- Centre for Biological Timing, Faculty of Biology Medicine and Health, University of Manchester, Manchester, United Kingdom
The gut microbiome plays a critical role in regulating host immunity and can no longer be regarded as a bystander in human health and disease. In recent years, circadian (24 h) oscillations have been identified in the composition of the microbiota, its biophysical localization within the intestinal tract and its metabolic outputs. The gut microbiome and its key metabolic outputs, such as short chain fatty acids and tryptophan metabolites contribute to maintenance of intestinal immunity by promoting barrier function, regulating the host mucosal immune system and maintaining the function of gut-associated immune cell populations. Loss of rhythmic host-microbiome interactions disrupts host immunity and increases risk of inflammation and metabolic complications. Here we review factors that drive circadian variation in the microbiome, including meal timing, dietary composition and host circadian clocks. We also consider how host-microbiome interactions impact the core molecular clock and its rhythmic outputs in addition to the potential impact of this relationship on circadian control of immunity.
Introduction
The circadian clock is a critical regulator of immunity, including homeostatic processes and responses to immune challenge (1–3). It is now very well established that intrinsic clocks within cells of the immune system regulate their function, influencing the manner in which they respond to a pathogen. However, extrinsic rhythmic signals also drive rhythmic behavior in immune cells. The gut microbiome is recognized as a major influence over the immune system, impacting on its development and daily function. Work over the last decade has established that this bacterial community exhibits 24 h oscillations in composition, biophysical localization and function. Furthermore, these oscillations are critical for driving rhythmic metabolic processes both within the gut and further afield in organs such as the liver. Here we examine the potential impact of circadian rhythms in the microbiome on immunity, and pose the question might rhythms in the microbiome influence circadian control of immunity?
The Circadian Clock
Entrainment to the 24 h environment is vital to allow organisms to temporally arrange their daily functions (such as feeding, metabolism and sleeping) to align with optimal conditions. Nearly every cell of the body (including those of the immune system) contains the molecular machinery required to keep the clock ticking (4). However, organisms cannot set the time correctly without the help of environmental cues termed “zeitgebers” (meaning time-givers). The most important zeitgeber is the light-dark cycle, which entrains the central clock within the suprachiasmatic nucleus (SCN). Other zeitgebers, such as food availability (5), hormones (6), and body temperature (7) are also important, particularly for influencing peripheral pacemakers (8). Peripheral clocks are synchronized by the SCN, but continue to oscillate when uncoupled from the central clock, highlighting their ability to respond autonomously to changes in the local micro-environment (3, 9, 10).
The molecular clockwork machinery relies on transcriptional-translational feedback loops (TTFL), the duration of which dictates the period of the circadian rhythm. The primary TTFL involves activator proteins circadian locomotor output cycles kaput (CLOCK) and brain muscle arnt-like 1 (BMAL1) interacting with repressor proteins PERIOD and CRYPTOCHROME (4). CLOCK and BMAL1 heterodimerise and bind Enhancer-box (E-box) sequences on target genes for PERIOD and CRYPTOCHROME, which accumulate in the cytoplasm before translocating to the nucleus to inhibit further CLOCK-BMAL1 activity by negative feedback (11). The main TTFL is stabilized by an auxiliary feedback loop involving REV-ERBα and retinoid-related orphan receptor (ROR)α, which repress and activate BMAL1, respectively (12). In this review, we consider bi-directional interactions between the circadian timing system and the gut microbiome and how this influences immune function.
The Microbiome
The microbiome is defined as an environment containing a heterogeneous community of microbes (including bacteria, fungi, and viruses) and the functional consequences of these microbes, such as their metabolic outputs and interaction with the host. By contrast, microbiota solely describes the community of organisms in isolation (13). It is generally accepted that the fetus develops in a sterile environment and first contact with microbes occurs during birth. An infant's microbiome is influenced by delivery route and method of feeding (14). The first 2–3 years of life oversee significant flux in the composition of the gut microbiome before assuming an adult-like profile, influenced by cessation of breast-feeding, increasing complexity of dietary nutrients and environmental exposures, including antibiotics (15, 16).
The advent of high throughput genome-sequencing technology and large scale projects such as the National Institute for Health's Human Microbiome Project (17) and the European Commission's Metagenomics of the Human Intestinal Tract (18) have dramatically improved our understanding of the phylogenic composition and variability of the microbiota, which in humans has over 150 genera from three main phyla: Firmicutes, Bacteroidetes and Actinobacteria (19). The abundance of these phyla varies significantly between individuals (20, 21).
Our understanding of microbiome function is growing constantly, with main areas of interest in development, immunity and metabolism. Here we focus on the impact of gut microbiota rhythmicity in directing immunity.
The Gut Microbiome Shapes Immunity
The gut microbiota contribute to the development, maturation and regulation of the host immune system. The importance of the microbiome in shaping the immune system is perhaps best demonstrated by studies with germ free (GF) animals. GF mice have underdeveloped gut-associated lymphoid tissues, fewer and smaller Peyer's patches and mesenteric lymph nodes, and impaired development of isolated lymphoid follicles (22, 23). GF mice have reduced levels of secretory IgA in the intestine (24), and the morphology of the intestinal epithelial cells (IECs), which are normally in direct contact with the microbiota, is modified, with altered microvilli formation and slower cell turnover (25). Furthermore, the absence of a microbiota is associated with arrested capillary network formation within the intestines (26, 27).
Studies with GF mice elegantly demonstrate that commensal bacteria are essential for the development of immune cell subsets, both within the gut lamina propria and further afield. The absence of gut commensals leads to defects in circulating innate immune cell populations (including neutrophils, monocytes and macrophages) and cells within systemic immune sites (including the spleen, bone marrow and liver) as well as delayed neutrophil aging (28–30). Much focus in the field has been surrounding the influence of the microbiota on shaping T cell populations, although it is clear that this influence extends much further to include B cells and innate immune cells. Work over the last decade has assigned roles for individual commensal species in influencing the composition of the lamina propria T lymphocyte subsets. For example, in rodents, segmented filamentous bacteria (SFB) induce intestinal T helper 17 (Th17) cells (31, 32), and in humans Bifidobacterium adolescentis plays a similar role (33). GF mice have reduced numbers of regulatory T cells (Tregs) in the lamina propria, which can be rescued by re-colonization with strains of clostridium (34). These studies highlight the importance of the microbiota for maintenance of the Th17/Treg axis. In support, outcomes of mouse models of autoimmunity are often dependent on colonization status. For example, GF mice exhibit marked attenuation in murine models of experimental arthritis (35), experimental autoimmune encephalomyelitis (EAE) (36) and uveitis (37). In the case of EAE (a model of CNS inflammation), the absence of the microbiota is associated with loss of susceptibility as a consequence of perturbations in the balance of Tregs and Th17 cells. This balance is restored after re-colonization with SFB and susceptibility returns (36). Similarly the reduction in Th17 cells associated with GF mice results in an attenuated arthritis in the K/BxN model of experimental arthritis (35). Intriguingly, in uveitis, retina-specific T cells have been detected in the intestine. Given that the eye is immunologically privileged, it may be that the gut microbiome is playing a role in antigen-mimicry, whereby T cells are triggered by a surrogate antigen present in the gut environment, mimicking a retinal antigen (38). The existence of a gut-retina axis is supported by the presence of uveitis in a subset of patients with inflammatory bowel disease, although further work is required to implicate the microbiome directly (39).
Murine models of autoimmune-driven Type 1 diabetes mellitus, such as non-obese diabetic (NOD) mice are driven in part by activation of innate immune component toll-like receptors (TLR) (40). NOD mice with genetic deletion of MyD88, a common adaptor protein for TLR signaling, are protected from type 1 diabetes, but this protection is lost in GF conditions (41). In keeping, NOD mice treated with antibiotics from an early age demonstrate an increased incidence of type 1 diabetes mellitus (42, 43). Together, these data suggest the microbiome modulates innate immunity through TLR signaling to impact risk of autoimmune metabolic diseases such as diabetes in mice.
Metabolic Outputs of the Microbiota Direct Immunity
Many of the effects the microbiome has on immunity are attributable to the metabolic outputs of the gut microbiota (44, 45). Intestinal bacteria produce a wide range of metabolites, including short chain fatty acids (SCFAs), tryptophan metabolites, essential vitamins, phenolic acids, polyamines and bile acids, which act to modulate host metabolism and immunity (Figure 1).
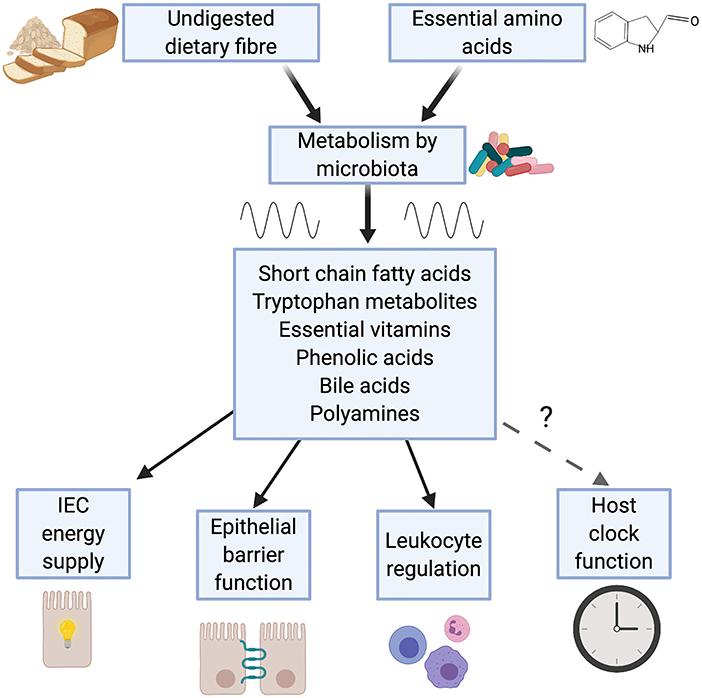
Figure 1. Metabolic outputs from the microbiome influence immunity. Bacteria within the gut breakdown substances derived from the diet. Emerging evidence demonstrates that production of a number of these metabolites is rhythmic, a consequence of feeding rhythms and rhythmic function of the microbiome. Many of these microbial metabolites (e.g. short chain fatty acids, tryptophan metabolites and bile acids) play a key role in immunity, contributing to: intestinal epithelial cell (IEC) function; maintenance of leukocyte populations such as regulatory T cells and macrophages and intestinal tolerance.
Short Chain Fatty Acids
SCFAs [for example, acetate (C2), proprionate (C3), and butyrate (C4)] are volatile fatty acids composed of a backbone of between 1 and 6 carbon atoms. Derived from bacterial anaerobic fermentation of dietary fibers in the colon, they promote intestinal epithelial barrier function and regulate the host mucosal immune system. SCFAs are important for maintenance of IEC turnover and barrier function (46–48). Butyrate is a critical source of energy for IECs (49), and also regulates expression of the transcription factor hypoxia-inducible factor (HIF)1α, which acts to co-ordinate barrier function (46) and induces IL10RA, which promotes expression of the tight junction protein Claudin 2 (48). In addition, SCFAs act on multiple gut resident immune cells to facilitate maintenance of intestinal immunity. The earliest observation of the role for SCFAs in maintenance of immune cell populations was made in Tregs. Butyrate and proprionate facilitate extrathymic generation of Tregs (50, 51) by enhancing histone H3 acetylation in the promoter and conserved non-coding sequence regions of the FoxP3 locus (52). More recently it has been established that the effects of SCFA extend far beyond these anti-inflammatory cells to influence the function of both adaptive and innate immune cell populations. These include macrophages (53, 54), B cells (55), CD8+T cells (56), type 3 innate lymphoid cells (ILC3s) (57, 58) and neutrophils (58). For example, butyrate alters the metabolic behavior of intestinal macrophages, promoting a state of alternative activation and thus promoting microbial tolerance (53).
Tryptophan Metabolites
Tryptophan is a dietary essential amino acid metabolized in the gastrointestinal tract via three different pathways: the kynurenine pathway; the serotonin pathway; or via direct metabolism by microbiota (59). Whilst the former two metabolic pathways are regarded as host-dependent, the microbiota has been shown to influence them (59). Tryptophan metabolism by the microbiota includes the transformation of tryptophan to indole and its derivatives, many of which are ligands for the aryl hydrocarbon receptor (AhR). This includes: indole-3-aldehyde (IAId), indole-3-acetate (I3A), indole 3-proprionic acid (IPA), indole-3-acetaldehyde (IAAId), 5-hydroxyindole-3-acetic acid (5-HIAA) and indoleacrylic acid.
AhR is a Per/Arnt/Sim (PAS) domain protein and a fundamental modulator of immunity, controlling the differentiation and inflammatory potential of innate and adaptive immune cells at the gut barrier and systemically (60, 61). AhR shares significant sequence homology to the core clock protein CLOCK and as such offers itself as a nodal point between the gut microbiota, circadian clocks and immunity (Figure 2). Ligand bound AhR heterodimerises with aryl hydrocarbon receptor nuclear translocator (ARNT) in the nucleus, but can also heterodimerise with BMAL (62) to disrupt normal binding of CLOCK/BMAL to the Per1 promoter (63). AhR plays a role in regulating circadian rhythms in behavior and physiology, as loss of one allele alters responses to changes in the light dark cycle, increases the amplitude of hepatic core clock genes and alters rhythms in glucose and insulin (64). Conversely, AhR activation alters rhythmic expression of core circadian regulators within the periphery; for example chronic administration of the AhR agonist 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) dampens rhythmic expression of core clock genes and clock-controlled hepatic genes disrupting circadian regulation of hepatic metabolism (65). To date there is no direct evidence that microbiota-derived tryptophan metabolites affect timing mechanisms, but this is a certainly a possibility.
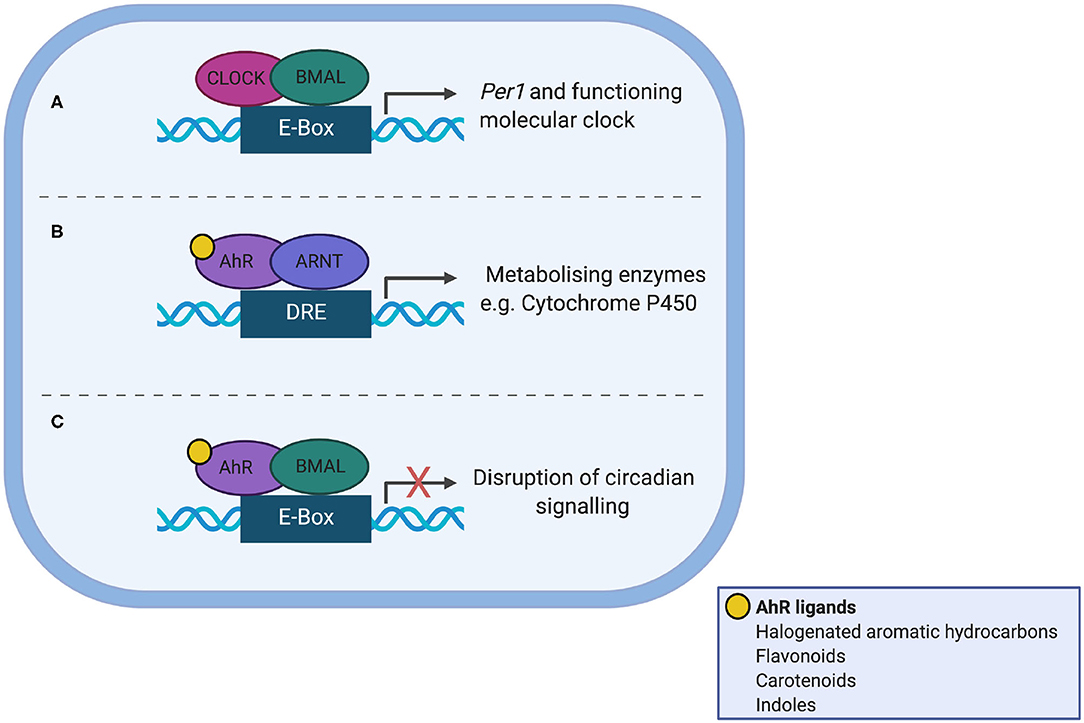
Figure 2. Interaction between the aryl hydrocarbon receptor (AhR) and the molecular clock. (A) In normal circadian clock function, CLOCK and BMAL heterodimerise and bind the E-box promotor, to transcribe Per1. (B) AhR shares sequence homology with CLOCK. Ligand-bound AhR heterodimerises with aryl hydrocarbon receptor nuclear translocator (ARNT) in the nucleus leading to transcription of enzyme superfamilies. (C) Ligand-bound AhR can also heterodimerise to BMAL1, which disrupts normal binding of CLOCK/BMAL to the Per1 promoter, leading to circadian disruption.
Tryptophan metabolites produced by the microbiota are important for regulation of immunity. For example, IAId produced by lactobacilli induces IL22 production to support antifungal resistance and ameliorate colitis in mouse models (66). I3A reduces the production of pro-inflammatory cytokines by macrophages and slows their migration toward a chemotactic signal in an AhR dependent manner (67). Additionally, tryptophan metabolites such as IPA act via pregnane X receptors (PXR) to regulate intestinal barrier function through TLR4 activity (68, 69). Recent work has uncovered a mechanism whereby bacteria derived butyrate enhances production of 5-HIAA which acts via AhR to support regulatory B cell function (70). This highlights the importance of interactions between different microbial species to promote immunity.
Bile Acids
Bile acids (BAs) are cholesterol-derived molecules produced in the liver and secreted into the duodenum. Whilst the vast majority (~95%) are re-absorbed through the terminal ileum, several different phyla of gut bacteria can transform the remainder to secondary bile acids, with immune modulatory effects. Recently there has been a flurry of activity in this field ascribing a role for secondary BAs in T cell biology. The lithocholic acid (LCA) derivatives 3-oxoLCA and isoalloLCA regulate Th17 and Treg cell differentiation in the lamina propria (71) and RORγ+ Tregs in the colon (72). Furthermore, isodeoxycholic acid (isoDCA) promotes the generation of peripheral Tregs (73).
Evidence for Intrinsic Circadian Clocks Within Gut Bacteria
The existence of intrinsic circadian clocks has rarely been demonstrated in organisms outside of the eukaryotic kingdom, with the exception of certain photosynthetic cyanobacteria. Synechococcus elongatus utilizes 3 clock proteins (KaiA, KaiB and KaiC) in a post-translational oscillator cycle of phosphorylation and de-phosphorylation to generate 24 h rhythms (74). The S. elongatus clock can synchronize to 24 h temperature cycles and directs clock-regulated gene expression of almost the entire genome (75, 76). It appears that it is not necessary to have all three Kai proteins in order to exhibit rhythmicity. Rhodobacter sphaeroides are purple photosynthetic bacteria which express self-sustaining rhythmicity under aerobic condition but lack KaiA (77), suggesting that either KaiB and KaiC function differently from those in Cyanobacteria, or that there may be novel components regulating the clock mechanisms in place here.
The existence of timekeeping components in non-photosynthetic bacteria has been addressed in part by exploring whether homologs of the Kai genes exist in other bacteria. Whilst KaiB and KaiC are quite highly conserved, KaiA is missing in many other prokaryotes (78). However, like R. Sphaeroides, circadian cycles can still be still displayed as in the case of the marine cyanobacterium Prochlorococcus (77). Enterobacter aerogenes is the first gut commensal bacteria found to express 24 h rhythms outside the host (79). This gram-negative motile bacterium from the phylum Proteobacteria shows circadian rhythmic swarming behavior in the presence of melatonin, which was unreproducible in other gram-negative bacteria such as Escherichia coli or Klebsiella pneumoniae. Furthermore, transformation with a flagellar motor-protein-driven luciferase reporter reveals temperature-compensated circadian rhythm of luciferase activity synchronized by melatonin (80). Given the abundance of extrapineal melatonin secreted within the gut (81), these studies present this diurnal hormone as a potential zeitgeber for the gut microbiome and may present one mechanism of host control of microbiome rhythmicity. Further studies are required to understand the intrinsic circadian mechanisms of E. aerogenes and explore the presence of intrinsic rhythms within other species of the gut microbiota. One possibility is that gut commensals have evolved to respond rhythmically to environmental signals through horizontal gene transfer with human hosts (80, 82). The presence of intrinsic circadian machinery in gut microbiota would have far-ranging implications for understanding host-microbiome interactions.
Rhythmicity of the Microbiome
With the discovery of resident intestinal clocks (83–85) and rhythmicity in multiple immune cells, interest is growing in the circadian influence on host-microbiome interaction and immunity. 16S rRNA gene sequencing of the fecal microbiome has demonstrated diurnal oscillations in the relative abundance of roughly 15% of OTUs, accounting for around 60% of the murine microbiome, with corresponding oscillation in 10% of OTUs in humans (86, 87). In addition rhythmicity in total microbiota has been described, driven predominantly by Bacteroidetes, which peak during the murine active phase (88). Diurnal oscillations in abundance also exist robustly in healthy humans despite marked inter-human variation in gut microbiome composition (20).
The potential functional consequences of a rhythmic microbiome have been highlighted via metagenomic studies showing oscillations in up to a quarter of total microbiome genetic material in both mice and humans (86). KEGG pathway analysis identified time-of-day grouping of functions such as cell growth and energy metabolism during the murine dark phase, with environmental sensing and flagellar assembly during the light phase (86). Human metagenome analysis demonstrated inversion of peaks in pathway activity compared to nocturnal mice (86). Interestingly, in a cohort of patients with Type 2 diabetes mellitus, diurnal oscillations in microbiome composition were dampened with metagenomic analysis highlighting impact on microbial pathways that process amino acids and fatty acids (89). This is notable as one of the first studies to associate an arrhythmic microbiome with a human disease state.
Diurnal oscillations have been detected in the biophysical distance between mucosal bacteria and IECs, with closest proximity during the murine active, dark phase (90). In mice with global knockout of RegIIIγ, an antimicrobial peptide (AMP) secreted by IECs into the mucus layer, rhythms in the mucosal microbiome were attenuated, suggesting AMPs have a part to play in control of mucosal rhythmicity as well as epithelial cell-microbe distance (90, 91). The mucosal microbiome in closest proximity to the epithelial barrier is most likely to be sampled by mucosal immune components and co-ordinated rhythmicity between the host and microbiome will likely produce the most effective balance in gut homeostasis between tolerance and activated immune response. Thus, whilst the fecal microbiome is easy to sample, the mucosal microbiome may provide more answers to the links in circadian host-microbiome interactions.
Metabolic outputs of the microbiota also exhibit diurnal rhythmicity (Table 1). This is heavily influenced by feeding rhythms of the host providing targets for metabolism, but rhythms in microbial activity clearly also contribute. SCFA levels show diurnal fluctuations in concentration in feces, caecal samples and plasma (92–94). These oscillations are regulated by feeding time as rhythmicity in caecal SCFA are abolished in arrhythmic Bmal1−/− mice, but restored by a restricted feeding regimen (94). Oscillations are also sensitive to dietary composition as a high-fat diet (HFD) abolishes rhythms in fecal butyrate (92). While levels of liver-derived conjugated bile acids exhibit post-prandial fluxes during the day in humans, levels of unconjugated bile acids peak late at night, likely reflecting their dependence on microbial activity (95). Many other microbiota-derived or microbiota-modulated metabolites exhibit diurnal rhythms in the intestines and serum, including the polyamine ornithine and the amino acid proline (90, 96). To date, the impact of these rhythms on downstream immunological processes has not been explored, but the importance of microbial metabolites in host immunity makes this an important area to explore.
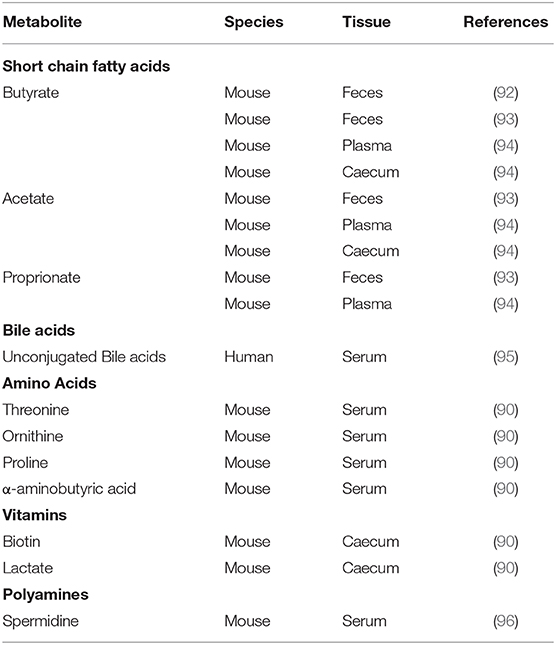
Table 1. Microbial metabolites with demonstrated daily rhythmicity in tissue or circulating concentrations.
Timing of food intake is a circadian process centrally controlled by the host and influenced by hunger, food availability and social and cultural norms (97). Mice with disrupted circadian light entrainment show a loss of fecal microbiome diversity, loss of microbiome oscillations, increased weight gain and impaired glucose tolerance despite similar calorie intake and energy expenditure to controls on a standard light:dark cycle (98–100). In a “jet-lag” model, mice phase-shifted by 8 h every 3 days for 4 weeks demonstrate attenuated diurnal oscillations in microbiome abundance, likely driven by loss of diurnal food intake (86).
In addition to timing of food, dietary composition also impacts the microbiome rhythmicity. The availability of high-fat diet ad libitum drives mice to lose their diurnal eating habits and spread their intake through the day (101). This dampens diurnal oscillations in the microbiome (which are restored when time-restricted feeding is applied) and results in persistent microbial dysbiosis (87). Jet-lagged mice fed a high-fat diet experience greater weight gain compared to non-shifted controls, despite equal food intake; a phenotype attenuated by concurrent antibiotic administration or provision of a high-fat diet to GF mice (86, 92). Fecal microbial transplantation (FMT) from mice with jet lag to GF, non-shifted mice conferred an obesity phenotype not seen in FMT of non-jet-lagged mice (86). This suggests that hosts with a disrupted clock are more susceptible to harboring a dysbiotic microbiome, which is sufficient to drive adverse metabolic consequences for the host (Figure 3).
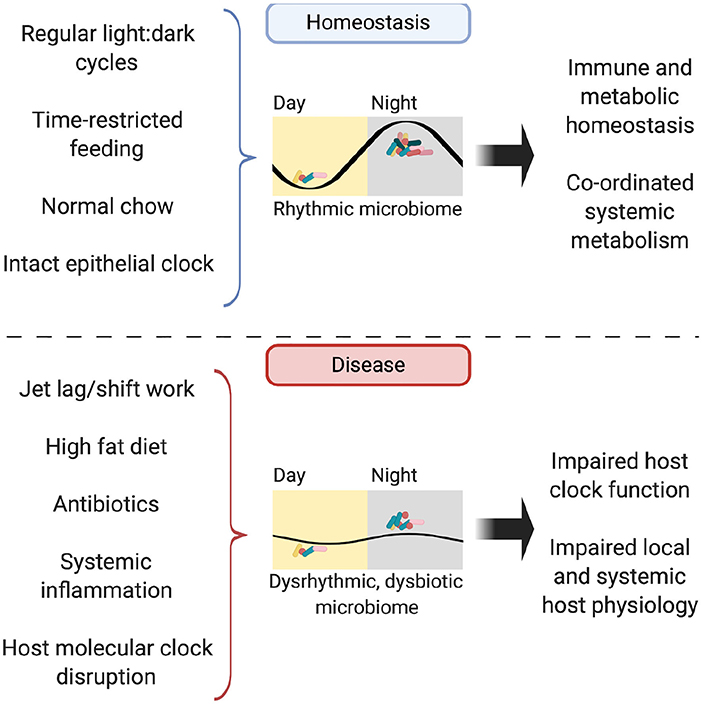
Figure 3. The rhythmic microbiome. In homeostasis, the microbiome is rhythmic in its composition, abundance and biogeographic distance from the intestinal mucosa, with its abundance peaking in the murine dark, active phase. Possible entraining factors include light:dark cycles, timing and composition of food intake and host molecular clock function. Functioning oscillations in the microbiome drive homeostasis in both immunity and metabolism, both locally in the intestine and further afield in organs such as the liver. In disease, disruption to entrainment via jet lag, high fat diet, illness and treatments such as antibiotics lead to perturbations in microbiome rhythmicity and subsequent impairment of local and systemic immune-metabolic homeostasis.
The Host Clock Shapes the Microbiome
Whilst feeding times clearly drive rhythms in the microbiome, the host circadian molecular clock also plays a role, with studies evidencing the importance of the biological clock in regulating the composition of the microbial community as well as the rhythmicity. Studies utilizing ClockΔ19 mice (as a genetic approach to model circadian disruption) report increased intestinal hyperpermeability (102) and reduced microbial diversity (103) of the gut microbiome. It is feasible, but as yet undemonstrated, that these changes to the microbiota influence barrier integrity (or vice versa). To address the contribution of the host clock in driving microbiome rhythmicity, studies often use transgenic mice lacking a functional clock. Most studies here have been undertaken using animals with global deletion of core clock genes, which complicates interpretation of results as this likely leads to arrhythmic feeding behavior. Mice with global Bmal1 knockout are arrhythmic and demonstrate attenuated compositional microbiome rhythmicity, whilst retaining circadian oscillations in total fecal microbiome abundance (88). Arrhythmic mice with global Per1/2 deletion lose rhythmicity in abundance of fecal and mucosal microbiomes as well as metagenomic and metabolic outputs (86, 90). In addition, the intestinal microbiome of global Per1/2 knockout mice is dysbiotic with less diversity compared to littermate controls (86). Instigation of a time-restricted feeding schedule to Per1/2 knockout mice to align food consumption with wildtypes recovers oscillations in the microbiome (86). Targeting the molecular clock in specific cells will further delineate the relative contributions of central host rhythms and circadian rhythms in local mucosal immunity to microbiome dysbiosis. Indeed, this approach has revealed a subset of ILCs as critical regulators of the rhythmic microbiome.
ILCs as Rhythmic Regulators of the Microbiome
ILCs are circadian rhythmic tissue-resident lymphoid cells. There are currently five subsets of ILCs, which align with their T helper cell counterparts comprised of natural killer cells, ILC1s, ILC2s, ILC3s and lymphoid tissue inducer cells (104). ILC3s are innate counterparts to Th17 cells, characterized by expression of RORγt and production of IL17, IL22, and GM-CSF and are the predominant ILC class found at mucosal sites such as the intestine. ILC3s regulate the composition of the microbiota by encouraging preferential commensal growth whilst contributing to removal of pathogens. ILC3-derived IL22 binds directly to IECs and acts through Signal Transducer and Activator of Transcription (STAT)3 to support a multitude of functions that bolster the luminal biophysical barrier including production of AMPs, goblet cell differentiation and subsequent mucus production (105, 106). In addition, IL22 improves colonization resistance by fucosylating IEC-associated proteins that commensal bacterial preferentially use as an energy source, thus helping commensals out-compete pathogens (107, 108).
Recent work has uncovered the importance of the circadian clock on the function of ILC3s (Figure 4). In homeostasis, ILC3s exhibit oscillating expression of core clock genes, transcription factors (such as Ahr) and functional genes (such as il17 and il22) (109–111). ILC3-specific knockout of either Bmal1 or Rev-erbα decreases intestinal ILC3 numbers, decreases gene expression of anti-microbial peptides and mucus production and disrupts oscillations of Proteobacteria and Bacteroidetes in the microbiome (109–111). Intriguingly, these genetic manipulations do not affect abundance of ILC3s in the spleen and lung, suggesting intestinal ILC3s may be uniquely reliant on Bmal1 (109).
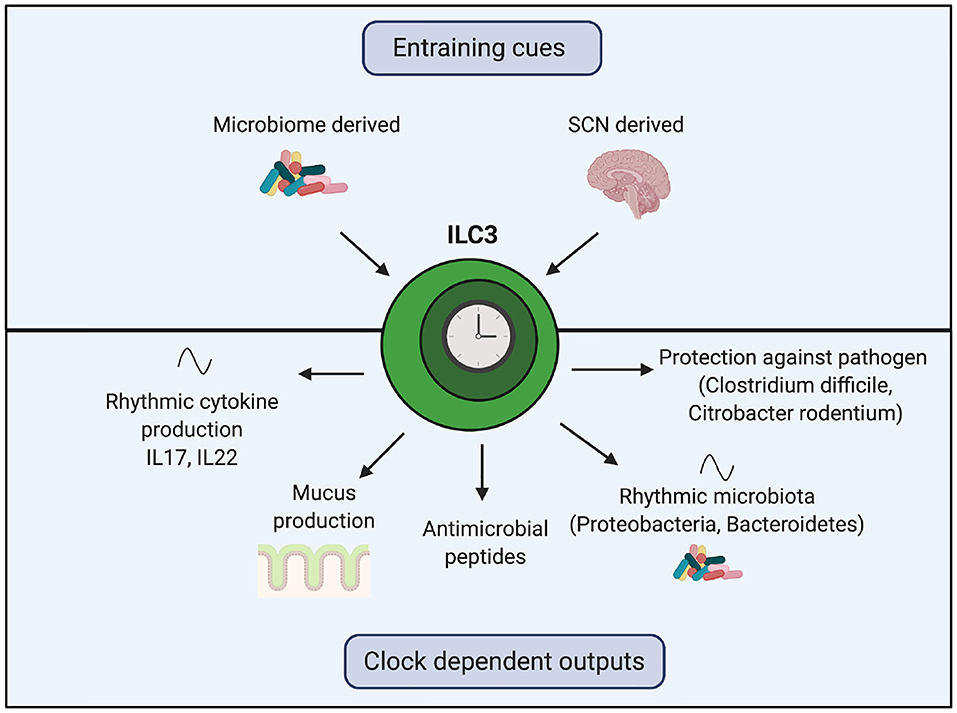
Figure 4. The role of the ILC3 clock in regulating the rhythmic microbiome and barrier function. Clocks within ILC3s are synchronized by the SCN and local microbiome-derived cues. Genetic manipulation of core clock genes within ILC3s demonstrates the importance of this intrinsic clock for regulating crosstalk between the host and the microbiome. Loss of the ILC3 clock results in decreased gene expression of anti-microbial peptides, reduced mucus production, disrupted oscillations in components of the microbiota and cytokines and impaired responses to harmful bacteria.
Mice with Rev-erbα-deficient ILC3s exposed to Clostridium difficile show an exaggerated IL17 response and a higher bacterial burden (111). Overproduction of IL17 and IL22 in Bmal1-deficient ILC3s is abrogated by antibiotic treatment (110). Lymphocyte-deficient mice with Bmal1-deficient ILC3s are more susceptible to gut inflammation induced by Citrobacter rodentium, with increased epithelial barrier permeability and reduced survival (109). In wildtype mice, oral antibiotic treatment shifts the timing of Per1 expression in ILC3s and dampens IL22 oscillations (109, 110). Together, these observations illustrate the importance of the ILC3-intrinsic circadian clock in regulation of crosstalk between host ILC3s and the microbiome. In humans with inflammatory bowel disease, there is a reduction in ILC3s within inflamed intestine compared to intra-individual non-inflamed tissue, with associated disruption of ILC3 circadian gene expression (110). Of interest, Rev-erbα gene expression was elevated in human ILC3s from inflamed tissue (110, 112). These perturbations to the ILC3 clockwork may have downstream effects on management of microbial rhythmicity.
Multiple components of mucosal immunity exhibit rhythmicity, including the microbiome, IECs, dendritic cells and ILC3s, which will likely need to be co-ordinated and entrained for maximal gut homeostasis. It will be pertinent to delineate the local power balance and determine which circadian clocks, local or central, drive rhythmicity in the gut microbiome during homeostasis and the consequences of disease (90, 113–115).
Rhythms in the Gut Microbiome Affect Rhythmic Processes Elsewhere
Communication between the host and microbiome is bi-directional. Whilst host clocks and rhythmic feeding drive daily variation in the composition of the microbiome, signals derived from the microbiome can influence circadian host physiology. We are now beginning to establish an understanding of how the microbiota and its metabolic outputs effects rhythmic processes.
A significant proportion of rhythmic transcripts within the intestines are influenced by the intestinal microbiota. The dependence of rhythmic expression of the core clock genes on the microbiome appears to be tissue dependent, with depletion of the microbiome affecting clock genes such as Rorα and Rev-erbα differently in the colon compared to the small intestines. Mukherji et al. mapped the rhythmic transcriptome in ileal IECs, which includes nuclear factor interleukin 3 regulated (Nfil3), tlr genes and clock genes Rorα and Rev-erbα. Depletion of the microbiome via administration of antibiotics results in the cessation of rhythmic gene expression and a fall in Tlr expression, whilst peroxisome proliferator activated receptor (Ppar)α becomes constitutively expressed, suppressing downstream circadian outputs including Nfil3, with subsequent impairment of host metabolism (113). Administration of LPS (as a surrogate for microbiome activation of toll-like receptors) reinstated RORα-driven circadian Tlr expression and recovered homeostatic IEC outputs including hypothalamic-pituitary-adrenal axis-independent corticosterone rhythmicity (113). NFIL3 is important in the development and regulation of immune cells including ILCs, macrophages and dendritic cells and may be one pathway for an arrhythmic microbiome to impact host immunity (116–118). In addition, lower NFIL3 levels have been detected in patients with active inflammatory pathologies such as colitis and arthritis, conditions that have been associated with microbiome dysbiosis (117, 119).
Transcriptomic examination of colonic IECs by Thaiss et al. demonstrated that depletion of the microbiota causes loss of rhythms in many pathways (including nucleotide metabolism and cell-cycle pathways) without affecting the core circadian clock genes (90). Intriguingly, this study also noted emerging de novo circadian rhythms in genes associated with metabolic pathways, suggesting the host may acquire compensatory oscillatory programmes in the absence of the microbiota. In keeping, depletion of the microbiome resulted in distinct changes to the temporal organization of the chromatin landscape within IECs. The regulation of transcriptional oscillations is dependent on rhythmic bacterial adherence, as mice lacking RegIIIγ (which have an abundant but non-rhythmic mucosal microbiome) showed an overlapping oscillating transcript with antibiotic-treated mice (90).
Further support for a role for the intestinal microbiota in programming daily rhythms in metabolic networks within the gut comes from a study, which found the gut microbiota drives rhythmic recruitment of HDAC3 to chromatin in IECs of the small intestine (120). Resultant HDAC3-NCoR complexes produce synchronized diurnal oscillations in histone acetylation, expression of metabolic genes, nutrient uptake and intestinal lipid absorption. Bacteroides thetaiotaomicron is critical here, as mono-colonization of GF mice with this Bacteroide is sufficient to restore rhythms in HDAC3 expression. Interestingly, the mechanism appears to be restricted to the small intestine; within the colon, genome wide acetylation rhythms persist in the absence of the microbiota (90, 120).
The microbiota also influences metabolic programmes within the gut via direct action on the clock. Wang et al. describe a signaling network by which the microbiota modulates clock activity with IECs to influence their metabolic programme (114) (Figure 5). This network integrates local immune cells which respond directly to the microbiota and signal to the IECs. Gram-negative bacteria are detected via toll-like receptors on myeloid cells within the intestinal lamina propria, via a number of potential routes (121). This prompts IL23 release which drives IL22 production by ILC3s. IECs respond to IL22 by up-regulating expression of STAT3, which binds to the promoter of Rev-erbα to repress expression. This reduction in REV-ERB levels allows enhanced transcription of Nfil3, a transcription factor that regulates a circadian transcription program of genes involved in lipid uptake, immunity and metabolism. In this way, the gut microbiota functions as an entraining cue for systemic processes and thus daily oscillations in the abundance of key bacterial species and/or the biogeography within the gut may be critical here. These studies highlight the ability of the local immune network to sense daily changes in the microbiota and raise the possibility that local immune networks may also be temporally responsive to the microbiota.
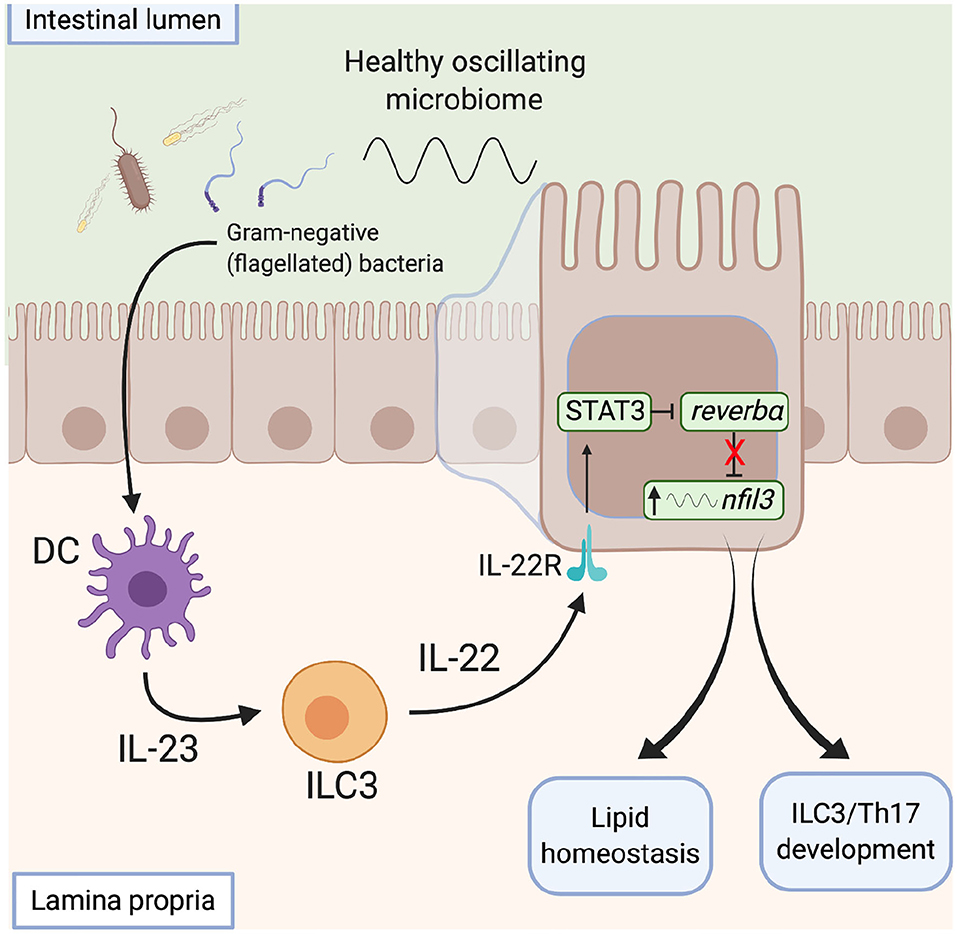
Figure 5. The microbiota influences metabolic programmes within the gut. Signals derived from gram-negative flagellated bacteria resident within a healthy microbiome activate toll-like receptors on lamina propria CD11c+ dendritic cells and induce IL23 secretion. In response to increased IL23 levels, ILC3s signal to intestinal epithelial cells via IL22, which activates a STAT3 signaling pathway to repress rev-erbα expression, with subsequent de-repression of nfil3 transcription. Increased nfil3 expression drives circadian lipid homeostasis and development of ILCs and Th17 cells, promoting intestinal homeostasis. DC, dendritic cell. ILC, innate lymphoid cell, STAT, signal transducer and activator of transcription.
It is evident from these studies that the microbiota itself can modulate the core clockwork machinery. It is also now becoming clear that metabolic outputs from the microbiota can directly alter the phase and amplitude of the circadian clock. Oral administration of SCFAs can induce phase changes in peripheral clocks (122). Additionally, metabolites produced by Clostridium sprogenes such as [3-(4-hydroxyphenyl) propionic acid (4-OH-PPA) and 3-phenylpropionic acid (PPA)] increase the amplitude and lengthen the period of PER2 oscillations in organotypic SCN slices and other tissues (123). Polyamines (putrescine, spermidine and spermine) can regulate the circadian period of cultured cells and alter the circadian period of mice, likely as a consequence of their ability to promote the interaction between PER2 and CRY1 (96). Finally, secondary bile acids (deoxycholic acid and chenodeoxycholic acid) alter clock gene expression in the ileum, colon and liver (124). Given that some secondary bile acids have been shown to be able to directly bind to RORγt (71), this represents a potential mechanism by which bile acids may modulate the core clock. Together, these findings highlight the possibility that microbiome-derived metabolites act as zeitgebers, perhaps fine-tuning the clock to feeding rhythms.
In addition to the microbiota driving rhythms within the gut, bacterial-derived signals also act further afield to programme circadian networks in the liver (90, 125). The gut microbiome has been identified as a potential driver of HFD-induced changes in liver circadian metabolism (125). The nutritional challenge afforded by HFD drives transcriptional and metabolic reprogramming within the liver, resulting in a gain of rhythms in previously non-circadian transcription factors such as peroxisome proliferative activated receptor γ (PPARγ) (126, 127). Murakami et al. demonstrated that fecal transfer from HFD-fed mice to recipient normal chow fed mice is sufficient to elicit the emergence of circadian rhythms in PPARγ leading to specific rewiring of circadian transcription. Thaiss et al. demonstrated in mice fed normal chow, depletion of the microbiome re-programmes the liver transcriptome, with both loss and gain of rhythmic pathways (90). They went on to identify polyamines and amino acids as contributing microbiome-regulated rhythmic metabolites that signal from the gut to the liver to regulate transcriptional programmes (90). These studies provide proof-of concept that systemic metabolic rhythms are reliant on the rhythmic microbial outputs. It will be interesting to explore the extent to which these rhythmic signals direct other physiological processes, with immune homeostasis being an obvious candidate.
Conclusions and Future Direction
The intestinal microbiome is very well established as an influential component of immune-metabolic homeostasis. A healthy microbiome exhibits rhythmicity in microbial composition, as well as rhythmicity in its biophysical location and metabolic outputs, each of which will influence immune homeostasis. Whilst evidence is emerging of gut commensals with potential intrinsic rhythmic capacity, it is likely that these microbiome rhythms will be heavily reliant on entrainment derived largely from the host. This may take the form of food timing, food composition, secreted host signals or non-dietary intake including antibiotics and immunomodulatory medication, as well as host disease state. Initial observations of microbiome rhythmicity in immune homeostasis and associated dysrhythmia in inflammatory disease states should drive scientific investigation into the mechanisms of host circadian regulation of the microbiome and vice versa, as it is not yet clear which immune components are directly sensitive to changes in microbiome rhythmicity and which host-microbiome pathways predominate in health and disease. Whilst current studies have predominantly focused on the metabolic implications of an arrhythmic microbiome, future work should expand on early work exploring the impact of microbiome rhythmicity on composition and function of immune subsets. Piecing together the relative contribution of each moving part in the microbiome, intestinal epithelium and circulating immune cells will help to understand the role of circadian rhythmicity in crosstalk between the microbiome and immune system.
Evidence of microbiome dysbiosis associated with chronic human inflammatory conditions is growing, however, addition of a circadian component to human immuno-microbiome trials is in its infancy. In humans, manipulation of the microbiome via fecal microbial transplantation is already established to modulate immune response in infective conditions such as Clostridium difficile infection and is under investigation in inflammatory conditions such as inflammatory bowel disease (128, 129). It is exciting to consider circadian influence on microbiome health as a pathway to further understanding of mucosal immunology and ultimately improve efficacy of microbiome modulation as a chronotherapy for human inflammatory and auto-immune disease.
Author's Note
Figures were created with QmlvcmVuZGVyLmNvbQ==.
Author Contributions
TB and JG wrote the manuscript and designed the figures. All authors contributed to the article and approved the submitted version.
Funding
Work in JG's laboratory is funded by the MRC (MR/S002715/1 and MR/P023576/1). TB is an MRC Clinical Research Training Fellow (MR/S02199X/1).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling Editor declared a past co-authorship with one of the authors JG.
Abbreviations
5-HIAA, 5-hydroxyindole-3-acetic acid; AHR, Aryl hydrocarbon receptor; AMP, Antimicrobial peptide; ARNT, Aryl hydrocarbon receptor nuclear translocator; BA, Bile acids; BMAL1, Brain muscle arnt-like 1; CLOCK, Circadian locomotor output cycles kaput; CNS, Central nervous system; EAE, Experimental autoimmune encephalomyelitis; FFAR, Free fatty acid receptor; FMT, Fecal microbial transfer; GF, Germ free; GM-CSF, Granulocyte macrophage colony stimulating factor; GPCR, G-protein coupled receptor; HCAR, hydroxycarboxylic acid receptor; HDAC, Histone deacetylase; HFD, high-fat diet; HIF, Hypoxia-inducible factor; I3A, Indole-3-acetate; IAId, Indole-3-aldehyde; IEC, Intestinal epithelial cell; IL, Interleukin; ILC, Innate lymphoid cell; IPA, Indole 3-proprionic acid; isoDCA, isodeoxycholic acid; LCA, lithocholic acid; LPS, lipopolysaccharide; NFIL3, nuclear factor interleukin 3 regulated; NOD, non-obese diabetic; OTU, Operational taxonomic unit; PAS, Per/Arnt/Sim; PPAR, peroxisome proliferator activated receptor; PXR, Pregnane X receptor; ROR, Retinoid related orphan receptor; SCFA, Short chain fatty acid; SCN, Suprachiasmatic nucleus; SFB, Segmented filamentous bacteria; STAT, Signal transducer and activator of transcription; TCCD, 2,3,7,8-tetrachlorodibenzo-p-dioxin; Th17, T helper 17; Treg, Regulatory T cell; TLR, toll-like receptor; TTFL, Transcriptional-translational feedback loop.
References
1. Curtis AM, Bellet MM, Sassone-Corsi P, O'neill LA. Circadian clock proteins and immunity. Immunity. (2014) 40:178–86. doi: 10.1016/j.immuni.2014.02.002
2. Man K, Loudon A, Chawla A. Immunity around the clock. Science. (2016) 354:999–1003. doi: 10.1126/science.aah4966
3. Scheiermann C, Gibbs J, Ince L, Loudon A. Clocking in to immunity. Nat Rev Immunol. (2018) 18:423–37. doi: 10.1038/s41577-018-0008-4
4. Carter SJ, Durrington HJ, Gibbs JE, Blaikley J, Loudon AS, Ray DW, et al. A matter of time: study of circadian clocks and their role in inflammation. J Leukoc Biol. (2016) 99:549–60. doi: 10.1189/jlb.3RU1015-451R
5. Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. (2000) 14:2950–61. doi: 10.1101/gad.183500
6. Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. (2000) 289:2344–7. doi: 10.1126/science.289.5488.2344
7. Brown SA, Zumbrunn G, Fleury-Olela F, Preitner N, Schibler U. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr Biol. (2002) 12:1574–83. doi: 10.1016/S0960-9822(02)01145-4
8. Scheiermann C, Kunisaki Y, Frenette PS. Circadian control of the immune system. Nat Rev Immunol. (2013) 13:190–8. doi: 10.1038/nri3386
9. Yoo S-H, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. (2004) 101:5339. doi: 10.1073/pnas.0308709101
10. Richards J, Gumz ML. Advances in understanding the peripheral circadian clocks. FASEB J. (2012) 26:3602–13. doi: 10.1096/fj.12-203554
11. Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. (2014) 24:90–9. doi: 10.1016/j.tcb.2013.07.002
12. Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. (2012) 485:123. doi: 10.1038/nature11048
13. Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. (2001) 292:1115. doi: 10.1126/science.1058709
14. Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. (2015) 17:690–703. doi: 10.1016/j.chom.2015.04.004
15. Tamburini S, Shen N, Wu HC, Clemente JC. The microbiome in early life: implications for health outcomes. Nat Med. (2016) 22:713–22. doi: 10.1038/nm.4142
16. Zmora N, Suez J, Elinav E. You are what you eat: diet, health and the gut microbiota. Nat Rev Gastroenterol Hepatol. (2019) 16:35–56. doi: 10.1038/s41575-018-0061-2
17. Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. (2012) 486:207–14. doi: 10.1038/nature11234
18. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. (2010) 464:59–65. doi: 10.1038/nature08821
19. Browne HP, Forster SC, Anonye BO, Kumar N, Neville BA, Stares MD, et al. Culturing of ‘unculturable' human microbiota reveals novel taxa and extensive sporulation. Nature. (2016) 533:543–6. doi: 10.1038/nature17645
20. Huse SM, Ye Y, Zhou Y, Fodor AA. A core human microbiome as viewed through 16S rRNA sequence clusters. PLoS ONE. (2012) 7:e34242. doi: 10.1371/journal.pone.0034242
21. Kho ZY, Lal SK. The human gut microbiome – a potential controller of wellness and disease. Front Microbiol. (2018) 9:1835. doi: 10.3389/fmicb.2018.01835
22. Bouskra D, Brezillon C, Berard M, Werts C, Varona R, Boneca IG, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. (2008) 456:507–10. doi: 10.1038/nature07450
23. Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. (2009) 9:313–23. doi: 10.1038/nri2515
24. Moreau MC, Ducluzeau R, Guy-Grand D, Muller MC. Increase in the population of duodenal immunoglobulin A plasmocytes in axenic mice associated with different living or dead bacterial strains of intestinal origin. Infect Immun. (1978) 21:532–9. doi: 10.1128/IAI.21.2.532-539.1978
25. Abrams GD, Bauer H, Sprinz H. Influence of the normal flora on mucosal morphology and cellular renewal in the ileum. A comparison of germ-free and conventional mice. Lab Invest. (1963) 12:355–64.
26. Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci USA. (2002) 99:15451–5. doi: 10.1073/pnas.202604299
27. Reinhardt C, Bergentall M, Greiner TU, Schaffner F, Ostergren-Lunden G, Petersen LC, et al. Tissue factor and PAR1 promote microbiota-induced intestinal vascular remodelling. Nature. (2012) 483:627–31. doi: 10.1038/nature10893
28. Deshmukh HS, Liu Y, Menkiti OR, Mei J, Dai N, O'leary CE. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med. (2014) 20:524–30. doi: 10.1038/nm.3542
29. Khosravi A, Yanez A, Price JG, Chow A, Merad M, Goodridge HS, et al. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe. (2014) 15:374–81. doi: 10.1016/j.chom.2014.02.006
30. Zhang D, Chen G, Manwani D, Mortha A, Xu C, Faith JJ, et al. Neutrophil ageing is regulated by the microbiome. Nature. (2015) 525:528–32. doi: 10.1038/nature15367
31. Ivanov Ii, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. (2009) 139:485–98. doi: 10.1016/j.cell.2009.09.033
32. Schnupf P, Gaboriau-Routhiau V, Sansonetti PJ, Cerf-Bensussan N. Segmented filamentous bacteria, Th17 inducers and helpers in a hostile world. Curr Opin Microbiol. (2017) 35:100–9. doi: 10.1016/j.mib.2017.03.004
33. Tan TG, Sefik E, Geva-Zatorsky N, Kua L, Naskar D, Teng F, et al. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc Natl Acad Sci USA. (2016) 113:E8141–50. doi: 10.1073/pnas.1617460113
34. Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. (2011) 331:337–41. doi: 10.1126/science.1198469
35. Wu HJ, Ivanov Ii, Darce J, Hattori K, Shima T, Umesaki Y, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. (2010) 32:815–27. doi: 10.1016/j.immuni.2010.06.001
36. Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA. (2011) 108 (Suppl. 1):4615–22. doi: 10.1073/pnas.1000082107
37. Horai R, Zarate-Blades CR, Dillenburg-Pilla P, Chen J, Kielczewski JL, Silver PB, et al. Microbiota-dependent activation of an autoreactive T cell receptor provokes autoimmunity in an immunologically privileged site. Immunity. (2015) 43:343–53. doi: 10.1016/j.immuni.2015.07.014
38. Horai R, Caspi RR. Microbiome and autoimmune uveitis. Front Immunol. (2019) 10:232. doi: 10.3389/fimmu.2019.00232
39. Vavricka SR, Schoepfer A, Scharl M, Lakatos PL, Navarini A, Rogler G. Extraintestinal manifestations of inflammatory bowel disease. Inflamm Bowel Dis. (2015) 21:1982–92. doi: 10.1097/MIB.0000000000000392
40. Paun A, Yau C, Danska JS. The influence of the microbiome on type 1 diabetes. J Immunol. (2017) 198:590–5. doi: 10.4049/jimmunol.1601519
41. Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. (2008) 455:1109–13. doi: 10.1038/nature07336
42. Candon S, Perez-Arroyo A, Marquet C, Valette F, Foray AP, Pelletier B, et al. Antibiotics in early life alter the gut microbiome and increase disease incidence in a spontaneous mouse model of autoimmune insulin-dependent diabetes. PLoS ONE. (2015) 10:e0125448. doi: 10.1371/journal.pone.0125448
43. Livanos AE, Greiner TU, Vangay P, Pathmasiri W, Stewart D, Mcritchie S, et al. Antibiotic-mediated gut microbiome perturbation accelerates development of type 1 diabetes in mice. Nat Microbiol. (2016) 1:16140. doi: 10.1038/nmicrobiol.2016.140
44. Levy M, Thaiss CA, Elinav E. Metabolites: messengers between the microbiota and the immune system. Genes Dev. (2016) 30:1589–97. doi: 10.1101/gad.284091.116
45. Garrett WS. Immune recognition of microbial metabolites. Nat Rev Immunol. (2020) 20:91–2. doi: 10.1038/s41577-019-0252-2
46. Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, et al. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe. (2015) 17:662–71. doi: 10.1016/j.chom.2015.03.005
47. Park JH, Kotani T, Konno T, Setiawan J, Kitamura Y, Imada S, et al. Promotion of intestinal epithelial cell turnover by commensal bacteria: role of short-chain fatty acids. PLoS ONE. (2016) 11:e0156334. doi: 10.1371/journal.pone.0156334
48. Zheng L, Kelly CJ, Battista KD, Schaefer R, Lanis JM, Alexeev EE, et al. Microbial-derived butyrate promotes epithelial barrier function through IL-10 receptor-dependent repression of claudin-2. J Immunol. (2017) 199:2976–84. doi: 10.4049/jimmunol.1700105
49. Roediger WE. Utilization of nutrients by isolated epithelial cells of the rat colon. Gastroenterology. (1982) 83:424–9. doi: 10.1016/S0016-5085(82)80339-9
50. Arpaia N, Campbell C, Fan X, Dikiy S, Van Der Veeken J, Deroos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. (2013) 504:451–5. doi: 10.1038/nature12726
51. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. (2013) 341:569–73. doi: 10.1126/science.1241165
52. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. (2013) 504:446–50. doi: 10.1038/nature12721
53. Scott NA, Andrusaite A, Andersen P, Lawson M, Alcon-Giner C, Leclaire C, et al. Antibiotics induce sustained dysregulation of intestinal T cell immunity by perturbing macrophage homeostasis. Sci Transl Med. (2018) 10:eaao4755. doi: 10.1126/scitranslmed.aao4755
54. Schulthess J, Pandey S, Capitani M, Rue-Albrecht KC, Arnold I, Franchini F, et al. The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity. (2019) 50:432–45 e437. doi: 10.1016/j.immuni.2018.12.018
55. Sanchez HN, Moroney JB, Gan H, Shen T, Im JL, Li T, et al. B cell-intrinsic epigenetic modulation of antibody responses by dietary fiber-derived short-chain fatty acids. Nat Commun. (2020) 11:60. doi: 10.1038/s41467-019-13603-6
56. Bachem A, Makhlouf C, Binger KJ, De Souza DP, Tull D, Hochheiser K, et al. Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated CD8(+) T cells. Immunity. (2019) 51:285–97 e285. doi: 10.1016/j.immuni.2019.06.002
57. Kim SH, Cho BH, Kiyono H, Jang YS. Microbiota-derived butyrate suppresses group 3 innate lymphoid cells in terminal ileal Peyer's patches. Sci Rep. (2017) 7:3980. doi: 10.1038/s41598-017-02729-6
58. Fachi JL, Secca C, Rodrigues PB, Mato FCP, Di Luccia B, Felipe JS, et al. Acetate coordinates neutrophil and ILC3 responses against C. difficile through FFAR2. J Exp Med. (2020) 217:jem.20190489. doi: 10.1084/jem.20190489
59. Agus A, Planchais J, Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe. (2018) 23:716–24. doi: 10.1016/j.chom.2018.05.003
60. Shinde R, Mcgaha TL. The Aryl hydrocarbon receptor: connecting immunity to the microenvironment. Trends Immunol. (2018) 39:1005–20. doi: 10.1016/j.it.2018.10.010
61. Trikha P, Lee DA. The role of AhR in transcriptional regulation of immune cell development and function. Biochim Biophys Acta Rev Cancer. (2020) 1873:188335. doi: 10.1016/j.bbcan.2019.188335
62. Hogenesch JB, Chan WK, Jackiw VH, Brown RC, Gu YZ, Pray-Grant M, et al. Characterization of a subset of the basic-helix-loop-helix-PAS superfamily that interacts with components of the dioxin signaling pathway. J Biol Chem. (1997) 272:8581–93. doi: 10.1074/jbc.272.13.8581
63. Xu CX, Krager SL, Liao DF, Tischkau SA. Disruption of CLOCK-BMAL1 transcriptional activity is responsible for aryl hydrocarbon receptor-mediated regulation of Period1 gene. Toxicol Sci. (2010) 115:98–108. doi: 10.1093/toxsci/kfq022
64. Jaeger C, Khazaal AQ, Xu C, Sun M, Krager SL, Tischkau SA. Aryl hydrocarbon receptor deficiency alters circadian and metabolic rhythmicity. J Biol Rhythms. (2017) 32:109–20. doi: 10.1177/0748730417696786
65. Fader KA, Nault R, Doskey CM, Fling RR, Zacharewski TR. 2,3,7,8-Tetrachlorodibenzo-p-dioxin abolishes circadian regulation of hepatic metabolic activity in mice. Sci Rep. (2019) 9:6514. doi: 10.1038/s41598-019-42760-3
66. Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. (2013) 39:372–85. doi: 10.1016/j.immuni.2013.08.003
67. Krishnan S, Ding Y, Saedi N, Choi M, Sridharan GV, Sherr DH, et al. Gut microbiota-derived tryptophan metabolites modulate inflammatory response in hepatocytes and macrophages. Cell Rep. (2018) 23:1099–111. doi: 10.1016/j.celrep.2018.03.109
68. Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet AP, et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity. (2014) 41:296–310. doi: 10.1016/j.immuni.2014.06.014
69. Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature. (2017) 551:648–52. doi: 10.1038/nature24661
70. Rosser EC, Piper CJM, Matei DE, Blair PA, Rendeiro AF, Orford M, et al. Microbiota-derived metabolites suppress arthritis by amplifying aryl-hydrocarbon receptor activation in regulatory B cells. Cell Metab. (2020) 31:837–51 e810. doi: 10.1016/j.cmet.2020.03.003
71. Hang S, Paik D, Yao L, Kim E, Trinath J, Lu J, et al. Bile acid metabolites control TH17 and Treg cell differentiation. Nature. (2019) 576:143–8. doi: 10.1038/s41586-019-1785-z
72. Song X, Sun X, Oh SF, Wu M, Zhang Y, Zheng W, et al. Microbial bile acid metabolites modulate gut RORγ(+) regulatory T cell homeostasis. Nature. (2020) 577:410–5. doi: 10.1038/s41586-019-1865-0
73. Campbell C, Mckenney PT, Konstantinovsky D, Isaeva OI, Schizas M, Verter J, et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature. (2020) 581:475–9. doi: 10.1038/s41586-020-2193-0
74. Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson CR, Tanabe A, et al. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. (1998) 281:1519–23. doi: 10.1126/science.281.5382.1519
75. Liu Y, Tsinoremas NF, Johnson CH, Lebedeva NV, Golden SS, Ishiura M, et al. Circadian orchestration of gene expression in cyanobacteria. Genes Dev. (1995) 9:1469–78. doi: 10.1101/gad.9.12.1469
76. Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. (2005) 308:414–5. doi: 10.1126/science.1108451
77. Min H, Guo H, Xiong J. Rhythmic gene expression in a purple photosynthetic bacterium, Rhodobacter sphaeroides. FEBS Lett. (2005) 579:808–12. doi: 10.1016/j.febslet.2005.01.003
78. Dvornyk V, Vinogradova O, Nevo E. Origin and evolution of circadian clock genes in prokaryotes. Proc Natl Acad Sci USA. (2003) 100:2495–500. doi: 10.1073/pnas.0130099100
79. Edgar RS, Green EW, Zhao Y, Van Ooijen G, Olmedo M, Qin X, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. (2012) 485:459–64. doi: 10.1038/nature11088
80. Paulose JK, Wright JM, Patel AG, Cassone VM. Human gut bacteria are sensitive to melatonin and express endogenous circadian rhythmicity. PLoS ONE. (2016) 11:e0146643. doi: 10.1371/journal.pone.0146643
81. Bubenik GA. Gastrointestinal melatonin: localization, function, and clinical relevance. Dig Dis Sci. (2002) 47:2336–48. doi: 10.1023/A:1020107915919
82. Aminov RI. Horizontal gene exchange in environmental microbiota. Front Microbiol. (2011) 2:158–158. doi: 10.3389/fmicb.2011.00158
83. Hoogerwerf WA, Hellmich HL, Cornelissen G, Halberg F, Shahinian VB, Bostwick J, et al. Clock gene expression in the murine gastrointestinal tract: endogenous rhythmicity and effects of a feeding regimen. Gastroenterology. (2007) 133:1250–60. doi: 10.1053/j.gastro.2007.07.009
84. Sladek M, Rybova M, Jindrakova Z, Zemanova Z, Polidarova L, Mrnka L, et al. Insight into the circadian clock within rat colonic epithelial cells. Gastroenterology. (2007) 133:1240–9. doi: 10.1053/j.gastro.2007.05.053
85. Moore SR, Pruszka J, Vallance J, Aihara E, Matsuura T, Montrose MH, et al. Robust circadian rhythms in organoid cultures from PERIOD2::LUCIFERASE mouse small intestine. Dis Model Mech. (2014) 7:1123–30. doi: 10.1242/dmm.014399
86. Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. (2014) 159:514–29. doi: 10.1016/j.cell.2014.09.048
87. Zarrinpar A, Chaix A, Yooseph S, Panda S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. (2014) 20:1006–17. doi: 10.1016/j.cmet.2014.11.008
88. Liang X, Bushman FD, Fitzgerald GA. Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc Natl Acad Sci USA. (2015) 112:10479–84. doi: 10.1073/pnas.1501305112
89. Reitmeier S, Kießling S, Clavel T, List M, Almeida EL, Ghosh TS, et al. Arrhythmic gut microbiome signatures for risk profiling of Type-2 Diabetes. Cell Host Microbe. (2020). doi: 10.1016/j.chom.2020.06.004. [Epub ahead of print].
90. Thaiss CA, Levy M, Korem T, Dohnalová L, Shapiro H, Jaitin DA, et al. Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell. (2016) 167:1495–510.e1412. doi: 10.1016/j.cell.2016.11.003
91. Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. (2012) 336:1268–73. doi: 10.1126/science.1223490
92. Leone V, Gibbons SM, Martinez K, Hutchison AL, Huang EY, Cham CM, et al. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe. (2015) 17:681–9. doi: 10.1016/j.chom.2015.03.006
93. Segers A, Desmet L, Thijs T, Verbeke K, Tack J, Depoortere I. The circadian clock regulates the diurnal levels of microbial short-chain fatty acids and their rhythmic effects on colon contractility in mice. Acta Physiol. (2019) 225:e13193. doi: 10.1111/apha.13193
94. Segers A, Desmet L, Sun S, Verbeke K, Tack J, Depoortere I. Night-time feeding of Bmal1-/- mice restores SCFA rhythms and their effect on ghrelin. J Endocrinol. (2020) 245:155–64. doi: 10.1530/JOE-20-0011
95. Al-Khaifi A, Straniero S, Voronova V, Chernikova D, Sokolov V, Kumar C, et al. Asynchronous rhythms of circulating conjugated and unconjugated bile acids in the modulation of human metabolism. J Intern Med. (2018) 284:546–59. doi: 10.1111/joim.12811
96. Zwighaft Z, Aviram R, Shalev M, Rousso-Noori L, Kraut-Cohen J, Golik M, et al. Circadian clock control by polyamine levels through a mechanism that declines with age. Cell Metab. (2015) 22:874–85. doi: 10.1016/j.cmet.2015.09.011
97. Asher G, Sassone-Corsi P. Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell. (2015) 161:84–92. doi: 10.1016/j.cell.2015.03.015
98. Fonken LK, Workman JL, Walton JC, Weil ZM, Morris JS, Haim A, et al. Light at night increases body mass by shifting the time of food intake. Proc Natl Acad Sci USA. (2010) 107:18664–9. doi: 10.1073/pnas.1008734107
99. Deaver JA, Eum SY, Toborek M. Circadian disruption changes gut microbiome taxa and functional gene composition. Front Microbiol. (2018) 9:737. doi: 10.3389/fmicb.2018.00737
100. Wu G, Tang W, He Y, Hu J, Gong S, He Z, et al. Light exposure influences the diurnal oscillation of gut microbiota in mice. Biochem Biophys Res Commun. (2018) 501:16–23. doi: 10.1016/j.bbrc.2018.04.095
101. Hatori M, Vollmers C, Zarrinpar A, Ditacchio L, Bushong EA, Gill S, et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. (2012) 15:848–60. doi: 10.1016/j.cmet.2012.04.019
102. Summa KC, Voigt RM, Forsyth CB, Shaikh M, Cavanaugh K, Tang Y, et al. Disruption of the circadian clock in mice increases intestinal permeability and promotes alcohol-induced hepatic pathology and inflammation. PLoS ONE. (2013) 8:e67102. doi: 10.1371/journal.pone.0067102
103. Voigt RM, Summa KC, Forsyth CB, Green SJ, Engen P, Naqib A, et al. The circadian clock mutation promotes intestinal dysbiosis. Alcohol Clin Exp Res. (2016) 40:335–47. doi: 10.1111/acer.12943
104. Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells: 10 years on. Cell. (2018) 174:1054–66. doi: 10.1016/j.cell.2018.07.017
105. Sonnenberg GF, Fouser LA, Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol. (2011) 12:383–90. doi: 10.1038/ni.2025
106. Penny HA, Hodge SH, Hepworth MR. Orchestration of intestinal homeostasis and tolerance by group 3 innate lymphoid cells. Semin Immunopathol. (2018) 40:357–70. doi: 10.1007/s00281-018-0687-8
107. Sonnenburg JL, Xu J, Leip DD, Chen C-H, Westover BP, Weatherford J, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. (2005) 307:1955–9. doi: 10.1126/science.1109051
108. Goto Y, Obata T, Kunisawa J, Sato S, Ivanov II, Lamichhane A, et al. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science. (2014) 345:1254009. doi: 10.1126/science.1254009
109. Godinho-Silva C, Domingues RG, Rendas M, Raposo B, Ribeiro H, Da Silva JA, et al. Light-entrained and brain-tuned circadian circuits regulate ILC3s and gut homeostasis. Nature. (2019) 574:254–8. doi: 10.1038/s41586-019-1579-3
110. Teng F, Goc J, Zhou L, Chu C, Shah MA, Eberl G, et al. A circadian clock is essential for homeostasis of group 3 innate lymphoid cells in the gut. Sci Immunol. (2019) 4:eaax1215. doi: 10.1126/sciimmunol.aax1215
111. Wang Q, Robinette ML, Billon C, Collins PL, Bando JK, Fachi JL, et al. Circadian rhythm–dependent and circadian rhythm–independent impacts of the molecular clock on type 3 innate lymphoid cells. Sci Immunol. (2019) 4:eaay7501. doi: 10.1126/sciimmunol.aay7501
112. Zeng B, Shi S, Ashworth G, Dong C, Liu J, Xing F. ILC3 function as a double-edged sword in inflammatory bowel diseases. Cell Death Dis. (2019) 10:315. doi: 10.1038/s41419-019-1540-2
113. Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. (2013) 153:812–27. doi: 10.1016/j.cell.2013.04.020
114. Wang Y, Kuang Z, Yu X, Ruhn KA, Kubo M, Hooper LV. The intestinal microbiota regulates body composition through NFIL3 and the circadian clock. Science. (2017) 357:912–6. doi: 10.1126/science.aan0677
115. Hopwood TW, Hall S, Begley N, Forman R, Brown S, Vonslow R, et al. The circadian regulator BMAL1 programmes responses to parasitic worm infection via a dendritic cell clock. Sci Rep. (2018) 8:3782. doi: 10.1038/s41598-018-22021-5
116. Kashiwada M, Pham NL, Pewe LL, Harty JT, Rothman PB. NFIL3/E4BP4 is a key transcription factor for CD8α+ dendritic cell development. Blood. (2011) 117:6193–7. doi: 10.1182/blood-2010-07-295873
117. Kobayashi T, Matsuoka K, Sheikh SZ, Elloumi HZ, Kamada N, Hisamatsu T, et al. NFIL3 is a regulator of IL-12 p40 in macrophages and mucosal immunity. J Immunol. (2011) 186:4649–55. doi: 10.4049/jimmunol.1003888
118. Seillet C, Rankin LC, Groom JR, Mielke LA, Tellier J, Chopin M, et al. Nfil3 is required for the development of all innate lymphoid cell subsets. J Exp Med. (2014) 211:1733–40. doi: 10.1084/jem.20140145
119. Halfvarson J, Brislawn CJ, Lamendella R, Vázquez-Baeza Y, Walters WA, Bramer LM, et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol. (2017) 2:17004. doi: 10.1038/nmicrobiol.2017.4
120. Kuang Z, Wang Y, Li Y, Ye C, Ruhn KA, Behrendt CL, et al. The intestinal microbiota programs diurnal rhythms in host metabolism through histone deacetylase 3. Science. (2019) 365:1428–34. doi: 10.1126/science.aaw3134
121. Mowat AM. To respond or not to respond — a personal perspective of intestinal tolerance. Nat Rev Immunol. (2018) 18:405–15. doi: 10.1038/s41577-018-0002-x
122. Tahara Y, Yamazaki M, Sukigara H, Motohashi H, Sasaki H, Miyakawa H, et al. Gut microbiota-derived short chain fatty acids induce circadian clock entrainment in mouse peripheral tissue. Sci Rep. (2018) 8:1395. doi: 10.1038/s41598-018-19836-7
123. Ku K, Park I, Kim D, Kim J, Jang S, Choi M, et al. Gut microbial metabolites induce changes in circadian oscillation of clock gene expression in the mouse embryonic fibroblasts. Mol Cells. (2020) 43:276–85. doi: 10.14348/molcells.2020.2309
124. Govindarajan K, Macsharry J, Casey PG, Shanahan F, Joyce SA, Gahan CG. Unconjugated bile acids influence expression of circadian genes: a potential mechanism for microbe-host crosstalk. PLoS ONE. (2016) 11:e0167319. doi: 10.1371/journal.pone.0167319
125. Murakami M, Tognini P, Liu Y, Eckel-Mahan KL, Baldi P, Sassone-Corsi P. Gut microbiota directs PPARgamma-driven reprogramming of the liver circadian clock by nutritional challenge. EMBO Rep. (2016) 17:1292–303. doi: 10.15252/embr.201642463
126. Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. (2007) 6:414–21. doi: 10.1016/j.cmet.2007.09.006
127. Eckel-Mahan KL, Patel VR, De Mateo S, Orozco-Solis R, Ceglia NJ, Sahar S, et al. Reprogramming of the circadian clock by nutritional challenge. Cell. (2013) 155:1464–78. doi: 10.1016/j.cell.2013.11.034
128. Cammarota G, Ianiro G, Tilg H, Rajilić-Stojanović M, Kump P, Satokari R, et al. European consensus conference on faecal microbiota transplantation in clinical practice. Gut. (2017) 66:569–80. doi: 10.1136/gutjnl-2016-313017
Keywords: microbiome, circadian, immunity, diurnal, short chain fatty acids
Citation: Butler TD and Gibbs JE (2020) Circadian Host-Microbiome Interactions in Immunity. Front. Immunol. 11:1783. doi: 10.3389/fimmu.2020.01783
Received: 03 April 2020; Accepted: 03 July 2020;
Published: 14 August 2020.
Edited by:
Christoph Scheiermann, Ludwig Maximilian University of Munich, GermanyReviewed by:
Eleni Beli, Queen's University Belfast, United KingdomChristoph Reinhardt, Johannes Gutenberg University Mainz, Germany
Copyright © 2020 Butler and Gibbs. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julie E. Gibbs, anVsaWUuZ2liYnNAbWFuY2hlc3Rlci5hYy51aw==
 Thomas D. Butler
Thomas D. Butler Julie E. Gibbs
Julie E. Gibbs