibiotics in the first year of life and celiac disease. Gastroenterology. (2019) 156:2217–29. doi: 10.1053/j.gastro.2019.02.039
49. Marild K, Kahrs CR, Tapia G, Stene LC, Stordal K. Infections and risk of celiac disease in childhood: a prospective nationwide cohort study. Am J Gastroenterol. (2015) 110:1475–84. doi: 10.1038/ajg.2015.287
50. Marild K, Ye W, Lebwohl B, Green PH, Blaser MJ, Card T, et al. Antibiotic exposure and the development of coeliac disease: a nationwide case-control study. BMC Gastroenterol. (2013) 13:109. doi: 10.1186/1471-230X-13-109
51. Kemppainen KM, Vehik K, Lynch KF, Larsson HE, Canepa RJ, Simell V, et al. Association between early-life antibiotic use and the risk of islet or celiac disease autoimmunity. JAMA Pediatr. (2017) 171:1217–25. doi: 10.1001/jamapediatrics.2017.2905
52. Kołodziej M, Patro-Gołab B, Gieruszczak-Białek D, Skórka A, Pieścik-Lech M, Baron R, et al. Association between early life (prenatal and postnatal) antibiotic administration and coeliac disease: a systematic review. Arch Dis Child. (2019) 104:1083–9. doi: 10.1136/archdischild-2019-317174
53. Leonard MM, Camhi S, Huedo-Medina TB, Fasano A. Celiac disease genomic, environmental, microbiome, and metabolomic (CDGEMM) study design: approach to the future of personalized prevention of celiac disease. Nutrients. (2015) 7:9325–36. doi: 10.3390/nu7115470
54. Caminero A, Herran AR, Nistal E, Perez-Andres J, Vaquero L, Vivas S, et al. Diversity of the cultivable human gut microbiome involved in gluten metabolism: isolation of microorganisms with potential interest for coeliac disease. FEMS Microbiol Ecol. (2014) 88:309–19. doi: 10.1111/1574-6941.12295
55. Olivares M, Laparra M, Sanz Y. Influence of Bifidobacterium longum CECT 7347 and gliadin peptides on intestinal epithelial cell proteome. J Agric Food Chem. (2011) 59:7666–71. doi: 10.1021/jf201212m
56. Caminero A, Galipeau HJ, McCarville JL, Johnston CW, Bernier SP, Russell AK, et al. Duodenal bacteria from patients with celiac disease and healthy subjects distinctly affect gluten breakdown and immunogenicity. Gastroenterology. (2016) 151:670–83. doi: 10.1053/j.gastro.2016.06.041
57. Heyman M, Abed J, Lebreton C, Cerf-Bensussan N. Intestinal permeability in coeliac disease: insight into mechanisms and relevance to pathogenesis. Gut. (2012) 61:1355–64. doi: 10.1136/gutjnl-2011-300327
58. Vorobjova T, Raikkerus H, Kadaja L, Talja I, Uibo O, Heilman K, et al. Circulating zonulin correlates with density of enteroviruses and tolerogenic dendritic cells in the small bowel mucosa of celiac disease patients. Dig Dis Sci. (2017) 62:358–71. doi: 10.1007/s10620-016-4403-z
59. Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. (2012) 336:1268–73. doi: 10.1126/science.1223490
60. Serena G, Yan S, Camhi S, Patel S, Lima RS, Sapone A, et al. Proinflammatory cytokine interferon-γ and microbiome-derived metabolites dictate epigenetic switch between forkhead box protein 3 isoforms in coeliac disease. Clin Exp Immunol. (2017) 187:490–506. doi: 10.1111/cei.12911
61. Lindfors K, Blomqvist T, Juuti-Uusitalo K, Stenman S, Venalainen J, Maki M, et al. Live probiotic Bifidobacterium lactis bacteria inhibit the toxic effects induced by wheat gliadin in epithelial cell culture. Clin Exp Immunol. (2008) 152:552–8. doi: 10.1111/j.1365-2249.2008.03635.x
62. Medina M, De Palma G, Ribes-Koninckx C, Calabuig M, Sanz Y. Bifidobacterium strains suppress in vitro the pro-inflammatory milieu triggered by the large intestinal microbiota of coeliac patients. J Inflamm. (2008) 5:19. doi: 10.1186/1476-9255-5-19
63. Laparra JM, Olivares M, Gallina O, Sanz Y. Bifidobacterium longum CECT 7347 modulates immune responses in a gliadin-induced enteropathy animal model. PLoS ONE. (2012) 7:e30744. doi: 10.1371/journal.pone.0030744
64. Zyrek AA, Cichon C, Helms S, Enders C, Sonnenborn U, Schmidt MA. Molecular mechanisms underlying the probiotic effects of Escherichia coli Nissle 1917 involve ZO-2 and PKCzeta redistribution resulting in tight junction and epithelial barrier repair. Cell Microbiol. (2007) 9:804–16. doi: 10.1111/j.1462-5822.2006.00836.x
65. D'Arienzo R, Stefanile R, Maurano F, Mazzarella G, Ricca E, Troncone R, et al. Immunomodulatory effects of Lactobacillus casei administration in a mouse model of gliadin-sensitive enteropathy. Scand J Immunol. (2011) 74:335–41. doi: 10.1111/j.1365-3083.2011.02582.x
66. Nadal I, Donat E, Donant E, Ribes-Koninckx C, Calabuig M, Sanz Y. Imbalance in the composition of the duodenal microbiota of children with celiac disease. J Med Microbiol. (2008) 56:1669–74. doi: 10.1099/jmm.0.47410-0
67. De Palma G, Nadal I, Medina M, Donat E, Ribes-Koninckx C, Calabuig M, et al. Intestinal dysbiosis and reduced immunoglobulin-coated bacteria associated with coeliac disease in children. BMC Microbiol. (2010) 10:63. doi: 10.1186/1471-2180-10-63
68. Collado MC, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. Imbalances in faecal and duodenal Bifidobacterium species composition in active and non-active coeliac disease. BMC Microbiol. (2008) 8:232. doi: 10.1186/1471-2180-8-232
69. Collado MC, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. Specific duodenal and faecal bacterial groups associated with paediatric coeliac disease. J Clin Pathol. (2009) 62:264–9. doi: 10.1136/jcp.2008.061366
70. Ou G, Hedberg M, Horstedt P, Baranov V, Forsberg G, Drobni M, et al. Proximal small intestinal microbiota and identification of rod-shaped bacteria associated with childhood celiac disease. Am J Gastroenterol. (2009) 104:3058–67. doi: 10.1038/ajg.2009.524
71. Schippa S, Iebba V, Barbato M, Di Nardo G, Totino V, Checchi MP, et al. A distinctive 'microbial signature' in celiac pediatric patients. BMC Microbiol. (2010) 10:175. doi: 10.1186/1471-2180-10-175
72. Di Cagno R, De Angelis M, De Pasquale I, Ndagijimana M, Vernocchi P, Ricciuti P, et al. Duodenal and faecal microbiota of celiac children: molecular, phenotype and metabolome characterization. BMC Microbiol. (2011) 11:219. doi: 10.1186/1471-2180-11-219
73. Wacklin P, Laurikka P, Lindfors K, Collin P, Salmi T, Lahdeaho ML, et al. Altered duodenal microbiota composition in celiac disease patients suffering from persistent symptoms on a long-term gluten-free diet. Am J Gastroenterol. (2014) 109:1933–41. doi: 10.1038/ajg.2014.355
74. Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. (2014) 11:506–14. doi: 10.1038/nrgastro.2014.66
75. De Angelis M, Rizzello CG, Fasano A, Clemente MG, De Simone C, Silano M, et al. VSL#3 probiotic preparation has the capacity to hydrolyze gliadin polypeptides responsible for Celiac Sprue. Biochim Biophys Acta. (2006) 1762:80–93. doi: 10.1016/j.bbadis.2005.09.008
76. Rizzello CG, De Angelis M, Di Cagno R, Camarca A, Silano M, Losito I, et al. Highly efficient gluten degradation by lactobacilli and fungal proteases during food processing: new perspectives for celiac disease. Appl Environ Microbiol. (2007) 73:4499–507. doi: 10.1128/AEM.00260-07
77. Francavilla R, De Angelis M, Rizzello CG, Cavallo N, Dal Bello F, Gobbetti M. Selected probiotic lactobacilli have the capacity to hydrolyze gluten peptides during simulated gastrointestinal digestion. Appl Environ Microbiol. (2017) 83:e00376–17. doi: 10.1128/AEM.00376-17
78. Smecuol E, Hwang H, Sugai E, Corso L, cherñavsky A, Bellavite F, et al. Exploratory, randomized, double-blind, placebo-controlled study on the effects of bifidobacterium infantis natren life start strain super strain in active celiac disease. J Clin Gastroenterol. (2013) 47:139–47. doi: 10.1097/MCG.0b013e31827759ac
79. Klemenak M, Dolinšek J, Langerholc T, Di Gioia D, Mičetić-Turk D. Administration of bifidobacterium breve decreases the production of TNF-α in children with celiac disease. Dig Dis Sci. (2015) 60:3386–92. doi: 10.1007/s10620-015-3769-7
80. Harnett J, Myers SP, Rolfe M. Probiotics and the microbiome in celiac disease: a randomised controlled trial. Evid Based Complement Alternat Med. (2016) 2016:1–16. doi: 10.1155/2016/9048574
81. Quagliariello A, Aloisio I, Bozzi Cionci N, Luiselli D, D'Auria G, Martinez-Priego L, et al. Effect of bifidobacterium breve on the intestinal microbiota of coeliac children on a gluten free diet: a pilot study. Nutrients. (2016) 8:660. doi: 10.3390/nu8100660
82. Savilahti EM, Ilonen J, Kukkonen AK, Savilahti E, Kuitunen M. Celiac disease by the age of 13 years is not associated with probiotics administration in infancy. J Pediatr Gastroenterol Nutr. (2018) 66:937–40. doi: 10.1097/MPG.0000000000001846
83. Uusitalo U, Andren Aronsson C, Liu X, Kurppa K, Yang J, Liu E, et al. Early probiotic supplementation and the risk of celiac disease in children at genetic risk. Nutrients. (2019) 11:1790. doi: 10.3390/nu11081790
84. Primec M, Klemenak M, Di Gioia D, Aloisio I, Bozzi Cionci N, Quagliariello A, et al. Clinical intervention using Bifidobacterium strains in celiac disease children reveals novel microbial modulators of TNF-α and short-chain fatty acids. Clin Nutr. (2019) 38:1373–81. doi: 10.1016/j.clnu.2018.06.931
85. Håkansson Å, Andrén Aronsson C, Brundin C, Oscarsson E, Molin G, Agardh D. Effects of Lactobacillus plantarum and Lactobacillus paracasei on the peripheral immune response in children with celiac disease autoimmunity: a randomized, double-blind, placebo-controlled clinical trial. Nutrients. (2019) 11:1925. doi: 10.3390/nu11081925
86. Cook LC, Munier ML, Seddiki N, van Bockel D, Ontiveros N, Hardy MY, et al. Circulating gluten-specific FOXP3(+)CD39(+) regulatory T cells have impaired suppressive function in patients with celiac disease. J Allergy Clin Immunol. (2017) 140:1592–603.e8. doi: 10.1016/j.jaci.2017.02.015
87. Cinova J, De Palma G, Stepankova R, Kofronova O, Kverka M, Sanz Y, et al. Role of intestinal bacteria in gliadin-induced changes in intestinal mucosa: study in germ-free rats. PLoS ONE. (2011) 6:e16169. doi: 10.1371/journal.pone.0016169
88. Sanchez E, Laparra JM, Sanz Y. Discerning the role of Bacteroides fragilis in celiac disease pathogenesis. Appl Environ Microbiol. (2012) 78:6507–15. doi: 10.1128/AEM.00563-12
89. Papista C, Gerakopoulos V, Kourelis A, Sounidaki M, Kontana A, Berthelot L, et al. Gluten induces coeliac-like disease in sensitised mice involving IgA, CD71 and transglutaminase 2 interactions that are prevented by probiotics. Lab Invest. (2012) 92:625–35. doi: 10.1038/labinvest.2012.13
90. Jeon SG, Kayama H, Ueda Y, Takahashi T, Asahara T, Tsuji H, et al. Probiotic Bifidobacterium breve induces IL-10-producing Tr1 cells in the colon. PLoS Pathog. (2012) 8:e1002714. doi: 10.1371/journal.ppat.1002714
91. Zheng B, van Bergenhenegouwen J, Overbeek S, van de Kant HJG, Garssen J, Folkerts G, et al. Bifidobacterium breve attenuates murine dextran sodium sulfate-induced colitis and increases regulatory T cell responses. PLoS ONE. (2014) 9:e95441. doi: 10.1371/journal.pone.0095441
Keywords: celiac disease, dysbiosis, gut microbiota, microbiome, probiotics
Citation: Pecora F, Persico F, Gismondi P, Fornaroli F, Iuliano S, de'Angelis GL and Esposito S (2020) Gut Microbiota in Celiac Disease: Is There Any Role for Probiotics? Front. Immunol. 11:957. doi: 10.3389/fimmu.2020.00957
Received: 04 March 2020; Accepted: 23 April 2020;
Published: 15 May 2020.
Edited by:
Veena Taneja, Mayo Clinic, United StatesReviewed by:
Luis Vitetta, University of Sydney, AustraliaStefano Guandalini, The University of Chicago Pritzker School of Medicine, United States
Copyright © 2020 Pecora, Persico, Gismondi, Fornaroli, Iuliano, de'Angelis and Esposito. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Susanna Esposito, c3VzYW5uYS5lc3Bvc2l0b0B1bmltaS5pdA==
 Francesco Pecora
Francesco Pecora Federica Persico1
Federica Persico1 Gian Luigi de'Angelis
Gian Luigi de'Angelis Susanna Esposito
Susanna Esposito