- 1Institute of Translational Medicine, Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 2Institute for Diabetes, Obesity, and Metabolism, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, United States
- 3Division of Endocrinology, Diabetes, and Metabolism, Department of Medicine, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, United States
Circular RNAs (circRNAs) are covalently closed RNA molecules in eukaryotes with features of high stability, tissue-specific and cell-specific expression. According to their biogenesis, circRNAs are mainly classified into five types, i.e. exonic circRNAs (EciRNAs), exon-intron circRNAs (EIciRNAs), intronic RNAs (CiRNAs), fusion circRNAs (f-circRNAs), and read-through circRNAs (rt-circRNAs). CircRNAs have been emerging as important non-coding regulatory RNAs in a variety of human cancers. CircRNA4s were revealed to exert regulatory function through multiple mechanisms, such as sponges/decoys of miRNAs and proteins, enhancers of protein functions, protein scaffolds, protein recruitment, or protein translation templates. Furthermore, some circRNAs are intensively associated with immune cells in tumor immune microenvironment (TIME), e.g. circARSP91 and natural killer cells. Through regulating immune checkpoint genes, circRNAs are demonstrated to modulate the immune checkpoint blockade immunotherapy, e.g. circCPA4 could up-regulate PD-L1 expression. In summary, we reviewed the molecular features of circRNAs and mechanisms how they exert functions. We further summarized functional implications of circRNA regulations in tumor immunology and immunotherapy. Further understanding of the regulatory roles of circRNAs in tumor immunology and immunotherapy will benefit tumor treatment.
Introduction
Circular RNAs (circRNAs) are single-stranded circularized RNA molecules produced from back-splicing. Accumulating evidence has shown that circRNA dysregulations are involved in a variety of human disorders, including viral infection (1), cardiac fibrosis (2), diabetes (3), and cancer (4). Advances in high-throughput sequencing technologies and computational algorithms have driven the systematic detection and investigation of circRNAs. Through diverse mechanisms, circRNAs have shown important roles in tumor immunology and immunotherapy. In this review, we summarized the molecular characteristics of circRNAs and how they exert functions through various mechanisms. We further reviewed and discussed the prospective of circRNAs utilities in tumor immunology and immunotherapy.
The Regulatory Roles of circRNAs in Human Cancers
Molecular Properties of circRNAs
Circular RNAs (circRNAs) are single-stranded covalently closed RNA molecules, which are generated by “back-splicing” where the spliceosome joins the 3’ end of an exon with an upstream 5’ end of the same or different exons (5). Briefly, the length of circRNAs can be ranging from hundreds of nucleotides to more than 1,000 nucleotides dependent on their host genes. They are highly stable in general due to their covalently closed ring structure. While the turnover of circRNAs are still in investigation, one report indicated that circRNAs with N6-methyladenosine (m6A) modification could be cleaved by the ribonuclease P (RNase P) multi-drug resistance-associated protein 1 (MRP) complex in a way that dependent on YTH domain-containing family protein 2 (YTHDF2) and heart-responsive protein 12 (HRSP12) (6). In another report, CDR1AS could be cleaved by protein argonaute 2 (AGO2) which plays an important role in RNA interference (7). It was initially considered as byproducts generated from aberrant splicing events (8–11). In recent years, the rapid development of high-throughput RNA sequencing (RNA-seq) and bioinformatics methods has promoted the extensive identification of circRNAs in eukaryotes (12–16). CircRNAs are characterized by high stability, widespread expression in diverse species, and high specificity among different species.
CircRNA Classification
Most circRNAs are generated from protein-coding genes, which are processed in the exon skipping during pre-messenger RNA (pre-mRNA) transcription to form a lariat structure containing single or multiple exons. This is called exonic circRNAs (EciRNAs) (17–19). Some circRNAs contain both exonic and intronic sequences that are derived from internal intron retention, which are called exon-intron circRNAs (EIciRNAs) (20). Circular intronic RNAs (CiRNAs) are generated from intronic lariats that are kept during canonical splicing process (21). In addition, circRNAs can also be produced from exon joint of different genes located in different or the same chromosomes, which are called fusion circRNAs (f-circRNAs) (22) and read-through circRNAs (rt-circRNAs) (4), respectively.
CircRNA Biogenesis
CircRNAs are generated from canonical splice sites from back-splicing, which is partly dependent on the canonical splicing machinery (14, 23) and have been shown to compete with linear RNAs (24). The biogenesis of most circRNAs is affected by cis-acting elements and trans-acting factors (17). In general, circRNAs are produced from looping intron sequences flanking the downstream splice-donor site and upstream splice-acceptor site. This process could be mediated by base pairing between inverted repeat elements or RNA-binding proteins (RBPs), such as QKI and FUS (25, 26). Additionally, during the process of exon skipping, some of excised lariats could undergo internal back-splicing, which would lead to circRNA formation (27). CiRNAs could also be produced from intronic lariats that escape from debranching (21). Other factors influencing circRNA biogenesis include epigenetic changes, such as histone modification and DNA methylation status variations within gene bodies (28, 29).
The Functional Mechanisms of circRNA Regulation in Human Cancers
Accumulating studies have shown that perturbations of circRNAs is prevalent in human cancers (4, 30), including thyroid cancer (31), ovarian cancer (32), and gastrointestinal cancers (33). Recently, a global analysis of circRNA landscape using clinical tumor samples (>2,000) was performed across more than 40 cancer types (4). Notably, this study identified over 160,000 circRNAs that showed expression in at least one cancer type. Another recent study, specifically in localized prostate cancer, identified 76,311 circRNAs through analyzing RNA-seq data derived from prostate tumor specimens (34). Furthermore, they also found a variety of circRNAs were functionally dysregulated in cancer. In particular, Chen et al. identified 171 circRNAs that were essential to prostate cancer cell proliferation. Collectively, these studies demonstrated the high prevalence of circRNA expression and their perturbations in cancers.
CircRNAs can exert their regulatory roles in cancer via different ways (17, 35) (Figure 1), i.e. protein sponges/decoys (Figure 1B) (23), protein recruitment (Figure 1C) (36), and templates for translation (Figure 1D) (37), miRNA sponges/decoys (Figure 1E) (38), protein scaffolding (39), and enhancer of protein function (Figure 1F) (20).
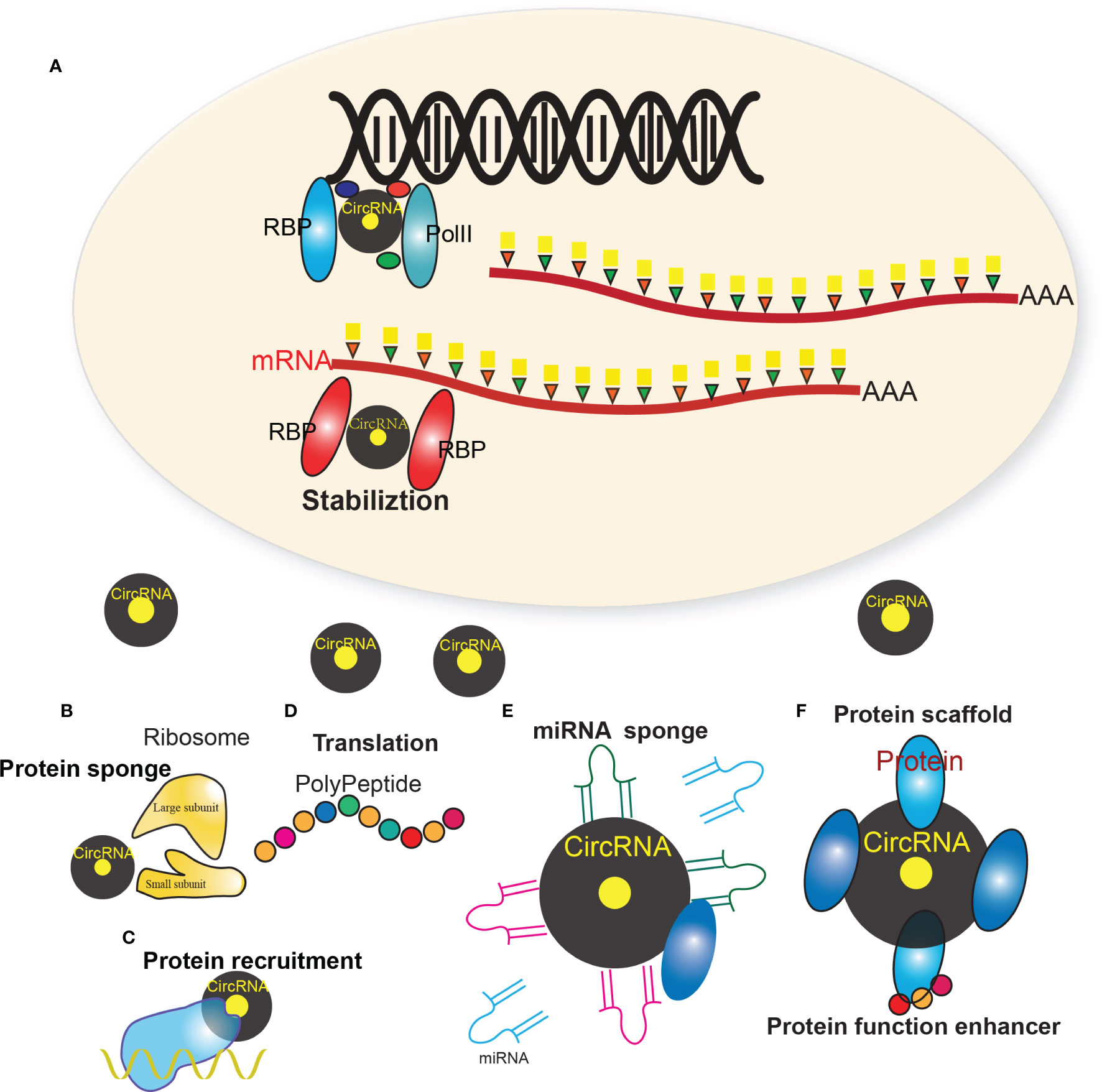
Figure 1 Multiple regulatory mechanism of circRNAs. (A) The biogenesis of circRNAs mainly involves the complex of PolII, RBPs and other factors. (B) CircRNAs that harbor RNA binding protein (RBP) binding motifs may serve as sponges/decoys of the corresponding proteins and regulate their functions. (C) CircRNAs may also recruit specific proteins to certain loci or subcellular compartments. (D) Some circRNAs harbor internal ribosome entry site elements and AUG sites, which enables circRNAs to be translated to unique peptides under certain circumstances. (E) Many circRNAs were found to act as miRNAs sponges, which sequester miRNAs via complementary RNA base-pairing and thus prevent miRNAs from binding their target. (F) Some circRNAs have been shown to facilitate the colocalization of enzymes and their substrates through acting as protein scaffolds. CircRNAs may also enhance the function of particular proteins through circRNA-protein interactions.
CircRNAs as miRNA Sponges
Many circRNAs were found to act as miRNAs sponges, which sequester miRNAs via complementary RNA base-pairing and thus prevent miRNAs from binding their target. For example, circTP63, a cell cycle related circRNA, is up-regulated in lung squamous cell carcinoma (LUSC) tissues and its up-regulation is correlated with larger tumor size and higher TNM stage in LUSC patients. Mechanically, circTP63 competitively binds to miR-873-3p and prevents miR-873-3p to decrease the level of FOXM1, which up-regulates CENPA and CENPB, and finally facilitates cell cycle progression (40). In colorectal cancer (CRC), circHIPK3 acts as the sponge of miR-7, which is a well-known tumor suppressor, to promote colorectal cancer growth and metastasis (41). In breast cancer, circFBXW7 acts as the sponge of miR-197-3p, which induces c-Myc degradation by up-regulating FBXW7 expression, to inhibit the malignant progression of triple-negative breast cancer (42). In bladder cancer, circ-ACVR2A could directly interact with miR-626 and acts as a miRNA sponge to regulate EYA4 expression, thus inhibiting bladder cancer cell proliferation and metastasis (43).
CircRNAs as Protein Decoys
CircRNAs that harbor RNA binding protein (RBP) binding motifs may serve as sponges/decoys of the corresponding proteins and regulate their functions. For example, Abdelmohsen et al. identified circPABPN1 in human cervical carcinoma HeLa cells, which suppressed the translation of nuclear poly(A) binding protein 1 (PABPN1) mRNA through sequestering the RBP Hu-antigen R (HUR) (44). By binding prescadillo homologue 1 (PES1, an essential 60S pre-ribosomal assembly factor), circANRIL was found to impair pre-rRNA processing and ribosome biogenesis, which further led to nucleolar stress and p53 activation (45). YAP (yes-associated protein), a key component of Hippo pathway which plays crucial roles in tumorigenesis, can inhibit apoptosis and promote proliferation and metastasis of cancer cells. Wu et al. showed that circYAP could bind with YAP mRNA and translation-associated protein eIF4G and PABP to negatively regulate the expression of YAP. Moreover, the malignant phenotypes can be reversed by the ectopic expression of circYAP, which is similar to silencing endogenous Yap (46).
CircRNAs Enhancing Protein Functions
CircRNAs may also enhance the function of particular proteins through circRNA-protein interactions. Sun et al. demonstrated that circMYBL2, generated from the cell cycle check point gene MYLB2, could promote the proliferation of FLT3-ITD+ cells in vitro and in vivo through enhancing the translational efficiency of FLT3 kinase via increasing the binding of polypyrimidine tract-binding protein 1 (PTBP1) to FLT3 messenger RNA (47). In addition, circEIF3J and circPAIP2 were demonstrated to be able to positively regulate the expression their parental genes through enhancing the function of transcription factors (20). Both circEIF3J and circPAIP2 were EIciRNAs residing in the nucleus, they could promote RNA polymerase II (Poll II)-mediated transcription by interacting with the U1 small nuclear ribonuleoprotein (snRNP)
CircRNAs as Protein Scaffolds or Recruitment
Some circRNAs have been shown to facilitate the colocalization of enzymes and their substrates through acting as protein scaffolds. Circ-Foxo3 physically binds to mouse double-minute 2 (MDM2) and mutant p53 through acting as protein scaffolds (48). Circ-Foxo3 was further demonstrated to facilitate MDM2-mdiated ubiquitylation of mutant p53, leading to proteasome-mediated degradation. CircRNAs may also recruit specific proteins to certain loci or subcellular compartments. For instance, Chen et al. found that circ-FECR1 could induce demethylation of CpG sites and promote transcription of FLI1 through recruiting TET1 to the promoter region of FLI1 (36).
CircRNAs Translating Into Peptides
Some circRNAs harbor internal ribosome entry site elements and AUG sites, which enables circRNAs to be translated to unique peptides under certain circumstances. Zhao et al. suggested that circE7, generated by oncogenic human papillomaviruses (HPVs), can be translated to produce E7 oncoprotein which is biologically functional and linked to the transforming properties of some HPV (49). Zhang et al. suggested that an 87-animo-acid peptide, encoded by circular form of the long intergenic non-protein-encoding RNA p53-induced transcript (LINC-PINT), directly interact with polymerase associated factor complex (PAF1c) and inhibited the transcriptional elongation of multiple oncogenes to suppresses glioblastoma cell proliferation in vitro and in vivo (50). Liang et al. showed that a novel 370-amino acid β-catenin isoform encoded by circRNA circβ-catenin could stabilize full-length β-catenin by antagonizing GSK3β-induced β-catenin phosphorylation and degradation, leading to activation of Wnt pathway, thus promoting liver cancer cell growth (51).
Interactions Between circRNAs and m6A Modification
N6-methyladenosine (m6A), which has been discovered in the early 1970s, and whose predominant accumulations around stop codons and 3’ untranslated regions (3’ UTRs) of mRNA with a typical consensus sequence RRACH (R = G or A and H = A, C, or U) have been reported (52–57), is one of the most common RNA modifications. Accumulating studies show that m6A play crucial roles in many different aspects including circadian rhythm, gene expression, cell differentiation, stress response, tumorigenesis, development, and inflammatory response (54, 58–63). Recently, Zhou et al. defined thousands of m6A circRNAs that showed cell-type-specific expression patterns (64). These circRNAs interact with m6A reader proteins YTHDF1 and YTHDF2, and m6A writer protein METTL3. Besides, Chen et al. also presented that the m6A reader YTHDC1 increase the cytoplasmic export of circRNA NOP2/Sun RNA methyltransferase 2 (circNSUN2), forming a circNSUN2-IGF2BP2-HMGA2 RNA-protein ternary complex in the cytoplasm contributing the stabilization of HMGA2 mRNA and the enhancement of colorectal liver metastasis (65). In addition, m6A writer protein METTL3 was reported to impact m6A modification of circZNF609, and the m6A reader proteins YTHDF1 was reported to regulate the backsplicing of circZNF609, suggesting the role of m6A in the biogenesis of circZNF609 (66). Besides, the translation of circRNAs was affected by m6A methylation (64), and the m6A modification on circRNAs can be recognized by mammalian cells to inhibit innate immunity by abrogating immune gene activation (67). All these studies expand our knowledge on the complex interactions between m6A modification and circRNAs.
In conclusion, circRNAs are prevalently expressed in human cancers and can exert its regulatory roles by acting as sponges/decoys for miRNAs and proteins, enhancing protein functions, protein scaffolds, recruiting proteins, or protein translation templates.
Tumor Immunology and Immunotherapy
Immune System in Human Cancers
The first indication of the immune system involvement in cancer was discovering the links between inflammation and cancer in 1863 (68). Endeavors have focused on how the immune system can be able to recognize and ultimately destroy cancer, which is made up of tumor and “self” cells. Cancer cells can express two types of tumor antigens: tumor-specific antigen (TSAs) and tumor-associated antigens (TAAs). TSAs are highly tumor-specific and are expressed only in tumor cells, while TAAs are more widely expressed in both tumor and normal cells (69). The immune system can respond to cancer cells in two ways, i.e. against TSAs or against TAAs. In immunosurveillance hypothesis, immune system recognizes malignant tumor cells as foreign agents and eliminates them (70). However, in the past decades, scientists found that cancer could actively deploy various tactics, which collectively termed “immune evasion mechanisms” and continuously develop diversity and complexity in late-stage, to delay, alter, or even stop anti-tumor immunity.
Interference on tumor immunology to intensify the immune response to eliminate tumor cells has provide novel insights on tumor therapy, i.e. immunotherapy, which includes immune checkpoint blockade (ICB) therapy, CAR T cell adoptive therapy, cancer vaccines, and oncolytic virus therapy.
Immune Checkpoint Blockade Therapy
Tumor cells can overexpress some specific molecules such as PD-L1 and CTLA-4 to silence the immune response and these molecules are collectively termed as “immune checkpoints.” Programmed cell death (PD) pathway is the first and most characterized “immune checkpoints.” PD-1, a co-inhibitory receptor, is highly expressed on activated T cells, B lymphocytes, natural killer cells, and MDSCs. PD-1 expression can be induced by TCR-antigen engagement and common γ-chain cytokines like interleukin (IL)-2, IL-7, IL-15, and IL-21 in the effector phase of the immune response (71). PD-L1 and PD-L2 are two known ligands of PD-1 and their expression on healthy tissues are relatively low (72). Effector T cell exhausted and could be triggered apoptosis upon engaging with PD-L1 (73). Multiple cancers including melanoma, non-small cell lung cancer (NSCLC), breast cancer, and squamous cell head and neck cancer has been documented with up-regulated expression of PD-L1 (74–76). Therapeutic monoclonal antibodies directly against PD-1 and PD-L1, with avelumab for PD-L1 and nivolumab for PD-1, respectively, have been proven effective for treating multiple solid tumors. CTLA-4, mainly expressed on T cells, acts as a negative regulatory receptor of T cells. Upon the TCR engaging with antigens, the expression of CTLA-4 rapidly up-regulates. CTLA-4 can compete with CD28, a key co-stimulatory receptor on T cells, for the binding of the same ligands, CD80 and CD86. And CTLA-4 has a higher affinity than CD28 for both ligands, resulting in interference with the immune synapse and T-cell inactivation. Therapeutic anti-CTLA-4 monoclonal antibodies, such as ipilimumab, have achieved promising clinical outcomes in advanced melanoma. Although ICB are considered as a revolution of cancer treatment, many patients including microsatellite stable colorectal cancer (CRC), ovarian cancer, prostate cancer, and pancreatic ductal adenocarcinoma (PDA) rarely exhibit objective responses to ICB (77). There must be other mechanisms leveraged by tumor cells to suppress the immune response.
Chimeric Antigen Receptor T Cell Adoptive Therapy
Chimeric antigen receptor T (CAR T) technique was first reported to transduce T cells with chimeric genes encoding single-chain antibodies that are linked to a transmembrane region and an intracellular domain encoding the signaling adaptor for the T cell receptor (78). CAR T cells could recognize tumor antigens independent of MHC presentation. By adopting CAR T technology, CAR T therapeutics genetically modified autologous T cells isolated from patients to express the CAR construct. After expansion, patient-derived genetically modified T cells were returned to patients to kill malignant cells. CAR T immunotherapy was demonstrated to redirect T cell killing to cells that express the antibody’s cognate antigen (79–81). Currently, the major targets of CAR T therapy is CD19, the B cell costimulatory receptor widely expressing on B cell leukemias and lymphomas (80, 82), showing highly therapeutic effects even in otherwise refractory diseases and can induce durable remissions (79, 83). Despite of impressive clinical outcomes in treating tumor patients, especially for those with lymphoma, CAR T therapy still have many problems, particularly for solid tumors (84). As CAR T therapy is based on direct recognition of tumor cells expressing CD19, deletion of the CAR-binding epitope frequently induces disease relapse (85). Beyond CD19, next-generation CAR T therapy will include novel targets such as CD22, which is a B cell regulatory receptor expressed by many B cell malignancies (83, 86). CD70 is also considered as a novel CAR T therapy target (87, 88).
Cancer Vaccines
Efforts of cancer vaccines have been made to promote cancer-specific immune response, which generate antitumor immunity, especially cytotoxic CD8+ T cells that are specific to tumor antigens. Administration of tumor antigens (e.g. overexpressed antigens, cancer-testis antigens, oncofetal antigens, and mutated antigens) with antigen-presenting cells (APC, e.g. DCs, B cells, and monocytes) shows therapeutic promise (83, 89–91). Current cancer vaccination includes four major types, i.e. peptide-based vaccines, APC-based vaccines, tumor-based vaccines, and virus-based vaccines. The most commonly used cancer vaccine is the MHC class I restricted peptide epitopes that are from shared TAAs aiming to activate rare specific CD8+ T cells, which has shown substantial therapeutic effects (92–94). Peptide vaccines with adjuvant formulation, such as cytokines and toll-like receptor (TLR) ligands, showed significant clinical benefits (94–96). Multiple peptides can be given at the same time (97, 98) and combinations of multi-peptide vaccines and chemotherapy also indicated benefits (99). Among various types of APCs (e.g. peripheral blood mononuclear cells, activated B cells and dendritic cells), the heterogeneous populations of dendritic cells could efficiently process and present antigens to CD4+ and CD8+ T cells. Application of dendritic cells vaccines offered clinical efficiency (100–102). Tumor cells from killed mice could be used to immunize other mice by expressing immune stimulatory cytokines (e.g. GM-CSF) (103, 104). These findings offered the possibility of tumor cell- based immunotherapy. Tests using allogeneic cell lines or autologous tumor cells exhibited capability to activate immunity killing tumor cells (105, 106). Although huge endeavors have been made in cancer vaccinations, the effects of cancer vaccines are limited due to the difficulty in target antigens selection and immunosuppression from tumor microenvironment. Improvements in antigen choice and vaccine design will obtain better clinical outcomes.
Oncolytic Virus Therapy
In the context of presenting tumor antigens, pathogens involvement can largely increase immune stimulation of tumor patients. Oncolytic viruses selectively replicate in tumor cells and kill them without harming normal cells, which can be genetically engineered or naturally occur (107). The most widely known virus vaccines for cancer are the human papillomavirus vaccines that are designed to prevent human papillomavirus (HPV) mediated cervical cancers. The oncolytic viruses have been used for decades, including adenoviruses, vaccinia viruses, and herpesviruses (91). Adenoviruses drive the transactivator early genes E1a and E1b and viral replication from tumor specific promoters. Taking advantage of natural infectivity, adenoviruses have been used to directly immunize with tumor antigens, where they are injected into muscle tissue as vectors (108, 109). The therapeutic efficacy of vaccinia viruses can be improved by genetically engineering with chemokine genes or combinations with costimulation (110). Oncolytic virus therapy has shown promising therapeutic values, but needs further exploration.
In conclusion, tumor immunology advances our understanding of the development of malignant tumor cells and their interactions with host, and immunotherapy has achieved great efforts in some malignant cancers. But tumor are a highly heterogeneous disease and tumor micro-environment are also complicated in different cancers. In the future, we should get more insights into complicated interplay between immune cells and tumor microenvironment, which will develop strategies to break the tumor evasion from immunosurveillance.
CircRNAs and Tumor Immune Microenvironment
Tumor Immune Microenvironment
Studies over several years has demonstrated that tumor cells could evade immune surveillance by establishing an immune-privileged microenvironment, which is functionally analogous to that of certain normal tissue (111, 112). Tumor immune microenvironment (TIME) is composed of various cell types except cancer cells, including tumor-associated macrophages, myeloid-derived suppressor cells (MDSCs), dendritic cells, cancer-associated fibroblasts cells, NK cells, tumor-associated neutrophils, and tumor-infiltrating lymphocytes. Over the past years scientists has realized that tumor cells could turn these cells to favor their progression and contribute to the immune escape. Firstly, accumulating evidences suggested that the vessels of TIME could control the extravasation of effector T cells from the circulatory system into tumors. Upon activated by the antigen present cells, the effector T cells traffic to the tumor via the circulatory system. For example, the apoptosis inducer Fas ligand (FAS-L) is highly expressed in the tumor vasculature of multiple tumor types including ovarian, colon, prostate, breast, bladder, and renal cancer, which substantially reduces the number of CD8+T cell infiltration into tumors. Accordingly, inhibition of Fas-L in preclinical models resulted in a significant increase of effector T cells in tumors and led to T cell dependent tumor suppression (113). Secondly, dendritic cells and MDSCs within the TIME can inhibit the immune response within the tumor via multiple mechanisms. After extravasation of cancer-specific T cells into tumor, it must locally replicate to further increase their numbers to kill the tumor cells effectively and also overcome barriers that restrict their distribution and the hostile elements of the TIME. MDSCs, key components of TIME, are broadly defined as myeloid cells and are different from mature myeloid cells (i.e. macrophages, DCs, neutrophils), which are terminally differentiated. MDSCs are consisted of myeloid progenitors, immature mononuclear cells (M-MDSCs), which are morphologically and phenotypically similar to monocytes, and immature polymorphonuclear (PMN-MDSCs), which are morphologically and phenotypically similar to neutrophils. M-MDSCs and PMN-MDSCs utilize different mechanisms to inhibit tumor immune response. M-MDSCs primarily utilize mechanisms associated with production of NO and cytokines to suppress both antigen-specific and nonspecific T cell response and have stronger suppressive activity than PMN-MDSCs (114). Other studies indicated that MDSCs and tumor-associated macrophage cells (TAMs) could also produce arginase-1, inducible nitric oxide synthase (iNOS) and reactive oxygen species (ROS) to suppress the proliferation of T cells within the tumor. Arginase-1 produced by MDSCs and TAMs within the TIME converts L-arginine, essential for the proliferation of T cells, to urea and L-ornithine, exhausting the pool of L-arginine within the TIME and thus impairing the proliferation of T cells. And MDSC-derived iNOS converts L-arginine to citrulline and NO, which suppresses T cell function by inhibiting JAK/STAT signaling, reducing MHC class II expression and inducing T cell apoptosis. While ROS and NO produced by MDSCs and TAMs result in nitration of the T cell receptor, which impairs the recognition of peptide antigens presented by MHC. Additionally, MDSCs can directly inhibit T cell response in a contact-dependent manner via membrane-bound TGF-β (115, 116). Furthermore, cancer associated-fibroblasts (CAFs) can prevent the effector T cells from accumulating in the vicinity of cancer cells within the TIME. Cancer-associated fibroblasts can leverage two methods to mediate this restriction. For the one hand, CAFs can produce extracellular matrix to exclude effector T cell. And studies found increased T cells movement out of the stromal regions and into contact with cancer cells when collagenase was added to reduce matrix rigidity. CAFs can also produce CXCL12 and IL-6 to exclude the effector T cells. Accordingly, administering an inhibitor of CXCR4, the receptor for CXCL12, to the PDA-bearing mice led to the rapid accumulation of effector T cells within the tumor and blockage IL-6 could improve T-cell trafficking, migration and tumor immunosuppression (117).
The Relation Between circRNAs and Tumor Immune Microenvironment
Emerging studies showed that circRNAs played an important role in key components of tumor immune microenvironment (89, 118). Here we briefly review the interactions between key components of tumor immune microenvironment and circRNAs. One study suggested that circSLC8A1, derived from the SLC8A1 gene, could act as a sponge of miR-494, which is crucial for migration of MDSCs into tumor site and regulation of the production of ARG1 and iNOS, thus enhancing the tumor immune response. This evidence indicated that circRNAs could serve as potential targets by modulating the MDSC-mediated immune response. Natural killer cells (NK cells) play an important role in tumor immune surveillance, which possesses the ability to direct against tumor and infected cells without stimulation like B or T lymphocytes. They simultaneously express activating and inhibitory receptors that encounter target cells by the subtle balance of transmitted signals for activation or inhibition. NK cells primarily leveraged two tactics to against tumors. One is that NK cell can express apoptotic ligands such as TNF-related apoptosis-inducing ligand (TRAIL) and tumor necrosis factor (TNF) family members Fas-L and then interact with their related receptor on tumor cells which inducing the apoptosis of tumor cells. The other is that NK cell secrete perforin and granzyme to direct lyse tumor cells. In addition to direct against tumors, NK cells can exert “helper role” to against tumor cells. It was reported that NK cells could contribute to the accumulation of T-bet+ CD4+ T cells in the tumor site, promote the production of TNF-αand IFN-γ by tumor infiltrating CD8+T cells, suppress the expression of exhaustion marker PD-1 on these CD8+ T cells and promote the induction of tumor-specific T cell memory in the mouse model. Dysfunction of NK cells has been documented over the several years in TIME (119–122). And some studies suggested that circRNAs can enhance or attenuate the function of NK cells. Ma et al. showed that overexpression of circARSP91 (circRNA of AR-suppressed PABPC1 91bp) could enhance cytotoxicity of NK cells to HCC cells via up-regulating UL16 binding protein 1 (ULBP1) expression in HCC cells at the mRNA and protein levels (123). Interestingly, in the another study, Zhang et al. demonstrated that HCCs could secrete circUHRF1 in exosomal manner to inhibit NK cells-derived IFN-γ and TNF-α secretion by up-regulating the expression of TIM-3 via degradation of miR-449c-5p. Additionally, circUHRF1 could mediate the resistance to anti-PD1 immunotherapy in HCC patients (124). Macrophages, key components of innate immunity, mainly derived from embryonically and seeded in tissues, serve as the first-line defense against pathogens and antigen-presenting cells for cellular immunity (125). Macrophages can be divided into M1 and M2 macrophages based their catabolism of L-arginine. The M1 macrophages has a function of eliciting inflammation, while the M2 macrophages show anti-inflammatory effect. This concept may explain the heterogeneity of macrophages. M2 phenotype macrophages, primarily derived from circulating bone marrow monocytes, is induced by soluble factors secreted by cancer stem cells (CSCs) (126, 127). Studies over human tumor samples have indicated that a high number of macrophages, especially M2 macrophages, is closely related to worse clinical prognosis in numerous malignant cancers (128). Accumulating evidence suggested that tumor-associated macrophages could inhibit cytotoxic T cell response by the following mechanisms: depletion of essential metabolite required for T cell proliferation, inhibition of T cell functions via producing anti-inflammatory cytokines and activation of T cell checkpoint blockade via inhibitory receptors. One study indicated that the expression levels of circRNA-003780, circRNA-010056, and circRNA-010231 were high in M1 cells, while the expression levels of circRNA-003424, circRNA-013630, circRNA-001489, and circRNA-018127 were five times than those of M1 macrophage. Different expression levels of specific circRNAs in different polarization state of macrophages suggested that circRNAs may play an important role in the polarization of macrophages (129). Additionally, another study demonstrated that high expression level of circ-CDR1as was correlated with higher ratio of M2 macrophage, suggesting circ-CDR1as may be involved in the polarization of macrophages in TIME (130). Tumor infiltrating lymphocytes, considered as selected population of T-cell with a higher specific immunological reactivity against tumor cells than the non-infiltrating lymphocytes, are mainly composed of CD3+CD4+T cells and CD3+CD8+T cells. Multiple studies have showed that higher proportion of tumor infiltrating lymphocytes (TILs) predicted a better prognostics (131). It was reported that HCC patients with higher percentage of TILs displayed better clinical outcomes, suggesting the prognostic value of TILs for HCC patients. In addition, Weng et al., performed global circRNA microarray between plasma of HCC patients with high TILs and low TILs. The results suggested HCC patients with high TILs had lower expression of hsa_circ_0064428 and was negatively correlated with overall survival, tumor size and metastasis. It can be concluded that hsa_circ_0064428 functioned as a novel immune-associated prognostic biomarker for HCC patients (132).
In conclusion, reprogrammed TIME provides a “shield” for tumor cell and contributes immune therapy resistance. CircRNAs play an important role in tumor progression. Aberrant functions in the TIME caused by circRNAs can be valuable new targets to treat cancer or become novel biomarkers for immunotherapy.
CircRNAs and Immunotherapy
Cancer immunotherapy have achieved therapeutic advances in recent years and was named as 2013’s breakthrough of the year by Science. It highlights the importance of human immune system in treating cancer. Immune checkpoint inhibitors, using therapeutic antibodies including anti-CTLA4 and anti-PD1/L1 mAb, to unleash cytotoxic T cells in tumor microenvironment, has achieved great success in clinical practices. Accumulating studies suggested that B7-H1/PD-1 interaction was the major ways used by tumor cells to suppress immune response in both preclinical and clinical settings (133, 134). Anti-PD1 therapy have achieved higher objective response rates in patients and with much fewer immune-related adverse events (irAEs), which is the most characterized feature of this approach. It is effective in more than 25 different types of solid tumor and has favorable response-to-toxicity profile, with a 40% objective tumor response rate and a 7–12% grade 3–5 irAEs immune across multiple tumor types (135). Although anti-PD1 therapy has achieved great clinical success in most solid cancers, a considerable portion of patients did not benefit from anti-PD1 therapy. The reason is that the tumor microenvironment of different cancers of different patients are heterogeneous (136). For a successful immune-mediated elimination, it requires substantial leukocytes to infiltrate into tumor tissue and recognize the malignant cells. However, there exists significant difference across numerous tumors. For example, it was reported that effector T cells infiltrated in melanoma and breast cancer, but rarely infiltrated in pancreatic ductal adenocarcinoma. So pancreatic ductal adenocarcinoma did not response to anti-PD1 therapy. Clinically, cancer patients are broadly classified into three categories based on the level of tumor infiltrating leukocytes and B7-H1 expression level: (1) Types I and IV: There is lack of significant TILs in the TIME. (2) Types II: There exist many TILs in the TME and these TILs were over-regulated due to the effect of the B7-H1/PD-1 pathway. (3) Type III: there are many TILs in the TIME which are dysfunctional due to suppression by other molecular pathways (non-B7-H1/PD-1). Only patients classified as Types II can benefit from the anti-PD1 therapy (135). Thus understanding the dominant immune defects in TIME of patients is critical for cancer immunotherapy. And immune defects induced by tumor cells are highly heterogeneous (136). With the rapid development of cancer immunotherapy, there are many novel immune checkpoints including LAG3 (CD233), TIM, TIGIT (T cell immunoglobulin and ITIM domain), VISTA, B7-H3, BTLA, and siglec-15 emerging (137). And inhibitors targeting these checkpoints are on clinical trials. Interestingly, a study indicated that when the mice treated with anti-PD-1 mAb, the level of LAG-3 and CTLA-4 increased and treated with anti-LAG-3 mAb, the level of PD-1 were up-regulated. This suggested that blockade of a single immune checkpoint targets may led to the compensatory up-regulation of other checkpoint receptors in TME (138). This compensatory mechanism of immune checkpoints may be another mechanism of anti-PD1 therapy resistance in patients and also indicated that there existed common compensatory mechanism across different types of cancers. CircRNAs, as novel non-coding RNAs in the past few years, have been implicated in multiple physiological and pathophysiological conditions. And they can also play a regulatory role of immune checkpoints and have the potential to serve as a predictive biomarker of immune checkpoint therapy (Figure 2). One study suggested that hsa_circ_0020397 can regulate CRC cell viability, apoptosis, and invasion by promoting the expression of miR-138 target genes, telomerase reverse transcriptase (TERT), and programmed death-ligand 1 (PD-L1) (139). CircCDR1-AS is representative circular RNA that is associated with poor prognosis in gastrointestinal cancers including colon, liver, and pancreatic cancers. Tanaka et al. demonstrated that CircCDR1-AS can significantly increase the expression of PD-L1 at the surface of colon cancer cells via CMTM4 and CMTM6 and led to the poor prognosis of CRC cancer patients (140). In another study, Hong et al. indicated that circ-CPA4 could promote cell growth, mobility and epithelial-mesenchymal transition and inhibited cell deaths of NSCLC cells by up-regulating expression levels of PD-L1 via acting as an RNA sponge for let-7 miRNA. Moreover, circ-CPA4 could positively regulate exosomal PD-L1 derived from NSCLC cells, which promoted cell stemness and inactivated CD8+ T cells (141). Additionally, Li et al. found that circ_0000284 could up-regulate the expression of PD-L1 via binding miR-377-3p and thus promoting the progression of NSCLC (142). TIM-3, also called hepatitis A virus cellular receptor 2 (HAVCR2), is another intriguing immune checkpoint. The gene TIM, located on human chromosome 5q33.2, expresses a protein of 302 amino acid which belongs to Ig superfamily (IgSF). It was expressed on different immune cells including B cells, T cells, NK cells, DCs, Tregs, monocytes, and macrophages. And higher expression of TIM-3 was closely associated with poor prognosis in solid malignant. More importantly, accumulating evidence have verified that therapeutic benefit of TIM-3 blockade and inhibit tumor growth especially combined with anti-PD therapy. For the time being, there are at least eight anti-TIM-3 mAbs has been registered on clinicaltrials.gov. Anti-TIM-3 therapy combined with TSR-042 are already on phase II clinical trial for liver cancer (137). Zhang et al. reported circUHRF1, circular ubiquitin-like with PHD and ring finger domain 1 RNA (circUHRF1), is highly expressed in human HCC tissues and closely related to poor clinical prognosis of HCC patients. Mechanically, hepatocellular carcinoma (HCC) can secreted circUHRF1 in an exosomal manner, inhibiting the secretion of NK cell-derived IFN-γ and TNF-α and inhibiting NK cell function by up-regulating the expression of TIM-3 through degradation of miR-449c-5p (124).
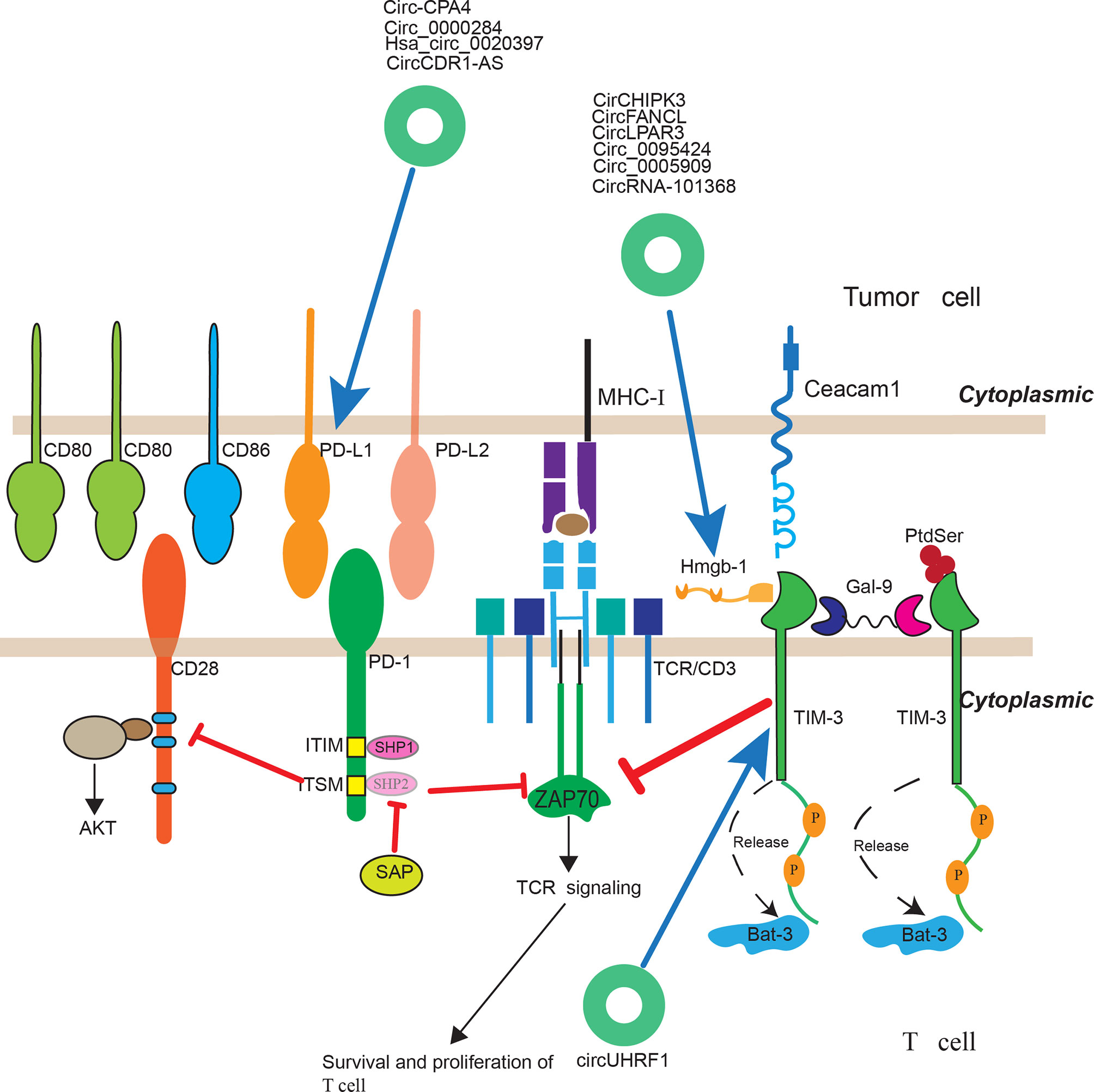
Figure 2 The regulatory roles of circRNAs in immunotherapy. CircRNAs have been implicated to play a regulatory role of immune checkpoints and have the potential to serve as a predictive biomarker of immune checkpoint therapy.
One challenge for cancer immunotherapy is that tumor-induced immune defects not only occur among different patients but also extend to different areas in a single tumor lesion. It is important to evaluate immune response at the TIME level which require sequential tumor tissue collection and analyses. CircRNAs, presented as a stable covalently closed single RNA and can also be secreted in exosomal manner, can play a regulatory role in some of these immune checkpoints and have the potential to serve as a biomarker of immune checkpoint therapy response. Hence, more regulatory roles of circRNAs playing in these immune checkpoints need further investigation.
Conclusions and Future Perspective
Due to the rapid progress of high-throughput sequencing and bioinformatics methodologies, researchers have unveiled the biogenesis and biological characteristics of circRNAs. Accumulating evidences exhibited that circRNAs are closely linked to immune cells in the tumor immune environment and are potentially to modulate or mark the infiltrating abundance of immune cells. Through regulating immune checkpoint genes, circRNAs are potentially capable to mediate therapeutic efficacy of immune checkpoint blockade therapy. The regulatory roles of circRNAs in cancer biology and therapy, especially tumor immunology and immunotherapy, need to be further explored. Several FDA-approved anti-cancer drugs were reported to be able target non-coding oncogenic RNAs. For example, miR-21 could be inhibited by compounds and its function could be ablated upon overexpressing pre-miR21. This finding indicates that onco-ncRNAs are druggable. However, there are no drugs specifically targeting onco-ncRNAs been clinically approved by FDA. Accumulating endeavors have been made to find small molecules targeting onco-ncRNAs. In terms of circRNA treatment, although no clinical trials have been reported, siRNAs targeting their unique conjunction cite, which can avoid targeting host gene, could be used to down-regulate specific onco-circRNAs to treat cancers. The major problem would be how to construct an efficient vector to deliver these siRNAs to tumor. Thus, ncRNAs treatment, we believe, will come true with the development of vector deliver system in the near future. To our best knowledge, the major possible negative effect of circRNA treatment could be mis-targeting. And this problem could be addressed by carefully designed siRNAs targeting the conjunction sites of circRNAs. In speaking of translational study of circRNAs, one study suggested that artificially designed circRNA could act as a miRNA sponge to sequester miR-122, which plays an important role in the transfer of hepatitis, to inhibit the replication of HCV (143). And another study showed that artificially designed circRNA sponging miR-132 and miR-212 could attenuate pressure overloaded-induced cardiac hypertrophy (144). These studies show a great potential for translational application of circRNAs. As we all known, there are many miRNAs associated with human diseases has been unraveled in the past years and one major functions of circRNA is to act as miRNA sponge to sequester miRNAs. In the future, not only circRNAs could be “druggable,” but also it could be “drug.” CircRNAs are characterized by their covalently closed loop structures and conjunction cites. When a siRNA specifically targeting conjunction cites of onco-circRNAs are used to treat cancer, it wouldn’t target mRNAs of host genes and this could be more precise than traditional ones. In addition, not only circRNAs could be targeted by siRNAs, but also artificially designed circRNAs could be delivered into cells to sponge onco-miRNAs to treat cancers.
Utilization of cutting-edge biotechnologies, such as single cell sequencing, the regulatory roles of circRNAs in tumor progression and immunology will be clearer. Thus, circRNAs will be employed in modulating the abundance or activity of immune cells and immune checkpoints expression, which will enhance the clinical efficiency of tumor immunotherapy.
Author Contributions
ZF, SL, and CJ collected the materials. ZF and SL draw the figures and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study is supported by the Shanghai General Hospital Startup Funding (02.06.01.20.06).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
CircRNAs, circular RNAs; EciRNAs, exonic circRNAs; EIciRNAs, exon-intron circRNAs; CiRNAs, intronic RNAs; f-circRNAs, fusion circRNAs; rt-circRNAs, read-through circRNAs; TIME, tumor immune microenvironment; RNA-seq, RNA sequencing; pre-mRNA, pre-messenger RNA; LUSC, lung squamous cell carcinoma; CRC, colorectal cancer; RBP, RNA binding protein; PABPN1, poly(A) binding protein 1; HUR, Hu-antigen R; PES1, prescadillo homologue 1; YAP, yes associated protein; PTBP1, polypyrimidine tract-binding protein 1; MDM2, mouse double-minute 2; HPV, human papillomaviruses; PAF1c, polymerase associated factor complex; TSA, tumor-specific antigen; TSA, tumor-specific antigen; TAA, tumor-associated antigens; PD, programmed cell death; IL, interleukin; NSCLC, non-small cell lung cancer; PDA, pancreatic ductal adenocarcinoma; ICB, immune checkpoint blockade; snRNP, small nuclear ribonuleoprotein; Pol II, polymerase II.
References
1. Nahand JS, Jamshidi S, Hamblin MR, Mahjoubin-Tehran M, Vosough M, Jamali M, et al. Circular RNAs: New Epigenetic Signatures in Viral Infections. Front Microbiol (2020) 11:1853. doi: 10.3389/fmicb.2020.01853
2. Yousefi F, Shabaninejad Z, Vakili S, Derakhshan M, Movahedpour A, Dabiri H, et al. TGF-β and WNT signaling pathways in cardiac fibrosis: Non-coding RNAs come into focus. Cell Commun Signal (2020) 18:1–16. doi: 10.1186/s12964-020-00555-4
3. Abbaszadeh-Goudarzi K, Radbakhsh S, Pourhanifeh MH, Khanbabaei H, Davoodvandi A, Fathizadeh H, et al. Circular RNA and Diabetes: Epigenetic Regulator with Diagnostic Role. Curr Mol Med (2020) 20:516–26. doi: 10.2174/1566524020666200129142106
4. Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, et al. The Landscape of Circular RNA in Cancer. Cell (2019) 176:869–81. doi: 10.1016/j.cell.2018.12.021
5. Starke S, Jost I, Hung L, Bindereif A. Exon Circularization Requires Canonical Splice Signals. CellReports (2015) 10:103–11. doi: 10.1016/j.celrep.2014.12.002
6. Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, et al. Endoribonucleolytic Cleavage of m6A-Containing RNAs by RNase P/MRP Complex. Mol Cell (2019) 74:494–507.e8. doi: 10.1016/j.molcel.2019.02.034
7. Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, et al. MiRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J (2011) 30:4414–22. doi: 10.1038/emboj.2011.359
8. Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J Vol (2016) 7:155–60. doi: 10.1096/fasebj.7.1.7678559
9. Pasman Z, Been DM, Garcia-Blanco AM. Exon circularization in mammalian nuclear extracts. RNA (1996) 2:603–10.
10. Cocquerelle C, Daubersies P, Majerus MA, Kerckaert JP, Bailleul B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J (1992) 11:1095–8. doi: 10.1002/j.1460-2075.1992.tb05148.x
11. Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, et al. Scrambled exons. Cell (1991) 64:607–13. doi: 10.1016/0092-8674(91)90244-S
12. Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, et al. Circular RNA is expressed across the eukaryotic tree of life. PloS One (2014) 9:e90859. doi: 10.1371/journal.pone.0090859
13. Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep (2015) 10:170–7. doi: 10.1016/j.celrep.2014.12.019
14. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA (2013) 19:426. doi: 10.1261/rna.035667.112
15. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-Type Specific Features of Circular RNA Expression. PloS Genet (2013) 9:e1003777. doi: 10.1371/annotation/f782282b-eefa-4c8d-985c-b1484e845855
16. Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, et al. Genome-wide Analysis of Drosophila Circular RNAs Reveals Their Structural and Sequence Properties and Age-Dependent Neural Accumulation. Cell Rep (2014) 9:1966–80. doi: 10.1016/j.celrep.2014.10.062
17. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet (2019) 20:675–91. doi: 10.1038/s41576-019-0158-7
18. Ma S, Kong S, Wang F, Ju S. CircRNAs : biogenesis, functions, and role in drug-resistant Tumours. Mol Cancer (2020) 19:119. doi: 10.1186/s12943-020-01231-4
19. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol (2014) 15:1–14. doi: 10.1186/s13059-014-0409-z
20. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol (2015) 22:256–64. doi: 10.1038/nsmb.2959
21. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular Intronic Long Noncoding RNAs. Mol Cell (2013) 51:792–806. doi: 10.1016/j.molcel.2013.08.017
22. Jeong JC, Lo-Coco F, Guarnerio J, Bezzi M, Naldini MM, Berry K, et al. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell (2016) 165:289–302. doi: 10.1016/j.cell.2016.03.020
23. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. CircRNA Biogenesis competes with Pre-mRNA splicing. Mol Cell (2014) 56:55–66. doi: 10.1016/j.molcel.2014.08.019
24. Liang D, Tatomer DC, Luo Z, Wu H, Yang L, Chen LL, et al. The Output of Protein-Coding Genes Shifts to Circular RNAs When the Pre-mRNA Processing Machinery Is Limiting. Mol Cell (2017) 68:940–54. doi: 10.1016/j.molcel.2017.10.034
25. Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell (2014) 159:134–47. doi: 10.1016/j.cell.2014.09.001
26. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell (2015) 160:1125–34. doi: 10.1016/j.cell.2015.02.014
27. Eger N, Schoppe L, Schuster S, Laufs U, Boeckel JN. Circular RNA splicing. Adv Exp Med Biol (2018) 1087:41–52. doi: 10.1007/978-981-13-1426-1_4
28. Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature (2011) 479:74–9. doi: 10.1038/nature10442
29. Rinaldi L, Datta D, Serrat J, Morey L, Solanas G, Avgustinova A, et al. Dnmt3a and Dnmt3b Associate with Enhancers to Regulate Human Epidermal Stem Cell Homeostasis. Cell Stem Cell (2016) 19:491–501. doi: 10.1016/j.stem.2016.06.020
30. Li S, Han L. Circular RNAs as promising biomarkers in cancer : detection, function, and beyond. Genome Med (2019) 11:15. doi: 10.1186/s13073-019-0629-7
31. Borran S, Ahmadi G, Rezaei S, Anari MM, Modabberi M, Azarash Z, et al. Circular RNAs: New players in thyroid cancer. Pathol Res Pract (2020) 216:153217. doi: 10.1016/j.prp.2020.153217
32. Shabaninejad Z, Vafadar A, Movahedpour A, Ghasemi Y, Namdar A, Fathizadeh H, et al. Circular RNAs in cancer: New insights into functions and implications in ovarian cancer. J Ovarian Res (2019) 12:1–12. doi: 10.1186/s13048-019-0558-5
33. Naeli P, Pourhanifeh MH, Karimzadeh MR, Shabaninejad Z, Movahedpour A, Tarrahimofrad H, et al. Circular RNAs and gastrointestinal cancers: Epigenetic regulators with a prognostic and therapeutic role. Crit Rev Oncol Hematol (2020) 145:102854. doi: 10.1016/j.critrevonc.2019.102854
34. Hua JT, Chen S, He HH. Landscape of Noncoding RNA in Prostate Cancer. Trends Genet (2019) 35:840–51. doi: 10.1016/j.tig.2019.08.004
35. Yang L, Fu J, Zhou Y. Circular RNAs and Their Emerging Roles in Immune Regulation. Front Immunol (2018) 9:2977. doi: 10.3389/fimmu.2018.02977
36. Chen N, Zhao G, Yan X, Lv Z, Yin H, Zhang S, et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol (2018) 19:1–14. doi: 10.1186/s13059-018-1594-y
37. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol Cell (2017) 66:9–21. doi: 10.1016/j.molcel.2017.02.021
38. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature (2013) 495:333–8. doi: 10.1038/nature11928
39. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res (2016) 44:2846–58. doi: 10.1093/nar/gkw027
40. Cheng Z, Yu C, Cui S, Wang H, Jin H, Wang C, et al. circTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nat Commun (2019) 10:3200. doi: 10.1038/s41467-019-11162-4
41. Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7 article. Cell Death Dis (2018) 9:417. doi: 10.1038/s41419-018-0454-8
42. Ye F, Gao G, Zou Y, Zheng S, Zhang L, Ou X, et al. circFBXW7 Inhibits Malignant Progression by Sponging miR-197-3p and Encoding a 185-aa Protein in Triple-Negative Breast Cancer. Mol Ther - Nucleic Acids (2019) 18:88–98. doi: 10.1016/j.omtn.2019.07.023
43. Dong W, Bi J, Liu H, Yan D, He Q, Zhou Q, et al. Circular RNA ACVR2A suppresses bladder cancer cells proliferation and metastasis through miR-626/EYA4 axis. Mol Cancer (2019) 18:1–16. doi: 10.1186/s12943-019-1025-z
44. Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol (2017) 14:361–9. doi: 10.1080/15476286.2017.1279788
45. Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun (2016) 7:12429. doi: 10.1038/ncomms12429
46. Wu N, Yuan Z, Du KY, Fang L, Lyu J, Zhang C, et al. Translation of yes-associated protein (YAP) was antagonized by its circular RNA via suppressing the assembly of the translation initiation machinery. Cell Death Differ (2019) 26:2758–73. doi: 10.1038/s41418-019-0337-2
47. Sun YM, Wang WT, Zeng ZC, Chen TQ, Han C, Pan Q, et al. CircMYBL2, a circRNA from MYBL2, regulates FLT3 translation by recruiting PTBP1 to promote FLT3-ITD AML progression. Blood (2019) 134:1533–46. doi: 10.1182/blood.2019000802
48. Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ (2017) 24:357–70. doi: 10.1038/cdd.2016.133
49. Zhao J, Lee EE, Kim J, Yang R, Chamseddin B, Ni C, et al. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat Commun (2019) 10:2300
50. Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei P, et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun (2018) 9:4475. doi: 10.1038/s41467-018-06862-2
51. Liang W, Wong C, Liang P, Shi M, Cao Y, Rao S, et al. Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol (2019) 20:84. doi: 10.1186/s13059-019-1685-4
52. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature (2012) 485:201–6. doi: 10.1038/nature11112
53. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell (2012) 149:1635–46. doi: 10.1016/j.cell.2012.05.003
54. Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol (2016) 18:31–42. doi: 10.1038/nrm.2016.132
55. Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature (2015) 519:482–5. doi: 10.1038/nature14281
56. Lee M, Kim B, Kim VN. Emerging roles of RNA modification: M6A and U-Tail. Cell (2014) 158:980–7. doi: 10.1016/j.cell.2014.08.005
57. Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA (1974) 71:3971–5. doi: 10.1073/pnas.71.10.3971
58. Edens BM, Vissers C, Su J, Arumugam S, Xu Z, Shi H, et al. FMRP Modulates Neural Differentiation through m6A-Dependent mRNA Nuclear Export. Cell Rep (2019) 28:845–54.e5. doi: 10.1016/j.celrep.2019.06.072
59. Engel M, Eggert C, Kaplick PM, Eder M, Röh S, Tietze L, et al. The Role of m6A/m-RNA Methylation in Stress Response Regulation. Neuron (2018) 99:389–403.e9. doi: 10.1016/j.neuron.2018.07.009
60. Yu R, Li Q, Feng Z, Cai L, Xu Q. m6A reader YTHDF2 regulates lps-induced inflammatory response. Int J Mol Sci (2019) 20:1323. doi: 10.3390/ijms20061323
61. Lan Q, Liu PY, Haase J, Bell JL, Huttelmaier S, Liu T. The critical role of RNA M6A methylation in cancer. Cancer Res (2019) 79:1285–92. doi: 10.1158/0008-5472.CAN-18-2965
62. Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M. XRNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell (2013) 155:793. doi: 10.1016/j.cell.2013.10.026
63. Dai D, Wang H, Zhu L, Jin H, Wang X. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis (2018) 9:124. doi: 10.1038/s41419-017-0129-x
64. Zhou C, Molinie B, Daneshvar K, Pondick JV, Wang J, Van Wittenberghe N, et al. Genome-Wide Maps of m6A circRNAs Identify Widespread and Cell-Type-Specific Methylation Patterns that Are Distinct from mRNAs. Cell Rep (2017) 20:2262–76. doi: 10.1016/j.celrep.2017.08.027
65. Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, et al. N 6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun (2019) 10:1–15. doi: 10.1038/s41467-019-12651-2
66. Di Timoteo G, Dattilo D, Centrón-Broco A, Colantoni A, Guarnacci M, Rossi F, et al. Modulation of circRNA Metabolism by m6A Modification. Cell Rep (2020) 31:107641. doi: 10.1016/j.celrep.2020.107641
67. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res (2017) 27:626–41. doi: 10.1038/cr.2017.31
68. Galon J, Bruni D. Tumor Immunology and Tumor Evolution: Intertwined Histories. Immunity (2020) 52:55–81. doi: 10.1016/j.immuni.2019.12.018
69. Calabrese L, Velcheti V. Checkpoint immunotherapy: Good for cancer therapy, bad for rheumatic diseases. Ann Rheum Dis (2017) 76:1–3. doi: 10.1136/annrheumdis-2016-209782
70. Melief CJM, Finn OJ. Cancer immunology. N Engl J Med (2008) 358:2704–15. doi: 10.1056/NEJMra072739
71. Good-Jacobson KL, Szumilas CG, Chen L, Sharpe AH, Tomayko MM, Shlomchik MJ. PD-1 regulates germinal center B cell survival and the formation and affinity of long-lived plasma cells. Nat Immunol (2010) 11:535–42. doi: 10.1038/ni.1877
72. Ghiotto M, Gauthier L, Serriari N, Pastor S, Truneh A, Nunès JA, et al. PD-L1 and PD-L2 differ in their molecular mechanisms of interaction with PD-1. Int Immunol (2010) 22:651–60. doi: 10.1093/intimm/dxq049
73. Latchman Y, Wood CR, Chernova T, Chaudhary D, Borde M, Chernova I, et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol (2001) 2:261–8. doi: 10.1038/85330
74. Velcheti V, Schalper KA, Carvajal DE, Anagnostou VK, Syrigos KN, Sznol M, et al. Programmed death ligand-1 expression in non-small cell lung cancer. Lab Invest (2014) 94:107–16. doi: 10.1038/labinvest.2013.130
75. McLaughlin J, Han G, Schalper KA, Carvajal-Hausdorf D, Pelekanou V, Rehman J, et al. Quantitative assessment of the heterogeneity of PD-L1 expression in non-small-cell lung cancer. JAMA Oncol (2016) 2:46–54. doi: 10.1001/jamaoncol.2015.3638
76. Vassilakopoulou M, Avgeris M, Velcheti V, Kotoula V, Rampias T, Chatzopoulos K, et al. Evaluation of PD-L1 Expression and Associated Tumor-Infiltrating Lymphocytes in Laryngeal Squamous Cell Carcinoma. Clin Cancer Res (2016) 22:704–13. doi: 10.1158/1078-0432.CCR-15-1543
77. Brahmer JR, Tykodi SS, Chow LQM, Hwu WJ, Topalian SL, Hwu P, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med (2012) 366:2455–65. doi: 10.1056/NEJMoa1200694
78. Yang Y. Cancer immunotherapy : harnessing the immune system to battle cancer. J Clin Invest (2015) 125:3335–7. doi: 10.1172/JCI83871
79. Park JH, Rivière I, Gonen M, Wang X, Sénéchal B, Curran KJ, et al. Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N Engl J Med (2018) 378:449–59. doi: 10.1056/NEJMoa1709919
80. Neelapu SS, Locke FL, Bartlett NL, Lekakis LJ, Miklos DB, Jacobson CA, et al. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-Cell lymphoma. N Engl J Med (2017) 377:2531–44. doi: 10.1056/NEJMoa1707447
81. Schuster SJ, Svoboda J, Chong EA, Nasta SD, Mato AR, Anak Ö, et al. Chimeric antigen receptor T Cells in refractory B-Cell lymphomas. N Engl J Med (2017) 377:2545–54. doi: 10.1056/NEJMoa1708566
82. Maude SL, Laetsch TW, Buechner J, Rives S, Boyer M, Bittencourt H, et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N Engl J Med (2018) 378:439–48. doi: 10.1056/NEJMoa1709866
83. Dougan M, Dranoff G, Dougan SK. Cancer immunotherapy: Beyond checkpoint blockade. Annu Rev Cancer Biol (2019) 3:55–75. doi: 10.1146/annurev-cancerbio-030518-055552
84. Xia A-L, Wang X-C, Lu Y-J, Lu X-J, Sun B. Chimeric-antigen receptor T (CAR-T) cell therapy for solid tumors: challenges and opportunities. Oncotarget (2017) 8:90521–31. doi: 10.18632/oncotarget.19361
85. Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med (2014) 371:1507–17. doi: 10.1056/NEJMoa1407222
86. Fry TJ, Shah NN, Orentas RJ, Stetler-Stevenson M, Yuan CM, Ramakrishna S, et al. CD22-targeted CAR T cells induce remission in B-ALL that is naive or resistant to CD19-targeted CAR immunotherapy. Nat Med (2018) 24:20–8. doi: 10.1038/nm.4441
87. Shaffer DR, Savoldo B, Yi Z, Chow KKH, Kakarla S, Spencer DM, et al. T cells redirected against CD70 for the immunotherapy of CD70-positive malignancies. Blood (2011) 117:4304–14. doi: 10.1182/blood-2010-04-278218
88. Jin L, Ge H, Long Y, Yang C, Chang YE, Mu L, et al. CD70, a novel target of CAR T-cell therapy for gliomas. Neuro Oncol (2018) 20:55–65. doi: 10.1093/neuonc/nox116
89. Xu Z, Li P, Fan L, Wu M. The potential role of circRNA in tumor immunity regulation and immunotherapy. Front Immunol (2018) 9:1–11. doi: 10.3389/fimmu.2018.00009
90. Sahin U, Türeci Ö. Personalized vaccines for cancer immunotherapy. Sci (80- ) (2018) 359:1355–60. doi: 10.1126/science.aar7112
92. Zhao X, Bose A, Komita H, Taylor JL, Chi N, Lowe DB, et al. Vaccines Targeting Tumor Blood Vessel Antigens Promote CD8 + T Cell-Dependent Tumor Eradication or Dormancy in HLA-A2 Transgenic Mice. J Immunol (2012) 188:1782–8. doi: 10.4049/jimmunol.1101644
93. Komita H, Zhao X, Taylor JL, Sparvero LJ, Amoscato AA, Alber S, et al. CD8+ T-cell responses against hemoglobin-β prevent solid tumor growth. Cancer Res (2008) 68:8076–84. doi: 10.1158/0008-5472.CAN-08-0387
94. Kirkwood JM, Lee S, Moschos S, Albertini MR, Michalak JC, Sander C, et al. Immunogenicity and antitumor effects of vaccination with peptide vaccine +/- granulocyte-monocyte colony-stimulating factor and/or IFIN-α2b in advanced metastatic melanoma: Eastern cooperative oncology group phase II trial E1696. Clin Cancer Res (2009) 15:1443–51. doi: 10.1158/1078-0432.CCR-08-1231
95. Slingluff CL, Lee S, Zhao F, Chianese-Bullock KA, Olson WC, Butterfield LH, et al. A randomized phase II trial of multiepitope vaccination with melanoma peptides for cytotoxic T cells and helper T cells for patients with metastatic melanoma (E1602). Clin Cancer Res (2013) 19:4228–38. doi: 10.1158/1078-0432.CCR-13-0002
96. Pollack IF, Jakacki RI, Butterfield LH, Hamilton RL, Panigrahy A, Potter DM, et al. Antigen-specific immune responses and clinical outcome after vaccination with glioma-associated antigen peptides and polyinosinic-polycytidylic acid stabilized by lysine and carboxymethylcellulose in children with newly diagnosed malignant brainstem and n. J Clin Oncol (2014) 32:2050–8. doi: 10.1200/JCO.2013.54.0526
97. Slingluff CL, Petroni GR, Chianese-Bullock KA, Smolkin ME, Ross MI, Haas NB, et al. Randomized multicenter trial of the effects of melanoma-associated helper peptides and cyclophosphamide on the immunogenicity of a multipeptide melanoma vaccine. J Clin Oncol (2011) 29:2924–32. doi: 10.1200/JCO.2010.33.8053
98. Walter S, Weinschenk T, Stenzl A, Zdrojowy R, Pluzanska A, Szczylik C, et al. Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival. Nat Med (2012) 18:1254–61. doi: 10.1038/nm.2883
99. Disis ML, Gad E, Herendeen DR, Phan-Lai V, Park KH, Cecil DL, et al. A multiantigen vaccine targeting neu, IGFBP-2, and IGF-IR prevents tumor progression in mice with preinvasive breast disease. Cancer Prev Res (2013) 6:1273–82. doi: 10.1158/1940-6207.CAPR-13-0182
100. Giordano FJ, Ping P, Mckirnan MD, Nozaki S, Demaria AN, Dillmann WH, et al. Vaccination of patients with B-cell lymphoma using autologous antigen-pulsed dendritic cells. Nat Med (1996) 2:534–9. doi: 10.1038/nm0596-534
101. Dummer R, Burg G, Schadendorf D. Vaccination of melanoma patients with peptide- or tumor lysate-pulsed dendritic cells. Nat Med (1998) 4:328–32. doi: 10.1038/nm0398-328
102. Banchereau J, Palucka AK, Dhodapkar M, Burkeholder S, Taquet N, Rolland A, et al. Immune and clinical responses in patients with metastatic melanoma to CD34+ progenitor-derived dendritic cell vaccine. Cancer Res (2001) 61:6451–8.
103. Dranoff G, Jaffee E, Lazenby A, Golumbek P, Levitsky H, Brose K, et al. Vaccination with irradiated tumor cells engineered to secrete murine granulocyte-macrophage colony-stimulating factor stimulates potent, specific, and long-lasting anti-tumor immunity. Proc Natl Acad Sci USA (1993) 90:3539–43. doi: 10.1073/pnas.90.8.3539
104. McBride WH, Thacker JD, Comora S, Economou JS, Kelley D, Hogge D, et al. Genetic Modification of a Murine Fibrosarcoma to Produce Interleukin 7 Stimulates Host Cell Infiltration and Tumor Immunity. Cancer Res (1992) 52:3931–7.
105. Le DT, Brockstedt DG, Nir-Paz R, Hampl J, Mathur S, Nemunaitis J, et al. A live-attenuated listeria vaccine (ANZ-100) and a live-attenuated listeria vaccine expressing mesothelin (CRS-207) for advanced cancers: Phase I studies of safety and immune induction. Clin Cancer Res (2012) 18:858–68. doi: 10.1158/1078-0432.CCR-11-2121
106. Kim TS, Chopra A, O-Sullivan IS, Cohen EP. Enhanced immunity to breast cancer in mice immunized with fibroblasts transfected with a complementary DNA expression library from breast cancer cells: Enrichment of the vaccine for immunotherapeutic cells. J Immunother (2006) 29:261–73. doi: 10.1097/01.cji.0000197097.46100.bb
107. Fukuhara H, Ino Y, Todo T. Oncolytic virus therapy: A new era of cancer treatment at dawn. Cancer Sci (2016) 107:1373–9. doi: 10.1111/cas.13027
108. Meng WS, Butterfield LH, Ribas A, Dissette VB, Heller JB, Miranda GA, et al. α-Fetoprotein-specific tumor immunity induced by plasmid prime-adenovirus boost genetic vaccination. Cancer Res (2001) 61:8782–6.
109. Butterfield LH, Economou JS, Gamblin TC, Geller DA. Alpha fetoprotein DNA prime and adenovirus boost immunization of two hepatocellular cancer patients. J Transl Med (2014) 12:1–9. doi: 10.1186/1479-5876-12-86
110. John LB, Howland LJ, Flynn JK, West AC, Devaud C, Duong CP, et al. Oncolytic virus and anti-4-1BB combination therapy elicits strong antitumor immunity against established cancer. Cancer Res (2012) 72:1651–60. doi: 10.1158/0008-5472.CAN-11-2788
111. Vinay DS, Ryan EP, Pawelec G, Talib WH, Stagg J, Elkord E, et al. Immune evasion in cancer: Mechanistic basis and therapeutic strategies. Semin Cancer Biol (2015) 35:S185–98. doi: 10.1016/j.semcancer.2015.03.004
112. Cervantes-Villagrana RD, Albores-García D, Cervantes-Villagrana AR, García-Acevez SJ. Tumor-induced neurogenesis and immune evasion as targets of innovative anti-cancer therapies. Signal Transduct Target Ther (2020) 5:99. doi: 10.1038/s41392-020-0205-z
113. Motz GT, Santoro SP, Wang LP, Garrabrant T, Lastra RR, Hagemann IS, et al. Tumor endothelium FasL establishes a selective immune barrier promoting tolerance in tumors. Nat Med (2014) 20:607–15. doi: 10.1038/nm.3541
114. Marvel D, Gabrilovich DI. Myeloid-derived suppressor cells in the tumor microenvironment: Expect the unexpected. J Clin Invest (2015) 125:3356–64. doi: 10.1172/JCI80005
115. Brown JM, Recht L, Strober S. The Promise of Targeting Macrophages in Cancer Therapy. Clin Cancer Res (2017) 23:3241–50. doi: 10.1158/1078-0432.CCR-16-3122
116. Veglia F, Perego M, Gabrilovich D. Myeloid-derived suppressor cells coming of age review-article. Nat Immunol (2018) 19:108–19. doi: 10.1038/s41590-017-0022-x
117. Feig C, Jones JO, Kraman M, Wells RJB, Deonarine A, Chan DS, et al. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc Natl Acad Sci USA (2013) 110:20212–7. doi: 10.1073/pnas.1320318110
118. Song H, Liu Q, Liao Q, Circular RNA. and tumor microenvironment. Cancer Cell Int (2020) 20:1–15. doi: 10.1186/s12935-020-01301-z
119. Sun C, Sun HY, Xiao WH, Zhang C, Tian ZG. Natural killer cell dysfunction in hepatocellular carcinoma and NK cell-based immunotherapy. Acta Pharmacol Sin (2015) 36:1191–9. doi: 10.1038/aps.2015.41
120. Cong J, Wang X, Zheng X, Wang D, Fu B, Sun R, et al. Dysfunction of Natural Killer Cells by FBP1-Induced Inhibition of Glycolysis during Lung Cancer Progression. Cell Metab (2018) 28:243–55.e5. doi: 10.1016/j.cmet.2018.06.021
121. Merino A, Miller JS, Cichocki F, Merino A, Zhang B, Dougherty P, et al. Chronic stimulation drives human NK cell dysfunction and epigenetic reprograming Find the latest version : Chronic stimulation drives human NK cell dysfunction and epigenetic reprograming. J Clin Invest (2019) 129:3770–85. doi: 10.1172/JCI125916
122. Piątkiewicz P, Miłek T, Bernat-Karpińska M, Ohams M, Czech A, Ciostek P. The dysfunction of nk cells in patients with type 2 diabetes and colon cancer. Arch Immunol Ther Exp (Warsz) (2013) 61:245–53. doi: 10.1007/s00005-013-0222-5
123. Ma Y, Zhang C, Zhang B, Yu H, Yu Q. circRNA of AR-suppressed PABPC1 91 bp enhances the cytotoxicity of natural killer cells against hepatocellular carcinoma via upregulating UL16 binding protein 1. Oncol Lett (2019) 17:388–97. doi: 10.3892/ol.2018.9606
124. Zhang PF, Zhang PF, Zhang PF, Gao C, Gao C, Huang XY, et al. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer (2020) 19:1–15. doi: 10.1186/s12943-020-01222-5
125. Pathria P, Louis TL, Varner JA. Targeting Tumor-Associated Macrophages in Cancer. Trends Immunol (2019) 40:310–27. doi: 10.1016/j.it.2019.02.003
126. Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature (2011) 475:222–5. doi: 10.1038/nature10138
127. Komohara Y, Fujiwara Y, Ohnishi K, Shiraishi D, Takeya M. Contribution of macrophage polarization to metabolic diseases. J Atheroscler Thromb (2016) 23:10–7. doi: 10.5551/jat.32359
128. Komohara Y, Jinushi M, Takeya M. Clinical significance of macrophage heterogeneity in human malignant tumors. Cancer Sci (2014) 105:1–8. doi: 10.1111/cas.12314
129. Zhang Y, Zhang Y, Li X, Zhang M, Lv K. Microarray analysis of circular RNA expression patterns in polarized macrophages. Int J Mol Med (2017) 39:373–9. doi: 10.3892/ijmm.2017.2852
130. Zou Y, Zheng S, Deng X, Yang A, Xie X, Tang H, et al. The role of circular RNA CDR1as/cirs-7 in regulating tumor microenvironment: A pan-cancer analysis. Biomolecules (2019) 9:1–13. doi: 10.3390/biom9090429
131. Badalamenti G, Fanale D, Incorvaia L, Barraco N, Listì A, Maragliano R, et al. Role of tumor-infiltrating lymphocytes in patients with solid tumors: Can a drop dig a stone? Cell Immunol (2019) 343:103753. doi: 10.1016/j.cellimm.2018.01.013
132. Weng Q, Chen M, Li M, Zheng YF, Shao G, Fan W, et al. Global microarray profiling identified hsa-circ-0064428 as a potential immune-associated prognosis biomarker for hepatocellular carcinoma. J Med Genet (2019) 56:32–8. doi: 10.1136/jmedgenet-2018-105440
133. Zou W, Wolchok JD, Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci Transl Med (2016) 8:328rv4. doi: 10.1126/scitranslmed.aad7118
134. Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Sci (80- ) (2018) 359:1350–5. doi: 10.1126/science.aar4060
135. Sanmamed MF, Chen L. A Paradigm Shift in Cancer Immunotherapy: From Enhancement to Normalization. Cell (2018) 175:313–26. doi: 10.1016/j.cell.2018.09.035
136. Schalper KA, Carvajal-Hausdorf D, McLaughlin J, Altan M, Velcheti V, Gaule P, et al. Differential expression and significance of PD-L1, IDO-1, and B7-H4 in human lung cancer. Clin Cancer Res (2017) 23:370–8. doi: 10.1158/1078-0432.CCR-16-0150
137. Qin S, Xu L, Yi M, Yu S, Wu K, Luo S. Novel immune checkpoint targets: Moving beyond PD-1 and CTLA-4. Mol Cancer (2019) 18:1–14. doi: 10.1186/s12943-019-1091-2
138. Huang RY, Francois A, McGray AR, Miliotto A, Odunsi K. Compensatory upregulation of PD-1, LAG-3, and CTLA-4 limits the efficacy of single-agent checkpoint blockade in metastatic ovarian cancer. Oncoimmunology (2017) 6:e1249561. doi: 10.1080/2162402X.2016.1249561
139. Zhang X, Xu L, Wang F. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis, and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol Int (2017) 41:1056–64. doi: 10.1002/cbin.10826
140. Tanaka E, Miyakawa Y, Kishikawa T, Seimiya T, Iwata T, Funato K, et al. Expression of circular RNA CDR1-AS in colon cancer cells increases cell surface PD-L1 protein levels. Oncol Rep (2019) 42:1459–66. doi: 10.3892/or.2019.7244
141. Hong W, Xue M, Jiang J, Zhang Y, Gao X. Circular RNA circ-CPA4/ let-7 miRNA/PD-L1 axis regulates cell growth, stemness, drug resistance and immune evasion in non-small cell lung cancer (NSCLC). J Exp Clin Cancer Res (2020) 39:1–19. doi: 10.1186/s13046-020-01648-1
142. Li L, Zhang Q, Lian K. Circular RNA circ_0000284 plays an oncogenic role in the progression of non-small cell lung cancer through the miR-377-3p-mediated PD-L1 promotion. Cancer Cell Int (2020) 20:1–11. doi: 10.1186/s12935-020-01310-y
143. Jost I, Shalamova LA, Gerresheim GK, Niepmann M, Bindereif A, Rossbach O. Functional sequestration of microRNA-122 from Hepatitis C Virus by circular RNA sponges. RNA Biol (2018) 15:1032–9. doi: 10.1080/15476286.2018.1435248
Keywords: circular RNA, regulatory roles, tumor immunology, immunotherapy, immune checkpoint
Citation: Fang Z, Jiang C and Li S (2021) The Potential Regulatory Roles of Circular RNAs in Tumor Immunology and Immunotherapy. Front. Immunol. 11:617583. doi: 10.3389/fimmu.2020.617583
Received: 15 October 2020; Accepted: 14 December 2020;
Published: 03 February 2021.
Edited by:
Alejandro Villagra, George Washington University, United StatesReviewed by:
XiuKai Cao, Yangzhou University, ChinaHamed Mirzaei, Kashan University of Medical Sciences, Iran
Chengchuang Song, Northwest A and F University, China
Xiang Li, Virginia Tech, United States
Copyright © 2021 Fang, Jiang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shengli Li, c2hlbmdsaS5saUBzaHNtdS5lZHUuY24=; Chunjie Jiang, Y2h1bmppZS5qaWFuZzkxN0BvdXRsb29rLmNvbQ==
 Zhixiao Fang1
Zhixiao Fang1 Chunjie Jiang
Chunjie Jiang Shengli Li
Shengli Li